Abstract
Temperature is considered to affect the activity of drug-metabolizing enzymes; however, no previous studies have compared temperature dependency among cytochrome P450 genetic variants. This study aimed to analyse warfarin 7-hydroxylation by CYP2C9 variants; omeprazole 5-hydroxylation by CYP2C19 variants; and midazolam 1-hydroxylation by CYP3A4 variants at 34 °C, 37 °C, and 40 °C.
Compared with that seen at 37 °C, the intrinsic clearance rates (Vmax/Km) of CYP2C9.1 and .2 were decreased (76~82%), while that of CYP2C9.3 was unchanged at 34°C. At 40 °C, CYP2C9.1, .2, and .3 exhibited increased (121%), unchanged and decreased (87%) intrinsic clearance rates, respectively. At 34 °C, the clearance rates of CYP2C19.1A and .10 were decreased (71~86%), that of CYP2C19.1B was unchanged, and those of CYP2C19.8 and .23 were increased (130~134%). At 40 °C, the clearance rates of CYP2C19.1A, .1B, .10, and .23 remained unaffected, while that of CYP2C19.8 was decreased (74%). At 34°C, the clearance rates of CYP3A4.1 and .16 were decreased (79~84%), those of CYP3A4.2 and .7 were unchanged, and that of CYP3A4.18 was slightly increased (112%). At 40 °C, the clearance rate of CYP3A4.1 remained unaffected, while those of CYP3A4.2, .7, .16, and .18 were decreased (58~82%).
These findings may be clinically useful for dose optimisation in patients with hypothermia or hyperthermia.
Keywords: Cytochrome p450, genetic polymorphism, CYP allele, temperature sensitivity, body temperature, hypothermia, hyperthermia
Introduction
In humans, body temperature is physiologically maintained around 36.5 °C; therefore, the activity of drug-metabolizing enzymes is usually examined at 37 °C in in vitro experiments. However, in the clinical setting some patients experience high fevers of ≥39 °C, while others with therapeutic hypothermia are treated at 32–34 °C (Mody et al. 2019). Therefore, drugs are administered to patients with a wide range of body temperatures.
Cytochrome P450 (CYP) is one of the most important enzyme families involved in drug metabolism. Both the CYP2C and CYP3A subfamilies are expressed in the liver and small intestine in humans (Zanger and Schwab 2013). The CYP2C subfamily is involved in the metabolism of approximately 20% of clinical CYP substrates (Chiba 1998). CYP3A4, the primary member of the CYP3A subfamily, is involved in the metabolism of >50% of drugs used in the clinical setting (Murayama et al. 2002).
Some in vitro and in vivo studies have suggested that temperature affects the activity of CYPs. Soons et al. reported that in febrile, infected patients (39 °C) the oral clearance rate of nitrendipine (a CYP3A4 substrate) was decreased to 53% of that seen in afebrile patients (Soons et al. 1992), although pathophysiological effects of the infections on such enzyme activity cannot be ruled out. Shedlofsky et al. reported that the administration of lipopolysaccharide (LPS) (an endotoxin derived from Gram-negative bacteria) to healthy subjects led to an increase in body temperature to 38 °C along with a significant reduction in the oral clearance rates of antipyrine (a substrate of CYP1A2 and CYP2C), hexobarbital (a CYP2C substrate), and theophylline (a CYP1A2 substrate) to 65%, 73%, and 78%, respectively (Shedlofsky et al. 1994), although it remains unclear whether the increase in temperature directly caused the reduction in metabolic activity because LPS-induced inflammatory cytokines may have affected metabolic activity independently of body temperature.
Regarding hypothermic conditions, Empey et al. reported in hypothermia-treated paediatric patients (32–33 °C) with severe traumatic brain injuries that the maximum clearance rate (mg/h) of phenytoin (a CYP2C9 substrate) was decreased to 21.8% of that seen at 36.5–37.9 °C (Empey et al. 2013). A similar observation was reported in an animal study. Li et al. measured the total clearance rates of diclofenac (a CYP2C9 substrate) and glibenclamide (a substrate of CYP2C9 and CYP3A4) in rats whose body temperatures had been lowered to 33 °C and found that they decreased significantly to 70% and 42%, respectively, of those seen at 37 °C (Li et al. 2016).
In vitro kinetic analysis of CYP3A4 activity, as assessed by midazolam (MDZ) metabolism, at different temperatures revealed a reduction in the maximum reaction velocity (Vmax) to 75% and 60% at 32 °C and 28 °C, respectively, without a significant change in the value of the Michaelis constant (Km) (Miyamoto et al. 2015). However, the temperature range of 28–32 °C is not physiologically relevant, and they did not investigate enzymatic kinetics at temperatures higher than 37 °C.
As genetic variants of CYP2C9, CYP2C19, and CYP3A4 are known to exhibit altered properties (e.g. differences in enzyme kinetics and sensitivity to inhibitors) (Lin and Lu 2001; Zhou et al. 2009; Yamaguchi et al. 2021), it is of interest whether the genetic variants of these enzymes also exhibit altered temperature sensitivity. To the best of our knowledge, only one previous study has examined the influence of temperature on CYP metabolic activity in genotyped subjects. Poope et al. measured the total clearance of pantoprazole (a CYP2C19 substrate) in patients that underwent hypothermia treatment (<34 °C) after suffering cardiac arrests. Although they found that the overall mean total clearance rate decreased to 53% of that seen under normothermic conditions (36.4 °C) after the patients regained consciousness, they failed to detect an influence of body temperature among the genotypes (Poppe et al. 2022).
No previous studies have systematically compared the effects of temperature on metabolic activity among genetic variants of multiple CYP isoforms.
The aim of this study was to compare the in vitro enzyme kinetics of genetic variants of CYP2C9, CYP2C19 and CYP3A4 (CYP2C9.1, .2, and .3; CYP2C19.1A, .1B, .8, .10, and .23; and CYP3A4.1, .2, .7, .16, and .18) among three different temperatures (34 °C, 37 °C, and 40 °C).
Materials & methods
Chemicals and materials
Warfarin sodium (WF) was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). 7-Hydroxywarfarin (7-OH WF) and 5-hydroxyomeprazole (5-OH OPZ) were purchased from Toronto Research Chemicals, Inc. (Toronto, Canada). Naproxen was purchased from LKT Laboratories, Inc. (St Paul, MN). Omeprazole (OPZ) was purchased from Fujifilm Wako Pure Chemical Corporation (Osaka, Japan). Phenacetin was purchased from Nacalai Tesque (Kyoto, Japan). MDZ and nitrazepam were purchased from Wako Pure Chemical Industries (Osaka, Japan). 1-Hydroxymidazolam (1-OH MDZ) was purchased from Biosciences-Discovery Labware (Tokyo, Japan). All other reagents were of reagent or high-performance liquid chromatography (HPLC) grade and were obtained commercially.
Preparation of the Escherichia coli membrane fractions containing CYPs
Recombinant CYP2C9.1, .2 and .3 were purchased from Cypex Ltd. (Dundee, UK) as bactosomes; EasyCYP CYP2C9R + cytochrome b5, EasyCYP CYP2C9*2R + cytochrome b5 and EasyCYP CYP2C9*3R + cytochrome b5 (catalog number: CYP/EZ038, CYP/EZ042 and CYP/EZ044). The E. coli membrane fractions of CYP2C19.1A, .1B, .8, .10, and .23 were prepared as described below. The expression plasmid for E. coli, pCW2C19/hCPR, was developed by Dr. Yamazaki, Showa Pharmaceutical University. The pCW2C19/hCPR plasmid carries a gene for CYP2C19* 1B. We used it as the first polymerase chain reaction (PCR) template to obtain a fragment containing CYP2C19* 1B cDNA. A pBluescript II vector SK (+) carrying a CYP2C19 *1B cDNA was modified for all variant constructs to avoid non-objective mutation generation during the PCR.
Information about the variants was obtained from the “PharmVar” website in spring 2017. All objective mutations in the variants were generated by Quick Change II site-directed mutagenesis (Agilent Technologies, Santa Clara, CA).
After the expression of CYP2C19 proteins in E. coli DH5α cells and the preparation of the membrane fraction, the P450 content of the fractions was determined using the CO difference spectra method of Omura and Sato (Omura and Sato 1964), as modified by Iwata et al. (Iwata et al. 1998), after diluting the suspension with 100 mM potassium phosphate buffer (pH 7.4) containing 20% (v/v) glycerol and 0.2% (w/v) Emulgen 911. The E. coli membrane fractions of CYP3A4.1, .2, .7, .16, and .18 were prepared as described previously (Gillam et al. 1993; Miyazaki et al. 2008).
Study of the effects of temperature on metabolism by each CYP
In all experiments, an SN-100SD shaking thermostatic bath (NISSIN, Tokyo, Japan) was used to control the experimental temperature. The temperature of the water bath was set at 34 °C, 37 °C, or 40 °C and continuously monitored using an SPT-800PT digital platinum thermometer (SANSYO, Tokyo, Japan). Metabolic reactions were carried out by incubating the CYP protein with the relevant substrates in potassium phosphate buffer (pH 7.4) containing an NADPH regenerating system at the designated temperature in the experimental conditions shown in Table 1.
Table 1.
Experimental conditions for the metabolic reactions.
| CYP variants | CYP concentration nmol P450/mL | Substrate | Substrate concentration μM | Incubation time min |
|---|---|---|---|---|
| 2C9 | ||||
| .1, .2 | 30 | |||
| 0.008 | Warfarin | 0, 1, 3, 10, 30, 100, 300 | ||
| .3 | 90 | |||
| 2C19 | ||||
| .1A, .1B, .10, .23 | 0, 0.3, 1, 3, 10, 30 | |||
| 0.005 | Omeprazole | 10 | ||
| .8 | 0, 1, 3, 10, 30, 50 | |||
| 3A4 | ||||
| .1, .2, .7, .16, .18 | 0.0125 | Midazolam | 0, 0.5, 1, 3, 10, 30, 50 | 5 |
A 25-μL of a methanol solution of WF was added to a tube, and the methanol was removed under a dry nitrogen stream at 40 °C. A 400-μL of aliquot of 50 mM K+ phosphate buffer (pH 7.4) containing ethylenediaminetetraacetic acid (EDTA) (0.1 mM in the incubation mixture) and a 50-μL aliquot of a CYP2C9 bactosome (0.008nmol P450/mL in the incubation mixture) were added and preincubated at the relevant temperature for 10min. The reaction was initiated by adding a 50-μL aliquot of an NADPH regenerating system (0.2 mM NADPH, 0.5 mM NADP+, 5 mM glucose-6-phosphate, 1 U/mL glucose-6-phosphate dehydrogenase, and 5 mM MgCl2 in the incubation mixture). The final volume of the reaction mixture was 500 μL. After the reaction mixture had been incubated, the reaction was terminated by adding 500μL of ice-cold methanol containing naproxen (4.5μM after the reaction terminated) as an internal standard (IS). Subsequently, the reaction mixture was centrifuged at 7000×g at 4°C for 20min. The concentration of 7-OH WF in the supernatant was determined using the method described below.
A 350-μL of aliquot of 50 mM K+ phosphate buffer (pH 7.4) containing EDTA (0.1 mM in the incubation mixture), a 50-μL aliquot of a recombinant CYP2C19 membrane fraction (0.005nmol P450/mL in the incubation mixture), and a 50-μL aliquot of an NADPH regenerating system (0.2 mM NADPH, 0.5 mM NADP+, 5 mM glucose-6-phosphate, 1 U/mL glucose-6-phosphate dehydrogenase, and 1.25 mM MgCl2 in the incubation mixture) were preincubated at the relevant temperature for 5min. The reaction was initiated by adding 50μL of OPZ solution. The final volume of the reaction mixture was 500μL. The OPZ was dissolved in the buffer using methanol. The final concentration of methanol in the reaction mixture did not exceed 1% (v/v). After the reaction mixture had been incubated, the reaction was terminated by adding 500μL of ice-cold methanol containing phenacetin (10μM after the reaction terminated) as an IS. Subsequently, the reaction mixture was centrifuged at 12 000×g at 4°C for 20min. The concentration of 5-OH OPZ in the supernatant was determined using the method described below.
A 350-μL aliquot of 300 mM K+ phosphate buffer (pH 7.4) containing EDTA (1 mM in the incubation mixture), a 50-μL aliquot of a recombinant CYP3A4 membrane fraction (0.0125nmol P450/mL in the incubation mixture), and a 50-μL aliquot of an NADPH regenerating system (0.2 mM NADPH, 0.5 mM NADP+, 5 mM glucose-6-phosphate, 1 U/mL glucose-6-phosphate dehydrogenase, and 3 mM MgCl2 in the incubation mixture) were preincubated at the relevant temperature for 10min. The reaction was initiated by adding 50μL of MDZ solution. The final volume of the reaction mixture was 500μL. The MDZ was dissolved in the buffer using both ethanol and methanol. The final concentrations of ethanol and methanol in the reaction mixture did not exceed 1% and 2.5% (v/v), respectively. After the reaction mixture had been incubated, the reaction was terminated by adding 500μL of ice-cold methanol containing nitrazepam (0.1μM after the reaction terminated) as an IS. Subsequently, the reaction mixture was centrifuged at 7000×g at 4°C for 20min. The concentration of 1-OH MDZ in the supernatant was determined using the method described below.
Under a higher temperature of 40 °C, the enzymatic activity may be rapidly inactivated. Therefore, we investigated the time course of metabolite formation by all the tested variants of CYP2C9, CYP2C19, and CYP3A4 at 40 °C. The substrate concentrations were set close to the respective Km value as follows: CYP2C9; 10μM for CYP2C9.1, .2, and 30μM for CYP2C9.3, CYP2C19; 3μM for CYP2C19.1A, .1B, 10μM for CYP2C19.10, .23, and 30μM for CYP2C19.8, CYP3A4; 3μM for CYP3A4.1, .18, and 10μM for CYP3A4.2, .7, .16. The incubation times were set to cover the time to investigate the effect of temperature (Supplemental Table).
Determination of the 7-OH WF concentration using the HPLC-FL method
The HPLC system consisted of a pump (LC-2030C, Shimadzu, Kyoto, Japan), a fluorescence detector (RF-20A, Shimadzu), and an octadecylsilane reversed-phase column (COSMOSIL, 5C18-MS-II, 4.6 ID × 250mm; Nacalai Tesque). The column temperature was set at 40 °C. The mobile phase was a mixture of 0.5% (w/v) KH2PO4 (pH 3.5 adjusted with H3PO4) and acetonitrile (63:37, v/v). The flow rate and excitation/emission wavelengths were set at 1.5mL/min and 320/415nm, respectively. The concentration of 7-OH WF was determined based on the ratio of the peak area of 7-OH WF to that of the IS. The limit of quantification was 1nM.
Determination of the 5-OH OPZ concentration using the HPLC-UV method
The HPLC system consisted of a pump and an ultraviolet (UV) detector (LC-2030C, Shimadzu) and an octadecylsilane reversed-phase column (COSMOSIL, 5C18-MS-II, 4.6 ID × 150 mm; Nacalai Tesque). The column temperature was set at 25 °C. The mobile phase was a mixture of 20 mM potassium phosphate buffer (pH 6.8) and methanol (65:35, v/v). The flow rate and detection wavelength were set at 1.5mL/min and 302nm, respectively. The concentration of 5-OH OPZ was determined based on the ratio of the peak area of 5-OH OPZ to that of the IS. The limit of quantification was 0.01nM.
Determination of the 1-OH MDZ concentration using the HPLC-UV method
The HPLC system consisted of a pump (LC-20AD, Shimadzu), a UV detector (SPD-20A, Shimadzu), and an octadecylsilane reversed-phase column (COSMOSIL, 5C18-MS-II, 4.6 ID × 150 mm; Nacalai Tesque). The column temperature was set at 42 °C. The mobile phase was a mixture of purified water and methanol (50:50, v/v). The flow rate and detection wavelength were set at 1.8mL/min and 245nm, respectively. The concentration of 1-OH MDZ was determined based on the ratio of the peak area of 1-OH MDZ to that of the IS. The limit of quantification was 0.02nM.
Calculation of kinetic parameters
The Michaelis-Menten equation (Equation 1) was fitted to the relationship between the substrate concentration ([S]) and the reaction rate (v) to obtain the maximum reaction velocity (Vmax) and the Michaelis constant (Km) using the non-linear least squares program (Multi (Yamaoka et al. 1981)).
| (1) |
Statistical analysis
The statistical significance of the differences in Vmax, ln Km, and Vmax/Km values among different temperatures was determined by one-way analysis of variance followed by Dunnett’s multiple comparisons test. P-values of <0.05 were considered statistically significant.
Results
3.1. Comparison of the metabolic kinetics of CYP2C9 genetic variants among temperatures
The metabolic activities of CYP2C9.1, .2, and .3, as assessed based on the 7-hydroxylation of WF, showed saturable kinetics at 34 °C, 37 °C, and 40 °C (Figure 1, Table 2). At 34 °C, the metabolic activity levels (measured based on Vmax/Km values) of CYP2C9.1 and .2 were decreased to 76% and 82%, respectively, of those seen at 37 °C, while that of CYP2C9.3 was almost unchanged (92%). At 40 °C, the metabolic activity levels of CYP2C9.1, .2, and .3 were increased (121%), unchanged, and decreased (87%), respectively, compared with those seen at 37 °C. The higher the temperature, the larger the Vmax was for all CYP2C9 genetic variants. While the Km value for CYP2C9.1 was almost unaffected by temperature, those for CYP2C9.2 and .3 increased as the temperature rose. The metabolite formation by all CYP2C9 variants were almost linear until the time to evaluate the effect of temperature (Supplemental Figure 1).
Figure 1.
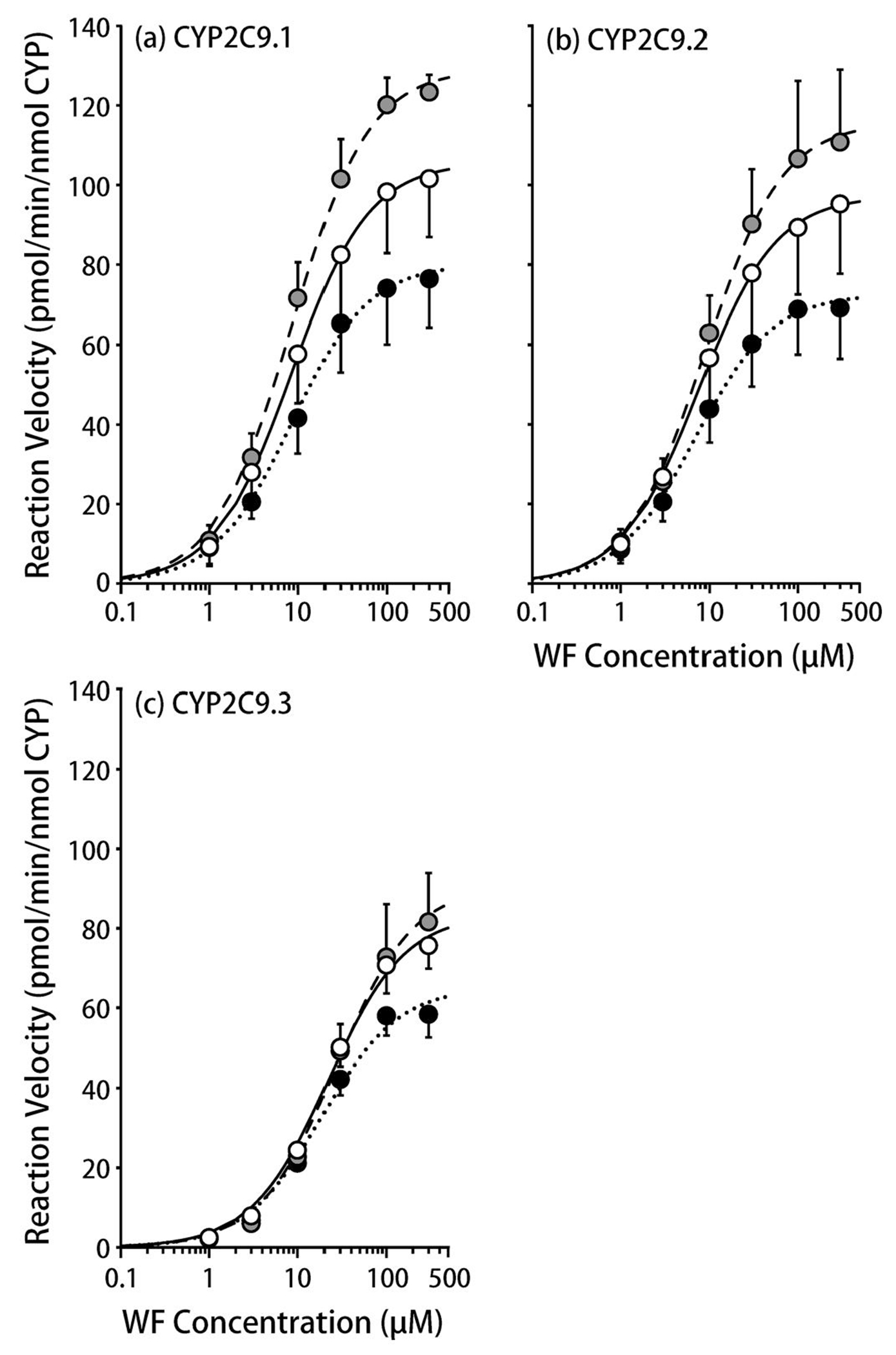
The influence of temperature on warfarin 7-hydroxylation by CYP2C9 variants.
(a) CYP2C9.1, (b) CYP2C9.2, (c) CYP2C9.3.
The solid (open), dotted (closed), and dashed lines (gray symbols) represent 37 °C, 34 °C, and 40 °C, respectively.
Each symbol represents a mean ± S.D. value derived from five independent experiments.
Table 2.
Kinetic parameters of WF metabolism by CYP2C9 variants.
|
Vmax/Km |
|||||
|---|---|---|---|---|---|
| CYP variants | Temperature | Vmax pmol/min/nmol P450 | Km μM | μL/min/nmol P450 | 37 °C ratio |
| CYP2C9.1 | |||||
| 34 °C | 80.2 ± 14.0* | 8.41 (6.68–10.6) | 9.72 ± 2.55 | 0.76 | |
| 37 °C | 106 ± 15.3 | 8.53 (6.94–10.5) | 12.8 ± 3.97 | (1) | |
| 40 °C | 129 ± 5.41* | 8.55 (7.18–10.2) | 15.4 ± 3.57 | 1.21 | |
| CYP2C9.2 | |||||
| 34 °C | 72.6 ± 12.5 | 6.88 (6.01–7.88) | 10.7 ± 2.76 | 0.82 | |
| 37 °C | 97.3 ± 18.2 | 7.63 (6.55–8.89) | 13.0 ± 3.31 | (1) | |
| 40 °C | 116 ± 20.3 | 8.91 (7.60–10.5) | 13.0 ± 2.46 | 1.00 | |
| CYP2C9.3 | |||||
| 34 °C | 65.2 ± 6.27* | 18.5 (17.4–19.7) | 3.52 ± 0.29 | 0.92 | |
| 37 °C | 83.6 ± 6.93 | 22.0 (20.7–23.4) | 3.81 ± 0.41 | (1) | |
| 40 °C | 90.8 ± 15.4 | 27.2 (25.2–29.4)* | 3.32 ± 0.41 | 0.87 | |
Vmax, Vmax/Km: arithmetic mean ± S.D., Km: geometric mean (−1S.D. ~ +1S.D.), n = 5
: p < 0.05 vs. 37 °C
Comparison of the metabolic kinetics of CYP2C19 genetic variants among temperatures
The metabolic activity levels of CYP2C19.1A, .1B, .8, .10, and .23, as assessed based on the 5-hydroxylation of OPZ, showed saturable kinetics at 34 °C, 37 °C, and 40 °C (Figure 2, Table 3). At 34 °C, the metabolic activity levels of CYP2C19.1A and .10 were decreased to 71% and 86%, respectively, of those seen at 37 °C, while that of CYP2C19.1B were almost unchanged (97%), and those of CYP2C19.8 and .23 were increased to 130% and 134%, respectively, of those seen at 37 °C. At 40 °C, the metabolic activity levels of CYP2C19.1A, .1B, .10, and .23 were almost unchanged (92-108%), while that of CYP2C19.8 decreased to 74% of that seen at 37 °C because only the Km value of CYP2C19.8 increased as the temperature rose. The metabolite formation by all CYP2C19 variants was almost linear until the time to evaluate the effect of temperature (Supplemental Figure 1).
Figure 2.
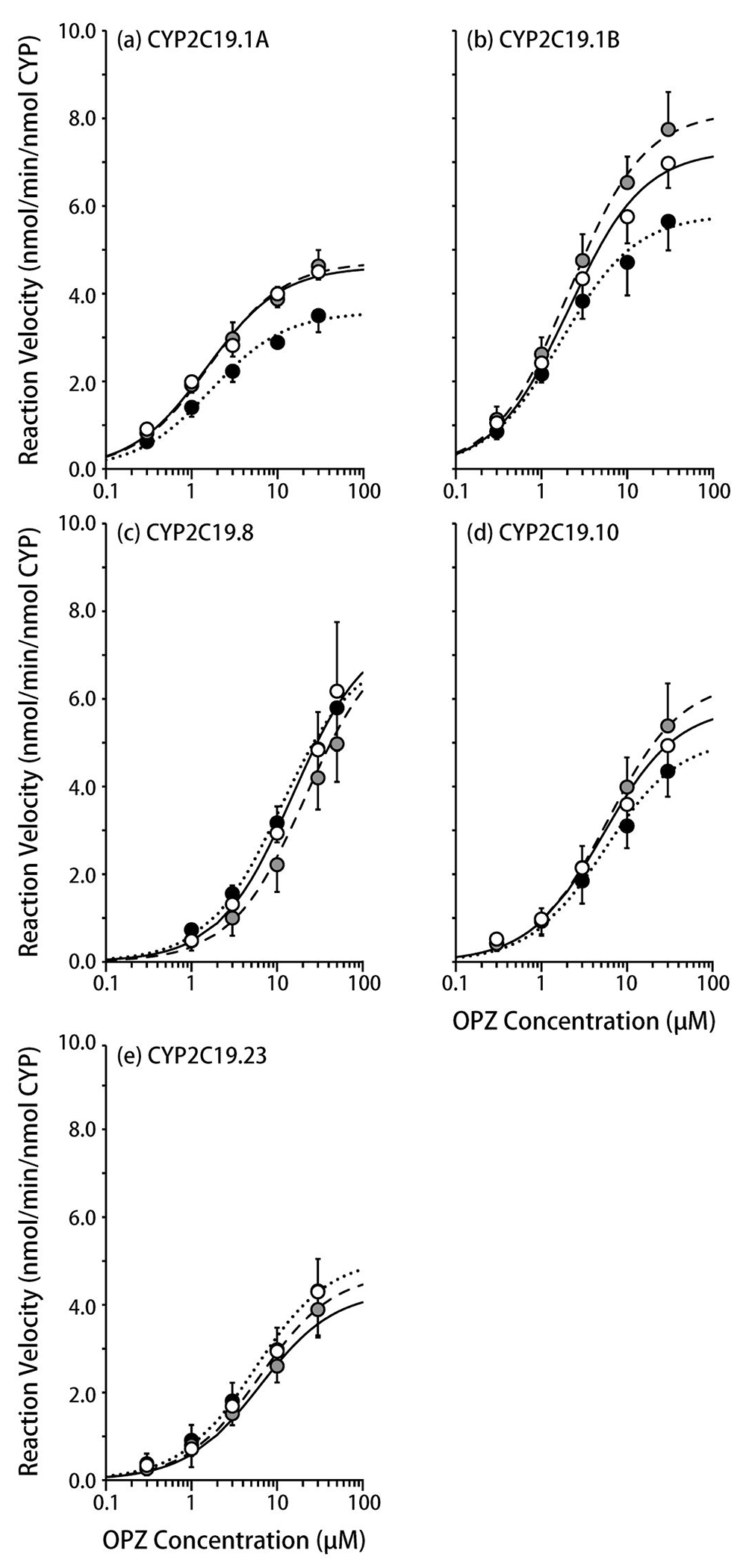
The influence of temperature on omeprazole 5-hydroxylation by CYP2C19 variants.
(a) CYP2C19.1A, (b) CYP2C19.1B, (c) CYP2C19.8, (d) CYP2C19.10, (e) CYP2C19.23
The solid (open), dotted (closed), and dashed lines (gray symbols) represent 37 °C, 34 °C, and 40 °C, respectively.
Each symbol represents a mean ± S.D. value derived from five independent experiments.
Table 3.
Kinetic parameters of OPZ metabolism by CYP2C19 variants.
| CYP variants | Temperature | Vmax nmol/min/nmol P450 | Km μM | Vmax/Km mL/min/nmol P450 | 37 °C ratio |
|---|---|---|---|---|---|
| CYP2C19.1A | |||||
| 34 °C | 3.58 ± 0.391* | 1.66 (1.17–2.34) | 2.23 ± 0.695 | 0.71 | |
| 37 °C | 4.61 ± 0.259 | 1.49 (1.22–1.82) | 3.15 ± 0.715 | (1) | |
| 40 °C | 4.72 ± 0.374 | 1.61 (1.27–2.05) | 2.98 ± 0.634 | 0.95 | |
| CYP2C19.1B | |||||
| 34 °C | 5.81 ± 0.793* | 1.67 (1.56–1.80) | 3.45 ± 0.217 | 0.97 | |
| 37 °C | 7.27 ± 0.600 | 2.07 (1.77–2.43) | 3.56 ± 0.737 | (1) | |
| 40 °C | 8.16 ± 0.865 | 2.13 (1.86–2.43) | 3.85 ± 0.599 | 1.08 | |
| CYP2C19.8 | |||||
| 34 °C | 7.08 ± 1.72 | 11.1 (7.21–17.1) | 0.655 ± 0.230 | 1.30 | |
| 37 °C | 7.61 ± 1.44 | 15.2 (9.44– 24.5) | 0.504 ± 0.267 | (1) | |
| 40 °C | 7.20 ± 1.61 | 20.5 (12.6– 33.5) | 0.373 ± 0.170 | 0.74 | |
| CYP2C19.10 | |||||
| 34 °C | 5.10 ± 0.652 | 5.43 (3.60–8.20) | 1.02 ± 0.507 | 0.86 | |
| 37 °C | 5.83 ± 0.788 | 5.28 (3.36–8.31) | 1.19 ± 0.595 | (1) | |
| 40 °C | 6.42 ± 1.11 | 5.90 (5.13–6.79) | 1.10 ± 0.272 | 0.92 | |
| CYP2C19.23 | |||||
| 34 °C | 5.08 ± 1.10 | 5.50 (3.48–8.69) | 0.988 ± 0.526 | 1.34 | |
| 37 °C | 4.31 ± 2.48 | 6.33 (4.08–9.80) | 0.735 ± 0.411 | (1) | |
| 40 °C | 4.74 ± 1.15 | 6.29 (3.47–9.96) | 0.787 ± 0.373 | 1.07 |
Vmax, Vmax/Km: arithmetic mean ± S.D., Km: geometric mean (−1S.D. ~ +1S.D.), n = 5
: p < 0.05 vs. 37 °C
Comparison of the metabolic kinetics of CYP3A4 genetic variants among temperatures
The metabolic activities of CYP3A4.1, .2, .7, .16, and .18, as assessed based on the 1-hydroxylation of MDZ, showed saturable kinetics at 34 °C, 37 °C, and 40 °C (Figure 3, Table 4). At 34 °C, the metabolic activity levels of CYP3A4.1 and .16 were decreased to 79% and 84%, respectively, compared with those seen at 37 °C, while those of CYP3A4.2 and .7 were almost unchanged (106% and 107%) and that of CYP3A4.18 was slightly increased to 112% of that seen at 37 °C. At 40 °C, the metabolic activity level of CYP3A4.1 was almost unchanged (95%), while those of CYP3A4.2, .7, .16, and .18 decreased to 72%, 58%, 82%, and 82%, respectively, of those seen at 37 °C. In particular, at 40 °C the Km value for CYP3A4.7 increased to 170% of that seen at 37 °C. Regarding four out of five CYP3A4 variants (CYP3A4.1, .2, .7, and .18), slight (2-20%) decreases in the metabolite formation were observed at the time to evaluate the effect of temperature (5 min) (Supplemental Figure 1).
Figure 3.
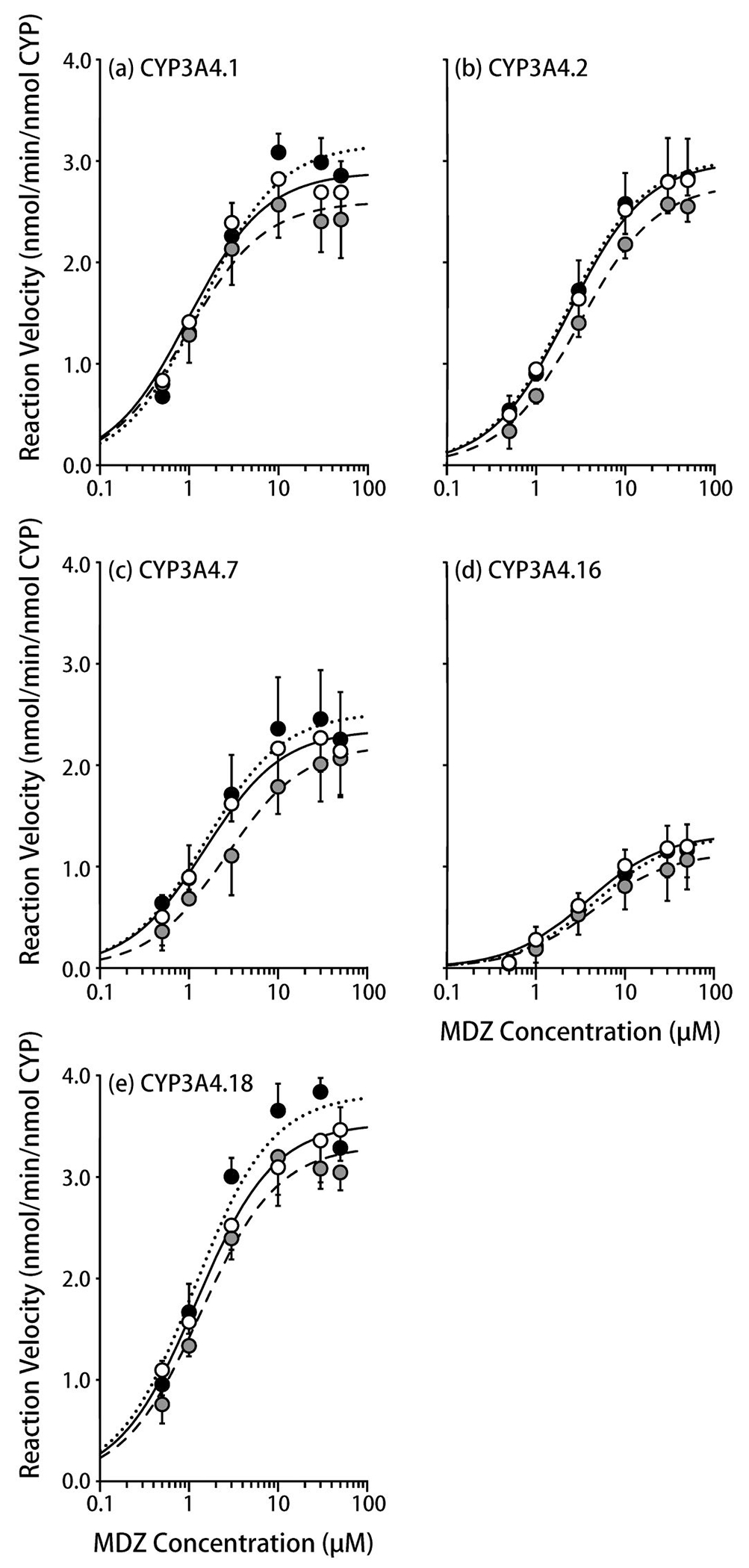
The influence of temperature on midazolam 1-hydroxylation by CYP3A4 variants.
(a) CYP3A4.1, (b) CYP3A4.2, (c) CYP3A4.7, (d) CYP3A4.16, (e) CYP3A4.18
The solid (open), dotted (closed), and dashed lines (gray symbols) represent 37 °C, 34 °C, and 40 °C, respectively.
Each symbol represents a mean ± S.D. value derived from five independent experiments.
Table 4.
Kinetic parameters of MDZ metabolism by CYP3A4 variants.
| CYP variants | Temperature | Vmax nmol/min/nmol P450 | Km μM | Vmax/Km mL/min/nmol P450 | 37 °C ratio |
|---|---|---|---|---|---|
| CYP3A4.1 | |||||
| 34 °C | 3.17 ± 0.19 | 1.37 (1.10 – 1.71) | 2.37 ± 0.49 | 0.79 | |
| 37 °C | 2.89 ± 0.24 | 0.97 (0.88 – 1.06) | 3.00 ± 0.39 | (1) | |
| 40 °C | 2.60 ± 0.36 | 0.94 (0.73 – 1.23) | 2.86 ± 0.91 | 0.95 | |
| CYP3A4.2 | |||||
| 34 °C | 3.02 ± 0.44 | 2.19 (1.85 – 2.59) | 1.38 ± 0.17 | 1.06 | |
| 37 °C | 3.00 ± 0.22 | 2.31 (2.12 – 2.53) | 1.30 ± 0.17 | (1) | |
| 40 °C | 2.78 ± 0.07 | 3.02 (2.59 – 3.52)* | 0.93 ± 0.15* | 0.72 | |
| CYP3A4.7 | |||||
| 34 °C | 2.52 ± 0.49 | 1.56 (1.37 – 1.77) | 1.67 ± 0.45 | 1.07 | |
| 37 °C | 2.35 ± 0.39 | 1.52 (1.43 – 1.61) | 1.56 ± 0.29 | (1) | |
| 40 °C | 2.20 ± 0.37 | 2.59 (1.68 – 4.00) | 0.91 ± 0.32 | 0.58 | |
| CYP3A4.16 | |||||
| 34 °C | 1.31 ± 0.24 | 4.67 (3.12 – 6.97) | 0.32 ± 0.14 | 0.84 | |
| 37 °C | 1.32 ± 0.30 | 3.76 (2.88 – 4.91) | 0.38 ± 0.15 | (1) | |
| 40 °C | 1.14 ± 0.28 | 4.40 (2.76 – 7.02) | 0.31 ± 0.19 | 0.82 | |
| CYP3A4.18 | |||||
| 34 °C | 3.82 ± 0.13 | 1.15 (0.96 – 1.38) | 3.38 ± 0.66 | 1.12 | |
| 37 °C | 3.53 ± 0.26 | 1.20 (0.87 – 1.67) | 3.03 ± 0.71 | (1) | |
| 40 °C | 3.31 ± 0.24 | 1.34 (1.23 – 1.47) | 2.47 ± 0.19 | 0.82 |
Vmax, Vmax/Km: arithmetic mean ± S.D., Km: geometric mean (−1S.D. ~ +1S.D.), n = 5
: p < 0.05 vs. 37 °C
Discussion
Under the normothermic conditions of 37 °C, the observed differences in the metabolic activity among the genetic variants of each CYP isoform were consistent with previous findings. Compared with CYP2C9.1, the metabolic activity (assessed based on the 7-hydroxylation of WF) of CYP2C9.3 is known to be decreased, whereas that of CYP2C9.2 is unaltered (Sullivan-Klose et al. 1996), and these observations are consistent with the present results. Similarly, the results obtained in the present study for CYP2C19.1B, .10, and .23 were consistent with the previous findings that compared with the metabolic activity level of CYP2C19.1A (assessed based on the 5-hydroxylation of OPZ) that those of CYP2C19.10 and .23 were decreased, but that of CYP2C19.1B was unaltered (Wang et al. 2011; Lau et al. 2017). The result for CYP2C19.8 was also consistent with the previous finding obtained using another CYP2C19 substrate, nebivolol, that the metabolic activity of CYP2C19.8 was lower than that of CYP2C19.1A (Zhou et al. 2018). The rank order of CYP3A4 activity among CYP3A4 variants, as assessed based on the 1-hydroxylation of MDZ, was reported to be CYP3A4.1 = .18 > .7 > .2 > .16 (Miyazaki et al. 2008), which is consistent with the present results, although not completely identical. Taken together, the present study is considered to have successfully reproduced the effects of genetic variations at 37 °C.
For CYP2C9, Vmax was higher at higher temperatures. Based on the conventional enzyme reaction model (Equation 2), Vmax is expressed as the product of the total amount of enzyme and the rate constant k2 of the reaction process after substrate binding to the enzyme.
| (2) |
where E is the enzyme, S is the substrate, ES is the enzyme-substrate complex, and P is the product.
Razavi et al. reported that for various enzymes, such as β-glucosidase, cellobiohydrolase, and xylanase, the relationship between temperature and Vmax can be explained by the Arrhenius equation (Razavi et al. 2015). In the present study, Vmax increased as the temperature rose for all of the examined genetic variants of CYP2C9, and the temperature dependence of Vmax was well explained by the Arrhenius equation (Supplemental Figure 2). For CYP2C19, Vmax increased as the temperature rose for CYP2C19.1A, .1B, and .10. The temperature dependence of Vmax for CYP2C19.1B and .10 was again well explained by the Arrhenius equation (Supplemental Figure 2). In contrast, the temperature dependence of Vmax for other CYP2C19 variants and CYP3A4 were not appropriately explained by the Arrhenius equation (data not shown). In other words, while molecular thermodynamics can simply explain the temperature dependency of Vmax for CYP2C9 and certain variants of CYP2C19, other factors, such as conformational changes in the enzyme, may be crucial determinants of the temperature dependency of Vmax for other enzyme molecules.
The Km value, which represents the affinity between the enzyme and the substrate, is expressed as the ratio of k−1 to k−1 in Equation 3, which is based on the assumption that k−1 ⪢ k2 in Equation 2.
| (3) |
Thus, the Km values are considered useful for approximating the equilibrium constant in the reversible reaction of enzyme-substrate complex formation for use in the Van’t Hoff equation. For CYP2C9.2, CYP2C9.3, CYP2C19.1B, CYP2C19.8, CYP3A4.2, and CYP3A4.18, the Km values increased with an increase in the temperature. For three out of these six variants, CYP2C9.2, CYP2C9.3, and CYP2C19.8, the Van’t Hoff equation, based on molecular thermodynamics, could explain the relationship between the Km value and the temperature (Supplemental Figure 3). On the other hand, the Van’t Hoff equation failed to explain the relationship of the other variants, implying that the effect of temperature on the Km value is not fully explained by molecular thermodynamics. Other factors, such as the conformational changes of the enzyme, may be crucial for determining the temperature dependence of Km for these variants.
For CYP2C9, the Km values for CYP2C9.2 and .3, but not that for CYP2C9.1, were temperature-dependent. Based on docking simulations of CYP2C9 and substrates, including WF, Sano et al. suggested that the substrate-binding pockets of CYP2C9.2 and .3 are larger than that of CYP2C9.1 so the substrates bind less closely to haem, leading to reduced metabolic activity (Sano et al. 2010). According to their binding model, the hydrogen bonds between Q214 and two amino acid residues, N474 and F476, are weaker in CYP2C9.3 than in CYP2C9.1. On the other hand, CYP2C9.2 has an R144C amino acid substitution, which results in decreased interaction with the V177-D188 domain. Therefore, there may be differences in the temperature dependence of these hydrogen bonds among CYP2C9 genetic variants. CYP2C19.8 has an amino acid substitution at W120, which is well conserved in the CYP2C subfamily and contributes to the interaction of haem iron with substrates in CYP molecules (Lewis 1998). The suppressive effect of the W120R substitution on substrate-haem interactions may be more substantial at higher temperatures, leading to a temperature-dependent increase in Km and a temperature-dependent reduction in Vmax/Km. In contrast, in comparison with those seen at 37 °C, the Km values of CYP2C19.1A, .1B, and .23 were almost unchanged (99-108%) at the higher temperature of 40 °C, and that of CYP2C19.10 was slightly increased (112%). On the other hand, the Km value for CYP2C19.8, which contains W120R, was more temperature-dependent (it exhibited a 35% increase at 40 °C). The substitution I331V, which is common to CYP2C19.1B, .10, and .23, is located outside of the substrate-binding region (Takahashi et al. 2015), so it is less likely to affect the temperature dependence of substrate binding (although Km should not be simply equated with substrate binding).
One of the limitations of the present study is the inactivation of the enzyme at a higher temperature of 40 °C. Unlike CYP2C9 and CYP2C19 variants, some CYP3A4 variants (.1, .2, .7, and .18) did not show linear kinetics of metabolite formation even within 5min (Supplemental Figure 1). As the substrate has not been depleted, this observation may imply the time-dependent inactivation of these four variants at the higher temperature of 40 °C. Therefore, the effects of temperature shown in Figure 3 may be, at least in part, attributable to the time-dependent inactivation, conceivably through oxidative damage.
The in vitro data obtained in the present study may explain, at least qualitatively, previously reported in vivo findings. For example, under the hypothermic conditions of 34 °C, the metabolic activity levels (Vmax/Km) of CYP2C9.1 and CYP3A4.1 were decreased to 76% and 79% of those seen at 37 °C, respectively. These reductions in metabolic activity are consistent with the findings obtained in rats by Li et al. that under hypothermic conditions (33 °C) the total body clearance rates of diclofenac (a CYP2C9 substrate) and glibenclamide (a substrate of CYP2C9 and CYP3A4) were significantly decreased to 70% and 42% of those seen at 37 °C, respectively (Li et al. 2016). Empey et al. also reported a similar result in rats; i.e. under the hypothermic conditions of 33 °C the total body clearance rate of MDZ was decreased to 83% of that seen at 37 °C (Empey et al. 2012). As the reduction in substrate clearance observed in rats in vivo was quantitatively comparable to the reduction in Vmax/Km seen in the present study, the decreased in vivo clearance observed under hypothermic conditions may be primarily explained by reductions in the activity of CYPs under low-temperature conditions.
In the clinical setting, various drugs, including phenytoin, are administered to patients during hypothermia treatment performed at 32–34 °C after a cardiac arrest or traumatic brain injury (Carney et al. 2017; Mody et al. 2019). In addition, body temperature is unintentionally affected by various factors in the clinical setting. Furthermore, the physiological body temperature of elderly individuals is lower than that of younger individuals. While infections or inflammation may lead to a high fever of ≥39 °C, such patients need to be treated with a variety of medications, including antibiotics and anti-inflammatory drugs. The present study provides clinically significant information regarding the metabolism of drugs under high or low body temperatures that are encountered in the clinical setting. Furthermore, genetic variations in CYP isoforms are known to affect metabolic activity, sensitivity to inhibitors, and the mode of inhibition (Lin and Lu 2001; Zhou et al. 2009; Yamaguchi et al. 2021). This is the first study to show that the temperature dependency of metabolic activity differs among genetic variants of CYP. Although the in vitro data obtained in the present study may provide clinically useful information for optimising pharmacotherapy in patients with altered body temperatures, there are some other factors that need to be taken into account. First, hepatic clearance is determined not only by intrinsic metabolic activity, but also by drug uptake into hepatocytes, hepatic blood flow, and plasma protein binding, which are also known to be affected by changes in body temperature (Anderson et al. 2016). Furthermore, although inflammation is one of the major causes of fever, it also alters physiological conditions and pathways, such as cytokine cascades. Shedlofsty et al. reported that the oral clearance rates of antipyrine, hexobarbital, and theophylline were significantly decreased in healthy subjects with LPS-induced fevers of 38 °C (Shedlofsky et al. 1994). The authors concluded that inflammatory cytokines, such as tumour necrosis factor α and interleukin-6, whose expression was induced by LPS were likely to be responsible for the reduction in metabolic activity. Thus, changes in body temperature are not independent of other physiological factors. Therefore, when predicting the changes in vivo metabolic clearance seen under altered body temperature, factors other than the temperature dependence of metabolic activity should be taken into account. By performing an in silico analysis of population-based in vivo physiological data for subjects with altered body temperatures, it may be possible to extrapolate the results of the present study in order to predict changes in vivo drug clearance.
Conclusion
In this study, we have shown that the metabolic activity levels of all of the examined genetic variants of CYP2C9, CYP2C19, and CYP3A4 were affected by temperature, while the trends in temperature dependence differed among the genetic variants. Furthermore, the temperature dependences of Vmax and Km differed among the isoforms and among the genetic variants of each isoform. This study highlights the importance of the temperature dependence of enzymes and how it differs among genetic variants for the optimisation of pharmacotherapy in patients with hypothermia or hyperthermia in the clinical setting.
Supplementary Material
Acknowledgements
We would like to thank Medical English Service (http://www.med-english.com) for English language editing.
Funding
This study was supported in part by the Japan Research Foundation for JSPS Kakenhi (OH; Grant Number 18K06758) and by the United States National Institutes of Health (FPG; Grant Number R01 GM118122) FP Guengerich was funded by the United States National Institutes of Health (Grant Number R01 GM118122).
Abbreviations:
- CYP
cytochrome P450
- WF
warfarin
- 7-OH WF
7-hydroxywarfarin
- OPZ
omeprazole
- 5-OH OPZ
5-hydroxyomeprazole
- MDZ
midazolam
- 1-OH MDZ
1-hydroxymidazolam
- Vmax
maximal rate of metabolism
- Km
Michaelis-Menten constant, or the concentration of the substrate that gives the half-maximal rate of metabolism
- Vmax/Km
intrinsic clearance
- HPLC-FL
high-performance liquid chromatography with fluorescence detection
- HPLC-UV
high-performance liquid chromatography with ultraviolet detection
- LPS
lipopolysaccharide
- IS
internal standard
- PCR
polymerase chain reaction
Footnotes
Supplemental data for this article can be accessed online at https://doi.org/10.1080/00498254.2023.2248498.
Disclosure statement
The authors report no declarations of interest.
References
- Anderson KB, Poloyac SM, Kochanek PM, Empey PE. 2016. Effect of hypothermia and targeted temperature management on drug disposition and response following cardiac arrest: a comprehensive review of pre-clinical and clinical investigations. Ther Hypothermia Temp Manag. 6(4):169–179. doi: 10.1089/ther.2016.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney N, Totten AM, O’Reilly C, Ullman JS, Hawryluk GW, Bell MJ, Bratton SL, Chesnut R, Harris OA, Kissoon N, et al. 2017. Guidelines for the management of severe traumatic brain injury, fourth edition. Neurosurgery. 80(1):6–15. doi: 10.1227/NEU.0000000000001432. [DOI] [PubMed] [Google Scholar]
- Chiba K. 1998. Genetic polymorphism of the CYP2C subfamily. Nihon Yakurigaku Zasshi. 112(1):15–21. doi: 10.1254/fpj.112.15. [DOI] [PubMed] [Google Scholar]
- Empey PE, Miller TM, Philbrick AH, Melick JA, Kochanek PM, Poloyac SM. 2012. Mild hypothermia decreases fentanyl and midazolam steady-state clearance in a rat model of cardiac arrest. Crit Care Med. 40(4):1221–1228. doi: 10.1097/CCM.0b013e31823779f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empey PE, Velez de Mendizabal N, Bell MJ, Bies RR, Anderson KB, Kochanek PM, Adelson PD, Poloyac SM, 2013. Therapeutic hypothermia decreases phenytoin elimination in children with traumatic brain injury. Crit Care Med. 41(10):2379–2387. doi: 10.1097/CCM.0b013e318292316c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam EMJ, Baba T, Kim BR, Ohmori S, Guengerich FP. 1993. Expression of modified human cytochrome P450 3A4 in Esherichia coli and purification and reconstitution of the enzyme. Arch Biochem Biophys. 305(1):123–131. doi: 10.1006/abbi.1993.1401. [DOI] [PubMed] [Google Scholar]
- Iwata H, Fujita K, Kushida H, Suzuki A, Konno Y, Nakamura K, Fujino A, Kamataki T. 1998. High catalytic activity of human cytochrome P450 Co-expressed with human NADPH-cytochrome P450 reductase in Escherichia coli. Biochem Pharmacol. 55(8):1315–1325. doi: 10.1016/s0006-2952(97)00643-6. [DOI] [PubMed] [Google Scholar]
- Lau PS, Leong KVG, Ong CE, Dong ANHM, Pan Y. 2017. In vitro functional characterisation of cytochrome P450 (CYP) 2C19 allelic variants CYP2C19* 23 and CYP2C19* 24. Biochem Genet. 55(1):48–62. doi: 10.1007/s10528-016-9771-8. [DOI] [PubMed] [Google Scholar]
- Lewis DF. 1998. The CYP2 family: models, mutants and interactions. Xenobiotica. 28(7):617–661. doi: 10.1080/004982598239236. [DOI] [PubMed] [Google Scholar]
- Li X, Ji Z, Gu Y, Hu Y, Huang K, Pan S. 2016. Mild hypothermia decreases the total clearance of glibenclamide after low dose administration in rats. Neurosci Lett. 614:55–59. doi: 10.1016/j.neulet.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Lin JH, Lu AY. 2001. Interindividual variability in inhibition and induction of cytochrome P450 enzymes. Annu Rev Pharmacol Toxicol. 41:535–567. doi: 10.1146/annurev.pharmtox.41.1.535. [DOI] [PubMed] [Google Scholar]
- Miyamoto H, Matsueda S, Moritsuka A, Shimokawa K, Hirata H, Nakashima M, Sasaki H, Fumoto S, Nishida K. 2015. Evaluation of hypothermia on the in vitro metabolism and binding and in vivo disposition of midazolam in rats. Biopharm Drug Dispos. 36(7):481–489. doi: 10.1002/bdd.1960. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Nakamura K, Fujita Y, Guengerich FP, Horiuchi R, Yamamoto K. 2008. Defective activity of recombinant cytochromes P450 3A4.2 and 3A4.16 in oxidation of midazolam, nifedipine, and testosterone. Drug Metab Dispos. 36(11):2287–2291. doi: 10.1124/dmd.108.021816. [DOI] [PubMed] [Google Scholar]
- Mody P, Kulkarni N, Khera R, Link MS. 2019. Targeted temperature management for cardiac arrest. Prog Cardiovasc Dis. 62(3):272–278.; doi: 10.1016/j.pcad.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Murayama N, Nakamura T, Saeki M, Soyama A, Saito Y, Sai K, Ishida S, Nakajima O, Itoda M, Ohno Y, et al. 2002. CYP3A4 gene polymorphisms influence teststerone 6β-hydroxylation. Drug Metab Pharmacokinet. 17(2):150–156. doi: 10.2133/dmpk.17.150. [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem. 239(7):2379–2385. doi: 10.1016/S0021-9258(20)82245-5. [DOI] [PubMed] [Google Scholar]
- PharmVar. https://www.pharmvar.org/gene/CYP2C19. last accessed: 2023/3/06
- Poppe M, Clodi C, Schriefl C, Mueller M, Sunder-Plaßmann R, Reiter B, Rechenmacher M, van Os W, van Hasselt JGC, Holzer M, et al. 2022. Targeted temperature management after cardiac arrest is associated with reduced metabolism of pantoprazole - A probe drug of CYP2C19 metabolism. Biomed Pharmacother. 146:112573. Feb; doi: 10.1016/j.biopha.2021.112573. [DOI] [PubMed] [Google Scholar]
- Razavi BS, Blagodatskaya E, Kuzyakov Y. 2015. Nonlinear temperature sensitivity of enzyme kinetics explains canceling effect-a case study on loamy haplic Luvisol. Front Microbiol. 6:1126. doi: 10.3389/fmicb.2015.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano E, Li W, Yuki H, Liu X, Furihata T, Kobayashi K, Chiba K, Neya S, Hoshino T. 2010. Mechanism of the decrease in catalytic activity of human cytochrome P450 2C9 polymorphic variants investigated by computational analysis. J Comput Chem. 31(15):2746–2758. doi: 10.1002/jcc.21568. [DOI] [PubMed] [Google Scholar]
- Shedlofsky SI, Israel BC, McClain CJ, Hill DB, Blouin RA. 1994. Endotoxin administration to humans inhibits hepatic cytochrome P450-mediated drug metabolism. J Clin Invest. 94(6):2209–2214. doi: 10.1172/JCI117582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soons PA, Grib C, Breimer DD, Kirch W. 1992. Effects of acute febrile infectious diseases on the oral pharmacokinetics and effects of nitrendipine enantiomers and of bisoprolol. Clin Pharmacokinet. 23(3):238–248. doi: 10.2165/00003088-199223030-00006. [DOI] [PubMed] [Google Scholar]
- Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. 1996. The role of the CYP2C9 Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 6(4):341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Saito T, Ito M, Tsukada C, Katono Y, Hosono H, Maekawa M, Shimada M, Mano N, Oda A, et al. 2015. Functional characterization of 21 CYP2C19 allelic variants for clopidogrel 2-oxidation. Pharmacogenomics J. 15(1):26–32. doi: 10.1038/tpj.2014.30. [DOI] [PubMed] [Google Scholar]
- Wang H, An N, Wang H, Gao Y, Liu D, Bian T, Zhu J, Chen C. 2011. Evaluation of the effects of 20 nonsynonymous single nucleotide polymorphisms of CYP2C19 on S-Mephenytoin 4-hydroxylation and omeprazole 5-Hydroxylation. Drug Metab Dispos. 39(5):830–837. doi: 10.1124/dmd.110.037549. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Akiyoshi T, Kawamura G, Imaoka A, Miyazaki M, Guengerich FP, Nakamura K, Yamamoto K, Ohtani H. 2021. Comparison of the inhibitory effects of azole antifungals on cytochrome P450 3A4 genetic variants. Drug Metab Pharmacokinet. 38: 100384.; doi: 10.1016/j.dmpk.2021.100384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka K, Tanigawara Y, Nakagawa T, Uno T. 1981. A pharmacokinetic analysis program (multi) for microcomputer. Journal of Pharmacobio-Dynamics. 4(11):879–885. doi: 10.1248/bpb1978.4.879. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Schwab M. 2013. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Zhou SF, Liu JP, Chowbay B. 2009. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 41(2):89–295. doi: 10.1080/03602530902843483. [DOI] [PubMed] [Google Scholar]
- Zhou X-Y, Hu X-X, Li M-F, Wang H, Zhang L-Q, Hu G-X, Cai J-P. 2018. Functional characterization of CYP2C19 variants in nebivolol 4-hydroxlation in vitro. Drug Test Anal. 10(5):807–813. doi: 10.1002/dta.2334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


