Abstract
During growth, cells need to synthesize and expand their envelope, a process that requires careful regulation. Here, we show that the GTPase ObgE of E. coli contributes to the regulation of lipopolysaccharide (LPS) synthesis, an essential component of the Gram-negative outer membrane. Using a dominant-negative mutant (named ‘ObgE*’), we show a direct interaction between ObgE and LpxA, which catalyzes the first step in LPS synthesis. This interaction is enhanced by the mutation in ObgE* which, when bound to GTP, leads to inhibition of LpxA, decreased LPS synthesis, and cell death. Although wild-type ObgE does not exert the same strong effects as ObgE* on LpxA or LPS synthesis, our data indicate that ObgE participates in the regulation of cell envelope synthesis in E. coli. Because ObgE also influences other cellular functions (i.e., ribosome assembly, DNA replication, etc.), it seems increasingly plausible that this GTPase coordinates several processes to finetune cell growth.
Subject terms: Cellular microbiology, Bacterial physiology, Bacteriology
During growth, cells need to synthesize and expand their envelope, a process that requires careful regulation. Here, Dewachter et al. show that the GTPase ObgE of E. coli contributes to regulation of the synthesis of lipopolysaccharide, an essential component of the Gram-negative outer membrane.
Introduction
Obg is a small GTPase that is conserved and essential within the bacterial kingdom1. This protein has been implicated in many different processes, but, despite its obvious importance, most cellular roles of Obg have not been thoroughly characterized. Obg is mostly known to facilitate ribosome assembly by interacting with the 50S ribosomal subunit and acting as an anti-association factor2–6. The strength of Obg’s ribosome anti-association activity appears to be regulated by its nucleotide binding state2.
Since Obg is a GTPase, it can interact with different guanine nucleotides and is expected to change its conformation and activity in response1. Indeed, Obg has been found to bind GTP, GDP, and the stringent response alarmone ppGpp and is able to hydrolyze GTP, albeit inefficiently7–9. Since Obg’s GTP hydrolysis rate is low and nucleotide exchange occurs very fast, Obg is generally thought to act as a sensor for cellular guanine nucleotide concentrations2,7–9.
Apart from Obg’s role in translation, several other functions have been attributed to this enigmatic protein. Obg has been shown to influence a large variety of processes, including DNA replication10–14, chromosome segregation11,15, several stress responses12,16,17, antibiotic persistence18, sporulation19, etc. More recently, a connection between Obg and the gram-negative outer membrane was reported. In Acinetobacter baumannii, a synthetically sick phenotype was obtained by combining a mutant obg allele (obgN258I) with a defect in the Mla pathway that functions in maintaining outer membrane asymmetry20.
The outer membrane is an essential part of the gram-negative cell envelope that acts as a permeability barrier and a load-bearing and osmoprotective structure21–23. Lipopolysaccharides (LPS) make up a large part of the outer leaflet of this asymmetric membrane24. LPS consists of the hydrophobic membrane anchor lipid A coupled to a core oligosaccharide which is additionally decorated with an O-antigen polysaccharide25. Whereas the O-antigen and core oligosaccharide are important for protection against environmental stresses and outer membrane stability, they are not strictly essential in Escherichia coli24,26. In fact, E. coli K12 lab strains do not produce O-antigen at all27,28. The minimal LPS structure that is required for E. coli survival under standard growth conditions is the Kdo-modified lipid IVA24,29, while at low temperatures, E. coli can survive with only lipid IVA30.
Kdo2-lipid A is synthesized in nine enzymatic steps that collectively form the Raetz pathway24,25. In the first step, the essential LpxA enzyme transfers an acyl group from acyl-ACP onto uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) to form UDP-3-O-acyl-GlcNAc24,25. This reaction is reversible and thermodynamically unfavorable, meaning that the committed step of the pathway is performed by the second enzyme, LpxC24,25,31. The LpxC enzyme has been established as a major point of regulation in lipid A and LPS synthesis. In E. coli, LpxC activity is mostly regulated at the posttranslational level by the protease FtsH32,33. Degradation of LpxC by FtsH is modulated by accessory proteins LapB (also known as YciM) and YejM (also known as PbgA) and depends on factors such as growth rate, fatty acid synthesis, and others34–43. LpxA, on the other hand, was widely believed not to be subjected to any kind of regulation. However, recent work demonstrated that LpxA activity can be altered by RnhB and ppGpp44, thereby indicating that additional levels of regulation in LPS synthesis exist. The reaction catalyzed by LpxA is an interesting one since it consumes substrates that can alternatively be used for the synthesis of phospholipids or peptidoglycan25,31,45. LpxA can, therefore, be seen as a hub in cell envelope synthesis, and changes in its activity could potentially affect the production of three major cell envelope components: LPS, phospholipids, and peptidoglycan. Because of its importance for the survival of most Gram-negative bacteria and its complete absence from humans, lipid A synthesis is seen as an interesting target for the development of new antibiotics46. In this respect, most attention is focused on the identification and optimization of compounds that inhibit LpxC, while other enzymes of the lipid A synthesis pathway are not explored as much46–48.
Here, we demonstrate that LPS synthesis in Escherichia coli is also regulated at the level of LpxA by the GTPase ObgE (Obg of E. coli). We have reached this conclusion by characterizing the toxic effects of ObgE*, a mutant isoform of ObgE that contains one amino acid substitution, K268I. This small variation in the protein results in a drastic change in functionality. In contrast to wild-type (wt) ObgE, which is essential for bacterial viability, expression of obgE* leads to E. coli cell death even in the presence of a wt chromosomal obgE copy49. We hypothesized that the dominant-negative phenotype caused by ObgE* stems from dysregulation of ObgE’s normal function and that by studying the cell death pathway triggered by ObgE*, we could get more insight into the cellular role of wt ObgE. We here show that ObgE* toxicity is caused by direct inhibition of LpxA catalytic activity, which leads to a decrease in LPS synthesis and subsequent cell death. Similarly, we show that wt ObgE interacts with LpxA. Even though this interaction did not strongly alter LpxA activity in vitro, upregulation of wt ObgE did sensitize E. coli to LPS inhibitors, demonstrating a regulatory role for ObgE in LPS synthesis in vivo. Taken together, our data show that ObgE* is a very potent inhibitor of LpxA and that wt ObgE likely regulates LpxA activity in vivo, although the conditions under which it does so remain to be further investigated. Our results, therefore, point toward a novel regulatory mechanism that controls LPS synthesis and that operates at the level of LpxA, a hub in cell envelope synthesis previously thought to not be subjected to any kind of regulation.
Results
ObgE*, a toxic mutant isoform of ObgE, activates the Rcs stress response
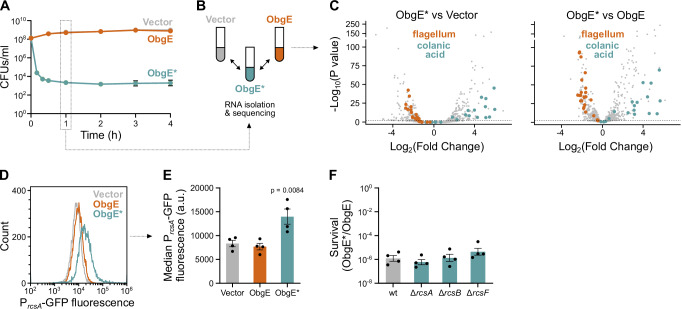
The amino acid substitution that is present in ObgE*, K268I, turns this essential GTPase into a highly toxic one that exerts a dominant negative effect on E. coli cell survival (Fig. 1A). To reveal how ObgE* causes cell death, we performed RNA sequencing and identified all genes that are up- or downregulated upon expression of the toxic obgE* allele. To do so, we expressed obgE* from a plasmid (pBAD33Gm-obgE*) in an E. coli strain that still contains the genomic wild-type (wt) obgE gene and compared the results to a vector control (pBAD33Gm) and overexpression of wt obgE (pBAD33Gm-obgE) (Fig. 1B). Results show that ObgE* causes a massive disturbance of gene expression (Fig. 1C, Supplementary Fig. S1A and Supplementary data 1), with 359 or 1045 genes (8% and 23% of all genes) significantly up- or downregulated compared to the vector control or overexpression of wt obgE, respectively.
Fig. 1. The toxic ObgE isoform, ObgE*, induces the Rcs stress response.
A Expression of obgE* (obgEK268I) causes a dominant-negative effect on cell survival. Cultures of E. coli containing pBAD33Gm (Vector), pBAD33Gm-obgE (ObgE), or pBAD33Gm-obgE* (ObgE*) were induced with arabinose, and cell survival was measured over time by determining CFUs/ml. Data are represented as the mean ± SEM, number of biological replicates n = 3, except for 2 h where n = 4. B RNA-seq analysis was performed on the Vector, ObgE, and ObgE* samples after 1 h of induction with arabinose. C ObgE* causes a massive disturbance in gene expression. Volcano plots of RNA-seq data reveal genes that are significantly up- or downregulated by ObgE* compared to the Vector (left) or wt ObgE (right) control. P-values were obtained using a two-sided Wald test and corrected for multiple testing using the Benjamini and Hochberg method. The gray dotted line marks an adjusted p-value of 0.01. Genes belonging to the GO category “Colanic Acid Biosynthetic Process” (GO:0009242) or “Bacterial-type Flagellum Organization” (GO:0044781) are highlighted in blue and orange, respectively. D ObgE* activates the Rcs stress response. A representative flow cytogram shows that ObgE* upregulates the rcsA promotor, which is part of the Rcs regulon. E From flow cytograms such as shown in (D), the median GFP fluorescence was recorded and plotted separately. Bar graphs and error bars represent the mean ± SEM, number of biological replicates n = 4. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test was performed against the Vector control condition. F ObgE* toxicity is not altered in the absence of a functional Rcs stress response. E. coli wt and Rcs stress response mutant cultures carrying pBAD33Gm-obgE or pBAD33Gm-obgE* were induced with arabinose to activate obgE(*) expression. Two hours after induction, CFUs/ml were determined, and the level of survival was calculated by dividing CFUs/ml upon obgE* expression by those recorded upon wt obgE overexpression. Bar graphs and error bars represent the mean ± SEM, number of biological replicates n = 4. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test was performed against the wt control condition, and no significant differences were detected.
To gain a more high-level overview of the cellular effects of ObgE*, we performed Gene Ontology (GO) enrichment analyses on genes that are significantly up- or downregulated by ObgE* compared to both the vector and wt ObgE control (Supplementary Fig. S1B and Supplementary data 1). These analyses revealed that genes involved in colanic acid synthesis, a protective exopolysaccharide involved in biofilm formation50, are strongly enriched among upregulated genes. Flagellar genes, on the other hand, are strongly downregulated (Fig. 1C). Since both colanic acid production and flagellar genes are controlled by the Rcs response51,52, our results hint at the activation of this cell envelope stress response by ObgE*. Indeed, our RNA-seq data confirm that rcsA, an Rcs auxiliary gene that is under the direct control of this stress response52, is strongly upregulated by ObgE* (14- and 9-fold upregulated compared to the vector and wt ObgE controls, respectively (Supplementary data 1)). These results were further confirmed by showing that ObgE* activates a PrcsA-GFP promoter fusion (Fig. 1D, E).
Strong activation of the Rcs response was shown to be toxic under certain conditions51. However, because ObgE* toxicity remained unchanged in E. coli mutants that are unable to launch the Rcs stress response51–53 (Fig. 1F), we conclude that Rcs activation does not contribute to the toxic ObgE* phenotype. Likewise, eliminating the production of colanic acid synthesis upon obgE* expression did not increase survival54,55 (Supplementary Fig. S1C). These findings demonstrate that neither the Rcs response itself nor the activation of the colanic acid synthesis pathway can explain ObgE*-mediated toxicity. Rather, ObgE* likely causes defects in the cell envelope that trigger the Rcs stress response and ultimately lead to cell death.
Toxicity in L-forms reveals that ObgE* primarily targets the E. coli outer membrane
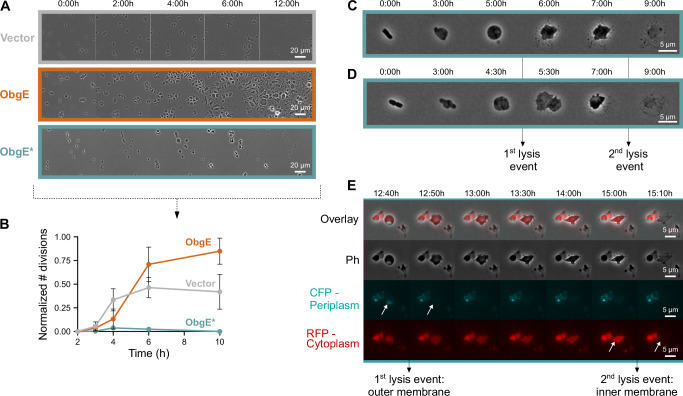
The Rcs response can be activated by disturbances in both the Gram-negative outer membrane and the peptidoglycan cell wall51,52. To investigate whether ObgE* exerts its toxic effect by targeting the cell wall, we used E. coli L-forms that are devoid of peptidoglycan56–58. As can be seen in Fig. 2A, B, and Supplementary Movie S1, ObgE* remains toxic in L-forms as it negatively affects growth and strongly inhibits L-form proliferation, which depends on membrane synthesis59.
Fig. 2. ObgE* toxicity in E. coli L-forms hints at an effect of ObgE* on the outer membrane.
A ObgE* remains toxic in E. coli L-forms that are devoid of peptidoglycan. E. coli pBAD33Gm (Vector), pBAD33Gm-obgE (ObgE), or pBAD33Gm-obgE* (ObgE*) were grown on agarose pads that contain arabinose, the inducer of obgE(*) expression, and fosfomycin, which triggers the transition into the L-form state. Snapshot images of time-lapse microscopy recordings are shown. B Quantitative analysis of these time-lapse images reveals that ObgE* strongly inhibits L-form proliferation. The number of successful L-form divisions was recorded and normalized to the number of intact L-forms present in the field of view. Data are represented as the mean ± SEM, number of biological replicates n = 3, where each repeat contains > 40 L-forms. C, D Time-lapse images of single (C) or dividing (D) E. coli cells that express obgE* are shown as they transition into the L-form stage and subsequently undergo cell lysis that proceeds in two distinct steps. E Time-lapse images of E. coli L-forms that express obgE* and contain a blue periplasmic and red cytoplasmic marker reveals how ObgE* triggers two-stage L-form lysis. ObgE* initially causes rupture of the outer membrane and loss of periplasmic content, while the second stage of lysis corresponds to the loss of inner membrane integrity and cytoplasmic content.
Because L-forms are known to be highly susceptible to reactive oxygen species (ROS)56, we verified that ObgE* toxicity in L-forms does not depend on ROS. In contrast to walled E. coli cells60, ObgE* did not lead to increased ROS levels in L-forms (Supplementary Fig. S2A). Moreover, a combination of scavengers that lowered ROS levels did not alter ObgE* L-form toxicity (Supplementary Fig. S2A, B and Supplementary Movie S2). Our data, therefore, indicate that ObgE* toxicity in E. coli L-forms is unrelated to ROS production.
Because ObgE* remains toxic in E. coli L-forms independently of ROS production, we hypothesize that ObgE* targets the outer membrane. Indeed, close inspection of L-form phenotypes upon obgE* expression hints at an outer membrane defect. More specifically, ObgE* causes lysis of L-forms in two distinct steps (Fig. 2C and Supplementary Movie S3). In the first lysis step, the cell abruptly expands and loses its smooth edge, but is able to retain cellular content. In the second lysis step, the cell bursts and dies. These two lysis steps correspond to the rupture of first the outer and then the inner membrane as demonstrated by (1) cells that were in the process of dividing prior to adopting the L-form state where the first lysis step liberates two separated protoplasts (Fig. 2D and Supplementary Movie S4), and (2) differentially labeling the cytoplasm with mCherry and the periplasm with superfolder mTurquoise2ox21,61 (Fig. 2E and Supplementary Movie S5). This 2-stage lysis process occurs in the vast majority of L-forms that express obgE*, and the delay between the first and second steps can range anywhere from minutes to hours (Supplementary Fig. S2C). These results indicate that ObgE*’s primary effect is to weaken the E. coli outer membrane, eventually resulting in L-form lysis. We hypothesize that ObgE*’s negative effect on the outer membrane also causes cell death in walled E. coli cells and serves as the trigger for the Rcs response.
Suppressor mutations in lpxA provide resistance to ObgE*
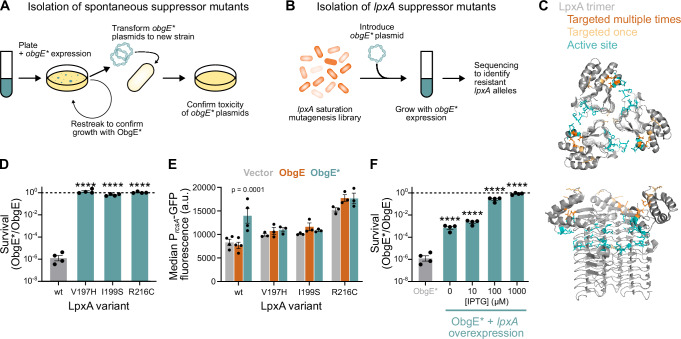
To be able to determine how ObgE* affects the E. coli outer membrane, we generated spontaneous suppressor mutants that have become resistant to the toxic action of ObgE* (Fig. 3A). To limit the isolation of mutants that no longer produce ObgE* (e.g., due to promoter or stop codon mutations), we expressed obgE* from two independent promoters; PBAD and Plac, and confirmed full-length ObgE* production using C-terminal fluorescent fusions to Venus and mCherry. For selected colonies, the toxicity of their obgE* alleles was confirmed by transforming the isolated plasmids to fresh genetic backgrounds. This way, we isolated 34 suppressor mutants from 22 independent overnight cultures. Whole genome sequencing was performed on these suppressor strains, and mutations are listed in Supplementary Data 2. Of note, 30 out of the 34 suppressors carried mutations in lpxA (Table 1). The essential LpxA enzyme is a UDP-N-acetylglucosamine acyltransferase that catalyzes the first step in the synthesis of lipid A, the membrane anchor of LPS24,25.
Fig. 3. Suppressor mutations in lpxA provide resistance to ObgE*.
A The workflow used to isolate spontaneous suppressor mutants resistant to ObgE* is shown. Colonies of E. coli pBAD33Gm-obgE*-venus pQE80L-obgE*-mCherry that formed despite induction of obgE* and that retained yellow (ObgE*-Venus) and red (ObgE*-mCherry) fluorescence were restreaked to confirm ObgE* resistance. Next, plasmids were isolated and transformed into a fresh parental strain to confirm the toxicity of their obgE* alleles. B Additional lpxA suppressors were isolated from a lpxA saturation mutagenesis library that was transformed with pBAD33Gm-obgE* and grown overnight with obgE* expression. Surviving cells were sequenced to identify lpxA alleles that provide ObgE* resistance. C The trimeric LpxA protein structure is shown (PDB 2jf3). Residues substituted in resistant lpxA mutants are highlighted in orange. The active site is shown in blue. D ObgE* toxicity is completely neutralized by selected lpxA mutations. E. coli wt and lpxA mutant cultures carrying pBAD33Gm-obgE or pBAD33Gm-obgE* were induced with arabinose for two hours. CFUs/ml were determined, and survival was calculated by dividing CFUs/ml with ObgE* by those with ObgE. Bar graphs and error bars represent the mean ± SEM, number of biological replicates n = 4. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test was performed against the wt control condition, ****p < 0.0001. E Based on flow cytometry data, the median GFP fluorescence from PrcsA-GFP was recorded and plotted. Bar graphs and error bars represent the mean ± SEM of these median values, number of biological replicates n ≥ 3. Ordinary one-way ANOVA with Sidak’s multiple comparisons test was performed to compare the ObgE and ObgE* condition of each mutant strain to their Vector control. F lpxA overexpression can neutralize ObgE* toxicity. E. coli carrying pBAD33Gm-obgE or pBAD33Gm-obgE* and pCA24N-lpxA were induced with arabinose and different concentrations of IPTG to induce different levels of lpxA expression. Two hours after induction, CFUs/ml were determined, and survival was calculated by dividing CFUs/ml with ObgE* by those with ObgE. Bar graphs and error bars represent the mean ± SEM, number of biological replicates n = 4. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test was performed to compare against the wt control condition without pCA24N-lpxA (thereby also eliminating leaky lpxA expression), ****p < 0.0001.
Table 1.
lpxA suppressor mutations that provide resistance to ObgE*
| Mutated Amino Acids | ||
|---|---|---|
| Targeted Residue | Spontaneous suppressors | Saturation mutagenesis |
| LpxA N120 | / | G |
| LpxA V197 | / | H, W |
| LpxA I199 | S | A, K, S |
| LpxA R216 | C, S | A, G, K, M, S, W, Y |
| LpxA K220 | N | / |
This table shows lpxA mutations (at the protein level) that are expected to provide resistance to ObgE*. These mutations were either identified in spontaneous suppressor mutants (‘Spontaneous suppressors’) or were isolated from the lpxA saturation mutagenesis library (‘Saturation mutagenesis’).
Because our data hinted at a pivotal role for LpxA in neutralizing ObgE* toxicity, we set out to identify all possible lpxA mutations that could provide resistance against ObgE*. To do so, we performed saturation mutagenesis on lpxA. Using high-throughput CRISPR-Cas editing62, we introduced genomic mutations so that, at the protein level, each amino acid of LpxA would be replaced by every other amino acid (Fig. 3B). The more than 5000 genomic mutants of this pooled library were transformed with pBAD33Gm-obgE* and grown overnight in triplicate while inducing obgE* expression. After a growth period of 20 h, PacBio sequencing of the lpxA gene was performed (Supplementary Data 2). This way, 14 putative resistance-conferring lpxA alleles were identified (Table 1). These 14 identified mutations, together with the 4 isolated spontaneous suppressor mutations, target only six different LpxA residues. Interestingly, 5 out of the 6 identified residues are clustered closely together within the LpxA protein structure (Fig. 3C).
For the three LpxA residues that were targeted multiple times, we chose representative mutations for further analysis; V197H, I199S, and R216C. We introduced these mutations into a clean parental strain and assessed their ability to counteract ObgE* toxicity. All three mutations provide full resistance to ObgE* (Fig. 3D) and prevent the activation of the Rcs stress response (Fig. 3E). Similarly, overexpression of lpxA was also found to counteract ObgE* toxicity in a dose-dependent manner (Fig. 3F).
In the next step, we characterized the effects of the selected lpxA mutations both in vitro and in vivo. Whereas the R216C mutation caused a minor growth defect and produced slightly elongated exponential-phase cells, neither the V197H nor I199S mutations affected growth or morphology (Supplementary Fig. S3A, B). Similarly, LPS synthesis is significantly decreased to 79% of wt levels in E. coli lpxAR216C, while it appears unchanged in the other lpxA mutant strains (Supplementary Fig. S3C). In line with these findings, sensitivity assays show that lpxAR216C strains are hypersensitive to the LPS inhibitor PF-04753299, which targets the LpxC enzyme, and also shows an increased sensitivity to vancomycin, an antibiotic that cannot penetrate an intact outer membrane (Supplementary Fig. S3D). The other lpxA mutant alleles (V197H and I199S) remain insensitive to vancomycin, and only lpxAV197H is slightly sensitized to LPS inhibition by PF-04753299 (Supplementary Fig. S3D). These data are confirmed by in vitro LpxA activity assays that show that LpxAI199S is almost as active as wt LpxA, but that LpxAR216C displays lowered catalytic activity (Supplementary Fig. S3E). Unfortunately, purified LpxAV197H proved to be unstable and its in vitro activity could therefore not be tested.
ObgE and ObgE* directly interact with LpxA
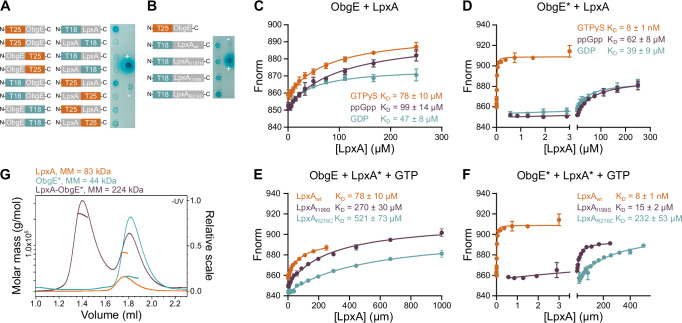
Because several LpxA amino acid substitutions can neutralize the negative effect of ObgE*, we hypothesized that ObgE* influences LpxA activity. We, therefore, first checked whether ObgE interacts with LpxA in vivo using the Bacterial Adenylate Cyclase-Based Two-Hybrid (BACTH) assay63. As shown in Fig. 4A, ObgE and LpxA indeed interact, but only when their C-terminal domains are free and available for binding. In contrast to our expectations, also mutant LpxA proteins that provide resistance against ObgE* (LpxAV197H, LpxAI199S, and LpxAR216C) interact with wt ObgE in vivo (Fig. 4B).
Fig. 4. ObgE and ObgE* directly interact with LpxA.
A Bacterial two-hybrid (BACTH) assays with ObgE and LpxA in all possible orientations show that these proteins interact when both are fused at their N terminus. ‘+’ and ‘−’ indicate positive and negative controls, respectively. B BACTH assays show that ObgE not only interacts with LpxA wt but also with LpxA mutant proteins, LpxAV197H, LpxAI199S, and LpxAR216C. C–F Microscale thermophoresis (MST) was performed to measure binding affinities between wt ObgE and wt LpxA in the presence of different nucleotides (C), between ObgE* and wt LpxA in the presence of different nucleotides (D), between wt ObgE and mutant LpxA proteins in the presence of GTPγS (E) and between ObgE* and mutant LpxA proteins in the presence of GTPγS (F). Data points and error bars represent the mean ± SEM, n = 3. G SEC-MALS confirms a 1:1 stoichiometry of the ObgE*-LpxA complex where three ObgE* monomers bind per LpxA trimer. LpxA concentrations are given as subunit concentrations.
The ObgE-LpxA interaction that was detected in vivo by BACTH was subsequently confirmed in vitro via chemical crosslinking mass spectrometry (XL-MS) using the purified LpxA and ObgE or ObgE* proteins and using the amine-reactive crosslinker disuccinimidyl suberate (DSS) (Supplementary Fig. S4) and was subsequently quantified via Microscale Thermophoresis (MST) (Fig. 4C–F). Hereto, fixed concentrations of randomly Cy5-labeled ObgE or ObgE* were titrated with a concentration gradient of LpxA. Since ObgE and ObgE* are GTPases that bind GTP, GDP, and ppGpp1,8, they are expected to change conformation and activity based on their nucleotide-binding state1,8. We, therefore, studied the interaction between LpxA and ObgE(*) in the presence of these different nucleotides. For wt ObgE, we observe a low-affinity interaction with LpxA that is nearly independent of the ObgE nucleotide binding state, with equilibrium dissociation constants (KD) of 47 ± 8 µM, 99 ± 14 µM and 78 ± 10 µM for the GDP-, ppGpp- and GTPγS-bound states of ObgE, respectively. Interestingly, while ObgE* displays similar low binding affinities in the presence of GDP and ppGpp, with KD’s of 39 ± 9 µM and 62 ± 8 µM respectively, a drastic increase in binding affinity is observed in the presence of GTPγS. When bound to this non-hydrolyzable GTP analog, the affinity of ObgE* for LpxA is increased more than 1000-fold, with a KD of 8 ± 1 nM. However, GTPγS-bound ObgE* no longer displays this tight interaction with the mutant LpxA proteins, with a KD of 15 ± 2 µM for LpxAI199S and a KD of 232 ± 53 µM for LpxAR216C. These data suggest that ObgE* toxicity is suppressed in these mutants by weakening the interaction with ObgE* in its GTP-bound state. A similar trend in the effect of the LpxA mutations is observed for the interaction with wt ObgE, albeit much less pronounced. For GTPγS-bound wt ObgE, the KD value increases from 78 ± 10 µM for wt LpxA to 270 ± 30 µM for LpxAI199S and 521 ± 73 µM for LpxAR216C.
Since ObgE and LpxA behave as a monomer and homotrimer in solution, we next wondered whether three molecules of ObgE* bind the LpxA trimer. To assess the binding stoichiometry, an SEC-MALS experiment was performed for the LpxA-ObgE* complex in excess of GTPγS, while the individual proteins were also analyzed as a reference (Fig. 4G). The molar masses obtained for ObgE* (44 ( ± 0.3%) kDa) and the LpxA trimer (83 ± (0.3%) kDa) correspond well to the expected molar masses for an ObgE* monomer (45 kDa) and LpxA trimer (90 kDa). The molar mass obtained for the ObgE*-LpxA complex (224 ( ± 0.1%) kDa) is in very good agreement with the expected molar mass for a LpxA trimer bound to three ObgE* monomers (225 kDa), confirming the 1:1 stoichiometry of the complex.
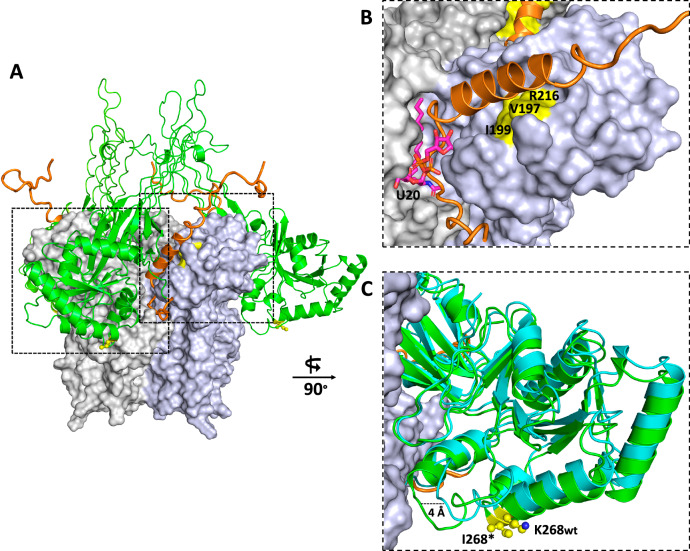
To obtain a better understanding of the molecular mechanisms underlying the interaction of ObgE* and LpxA, AlphaFold Multimer was used to predict a model of the ObgE*-LpxA complex64,65. Based on our SEC-MALS results, a 3:3 subunit ratio was used to model three ObgE* proteins binding to a LpxA trimer66. The top five solutions generated by AlphaFold Multimer fall apart in two distinct interaction modes, represented by models 1 and 2 in Supplementary Fig. S5, with model 1 being the most probable according to the AlphaFold Multimer ranking. The two models drastically differ in the way ObgE* interacts with LpxA: while model 1 mainly relies on interactions with the ObgE* G domain and C-terminal domain, model 2 relies on interactions with the N-terminal Obg domain and C-terminal domain. Considering that in model 1 the G domain of ObgE*, which contains the K268I mutation, is involved in the interaction with LpxA, we consider model 1 also as the most biologically relevant, and will use this model for further interpretation. Nevertheless, an experimental ObgE*-LpxA structure would be required to unambiguously validate this model. In our preferred model 1, the G domain and β-barrel region of the N-terminal domain of ObgE* are inserted in between two adjacent subunits of the LpxA trimer, hence partially covering the LpxA active site pocket (Fig. 5A). In particular, the G domain is within close distance to both the N-terminal left-handed parallel β-helix (LβH) domain and C-terminal α-helical domain of an LpxA subunit66–68. Interestingly, according to the AlphaFold model, extensive interactions are mediated via the ObgE* C-terminal intrinsically disordered domain (aa 340-390)8. A part of this domain (aa 344 – 356) adopts a coil conformation that inserts deep into the LpxA active site and significantly overlaps with the UDP-GlcNAc-binding site while also covering the R-3-hydroxymyristoyl binding groove, thus providing a structural explanation for the observed inhibition of LpxA activity (Fig. 5B)68. A second part of the ObgE* C-terminal domain adopts an α-helical conformation that directly stacks to a surface patch of LpxA containing residues V197, I199, and R216. Mutation of either of these latter 3 residues confers resistance to ObgE* toxicity, hence providing additional credibility to the proposed model. A remaining question is how the K268I mutation, located within the ObgE G domain, can drastically influence the binding affinity toward LpxA, especially considering that the site of mutation is located relatively far away from the ObgE*-LpxA binding interface. Part of the answer to this question is provided by comparing the AlphaFold models of the complexes formed between LpxA and ObgE* versus ObgE. Although the exact mechanism underlying this observation is not entirely clear, this comparison suggests that the K268I mutation of ObgE* induces a conformational change in the G domain that brings this domain in closer contact with LpxA, hence potentially leading to a stronger overall interaction between both proteins (Fig. 5C). An additional AlphaFold multimer modeling of complexes formed between several representatives of homologs of Obg and LpxA belonging to the class of the Gammaproteobacteria shows that in all tested cases a structure very close to Model 1 is obtained among the top 5-ranked predictions (Supplementary Fig. S6). This suggests that the binding of Obg to LpxA is more widely conserved among the Gammaproteobacteria.
Fig. 5. AlphaFold model of the LpxA-ObgE* complex.
A One of the two AlphaFold models (“model 1”, see Supplementary Fig. S5 for a comparison of the two generated models) predicts the binding interaction mode of 3 ObgE* molecules to an LpxA trimer. LpxA is shown in surface representation with each of the subunits in a different shade of gray, while ObgE* is shown in green (G domain and N-terminal domain) and orange (C-terminal domain). The position of the K268I mutation of ObgE* is indicated in yellow sticks. B Zoom-in on the interaction area of LpxA with the C-terminal domain of ObgE*. The same color code as in (A) is used. The position of the resistance-conferring LpxA residues (V197, I199, R216) is shown in yellow. The position of the LpxA active site is indicated by the presence of U20 (uridine-5’-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-D-glucosamine), represented in magenta sticks and obtained by superposing the AlphaFold model on PDB 2QIA68. C Superposition of the AlphaFold models predicting the binding to LpxA of ObgE* (green) and ObgE wild-type (cyan), respectively, focusing on the observed conformational changes in their G domains. The K268 (ObgE wild-type) and I268 (ObgE*) residues are shown in yellow sticks.
The GTPase ObgE* inhibits LpxA activity when bound to GTP
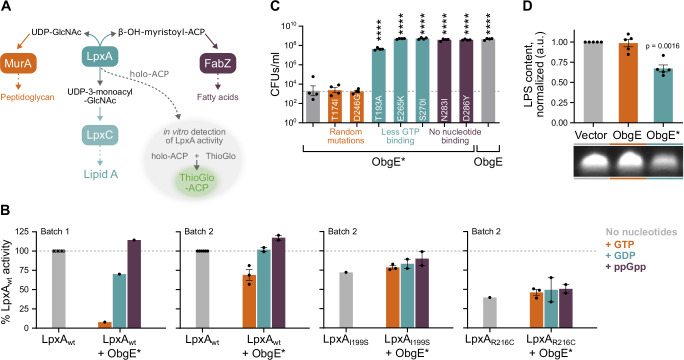
We next tested if and how the observed interaction between ObgE or ObgE* and LpxA would influence LpxA’s enzymatic activity. LpxA catalyzes a reversible reaction that, in the forward direction, transfers an acyl group from acyl-ACP onto UDP-GlcNAc24,25. When the reaction is performed in vitro in the presence of ThioGlo, LpxA activity can be detected by the production of a fluorescent ThioGlo-ACP conjugate (Fig. 6A)44,69. This assay was performed with two separately produced substrate batches and purified proteins. Like others44, we noticed strong batch-to-batch variability in our results, which necessitated the use of different protein concentrations in different assays and prevented us from precisely quantifying the observed effects. However, because general trends are conserved across batches, we can draw conclusions on the overall effect of the tested conditions on LpxA activity, rather than the magnitude of the effect.
Fig. 6. ObgE* influences LpxA activity and LPS synthesis.
A LpxA catalyzes a reversible reaction that consumes substrates that can also be used for peptidoglycan or fatty acid synthesis and produces a compound that can be further processed into lipid A, the membrane anchor of LPS. LpxA activity can be measured in vitro by monitoring the production of fluorescent ThioGlo-ACP, a compound that is produced when the liberated ACP molecule reacts with ThioGlo. B When bound to GTP, ObgE* reduces LpxA activity in vitro. The effect of different concentrations of ObgE* on LpxA activity was tested. Assays were performed with two different batches of substrate and purified proteins (indicated in the figure). In batch 1, the influence of 125 nM ObgE* on 10 nM LpxA was tested. In batch 2, LpxA and ObgE* were used at concentrations of respectively 3 and 7 nM. Results were normalized to the activity of LpxAwt without the addition of ObgE* or nucleotides. Bar graphs and error bars represent the mean ± SEM, number of biological replicates n = 1–3. C Combining the K268I amino acid substitution of ObgE* with other mutations that were shown to affect nucleotide binding can strongly decrease toxicity in vivo. CFUs/ml were determined upon expression of obgE*, obgE*T174I, obgE*D246G, obgE*T193A, obgE*E265K, obgE*S270I, obgE*N283I, obgE*D286Y or wt obgE from pBAD33Gm. Data are represented as the mean ± SEM, number of biological replicates n = 4. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test was performed against the ObgE* sample, ****p < 0.0001. D ObgE* decreases cellular LPS content. The effect of ObgE and ObgE* on the amount of LPS found in the cell was measured using a gel-based assay. Quantitative interpretation of these signals, normalized to the Vector control sample, shows that ObgE* leads to a decrease in cellular LPS content. Bar graphs and error bars represent the mean ± SEM, number of biological replicates n = 5. A one-sample, two-sided t test was performed to assess which samples display a normalized LPS content that deviates from one. ACP acyl carrier protein, GlcNAc N-acetylglucosamine, UDP uridine diphosphate.
Considering that the affinity of the interaction between LpxA and ObgE* is strongly increased in the presence of GTP, we assessed the effect of ObgE* on LpxA catalytic activity when bound to GTP, GDP or ppGpp (Fig. 6B and Supplementary Fig. S7). Despite strong batch-to-batch variability, all our repeats demonstrate that ObgE* inhibits LpxA catalytic activity when bound to GTP (Fig. 6B). Addition of other nucleotides (GDP or ppGpp) did not consistently alter LpxA activity (Fig. 6B). In contrast to wt LpxA, the activity of LpxA mutant proteins LpxAI199S and LpxAR216C in the presence of ObgE* was not significantly changed by the addition of GTP or any other nucleotide tested (Fig. 6B). These results indicate that the increased affinity of GTP-bound ObgE* for wt LpxA and the associated inhibition of LpxA activity is causal to toxicity. Wt ObgE did not strongly alter the activity of the LpxA wt or mutant proteins with any of the nucleotides tested (Supplementary Fig. S7).
Based on these in vitro results, we hypothesized that the GTP-bound form of ObgE* is responsible for toxicity and cell death in vivo. To confirm this hypothesis, we introduced mutations into obgE* that are known to change nucleotide binding affinities70,71 (Supplementary Table S1). As shown in Fig. 6C, preventing all nucleotide binding by amino acid substitutions N283I and D286Y completely eliminated toxicity. The apo-form of ObgE* is therefore not harmful to E. coli. Similarly, decreasing the relative affinity for GTP compared to other nucleotides (GDP and ppGpp) with amino acid substitutions T193A, E265K, and S270I allows for strong increases in cell survival (Fig. 6C). Although we cannot fully exclude that these mutations eliminate toxicity by effects other than their altered nucleotide affinities, we previously verified that these non-toxic ObgE* double mutants can substitute for wt ObgE in supporting E. coli viability, thereby confirming that they are properly folded and functional70. In addition, we show that mutations T174I and D246G that do not change the relative affinity for GTP also did not change ObgE* toxicity. Collectively, these results corroborate our in vitro findings and confirm that, in vivo, ObgE* needs to be bound to GTP in order to reduce LpxA activity to an extent that is toxic to E. coli. Isolated LpxA variants that provide resistance to ObgE* likely do so by weakening the interaction with GTP-bound ObgE* and thereby alleviating the inhibition of LpxA catalytic activity.
We next investigated the cellular effects of inhibition LpxA catalytic activity by GTP-bound ObgE*, which is expected to cause defects in LPS production (Fig. 6A). Indeed, after 2 h of expression, ObgE* has reduced the amount of LPS detected to 66% of normal concentrations (Fig. 6D). As expected, this negative effect of ObgE* on LPS was eliminated by each of the selected lpxA mutations (Supplementary Fig. S8A). The E. coli BW25113 lab strain that was used for all previous experiments72 is a K-12 derivative that is defective in O-antigen synthesis and only contains the LPS lipid A-core structure27,28. We therefore verified that ObgE* retains toxicity in the uropathogenic E. coli strain CFT073 (O6:K2:H1) that produces LPS with O-antigen polysaccharide73. Indeed, very strong toxicity was detected in E. coli CFT073, albeit slightly decreased compared to the E. coli BW25113 lab strain (Supplementary Fig. S8B). The absence of LPS O-antigen in E. coli BW25113 can, therefore, likely not explain the toxic ObgE* phenotype.
In order to produce lipid A precursors, LpxA consumes UDP-GlcNAc and β-hydroxymyristoyl-ACP, substrates that can also be used for respectively the production of peptidoglycan or fatty acids25,31,45. The LpxA enzyme can, therefore, be seen as a hub in the synthesis of three vital components of the Gram-negative cell envelope; peptidoglycan, phospholipids, and LPS. Because GTP-bound ObgE* blocks LpxA activity in the forward direction (Fig. 6B), it is expected to free up precursors for the synthesis of peptidoglycan and fatty acids. However, we could not detect any increases in peptidoglycan or fatty acid production upon obgE* expression (Supplementary Fig. S8C, D). On the contrary, the amount of cellular fatty acids is decreased by ObgE*.
Wt ObgE is involved in the regulation of cell envelope synthesis through LpxA
Even though we did not detect strong inhibitory effects of wt ObgE on either LpxA activity or LPS content, this GTPase was shown to interact with LpxA (both in vitro and upon overexpression in vivo). To investigate whether this interaction is physiologically relevant, we performed genome-wide CRISPRi screens upon obgE overexpression. In these screens, we made use of a previously validated pooled E. coli sgRNA plasmid library74 and transformed this library into E. coli strains with a chromosomal dcas9 gene under the control of the tight inducible Ptet promoter and carrying either pBAD33Gm or pBAD33Gm-obgE.
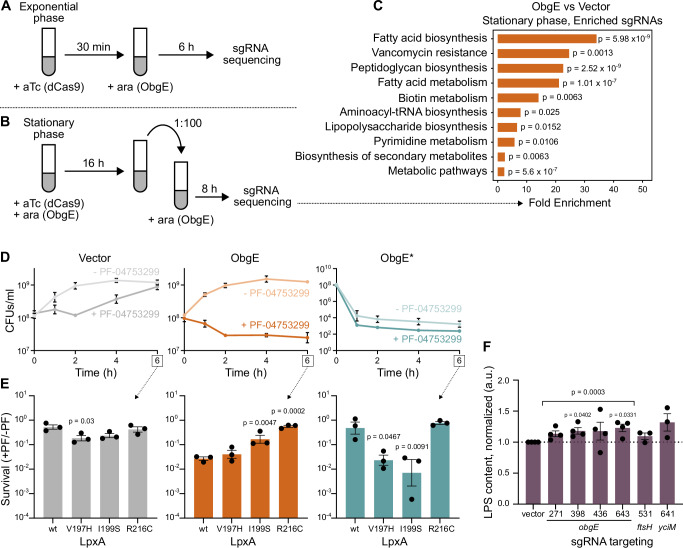
We initially induced dcas9 expression in the early exponential phase and added the inducer of obgE expression 30 min later (Fig. 7A). Six hours after inducing obgE expression, cell numbers were determined, the sgRNAs present in the population were sequenced, and sgRNA frequencies upon obgE overexpression were compared to a vector control (Supplementary Fig. S9A, B, and Supplementary data 3). The results from this exponential phase CRISPRi screen showed that, upon obgE overexpression, a variety of sgRNAs are significantly enriched or depleted. However, apart from ftsH, which encodes the LpxC-degrading protease FtsH32,33, no LPS-related genes were found among the significant hits (Supplementary data 3). Indeed, a KEGG enrichment analysis failed to show any link between ObgE and the E. coli cell envelope under these conditions and only identified “oxidative phosphorylation” as a process that was significantly enriched among genes that cause a fitness defect in the presence of excess ObgE (fold enrichment = 20, p = 0.0045).
Fig. 7. The wt ObgE protein is involved in the regulation of E. coli cell envelope synthesis.
A Schematic representation of the set-up of the CRISPRi screen performed in exponential phase. B Schematic representation of the set-up of the CRISPRi screen performed in the stationary phase. C KEGG pathway enrichment results are shown for the comparison of ObgE to vector control in the stationary phase CRISPRi screen. As input, genes were used for which our gene-level analysis showed significant enrichment of sgRNAs targeting this gene. Enrichment was determined by Fisher’s exact test. Two-sided p-values were FDR adjusted. Ara, arabinose. D Both ObgE and ObgE* synergize with LPS inhibitor PF-04753299. E. coli carrying pBAD33Gm, pBAD33Gm-obgE, or pBAD33Gm-obgE* were induced with arabinose, and PF-04753299 was added at a concentration of 1/8x MIC (0.03125 µg/ml). CFUs/ml were monitored for a growth period of 6 h. Data are represented as the mean ± SEM, number of biological replicates n = 3. E E. coli wt and lpxA mutants carrying pBAD33Gm, pBAD33Gm-obgE, or pBAD33Gm-obgE* were induced with arabinose and treated with PF-04753299 at a concentration of 1/8x MIC (0.03125 µg/ml for lpxAwt, lpxAV197H, lpxAI199S, and 0.0078 µg/ml for lpxAR216C). After 6 h, CFUs/ml were determined, and survival was calculated by dividing CFUs/ml with PF-04753299 treatment by those without PF-04753299. Bar graphs and error bars represent the mean ± SEM, number of biological replicates n = 3. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test was performed against the lpxAwt control condition. F CRISPRi depletion of ObgE using four different sgRNAs leads to slightly increased LPS levels, similar to depletion of known LPS inhibitors FtsH and YciM with CRISPRi74. Quantitative interpretation of this gel-based assay was performed and values were normalized to the vector control sample. Bar graphs and error bars represent the mean ± SEM, number of biological replicates n ≥ 3. A one-sample, two-sided t test was performed to assess if samples display a normalized LPS content that deviates from one. Samples were tested individually, and a sample where all obgE-targeting sgRNAs were taken together was also considered.
However, because an effect of obgE overexpression on cell envelope synthesis might be apparent only under specific growth conditions, we decided to expand our CRISPRi screening efforts beyond the exponential growth phase. In our stationary phase CRISPRi screen, we induced the expression of both dcas9 and obgE in the early stationary phase and maintained this expression overnight. Then, cultures were diluted into a fresh growth medium containing the inducer of obgE but not dcas9 (Fig. 7B). dcas9 expression was omitted at this point to not fully inhibit the growth of strains where expression of essential genes (including LPS synthesis genes) are targeted. After eight hours of growth, cell numbers were determined, sgRNAs were sequenced, and their frequencies were compared (Supplementary Fig. S9C, D and Supplementary Data 3). This setup allows us to assess fitness benefits or defects that occur during the stationary phase, lag phase, and/or very early exponential phase when dCas9 levels are still high. In this screen, only a small number of sgRNAs are depleted. These few sgRNAs correspond to genes whose expression is important for proliferation when ObgE is present in excess. These genes are involved in protein secretion and lead to the identification of “protein export” (fold enrichment = 77, p = 0.0376) and “bacterial secretion system” (fold enrichment = 46, p = 0.0487) in a KEGG enrichment analysis. On the other hand, a large number of sgRNAs was found to be significantly enriched, meaning that inhibiting the expression of corresponding genes increases fitness upon obgE overexpression. KEGG-based pathway analysis reveals that the majority of these genes are involved in cell envelope synthesis since categories such as “fatty acid biosynthesis”, “peptidoglycan biosynthesis”, and “lipopolysaccharide biosynthesis” are significantly enriched among detected hits (Fig. 7C). These results uncover an undeniable link between the GTPase ObgE and E. coli cell envelope synthesis.
Encouraged by the detected link between wt ObgE and the E. coli cell envelope, we set out to interrogate a potential role for wt ObgE in LPS synthesis in more detail. To do so, we assessed potential growth defects upon overexpression of wt obgE or obgE* while simultaneously applying sub-inhibitory concentrations of the LPS inhibitor PF-04753299 (Fig. 7D). As expected, the addition of PF-04753299 increases toxicity caused by ObgE*, albeit only slightly likely because the detection of synergy is limited by the emergence of suppressor mutants that no longer express obgE* (i.e., frameshifts, promoter and/or stop codon mutations). However, also for wt ObgE, synergy with the LPS inhibitor can be observed. Importantly, this synergy is dependent on LpxA since it is significantly weaker in two out of the three tested lpxA suppressor mutants (Fig. 7E). This clear LpxA-dependent synergy indicates that like ObgE*, wt ObgE has the potential to negatively influence LPS synthesis. However, unlike ObgE*, this effect appears to be minor and may be subjected to additional levels of control that are bypassed by the mutation present in ObgE*.
Finally, whereas overexpression of wt obgE did not significantly alter LPS levels (Fig. 6D), we also assessed the effect of ObgE depletion imposed by CRISPRi, using four different obgE-targeting sgRNAs74. dcas9 expression was induced in the exponential phase, and LPS levels were measured 4 h later. Two out of the four sgRNAs tested led to significantly increased LPS levels. For the other two sgRNAs, the observed increase is not statistically significant (Fig. 7F). Under the same conditions (activation of CRISPRi for 4 h starting in exponential phase), targeting the known LPS inhibitors FtsH or YciM37 leads to increases in LPS levels that are highly similar to what is observed for obgE depletion (Fig. 7F). Because decreasing cellular ObgE levels appear to increase LPS production, these results indicate that wt ObgE might be capable of negatively regulating LPS production like ObgE* does. Collectively, our results, therefore, support a role for wt ObgE in the regulation of LPS synthesis through modulating LpxA activity, although further research is needed to firmly establish this type of LPS regulation.
Discussion
Collectively, our results point to the existence of a previously unknown mechanism for the regulation of LPS synthesis in E. coli. More specifically, we suggest that the GTPase ObgE regulates LPS synthesis by directly interacting with LpxA and thereby modulating LpxA’s enzymatic activity under specific conditions. The ObgE amino acid K268 appears to be crucial in this regulatory mechanism since mutating this lysine residue to isoleucine in ObgE* leads to constitutive inhibition of LpxA when ObgE* is bound to GTP.
Our data demonstrate that ObgE directly interacts with LpxA both in vitro and in vivo. This protein-protein interaction is characterized by a KD of 50–100 µM, depending on the nucleotide binding state of ObgE. However, we show that the K268I amino acid substitution in ObgE* strongly impacts this interaction, leading to affinities that are increased more than 1000-fold, but only when ObgE* is bound to GTP. In the presence of other nucleotides (GDP or ppGpp), the affinity of ObgE* for LpxA remains unaltered compared to wt ObgE. The observed changes in the ObgE*-LpxA interaction can explain why ObgE* is highly toxic to E. coli. The tight interaction with GTP-bound ObgE* prevents LpxA catalytic activity and leads to a decrease in LPS synthesis. This LPS deficit triggers the Rcs cell envelope stress response and also causes cell death independently of Rcs activation.
Resistance to ObgE* toxicity can be obtained in a variety of ways. First, decreasing GTP binding strongly increases survival, thereby highlighting the impact of GTP binding on the function and characteristics of ObgE*. Second, resistance can be obtained by overexpressing lpxA, likely because of titration of the toxic ObgE* and liberation of unbound and, therefore, active LpxA proteins. Third, several specific mutations in lpxA can provide resistance. Although we tested the impact of physiological levels of over 5000 LpxA variants carrying all possible single amino acid substitutions, only a very limited number was able to counteract ObgE* toxicity. We believe it is likely that some resistant LpxA variants were missed by our analysis since the sequencing depth provided by PacBio was likely insufficient to pick up all relevant mutations. Nonetheless, it is clear that only a small number of lpxA alleles can rescue cells from ObgE*. Interestingly, all but one of the detected mutations are clustered together in the LpxA protein structure at a location that is predicted to be important in the interaction with ObgE*. We thoroughly characterized three representative LpxA variants and show that they provide resistance to ObgE* by significantly weakening the interaction with the GTP-bound version of this mutant GTPase. As a result, the cell can maintain normal LPS levels and survive, even in the presence of GTP-bound ObgE*. Interestingly, these LpxA mutant proteins also show a decreased binding affinity to wt ObgE, indicating that the ObgE* and ObgE interaction with LpxA is highly similar.
Intriguingly, none of the LpxA variants characterized here provide resistance to ObgE* by completely eliminating the ObgE-LpxA interaction. Rather, they reduce the increased affinity for GTP-bound ObgE* back to levels comparable to the interaction with wt ObgE. Although further investigation is necessary to draw strong conclusions, it is tempting to speculate that completely abolishing the interaction between ObgE and LpxA is not a viable option for E. coli and that regulation of LpxA activity by ObgE is needed to properly control LPS synthesis. Because both up- and downregulation of LPS synthesis are detrimental to the cell37, we believe it is possible that ObgE is needed to prevent overproduction of LPS by limiting LpxA activity under appropriate conditions. However, other explanations can be put forward. For example, it is possible that mutations in lpxA that abolish the interaction with ObgE also directly interfere with substrate binding, catalysis, and/or protein stability.
Although further investigation is necessary to conclusively show if and under which conditions wt ObgE regulates LpxA activity, we here present strong indications for the involvement of this universally conserved GTPase in the regulation of cell envelope synthesis in E. coli. Apart from the direct interaction between ObgE and LpxA, we show that overexpression of wt obgE increases the sensitivity of E. coli to the LPS inhibitor PF-04753299 in an LpxA-dependent manner. In addition, depleting ObgE leads to small increases in cellular LPS levels. Both results hint at a biological role for ObgE in limiting LPS production in vivo. Moreover, our genome-wide screening efforts tightly link wt ObgE to cell envelope synthesis. Indeed, we show that when ObgE is present in excess, cells experience a fitness benefit when downregulating genes involved in the synthesis of fatty acids, peptidoglycan, and LPS. Although these findings strongly implicate ObgE in the regulation of cell envelope synthesis, they are somewhat contrary to our expectations that ObgE, like ObgE*, could block LPS synthesis by inhibiting LpxA activity. On the other hand, the fact that ObgE is linked to the production of phospholipids, peptidoglycan, and LPS is in line with a potential effect on LpxA, which serves as a hub in the synthesis of these three envelope components25,31,45.
Whatever the effect of ObgE on the cell envelope may be, it is likely subtle, transient, and/or condition-dependent, thereby preventing us from detecting large changes in LPS content in standard growth conditions. In addition, when studying the effect of wt ObgE on LpxA activity in vitro, we could not detect any significant changes. We, therefore, suspect that regulation of LpxA by ObgE in vivo is more complex and takes into account additional regulatory input besides only the nucleotide-binding state of ObgE (e.g., additional interaction partners, ligands, etc.). In this regard, it is interesting to note that LpxA was recently found to be anchored to the membrane by LapB in a multi-component network that also contains the LPS biosynthesis enzymes LpxC and LpxD and the phospholipid biosynthesis enzyme FabZ75. This finding indicates that additional input regulating the nature of the interaction between LpxA and ObgE may very well exist. Our CRISPRi screens indeed indicate that the link between ObgE and the cell envelope can only be detected under specific conditions. Clearly, further research will be necessary to confirm if, when, and how ObgE influences cell envelope synthesis. In addition, since ObgE is mostly known to affect ribosome assembly and activity2,3,76, it will be interesting to determine whether both functions of this GTPase are related. If so, ObgE could function in coupling cell expansion to protein synthesis and thereby bridge these two vital aspects of cellular growth.
The reaction catalyzed by LpxA is thermodynamically unfavorable and reversible24,25,31. LpxA activity can be nudged towards the production of lipid A by the next step in the synthesis pathway, which is catalyzed by LpxC24,25. Since LpxC catalyzes the committed step and is subjected to tight regulation37, it is generally assumed that LpxC represents the first point of control in the LPS synthesis pathway. However, it was recently demonstrated that LpxA activity can be altered by RnhB and ppGpp44. We here now show that LpxA activity can also be drastically affected by ObgE* and is likely also altered by wt ObgE under appropriate conditions. Our data, therefore, highlight the cellular potential to regulate LPS synthesis at the level of LpxA prior to the committed step executed by LpxC, and thereby reveal a novel point of control in this biosynthetic pathway that is essential to the vast majority of Gram-negative bacteria.
Methods
Bacterial strains and growth conditions
Experiments were performed with E. coli BW2511377 and derivatives unless mentioned otherwise. obgE and obgE* were expressed from the PBAD promoter of pBAD33Gm49. Whenever single-gene deletion mutants from the Keio collection were used77, the kanamycin resistance cassette was first removed by transformation of pCP2072, which in turn was cured prior to performing further experiments. CRISPRi experiments were performed with E. coli MG1655 dcas9, which was constructed using the integrative pLC143 plasmid78. This plasmid contains an FLP-excisable integration module consisting of an integrase-coding region and a kanamycin-resistance cassette. Upon transformation into E. coli MG1655 ΔaraBAD and growth at 37 °C, the plasmid integrated into the chromosome at the ileY attB site. Afterwards, the integration module was excised by expressing the FLP recombinase from the temperature-sensitive pE-FLP plasmid, by transforming pE-FLP into the strain and culturing at 30 °C. Finally, the pE-FLP plasmid was cured by culturing at 42 °C79. All strains and plasmids used in this study are listed in Supplementary Table S2.
Strains were grown in lysogeny broth (LB) containing the appropriate antibiotics (ampicillin 100 µg/ml, carbenicillin 100 µg/ml, chloramphenicol 35 µg/ml, gentamicin 25 µg/ml, kanamycin 40 µg/ml) and incubated at 37 °C. Liquid cultures were incubated with continuous shaking at 200 rpm. Plates were supplemented with 1.5% agar. Expression from the PBAD promoter was induced with 0.2% w/v arabinose. Expression from Plac promoters was done with 0 mM IPTG (pQE80L and pTargetF_lac_sgRNA, leaky expression), 10 µM IPTG (pMDeg02), 0.5 mM IPTG (pKT25, pKNT25, pUT18, pUT18C), 1 mM IPTG (pET28a) or various IPTG concentrations (pCA24N). Expression from the Ptet promoter was induced with 100 ng/ml aTc.
L-forms were created in an osmoprotective environment provided by pads of LM medium (100 g sucrose, 1 g MgSO4, 18.5 g brain heart infusion, 500 ml dH2O) with 1% agarose80. LM pads were supplemented with 400 µg/ml fosfomycin to trigger the transition into the L-form state by degrading the cell wall and simultaneously stimulating excess membrane synthesis57–59. Also, 25 µg/ml gentamicin was added to LM pads to prevent plasmid loss. Where appropriate, additional supplements were added as indicated in the text. Pads containing L-forms were incubated at 30 °C.
Plasmids generated in this study were constructed in the following ways. pET28a-lpxA was constructed by amplifying the lpxA ORF through PCR with primers BD1&2, using a pBAD-lpxA plasmid as a template. The PCR product was subsequently inserted within the NdeI and EcoRI restriction sites of a pET28a vector. pQE80L-obgE*-mCherry was constructed by restriction with SacI-HF HindIII-HF and T4 ligation. The obgE*-mCherry insert was obtained by PCR amplification with primers SPI10499&10500 from pBAD/His A-obgE*-mCherry. The bacterial-two-hybrid plasmids pKT25-obgE, pKNT25-obgE, pUT18-obgE, and pUT18C-obgE were constructed by restriction with BamHI and KpnI and T4 ligation. The obgE insert was obtained by PCR amplification from the E. coli BW25113 genome using primer pairs P209&210 (for pKT25 and pUT18C) or P211&212 (for pKNT25 and pUT18). The K268I mutation was introduced into pET28a-obgE* by PCR with mismatch primers SPI10930&10931 containing the desired mutation. Similarly, pET28a-lpxAV197H, pET28a-lpxAI199S, pET28a-lpxAR216C, pUT18C-lpxAV197H, pUT18C-lpxAI199S and pUT18C-lpxAR216C were constructed by PCR with mismatch primers starting from either pET28a-lpxA or pUT18C-lpxA. Primer pairs used were P213&214 for lpxAR216C, P215&216 for lpxAI199S, and P217&218 for lpxAV197H. All primer sequences are shown in Supplementary Table S3.
CFU measurements
The numbers of CFUs per ml were measured by preparing serial dilutions in 10 mM MgSO4 that were plated on solid LB medium using the Eddy Jet spiral plater (IUL Instruments). After overnight incubation at 37 °C, colonies were counted, and the number of CFUs per ml was calculated by the Flash and Grow Automatic Colony Counter (IUL Instruments).
For most ObgE* toxicity measurements, overnight cultures were diluted 100 times and grown for 2 h prior to induction of obgE or obgE*. Induction was maintained for 2 h, after which CFUs were determined. Survival was calculated by dividing the number of CFUs per ml obtained upon obgE* expression by those obtained upon overexpression of wt obgE. For Fig. 1A, CFUs per ml were determined at several time points after induction was performed. To assess potentially small differences in growth caused by lpxA mutant alleles, overnight cultures were diluted 1000x, and CFUs were measured at several time points after this dilution step (Supplementary Fig. S3A). To assess the potentially small effects of obgE overexpression on LPS synthesis, overnight cultures were diluted 100x and grown for 2 h before expression of obgE or obgE* was induced with 0.2% arabinose. At this time point, the LPS inhibitor PF-04753299 was or was not added to cultures at a concentration of 1/8x MIC (0.03125 µg/ml for lpxAwt, lpxAV197H, lpxAI199S and 0.0078 µg/ml for lpxAR216C). At several time points after induction, CFUs were measured (Fig. 7D, E).
RNA-sequencing
RNA-sequencing was performed on E. coli ΔrecA carrying plasmids pBAD33Gm, pBAD33Gm-obgE, or pBAD33Gm-obgE*. A ΔrecA mutant strain was initially used to prevent any potential interference of RecA and the SOS response, which we later found do not play a role in toxicity (data not shown). Overnight cultures were diluted 100x, grown for 2 h, and then induced with 0.2% arabinose for 1 h. All growth steps were performed at 37 °C. 20 ml of each culture was added to 4 ml fixation fluid (5:100 phenol in ethanol) and immediately frozen by submersion in liquid nitrogen. Frozen samples were stored at − 80 °C. After thawing on ice, samples were centrifuged (3750 ×g, 30 min, 4 °C), and 5 ml TE buffer with 100 mM NaCl was added to cell pellets.
After vortexing, cells were collected by centrifugation (3750 × g, 5 min, 4 °C), and pellets were dissolved in 1 ml trizol. Cell suspensions were transferred to screw cap microcentrifuge tubes filled with sterile sand and small glass beads. Mechanical lysis was performed by the Precellys®24 (Bertin Corp.) in 2 cycles of 45 sec at 6500 rpm with a 30 sec break, after which cells were incubated for 10 min at room temperature. Interphase-protein contamination was removed by Phase Lock GelTM (QuantaBio, VWR). Spin cartridges of the PureLink® RNA Mini kit (ThermoFisher Scientific) were used for RNA purification, and the remaining DNA was degraded by the TURBO DNA-freeTM kit (ThermoFisher Scientific). The concentration and purity of RNA samples were determined using Nanodrop (ThermoFisher Scientific), Qubit (ThermoFisher Scientific), and ExperionTM RNA StdSens analysis (Bio-Rad). rRNA was depleted, and libraries were prepared using the TruSeq® Stranded Total RNA kit (Illumina) at the EMBL GeneCore (Heidelberg, Germany). The cDNA libraries were multiplexed and subjected to 50-cycle single-end massive parallel sequencing with the Illumina HiSeq2000 (GeneCore, EMBL, Heidelberg). The sequencing data were uploaded to the Galaxy web platform, and we used the public server at galaxy.be for analyses81. Reads were trimmed by Trimmomatic82 using sliding window trimming and a PHRED threshold score of 20. Quality assessment was done with FastQC83. Reads were then mapped on the E. coli K12 MG1655 reference genome (NC_000913.3) using Bowtie284 with standard settings. The quality of mapping was evaluated using Qualimap85. HTSeq-count86 was used to get read counts per gene, after which differential expression was determined by DESeq287 in R. Gene ontology enrichment analyses were performed using the online Gene Ontology Resource platform that is coupled to the PANTHER classification system analysis tool88–90. Genes that were significantly (p < 0.01) up- or downregulated by ObgE* in comparison to both the Vector control and overexpression of wt obgE were used as input. As a reference gene list, all genes that generated non-NA values in the DESeq2 comparisons were used. A PANTHER analysis was performed to identify biological processes that are overrepresented as defined by Fisher’s exact test using FDR-corrected p values. The significance cut-off was set at an adjusted p value < 0.05.
Measurements of expression levels
To determine the PrcsA promoter activity (Figs. 1D, E, and 3E), we made use of the pMS201-PrcsA-gfp plasmid from the E. coli promoter fusion library91. E. coli BW25113 carrying plasmid pBAD33Gm, pBAD33Gm-obgE or pBAD33Gm-obgE* was co-transformed with pMS201-PrcsA-gfp. Resulting colonies were grown overnight, diluted 100x in selective LB medium, grown for 2 h, induced with 0.2% w/v arabinose and incubated for 2 h. Cultures were then diluted 1000x in PBS and measured using a CytoFLEX S instrument (Beckman Colter Life Sciences) equipped with 405 nm, 488 nm, and 561 nm lasers. To generate Figs. 1E and 3E, the population-wide median GFP value was recorded for ≥ 3 independent biological repeats. The mean ± SEM of these median values is shown.
Microscopy experiments and analyses
All microscopy analyses were performed using a Nikon Ti-E inverted microscope equipped with a DS-Qi2 CMOS camera and temperature-controlled cage incubator. The Objective Plan Apo λ 100x oil Ph3 DM with NA 1.45 was used. mCherry was recorded with the mCherry-B filter cube with an exposure time of 100 msec and a gain of 74.1x. GFP was recorded with the GFP-4050B filter cube with an exposure time of 200 msec and gain 13.9x. mTurquoise2ox was recorded with the CFP-2432C filter cube with an exposure time of 100 msec and gain of 20.9x.
For snapshot analyses of walled E. coli cells performed at discrete time points (Supplementary Fig. S3B), cells were spotted onto pads of 10 mM MgSO4 with 2% w/v agarose. Quantitative analysis of cell length was performed using MicrobeJ92.
For analyses of E. coli L-forms (Fig. 2 and Supplementary. Fig.S2), cells from overnight cultures were spotted onto pads of LM medium supplemented with 400 µg/ml fosfomycin, 25 µg/ml gentamicin and 0.2% w/v arabinose. When using cytoplasmic and periplasmic fluorescent markers (Fig. 2E), overnight cultures were first diluted into LB medium with appropriate antibiotics and supplemented with 10 µM IPTG to allow for the expression of the fluorescent markers. After incubating these cultures for 3 h at 37 °C with continuous shaking, cells were spotted on LM pads as described above. In this case, 10 µM IPTG was added to the pads. For time-lapse experiments (Fig. 2A, C, D, E, and Supplementary Fig. S2B), L-forms were grown for 18 h at 30 °C and pictures were taken every 10 min. Quantitative analyses of L-form proliferation were performed manually (Fig. 2B). For a growth period of 10 h, the number of successful L-form divisions per time window of 2 h was counted. A successful L-form division was defined as a fission event where at least one of the progeny L-forms was able to survive for a minimum of 20 min after the division event. For each time window, the number of successful divisions was normalized to the number of intact L-forms present at the start of the time window. For quantitative measurements of ROS production in L-forms (Supplementary Fig. S2A), cells were spotted onto control pads and pads supplemented with the following mix of scavengers: 100 µM MnTBAP, 0.5% v/v DMSO, 10 mM sodium pyruvate. Cells were incubated for 4 h at 30 °C after which GFP fluorescence (expressed from Pdps) was recorded. Using MicrobeJ, the mean GFP fluorescence per L-form (i.e., corrected for cell size) was corrected for background fluorescence. The 2-stage lysis phenotype was quantified manually (Supplementary Fig. S2C). Only cells that successfully transitioned into L-forms, i.e., cells that adopted the characteristic round morphology, were taken into account.
For the analysis and statistical comparison of microscopic properties (i.e., GFP fluorescence (Supplementary Fig. S2A) or cell size (Supplementary Fig. S3B)) across different mutants and/or conditions, the following approach was used93. For every mutant or condition, the relevant property was measured in 3 biologically independent repeats. For every repeat, at least 50 cells were recorded and the median property value of these > 50 cells was determined in each biological repeat. These median values were used to calculate the mean and SEM values that are shown in Supplementary Figs. S2A and S3B. The median values of each repeat were also used to determine any statistically significant differences using ordinary one-way ANOVA with Dunnett’s multiple comparisons test.
Suppressor mutants
Spontaneous suppressor mutants were generated starting from E. coli BW25113 carrying pBAD33Gm-obgE*-venus and pQE80L-obgE*-mCherry. Overnight cultures were plated on a selective medium containing 0.2% w/v arabinose and 0 µM IPTG (for production of obgE*-mCherry from pQE80L, we relied on leaky expression). After overnight incubation, colonies that displayed both yellow and red fluorescence were transferred to new selective LB plates with 0.2% w/v arabinose. Plasmids from strains that were able to grow and that retained fluorescence were purified and transformed into a fresh E. coli BW25113 background. Transformants were first plated on a selective LB medium without arabinose and were then transferred to a selective medium with 0.2% w/v arabinose. Plasmids that, in this fresh E. coli background, still confer toxicity in the presence of arabinose, were isolated from strains that are resistant to ObgE* due to genomic alterations. These strains were selected for further analysis by whole-genome sequencing. Genomic DNA was isolated from selected strains by the DNeasy Blood & Tissue kit (Qiagen), and the concentration and purity of DNA were determined. Libraries were prepared at the EMBL GeneCore (Heidelberg, Germany) using the NEBNext kit with an average insert size of 200 bp. The DNA libraries were multiplexed and subjected to 100-cycle paired-end massively parallel sequencing with the Illumina HiSeq2000 (GeneCore, EMBL, Heidelberg). CLC Genomics Workbench version 7.6 (www.qiagenbioinformatics.com) was used for analysis of the sequences. Following quality assessment of the raw data, reads were trimmed and mapped to the E. coli MG1655 reference genome (NC 000913.1) using the CLC Assembly Cell 4.0 algorithm yielding an average coverage of ~ 150x. To obtain sufficient coverage, some samples were sequenced twice, and both fastq files were used for mapping. Finally, mutations in all samples were detected using the CLC Fixed Ploidy Variant Detector. We focused on variant calling for non-synonymous changes in open reading frames and did not report indels in our supplementary data set.
To identify additional lpxA mutations that confer resistance to ObgE*, we selected suppressor mutations from an lpxA saturation mutagenesis library. This saturation mutagenesis library was constructed as done previously, using high-throughput CRISPR-based editing provided by the Onyx® Digital Genome Engineering platform62. Briefly, repair templates were designed using Inscripta’s Designer software (development version) so that each amino acid would be replaced by every other amino acid and so that every codon would be replaced by a synonymous codon (if a synonymous codon exists). Besides the desired mutation, each oligo may also contain one or more synonymous edits that prevent re-cutting by eliminating the PAM site and/or introducing edits that interfere with cutting. For each mutation present in the repair template, the most frequently used available codon was chosen. The repair template and corresponding sgRNA were cloned in bulk into a high-copy plasmid backbone. Saturation mutagenesis was performed on the Onyx® Digital Genome Engineering Platform, a fully automated instrument that uses the MAD7 nuclease, a type V CRISPR nuclease, to generate multiplexed genome-engineered libraries. 1 ml of an overnight E. coli culture was prepared using the Onyx® E. coli Edit Competency Kit and placed into the Onyx® instrument that performed the genomic editing using the OnyxWare program K-strain v1.1. The resulting pool of edited cells was collected from the instrument and transformed with pBAD33Gm-obgE*. Transformed mutant pools were grown in the presence of 0.2% w/v arabinose for 20 h in triplicate. The lpxA genes present after selection were amplified using forward and reverse primers LD1 and LD2 and subjected to PacBio sequencing. This resulted in ± 18,000 reads per library, which, considering an editing efficiency of 5% and ± 5700 edits, corresponds to ± 0.16 expected reads per mutant. This limiting read depth indicates that low-frequency variants might be missed. However, lpxA alleles that provide resistance against ObgE* are expected to strongly increase in frequency during our selection step, thereby enabling their identification using PacBio sequencing. Those lpxA alleles that were detected with a summed read count of 5 or higher across the three replicates were identified as potential suppressor mutations.
Selected lpxA suppressor mutations (i.e., V197H, I199S, and R216C) were introduced into a fresh E. coli BW25113 background for further analyses. To do so, lpxA mutant alleles were amplified using primers LD3 and LD4 and introduced into E. coli ΔrnhB using CRISPR-FRT94. Successful mutant construction was confirmed by Sanger sequencing of the lpxA gene. Because the lpxAV197H mutant allele was isolated from the lpxA saturation mutagenesis library, also synonymous PAM mutations were transferred (see above).
Spot assays
To determine sensitivities to different compounds (Supplementary Fig. S3D), serial dilutions of overnight cultures were prepared in 10 mM MgSO4. 10 µl of dilutions 10−1 to 10−7 were spotted onto LB agar plates supplemented with 0.05 µg/ml PF-04753299, 25 µg/ml vancomycin or no additional compounds (control). After overnight incubation at 37 °C, pictures were taken.
Bacterial Adenylate Cyclase-Based Two-Hybrid (BACTH) assays
Bacterial two-hybrid assays were performed as described previously75. Expression plasmids pKT25, pKNT25, pUT18, and pUT18C with different gene inserts were transformed to E. coli DHM1 in several different combinations that allow for testing protein-protein interactions in distinctive conformations. Single colonies were dissolved in 50 µl 0.9% NaCl, and 3 µl of cell suspension was spotted onto agar plates of M63 minimal medium supplemented with 0.5 mM IPTG, 40 μg/ml X-Gal, 50 mg/ml ampicillin and 25 mg/ml kanamycin. Plates were incubated at 30 °C for up to 7 days. As a negative control, the combination of empty plasmids pKT25 and pUT18C was tested. As a positive control, plasmids pKT25-zip and pUT18C-zip that together encode the leucine zipper of GCN4 were combined.
Protein purification
The expression and purification of the N-terminally His6-tagged ObgE and ObgE* proteins were performed as previously described95. Briefly, the proteins were expressed from a pET28a vector (Novagen) in E. coli BL21 (DE3) pLysS cells or E. coli BL21(DE3) lpxAR216C pLysS cells in the case of ObgE*. The proteins were purified on a HisTrap FF column (5 ml, Cytiva), after which an alkaline phosphatase treatment was performed to obtain nucleotide-free protein. Subsequently, the alkaline phosphatase was removed through anion exchange using a HiTrap Q HP column (5 ml, Cytiva). Finally, the protein was dialyzed to storage buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2 mM DTT, 5 % glycerol), flash frozen in liquid nitrogen, and stored at − 80 °C.
All LpxA protein constructs were expressed in E. coli BL21 (DE3) pLysS cells in Terrific Broth (TB) medium at 37 °C and 120 rpm. After reaching an OD600nm of 0.6, protein expression was induced by adding 1 mM IPTG, while the cultures continued to grow overnight at 25 °C and 120 rpm. The cells were then collected by centrifugation and resuspended in lysis buffer (20 mM HEPES (pH 7.5), 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol, 5% glycerol) containing 50 µg/ml DNase, 0.1 mg/ml AEBSF and 1 µg/ml leupeptin. After lyzing the cells using a cell disruptor system (Constant Systems) and clearing the lysate through centrifugation, the lysate was applied to a HisTrap FF column (5 ml, Cytiva) equilibrated in buffer A (20 mM HEPES (pH 7.5), 1 M NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol, 5% glycerol). The column was extensively washed with buffer A before eluting the protein with a linear gradient of buffer B (20 mM HEPES (pH 7.5), 300 mM NaCl, 500 mM imidazole, 5 mM β-mercaptoethanol, 5% glycerol). Size exclusion chromatography was performed as a final purification step using a Superdex 200 26/60 column (GE Healthcare) equilibrated in 20 mM HEPES (pH 7.5), 200 mM NaCl, 1 mM DTT, and 5% glycerol. The purified LpxA proteins were flash-frozen in liquid nitrogen and stored at − 80 °C.
Chemical crosslinking and MALDI-TOF MS/MS analysis
ObgE, ObgE*, and LpxA were dialyzed in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 5% glycerol, and 2 mM DTT. Next, 20 µM LpxA was mixed with 40 µM of ObgE or ObgE*, in presence of either 1 mM of GDP or GTPγS and crosslinked with 1 mM of disuccinimidyl suberate (DSS). As a control, a similar crosslinking reaction was performed using each of the proteins individually. After 10 min, the reactions were stopped by adding Tris (pH 7.5) at a final concentration of 50 mM, followed by dialysis to remove the reagents. The samples were loaded on SDS-PAGE, and, after excision, the bands of interest were subjected to in-gel digestion with trypsin and ProteaseMax™ surfactant, both obtained from Promega (Maddison, WI, USA), following the manufacturer’s instructions. The samples were measured two times: once immediately after digestion and once after ZipTIPC18 purification. Matrix Assisted Laser Desorption Ionization – Time of Flight MS and MSMS data were acquired on an Ultraflextreme enhanced MALDI TOF/TOF-MS system (Bruker Daltonics, Bremen, Germany) using FlexControl 3.4 acquisition software (Bruker). In MS mode, spectra were measured in the positive reflector mode within a mass range of 700 to 4000 m/z. Up to 5000 shots were acquired with a laser repetition rate of 2000 Hz and 200 shots per raster spot. All MS spectra were analyzed and processed using FlexAnalysis 3.4 (Bruker). Peaks were detected by means of the Snap peak detection algorithm and a signal-to-noise ratio of at least 3. All spectra were externally calibrated by means of the cubic enhance mode (min 6 points) within a range of 757 to 3147 Da. The acquired peak lists were subjected to a peptide mass fingerprint (PMF) search with BioPharma Compass 4.0.1 (Bruker) and the MASCOT server 2.8.2 (MatrixScience). In the MS-MS mode, spectra were acquired using the LIFT method provided by the manufacturer. Concretely, up to 4000 shots were acquired with a laser repetition rate of 1000 Hz for the fragment ions and 1000 shots at 2000 Hz for the precursor ion. All MSMS spectra were baseline subtracted by means of the TopHat Algorithm and smoothed. Peak finding was performed with the Snap algorithm. Subsequently, MS/MS-spectra acquired from three sample spots (technical replicates) were compiled and subjected to database searches using BioPharma Compass 4.0.1 (Bruker) and the MASCOT server 2.8.2 (MatrixScience).
Microscale thermophoresis (MST)
The ObgE and ObgE* proteins were fluorescently labeled using sulfo-Cyanine5 NHS ester (Lumiprobe), by incubating 1 mg/ml of protein with an 8-fold molar excess of the sulfo-Cyanine5 NHS ester at room temperature in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 5 % glycerol, and 2 mM DTT. After 30 minutes, the reaction was stopped by adding 1 M of Tris (pH 7.5). The labeled protein was separated from the excess of the fluorophore by size exclusion chromatography using a Superdex 200 10/30 column (GE Healthcare) equilibrated in MST buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 mM DTT & 0.1% Tween-20).
All MST measurements were performed using a Monolith NT.115 instrument (NanoTemper Technologies) at 25 °C, Monolith Premium Capillaries (NanoTemper Technologies), and MST buffer supplemented with 0.2% BSA. The LED and Laser power were both fixed at 50%, while the laser on and off times were set at 30 s and 5 s, respectively. To determine the affinity between ObgE and LpxA WT in the presence of different nucleotides, 50 nM of Cy5-labeled ObgE was incubated with a 2:1 serial dilution of LpxA WT starting at a maximum concentration of 250 µM. This was done in the presence of 200 µM GDP, ppGpp, or GTPγS. In the case of ObgE*, the affinity for LpxA WT in the presence of 200 µM GDP and ppGpp was determined by incubating 60 nM of Cy5-labeled ObgE* with a 2:1 serial dilution of LpxA WT starting at 250 µM (LpxA subunit concentration). To determine the affinity in the presence of 200 µM GTPγS, 20 nM of Cy5-labeled ObgE* was incubated with a 1:1 serial dilution of LpxA WT starting at 3 µM (subunit concentration). The affinity of ObgE for the LpxA I199S and LpxA R216C mutants in the presence of GTPγS was determined by incubating 75 nM of Cy5-labeled ObgE with a 2:1 serial dilution of the LpxA mutants starting at 1000 µM (subunit concentration). The affinity of ObgE* for the LpxA I199S and LpxA R216C mutants in the presence of GTPγS was determined by incubating 60 nM of Cy5-labeled ObgE* with a 2:1 serial dilution of LpxA I199S starting at 250 µM (subunit concentration) or a 4:1 serial dilution starting at 500 µM (subunit concentration) in case of LpxA R216C. All measurements were performed in triplicate.
The MST traces were analyzed at the 2.5 s time point after the laser was turned on. The normalized fluorescence values (Fnorm) were plotted against the LpxA subunit concentration and fitted on the quadratic binding curve to obtain a value for the equilibrium dissociation constant (KD).
Multi-angle light scattering (MALS)
For SEC-MALS, a Superdex 200 5/150 GL increase column (Cytiva) was coupled to an HPLC Alliance system (Waters) equipped with a 2998 PDA detector (Waters), a TREOS II MALS detector (Wyatt Technology) and a RI-501 refractive index detector (Shodex). Samples of ObgE* and LpxA separately were prepared at a final concentration of 26 µM. For the LpxA-ObgE* complex, the proteins were pre-mixed at respective (subunit) concentrations of 26 µM and 39 µM and incubated on ice in the presence of 500 µM GTPγS before injection. For each sample, 50 µl was injected, and 20 mM HEPES pH 7.5, 200 mM NaCl, 5 mM MgCl2, 5% glycerol, and 2 mM DTT were used as running buffer at a flow rate of 0.2 ml/min. The Astra 7.3.0 software (Wyatt Technology) was used to analyze the data. A BSA sample (1 mg/ml) was used to normalize and align the signals of the different detectors and to account for any band-broadening effects before further analyzing the other runs. A dn/dc value of 0.1850 ml/g was used.
In vitro LpxA activity measurements
A previously described in vitro fluorescent LpxA activity assay was performed to measure enzymatic activity in the presence of ObgE, ObgE* and different nucleotides44,69. To do so, several substrates were synthesized in house as described previously and detailed below44.
AasSHis6 production and purification
To obtain Vibrio harveyi AasSHis6, E. coli TOP10 cells were transformed with pBO4874 (pBAD24 + aasSHis6). An overnight culture was used to inoculate the main culture (1 l LB + ampicillin) at an OD600 of 0.05. Cells were cultivated at 37 °C until they reached an OD600 of 0.6–0.8 and protein overproduction was induced by adding 0.2 % (w/v) of L-arabinose. Cultivation continued at 30 °C for 4 h. Cells were harvested and stored as described above.
Cell pellets were resuspended in 10 ml lysis buffer consisting of 20 mM HEPES/NaOH (pH 8.0), 500 mM NaCl, 10% (v/v) glycerol, 10 mM imidazole, and 1 x cOmplete™ EDTA-free, with the addition of DNase I, RNase A, and lysozyme (0.1 mg/ml). The cells were lysed using a French press. The lysate was centrifuged (10 min, 16000 × g, 4 °C), and the supernatant was passed through a Ni-NTA agarose column (Bio-Rad) after equilibration with lysis buffer (without enzymes). Three wash steps with 10 column volumes (CVs) each, with wash buffers I-III, were performed to remove unspecifically bound proteins (20 mM HEPES/NaOH pH 8.0, 500/300/150 mM NaCl (I/II/III), 10% (v/v) glycerol, 50 mM imidazole). AasSHis6 was eluted using wash buffer III supplemented with 250 mM imidazole. The buffer was exchanged by a PD-10 column to store the protein in 20 mM Tris-HCl pH 7.5, 10% (v/v) glycerol, 1 mM EDTA, 0.1 mM TCEP, and 0.002% (v/v) Triton X-10096. Protein aliquots were stored at − 80 °C. Protein concentration was determined with the Roti®-Quant reagent (Carl Roth) and BSA as a standard protein.
Holo-ACP production, purification, and acylation
E. coli BL21(DE3) was transformed with pBO4886 (pACYCDuet-1 + acpS-his6 + acpP). An overnight culture was grown in an LB medium supplemented with chloramphenicol and used to inoculate a 1 l main culture to an initial optical density (OD600) of 0.05. The bacteria were allowed to grow at 37 °C until they reached an OD600 of 0.6–0.8. Protein overproduction was induced by adding 1 mM IPTG. The temperature was shifted to 18 °C, and the culture was incubated overnight. Afterward, the cells were sedimented by centrifugation (10 min, 4000 × g, 4 °C), washed in 20 mM HEPES/NaOH pH 8.0, and cell pellets were stored at − 20 °C.
Cell pellets were resuspended in 10 ml of buffer A (20 mM HEPES/NaOH pH 8.0, 1 mM TCEP pH 7.0), supplemented with lysozyme, DNase I, and RNase A at a concentration of 0.1 mg/ml each. Following cell lysis via French press, the clarified lysate was applied to a Ni-NTA gravity-flow column (Bio-Rad) pre-equilibrated with buffer A. The resin was washed twice with five CVs of buffer A. AcpSHis6 was eluted through an imidazole gradient (10–1000 mM). The elution fractions containing imidazole concentrations of 50–1000 mM were combined, and an equal volume of isopropanol was slowly added with gentle stirring at 4 °C. After 1 h at 4 °C, aggregated proteins were removed by centrifugation (30 min, 16000 × g, 4 °C), and buffer A was added in equal volume.
AcpSHis6 and holo-ACP were separated using a HiScreen™ QHP column (4.7 ml CV; Cytiva) that was equilibrated with buffer A. The supernatant from the previous step was applied using an AEKTA sample pump. Following a five-CV wash with buffer A, an 80 ml linear gradient of NaCl (0–500 mM) in buffer A was employed to elute holo-ACP. Holo-ACP was detected at ~ 300 mM NaCl, and fractions were identified using SDS-PAGE, Coomassie, and SYPRO™ Orange staining (Sigma). Holo-ACP was only visible in SYPRO™ Orange-stained SDS-PAGE, while AcpSHis6 was only visible with Coomassie staining. The fractions containing holo-ACP were combined, subjected to buffer exchange, and concentrated in 20 mM HEPES/NaOH (pH 8.0) utilizing an Amicon® with a 3-kDa cutoff. Protein concentration was quantified via absorbance at 280 nm (A280).
Holo-ACP was reduced by adding two molar equivalents of TCEP (pH 7.0) and incubation for 1 h at 21 °C. The acylation of holo-ACP was conducted in a 10 ml reaction mixture comprising 70 µM reduced holo-ACP, 100 mM Tris/HCl (pH 7.5), 5 mM ATP, 5 mM MgCl2, 100 µM TCEP (pH 7.0), 0.01% (v/v) Triton X-100, 100 µg AasSHis6, and 300 µM R-3-hydroxymyristic acid. The reaction proceeded for 45 min at 30 °C, followed by adding 50 µg of AasSHis6 and an additional 20-min incubation at 30 °C. The reaction mixture was then cooled and directly applied to a HiScreen™ QHP column (4.7 ml CV; Cytiva) pre-equilibrated with 20 mM HEPES/NaOH (pH 8.0). After a three-CV wash, an 80 ml linear NaCl gradient (0–500 mM) was employed to elute acyl-ACP. Acylation was confirmed by SDS-PAGE and SYPRO™ Orange staining. Acyl-ACP exhibited faster migration through the gel matrix.
Due to the inability to separate holo- and acyl-ACP, the fractions containing acyl-ACP were pooled, subjected to buffer exchange, and concentrated in 20 mM HEPES/NaOH (pH 7.0) using an Amicon® with a 3-kDa cutoff. An excess (~ 20 mg) of N-ethylmaleimide (NEM; Sigma) was added to block free thiol groups from remaining holo-ACP. Following a 2-hour incubation at 21 °C and 16 h at 4 °C, NEM was removed through buffer exchange in 20 mM HEPES/NaOH (pH 7.0). The NEM-blocked acyl-ACP preparation was then buffer exchanged and concentrated in 20 mM HEPES/NaOH (pH 8.0). Visualization and quantification of acyl-ACP were achieved via SDS-PAGE, SYPRO™ Orange staining, and densitometric analysis using the Image QuantTM software (Bio-Rad). Protein concentration was determined using A280 measurements. The acyl-ACP preparation of batch 1, stored at − 20 °C, had a total concentration of 35.8 µM, with acyl-ACP constituting 33.9% or 12.1 µM. Batch 2 had a total concentration of 38.5 µM, with acyl-ACP constituting 7.8% or 3 µM.
Fluorescent assay for LpxA activity
For each reaction, 20 µl of the NEM-blocked acyl-ACP preparation was mixed with 20 µl of 20 mM UDP-GlcNAc, 30 µl of 33.33 µM ThioGlo® and 10 µl 20 mM HEPES/NaOH [pH 8.0] into the wells of a black 96-well half-area plate (Corning). All compounds were prepared in 20 mM HEPES/NaOH [pH 8.0]. The plate was equilibrated in a Tecan Infinite® M Plex in the dark at 25 °C for five minutes prior to adding LpxA, LpxA mutant proteins, ObgE, ObgE*, and/or different nucleotides with a total volume of 20 µl. The reaction was mixed by pipetting up and down, and the fluorescence intensity was monitored for ten minutes at λex = 379 nm and λem = 513 nm at 10-sec intervals. When working with batch 1 of synthesized substrates and purified proteins, LpxA and LpxA mutant proteins were always added at a trimeric concentration of 10 nM. ObgE and ObgE* were added at a fixed concentration of 125 nM. When working with batch 2 of synthesized substrates and purified proteins, increased background signals forced us to lower the protein and nucleotide concentrations added to the reactions. In this case, reactions were performed with a trimeric LpxA or LpxA mutant concentration of 3 nM and an ObgE or ObgE* concentration of 7 nM. To test the effect of ObgE or ObgE* on LpxA or LpxA variants, both proteins were pre-incubated together at a 5x concentration for 30 min at room temperature on a shaking platform. If needed, nucleotides (GTP, GDP, ppGpp) were added during the pre-incubation step at a concentration of 800 µM (batch 1) or 40 µM (batch 2).
LpxA activity was determined as the initial reaction rate, represented by the slope of the linear regression fitted onto the linear range of the measurement curve. To account for differences in signal intensities between different experiments, all values were normalized to the wt LpxA control (no ObgE(*), no nucleotides) that was included in each experiment.
Determining cellular protein concentrations
To allow for accurate comparisons of cellular LPS levels and fatty acid concentrations between different samples, these measurements were normalized to total cellular protein concentrations. To determine the latter, 1 ml culture volume was pelleted by centrifugation (20,000 × g, 5 min). Pellets were dissolved in 100 µl dH2O and incubated at 95 °C for 10 min with continuous shaking. Next, protein concentrations were determined using the QubitTM protein assay kit (ThermoFisher Scientific) following the manufacturer’s instructions.
Quantification of LPS
Cellular LPS was quantified using the Pro-Q Emerald 300 Lipopolysaccharide Gel Stain Kit (ThermoFisher Scientific) as described previously97. For this protocol, approximately 5 × 108 cells were collected. To do so, overnight cultures were diluted 100x and either grown for 4 h (Supplementary Fig. S3C) or grown for 2 h and again 2 h after adding 0.2% w/v arabinose (Fig. 6D and Supplementary Fig. S8A) or grown for 2 h and 4 more hours after adding 100 ng/ml aTc (Fig. 7F). Cells from 2 ml of culture volume were collected by centrifuging (20,000 × g, 5 min) and resuspended in 100 µl of 1x NuPAGETM LDS sample buffer (ThermoFisher Scientific) with 4% v/v β-mercaptoethanol. Samples were incubated for 10 min at 95 °C with continuous shaking. After cooling down, they were then treated with 125 µg/ml proteinase K at 55 °C overnight (16 to 20 h). Proteinase K was inactivated at 100 °C for 5 min, and samples were loaded onto a 4–12% bis-Tris NuPAGETM gradient gel (ThermoFisher Scientific) and run with MES buffer. The loaded sample volume was normalized to total protein content so that an equivalent of 270 µg of protein was loaded for each sample. The gel was fixed, washed, oxidized, and stained according to the manufacturer’s instructions and visualized by UV transillumination with the E-box Gel Documentation Imager (Vilber). Two bands were visualized per sample. Based on personal correspondence with a number of research groups that are experts in this assay, we quantified signal intensities of the top band that corresponds to the LPS species of the E. coli lab strain used here. Signal intensities were quantified using ImageJ98. In case not all repeats fitted onto the same gel, signals were normalized to a control signal present on the same gel to account for differences in staining intensities (Fig. 6D, Supplementary Fig. S8A, and 7F, control signal = E. coli wt pBAD33Gm or E. coli dcas9 pTargetF_lac_sgRNA).
Quantification of peptidoglycan synthesis
To determine the rate of peptidoglycan synthesis, incorporation of the radioactive marker (DL + meso)-2,6-Diaminopimelic acid, [2,6-3H] (3H-DAP) was measured. Overnight cultures of E. coli BW25113 carrying plasmid pBAD33Gm, pBAD33Gm-obgE, or pBAD33Gm-obgE* were diluted 100x and grown for 2 h. Then, cultures were induced with 0.2% w/v arabinose, and 1 µl of 1 mCi/ml 3H-DAP was added. 2 h later, 100 µl of each culture was added to 3.5 ml ice-cold 10% TCA, and precipitates were collected under vacuum on 25 mm glass microfiber filters (Whatman® Grade GF/C). Filters were washed twice with 4 ml ice-cold dH2O and added to 3.5 ml scintillation liquid (Ultima-Flo M, Perkin Elmer). The incorporation of the radiolabel was assessed using a Hidex 300SL scintillation counter, and resulting Counts Per Minute (CPM) were used to evaluate the incorporation of radiolabeled 3H-DAP.
Quantification of fatty acids
To determine the fatty acid composition, overnight cultures were diluted 100x and grown for 2 h. At this point, the inducer 0.2% w/v arabinose was added, and cultures were incubated for 2 more hours. 10 ml of each culture was spun down (3750 g, 20 min, 4 °C), and pellets were dissolved in 1 ml dH2O. This cell suspension was used to extract the fatty acids and derivate them to fatty acid methyl esters. The protocol was obtained and adapted from literature99. First, the samples were spiked with 10 µL of a pentadecanoic fatty acid standard (15.6 mg/mL). Next, 50 µL of acetic acid was added, followed by 2 mL of a 1:1 chloroform:methanol mixture. The suspension was left at room temperature overnight. The lower organic phase was taken and dried using a continuous nitrogen gas flow. The dried samples were re-suspended in 2 mL of 5 % H2SO4 in methanol and incubated for 3 h at 80 °C. Next, 500 µL of a 0.9 % NaCl solution and 500 µL hexane were added. After vortexing, the upper phase was collected for GC analysis. 1 µL of the sample was measured for fatty acid methyl esters and injected into a 59:1 split injection system of a Hewlett Packard HP6890 gas chromatograph, equipped with a CP-Sil 88 column (100 m length, 0.25 mm diameter, and 0.2 µm film thickness). Based on retention time, peaks were matched to specific fatty acids. Peak areas were converted to concentrations based on the C15 standard. Furthermore, fatty acid quantities were normalized to protein concentrations measured in each sample.
Alphafold-multimer modeling
The modeling of the ObgE*-LpxA and Obg-LpxA complexes was performed with AlphaFold-multimer v3 running on a local server. The sequences for all Obg and LpxA orthologues were retrieved from the UniProt database website, and the inputs for both complexes were generated by the AlphaFold-Multimer pipeline using default parameters64,65. Nine cycles of Amber relaxation were used on the top 5 models, ranked based on the iptm + ptm scores. The top-ranked predictions, along with the pLDDT scores and PAE plots, are deposited on Zenodo as record 13902471. Visualization of the three-dimensional structures and analysis was done using pymol (v 2.5.2).
CRISPRi depletions
Genome-wide CRISPRi screens were performed using previously validated pooled E. coli sgRNA plasmid libraries 1-474. Libraries were transformed into E. coli MG1655 dcas9 carrying pBAD33Gm or pBAD33Gm-obgE by electroporation. Briefly, mid-exponential phase cultures were pelleted by centrifugation (10 min, 4.750 rpm), and pellets were washed four times in chilled 10% glycerol. After the final wash steps, cultures were concentrated 100x in 10% glycerol, and an aliquot of 100 µl was electroporated with 50 ng of the pooled sgRNA libraries. After electroporation, cells were allowed to recover in 1 ml SOC medium for 45 min and were then transferred to 100 ml selective LB medium. After overnight growth, libraries were stored at − 80 °C for later use in CRISPRi experiments. Coverage of the libraries was determined right after recovery in SOC medium and again before storage at – 80 °C. This coverage exceeded our pre-set threshold of 100x, which was also maintained as a lower limit for all experimental steps.
For the exponential phase CRISPRi screen, libraries were grown overnight in a selective LB medium and diluted 100x into fresh medium in 2-fold. After 2 h of growth, 100 ng/ml aTc (the inducer of dcas9 expression) was added to one of the duplicate culture tubes, while the CRISPRi system was not activated in the second duplicate. 30 min later, 0.2% arabinose was added to all tubes (both with and without aTc) to induce obgE expression. Cultures were grown for 6 hours before sampling. In the stationary phase screen, libraries were grown overnight in a selective LB medium and diluted 100x into fresh medium in 2-fold. After 8 h of growth, 100 ng/ml aTc was added to one of the duplicate culture tubes and 0.2% arabinose was added to all tubes. Cultures were further incubated overnight for 16 hrs. Next, cultures were diluted 100x into fresh selective LB medium with 0.2% arabinose but without aTc. After 8 hours of growth, cultures were sampled.
Sampling was done by centrifuging 1 ml of culture for 1 min at 13.000 × g. Pellets were stored at − 20 °C prior to isolating the sgRNA-encoding plasmids using the NucleoSpin Plasmid EasyPure kit (Machery-Nagel). Plasmid concentrations were measured by Qubit and adjusted to 0.5 ng/µl to serve as input for a first PCR reaction that amplifies the sgRNA N20 region and introduces i5 and i7 adapters. PCR products of appropriate size (149 bp) were selected with the Mag-Bind TotalPurge NGS kit (Omega Bio-Tek), their concentration was adjusted to 1 ng/µl and they were used as input for a second PCR reaction in which sample-specific indices were introduced. Again, PCR products of the correct size (218 bp) were selected with the Mag-Bind TotalPurge NGS kit, and their size was confirmed using the QIAxcel fragment analyzer. Samples were pooled at a concentration of 20 nM. This pool was sequenced using an Element Bio AVITI instrument (2×75 cloudbreak high sequencing kit single-end reads) with a custom sequencing primer (CGACGCTCTTCCGATCTGGATAACAAGATACTGAGCAC) and 2% PhiX DNA. After demultiplexing, raw read counts were extracted and used to detect significantly enriched or depleted sgRNAs by DESeq287. Our DESeq2 analysis made use of the interaction term (strain:inducer) and was therefore designed to compare the changes that occur with and without aTc in our vector sample to the changes that occur with and without aTc upon obgE overexpression. As output, a shrunken log2 fold change was calculated, and a Wald test was performed to obtain p-values. FDR-adjusted p-values were calculated using the Benjamini-Hochberg correction for multiple hypothesis testing. Next, to obtain results at the gene level, the α Robust Rank Aggregation (α-RRA) method from the MAGeCK pipeline was applied in R100. Here, a gene-level average log2 fold change was calculated based on the shrunken log2 fold change values of significantly enriched or depleted sgRNAs (FDR-adjusted p-value < 0.05). For each gene, this calculation was done twice: once for sgRNAs with a positive log2 fold change and once for sgRNAs with a negative log2 fold change. Another Wald test was applied to obtain gene-level p-values that were once again FDR-adjusted for multiple testing. This results in two log2 fold changes and corresponding p-values per gene, one for enriched sgRNAs and one for depleted sgRNAs. Finally, significant hit genes were used as input for an enrichment analysis based on KEGG pathways which was performed with the STRINGdb R package101,102.
For individual gene depletions (Fig. 7F), overnight cultures were diluted 100x, grown for 2 h, and then induced with 100 ng/ml aTc. 4 h after induction, cultures were sampled.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We are thankful to Maarten Fauvart and Natalie Verstraeten for the valuable discussions we had surrounding this project. We also thank Steven Janvier for performing and interpreting the MALDI-TOF/TOF analysis, Aaron N. Brooks and Nandini Krishnamurthy from Inscripta, Inc., for their assistance in the construction and analysis of the lpxA saturation mutagenesis library and the VIB nucleomics core for their efforts in next-generation sequencing of our samples. We also thank the labs of David Bikard and Jean-François Collet for kindly sharing useful plasmids and Wieland Steinchen and Gert Bange (University Marburg) for providing ppGpp. All schematics were drawn with Inkscape, except for the DNA plasmid icon shown in Fig. 3A, B, which was created by Tim Madle and made available through the Noun Project (Creative Commons Attribution License CC BY 3.0). This research was supported by projects from the Research Foundation Flanders (FWO) G0B0420N, G0C4322N, EOS project 40007496, KU Leuven project C16/17/006, VUB Strategic Research Program SRP50 and SRP95 and VIB. In addition, FWO PhD fellowships were awarded to B.D. (11D4621N) and E.L. (1S88319N). L.D. is currently supported by the F.R.S.-FNRS (Research Associate) and previously received an FWO junior postdoctoral fellowship (12Q4319N). F.N. was supported by the German Research Foundation (DFG; Research Training Group GRK2341 “Microbial Substrate Conversion (MiCon)” and Priority Program SPP1879 “Nucleotide second messenger signaling in bacteria”).
Author contributions
Conceptualization: L.D., B.D., W.V., and J.M.; Data analysis: L.D., B.D., I.M.M., S.C.V., and W.V.; Investigation: L.D., B.D., I.M.M., E.L., S.V., P.M., S.B., and A.M.M.; Supervision: L.D., F.N., W.V., and J.M.; Writing – original draft: L.D., B.D., and W.V.; Writing – Review & Editing: L.D., B.D., W.V., and J.M.; Visualization: L.D., B.D., and W.V.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All raw data included in this manuscript are available and stored in the following repositories. The sequencing data generated in this study have been deposited in the Sequence Read Archive (SRA) with BioProject accession number PRJNA1061367, and microscopy images and videos have been deposited in the EMBL-EBI BioImages Archive with accession number S-BIAD1002. The output of AlphaFold-Multimer and all other data presented here (including raw gel images and MS data) are deposited on Zenodo as record 13902471 with 10.5281/zenodo.13902471.
Code availability
Code for analysis of our CRISPRi screening data has been deposited on Zenodo as record 13902656 with 10.5281/zenodo.13902656.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wim Versées, Jan Michiels.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53980-1.
References
- 1.Verstraeten, N., Fauvart, M., Versées, W. & Michiels, J. The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev.75, 507–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng, B. et al. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol.12, e1001866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang, M. et al. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol.188, 6757–6770 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin, B., Thayer, D. A., Maddock, J. R. & Al, L. I. N. E. T. The caulobacter crescentus CgtA C protein cosediments with the free 50S ribosomal subunit. J. Bacteriol.186, 481–489 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott, J. M., Ju, J., Mitchell, T. & Haldenwang, W. G. The bacillus subtilis GTP binding protein Obg and regulators of the sigma B stress response transcription factor cofractionate with ribosomes. J. Bacteriol.182, 2771–2777 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan, J., Jakob, U., Bardwell, J. C. A. & Court, D. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol.184, 2692–2698 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buglino, J., Shen, V., Hakimian, P. & Lima, C. D. Structural and biochemical analysis of the Obg GTP binding protein. Structure10, 1581–1592 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Gkekas, S. et al. Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence. J. Biol. Chem.292, jbc.M116.761809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, B., Covalle, K. L., Maddock, J. R. & Lin, B. I. N. The caulobacter crescentus CgtA protein displays unusual guanine nucleotide binding and exchange properties. J. Bacteriol.181, 5825–5832 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta, K., Skidmore, J. M., Pu, K. & Maddock, J. R. The caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels. Mol. Microbiol.54, 1379–1392 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Dutkiewicz, R., Slomińska, M., Wegrzyn, G. & Czyz, A. Overexpression of the cgtA (yhbZ, obgE) gene, coding for an essential GTP-binding protein, impairs the regulation of chromosomal functions in Escherichia coli. Curr. Microbiol.45, 440–445 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Foti, J. J., Schienda, J., Sutera, V. A. & Lovett, S. T. A bacterial G protein-mediated response to replication arrest. Mol. Cell17, 549–560 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Sikora, A. E., Zielke, R., Datta, K. & Maddock, J. R. The vibrio harveyi GTPase CgtA V is essential and is associated with the 50S ribosomal subunit. J. Bacteriol.188, 1205–1210 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulanowska, K., Sikora, A., Węgrzyn, G. & Czyż, A. Role of the cgtA gene function in DNA replication of extrachromosomal elements in Escherichia coli. Plasmid50, 45–52 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Foti, J. J., Persky, N. S., Ferullo, D. J. & Lovett, S. T. Chromosome segregation control by Escherichia coli ObgE GTPase. Mol. Microbiol.65, 569–581 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Persky, N. S., Ferullo, D. J., Cooper, D. L., Moore, H. R. & Lovett, S. T. The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol.73, 253–266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wout, P. et al. The Escherichia coli GTPase CgtA E cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase / hydrolase. J. Bacteriol.186, 5249–5257 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verstraeten, N. et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell59, 9–21 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kok, J. A. N., Trach, K. A. & Hoch, J. A. Effects on bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. J. Bacteriol.642, 7155–7160 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powers, M. J., Simpson, B. W. & Stephen Trent, M. The mla pathway in acinetobacter baumannii has no demonstrable role in anterograde lipid transport. ELife9, 1–21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deghelt, M. et al. The outer membrane and peptidoglycan layer form a single mechanical device balancing turgor. Preprint at 10.1101/2023.04.29.538579 (2023).
- 22.Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev.67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojas, E. R. et al. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature559, 617–621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitfield, C. & Trent, M. S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem.83, 99–128 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Bertani, B. & Ruiz, N. Function and biogenesis of lipopolysaccharides. EcoSal Plus8, ESP-0001-2018 (2018). [DOI] [PMC free article] [PubMed]
- 26.Whitfield, C., Williams, D. M. & Kelly, S. D. Lipopolysaccharide O-antigens—bacterial glycans made to measure. J. Biol. Chem.295, 10593–10609 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, D. & Reeves, P. R. Escherichia coli K12 regains its O antigen. Microbiology140, 49–57 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Qin, J., Hong, Y., Morona, R. & Totsika, M. O antigen biogenesis sensitises Escherichia coli K-12 to bile salts, providing a plausible explanation for its evolutionary loss. PLOS Genet.19, e1010996 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brabetz, W., Muller-Loennies, S., Holst, O. & Brade, H. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia Coli K-12 results in an re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur. J. Biochem.247, 716–724 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Klein, G., Lindner, B., Brabetz, W., Brade, H. & Raina, S. Escherichia coli K-12 suppressor-free mutants lacking early glycosyltransferases and late acyltransferases: Minimal lipopolysaccharide structure and induction of envelope stress response. J. Biol. Chem.284, 15369–15389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson, M. S. et al. UDP-N-acetylglucosamine acyltransferase of Escherichia coli: The first step of endotoxin biosynthesis is thermodynamically unfavorable. J. Biol. Chem.268, 19858–19865 (1993). [PubMed] [Google Scholar]
- 32.Führer, F. et al. Sequence and length recognition of the C-terminal turnover element of LpxC, a soluble substrate of the membrane-bound FtsH protease. J. Mol. Biol.372, 485–496 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Ogura, T. et al. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol.31, 833–844 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Clairfeuille, T. et al. Structure of the essential inner membrane lipopolysaccharide–PbgA complex. Nature584, 479 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Emiola, A., George, J. & Andrews, S. S. A complete pathway model for lipid a biosynthesis in Escherichia coli. PLoS ONE10, 1–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fivenson, E. M. & Bernhardt, T. G. An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in escherichia coli. mBio11, e00939–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guest, R. L., Rutherford, S. T. & Silhavy, T. J. Border control: Regulating LPS biogenesis. Trends Microbiol.29, 334–345 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guest, R. L., Guerra, D. S., Wissler, M., Grimm, J. & Silhavy, T. J. YejM modulates activity of the YciM/FtsH protease complex to prevent lethal accumulation of lipopolysaccharide. mBio11, e00598–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein, G., Kobylak, N., Lindner, B., Stupak, A. & Raina, S. Assembly of lipopolysaccharide in escherichia coli requires the essential LapB heat shock protein. J. Biol. Chem.289, 14829–14853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahalakshmi, S., Sunayana, M. R., Saisree, L. & Reddy, M. YciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol. Microbiol.91, 145–157 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Nguyen, D., Kelly, K., Qiu, N. & Misra, R. YejM Controls LpxC levels by regulating protease activity of the FtsH/YciM complex of Escherichia coli. J. Bacteriol.202, e00303–e00320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schäkermann, M., Langklotz, S. & Narberhaus, F. FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J. Bacteriol.195, 1912–1919 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shu, S. & Mi, W. Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli. Nat. Commun.13, 4576 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brückner, S. et al. (p)ppGpp and moonlighting RNases influence the first step of lipopolysaccharide biosynthesis in Escherichia coli. microLife4, uqad031 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trent, M. S. Biosynthesis, transport, and modification of lipid A. Biochem. Cell. Biol. Biochim. Biol. Cell.82, 71–86 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Theuretzbacher, U., Blasco, B., Duffey, M. & Piddock, L. J. V. Unrealized targets in the discovery of antibiotics for Gram-negative bacterial infections. Nat. Rev. Drug Discov.22, 957–975 (2023). [DOI] [PubMed] [Google Scholar]
- 47.Erwin, A. L. Antibacterial drug discovery targeting the lipopolysaccharide biosynthetic enzyme LpxC. Cold Spring Harb. Perspect. Med.6, a025304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalinin, D. V. & Holl, R. LpxC inhibitors: a patent review (2010-2016). Expert Opin. Ther. Pat.27, 1227–1250 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Dewachter, L. et al. A mutant isoform of ObgE causes cell death by interfering with cell division. Front. Microbiol.8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Limoli, D. H., Jones, C. J. & Wozniak, D. J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 3, MB-0011-2014 (2015). [DOI] [PMC free article] [PubMed]
- 51.Mitchell, A. M. & Silhavy, T. J. Envelope stress responses: balancing damage repair and toxicity. Nat. Rev. Microbiol.17, 417–428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wall, E., Majdalani, N. & Gottesman, S. The complex Rcs regulatory cascade. Annu. Rev. Microbiol.72, 111–139 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Cho, S. H. et al. Detecting envelope stress by monitoring β-barrel assembly. Cell159, 1652–1664 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Patel, K. B. et al. Functional characterization of UDP-glucose:Undecaprenyl-Phosphate Glucose-1-phosphate transferases of Escherichia coli and caulobacter crescentus. J. Bacteriol.194, 2646–2657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott, P. M., Erickson, K. M. & Troutman, J. M. Identification of the functional roles of six key proteins in the biosynthesis of Enterobacteriaceae colanic acid. Biochemistry58, 1818–1830 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawai, Y. et al. Cell growth of wall-free L-form bacteria is limited by oxidative damage. Curr. Biol.25, 1613–1618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mercier, R., Kawai, Y. & Errington, J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife3, e04629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mickiewicz, K. M. et al. Possible role of L-form switching in recurrent urinary tract infection. Nat. Commun.10, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mercier, R., Kawai, Y. & Errington, J. Excess membrane synthesis drives a primitive mode of cell proliferation. Cell152, 997–1007 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Dewachter, L., Herpels, P., Verstraeten, N., Fauvart, M. & Michiels, J. Reactive oxygen species do not contribute to ObgE*-mediated programmed cell death. Sci. Rep.6, 33723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meiresonne, N. Y. et al. Superfolder mTurquoise2ox optimized for the bacterial periplasm allows high efficiency in vivo FRET of cell division antibiotic targets. Mol. Microbiol.111, 1025–1038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dewachter, L. et al. Deep mutational scanning of essential bacterial proteins can guide antibiotic development. Nat. Commun.14, 241 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karimova, G., Gauliard, E., Davi, M., Ouellette, S. P. & Ladant, D. Protein–protein interaction: Bacterial two-hybrid. in Bacterial Protein Secretion Systems: Methods and Protocols (eds. Journet, L. & Cascales, E.) 159–176 (Springer, New York, NY, 2017). [DOI] [PubMed]
- 64.Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at 10.1101/2021.10.04.463034 (2022).
- 65.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raetz, C. R. & Roderick, S. L. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science270, 997–1000 (1995). [DOI] [PubMed] [Google Scholar]
- 67.Smith, E. W. et al. Structures of Pseudomonas aeruginosa LpxA reveal the basis for its substrate selectivity. Biochemistry54, 5937–5948 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Williams, A. H. & Raetz, C. R. H. Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase. Proc. Natl. Acad. Sci. USA104, 13543–13550 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jenkins, R. J. & Dotson, G. D. A continuous fluorescent enzyme assay for early steps of lipid A biosynthesis. Anal. Biochem.425, 21–27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dewachter, L. et al. GTP Binding is necessary for the activation of a toxic mutant isoform of the essential GTPase ObgE. Int. J. Mol. Sci.21, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verstraeten, N. et al. Biochemical determinants of ObgE‐mediated persistence. Mol. Microbiol.112, 1593–1608 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarkar, S., Ulett, G. C., Totsika, M., Phan, M.-D. & Schembri, M. A. Role of capsule and O antigen in the virulence of uropathogenic Escherichia coli. PloS ONE9, e94786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, T. et al. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat. Commun.9, 2475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Möller, A.-M. et al. LapB (YciM) orchestrates protein–protein interactions at the interface of lipopolysaccharide and phospholipid biosynthesis. Mol. Microbiol.119, 29–43 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Sato, A. et al. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells10, 393–408 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol.2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui, L. et al. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun.9, 1912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.St-Pierre, F. et al. One-step cloning and chromosomal integration of DNA. ACS Synth. Biol.2, 537–541 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Davison, F., Chapman, J. & Mickiewicz, K. Isolation of L-form bacteria from urine using filtration method. J. Vis. Exp. 160, e61380 (2020). [DOI] [PubMed]
- 81.The galaxy community. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 50, W345–W351 (2022). [DOI] [PMC free article] [PubMed]
- 82.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. (2010).
- 84.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.García-Alcalde, F. et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics28, 2678–2679 (2012). [DOI] [PubMed] [Google Scholar]
- 86.Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet.25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed]
- 90.Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res.47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zaslaver, A. et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods3, 623–628 (2006). [DOI] [PubMed] [Google Scholar]
- 92.Ducret, A., Quardokus, E. & Brun, Y. MicrobeJ, a high throughput tool for quantitative bacterial cell detection and analysis. Nat. Microbiol.1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lord, S. J., Velle, K. B., Mullins, R. D. & Fritz-Laylin, L. K. SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol.219, e202001064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swings, T. et al. CRISPR-FRT: Targeting shared sites in a knock-out collection for off-the-shelf genome editing. Nat. Commun.9, 2331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deckers, B. et al. YbiB: a novel interactor of the GTPase ObgE. Nucleic Acids Res.51, gkad127 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang, Y., Chan, C. H. & Cronan, J. E. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry45, 10008–10019 (2006). [DOI] [PubMed] [Google Scholar]
- 97.Sutterlin, H. A. et al. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. USA113, E1565–E1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Voelker, T. A. & Davies, H. M. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J. Bacteriol.176, 7320–7327 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol.15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res.51, D638–D646 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res.47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All raw data included in this manuscript are available and stored in the following repositories. The sequencing data generated in this study have been deposited in the Sequence Read Archive (SRA) with BioProject accession number PRJNA1061367, and microscopy images and videos have been deposited in the EMBL-EBI BioImages Archive with accession number S-BIAD1002. The output of AlphaFold-Multimer and all other data presented here (including raw gel images and MS data) are deposited on Zenodo as record 13902471 with 10.5281/zenodo.13902471.
Code for analysis of our CRISPRi screening data has been deposited on Zenodo as record 13902656 with 10.5281/zenodo.13902656.