Abstract
Although >98% of our genome is noncoding1, nearly all drugs on the market target one of ~700 disease-related proteins. The historical reluctance to invest in noncoding RNA stems partly from requirements for drug targets to adopt a single stable conformation2. Most RNAs can adopt several conformations of similar stabilities. RNA structures also remain challenging to determine3. Nonetheless, an increasing number of diseases is now being attributed to noncoding RNA4 and the ability to target them would vastly expand the chemical space for drug development. Here we devise a screening strategy and identify small molecules that hit the non-coding RNA prototype, Xist5. The X1 compound has drug-like properties and binds specifically to Xist’s RepA motif6 in vitro and in vivo. SAXS analysis reveals that RepA can adopt multiple conformations but favors one structure in solution. X1 binding reduces RepA’s conformational space, displaces cognate interacting protein factors (PRC2, SPEN), suppresses H3K27 trimethylation, and blocks initiation of X-inactivation. X1 inhibits cell differentiation and growth in a female-specific manner. Thus, RNA can be systematically targeted by drug-like compounds that disrupt RNA structure and epigenetic function.
The large non-coding transcriptome and its potential to encode disease variants make RNAs appealing drug discovery targets. The quest to find RNA-binding small molecules has intensified in recent years7,8,3. Early examples did not have good drug-like properties, as they tend to be positively charged compounds with low selectivity, poor cellular uptake, and high toxicity3. More promising have been recent examples such as Ribocil (flavin mononucleotide riboswitch inhibitor)9 and Evrysdi (splice modulator for spinal muscular atrophy)10. However, these examples were discovered in phenotypic screens and only later determined to be RNA interactors. Currently lacking are scalable methods to screen RNA-targeting compounds. Although rational design by exploiting structural motifs is becoming more feasible11, tridimensional structures for RNA are rarely available. To overcome conventional limitations, we devised an unbiased screen based on affinity-selection mass spectrometry. The “Automated Ligand Identification System” (ALIS)12 is a biophysical binding technology that identifies reversibly binding ligands with better than 10 μM affinities. ALIS includes a fast size-exclusion chromatography (SEC) step to isolate the target-ligand complex away from unbound components and then employs LC-MS to release, separate, and identify the bound ligand (Fig. 1b, bottom). Although ALIS has been used to identify protein hits, its potential for RNA drug discovery is under-explored13,14.
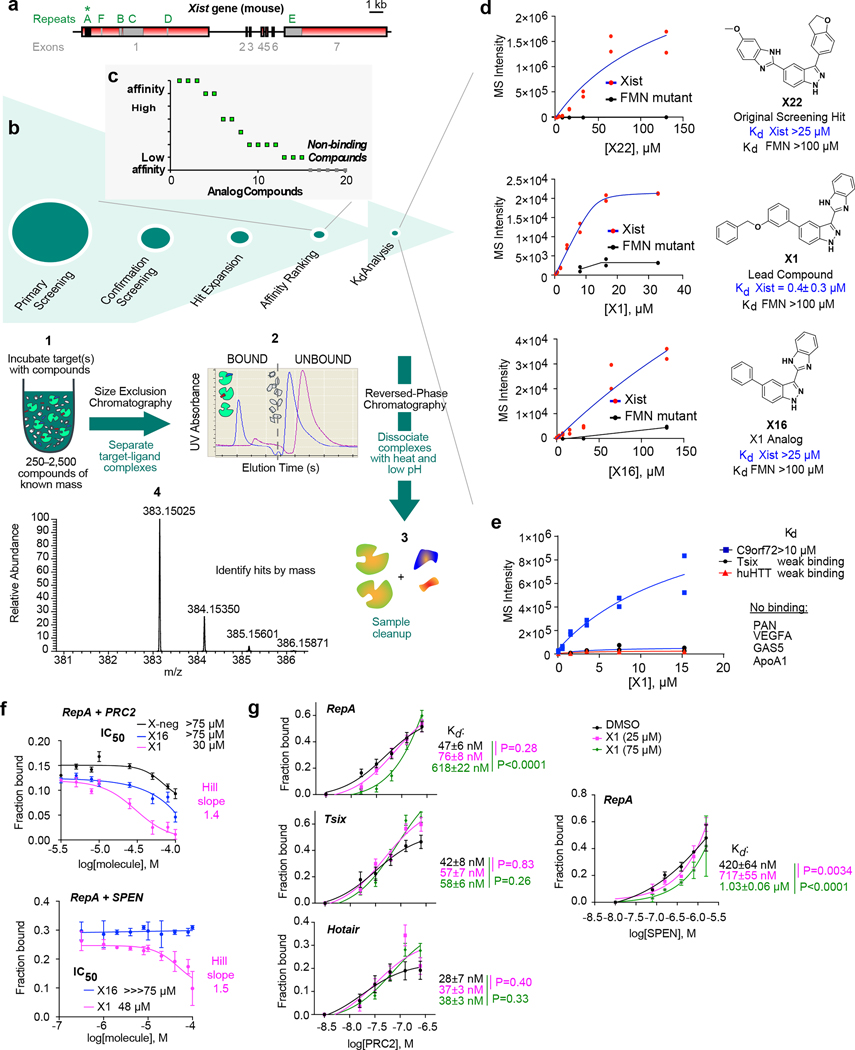
Figure 1: X1 binds Xist RepA and weakens RNA binding to interacting protein partners in vitro.
a, The mouse Xist gene with conserved repeat motifs (A-F) and exons (1–7).
b, ALIS screening of 50,000 diverse small molecule compounds against RepA. (1) Compound mixture is equilibrated with target, (2) target-ligand complex is separated from unbound ligands by SEC, (3) RP-HPLC dissociates bound ligands and purifies sample, (4) MS identifies ligand by mass. Confirmation Screening: Single hit identified (X22) and verified for target binding in ALIS.
c, Hit Expansion around the original hit, X22. 20 analogs were identified with 70% similarity from the complete Merck collection, and Affinity Ranking was performed. y axis represents the relative binding affinity of each compound to RepA as determined by ALIS following the MS step.
d, The original hit X22, X1 from expansion series, and X16 from synthetic analogs shown with affinities for RepA. Negative control: FMN Δ94–102 riboswitch9. Data are represented as mean +/− S.E.M. n=2 independent experiments.
e, X1 affinity for PAN, VEGFA, GAS5, ApoA1, C9orf72, Tsix, and huHTT RNAs.
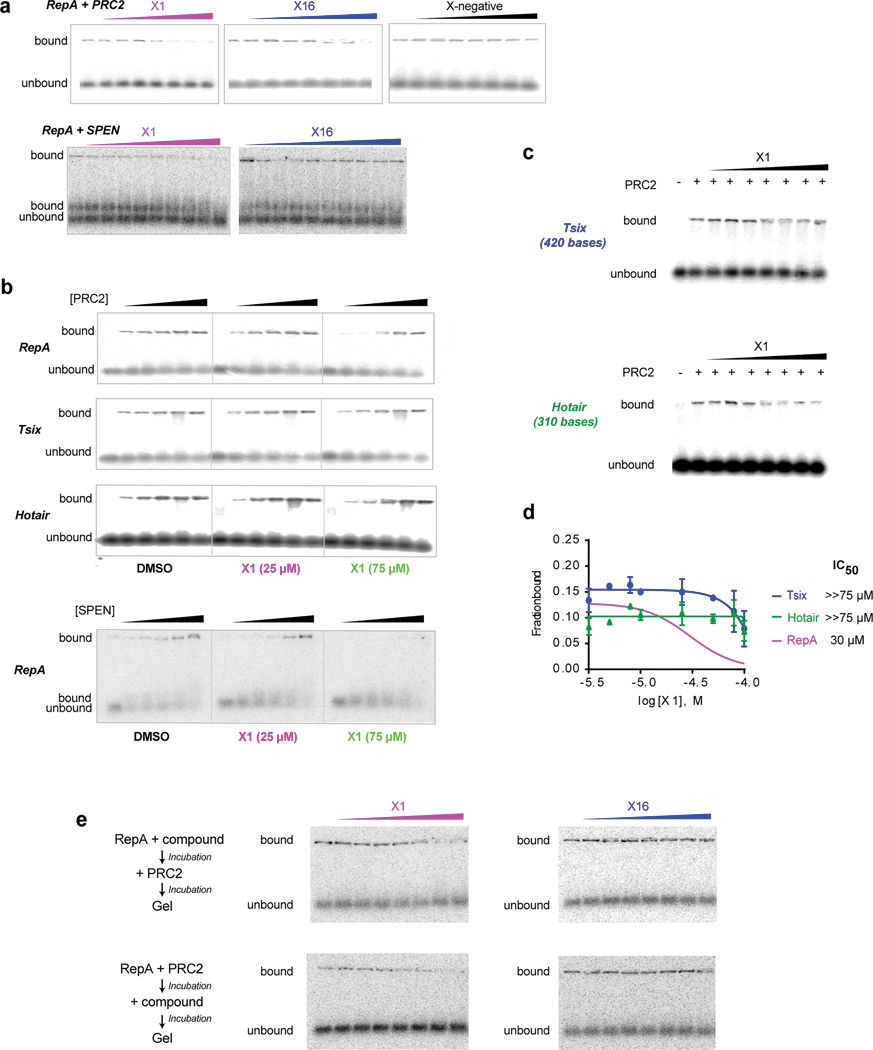
f, X1 weakens RepA-PRC2 and RepA-SPEN interactions. Densitometric analysis of EMSAs (Ext. Data-Fig. 2a) to determine IC50 and Hill coefficient. Increasing concentrations of indicated compounds (0, 5, 7.5, 10, 25, 50, 75, 100 μM) were titrated against 0.5 nM RNA and 15.6 nM PRC2, or 0.1 nM RNA and 158 nM SPEN-RRM. Mean +/− S.D. shown. n=3 independent experiments.
g, Quantitation of RNA-EMSAs (Ext. Data-Fig. 2b) titrating PRC2 (0, 15.6, 31.2, 62.4, 124.9, 250 nM) or SPEN-RRM (0, 79.2, 158, 396, 792, 1580 nM) against 25 or 75 μM X1. [RNA], 0.5 nM. Mean +/− S.D. shown. P-values: one-way ANOVA with Dunnet’s post-test comparing all conditions to control. n=3 independent experiments.
To test applicability to RNA, we conducted a screen using Xist, the 17 kb transcript (Fig. 1a) that initiates X-chromosome inactivation (XCI) as it spreads along the inactive X chromosome (Xi)6,5. Xist’s “Repeat A” (RepA) is a 431-nucleotide domain consisting of 8.5 units of a GC-rich motif responsible for gene silencing6. We synthesized murine RepA RNA15 and performed purification under native conditions to retain secondary structures16 (Ext. Data-Fig. 1). Using ALIS, we screened 50,000 compounds representing Merck’s chemical diversity collection (Fig. 1b) and obtained one positive hit, X22, with a molecular weight of 382 Da. X22 exhibited reasonable pharmacological properties and “drug-likeness”17. None of 42 control RNAs14 of varying lengths and GC content demonstrated binding to X22, including multiple other GC-rich RNAs (Ext. Data-Table 1). We carried out an expansion series around X22 for compounds with >70% structural similarity using Tanimoto scoring and identified 20 analogs (Fig. 1c; Ext. Data-Table 2). ALIS confirmed binding via competition studies. We rank-ordered their relative affinities for RepA in order to prioritize candidates. Among the 20 expanded compounds, 5 compounds did not bind appreciably (Fig. 1d, grey boxes) and were not pursued further. The remaining 15 demonstrated a spectrum of binding affinities (Fig. 1d; Ext. Data-Table 2).
One compound, X1, has a unique indazole-benzimidazole scaffold and emerged as the highest affinity binder (Kd = 0.4 ± 0.3 μM; Fig. 1d). Like X22, X1 displayed drug-likeness, with a molecular weight of 416 Da, 4 hydrogen bond donors, and 1 hydrogen bond acceptor. X1 has a low percent effect of −11.7 at 40 μM in a Thiazole Orange displacement assay, indicating that it is not a nucleic acid intercalator. X1 did not appreciably bind other tested RNAs, including bacterial FMN Δ94–102 mutant riboswitch RNA9 (Kd >>100 μM; Fig. 1d, black line) and various mammalian RNAs of similar size such as PAN, VEGFA, GAS5, huHTT, and APOA1 (Fig. 1e; Ext. Data-Table 1). Xist’s reverse complement (Tsix) and CG-rich C9orf72 (which nonspecifically binds a range of compounds14) also showed weak affinity for X1 (Kd >10 μM; Fig. 1e). Thus, X1 has high selectivity for RepA.
To determine if X1 affects RepA function, we examined its interaction with cognate protein partners that facilitate Xist spreading and gene silencing. RepA binds Polycomb repressive complex 2 (PRC2), an epigenetic complex that trimethylates histone H3 at lysine 27 (H3K27me3)15, and the RNA recognition motif (RRM) of SPEN, a megadalton RNA-binding protein that interacts with histone deacetylases18. Using purified PRC2 (subunits EZH2, SUZ12, EED, and RBAP48) and SPEN-RRM, we assessed whether X1 could disrupt RepA-PRC2 and RepA-SPEN interactions in electrophoretic mobility shift assays (EMSAs). In an X1 titration series (0–100 μM) against fixed concentrations of RepA RNA, PRC2, and SPEN-RRM, we observed progressively disruptive effects on RepA-PRC2 interactions, with an IC50 of 30 μM; similarly, X1 also disrupted RepA-SPEN interactions, but with an IC50 of 48 μM (Fig. 1f; Ext. Data-Fig. 2a). With Hill coefficients of 1.4–1.5, X1 may exert cooperative binding, suggesting that more than one X1 molecule binds one RepA.
To perform structural activity relationship (SAR) studies, we synthesized analogs around X1. Among them, X16 lacks the benzyloxy group but still bound RepA. Consistent with its much lower affinity of 25 μM (Fig. 1d), X16 was also able to reduce RepA-PRC2 and RepA-SPEN interactions, but a higher compound concentration was required (IC50 > 75 μM)(Fig. 1f). We also tested a negative control compound (“X-negative”) known to bind influenza A virus RNA promoter19. Although X-negative bound similar size RNAs in other screenings14, it could not bind RepA and, accordingly, could not disrupt RepA-PRC2 interactions (Fig. 1f; Ext. Data-Fig. 2a).
To quantify disruptive effects, we measured Kd changes of RepA for PRC2 or SPEN in the presence of X1. Without X1, RepA bound PRC2 at 47 nM, whereas RepA bound SPEN with a 10-fold lower affinity of 420 nM (Fig. 1g; Ext. Data-Fig. 2b). Addition of 25 μM X1 only modestly reduced the RepA-PRC2 affinity to 76 nM, and likewise modestly reduced the RepA-SPEN affinity to 717 nM (Fig. 1g; Ext. Data-Fig. 2b). However, addition of 75 μM X1 caused a significant reduction of RepA’s affinity to 618 nM for PRC2 and 1.3 μM for SPEN. On the other hand, X1 had no significant effect on binding to other PRC2-interacting RNAs, including Hotair and Tsix, even at 75 μM X1 (Fig. 1g; Ext. Data-Fig. 2b–d). X1’s effect was not affected by order of addition, as preincubating RepA with either X1 or PRC2 did not change the results (Ext. Data-Fig. 2e). Thus, X1 selectively inhibits RepA’s interactions with binding partners in vitro.
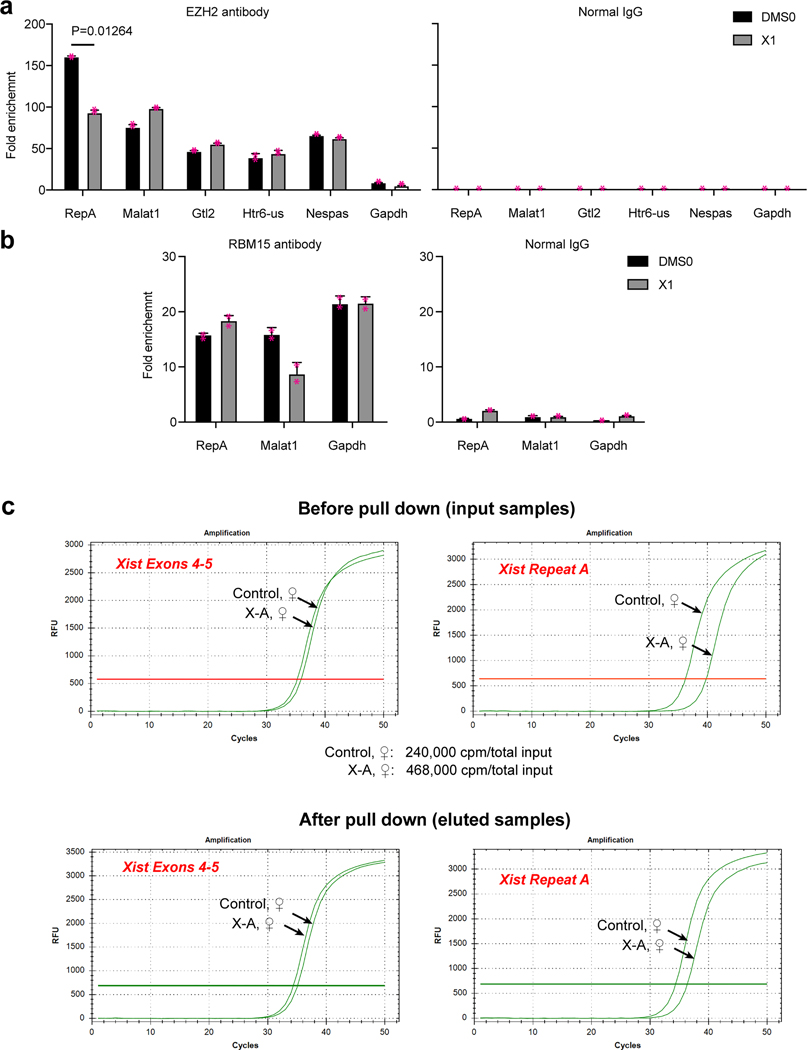
To assessed X1 effects inside cells, we performed RNA immunoprecipitation (RIP) using anti-SUZ12 or -EZH2 antibodies to pull down PRC2-interacting RNAs and conducted RT-qPCR to quantitate co-eluted RNAs. The female mouse embryonic stem (ES) cell model (TsixTST/+, henceforth TST20) initially carry two active X’s (Xa) but recapitulate XCI when induced to differentiate into embryoid bodies (EB) for 4 days (Fig. 2a). We added 10 μM X1 on day 2, performed RIP on day 4, and observed that, Xist recovery was reduced to 30–40% of control in multiple biological replicates (Fig. 2b). In contrast, other PRC2 interactors21 such as Malat1, Gtl2, Htr-us, or Nespas were not affected by X1 (Fig. 2b, Ext. Data-Fig. 3a). Non-interacting RNAs such as Gapdh21 were not immunoprecipitated, and X1 treatment had no effect on this outcome. Moreover, RIP using negative control IgG antibodies gave undetectable signals (Fig. 2b,c and Ext. Data-Fig.3a). We also performed RIP using anti-SPEN antibodies and observed that X1 reduced RepA–SPEN interaction in vivo (Fig. 2b). Notably, X1 exerted a stronger inhibitory effect on SPEN than PRC2, consistent with RepA’s differential affinities for X1 (Kd = 400 nM), PRC2 (47 nM), and SPEN (420 nM). Because RepA binds PRC2 more avidly than it does SPEN or X1, RepA-PRC2 interactions are less inhibited by X1 than RepA-SPEN interactions. Xist RNA interacts with other protein partners through other domains, including CIZ1 and RBM1522,23. The m(6)A RNA methyl-transferase, RBM15, binds near RepA22 and, interestingly, X1 did not disrupt RMB15-RepA interactions (Ext. Data-Fig.3b). Nor did X1 affect Xist-CIZ1 interactions (Fig. 2b), which occur via Xist exon 7 within Repeat E23. Non-interacting RNAs such as Malat1 and Gapdh were not pulled down by CIZ1 or RBM15 antibodies. Treating cells with X-negative also had no effect on Xist-PRC2 interactions (Fig. 2c). X1 can therefore selectively disrupt Xist’s interaction with cognate proteins in vitro and inside cells.
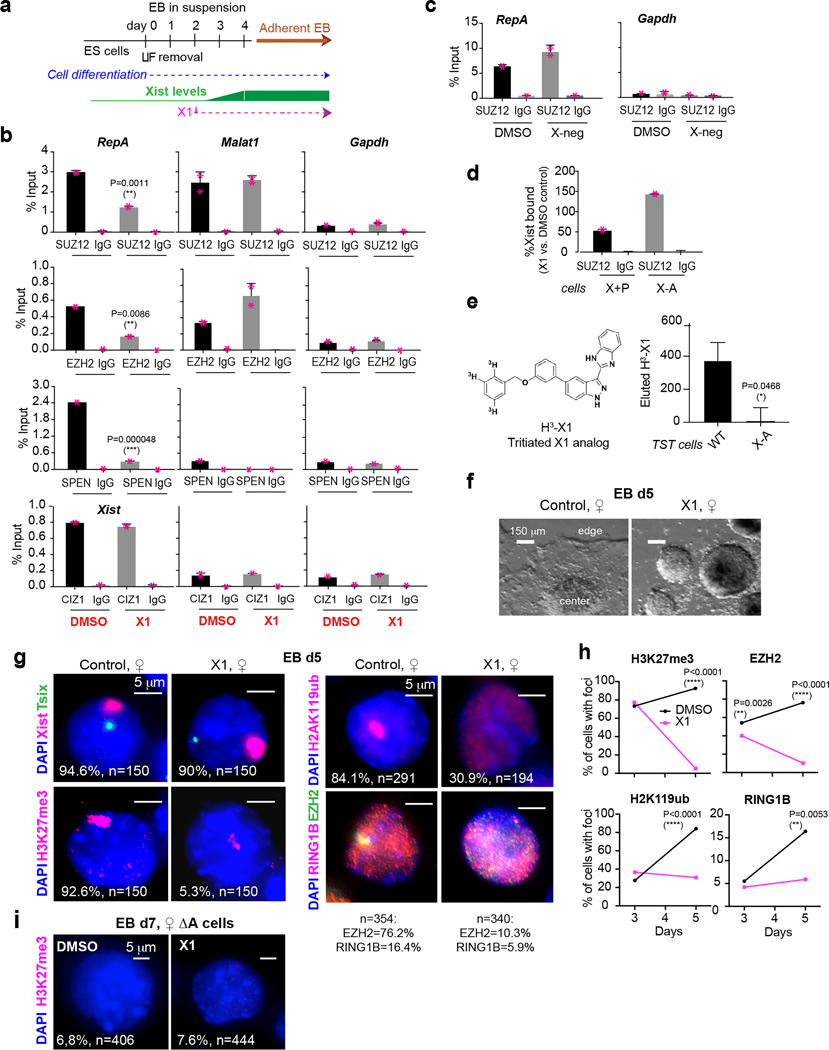
Figure 2: X1 weakens Xist’s interaction with PRC2 and SPEN in a RepA-dependent manner inside cells.
See also Ext. Data-Fig. 4–6.
a, Analysis of XCI effects using the schematic shown. 10 μM X1 was added at day 2 and replenished daily with medium change. EBs grown in suspension, followed by adherent culture. All experiments performed in 3 biological replicates (n=3) unless otherwise specified.
b,c, RIP-qPCR in d4 female TST ES cells to evaluate Xist binding to SUZ12, EZH2, SPEN, and CIZ1, as indicated, +/− X1 (b) or X-negative (X-neg) control compound (c). IgG, control antibodies. Bars: %input (mean ± S.D). P-values: two-tailed Student’s t-test. Individual data points shown. Malat1 and Gapdh: negative control RNAs.
d, RIP-qPCR in fibroblasts over-expressing transgenic Xist (X+P) or a RepA-deleted version (X-A). Bars: %Xist (mean +/− S.D.) recovered with X1-treatment relative to DMSO. n=2 biological replicates.
e, Validation of target engagement using [3H]X1. Bars: cpm (mean +/− S.D.). P-value: two-tailed Student’s t-test.
f, X1 inhibits growth of day 5 differentiating ♀-TST cells. Scale bar, 150 μm. One representative field shown.
g, Xist/Tsix RNA-FISH and immunostain for H3K27me3, H2AK119ub, EZH2, and RING1B in ♀-TST EB at day 5. One representative nucleus is shown. %cells with Xist foci is indicated. n, sample size. Scale bar, 5 μm.
h, Enrichment of indicated protein epitopes across time (immunostain). P-values: two-tailed Chi-square test.
i, X1 inhibition depends on RepA, shown by H3K27me3 immunostaining in day 7 ♀-TST-ES cells lacking RepA (ΔA). %cells with H3K27me3 enrichment is indicated. n, sample size from two biological replicates. Scale bar, 5 μm.
To validate target engagement inside cells, we reasoned that if X1’s effect was mediated by interaction with RepA, deleting RepA would eliminate the effect. We compared RIP results in fibroblasts expressing either a wildtype Xist transgene (X+P cells)24 or a version lacking RepA (X-A cells)25. In 10 μM X1, SUZ12 RIP recovered significantly less Xist RNA from X+P cells relative to DMSO-treated cells (Fig. 2d; incomplete abrogation is consistent with there being other Xist domains (e.g., Repeat B) involved in recruiting PRC225). X1’s inhibitory effect was lost entirely when RepA was deleted (Fig. 2d), indicating that X1 indeed acts through RepA. To further validate target engagement, we synthesized a tritiated analog of X1 (Fig. 2e, [3H]X1) and performed RNA pulldown assays using biotinylated probes to capture Xist in TST cells expressing either a wildtype Xist allele (WT) or one lacking RepA (X-A)25. When [3H]X1 was added on day 3, pulldown assays on day 4 showed that radioactive X1 was recovered only from WT cells (Fig. 2e). Because X1 treatment had little effect on steady state Xist levels before or after pulldown (Ext. Data-Fig. 3c), the absence of [3H]X1 counts in the X-A cells could not be attributed to absence of Xist RNA. The overall RepA-dependence argues that X1 selectively binds RepA inside cells.
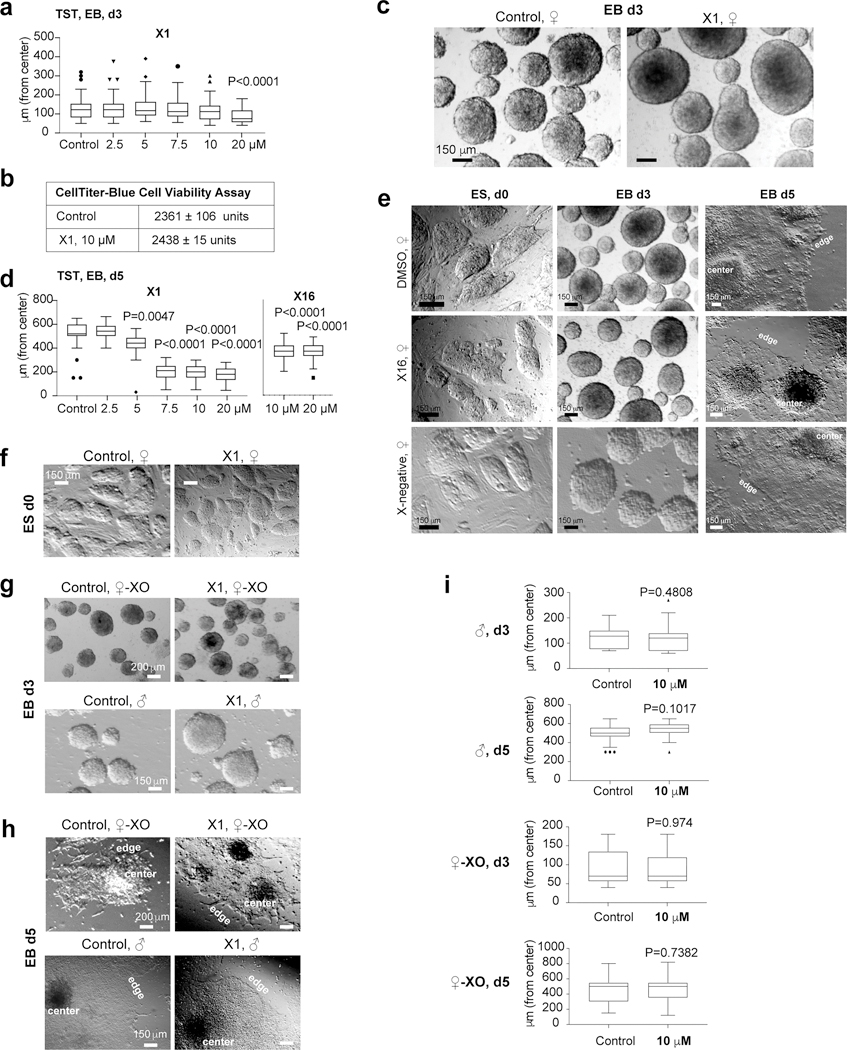
We asked whether X1’s interaction with RepA affected XCI. To determine appropriate compound concentrations, we conducted dose-response assays (0–20 μM). Female ES cells showed no growth retardation or inviability when treated with ≤10 μM X1 for 24 hours (Ext. Data-Fig. 4a,b). We therefore performed phenotypic assays at 10 μM. When differentiated in the presence of 10 μM X1 for 24 hours (day 3), female EB remained healthy (Ext. Data-Fig. 4c). However, prolonged exposure to day 5 resulted in poor EB outgrowth (Fig. 2f). Reducing X1 to 7.5 μM yielded the same result (Ext. Data-Fig. 4d). The less potent analog, X16, also suppressed EB outgrowth at 10 μM, but to a lesser extent, whereas X-neg had no effect at all (Ext. Data-Fig. 4d,e), thereby correlating compound potency with EB growth suppression. X1’s effect is Xist-dependent, as pre-XCI (day 0) female ES cells, as well as days 3–5 XY male and XO female cells, all grew normally when treated with X1 (Fig. 2g; Ext. Data-Fig. 4f,g and 5a). Thus, X1 exerts a phenotypic effect on EB outgrowth. This effect is Xist-dependent and observed only in XX female ES cells.
To determine if female-specific effects resulted from aberrant XCI, we performed RNA fluorescence in situ hybridization (FISH). In both control and X1-treated cells, Tsix RNA was observed from the Xa5 and Xist RNA (red) was properly upregulated from the Xi at day 3, indicating initiation of XCI (Ext. Data-Fig. 5b,6). Immunostaining for EZH2 and the repressive histone mark, H3K27me3, showed proper PRC2 enrichment on the Xi in 40% cells and H3K27me3 enrichment in 70–80% at 24 hours of X1 treatment, similar to those for controls (Ext. Data-Fig. 5b,6). However, X1 exposure for 72 hours to day 5 resulted in a strong effect on PRC2 recruitment. Whereas Xist foci continued to be observed in X1-treated cells, PRC2 and H3K27me3 enrichment was severely blunted (P<0.01; Fig. 2g,h). Only 10.3% and 5% of treated cells showed enrichment, respectively, while >76.2% and >92% of control cells displayed enrichment. These data indicate that X1 requires 1–3 days to take effect, as is characteristic of some pharmacological agents. A 3-way interaction between Xist RNA, PRC2, and Polycomb repressive complex 1 (PRC1) is required for spreading along the Xi25. Interestingly, X1 treatment also did not affect enrichment of PRC1 (RING1B subunit) and associated H2AK119ub histone marks at day 3, but significantly compromised their enrichment at day 5 (Fig. 2g,h; Ext. Data-Fig. 5b,6). Notably, in female ES cells lacking RepA (ΔA)25, immunostaining showed that H3K27me3 enrichment on the Xi was severely blunted in untreated cells and remained low in X1-treated cells (Fig. 2i; Ext. Data-Fig. 6b), further affirming that X1 action depends on RepA. Importantly, unlike EPZ-6438 and PF-06821497 (two known EZH2 inhibitors), X1 is not a general PRC2 inhibitor, as bulk H3K27me3 in cells was unchanged (Ext. Data-Fig. 6c). Altogether, these findings demonstrate that X1 does not affect Xist expression but instead affects Xist’s ability to recruit interacting proteins. X1 thereby uncouples Xist expression from Polycomb function in cells.
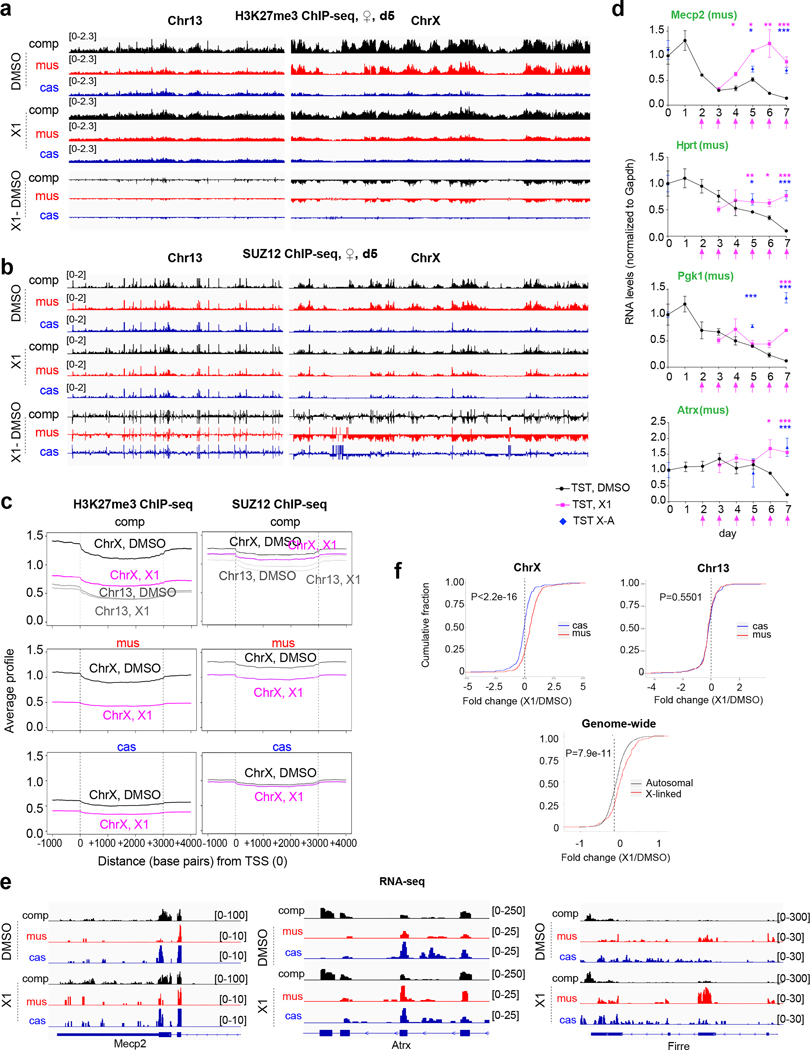
To obtain a high-resolution view and assess potential off-target effects, we performed allele-specific ChIP-seq in hybrid ES cells to distinguish Xa from Xi. Due to a Tsix mutation20, TST cells invariably inactivate the X-chromosome of Mus musculus (Xmus) origin and maintain the X of Mus castaneus (Xcas) origin as Xa. Whereas DMSO-treated cells showed PRC2 and H3K27me3 enrichment on Xmus, X1-treated cells (10 μM) failed to do so (Fig. 3a,b; Ext. Data-Fig. 7a,b). Specific X-linked genes, including Xist, displayed reduced H3K27me3 (Ext. Data-Fig. 7c,d). In contrast, genes that escape XCI and autosomal genes were not affected (Ext. Data-Fig. 7e,f). Metagene analysis showed that X1 treatment resulted in strongly inhibited H3K27me3 and PRC2 accumulation specifically on the Xi (mus), while the Xa and autosomes were minimally affected (Fig. 3c; Ext. Data-Fig. 7g). Notably, although 30–48 μM (IC50) of X1 was required to disrupt RepA-protein interactions in vitro (Fig. 1f), cellular effects were observed at 5 μM. Cells may concentrate X1 intracellularly, as quantitation using radio-labelled X1 (Fig. 2e) showed 180-fold higher concentrations inside cells (Ext. Data-Table 3). Thus, X1 shows strong on-target effects on PRC2 recruitment to the Xi.
Figure 3: X1 treatment leads to Xi-specific loss of PRC2 and H3K27me3 enrichment and failure of XCI.
a,b, Allele-specific H3K27me3 (a) and SUZ12 (b) ChIP-seq analyses of day 5 female EB treated with 10 μM X1 or DMSO (control) for 72 h. Tracks for all reads (composite, “comp”), mus (Xi), and cas (Xa). X1–DMSO, subtraction of X1 reads from DMSO reads. Chr13 and ChrX shown, with a sliding window of 100 kb, step size 50 kb. FPM scale shown in brackets.
c, Metagene analysis of the average Xi gene, Xa gene, and Chr13 gene with or without X1 treatment.
d, Time course allele-specific RT-qPCR of indicated X-linked genes in DMSO- or X1-treated female EB. TST X-A cells analyzed on days 5 and 7 phenocopy X1-treatment. Mus allele (Xi) shown. X1 added on indicated days (pink arrows). P-values from two-tailed Student’s t-test compared to DMSO control. *, P<0.05. **, P<0.01. ***, P<0.001. Mean and S.D. shown for 3 biological replicates.
e, RNA-seq analyses of day 5 DMSO- or X1-treated female EB. Tracks shown for all reads (comp), mus (Xi), and cas (Xa). FPM scale shown in brackets.
f, Cumulative distribution plots of fold-changes in gene expression in X1- versus DMSO-treated EB. Mus, Xi. Cas, Xa. ChrX and Chr13 shown. Genes with RPKM>1 were included in analysis. P-values: two-tailed Wilcoxon test.
Allele-specific RT-qPCR showed that X1 did not affect proper Tsix downregulation (Xist repressor5), upregulation of Jpx (Xist activator5), or upregulation of Xist (Ext. Data-Fig. 8a). Nor did it affect downregulation of pluripotency markers (e.g., Klf4 and Sox2), suggesting normal ES cell differentiation. However, while there was also no effect on expression of Xa (Xcas) genes, there was a major effect on silencing of Xi (Xmus) genes between days 0–7 (Fig. 3d, pink line; Ext. Data-Fig. 8b). The effect was dose-dependent between 0–10 μM X1, with 10 μM resulting in full inhibition (Ext. Data-Fig. 8c). Interestingly, deleting RepA phenocopied X1 treatment (Fig. 3d, blue dots), further arguing that X1 effects are mediated through RepA. The effects were also reversible, as Xi silencing and EB growth resumed when X1 was withdrawn (Ext. Data-Fig. 8d,e). We conclude that X1 blocks PRC2 recruitment and Xi silencing in a RepA-dependent manner.
Allele-specific RNA-seq analysis verified the loss of Xi silencing. While control cells showed Xi silencing by day 5, X1-treated cells retained high-level Xi expression despite proper Xist upregulation (Fig. 3e; Ext. Data-Fig. 9a,b). Escapees and autosomal genes were not affected (Ext. Data-Fig. 9c,d). Cumulative distribution plots (CDPs) confirmed a significant shift in Xi expression relative to Xa expression in X1-treated cells (Fig. 3f, upper left panel). Increased expression of Xi genes was also reflected in the right shift for X-linked genes relative to autosomal genes and to the genome (Fig. 3f bottom). In contrast, there was no allele-specific effect on a representative autosome (Fig. 3f upper right panel). These data further support X1’s on-target effect on Xi genes.
Although autosomes were unaffected in bulk, off-target changes could occur at individual genes. We therefore performed DE-seq analysis and found that 197 autosomal genes showed significant changes, of which 141 were upregulated and 56 downregulated (P<0.05). These changes could result directly from X1 treatment (true “off-target” effect) or indirectly from X-linked changes (“secondary effect”). To distinguish between them, we correlated the changes with H3K27me3 alterations, reasoning that direct effects would be accompanied by H3K27me3 decreases. Among the 197 affected autosomal genes, only 19 showed upregulation accompanied by H3K27me3 decrease (Quadrant II: Ext. Data-Fig. 9e, Ext. Data-Table 4), and only 9 showed downregulation with increased H3K27me3 (Quadrant IV). Quadrants I and III also contained only 14 and 1 genes, respectively. Therefore, autosomal effects were small relative to Xi effects (190 of 335 total X-linked genes expressed in ES cells)(Ext. Data-Table 5) and most may be secondary effects of Xi changes.
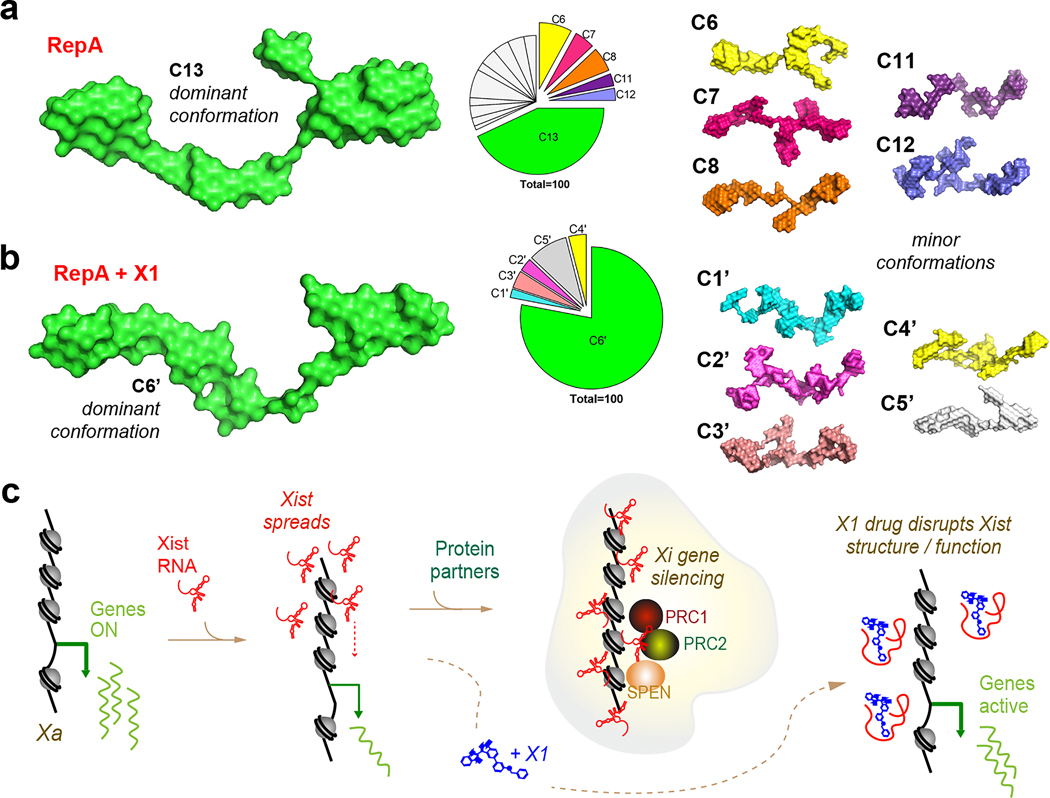
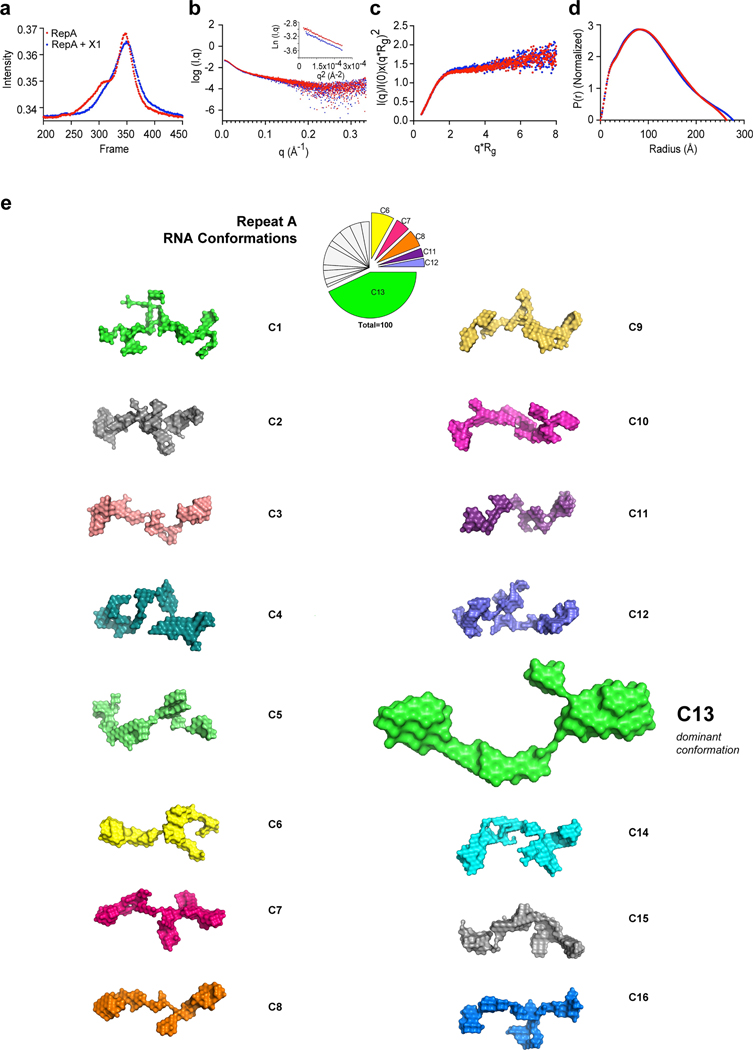
A priori, X1’s effect on RepA could occur either through steric blockade or a change in RNA conformation. Because X1 affects RepA’s interaction with both PRC2 and SPEN, we favored the latter hypothesis. RNA can generally fold into many stable conformers, perhaps explaining why chemical probing of RepA’s structure has yielded no consensus so far26. Here, we employed small-angle X-ray scattering (SAXS), which uses nanoscale density differences to determine size and shape distributions of biomolecules27, in line with HPLC to collect monodispersed samples without aggregation and degradation (Ext. Data-Fig. 10a). Quality analysis in PRIMUS showed absence of upturned or downturned profile at low q regions indicative that RNAs were free of aggregation or interparticulate interference (Ext. Data-Fig. 10b). The lower q region was used to generate the Guinier plot (Ext. Data-Fig. 10b, inset) and estimate the Radius of Gyration (Rg). Free RepA demonstrated an Rg of 83.11±0.37 Å, and X1-treated RepA showed an Rg of 83.63±2.67 Å, suggesting similar overall dimensions and agreeing with HPLC data where RepA yielded similar elution profiles with or without X1 (Ext. Data-Fig. 10a). Dimensionless Kratky analysis showed a sharply bent profile (Ext. Data-Fig. 10c), implying that the RNA is folded in solution, similar to previous reports28. Pairwise distance distribution profile (P(r)) yielded a real space Rg for RepA versus RepA+X1 of 84.58 ± 0.17 and 85.66 ± 0.25 Å, respectively (Ext. Data-Fig.10d), in close agreement with Guinier analysis (Ext. Data-Fig. 10b, inset). P(r) also estimated the maximum dimension (Dmax) of 263 and 277 Å for RepA and RepA+X1, respectively (Ext. Data-Fig. 10d), hinting at an underlying conformational change.
To determine the tridimensional structure of RepA in solution with and without X1, we used ab initio modeling package DAMMIN. To reveal all possible conformations, 100 models were generated independently for each condition, followed by clustering of similar conformations using DAMCLUST. For RepA alone, although 16 conformation clusters were identified (C1-C16, Fig. 4a, Ext Data-Fig. 10e), C13 was dominant (Video 1). These structures are distinct from prior models derived from population-averaging in silico and/or chemical probing methods, which ranged from simple two-hairpin motifs 6 to complex structures involving inter-repeat duplexes26. An advantage of SAXS is the ability to account for conformational polydispersity27. The existence of 16 RepA conformers may explain why existing chemical studies have reached no consensus for the structure of RepA26. Intriguingly, addition of X1 reduced RepA’s conformational space to 6, with one dominant form, C6’, that is distinct from all prior RepA structures (Fig. 4b; Video 2). C6’ accounted for 78% of all possible conformations. Thus, SAXS analysis has enabled a first 3D solution-based visualization of RepA and revealed that X1 reduces its conformational heterogeneity and induces formation of new structures. We propose that X1 acts on RepA by stabilizing a de novo conformation with reduced affinity for cognate interactors, explaining the loss of XCI in X1-treated cells.
Figure 4: 3D structure of RepA, with and without X1.
a, 16 representative conformations of RepA (C1-C16). C13 (43% of species) is enlarged. Pie-chart shows relative abundance of structural clusters. See Ext. Data-Fig.10 for all 16 conformations.
b, 6 representative conformations of RepA with X1 (C1’-C6’). C6’ (78% of species) is magnified. Pie-chart shows relative abundance of structural clusters.
c, XCI and the effects of X1 treatment.
Here, we have shown that non-coding RNA can be systematically targeted by a small molecule for phenotypic impact. X1 engages Xist’s RepA motif, disrupts its conformation, displaces interacting proteins (PRC2, PRC1, SPEN), and abrogates XCI, intriguingly without affecting Xist expression or localization (Fig. 4c). Medicinal chemistry of X1 could further increase potency and specificity, potentially for Xi-reactivation and treatment of X-linked diseases such as Rett syndrome29. Unlike prior compounds shown retrospectively to bind RNA9,10, our hits arose not from phenotypic screens but from a direct RNA-binding screen. ALIS is agnostic to RNA’s mechanism of action and is potentially applicable to any RNA, including the vast majority which lacks detailed structural information despite most efforts being focused on sequence- or structure-based design30,11. Xist’s large size, multiple modular domains, likelihood of being mostly unstructured, and coexisting structural subpopulations present a unique challenge that was circumvented by ALIS. Interestingly, total hits to RepA were lower than to typical protein targets using ALIS, possibly reflecting that the diversity set was originally designed to hit proteins13. A goal of future research will be development of curated sets with diverse RNA-binding properties to enhance identification of RNA-targeting drugs.
Extended Data
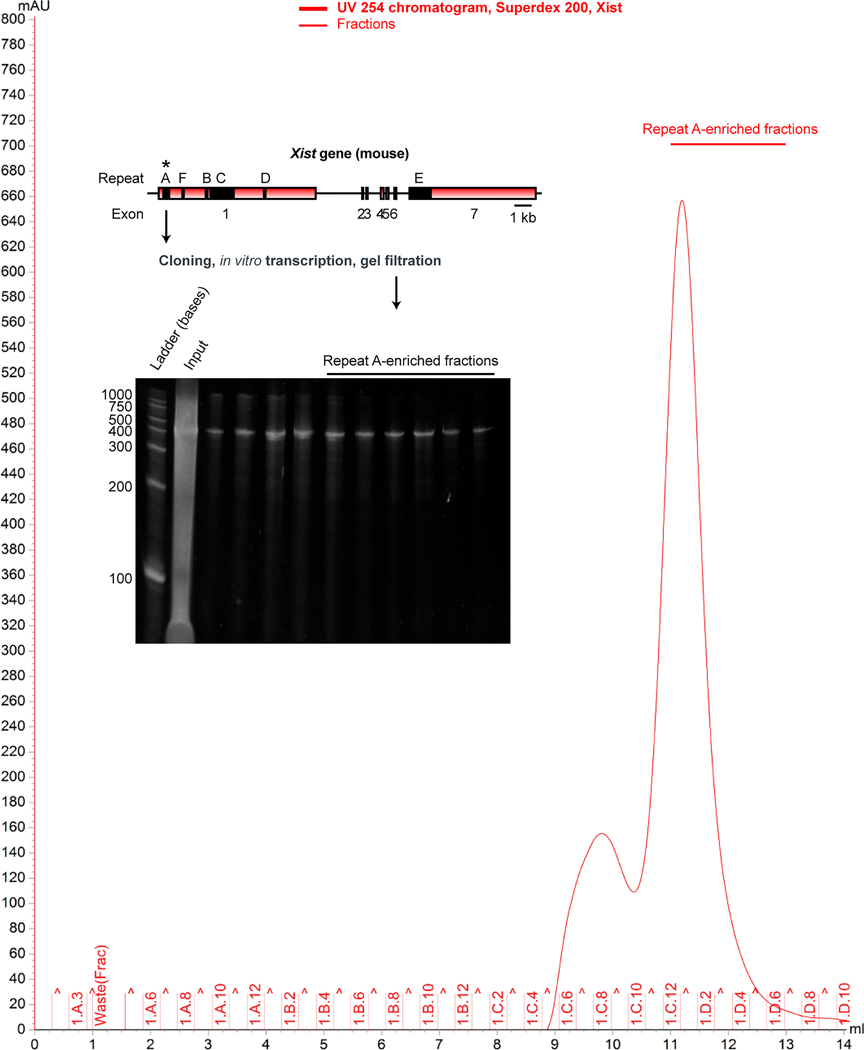
Figure 1: Purification of Xist RepA RNA.
A 431 Repeat A fragment of Xist RNA was in vitro transcribed and purified under native conditions by FPLC. A representative chromatogram is shown. To confirm size and stability of the sample just prior to ALIS, we visualized the RNA in a denaturing urea-PAGE
Figure 2: X1 inhibits interaction of Xist RepA with cognate interacting proteins in vitro.
a, RNA EMSAs show that X1 weakens interaction between RepA and PRC2, and RepA and SPEN-RRM. Increasing concentrations of the compounds (0, 5, 7.5, 10, 25, 50, 75, 100 μM) were titrated against 0.5 nM RNA and 15.6 nM PRC2, or 0.1 nM RNA and 158 nM SPEN-RRM. Two replicates showed similar results.
b, RNA EMSAs titrating PRC2 (0, 15.6, 31.2, 62.4, 124.9, 250 nM) or SPEN-RRM (0, 79.2, 158, 396, 792, 1580 nM) against a fixed concentration of X1 (25 or 75 μM) and 0.5 nM RepA, Tsix (reverse complement of RepA), or Hotair RNA—all of which are known PRC2 interactors. For SPEN, RNA was 0.1 nM. Two or more replicates showed similar results.
c, Increasing concentrations of X1 (0, 5, 7.5, 10, 25, 50, 75, 100 μM) was titrated against 0.5 nM RNA (Tsix, Hotair) and 15.6 nM PRC2. One representative gel of two replicates is shown.
d, Densitometric analysis to determine IC50, which were too high to be measured for Tsix and Hotair. Data are represented as mean +/− SD. n=2 independent experiments. RepA result from Fig. 1f is shown as reference.
e, Order of addition does not affect X activity. Increasing concentrations of the compounds (0, 5, 7.5, 10, 25, 50, 75, 100 μM) was titrated against 0.5 nM RepA and 15.6 nM PRC2. One representative gel of two replicates is shown. Top, PRC2 was added to a RepA-molecule pre-incubated mix. Bottom, Molecule was added to a RepA-PRC2 pre-incubated mix.
Figure 3: X1 also inhibits interaction of Xist RepA with cognate interacting proteins in vivo.
a-b, RIP-qPCR analysis in d4 female TST ES cells to evaluate Xist binding to EZH2 (a) and RBM15 (b) in 10 μM X1. IgG, negative control antibody. Other EZH2 interactors Malat1, Gtl2, Htr6-us and Nespas are shown. Gapdh, negative control RNA. Bars: mean ± S.D. P-value: two-tailed Student’s t-test. Individual data points included. n=2 biologically independent experiments.
c, Top: RT-qPCR confirms similar quantities of Xist RNA in control and X-A samples prior to Xist RNA pulldown. Xist exons 4–5 primers were used. Bottom: Similar quantities were also present following Xist RNA pulldown, thereby ruling out unequal Xist expression as a cause of unequal H3 radioactive counts. X-A cells amplified poorly with RepA primers, consistent with deletion of RepA.
Figure 4: X1 effects on EB outgrowth in ♀- TST-XX, ♀-XO, and ♂-XY EB cells.
a, Growth of differentiating ♀-TST cells at day 3, or 24 h post-X1 treatment, up to 10 μM X1. Data are represented as Tukey box plots. Lower whisker: 25th percentile minus 1.5xInterquartile Range (IQR). Higher whisker: 75th percentile plus 1.5xIQR. Box range: 25th (bottom) to 75th (top) percentile. Line within box: median. Points beyond higher whisker are shown. P-values: one-way ANOVA with respect to control cells. n=150 colonies combined from 3 independent experiments.
b, Viability of d5 cells. n=3 biologically independent experiments
c, No obvious effect on day 3 female EB growth after 24 hours X1 treatment.
d, Quantitation of EB outgrowth at day 5 (72 h post-drug application). The distance from EB center to edge of outgrowth was measured in 100 d3 or 30 d5 EBs combined from 3 independent experiments. Data presented as in panel (a). P-values: one-way ANOVA with respect to control cells.
e, Weaker effect of X16 on ♀-TST EB outgrowth. No obvious effect of X-negative.. One representative brightfield microscopy from 3 independent cultures is shown. Center of the EB and edge of outgrowth as marked. Scale as indicated.
f, X1 had no effect on growth of pre-XCI (d0) female cells.
g,h, X1 also did not inhibit ♀-TST-XO and ♂-XY ES cells at day 3 (g) or day 5 (h). Neither cell line expresses Xist or undergoes XCI. One representative field is shown. Scale bar, 150 μm.
i, Quantitation of EB outgrowth in XY male and XO female EBs at days 3 and 5. Distance from the EB center to the edge of outgrowth was measured. Day 3: n=136, XO colonies; n=112, XY colonies. Day 5: n=40, XO colonies; n=60, XY colonies). Data presented as in panel (a).
Figure 5. Karyotype analysis of ES cells and RNA immunoFISH analysis of day 3 X1-treated cells.
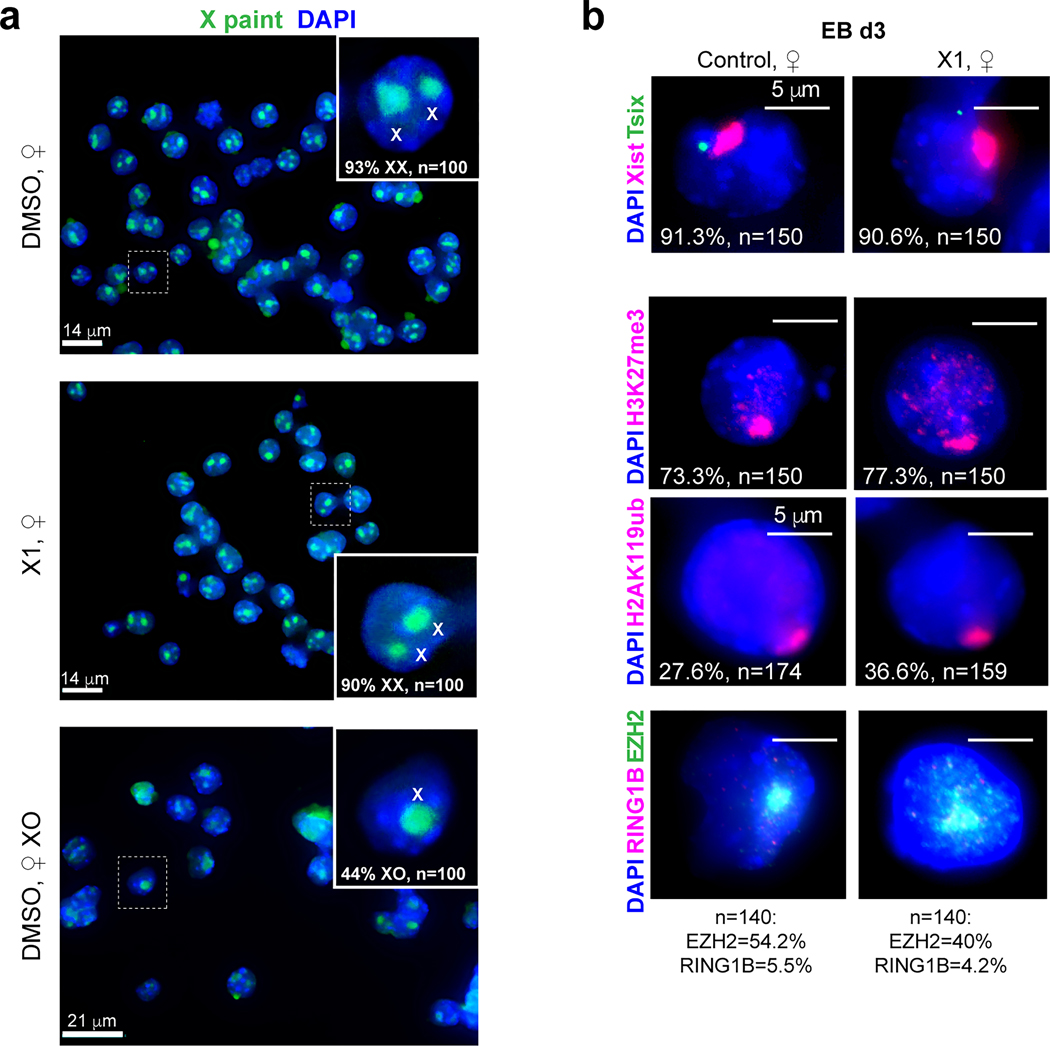
a, X-chromosome painting DNA FISH of DMSO- and X1-treated XX TST cells, and a DMSO-treated XO clone that spontaneously arose from the XX TST cells. Scale as shown. Inset: magnification of representative nucleus. %nuclei with indicated X chromosome number shown. n, sample size combining from 3 biologically independent experiments.
b, Xist/Tsix RNA-FISH and immunostaing for H3K27me3, H2AK119ub, EZH2, and RING1B in ♀-TST EB at day 3. One representative nucleus is shown. %cells with Xist foci is indicated. n, sample size. Scale bar, 5 μm.
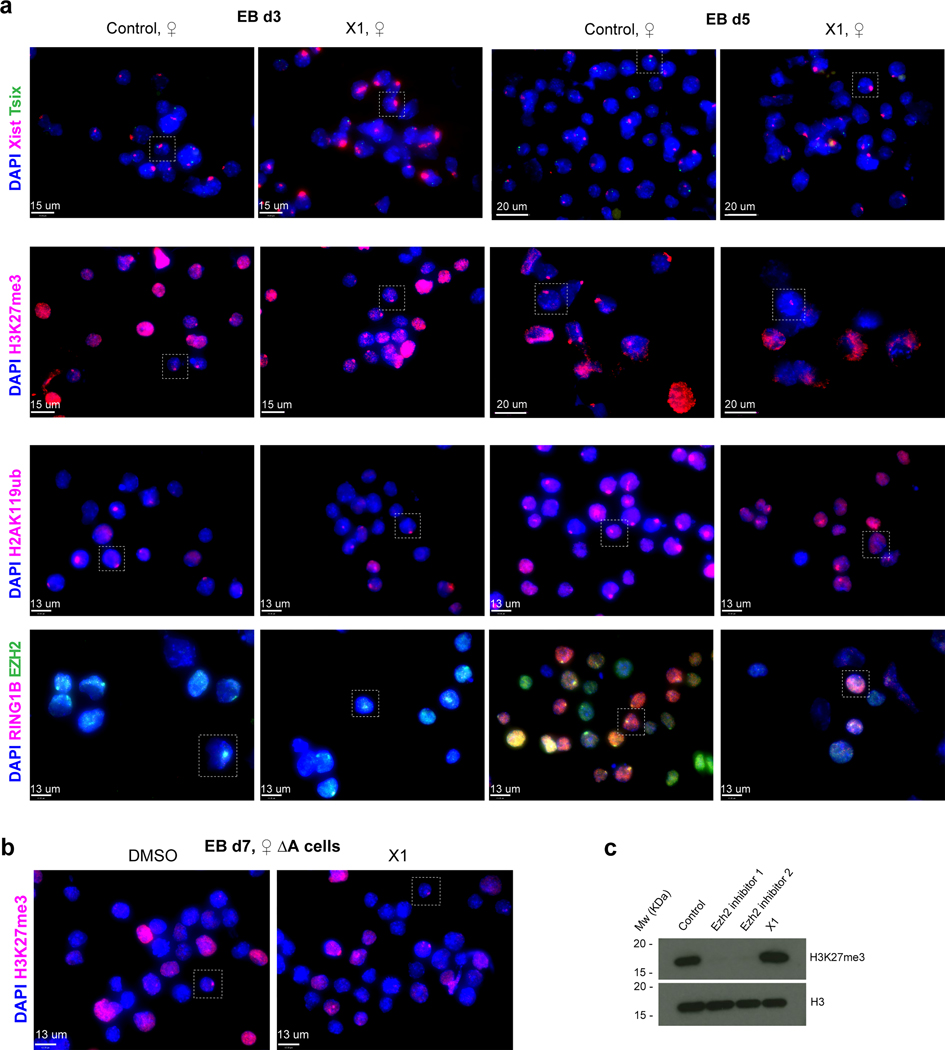
Figure 6: Full fields for RNA immunoFISH experiments of Figure 2 and Extended Data-Figure 5.
a, Full fields for the RNA FISH and Immunofluorescence experiments, with boxed nuclei presented in Figure 2g and Ext. Data-Fig. 5b.
b, Full fields for H3K27me3 immunostaining of DMSO- or X1-treated ♀ TST-A cells, with boxed nuclei presented in Figure 2h. %cells with foci on the Xi as indicated (sample size, n, from two biologically independent experiments combined).
c, X1 does not inhibit PRC2’s catalytic activity. Western blot using H3K27me3 and total histone H3 antibodies. Total cell extracts were obtained from day 7 female EB cells after treating with 10 μM of various compounds from day 2. Compounds: EZH2 inhibitor 1 (EPZ-6438, MedChem Express), EZH2 inhibitor 2 (PF-06821497, Pfizer), or X1. One representative film of two replicates is shown.
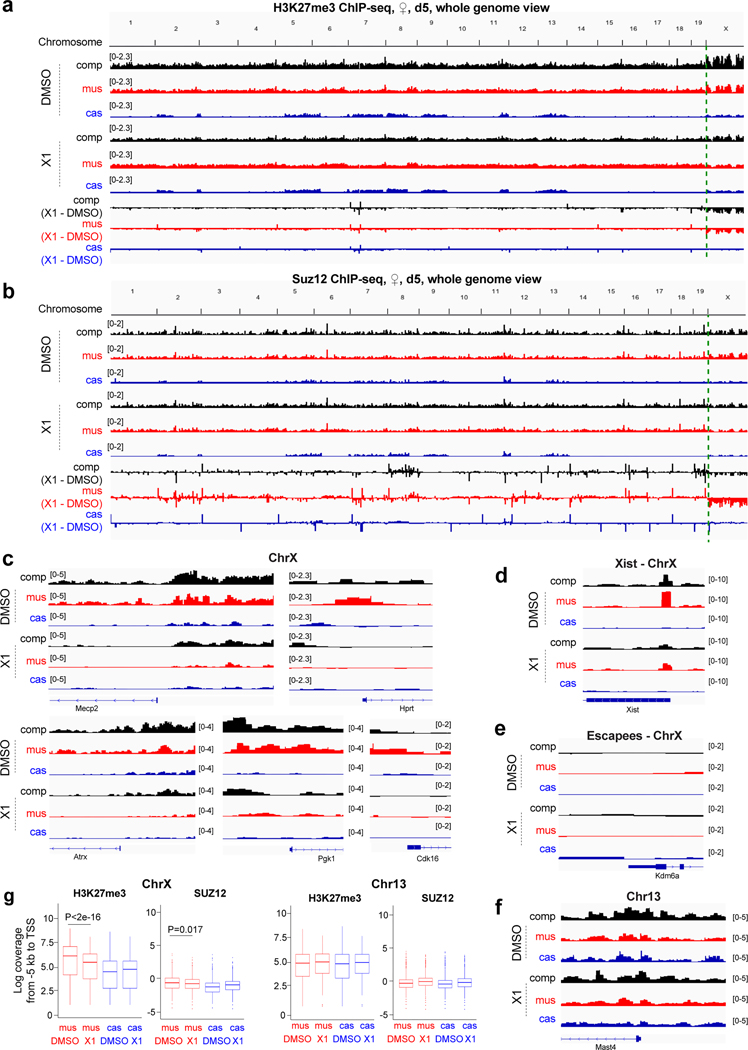
Figure 7: Epigenomic analyses of PRC2 and H3K27me3 enrichment in the presence of X1.
a-b, Allele-specific H3K27me3 (a) and SUZ12 (b) ChIP-seq analyses of day 5 female EB treated with 10 μM X1 or DMSO (control) for 72 h. Tracks for all reads (composite, “comp”), mus (Xi), and cas (Xa). Dotted green lines separate ChrX.
c-f, Zoom-ins for allele-specific H3K27me3 ChIP-seq analyses of day 5 female EB treated with 10 μM X1 or DMSO (control) for 72 h. Browser shots shown with sliding window 1 kb, step size 0.5 kb. Scale shown in brackets. c, X-linked genes subjected to XCI. d, the Xist gene. e, Escapees. f, Representative control autosomal gene on Chr13.
g, Box plot of normalized read densities for the −5000 to +1 region of ChrX and Chr13 refSeq genes, parsed into mus and cas alleles. Lower whisker: 10th percentile. Higher whisker: 90th percentile. Box range: 25th (bottom) to 75th (top) percentile. Line within box: median. Points beyond whiskers are shown. P-values: two-tailed Wilcoxon test from data gathered from individual H3K27me3 and Suz12 ChIP-seq experiments.
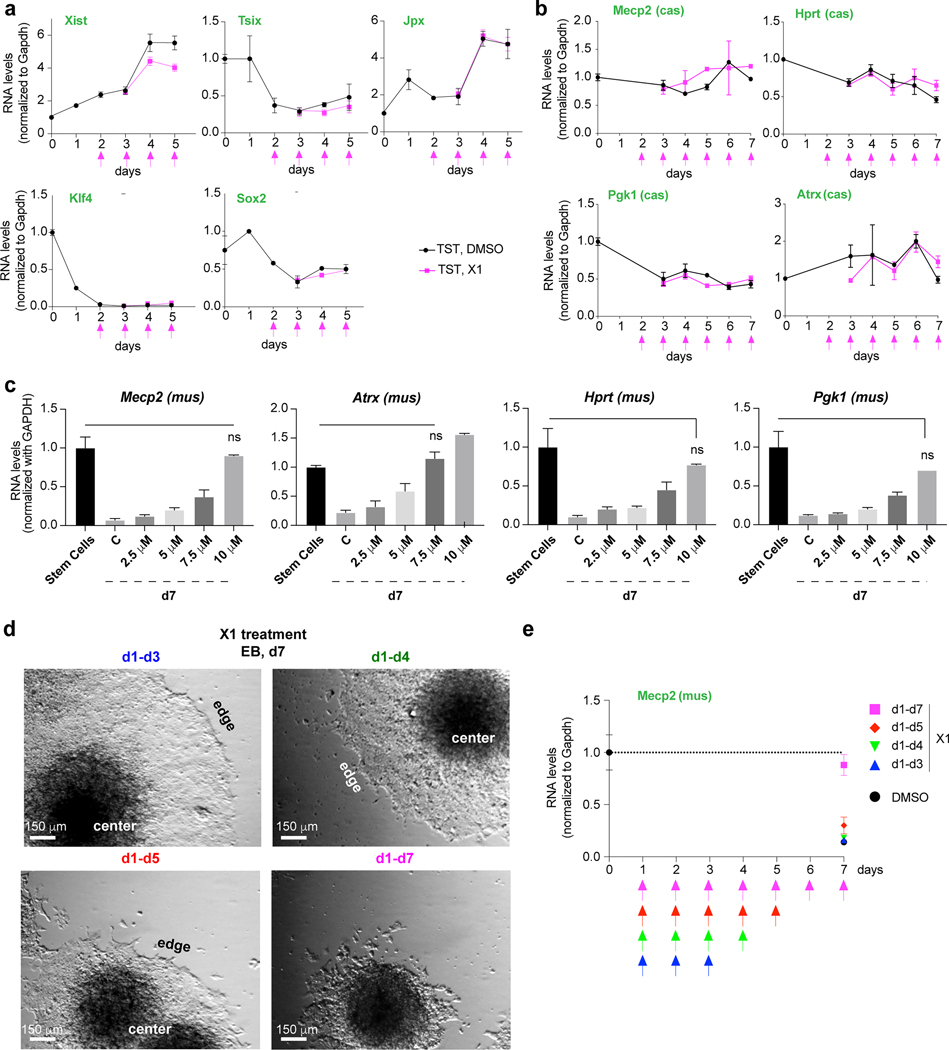
Figure 8: Analysis of gene expression and reversibility of X1’s effect.
a, Time course RT-qPCR of indicated control genes in DMSO- or X1-treated female EB. X1 added on indicated days (pink arrows). Mean and S.D. shown for 3 biological replicates.
b, Time course allele-specific RT-qPCR of indicated Xa genes in DMSO- or X1-treated female EB. X1 added on indicated days (pink arrows). Mean and S.D. shown for 3 biological replicates.
c, Dose-response analysis in the range of 0–10 μM X1 compound. Allele-specific RT-qPCR of indicated X-linked genes in DMSO- or X1-treated female EB. Mus allele (Xi) shown. X1 was added on d2. P, two-tailed Student’s t-test with respect to DMSO-treated TST control. Mean and S.D. shown for 2 replicates. At 10 μM X1, the Student’s t-test reveal no significant difference between d7 cells and expression found in control ES cells.
d-e, X1 effect is reversible upon drug withdrawal. Female EB were grown from d1 in 10 μM X1 and the treatment was either suspended on day 3, 4, 5, or maintained up to day 7. The growth morphology (d) and Mecp2 expression from the Xi is evaluated at d7 (e). One representative brightfield microscopy from 3 independent cultures is shown.
Figure 9: Transcriptomic studies of on- and off-target effects.
a-d, RNA-seq analyses of day 5 DMSO- or X1-treated female EB. Zoom-ins to representative X-linked genes subjected to XCI (a), Xist (b), escapee gene (c) and autosomal gene (d). Tracks for all reads (comp), mus reads (Xi), and cas reads (Xa). FPM scale shown in brackets.
e, Differentially expressed autosomal genes (y axis) and their corresponding changes in H3K27me3 enrichment (x axis). Each dot represents a gene. Number of genes on each of the nine sections as shown. Comp tracks were sampled to the smallest library, then MultiTesting and IndependentFiltering DESeq2 filtering was performed reporting significance below 0.05 (Wald test) after Benjamini and Hochberg correction with the application of independent intensity filtering.
Figure 10: Sixteen conformational clusters identified for native RepA RNA without X1 treatment.
a, HPLC-SEC profile of the purified RepA RNA with or without X1 previous to SAXS data collection.
b, PRIMUS analysis for initial data quality analysis. Inset: Guinier plot to determine the Radius of Gyration (Rg).
c, Dimensionless Kratky analysis [q*Rg vs. I(q)/I(0)x(q*Rg)2] of samples
d, Pairwise distance distribution profile (P(r)) to estimate the real space dimensions of the molecule in Å
e, 16 clusters (C1-C16) of RepA are presented in their native state without X1. C13 is the dominant conformation. Pie-chart shows relative abundance of structural clusters. See also Fig. 4.
Supplementary Material
Table 2: Analog compounds from Ext. Data - Figure 1c that demonstrated binding in affinity ranking experiments.
Table 5: List of upregulated genes on the Xi following X1 treatment.
Left: All active genes encoded in the X chromosome with FKPM > 0.5 are listed. Right: genes encoded on Xmus, up-regulated with X1. Results are expressed as the relative change (%) in the fraction of reads transcribed from the mus allele with respect to the overall (mus+cas) expression.
Table 4: Further demonstration of selectivity of X1 for Xist and X-inactivation.
List of autosomal genes where relative changes in RNA expression are > 2 or < −0.5, and relative changes in H3K27me3 enrichment are > 4 or < −0.25. All values are logarithmic (base 2).
Table 3: Cellular uptake and concentration of X1 molecule by up to 180-fold.
Here we addressed whether cells concentrated the X1 molecule following uptake. Briefly, we added a known amount of tritiated X1 molecule to Embryonic Stem (ES) cells, differentiation day 7 Embryoid Bodies (EB), or fibroblasts (T4)(Row 1). After 24h, radioactivity was measured as counts per minute (cpm) from a total cell lysate using a scintillation counter (Row 2). Knowing the cellular volume and the number of total cells in each assay (Rows 3 and 4), we calculated the total volume of cells (Row 5) into which X1 molecule was diluted (cpm/μL) (Row 6). The cpm/μL value was interpolated to a standard curve (Sheet 2) constructed with increasing concentrations of the tritiated molecule: [cpm/μL] = 33.954 x [nM molecule] - 898.7. The concentration factor could then be derived by comparing the concentration of the molecule found in cells (Row 7) with the original amount added to the culture (Row 1).
Table 1: RNAs tested in ALIS surveys.
The 42 transcripts tested against X22 or X1 are listed. The murine Xist RepA RNA (the only X22 binder) is highlighted in yellow. RNAs with >60% GC content are highlighted in pink.
ACKNOWLEDGEMENTS
We thank all members of the Lee Lab, Patel Lab, Charles Lesberg and other Merck team members for stimulating scientific discussions. We also acknowledge the Merck MINt Award and funding from the Howard Hughes Medical Institute to J.T.L., the Pew Charitable Trust Latin American Fellows Program to R.A. and C.R. and the MGH Fund for Medical Discovery to R.A. M.D.B. and T.M. acknowledge MITACS and NSERC PGS-D fellowships respectively. T.R.P. is Canada Research Chair in RNA and Protein Biophysics. We thank DIAMOND Light Source B21 beamline staff for their help with data collection (BAG SM22113).
Footnotes
COMPETING INTEREST DECLARATION
All authors, except RA, BK, CR, TM, MDB, TRP, and JTL, are current or former employees of Merck & Co., Inc., and may hold stock or other financial interests in Merck & Co. JTL. is a cofounder of Translate Bio and Fulcrum Therapeutics and is also a scientific advisor to Skyhawk Therapeutics.
DATA AVAILABILITY
Sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) repository with accession number GSE141683: “Targeting Xist with compounds that disrupt RNA structure and X-inactivation” https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141683
REFERENCES
- 1.Consortium EP et al. , Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447 (7146), 799–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos R. et al. , A comprehensive map of molecular drug targets. Nat Rev Drug Discov 16 (1), 19–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warner KD et al. , Principles for targeting RNA with drug-like small molecules. Nat Rev Drug Discov 17 (8), 547–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F. & Lupski JR, Non-coding genetic variants in human disease. Hum Mol Genet 24 (R1), R102–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JT, Epigenetic regulation by long noncoding RNAs. Science 338 (6113), 1435–1439 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Wutz A. et al. , Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30 (2), 167–174 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Rizvi NF & Smith GF, RNA as a small molecule druggable target. Bioorg Med Chem Lett 27 (23), 5083–5088 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Disney MD et al. , Drugging the RNA World. Cold Spring Harb Perspect Biol 10 (11) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe JA et al. , Selective small-molecule inhibition of an RNA structural element. Nature 526 (7575), 672–677 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Palacino J. et al. , SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat Chem Biol 11 (7), 511–517 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Disney MD et al. , Inforna 2.0: A Platform for the Sequence-Based Design of Small Molecules Targeting Structured RNAs. ACS Chem Biol 11 (6), 1720–1728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen Annis D. et al. , An affinity selection-mass spectrometry method for the identification of small molecule ligands from self-encoded combinatorial libraries - Discovery of a novel antagonist of E-coli dihydrofolate reductase. International Journal of Mass Spectrometry 238, 77–83 (2004). [Google Scholar]
- 13.Rizvi NF et al. , Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry. ACS Chem Biol 13 (3), 820–831 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Rizvi NF et al. , Targeting RNA with Small Molecules: Identification of Selective, RNA-Binding Small Molecules Occupying Drug-Like Chemical Space. SLAS Discov, 2472555219885373 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Cifuentes-Rojas C. et al. , Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 55 (2), 171–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chillon I. et al. , Native Purification and Analysis of Long RNAs. Methods Enzymol 558, 3–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipinski CA, Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1 (4), 337–341 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Monfort A. et al. , Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells. Cell Rep 12 (4), 554–561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MK et al. , A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication. Chem Commun (Camb) 50 (3), 368–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa Y. et al. , Intersection of the RNA interference and X-inactivation pathways. Science 320 (5881), 1336–1341 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J. et al. , Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40 (6), 939–953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patil DP et al. , m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunwoo H. et al. , Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc Natl Acad Sci U S A 114 (40), 10654–10659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeon Y. & Lee JT, YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146 (1), 119–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colognori D. et al. , Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X. Mol Cell 74 (1), 101–117 e110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F. et al. , Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat Chem Biol 13 (3), 282–289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikhney AG & Svergun DI, A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett 589 (19 Pt A), 2570–2577 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Kim DN et al. , Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution. Nat Commun 11 (1), 148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrette LLG et al. , A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc Natl Acad Sci U S A (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stelzer AC et al. , Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol 7 (8), 553–559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 2: Analog compounds from Ext. Data - Figure 1c that demonstrated binding in affinity ranking experiments.
Table 5: List of upregulated genes on the Xi following X1 treatment.
Left: All active genes encoded in the X chromosome with FKPM > 0.5 are listed. Right: genes encoded on Xmus, up-regulated with X1. Results are expressed as the relative change (%) in the fraction of reads transcribed from the mus allele with respect to the overall (mus+cas) expression.
Table 4: Further demonstration of selectivity of X1 for Xist and X-inactivation.
List of autosomal genes where relative changes in RNA expression are > 2 or < −0.5, and relative changes in H3K27me3 enrichment are > 4 or < −0.25. All values are logarithmic (base 2).
Table 3: Cellular uptake and concentration of X1 molecule by up to 180-fold.
Here we addressed whether cells concentrated the X1 molecule following uptake. Briefly, we added a known amount of tritiated X1 molecule to Embryonic Stem (ES) cells, differentiation day 7 Embryoid Bodies (EB), or fibroblasts (T4)(Row 1). After 24h, radioactivity was measured as counts per minute (cpm) from a total cell lysate using a scintillation counter (Row 2). Knowing the cellular volume and the number of total cells in each assay (Rows 3 and 4), we calculated the total volume of cells (Row 5) into which X1 molecule was diluted (cpm/μL) (Row 6). The cpm/μL value was interpolated to a standard curve (Sheet 2) constructed with increasing concentrations of the tritiated molecule: [cpm/μL] = 33.954 x [nM molecule] - 898.7. The concentration factor could then be derived by comparing the concentration of the molecule found in cells (Row 7) with the original amount added to the culture (Row 1).
Table 1: RNAs tested in ALIS surveys.
The 42 transcripts tested against X22 or X1 are listed. The murine Xist RepA RNA (the only X22 binder) is highlighted in yellow. RNAs with >60% GC content are highlighted in pink.
Data Availability Statement
Sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) repository with accession number GSE141683: “Targeting Xist with compounds that disrupt RNA structure and X-inactivation” https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141683