Abstract
Although primarily studied in relation to photorespiration, serine metabolism in chloroplasts may play a key role in plant CO2 fertilization responses by linking CO2 assimilation with growth. Here, we show that the phosphorylated serine pathway is part of a ‘photosynthetic C1 pathway’ and demonstrate its high activity in foliage of a C3 tree where it rapidly integrates photosynthesis and C1 metabolism contributing to new biomass via methyl transfer reactions, imparting a large natural 13C-depleted signature. Using 13CO2-labelling, we show that leaf serine, the S-methyl group of leaf methionine, pectin methyl esters, and the associated methanol released during cell wall expansion during growth, are directly produced from photosynthetically-linked C1 metabolism, within minutes of light exposure. We speculate that the photosynthetic C1 pathway is highly conserved across the photosynthetic tree of life, is responsible for synthesis of the greenhouse gas methane, and may have evolved with oxygenic photosynthesis by providing a mechanism of directly linking carbon and ammonia assimilation with growth. Although the rise in atmospheric CO2 inhibits major metabolic pathways like photorespiration, our results suggest that the photosynthetic C1 pathway may accelerate and represents a missing link between enhanced photosynthesis and plant growth rates during CO2 fertilization under a changing climate.
Subject terms: Rubisco, Cell growth, Metabolomics
A photosynthetic C1 pathway starting with CO2 and NH3 assimilation and ending with methionine synthesis is highly active in foliage of a C3 tree, where it rapidly integrates photosynthesis and C1 metabolism contributing to new biomass via methyl transfer reactions.
Introduction
Terrestrial and marine ecosystems assimilate atmospheric CO2 into biomass through photosynthesis1 serving as a foundation for Earth’s carbon cycle. Since pre-industrial times (i.e. before 1850), global terrestrial gross primary production (GPP) is estimated to have increased by 31 ± 5%, largely driven by an increase in atmospheric CO2 concentrations2,3. However, while the mechanisms involving enhanced plant growth under elevated CO2 have primarily focused on increased carboxylation and decreased oxygenation rates of ribulose bisphosphate carboxylase/oxygenase (RuBisCO) and enhanced water use efficiency related to decreased stomatal conductance, these processes alone cannot fully explain the dynamic changes in plant growth to elevated CO24.
Plants assimilate CO2 via gross photosynthesis (carboxylation) but also produce CO2 via photorespiration and respiration. Photorespiration involves a series of reactions linked to C1 metabolism during the conversion of glycine to serine active during abiotic stress responses5. However, although the majority of research on serine metabolism in plants has focused on the role of photorespiration, the presence of a phosphorylated serine pathway has been established in cyanobacteria6 and higher plants7 and appears to be well conserved across prokaryotic and eukaryotic species8. High expression levels of key enzymes of the phosphorylated serine pathway have been documented in light-grown shoots9 with the phosphorylated serine pathway directly linked to nitrogen assimilation by providing 2-oxogluterate for ammonia fixation10.
Disruption of the phosphorylated serine pathway leads to a reduction in nitrogen and sulfur contents in shoots and a general transcriptional response to nutrient deficiency11. While the in vivo activity of the phosphorylated serine pathway, including its potential links with photosynthesis remains poorly characterized, genetic and molecular evidence revealed a critical role of the phosphorylated serine pathway for proper embryo and pollen development and root growth12. In the model plant Arabidopsis thaliana, mutagenesis studies demonstrated its essential role in light and sugar-dependent growth promotion; A downregulation of the phosphorylated serine pathway led to a severe inhibition of shoot and root growth8 and mutants of the key enzymes phosphoglycerate dehydrogenase (PGDH) and phosphoserine phosphatase (PSP) resulted in an embryo-lethal phenotype10,13,14. In addition, the phosphorylated serine pathway was shown to be critical for pollen development and metabolomics studies suggested that it directly integrates central carbon and energy metabolism with growth and development by affecting ammonia assimilation, glycolysis, the tricarboxylic acid cycle, and the biosynthesis of amino acids such as tryptophan10,13. While most of what is known about leaf serine derives from studies on photorespiration7, the importance of the phosphorylated serine pathway as a source of serine was found to be particularly important under elevated ambient CO2 concentrations associated with enhanced growth rates. Wild-type A. thaliana plants grown under elevated CO2 concentrations showed enhanced leaf growth rates together with increased expression of PGDH1. In contrast, leaf serine content and growth rates of mature PGDH1-silenced plants were severely impaired while increased ammonia and some amino acid concentrations were observed10. These observations suggest that the phosphorylated serine pathway is particularly important under high CO2 conditions that promote photosynthesis while suppressing photorespiration. Consistent with this view, a recent analysis of cell proliferation and elongation in A. thaliana revealed that the phosphorylated serine pathway is indispensable for plant growth and its loss cannot be compensated by photorespiratory serine biosynthesis15.
Although the potential direct connection between the phosphorylated serine pathway and the Calvin-Benson cycle for C3 substrate linked with photosynthesis has not been investigated, a major role in plant growth and development and C1 metabolism has been discussed, with serine involved in methyl group transfer reactions by supplying tetrahydrofolate (THF) metabolism C1 units16,17. Thus, while the mechanisms behind this critical and indispensable role of the phosphorylated serine pathway for growth and development remain under investigation, C1 methyl transfer reactions are likely involved. In A. thaliana, chloroplasts were shown to be autonomous for the de novo synthesis of methionine, with the phosphorylated serine pathway the likely source of the serine used in the methylation of homocysteine to methionine18. Mediated by a chloroplast serine hydroxymethyltransferase (SHMT), serine from the phosphorylated serine pathway may be a major source of activated one-carbon units for plant C1 metabolism19, especially under conditions like elevated CO2 which suppresses photorespiration while enhancing photosynthesis.
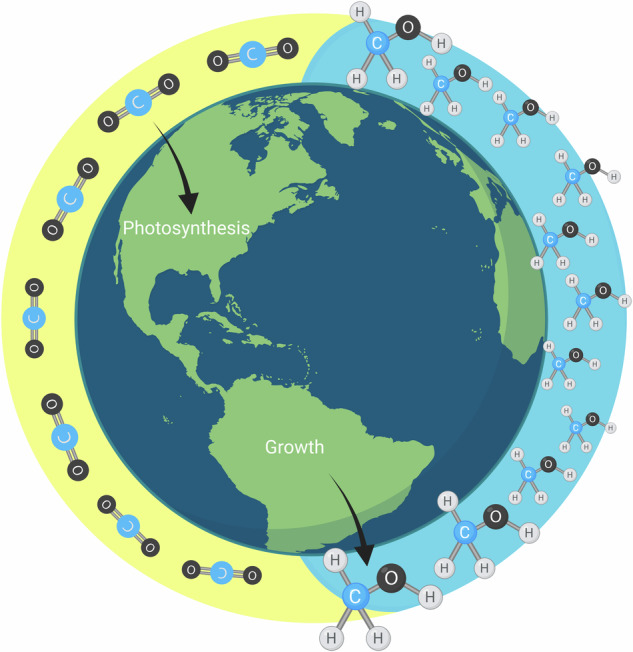
Following the export of methionine from chloroplasts to the cytosol and its activation to the universal methyl group donor S-adenosylmethionine (AdoMet or SAM), AdoMet is subsequently imported into numerous organelles where it is utilized by diverse methyltransferase enzymes during biosynthetic and regulatory processes including the methylation of proteins, carbohydrates, nucleic acids, and metabolites20. A large sustained turnover of methionine in plant tissues may be possible with AdoMet formation and regeneration, without net consumption of methionine21. AdoMet metabolism may be intimately involved in plant growth by sustaining nitrogen assimilation10 and a large, but poorly characterized flux of C1 methyl transfers during new biopolymer and metabolite synthesis including regulatory components of plant growth such as nucleic acids20, the growth hormones auxins10 and ethylene2 and methylated pectin22. While previously considered a byproduct of growth, the release of methanol from methylated pectin is now recognized to dramatically alter the elasticity of the primary cell wall, a critical parameter controlling initiation and propagation of tissue morphogenesis and growth22–24 (see Supplementary Note 1: ‘pectin demethylation, methanol emission, and growth’).
In this study, we hypothesize that light-dependent methionine synthesis and metabolism are tightly linked with growth by providing a potentially large flux of C1 transfers during biopolymer and metabolite synthesis and regulation via methylation. Moreover, synthesis of methylated pectin influences tissue morphogenesis and growth via subsequent demethylation on the primary cell wall, leading to changes in mechanical properties like elasticity and the associated release of methanol into the apoplast. We hypothesize that carbon for the activated methyl donor synthesis of AdoMet is directly produced by a ‘photosynthetic C1 pathway’ in chloroplasts initiated by the carboxylation reaction catalyzed by RuBisCO which generates 3-phosphoglyceric acid (PGA) then utilized by the plastidic phosphorylated serine pathway. Serine then transfers C1 units to THF which subsequently transfers C1 units to homocysteine during methionine synthesis. Followed by the export of methionine to the cytosol and its activation to AdoMet, the universal methyl group donor20, a high flux of methyl transfer reaction might be sustained through an AdoMet recycling pathway in the cytosol involving the regeneration of homocysteine, independently of photorespiration. To test these hypotheses, we employed dynamic 13CO2-labeling of California poplar (Populus trichocarpa) leaves under controlled environmental conditions and whole branches under day/night light cycles together with analysis of stable carbon isotope ratios (13C/12C, δ13C values) of leaf phosphoglyceric acid (PGA), serine, and methionine, cell wall methyl esters (OCH3), and leaf methanol (CH3OH), acetaldehyde (CH3CHO), and isoprene (C5H8) emissions (that is, C1, C2, C3 and C5 metabolism products). We compared 13CO2-branch labeling patterns under a 21% and 1% O2 atmosphere to evaluate the potential roles of C1-photosynthesis versus photorespiration in generating new cell wall methyl esters and the release of methanol during growth and development. We also evaluated P. tricocharpa leaf and branch gas exchange patterns in a field experiment to gain insight into potential temperature and hydraulic controls over diurnal growth and methanol emissions during the 2023 growing season in Berkeley, CA, USA. Finally, using comparative genomics, we examine the co-occurrence of proteins in the photosynthetic C1 pathway and trace the evolutionary origin of methionine synthase through cyanobacteria and algae to land plants.
The dynamic labeling of 13CO2 in California poplar demonstrated a strong connection between photosynthetic carbon assimilation and methanol emissions. Methanol emissions were highly responsive to light, with 13C-labeled pectin methyl esters (PMEs) observed within 5 min of light exposure and branch methanol emissions reaching 36% after two diurnal cycles. Methionine and PMEs were found to be primary recipients of 13C-labeled carbon from the Calvin cycle, confirming the involvement of the photosynthetic C1 pathway in methyl group transfer and cell wall expansion during plant growth. This study reveals that the photosynthetic C1 pathway plays a critical role in linking carbon assimilation with growth processes in California poplar by driving light-dependent methionine synthesis and methyl group transfer reactions. The observed diurnal fluctuations in methanol emissions and their correlation with photosynthesis underscore the importance of this pathway in regulating cell wall dynamics and overall plant growth. These findings provide new insights into the mechanisms by which elevated CO2 and photosynthesis enhance plant growth, potentially representing a critical component of plant responses to changing environmental conditions.
Results
13C abundance in methanol, isoprene and acetaldehyde emitted by branches depends on light and % O2 during diurnal 13CO2 labeling
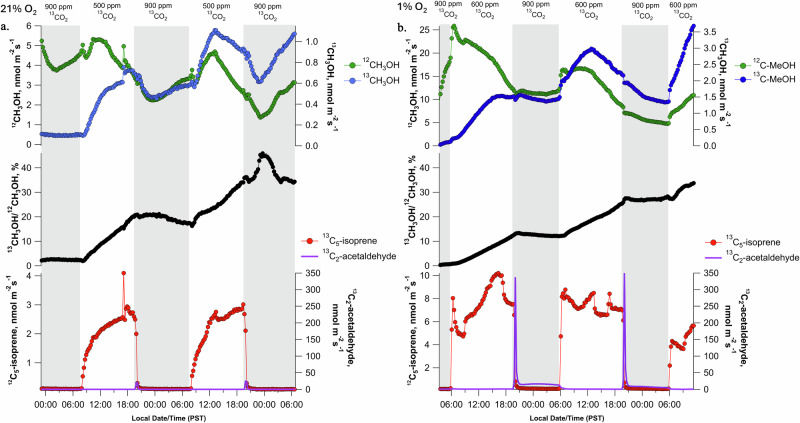
When a gas-exchange enclosure with elevated 13CO2 (900 µmol mol−1) was installed on an intact poplar branch at night in the dark under a 21% O2 atmosphere, methanol emissions displayed a typical nocturnal pattern of initially decreasing in the evening, reaching a minimum around midnight, followed by an increase throughout the night (Fig. 1a). Despite continued branch methanol emissions in the dark, 13C-labeling was not observed, with a 13C/12C isotope ratio of 1.0–2.5%, similar to natural abundance (1.1%). Moreover, emissions of isoprene in the dark could not be detected. However, upon switching to the light in the morning, emissions of 13C5-isoprene (i.e., fully 13C-labeled isotopologue) was immediately apparent and dominated other isoprene isotopologues (13C5-isoprene emissions accounting for > 95% of total isoprene emissions). In the light, the 13C/12C isotope ratio in methanol continuously increased, reaching 36% after two diurnal cycles (Fig. 1a). In contrast, at night, the isotope ratio in methanol showed some variation but did not increase. Interestingly, branch emissions of both 12C- and 13C-methanol started to increase at midnight, i.e. when leaf water potential recovered (−0.2 ± 0.1 MPa) from midday values (−1.0 ± 0.1 MPa). Replicate 2-day experiments showed similar diurnal patterns and maximum isotope ratio in methanol after 2 light periods (Supplementary Figs. S1–S4). A single long-term 13CO2 branch labeling experiment (5 days) demonstrated that the pattern of light-dependent 13C-methanol labeling continued to increase, with an isotope ratio in methanol reaching 80% by the end of day 5 (Supplementary Fig. S5).
Fig. 1. Diurnal 13C-labeling dynamics of branch methanol emissions during light-dark cycles under an elevated 13CO2 atmosphere.
a Example 2-day time series in a 21% and (b) 1% O2 atmosphere showing dynamic branch labeling of methanol (CH3OH) emissions via the photosynthetic-C1 pathway under elevated 13CO2. Night periods are shaded in gray. Also shown are branch emissions of 13C5-isoprene during the day and 13C2-acetaldehyde bursts at the beginning of the dark period.
Under a 1% O2 atmosphere used to suppress photorespiration (Fig. 1b), branch 13CO2 assimilation and O2 production continued in the light (Supplementary Fig. S6) and the isotope ratio in methanol increased to 28% after two light periods. This value was slightly lower than that under 21% O2 (36%). The branch also emitted 13C5-isoprene. Compared to 21% O2, there was a strong increase in fermentation-type metabolism in 1% O2. In fact, the burst of acetaldehyde emission at the light-to-dark transition increased considerably (nearly 20-fold) under 1% O2 (purple line, Fig. 1a, b). Moreover, acetaldehyde emission lasted longer (2–3 h) and was followed by a period of elevated, but gradually decreasing 13C2-acetaldehyde emissions for the entire night (Fig. 1b). Collectively, it shows that C1 metabolism producing methanol was mostly fed by photosynthetic carbon (and only marginally by photorespiration), with a relatively slow turn-over (compared to, e.g., isoprene).
Instantaneous leaf 13C-methanol emission in the light correlates with cumulative 13CO2 assimilation
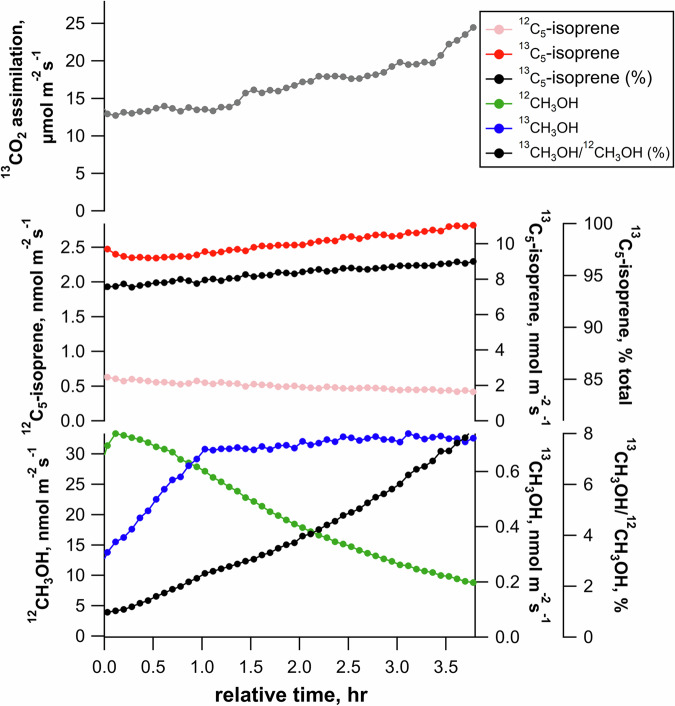
A labeling experiment was carried out with a leaf placed in the chamber in the light under elevated 13CO2 (750 µmol mol−1). As expected, there was a progressive rise in assimilation rate (from 13 to 24 mol m−2 s−1, Fig. 2). Light-dependent isoprene emissions were dominated by the 13C5-isotopologue (>95% as determined by PTR-MS) and slowly increased with time, while the monoisotopic species (12C5-isoprene) decreased concurrently. As a result, after 3 h in the light, isoprene was more than 95% 13C-labeled. Similarly, 13C-labeling in methanol increased, reaching 8% 13C after 3.5 h (Fig. 2). 13C-labeling in methanol was verified using thermal desorption-gas chromatography-mass spectrometry (TD-GC-MS) (Supplementary Fig. S7). When individual leaves were illuminated for 1–5 h under elevated 13CO2 (at 21% O2), we found a strict linear correlation (R2 > 0.96) between cumulative 13CO2 assimilation in the light and instantaneous %13C in methanol emissions (Supplementary Fig. S8). Also, with increasing labeling time from 1 to 2, 3, 4, or 5 h, the maximum 13C/12C ratio in methanol increased proportionally to 2.4%, 2.8%, 5%, 9%, and 11%, respectively. Interestingly, the slope of linear regressions, representing increase in 13C/12C ratio per µmol of assimilated 13C per m−2, varied from leaf to leaf with a mean of 2.7 × 10−5 ± 1.7 × 10−5 ( ± 1 SD). Overall, it suggests that the carbon source used to synthesize methanol was fed directly by photosynthetic assimilation.
Fig. 2. Example time series showing the 13C-labeling dynamics of leaf isoprene and methanol in the light under constant environmental conditions including optimal conditions for photosynthesis (1200 µmol m−2 s−1 PAR, 32 °C leaf temperature, and 900–1000 ppm reference 13CO2).
Note the rapid and near complete 13C-labeling of isoprene emissions, and the comparably slower and incomplete labeling of 13C-methanol emissions.
Leaf 13C-methanol emission strongly depends on temperature upon 13CO2 labeling
Selected branches from the 2-day 13CO2 diurnal experiment (above) were detached from the tree with the stem recut under water, and one mature leaf placed in a gas exchange system under a 12CO2 atmosphere (400 µmol mol−1). There was a strong increase in both 13C-methanol and 12C-methanol emission with temperature, despite a rather constant 13C/12C isotope ratio, near 20% (Supplementary Fig. S9). Thus, temperature led to an increase in the rate of, but no change in the carbon pool used for methanol production.
13C-pectin methyl esters are synthesized from assimilated 13CO2 even without photorespiration
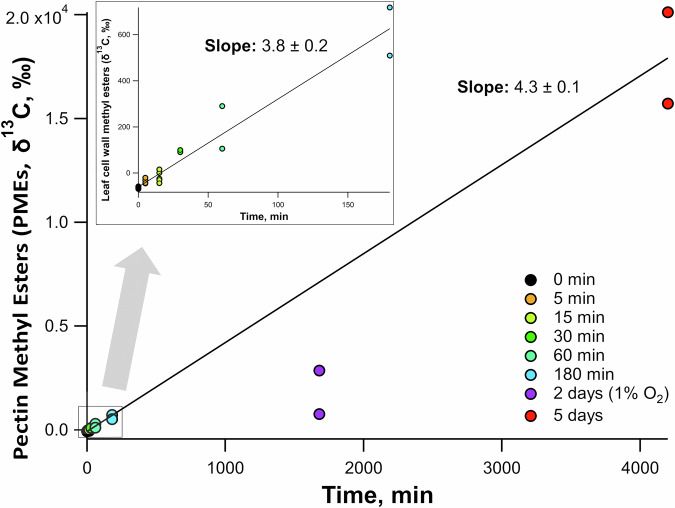
While measurement of a small % changes in 13C/12C isotope ratio in leaf-emitted methanol required 1 h or more, we used more sensitive measurements of changes of the 13C/12C ratio of pectin methyl esters (PMEs) using gas chromatography-combustion-isotope ratio mass spectrometry (GC-C-IRMS) and present results as δ13C values (‰, see Methods). PMEs were naturally 13C-depleted and thus typical of C1 carbon units of a C3 plant. In effect, values obtained from control experiments confirmed the large natural 13C-anomaly of plant C1 carbon (i.e. down to –60‰, Fig. 3). Upon 13CO2 labeling in the light (1000 µmol mol−1 13CO2, 21% O2), the 13C-enrichment in leaf PMEs could be detected after the shortest possible time experimentally (5 min) to place a leaf in the chamber and reach steady state gas exchange (−35‰) and after 180 min, reached +614‰ (i.e. about 1.8% 13C). The δ13C value increased after 5 d, reaching ca. +18000‰ (i.e. about 17.6% 13C), thus showing a consistent rate of increase throughout (3.8–4.3‰ min−1). Interestingly, 13CO2 labeling for 2 days under 1% O2 led to an average 13C-enrichment in PMEs of +2860‰ (purple circles, Fig. 3). Although slightly lower than expected compared to the situation under 21% O2, it shows that the suppression of photorespiration did not stop the synthesis of 13C-labeled PMEs. Collectively, PMEs synthesis appeared to be consistent with methanol emission, that is, methyl groups of PMEs came, at least partly, from non-photorespiratory, recently assimilated photosynthetic carbon.
Fig. 3. δ13C values of bulk leaf cell wall methyl esters by GC-C-IRMS plotted versus leaf 13CO2 labeling period.
Note the strong natural 13C-depletion of control (0 min) leaf samples (δ13C of −60‰). All leaves were under a 21% O2 atmosphere except the 2-day samples which were under a 1% O2 atmosphere. Trendlines only include data under a 21% O2 atmosphere. Short labeling durations 0–180 min shown in inset.
13C-labeling in metabolites suggests the involvement of the Calvin–Benson cycle and P-serine pathway
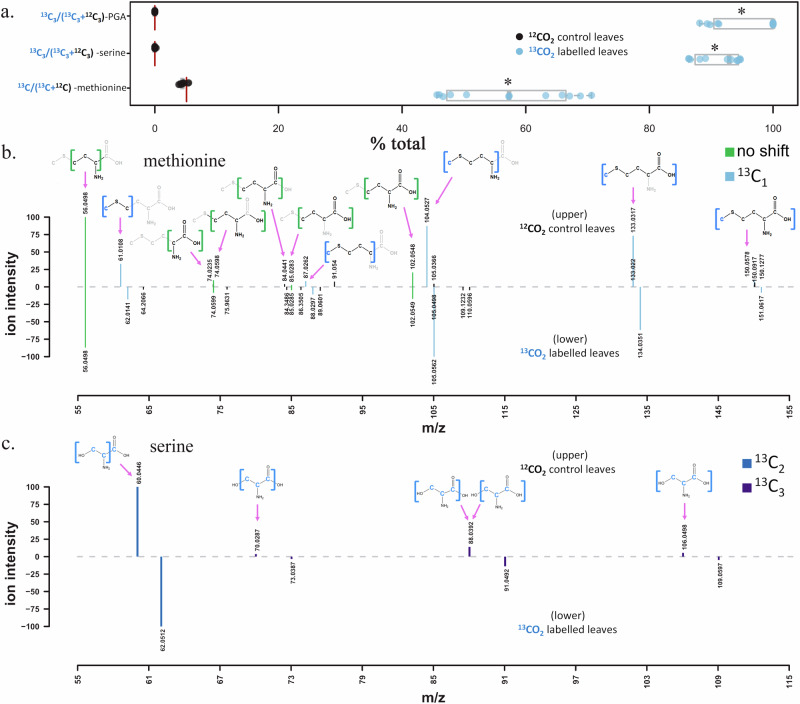
Following 13CO2-labeling period under a 21% O2 atmosphere, liquid chromatography mass spectrometry (LC-MS/MS) analysis of extracted bulk leaf phosphoglyceric acid (3-PGA and/or 2-PGA), serine, and methionine showed significant 13C-incorporation relative to control leaves exposed to 12CO2 (Fig. 4a). PGA, an intermediate of the Calvin-Benson cycle25, and serine, a product of the plastidic phosphorylated serine pathway9, showed near complete labeling on all three carbon atoms (total relative abundance 13C3/(13C3 + 12C3) of 95.8 ± 1.5% and 91.2 ± 1.0%, respectively). In addition, in leaves exposed to 13CO2, the 13C/12C-methionine abundance ratio of singly 13C-labeled methionine to unlabeled methionine was highly elevated (57.2 ± 2.8%). In contrast, the 13C/12C methionine abundance ratio from control leaves exposed to 12CO2 was 4.55 ± 0.17%, consistent with the value expected from natural 13C-abundance (5.13% as 13C1/(13C1 + 13C0)). MS/MS analysis of methionine extracted from leaf samples identified 10 fragments observed in the natural abundance (12C-methyl)-methionine and (13C-methyl)-methionine standards (Supplementary Fig. S10), also previously reported in the literature26. The molecular ion and all four fragments containing the sulfur-bonded methyl group (CH3-S) showed 13C-incoproration (m/z 61.0108, 87.0262, 104.0527, 133.0317). In contrast, all 6 fragments without the sulfur-bonded methyl group were not 13C-labeled (Fig. 4b and Supplementary Fig. S10), also previously reported in the literature26. The molecular ion and all four fragments containing the sulfur-bonded methyl group (CH3-S) showed 13C-incoproration (m/z 61.0108, 87.0262, 104.0527, 133.0317). In contrast, all 6 fragments without the sulfur-bonded methyl group were not 13C-labeled. For example, the methionine [CH3SCH2]+ fragment containing the sulfur-bonded methyl group showed 13C-incorporation (m/z 61.0106 → 62.0105) whereas the [C4H8NO2]+ fragment, which does not contain the sulfur-bonded methyl group, did not (m/z 102.0549) (Fig. 4b and Supplementary Fig. S10). These observations reveal that the 13C-incorporation in methionine during photoassimilaton of 13CO2 occurred in the sulfur bonded methyl group, (13C-methyl)-methionine. All serine fragments showed expected shifts in the fragmentation spectrum of the 13CO2 labeled leaves based on the predicted fragment structures and 13C-labeling of all 3 carbon atoms (Fig. 4c), while no shifts were observed in leaves exposed to 12CO2 (Supplementary Fig. S10). 2-phosphoglyceric acid is known to elute on our methods at a similar retention time as 3-phosphoglyceric acid, and produce similar fragmentation spectra, thus we could not differentiate them in these samples, and no fragmentation spectra were collected for PGA due to the low abundance in the samples. However, mediated by a plastidic phosphoglycerate mutase, photosynthetically generated 3-PGA is known to be converted to 2-PGA in the light27.
Fig. 4. LC-MS/MS 13C-labeling analysis of phosphoglyceric acid (PGA), serine, and methionine extracted from P. trichocarpa leaves following 2-day gas exchange studies under a 13CO2 and 21% O2 atmosphere.
a % total abundance ratio plot of [13C3/(13C3 + 12C3)]-PGA, [13C3/(13C3 + 12C3)]-serine, and [13C/12C]-methionine from control leaves exposed to a natural abundance 12CO2 atmosphere (n = 10), and leaves exposed to 13CO2 over two diurnal cycles (n = 12). The red vertical lines are the theoretical natural abundance ratios and * indicates significant difference by one tailed t test at P < 0.001. PGA was confirmed by RT, and m/z comparison with pure reference standard, while serine and methionine were confirmed with RT, m/z and MSMS. Also shown are the mirror plots comparing fragmentation patterns of leaf methionine (b) and serine (c) from 12CO2 and 13CO2 exposed leaves. Serine and methionine carbon atoms within each fragment are shown as previously determined26. Green fragments indicate matching ions within 15 ppm and blue and purple fragments indicate that the lower ion is within 15ppm of the expected 13C-labeled fragment isotopologue (see legend).
The molecular ion and all four fragments containing the sulfur-bonded methyl group (CH3-S) showed strong 13C-incoproration (m/z 61.0108, 87.0262, 104.0527, 133.0317). In contrast, all 6 fragments without the sulfur-bonded methyl group were not 13C-labeled (Fig. 4b and Supplementary Fig. S10). For example, the methionine [CH3SCH2]+ fragment containing the sulfur-bonded methyl group showed strong 13C-incorporation (m/z 61.0106 → 62.0105) whereas the [C4H8NO2]+ fragment, which does not contain the sulfur-bonded methyl group, did not (m/z 102.0549) (Fig. 4b). These observations reveal that the majority of 13C-incorporation in methionine during photoassimilaton of 13CO2 occurred in the sulfur bonded methyl group, (13C-methyl)-methionine.
In situ methanol emission rate in the field is mostly linked to air temperature
Diurnal branch gas exchange on P. trichocarpa individuals in the field showed a tight temporal relationship between air temperature and sap velocity, as well as branch gas exchange rates including net photosynthesis, transpiration, and methanol emissions during the early, mid, and late-growth season (April, May, and June 2023, Supplementary Figs. S11–S13, respectively). In April, in early growth season, night temperature was relatively low (i.e. 10 °C) and little to no methanol emission was observed. However, significant night-time methanol emission could be observed in May and June, although night temperatures kept relatively low (i.e. 3.0 nmol m−2 s−1 at 12 °C). Methanol emissions tended to peak around midday (i.e. 13:00) whereas net photosynthesis, transpiration, and sap flow continued at elevated rates until later in the day (i.e. showing a relatively broad peak centered at 16:00). Therefore, during late afternoons, methanol emissions were low while photosynthesis remained elevated (along with sap velocity and air temperature). When put together, the dynamic branch gas exchange datasets showed that methanol emission increased exponentially with air temperature (Supplementary Fig. S14). Similar results were obtained when diurnal leaf gas exchange time series under controlled environmental conditions were compiled during April and June as bar plots (Supplementary Figs. S15 and S16). It was visible that methanol emission increased with temperature (and thus transpiration) throughout the mornings, peaked and declined in the late afternoon while temperature and transpiration remained high.
Discussion
To date, in vivo studies on plant serine and C1 metabolism have focused on mitochondrial glycine-to-serine conversion during photorespiration, associated with THF-mediated methyl transfer28,29. For example, in a previous study on photorespiration and sulfur metabolism using 13CO2 labeling of sunflower (Helianthus annuus) leaves in the light30, high-resolution 33S and 13C tracing with nuclear magnetic resonance (NMR) spectroscopy and LC-MS demonstrated a direct and rapid (2 h) 13C-incorporation mostly in the methyl group (C-5 atom) of methionine; using various CO2/O2 ratios, it was shown that methionine C-5 labeling was directly dependent on RuBisCO-catalyzed oxygenation29. However, although the methyl group of methionine can be redistributed to other compounds and thus participate in cellular C1 metabolism, products of C1 metabolism were not followed in that study. These results are consistent with studies using metabolic flux analysis, modeling, and gas exchange measurements that suggest a fraction of photorespiratory carbon can be exported as serine under photorespiratory conditions31–33. Moreover, recent findings show that the photorespiratory pathway could contribute to C1 metabolism; Disruption of mitochondrial SHMT1 boosted glycine and C1 carbon (in the form of 5,10-methylene-THF) flux out of the photorespiratory cycle11. Thus, it is important to note that photorespiratory C1 carbon flux could be a substantial source of C1 units, especially during abiotic stress conditions like heat waves and droughts that stimulate photorespiration while suppressing photosynthesis.
Here, we used 13CO2-labeling to monitor the isotope enrichment not only in leaf methionine, but also in PGA, serine, PMEs, and methanol, acetaldehyde, and isoprene emissions under conditions that promoted high rates of photosynthesis while suppressing photorespiration (e.g. moderate light, optimal leaf temperatures, elevated 13CO2, and 1% O2). Our results show that PMEs and methanol are 13C-labeled even in the absence of photorespiration, suggesting that the plastidic phosphorylated serine pathway7,9 rather than photorespiration, plays the role of a light-dependent ‘photosynthetic C1’ pathway initiated by RuBisCO-catalyzed carboxylation. That is, CO2 fixation in the light can be a direct carbon source for cellular methyl transfers mediated by the methyl group of methionine. We nevertheless recognize that despite the strong 13C-labeling in both PMEs and methanol in the light under non-photorespiratory conditions (1% O2), it was slightly lower than that obtained under 21% O2 (i.e. 28% compared with 32-42% after 2 diurnal cycles, Fig. 1). This effect was likely caused by (i) a slight inhibition of photosynthetic 13CO2 fixation by O2 mole fraction below 2%34, (ii) an acceleration of fermentative metabolism at night (here, elevated nocturnal 13C2-acetaldehyde emissions)35, and (iii) reduced methanol dynamics (oscillations) at night-time, perhaps suggesting reduced growth rates (Fig. 1b).
The results demonstrate that the synthesis of PMEs via the light-dependent ‘photosynthetic C1 pathway’ is independent of the release of methanol from PMEs during temperature-stimulated (day) as well as hydraulically-driven (night) growth processes. Thus, methanol emissions require light-dependent processes of C1 photosynthesis leading to synthesis of new PMEs on the primary cell wall, and light-independent growth processes involving cell wall expansion, pectin demethylation, and methanol production. This implies that PME synthesis and export and incorporation into the primary cell wall occurs only during the day while PME demethylation and methanol production occurs during both the day and night, impacted by temperature and plant hydraulics.
Metabolic route from photosynthetic carbon to methanol
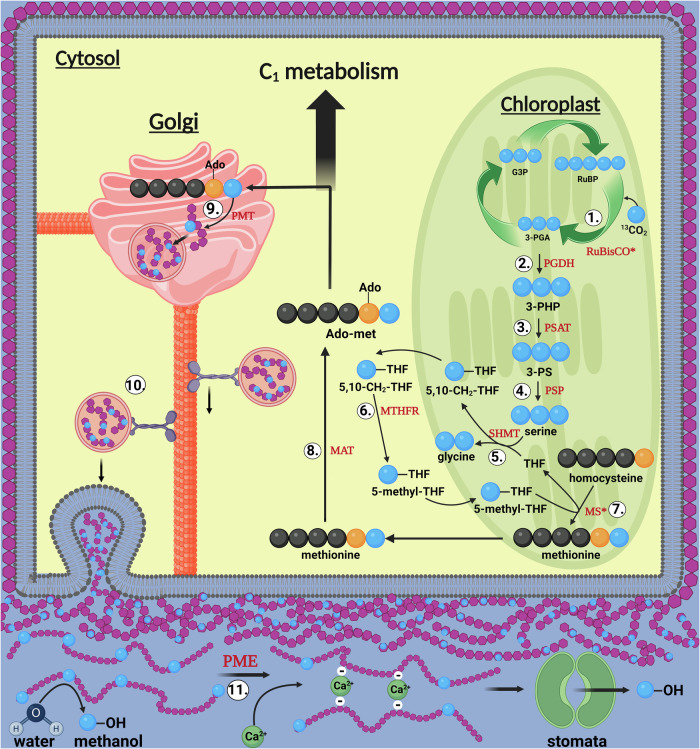
Methionine 13C-labeling was strong only in the C-5 atom position (methyl group, Fig. 4a, b and Supplementary Fig. S10), demonstrating the importance of THF-mediated C1 metabolism. There was a dominance of 13C3 isotopologues in both PGA and serine (more than 90% of total, Fig. 4a, c), consistent with the conversion of 3-PGA from the Calvin-Benson cycle to serine as the source of the methyl group (C-5 atom) of methionine1,9. The very high 13C-enrichment in PGA and its direct products was also suggested by isoprene 13C-enrichment. As observed in other studies36,37, more than 95% of isoprene emitted was fully 13C-labeled (13C5 isotopologue) within minutes of placing the leaf into a 13CO2 atmosphere in the light under elevated 13CO2 (Fig. 2). Isoprene is not stored in plants and is produced via the isoprenoid pathway in chloroplasts which requires C3 carbon skeletons synthesized by the Calvin-Benson cycle (i.e. 3-PGA)36. A rapid conversion of 3-PGA to 2-PGA via plastidic phosphoglycerate mutase in the light27 supports substrate flow to pyruvate and acetyl-CoA during light-dependent isoprenoid38 and fatty acid synthesis39, respectively. Moreover, the observation of strong near complete 13C2-labeling of acetaldehyde emissions during light-dark transitions supports the previous suggestions of a pyruvate overflow mechanism where photosynthetically generated pyruvate in the light is decarboxylated to acetaldehyde via aerobic fermentation upon sudden darkening, mediated by pyruvate decarboxylase40–42. Under the 1% O2, this process was amplified resulting in nearly 20-fold increase relative to 21% O2 with highly elevated 13C2-acetaldehyde persisting the entire night. Acetaldehyde oxidation to acetate is considered an effective alternative route to acetyl-CoA synthesis used in respiration as a part of a so called pyruvate dehydrogenase bypass pathway43, and O2 concentrations of 2% or lower negatively impact aerobic respiration, with typical symptoms of hypoxia44,45 such as an upregulation of fermentation metabolism41. Also, the 13C-enrichment in PMEs (including under 1% O2) is consistent with the involvement of the phosphorylated serine pathway since the methyl group of methionine (and thus AdoMet) and the oxygen-bonded methyl group of PMEs are directly linked via a series of reactions (Fig. 5). A proposed biochemical model of the ‘Photosynthetic C1 pathway’ involves seven distinct enzymatic steps followed by AdoMet regeneration in the cytosol as a major source of cellular methyl transfer reactions in the light (see Fig. 6 and Supplementary Note 2: Biochemical steps of the ‘Photosynthetic C1 pathway’). Here, chloroplast-localized methionine synthase18 (step 7) is crucial to transfer a C1 unit from 5-methyl-THF to methionine. After activation to AdoMet (step 8), methyl esterification of new pectin monomers can occur via pectin methyltransferase46 (step 9). Further utilization (transport, export, and incorporation to new PMEs in growing primary cell walls47, step 10) and methanol production during pectin demethylation (step 11)46 can then take place. Our results effectively show that it was possible to trace photosynthetic carbon (13C) utilization through C1 metabolism including methanol liberation (Fig. 1).
Fig. 5. Simplistic model of the ‘Photosynthetic C1’ pathway and methanol emission during growth.
The ‘Photosynthetic C1’ pathway (steps 1–7) results in light-dependent methionine synthesis followed by the activation of methionine to AdoMet in the cytosol (step 8) and its utilization as a major source of cellular methyl transfer reactions including in the Golgi during the methylation of newly synthesized pectin polysaccharides (step 9). Following the export and incorporation of newly synthesized, highly methylated pectin into the primary cell wall (step 10), cell wall expansion during growth processes is regulated by pectin demethylation (step 11). Pectin methyl ester hydrolysis results in the change in cell wall elasticity via formation of calcium pectinate “egg-box”99 and release of methanol into the apoplast. Methanol partitions between the plant aqueous and gas phases and rapidly escapes the plant via stomatal emission to the atmosphere. Blue balls represent carbon atoms recently assimilated by the Calvin–Benson cycle during photosynthesis as CO2; Black balls represent stored or non-recently assimilated carbon atoms; Orange balls represent the sulfur atom of homocysteine, methionine and AdoMet. For abbreviations of metabolites and enzymes, see Fig. 6 for a more detailed schematic of the ‘Photosynthetic C1’ pathway as well as Note 2 in the supplementary information document. Created in BioRender. K. Jardine (2023) BioRender.com/p03i521.
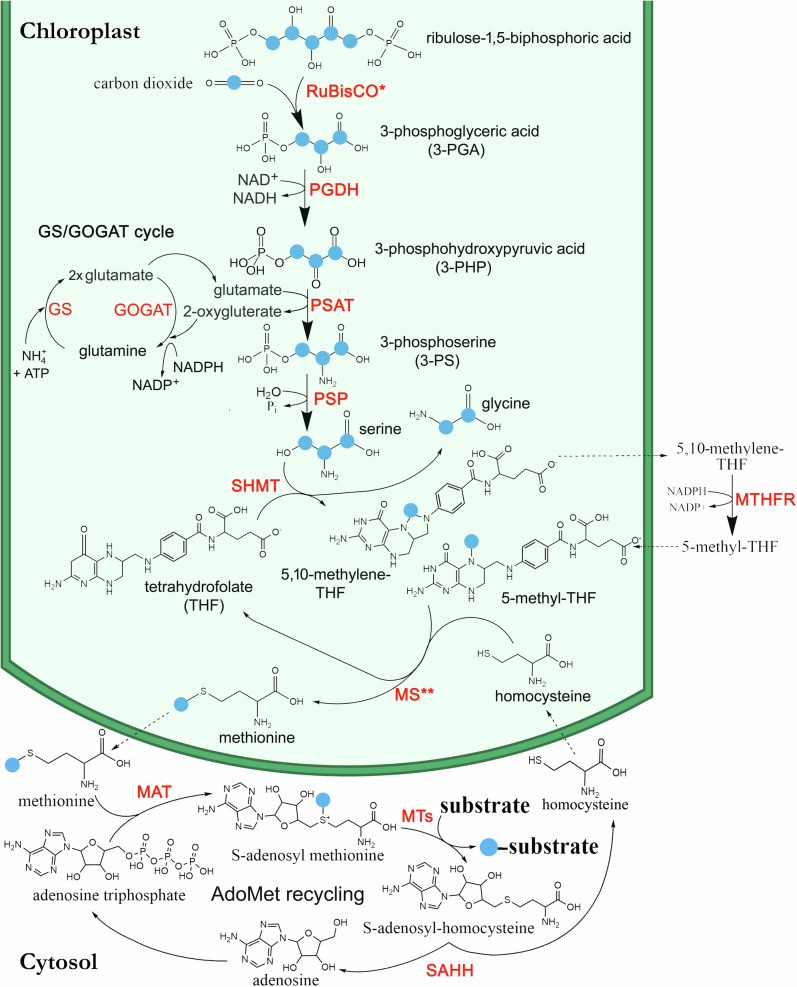
Fig. 6. Proposed biochemical model of the ‘Photosynthetic C1 pathway’ in California poplar.
Photosynthetic C1 pathway initiated in chloroplasts by the Calvin-Benson cycle linked to NH4+ assimilation via the glutamine synthetase/glutamine oxoglutarate aminotransferase (GS/GOGAT) cycle11. Enzymes abbreviations shown in red include (1) ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), (2) 3-phosphoglycerate dehydrogenase (PGDH), (3) 3-phosphoserine aminotransferase (PSAT), (4) 3-phosphoserine phosphatase (PSP), (5) Serine hydroxymethyltransferase (SHMT), (6) methylenetetrahydrofolate reductase (MTHFR), and (7) methionine synthase (MS). PGDH, PSAT and PSP are enzymes of the phosphorylated serine pathway9. All enzymes of the photosynthetic C1 pathway and GS/GOAGT cycle are chloroplast localized, except MTHFR which is thought to be in the cytosol. Also shown is the AdoMet recycling pathway in the cytosol as a source of cellular methylation reactions with enzymes including methionine adenosyltransferase (MAT), methyltransferases (MT), and S-adenosyl-homocysteine hydrolase (SAHH). The blue balls represent carbon atoms recently assimilated by RuBisCO via the Calvin-Benson cycle during photosynthesis as CO2. Note, the regeneration of THF and homocysteine intermediates, and the lack of ATP and NADPH requirements for methionine synthesis in the chloroplast. However, the GS/GOAGT cycle for NH4+ assimilation requires both ATP and NAPDH. * Large kinetic isotope effect leading a natural 13C-depletion of C3 biomass relative to CO2. ** Large kinetic isotope effect leading to natural 13C-depletion of C1 carbon pools relative to C3 biomass.
Interestingly, there was a considerable difference in %13C between metabolic end products examined here, namely PGA, serine, isoprene, acetaldehyde, PMEs and methanol. This effect was simply due to isotopic dilution (and pool size) along metabolic pathways. In particular, PGA, serine, isoprene and acetaldehyde were highly labeled while methanol and PMEs showed some isotopic dilution. The methyl group of methionine itself was at about 60% 13C under our conditions (Fig. 4a and Supplementary Fig. S10) and preexisting methyl groups in PMEs also participated in methanol production (Fig. 1). Although six of seven enzymes of the ‘photosynthetic C1 pathway’ are chloroplast localized, 5-methyl-THF synthesis catalyzed by MTHFR is thought to occur in the cytosol, with the export of 5,10-methylene-THF out of chloroplasts together with 5-methyl-THF import into chloroplasts likely mediated by one or more chloroplastic folate transporters48. Thus, export of strongly 13C-labeled 5,10-methylene-THF may be diluted by pre-existing 5,10-methylene-THF in the cytosol, resulting in a decrease in 13C-lableing of the methyl group of methionine (~60%) relative to serine (>90%). Methanol emission could continue at night in the absence of photosynthesis due to the large reservoir represented by PMEs in the primary cell wall49 and continued growth processes at night. Thus, 13CO2 photosynthetic fixation during the day allowed for the 13C-label to rapidly enter the PME pool (within 5 min), detectable in methanol emissions within 1 h (the initial, naturally 13C-depleted signature of PMEs is discussed further in Supplementary Note 3: Large, natural, and universal 13C depletion of plant C1 pools). While the cause of the natural 13C-isotopic anomaly in the leaf C1 pool remains unclear, we propose that the sequential events of natural discrimination against 13C during CO2 diffusion and carboxylation by RuBisCO followed by methionine synthesis50 as the last step of the photosynthetic C1 pathway imparts an extreme 13C-depleted carbon isotope signature on the S-methyl group of methionine, and consequently all substrates that receive C1 carbon during their synthesis or regulation via methyl transfer from AdoMet (see Fig. 6 and Note 3). Thus, the photosynthetic C1 pathway may be responsible for the large carbon isotope anomaly observed in major leaf C1 carbon pools51. However, due to the large pool size of preexisting 12C-PMEs at the beginning of the labeling experiments, 13C-labeling was still incomplete after 5 d with 13C/12C-methanol emission ratios reaching up to 80% (13C-methanol emissions representing 45% of total methanol emissions). This is in strong contrast to isoprene emissions, a known photosynthetic product that is not stored in plants36. Within minutes with elevated 13CO2, leaf isoprene emissions were dominated by the fully labeled (13C5-isotopologue) isoprene molecular species (Fig. 2).
Relationship between PMEs, methanol production and eco-physiological parameters
We found a high resilience of the 13C/12C isotope ratio in emitted methanol with respect to time (i.e. rather stable ratio at night) (Fig. 1 and Supplementary Figs. S1–5) and temperature (Supplementary Fig. S9), suggesting that the metabolic pool feeding methanol production (i.e. PMEs on the primary cell wall) was large enough to dampen variations due to new PME synthesis in the light. In fact, after the first day of 13CO2 labeling, methanol emission rates at night were not associated with a strong decline in the isotope ratio in methanol during the night (Fig. 1a and Supplementary Figs. S1–5), as would be expected if methanol emission represented a significant fraction of total active PMEs pool. Moreover, in leaves previously exposed to 13CO2 and placed in a 12CO2 atmosphere, the isotope ratio in methanol remained elevated and stable despite the considerable increase in individual 13C- and 12C-methanol emission rate with temperature (Supplementary Fig. S9). The dependence of methanol emission rate on temperature (Supplementary Figs. S9 and S14) is consistent with previous observations of high temperature-sensitivity of foliar methanol emissions in many plant species52 as well as direct observations of the temperature sensitivity of PMEs hydrolysis and methanol release in hydrated leaf cell wall extracts49. Our results also show that methanol emission occurs at night, likely possible due to high night-time stomatal conductance in P. trichocarpa leaves, and enhanced methanol emission starting at around midnight was associated with a recovery in leaf water potential; this was followed by an increase until midday and then reduction of afternoon methanol emission associated with increased leaf water stress (Fig. 1a, Supplementary Figs. S1–S5, S11–S13, and S15, S16).
Importantly, our results suggest that methanol emission is connected to photosynthesis metabolically (via 3-PGA and the phosphorylated serine pathway, see above) but also numerically, with a linear response of 13C-content in emitted methanol to cumulated fixed 13CO2 (Fig. S8). This is in contrast with previous studies that suggested little direct connection between photosynthesis and methanol emission, considering the large reservoir of methylated pectin potentially independent of photosynthesis, and continued methanol emissions at night49. Here, 13CO2 labeling showed a rapid (within 5 min) metabolic connection between PMEs and CO2 assimilation (Fig. 3).
Methanol emissions observed in the present study in poplar leaves are reminiscent of another C1 volatile molecule produced during photosynthesis, methane. Light-dependent methane production and emission from 13C fixed photosynthetically have recently been observed in cyanobacteria53. In that study, stable isotope techniques were used to demonstrate methane production during oxygenic photosynthesis in the light directly from 13CO2. In land plants, while metabolic mechanisms are still under investigation, previous studies demonstrated that methane production and emission can be linked to the metabolism of the methyl group of methionine54–56. Given that cyanobacteria also possess the phosphorylated serine pathway6, one possibility is that in both land plants and cyanobacteria, methane production is linked to photosynthesis via the ‘photosynthetic C1’ pathway and light-dependent methionine biosynthesis. Since methane is not readily soluble and thus expected to be liberated, methane emission should be dependent on photosynthetic carbon fixation in the light. Accordingly, a recent study of methane emission by shoots of Scotts pine trees showed a pronounced diurnal cycle that closely followed incident photosynthetically active radiation57.
Potential implications of the “photosynthetic C1” pathway
Since C1 metabolism and methanol emission was mostly independent of photorespiration and reflected photosynthetic carbon fixation as well as primary cell wall expansion, one may hypothesize that methanol production and emission can be used as an indirect indicator of growth. After mid-night, branch emissions of both 12C-methanol and 13C-methanol under a 21% O2 atmosphere increased steadily throughout the night (Fig. 1a and Supplementary Figs. S1–5), suggesting an important link to tissue water status (see graphical representation in Supplementary Fig. S17). This is consistent with PME hydrolysis liberating methanol linked to changes in primary cell wall elasticity and growth, which may be downregulated during tissue water stress, possibly via enhanced activity of pectin methyl esterase inhibitors58. In other words, it highlights the potential for methanol emission as a non-invasive chemical tracer of plant water status and growth processes across temporal and spatial scales (this is further discussed in Supplementary Note S4: Methanol emission: a metabolic biomarker of plant physiological status?).
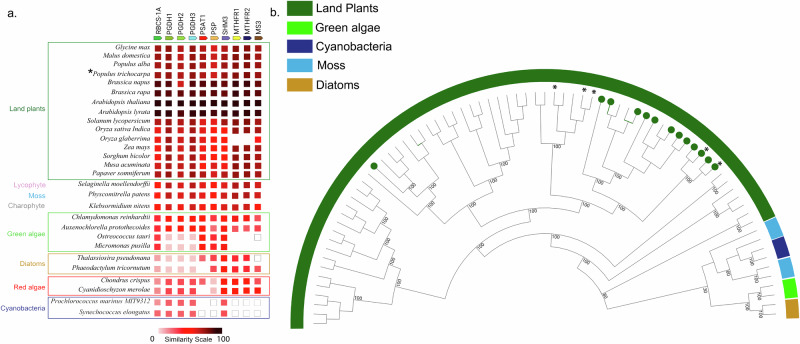
Considering the central role of the ‘photosynthetic C1’ pathway in synthesis of the methyl donor group of AdoMet, our results raise the question as to whether enzymes of this pathway are well-represented in the green lineage. A comparative genomics approach was used to explore the molecular evolution of the seven genes comprising the ‘photosynthetic C1’ pathway leading to light-dependent methionine synthesis across photosynthetic lineages. Representative orthogroups for enzymes of the model plant A. thaliana C1 pathway were identified using the STRING-DB59. The co-occurrence analysis reveals that, within land plants, proteins in the C1 pathway are present with high similarity (~60–100%) (Fig. 7a). Beyond land plants, photosynthetic C1 proteins were present throughout photosynthetic organisms including in a lycophyte, a moss, a charophyte, green algae, diatoms, red algae, and cyanobacteria. Availability of more genomes will improve the understanding of this pathway, especially in lycophytes, mosses and charophytes where only a single species was represented in the database. Intriguingly, some green algae, which are at the base of Viridiplantae (the green lineage) and ancestral to land plants, do not have methylene tetrahydrofolate reductase (MTHFR) nor methionine synthase (MS) detected in their genomes, and we confirmed these absences manually (see Methods). It suggests that in the green lineage, enzymes of chloroplastic C1 metabolism may have a shared common ancestor that was lost in the early diverging green algae (e.g. Ostreococcus) but was retained in the later diverging chlorophytes (e.g. Chlamydomonas). Surprisingly, we also observed a gene fusion event of serine hydroxymethyltransferase (SHMT) and MS sequences in the green alga Chlorella sorokiniana suggesting that the two sequences are translated into a single bifunctional polypeptide and act in the same pathway. Many cyanobacterial C1 proteins had to be extracted from Cyanobase (white boxes) because they did not belong to the same orthogroup as algae and land plants suggesting that the domain was not conserved. The absence of a protein may be due to improper genome annotation or an incomplete genome (e.g. MTHFRs in Oryza glaberrima).
Fig. 7. Photosynthetic C1 pathway protein phylogeny and evolution across cyanobacteria and algae to land plants.
a A co-occurrence plot for the presence of orthologs for the seven proteins of the C1 pathway from representative species (see Fig. 6). Similarity between orthologous proteins is represented by color with a color scale ranging from white (low similarity) to black (high similarity) (see scale bar). A white box indicates that the sequence was detected or reported in literature but did not belong to the same orthogroup whereas a lack of a box indicates that the ortholog was not detected. Asterisk highlights P. trichocarpa. b Phylogenetic tree of methionine synthase (MS) sequences from representative species. This tree is based on 76 amino acid sequences from 22 species and constructed using the maximum likelihood method (see Supplementary Data 2 for species and sequences). Sequences with predicted chloroplast localization based on transit peptide prediction have a green circle at the edge of the nodes. Asterisk indicates nodes of P. trichocarpa MS sequences. Bootstrap values (percentage of 1000 bootstraps) for selected nodes are indicated at the branch point.
Because MS is the key enzyme responsible for generating methionine for the ‘photosynthetic C1’ pathway (with a large kinetic isotope effect, contributing to explain the natural 13C depletion50, see Supplementary Notes S3), we performed a phylogenetic analysis of MS from representative species across photosynthetic lineages. MS of land plants form a distinct clade that branch from the basal clade which includes diatoms, green algae, and cyanobacteria (Fig. 7b). Land plants have multiple copies of MS and more than half (8 of 15 species) had a copy with predicted chloroplast localization. Since the ‘photosynthetic C1’ pathway is predicted to occur in the chloroplast, the phylogenetic clustering of most chloroplastic MS sequences may suggest a common, cyanobacterial origin of the ‘photosynthetic C1’ pathway in land plants. In A. thaliana, all proteins of the pathway except MTHFR have been found in the chloroplast proteome60,61. Altogether, these results suggest that most enzymes of the ‘photosynthetic C1’ pathway are found across photosynthetic lineages and that those within Angiosperms have high protein similarity, suggesting a conserved function in generating light-dependent synthesis of methionine.
Conclusions and perspectives
Our results show that C1 metabolism at the origin of PMEs and methanol depends on RuBisCO-catalyzed carboxylation via the phosphorylated serine pathway. As such, methanol is linked to photosynthesis and partly independent of photorespiration. Considering the value of C1 metabolism in support of biopolymer and metabolite synthesis and regulation, we presume that the carbon flux through the ‘photosynthetic C1’ pathway may be significant relative to the rate of carboxylation. Nevertheless, future studies should focus on characterizing this photosynthetic C1 flux in relation to carboxylation and how it varies with instantaneous environmental variables that impact photosynthesis (i.e. CO2, light, temperature, soil moisture, etc.) and biological variables such as the maximum carboxylation velocities and growth rates as they vary across plant development and early and late successional species, as well as relationships with gene expression patterns. Although limited information is available on the expression patterns of the seven genes of the photosynthetic C1 pathway in leaves, a study of leaf gene expression in natural P. trichocharpa trees in the field during the growing season showed that most genes of the photosynthetic C1 pathway have a ~4X increase during the growth phase (May)62. Although we used conditions that promote high rates of photosynthesis while suppressing photorespiration, we were not able to discriminate serine produced form photorespiration versus the phosphorylated serine pathway. Assuming that serine can move freely between organelles, one important area of research is the allocation of serine from photorespiration versus the phosphorylated serine pathway to C1 metabolism and other major sinks like protein synthesis15. Given that the SHMT reactions operate in opposite directions in photorespiration (glycine and 5,10-methylene-THF converted to serine) versus C1 photosynthesis (serine converted to glycine and 5,10-methylene-THF), exchange of substrates and products between these pathways should be investigated. This includes CO2 and NH4+ generated from photorespiration which may be re-assimilated by C1 photosynthesis linked to the GS/GOGAT cycle in chloroplasts (Fig. 6). Even though photorespiration generates high amounts of serine in plants, serine derived from the phosphorylated serine pathway appears to be more important for plant growth and its deficiency triggers the induction of nitrogen assimilation as an amino acid starvation response15. This may be because the photosynthetic C1 pathway is a critical biosynthetic pathway required for the synthesis of numerous biopolymers and metabolites leading to the gain of carbon and nitrogen, whereas photorespiration is largely a recycling pathway for Calvin-Benson cycle intermediates important for abiotic stress signaling (via H2O2) associated with the loss of carbon (CO2) and nitrogen (NH4+)5. It is important to point out that the production of methanol and other C1 pools via the ‘photosynthetic C1’ pathway proposed here is distinct from the previously described oxidative C1 pathway17,63. The latter is initiated by methanol oxidation to formaldehyde and formic acid which can then integrate into photorespiration via 5,10-methylene-THF formation64. Moreover, formic acid can be oxidized to CO2 which can be refixed and thus support photosynthesis65. It is possible that the synthesis of AdoMet via the ‘photosynthetic C1’ pathway, ammonia assimilation via the glutamine synthetase/glutamine oxoglutarate aminotransferase (GS/GOGAT) cycle15, photorespiration, the release of methanol during cell wall expansion, and methanol recycling mechanisms via the oxidative C1 pathway, operate concurrently during the day (involving photosynthesis and photorespiration) and/or separately during the night (methanol emissions at night).
Another important question that warrants more research work is the way the ‘photosynthetic C1’ pathway changes (and acclimates) with rising CO2 mole fraction. Depending on stomatal conductance and photosynthetic capacity, elevated CO2 tends to enhance photosynthesis and decrease photorespiration. Therefore, the provision of C1 units including for sulfur assimilation to methionine should decline and perhaps, the ‘photosynthetic C1’ pathway plays an increasingly important role in both carbon and nitrogen assimilation15. Potentially mediated by methionine phloem loading and transport, we also note that it may have consequences for whole plant growth responses as well as microbial methanotrophy and methylotrophy above ground in the phylosphere66 and below ground in the rhizosphere67. It is generally understood that elevated CO2 tends to suppress biosynthetic pathways like the isoprenoids synthesis in chloroplasts that are unable to compete with the Calvin-Benson cycle for ATP and NADPH68,69. Importantly, the ‘photosynthetic C1’ pathway from RuBisCO-catalyzed carboxylation to methionine synthesis by MS has no requirement for ATP and generates one NADH. Although the MTHFR reaction in the cytosol consumes NADPH, the lack of ATP and NADPH requirements in the chloroplast means that in principle, the flux of methionine synthesis could increase together with the Calvin-Benson cycle as atmospheric CO2 mole fraction increases. However, it is important to note that ammonia assimilation in chloroplasts via the GO/GOGAT cycle, which directly links with C1 photosynthesis by providing glutamate for the formation of 3-phosphoserine (3-PSP) while consuming 2-oxygluterate, requires ATP and NADPH (catalyzed by PSAT, see Fig. 6). Our observations suggest that the ‘photosynthetic C1’ pathway, which appears to be highly conserved in land plants (Fig. 7) may play a critical, yet poorly understood role in enhancing net primary productivity of C3 plants as atmospheric CO2 continues to rise, and therefore might represent a missing link between photosynthesis and growth of natural and managed forests under a changing climate (see Supplementary Video 1, The global photosynthetic C1 Pathway and plant growth).
It should be kept in mind that while the total flux associated with carbon utilization from 3-PGA to overall C1 metabolism may be large under elevated CO2, it remains uncharacterized. Future research should quantify the carbon flux through the photosynthetic C1 pathway in relation to rates of RuBisCO carboxylation, carbohydrate, isoprenoid, and fatty acid synthesis, and its impact on 3-PGA and RuBP regeneration. Efforts to improve photosynthesis have shown that overexpression of a plastidic glyceraldehye-3-phosphate dehydrogenase (GAPDH), which dedicates triosphosphates to glycolysis, limits the regeneration of RuBP, but slightly increases photosynthesis rates under elevated CO2 in rice70. RuBisCO overexpression has also been considered as method to improve photosynthesis rates, but has generally been shown to not lead to substantial photosynthesis improvement, possibly due to over-accumulation of 3-PGA71. However, simultaneous overexpression of RuBisCO and the glycolytic enzymes GAPDH and triosephosphate isomerase (TPI) only slightly improved photosynthesis at elevated CO2, It was concluded that these modifications did not alleviate over-accumulation of 3-PGA71. Given its role in both CO2 assimilation and utilization of 3-PGA, future work could evaluate if the overexpression of the seven enzymes of the ‘photosynthetic C1 pathway’ under elevated CO2 leads to more substantial improvements in photosynthesis by more effectively alleviating 3-PGA over-accumulation. Indeed, recent studies have shown that in most eukaryotic algae, RuBisCO is located in a microcompartment known as the pyrenoid in association with CO2-concentrations mechanisms that promote photosynthesis over photorespiration72. Moreover, the second enzyme of the photosynthetic C1 pathway, PGDH which catalyzes the commitment step of serine synthesis, is located to puncta directly adjacent to the pyrenoid, likely acting as metabolic channel to enhance serine biosynthesis by capturing 3-PGA exiting the pyrenoid73.
Given that the demonstrated role of the methyl group of methionine in methane production and emission in plants, the photosynthetic C1 pathway provides a biochemical mechanism for light-dependent production and emission of methane observed from cyanobacteria53 and trees57 and may be expanded with additional studies to other C1 volatiles like methanethiol and dimethyl sulfide74. Thus, this resolves the controversy for many years that plants produce methane75–77 versus the widespread view that vascular plants do not possess a biochemical pathway to produce methane78, but rather only act as a conduit for microbially produced methane in soils to the atmosphere79. Thus, the presence of a ‘photosynthetic C1 pathway’ across the photosynthetic tree of life may have major implications for not only understanding the evolution of oxygenic photosynthesis80, but also represents the basis for the development of predictive Earth system models that simulate natural greenhouse gas emissions from marine and terrestrial ecosystems to the atmosphere and their feedback with the climate system.
Methods
Greenhouse plants
We used 15 potted California poplar (Populus trichocarpa) saplings (average height of 2 meter) obtained from Plants of the Wild (Washington State, USA) and maintained for three years in the South Greenhouse at the Oxford Tract Experimental Facility in Berkeley, CA, USA, where they were regularly watered and subject to standard pest control practices. Water was delivered to each individual using an automated watering system in 15 gal pots containing Supersoil planting media (Scotts Co., Marysville, Ohio, USA). Ambient natural light was supplemented with LED lighting using an Argus Titan environmental control system with photocell (Argus Controls, British Columbia, Canada). The controller was programmed to turn LED lights off when detecting exterior light levels above 850 μmol m−2 s−1 during the 16-h photoperiod (6:00 to 22:00).
Methanol, acetaldehyde, and isoprene 13C-labeling analysis by PTR-MS
For all leaf and branch gas exchange experiments, a high sensitivity quadrupole proton transfer reaction- mass spectrometer (PTR-MS with QMZ 422 quadrupole, Balzers, Switzerland) was utilized for real-time flux and stable carbon isotope analysis of methanol, acetaldehyde and isoprene emissions as previously described36,42,81. The PTR-MS was operated with a drift tube voltage of 440 V and pressure of 1.8 mbar. For each measurement cycle lasting 24 s, the following mass to charge (m/z) ratios were monitored including m/z 21 (H318O+), 32 (O2+), m/z 37 (H3O+-H2O), m/z 33 (H+-12C-methanol), m/z 34 (H+-13C-methanol), m/z 45 (12C2-acetaldehyde), m/z 47 (13C2-acetaldehyde), m/z 69 (H+-12C5-isoprene), m/z 70 (H+-13C1-isoprene), m/z 71 (H+-13C2-isoprene), m/z 72 (H+-13C3-isoprene), m/z 73 (H+-13C4-isoprene), and m/z 74 (H+-13C5-isoprene). Quantification of the methanol, acetaldehyde, and isoprene concentrations was based on dynamic dilution of a primary gas standard (1,000 ppb methanol and isoprene in nitrogen, Restek Corporation). Calibration curves were generated for m/z 33 (methanol), m/z 45 (acetaldehyde), and m/z 69 (isoprene) for 0-45 ppb generated by dynamic dilution of the primary standard (1000 ppb, Restek, Inc.) with hydrocarbon-free air. 13C/12C-methanol emission ratios were calculated as the ratio of 13C-methanol/12C-methanol and expressed as a % whereas 13C5-isoprene emissions were expressed as a % of total isoprene emissions (12C5-isoprene + 13C5-isoprene) emissions.
13CO2 leaf labeling under optimal environmental conditions for photosynthesis
In order to study 13CO2 labeling of the putative photosynthetic-C1 pathway in poplar leaves under constant environmental conditions and short time periods, a portable photosynthesis system (Li6800, Li-COR Biosciences) with a 36 cm2 leaf chamber and light source (6800-03, Li-COR Biosciences) was coupled to a high precision cavity ringdown spectrometer (CRDS) for 13CO2 (G2131-i, Picarro Inc.) and quadrupole PTR-MS inside the greenhouse laboratory (see configuration diagram in Supplementary Fig. S18a). Leaves were maintained under constant environmental conditions of 1200 µmol m−2 s−1 photosynthetically active radiation (PAR), 32 °C leaf temperature, and an air flow rate entering the leaf chamber of 538 ml min−1 maintained with humidity between 4 and 10 mmol mmol−1. A small fraction of air exiting the chamber was diverted to a CRDS for determination of 13CO2 concentrations (25 ml min−1) and a PTR-MS (75 ml min−1) for determination of 12C-methanol and 13C-methanol leaf emissions. CO2-free zero air (ultra-zero air, CAS No: 132259-10-0, Linde Gas) was delivered to the Li6800 system after passing through a catalytic converter system (ZA30 catalyst, Aadco instruments, USA) to oxidize any trace volatile organic compounds present. A 13CO2 cylinder (99% 13CO2, CAS No: 1111-72-4, Sigma-Aldrich Inc., USA) was used to supply the internal CO2 injector using the CO2 tank adapter kit (part number 9968-109, Li-COR-Biosciences). An elevated 13CO2 concentration in the leaf chamber was maintained between 900 and 1100 ppm 13CO2 (determined by CRDS) throughout each leaf labeling experiment in the light. Net fluxes of 13CO2 photoassimilation (µmol 13CO2 m−2 s−1) were calculated as a function of time in the leaf chamber as A (t) based on Eq. (1) where µ is the constant air flow rate entering the leaf chamber (400 mol air s−1), Δ13CO2 (t) is the difference in 13CO2 mole fraction between leaf chamber and reference air (µmol mol−1) as a function of time, and S is leaf surface area (0.0036 m2) placed inside the chamber.
| 1 |
Leaf 13CO2 labeling studies lasting 1 h (n = 5), 2 h (n = 2), 3 h (n = 1), 4 h (n = 2), and 5 h (n = 2) were conducted together with 13C/12C-methanol emission ratios analysis. On the morning of the experiment, poplar branches were detached from one of the 15 potted California poplar trees in the greenhouse in the morning (9:00–11:00) and immediately immersed and recut under water and transferred to the nearby greenhouse laboratory. A selected leaf from the branch was placed inside the leaf chamber. To minimize branch water loss through transpiration and maintain hydrated leaves, a mylar sheet was placed over the branch outside of the leaf chamber and moist paper towels were placed inside around the branch. This was found to be important for minimizing leaf water stress where high leaf transpiration rates occurred over long periods of time (e.g. up to 5 h). For each leaf 13CO2 labeling experiment, 13C/12C-methanol emission ratios were plotted versus cumulative 13CO2 photoassimilation (µmol 13CO2 m−2). For a single leaf sample (5 h), 13C-methanol labeling was verified using TD-GC-MS (see supplementary methods, ‘TD-GC-MS verification of leaf methanol 13C-labeling during 13CO2 photosynthesis’, for experimental details).
Diurnal 13CO2 branch labeling of intact potted saplings
Diurnal branch 13CO2 labeling occurred over a 2–5-day period in the plant growth area in the real-time volatile metabolomics laboratory at the Oxford greenhouse facilities in Berkeley, CA (see configuration diagram in Supplementary Fig. S18b). Potted P. trichocarpa trees were transported to the nearby laboratory from the greenhouse and placed under a full spectrum grow light (Bestva Diammble Pro 4000 W, USA) approximately 1 m above the top branches of the tree. The light intensity was maintained constant (700–900 µmol m−2 s−1 of photosynthetically active radiation, depending on the tree) during the light period (6:00–20:00). Tap water was automatically added to the soil every 1 h for 20 s (200 ml) using an automated watering system (smart watering device, Model: Sw-C03-KA, sPlant Technology Co., Ltd.). A transparent branch chamber (Restek, 10 L ALTEF gas sampling bag, Catalog#: 22962) was continuously supplied with 2.75 l min−1 of CO2-free zero air (ultra-zero air, CAS No: 132259-10-0, Linde Gas) which first passed through a catalytic converter system (ZA30 catalyst, Aadco instruments, USA) to oxidize any trace volatile organic compounds present in the high purity air. A small flow (3.0 ml min−1) of 99% 13CO2 (Sigma-Aldrich Inc., CAS No:1111-72-4, USA) was mixed into the 2.75 l min−1 dilution flow of zero air just prior to the catalytic converter to generate an expected 13CO2 concentration in the empty branch chamber of 1090 ppm. CO2 as well as isoprene and methanol concentrations and 13C/12C ratios were monitored in real-time inside the branch chamber by CRDS and PTR-MS, respectively. While the majority of the air flow entering the chamber passed through the enclosure (2653 ml min−1), a small fraction was diverted through a 1/8” O.D. Teflon PFA tubing to the CRDS (25 ml min−1) and PTR-MS (75 ml min1). Following the initiation of the 13CO2 flow, background CRDS and PTR-MS data were collected for several hours without a branch inside the enclosure. A branch near the top of the tree canopy was subsequently placed carefully inside the chamber during the night period, and branch gas exchange measurements were initiated. In the light, 13CO2 was drawn down inside the branch chamber between 700 and 900 ppm, depending on the branch studied. The experiment was repeated on five different potted poplar trees.
For all but one diurnal branch 13CO2 labeling experiment, a 21% O2 atmosphere was used and occurred over 2 days (5 different trees, 1 branch per tree) and 5 days (1 tree, 1 branch). For a single 2-day branch 13CO2 labeling experiment, a 1% O2 atmosphere was used in order to suppress photorespiration34. During the single 2-day 13CO2 branch label experiment under a 1% O2 atmosphere, a high precision O2 cavity ringdown laser spectrometer (G2207-i, Picarro Inc.) was used to both verify the 1% O2 concentrations in the dynamic branch headspace atmosphere and to demonstrate net O2 production during photosynthesis in the light.
Verification of leaf methanol 13C-labeling during 13CO2 photosynthesis by TD-GC-MS
During branch 13CO2 labeling of potted P. trichocarpa plants, 13C-labeling of methanol was verified using thermal desorption gas chromatography-mass spectrometry (TD-GC-MS). Briefly, roughly 1.0 l of air sample from inside the dynamic branch enclosure was transferred to an empty Tedlar gas sample bag using a diaphragm pump set to 100 ml min-1. The gas sample bags were connected to the air server-xr system (Markes International, UK) via tubing to allow the TD-GC-MS system to collect and analyze each sample for methanol 13C/12C analysis. Air samples (25 ml min−1 × 10 min: 0.25 l) were first dried by passing the air sample through the Kori-xr held at −20 oC before the methanol was quantitatively pre-concentrated onto the cold trap (Air toxics, Markes International, UK) held at −30 oC with the sample flow path maintained at 150 oC. Following the sample pre-concentration step, the collected volatiles were injected onto the analytical column, by rapidly heating the cold trap to 280 oC for 3 min while back-flushing with carrier gas at a flow of 7.5 ml min−1. In order to improve peak shape and further reduce the amount of water introduced into the GC-MS, 6.0 ml min−1 of this flow was vented through the split while the remaining 1.5 ml min−1 was directed to the column, temperature programmed with an initial hold at 40 °C for 4 min followed by an increase to 70 °C at 4 °C min−1. A post run temperature of 230 °C was applied for 5 min. The mass spectrometer was configured for trace analysis (SIM Mode and 10X detector gain factor) with 25 ms dwell times for the target ions of methanol (m/z 15, 16, 28, 29, 30, 31, 33). Methanol showed a retention time of 7.8 min, verified using a standard.
13C-labeling analysis of metabolites by LC-MS/MS
For three of the 2-day P. trichocarpa branch 13CO2 experiments as well as control branches not exposed to 13CO2, 13C-labeling patterns of leaf metabolites were determined using hydrophilic liquid interaction chromatography and tandem mass spectrometry (LC-MS/MS). Following the 13CO2 labeling period, the leaves were removed from the plant, flash frozen and ground with mortar and pestle under liquid nitrogen, and stored at −80 °C. Frozen partially homogenized leaf tissues were freeze-dried (Labconco) overnight and then bead-milled using methanol washed 3.2 mm stainless steel beads in a Mini-Beadbeater-96 (BioSpec Products). Milling was done for a total of 45 s total in 5 s intervals separated by 10 s breaks to prevent overheating of sample; samples were re-cooled every 3 milling intervals. Powderized samples were refrozen and then suspended in 70% methanol with internal standards (see details in Supplementary Data 1) at 5 µl/mg dry tissue. Samples were then vortexed 2 ×10 sec, bath sonicated in ice water for 15 min, and centrifuged at 10,000 G (5 min, 10 °C); supernatants were 0.22-µm filtered in microcentrifuge filter tubes, and filtrates were transferred to amber glass vials for LC-MS/MS analysis. Metabolites were separated on an Agilent 1290 HPLC equipped with a hydrophilic liquid interaction column and detected with a Thermo QExactive Hybrid Orbitrap Mass Spectrometer; method parameters are described in Supplementary Data 1. Raw files were converted to MZML using ProteoWizard MSConvert 3.0.2126582 or ThermoRawFileParser version 1.4.2. Data were evaluated and peak heights for methionine and singly 13C-labeleled methionine were extracted using MZmine 2.083. MS2 spectra were extracted using the RaMS package version 1.3.184 and plotted in base R 4.3.085; vector-based PDFs were generated using Cairo 1.6. From a centroided mzml file, the most intense MS2 spectrum within a 5-ppm window of the precursor m/z for each file was selected and then filtered to remove background intensity fragment ions; a 15-ppm window was used to determine if sample fragment ions matched when comparing MS2 spectra (for example between leaf tissue and standard reference compound). Samples metabolite retention times, mass-to-charge (m/z) ratios and fragmentation spectra were compared with authentic reference standards when available.
13C-labeling analysis of methyl esters from purified leaf cell wall extracts
Following a period of cumulative photosynthesis under a 13CO2 atmosphere with individual leaves under optimal environmental conditions for photosynthesis and branches under diurnal light/dark periods, leaves were detached and immediately flash frozen in liquid nitrogen. For cell wall methyl ester 13C-labeling analysis, two replicate leaves were labeled with 13CO2 for 5 min, 15 min, 30 min, 1 h, and 3 h before flash freezing. In addition, leaves of two intact branches were labeled with 13CO2 for 2 days and 5 days, respectively, before flash freezing all mature leaves (5–8 leaves) on the branch together in liquid nitrogen (leaf samples pooled into a single average branch sample). Two replicate leaves never exposed to the 13CO2 atmosphere were also flash frozen as controls. Whole leaf cell walls were isolated from the frozen leaf tissues through the generation of alcohol insoluble residues (AIR) as previously described86. Briefly, control and 13CO2-labeled frozen poplar leaves were kept frozen on dry ice and ground with a pestle and mortar before placing one ground frozen leaf (0.2–0.3 g) in a 50 ml Falcon tube with 96% ethanol at 70 °C. The Falcon tube was then immediately placed in a 70 °C water bath for 60 min before cooling to room temperature. Homogenized tissue was allowed to settle, and the excess solvent was removed by pipetting and ~250 μl of the resulting semi-solid ‘sludge’ was transferred to a 2-ml microcentrifuge tube and washed with 96% ethanol by vortexing for 1 min and centrifuging for 15 min at max speed (10,000–15,000 G). The supernatant was removed and the same solvent washing procedure was performed on all samples using 100% ethanol before washing with methanol:chloroform, 2 v:3 v two times in a shaker held at 4 °C for 1 h. Leaf cell wall extracts were then washed with 100%, 65%, 80%, and again 100% ethanol before being dried at room temperature for 4 h using a speedvac to generate the AIR samples. Dried whole cell wall samples (AIR) where then stored at room temperature and shipped to Heidelberg University, Germany for analysis of methyl ester 13C-labeling patterns as described in the methods section below, ‘Methyl ester 13C-labeling analysis of isolated whole leaf cell walls by GC-C-IRMS’.
Methyl ester 13C-labeling analysis of isolated whole leaf cell walls by GC-C-IRMS
Methyl ester 13C-labeling analysis of leaf AIR samples was achieved by conversion to CH3I using a 57% aqueous solution of hydriodic acid (Acros, Thermo Fisher Scientific, Geel, Belgium) followed by stable carbon isotope ratio analysis of CH3I with gas chromatography-combustion-isotope ratio mass spectrometry (GC-C-IRMS) as previously described87,88. Hydriodic acid (0.25 ml) was added to AIR samples (3-5 mg) in crimp-top glass vials (1.5 ml; IVA Analysentechnik, Meerbusch, Germany). The vials were sealed with crimp caps containing PTFE-lined butyl rubber septa (thickness 0.9 mm) and incubated for 30 min at 130 °C. After heating, the samples were allowed to equilibrate at room temperature (22 ± 0.5 °C) for at least 30 min before 30–50 µl of the headspace were directly injected into the GC using a 100 μl gas-tight syringe (SGE Analytical Science). δ13C values of CH3I were measured using an HP 6890 N gas chromatograph (Agilent, Santa Clara, USA) equipped with an auto sampler A200S (CTC Analytics, Zwingen, Switzerland), coupled to a MAT253 isotope ratio mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) via an oxidation reactor [ceramic tube (Al2O3), length 320 mm, 0.5 mm i.d., with Cu/Ni/Pt wires inside (activated by oxygen), reactor temperature 960 °C and a GC Combustion III Interface (ThermoQuest Finnigan, Bremen, Germany). The GC was fitted with a Zebron ZB-5MS capillary column (Phenomenex, Torrance, USA) (30 m × 0.25 mm i.d., df = 1 μm) and the following GC conditions were employed: split injection (10:1), initial oven temperature at 40 °C for 3.8 min, ramp at 50 °C/min to 110 °C. High-purity helium (5 N) was used as carrier gas at a constant flow of 1.8 ml min−1. A tank of high-purity carbon dioxide (99.995% or N45, Air Liquide, Düsseldorf, Germany) was used as the monitoring gas. All stable carbon isotope ratios of methyl esters are expressed in the conventional ‘delta’ (δ) notation, meaning the relative difference of the isotope ratio of a substance compared to the standard substance Vienna Peedee Belemnite (VPDB). The δ13C values were calibrated with linear regression using reference materials HUBG1 and HUBG289. Both materials were measured by EA‐IRMS and calibrated to the VPDB scale using the reference material IAEA‐603 (+2.474 ± 0.05‰) and an in‐house standard (acetanilide; −30.06 ± 0.20‰), which, in turn, was calibrated against the two reference materials NBS22 (−30.03 ± 0.08‰) and USGS44 (−42.21 ± 0.10‰). The calibrated values for HUBG1 and HUBG2 are −50.21 ± 0.08‰ (N = 14) and +1.61 ± 0.05‰ (N = 16), respectively.
Diurnal observations of leaf water potential and leaf and branch gas exchange in field grown P. trichocarpa in Berkeley, CA, USA during the 2023 growing season
During three individual days during the growing season (25-April-2023, 19-May-2023, and 30-June-2023) diurnal measurements of leaf water potential occurred together with leaf gas exchange including CO2/H2O fluxes with methanol and isoprene emissions. Leaf water potential (LWP, KPa) was manually measured from one leaf from five different California poplar trees during midnight (23:30 and 00:30), pre-dawn (5:00–6:00), mid-day (11:30–12:30), and afternoon (14:00–15:00) periods. The leaf was removed from the tree by cutting the petiole with scissors as long as possible. After cutting the petiole, LWP was immediately determined using a Scholander pressure chamber with nitrogen (Model 1005 with digital display, PMS instrument company). As soon as water emerged from the petiole top, the nitrogen pressure increase was immediately stopped, and the chamber pressure value (KPa) noted. LWP was recorded as negative value of the determined pressure. During the same three days, diurnal measurements of leaf gas exchange fluxes in the field were collected manually using the small leaf chamber (6 cm2). The Li6800 console was supplied with hydrocarbon free air generated by passing ambient air through a catalytic converter (ZA30-120, Aadco Inc.). Environmental conditions maintained in the leaf chamber throughout the day include constant air flow through the chamber (470 ml min−1), leaf chamber CO2 concentrations (400 ppm), and photosynthetically active radiation (red: 990 µmol m−2 s−1, blue 10 µmol m−2 s−1). Reference humidity was not regulated, and varied slightly based on the lab conditions between 6 and 12 mmol mol−1. A fraction of the air exiting the chamber (75 ml min−1) in the field was diverted to the PTR-MS in the laboratory through 1/8” O.D. Teflon PTFA tubing for continuous analysis of leaf methanol and isoprene concentrations. Leaf temperature was increased by 1 °C for each leaf inside the leaf chamber and ranged from a minimum of 20 °C for measurement at sunrise and up to 38 °C for the afternoon measurement in 30-June-2023. Leaf gas exchange was measured between 6:00 and 17:00 from a single California poplar tree and consisted of setting the leaf temperature without a leaf inside the chamber and collecting background isoprene and methanol concentration data (10 min) before placing a sun-exposed leaf inside the chamber and waiting for leaf gas exchange to stabilize (10 min) and logging net photosynthesis (Anet, µmol m−2 s−1), transpiration (E, mmol m−2 s−1), and stomatal conductance (mol m−2 s−1) data together with isoprene and methanol concentration data (ppb) for 15 min. Following this, the leaf was removed and the procedure was repeated on another sunlit leaf on the same tree. Leaf methanol and isoprene concentration changes were calculated by subtracting background concentrations (empty leaf chamber) from concentrations with a leaf in the chamber. Isoprene and methanol fluxes (nmol m−2 s−1) were calculated from the concentration changes, air flow rate through the chamber, and leaf area (6 cm2).
Branch gas exchange measurements of CO2, H2O, methanol, and isoprene fluxes from a single California poplar tree in the field were continuously quantified under ambient environmental conditions of light and temperature during the early (13–24 April), mid (25–31 May), and late (13–28 June) 2023 growing season. A dynamic branch enclosure (5 Liter Tedlar Bag, CEL Scientific Corporation, Los Angeles, CA, USA) was installed around a poplar tree branch together with a second empty Tedlar enclosure installed near the tree as a control. 5.0 l min−1 hydrocarbon free air continuously passed through both branch and control enclosures with ambient levels of CO2 and H2O, with trace volatile organic compounds (VOCs) removed by catalytic oxidation of laboratory air (ZA30-120, Aadco Inc.). The enclosure was secured around the stem at the base with air entering the top through the ¼” valve port and venting at the bottom where the enclosure was secured to the stem (4875 ml min−1), with 125 ml min−1 diverted to two real-time gas sensors for quantification of (1) CO2 and H2O branch headspace concentrations (Li7000, Li-Cor Biosciences, Lincoin, NE, USA), (2) methanol and isoprene branch headspace concentrations via high sensitivity quadrupole Proton Transfer Reaction-Mass Spectrometer (PTR-MS, Ionicon, Austria). The Li7000 was operated in absolute mode with the reference cell continuously provided 50 ml min−1 of ultra-high purity nitrogen (5.0 purity, Praxair, Danbury, CT, USA) and designated as 0 ppm CO2 and 0 mmol mol−1 H2O while 50 ml min−1 of air from inside the branch enclosure was directed to the Li7000 sample cell with average CO2 and H2O concentrations recorded every min. Using an automated solenoid valve, concentration measurements were made every hour of the empty branch enclosure (20 min) and the enclosure with the poplar branch inside (20 min). CO2, H2O, isoprene, and methanol fluxes each hour were determined by the difference in concentrations between the dynamic branch enclosure and empty chamber, the air flow through the chambers, and the enclosed leaf area, estimated by tracing the leaves paper and using the determined ratio of the paper area/weight ratio.
Co-occurrence analysis
We collected the protein sequences from Phytozome V1390 for the following A. thaliana enzymes AtRBCS1A (RuBisCo small subunit), AtPGDH1, AtPGDH2, AtPGDH3, AtPSAT1, AtPSP, AtSHMT3, AtMTHFR1/2, and AtMS3 (Supplementary Data 2). All the genes except the AtMTHFR have a known/predicted chloroplast transit peptide and have proteomic experimental evidence confirming chloroplast localization60,61. However, it should be noted that depending on specific gene expression patterns, proteins could be plastid-localized in heterotrophic cells and/or chloroplast-localized in autotrophic cells. Consistent with the 60-amino acid leader sequence acting as a transit peptide for transportation of A. thaliana PGDH to plastids91, all three proteins (AtPGDH1, AtPGDH2, AtPGDH3) were found to be present in the A. thaliana chloroplast proteome61 and are therefore included in the co-occurance analysis. Using STRING-DB V12.0 (Search Tool for Retrieval of Interacting Genes/Proteins DataBase)59, orthogroups of proteins of the above-listed genes in the C1 pathway were identified across all organisms available in STRING-DB. The STRING-DB classified orthologous sequences into specific orthogroups and proteins from other species were extracted based on their specific groups. STRING-DB uses genomes available in the NCBI database and has significant representation of angiosperm genomes; however, only a single lycophyte, moss, and charophyte species was currently present in the STRING-DB. Based on their similarity score, a co-occurrence figure was made for selected species. For each protein, all sequences were manually confirmed to belong to a specific orthogroup. Protein absences of PSAT1, MTHFR, and MS in green algae, diatoms and red algae were confirmed manually using BLAST of A. thaliana proteins in Phycocosm92. Absences of cyanobacterial proteins were validated in Cyanobase93 and sequences for present enzymes were extracted.
Phylogenetic analysis
To construct a phylogenetic tree, all protein sequences from the methionine synthase (MS) orthogroup were extracted from the STRING-DB59 and manually curated to filter sequences specific to two diatoms, two green algae, one moss and 15 land plants (dicots and monocots). Two cyanobacterial MS sequences were extracted from Cyanobase93. In total, 76 sequences from 22 species were used to construct the tree (see Supplementary Data 2). A multiple sequence alignment (MSA) of the selected protein sequences was performed using the msa() function94 with the ClustalW method in R version 4.2.2. A maximum likelihood (ML) tree was generated using the phangorn package in R95. Based on the MSA, various models were tested for the estimation of likelihood to the provided sequences with the modelTest() function and the best available model was selected to construct a ML tree using the pml_bb() function. To improve confidence, the tree was bootstrapped a 1000 times using the default ultrafast bootstrapping in the phangorn() package. The tree was plotted using the ggtree package in R96. The sequences were checked for localization within the chloroplast by investigating the presence of transit peptide in TargetP97 and prediction of plastid localization using DeepLoc2.0 (0.9 cutoff)98. Sequences with either transit peptides or predicted plastid localization were manually annotated on the tree.
Field experiment
A field experiment was carried out throughout the 2023 growing season to explore the seasonal pattern in diurnal growth, methanol emissions, transpiration, and temperature in an established field plot adjacent to the analytical trace gas laboratory. Detailed protocols used for ‘Diurnal observations of leaf water potential and leaf and branch gas exchange’ can be found in the supplementary information.
Field plants
Twenty California poplar saplings (Populus trichocarpa) were planted on May 24, 2021 in the Oxford Tract Experimental Facility field site. California poplar saplings were obtained from Plants of the Wild, WA, USA. Automated watering was configured during the growing season (April-December), such that 0.5 gal h−1 was delivered to each tree for 1 h each day at 8:00. However, during the warm summer months (June through October) watering duration increased to 1.5–3 h in order to maintain relatively constant soil moisture. Prior to the commencement of experimentation in April 2023, the trees were grown in the field for 689 days (1 year, 10 months, and 20 days) yielding a height of 2.0–2.6 m.
Environmental monitoring of air temperature, humidity, and sap velocity
Starting on 31-Jan-2023 in the poplar tree field site, air temperature was continuously recorded at 1.5–2.0 m above the ground every 5 min using type-T thermocouples and a temperature data logger (OM-CP-OCTTCTEMP-A2, Omega Engineering). Moisture of the soil (0-50%) was determined weekly throughout the field experiment manually for each tree using a Extech MO750 Soil Moisture Meter with Integrated 8” Heavy Duty Probe inserted 10 cm into the soil. In addition, ambient air relative humidity and temperature were continuously recorded every 10 min at 1.0 m height above the ground with a HOBO MX2300 Series Temp, RH data logger in the center of the field plot adjacent to the ambient air temperature thermocouple. Sap velocity was continuously monitored at 50 cm height on the main stem of one California poplar (CP02) using the heat ratio method (SFM1 sap flow meter, ICT International). The SFM1 sensor was configured as previously described39. Briefly, heat pulses (30 J) were programmed every 15 min and stem sap velocity (sapwood) was recorded.
Software and Code
Commercial software used to control and collect data form the following instruments were used as follows;
• Online volatile organic compound concentrations were collected the commercial PTR-MS software for quadrupole mass spectrometers (PTR-MS Control software, Ionicon Analytic)
• Online 12CO2, 13CO2 (isotopic CRDS) and high precision O2 CRDS concentrations data were collected using the commercial Picarro Surveyor software (Picarro Inc.)
• Online leaf photosynthesis, transpiration, and stomatal conductance data as well as controlled environmental variables were collected using Bluestem OS. Version 2.1.13 on a Li6800 portable photosynthesis system (Licor, Inc.)
• Branch level fluxes of leaf photosynthesis and transpiration were acquired using a Li7000 under computer control using the Li-7000 Control software (Licor, Inc.)
• Extracted leaf metabolite data using LC-MS/MS were collected using Thermo Xcalibur software.
• Verification of leaf methanol 13C-labeling by GC-MS was performed using Masshunter version 11.1 (Agilent)
• Data for analysis of methyl ester 13C-labeling patterns of isolated whole leaf cell walls by GC-C-IRMS was collected using Isodat (Thermo)
• STRING-DB V12.0 (Search Tool for Retrieval of Interacting Genes/Proteins DataBase) was used to identify orthogroups of proteins of the seven genes in the C1 pathway were identified across all organisms available in STRING-DB.
• Phylogenetic tree construction: Protein sequences from the methionine synthase (MS) orthogroups were extracted from the STRING-DB and manually curated to filter sequences specific to two diatoms, two green algae, one moss and 15 land plants (dicots and monocots). Two cyanobacterial MS sequences were extracted from Cyanobase. In total, 76 sequences from 22 species were used to construct the tree (see Supplementary Data 2).
• Environmental data (air temperature) during field studies were collected using Omega Data Logger Software (ODLS): A software platform for configuring, downloading, and analyzing data from the OM-CP-OCTTCTEMP-A2 thermocouple data logger.
• Sap flow data was configured and downloaded from SFM1 sap flow sensors using Sap Flow Tool version 3.1.4 (ICT International).
Data analysis
Leaf and branch gas exchange time series data were analyzed using Igor Pro software (Version 8.0).
Leaf metabolite analysis: Raw LC-MS files were converted to MZML format using either ProteoWizard MSConvert (Version 3.0.21265) or ThermoRawFileParser (Version 1.4.2). Data were evaluated, and peak heights for methionine and singly 13C-labeled methionine were extracted using MZmine (Version 2.083). MS2 spectra were extracted using the RaMS package (Version 1.3.184) and plotted in base R (Version 4.3.0). Vector-based PDFs were generated using the Cairo package (Version 1.6).
Statistical analysis of leaf metabolites (LC-MS/MS) was performed using R (Version 4.3.0).
Phylogenetic tree construction for methionine synthase (MS): A multiple sequence alignment (MSA) of the selected protein sequences was performed using the msa() function with the ClustalW method in R (Version 4.2.2). A maximum likelihood (ML) tree was generated using the phangorn package. Various models were tested for likelihood estimation using the modelTest() function, and the best-fitting model was selected to construct the ML tree using the pml_bb() function. The tree was bootstrapped 1000 times to improve confidence using the ultrafast bootstrapping default in phangorn. The tree was plotted using the ggtree package in R. Protein sequences were further examined for chloroplast localization by identifying transit peptides with TargetP and plastid localization predictions using DeepLoc2.0 (0.9 cutoff). Sequences with either transit peptides or predicted plastid localization were manually annotated on the tree.
Statistics and Reproducibility
For all leaf and branch experiments involving replicates, biological replicates were defined as independent samples derived from different individuals or experimental units (e.g., separate leaves or branches on a tree). All replicate studies were successful, and no data were excluded from the field study. Reproducibility was confirmed by performing independent repeats of experiments on different days under identical conditions or in the field by performing repeat experiments at different times throughout the growing season.
15 potted poplar trees were used for leaf (1–5 h) and branch (2–5) day 13CO2 labeling under conditions which promoted high rates of net photosynthesis while suppressing photorespiration. The group of 15 potted poplar trees grown in the greenhouse and used for leaf and branch 13CO2 labeling studies were randomly selected for either leaf gas exchange and cell wall studies (detached branches) or branch gas exchange (intact branches) coupled with leaf metabolite analysis. During the branch gas exchange studies, a random potted tree was selected and moved into the analytical laboratory and placed under an LED grow light and automated watering system (and allowed to acclimate for several days). The branch selected for continuous 2-5 day gas exchange studies during 13CO2 labeling was also randomly selected, but was required to be in the upper canopy so that it was not shaded by branches above it. All branches studied had a mix of young light green immature leaves (near the end of the branch) and darker green maturing leaves.
Leaf 13CO2 labeling studies under constant environmental conditions for short time periods (up to 5 h) promoting high rates of photosynthesis lasting 1 h (n = 5), 2 h (n = 2), 3 h (n = 1), 4 h (n = 2), and 5 h (n = 2) were conducted together with online 13CO2 assimilation quantification and 13C/12C-methanol emission ratios analysis. In addition, short term (0, 5, 15, 30, 60, and 180 min) 13CO2 leaf labeling (replicated 2x for each time point) was conducted followed by flash freezing leaves at the end of the experiment for bulk cell wall pectin methyl ester 13C-analysis. For cell wall methyl ester 13C-labeling analysis, two replicate leaves (n = 2) were labeled with 13CO2 for 5 min, 15 min, 30 min, 1 h, and 3 h before flash freezing. In addition, following 13CO2 branch labeling for 2 days and 5 days, mature leaves (5-8 leaves) on each of the branches were flash frozen in liquid nitrogen for PME 13C-labeling.
Branch 13CO2 labeling studies were performed under diurnal light/dark cycles and 21% O2 over 2 days with online 13CO2 assimilation quantification and 13C/12C-methanol emission ratios analysis and flash frozen at the end of the experiment for bulk cell wall pectin methyl ester and leaf metabolomics 13C-analysis. A branch 13CO2 labeling study with gas exchange and leaf PME 13C-labeling analysis was conducted over 5 days (n = 1) as well as under a 1% O2 atmosphere over 2 days (n = 1).
For leaf metabolomics 13C-labeling analysis of PGA, serine, and methionine were compared between control leaves exposed to a natural abundance 12CO2 atmosphere (n = 10), and leaves exposed to 13CO2 over two diurnal cycles (n = 12). One tailed t test (P < 0.001) was used for comparisons between 13C/12C leaf metabolite labeling patterns among the 12CO2 and 13CO2 treated leaf samples and all statistical analyses of leaf metabolites.
A population of 20 field grown poplar trees was used to study the temperature and hydraulic effects of methanol emissions. One tree was randomly selected for 1–2 week continuous branch gas exchange studies during April, June, and July 2023 as well as a single-day experiment at the leaf level on a separate individual to quantify sap velocity and leaf gas exchange together with leaf water potential (from 10 trees) at midnight, predawn, mid-day, and early afternoon.
For the co-occurrence analysis of proteins of the photosynthetic C1 pathway and phylogenetic analysis of methionine synthase across the photosynthetic tree of life, all orthogroups of proteins of the listed genes in the C1 pathway were identified across all organisms available in STRING-DB.
To construct a phylogenetic tree, all protein sequences from the methionine synthase (MS) orthogroup were extracted from the STRING-DB and manually curated to filter sequences specific to two diatoms, two green algae, one moss and 15 land plants (dicots and monocots). Two cyanobacterial MS sequences were extracted from Cyanobase. In total, 76 sequences from 22 species were used to construct the tree.
Sampling strategy
For all leaf and branch experiments involving replicates, biological replicates were defined as independent samples derived from different individuals or experimental units (e.g., separate leaves or branches on a tree). All replicate studies were successful, and no data were excluded from the field study. Reproducibility was confirmed by conducting independent repeats of experiments on different days under identical conditions or in the field at different times throughout the growing season.
For each of the leaf and branch 13CO2 labeling experiments, one out of 15 potted trees maintained in the greenhouse was randomly selected for study. These 15 trees were grown in the greenhouse and randomly chosen for either leaf gas exchange and cell wall studies (detached sun-exposed branches in the upper canopy) or branch gas exchange (intact branches) coupled with leaf metabolite analysis. During the branch gas exchange studies, a random potted tree was selected, moved into the analytical laboratory, and placed under an LED grow light and automated watering system (allowed to acclimate for several days). The branch selected for continuous 2–5 day gas exchange studies during 13CO2 labeling was also randomly selected but required to be in the upper canopy to avoid shading. All branches studied contained a mix of immature light-green leaves near the tip and more mature darker green leaves. All replicate leaf and branch 13CO2 labeling experiments showed a strict- light-dependence of 13C-labeling of pectin methyl esters in primary cell walls, leaf methanol emissions, and targeted leaf metabolites.
Leaf 13CO2 labeling studies under constant environmental conditions for short time periods promoting high rates of photosynthesis lasting 1 h (n = 5), 2 h (n = 2), 3 h (n = 1), 4 h (n = 2), and 5 h (n = 2) were conducted together with online 13CO2 assimilation quantification and 13C/12C-methanol emission ratios analysis. All leaf samples showed a very tight linear correlations (r2 > 0.97) between cumulative 13CO2 photoassimilation and the instantaneous 13C/12C-methanol emission ratio.
For cell wall methyl ester 13C-labeling analysis, leaves were labeled with 13CO2 for 5 min, 15 min, 30 min, 1 h, and 3 h under optimal conditions for net photosynthesis before flash freezing (replicated 2X per time point). In addition, leaves of two intact branches were labeled with 13CO2 for 2 days and 5 days, respectively, before flash freezing all mature leaves (5-8 leaves) on the branch together in liquid nitrogen (leaf samples pooled into a single average branch sample). Leaves on branches never exposed to the 13CO2 atmosphere were also flash frozen as controls for bulk cell wall pectin methyl ester 13C-analysis (n = 8). All leaf samples showed a tight linear correlation (r2 > 0.97) between δ13C of leaf PMEs and light period duration.
Long-term branch 13CO2 labeling studies (2 days) under diurnal light/dark cycles were repeated five times using randomly selected potted poplar trees. These experiments occurred under 21% O2, with conditions favoring photosynthesis over photorespiration (i.e. moderate light and temperature, high soil moisture, and elevated 13CO2). RuBisCO oxygenation (Vo) was estimated to be less than 5% of carboxylation (Vc) under these conditions. Each replicate showed similar light-dependent 13C-labeling patterns of methanol emissions, bulk leaf cell wall PMEs, as well as targeted leaf metabolites. To assess extended labeling, a single 5-day branch 13CO2 labeling study was performed on a randomly selected potted tree.
A 2-day branch 13C-labeling study under 1% O2 was conducted to effectively eliminate photorespiration (Vo < 0.5% Vc). Branch headspace was verified to contain 1% O2, and net O2 production was confirmed using CRDS during the light period. Light-dependent 13C-labeling patterns and methanol emissions, as well as 13C-labeling of bulk leaf cell wall pectin methyl esters, were only slightly reduced relative to branches exposed to 21% O2. This slight decrease was attributed to reduced photosynthesis under 1% O2 and the activation of fermentation metabolism.
In the field experiment, 1–2 weeks of continuous branch gas exchange studies were conducted on a single randomly selected tree. These branch gas exchange studies were repeated in April, June, and July 2023 (n = 3). When branch data from each period were combined, a clear exponential dependence of methanol emissions on temperature was observed. During April and June 2023, a second tree was randomly selected for high temporal resolution observations of leaf gas exchange and sap velocity, with leaf water potential measurements taken across 10 individuals at midnight, predawn, mid-day, and early afternoon.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors would like to thank Christina Wistrom for maintaining optimal growth conditions of poplar trees in the UC Berkeley Oxford Tract greenhouse and facilitating the integration of a real-time volatile metabolomics laboratory. We would also like to thank Christina Procopiou for the assistance with the C1 photosynthesis animation development and helpful discussions with Alister Rogers on AdoMet recycling mechanisms, and Dan Feldman on methanol remote sensing. This research was supported by the DOE Office of Science, Office of Biological and Environmental Research (BER), Early Career Research Project (ECRP) grant no. FP00007421 and the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by contract DE-AC02-05CH11231. Additional DOE funding for the research came from the Next Generation Ecosystem Experiments-Tropics (NGEE-Tropics) through contract no. DE-AC02-05CH11231 as part of DOE’s Terrestrial Ecosystem Science Program. The metabolomics analysis was supported by m-CAFEs Microbial Community Analysis & Functional Evaluation in Soils, (m-CAFEs@lbl.gov) a Science Focus Area at Lawrence Berkeley National Laboratory funded by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research DE-AC02-05CH11231. TFD received financial support from the Brazilian National Council for Scientific and Technological Development (CNPq) grant 312589/2022-0 (Bolsa de produtividade em Pesquisa). LBG received financial support from AUSPIN Edital 001/2022 – Programa de Bolsas de Intercâmbio Internacional.
Author contributions
These authors contributed equally to the development of the C1 photosynthesis metabolic illustrations and associated animation (K. Jardine and L. Gallo) and these authors contributed equally to collecting and/or analyzing the data and assisted in manuscript preparation and revising the work critically for important intellectual content (K. Jardine, L. Gallo, A. Eudes, M. Roth, S. Upadhyaya, T. Northen, S. Kosina, G. Tcherkez, T. Dominges, M. Greule, S. Som, and F. Keppler).
Peer review
Peer review information
Communications Biology thanks Roc Ros, Berkley Walker and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Xiaoling Xu and David Favero. A peer review file is available.
Data availability
All data needed to evaluate the conclusions in this paper including the source data of every figure and Supplementary Fig. is available for download as a supplementary file. All raw LC-MS/MS data For LC-MS data used in Fig. 4 and supplementary Fig. S10 are available in the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=aabec0a91ce449e58e843e60ff3e5021). Supplementary Data 1 (.xlsx file): Method details of the leaf methionine analysis from frozen leaf extracts separated using hydrophilic liquid interaction chromatography. Supplementary Data 2 (.xlsx file): 76 protein sequences from 22 species used to construct the phylogenetic tree shown in Fig. 7 with protein ID, Genus and species, and plant category. Supplementary Data 3 (.zip file): Data set for all data-based figures including Fig. 1a, Fig. 1b, Fig. 2, Fig. 3, and supplementary Figs. S1, S2, S3, S4, S5, S6, S7, S8, S9, S11, S12, S13, S14, S15, and S16.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07142-0.
References
- 1.Benson, A. & Calvin, M. The dark reductions of photosynthesis. Science105, 648–649 (1947). [DOI] [PubMed] [Google Scholar]
- 2.Haverd, V. et al. Higher than expected CO2 fertilization inferred from leaf to global observations. Glob. Change Biol.26, 2390–2402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pachauri, R. K. & Reisinger, A. IPCC Fourth Assessment Report 044023 (IPCC, 2007) (2007).
- 4.Gamage, D. et al. New insights into the cellular mechanisms of plant growth at elevated atmospheric carbon dioxide concentrations. Plant Cell Environ.41, 1233–1246 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Voss, I., Sunil, B., Scheibe, R. & Raghavendra, A. Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol.15, 713–722 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Colman, B. & Norman, E. G. Serine synthesis in cyanobacteria by a nonphotorespiratory pathway. Physiologia Plant.100, 133–136 (1997). [Google Scholar]
- 7.Ros, R., Muñoz-Bertomeu, J. & Krueger, S. Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci.19, 564–569 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Wulfert, S. & Krueger, S. Phosphoserine Aminotransferase1 is part of the phosphorylated pathways for serine biosynthesis and essential for light and sugar-dependent growth promotion. Front. Plant Sci.9, 1712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, C.-L. & Saito, K. Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids20, 243–259 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Benstein, R. M. et al. Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell25, 5011–5029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosa-Téllez, S. et al. The serine–glycine–one-carbon metabolic network orchestrates changes in nitrogen and sulfur metabolism and shapes plant development. Plant Cell36, 404–426 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz-Bertomeu, J. et al. The essential role of the phosphorylated pathway of serine biosynthesis in Arabidopsis. Plant Signal. Behav.8, e27104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cascales-Miñana, B. et al. The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell25, 2084–2101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toujani, W. et al. Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol.163, 1164–1178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmermann, S. E. et al. The phosphorylated pathway of serine biosynthesis links plant growth with nitrogen metabolism. Plant Physiol.186, 1487–1506 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson, A. D. & Gregory, J. F. III Folate biosynthesis, turnover, and transport in plants. Annu. Rev. Plant Biol.62, 105–125 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Hanson, A. D. & Roje, S. One-carbon metabolism in higher plants. Annu. Rev. Plant Biol.52, 119–137 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Ravanel, S. et al. Methionine metabolism in plants: chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem.279, 22548–22557 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Ruszkowski, M., Sekula, B., Ruszkowska, A. & Dauter, Z. Chloroplastic serine hydroxymethyltransferase from Medicago truncatula: a structural characterization. Front. Plant Sci.9, 584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roje, S. S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry67, 1686–1698 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Amir, R & Hacham, Y. in A Missing Link Between Soils, Crops and Nutrition Vol. 50 (ed. Jez, J.) 251–279 (Wiley, 2008). .
- 22.Zhang, D. & Zhang, B. Pectin drives cell wall morphogenesis without turgor pressure. Trends Plant Sci.25, 719–722 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Haas, K. T., Wightman, R., Meyerowitz, E. M. & Peaucelle, A. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science367, 1003–1007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peaucelle, A., Braybrook, S. & Höfte, H. Cell wall mechanics and growth control in plants: the role of pectins revisited. Front. plant Sci.3, 121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tommasi, I. C. The mechanism of Rubisco catalyzed carboxylation reaction: chemical aspects involving acid-base chemistry and functioning of the molecular machine. Catalysts11, 813 (2021). [Google Scholar]
- 26.Zhang, P. et al. Revisiting fragmentation reactions of protonated α-amino acids by high-resolution electrospray ionization tandem mass spectrometry with collision-induced dissociation. Sci. Rep.9, 6453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulze-Siebert, D., Heintze, A. & Schultz, G. Substrate flow from photosynthetic carbon metabolism to chloroplast isoprenoid synthesis in spinach evidence for a plastidic phosphoglycerate mutase. Z. Naturforsch. C42, 570–580 (1987). [Google Scholar]
- 28.Timm, S. et al. Serine acts as a metabolic signal for the transcriptional control of photorespiration-related genes in Arabidopsis. Plant Physiol.162, 379–389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siqueira, J. A., Zhang, Y., Nunes-Nesi, A., Fernie, A. R. & Araújo, W. L. Beyond photorespiration: the significance of glycine and serine in leaf metabolism. Trends Plant Sci.28, 1092–1094 (2023). [DOI] [PubMed]
- 30.Abadie, C. & Tcherkez, G. Plant sulphur metabolism is stimulated by photorespiration. Commun. Biol.2, 379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abadie, C., Boex-Fontvieille, E. R., Carroll, A. J. & Tcherkez, G. In vivo stoichiometry of photorespiratory metabolism. Nat. Plants2, 1–4 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Busch, F. A., Sage, R. F. & Farquhar, G. D. Plants increase CO2 uptake by assimilating nitrogen via the photorespiratory pathway. Nat. Plants4, 46–54 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Fu, X., Gregory, L. M., Weise, S. E. & Walker, B. J. Integrated flux and pool size analysis in plant central metabolism reveals unique roles of glycine and serine during photorespiration. Nat. Plants9, 169–178 (2023). [DOI] [PubMed] [Google Scholar]
- 34.Björkman, O. The effect of oxygen concentration on photosynthesis in higher plants. Physiologia Plant.19, 618–633 (1966). [Google Scholar]
- 35.Abadie, C., Blanchet, S., Carroll, A. & Tcherkez, G. Metabolomics analysis of postphotosynthetic effects of gaseous O2 on primary metabolism in illuminated leaves. Funct. Plant Biol.44, 929–940 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Jardine, K. et al. Dynamic balancing of isoprene carbon sources reflects photosynthetic and photorespiratory responses to temperature stress. Plant Physiol.166, 2051–2064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karl, T. et al. On-line analysis of the 13 CO 2 labelling of leaf isoprene suggests multiple subcellular origins of isoprene precursors. Planta215, 894–905 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Heintze, A. et al. Plastidic isoprenoid synthesis during chloroplast development: change from metabolic autonomy to a division-of-labor stage. Plant Physiol.93, 1121–1127 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tovar‐Méndez, A., Miernyk, J. A. & Randall, D. D. Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur. J. Biochem.270, 1043–1049 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Karl, T., Curtis, A., Rosenstiel, T., Monson, R. & Fall, R. Transient releases of acetaldehyde from tree leaves− products of a pyruvate overflow mechanism? Plant Cell Environ.25, 1121–1131 (2002). [Google Scholar]
- 41.Jardine, K. J. & McDowell, N. Fermentation‐mediated growth, signaling, and defense in plants. New Phytol.239, 839–851 (2023). [DOI] [PubMed]
- 42.Jardine, K. et al. Green leaf volatiles and oxygenated metabolite emission bursts from mesquite branches following light–dark transitions. Photosynthesis Res.113, 321–333 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Wei, Y., Lin, M., Oliver, D. J. & Schnable, P. S. The roles of aldehyde dehydrogenases (ALDHs) in the PDH bypass of Arabidopsis. BMC Biochem.10, 1–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tcherkez, G. et al. Leaf day respiration: low CO2 flux but high significance for metabolism and carbon balance. N. Phytologist216, 986–1001 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Tcherkez, G. et al. Short‐term effects of CO2 and O2 on citrate metabolism in illuminated leaves. Plant, Cell Environ.35, 2208–2220 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Scheller, H. V., Jensen, J. K., Sørensen, S. O., Harholt, J. & Geshi, N. Biosynthesis of pectin. Physiologia Plant.129, 283–295 (2007). [Google Scholar]
- 47.Harholt, J., Suttangkakul, A. & Vibe Scheller, H. Biosynthesis of pectin. Plant Physiol.153, 384–395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bedhomme, M. et al. Folate metabolism in plants: an Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J. Biol. Chem.280, 34823–34831 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Jardine, K. J. et al. Cell wall ester modifications and volatile emission signatures of plant response to abiotic stress. Plant, Cell Environ.45, 3429–3444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romek, K. M. et al. Insights into the role of methionine synthase in the universal 13C depletion in O-and N-methyl groups of natural products. Arch. Biochem. Biophys.635, 60–65 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Keppler, F., Kalin, R., Harper, D., McRoberts, W. & Hamilton, J. T. Carbon isotope anomaly in the major plant C 1 pool and its global biogeochemical implications. Biogeosciences1, 123–131 (2004). [Google Scholar]
- 52.Harley, P., Greenberg, J., Niinemets, Ü. & Guenther, A. Environmental controls over methanol emission from leaves. Biogeosciences4, 1083–1099 (2007). [Google Scholar]
- 53.Bižić, M. et al. Aquatic and terrestrial cyanobacteria produce methane. Sci. Adv.6, eaax5343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lenhart, K., Althoff, F., Greule, M. & Keppler, F. Methionine, a precursor of methane in living plants. Biogeosciences12, 1907–1914 (2015). [Google Scholar]
- 55.Ernst, L. et al. Methane formation driven by reactive oxygen species across all living organisms. Nature603, 482–487 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Althoff, F. et al. Abiotic methanogenesis from organosulphur compounds under ambient conditions. Nat. Commun.5, 4205 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Kohl, L. et al. Radiation and temperature drive diurnal variation of aerobic methane emissions from Scots pine canopy. Proc. Natl Acad. Sci.120, e2308516120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wormit, A. & Usadel, B. The multifaceted role of pectin methylesterase inhibitors (PMEIs). Int. J. Mol. Sci.19, 2878 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res.51, D638–D646 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang, M. et al. Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. J. Proteome Res.12, 491–504 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Rowland, E. et al. The CLP and PREP protease systems coordinate maturation and degradation of the chloroplast proteome in Arabidopsis thaliana. N. Phytol.236, 1339–1357 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Lu, H., Gordon, M. I., Amarasinghe, V. & Strauss, S. H. Extensive transcriptome changes during seasonal leaf senescence in field-grown black cottonwood (Populus trichocarpa Nisqually-1). Sci. Rep.10, 6581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nonomura, A. & Benson, A. The path of carbon in photosynthesis: improved crop yields with methanol. Proc. Natl Acad. Sci.89, 9794–9798 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jardine, K. J. et al. Integration of C1 and C2 metabolism in trees. Int. J. Mol. Sci.18, 2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alekseeva, A., Savin, S. & Tishkov, V. NAD+-dependent formate dehydrogenase from plants. Acta Nat. (англоязычная версия)3, 38–54 (2011). [PMC free article] [PubMed] [Google Scholar]
- 66.Yurimoto, H. & Sakai, Y. Interaction between C1-microorganisms and plants: contribution to the global carbon cycle and microbial survival strategies in the phyllosphere. Biosci. Biotechnol. Biochem.87, 1–6 (2023). [DOI] [PubMed] [Google Scholar]
- 67.Iguchi, H., Yurimoto, H. & Sakai, Y. Interactions of methylotrophs with plants and other heterotrophic bacteria. Microorganisms3, 137–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niinemets, Ü., Tenhunen, J., Harley, P. C. & Steinbrecher, R. A model of isoprene emission based on energetic requirements for isoprene synthesis and leaf photosynthetic properties for Liquidambar and Quercus. Plant Cell Environ.22, 1319–1335 (1999). [Google Scholar]
- 69.Morfopoulos, C. et al. A model of plant isoprene emission based on available reducing power captures responses to atmospheric CO 2. N. Phytol.203, 125–139 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Suzuki, Y. et al. Overproduction of chloroplast glyceraldehyde-3-phosphate dehydrogenase improves photosynthesis slightly under elevated [CO2] conditions in rice. Plant Cell Physiol.62, 156–165 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Suzuki, Y. et al. Effects of simultaneous Rubisco, glyceraldehyde-3-phosphate dehydrogenase, and triosephosphate isomerase overexpression on photosynthesis in rice. Soil Sci. Plant Nutr. 1–8. 10.1080/00380768.2023.2255213 (2023).
- 72.Atkinson, N., Mao, Y., Chan, K. X. & McCormick, A. J. Condensation of Rubisco into a proto-pyrenoid in higher plant chloroplasts. Nat. Commun.11, 6303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, L. et al. A chloroplast protein atlas reveals punctate structures and spatial organization of biosynthetic pathways. Cell186, 3499–3518.e3414 (2023). [DOI] [PubMed] [Google Scholar]
- 74.Lawson, S. J. et al. Methanethiol, dimethyl sulfide and acetone over biologically productive waters in the southwest Pacific Ocean. Atmos. Chem. Phys.20, 3061–3078 (2020). [Google Scholar]
- 75.Keppler, F., Hamilton, J. T., Braß, M. & Röckmann, T. Methane emissions from terrestrial plants under aerobic conditions. Nature439, 187–191 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Keppler, F. et al. Methane formation in aerobic environments. Environ. Chem.6, 459–465 (2009). [Google Scholar]
- 77.Parsons, A. J., Newton, P. C., Clark, H. & Kelliher, F. M. Scaling methane emissions from vegetation. Trends Ecol. Evol.21, 423–424 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Nisbet, R. et al. Emission of methane from plants. Proc. R. Soc. B: Biol. Sci.276, 1347–1354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bastviken, D. et al. The importance of plants for methane emission at the ecosystem scale. Aquat. Bot.184, 103596 (2023). [Google Scholar]
- 80.Blankenship, R. E. & Hartman, H. The origin and evolution of oxygenic photosynthesis. Trends Biochem. Sci.23, 94–97 (1998). [DOI] [PubMed] [Google Scholar]
- 81.Jardine, K. J. et al. Methanol and isoprene emissions from the fast growing tropical pioneer species Vismia guianensis (Aubl.) Pers.(Hypericaceae) in the central Amazon forest. Atmos. Chem. Phys.16, 6441–6452 (2016). [Google Scholar]
- 82.Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol.30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pluskal, T., Castillo, S., Villar-Briones, A. & Orešič, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform.11, 1–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumler, W., & Ingalls, A. E. Tidy Data Neatly Resolves Mass-Spectrometry's Ragged Arrays. The R Journal, 14, 193–202 (2022).
- 85.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
- 86.Dewhirst, R. A., Afseth, C. A., Castanha, C., Mortimer, J. C. & Jardine, K. J. Cell wall O-acetyl and methyl esterification patterns of leaves reflected in atmospheric emission signatures of acetic acid and methanol. PLoS ONE15, e0227591 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greule, M., Mosandl, A., Hamilton, J. T. & Keppler, F. A simple rapid method to precisely determine 13C/12C ratios of plant methoxyl groups. Rapid Commun. Mass Spectrom.23, 1710–1714 (2009). [DOI] [PubMed] [Google Scholar]
- 88.Greule, M., Moossen, H., Geilmann, H., Brand, W. A. & Keppler, F. Methyl sulfates as methoxy isotopic reference materials for δ13C and δ2H measurements. Rapid Commun. Mass Spectrom.33, 343–350 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Greule, M. et al. Three wood isotopic reference materials for δ2H and δ13C measurements of plant methoxy groups. Chem. Geol.533, 119428 (2020). [Google Scholar]
- 90.Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res.40, D1178–D1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ho, C.-L., Noji, M., Saito, M. & Saito, K. Regulation of serine biosynthesis in Arabidopsis: crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J. Biol. Chem.274, 397–402 (1999). [DOI] [PubMed] [Google Scholar]
- 92.Grigoriev, I. V. et al. PhycoCosm, a comparative algal genomics resource. Nucleic acids Res.49, D1004–D1011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fujisawa, T. et al. CyanoBase: a large-scale update on its 20th anniversary. Nucleic Acids Res.45, D551–D554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bodenhofer, U., Bonatesta, E., Horejš-Kainrath, C. & Hochreiter, S. msa: an R package for multiple sequence alignment. Bioinformatics31, 3997–3999 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Schliep, K. P. phangorn: phylogenetic analysis in R. Bioinformatics27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T. Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol.8, 28–36 (2017). [Google Scholar]
- 97.Armenteros, J. J. A. et al. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance2, e201900429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thumuluri, V., Almagro Armenteros, J. J., Johansen, A. R., Nielsen, H. & Winther, O. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res.50, W228–W234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cao, L., Lu, W., Mata, A., Nishinari, K. & Fang, Y. Egg-box model-based gelation of alginate and pectin: a review. Carbohydr. Polym.242, 116389 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data needed to evaluate the conclusions in this paper including the source data of every figure and Supplementary Fig. is available for download as a supplementary file. All raw LC-MS/MS data For LC-MS data used in Fig. 4 and supplementary Fig. S10 are available in the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=aabec0a91ce449e58e843e60ff3e5021). Supplementary Data 1 (.xlsx file): Method details of the leaf methionine analysis from frozen leaf extracts separated using hydrophilic liquid interaction chromatography. Supplementary Data 2 (.xlsx file): 76 protein sequences from 22 species used to construct the phylogenetic tree shown in Fig. 7 with protein ID, Genus and species, and plant category. Supplementary Data 3 (.zip file): Data set for all data-based figures including Fig. 1a, Fig. 1b, Fig. 2, Fig. 3, and supplementary Figs. S1, S2, S3, S4, S5, S6, S7, S8, S9, S11, S12, S13, S14, S15, and S16.