Abstract
Oral squamous cell carcinoma (OSCC) is known to be driven by multiple intricated receptor tyrosine kinases (RTKs) including EGFR, PI3K/AKT and MAPK signaling pathways. However, whilst targeting EGFR with cetuximab has been approved for the treatment of OSCC, other single-agent inhibitors of the RTKs have shown modest effects in improving survival. From the genome-wide CRISPR/Cas9 screen on 21 OSCC cell lines, we have identified PTPN11 among the top essential genes in OSCC. PTPN11 encodes for SHP2, a phosphatase that acts as a master signal transducer, downstream of various RTKs. Although PTPN11 overexpression has been reported in OSCC, little is known about its role as an essential gene for OSCC survival and its potential as a therapeutic target. Herein, we confirmed that PTPN11 is an essential gene in OSCC where its deletion significantly impacted cell survival. We evaluated three SHP2 inhibitors on 21 OSCC cell lines and found TNO155 to be significantly associated with CRISPR dependency score. We showed that TNO155 caused dose-dependent suppression on p-ERK and p-MEK, and suppresses the JAK/STAT pathway via downregulating p-JAK1, p-STAT1, p-STAT3. Furthermore, we confirmed that the combination of the mTOR inhibitor, everolimus with TNO155 is synergistic in OSCC. In summary, PTPN11 is a promising therapeutic target in OSCC that can be selectively targeted by SHP2 inhibitor such as TNO155. Our findings on the use of mTOR inhibitor, everolimus to overcome resistance to TNO155 are essential to inform on next phases of clinical trials which is warranted for the treatment of OSCC.
Keywords: RTK signaling, OSCC, Drug resistance, Essential genes, PTPN11, SHP2, Protein tyrosine phosphatase, Allosteric inhibitor
1. Introduction
Oral squamous cell carcinoma (OSCC) is the commonest subtype of head and neck squamous cell carcinoma (HNSCC) and accounts for 90 % of all oral malignancies. Globally, there are more than 300,000 cases of OSCCs reported annually, with approximately 145,000 associated deaths, resulting in a mortality rate of nearly 50 % [1]. The disease is especially prevalent in Asia, with more than 70 % of the deaths occurring among Asians [2]. Despite the characterization of the OSCC genomics landscape, the development of targeted therapies remain limited and treatment resistance continues to be a major challenge [3]. Cetuximab, the monoclonal antibody targeting EGFR remained as the only molecular targeted therapy approved for advanced stage OSCC, but only a subset of OSCC patients are benefitting [4]. To identify novel targets that govern the OSCC survival, we performed the genome-wide CRISPR/Cas9 screen in 21 OSCC lines to identify potentially druggable genetic vulnerabilities [5]. One of the top essential genes is the Protein Tyrosine Phosphatase Non-receptor 11 (PTPN11).
PTPN11 is a unique proto-oncogene that encodes for the Src homology-2-containing protein tyrosine phosphatase 2 (SHP2), that contains two SH2 domains [6]. These SH2 domains play a crucial role in recognizing phosphorylated molecules and are found in many intracellular signal-transducing and adaptor proteins, downstream of many receptor tyrosine kinases (RTKs) such as EGFR. As a result, SHP2 serves as a master regulator of multiple intracellular oncogenic signaling pathways, such as RAS/RAF/MAPK, PI3K/AKT, JAK/STAT, TGF-β and PD-1/PD-L1 pathways, making SHP2 a promising drug target for simultaneous inhibition of multiple oncogenic pathways [[7], [8], [9], [10]]. In recent years, the development of SHP2 inhibitors afforded an opportunity to target SHP2, these include the allosteric inhibitor SHP099 [8], TNO155 [11], RMC4550 [12], RMC4630 and JAB-3068 [13]. SHP2 inhibitors such as TNO155 [14], RMC4630 and JAB-3068 are currently in early phase clinical trials (NCT03114319, NCT03634982, NCT03518554 and NCT03565003), where generally patients with advanced solid tumors are recruited. However, the efficacy of targeting PTPN11 dependency with these SHP2 inhibitors has not been studied in the specific context of OSCC. Hence, the sensitivity profile of OSCC lines towards these SHP2 inhibitors and their potential mechanism of action remains unknown. Preclinical mechanistic study of SHP2 inhibitors in OSCC could help inform their future clinical development, in identifying possible biomarkers of response or resistance and suggesting rational combinations.
Aberrant activation of the mitogen-activating protein kinase (MAPK) pathway has been reported in 50 % of OSCC [15], with ∼18 % due to mutations. Drugs targeting different RTK pathways have been tested in clinical trials [16], but mostly failed due to limited clinical benefit and dose-limiting toxicities [16]. The reason behind the lack of efficacy of drugs targeting the RAS/RAF/MAPK pathway is likely caused by the high plasticity in the rewiring of oncogenic signaling such as the feedback or cross talking with the PI3K/AKT/mTOR pathways. However, the exact mode of action and mechanism of resistance in OSCC towards these new SHP2 inhibitor, which will be essential to inform on next phases of clinical trials have yet to be explored.
In this study, we aimed to validate PTPN11 as an essential gene in OSCC and evaluate the efficacy of the SHP2 inhibitors on multiple downstream signaling pathways. Additionally, we used our CRISPR/Cas9 screen to identify other dependencies that could inform on resistant mechanisms towards PTPN11 inhibition and suggest drug combinations that are synergistic in killing OSCC.
2. Materials and methods
2.1. Cell lines
All the OSCC cell lines (ORL-48: CVCL_S692, ORL-115: CVCL_S690, ORL-136: CVCL_S691, ORL-150: CVCL_VJ37, ORL-153: CVCL_VJ38, ORL-156: CVCL_VJ39, ORL-166: CVCL_VJ40, ORL-174: CVCL_VJ41, ORL-188: CVCL_VJ42, ORL-195: CVCL_VJ43, ORL-204: CVCL_VJ45, ORL-207: CVCL_VJ46, ORL-214: CVCL_VJ47, ORL-215: CVCL_VJ48, BICR10: CVCL_2307, HSC-2: CVCL_1287, HSC-4: CVCL_1289, Ho-1-u-1: CVCL_2784, PE/CA-PJ15: CVCL_2678, SAS: CVCL_1675, SCC-9: CVCL_1685) used were cultured in Dulbecco's Modified Eagle's Medium (DMEM)/Nutrient Mixture F-12 medium (Gibco) (DMEM/F12) containing 10 % (v/v) heat inactivated fetal bovine serum (Gibco), and 100 IU penicillin/streptomycin (Gibco), as previously described [5]. All lines were grown in humidified incubator at 37 °C with 5 % carbon dioxide. The authenticity of cell lines was confirmed by short tandem repeat (STR) profiling, and routine tests were conducted to ensure mycoplasma-free status.
2.2. Compounds
SHP099, RMC4550 and TNO155 were purchased from Chemietek, USA. Everolimus was purchased from MedChemExpress. All drugs were dissolved in Dimethylsulfoxide (DMSO) at a stock concentration of 50 mmol/L.
2.3. CRISPR/Cas9 knockout
Cas9-expressing cell lines were transduced with lentivirus carrying PTPN11 sgRNA in pKLV2-U6gRNA5(BbsI)-PGKpuro2ABFP-W as described previously [5]. Two sgRNA were used for PTPN11 -"sg1k” (P1k) from Kosuke Yusa's CRISPR Library v1 and another independently designed sgRNA using Broad's sgRNA-designer tool - “sg2b” (P2b). Non-targeting (NT) sgRNA was included as control. Transduced cells were enriched for by selection with puromycin at 2 μg/mL the next day. Sequences of the sgRNAs used can be found in Supplementary Table 1.
2.4. Co-competition assay
The co-competition assays were used to validate the CRISPR screen result, as described previously [5]. The percentage of BFP-positive sgRNA-transduced cells was measured on day 4/6, 8, 11, 15 and 18 post-transduction. The relative growth rate of sgRNA-transduced and non-transduced cells at each timepoint was measured by comparing and normalizing with the percentage of BFP-positive transduced cells on day 4/6.
2.5. Cell viability assay
The half maximal inhibitory concentration (IC50) was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. OSCC cells were seeded at optimized density in triplicates. The next day, seven varying concentrations of PTPN11 inhibitors at 10× fold dilution ranging from 0.1 μM to 100 μM were added. After 72 h, MTT was added to the cells and incubated for 4 h at 37 °C. Subsequently, DMSO was used to dissolve the formazan crystal after medium removal. Synergy H1M micro-plate reader (BioTek Instruments, USA) was used to measure the optical density at 570 nm. Normalized cell viability data was entered into GraphPad Prism (version 9.2.0; RRID:SCR_002798; GraphPad Software, Inc.) which was used to plot the dose-response curve and IC50 was determined using the nonlinear regression analyses function, with the equation “log (inhibitor) vs. normalized response”.
For PTPN11-knockout experiments, day 4 post-transduced cells were seeded in triplicates into 96-well plates at 2,000 cells per well and were processed as above on day 1, 3 and 5 post seeding.
2.6. Apoptosis assay
Cells were pelleted and washed with PBS. For detection of apoptotic cells, the cell pellet was suspended in Annexin V buffer containing Annexin V solution and propidium iodide from FITC Annexin V Apoptosis Detection Kit I (BD Biosciences; Cat. No. 556547) as described in Ref. [5]. The apoptotic cells were analyzed using the LSR Fortessa X-20 cell analyzer (BD Biosciences) and FlowJo (version 10.8.1, BD Biosciences, RRID:SCR_008520), considering all single- and double-stained cells as apoptotic cells.
2.7. Lysate preparation and western blotting
Total cell lysates were extracted with RIPA buffer 50 mM Tris pH 8, 1 % (v/v) NP-40, 0.5 % (w/v) sodium deoxycholate, 0.1 % (w/v) SDS, and 150 mM NaCl supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Pierce Biotechnology, Rockford, IL, USA) on ice. TCL was collected by centrifugation and quantified using the BCA method (Thermo Fisher Scientific, USA). Total protein (15 or 30 μg) was resolved by SDS-PAGE and analyzed by western blotting as previously described [1]. The resolved proteins were detected by Immobilon Forte Western HRP Substrate (Merck, Germany) and visualized using Azure c300 imaging system (Azure Biosystems, USA). To normalize for loading, the blots were subsequently probed for house-keeping proteins (vinculin, α-tubulin, actin) and processed as described above. List of primary and secondary antibodies used is found in Supplementary Table 2. Uncropped Western blot images can be found in Supplementary Fig. 2.
2.8. Colony formation assay
On day-4 post transduction, PTPN11-knockout cells were seeded into 24-well plates at 2000–4000 cells per well and were harvested after a week. To assess the drug's effects, OSCC cells were seeded and treated the next day with the indicated drugs. A week later, cells were fixed using ice-cold methanol and stained with 0.5 % crystal violet in 25 % methanol and 75 % water.
2.9. Phosphorylation pathway profiling array
The phosphorylation pathway profiling array was performed using the Human Phosphorylation Pathway Profiling Array C55 kit (Raybiotech, Inc. AAH-PPP-1-4) following the manufacturer's instructions. The array was used to analyse the differential phosphorylation levels of 55 proteins in five oncogenic pathways (MAPK, AKT, JAK/STAT, NF-κB, and TGF-β) in response to TNO155 treatment. Lysates used were OSCC cells treated with 3 μM TNO155 for 1 h, with 3 μM equivalent DMSO-treated lysates as control. The membranes were incubated with blocking buffer at room temperature (RT) for 30 min and then incubated overnight at 4 °C with 1 mL of cell lysates (1/3 dilution). Subsequently, the membranes were incubated with Detection Antibody Cocktail for 2 h at RT and with HRP-Anti-Rabbit IgG for 2 h at RT. Signals were detected using Azure c300 imaging system (Azure Biosystems, USA). Densitometric analysis of the array spots was performed using Carpentier G. Protein Array Analyzer (2010) [17] for ImageJ software (v.1.53, RRID:SCR_003070). Markers that did not reach the minimum intensity threshold of 4000 were excluded from the analysis. The pixel density of the negative control served as a background value and the positive control spots served as a normalizing factor. The threshold of the positive or negative fold-change was defined as |≥15 %|.
2.10. Drug combination synergism via combination index estimation
For the evaluation of synergistic effect between TNO155 and everolimus, the relative cell growth inhibition effect at each drug combination doses were used as input for the CompuSyn software (v1.0) [18]. Fraction affected (Fa) and combination index (CI) was computed by the software based on Chou-Tatalay's median-effect equation, where a value less than 1 is indicative of a synergistic effect, while CI = 1 is addictive and CI > 1 reflects antagonism [18,19].
2.11. Statistical analyses
Statistical analyses were performed using an unpaired parametric two-tailed t-test in GraphPad Prism (version 9.2.0; RRID:SCR_002798; GraphPad Software, Inc.), unless otherwise stated.
2.12. Data availability
Raw data for the phosphorylation array can be accessed from Figshare - dx.doi.org/10.6084/m9.figshare.26,342,137.
3. Results
3.1. PTPN11 is an essential gene in OSCC cell lines
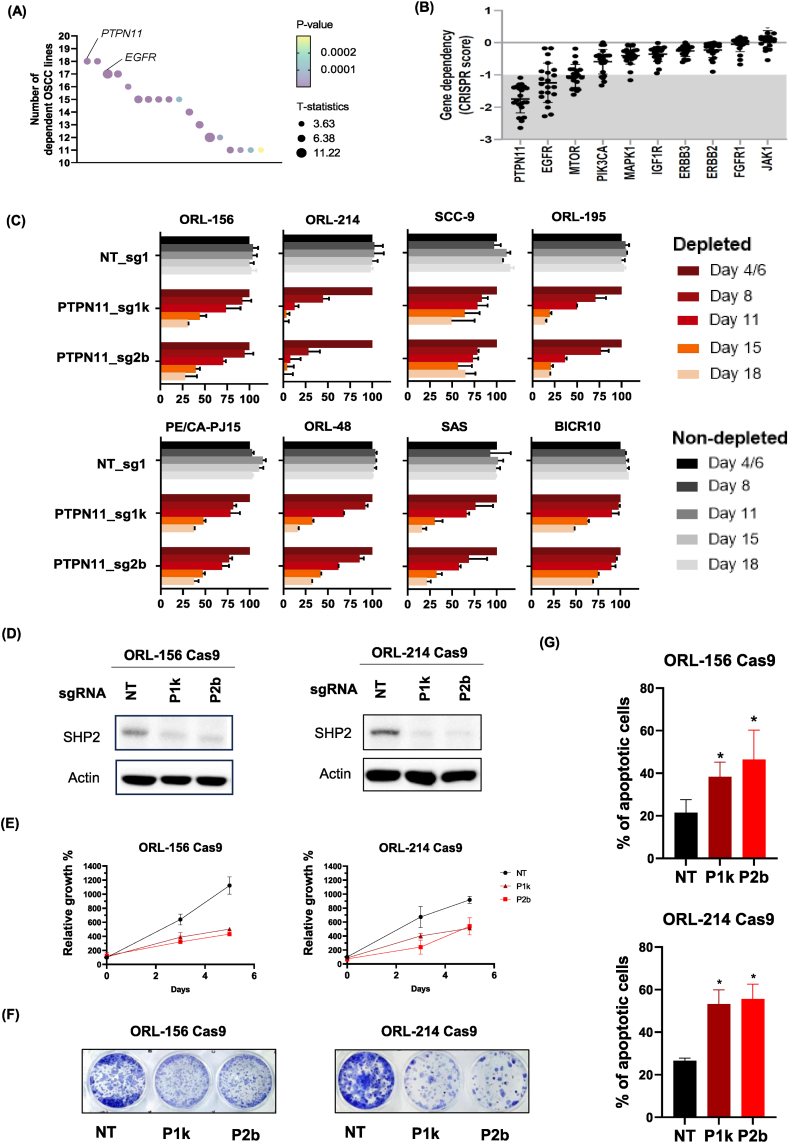
Our genome-wide CRISPR screens on 21 OSCC cell lines revealed that PTPN11 is among the top essential genes of the 918 identified [5], where most OSCC lines are dependent on PTPN11. Among those 918 genes that have at least ten dependent OSCC lines, eighteen genes were also found to be enriched in the lineage of upper aerodigestive tract (DepMap Public 22Q4, https://depmap.org/portal/). Among which, PTPN11 appeared as the top candidate with enrichment of essentiality in OSCC [Fig. 1A, Supplementary Table 1].
Fig. 1.
Confirmation of PTPN11 as an essential gene for OSCC survival. (A) Plot of number of dependent OSCC cell lines based on our previous CRISPR/Cas9 screen of 21 OSCC lines (Chai et al., 2020), for the essential genes that are enriched in the lineage of upper aerodigestive tract (DepMap Public 22Q4, https://depmap.org/portal/). (B) Scatter plots of CRISPR scores of PTPN11 and other genes that are targets of drugs being tested for OSCC. (C) Co-competition assays between non-transduced cells with either Non-targeting (NT) or PTPN11 sgRNAs-transduced OSCC to validate dependency on PTPN11. The percentage of BFP-positive transduced cells obtained at different time points were normalized to the day 4 or 6 readings for respective sgRNAs. (D) Western blots showing depletion of the SHP2 level, the protein encoded by PTPN11 gene upon sgRNA-mediated knockout. Actin is used as loading control. NT – Non-targeting sgRNAs; “P1k” – PTPN11_sg1k”; “P2b” – PTPN11_sg2b. (E) Cell viability assay of OSCC cells upon PTPN11 knockout with P1k and P2b. (F) Colony forming assay of OSCC cells upon PTPN11 knockout with P1k and P2b. (G) Apoptosis assay of OSCC cells upon PTPN11 knockout with P1k and P2b.
When comparing the PTPN11 with putative targets of several anti-cancer drugs being tested for HNSCC treatment, PTPN11 has the lowest average CRISPR scores, and is the only gene with all OSCC lines showing dependency (CRISPR score ≤ −1), suggesting that it could be a better therapeutic target to inhibit over other targets [Fig. 1B]. Besides, the survival analysis performed using the Cancer Genome Atlas (TCGA) cohort of head and neck cancer patients revealed that high PTPN11 expression is associated with poorer overall survival (Hazard Ratio (HR) = 1.4455, p = 0.0156) [Supplementary Fig. 1] [20]. Furthermore, given the previously reported role of PTPN11-encoded SHP2 protein in promoting proliferation, invasion and metastasis of OSCC [21,22], we shortlisted PTPN11 as a promising therapeutic target for OSCC.
To validate the results from the genome-wide CRISPR screens [5], we used two sgRNAs (PTPN11_sg1k as “P1k”; PTPN11_sg4b as “P2b”), to knockout PTPN11 and evaluated the essentiality of PTPN11 in various OSCC lines. Co-competition assay was performed on a panel of eight OSCC cell lines [Fig. 1C]. Non-targeting (NT) sgRNA was included as control, while we also included PLK1 (a core essential gene) as positive control, and CHAT as negative control [Supplementary Fig. 2]. Depletion of PTPN11-sgRNA transduced, BFP-positive cells over time serve as a validation that PTPN11 is an essential gene in the OSCC lines of diverse genomic background, for example, ORL-214 has HRAS, TP53, FAT1 and NOTCH1 mutations, while BICR10 is a PIK3CA-mutated cell line [5]. This suggests that PTPN11 dependency is ubiquitous in OSCC.
To confirm and characterize the impact of PTPN11 knockout in OSCC, cell viability assay, apoptosis assay and clonogenic assay were performed. With CRISPR/Cas9-mediated depletion of PTPN11-encoding protein – SHP2 [Fig. 1D], the relative cell growth was found to be inhibited [Fig. 1E] and likewise, colony formation ability was drastically impaired [Fig. 1F]. Furthermore, the number of apoptotic cells was significantly increased upon PTPN11 depletion [Fig. 1G]. These indicate that PTPN11 is essential for OSCC viability and growth whereby the knockout of PTPN11 causes apoptotic cell death in OSCC.
3.2. Evaluation of SHP2 inhibitors efficacy in OSCC
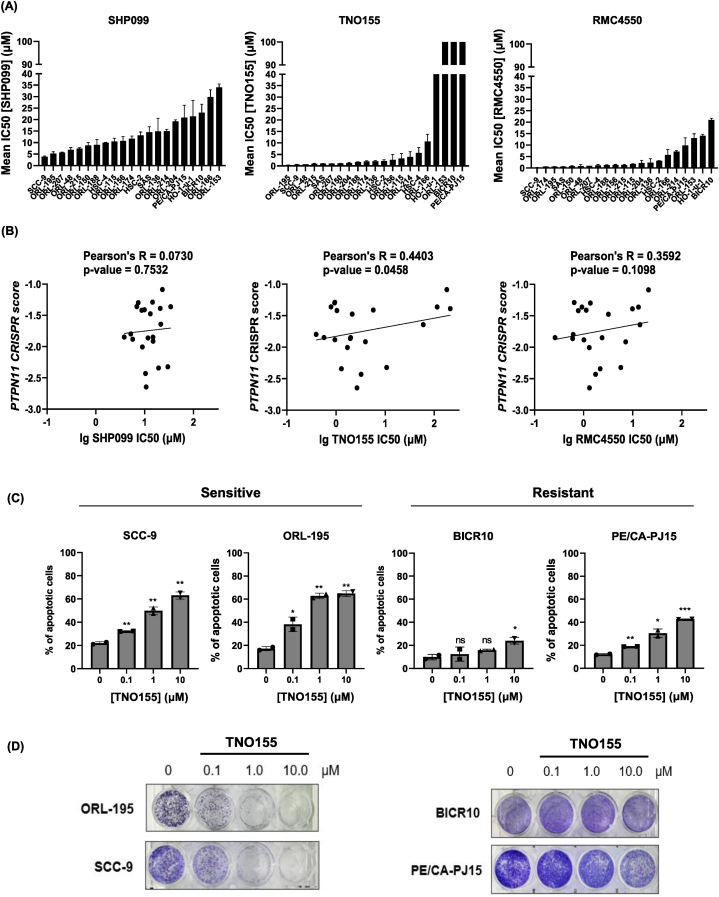
Given the dependency to PTPN11, we proceeded to assess the potency of SHP2 inhibitors in OSCC, and determined the half-maximal inhibitory concentration (IC50) of three SHP2 inhibitors SHP099, TNO155 and RMC4550 on 21 OSCC lines. Among the three inhibitors, TNO155 (IC50 ranging from 0.39 μM to 211.1 μM) and RMC4550 (IC50 ranging from 0.261 μM to 20.9 μM) are more efficacious in majority of the OSCC cell lines (with 6 cell lines having IC50 < 1 μM, 16 and 18 cell lines having IC50 < 10 μM respectively), compared to SHP099 (IC50 ranging from 3.822 μM to 34.0 μM) (only 8 cell lines with IC50 < 10 μM) [Fig. 2A]. We also showed that the mean IC50 of TNO155 showed the best and statistically significant correlation with PTPN11 CRISPR dependency score (Pearson's R = 0.4403, p-value = 0.0458) [Fig. 2B]. Considering also that TNO155 is the most clinically advanced SHP2 inhibitor, TNO155 was subsequently selected for further investigation. TNO155 also has the widest range of IC50 in OSCC lines that could help identify mechanism of resistance (those with IC50 > 10 μM). For subsequent analyses, the two most sensitive (ORL-195 and SCC-9) and two most resistant cell lines (BICR10 and PE/CA-PJ15) towards TNO155, were used. These lines are also consistently amongst the top five most sensitive and resistant lines respectively for the other two inhibitors.
Fig. 2.
TNO155, an inhibitor of SHP2 exhibit selective sensitivity in OSCC. (A) IC50 of three SHP2 inhibitors (SHP099, TNO155 and RMC4550), were determined for 21 OSCC cell lines using MTT assay. Data shown is mean ± standard deviation (in μM). (B) Correlation plot of PTPN11 CRISPR scores against log10 (lg) IC50 of SHP099/TNO155/RMC4550 IC50 (μM). (C) Dose-dependent effects of TNO155 on two selective “sensitive” lines – ORL-195 and SCC-9; and two “resistant” lines – BICR10 and PE/CA-PJ15 were measured by apoptosis assay. Bar charts show the percentage of apoptotic cells at increasing concentration of TNO155. Error bars are mean±SD of two biological repeats with technical triplicates. Unpaired student's t-test was used with untreated control (0 μM TNO155) as comparator. ∗p: <0.05, ∗∗: p < 0.01, ∗∗∗: not significant (ns). (D) Dose-dependent effect of TNO155 on colony formation assay in sensitive and resistant OSCC cell lines.
To evaluate the cytotoxicity effect of TNO155 on OSCC, the selected cell lines were subjected to increasing concentrations of TNO155. We showed that TNO155 treatment led to dose-dependent increase in apoptotic cells in the sensitive cell lines [Fig. 2C]. In addition, TNO155 significantly suppressed the colony-forming ability in a dose-dependent manner in sensitive cell lines [Fig. 2D]. Among the resistant lines, the effect of TNO155 was less drastic, indicating that although TNO155 is an effective agent for inhibiting cell viability and colony formation ability, it nevertheless has a heterogenous effect across OSCC lines.
3.3. Dose-dependent effect of TNO155 treatment on downstream signaling in OSCC
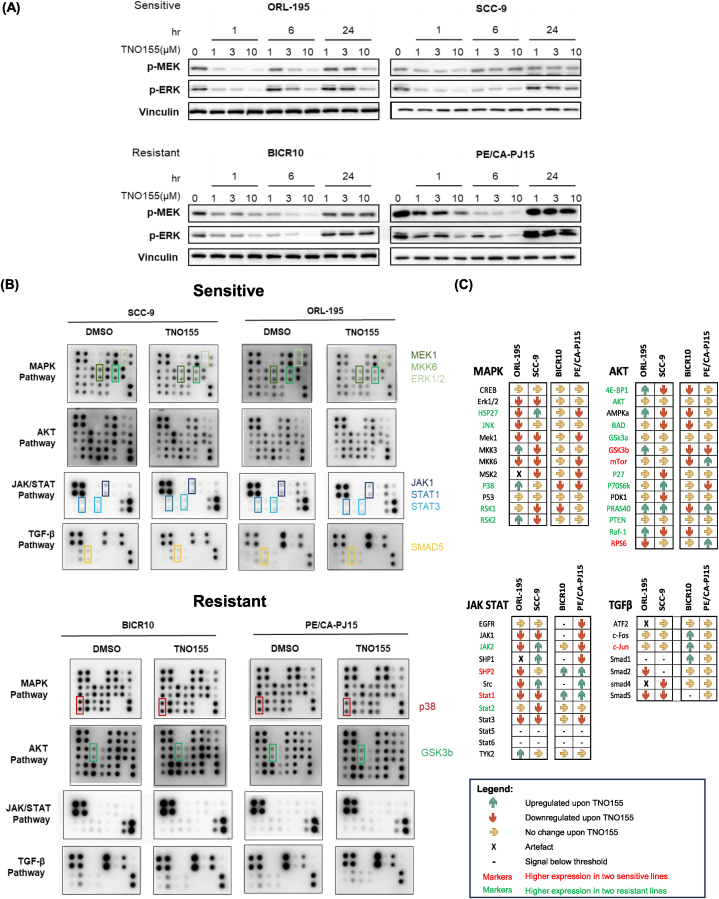
Next, we determined if the TNO155 exhibits a dose-dependent suppression effect on SHP2 signaling by looking at p-MEK and p-ERK levels of the MAPK pathway. Both p-MEK and p-ERK showed a dose-dependent down-regulation as early as 1-h post-treatment [Fig. 3A]. The effect of downregulation is more obvious in the two sensitive lines, ORL-195 and SCC-9. The inhibition effect on p-MEK and p-ERK were also more durable in the sensitive lines, while the effect was already abrogated in the resistant lines, BICR10 and PE/CA-PJ15, after 24 h of 10 μM TNO155 treatment.
Fig. 3.
Effect of TNO155 on phosphorylation levels of kinases in its downstream and parallel signaling pathways. (A) Western blot shows the effect of various TNO155 concentration on MEK and ERK phosphorylation levels upon 1-, 6- and 24-h treatment, in sensitive and resistant OSCC lines. (B) Phosphorylation arrays of DMSO-treated (control) and TNO155-treated OSCC lines reveal the overall effects of TNO155 on the downstream (MAPK) and other related phosphorylation signaling pathways (AKT, JAK/STAT, and TGFβ) and identify potential markers that are distinctive between sensitive and resistant lines. (C) Graphical representation of the quantified changes on the phosphorylated markers upon TNO155 treatment in the sensitive and resistant lines. Icons in the boxes represent whether the markers expression was upregulated (green upward arrow), downregulated (red downward arrow), or unchanged (yellow rightward arrow) upon TNO155 treatment. Differences in baseline expression were also indicated by the font colours of the markers, by comparing the signal intensities between the average of sensitive lines and average of resistant lines. Red font indicate that the markers have significantly higher level among sensitive lines than resistant lines; while green font indicate that the markers have significantly higher level among resistant lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Consistent with our Western blot analysis, TNO155 treatment resulted in the downregulation of phosphorylated proteins of the MAPK pathway, particularly affecting the key downstream components of the ERK pathway, including MEK1 and ERK1/2. This observation is coherent with the Western blot results in Fig. 3A. Notably, we observed a downregulation trend of essential JAK/STAT elements consistently in both the sensitive lines, this includes p-JAK1 (Tyr1022), p-STAT1 (Ser727), p-STAT3 (Tyr705). By contrast, TNO155 treatment did not result in the significant downregulation of those markers mentioned in the two resistant lines.
We are interested in the mechanism by which some lines are intrinsically resistant to TNO155. We found that both 4E-BP1 and p-70S6K (both are key downstream effectors/indicators of mTOR signaling activation) levels were on average higher among the two resistant lines. Interestingly, we also found that the MTOR-dependent OSCCs were significantly less dependent on PTPN11 [Supplementary Fig. 3]. These TNO155-resistant lines such as BICR10 might be more addicted to the mTOR pathway and show less dependency on PTPN11, and therefore the lack of sensitivity towards TNO155.
3.4. Combination of TNO155 with mTOR inhibitor is synergistic in OSCC
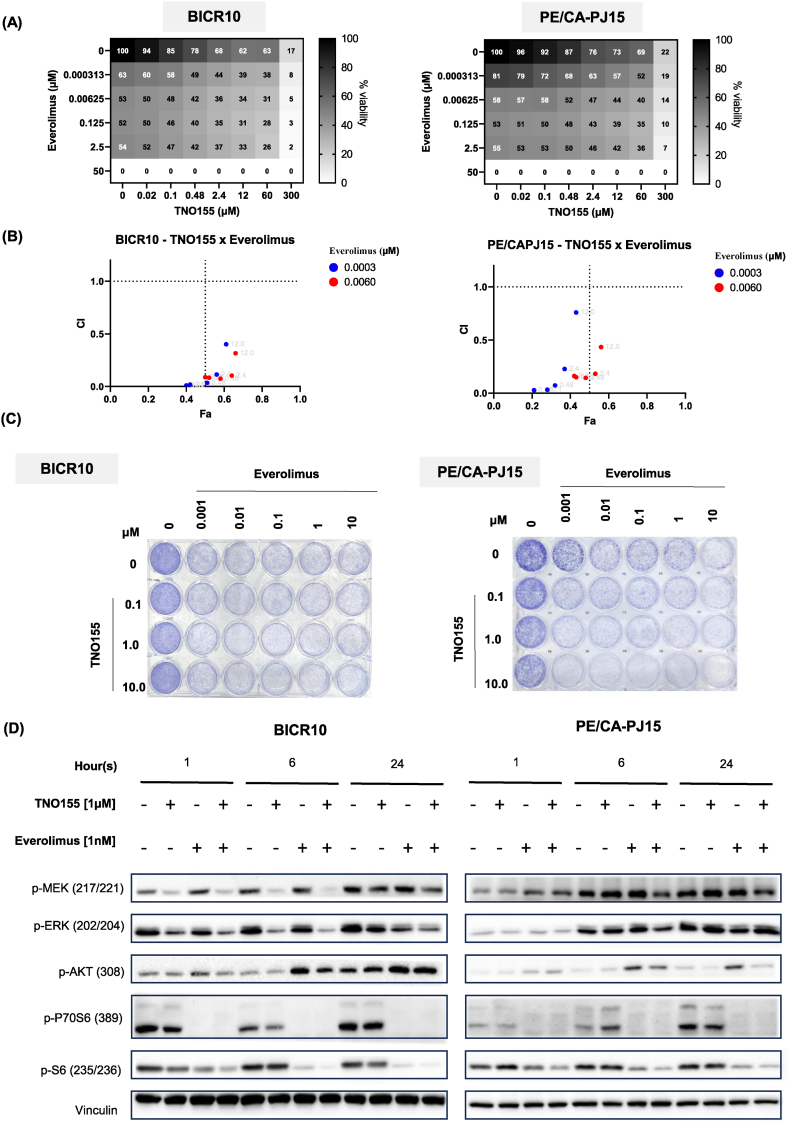
Given the dependency of mTOR and the higher levels of mTOR activation markers (p-4E-BP1 and p70S6K) in the TNO155-resistant lines, we hypothesize that mTOR inhibition can overcome the resistance of these OSCC towards TNO155. We then investigated if there is any synergistic effect in the combination of TNO155 with everolimus, an mTOR inhibitor that is FDA-approved for several cancer indications. In both BICR10 and PE/CA-PJ15, we tested the combination of 0.3 nM–50 μM everolimus with 0.02 μM–300 μM TNO155, at different ratios [Fig. 4A]. For BICR10 which has an IC50 estimated to be above 100 μM, we can clearly observe synergistic effect with everolimus, where 6.25 nM everolimus in combination with 0.02 μM TNO155 resulted in 50 % viability reduction. The combination indexes (CI) were plotted against fraction affected (Fa) for BICR10 and PE/CA-PJ15 [Fig. 4B]. In both lines, addition of very low dose of everolimus (0.3 nM and 6 nM) could result in synergistic effect (CI < 1) across all doses of TNO155.
Fig. 4.
Combination of TNO155 and everolimus are synergistic in OSCC lines. (A) Matrix of cell viability across various combinations of TNO155 (SHP2 inhibitor) and everolimus (mTOR inhibitor) in BICR10 and PE/CA-PJ15, suggesting a synergistic effect. (B) Plot of combination index (CI) against fraction affected (Fa) of TNO155 (0.02 μM, 0.1 μM, 0.48 μM, 2.4 μM and 12 μM) and everolimus (0.0003 μM and 0.006 μM) combinations in BICR10 and PE/CA-PJ15. Only doses that are lower than single-agent IC50 is plotted here. (C) Colony formation assays at various combinations of TNO155 and everolimus showing the synergistic effect of the combinations. (D) Western blot showing the effect of the TNO155 and everolimus combinations on the phosphorylation levels of key downstream markers.
The combinations of TNO155 and everolimus were also tested in colony formation assay [Fig. 4C]. The synergistic effect was observed in low-doses combination as low as 1 μM TNO155 and 1 nM everolimus. Subsequently, to characterize the effect of the combination of TNO155 and everolimus in SHP2 downstream signal pathways, the resistant cell lines were treated with the inhibitors at these indicated concentrations. In both cell lines, synergistic effect can be seen with the combination of TNO155 and everolimus, especially when looking at the downstream key effector marker of the mTOR pathway, p-S6 [Fig. 4D]. Interestingly, we observed an upregulation of p-AKT in samples treated with everolimus, which is evident in both cell lines at 6- and 24-h post-treatment. But in samples treated with everolimus and TNO155, the p-AKT suppression effect is more prominent, as exemplified in the case of PE/CA-PJ15. Overall, our data suggest that combination of everolimus and TNO155 is synergistic whilst mitigating the p-AKT activation feedback loop that may otherwise lead to acquired resistance towards everolimus in these OSCCs.
4. Discussion
Targeting of the RTK signaling pathway has been a logical approach for the treatment of cancer but this has been challenging due to the intricate crosstalk and feedback mechanisms that underlie intrinsic and acquired resistance towards single-agent RTK inhibitor [23,24]. The heterogeneity in OSCCs’ dependency on RTK signaling pathways necessitates targeting of a central node that can allow co-inhibition of multiple oncogenic pathways [25]. One such target is SHP2, encoded by the PTPN11 gene [13,[25], [26], [27]]. Importantly, our genome-wide CRISPR/Cas9 screen revealed that a vast majority of OSCCs are dependent on PTPN11. Previous literature has also shown that the use of RNA interference to knockdown PTPN11 inhibits cell viability, invasion, and metastasis of oral cancer cells [21,22]. Herein, we showed that CRISPR/Cas9-mediated knockout of PTPN11 gene resulted in the inhibition of cell viability and colony forming ability, and increased apoptosis. Furthermore, significant upregulation of the SHP2 protein has been reported in clinical specimens of OSCC and its overexpression was found to be associated with advanced clinical stages [21], and significantly poorer overall survival. Taken together, targeting SHP2 is a promising therapeutic strategy.
SHP099 is the first selective allosteric inhibitor of SHP2, developed by Novartis in 2016 [28]. Subsequently, RMC4550 was developed, demonstrating a similar mode of action but higher potency compared to SHP099 [12]. Initial findings indicated that these compounds would be particularly potent in treating cancers driven by various aberrantly regulated RTKs [8,12,28]. However, the first SHP2 inhibitor in clinical testing is TNO155, from Novartis [11]. TNO155 is a structure-optimized, potent inhibitor of SHP2 that binds to the allosteric tunnel site of SHP2 [11]. The selectivity of TNO155 has been demonstrated in vitro [11] and the TNO155's dose-escalation Phase I study showed favorable pharmacokinetics and tolerable safety profile [14]. Given TNO155's high potency and direct clinical relevance, its effect in modulating downstream phosphorylation-activated signaling was further investigated, for the first time in OSCC.
Despite OSCC patients being eligible for the on-going Phase I clinical trials of TNO155 (8 %, n = 9 of NCT03114319 were patients with head and neck cancers) [14], information on the pathways inhibited by TNO155 remains unclear since no pre-clinical studies have been reported in OSCC. Using the phosphorylation array, we investigated the effect of TNO155 simultaneously on multiple downstream effectors signaling pathways, to delineate its mechanism of actions and potential biomarkers of response/resistance. Importantly, we showed consistently that OSCC that are resistant to TNO155 exhibit significant upregulation of p-SHP2 due to positive-feedback mechanism. Similar phenomenon of p-SHP2 induction upon SHP2 inhibitor treatment has also been reported in two SHP-099-resistant liver cancer cell lines (JHH-7 and Hep3B) [29]. Upregulation of p-SHP2 upon SHP2 inhibition could therefore represent a marker of resistance towards SHP2 inhibitor and could be of prognostic significance to help plan for next treatment.
In OSCC, we showed the expected inhibition effects on p-ERK and p-MEK directly downstream of the SHP2 in the MAPK pathway [11], which in turn resulted in suppression of OSCC cell survival and proliferation. Besides, we also discovered that the JAK/STAT pathway can be inhibited by TNO155, specifically p-STAT1 and p-STAT3 downregulation can be seen among the sensitive OSCC lines. Increasing evidence supports that the JAK/STAT signaling pathway is related to OSCC development [30] and inhibition of STATs, specifically STAT3 could lead to promising anti-cancer effects [[31], [32], [33]]. In the past, SHP2 has been reported to both positively and negatively regulate the JAK/STAT pathway and this is likely context-dependent, and different functional domains of the SHP2 can exert different effects both in catalytic and non-catalytic dependent manner [34]. However, the role of SHP2 and consequences of its inhibition on the JAK/STAT pathway has been unclear. In this study, we confirmed for the first time that in OSCCs, inhibition of SHP2 using TNO155 can lead to the downregulation of the JAK/STAT pathways, potentiating the anti-proliferative effect.
The use of SHP2 inhibitors has been extensively investigated for use in combinations with RAS inhibitors in malignancies driven by RAS mutations. However, the utility of SHP2 inhibitors in HNSCC should not be undermined although the mutation rate of RAS is very rare. We and others have shown that consistent with our finding that PTPN11 is among the most enriched essential gene in OSCCs, a recent drug screen also revealed that HNSCC (comprising mainly of OSCC) is the most sensitive subtype of adult cancer towards SHP099 [9]. Nevertheless, with the quick rebound of the suppression effect on p-ERK and p-MEK as seen in the 24 h-treated samples, it is likely that single agent SHP2 inhibitors may still not be effective in giving a durable anti-cancer effect and combination therapies would warrant further investigation. Notably, combinations of SHP2 inhibitors have been shown to overcome drug resistance to a variety of drugs including RTK inhibitors, such as BYL719 (PI3K inhibitor), poziotinib (HER2 inhibitor) and osimertinib (EGFR inhibitor) [25,35,36], and will likely be the preferred mode forward for maximal and durable tumor growth inhibition [25,37,38].
Our comprehensive phospho-array enabled us to identify potential mechanisms underlying TNO155 resistance. We found that the level of p-4E-BP1 and p-P70S6 K to be generally higher among the TNO155-resistant lines, suggestive of mTOR pathway activation [Fig. 3C]. A recent study by Kurupi et al. using SHP099 suggested that its efficacy is achieved via co-inhibition of PI3K and MEK signaling that eventually converged to downregulation of mTOR [9]. We hypothesized that a small subset of OSCCs may have constitutive activation of the mTOR pathway that rescue the cells from SHP2 inhibition. Interestingly, our previous CRISPR screen data also showed corroborating results where MTOR-dependent OSCC is significantly less dependent on PTPN11 [Supplementary Fig. 3]. Our study is the first to evaluate the rational combination of TNO155 with the mTOR inhibitor, everolimus in OSCC and confirmed that everolimus could significantly lower the IC50 of TNO155 with synergistic effects seen across various low doses combinations. Everolimus is an FDA-approved drug for few other indications and recent Phase II trial with head and neck cancer (HNC) patients using everolimus as adjuvant therapy have yielded promising results in terms of improving progression free survival (PFS) [39]. While for TNO155, early data from its first-in-human (FIH) trial also showed safety and favorable pharmacokinetics. However, limited single-agent activity was observed [14], and novel combinations are still being explored. Moving forward, challenges lie in the identification of optimal doses for the combination therapy of TNO155 with everolimus, for minimal toxicities and maximal synergy. An on-going study using a novel SHP2 inhibitor, PF-07284892 have demonstrated proof-of-concept evidence that a novel Phase I trial design which allowed for evaluation of effective combinations during dose escalation study is feasible, more ethical and patient-centric [40]. Investigations on the clinical benefit on OSCC patients treated with TNO155 combined with everolimus should ideally follow this novel design [40] to accelerate testing of this combinations. In addition to the combination with everolimus, an area of focus in the future is to explore the combination of TNO155 with other mainstay treatments for OSCC such as chemotherapy or radiotherapy. Additionally, the long-term effects and potential development of resistance from using the TNO155 and everolimus combination should be investigated. This research will help identify mechanisms of resistance and potential biomarkers that could anticipate refractory disease.
In summary, our study reports that targeting the enriched PTPN11 dependency using the SHP2 inhibitor TNO155 is promising, and provides novel insights on the effect of TNO155 on MAPK and JAK/STAT signaling pathway. We have uncovered that TNO155 treatment results in the upregulation of p-SHP2 among resistance lines and confirmed that TNO155 downregulates p-STAT1 and p-STAT3 exclusively among sensitive lines. From these observations our drug combination studies also demonstrated TNO155's synergistic interaction with everolimus in OSCC. The clinical development of SHP2 inhibitors as single agent or in combination with other targeted therapy and immunotherapy are actively on-going. Our study provides insights into potential mechanisms of resistance that could be further investigated in patients enrolled in clinical studies and suggest that drug combinations especially with mTOR inhibitors like everolimus warrants further investigation in this tumor type to overcome intrinsic resistance.
CRediT authorship contribution statement
Annie Wai Yeeng Chai: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Yee Hua Tan: Visualization, Validation, Methodology, Formal analysis, Data curation. Shiyin Ooi: Visualization, Validation, Methodology, Formal analysis, Data curation. Pei San Yee: Visualization, Validation, Methodology, Formal analysis, Data curation. Shi Mun Yee: Visualization, Validation, Methodology, Formal analysis, Data curation. Sok Ching Cheong: Writing – review & editing, Supervision, Investigation, Conceptualization.
Funding statement
This project is funded by the Dr. Ranjeet Bhagwan Singh (RBS) Research Grant 2021–2023 awarded to Dr. Annie Chai, administered by the Academy Sains Malaysia (ASM).
Acknowledgments
The CRISPR screen data is generated from a previous work funded by the Newton Ungku Omar Fund to Dr. Mathew Garnett (Wellcome Sanger Institute) and Prof. Dr. Sok Ching Cheong (Chai et al., 2020, eLife). We also thank Dr. Hazwani Mat Saad and Ms. Megan Lian Jia Wen in their help with some of the technical replicates contributing to Fig. 1. This work was supported by donors of Cancer Research Malaysia, a non-profit research organization, committed to an understanding of cancer prevention, diagnosis and treatment of cancers common in Asia.
Footnotes
The authors declare no potential conflicts of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e39677.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Torre L.A., et al. Global cancer statistics, 2012. CA A Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.J F., et al. International Agency for Research on Cancer; Lyon, France: 2020. Global Cancer Observatory: Cancer Today. [Google Scholar]
- 3.Chai A.W.Y., Lim K.P., Cheong S.C. Translational genomics and recent advances in oral squamous cell carcinoma. Semin. Cancer Biol. 2019;61:71–83. doi: 10.1016/j.semcancer.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Bonner J.A., et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11(1):21–28. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 5.Chai A.W.Y., et al. Genome-wide CRISPR screens of oral squamous cell carcinoma reveal fitness genes in the Hippo pathway. Elife. 2020;9 doi: 10.7554/eLife.57761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentires-Alj M., et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64(24):8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 7.Liu Q., et al. Targeting SHP2 as a promising strategy for cancer immunotherapy. Pharmacol. Res. 2020;152 doi: 10.1016/j.phrs.2019.104595. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y.N., et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535(7610):148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 9.Kurupi R., et al. Pharmacologic inhibition of SHP2 blocks both PI3K and MEK signaling in low-epiregulin HNSCC via GAB1. Cancer Res Commun. 2022;2(9):1061–1074. doi: 10.1158/2767-9764.CRC-21-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai X., et al. SHP2 inhibitors maintain TGFbeta signalling through SMURF2 inhibition. npj Precis. Oncol. 2023;7(1):136. doi: 10.1038/s41698-023-00486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaMarche M.J., et al. Identification of TNO155, an allosteric SHP2 inhibitor for the treatment of cancer. J. Med. Chem. 2020;63(22):13578–13594. doi: 10.1021/acs.jmedchem.0c01170. [DOI] [PubMed] [Google Scholar]
- 12.Nichols R.J., et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol. 2018;20(9):1064–1073. doi: 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong J., Long Y.Q. Recent advances in the discovery of protein tyrosine phosphatase SHP2 inhibitors. RSC Med. Chem. 2022;13(3):246–257. doi: 10.1039/d1md00386k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brana I., et al. Initial results from a dose finding study of TNO155, a SHP2 inhibitor, in adults with advanced solid tumors. J. Clin. Oncol. 2021;39(15_suppl) 3005-3005. [Google Scholar]
- 15.Cheng Y., et al. MAPK signaling pathway in oral squamous cell carcinoma: biological function and targeted therapy. Cancers. 2022;14(19) doi: 10.3390/cancers14194625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngan H.L., et al. Precision drugging of the MAPK pathway in head and neck cancer. NPJ Genom Med. 2022;7(1):20. doi: 10.1038/s41525-022-00293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpentier G. Dot blot analyzer. 2010. http://rsb.info.nih.gov/ij/macros/toolsets/ProteinArrayAnalyzer.txt Available from:
- 18.Chou T.-C. vol. 7. Synergy; 2018. pp. 49–50. (The Combination Index (CI < 1) as the Definition of Synergism and of Synergy Claims). [Google Scholar]
- 19.Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 20.Zhang G., et al. OShnscc: a novel user-friendly online survival analysis tool for head and neck squamous cell carcinoma based on RNA expression profiles and long-term survival information. J. Zhejiang Univ. - Sci. B. 2022;23(3):249–257. doi: 10.1631/jzus.B2100512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie H., et al. Upregulation of Src homology phosphotyrosyl phosphatase 2 (Shp2) expression in oral cancer and knockdown of Shp2 expression inhibit tumor cell viability and invasion in vitro. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117(2):234–242. doi: 10.1016/j.oooo.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 22.Wang H.-C., et al. Src-homology 2 domain-containing tyrosine phosphatase 2 promotes oral cancer invasion and metastasis. BMC Cancer. 2014;14(1):442. doi: 10.1186/1471-2407-14-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L., Fu L. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm. Sin. B. 2015;5(5):390–401. doi: 10.1016/j.apsb.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadhan R., Srinivas P., Pillai M.R. RTKs in pathobiology of head and neck cancers. Adv. Cancer Res. 2020;147:319–373. doi: 10.1016/bs.acr.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 25.Liu C., et al. Combinations with allosteric SHP2 inhibitor TNO155 to block receptor tyrosine kinase signaling. Clin. Cancer Res. 2021;27(1):342–354. doi: 10.1158/1078-0432.CCR-20-2718. [DOI] [PubMed] [Google Scholar]
- 26.Grossmann K.S., et al. The tyrosine phosphatase Shp2 in development and cancer. Adv. Cancer Res. 2010;106:53–89. doi: 10.1016/S0065-230X(10)06002-1. [DOI] [PubMed] [Google Scholar]
- 27.Prahallad A., et al. PTPN11 is a central node in intrinsic and acquired resistance to targeted cancer drugs. Cell Rep. 2015;12(12):1978–1985. doi: 10.1016/j.celrep.2015.08.037. [DOI] [PubMed] [Google Scholar]
- 28.Garcia Fortanet J., et al. Allosteric inhibition of SHP2: identification of a potent, selective, and orally efficacious phosphatase inhibitor. J. Med. Chem. 2016;59(17):7773–7782. doi: 10.1021/acs.jmedchem.6b00680. [DOI] [PubMed] [Google Scholar]
- 29.Lu H., et al. Resistance to allosteric SHP2 inhibition in FGFR-driven cancers through rapid feedback activation of FGFR. Oncotarget. 2020;11(3):265–281. doi: 10.18632/oncotarget.27435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qureshy Z., Johnson D.E., Grandis J.R. Targeting the JAK/STAT pathway in solid tumors. J Cancer Metastasis Treat. 2020;6:27. [PMC free article] [PubMed] [Google Scholar]
- 31.Lai S.Y., Johnson F.M. Defining the role of the JAK-STAT pathway in head and neck and thoracic malignancies: implications for future therapeutic approaches. Drug Resist. Updates. 2010;13(3):67–78. doi: 10.1016/j.drup.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Mali S.B. Review of STAT3 (signal transducers and activators of transcription) in head and neck cancer. Oral Oncol. 2015;51(6):565–569. doi: 10.1016/j.oraloncology.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Jiang M., Li B. STAT3 and its targeting inhibitors in oral squamous cell carcinoma. Cells. 2022;11(19) doi: 10.3390/cells11193131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu D., Qu C.K. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 2008;13:4925–4932. doi: 10.2741/3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heynen G.J.J.E., et al. Targeting SHP2 phosphatase in breast cancer overcomes RTK-mediated resistance to PI3K inhibitors. Breast Cancer Res. 2022;24(1):23. doi: 10.1186/s13058-022-01521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reischmann N., et al. Overcoming MET-mediated resistance in oncogene-driven NSCLC. iScience. 2023;26(7) doi: 10.1016/j.isci.2023.107006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M., et al. Strategies to overcome drug resistance using SHP2 inhibitors. Acta Pharm. Sin. B. 2021;11(12):3908–3924. doi: 10.1016/j.apsb.2021.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris S. SHP2 inhibitors undergo exploration in combinations. Targeted Therapies in Oncology. 2023;12:56. [Google Scholar]
- 39.Nathan C.O., et al. A randomized multi-institutional phase II trial of everolimus as adjuvant therapy in patients with locally advanced squamous cell cancer of the head and neck. Clin. Cancer Res. 2022;28(23):5040–5048. doi: 10.1158/1078-0432.CCR-21-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drilon A., et al. SHP2 inhibition sensitizes diverse oncogene-addicted solid tumors to Re-treatment with targeted therapy. Cancer Discov. 2023;13(8):1789–1801. doi: 10.1158/2159-8290.CD-23-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for the phosphorylation array can be accessed from Figshare - dx.doi.org/10.6084/m9.figshare.26,342,137.