Abstract
Fibronectin type III domain-containing (FNDC) proteins play critical roles in cellular homeostasis and cardiac injury, and our recent findings define FNDC5 as a promising cardioprotectant against doxorubicin- and aging-related cardiac injury. FNDC4 displays a high homology with FNDC5; however, its role and mechanism in cardiac ischemia/reperfusion (I/R) injury remain elusive. Here, we show that cardiac and plasma FNDC4 levels are elevated during I/R injury in a hypoxia-inducible factor 1α (HIF1α)-dependent manner. Cardiac-specific FNDC4 overexpression facilitates, while cardiac-specific FNDC4 knockdown inhibits cardiomyocyte survival and angiogenesis in I/R-stressed hearts of male mice through regulating the proteasomal degradation of HIF1α. Interestingly, FNDC4 does not directly stimulate angiogenesis of endothelial cells, but increases the expression and secretion of fibroblast growth factor 1 from cardiomyocytes to enhance angiogenesis in a paracrine manner. Moreover, therapeutic administration of recombinant FNDC4 protein is sufficient to alleviate cardiac I/R injury in male mice, without resulting in significant side effects. In this work, we reveal that FNDC4 alleviates cardiac I/R injury through facilitating HIF1α-dependent cardiomyocyte survival and angiogenesis, and define FNDC4 as a promising predictive and therapeutic target of cardiac I/R injury.
Subject terms: Cardiovascular diseases, Ubiquitylation
Cardiac ischemia/reperfusion injury is a deleterious consequence of reperfusion therapy for myocardial infarction. Here, the authors show that FNDC4 alleviates cardiac ischemia/reperfusion injury through facilitating HIF1α-dependent cardiomyocyte survival and angiogenesis.
Introduction
Coronary recanalization is a pivotal strategy to rescue the viable cardiomyocytes during acute myocardial injury; however, it also induces a second wave of cardiac damage, termed ischemia/reperfusion (I/R) injury1. In addition, cardiac I/R injury also results in no-reflow phenomenon after coronary recanalization, thereby exacerbating acute cardiac injury and chronic cardiac remodeling2. Therapeutic angiogenesis accelerates the formation of collateral circulation, improves the blood flow of the ischemic myocardium and subsequently alleviates cardiac I/R injury. Hypoxia-inducible factor 1α (HIF1α) emerges as an oxygen-sensitive transcription factor that orchestrates the adaptive response to hypoxia and ischemia through increasing the expression of various pro-angiogenic factors, including vascular endothelial growth factor (VEGF), angiopoietin (ANGPT), angiopoietin-like (ANGPTL), insulin like growth factor (IGF), slit guidance ligand (SLIT) and fibroblast growth factor (FGF), etc3,4. Under normoxic conditions, HIF1α is hydroxylated by prolyl hydroxylase domain (PHD) proteins, bound and ubiquinated by E3 ubiquitin ligase von Hippel-Lindau protein (VHL), and eventually delivered to the proteasome for degradation. In response to hypoxia, HIF1α is stabilized and accumulated to the nucleus to activate an angiogenic transcription program5,6. Moreover, HIF1α activation also directly prevents cardiomyocyte apoptosis during cardiac I/R injury, and facilitates functional recovery of the heart5,7,8. In addition, our recent studies revealed that inhibiting cardiomyocyte apoptosis dramatically mitigated I/R-induced acute cardiac injury, chronic cardiac remodeling and cardiac dysfunction9,10. These findings identify HIF1α as a promising therapeutic target to treat cardiac I/R injury.
Fibronectin type III domain-containing (FNDC) proteins, distinguished by the conserved fibronectin type III domains, belong to a unique extracellular matrix protein family and play critical roles in cellular homeostasis and cardiac injury11–13. Sato et al. found that FNDC1 expression was elevated by ischemic stimuli in isolated adult ventricular cardiomyocytes but not in other cell types, and that FNDC1 knockdown completely prevented hypoxia-induced cardiomyocyte apoptosis14,15. You and colleagues determined that hepatocyte-restrict FNDC3B ablation exacerbated alcohol-induced liver steatosis and ferroptosis16. Intriguingly, overexpression of circFndc3b, a non-coding RNA of FNDC3B gene, in endothelial cells dramatically enhanced their angiogenic activity and reduced cardiomyocyte apoptosis, eventually promoting cardiac repair after myocardial infarction17. FNDC5 is a well-known myokine to regulate energy expenditure, and also functions as the most studied FNDC proteins in cardiovascular diseases13,18. Lecker et al. demonstrated that FNDC5 expression positively correlated with aerobic exercise performance in patients with heart failure19. Further findings by Li et al. suggested that supplementation of irisin, a cleaved and secreted fragment of FNDC5, could prevent pressure overload-induced cardiac hypertrophy and dysfunction20. Meanwhile, our recent studies revealed that FNDC5 overexpression could attenuate inflammation, oxidative stress and cardiomyocyte apoptosis, thereby diminishing aging- or doxorubicin-induced cardiac dysfunction21,22. FNDC4 is a type I transmembrane glycoprotein and displays a high homology with FNDC5. Bosma et al. previously revealed that administration of recombinant FNDC4 (rFNDC4) protein could suppress the disease severity of colitis in mice, and that FNDC4-deficient mice developed more severe colitis23. In addition, liver-derived soluble FNDC4 promoted insulin signaling and insulin-mediated glucose uptake in white adipocytes, and subsequently improved glucose tolerance in pre-diabetic mice24. Yet, the role and molecular basis of FNDC4 in cardiovascular diseases, especially in cardiac I/R injury, remain unclear.
Here, we show that cardiac and plasma FNDC4 levels are elevated during I/R injury in a HIF1α-dependent manner, and that higher FNDC4 expression predicts better cardiac function and prognosis. Cardiac-specific FNDC4 overexpression facilitates, while cardiac-specific FNDC4 knockdown inhibits cardiomyocyte survival and angiogenesis in I/R-stressed hearts. The unbiased transcriptome analysis reveals that FNDC4 blocks the proteasomal degradation of HIF1α and subsequently activates HIF1α signaling pathway to exert the cardioprotective effects. Meanwhile, FNDC4 does not directly stimulate angiogenesis of endothelial cells, but increases the expression and secretion of FGF1 from cardiomyocytes to enhance angiogenesis in a paracrine manner. Moreover, therapeutic administration of rFNDC4 protein is sufficient to alleviate cardiac I/R injury, without resulting in significant side effects.
Results
FNDC4 expression is elevated during cardiac I/R injury in a HIF1α-dependent manner
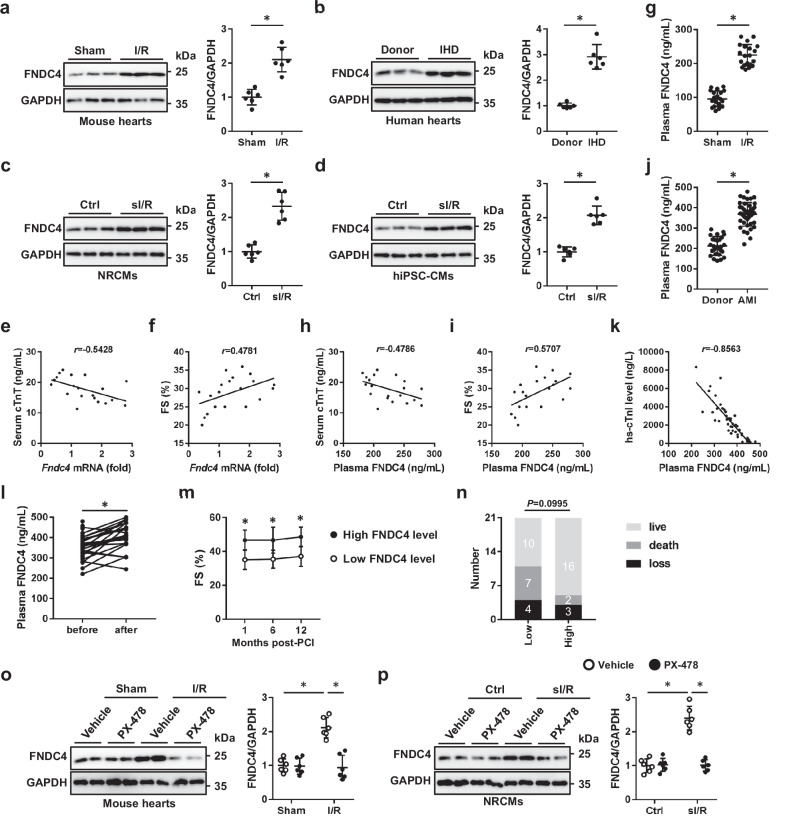
Previous studies by us and the others have identified the cardioprotective role of FNDC5 in response to different stresses, including doxorubicin, aging and pressure overload, etc20–22. FNDC4 displays a high homology with FNDC5; however, its role and mechanism in cardiac I/R injury remain unclear. To address this issue, we firstly measured the expression of FNDC4 during cardiac I/R injury. As shown in Fig. 1a, FNDC4 protein level was dramatically elevated in I/R-stressed hearts, compared with that in sham-operated hearts. Meanwhile, we found that the hearts from ischemic heart disease (IHD) patients also displayed higher FNDC4 expression (Fig. 1b). Our recent study revealed that cardiac FNDC4 was abundantly localized to cardiomyocytes, so we measured the alteration of FNDC4 expression in cardiomyocytes in vitro. Consistent with the in vivo findings, FNDC4 protein in neonatal rat cardiomyocytes (NRCMs) was also elevated by simulated I/R (sI/R) stimulation (Fig. 1c). To further confirm the results, human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) were prepared and exposed to sI/R stimulation as we recently described9. As shown in Fig. 1d, FNDC4 expression was dramatically increased in sI/R-stimulated human cardiomyocytes. Interestingly, Fndc4 mRNA level in I/R-stressed murine hearts negatively correlated with serum cardiac isoform of tropnin T (cTnT) level, a biomarker of cardiomyocyte injury (Fig. 1e). In contrast, Fndc4 mRNA level positively correlated with the systolic function of I/R-stressed hearts (Fig. 1f). FNDC4 is a type I transmembrane glycoprotein and can be cleaved to a soluble bioactive fragment, and we thus detected plasma FNDC4 in I/R- or sham-operated mice. As shown in Fig. 1g, plasma FNDC4 level in mice was dramatically increased by I/R surgery. Interestingly, plasma FNDC4 level negatively correlated with serum cTnT level, but positively correlated with fractional shortening (FS) in I/R-stressed mice (Fig. 1h, i). To examine the clinical relevance of elevated plasma FNDC4 in I/R-injured mice, we first measured plasma FNDC4 level in acute myocardial infarction (AMI) patients, and found that plasma FNDC4 level at admission was dramatically increased in AMI patients in comparison to healthy controls (Fig. 1j). In addition, AMI patients with higher plasma FNDC4 level displayed lower plasma high-sensitivity tropnin I (hs-TnI) level, a biomarker of cardiomyocyte injury (Fig. 1k). Moreover, plasma FNDC4 level in AMI patients at admission was further increased 24 h after percutaneous coronary intervention (PCI) surgery (Fig. 1l). Next, we divided AMI patients into low FNDC4 and high FNDC4 groups using the median of plasma FNDC4 post-PCI as the threshold. As shown in Fig. 1m, AMI patients with higher plasma FNDC4 level post-PCI displayed better cardiac function, as evidenced by the increased FS. Of note, higher plasma FNDC4 level also predicted a trend of lower mortality rate, with comparable ratio of loss to follow-up (Fig. 1n). Previous studies by us and the others have shown that the oxygen-dependent transcription factor HIF1α contributes to cardioprotection against ischemic stimuli through controlling the expressions of various proteins10,25. We next investigated whether FNDC4 upregulation in response to cardiac I/R injury was secondary to the activation of HIF1α. Various putative binding sites of HIF1α (5′-RCGTG-3′) were identified in the promoter of FNDC4. As shown in Fig. 1o, p, HIF1α inhibition dramatically suppressed the increases of FNDC4 expression in I/R-stressed hearts and sI/R-stimulated NRCMs. Collectively, we demonstrate that cardiac and plasma FNDC4 levels are elevated during I/R injury in a HIF1α-dependent manner, and that higher FNDC4 expression predicts better cardiac function and prognosis.
Fig. 1. FNDC4 expression is elevated during cardiac I/R injury in a HIF1α-dependent manner.
a The hearts with or without I/R injury were harvested for western blot (n = 6). b The left ventricles from ischemic heart disease (IHD) patients or donors were prepared for western blot (n = 6). c, d Neonatal rat cardiomyocytes (NRCMs) and human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) with or without sI/R injury were prepared for western blot (n = 6 independent experiments). e, f Correlations between cardiac Fndc4 mRNA and serum cardiac isoform of tropnin T (cTnT) or fractional shortening (FS) in I/R-stressed mice (n = 20). g Plasma FNDC4 levels in sham- or I/R-stressed mice (n = 20). h, i Correlations between plasma FNDC4 and serum cTnT or FS in I/R-stressed mice (n = 20). j Plasma FNDC4 levels in acute myocardial infarction (AMI) patients or healthy controls (n = 26 for donors and n = 42 for AMI patients). k Correlation between plasma FNDC4 and high-sensitivity tropnin I (hs-TnI) in AMI patients (n = 42). l Plasma FNDC4 levels in AMI patients at admission and after PCI surgery (n = 42). m FS in AMI patients with low or high FNDC4 levels at 1, 6 and 12 months post-PCI surgery (n = 10 for low FNDC4 group and n = 16 for high FNDC4 group). n The clinical outcome of AMI patients with low or high FNDC4 levels after PCI surgery (n = 21). o The hearts with or without HIF1α inhibition were prepared for western blot (n = 6). p NRCMs with or without HIF1α inhibition were prepared for western blot (n = 6 independent experiments). Data were presented as the mean ± S.D., and analyzed using an unpaired two-tailed Student′s t-test. For the analysis in (e–k), Pearson’s correlation analysis was used. For the analysis in (l), a paired two-tailed Student′s t-test was performed. For the analysis in (m), repeated measures ANOVA was conducted. For the analysis in (n), a two-tailed χ2 test with Yates’ correction was used, χ2 = 2.713, df = 1, z = 1.647. For the analysis in (o, p), one-way ANOVA followed by Tukey post hoc test was used. *P < 0.0001. Source data are provided as a Source Data file.
Cardiac-specific FNDC4 overexpression facilitates cardiomyocyte survival and angiogenesis during cardiac I/R injury
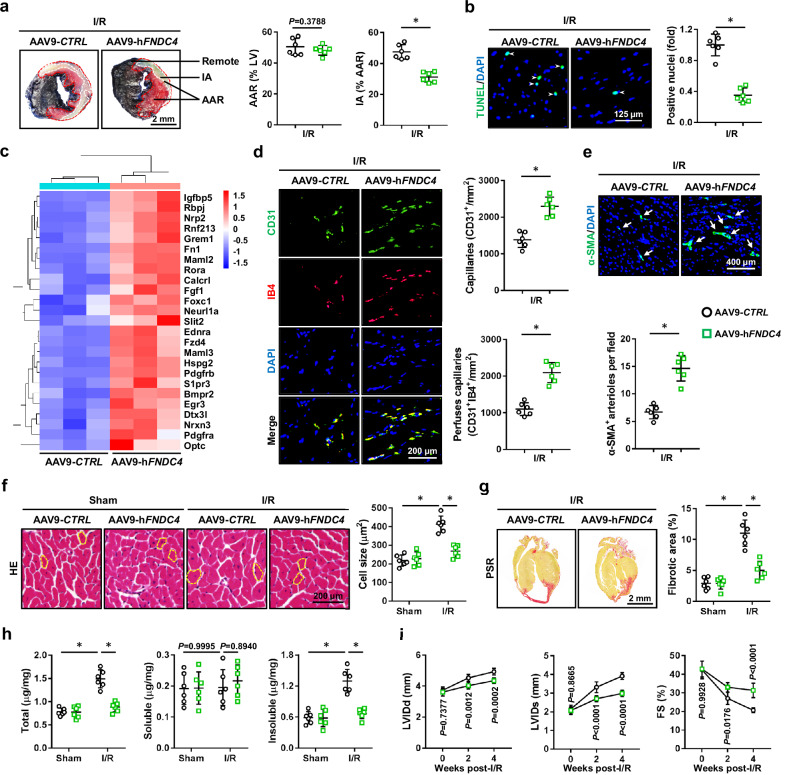
To investigate the cardiomyocyte-autonomous role of FNDC4 during cardiac I/R injury, mice were intravenously injected with AAV9-hFNDC4 to specifically overexpress FNDC4 in the myocardium. Intriguingly, FNDC4 overexpression did not affect the body weight in mice (28.3 ± 2.44 g versus 27.6 ± 3.28 g), which was consistent with a previous study by Georgiadi et al.24. As shown in Fig. 2a, FNDC4-overexpressed mice exhibited smaller infarct area (IA) in response to cardiac I/R injury, despite a similar surgical injury as the control mice. Accordingly, serum levels of cTnT, creatine kinase isoenzymes (CK-MB) and lactate dehydrogenase (LDH) were decreased in I/R-stressed mice with FNDC4 overexpression (Supplementary Fig. 1a). Meanwhile, I/R-induced cell apoptosis in the myocardium was also mitigated by FNDC4 overexpression, as determined by the decreased levels of TdT-mediated dUTP nick end-labeling (TUNEL)+ nuclei and DNA fragments (Fig. 2b and Supplementary Fig. 1b). Consistent with the phenotypic alteration, B-cell lymphoma-2 (BCL-2) protein was elevated, while BCL-2-associated X protein (BAX) expression was reduced in FNDC4-overexpressed hearts upon I/R injury, accompanied with a decreased caspase3 activity (Supplementary Fig. 1c, d). Transcriptome analysis also revealed that cell death-related genes and GO terms in I/R-stressed hearts were dramatically regulated by FNDC4 overexpression (Supplementary Fig. 1e, f). As shown in Supplementary Fig. 2a, FNDC4 overexpression also regulated angiogenic pathways during cardiac I/R injury, including endothelial cell differentiation, vascular endothelial growth factor binding, angiogenesis, Notch signaling pathway and vascular endothelial growth factor receptor activity, etc. Similarly, the pro-angiogenic genes in I/R-stressed hearts were dramatically upregulated by FNDC4 overexpression (Fig. 2c). As expected, FNDC4 overexpression dramatically increased the densities of capillaries and arterioles in the myocardium 4 weeks post-I/R surgery (Fig. 2d, e). We also investigated whether cardiac-specific FNDC4 overexpression elicited long-term structural or functional protections against cardiac I/R injury. As shown in Fig. 2f, g, histological staining revealed that I/R-induced cardiac hypertrophy and fibrotic remodeling were effectively inhibited by FNDC4 overexpression. The dysregulated mRNA levels of hypertrophic and fibrotic markers in I/R-stressed hearts were also restored by FNDC4 overexpression, as evidenced by the decreased mRNA levels of β-Mhc, Col1α1, Col3α1, and increased mRNA level of α-Mhc (Supplementary Fig. 2b). In addition to the level of collagen deposition, the extent of collagen cross-linking in the heart also correlated with cardiac function. Interestingly, we found that cardiac-specific FNDC4 overexpression dramatically reduced the amount of insoluble cross-linked collagen, without affecting the level of soluble collagen (Fig. 2h). Consistent with the improvements of acute cardiac injury and long-term structural remodeling, echocardiographic analysis identified an alleviative cardiac hypertrophy and dysfunction in I/R-injured mice with FNDC4 overexpression, as determined by the decreased left ventricle internal diameters at diastole (LVIDd), left ventricle internal diameters at systole (LVIDs), interventricular septal thickness at diastole (IVSd), interventricular septal thickness at systole (IVSs), and increased FS (Fig. 2i and Supplementary Fig. 2c). Of note, no difference in heart rate (HR) was identified between groups before- or post-I/R surgery (Supplementary Fig. 2d). Collectively, we demonstrate that cardiac-specific FNDC4 overexpression facilitates cardiomyocyte survival and angiogenesis, thereby preventing cardiac remodeling and dysfunction during cardiac I/R injury.
Fig. 2. Cardiac-specific FNDC4 overexpression facilitates cardiomyocyte survival and angiogenesis during cardiac I/R injury.
a Evans blue and TTC double staining was performed to identify the infarct area (IA), area at risk (AAR) and remote area of heart samples 24 h after I/R surgery (n = 6). b Heart samples with or without FNDC4 overexpression were collected 24 h after I/R surgery and subjected to TUNEL staining, and TUNEL+ nuclei were quantified. Arrows indicate TUNEL+ nuclei (n = 6). c Heart samples with or without FNDC4 overexpression were collected 24 h after I/R surgery and subjected to unbiased transcriptome analysis, and the expressions of angiogenesis-related genes were presented using a heatmap (n = 3). d, e Heart samples with or without FNDC4 overexpression were collected 4 weeks after I/R surgery and subjected to immunofluorescence staining, and the numbers of capillaries as well as arterioles were quantified. Arrows indicate α-SMA+ arterioles (n = 6). f, g Heart samples with or without FNDC4 overexpression were collected 4 weeks after I/R surgery and subjected to hematoxylin-eosin (HE) and picrosirius red (PSR) staining, and cell size as well as fibrotic area were quantified. Circles indicate the cross-sectional area of cardiomyocytes (n = 6). h Total, soluble and insoluble collagen content in the heart 4 weeks post-I/R surgery (n = 6). i Cardiac function of FNDC4-overexpressed and control mice was analyzed by transthoracic echocardiography at the indicated time points (n = 6). Data were presented as the mean ± S.D., and analyzed using an unpaired two-tailed Student′s t-test. For the analysis in (f–h), one-way ANOVA followed by Tukey post hoc test was used. For the analysis in (i), repeated measures ANOVA was performed. *P < 0.0001. Source data are provided as a Source Data file.
Cardiac-specific FNDC4 knockdown inhibits cardiomyocyte survival and angiogenesis during cardiac I/R injury
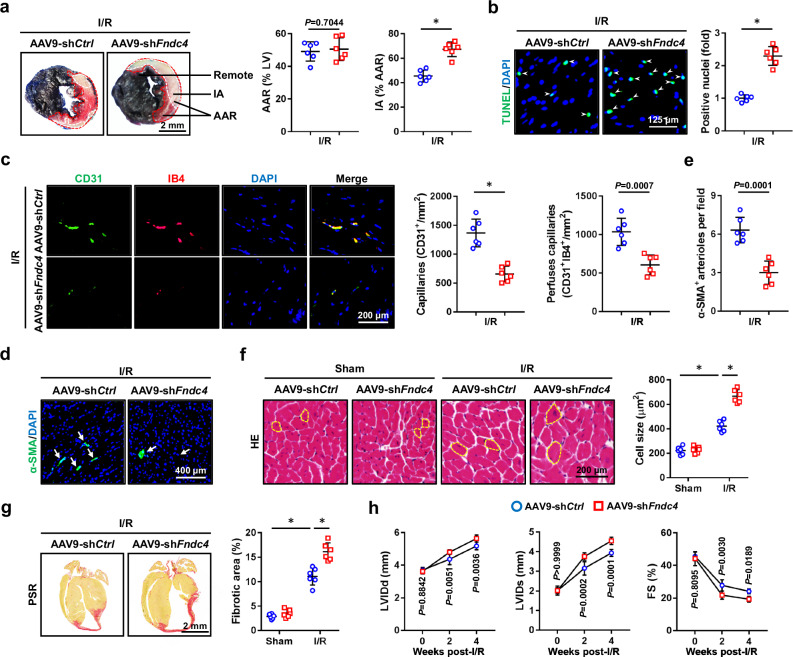
To further decipher the role of FNDC4, we specifically knocked down its expression in the myocardium using AAV9 vectors. As expected, the body weight was unaltered by FNDC4 knockdown (29.1 ± 3.76 g versus 29.5 ± 5.03 g). As shown in Fig. 3a and Supplementary Fig. 3a, cardiac-specific FNDC4 knockdown dramatically increased IA in I/R-stressed hearts, accompanied with elevated levels of serum cTnT, CK-MB and LDH. Meanwhile, I/R-induced cell apoptosis in the myocardium was also aggravated by FNDC4 knockdown, as determined by the increased levels of TUNEL+ nuclei and DNA fragments (Fig. 3b and Supplementary Fig. 3b). Consistently, the pro-apoptotic protein, BAX, was upregulated, while the anti-apoptotic protein, BCL-2, was downregulated in I/R-stressed hearts by FNDC4 knockdown, accompanied with an increased caspase3 activity (Supplementary Fig. 3c, d). In addition, cardiac-specific FNDC4 knockdown also inhibited angiogenesis in the myocardium under I/R stresses (Fig. 3c–e). I/R-induced increases of cardiomyocyte hypertrophy, interstitial fibrosis as well as collagen cross-linking were further exacerbated by FNDC4 knockdown (Fig. 3f, g and Supplementary Fig. 3e). Accordingly, the increased mRNA levels of β-Mhc, Col1α1, Col3α1 were further elevated, while the decreased mRNA level of α-Mhc was reduced in I/R-stressed hearts by FNDC4 knockdown (Supplementary Fig. 3f, g). Echocardiographic results showed that cardiac-specific FNDC4 knockdown further exacerbated I/R-induced ventricular dilation, hypertrophic growth and systolic dysfunction, as determined by the increased LVIDd, LVIDs, IVSd, IVSs, and decreased FS (Fig. 3h and Supplementary Fig. 3h). As expected, HR was unaffected by FNDC4 knockdown either at baseline or under I/R stresses (Supplementary Fig. 3i). Collectively, we demonstrate that cardiac-specific FNDC4 knockdown inhibits cardiomyocyte survival and angiogenesis, thereby facilitating cardiac remodeling and dysfunction during cardiac I/R injury.
Fig. 3. Cardiac-specific FNDC4 knockdown inhibits cardiomyocyte survival and angiogenesis during cardiac I/R injury.
a Evans blue and TTC double staining was performed to identify the IA, AAR and remote area of heart samples 24 h after I/R surgery (n = 6). b Heart samples with or without FNDC4 knockdown were collected 24 h after I/R surgery and subjected to TUNEL staining, and TUNEL+ nuclei were quantified. Arrows indicate TUNEL+ nuclei (n = 6). c–e Heart samples with or without FNDC4 knockdown were collected 4 weeks after I/R surgery and subjected to immunofluorescence staining, and the numbers of capillaries as well as arterioles were quantified. Arrows indicate α-SMA+ arterioles (n = 6). f, g Heart samples with or without FNDC4 knockdown were collected 4 weeks after I/R surgery and subjected to HE and PSR staining, and cell size as well as fibrotic area were quantified. Circles indicate the cross-sectional area of cardiomyocytes (n = 6). h Cardiac function of FNDC4-silenced and control mice was analyzed by transthoracic echocardiography at the indicated time points (n = 6). Data were presented as the mean ± S.D., and analyzed using an unpaired two-tailed Student′s t-test. For the analysis in (f, g), one-way ANOVA followed by Tukey post hoc test was used. For the analysis in (h), repeated measures ANOVA was performed. *P < 0.0001. Source data are provided as a Source Data file.
FNDC4 overexpression prevents, while FNDC4 knockdown exacerbates sI/R-induced cardiomyocyte injury in vitro
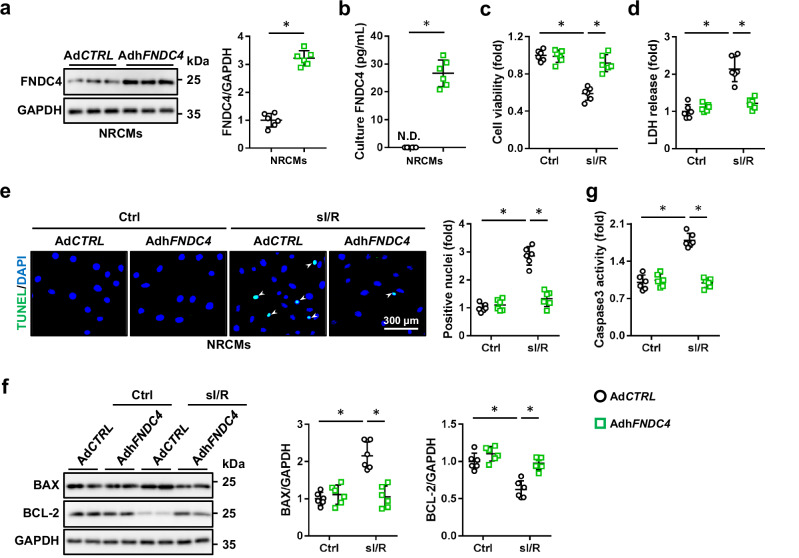
Next, NRCMs were overexpressed with FNDC4 to further confirm its cellular protection against I/R injury in vitro, and FNDC4 protein level was dramatically elevated in the lysates of AdhFNDC4-infected NRCMs (Fig. 4a). FNDC4 can be released as a soluble bioactive fragment to exert its biological functions, and we thus measured extracellular FNDC4 in cell medium. As shown in Fig. 4b, human FNDC4 protein was not detectable (N.D.) in the medium of AdCTRL-infected rat cardiomyocytes, but increased in the medium of AdhFNDC4-infected NRCMs. Consistent with the in vivo findings, we determined that FNDC4 overexpression dramatically attenuated sI/R-induced cardiomyocyte injury and death, as evidenced by the increased cell viability and decreased LDH releases (Fig. 4c, d). Results of TUNEL staining also identified an anti-apoptotic effect of FNDC4 (Fig. 4e). Accordingly, the increased levels of BAX protein and caspase3 activity were reduced, while the decreased BCL-2 protein level was elevated by FNDC4 overexpression in sI/R-treated NRCMs (Fig. 4f, g). In addition, NRCMs were also infected with shFndc4 to knock down the expression of FNDC4, and FNDC4 level in cell medium was also reduced by FNDC4 knockdown (Supplementary Fig. 4a, b). As shown in Supplementary Fig. 4c, d, sI/R-induced cardiomyocyte injury and death were further enhanced in shFndc4-infected NRCMs, as evidenced by the decreased cell viability and increased LDH releases. Meanwhile, FNDC4-silenced NRCMs also displayed a higher apoptotic rate in response to sI/R stimulation, compared with control cells (Supplementary Fig. 4e, f). Collectively, we demonstrate that FNDC4 overexpression prevents, while FNDC4 knockdown exacerbates sI/R-induced cardiomyocyte injury in vitro.
Fig. 4. FNDC4 overexpression prevents sI/R-induced cardiomyocyte injury in vitro.
a NRCMs with or without FNDC4 overexpression were harvested for western blot (n = 6 independent experiments). b FNDC4 level in the medium of NRCMs with or without FNDC4 overexpression (n = 6 independent experiments). c Cell viability was determined using the cell counting kit-8 (CCK-8) method (n = 6 independent experiments). d Lactate dehydrogenase (LDH) releases were calculated as (LDH level in ischemia medium + LDH in reperfusion medium)/(LDH in ischemia medium + LDH in reperfusion medium + LDH in cell lysate) (n = 6 independent experiments). e Representative TUNEL staining images of cell coverslips and quantitative results. Arrows indicate TUNEL + nuclei (n = 6 independent experiments). f NRCMs were collected for western blot (n = 6 independent experiments). g Caspase3 activity in NRCMs (n = 6 independent experiments). Data were presented as the mean ± S.D., and analyzed using one-way ANOVA followed by Tukey post hoc test. For the analysis in (a, b), an unpaired two-tailed Student′s t-test was used. *P < 0.0001. Source data are provided as a Source Data file.
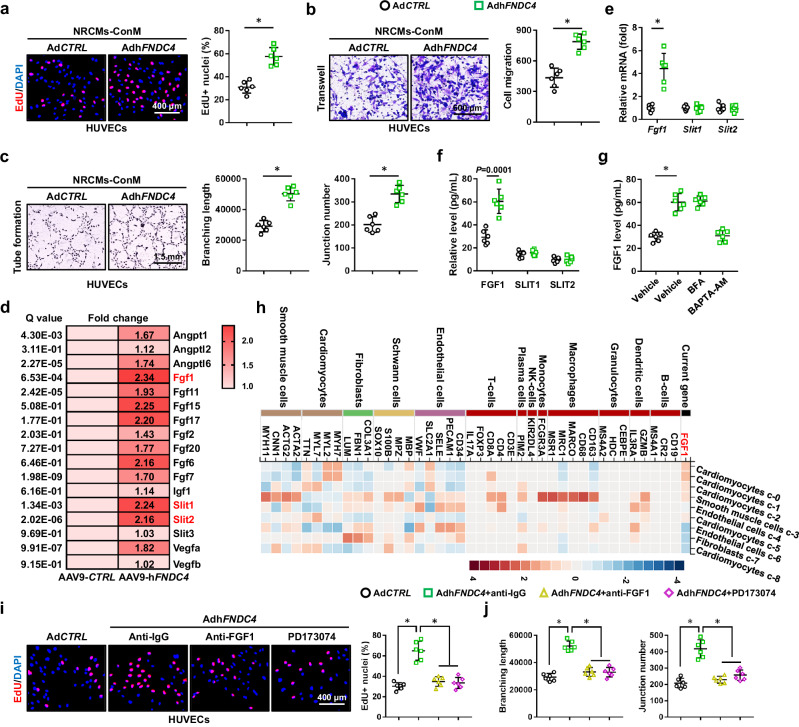
FNDC4 promotes angiogenesis of endothelial cells through increasing FGF1 secretion from cardiomyocytes
Given the angiogenic role of FNDC4 in I/R-stressed hearts, we investigated whether FNDC4 overexpression could directly enhance the angiogenic ability of human umbilical vein endothelial cells (HUVECs) in vitro. The abilities of proliferation, migration and tube formation are essential for angiogenesis of endothelial cells. Unexpectedly, FNDC4 overexpression in HUVECs did not affect their proliferative capacities, as evidenced by the unaltered cell viability and EdU+ nuclei (Supplementary Fig. 5a, b). Meanwhile, wound scratch assay and transwell assay also identified an unaltered migrative capacity of AdhFNDC4-infected HUVECs (Supplementary Fig. 5c, d). Compared with AdCTRL-infected HUVECs, FNDC4-overexpressed HUVECs displayed similar formations of vascular tubes on Matrigel (Supplementary Fig. 5e, f). Western blot results showed that FNDC4 protein level was dramatically increased in the lysates of AdhFNDC4-infected HUVECs (Supplementary Fig. 5g). FNDC4 is released as a soluble bioactive fragment to exert its biological functions, and we wondered whether FNDC4 release in HUVECs was blocked due to the lack of a secretory mechanism, which might be complete in cardiomyocytes. Yet, FNDC4 level was detectable in the medium of HUVECs at baseline, which was further increased by AdhFNDC4 overexpression (Supplementary Fig. 5h). Interestingly, the mRNA and protein levels of FNDC4 in endothelial cells were also unaffected by I/R stimulation in vivo and in vitro (Supplementary Fig. 5i, j). The above findings showed that FNDC4 overexpression did not directly enhance the angiogenic ability of HUVECs in vitro. Next, HUVECs and NRCMs were treated with rFNDC4 to directly evaluate the role of extracellular FNDC4. As shown in Supplementary Fig. 6, the angiogenic ability of HUVECs was unaffected by rFNDC4; however, treatment with rFNDC4 dramatically prevented sI/R-induced cardiomyocyte injury and apoptosis in vitro.
These results suggested that it was cardiomyocytes, instead of endothelial cells, that were the primary source and direct target of FNDC4. For this reason, we speculated that FNDC4 might promote the secretion of a certain angiogenic factor from cardiomyocytes to enhance angiogenesis of endothelial cells in a paracrine manner. To address this hypothesis, we firstly treated HUVECs with the conditioned medium from AdCTRL- or AdhFNDC4-infected NRCMs. As shown in Fig. 5a and Supplementary Fig. 7a, the conditioned medium from AdhFNDC4-infected NRCMs dramatically enhanced the proliferative capacity of HUVECs. Wound scratch assay and transwell assay also identified an increased migrative capacity of HUVECs that were cultured in the conditioned medium from AdhFNDC4-infected NRCMs (Fig. 5b and Supplementary Fig. 7b). In addition, the conditioned medium from cardiomyocytes overexpressing FNDC4 dramatically facilitated the vascular tube formation of HUVECs in vitro (Fig. 5c). In contrast, the conditioned medium from shFndc4-infected NRCMs suppressed the proliferative and migrative capacities of HUVECs (Supplementary Fig. 7c–f). Exposing HUVECs to the conditioned medium from shFndc4-infected NRCMs blocked their angiogenic behavior in vitro (Supplementary Fig. 7g). Due to the lack of FNDC4 neutralizing antibodies, to exclude the potential involvement of FNDC4 in the conditioned medium from cardiomyocytes overexpressing FNDC4, NRCMs were pre-treated with rFNDC4, and then washed and cultured in fresh medium to prepare the conditioned medium. We could not detect human FNDC4 in the conditioned medium from rat cardiomyocytes, and the level of rat FNDC4 was also comparable in the conditioned medium from vehicle- or rFNDC4-treated NRCMs (Supplementary Fig. 8a). As shown in Supplementary Fig. 8b–f, the conditioned medium from rFNDC4-treated NRCMs induced the proliferation, migration and vascular tuber formation of HUVECs.
Fig. 5. FNDC4 promotes angiogenesis of endothelial cells through increasing FGF1 secretion from cardiomyocytes.
a Human umbilical vein endothelial cells (HUVECs) were cultured with the conditioned medium (NRCMs-ConM) for 24 h, and then EdU+ nuclei were quantified using a commercial kit (n = 6 independent experiments). b HUVECs were cultured with NRCMs-ConM for 24 h, and then exposed to transwell assay. After 12 h, cells in the lower chamber were stained with crystal violet to quantify the migrated cells (n = 6 independent experiments). c HUVECs were cultured with NRCMs-ConM for 24 h, and then exposed to tube formation assay. After 8 h, the branching length and junction number were quantified (n = 6 independent experiments). d The upregulated angiogenic factors in FNDC4-overexpressed hearts were analyzed using the transcriptome data (n = 3). e The mRNA levels of fibroblast growth factor 1 (Fgf1), slit guidance ligand 1 (Slit1) and Slit2 in NRCMs with or without FNDC4 overexpression (n = 6 independent experiments). f The levels of FGF1, SLIT1 and SLIT2 in the medium of NRCMs with or without FNDC4 overexpression (n = 6 independent experiments). g The level of FGF1 in the medium of NRCMs with brefeldin A (BFA) or BAPTA-AM treatment (n = 6 independent experiments). h Single-cell sequencing data of FGF1 in human hearts. i HUVECs were cultured with NRCMs-ConM in the presence of anti-FGF1 or PD173074 for 24 h, and then EdU+ nuclei were quantified using a commercial kit (n = 6 independent experiments). j HUVECs were cultured with NRCMs-ConM in the presence of anti-FGF1 or PD173074 for 24 h, and then exposed to tube formation assay (n = 6 independent experiments). Data were presented as the mean ± S.D., and analyzed using an unpaired two-tailed Student′s t-test. For the analysis in (g–j), one-way ANOVA followed by Tukey post hoc test was used. *P < 0.0001. Source data are provided as a Source Data file.
Several known angiogenic factors, including VEGF, ANGPT, ANGPTL, IGF, SLIT and FGF, could induce angiogenesis in the heart. By analyzing the upregulated genes of the transcriptome data, we found that the mRNA levels of Fgf1, Slit1 and Slit2 were dramatically elevated in the myocardium with FNDC4 overexpression (Fig. 5d). To clarify the specific factors mediating cardiomyocytes-endothelial cells crosstalk, we measured the levels of FGF1, SLIT1 and SLIT2 in NRCMs with or without FNDC4 overexpression. Interestingly, we found that only the mRNA level and release of FGF1 were elevated in FNDC4-overexpressed NRCMs (Fig. 5e, f). Consistent with previous findings that FGF1 is released from cells in a non-classical manner by direct translocation through the plasma membrane, FNDC4-dependent FGF1 release from NRCMs was not affected by the vesicle formation and transport inhibitor brefeldin A (BFA), but was blocked by chelating intracellular Ca2+ using BAPTA-AM (Fig. 5g)26. As shown in Supplementary Fig. 9a, b, FGF1 gene was abundantly expressed in both human and mouse hearts27,28. Single-cell sequencing data also revealed that FGF1 in the heart was mainly localized to the cardiomyocytes (Fig. 5h and Supplementary Fig. 9c). To validate the necessity of FGF1 signaling pathway in mediating cardiomyocytes-endothelial cells crosstalk, FGF1 neutralizing antibody (ati-FGF1) and PD173074 were used to immunodeplete FGF1 or block FGF receptor, respectively. As shown in Fig. 5i, j and Supplementary Fig. 9d–g, the increased proliferation, migration and vascular tuber formation of HUVECs treated with the conditioned medium from cardiomyocytes overexpressing FNDC4 were dramatically blocked by inhibiting FGF1 signaling pathway. Interestingly, FGF1 neutralization did not affect the anti-apoptotic role of FNDC4 in sI/R-stimulated NRCMs (Supplementary Fig. 10a, b). Collectively, we demonstrate that FNDC4 does not directly stimulate angiogenesis of endothelial cells, but increases the expression and secretion of FGF1 from cardiomyocytes to enhance angiogenesis in a paracrine manner.
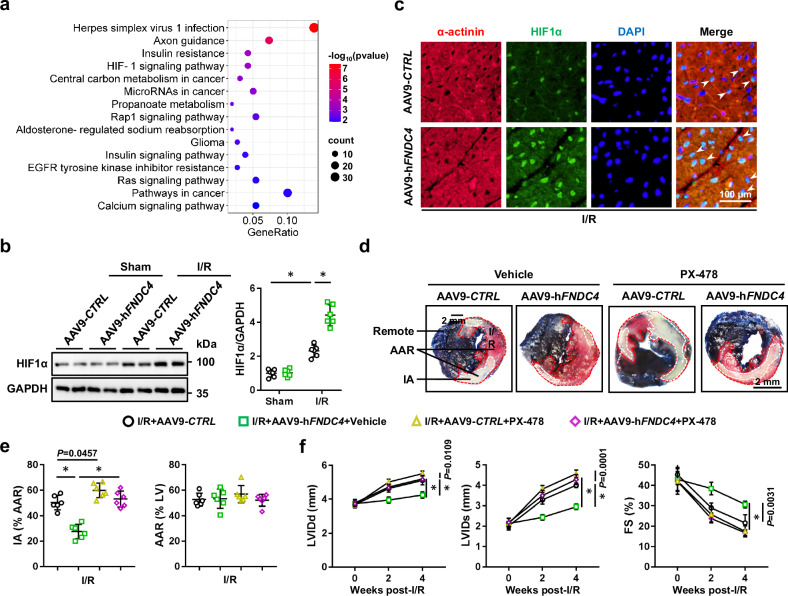
FNDC4 ameliorates cardiac I/R injury through activating HIF1α in vivo and in vitro
The signaling pathway of HIF1α, an oxygen-sensitive transcription factor that orchestrates the adaptive response to hypoxia and ischemia through increasing the expression of various pro-angiogenic factors, was dramatically activated in I/R-stressed hearts by FNDC4 overexpression; therefore, we determined whether FNDC4 attenuated cardiac I/R injury through preserving HIF1α signaling pathway (Fig. 6a and Supplementary Fig. 11). As shown in Fig. 6b, HIF1α protein level was increased by FNDC4 overexpression in I/R-stressed hearts. By isolating endothelial cells and cardiomyocytes from adult hearts, we found that FNDC4 overexpression in the myocardium mainly elevated HIF1α protein expression in I/R-stressed cardiomyocytes, but not in endothelial cells (Supplementary Fig. 12a). Immunofluorescence staining of I/R-stressed hearts and sI/R-stimulated NRCMs revealed that both the expression and nuclear accumulation of HIF1α were enhanced by FNDC4 overexpression (Fig. 6c and Supplementary Fig. 12b). To clarify the role of HIF1α, sI/R-stimulated NRCMs with FNDC4 overexpression were treated with 2-methoxyestradiol (2ME2) or PX-478 to inhibit HIF1α. As shown in Supplementary Fig. 12c, d, the protective role of FNDC4 against sI/R-induced cardiomyocyte injury was blocked by both 2ME2 and PX-478, as evidenced by the increased TUNEL+ nuclei and LDH releases. In addition, the elevated FGF1 level in the conditioned medium of cardiomyocytes overexpressing FNDC4 was reduced by HIF1α inhibitors (Supplementary Fig. 12e). Accordingly, the increased proliferation, migration and vascular tuber formation of HUVECs treated with the conditioned medium from cardiomyocytes overexpressing FNDC4 were also blocked by inhibiting HIF1α signaling pathway (Supplementary Fig. 12f–l). Consistent with the in vitro findings, we found that the decreased IA in FNDC4-overexpressed hearts after I/R surgery was dramatically increased by PX-478 treatment (Fig. 6d, e). In addition, HIF1α inhibition also abolished FNDC4 overexpression-mediated preservations of cardiomyocyte survival and angiogenesis in I/R-stressed hearts (Supplementary Fig. 13a). Consistently, the anti-hypertrophic and anti-fibrotic roles of FNDC4 were blunted in mice with PX-478 treatment (Supplementary Fig. 13b). Compared with the control mice, mice treated with PX-478 displayed worse ventricular dilation and systolic function in response to I/R surgery after FNDC4 overexpression, as evidenced by the increased LVIDd, LVIDs and decreased FS (Fig. 6f). Collectively, we demonstrate that FNDC4 ameliorates cardiac I/R injury through activating HIF1α in vivo and in vitro.
Fig. 6. FNDC4 ameliorates cardiac I/R injury through activating HIF1α in vivo.
a Heart samples with or without FNDC4 overexpression were collected 24 h after I/R surgery and subjected to unbiased transcriptome analysis, and the KEGG analysis was performed using the upregulated differentially expressed genes (DEGs) (n = 3). b Heart samples with or without FNDC4 overexpression were collected 24 h after I/R surgery and subjected to western blot (n = 6). c Heart samples with or without FNDC4 overexpression were collected 24 h after I/R surgery and subjected to immunofluorescence staining of HIF1α (green) and α-actinin (red). Arrows indicate HIF1α expression in the nuclei of cardiomyocytes (n = 6). d, e Mice with or without FNDC4 overexpression were treated with PX-478 to inhibit HIF1α, and heart samples were collected 24 h after I/R surgery for Evans blue and TTC double staining (n = 6). f Cardiac function of FNDC4-overexpressed mice with or without PX-478 treatment was analyzed by transthoracic echocardiography at the indicated time points (n = 6). Data were presented as the mean ± S.D., and analyzed using one-way ANOVA followed by Tukey post hoc test. *P < 0.0001. Source data are provided as a Source Data file.
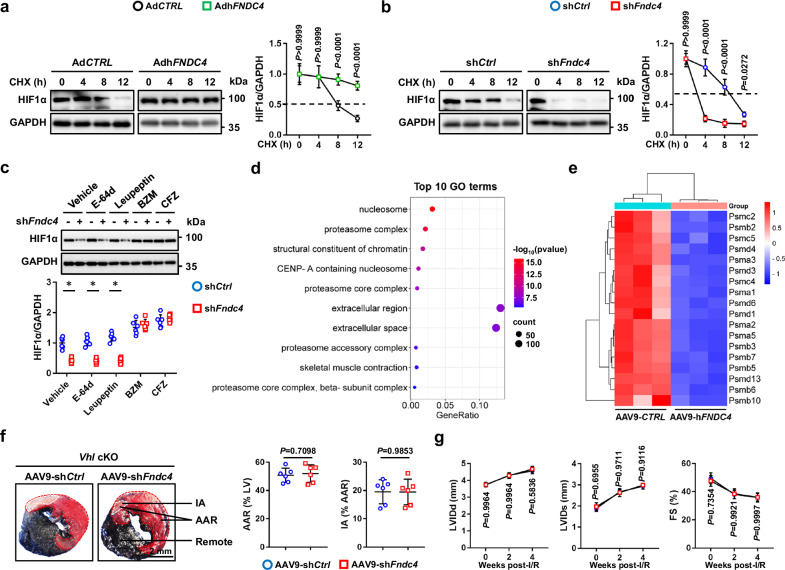
FNDC4 elevates HIF1α protein level through suppressing its proteasomal degradation
We then explored the underlying mechanism through which FNDC4 elevated HIF1α protein level. As shown in Supplementary Fig. 14a, Hif1α mRNA levels in I/R-stressed hearts, cardiomyocytes and endothelial cells were not affected by FNDC4 overexpression. We speculated that FNDC4 elevated HIF1α protein level through inhibiting its protein degradation. As expected, FNDC4 overexpression delayed, while FNDC4 knockdown facilitated HIF1α protein degradation (Fig. 7a, b). HIF1α is mainly degraded through the proteasomal pathway; however, emerging studies have also identified a lysosomal degradation of HIF1α29,30. To elucidate the specific mechanism mediating HIF1α degradation, FNDC4-silenced NRCMs were treated with either proteasomal inhibitors, bortezomib (BZM) and carfilzomib (CFZ), or lysosomal inhibitors (E-64d and leupeptin). As shown in Fig. 7c, the accelerated HIF1α degradation in NRCMs by FNDC4 knockdown was blocked by both BZM and CFZ, instead of E-64d or leupeptin, indicating that the increased expression and nuclear accumulation of HIF1α in FNDC4-overexpressed NRCMs were probably secondary to the inhibition of its proteasomal degradation. As expected, the hydroxylated and ubiquinated HIF1α levels were dramatically increased in FNDC4-overexpressed NRCMs under sI/R stimulation (Supplementary Fig. 14b). Interestingly, transcriptome data revealed that FNDC4 overexpression dramatically inhibited the expressions of proteasomal components in the myocardium (Fig. 7d, e). Next, we randomly selected five genes encoding the proteasomal components (Psma1, Psmb5, Psmb10, Psmc2 and Psmd3) based on the transcriptome data, and measured whether FNDC4 could regulate the expression of these genes using PCR analysis. As shown in Supplementary Fig. 14c, FNDC4 knockdown increased, while FNDC4 overexpression decreased the expressions of Psma1, Psmb5, Psmb10, Psmc2 and Psmd3 in I/R-stressed hearts. Consistent with the mRNA levels, the protein levels of PSMA1, PSMB5, PSMB10, PSMC2 and PSMD3 were also increased by FNDC4 knockdown, but decreased by FNDC4 overexpression (Supplementary Fig. 14d, e). Meanwhile, the proteasomal activity was also restored in I/R-stressed hearts by FNDC4 knockdown, but further compromised by FNDC4 overexpression (Supplementary Fig. 14f). During proteasomal degradation, hydroxylated HIF1α is ubiquinated by VHL and then delivered to the proteasome for degradation. Previous studies have shown that VHL ablation could block the proteasomal degradation of HIF1α, and subsequently activated HIF1α signaling pathway6,31. Next, we generated cardiac-specific VHL knockout (cKO) mice, and found that HIF1α protein level was dramatically elevated in the myocardium of Vhl cKO mice (Supplementary Fig. 14g). As shown in Fig. 7f, cardiac-specific FNDC4 knockdown failed to increase IA in I/R-stressed Vhl cKO mice. Accordingly, HIF1α activation in Vhl cKO mice also blocked FNDC4 knockdown-mediated aggravation of cardiac function in response to I/R surgery, as evidenced by the unaltered LVIDd, LVIDs and FS (Fig. 7g). Collectively, we demonstrate that FNDC4 elevates HIF1α protein level through suppressing its proteasomal degradation.
Fig. 7. FNDC4 elevates HIF1α protein level through suppressing its proteasomal degradation.
a, b NRCMs with FNDC4 overexpression or knockdown were treated with cycloheximide (CHX) to inhibit protein synthesis, and western blot was performed to analyze HIF1α protein levels at the indicated time points (n = 6 independent experiments). c NRCMs with FNDC4 knockdown were treated with proteasomal inhibitors (BZM and CFZ) or lysosomal inhibitors (E-64d and leupeptin), and western blot was performed to measure HIF1α protein level (n = 6 independent experiments). d The top 10 downregulated GO terms of the transcriptome analysis (n = 3). e The expressions of proteasome-related genes were presented using a heatmap (n = 3). f Heart samples were collected from von Hippel-Lindau protein (Vhl) cKO mice with or without FNDC4 knockdown 24 h after I/R surgery, and then exposed to Evans blue and TTC double staining (n = 6). g Cardiac function of Vhl cKO mice with or without FNDC4 knockdown was analyzed by transthoracic echocardiography at the indicated time points (n = 6). Data were presented as the mean ± S.D., and analyzed using repeated measures ANOVA. For the analysis in (c), one-way ANOVA followed by Tukey post hoc test was used. For the analysis in (f), an unpaired two-tailed Student′s t-test was used. *P < 0.0001. Source data are provided as a Source Data file.
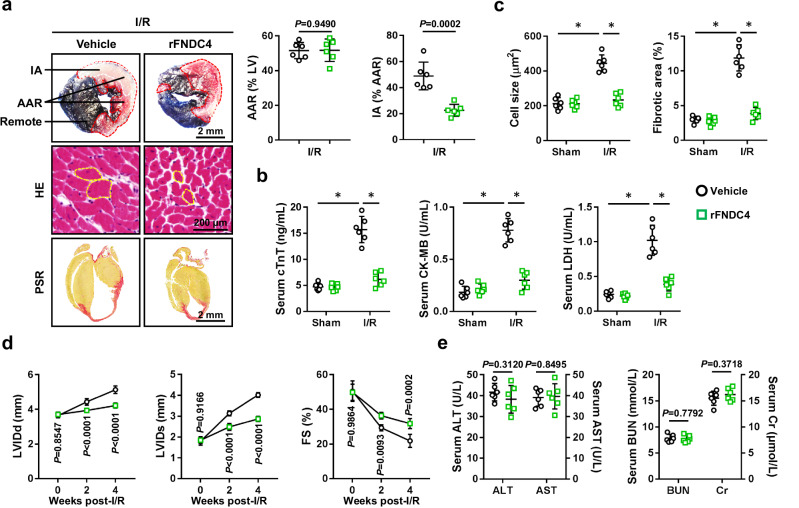
Therapeutic administration of rFNDC4 protein is sufficient to attenuate cardiac I/R injury
To enhance the clinical impact and translational value of our findings, we finally investigated whether administration of rFNDC4 protein could provide therapeutic effects against cardiac I/R injury. As shown in Fig. 8a, b, rFNDC4 treatment dramatically reduced IA in I/R-stressed hearts, accompanied with decreased levels of serum cTnT, CK-MB and LDH. Meanwhile, I/R-induced cardiac hypertrophy and fibrosis were also attenuated in mice with rFNDC4 treatment (Fig. 8c). Accordingly, ventricular dilation and systolic dysfunction in I/R-stressed hearts were ameliorated by rFNDC4 administration (Fig. 8d). Moreover, therapeutic administration of rFNDC4 protein did not result in any significant hepatic or renal toxicities (Fig. 8e). Of note, the body weight was comparable between vehicle-treated and rFNDC4-treated mice (27.9 ± 4.14 g versus 28.6 ± 3.88 g). Collectively, we demonstrate that therapeutic administration of rFNDC4 protein is sufficient to attenuate cardiac I/R injury, and that it may be a promising therapeutic target for the treatment of cardiac I/R injury.
Fig. 8. Therapeutic administration of rFNDC4 protein is sufficient to attenuate cardiac I/R injury.
a Evans blue and TTC double staining was performed to identify the IA, AAR and remote area of heart samples 24 h after I/R surgery. Circles indicate the cross-sectional area of cardiomyocytes (n = 6). b Serum levels of cTnT, creatine kinase isoenzymes (CK-MB) and LDH in I/R-stressed mice with or without recombinant FNDC4 (rFNDC4) protein treatment (n = 6). c Heart samples with or without rFNDC4 protein treatment were collected 4 weeks after I/R surgery and subjected to HE and PSR staining, and cell size as well as fibrotic area were quantified (n = 6). d Cardiac function of rFNDC4- or vehicle-treated mice was analyzed by transthoracic echocardiography at the indicated time points (n = 6). (e) Serum levels of alanine transaminase (ALT), aspartate transaminase (AST), blood urea nitrogen (BUN) and creatinine (Cr) (n = 6 biological samples). Data were presented as the mean ± S.D., and analyzed using one-way ANOVA followed by Tukey post hoc test. For the analysis in (a–e), an unpaired two-tailed Student′s t-test was used. For the analysis in (d), repeated measures ANOVA was performed. *P < 0.0001. Source data are provided as a Source Data file.
Discussion
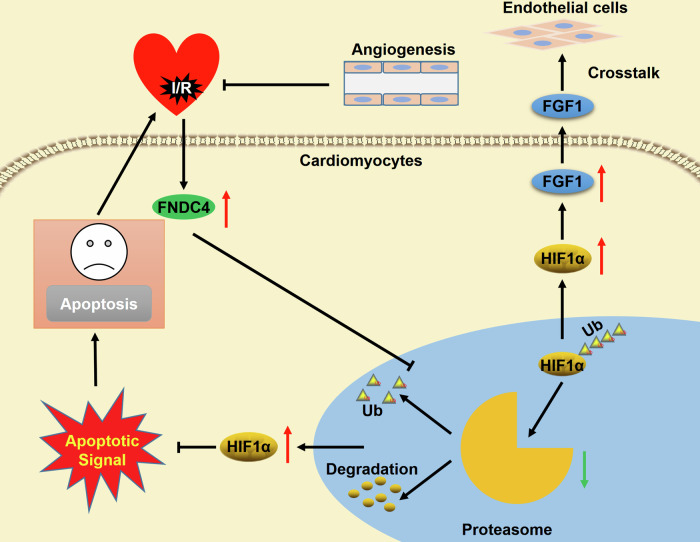
Therapeutic benefits from coronary recanalization of AMI patients are extremely impeded by cardiac I/R injury, and we herein identify a protective molecule and the underlying mechanism that limit cardiac I/R injury. In the present study, we determine that cardiac and plasma FNDC4 levels are elevated during I/R injury in a HIF1α-dependent manner, and that higher FNDC4 expression predicts better cardiac function and prognosis. Cardiac-specific FNDC4 overexpression facilitates, while cardiac-specific FNDC4 knockdown inhibits cardiomyocyte survival and angiogenesis in I/R-stressed hearts. The unbiased transcriptome analysis reveals that FNDC4 blocks the proteasomal degradation of HIF1α and subsequently activates HIF1α signaling pathway to exert the cardioprotective effects. Meanwhile, FNDC4 does not directly stimulate angiogenesis of endothelial cells, but increases the expression and secretion of FGF1 from cardiomyocytes to enhance angiogenesis in a paracrine manner (Fig. 9). Moreover, therapeutic administration of rFNDC4 protein is sufficient to alleviate cardiac I/R injury, without resulting in significant side effects. In summary, our study defines FNDC4 as a promising predictive and therapeutic target of cardiac I/R injury.
Fig. 9. Schematic diagram of the molecular mechanisms underlying FNDC4-mediated protection against cardiac I/R injury.
FNDC4 is elevated during cardiac I/R injury, which blocks the proteasomal degradation of HIF1α and subsequently activates HIF1α signaling pathway to inhibit cardiomyocyte apoptosis. In addition, FNDC4-mediated HIF1α activation increases the expression and secretion of FGF1 from cardiomyocytes to enhance the angiogenic capacity of endothelial cells in a paracrine manner to facilitate cardiac repair during I/R stress.
HIF1α is an oxygen-sensitive transcription factor and orchestrates the adaptive response to hypoxia as well as ischemia, whose activation provides significant cardioprotections against cardiac I/R injury5. Cai et al. previously demonstrated that HIF1α was required for intermittent hypoxia- or erythropoietin-mediated protection against cardiac I/R injury32. Meanwhile, HIF1α activation by PHD2 knockdown dramatically reduced infarct size in I/R-stressed hearts33. Emerging studies have suggested that stabilizing HIF1α levels could directly protect cardiomyocytes from apoptosis, and subsequently led to smaller infarct size during acute cardiac I/R surgery7,34. Ong et al. showed that acute HIF1α stabilization promoted aerobic glycolysis, decreased mitochondrial oxidative stress and prevented the opening of the mitochondrial permeability transition pore, thereby reducing acute cardiac I/R injury35. Further results implied that HIF1α-dependent frataxin expression in cardiomyocytes directly mitigated mitochondrial iron overload and overproduction of free radicals, and eventually preserved mitochondrial membrane integrity and viability of cardiomyocytes36. Intriguingly, our recent study showed that HIF1α-dependent upregulation of isthmin-1 could prevent cardiomyocyte apoptosis and cardiac I/R injury in mice10. In addition to the direct regulation of cardiomyocyte death, HIF1α is well-known as a major determinant of angiogenesis, and helps to restore blood supply to the infarct and border zone, which prolongs the survival of ischemic myocardium and improves cardiac remodeling as well as dysfunction following long-term observations. Accordingly, our previous study also showed that enhancing angiogenesis could prevent cardiomyocyte apoptosis and facilitate wound healing of the infarcted myocardium37. Findings from Kido et al. revealed that constitutive overexpression of HIF1α in cardiomyocytes dramatically increased capillary densities in infarct and border regions of the ischemic hearts25. Huang et al. also showed that HIF1α upregulation increased neovascularization in injured myocardium and subsequently improved cardiac function38. The proliferation and migration of endothelial cells contribute to the formation of vascular network, and subsequently increase the blood flow of the ischemic myocardium. Previous studies indicated that HIF1α activation was required for the expression and release of various angiogenic factors (VEGF, ANGPT1 and SLIT1, etc), and these angiogenic factors provoked angiogenesis of endothelial cells. Kelley et al. showed that HIF1α activation was sufficient to induce angiogenesis through elevating the expressions of ANGPT1, ANGPT2, VEGF and other angiogenic factors39. Cardiomyocytes are the primary cell population in mammalian hearts, and the crosstalk between cardiomyocytes and endothelial cells further enhances angiogenesis. Rose et al. showed that p38-dependent release of VEGF from cardiomyocytes promoted paracrine signaling-mediated pro-angiogenic activity40. Meanwhile, cardiomyocyte-derived small extracellular vesicles also regulated the angiogenic capacity of cardiac endothelial cells, thereby modulating cardiac I/R injury41,42. Moreover, Gladka et al. determined that Thymosin β4 and Prothymosin α released by cardiomyocytes in the infarcted hearts enhanced angiogenesis and prevented cardiac dysfunction after ischemic injury43. In our study, we found that FNDC4 facilitated the interplay between cardiomyocytes and endothelial cells, and subsequently increased angiogenesis in I/R-stressed hearts. In addition, treatment with rFNDC4 (an extracellular domain of FNDC4) directly prevented sI/R-induced cardiomyocyte injury and apoptosis in vitro, indicating that FNDC4 in the secretion medium was sufficient to confer the cardioprotection. However, whether intracellular FNDC4 might also have protective effects against I/R injury deserves further investigations. Moreover, FNDC4 was identified as an anti-inflammatory factor on macrophages, and subsequently prevented tissue injury under different disease contexts23,44. FNDC4 also played critical roles in metabolic control, which reduced lipogenesis, promoted fat browning and improved glucose tolerance24,45. GPR116 is highly expressed in the heart and in endothelial cells, which plays critical roles in angiogenesis46. Meanwhile, Georgiadi et al. previously identified GPR116 as a receptor of FNDC4 in the white adipose tissue24. However, our study revealed that endothelial cells were neither the origin nor the direct target of FNDC4. Moreover, Niaudet et al. found that GPR116 knockout dramatically increased vascular density, which was contrary to our findings showing that FNDC4 facilitated angiogenesis in I/R-stressed hearts47. Whether the cardioprotective effects of FNDC4 involve these mechanisms should be determined in further studies.
FGF proteins belong to a family of pleiotropic heparin-binding growth factors, and play critical roles in regulating embryonic development, organ morphogenesis and tissue repair, etc. Various FGF proteins have been implicated in the initiation and progression of angiogenesis and cardiac homeostasis48. Specially, recombinant FGF2 was used as an angiogenic protein for the treatment of ischemic diseases in clinic trials49,50. FGF1 is a multifunctional peptide and contributes to the regulation of cardiac morphogenesis51. Within the heart, FGF1 is primarily expressed by cardiomyocytes, and the protein is located in myocytes and in the extracellular matrix52. Consistent with the role of other FGF proteins, FGF1 is also related with angiogenesis and cardiac function. Fernandez et al. found that cardiac-specific FGF1 overexpression dramatically increased the density of coronary arteries, accompanied with an enhanced coronary flow53. Meanwhile, FGF1 coacervate promoted the proliferation of endothelial and mural cells, generated stable arterioles and capillaries in infarcted hearts, and improved cardiac function54. Accordingly, FGF1 overexpression was sufficient to improve cardiac functional recovery and enhance cell survival after cardiac I/R injury52. In the present study, we found that FNDC4 increased the expression and secretion of FGF1 from cardiomyocytes in a HIF1α-dependent manner, which drove the proliferation, migration and tube formation of endothelial cells, thereby preventing cardiac I/R injury. In contrast, neutralizing FGF1 or blocking FGF receptors effectively reduced the angiogenic role of the conditioned medium from FNDC4-overexpressed cardiomyocytes. FGF1 also provided cardioprotection within the cardiomyocyte, independent from angiogenesis; however, we found that the decreased cardiomyocyte injury by FNDC4 was not dependent on FGF155,56.
In summary, our findings reveal that FNDC4 alleviates cardiac I/R injury through facilitating HIF1α-dependent cardiomyocyte survival and angiogenesis, and define FNDC4 as a promising predictive and therapeutic target of cardiac I/R injury.
Methods
Ethics statement
All animal studies were approved by the Animal Care and Use Committee of Renmin Hospital of Wuhan University, and were performed in strict accordance with the Guidelines for the Care and Use of Laboratory Animals published by the US National Institutes of Health. All experimental procedures involving human samples in this study were approved by the Review Board of Renmin Hospital of Wuhan University.
Reagents
PX-478 (#HY-10231), a specific inhibitor of HIF1α, was purchased from MedChemExpress (Monmouth Junction, NJ, USA). FGF1 neutralizing antibody (anti-FGF1, #ab9588), LDH Assay Kit (#ab102526) and Mouse/Rat FGF1 ELISA Kit (#ab223587) were purchased from Abcam (Cambridge, UK). PD173074 (#P2499), BFA (#203729), 2ME2 (#454180), cycloheximide (CHX, a protein synthesis inhibitor, #01810), E-64d (a lysosomal inhibitor, #E8640) and leupeptin (a lysosomal inhibitor, #L2884) were purchased from Sigma-Aldrich (St. Louis, MO, USA). BAPTA-AM (#B1205) and EnzChek caspase3 assay kit (#E13183) were purchased from Invitrogen (Carlsbad, CA, USA). ApopTag Plus In Situ Apoptosis Fluorescein Detection Kit (#S7111) was purchased from Millipore (Billerica, MA, USA). Cell Counting Kit-8 (CCK-8, #C0037) was purchased from Beyotime (Shanghai, China). Click-iT EdU-594 Cell Proliferation Assay Kit (#G1603) and crystal violet (#G1014) were purchased from Wuhan Servicebio Technology Co., Ltd. (Wuhan, China). LDEV-free Matrigel® Basement Membrane Matrix (#356234) was purchased from Corning® (Corning, NY, USA). BZM (a reversible proteasomal inhibitor, #S1013) and CFZ (an irreversible proteasomal inhibitor, #S2853) were purchased from Selleck Chemicals (Houston, TX, USA). Mouse FNDC4 EIA Kit (#EK-067-90) was purchased from Phoenix Pharmaceuticals, Inc. (Burlingame, CA, USA). Human FNDC4 ELISA Kit (#MBS9332722), Rat FNDC4 ELISA Kit (#MBS9399687), Rat SLIT1 ELISA Kit (#MBS9711860) and Rat SLIT2 ELISA Kit (#MBS9711859) were purchased from MyBioSource, Inc. (San Diego, CA, USA). Cell Death Detection ELISAPLUS kit (#11774425001) was purchased from Roche Applied Science (Basel, Switzerland). Proteasome Activity Fluorometric Assay Kit I (#J4110), Suc-Leu-Leu-Val-Tyr-AMC (#G1100), Z-Leu-Leu-Glu-AMC (#G2100) and Boc-Leu-Arg-Arg-AMC (#G3100) were obtained from UBPBio (Aurora, USA). Plasmids encoding full-length human FNDC4 cDNA (#W2481) were obtained from GeneCopoeia, Inc. (Rockville, MD, USA), and then packaged to the cardiotropic AAV9 vectors under a cTnT promoter to produce AAV9-hFNDC4, while AAV9-CTRL was used as the negative control. Mouse FNDC4 shRNA plasmids were purchased from GeneCopoeia, Inc., and sequences were cloned under a cTnT promoter to produce AAV9-shFndc4, while AAV9-shCtrl was used as the negative control.
Animal studies
Male C57BL/6 mice were bred in a temperature- and humidity-controlled SPF condition (20-25 °C and 45-55% humidity) under 12/12 h light/dark cycles with free access to food and water57,58. Mice were acclimated at least for 1 week, and then those at 10–12 weeks of age received a single intravenous injection of AAV9-hFNDC4 or AAV9-shFndc4 at a dose of 1 × 1011 viral genome per mouse to specifically overexpress or knock down FNDC4 in the myocardium 4 weeks prior to I/R surgery22,59. To induce I/R injury, mice were anesthetized by 3% pentobarbital sodium, and the left thoracotomy was performed to expose the heart. Next, the left anterior descending (LAD) artery was ligated with a PE-10 tubing at 1 mm below the tip of the left atrial appendage, and cardiac ischemia was verified by both ST elevation on surface electrocardiogram and visual blanching. After 45 min of ischemia, the tubing was removed and the ligature was released to achieve reperfusion9. To inhibit HIF1α, mice were intraperitoneally injected with PX-478 (50 mg/kg) as previously described60. Vhl floxed (Vhlfl/fl) mice (#004081) were provided by Jackson Laboratories (Bar Harbor, ME, USA), and backcrossed to C57BL/6 strain over ten generations before used. Genotyping was performed using the following primers: primer oIMR1555: CTCAGGTCATCTTCTGCAACC; primer oIMR1556: TCTGTCTTGGCCTCCTGAGT. Cardiac-specific VHL knockout mice were generated by mating Vhlfl/fl mice with α-myosin heavy chain (Mhc)-MerCreMer transgenic mice (#005657, Jackson Laboratories), which were then intraperitoneally injected with tamoxifen (25 mg/kg/day, dissolved in corn oil) for consecutive 5 days to induce VHL ablation61,62. Two weeks post-tamoxifen injection to allow its clearance, Vhl cKO mice were intravenously injected with AAV9-shFndc4 or AAV9-shCtrl, and kept for 4 weeks before I/R surgery. To estimate the therapeutic value of FNDC4, mice were intraperitoneally injected with 0.2 mg/kg rFNDC4 protein every other day for 4 weeks, and then exposed to I/R surgery. The rFNDC4 protein was prepared by cloning the extracellular fragment of FNDC4 according to previous studies, and the protein was produced in mammalian cells and as such being free of endotoxin23,24. In this study, the investigators were blinded to group allocation during data collection.
Echocardiography
Mice were lightly anesthetized by 1.5% isoflurane, and then were exposed to echocardiographic examination at baseline, 2 and 4 weeks post-I/R surgery using a Vevo® 3100 high-resolution Preclinical Imaging System (FUJIFILM VisualSonics, Toronto, Canada) with a 30 MHz MX 400 linear ultrasound transducer9,21. Two-dimension-guided M mode measurements of ventricular diameter were obtained from at least five cardiac cycles, and the LVIDd, LVIDs, IVSs and IVSd were recorded. FS was calculated as (LVIDd-LVIDs)/LVIDd × 100%.
Histology
To measure infarct size, mice were re-anesthetized 24 h after reperfusion, and the LAD was isolated for re-ligation at the same location of I/R surgery. Subsequently, 2% Evans blue dye was injected into the jugular vein to demarcate the ischemic area at risk (AAR), and then heart samples were harvested, quickly froze and sectioned into 4–5 μm slices, which were incubated with 1% TTC solution at 37 °C for 15 min to distinguish IA. IA (pale), AAR (not blue) and LV area were measured by Media Cybernetics Image-Pro Plus 6.0 software (Bethesda, MD, USA)9. To measure cell size, hearts were harvested, fixed with 10% neutral formalin, dehydrated, embedded in paraffin and processed to 5 μm slices, which were then exposed to hematoxylin-eosin (HE) staining following standard protocols. More than 30 fields were included for calculating the cross-sectional area of cardiomyocytes each group, with at least 5 cardiomyocytes per field analyzed57. To measure fibrotic area, cardiac slices were subjected to picrosirius red (PSR) staining, and >60 fields per group were included for the calculation of collagen deposition63,64.
Collagen content analysis
Collagen content was analyzed by measuring the levels of hydroxyproline9. Briefly, fresh ventricles were homogenized with hydrochloric acid to obtain the total collagen content, and pepsin-acid buffer was used for the calculation of soluble collagen. The insoluble collagen content was determined by subtracting the amount of soluble collagen from total collagen.
Cell experiments
NRCMs and HUVECs (#NC006, YRGene, Changsha, China) were prepared and cultured as we previously described64,65. In addition, hiPSC-CMs were used to further enhance the translational value of our findings, and they were kindly provided by Dr. Si-Chi Xu in Peking Union Medical College Hospital. Written informed consent was obtained from the donors of the cells that were used to obtain hiPSCs, and the use of hiPSCs was also approved by the Review Board of Renmin Hospital of Wuhan University. The specific process of hiPSC-CMs induction was provided in our recent study9. Briefly, hiPSCs were cultured in mTeSR™1 medium on LDEV-free Corning® Matrigel® hESC-Qualified Matrix (Corning, NY, USA)-coated 6-well plates, and then, 3.5 × 105 single hiPSCs were prepared and seeded onto pre-coated 12-well plates with 10 µmol/L Y-27632. Next, the medium was replaced with fresh mTeSR™1 without Y-27632, and hiPSCs were cultured for an additional 24 h. After grown to 95% confluency, hiPSCs were cultured according to the manufacturer’s instructions with STEMdiff™ Cardiomyocyte Differentiation Medium containing Supplement A, B or C, respectively. On 8th day, hiPSCs were cultured with STEMdiff™ Cardiomyocyte Maintenance Medium, and the medium was replaced every 2 days. On 15th day, these hiPSCs were used as hiPSC-CMs and subjected to sI/R in vitro. Endothelial cells and cardiomyocytes were isolated from adult hearts as we previously described61,66. For sI/R, cells were cultured under hypoxia (95% N2, 5% CO2) with the Esumi ischemic buffer for 4 h, and then transferred to a normoxic environment (95% air, 5% CO2) with fresh medium containing 10% fetal bovine serum (FBS) overnight9. Cells cultured with fresh medium under normoxia were used as Ctrl. To overexpress of knock down FNDC4, cells were pre-infected with AdhFNDC4, shFndc4 or matched controls (AdCTRL for AdhFNDC4, shCtrl for shFndc4) for 4 h, and then cultured with fresh medium containing 10% FBS for an additional 24 h before sI/R injury9,22. To clarify the role of extracellular FNDC4, cells were also treated with 100 nmol/L rFNDC4 or an equal volume of vehicle for 24 h, and then stimulated with sI/R23. To determine the crosstalk between cardiomyocytes and endothelial cells, AdhFNDC4- or shFndc4-infected NRCMs were cultured in fresh medium for 24 h, and then supernatants were collected, centrifuged and filtered with a 0.22 μm filter to obtain the conditioned medium (NRCMs-ConM)67. Next, HUVECs were cultured with NRCMs-ConM for indicated times. To neutralize FGF1 in the conditioned medium, NRCMs-ConM from AdhFNDC4-infected NRCMs was incubated with 50 ng/mL anti-FGF1 or isotypic IgG at 37 °C for 1 h to immunodeplete FGF168. Meanwhile, 10 μmol/L PD173074 was added to the medium to block FGF receptor in endothelial cells67. Moreover, the conditioned medium was also obtained from NRCMs with or without rFNDC4 treatment. To reveal the specific mechanism mediating FGF1 release, NRCMs were pre-treated with 2.5 μmol/L BFA for 4 h to block classic secretory pathway or 75 μmol/L BAPTA-AM for 1 h to eliminate intracellular Ca2+ 61,69. For the inhibition of HIF1α, cells were treated with 10 μmol/L 2ME2 or PX-4784.
TUNEL staining
TUNEL staining was performed to evaluate cell apoptosis following the manufacturer’s instructions, and positive nuclei was calculated as the ratio of TUNEL positive nuclei to total nuclei9,70.
Immunofluorescence staining
Deparaffinized cardiac slices and fixed cell coverslips were blocked with 10% goat serum at room temperature for 1 h, incubated with the primary antibodies (Supplementary Table I) at 4 °C overnight and then reacted with goat anti-mouse IgG Alexa Fluor 488 or goat anti-rabbit IgG Alexa Fluor 568 secondary antibodies at 37 °C for an additional 1 h. SlowFade™ gold antifade reagent with DAPI was used for the visualization of cell nuclei, and the images were recorded with a DP74 fluorescence microscope (OLYMPUS, Tokyo, Japan)64,71.
Cell viability and proliferation
Cell viability at indicating times was measured using the CCK-8 method following the manufacturer’s instructions22,58. For cell proliferation analysis, Click-iT EdU-594 Cell Proliferation Assay Kit was used following the manufacturer’s instructions. Briefly, cells were incubated with 10 μmol/L EdU working solution at 37 °C for 2 h after the stimulation with NRCMs-ConM for 24 h, and then reacted with Click-iT reaction cocktail at room temperature for 30 min after fixation and permeabilization. Finally, SlowFade™ gold antifade reagent with DAPI was used for nuclear staining, and >30 fields were included for calculating EdU+ nuclei each group.
Wound scratch assay and transwell assay
Cell migration was evaluated using both wound scratch assay and transwell assay as previously described by us and the others57,72. For wound scratch assay, cells were seeded in 6-well plates to grow to a monolayer, which were then scraped using a 200 μL sterilized micropipette tip. Next, the detached cells were removed, and the wound sizes at 0 h together with 24 h were recorded immediately with an inverted microscope. Scratch closure (%) was calculated as [(wound at 0 h-wound at 24 h)/wound at 0 h] × 100 using ImageJ software. For transwell assay, HUVECs in 200 μL serum-free medium were seeded onto the upper chamber of the inserts (8.0 μm pore size) at a density of 2 × 104 cells per well, and the lower chamber was added with 400 μL complete medium. After 12 h, cells in the upper chamber were carefully removed with a cotton swab, while the migrated cells in the lower chamber were stained with 0.1% crystal violet at room temperature for 20 min before observation.
Tube formation assay
To perform tube formation assay, 96-well plates were pre-coated with 50 μL LDEV-free Matrigel® Basement Membrane Matrix at 37 °C for 1 h. Next, HUVECs were seeded onto at a density of 3 × 104 cells per well, and tube formation at 8 h was recorded for analysis72.
Western blot
Total proteins were extracted and separated by SDS-PAGE, which were then transferred onto PVDF membranes. Next, the membranes were incubated with indicating primary antibodies (Supplementary Table I) at 4 °C overnight, followed by the identification with HRP-conjugated secondary antibodies at room temperature for an additional 1 h. Finally, the membranes were visualized by a Bio-Rad ChemiDoc™ XRS+ System using the electrochemiluminescence reagent59,66.
Quantitative real-time PCR
Total RNA was extracted and reversely transcribed to cDNA using a Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland) following the manufacturer’s instructions. Next, SYBR Green I Master Mix (Roche) was used for the quantification of gene expression on the Roche LightCycler 480 system, and the primer sequences were provided in Supplementary Table II63,64.
Protein degradation assay
To clarify the degradation of HIF1α protein, NRCMs with FNDC4 overexpression or knockdown were treated with CHX (10 µmol/L) for indicated times to inhibit protein synthesis. Meanwhile, FNDC4-silenced NRCMs were treated with BZM (0.1 μmol/L), CFZ (1 μmol/L), E-64d (100 nmol/L) or leupeptin (100 µmol/L) to inhibit proteasome- or lysosome-mediated degradation as we recently described21.
Transcriptome analysis
I/R-stressed hearts with or without FNDC4 overexpression were harvested for total RNA purification and cDNA synthesis, and sequencing libraries were constructed using the Illumina Novaseq 6000 platform (San Diego, CA, USA). Raw reads were filtered, and mapped to the GRCm38/mm10 mouse genome using HISAT2 software. The fragments per kilobase million (FPKM) values of all mapped genes were calculated, and then the differential expression analysis was performed by DESeq2, and the differentially expressed genes (DEGs) were defined as those with |Fold Change | ≥ 2 and an adjusted P-value < 0.05. In addition, KEGG and GO enrichment analysis were performed for functional measurements.
Biochemical analysis
Plasma FNDC4 levels in mice and human were measured with commercial kits following the manufacturer’s instructions. Serum levels of cTnT, CK-MB, LDH, alanine transaminase (ALT), aspartate transaminase (AST), blood urea nitrogen (BUN) and creatinine (Cr) were measured using an ADVIA® 2400 automatic biochemical analyzer (Siemens Healthcare Diagnostics, Tarrytown, NY, USA). The levels of caspase3 activity and DNA fragment were measured with commercial kit following the manufacturer’s instructions as we recently described9,61. LDH releases were detected using a commercial kit and calculated as (LDH level in ischemia medium + LDH in reperfusion medium)/(LDH in ischemia medium + LDH in reperfusion medium + LDH in cell lysate)9. The levels of FNDC4, FGF1, SLIT1 and SLIT2 in cell medium were measured using ELISA kits following the manufacturer’s instructions. The proteasomal activity was detected using the commercial fluorometric kits as we previously described65.
Human samples
Human heart samples were obtained from the left ventricular free wall or apex of IHD patients undergoing heart transplantation, and the control samples were obtained from donor hearts that were unsuitable for transplantation for non-cardiac diseases. The source and identity of these heart samples were provided in our previous studies9,66. For the analysis of human blood samples, AMI patients (aged > 18 years) with typical chest pain < 12 h and ST-segment elevation ≥ 0.1 mV in two or more contiguous electrocardiographic leads were included, and those with cardiogenic shock, severe infections, severe liver diseases, malignancies, end-stage renal diseases and surgery history within 3 months were excluded. The healthy controls without cardiovascular risk factors and coronary artery disease were recruited as the donors. Blood samples were collected from 26 donors (15 males and 11 females) and 42 AMI patients (25 males and 17 females), and the age of all participants is between 25 and 85. All experimental procedures involving human samples in this study were in accordance with the Declaration of Helsinki, and written informed consent was obtained from all donors or their legal guardians.
Statistical analysis
Data were presented as the mean ± standard deviation (S.D.), and analyzed with the GraphPad Prism (version 7.0). Unpaired two-tailed Student′s t-test was performed to compare differences between two groups with a normal distribution and homogeneity of variance, while one-way analysis of variance (ANOVA) in combination with Tukey’s post hoc test was performed for multiple comparisons. Differences between groups over time were evaluated by repeated measures ANOVA. Correlations were evaluated using Pearson’s correlation analysis. A P-value <0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by grants from the Natural Science Foundation of Hubei Province (No. 2023AFB099, 2024AFB092), the Fundamental Research Funds for the Central Universities (No. 2042023kf0046), the Open Project of Hubei Key Laboratory (No. 2023KFZZ028), Undergraduate Training Programs for Innovation and Entrepreneurship of Wuhan University (202410486110, S202410486326), Clinical Medicine+ Youth Talent Support Program of Wuhan University, Free Innovation Pre-Research Fund of Wuhan Union Hospital (2023XHYN034) and Excellent and new plan of Wuhan Union Hospital. The RNA libraries were sequenced by OE Biotech, Inc. (Shanghai, China), and we are grateful to OE Biotech, Inc. for assisting in sequencing and/or bioinformatics analysis.
Author contributions
X.Z. and C.H. conceived the hypothesis. X.Z., Y.P.G., W.S.D., K.L. and C.H. performed the experiments. X.Z., Y.X.H. and Y.J.Y. interpreted the data. X.Z. drafted the manuscript, and X.Z. together with C.H. revised the manuscript. All authors made intellectual contributions and agreed to the published version of the manuscript.
Peer review
Peer review information
Nature Communications thanks Ralf Brandes, Anastasia Georgiadi, Dongqi Tang and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data that support the findings of this study are available within the main text and its Supplementary Information file and Source data. Source data are provided with this paper. Additional data that support the findings in this study are available from the corresponding author upon reasonable request. The datasets generated for the transcriptome analysis are available through the Gene Expression Omnibus under accession code GSE277729. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xin Zhang, Email: dr.zhangxin@whu.edu.cn.
Can Hu, Email: dr_hucan@hust.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53564-z.
References
- 1.Davidson, S. M. et al. Multitarget strategies to reduce myocardial ischemia/reperfusion injury: JACC review topic of the week. J. Am. Coll. Cardiol.73, 89–99 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Reffelmann, T., Hale, S. L., Dow, J. S. & Kloner, R. A. No-reflow phenomenon persists long-term after ischemia/reperfusion in the rat and predicts infarct expansion. Circulation108, 2911–2917 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Bosch-Marce, M. et al. Effects of aging and hypoxia-inducible factor-1 activity on angiogenic cell mobilization and recovery of perfusion after limb ischemia. Circ. Res.101, 1310–1318 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Fu, C. et al. Inositol polyphosphate multikinase inhibits angiogenesis via inositol pentakisphosphate-induced HIF-1alpha degradation. Circ. Res.122, 457–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loor, G. & Schumacker, P. T. Role of hypoxia-inducible factor in cell survival during myocardial ischemia-reperfusion. Cell Death Differ.15, 686–690 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Majmundar, A. J., Wong, W. J. & Simon, M. C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell.40, 294–309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu, J. et al. SENP1 protects against myocardial ischaemia/reperfusion injury via a HIF1alpha-dependent pathway. Cardiovasc. Res.104, 83–92 (2014). [DOI] [PubMed] [Google Scholar]
- 8.An, W. G. et al. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature392, 405–408 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Zhang, X. et al. Tisp40 prevents cardiac ischemia/reperfusion injury through the hexosamine biosynthetic pathway in male mice. Nat. Commun.14, 3383 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, M. et al. Isthmin-1 alleviates cardiac ischaemia/reperfusion injury through cGMP-PKG signalling pathway. Cardiovasc. Res.120, 1051–1064 (2024). [DOI] [PubMed] [Google Scholar]
- 11.Daudon, M., Bigot, Y., Dupont, J. & Price, C. A. Irisin and the fibronectin type III domain-containing family: structure, signaling and role in female reproduction. Reproduction164, R1–R9 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Zhang, X., Hu, C., Yuan, Y. P., Ma, Z. G. & Tang, Q. Z. A brief overview about the physiology of fibronectin type III domain-containing 5. Cell. Signal.76, 109805 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Zhang, X., Hu, C., Wu, H. M., Ma, Z. G. & Tang, Q. Z. Fibronectin type III domain-containing 5 in cardiovascular and metabolic diseases: a promising biomarker and therapeutic target. Acta Pharmacol. Sin.42, 1390–1400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato, M. et al. Identification of a receptor-independent activator of G protein signaling (AGS8) in ischemic heart and its interaction with Gbetagamma. Proc. Natl Acad. Sci. USA.103, 797–802 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato, M. et al. Activator of G protein signaling 8 (AGS8) is required for hypoxia-induced apoptosis of cardiomyocytes: role of G betagamma and connexin 43 (CX43). J. Biol. Chem.284, 31431–31440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You, Y. et al. FNDC3B protects steatosis and ferroptosis via the AMPK pathway in alcoholic fatty liver disease. Free Radic. Biol. Med.193, 808–819 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Garikipati, V. et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun.10, 4317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bostrom, P. et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature481, 463–468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecker, S. H. et al. Expression of the irisin precursor FNDC5 in skeletal muscle correlates with aerobic exercise performance in patients with heart failure. Circ. Heart Fail.5, 812–818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, R. L. et al. Irisin alleviates pressure overload-induced cardiac hypertrophy by inducing protective autophagy via mTOR-independent activation of the AMPK-ULK1 pathway. J. Mol. Cell. Cardiol.121, 242–255 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Hu, C. et al. Fibronectin type III domain-containing 5 improves aging-related cardiac dysfunction in mice. Aging Cell.21, e13556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, X. et al. FNDC5 alleviates oxidative stress and cardiomyocyte apoptosis in doxorubicin-induced cardiotoxicity via activating AKT. Cell Death Differ.27, 540–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosma, M. et al. FNDC4 acts as an anti-inflammatory factor on macrophages and improves colitis in mice. Nat. Commun.7, 11314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgiadi, A. et al. Orphan GPR116 mediates the insulin sensitizing effects of the hepatokine FNDC4 in adipose tissue. Nat. Commun.12, 2999 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kido, M. et al. Hypoxia-inducible factor 1-alpha reduces infarction and attenuates progression of cardiac dysfunction after myocardial infarction in the mouse. J. Am. Coll. Cardiol.46, 2116–2124 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Prudovsky, I. et al. Folding of fibroblast growth factor 1 Is critical for its nonclassical release. Biochemistry55, 1159–1167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom.13, 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature515, 355–364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbi, M. E. et al. Cyclin-dependent kinases regulate lysosomal degradation of hypoxia-inducible factor 1alpha to promote cell-cycle progression. Proc. Natl Acad. Sci. USA.111, E3325–E3334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbi, M. E. et al. Chaperone-mediated autophagy targets hypoxia-inducible factor-1alpha (HIF-1alpha) for lysosomal degradation. J. Biol. Chem.288, 10703–10714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan, Y. et al. Profilin-1 phosphorylation directs angiocrine expression and glioblastoma progression through HIF-1alpha accumulation. Nat. Cell Biol.16, 445–456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai, Z. et al. Hearts from rodents exposed to intermittent hypoxia or erythropoietin are protected against ischemia-reperfusion injury. Circulation108, 79–85 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Natarajan, R., Salloum, F. N., Fisher, B. J., Kukreja, R. C. & Fowler, A. R. Hypoxia inducible factor-1 activation by prolyl 4-hydroxylase-2 gene silencing attenuates myocardial ischemia reperfusion injury. Circ. Res.98, 133–140 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Holscher, M. et al. Cardiomyocyte-specific prolyl-4-hydroxylase domain 2 knock out protects from acute myocardial ischemic injury. J. Biol. Chem.286, 11185–11194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong, S. G. et al. HIF-1 reduces ischaemia-reperfusion injury in the heart by targeting the mitochondrial permeability transition pore. Cardiovasc. Res.104, 24–36 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Nanayakkara, G. et al. Cardioprotective HIF-1alpha-frataxin signaling against ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol.309, H867–H879 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Liu, F. Y. et al. TLR9 is essential for HMGB1-mediated post-myocardial infarction tissue repair through affecting apoptosis, cardiac healing, and angiogenesis. Cell Death Dis.10, 480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang, M. et al. Double knockdown of prolyl hydroxylase and factor-inhibiting hypoxia-inducible factor with nonviral minicircle gene therapy enhances stem cell mobilization and angiogenesis after myocardial infarction. Circulation124, S46–S54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly, B. D. et al. Cell type-specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circ. Res.93, 1074–1081 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Rose, B. A. et al. Cardiac myocyte p38alpha kinase regulates angiogenesis via myocyte-endothelial cell cross-talk during stress-induced remodeling in the heart. J. Biol. Chem.292, 12787–12800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, G. et al. Cardiomyocyte-derived small extracellular vesicles can signal eNOS activation in cardiac microvascular endothelial cells to protect against Ischemia/Reperfusion injury. Theranostics10, 11754–11774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ottaviani, L. et al. Intercellular transfer of miR-200c-3p impairs the angiogenic capacity of cardiac endothelial cells. Mol. Ther.30, 2257–2273 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gladka, M. M. et al. Cardiomyocytes stimulate angiogenesis after ischemic injury in a ZEB2-dependent manner. Nat. Commun.12, 84 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv, Z. T. et al. FNDC4 inhibits RANKL-induced osteoclast formation by suppressing NF-kappaB activation and CXCL10 expression. Biomed. Res. Int.2018, 3936257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fruhbeck, G. et al. FNDC4, a novel adipokine that reduces lipogenesis and promotes fat browning in human visceral adipocytes. Metab. Clin. Exp.108, 154261 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Wallgard, E. et al. Identification of a core set of 58 gene transcripts with broad and specific expression in the microvasculature. Arterioscler. Thromb. Vasc. Biol.28, 1469–1476 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Niaudet, C. et al. Adgrf5 contributes to patterning of the endothelial deep layer in retina. Angiogenesis22, 491–505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoh, N. & Ohta, H. Pathophysiological roles of FGF signaling in the heart. Front. Physiol.4, 247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lederman, R. J. et al. Therapeutic angiogenesis with recombinant fibroblast growth factor-2 for intermittent claudication (the TRAFFIC study): a randomised trial. Lancet359, 2053–2058 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Simons, M. et al. Pharmacological treatment of coronary artery disease with recombinant fibroblast growth factor-2: double-blind, randomized, controlled clinical trial. Circulation105, 788–793 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Engelmann, G. L., Dionne, C. A. & Jaye, M. C. Acidic fibroblast growth factor and heart development. Role in myocyte proliferation and capillary angiogenesis. Circ. Res.72, 7–19 (1993). [DOI] [PubMed] [Google Scholar]
- 52.Palmen, M. et al. Fibroblast growth factor-1 improves cardiac functional recovery and enhances cell survival after ischemia and reperfusion: a fibroblast growth factor receptor, protein kinase C, and tyrosine kinase-dependent mechanism. J. Am. Coll. Cardiol.44, 1113–1123 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Fernandez, B. et al. Transgenic myocardial overexpression of fibroblast growth factor-1 increases coronary artery density and branching. Circ. Res.87, 207–213 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Wang, Z. et al. Fibroblast growth factor-1 released from a heparin coacervate improves cardiac function in a mouse myocardial infarction model. Acs Biomater. Sci. Eng.3, 1988–1999 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Buehler, A. et al. Angiogenesis-independent cardioprotection in FGF-1 transgenic mice. Cardiovasc. Res.55, 768–777 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Li, G. et al. MicroRNA-27b-3p down-regulates FGF1 and aggravates pathological cardiac remodelling. Cardiovasc. Res.118, 2139–2151 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, X. et al. Rosmarinic acid attenuates cardiac fibrosis following long-term pressure overload via AMPKalpha/Smad3 signaling. Cell Death Dis.9, 102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu, C. et al. Matrine attenuates oxidative stress and cardiomyocyte apoptosis in doxorubicin-induced cardiotoxicity via maintaining AMPKalpha/UCP2 pathway. Acta Pharm. Sin. B.9, 690–701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu, C. et al. Osteocrin attenuates inflammation, oxidative stress, apoptosis, and cardiac dysfunction in doxorubicin-induced cardiotoxicity. Clin. Transl. Med.10, e124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hua, X. et al. Single-cell RNA sequencing to dissect the immunological network of autoimmune myocarditis. Circulation142, 384–400 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Hu, C. et al. Meteorin-like protein attenuates doxorubicin-induced cardiotoxicity via activating cAMP/PKA/SIRT1 pathway. Redox Biol.37, 101747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma, Z. G. et al. IRX2 regulates angiotensin II-induced cardiac fibrosis by transcriptionally activating EGR1 in male mice. Nat. Commun.14, 4967 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, X. et al. Matrine attenuates pathological cardiac fibrosis via RPS5/p38 in mice. Acta Pharmacol. Sin.42, 573–584 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, X. et al. Endothelial ERG alleviates cardiac fibrosis via blocking endothelin-1-dependent paracrine mechanism. Cell Biol. Toxicol.37, 873–890 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Zhang, X. et al. Osteocrin, a novel myokine, prevents diabetic cardiomyopathy via restoring proteasomal activity. Cell Death Dis.12, 624 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma, Z. G. et al. C1q-tumour necrosis factor-related protein-3 exacerbates cardiac hypertrophy in mice. Cardiovasc. Res.115, 1067–1077 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Wang, S. et al. Adipocyte Piezo1 mediates obesogenic adipogenesis through the FGF1/FGFR1 signaling pathway in mice. Nat. Commun.11, 2303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krist, B. et al. miR-378a influences vascularization in skeletal muscles. Cardiovasc. Res.116, 1386–1397 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Kruskal, B. A., Shak, S. & Maxfield, F. R. Spreading of human neutrophils is immediately preceded by a large increase in cytoplasmic free calcium. Proc. Natl Acad. Sci. USA.83, 2919–2923 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, X. et al. Rosmarinic acid alleviates cardiomyocyte apoptosis via cardiac fibroblast in doxorubicin-induced cardiotoxicity. Int. J. Biol. Sci.15, 556–567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma, Z. G. et al. A77 1726 (leflunomide) blocks and reverses cardiac hypertrophy and fibrosis in mice. Clin. Sci.132, 685–699 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Yu, B. et al. Extracellular vesicles engineering by silicates-activated endothelial progenitor cells for myocardial infarction treatment in male mice. Nat. Commun.14, 2094 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the main text and its Supplementary Information file and Source data. Source data are provided with this paper. Additional data that support the findings in this study are available from the corresponding author upon reasonable request. The datasets generated for the transcriptome analysis are available through the Gene Expression Omnibus under accession code GSE277729. Source data are provided with this paper.