Abstract
The nucleus pulposus (NP) plays a vital role in intervertebral disc degeneration (IVDD). Previous studies have revealed cellular heterogeneity in NP tissue during IVDD progression. However, the cellular and molecular alterations of diverse cell clusters during IVDD remain to be fully elucidated. NP tissues were isolated from patients with different grades of IVDD undergoing discectomy, and then subjected to single-cell RNA sequencing (scRNA-seq). Cell subsets were identified based on unbiased clustering of gene expression profiles. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to determine the molecular features of diverse cell clusters. Monocle analysis was used to illustrate the differentiation trajectories of chondrocytes. Additionally, CellPhoneDB analysis revealed potential interactions between chondrocytes and other cells during IVDD. Based on the expression profiles of 47,610 individual cells, eight putative clusters including chondrocytes, endothelial cells, fibroblasts, macrophages, mural cells, osteoclasts, proliferating stromal cells and T cells were identified. The chondrocyte cluster was classified into three subsets, C1-C3, which were associated with stress-resistance, fibrosis and inflammatory responses, respectively. Pseudo-time trajectories suggested that chondrocytes gradually differentiated into fibroblasts during IVDD. Immune cells including cDC2s, macrophages and monocytes were identified. Further analysis showed that chondrocytes might communicate with immune cells via the MIF, TNFSF9, SPP1 and CCL4L2 signaling pathways. In addition, we found that invading endothelial cells might interact with chondrocytes through the COL4A1, CXCL12, VEGFA and SEMA3E signaling pathways. Our results reveal the cellular complexity and phenotypic characteristics of NP tissues at single-cell resolution, which will contribute to the in-depth investigation of preventative and regenerative strategies for IVDD.
Keywords: Intervertebral disc degeneration, Nucleus pulposus, Single-cell RNA sequencing
Subject terms: Transcriptomics, Orthopaedics
Introduction
Low back pain (LBP), a prevalent and chronic disease worldwide, is predominantly triggered by intervertebral disc degeneration (IVDD)1. It is estimated that around 20% of adolescents have mild-degenerated discs and that 80% of the population experience LBP at some point in their lives2. The annual direct and indirect costs associated with IVDD exceed $100 billion, imposing a heavy financial burden on society3. The etiology of IVDD is related to endogenous genetic susceptibility and exogenous stressors, such as aging, mechanical overload and nutritional deficiency4. IVDD encompasses structural and biochemical changes in discs leading to extracellular matrix (ECM) degradation, matrix metalloproteinase generation, and inflammatory cytokine production. Currently, the symptoms of IVDD are relieved primarily by conservative and surgical treatments, which are often accompanied by relapse and various complications5. Therefore, it is critical to gain insight into the pathological mechanisms driving IVDD, which may help to identify novel biotherapies to reverse IVDD and reconstruct the mechanical function of the spine.
IVD consists of annulus fibrosus (AF), nucleus pulposus (NP) and cartilage endplate (CEP). The normal NP tissue produces ECM components including collagen, aggrecan to maintain the flexibility of IVDs6. In the influence of environmental factors and ageing, the proteoglycan and hydration ability of ECM decreases gradually resulting in the progressive dehydration of IVDs7. Simultaneously, NPCs undergo apoptosis or pyroptosis which may further promote the pathogenesis of IVDD8,9. NP is considered as an avascular and immune-privileged structure surrounded by AF and CEP. However, both ECM disruption and AF fissures facilitate the infiltration of endothelial and immune cells during degeneration10,11. Given the complexity and multifactorial nature of IVDD, identifying the key regulatory genes that initiate the degenerative cascade is challenging.
In recent years, single cell RNA sequencing (scRNA-seq) has provided a powerful tool for analyzing the transcriptomic and functional heterogeneity of different cell populations within organs. Previous studies have performed scRNA-seq on healthy and degenerative IVDs to identify various or new cell subsets and determine the cellular heterogeneity12–15. For instance, Liu et al. identified six clusters of chondrocytes with corresponding markers and analysed the crosstalk between endothelial cells and chondrocytes in a goat IVDD model16. Su et al. revealed a novel human notochord cluster in the early stage of IVD formation and the role of notochord-secreted SPP1 in regulating chondrogenesis17. Local mesenchymal progenitor cells have been reported in human degenerated intervertebral discs18–22. Consistent with the previous findings, scRNA-seq supports the existence of NP progenitor/stem cells expressing PROCR, PRRX1, CD44, LepR, CD105, THY1, and CD7323,24. ScRNA-seq facilitates deep data mining of cellular characteristics and cell–cell interactions, which can shed light on the cellular status under physiological and pathological conditions.
In this study, we performed scRNA-seq on NP tissues with different degenerative grades and captured 47,610 individual cells. Based on the transcriptomic analysis, we defined eight putative clusters and comprehensively characterized the transcriptomic features of chondrocytes, fibroblasts and MPs. We demonstrated the relationship between chondrocyte and fibroblast differentiation. We also investigated the intercellular crosstalk between chondrocytes and immune cells, endothelial cells. Our study revealed in detail NP cellular heterogeneity and intercellular signaling during IVDD, providing insights into the molecular mechanisms of IVDD pathogenesis.
Materials and methods
Human NP tissue specimens
NP tissues were obtained from six patients at L4/L5 or L5/S1 level, who were diagnosed with lumbar disc herniation and undergoing endoscopic lumbar discectomy. All participants underwent MRI before surgery. The degree of IVDD was evaluated based on the Pfirrmann grading system. In this study, grades II and III were classified as mild degeneration (MI), and grade IV was classified as severe degeneration (SE). This study was approved by the Ethics Committee of Affiliated Hospital of Jining Medical University (Ethics number: 2021C159). Informed consent was obtained from all patients for inclusion in the study.
Preparation of human NP single-cell suspensions
The removed NP tissues were immediately placed in sCelLive Tissue Preservation Solution (Singleron Biotechnologies) and transported to the laboratory at 4℃. The specimens were dissociated into single cell suspensions with a Singleron PythoNTM Automated Tissue Dissociator and sCelLiveTM Tissue Dissociation Mix. 0.4% trypan blue (C0040, Solarbio, China) was mixed with 10 μL cell suspension and incubated at room temperature for 3 min. Apply a drop of the trypan blue/cell mixture to a hemacytometer and evaluate by a microscopy (CKX31, Olympus, Japan). Count the unstained (viable) and stained (nonviable) cells separately, and calculate the percentage of viable cells as follows: cell viability (%) = total number of viable cells/total number of cells × 100. Samples with > 80% cell viability were used for subsequent experiments.
ScRNA-seq
The concentration of single-cell suspension was adjusted to 1 × 105 cells/mL with PBS (P1000, Solarbio, China). Single-cell suspensions were loaded onto microfluidic chips to obtain a single cell per microwell. ScRNA-seq libraries were constructed with a GEXSCOPE Single-Cell Sequencing Kit and Singleron Matrix Automated single-cell processing system. Individual libraries were diluted to 4 ng/µL and pooled for sequencing on an Illumina NovaSeq6000 platform with 150 bp paired-end reads.
Primary analysis of raw read data
Raw reads were processed to generate gene expression profiles using CeleScope v1.10.0 with default parameters. Briefly, barcodes and UMIs were extracted from R1 reads and corrected. Adapter sequences and poly A tails were trimmed from R2 reads and aligned against the GRCh38 (hg38) transcriptome using STAR (v2.6.1a). Uniquely mapped reads were then assigned to exons with FeatureCounts (v2.0.1). An expression matrix is generated by combining barcodes and UMI sequences, assigning aligned reads to the appropriate cells, and then calculating the number of UMIs for each gene in each cell.
Quality control, dimension-reduction and clustering
Scanpy v1.8.1 was used for quality control, dimensionality reduction and clustering in Python 3.7. For each sample dataset, we filtered the expression matrix by the following criteria: (1) cells with gene count less than 200 or with top 2% gene count were excluded; (2) cells with top 2% UMI count were excluded; (3) cells with mitochondrial content of 20% were excluded; and (4) genes expressed in fewer than 5 cells were excluded. After filtering, 47,610 cells were retained for the downstream analyses, with an average of 1913 genes and 7438 UMIs per cell. The raw count matrix was normalized by total counts per cell and logarithmically transformed into a normalized data matrix. The top 2000 variable genes were selected by setting flavor = ‘seurat’. Principal Component Analysis (PCA) was performed on the scaled variable gene matrix, and the top 20 principal components were used for clustering and dimensional reduction. Cells were separated into 21 clusters by using the Louvain algorithm and setting the resolution parameter to 1.2. Cell clusters were visualized by using Uniform Manifold Approximation and Projection (UMAP) or t-Distributed Stochastic Neighbor Embedding (t-SNE).
Cell-type recognition with Cell-ID
Cell-ID is a multivariate approach that extracts gene signatures for individual cells and performs cell identity recognition using hypergeometric tests (HGTs). Dimensionality reduction was performed on a normalized gene expression matrix through multiple correspondence analysis, where both cells and genes were projected in the same low-dimensional space. Then a gene ranking was calculated for each cell to obtain the most featured gene sets. Cell types were manually annotated based on the canonical markers from the reference database SynEcoSys, which contains CellMakerDB, PanglaoDB etc.
Differentially expressed genes (DEGs) analysis
DEGs of each cell type were identified using the scanpy.tl.rank_genes_groups function. The significance of the differences in gene expression was determined using the Wilcoxon rank sum test with Benjamini–Hochberg correction. DEGs were identified under the following criteria: (1) log(fold change) > 1; (2) adjusted p value < 0.05; and (3) expression in more than 10% of the cells in the compared groups.
Pathway enrichment analysis
To investigate the potential functions of DEGs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were performed with the clusterProfiler (v 3.16.1) R package. Pathways with p value which was calculated by Benjamini–Hochberg correction less than 0.05 were considered significantly enriched. For GSVA pathway enrichment analysis, the average gene expression of each cell type was used as input data.
Pseudotime trajectory analysis
The trajectory analysis of cell differentiation was constructed with Monocle2 (v 2.10.0). Marker genes of the cell subsets after Seurat clustering and raw expression counts of filtered cells were selected for analysis. Dimensional reduction and cell ordering were performed using the DDRTree method and orderCells function. The plot_cell_trajectory function was applied for visualization.
Cell communication analysis
Cell–cell interactions were predicted by CellPhone DB (v2.1.0), a public repository of ligands, receptors and their interactions. Permutation number for calculating the null distribution of average ligand-receptor pair expression in randomized cell identities was set to 1000. The threshold for individual ligand or receptor expression was based on the cut-off value of average log gene expression in each cell type. Predicted interaction pairs with p value < 0.05 and mean log expression > 0.1 were considered significant and visualized by heatmap_plot and dot_plot.
Validation of COL1A1 and TGFB1 expressions in a rat IVDD model
Three-month-old male Sprague–Dawley rats were used to establish the IVDD model as previously reported25. Six rats remained intact and were used as negative controls. Six rats were placed in the prone position after anesthesia with 2% (w/v) pentobarbital (40 mg/kg). Two IVDs (Co6/7 and Co8/9) were punctured by a 20-gauge needle from the dorsal side. The needle was inserted vertically through the center of IVD to the opposite side, rotated 360° and held for 10 s. Rats were anesthetized with 2% pentobarbital 4 weeks after surgery. IVDs were fixed in 4% paraformaldehyde (P1110, Solarbio, China) and decalcified in EDTA (C1038, Solarbio, China) for 4 weeks. The discs were dehydrated, embedded in paraffin and then cut into 5 μm thick sections. The sections were blocked with 5% (w/v) BSA (ST023, Beyotime, China) and incubated with primary antibodies against COL1A1 (AF7001, Affinity Biosciences, Australia) and TGFB1 (21898-1-AP, proteintech, America), followed by incubation with secondary antibodies conjugated with Alexa Fluor 488 (A-11094, ThermoFisher, America). Finally, the sections were scanned by Zeiss Axio Observer 7 microscopy (Germany).
Results
Single cell profiling and clustering of human NP tissue with IVDD pathology
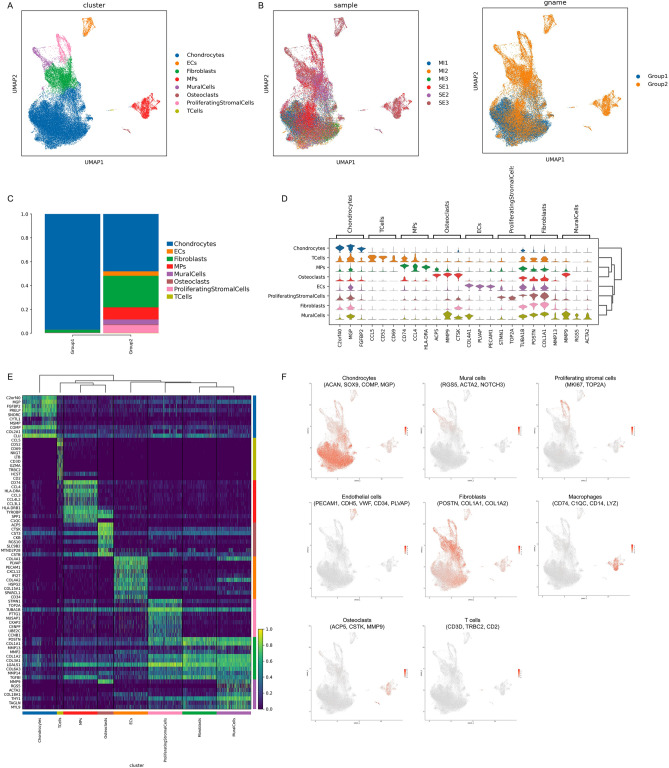
To understand the cellular heterogeneity and molecular characteristics of human NP tissue during IVDD, NP tissues from six human IVDD patients with different Pfirrmann degenerative grades [Pfirrmann II, III, and IV] were isolated and single-cell transcriptomic profiling (10X Genomics platform) was performed (Supplementary Fig. 1). A total of 47,610 individual cells meeting the quality control criteria were profiled. UMAP identified eight putative clusters, including chondrocytes, endothelial cells (ECs), fibroblasts (Fibros), mononuclear phagocytes (MPs), mural cells, osteoclasts (OCs), proliferating stromal cells and T cells (Fig. 1A). As the severity of IVDD increases, the cellular components become more complex (Fig. 1B). In addition to the decrease in the proportion of chondrocytes, other cells including ECs, Fibros, MPs, mural cells, OCs, proliferating stromal cells and T cells were upregulated (Fig. 1C). The heterogeneity of NPCs was classified by gene expression (Fig. 1D,E), and representative gene markers were shown with a feature plot map (Fig. 1F). The cartilage matrix specific markers ACAN, SOX9, COMP, MGP and COL2A1 were strongly expressed in chondrocytes. Mural cells highly expressed RGS5, ACTA2 and NOTCH3. A cluster of proliferating stromal cells expressing MKI67 and TOP2A was identified. EC cluster was characterized by the expression of feature genes PECAM1, CDH5, VWF, CD34 and PLVAP. Fibro cluster was identified by POSTN, COL1A1 and COL1A2. CD74, C1QC, CD14 and LYZ were mainly expressed in the MP cluster. ACP5, CTSK and MMP9 were strongly expressed in the OC cluster. CD3D, TRBC2 and CD2 were specifically expressed in the T cell cluster. Overall, the above results revealed the cellular heterogeneity and transcriptomic features of human NP tissue during IVDD pathogenesis.
Fig. 1.
Single-cell transcriptomic profiling of human nucleus pulposus (NP) with IVDD. (A) UMAP visualization of human NPCs identified eight different clusters. Each dot corresponds to a colored cell cluster. (B) UMAP plot depicting the proportions of cell types in NP tissues with different degenerative grading. MI: mildly degenerative, SE: severely degenerative. Group 1 represents MI tissues and Group 2 includes SE tissues. (C) Bar diagrams showing the grade distribution of each NP cell subset. (D) Violin diagrams displaying the top three highly expressed genes in each cell subset. (E) Heatmap revealing the scaled expression of differentially expressed genes (DEGs) for each cell cluster. (F) Feature plot map showing the expression and distribution of signature genes in each cell cluster.
Transcriptional characteristics and developmental trajectories of chondrocytes during IVDD
The ECM in NP tissue is composed of proteoglycans and collagen fibrils. Chondrocytes can recognize the loss of ECM and actively produce ECM components. Based on the pivotal role of chondrocytes in ECM homeostasis, it is essential to determine the changes in chondrocyte composition and gene expression during the progression of IVDD. The chondrocyte cluster was divided into three subclusters based on mRNA expression (Fig. 2A). Chondrocyte 1 (C1) preferentially expressed genes involved in resisting oxidative stress including SOD3, GPX3 and transcription factor NR4A2; subcluster chondrocyte 2 (C2) highly expressed fibrosis-related ECM genes, such as PRG4, COL1A1 and COL3A1; and subcluster chondrocyte 3 (C3) preferentially expressed the inflammation- and stress response-related genes ORM1/2, SLPI and TRIB3 (Fig. 2B,C). To analyse the signature alteration between the chondrocyte subclusters, DEGs were subjected to KEGG analysis. The results showed that C1 had high activity in ribosome and mitophagy; C2 was enriched in ECM-receptor interaction, focal adhesion, lysosome, PI3K-Akt signaling pathway and TGF-beta signaling pathway; and C3 exhibited strong enrichment in protein synthesis, ferroptosis, antigen processing and presentation, MAPK signaling pathway and cellular senescence (Fig. 2D). Next, the developmental trajectory of chondrocytes was investigated and it was found that C1 was located at the root of the trajectory, showing a fate of differentiation towards C2 and C3. The differentiation to fibrochondrocytes (C2) took the dominant position, and C2 became the major population involved in IVDD (Fig. 2E,F). Gene expression analysis showed that the progenitor cell marker TAGLN was widely expressed in both C1 and C2, PROCR and PRRX1 were preferentially expressed in C2, and TEK was expressed only in individual chondrocytes (Fig. 2G). Additionally, the molecular features during the differentiation trajectories of chondrocytes were analysed, and the results showed that the expression of stimulus responsive genes such as CHI3L1, CHI3L2, CEBPD and CLU was significantly down-regulated, while that of the fibrosis-related genes COL1A1, COL8A1, COL1A2 and ASPN was highly up-regulated (Fig. 2H,I).
Fig. 2.
Gene signatures and developmental trajectories of chondrocytes in IVDD. (A) UMAP plot of three chondrocyte clusters. (B) Dot plot showing the typically expressed genes in each chondrocyte cluster. (C) Heatmap displaying feature genes of each chondrocyte cluster. (D) Dot plot showing the enriched KEGG pathway of each chondrocyte cluster. (E,F) Pseudo-temporal differentiation trajectories of chondrocytes by Monocle2. (G) Violin plots showing TAGLN, TEK, PROCR and PRRX1 expression levels in three chondrocyte clusters. (H) Changes in relative expression of representative genes along the differentiation trajectories. (I) Fitting curves displaying the trend of key molecules along the pseudo time.
In summary, these results revealed the temporal transcriptomic variability of chondrocytes in IVDD, suggesting that the phenotype of chondrocytes shifted from resistance to stress and inflammation to fibrosis.
Intrinsic signatures of fibroblasts associated with IVDD
Fibrosis is a pathological feature of many chronic inflammatory diseases, including pulmonary fibrosis, rheumatoid arthritis and liver cirrhosis26. Recent studies have demonstrated the abnormal deposition of fibrotic proteins in the ECM and the presence of fibroblastic NP cells during IVDD27. The results showed that in NP tissues with severe degeneration, the proportion of fibroblast cluster was significantly increased (Fig. 1C). The fibroblast cluster was divided into four subclusters based on mRNA expression, with an increase in the proportion of fibroblasts 1 (Fib1) and Fib4, and a decrease in Fib2 and Fib3 during IVDD (Fig. 3A,B). Fib1 expressed high levels of fibrotic signature genes such as MMP13, IBSP, TAGLN, COL1A1 and COL3A1 (Fig. 3C,D). The inflammatory-related proteins NFKBIA, CXCL2 and stress responsive proteins HSPA1A, DNAJB1 were specifically highly expressed in Fib2 (Fig. 3C,D). Fib3 preferentially expressed FMOD, SCRG1, CHAD and PRELP to mediate the interaction with chondrocytes (Fig. 3C,D). FMOD is a small leucine-rich proteoglycan that is critical for collagen fibrillogenesis28. SCRG1 is a transcription factor that promotes chondrogenic gene expression and stimulates chondrogenesis29. CHAD and PRELP bind cell-surface proteoglycans, enhance focal adhesion formation and promote the attachment of fibroblasts and chondrocytes30,31. Fib4 specifically expressed the cell proliferation-related genes PCLAF, TK1 and MCM3 (Fig. 3C,D). To analyse the functional differences among the fibroblast subpopulations, GO and KEGG databases were used to investigate the associated biological processes and signaling pathways. Fib1 was enriched for processes related to extracellular matrix organization and ECM-receptor interaction (Fig. 3E,I), while Fib2 was involved in the cellular response to oxidative stress and regulation of the apoptotic signalling pathway (Fig. 3F). KEGG analysis suggested that Fib2 played a potential role by modulating the NOD-like receptor and MAPK signaling pathways (Fig. 3J). Fib3 was associated with glycosaminoglycan biosynthesis and positive regulation of collagen fibril organization (Fig. 3G,K). Consistent with the characteristic gene functions, GO and KEGG analyses revealed that Fib4 was enriched in terms associated with DNA replication and cell cycle (Fig. 3H,L).
Fig. 3.
Intrinsic signatures in fibroblasts associated with IVDD. (A) UMAP plot of four fibroblast clusters. (B) Histograms showing the proportion of each fibroblast cluster in different degenerative NP tissues. (C) Heatmap depicting characteristic genes of each fibroblast cluster. (D) Violin diagrams exhibiting the top three highly expressed genes in each fibroblast subset. (E–H) Histograms displaying the enriched GO terms including biological process (BP), cellular component (CC) and molecular function (MF) of each fibroblast cluster. (I–L) Dot plot showing the enriched KEGG pathway of each fibroblast cluster.
In brief, these findings pointed to an increase in the proportion and heterogeneity of fibroblasts, and predicted the potentially critical roles of each cell subset in IVDD process.
Pseudo-time trajectory of chondrocytes differentiation to fibroblasts in IVDD
To analyse the relationship between chondrocytes and fibroblasts during IVDD, chondrogenic trajectory was simulated using Monocle2. Chondrocytes lay in the root of trajectory and almost disappeared in the late period. In the severe degeneration group, chondrocytes differentiated into fibroblasts, and fibroblasts became the dominant population (Fig. 4A,B). Gene expression analysis showed that chondrocytes barely expressed CD146/MCAM, which determines cell migration, suggesting that they were resident original cells (Supplementary Fig. 2). Consistent with the trajectory of cell differentiation, the expression of chondrocyte markers ACAN, COL2A1 and APOD decreased, while that of the fibrosis-related genes COL1A1, COL1A2 and COL3A1 increased (Fig. 4C,D). To evaluate the molecular differences in chondrocytes between mild and severe degeneration groups, DEGs, GO and KEGG analyses were performed. In cluster C1, inflammatory-related proteins CHI3L2 and CXCL2 were significantly up-regulated in the severe group (Fig. 4E). GO and KEGG analyses revealed that C1 had higher scores for chemokine receptor binding and IL-17 signaling pathway (Supplementary Fig. 3 and Fig. 4H). In clusters C2 and C3, the expression of TGFB1 was notably elevated, which might stimulate progenitor/stem cells differentiation into chondrocytes and promote fibroblast proliferation at a later stage of the chondrogenic trajectory (Fig. 4F,G). In addition, heatmap results indicated that the expression of fibroblast markers COL1A1, COL1A2 and COL3A1 were up-regulated in the three clusters of the severely degenerative group (Fig. 4E–G). KEGG anlysis manifested that all three clusters were enriched in the AGE-RAGE pathway, which can induce the expression of COL1A1 and COL1A2 via NF-κB (Fig. 4I,J). Furthermore, the expression changes of COL1A1 and TGFB1 were determined using a puncture-induced IVDD rat model. The immunofluorescence data showed that COL1A1 and TGFB1 expression levels were increased in the puncture-induced IVDD, which was consistent with the scRNA-seq (Fig. 4K).
Fig. 4.
Pseudo-time trajectory of chondrocytes differentiation to fibroblasts in IVDD. (A,B) Monocle pseudotime trajectory displaying the progression of chondrocytes differentiation to fibroblasts in the severely degenerative NP tissues. (C) Changes in relative expression of feature genes along the differentiation trajectories. (D) Fitting curves showing the trend of key genes along the pseudo time. (E–G) Heatmap revealing DEGs of each chondrocyte cluster in different degenerative NP tissues. (H–J) Dot plot depicting the enriched KEGG pathways of each chondrocyte cluster. (K) IF staining of COL1A1 and TGFB1 in the rat tail discs of Co6/7 or Co8/9 at 4 weeks after needle puncture. Scale bar: 500 μm.
In conclusion, these data revealed changes in the transcriptome and signalling pathways of chondrocytes during IVDD pathogenesis, as well as potential trends in chondrocytes differentiation into fibroblasts.
Identification of MP infiltration in IVDD
The nucleus pulposus, which is surrounded by fibrous rings and cartilage endplates, is isolated from the immune system and considered an immune pardoning organ. During the degeneration process, the fibrous rings rupture and immune cells infiltrate. The results showed that the proportion of MPs was markedly increased (Fig. 1C). The MP cluster was divided into three subclusters including cDC2, macrophages and monocytes based on the feature genes (Fig. 5A). cDC2s were identified by CD1C, CLEC10A and FCER1A; macrophages were identified by CD68, CD163 and CD14; and monocytes were identified by FCN1, LYZ and VCAN (Fig. 5B). The heatmaps showed that the chemokine receptor CCR7 and HLA-DQA1/2 were highly expressed in cDC2s, lysosomal protease CTSD and ferroptosis-related gene HMOX1 were increasingly expressed in macrophages, while inflammatory-related genes CCL20 and IL1B were highly expressed in monocytes (Fig. 5C). GO and KEGG analyses indicated that the function of cDC2s was closely associated with ribosome and antigen processing and presentation, macrophages was related to lysosome and ferroptosis, whereas that of monocytes was related to TNF, IL-17 and NF-kappa B signaling pathways (Fig. 5D–F). Furthermore, the proportion of distinct MP clusters in mild vs. severe samples was analysed and it was found that the mild degeneration group rarely contained MPs except for MI1 (Fig. 5G). The MI1 sample only contained macrophages and monocytes, while the SE1, SE2 and SE3 samples included cDC2, macrophages and monocytes (Fig. 5H). To evaluate the molecular differences in macrophages between mild and severe degenerative groups, DEGs, GO and KEGG analyses were performed. The heatmap indicated that lysosomal proteases CTSB, LGMN, ASAH1 and antigen presenting molecules HLA-DRB5, HLA-DMA were significantly up-regulated in the severe group (Fig. 5I). Moreover, the expression of IL1B, a marker of M1-polarized macrophages, was also markedly increased. GO and KEGG analyses showed that DEGs were enriched in lysosome and antigen processing and presentation (Fig. 5J,K).
Fig. 5.
Identification of MPs infiltration in IVDD. (A) UMAP plot of three MP clusters. (B) Dot plot depicting the typically expressed genes in each MP cluster. (C) Heatmap showing characteristic genes of each MP cluster. (D–F) Dot plot displaying the enriched KEGG pathway of each MP cluster. (G) UMAP plot depicting the grade distribution of each MP subset in NP tissues. (H) Bar diagrams showing the proportions of each MP subset in NP tissues. (I) Heatmap revealing DEGs of macrophages between mild and severe degenerative NP tissues. (J) Lollipop chart depicting the enriched GO terms including BP, CC and MF of DEGs in (I). (K) Histograms showing the enriched KEGG pathway of of DEGs in (I).
Overall, these results revealed the expression patterns of immune cells in human NP tissue. A higher infiltration of MPs was observed in the severe group than in mild group, and M1-polarized macrophages, cDC2 and monocytes may play an important role in the inflammatory cascade during IVDD progression.
Intercellular crosstalks between MPs and chondrocytes
To explore the role of immune cells in the IVDD process, the cell–cell interaction network between chondrocytes and immune cells was analysed using CellPhone DB. Strong cellular interactions were found between MPs and chondrocytes (Fig. 6A). C1, C2 and C3 exhibited similar interactions with cDC2s, macrophages and monocytes, and they exhibited relatively high cellular communication with macrophages. Additionally, the three MP clusters had more interactions with the C2 subset than with the other subsets (Fig. 6B). Ligands and receptors with significant interactions were partially displayed. The SPP1, CCL4L2 and GRN signalling pathways were highly activated, with MPs as the signal senders and chondrocytes as the signal receivers (Fig. 6C). Notably, macrophage-secreted SPP1 was found to regulate chondrocytes via CD44, which is supported by the research that SPP1 suppresses hypertrophy and vascularization while stimulating chondrogenesis by interacting with CD4432. It had also been found that CCL4L2 (mainly secreted by monocytes) modulated chondrocytes via PGRMC2 and VSIR. Additionally, the CD74, OGN, ANXA1 and TNFSF9 signaling pathways were highly activated, with chondrocytes as the signal senders and MPs as the signal receivers (Fig. 6D). The CD74 pathway includes MIF_CD74, COPA_CD74 and APP_CD74 pairs; TNFSF9 pathway includes TNFSF9_HLA-DPA1 pair; OGN pathway includes OGN_HLA-DRB1 pair; and ANXA1 pathway includes ANXA1_FPR1 and ANXA1_FPR3 pairs. The data showed that MIF and TNFSF9, which play important roles in macrophage activation, were highly expressed in the C2 and C3 clusters, respectively. This suggests that C2-secreted MIF and C3-secreted TNFSF9 are potential factors mediating macrophage M1 polarization.
Fig. 6.
Intercellular crosstalks between MPs and chondrocytes. (A,B) Circle plot showing the interaction numbers between chondrocytes and MPs. (C,D) Dot plots revealing the ligand-receptor interactions of MPs on chondrocytes and chondrocytes on MPs.
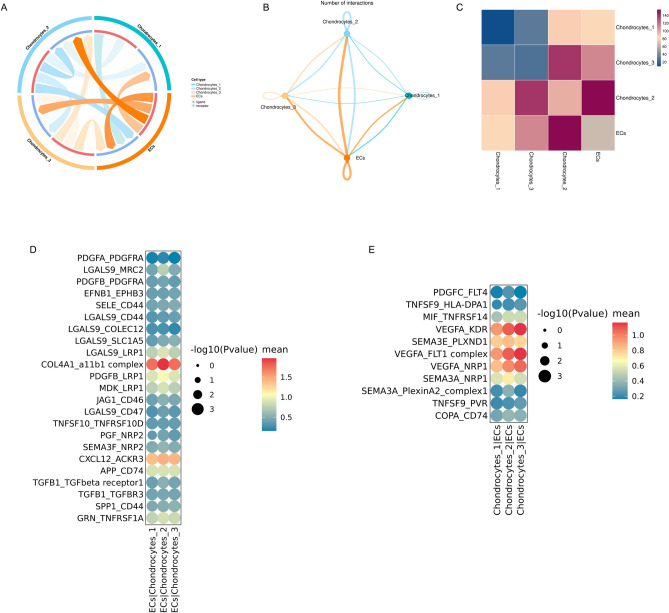
Characterization of endothelial cells-to-chondrocytes interactions involved in IVDD
Vessels invade IVDs during the degeneration process, leading to pain and dysfunction. Annulus fibrosus injury and proteoglycan reduction are intrinsically important factors in the vascular growth of degenerative intervertebral discs. The results also revealed a significant increase in the proportion of ECs during IVDD (Fig. 1C). To analyse the functions of EC penetration, CellPhone DB was used to establish a cellular communication network and to reveal the interactions between ECs and chondrocytes (Fig. 7A–C). The COL4A1 and CXCL12 signaling pathways were highly activated, with ECs as the signal senders and chondrocytes as the signal receivers (Fig. 7D). ECs might modulate chondrocytes through the interaction of COL4A1 with integrin a11b1 and CXCL12 with ACKR3. With condrocytes as the signal senders and ECs as the signal receivers, VEGFA signaling pathway, including VEGFA-KDR, VEGFA-FLT1 and VEGFA-NRP1 pairs, were preferentially activated (Fig. 7E). As the major angiogenic factor in IVD, VEGFA might bind to the receptors KDR, FLT1 and NRP1 on the surface of ECs to induce endothelial infiltration and vasculogenesis. The results also indicated that the SEMA3E-PLXND1 pathway was activated, suggesting that chondrocytes might secrete SEMA3E as a potential factor to inactivate ECs to maintain the avascular microenvironment of IVDs (Fig. 7E). These data pointed that the chondrocyte cluster might synergistically regulate EC penetration in IVDD via VEGFA and SEMA3E pathways.
Fig. 7.
Characterization of endothelial cells-to-chondrocytes interactions involved in IVDD. (A) Chord diagram showing potential ligand-receptor pairs between ECs and chondrocytes. (B) Circle plot displaying cellular crosstalks between ECs and chondrocytes. (C) Heatmap showing the interaction numbers between ECs and chondrocytes. (D) Dot plot depicting the ligand-receptor pairs of ECs on chondrocytes. (E) Dot plot exhibiting the ligand-receptor interactions of chondrocytes on ECs.
In summary, these results revealed the cellular crosstalks between ECs and chondrocytes. Chondrocytes secrete cytokines to affect EC penetration, while ECs can also negatively regulate chondrocyte functions in IVDD through ligand-receptor interactions.
Discussion
Due to overloaded mechanical stress and insufficient nutrient supply, NP tissues undergo parenchymal biological changes, including changes in cellular composition and molecular features during IVDD. Consistently, our scRNA-seq data indicated the cellular complexity of degenerative NP tissues. We performed scRNA-seq on NP tissues from mild and severe degenerative IVDD patients and identified eight putative clusters, including chondrocytes, endothelial cells, fibroblasts, MPs, mural cells, osteoclasts, proliferating stromal cells and T cells (Fig. 1A,B). In addition to chondrocytes, the proportion of other cell clusters significantly increased with an aggravation of degenerative degree (Fig. 1C). Mural cells, including smooth muscle cells and pericytes, are integral components of blood vessels that play important roles in angiogenesis and vessel maturation. We found that the numbers of ACTA2+ and RGS5+ mural cells were increased in degenerative NP tissue, indicating vascularization in IVDD (Fig. 1D–F). Osteoclasts can not only act as antigen- presenting cells, but also secrete chemokines and cytokines to promote the recruitment and activation of T cells33. A cluster of ACP5+ CTSK+ MMP9+ osteoclasts was discovered in the degenerative NP tissue, which might exacerbate the inflammatory microenvironment of IVD (Fig. 1D–F). LGALS1, encoding galectin-1, has immunomodulatory functions in a variety of immune cells and plays a role in regulating apoptosis, cell proliferation and differentiation34,35. LGALS1 was widely expressed in the eight cell clusters, with particularly high levels in proliferating stromal cells, suggesting that it might play a role in the progression of IVDD (Fig. 1E).
Chondrocytes are the dominant cell type in NP tissue and divided into three subsets based on the expression of characteristic genes. A population of SOD3+ GPX3+ NR4A2+ chondrocytes (C1) was identified to potentially function in resisting oxidative stress. NP is a type of avascular tissue that is chronically exposed to a hypoxic microenvironment. The mitochondrial function of NP cells is severely impaired during the degenerative process of IVD. DEGs in the C1 cluster were enriched in mitophagy, which suggested that C1 might degrade abnormal mitochondria to maintain mitochondrial homeostasis and resist oxidative stress. Moreover, we discovered a novel cluster of ORM1/2+ SLPI+ TRIB3+ chondrocytes (C3). ORM1/2 and SLPI modulate immune responses by regulating NF-kappa-B and interleukin-1 production36,37, while TRIB3 acts as a stress sensor in response to hypoxia, high glucose and AGEs38, indicating a potential role of C3 in the response to inflammation and cellular stress. Interestingly, we found that DEGs in the C3 cluster exhibited strong enrichment in ferroptosis, which is a novel type of cell death characterized by iron-dependent lipid peroxidation and contributes to the pathogenesis of IVDD. Therefore, we proposed that excessive oxidative stress and inflammation might trigger ferroptosis in the C3 cluster and exacerbate disc degeneration. Notably, a cluster of PRG4+ COL1A1+ COL3A1+ chondrocytes (C2), closely associated with fibrosis, was identified. Developmental trajectories suggested that C1 displayed two differentiation fates to C2 and C3, and that C2 took the dominant position in the late stage. Consistently, the proportion of fibroblasts significantly increased in severely degenerative NP tissue. Pseudo-time trajectories also suggested chondrocytes gradually differentiated into fibroblasts during IVDD.
Endogenous repair is an important self-repair mechanism in IVD to maintain long-term homeostasis. Progenitor cells within IVD play a crucial role in the endogenous repair39. Local mesenchymal progenitor cells have been reported in human degenerated intervertebral discs18–22. ScRNA-seq also supports the existence of NP progenitor/stem cells expressing PROCR, PRRX1, CD44, LepR, CD105, THY1, and CD7323,24. Progenitor cells exhibited the ability to delay the progression of disc degeneration40–42. Therefore, it is a promising strategy to activate endogenous progenitor cells or transplant exogenous progenitor cells for IVDD therapy. Interestingly, our results revealed that both C1 and C2 expressed the progenitor cell marker TAGLN, and C2 preferentially expressed PROCR and PRRX1, confirming the observation that progenitor cells are present in NP tissues. Notably, the expression of TGFB1 in C2 was notably elevated, which might stimulate progenitor/stem cells differentiation into chondrocytes and promote fibroblast proliferation43,44. CD146/MCAM distinguishes stem cell subpopulations with distinct migration and regenerative potential in degenerative intervertebral discs45,46. The CD146+ MSC subpopulation held greater migration potential towards degenerative IVDs, while the CD146- cells induced a stronger regenerative response in the resident IVD cells. We found that none of the three chondrocyte clusters expressed CD146, suggesting that these chondrocytes were resident original cells.
NP is considered an immune-privileged structure surrounded by AF and CEP. Structural destruction of AF and compression of spinal nerves by herniation induce immune cell activation and infiltration10,11. A variety of immune cells, including MPs and T cells, were detected in degenerated NP tissue. The MP cluster was divided into three subclusters: CD1C+ CLEC10A+ cDC2s, CD68+ macrophages and FCN1+ monocytes. We revealed that macrophages highly expressed lysosomal protease CTSD and ferroptosis-related gene HMOX1. HO-1 is a stress-inducible enzyme encoded by the HMOX1 gene involved in the breakdown of haem and production of ferrous iron, which promotes ferroptosis47,48. However, HO-1 also acts as an important immunoregulator in macrophages, which is not only highly expressed in M1-like or pro-inflammatory macrophages, but also associated with M2-like macrophage polarization49. Cellular interaction analysis revealed that the SPP1-CD44 signaling pathway was highly activated between macrophages and chondrocytes. SPP1, a secreted multifunctional glycoprotein, has been reported to suppress hypertrophy and vascularization while stimulating chondrogenesis by interacting with CD4432. Therefore, we predicted that macrophage-secreted SPP1 might modulate the activity of chondrocytes via CD44 during IVDD progression. In addition, our scRNA-seq analysis revealed that monocytes highly expressed the cytokines CCL20 and IL1B and were enriched in the TNF, IL-17 and NF-kappa B signaling pathways. Further cellular interaction analysis revealed that the CCL4L2-PGRMC2 and CCL4L2-VSIR signaling pathways were strongly up-regulated between monocytes and chondrocytes. CCL4L2 participates in the MIF-mediated glucocorticoid regulation and TGF-beta pathway, which is a central mediator of fibrosis50–52. These results indicated that cytokines secreted by monocytes might cause the inflammatory microenvironment of chondrocytes and promote their transition to a fibrotic state.
Healthy nucleus pulposus is avascular, whereas with the degeneration of NPCs and degradation of ECM, vessel ingrowth is observed in degenerated IVDs53. Consistently, our results also revealed a significant increase in the proportion of ECs during IVDD. As a major angiogenic factor, VEGF binds to receptors on the surface of ECs to trigger proliferation, migration and promote survival54. In this study, we found that the VEGFA signaling pathways, including VEGFA-KDR, VEGFA-FLT1 and VEGFA-NRP1 pairs, were preferentially activated, with condrocytes serving as the signal senders and ECs as the receivers. The KDR gene encoding VEGFR-2 is responsible for VEGF-stimulated EC proliferation and migration, whereas FLT-1 (encoding VEGFR-1) down-modulates this process. The axon guidance molecule SEMA3A plays a key role in vascular and neuronal development55,56. In this study, SEMA3E-PLXND1 pathway was found to be activated, which controls angiogenesis and vascular development by regulating VEGF signaling pathway57. CellPhone DB analysis also revealed that with ECs as the signal senders and chondrocytes as the signal receivers, the COL4A1-a11b1 and CXCL12-ACKR3 signaling pathways were highly activated. COL4A1 is mostly expressed in adjacent stromal cells, such as cancer-associated fibroblasts (CAFs) and endothelial cells. COL4A1 expressed on ECs might modulate chondrocytes by interacting with the collagen receptor-integrin a11b1. Moreover, ECs might reprogramme cellular metabolism of chondrocytes via CXCL12-ACKR3, which regulates the glycolysis and pentose phosphate pathway58. These results suggested that chondrocytes might intercrosstalk with ECs via multiple ligand-receptor interactions during IVDD.
There are several limitations and problems to be resolved in this study. First, due to clinical difficulties in human sample collection, normal NP tissues were not included in this study. Therefore, we compared only the mildly and severely degenerated nucleus pulposus, which might not fully demonstrate the changes in composition at single-cell resolution. IVD has the ability to regenerate spontaneously, as evidenced by the self-healing properties following disc degeneration, which may be due to the presence of in situ progenitor cells39. Progenitor cells exhibit certain ability to slow disc degeneration59,60. Tan et al. report that TAGLN+ cells located in the periphery of the NP are proliferative and characteristic of progenitors24. The TAGLN+ PeriNP cells become diminished during age and puncture-induced disc degeneration. In our research, we also found TAGLN+ cells in C1 and C2 clusters. Moreover, progenitor cell markers PROCR and PRRX1 were preferentially expressed in the C2 cluster. Gan et al. also identify an nucleus pulposus progenitor cells (NPPCs) cluster that highly expressed PROCR and PRRX1, which exhibited pluripotency with colony-formation capacity and osteochondrogenic potentials61. These results suggest that the progenitor cells exist in NP tissues and possess the plasticity. In the future study, we will try to collect normal NP tissues as controls. In addition, it is indeed a challenge to isolate adequate cells from severely degenerative nucleus pulposus, resulting in the absence of the most degenerated disc (Pfirrmann degenerative grade V) in this study. Finally, experiments to validate the scRNA-seq results using large-scale clinical samples are lacking. Hence, we intend to conduct further research in the future to overcome the above limitations and problems.
In summary, our scRNA-seq results allowed for the identification of NP cellular heterogeneity at the single-cell and transcriptomic levels, followed by the analysis of intrinsic signatures in cell subsets (i.e., chondrocytes, fibroblasts and immune cells), the developmental trajectories of chondrocytes, and the interactions between different cell clusters, including chondrocytes and immune cells, chondrocytes and ECs. These new findings provide new insights into the changes at the cellular and molecular levels and facilitate the in-depth investigation of treatments for IVDD.
Conclusions
In conclusion, the present study utilized single-cell RNA sequencing to elucidate in-depth the cellular ecosystem of NP tissues in IVDD patients. The biological heterogeneity of chondrocytes and fibroblasts was revealed. A unique evolutionary trajectory of chondrocytes differentiation into fibroblasts was identified. We further elucidated the biological features of immune cells and endothelial cells. Interactions between chondrocytes and immune cells, chondrocytes and ECs were comprehensively analysed. These findings enhance our understanding of NPC complexity and offer insights into potential novel therapeutic targets for IVDD.
Supplementary Information
Acknowledgements
We would like to thank Singleron Biotechnologies for scRNA-seq and data analysis.
Abbreviations
- NP
Nucleus pulposus
- IVDD
Intervertebral disc degeneration
- scRNA-seq
Single-cell RNA sequencing
- GO
Gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- ECM
Extracellular matrix
- AF
Annulus fibrosus
- CEP
Cartilage endplate
- PCA
Principle component analysis
- UMAP
Uniform manifold approximation and projection
- t-SNE
T-distributed stochastic neighbor embedding
- HGT
Hypergeometric tests
- DEGs
Differentially expressed genes
- ECs
Endothelial cells
- Fibros
Fibroblasts
- MPs
Mononuclear phagocytes
- OCs
Osteoclasts
- CAFs
Cancer-associated fibroblasts
Author contributions
SJ and CYM conceived this study. SJ, HML, TY, SG, ZFZ and XG analysed the scRNA-seq data. SJ wrote the manuscript. CYM reviewed the manuscript. DRL, ZYZ, YHL, XL and YXW collected the NP tissue samples. All authors reviewed and approved the final version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 82372477), Youth Project of Natural Science Foundation of Shandong Province (Grant No. ZR2021QH059), Key Research and Development Plan of Jining (Grant No. 2022YXNS001), Traditional Chinese Medicine Science and Technology Project of Shandong Province (Grant No. M-2022245).
Data availability
All the single-cell RNA sequencing data has been deposited in NCBI GEO databases under accession number: GSE251686.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Medical Science Research and Ethics Committee of Affiliated Hospital of Jining Medical University. All methods were performed in accordance with the relevant guidelines and regulations.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78675-x.
References
- 1.Knezevic, N. N., Candido, K. D., Vlaeyen, J. W. S., Van Zundert, J. & Cohen, S. P. Low back pain. Lancet398, 78–92 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Diwan, A. D. & Melrose, J. Intervertebral disc degeneration and how it leads to low back pain. Jor Spine6, e1231 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher, C., Underwood, M. & Buchbinder, R. Non-specific low back pain. Lancet389, 736–747 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Fine, N. et al. Intervertebral disc degeneration and osteoarthritis: A common molecular disease spectrum. Nat. Rev. Rheumatol.19, 136–152 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Bydon, M. et al. Lumbar fusion versus nonoperative management for treatment of discogenic low back pain a systematic review and meta-analysis of randomized controlled trials. J. Spinal Disord. Tech.27, 297–304 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Humzah, M. D. & Soames, R. W. Human intervertebral disc: Structure and function. Anat. Rec.220, 337–356 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Roughley, P. J. Biology of intervertebral disc aging and degeneration: Involvement of the extracellular matrix. Spine29, 2691–2699 (2004). [DOI] [PubMed] [Google Scholar]
- 8.He, R. J. et al. HIF1A Alleviates compression-induced apoptosis of nucleus pulposus derived stem cells via upregulating autophagy. Autophagy17, 3338–3360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo, J. Y., Yang, Y. X., Wang, X., Chang, X. Y. & Fu, S. B. Role of Pyroptosis in intervertebral disc degeneration and its therapeutic implications. Biomolecules12, 1804 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun, Z., Liu, B. & Luo, Z. J. The immune privilege of the intervertebral disc: Implications for intervertebral disc degeneration treatment. Int. J. Med. Sci.17, 685–692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye, F. B. A., Lyu, F. J., Wang, H. & Zheng, Z. M. The involvement of immune system in intervertebral disc herniation and degeneration. Jor Spine5, 1196 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, B. et al. Discovery and application of postnatal nucleus pulposus progenitors essential for intervertebral disc homeostasis and degeneration. Adv. Sci.9, 104888 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun, H., Wang, H., Zhang, W. D., Mao, H. J. & Li, B. Single-cell RNA sequencing reveals resident progenitor and vascularization-associated cell subpopulations in rat annulus fibrosus. J. Orthop. Transl.38, 256–267 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling, Z. M. et al. Single-Cell RNA-Seq analysis reveals macrophage involved in the progression of human intervertebral disc degeneration. Front. Cell Dev. Biol.9, 833420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, X. et al. Single-cell sequencing: New insights for intervertebral disc degeneration. Biomed. Pharmacother.165, 115224 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Lin, P. et al. Spatially multicellular variability of intervertebral disc degeneration by comparative single-cell analysis. Cell Proliferat.56, 13464 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou, T. F. et al. Spatiotemporal characterization of human early intervertebral disc formation at single-cell resolution. Adv. Sci.10, 6296 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriksson, H. et al. Identification of cell proliferation zones, progenitor cells and a potential stem cell niche in the intervertebral disc region: A study in four species. Spine34, 2278–2287 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Risbud, M. V. et al. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine32, 2537–2544 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Huang, S. et al. Coupling of small leucine-rich proteoglycans to hypoxic survival of a progenitor cell-like subpopulation in Rhesus Macaque intervertebral disc. Biomaterials34, 6548–6558 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Brisby, H. et al. The presence of local mesenchymal progenitor cells in human degenerated intervertebral discs and possibilities to influence these in vitro: A descriptive study in humans. Stem Cells Dev.22, 804–814 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Blanco, J. F. et al. Isolation and characterization of mesenchymal stromal cells from human degenerated nucleus pulposus: Comparison with bone marrow mesenchymal stromal cells from the same subjects. Spine35, 2259–2265 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Tu, J. et al. Single-cell transcriptome profiling reveals multicellular ecosystem of nucleus pulposus during degeneration progression. Adv. Sci. (Weinh.)9, e2103631 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan, Z. et al. Progenitor-like cells contributing to cellular heterogeneity in the nucleus pulposus are lost in intervertebral disc degeneration. Cell Rep.43, 114342 (2024). [DOI] [PubMed] [Google Scholar]
- 25.Zhang, H., La Marca, F., Hollister, S. J., Goldstein, S. A. & Lin, C. Y. Developing consistently reproducible intervertebral disc degeneration at rat caudal spine by using needle puncture. J. Neurosurg. Spine10, 522–530 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Weiskirchen, R., Weiskirchen, S. & Tacke, F. Organ and tissue fibrosis: Molecular signals, cellular mechanisms and translational implications. Mol. Aspects Med.65, 2–15 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Chen, C. et al. Autologous fibroblasts induce fibrosis of the nucleus pulposus to maintain the stability of degenerative intervertebral discs. Bone Res.8, 7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng, Z. et al. Fibromodulin reduces scar formation in adult cutaneous wounds by eliciting a fetal-like phenotype. Signal Transduct. Target2, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochi, K., Derfoul, A. & Tuan, R. S. A predominantly articular cartilage-associated gene, SCRG1, is induced by glucocorticoid and stimulates chondrogenesis in vitro. Osteoarthr. Cartilage14, 30–38 (2006). [DOI] [PubMed] [Google Scholar]
- 30.He, C. L. et al. Quantitative proteomic analysis of Bi Zhong Xiao decoction against collagen-induced arthritis rats in the early and late stages. Bmc Complement. Med.22, 5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bengtsson, E., Lindblom, K., Tillgren, V. & Aspberg, A. The leucine-rich repeat protein PRELP binds fibroblast cell-surface proteoglycans and enhances focal adhesion formation. Biochem. J.473, 1153–1164 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Frahs, S. M. et al. Prechondrogenic ATDC5 cell attachment and differentiation on graphene foam; modulation by surface functionalization with fibronectin. Acs Appl. Mater. Inter.11, 41906–41924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park-Min, K. H. & Lorenzo, J. Osteoclasts: Other functions. Bone165, 116576 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Alhabbab, R. et al. Galectin-1 is required for the regulatory function of B cells. Sci. Rep.8, 2725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong, L. et al. Comprehensive single-cell sequencing reveals the stromal dynamics and tumor-specific characteristics in the microenvironment of nasopharyngeal carcinoma. Nat. Commun.12, 1540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gemelli, C. et al. The Orosomucoid 1 protein is involved in the vitamin D—mediated macrophage de-activation process. Exp. Cell Res.319, 3201–3213 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Nugteren, S. & Samsom, J. N. Secretory Leukocyte Protease Inhibitor (SLPI) in mucosal tissues: Protects against inflammation, but promotes cancer. Cytokine Growth F R59, 22–35 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Izrailit, J., Jaiswal, A., Zheng, W., Moran, M. F. & Reedijk, M. Cellular stress induces TRB3/USP9x-dependent Notch activation in cancer. Oncogene36, 1048–1057 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Lyu, F. J. et al. IVD progenitor cells: A new horizon for understanding disc homeostasis and repair. Nat. Rev. Rheumatol.15, 102–112 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Pettine, K. A., Murphy, M. B., Suzuki, R. K. & Sand, T. T. Percutaneous injection of autologous bone marrow concentrate cells significantly reduces lumbar discogenic pain through 12 months. Stem Cells33, 146–156 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Mahla, R. S. Stem cells applications in regenerative medicine and disease therapeutics. Int. J. Cell Biol.2016, 6940283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, W. et al. Dual inhibition of HDAC and tyrosine kinase signaling pathways with CUDC-907 attenuates TGFβ1 induced lung and tumor fibrosis. Cell Death Dis.11, 765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia, P. et al. TGF-β1-induced chondrogenesis of bone marrow mesenchymal stem cells is promoted by low-intensity pulsed ultrasound through the integrin-mTOR signaling pathway. Stem Cell Res. Ther.8, 281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wangler, S. et al. CD146/MCAM distinguishes stem cell subpopulations with distinct migration and regenerative potential in degenerative intervertebral discs. Osteoarthr. Cartilage27, 1094–1105 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Papadimitriou, N. et al. Intradiscal injection of iron-labeled autologous mesenchymal stromal cells in patients with chronic low back pain: A feasibility study with 2 years follow-up. Int. J. Spine Surg.15, 1201–1209 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, Z. M. et al. HO-1-mediated ferroptosis as a target for protection against retinal pigment epithelium degeneration. Redox Biol.43, 101971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menon, A. V. et al. Excess heme upregulates heme oxygenase 1 and promotes cardiac ferroptosis in mice with sickle cell disease. Blood139, 936–941 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell, N. K., Fitzgerald, H. K. & Dunne, A. Regulation of inflammation by the antioxidant haem oxygenase 1. Nat. Rev. Immunol.21, 411–425 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Zhu, L. et al. Controlled release of TGF-β3 for effective local endogenous repair in IDD using rat model. Int. J. Nanomed.17, 2079–2096 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nie, C. H. et al. A comparative genomic database of skeletogenesis genes: From fish to mammals. Comp. Biochem. Phys. D38, 100796 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Zhu, H. et al. Identification of pathogenic immune cell subsets associated with checkpoint inhibitor-induced myocarditis. Circulation146, 316–335 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cornejo, M. C., Cho, S. K., Giannarelli, C., Iatridis, J. C. & Purmessur, D. Soluble factors from the notochordal-rich intervertebral disc inhibit endothelial cell invasion and vessel formation in the presence and absence of pro-inflammatory cytokines. Osteoarthr. Cartilage23, 487–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pulkkinen, H. H. et al. BMP6/TAZ-Hippo signaling modulates angiogenesis and endothelial cell response to VEGF. Angiogenesis24, 129–144 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moutal, A. et al. SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain162, 243–252 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, H. Y., Lu, Y., Wu, B. B. & Xia, F. Semaphorin 3A mitigates lipopolysaccharide-induced chondrocyte inflammation, apoptosis and extracellular matrix degradation by binding to Neuropilin-1. Bioengineered12, 9641–9654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, R. et al. Vascular Sema3E-Plexin-D1 signaling reactivation promotes post-stroke recovery through VEGF downregulation in mice. Transl. Stroke Res.13, 142–159 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luker, K. E. & Luker, G. D. The CXCL12/CXCR4/ACKR3 signaling axis regulates PKM2 and glycolysis. Cells-Basel11, 1775 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du, Y., Wang, Z., Wu, Y., Liu, C. & Zhang, L. Intervertebral disc stem/progenitor cells: A promising, “seed” for intervertebral disc regeneration. Stem Cells Int.2021, 2130727 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, Y. et al. Characterization of the nucleus pulposus progenitor cells via spatial transcriptomics. Adv. Sci. (Weinh.)11, e2303752 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gan, Y. et al. Spatially defined single-cell transcriptional profiling characterizes diverse chondrocyte subtypes and nucleus pulposus progenitors in human intervertebral discs. Bone Res.9, 37 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the single-cell RNA sequencing data has been deposited in NCBI GEO databases under accession number: GSE251686.