Abstract
Neurexin, a molecule associated with autism spectrum disorders, is thought to function mainly in neurons. Recently, it was reported that Neurexin is also present in muscle, but the role of Neurexin in muscle is still poorly understood. Here, we demonstrate that the overexpression of Neurexin in muscles effectively restored the locomotor function of Drosophila neurexin mutants, while rescuing effects are observed within the nervous. Notably, the defects in muscle structure and function caused by Neurexin deficiency were similar to those caused by mutations in dystroglycan, a gene associated with progressive muscular dystrophy. The absence of Neurexin leads to muscle attachment defects, emphasizing the essential role of Neurexin in muscle integrity. Furthermore, Neurexin deficiency reduces Dystroglycan glycosylation on the cell surface, which is crucial for maintaining proper muscle structure and function. Finally, Neurexin guides Dystroglycan to the glycosyltransferase complex through interactions with Rotated Abdomen, a homolog of mammalian POMT1. Our findings reveal that Neurexin mediates muscle development and function through Dystroglycan glycosylation, suggesting a potential association between autism spectrum disorders and muscular dystrophy.
Subject terms: Mechanisms of disease, Membrane proteins
The critical role of Neurexin in facilitating Dystroglycan glycosylation and its impact on muscle structure and function in Drosophila.
Introduction
Neurexin (NRX), a protein associated with autism spectrum disorders (ASDs), plays a critical role in synaptic morphology and function in both Drosophila and mammals1–5. In addition, NRX also functions in nonneuronal cells. α-NRX mediates Ca2+-induced exocytosis in pituitary melanotroph cells in mice6. α-NRX, β-NRX, and Neuroligins are expressed in the endothelial cells and smooth muscle cells of the mouse vascular wall7. NRX1-α plays a role in promoting insulin secretion in pancreatic β cells7, whereas β-NRX inhibits angiogenesis in the chick embryo chorioallantoic membrane8. Previous studies have reported that Drosophila Neurexin (DNRX) is expressed mainly in neurons in the central nervous system2,9. Subsequent studies have revealed that DNRX is also expressed in the embryonic and larval muscle cells10,11, and in the photoreceptor cells of adult flies12. These results indicate a potential function of DNRX in nonneuronal cells.
Muscular dystrophy is characterized by progressive muscle weakness and degeneration. Duchenne muscular dystrophy (DMD), one of the most severe muscular dystrophies, causes significantly decreased IQ scores, inadequate responses to bright light stimuli and electroretinogram measurements in patients13,14. Approximately 25% of patients with DMD meet the diagnostic criteria for ASDs15,16. The Dystrophin-Glycoprotein complex (DGC) is an essential component of the macromolecular structure linking the extracellular matrix to the actin cytoskeleton and provides mechanical support for the stability of the muscle cell membrane. Mutations in the DGC result in the most severe form of muscular dystrophy, such as Limb Girdle Muscular Dystrophy17,18. The central component of the DGC, Dystroglycan (DG), is responsible for the structural stability of the sarcolemma and has been reported to bind the 2nd LNS-domain of α-NRXs in an SS2-dependent manner19. Furthermore, NRX-3 binding to DG sustains inhibitory synaptic transmission, indicating that trans-synaptic DG and NRX binding is necessary and sufficient for a normal release probability at inhibitory synapses20. However, the physiological significance of the NRX-DG interaction in muscle cell remains inadequately understood.
DG is highly conserved among vertebrates and is commonly expressed in the brain and skeletal muscle21. Loss of DG causes defects in muscle attachment, contraction, and membrane resistance22. The extracellular mucin-like domain of DG undergoes extensive glycosylation, producing various glycans such as O-linked mannose and O-GalNAC structures. In addition to the Dg gene, a group of glycosyltransferases, which are located mainly in the endoplasmic reticulum (ER) and Golgi complex, play a role in the addition of O-mannosyl glycans to DG21,23,24, and mutations in these glycosyltransferases are associated with muscular dystrophy and neural migration defects21. The initial step in the generation of these glycans is the addition of mannose to serine or threonine residues of DG in an O-linked manner, which is carried out by two protein O-mannosyltransferases (POMTs), POMT1 and POMT225,26. These two components form a biochemically active complex, and mutation of either leads to muscular dystrophy27,28.
There are three DG isoforms in Drosophila, one of which contains the full mucin-like domain (DG-C) and is highly glycosylation29,30. Rotated Abdomen (RT) and Twisted (TW), the homologs of POMT1 and POMT2, respectively, in Drosophila, act as glycosyltransferases for DG31. Both single and double mutants of rt and tw results in defects in muscle attachment and contraction in Drosophila at the larval stage, mimicking the phenotypes caused by the loss of DG, suggesting the requirement of glycosylation for the normal function of Drosophila DG22. However, the mechanism underlying DG glycosylation by RT and TW is still unknown.
Here, we show that the loss of DNRX results in alterations in locomotion and muscle morphogenesis in Drosophila, which can be effectively rescued by DNRX overexpression in muscle. The muscle structural and functional defects induced by DNRX deficiency resemble those observed in Dg mutants. Moreover, DNRX facilitates DG localization through its N-terminal region. Furthermore, the loss of DNRX decreases DG glycosylation on the cell surface, which is essential for muscle structure and function. Overall, we demonstrate that DNRX bridges DG to the RT/TW glycosyltransferase complex, elucidating the crucial roles of DNRX in muscle structure and function.
Results
Absence of DNRX in muscle leads to musculature defects
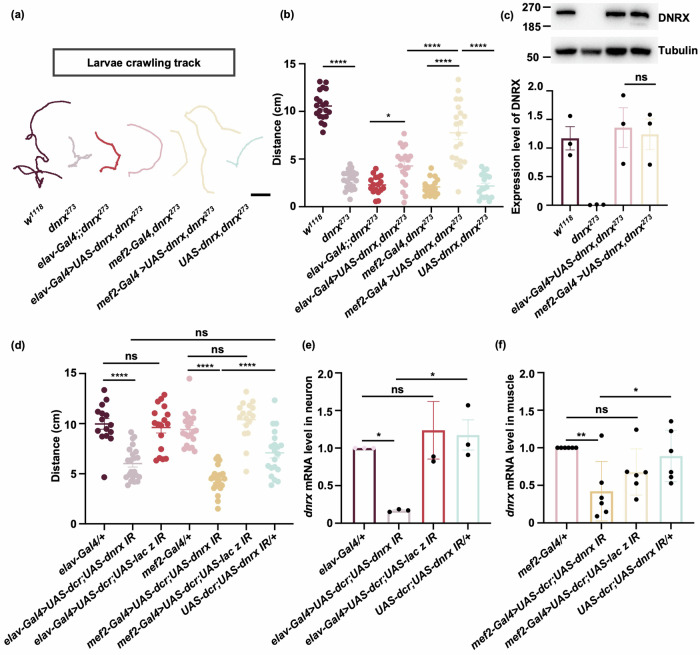
DNRX is expressed in Drosophila embryonic and larval muscle cells10,11. However, the function of DNRX in muscle is a mystery. We first assessed locomotor activity to evaluate the roles of DNRX in Drosophila in the larval stage. We assessed locomotor activity by recording the crawling trajectories of third-instar larvae for 3 min. dnrx273 homozygotes null mutants9,32 (hereafter referred to as dnrx273), which lack most of the coding sequences for the extracellular region of DNRX, presented a severe reduction in crawling ability, and the distance was decreased by approximately 80% compared to the w1118 larvae (Fig. 1a, b). Furthermore, we observed that when DNRX was overexpressed in dnrx273 mutants separately using muscle-specific mef2-Gal433 and neuron-specific elav-Gal434 driver, the crawling defects in third-instar larvae were partially restored (Fig. 1a, b). Western blot revealed that DNRX expression was the same when mef2-Gal4 and elav-Gal4 were used to overexpress DNRX (Fig. 1c), which excluded the possibility of different efficiencies of muscle and neuronal restoration as a result of DNRX expression. Additionally, the locomotor activity of dnrx273 larvae was improved significantly when DNRX was overexpressed in both neurons and muscle, but DNRX overexpression had no effect on the locomotor activity of wild-type larvae (Supplementary Fig. 1e). To further confirm the role of DNRX in the locomotion of Drosophila larvae, we knocked down DNRX expression in muscle and neurons separately. Consistent with the result of the rescue experiment, locomotor activity decreased significantly when DNRX was knocked down in muscle and neurons (Fig. 1d). However, it is worth noting that the UAS-dcr;UAS-dnrx IR flies displayed locomotor defects. Therefore, locomotor activity was significantly reduced in flies with DNRX knockdown in neurons compared to those with elav-Gal4-driven DNRX knockdown but not to UAS-dcr;UAS-dnrx IR flies (Fig. 1d). The efficiency of RNAi was determined via RT‒PCR, which revealed that RNAi significantly decreased dnrx mRNA expression in muscle and neurons (Fig. 1e, f). These findings suggest that DNRX is essential for proper locomotor activity in Drosophila larvae.
Fig. 1. DNRX in muscle and neurons is required for Drosophila locomotion.
a Examples of the crawling trajectories of Drosophila larvae of the indicated genotypes over 3 min. Scale bar, 10 mm. b The distance traveled by larvae of the indicated genotypes in 3 min. n = 20, 21, 17, 20, 17, 20, 17 for the different genotypes from w1118 to UAS-dnrx,dnrx273. The data are presented as the means ± SEMs. ****p < 0.0001, *p < 0.05; one-way ANOVA test. c Upper panel: Western blotting analysis of DNRX expression in neurons and muscle. Bottom panel: Quantitative analysis of DNRX expression levels from the data in the upper panel. The data are presented as the means ± SEMs of three independent experiments. ns, p > 0.05; one-way ANOVA test. d The distance traveled by larvae of DNRX knockdown within 3 min. n = 15, 22, 17, 20, 24, 17, 20 for the different genotypes from elav-Gal4/+ to UAS-dcr;UAS-dnrx IR/+. The data are presented as the means ± SEMs. ****p < 0.0001; ns, p > 0.05; one-way ANOVA test. e Quantification of dnrx mRNA levels in the neurons of DNRX-knockdown flies as indicated. The data are presented as the means ± SEMs of three independent experiments. *p < 0.05; ns, p > 0.05; one-way ANOVA test. f Quantification of dnrx mRNA levels in the muscles of DNRX-knockdown flies as indicated. The data are presented as the means ± SEMs of six independent experiments. **p < 0.01, *p < 0.05; ns, p > 0.05; one-way ANOVA test.
Next, to visualize DNRX in muscle, we used a knock-in line for DNRX via the CRISPR/CAS9 system. HA was introduced at the end of the open reading frame because of the low expression of endogenous DNRX in muscle and the limitations of anti-DNRX antibody. The line enabled us to observe DNRX signals indicated by HA staining, along the muscle fibers at the neuromuscular junction (NMJ) (Supplementary Fig. 1a, b’). Furthermore, we utilized dnrx-Gal435 to drive DNRX expression mimicking the endogenous expression pattern of DNRX and found that DNRX was present in muscle (Supplementary Fig. 1c, d’). These findings demonstrate that DNRX is expressed in muscle and at the NMJ in Drosophila larvae, suggesting that DNRX has a function in muscle tissue.
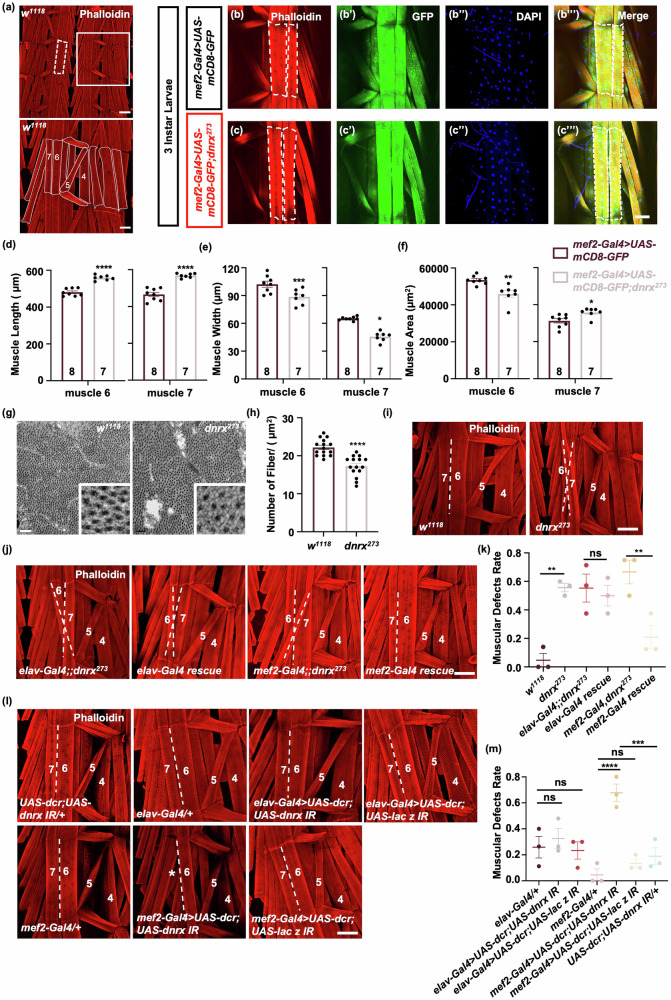
Given the crucial role of muscle integrity in facilitating proper locomotion, we further investigated whether the absence of DNRX impacts muscle structure. In w1118 larvae, muscle fibers are organized in a predictable pattern and establish connections with the underlying epidermis through interactions with tendon cells at specific locations36 (Fig. 2a). To visualize the muscle morphology of third-instar larvae, the expression of UAS-mCD8-GFP was driven by mef2-Gal4. Compared to control, muscle 6 and muscle 7 in segment A3 of the dnrx273 mutants appeared longer and narrower, with a significantly reduced muscle area specifically in muscle 6 (Fig. 2a–c”’, d–f). Similar morphological alterations were observed in second-instar larvae (Supplementary Fig. 2a–e). Further ultrastructural examination of muscle cross-sections via electron microscopy revealed a decrease in the number of muscle fibers per area in dnrx273 mutants (Fig. 2g–h). Additionally, we investigated muscle attachment, another crucial indicator of muscle integrity22. In w1118 larvae, muscles were connected to the underlying epidermis in a stereotypical pattern. However, one or more muscles were frequently improperly united or misattached in dnrx273 mutants (Fig. 2i). There was no consistent pattern in terms of which hemisegment or muscle was affected. Occasionally, we also observed the presence of additional muscle tissue (Supplementary Fig. 2f, g). Most instances of muscle mis-attachment occurred in muscle 6/7 crossover; in other cases, missing muscle was observed in the hemisegment. The ratio of samples with muscle attachment defects to the total number tested served as a crucial indicator of muscle integrity22, demonstrating that there were significantly more dnrx273 than in w1118 with muscle attachment defects (Fig. 2i, k). We also tested other dnrx null mutants, such as dnrx832, as well as trans-heterozygous null mutants, including dnrx273/dnrx1742 and dnrx273/dnrx83, and observed the same muscle attachment defects across these dnrx mutants (Supplementary Fig. 2f–k). Notably, these attachment defects were restored by overexpressing DNRX in muscle, but not in neurons (Fig. 2j, k). To further determine the role of DNRX in muscle integrity in Drosophila larvae, we knocked down DNRX expression specifically in muscle and neurons. Consistent with the results of the rescue experiment, the occurrence of muscle attachment defects increased significantly when DNRX was knocked down in muscle but not in neurons (Fig. 2l, m). These findings underscore the role of DNRX in modulating muscle function by influencing muscle structure and integrity.
Fig. 2. Ablation of DNRX causes defects in the muscle structure of Drosophila larvae.
a Upper panel: Muscle pattern diagram of third-instar Drosophila larvae. Scale bar, 200 μm. The dashed white box represents the area shown in (b, c”’). Bottom panel: The enlarged area is outlined in the white box in the upper panel for clarity. Scale bar, 100 μm. The arrangement of muscles in part of a hemisegment in w1118 files; the muscles referred to in the text are labeled by number according to standard nomenclature. b, c”’ Phalloidin-stained muscles of Drosophila larvae expressing mef2-Gal4 > UAS-mCD8-GFP (Control) and mef2-Gal4 > UAS-mCD8-GFP, dnrx273. Phalloidin (red) was used to visualize actin, GFP (green) was used to label the cell membrane, and DAPI (blue) was used to label the nucleus. Scale bar, 100 μm. d–f Characterization of larval muscle structure. Average muscle length (d), average muscle width (e), and muscle area (f) of muscle 6 or 7. mef2-Gal4 > UAS-mCD8-GFP (n = 8) and mef2-Gal4 > UAS-mCD8-GFP, dnrx273 (n = 7). The data are presented as the means ± SEMs. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05; two-tailed unpaired Student’s t-test. g Electron micrographs of larval muscle showing the muscle fibers of w1118 flies and dnrx273 mutants in transverse sections. Scale bar, 0.2 μm. h Quantification of the number of muscle fibers per unit area in w1118 flies (n = 15) and dnrx273 mutants (n = 15). The data are presented as the means ± SEMs. ****p < 0.0001; two-tailed unpaired Student’s t-test. i, j, l Phalloidin-stained muscles of Drosophila larvae showing muscle attachment in the indicated genotypes. The dashed lines represent the boundaries of muscles 6 and 7. The double dashed lines indicate misalignment of muscle attachment in muscles 6 and 7. * indicates muscle loss. Scale bar, 100 μm. k Quantification of the ratio of muscle defects. elav-Gal4 rescue (elav-Gal4 > UAS-dnrx, dnrx273), mef2-Gal4 rescue (mef2-Gal4 > UAS-dnrx, dnrx273). The data are presented as the means ± SEMs of three independent experiments. n = 22, 25, 22, 22, 24, 24 for the different genotypes from w1118 to mef2-Gal4 rescue. **p < 0.01; ns, p > 0.05; one-way ANOVA test. m Quantification of the ratio of muscle defects in Drosophila larvae in which DNRX was knocked down in muscle and in neurons. The data are presented as the means ± SEMs of three independent experiments. n = 24, 22, 24, 23, 21, 24, 20 for the different genotypes from elav-Gal4/+ to UAS-dcr;UAS-dnrx IR/+. ****p < 0.0001, ***p < 0.001; ns, p > 0.05; one-way ANOVA test.
DNRX interacts with DG through its extracellular region
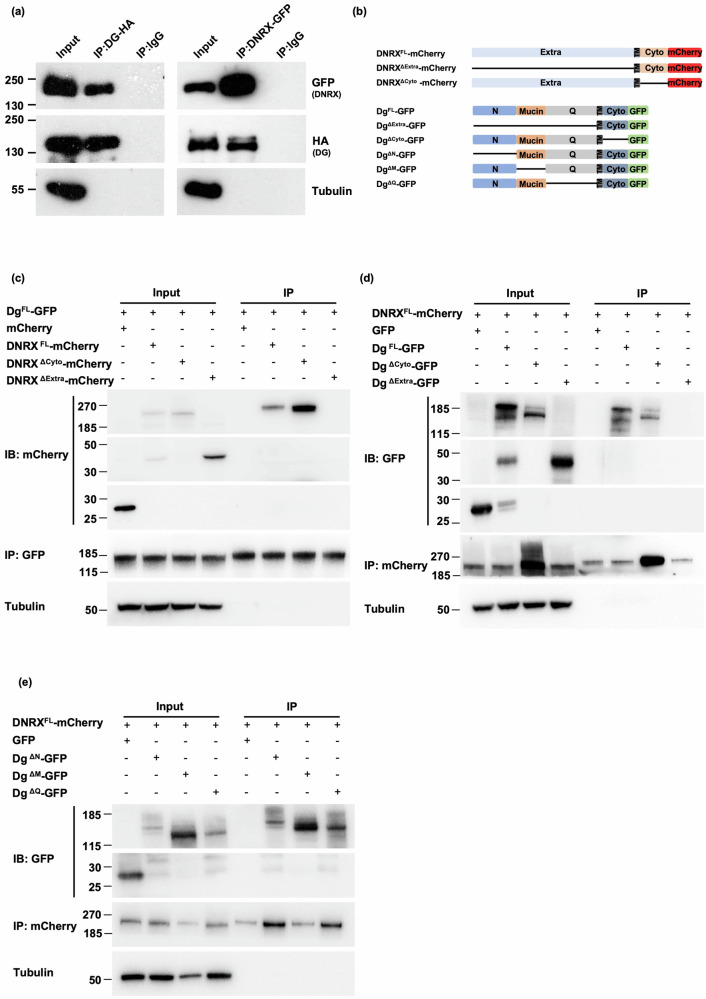
Given the resemblance of muscle attachment defects between dnrx mutants and Dg mutants22, we investigated the interaction between DNRX and DG in Drosophila, which has been demonstrated in mice19. Owing to the limitations of antibodies, we individually expressed DG tagged with HA or DNRX tagged with GFP via the mef2-Gal4 driver. Bidirectional coimmunoprecipitation (co-IP) experiments demonstrated that DNRX interacted with DG in vivo and that DNRX physically interacted with DG in Drosophila muscle (Fig. 3a).
Fig. 3. DNRX physically interacts with DG through its extracellular domain.
a Coimmunoprecipitation demonstrating the interaction between DNRX and DG in Drosophila muscle. An anti-GFP antibody was used to label DNRX, and an anti-HA antibody was used to label DG. Left: Immunoprecipitation with an anti-HA antibody. Right: Immunoprecipitation with an anti-GFP antibody. b Construction of plasmids with deletion of different regions of DNRX and the Dg-C isoform. c Coimmunoprecipitation demonstrating that DNRX interacts with DG through its extracellular region in S2 cells. An anti-GFP antibody was used for immunoprecipitation. d Coimmunoprecipitation demonstrating that DG interacts with DNRX through its extracellular region in S2 cells. An anti-mCherry antibody was used for immunoprecipitation. e Coimmunoprecipitation demonstrating that DNRX interacts with multiple regions of Dg-C in S2 cells. An anti-mCherry antibody was used for immunoprecipitation.
To map the region of DNRX that interacts with DG, we engineered multiple constructs encoding full-length DNRX or truncated DNRX fragments fused with mCherry, named DNRXFL, DNRXΔExtra (excluding Met1~Arg1693) and DNRXΔCyto (excluding Val1715~Val1840), and a construct encoding full-length DG tagged with GFP, named DGFL (Fig. 3b). We co-transfected the different DNRX constructs with DGFL into S2 cells and HEK293T cells, respectively. Co-IP experiments revealed that DG bound to DNRXFL and DNRXΔCyto but not DNRXΔExtra, suggesting that DNRX interacts with DG via its N-terminus (Fig. 3c and Supplementary Fig. 3a). Similarly, truncated DG fragments, namely, DgΔExtra (excluding Glu28~Thr1051), DgΔCyto (excluding Arg1077~Pro1179), DgΔN (excluding Glu28~Leu314), DgΔM (excluding Gly315~Leu592), and DgΔQ (excluding Pro593~Thr1051), were fused with GFP to map the region of DG that interacts with DNRX (Fig. 3b). We co-transfected the different DG constructs with DNRXFL into S2 cells and HEK293T cells, respectively. Co-IP experiments revealed that all of the DG constructs except DgΔExtra, i.e., DgΔCyto, DgΔN, DgΔM, and DgΔQ, bound to DNRX, suggesting that DG interacts with DNRX via its extracellular region (Fig. 3d, e and Supplementary Fig. 3b, c). These findings imply a slightly different binding mechanism between DNRX and DG in Drosophila in which the extracellular portions of DG potentially make significant contributions.
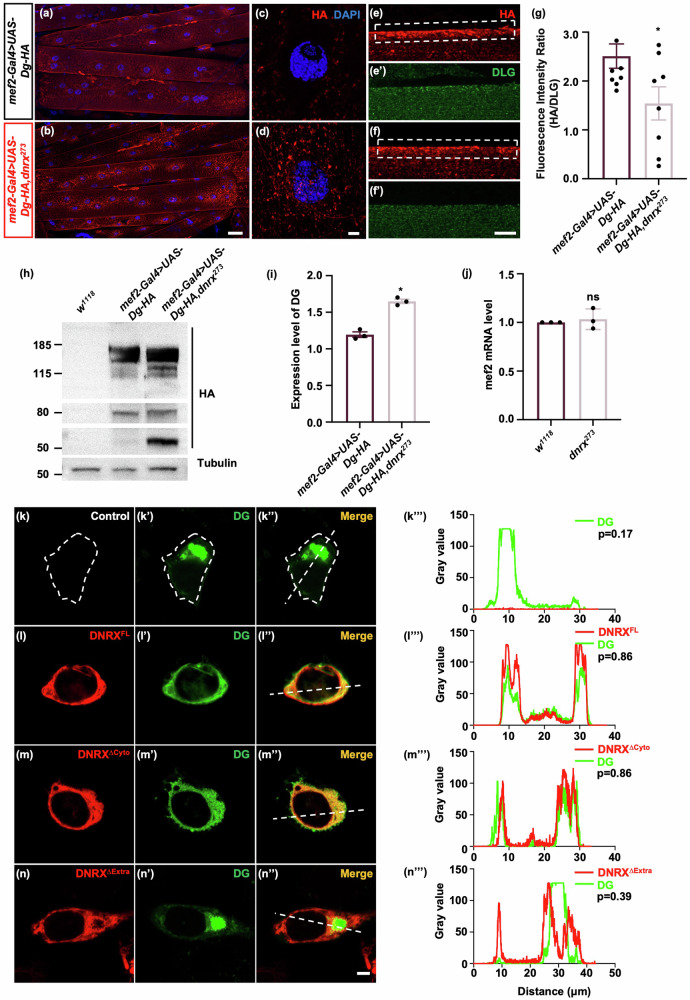
To explore the relationship between DNRX and DG in Drosophila muscle, we expressed DG tagged with HA via the mef2-Gal4 driver in both wild-type and dnrx273 mutant backgrounds. Compared with control, DG accumulated around nuclei in the muscle to a significantly greater extent in dnrx273 (Fig. 4a–d). Furthermore, compared with the control, the dnrx273 mutants presented a significant reduction in HA fluorescence intensity on the cell surface (Fig. 4e, f’). Quantitative analysis revealed that while the total amount of DG increased in dnrx273 mutants (Fig. 4h, i), the fluorescence intensity of DG on the cell surface significantly decreased (Fig. 4g), indicating that DG localization was impaired in dnrx273 mutants. Additionally, the level of DG isoforms with a molecular weight of approximately 50 kDa significantly increased in the dnrx273 mutants, whereas the level of isoforms with a molecular weight of approximately 80 kDa seemed to remain unchanged (Fig. 4h), suggesting the splicing or cleavage of DG via unknown mechanisms in Drosophila. The mRNA level of mef2 was assessed to rule out the possibility that increased MEF2 expression leads to increased DG expression (Fig. 4j). Then, the relationship between DNRX and DG accumulation was investigated in HEK293T cells. As expected, we observed DG accumulation around the nucleus following transfection of DGFL alone (Fig. 4k, k”). However, the aggregation of DG made observing its localization on the cell membrane challenging. Interestingly, abnormal DG accumulation was prevented when either DNRXFL or DNRXΔCyto was co-transfected with DGFL (Fig. 4l, m”), but not with DNRXΔExtra (Fig. 4n, n”). The results of colocalization analysis were consistent with the co-IP results in S2 cells (Figs. 4k”’–n”’ and 3c), suggesting that DNRX facilitates normal DG localization through its extracellular region.
Fig. 4. Loss of DNRX leads to abnormal accumulation of DG in vivo and in vitro.
a–d Immunostaining revealed DG localization in wild-type and dnrx273 mutants backgrounds. An anti-HA antibody was used to label DG (red), and DAPI (blue) was used to label the nucleus. Scale bars, 50 μm (a, b) and 5 μm (c, d). e, f ’ Immunostaining revealed that DG was localized on the muscle cell membrane in the wild-type and dnrx273 mutants backgrounds. An anti-HA antibody was used to label DG (red), and DLG (green) represents the postsynaptic muscle membrane. Scale bar, 10 μm. g Quantification of the fluorescence intensity ratio (HA/DLG) in the wild-type flies and dnrx273 mutant backgrounds. The data are presented as the means ± SEMs; mef2-Gal4 > UAS-Dg-HA, n = 9; mef-Gal42 > UAS-Dg-HA, dnrx273, n = 8. *p < 0.05; two-tailed unpaired Student’s t-test. h Western blotting revealed an increase in DG levels in the muscle of dnrx273 mutants. An anti-HA antibody used to label DG. The protein originated from larval body wall muscles. i Quantification of DG protein levels relative to tubulin levels. The data are presented as the means ± SDs of three independent experiments. *p < 0.05; two-tailed unpaired Student’s t-test. j Quantification of the mRNA levels of mef2 in the muscles of w1118 flies and dnrx273 mutants. The data are presented as the means ± SDs of three independent experiments. ns, p > 0.05; two-tailed unpaired Student’s t-test. k, k” Immunofluorescence images of HEK293T cells transfected with DG alone. l–l” Immunofluorescence images of HEK293T cells transfected with DG and DNRXFL. m–m” Immunofluorescence images of HEK293T cells transfected with DG and DNRXΔCyto. n–n” Immunofluorescence images of HEK293T cells transfected with DG and DNRXΔExtra. DAPI was used to stain the nucleus. Scale bar, 5 μm. k”’, l”’, m”’, n”’ Plots of pixel intensity along the straight dashed white line in the images to the left of each plot. p: Pearson’s coefficient.
DNRX facilitates DG glycosylation in Drosophila muscle
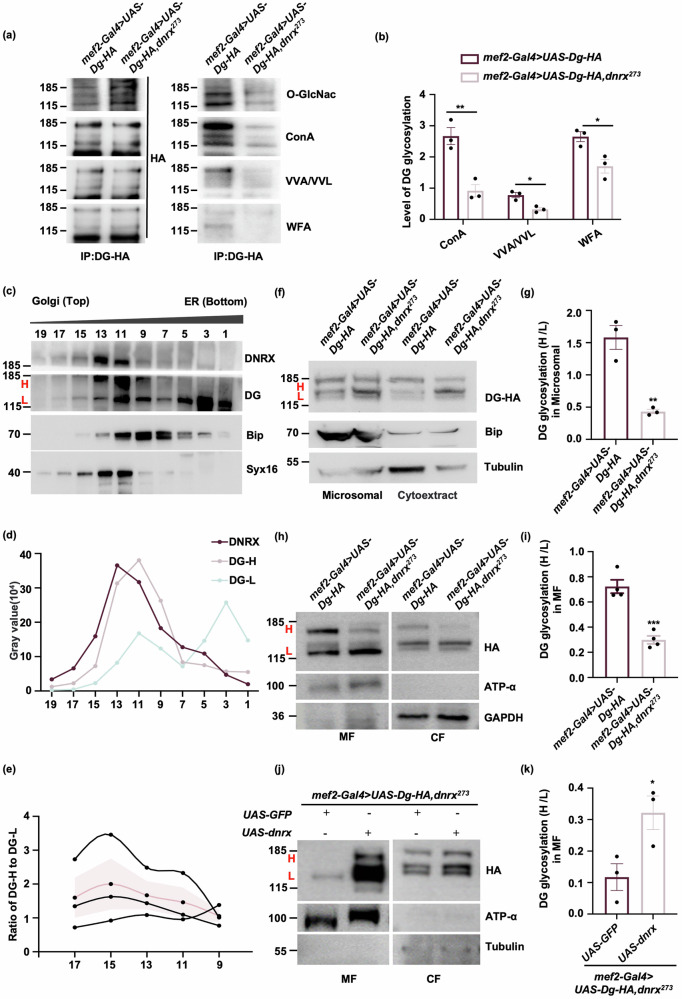
Extensive glycosylation of DG is crucial for normal muscle function, and aberrations in this process lead to various forms of muscular dystrophy37–40. To investigate the impact of DNRX deficiency on DG glycosylation, we expressed HA-tagged DG in both wild-type and dnrx273 mutant backgrounds via the mef2-Gal4. In comparison to control, the glycosylation of DG was significantly reduced in dnrx273 (Fig. 5a). Further analysis of the specific glycan structures of DG was conducted via lectin blotting. Compared to control, all the signals of Concanavalin A (Con A), representing α-linked mannose on DG, Vicia villosa (VVA), representing O-linked GalNAc, and floribunda agglutinin (WFA), representing terminal GalNAc were markedly diminished in dnrx273, indicating that the loss of DNRX results in a decrease in DG glycosylation (Fig. 5a, b).
Fig. 5. DG glycosylation is decreased by the absence of DNRX in Drosophila muscle.
a Decrease in DG glycosylation in the absence of DNRX. DG was labeled with an anti-HA antibody, and DG levels were subsequently analyzed via lectin blotting and western blotting. b Quantification of DG glycosylation via lectin blotting from the data in (a). The data are presented as the means ± SEMs of three independent experiments. **p < 0.01, *p < 0.05; two-tailed unpaired Student’s t-test. c Western blotting analysis that DNRX and increased DG glycosylation were present following density gradient centrifugation. The samples were separated into 19 fractions with varying densities through centrifugation, and the fractions were labeled 1 to 19. Bip was used as an ER marker. Syx16 was used as a Golgi marker. H: Heavily glycosylated DG, L: Lightly glycosylated DG. d The grayscale values of the bands corresponding to DNRX, DG-H and DG-L in each lane. e Quantification of the ratio of grayscale values of the DG-H to DG-L bands in lanes 9 to 17. The data are presented as the means ± SEMs of three independent experiments. f Western blot analysis revealed that loss of DNRX decreased DG glycosylation in the microsomal fraction. g Quantification of the ratio of DG-H to DG-L in the microsomal fraction. The data are presented as the means ± SEMs of three independent experiments. **p < 0.01; two-tailed unpaired Student’s t-test. h Western blot analysis revealed a decrease in DG glycosylation on the cell membrane in the absence of DNRX. MF: membrane fraction, CF: cytoplasmic fraction. An anti-HA antibody was used to label DG, ATP-α was used as a membrane fraction marker, and GAPDH was used as a cytoplasmic fraction marker. i Quantification of the ratio of DG-H to DG-L on the plasma membrane. The data are presented as the means ± SEMs of four independent experiments. ***p < 0.001; two-tailed unpaired Student’s t-test. j Western blot analysis revealed that DG glycosylation was rescued by the overexpression of full-length DNRX in muscle. Tubulin was used as a cytoplasmic fraction marker. k Quantification of the ratio of DG-H to DG-L on the plasma membrane. The data are presented as the means ± SEMs of three independent experiments. *p < 0.05, two-tailed unpaired Student’s t-test.
In eukaryotic cells, protein glycosylation most commonly begins in the ER and is completed in the Golgi. Protein glycosylation causes an observed shift in molecular weight, and glycosylated DG can be distinguished easily via gel electrophoresis, as the size of heavily glycosylated DG (H band, DG-H) is slightly greater than that of lightly glycosylated DG (L band, DG-L) (Fig. 5c, f, h, j). Organelle separation experiments utilizing density gradient centrifugation revealed that glycosylated DG was along with DNRX (Fig. 5c–e), revealing that DNRX is associated with the process of DG glycosylation. We quantified the grayscale values of the band corresponding to DNRX, DG-H, and DG-L and observed that the grayscale value of the DG-H band was consistent with that of the DNRX band (Fig. 5d). We performed quantitative analysis on the grayscale value ratio of DG-H band to DG-L band in lanes 9-17, as DG-H band surpasses DG-L band starting from lane 9 along with DNRX appearance prominently from lane 9, suggesting an increase in DG glycosylation (Fig. 5e). Furthermore, we isolated the microsomal fraction, which was rich in ER components and the Golgi apparatus, from Drosophila muscle, and western blotting revealed that DG glycosylation was decreased in the microsomal fraction of dnrx273 (Fig. 5f, g). These results imply that DNRX is essential for the glycosylation process of DG. To assess the glycosylation of DG on the cell membrane, we isolated the plasma membrane and the cytoplasmic components of Drosophila muscle. Compared with control, DG glycosylation at the plasma membrane was reduced of dnrx273 (Fig. 5h, i), and could be restored by overexpressing DNRXFL in muscle (Fig. 5j, k). Taken together, these findings suggest that DNRX is involved in promoting DG glycosylation, which is essential for the proper function of muscle cells.
RT/TW is essential for maintaining muscle structure and function
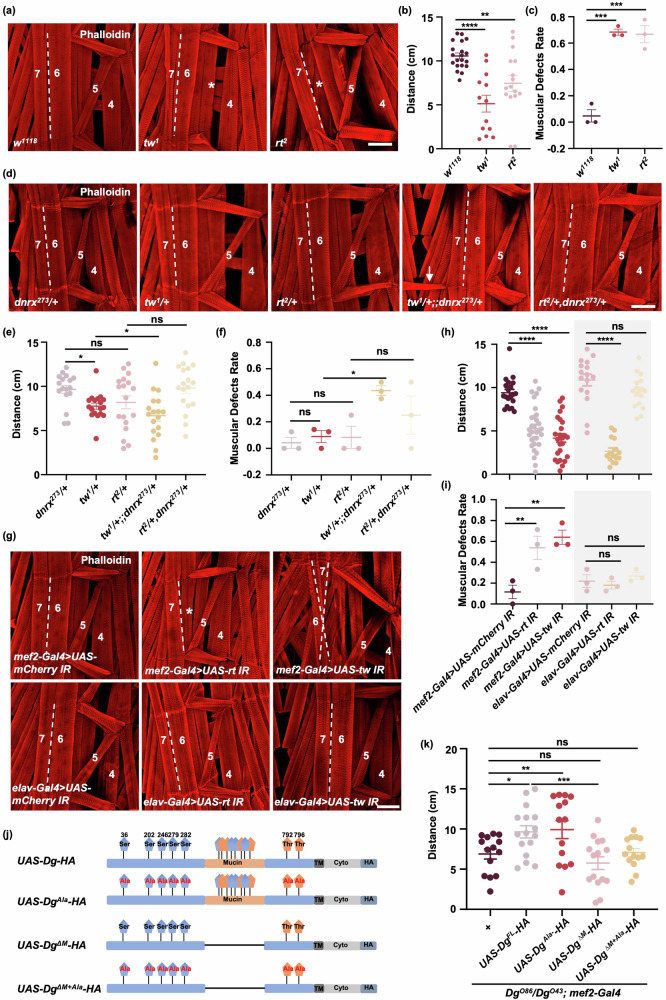
DG glycosylation plays a crucial role in connecting extracellular matrix proteins to muscle cells41. It has been reported that RT and TW are responsible for DG mannosylation in Drosophila31 and exhibit synergistic genetic interactions with DG glycosylation42,43. Consistent with previous study22, rt2 and tw1 mutants presented defects in muscle attachment (Fig. 6a, c), similar to dnrx mutants. Additionally, rt2 and tw1 mutants exhibited impaired locomotor ability (Fig. 6b). Furthermore, the genetic interactions between dnrx, rt and tw were investigated in heterozygous mutants. Interestingly, tw1/+ flies displayed a significant reduction in crawling ability but normal muscle attachment, and these deficits were exacerbated in tw1/+;;dnrx273/+ (Fig. 6d–f). However, rt2/+ and rt2/+,dnrx273/+ showed normal muscle attachment and locomotion (Fig. 6d–f), suggesting that partial retention of RT is sufficient to maintain muscle structure and function. Next, RT and TW were knocked down via mef2-Gal4 or elav-Gal4, and muscle morphology and function were evaluated. As expected, muscle defect rates and locomotor ability significantly decreased when RT or TW was knocked down in muscle, resembling what was seen in rt2 or tw1mutants (Fig. 6g–i). Surprisingly, locomotor ability also decreased significantly when RT was knocked down in neurons, although muscle morphology remained intact, suggesting that neuronal RT modulates locomotor ability independent of muscle function (Fig. 6g–i). DG glycosylation plays a crucial role in connecting extracellular matrix proteins to muscle cells. To determine whether extracellular matrix proteins are involved in muscle structure and function, we knocked down an extracellular matrix protein—Laminin—in muscle and neurons, respectively. Defects in muscle morphology and function were observed when Laminin A was specifically knocked down in muscle but not in neurons (Supplementary Fig. 5a–e). These findings indicate that RT/TW is essential for maintaining muscle structure and function.
Fig. 6. DG glycosylation is involved in muscle defects caused by DNRX deficiency.
a, d, g Phalloidin-stained muscles of Drosophila larvae showing muscle attachment defects in the indicated genotypes. The dashed lines represent the boundaries of muscles 6 and 7. The double dashed lines indicate misalignment of muscle attachment in muscles 6 and 7. * represents muscle loss. The arrow indicates muscle that is abnormal and should not be present, suggesting that muscle proliferated excessively. Phalloidin (red) was used to visualize actin. Scale bar, 100 μm. b The distance traveled by larvae of each genotype over 3 min. w1118, n = 20, tw1, n = 13, rt2, n = 16.The data are presented as the means ± SEMs. ****p < 0.0001, **p < 0.01; one-way ANOVA test. c Quantification of the ratio of flies of the indicated genotypes with muscle defects. w1118, n = 27, tw1, n = 25, rt2, n = 27. The data are presented as the means ± SEMs of three independent experiments. ***p < 0.001; one-way ANOVA test. e The distance traveled by larvae of each genotype over 3 min. n = 17, 18, 18, 18, 18 for the different genotypes from dnrx273/+ to rt2/+,dnrx273/+. *p < 0.05; ns, p > 0.05; one-way ANOVA test. f Quantification of the ratio of flies of the indicated genotypes with muscle defects. n = 23, 23, 22, 21, 21 for the different genotypes from dnrx273/+ to rt2/+,dnrx273/+. The data are presented as the means ± SEMs of three independent experiments. *p < 0.05; ns, p > 0.05; one-way ANOVA test. h The distance traveled by larvae of each genotype over 3 min. n = 20, 31, 25, 16, 15, 16 for the different genotypes from mef2-Gal4 > UAS-mCherry IR to elav-Gal4 > UAS-tw IR. ****p < 0.0001; ns, p > 0.05; one-way ANOVA test. i Quantification of the ratio of Drosophila larvae with knockdown of rt or tw in muscle or neurons with muscle defects. n = 22, 20, 23, 19, 22, 23 for the different genotypes from mef2-Gal4 > UAS-mCherry IR to elav-Gal4 > UAS-tw IR. The data are presented as the means ± SEMs of three independent experiments. **p < 0.01; ns, p > 0.05; one-way ANOVA test. j Schematic diagram of the DG plasmids constructed to generate transgenic Drosophila with different glycosylation site deletions. k The distance traveled by larvae of the indicated genotypes over 3 min. Glycosylation-deficient forms of DG were overexpressed by mef2-Gal4 in DgO86/DgO43 mutants. n = 14, 16, 14, 15, 14 for the different genotypes from Control (+) to UAS-Dg∆M+Ala-HA. The data are presented as the means ± SEMs. ***p < 0.001, **p < 0.01, *p < 0.05; ns, p > 0.05; one-way ANOVA test.
To simulate a lack of DG glycosylation, we mutated seven potential glycosylation sites (Ser36, Ser202, Ser246, Ser279, Ser282, Thr792 and Thr796) and the mucin-like domain on the basis of previous work31. We generated three transgenic flies: UAS-DgAla-HA (in which the seven glycosylation sites were mutated to alanine to emulate a non-glycosylated state), UAS-DgΔM-HA (in which the mucin-like domain was deleted), and UAS-DgΔM+Ala-HA (in which the glycosylation sites were mutated to alanine and the mucin-like domain was deleted) (Fig. 6j). To assess the impact of DG glycosylation on the locomotion of Drosophila larvae, we subsequently overexpressed wild-type DG (UAS-Dg-HA) as well as the aforementioned glycosylation-deficient DG mutants using mef2-Gal4 in Dg mutant background. DgO43 and DgO86 are nonsense mutations caused by single nucleotide changes that introduce premature stop codons into the sequence44. Owing to the lethality of the DgO86 homozygotes, DgO86/DgO43 trans-heterozygotes were used as controls in the experiment. As expected, overexpression of wild-type DG in muscle significantly alleviated the crawling deficiencies in DgO86/DgO43 (Fig. 6k), whereas overexpression of DgΔM and DgΔM+Ala in muscle failed to rescue the crawling defects in Dg mutants (Fig. 6k), highlighting the crucial role of DG glycosylation in muscle function. Furthermore, overexpression of DgAla in muscle also rescued the crawling defects, indicating that the elimination of isolated glycosylation sites alone does not impair DG function (Fig. 6k). The findings suggest that the mucin-like domain is essential for the crawling ability of Drosophila larvae. Given that this domain is highly glycosylated, it is hypothesized that DG glycosylation may partially contribute to the locomotor function of Drosophila larvae.
DNRX bridges DG to the glycosyltransferase complex through interactions with RT
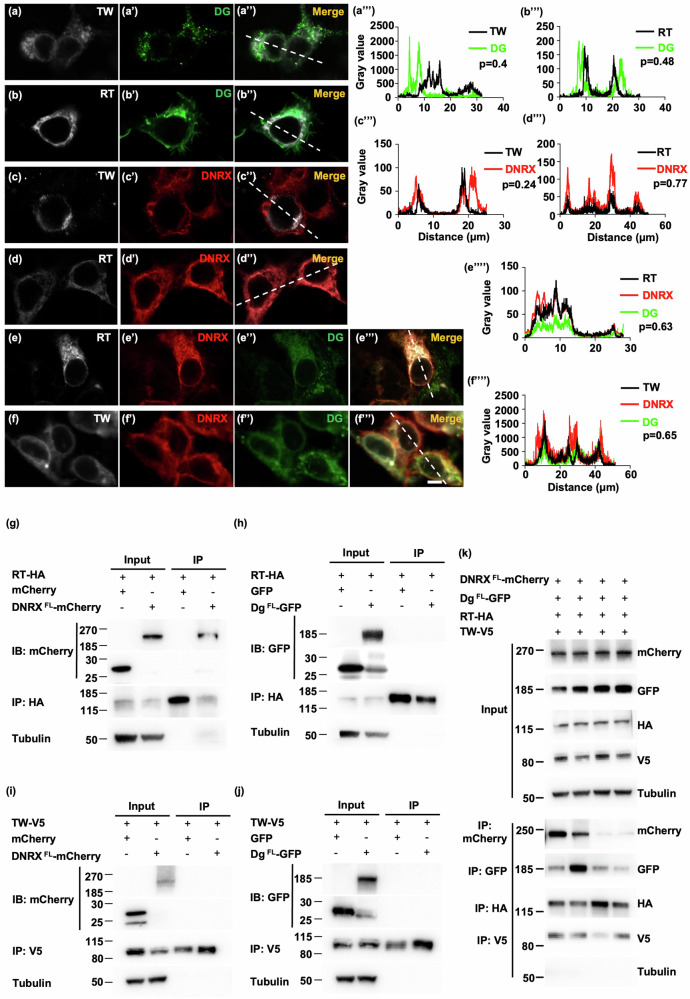
Next, we investigated how DNRX affects DG glycosylation in vitro. To visualize the fluorescence signals properly, we then transfected DG along with RT tagged with HA or TW tagged with V5 into HEK293T cell and observed the DG distribution within the cells. The results revealed that DG accumulated in the cells even in the presence of RT and TW (Fig. 7a, b”). Upon introduction of DNRX into these cells, the distribution of DG became more diffuse (Fig. 7e, f”’), indicating that the proper distribution of DG cannot be corrected by RT or TW alone and requires DNRX. Moreover, immunostaining demonstrated that DNRX colocalized with RT, but not with TW (Fig. 7c, d”). The colocalization of DNRX and RT suggested an interaction between these two proteins (Fig. 7d”’). Additionally, the fluorescence signals of RT, DNRX, and DG and those of TW, DNRX, and DG significantly overlapped and showed well-matched intensity profiles (Fig. 7e””, f””). This finding indicates a strong potential interaction among these proteins in the cells. Co-IP experiments revealed that, in both S2 cells and HEK293T cells, DNRX interacted with RT but not with TW. Additionally, DG exhibits no interactions with either RT or TW in either cell type (Fig. 7g–j and Supplementary Fig. 6a, d).
Fig. 7. DNRX links DG to the RT/TW complex.
a–a” Immunofluorescence images of HEK293T cells transfected with DG and TW. b–b” Immunofluorescence images of HEK293T cells transfected with DG and RT. c–c” Immunofluorescence images of HEK293T cells transfected with DNRX and TW. d–d” Immunofluorescence images of HEK293T cells transfected with DNRX and RT. e–e” Immunofluorescence images of HEK293T cells transfected with DG, DNRX and RT. f–f” Immunofluorescence images of HEK293T cells transfected with DG, DNRX and TW. Scale bar, 5 μm. a”’–d”’, e””–f”” Plots of the pixel intensity along the straight dashed white lines in the images (a”–d” and e”’–f”’). p: Pearson’s coefficient. g, h Immunoprecipitation demonstrating the interaction between RT and DNRX but not DG in S2 cells. i, j Immunoprecipitation showing that neither DNRX nor DG interacted with TW in S2 cells. k Immunoprecipitation results showing that DNRX, DG, RT, and TW formed a complex in S2 cells.
Given that DNRX interacts with both DG and RT, we hypothesized that DNRX might serve as a bridge linking DG to the RT/TW complex, forming a four-component structure that facilitates DG glycosylation through the RT/TW complex. To test this hypothesis, we co-transfected constructs of DNRX, DG, RT, and TW into S2 cells or HEK293T cells and evaluated the interactions among the components of the complex via co-IP. As expected, all of these proteins were able to coprecipitate with the other three, although the immunoprecipitation efficiency for TW was relatively low (Fig. 7k and Supplementary Fig. 5e). Taken together, these results suggest that DNRX links DG to RT/TW, forming a complex that is necessary for the proper physiological functions of DNRX and DG.
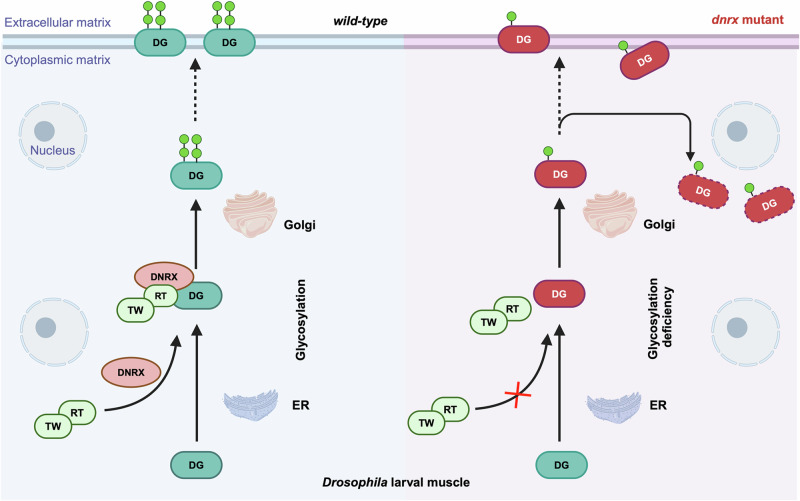
Here, we illustrate a working model in which DNRX plays a pivotal role in facilitating DG glycosylation and ensuring the proper localization of DG on the cell surface, which is essential for muscle structure and function, as shown in Fig. 8. Under normal circumstances, glycosylation modification of nascent DG occurs in the ER, matures in the Golgi apparatus, and finally glycosylated DG reaches the plasma membrane. DNRX acts as a bridge linking DG to the RT/TW complex, which is crucial for the glycosylation process, allowing glycosylated DG to be properly transported to the cell surface. However, glycosylation-deficient DG in dnrx mutant tends to accumulate around the nucleus, although some glycosylation-deficient DG can still reach the cell surface. DG localization on the cell surface is impaired, indicating disruptions in normal protein transport and localization. Our findings elucidate the role of DNRX in facilitating the proper glycosylation and localization of DG, thereby maintaining muscle structure and function in Drosophila. This understanding of the impact of DG glycosylation on muscle integrity could aid the development of new therapeutic strategies for muscular dystrophies.
Fig. 8. Model depicting how DNRX bridges DG to the heterodimeric glycosyltransferase complex RT/TW for its glycosylation.
In wild-type files, glycosylation modification of nascent DG (green DG) begins in the ER, matures (green DG modified by glycans) in the Golgi apparatus, and finally, glycosylated DG reaches the plasma membrane. DNRX recruits the RT/TW heterodimer to DG through its interaction with RT, facilitating the glycosylation of DG. Glycosylated DG is then transported to the cell surface, where it carries out its proper function. However, in dnrx mutants, lightly glycosylated DG (red DG) cannot reach the RT/TW heterodimer due to DNRX deficiency, resulting in reduced DG glycosylation and accumulation in the cytoplasm through an unknown mechanism (red dashed DG). Although some of the lightly glycosylated DG can be transported to the cell surface, decreased levels of DGs are present at the cell membrane.
Discussion
DNRX is essential for muscle development and locomotion in Drosophila larvae
Previous studies have shown that Neurexins, which are associated with ASDs3,45, are major participants in synapse development and plasticity at presynaptic sites1–5,7. However, our research, along with others, has gradually revealed that DNRX is also expressed in muscle during the embryonic and larval stages of Drosophila10,11. In this study, we demonstrate that locomotor defects in dnrx mutants can be partially rescued by overexpressing DNRX in muscle and neurons (Fig. 1a, b). This finding slightly differs from a previous report showing that the crawling ability of Drosophila larvae can be completely rescued by overexpressing DNRX in motor neurons via C380-Gal49. The reduced locomotor ability observed in our study might have been attributed to the dilution of DNRX function when its expression was driven by elav-Gal4, which has a broader expression spectrum than C380-Gal4. The transcription levels of DNRX may vary across different backgrounds due to their respective promoters, although it remains similar in motor neurons. Additionally, the dnrx273 homozygotes used in this study, which lack most of the coding sequence for the extracellular region of DNRX, were different from the dnrx273/Df flies used in the previous study9. For example, the dnrx273/Df mutants can survive to the adult stage, whereas dnrx273 mutants are lethal at the pupal stage. Consistent with the results of the rescue experiments, the results of the knockdown experiments demonstrated that DNRX functions in muscles (Fig. 1d–f). Our findings verify the crucial role of DNRX in muscle for the locomotor function of Drosophila larvae; however, the presynaptic role of DNRX cannot be entirely ruled out by the findings of this study.
Forward locomotion of larvae is achieved by the sequential contraction of muscles from the posterior to the anterior segments46–48, which is closely related to muscle development. Increasing muscle length is associated with the addition of new sarcomeres to each existing myofibril, whereas an increase in muscle diameter is promoted by the addition of parallel myofibrils49. Muscles were longer, and thinner in dnrx273 than in w1118, and the area of muscle 6 was significantly reduced in dnrx273 (Fig. 2a–f); moreover, the number of myofibers per unit area decreased in the absence of DNRX according to electron microscopy (Fig. 2g–i). In terms of the muscle structure, we observed defects similar to those of Dg mutants22, confirming the muscle specificity of these defects (Fig. 2). The attachment defects in dnrx273 mutants also resembled those observed in POMT mutants22, further supporting the role of DNRX in muscle integrity. These results suggest that DNRX in muscle has a regulatory effect on muscle morphology and development and thus maintains locomotor activity. Elucidating the molecular mechanisms by which DNRX regulates muscle development would be interesting. Understanding these pathways could provide deeper insights into how DNRX influences muscle structure and function.
DNRX modulates DG accumulation and glycosylation
Here, we demonstrate that DNRX interacts with DG in Drosophila muscle (Fig. 3a), which is consistent with a previous report that NRX interacts with DG through LNS domains in the brains of mice19. Loss of DNRX causes DG accumulation in muscle, primarily manifested by an increased amount of DG around the nucleus (Fig. 4a–d). Moreover, DG cluster indicated by puncta surrounding the cell membrane was disrupted in the absence of DNRX (Fig. 4e, f’), although some of DG are still transported to the cell surface. It has been reported that DNRX influences the organization and transport of axonal microtubules, as the loss of DNRX leads to transport failure and the aggregation of synaptotagmin on axonal microtubules50. These results suggest that DNRX can promote the accurate localization of protein in some cases.
Furthermore, there are other regulatory mechanisms. We detected DG isoforms with molecular weights of approximately 50 kDa and 80 kDa in Drosophila (Fig. 4h), which might represent spliced or truncated forms. Similarly, β-DG can be cleaved into a 30 kDa fragment51 and translocated to the nucleus via an endocytic pathway52, indicating that the accumulation of DG around the cell nucleus resulting from DNRX loss might be due to DG cleavage. Additionally, it is possible that overexpressed DG is stuck within the secretory pathway compartment, contributing to observed accumulation and functional disruptions. In conclusion, our study reveals a new physiological function of DNRX in regulating the localization of DG in muscle, highlighting the importance of DNRX in maintaining proper DG distribution and muscle integrity.
Given that a low level of glycosylated DG remains detectable at the ribbon synapse in POMGnT1 and myd mutant mice53, the absence of DG glycosylation could result in a diminished interaction with the extracellular matrix, ultimately leading to decreased stability of the cell membrane. In humans, aberrant glycosylation of α-DG is a key factor underlying dystroglycanopathies (DGPs)21, which are characterized by progressive muscular dystrophy. Our findings revealed that DG glycosylation is decreased in dnrx273, indicating that loss of DNRX results in reduced glycosylation of DG. Additionally, the glycosylation level of DG on the cell surface was reduced in dnrx273, but this could be rescued by the overexpression of DNRX (Fig. 5h–k). In the absence of DNRX, there was a slight increase in the overall level of DG and its accumulation both in vitro and in vivo (Fig. 4 and Supplementary Fig. 3d, e). This finding aligns with previous studies showing that the non-glycosylated PrP mutants in N181D/N197D are primarily localized in the cytoplasm and exhibit strong aggregation ability, whereas the glycosylation-deficient mutant in V180I shows impaired localization on the plasma membrane54. Investigating the relationship between DG glycosylation and localization in Drosophila muscle will be interesting.
DNRX bridges DG to the heterodimeric O-mannosyltransferase complex
Protein O-mannosylation is a prominent form of protein glycosylation with crucial biological functions38,55,56. POMT1 and POMT2 are conserved across all animals and function as a complex in the ER57,58. DG, the best-characterized O-mannosylation target, serves as a cell membrane-spanning bridge linking the extracellular matrix to the cytoskeleton. The muscle defects observed in dnrx mutants are identical to those associated with reduced DG or POMT function (Fig. 6), such as defects in muscle attachment22. These results highlight the importance of DG glycosylation in muscle structure and function, which is regulated by POMTs via DNRX. This is further supported by previous findings that glycosylated α-DG restores sarcolemma integrity in largemyd mice59. Taken together, these data reveal a central role for DG glycosylation in maintaining muscle integrity in Drosophila larvae, suggesting that DNRX may be involved not only in neurodegenerative diseases but also in muscular dystrophy.
Interestingly, DG does not bind directly to RT/TW but rather interacts with DNRX to form a complex with RT/TW. DNRX associates with multiple proteins that contain PDZ domains, thereby facilitating the formation of cytoplasmic scaffold complexes60,61. Additionally, DNRX interacts with Ephrin to promote its clustering62, mediates retinoid transport and rhodopsin maturation12, and forms a complex with Scribble and DPix to activate Rac1, stimulating presynaptic F-actin assembly and synaptic vesicle clustering32. Therefore, the linkage of DG to RT/TW through DNRX is likely critical for DG glycosylation. We hypothesize that DNRX guides DG to the POMT complex, facilitating DG glycosylation to maintain muscle structure and function in Drosophila (Fig. 8). While immunoprecipitation results suggest potential interactions between DNRX and DG, the functional significance of these complexes need to be validated in vivo, especially in Drosophila muscle cells. The mechanism underscores the multifaceted role of DNRX in muscle integrity and function, expanding our understanding of its contributions beyond neuronal functions to include potential implications for muscular dystrophy.
Limitations of the study
Here, we verified that DNRX facilitates the glycosylation of DG in postsynaptic muscle and plays a crucial role in larval locomotion. However, the evidence for DNRX localization in muscle is relatively insufficient owing to the low sensitivity and specificity of the DNRX antibodies and potentially low expression levels of DNRX. We are actively working on obtaining more robust evidence to validate DNRX expression in muscle through other methods, such as immunoelectron microscopy (immuno-EM). Given that DNRX associates with several proteins that mediate the assembly of cytoplasmic scaffold complexes and extracellular coupling, it is necessary to verify these direct interactions via GST pull-down assays. These findings help to elucidate the molecular mechanisms by which DNRX contributes to DG glycosylation and muscle function.
Methods
Drosophila strains
The flies were maintained on a standard medium at 25 °C with 60–80% relative humidity. The wild-type flies used in this study were w1118 flies. The Dg mutants DgO86 and DgO43, the rt mutants rt2 and the tw mutants tw1 were obtained from the Bloomington Stock Center. DgO43 and DgO86 flies carry nonsense mutations caused by single nucleotide changes that result in premature stop codons44. rt2 flies carry disruptions in the gene coding region caused by P-element insertion63. tw1 flies carry an amino acid substitution in the conserved POM domain43. rt2 and tw1 flies carry null mutations. UAS-GFP, UAS-dnrx IR, UAS-rt IR, UAS-tw IR, UAS-lanA IR, and UAS-lanB IR flies were purchased from the TsingHua Fly Center. dnrx-Gal411, dnrx273 (which lack most of the coding sequence for the extracellular region)9,32, dnrx83 (deletion of 4066 bp of the genome)2, dnrx174 (deletion of 1369 bp of the genome)2 and UAS-dnrx32 flies were generated in our previous work. The other flies used in this work, including UAS-mCD8-GFP, UAS-mCherry IR, UAS-dcr, elav-Gal4, and mef2-Gal4 flies, were obtained from the Bloomington Stock Center. UAS-lac Z IR flies were obtained from the Vienna Drosophila Resource Center.
Generation of transgenic and knock-in flies
UAS-dnrx-GFP and UAS-Dg-HA transgenic flies were generated via injection of a recombinant pUAST vector containing full-length dnrx cDNA and Dg-C cDNA into attP transgenic fly embryos. GFP was inserted at the intracellular end adjacent to the transmembrane region of DNRX, and HA was inserted at the C-terminus of DG. UAS-DgAla-HA, UAS-DgΔM-HA, and UAS-DgΔM+Ala-HA transgenic flies were generated via injection of a recombinant pUAST vector containing Dg-C cDNA into attP transgenic fly embryos. HA was inserted in the C-terminus of DG. Ala indicates the mutation of seven glycosylation sites (Ser36, Ser202, Ser246, Ser279, Ser282, Thr792, and Thr796 were mutant to ala). ΔM indicates deletion of the mucin region (Gly315~Leu592). ΔM+Ala indicates mutation of the seven glycosylation sites and deletion of the mucin region. The vas-phi-Zh2A-VK5 (75B1) flies were used to generate transgenic flies inserted at the 75B1 attP locus64. cDNAs were generated via overlap extension PCR of full-length Dg. dnrx-HA knock-in flies were generated by inserting 6 × HA just before the stop codon of the dnrx gene. yw; nos-Cas9 (BDSC: 51324) flies were used for the knock-in process.
Plasmid construction and purification
RNA was extracted from Drosophila via the TRIZOL kit (Sigma). The synthesis of full-length dnrx cDNA and Dg-C cDNA was carried out via reverse transcription via a Vazyme kit. To prepare plasmids for transfection into S2 cells, DNRXFL, DNRXΔExtra (excluding Met1~Arg1693) and DNRXΔCyto (excluding Val1715~ Val1840) cDNA was amplified via PCR, cloned and inserted into the pAC5.1- mCherry at Not I restriction sites to generate expressing plasmid. The DgFL, DgΔExtra (excluding Glu28~Thr1051), DgΔCyto (excluding Arg1077~Pro1179), DgΔN (excluding Glu28~Leu314), DgΔM (excluding Gly315~Leu592), and DgΔQ (excluding Pro593~Thr1051) cDNA was amplified via PCR, cloned and inserted into the pAC5.1- GFP at Not I restriction sites to generate an expressing plasmid. Tull-length rt and tw cDNA was amplified via PCR, tagged with HA or V5, and cloned and inserted into pAC5.1 V5-His at Not I restriction sites to generate an expressing plasmid. To prepare plasmids for transfection into HEK293T cells, the mCherry sequence was inserted into pCDNA3.1 at the BamH I restriction site to generate a tool plasmid (pCDNA3.1-mCherry). DNRX-expressing plasmids were generated by inserting constructs into pCDNA3.1-mCherry at the Hind III restriction site. DG-expressing plasmids were generated by inserting constructs into pCDNA3.1-GFP at the BstX I restriction site. Full-length rt and tw cDNA was amplified via PCR, tagged with HA or V5, cloned and inserted into pCDNA3.1 at EcoR I restriction sites to generate an expression plasmid.
Quantitative real‑time PCR
RNA was extracted from the heads of 20 third-instar larvae or the body wall muscles of 10 third-instar larvae via the TRIZOL kit (Sigma). cDNA synthesis was carried out via reverse transcription via a Vazyme kit. RT‒PCR was performed using AceQ qPCR SYBR Green Master Mix (Vazyme) on a QuantStudio 5 real-time PCR system. The following primers were used: mef2, F: 5’-CAACGACAGAGCCAGACAT-3’, R: 5’-TGCCAGACGCACATCC-3’, dnrx, F: 5’-GTACATGTACGATGGAGCGCTC-3’, R: 5’- TGCATCGATACTTTGTCAGACAACC-3’, rp49, F: 5’-TACAGGCCCAAGATCGTGAA-3’, R: 5’-ACCGTTGGGGTTGGTGGAG-3’.
Antibodies
DG antiserum was generated in rabbits against a synthetic KLH-conjugated peptide of the intracellular domain of DG. The sequence of the peptide synthesized was GKSPATPSYRKPPPYVSP-C, as has been reported30 (ABclonal).
Immunostaining and image acquisition
Immunostaining of dissected wandering third-instar larvae was performed as described previously65. In brief, after fixation in 4% paraformaldehyde, the samples were washed with PBST containing 0.3% Triton X-100 and blocked with 0.5% bovine serum albumin. The samples were then incubated with primary antibodies at 4 °C overnight. Following washes with PBST, the samples were incubated with Alexa Fluor-conjugated secondary antibodies at room temperature for 2 h. For HEK293T cell immunostaining, the samples were washed twice with precooled PBS and fixed in 4% paraformaldehyde and 4% sucrose for 20 min. The samples were then washed three times with 0.1% Triton X-100 and blocked with 1% bovine serum albumin for 1 h at room temperature. The samples were incubated with primary antibodies overnight at 4 °C, washed three times with PBST containing 0.1% Triton X-100, and incubated with secondary antibodies at room temperature for 2 h. Finally, the samples were washed four times with PBST. Images of the samples were acquired via a Zeiss LSM 900 confocal microscope and analyzed with ImageJ software. The primary antibodies used in this study were as follows: mouse anti-GFP (1:500, Santa Cruz Biotechnology), rabbit anti-GFP (1:500, Torrey Pines Biolabs), Texas Red-X phalloidin (1:300, Molecular Probes, Invitrogen), mouse anti-DLG (1:50, DSHB), rabbit anti-HA (1:1000, Cell Signaling Technology), rabbit anti-mCherry (1:500, Abcam), and mouse anti-V5 (1:300, Santa Cruz Biotechnology). Alexa Fluor 488-, Alexa Fluor 555-, or Alexa Fluor 647-conjugated secondary antibodies (1:500, Invitrogen) were used.
For the colocalization assay, images were captured via a 63× objective on a confocal microscope, and the maximum intensity projections of Z-series stacks were obtained. The background was removed via the Subtract Background plugin in ImageJ (rolling ball radius of 50 pixels). The fluorescence intensity along the cord was evaluated with the Plot Profile plugin. For each channel, the values were standardized to the maximum intensity value. The overlap of peaks in the intensity profiles suggested colocalization of the proteins. Pearson’s coefficients, obtained using ImageJ, were used to compare the distribution of fluorescence signals in each channel to evaluate their correlations, as depicted in Fig. 4k”–n” and Fig. 7a”–f”’.
Immunoprecipitation
Fly body wall muscles were homogenized via a glass homogenizer in ice-cold lysis buffer for Western blotting and IP (Beyotime) following the addition of 1% w/v protein inhibitor (Roche). The lysates were incubated on a shaker at 4 °C for 30 min and centrifuged twice at 12,000 rpm at 4 °C for 15 min. For cell extract preparation, cell pellets were lysed in precooled lysis buffer supplemented with 1% w/v protein inhibitor for 30 min and then centrifuged at 12,000 rpm for 10 min at 4 °C. The protein concentrations in the extracts were determined via a BCA protein assay kit (Thermo Fisher Scientific). The supernatant was incubated with 2 μl of preimmunized rabbit/mouse sera for 2 h at 4 °C, followed by 20 μl of protein A/G beads for another hour at 4 °C before centrifugation at 3000 × g for 5 min. The preabsorbed supernatants were incubated with beads and antibodies on a shaker for 2 h at 4 °C. After four washes at 3000 × g for 5 min with lysis buffer, the immune complexes were eluted with 2 × SDS sample buffer and subjected to SDS‒PAGE for Western blotting analysis.
Plasma membrane protein extraction
Plasma membrane fractions were extracted from fly body wall muscle via the Plasma Membrane Protein Extraction Kit (Abcam, ab65400). Briefly, approximately 500 fly body wall muscles were collected and thoroughly homogenized in 0.5 ml of homogenization buffer supplemented with protease inhibitors (Roche). After centrifugation at 700 × g for 10 min at 4 °C, the supernatant was further centrifuged at 10,000 × g for 30 min at 4 °C to collect the supernatant as the cytosolic fraction. The pellet, which contained total cellular membrane proteins (from both plasma membranes and cellular organelle membranes), was subjected to further extraction according to the kit protocol. The total membrane protein pellet was resuspended in 200 μl of Upper Phase Solution. Next, 200 μl of Lower Phase Solution was added, mixed well, and incubated on ice for 5 min. After centrifugation at 1000 × g for 5 min at 4 °C, the upper phase was carefully collected, and 100 μl of Lower Phase Solution and Upper Phase Solution were added again. After centrifugation again, the upper phase was diluted in 5 volumes of water and centrifuged at 15,000 × g for 10 min at 4 °C. The supernatant was then removed, leaving the pellet containing the plasma membrane protein fraction. The plasma membrane protein pellet was dissolved in 40–100 μl of 2 × SDS sample buffer for Western blotting.
Endoplasmic reticulum separation
The ER/Golgi was isolated from Drosophila as described previously66. Briefly, according to the manufacturer’s instructions for the ER Isolation Kit (Sigma-Aldrich, ER0100), approximately 500 wild-type fly body wall muscles were homogenized, and the lysates were subjected to differential centrifugation. The homogenized tissue was centrifuged at 12,000 × g for 15 min at 4 °C to obtain the post-mitochondrial fraction (PMF). The PMF was then centrifuged at 100,000 × g for 60 min at 4 °C. The resulting pellet—microsomal fraction—was rich in ER and Golgi and was used for further analysis. For density gradient centrifugation, the microsomal fraction was resuspended in isotonic extraction buffer. OptiPrep™ (60%) was added to achieve a final concentration of 20%. In a centrifuge tube, 30% OptiPrep™ was added to the bottom, followed by the addition of the 20% sample and then 15% OptiPrep™. The tube was subsequently centrifuged at 150,000×g for 3 h at 4 °C. After centrifugation, the samples were collected in equal-volume fractions from top to bottom for further experiments, such as Western blotting.
Western blotting
Fly body wall muscles were homogenized in ice-cold lysis buffer for Western blotting and IP (Beyotime) following the addition of 1% w/v protein inhibitor (Roche) and centrifuged at 12,000 rpm for 15 min at 4 C. The proteins were separated on SDS‒PAGE gels and subsequently transferred onto a PVDF membrane. To prevent nonspecific binding of antibodies, the membranes were blocked in PBST (phosphate-buffered saline solution with 0.05% Tween-20) containing 5% milk for 1 h at room temperature. The membranes were then incubated overnight with primary antibodies in antibody diluent (NCM Biotech). Following extensive washing, the membranes were incubated with appropriate HRP-conjugated secondary goat antibodies in PBST containing 5% milk for 2 h at room temperature. The blots were imaged via the Tanon 5200 imaging system. The primary antibodies used for Western blotting analyses in this study were as follows: mouse anti-HA (1:3000, Sigma), rabbit anti-O-GlcNac (1:1000, Cell Signaling), rabbit anti-DG (1:1000), mouse anti-DNRX (1:1000), mouse anti-tubulin (1:5000, Sigma‒Aldrich), mouse anti-GFP (1:1000, Santa Cruz Biotechnology), rabbit anti-GFP (1:2000, Torrey Pines Biolabs), mouse anti-mCherry (1:2000, ABclonal), mouse anti-ATPα (1:100, DSHB), rabbit anti-GAPDH (1:5000, GeneTex), rabbit anti-V5 (1:1000, Cell Signaling Technology), rat anti-Bip (1:3000, Abcam), and rabbit anti-syx16 (1:2000, Abcam).
Lectin blot
Proteins extracted from fly body wall muscle samples were separated on an 8% SDS–PAGE gel, and the separated proteins were then transferred onto a PVDF membrane. Nonspecific binding was blocked by incubating the membrane in 1× Carbo-Free™ Blocking Solution (SP-5040) for 30 min at room temperature. The membrane was then incubated with biotinylated lectins (2.5 μg/ml ConA or 10 μg/ml VVA or WFA, all from Vector) for 1 h at room temperature, followed by detection with a Vectastain ABC kit (Vector). The membrane was washed with TBST between steps. The blots were imaged via the Tanon 5200 imaging system.
Cell culture and cell transfection
S2 cells were obtained from the Drosophila Genomics Resource Center (DGRC). HEK293T cells were obtained from the American Type Culture Collection (ATCC). The cells routinely tested negative for mycoplasma using the MycoBlue Mycoplasma Detection Kit (Vazyme), and cell aliquots from early passages were used. S2 cells were maintained in Schneider’s Drosophila Medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) at a constant temperature of 28 °C in ambient air. Plasmid transfection was performed via X-treme GENE™ HP DNA Transfection Reagent (Roche) according to the manufacturer’s protocols. HEK293T cells were cultured in DMEM (Wisent Inc.) supplemented with 10% FBS (Gibco) in a 5% CO2 incubator at 37 °C. Plasmid transfection was carried out via Lipofectamine™ 3000 (Invitrogen) according to the manufacturer’s instructions. After 24 or 48 h of transfection, the cells were collected for further experiments, including Western blotting and immunostaining.
Locomotion activity detection
Larval locomotion assay was evaluated as previously described67. Larvae were placed in the center of 8.5 cm diameter dishes after being washed in PBS at room temperature. The transparent dishes, which contained 2% agar on the bottom, were colored dark purple by adding food colorant to the agar gel. The movement of the larvae was recorded with a video camera, and the trajectory over 3 min was tracked via tracker software written in MATLAB. The distance traveled was then calculated via ImageJ.
Muscle attachment defects analysis
Muscle attachment was assessed as previously described22. In wild-type larvae, muscle fibers are organized in a predictable pattern and establish connections with the underlying epidermis through interactions with tendon cells at specific locations. In mutant larvae, the lack of or incorrect attachment of one or more muscles was commonly observed, and the presence of extra muscle tissue was occasionally observed. For statistical analysis, the number of muscles with attachment defects was counted and compared to the total number of muscles studied to determine the defect ratio. This ratio served as a basis for assessing the frequency and severity of muscle defects.
Transmission electron microscopy analysis of larval muscle
TEM was performed according to the procedure described68. In brief, wandering third-instar larvae were dissected in precooled phosphate-buffered saline (PBS) and subsequently fixed using a mixed fixative containing 2% glutaraldehyde and 2% formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) at 4 °C overnight. The samples were rinsed multiple times with cacodylate buffer at 4 °C, post-fixed for 2 h 1% OsO4 0.1 M cacodylate buffer, and rinsed twice with distilled water. The samples were stained for 2 h with 2% uranyl acetate and rinsed twice with distilled water. The specimens were dehydrated through a graded series of ethanol, passed through propylene oxide two times, and embedded into a sheet in Epon812 (SPI Science). The embedded sectioned at 80 μm at the 6th/7th muscles of the A3 or A2 segment in one animal using a diamond specimen on a Leica UC7 ultrathin microtome. Each slice was 90 nm thick; 30–40 slices were gathered into a group and attached to a grid. Finally, each ultrathin slice was examined under a transmission electron microscope (Hitachi H-7650).
Statistical and reproducibility
The significance of differences among genotypes or conditions was determined via one-way ANOVA and two-tailed unpaired Student’s t-test. The data were analyzed using GraphPad Prism 10.0 software and Microsoft Excel. The data are presented as the means ± SDs or SEMs. All asterisks above a line indicate comparisons between the two groups specified: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank all members in Xie lab for the critical discussion of the manuscript, the core facility of Nanjing University for providing key equipment. We extend our sincere appreciation to the Bloomington Drosophila Stock Center, the Core Facility of Drosophila Resource and Technology at CEMCS, CAS, and the Tsinghua Fly Center for providing fly stocks. We thank BioRender for their support in creating and editing the scientific illustrations (Fig. 8, Created in BioRender. Zhao, Y. (2023) BioRender.com/o34o492) used in this work. This work was funded by the National Natural Science Foundation of China (31771592, 32171149 to W.X.; 32271013 to J.G.; 32070811 to G.G.).
Author contributions
W.X. and J.G. conceived of and supervised this project. Y.Z. and J.G. designed and performed the main experiments. Y.Z., Z.M., Y.S., M.O., L.X. and M.R. performed genetic experiments and morphological analysis; G.G. performed EM assay; M.R., M.L. and J.H. provided with resource and participated in data analysis. Y.Z., J.G. and W.X. interpreted data and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary handling editor: Christina Karlsson Rosenthal.
Data availability
The data that underpins the conclusions of this research are contained within the article and accompanying supplementary materials. P-values and numerical source data for all graphs can be found in Supplementary Data 1, while the original uncropped western blot images can be found in Supplementary Data 2. Drosophila strains and reagents can be found in Supplementary Data 3.
Competing interests
The authors declare no competing interests.
Ethical approval
The experiments described in this document adhere to the prevailing legislation of the countries where they were conducted. The research did not involve the study of individuals.
Consent to publish
The manuscript has been reviewed by all authors, who have provided their consent for the publication of its contents in Communications Biology.
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yu Zhao, Junhua Geng.
Contributor Information
Junhua Geng, Email: gengjunhua@seu.edu.cn.
Wei Xie, Email: wei.xie@seu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07191-5.
References
- 1.Dean, C. et al. Neurexin mediates the assembly of presynaptic terminals. Nat. Neurosci.6, 708–716 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng, X. et al. Neurexin-1 is required for synapse formation and larvae associative learning in Drosophila. FEBS Lett.581, 2509–2516 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Kim, H. G. et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet.82, 199–207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, T. et al. The neurexin/N-ethylmaleimide-sensitive factor (NSF) interaction regulates short term synaptic depression. J. Biol. Chem.290, 17656–17667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian, Y., Zhang, Z. C. & Han, J. Drosophila studies on autism spectrum disorders. Neurosci. Bull.33, 737–746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudanova, I. et al. Important contribution of α-neurexins to Ca2+-triggered exocytosis of secretory granules. J. Neurosci.26, 10599–10613 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottos, A. et al. The synaptic proteins neurexins and neuroligins are widely expressed in the vascular system and contribute to its functions. Proc. Natl Acad. Sci. USA106, 20782–20787 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosedale, M., Egodage, S., Calma, R. C., Chi, N. W. & Chessler, S. D. Neurexin-1α contributes to insulin-containing secretory granule docking. J. Biol. Chem.287, 6350–6361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, J., Ashley, J., Budnik, V. & Bhat, M. A. Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission. Neuron55, 741–755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, K. et al. Neurexin in embryonic Drosophila neuromuscular junctions. PLoS ONE5, e11115 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun, M., Zeng, X. & Xie, W. Temporal and spatial expression of Drosophila neurexin during the life cycle visualized using a DNRX-Gal4/UAS-reporter. Sci. China Life Sci.59, 68–77 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Tian, Y. et al. Neurexin regulates visual function via mediating retinoid transport to promote rhodopsin maturation. Neuron77, 311–322 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Tremblay, F., Laroche, R. G. & De Becker, I. The electroretinographic diagnosis of the incomplete form of congenital stationary night blindness. Vis. Res.35, 2383–2393 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Ricotti, V. et al. Ocular and neurodevelopmental features of Duchenne muscular dystrophy: a signature of dystrophin function in the central nervous system. Eur. J. Hum. Genet.24, 562–568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinton, V. J. et al. Association of autistic spectrum disorders with dystrophinopathies. Pediatr. Neurol.41, 339–346 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Alexander, M. S. et al. Reversal of neurobehavioral social deficits in dystrophic mice using inhibitors of phosphodiesterases PDE5A and PDE9A. Transl. Psychiatry6, e901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blake, D. J., Weir, A., Newey, S. E. & Davies, K. E. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol. Rev.82, 291–329 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, K. M. & Morgan, K. G. The importance of dystrophin and the dystrophin associated proteins in vascular smooth muscle. Front. Physiol.13, 1059021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugita, S. et al. A stoichiometric complex of neurexins and dystroglycan in brain. J. Cell Biol.154, 435–445 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trotter, J. H., Wang, C. Y., Zhou, P., Nakahara, G. & Südhof, T. C. A combinatorial code of neurexin-3 alternative splicing controls inhibitory synapses via a trans-synaptic dystroglycan signaling loop. Nat. Commun.14, 1771 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quereda, C., Pastor, A. & Martin-Nieto, J. Involvement of abnormal dystroglycan expression and matriglycan levels in cancer pathogenesis. Cancer Cell Int.22, 395 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haines, N., Seabrooke, S. & Stewart, B. A. Dystroglycan and protein O-mannosyltransferases 1 and 2 are required to maintain integrity of Drosophila larval muscles. Mol. Biol. Cell18, 4721–4730 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragni, E. et al. Protein O-mannosylation is crucial for human mesencyhmal stem cells fate. Cell Mol. Life Sci.73, 445–458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanagawa, M. & Toda, T. Ribitol-phosphate-a newly identified posttranslational glycosylation unit in mammals: structure, modification enzymes and relationship to human diseases. J. Biochem.163, 359–369 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Manya, H. et al. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl Acad. Sci. USA101, 500–505 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akasaka-Manya, K., Manya, H., Nakajima, A., Kawakita, M. & Endo, T. Physical and functional association of human protein O-mannosyltransferases 1 and 2. J. Biol. Chem.281, 19339–19345 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Akasaka-Manya, K., Manya, H. & Endo, T. Mutations of the POMT1 gene found in patients with Walker-Warburg syndrome lead to a defect of protein O-mannosylation. Biochem. Biophys. Res. Commun.325, 75–79 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Reeuwijk, J. et al. POMT2 mutations cause α-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J. Med. Genet.42, 907–912 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng, W. M. et al. Dystroglycan is required for polarizing the epithelial cells and the oocyte in Drosophila. Development130, 173–184 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Schneider, M. et al. Perlecan and Dystroglycan act at the basal side of the Drosophila follicular epithelium to maintain epithelial organization. Development133, 3805–3815 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura, N. et al. Drosophila Dystroglycan is a target of O-mannosyltransferase activity of two protein O-mannosyltransferases, Rotated Abdomen and Twisted. Glycobiology20, 381–394 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rui, M. et al. The neuronal protein Neurexin directly interacts with the Scribble-Pix complex to stimulate F-actin assembly for synaptic vesicle clustering. J. Biol. Chem.292, 14334–14348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banovic, D. et al. Drosophila Neuroligin 1 promotes growth and postsynaptic differentiation at glutamatergic neuromuscular junctions. Neuron66, 724–738 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Ogienko, A. A., Andreyeva, E. N., Omelina, E. S., Oshchepkova, A. L. & Pindyurin, A. V. Molecular and cytological analysis of widely-used Gal4 driver lines for Drosophila neurobiology. BMC Genet.21, 96 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun, M. et al. Genetic interaction between Neurexin and CAKI/CMG is important for synaptic function in Drosophila neuromuscular junction. Neurosci. Res.64, 362–371 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Bate, M. The embryonic development of larval muscles in Drosophila. Development110, 791–804 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Lauwereyns, J., Watanabe, K., Coe, B. & Hikosaka, O. A neural correlate of response bias in monkey caudate nucleus. Nature418, 413–417 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Yoshida-Moriguchi, T. & Campbell, K. P. Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology25, 702–713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Praissman, J. L. et al. The functional O-mannose glycan on α-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife5, e14473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheikh, M. O., Halmo, S. M. & Wells, L. Recent advancements in understanding mammalian O-mannosylation. Glycobiology27, 806–819 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dang, K., Jiang, S., Gao, Y. & Qian, A. The role of protein glycosylation in muscle diseases. Mol. Biol. Rep.49, 8037–8049 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Ichimiya, T. et al. The twisted abdomen phenotype of Drosophila POMT1 and POMT2 mutants coincides with their heterophilic protein O-mannosyltransferase activity. J. Biol. Chem.279, 42638–42647 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Lyalin, D. et al. The twisted gene encodes Drosophila protein O-mannosyltransferase 2 and genetically interacts with the rotated abdomen gene encoding Drosophila protein O-mannosyltransferase 1. Genetics172, 343–353 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christoforou, C. P., Greer, C. E., Challoner, B. R., Charizanos, D. & Ray, R. P. The detached locus encodes Drosophila Dystrophin, which acts with other components of the Dystrophin Associated Protein Complex to influence intercellular signalling in developing wing veins. Dev. Biol.313, 519–532 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Yan, J. et al. Neurexin 1α structural variants associated with autism. Neurosci. Lett.438, 368–370 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Fox, L. E., Soll, D. R. & Wu, C. F. Coordination and modulation of locomotion pattern generators in Drosophila larvae: effects of altered biogenic amine levels by the tyramine beta hydroxlyase mutation. J. Neurosci.26, 1486–1498 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iyengar, B. G. et al. Silencing synaptic communication between random interneurons during Drosophila larval locomotion. Genes Brain Behav.10, 883–900 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Hasegawa, E., Truman, J. W. & Nose, A. Identification of excitatory premotor interneurons which regulate local muscle contraction during Drosophila larval locomotion. Sci. Rep.6, 30806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luis, N. M. & Schnorrer, F. Mechanobiology of muscle and myofibril morphogenesis. Cells Dev.168, 203760 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Banerjee, S. & Riordan, M. Coordinated regulation of axonal microtubule organization and transport by Drosophila Neurexin and BMP Pathway. Sci. Rep.8, 17337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukai, Y. et al. Cleavage of β-dystroglycan occurs in sarcoglycan-deficient skeletal muscle without MMP-2 and MMP-9. Biochem. Biophys. Res. Commun.492, 199–205 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Gracida-Jiménez, V. et al. Retrograde trafficking of β-dystroglycan from the plasma membrane to the nucleus. Sci. Rep.7, 9906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanagawa, M. et al. Post-translational maturation of dystroglycan is necessary for pikachurin binding and ribbon synaptic localization. J. Biol. Chem.285, 31208–31216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi, C. W. et al. Glycosylation significantly inhibits the aggregation of human prion protein and decreases its cytotoxicity. Sci. Rep.8, 12603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marrone, A. K., Edeleva, E. V., Kucherenko, M. M., Hsiao, N. H. & Shcherbata, H. R. Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile. BMC Cell Biol.13, 26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells, L. The O-mannosylation pathway: glycosyltransferases and proteins implicated in congenital muscular dystrophy. J. Biol. Chem.288, 6930–6935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu, H. et al. Conditional knockout of protein O-mannosyltransferase 2 reveals tissue-specific roles of O-mannosyl glycosylation in brain development. J. Comp. Neurol.519, 1320–1337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freeze, H. H., Eklund, E. A., Ng, B. G. & Patterson, M. C. Neurological aspects of human glycosylation disorders. Annu. Rev. Neurosci.38, 105–125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han, R. et al. Basal lamina strengthens cell membrane integrity via the laminin G domain-binding motif of α-dystroglycan. Proc. Natl Acad. Sci. USA106, 12573–12579 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biederer, T. & Südhof, T. C. Mints as adaptors. Direct binding to neurexins and recruitment of munc18. J. Biol. Chem.275, 39803–39806 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Koroll, M., Rathjen, F. G. & Volkmer, H. The neural cell recognition molecule neurofascin interacts with syntenin-1 but not with syntenin-2, both of which reveal self-associating activity. J. Biol. Chem.276, 10646–10654 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Liu, L. et al. Neurexin restricts axonal branching in columns by promoting ephrin clustering. Dev. Cell41, 94–106.e104 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Martín-Blanco, E. & García-Bellido, A. Mutations in the rotated abdomen locus affect muscle development and reveal an intrinsic asymmetry in Drosophila. Proc. Natl Acad. Sci. USA93, 6048–6052 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bischof, J., Maeda, R. K., Hediger, M., Karch, F. & Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl Acad. Sci. USA104, 3312–3317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun, Y., Zhao, Y., Johnson, T. K. & Xie, W. Immunohistochemical analysis of the Drosophila larval neuromuscular junction. Methods Mol. Biol.2746, 201–211 (2024). [DOI] [PubMed] [Google Scholar]
- 66.Ge, L., Melville, D., Zhang, M. & Schekman, R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife2, e00947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, X. et al. Neuroligin 4 regulates synaptic growth via the bone morphogenetic protein (BMP) signaling pathway at the Drosophila neuromuscular junction. J. Biol. Chem.292, 17991–18005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guangming, G., Junhua, G., Chenchen, Z., Yang, M. & Wei, X. Neurexin and neuroligins maintain the balance of ghost and satellite boutons at the Drosophila neuromuscular junction. Front. Neuroanat.14, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data that underpins the conclusions of this research are contained within the article and accompanying supplementary materials. P-values and numerical source data for all graphs can be found in Supplementary Data 1, while the original uncropped western blot images can be found in Supplementary Data 2. Drosophila strains and reagents can be found in Supplementary Data 3.