Abstract
Testosterone deficiency can cause abnormal lipid metabolism in men, leading to hyperlipidemia. We identified the testosterone-degrading bacterium Pseudomonas nitroreducens in the fecal samples of male patients with hyperlipidemia. Gastric administration of P. nitroreducens in mice led to testosterone deficiency and elevated blood lipid levels. Whole-genome sequencing of P. nitroreducens revealed the presence of 3/17β-hydroxysteroid dehydrogenase (3/17β-HSD), a gene responsible for testosterone degradation, which is also associated with hyperlipidemia. Microbiota analysis of fecal samples collected from 158 patients with hyperlipidemia and 151 controls revealed that the relative abundance of P. nitroreducens and 3/17β-HSD in the fecal samples of patients with hyperlipidemia was significantly higher than that in controls. These results suggest that P. nitroreducens and 3/17β-HSD may be related to the onset of testosterone deficiency-induced hyperlipidemia. Therefore, treatments targeted at eradicating testosterone-degrading bacteria are a potential future option for patients with testosterone-induced hyperlipidemia and should thus be studied further.
Subject terms: Clinical microbiology, Cellular microbiology
Introduction
Hyperlipidemia is a prevalent chronic condition, with a staggering incidence rate of 40.4% among Chinese adults1. Moreover, it is associated with severe complications such as diabetes, coronary heart disease, obesity, and metabolic syndrome, posing a significant threat to human health2,3. Numerous cross-sectional and prospective observational studies have indicated that low endogenous testosterone levels in males are associated with high and low levels of low-density lipoprotein (LDL) and high-density lipoprotein (HDL), respectively4,5. Exogenous testosterone administration is associated with decreased levels of HDL, LDL, and total cholesterol (TC)6,7, suggesting a potential association between testosterone and the development of hyperlipidemia.
Testosterone is a steroid hormone that regulates endocrine homeostasis and is produced by the testes and adrenal glands8. It is extensively metabolized by multiple enzymes in the liver and in extrahepatic organs, and excreted in urine or feces as conjugated metabolites9. The transport of testosterone glucuronide metabolites is also a critical process for enterohepatic circulation. Testosterone excreted into the intestine through bile acids can be reactivated by gut microbiota via deglucuronidation, thereby promoting the reabsorption of testosterone into the bloodstream10,11. Previous studies have revealed differences in serum testosterone levels between germ-free (GF) and specific pathogen-free (SPF) mice12. Specifically, GF females displayed elevated testosterone relative to SPF females, while GF males had lower levels than SPF males. The colonization of the gut by filamentous bacteria in mice was correlated with increased testosterone levels13, suggesting that the gut microbiota regulate serum testosterone levels through certain pathways. However, the mechanism underlying male testosterone deficiency remains unclear.
The gut microbiota is considered important to the endocrine system. Recently, numerous studies have revealed that the intestinal microbiota can impact the endocrine status of the body through various pathways, thereby influencing the onset and progression of diseases14. An analysis of the gut microbiota of 893 individuals revealed that multiple bacterial genera were associated with serum lipid levels15. For example, Clostridiaceae/Lachnospiraceae correlated with LDL levels, whereas Pasteurellaceae, Coprococcus, and Collinsella correlated strongly with triglyceride (TG) levels. Notably, the abundance of Actinobacteria in the intestines of patients with hyperlipidemia is increased compared to healthy individuals. Concurrently, Actinobacteria have demonstrated the capability to metabolize testosterone within ecological niches16,17. However, the effect of the gut microbiota on the regulation of blood lipid metabolism via testosterone degradation has not been confirmed.
Therefore, we hypothesized that the gut microbiota can affect lipid metabolism via testosterone degradation.
Methods
Human participants
A total of 259 male patients (aged 20–65 years) with hyperlipidemia, admitted to the Renmin Hospital of Wuhan University from September 2020 to December 2021, were enrolled. Hyperlipidemia was diagnosed in accordance with the 2013 ACC/AHA diagnostic guidelines18. The exclusion criteria included5,19 the presence of diabetes, hypertension, coronary heart disease, and other metabolic diseases; severe liver and kidney function injury and cardiac insufficiency; and other diseases, such as tumors, infectious diseases, and depression; the history of antibiotic or yogurt use within 3 months. A total of 218 age-matched volunteers who failed to reach the diagnostic criteria for hyperlipidemia and underwent physical examination at the Renmin Hospital of Wuhan University were recruited as the control group. The clinical indicators of the control group, including chest radiography, urine and stool evaluation, routine blood examination, physical examination, and preoperative examination, were within the normal range. All participants were from central China and were of Asian descent. Serum and fecal samples were collected simultaneously from participants in the hyperlipidemia group and control group.
Detection of serum testosterone level using LC-MS/MS
Venous blood samples were collected from humans or mice after fasting for 8 h and centrifuged at 3500 rpm for 10 min to separate the serum. We placed 20 µL of serum sample (human or mouse) in a 1.5 mL Eppendorf tube and added 20 µL of testosterone internal standard and 200 µL of pure acetonitrile. The mixture was then vortexed for 5 min to ensure complete mixing. After high-speed centrifugation at 13,000 rpm at 4 °C for 5 min, 50 µL of the supernatant was mixed with 150 µL of molecular-grade water, and then placed in a 96-well plate. Testosterone levels were analyzed using an Ekspert ultra LC 100-XL system (AB SCIEX, Framingham, MA, USA) connected to an AB SCIEX 4500 QTRAP mass spectrometer on an ACQUITY UPLC BEH Shield RP18 column (particle size: 1.7 µm, 2.1 × 50 mm, Waters, Milford, MA, USA). For the mobile phase, methanol (A) and water (B) were used at a flow rate of 0.3 mL/min, and both eluents were enhanced with 0.2% formic acid The data were collected in the positive ion mode.
Detection of serum TC, TG, HDL-C, and LDL-C levels
Venous blood was collected from patients and mice after fasting for 8 h and then centrifuged at 3500 rpm for 10 min to collect the serum. Serum concentrations of TC, TG, HDL-C, and LDL-C were determined using a Siemens 2400 automatic biochemical analyzer (Siemens, Erlangen, Germany).
Stool sample collection
Stool samples were collected from the participants following normal defecation between 07:00 and 10:00. The specimens were stored at −80 °C within 1 h after collection for subsequent experiments.
Degradation of testosterone by fecal microbiota
Five patients with hyperlipidemia and five control participants were randomly selected. Fecal samples were weighed, and 1 g was placed in a sterile centrifuge tube and suspended in 10 mL of isotonic sodium chloride solution. Following the initial centrifugation at 2000 × g for 1 min at 4 °C, the bacteria-enriched supernatant was carefully transferred into a sterile tube and centrifuged at 15,000 × g for 5 min. After discarding the supernatant, testosterone (1 g/L) was added to a sterile tube, and 10 mL of normal saline was used to re-suspend the fecal microbiota of bacterial particles20. The suspension (0.5 mL) was collected every 24 h.
Thin-layer chromatography
After thawing at 22–25 °C, 500 µL of the sample was mixed with an equal volume of ethyl acetate. After centrifugation, 2 µL of the supernatant was added to the TLC plate (Qingdao Ocean Chemical Plant, Qingdao, China). Dissolve 1 milligram of testosterone or androstenedione in 1 milliliter of ethyl acetate to obtain a 1 g/L ethyl acetate solution of testosterone or androstenedione. Add 2 µL of ethyl acetate solution of testosterone and androstenedione to the TLC plate as a control. The product formation was observed after unfolding in ethyl acetate/petroleum ether (1:1).
Quantifying steroids using HPLC
We thawed 500 µL of the sample at 22–25 °C and extracted it with 500 µL of ethyl acetate. The extracted supernatant was detected using a utem3000 (Waltham, MA, USA) and a column Xterra RP-C18 (Milford, MA, USA). The mobile phase was methanol/water (80:20, v/v), with a column temperature of 30 °C and a flow rate of 8 mL/min. Testosterone and its products were detected at 254 nm using UV spectrophotometry.
Mice
In order to construct hyperlipidemia and atherosclerosis models, ApoE −/− mice were selected. SPF grade male C57BL/6J ApoE−/− mice, 6 weeks old and weighing 18–20 g, were purchased from Zhejiang Weitong Lihua Laboratory Animal Technology (Zhejiang, China). Adaptive feeding was performed in the experimental animal center of Renmin Hospital of Wuhan University (approval number: S0272012259A) with free access to drinking water. The animal room was maintained at 12 h of light/dark cycle, temperature of 22–25 °C, and relative humidity of 40–60%. After the mice were transported from their breeding grounds to the experimental animal center, they were adaptively fed for 1 week before the experiment to adapt to the environment. During this period, no experimental procedures were conducted on the mice, and their original dietary structure was maintained. After adaptive feeding for 1 week, the mice were administered broad-spectrum antibiotics (0.5 mg/mL vancomycin, 1 mg/mL neomycin, 1 mg/mL metronidazole, and 1 mg/mL ampicillin) in their drinking water for 1 week. Subsequently, they were given a high-fat diet (D12079B: 17% protein, 43% carbohydrate, 40% fat).
Fecal transplantation
Mice were treated with short-term antibiotics prior to bacterial gavage. For 7 days, the mice drank an antibiotic mixture containing 0.5 mg/mL vancomycin, 1 mg/mL neomycin, 1 mg/mL metronidazole, and 1 mg/mL ampicillin to eliminate intestinal microorganisms, followed by autoclaved water for 24 h21. Ten fecal samples were randomly selected from patients with hyperlipidemia and participants in the control group for microbiota transplantation. Then, 1 g of the participants’ fresh fecal particles were weighed and placed in sterile tubes, and 10 mL of normal saline was added to suspend them. After centrifugation at 2000 × g for 1 min at 4 °C, the supernatant was transferred into a sterile tube and centrifuged at 15,000 × g for 5 min. After discarding the supernatant, the bacterial pellet was re-suspended in 10 mL of saline to obtain a fecal microbiota suspension. Suspensions containing microbiota from patients or controls were mixed and used for transplantation. From the first day after antibiotic treatment, the mice were administered 100 µL of the microbiota suspension by gavage twice a week for 2 weeks. Subsequently, the mice were treated with the microbiota suspension by gavage once a week until the end of the experiment. Mice in the control group were treated with an equivalent volume of saline by gavage22.
Oil Red O staining
Oil red O staining of the whole aorta
The adipose tissue around the aortic vessels was removed, and the vessels were longitudinally dissected. After washing the blood vessels with water and immersing them in 60% isopropanol (Guoyao, China), they were placed in an Oil Red O staining solution (Solarbio, China) and stained in the dark for 60 min. Subsequently, they were immersed in 60% isopropanol until the fatty plaques in the lumen turned orange or bright red and then rinsed with distilled water. Photographs were taken, and images were analyzed.
Oil Red O staining of the aortic sinus
Sectioning was performed on the frozen heart tissue. Three aortic valves were cut at a similar level, and the sections were kept. The frozen slices were reheated, dried, fixed in a fixing solution for 15 min, rinsed with distilled water, and dried. The slices were then immersed in an Oil Red O staining solution and differentiated using 60% isopropanol. They were then washed with distilled water and re-stained with hematoxylin (Solarbio, China). They were soaked in distilled water again and differentiated with the differentiation solution (60% alcohol). Subsequently, the returning blue solution was used to return the color to blue. Finally, the slices were soaked, and the staining was observed under a microscope. A glycerol gelatin sealing agent was used to seal the film, and images were collected and analyzed under a microscope.
Isolation of testosterone-degrading bacteria
We placed 0.5 g of fecal sample from patients with hyperlipidemia in a conical flask containing 50 mL of testosterone-selective medium, and the conical flask was placed in a thermostatic shaker and incubated at 220 rpm and 37 °C for 3 days. Thereafter, 1 mL of the medium was placed in a new conical flask containing 49 mL of testosterone-selective medium and incubated at 220 rpm and 37 °C for 3 days. After repeating this process nine times, the ninth-generation medium liquid was placed on the Columbia blood plate, and the bacterial colonies were obtained after incubation at 37 °C for 12 h.
P. nitroreducens degradation of testosterone
P. nitroreducens was added to 5 mL LB medium and cultured at 37 °C. When OD600 was approximately 1.0, 3 mL of the bacterial culture was added to 50 mL of fermentation medium (3.5 g/L (NH4)2HPO4, 0.5 g/L K2HPO4, 0.5 g/L MgSO4·7H2O, 0.05 g/L ammonium ferric citrate, 10 g/L glucose, 2 g/L citric acid, pH 7.5). A methanol solution of testosterone was added to make the final testosterone concentration of the culture medium 1 g/L, after which 1 mL of the culture was collected every 24 h and stored at −80 °C for further analysis.
Drug sensitivity test
The disk diffusion method was used to detect the resistance of P. nitroreducens to aztreonam, cefoxitin, ampicillin, chloramphenicol, imipenem, and cefotaxime.
Transplantation of P. nitroreducens
The mice were treated with short-term antibiotics prior to bacterial gavage. The mice drank water mixed with an antibiotic mixture containing 0.5 mg/mL vancomycin, 1 mg/mL neomycin, 1 mg/mL metronidazole, and 1 mg/mL ampicillin for 7 days to eliminate intestinal microorganisms, followed by autoclaved water for 24 h. Sterile LB medium was inoculated with P. nitroreducens and incubated at 220 rpm and 37 °C in a thermostatic shaker. When the number of colonies in the culture solution was approximately 1 × 108 CFU, the mice in the P. nitroreducens group and imipenem treatment group were gavaged with 100 µL of the bacterial suspension thrice a week for 12 weeks. After 12 weeks of receiving P. nitroreducens gavage, the mice in the imipenem treatment group continuously received imipenem by gavage (50 mg/kg) once a day for 1 week. Mice in the control group were fed sterile LB medium by gavage (100 µL/mouse) thrice a week for 12 weeks. All the mice were fed a high-fat diet for 16 weeks.
Whole-genome sequencing of P. nitroreducens
The whole-genome DNA of P. nitroreducens was extracted using a genomic DNA extraction kit (Tiangen, Beijing, China). Utilizing a combination of PacBio Sequel IIe and Illumina sequencing platforms, DNA samples were prepared for sequencing library construction. For Illumina sequencing, genomic DNA was used for each strain in sequencing library construction. DNA samples were sheared into 400–500 bp fragments using a Covaris M220 Focused Acoustic Shearer following the manufacturer’s protocol. Illumina sequencing libraries were prepared from the sheared fragments using the NEXTFLEX Rapid DNA-Seq Kit. Briefly, 5’ prime ends were first end-repaired and phosphorylated. Next, the 3’ ends were A-tailed and ligated to sequencing adapters. The third step is to enrich the adapters-ligated products using PCR. The prepared libraries then were used for paired-end Illumina sequencing (2 × 150 bp) on Illumina Novaseq 6000 (Illumina Inc., San Diego, CA, USA). For PacBio sequencing, genomic DNA was fragmented at ~10 kb. DNA fragments were then purified, end-repaired and ligated with SMRT bell sequencing adapters following the manufacturer’s recommendations (Pacific Biosciences, CA). Next, the PacBio library was prepared and sequenced on one SMRT cell using standard methods.
The sequencing data underwent quality inspection, and a segment with an insertion length of ~400 bp was constructed. Paired-end 150 (PE150) sequencing was conducted on Illumina, encompassing both dual-end sequencing and single-end sequencing, with a read length of 150 base pairs. Each sample yielded raw sequencing data with a minimum coverage depth exceeding 100 times the genome coverage. Subsequently, a bacterial genome scanning map was generated using the short sequence assembly software SOAPdenovo 2 (http://soap.genomics.org.cn/). The map concatenated multiple Kmer parameters from the optimized sequence following second-generation sequencing to achieve the optimal contig assembly. The reads were then compared to the contig; based on the paid-end and overlay relationships of the reads, local assembly and optimization of the assembly results were performed to form the scaffolds. The assembly diagram integrates both second and third-generation sequencing methodologies, specifically employing Illumina and PacBio sequencing technologies. Each sample furnishes PacBio sequencing data, ensuring a minimum coverage depth exceeding 100 times that of the genome, along with Illumina sequencing data to further enhance assembly accuracy and completeness. This approach effectively mitigates the risk of losing information related to small plasmids (<15 kb) and guarantees the acquisition of comprehensive genomic data, inclusive of plasmid sequences. The bacterial genome completion map was assembled using the assembly software Unicycle for third-generation sequences, and during the assembly process, the Pilonjin software was used for sequence correction23. If there were overlays of a certain length or more at both ends of the final assembly sequence, the sequence was looped, and one end of the overlay sequence was truncated to obtain a complete chromosome and plasmid sequence. The analysis was conducted and assembled by the Majorbio cloud platform (www.majorbio.com).
Heterologous expression of 3/17β-HSD by E. coli
The pET28a plasmid was used as an overexpression vector for the expression of the 3/17β-HSD gene of P. nitroreducens in E. coli BL21 (DE3). Using the whole-genome DNA of P. nitroreducens as a template, 3/17β-HSD fragments were amplified by PCR (forward 5′- GGAATTCATATGAATCGAGTACAAGCAAG-3′; reverse 5′- CCCAAGCTTCAGATCCGCTGATCAG-3′). The amplified fragments were digested with NdeI and HindIII and ligated into the pET28a plasmid to construct pJT06. First-generation sequencing was used to confirm the construction of the correct pJT06 plasmid. Plasmid pJT06 was introduced into E. coli BL21 (DE3) to create E. coli BL21 (DE3)/pJT06, while the pET28a vector was introduced into E. coli BL21 (DE3) to establish E. coli BL21 (DE3)/pET28a as a control. E. coli BL21 (DE3)/pJT06 or E. coli BL21 (DE3)/pET28a was cultured in 30 mL LB medium at 37 °C on a rotating oscillator at a speed of 220 rpm. When the OD value of the culture medium was approximately 0.8 at 600 nm, 0.5 mM of IPTG was added to the medium to induce the expression of 3/17β-HSD.
Transplantation of E. coli BL21 (DE3)/pJT06 and E. coli BL21 (DE3)/pET28a
The mice were treated with short-term antibiotics prior to bacterial gavage. The mice drank water mixed with an antibiotic mixture containing 0.5 mg/mL vancomycin, 1 mg/mL neomycin, 1 mg/mL metronidazole, and 1 mg/mL ampicillin for 7 days to eliminate intestinal microorganisms, followed by autoclaved water for 24 h. E. coli BL21 (DE3)/pJT06 and E. coli BL21 (DE3)/pET28a were inoculated into a 10 mL sterile LB medium. When the OD600 value of the bacterial suspension was 0.6–0.8, 1 µL of isopropylthio-β-galactoside (IPTG) was added, which can induce the expression of recombinant E. coli BL21 (DE3)/pJT06 and E. coli BL21 (DE3)/pET28a. After 16–18 h of induced expression, 100 µL of bacterial suspension (approximately 2 × 108 CFU) was administered to the mice by gavage (thrice a week for 16 weeks).
Administration of androstenedione and testosterone
The mice in the AD treatment group were treated with AD gavage (0.5 mg/day for 16 weeks) after antibiotic treatment. Mice in the testosterone treatment group were treated by gavage with testosterone (0.5 mg/day, for 16 weeks) during the E. coli BL21 (DE3)/pJT06 gavage period.
Extraction of DNA from fecal samples
A QIAamp PowerFecal DNA Kit (Qiagen, Valencia, CA, USA) was used to extract DNA from 0.25 g of fecal samples following the manufacturer’s instructions.
Western blotting
Mice cecal contents (0.2 g) were placed in a 15 mL sterile centrifugal tube, suspended in 10 mL of physiological saline, and centrifuged at 2000 × g for 1 min; the supernatant was collected, placed in a new sterile centrifugal tube, and centrifuged at 15,000 × g for 5 min. The supernatant was discarded to obtain the bacterial precipitates. Subsequently, 250 μL of radioimmunoprecipitation assay lysis buffer was added. After ultrasound, it was centrifuged at 12,000 × g for 5 min. Next, 30 μg protein was added to 8% separation gel and 5% concentration gel for electrophoresis and imprinted on a polyvinylidene difluoride membrane. The membrane was sealed in 5% skim milk and incubated with His-Tag primary antibody. Finally, a freshly prepared enhanced chemiluminescence mixed solution was added dropwise to the protein side of the membrane. Luminescence was captured using a QuickChemi Imager, and photographs were taken.
Determining the prevalence of P. nitroreducens and 3/17β-HSD using qPCR
For P. nitroreducens-specific primers, we compared the P. nitroreducens JT2020 sequencing data from the NCBI database and the whole genome of P. nitroreducens with the Pseudomonas sequence from the NCBI database and selected the specific and conserved genes of P. nitroreducens. Specific primers targeting the conserved gas-producing P. nitroreducens sequences were designed using Premier 5.0 (Primer, Canada) (P. nitroreducens F: 5′-GGCTGTAGCGATGTTGGAGCA-3′; P. nitroreducens R: 5′-TCAGAGGCCGCCAGAAGGAT-3′). For 3/17β-HSD primers, specific primers targeting the 3/17β-HSD gene were designed using the same software (3/17β-HSD F: 5′- GCCGCTTCGTGTTCATCTGGG-3′; 3/17β-HSD R: 5′- CGCAAGGACAGGGCTCCATCA-3′). Additionally, the universal primer EUB was employed as an internal reference (EUB_F: 5′-ACTCCTACGGGAGGCAGCAGT-3′; EUB_R: 5′-ATTACCGCGGCTGCTGGC-3′). The relative abundances of gas-producing P. nitroreducens and 3/17β-HSD were determined using the 2−ΔΔct method24.
Statistical methods
Statistical analyses were performed with SPSS 23.0 and GraphPad Prism 7.0. Continuous variables that obey a normal distribution were expressed as the mean ± standard deviation. Continuous variables that did not obey a normal distribution were expressed as the median with the interquartile range. Comparison of differences between two groups was performed using a two-independent-sample t-test or Mann–Whitney U-test. Comparison of differences between more than two groups was via one-way analysis of variance or Kruskal–Wallis H-test. P-values < 0.05 indicate a statistical difference.
Results
Serum testosterone levels vary between patients with hyperlipidemia and controls
To investigate the prevalence of testosterone deficiency in individuals with hyperlipidemia, 259 patients diagnosed with hyperlipidemia according to the hyperlipidemia diagnostic criteria of the 2013 American College of Cardiology/American Heart Association (ACC/AHA) diagnostic guidelines18 were included in the experimental group, and 218 volunteers who did not meet the diagnostic criteria for hyperlipidemia were included in the control group.
After quantifying the serum testosterone levels of the participants in the two groups using liquid chromatography-tandem mass spectrometry (LC-MS/MS), we found that the serum testosterone level in the experimental group was 335.99 (295.44–375.76) ng/dL, which was significantly lower than that in the control group [402.39 (333.07–490.63) ng/dL (P < 0.001, Supplementary Fig. 1)].
Fecal microbiota can degrade testosterone
To explore whether the gut microbiota of individuals with hyperlipidemia can degrade testosterone (culturing intestinal microorganisms using a testosterone-containing solution), we collected the fecal samples of five patients in both the experimental and control groups. After performing thin-layer chromatography with regular sampling (24-h interval), we found that the fecal samples of patients in the experimental group had the degradation product, androstenedione present after 24 h, and testosterone was degraded completely within 96 h. However, 40% (2/5) of the fecal microbiota from the patients in the control group could not degrade testosterone, whereas the remaining 60% (3/5) weakly degraded testosterone. No degradation product was observed in the fecal sample of patients in the control group after 24 h, and testosterone was not degraded completely after 120 h (Supplementary Fig. 2).
Transplantation of fecal microbiota in patients with hyperlipidemia induced elevated serum lipid levels in mice
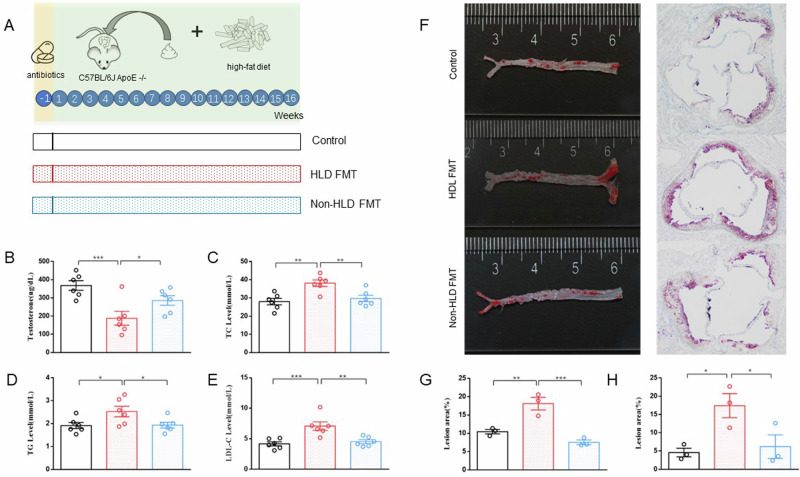
To verify the effects of gut microbiota on testosterone and lipid levels, we conducted fecal microbiota transplantation from humans into mice intestines. The mice were divided into three groups: (1) control group: orally administered with normal saline for 16 weeks (twice a week for the first 2 weeks, then once a week for the following 14 weeks); (2) HLD FMT group: orally administered with fecal microbiota suspension from patients with hyperlipidemia for 16 weeks (twice a week for the first 2 weeks, then once a week for the following 14 weeks); and (3) non-HLD FMT group: orally administered with fecal microbiota suspension from participants without hyperlipidemia for 16 weeks (twice a week for the first 2 weeks, then once a week for the following 14 weeks). Following the engraftment period, serological tests revealed that serum testosterone levels of mice in the HLD FMT group were significantly lower than those in both the control and non-HLD FMT groups. Additionally, serum TC, TG, and LDL-cholesterol (LDL-C) levels in the HLD FMT group were significantly higher than those in the control and non-HLD FMT groups. Furthermore, Oil Red O staining showed significantly increased lesion sizes in the whole aorta and aortic sinus of mice in the HLD FMT group compared with those in the controls and non-HLD FMT groups (Fig. 1A–H).
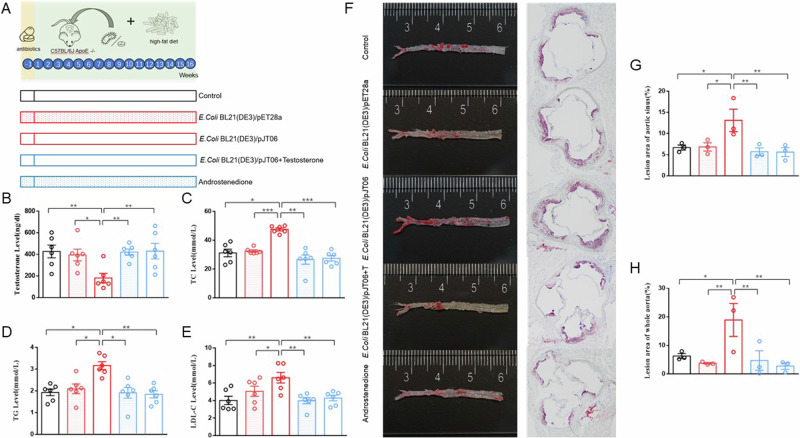
Fig. 1. Differences in serum testosterone, TC, TG, LDL-C, and HDL-C levels and atherosclerotic lipid deposits between the three groups of mice.
A Experimental design. B Serum testosterone levels (n = 6). C Serum TC levels (n = 6). D Serum TG levels (n = 6). E Serum LDL-C levels (n = 6). F Lipid deposit in mouse aorta and aortic valve plaque; G Lesion area of aortic sinus (n = 3). H Lesion area of whole aorta (n = 3). HLD hyperlipidemia, FMT transplantation of fecal microbiota, LDL-C low-density lipoprotein cholesterol, TC total cholesterol, TG triglyceride. Data are presented as means ± SEM, and one-way ANOVA with Tukey’s multiple comparisons tests was performed to calculate statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
Isolation of testosterone-degrading bacteria from patients with hyperlipidemia
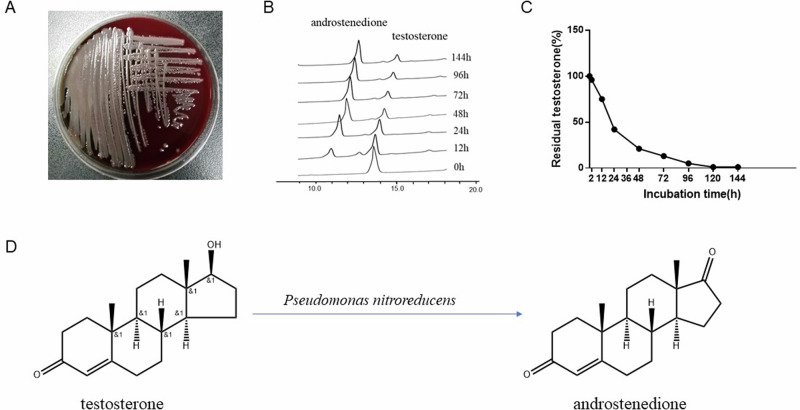
To isolate gut microbes that can degrade testosterone, we added the feces of the five patients with hyperlipidemia to a selective culture medium with testosterone as the sole carbon source. Finally, we isolated a gray white colony with a smooth surface (Fig. 2A). This strain was identified as P. nitroreducens by using MALDI–TOF mass spectrometry. In the phylogenetic tree, the 16S rRNA sequence of the strain was 99.50–99.64% similar to P. nitroreducens and 97.77–99.57% similar to other strains of Pseudomonas. To verify the testosterone degradation ability of P. nitroreducens, we added testosterone to the culture medium and incubated P. nitroreducens with it, and regularly measured the testosterone concentration in the culture medium. A high-performance liquid chromatography (HPLC) test showed that the testosterone level in the culture medium gradually decreased with time (Fig. 2B–D, Supplementary Fig. 3). The drug sensitivity test results showed that P. nitroreducens was sensitive to imipenem (Supplementary Fig. 4).
Fig. 2. Testosterone metabolism ability of Pseudomonas nitroreducens.
A Colony of P. nitroreducens. B Decreasing peak of testosterone displayed by HPLC. C Changes in testosterone concentration levels in culture medium. D The chemical formula for the degradation of testosterone to androstenedione. HPLC high-performance liquid chromatography.
Intragastric administration of P. nitroreducens in mice led to increased blood lipid levels
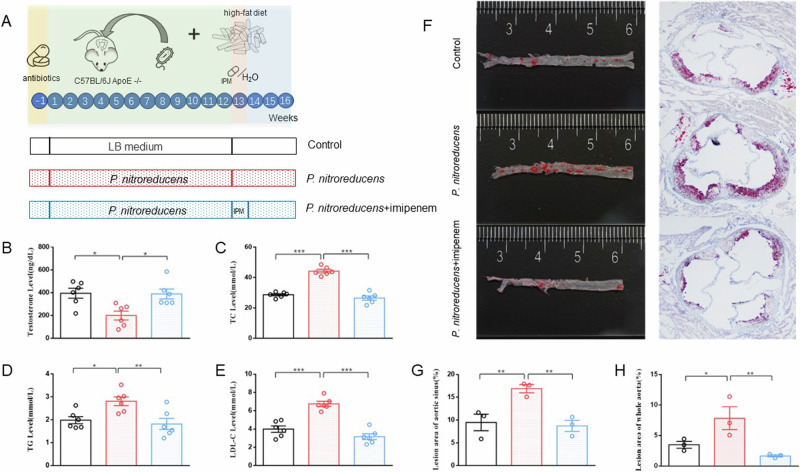
To investigate whether P. nitroreducens can affect testosterone and lipid levels, we introduced P. nitroreducens into the intestines of mice. We divided the mice into three groups: (1) P. nitroreducens group: P. nitroreducens was orally administered thrice a week for 12 weeks; (2) imipenem treatment group: After 12 weeks of P. nitroreducens gavage, imipenem was gavaged once a day for 1 week; (3) control group: LB medium was orally administered thrice a week for 12 weeks. Due to P. nitroreducens was suspended in LB medium for the other two groups, our control group chose to gavaged LB medium. After bacterial transplantation, the mice in the P. nitroreducens group showed significantly higher serum TC, TG, and LDL-C levels and lower testosterone levels than those in the control and imipenem treatment groups (P < 0.05, Fig. 3A–E). Oil Red O staining showed that lipid deposits in the aortic plaques of mice in the P. nitroreducens group were higher than those in the control and antibiotic-treatment groups (P < 0.05, Fig. 3F–H).
Fig. 3. Differences in serum testosterone, TC, TG, LDL-C, and HDL-C levels and atherosclerotic lipid deposits among the three groups of mice.
A Experimental design. B Serum testosterone levels (n = 6). C Serum TC levels (n = 6). D Serum TG levels (n = 6). E Serum LDL-C levels (n = 6). F Lipid deposit in mouse aorta and aortic valve plaque. G Lesion area of aortic sinus (n = 3). H Lesion area of whole aorta (n = 3). IPM imipenem, LDL-C low-density lipoprotein cholesterol, TC total cholesterol, TG triglyceride. Data are presented as means ± SEM, and one-way ANOVA with Tukey’s multiple comparisons tests was performed to calculate statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
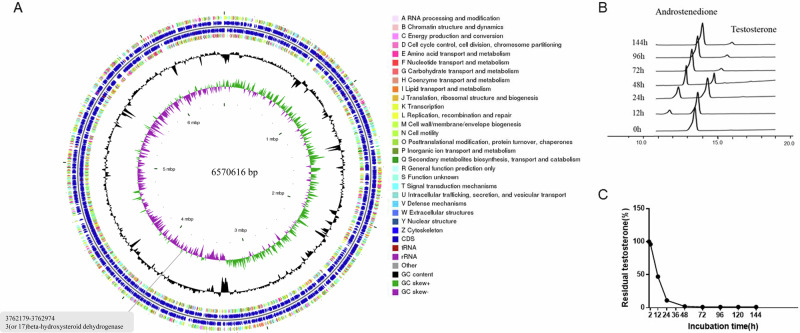
Identification of 3/17β-hydroxysteroid dehydrogenase (3/17β-HSD) gene in the P. nitroreducens genome
Whole-genome sequencing of P. nitroreducens was performed and compared to the Kyoto Encyclopedia of Genes and Genomes database, revealing the 3/17β-hydroxysteroid dehydrogenase (3/17β-HSD) gene (Fig. 4A). To construct a testosterone-degrading strain, the pET28a containing 3/17β-HSD gene was transformed into Escherichia coli BL21 (DE3) cells, and Escherichia coli BL21 (DE3)/pJT06 was obtained. HPLC combined with thin-layer chromatography showed that E. coli BL21 (DE3)/pJT06 transformed testosterone into androstenedione in a time-dependent manner (Fig. 4B, C, Supplementary Fig. 5A–C).
Fig. 4. Testosterone degradation ability of Escherichia coli BL21 (DE3)/pet28a and E. coli BL21 (DE3)/pjt06.
A 3/17β-HSD gene in P. nitroreducens. B Decreasing peak of testosterone displayed by HPLC. C Changes in testosterone concentration levels in culture medium.
E. coli BL21 (DE3)/pJT06 induced elevated serum lipid levels in mice
To explore whether E. coli BL21 (DE3)/pJT06 can affect testosterone and lipid levels, we gavaged mice with E. coli BL21 (DE3)/pJT06. The mice were divided into three groups: (1) control group: gavaged with LB medium thrice a week for 16 weeks; (2) E. coli BL21 (DE3)/pET28a group: gavaged with E. coli BL21 (DE3)/pET28a thrice a week for 16 weeks; (3) E. coli BL21 (DE3)/pJT06 group: gavaged with E. coli BL21 (DE3)/pJT06 thrice a week for 16 weeks. Compared with mice in the control group and E. coli BL21 (DE3)/pET28a group, serum TC, TG, LDL-C levels, and atherosclerotic lipid deposits in E. coli BL21 (DE3)/pJT06 group mice were significantly increased, whereas testosterone levels were significantly reduced (Fig. 5A–E).
Fig. 5. Differences in serum testosterone, TC, TG, LDL-C, and HDL-C levels and atherosclerotic lipid deposits among the five groups of mice.
A Experimental design. B Serum testosterone levels (n = 6). C Serum TC levels (n = 6). D Serum TG levels (n = 6). E Serum LDL-C levels (n = 6). F Lipid deposit in mouse aorta and aortic valve plaque. G Lesion area of aortic sinus (n = 3). H Lesion area of whole aorta (n = 3). T testosterone, LDL-C low-density lipoprotein cholesterol, TC total cholesterol, TG triglyceride. Data are presented as means ± SEM, and one-way ANOVA with Tukey’s multiple comparisons tests was performed to calculate statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
E. coli BL21 (DE3)/pJT06 had the capability to metabolize testosterone into androstenedione (AD). To explore the potential correlation between alterations in lipid levels and the rise in AD levels, we introduced a group of mice undergoing AD treatment, administered via gavage. Furthermore, to delve deeper into the impact of 3/17β-HSD on testosterone metabolism and lipid levels, we included a testosterone treatment group, where mice were supplemented with testosterone concurrently with the gavage of E. coli BL21 (DE3)/pJT06. Compared with the E. coli BL21 (DE3)/pJT06 group, mice in AD group and testosterone treatment group had significantly lower serum TC, TG, and LDL-C levels and a significantly higher serum testosterone level. Additionally, compared with the E. coli BL21 (DE3)/pJT06 group, lipid deposition in the aortic and aortic sinus plaques of mice in AD and testosterone treatment groups was significantly reduced (Fig. 5F–H).
To assess the presence of 3/17β-HSD in the intestinal tract of mice, we performed a western blotting experiment utilizing the cecal contents of the mice. No significant 3/17β-HSD bands were observed in the fecal samples of mice in the E. coli BL21 (DE3)/pET28a group, while 3/17β-HSD bands were detected in the fecal samples of the mice in the E. coli BL21 (DE3)/pJT06 and testosterone treatment groups, indicating that E. coli BL21 (DE3)/pJT06 induced 3/17β-HSD expression (Supplementary Fig. 5D–F).
Elevated relative abundances of P. nitroreducens and 3/17β-HSD gene in patients with hyperlipidemia
To investigate the relative abundance of P. nitroreducens and 3/17β-HSD in the gut microbiota of patients with hyperlipidemia and control populations, we conducted quantitative PCR (qPCR) on 158 patients with hyperlipidemia and 151 control populations using specific primers for P. nitroreducens and degenerate primers for 3/17β-HSD. The housekeeping gene approach was employed to measure relative abundance.
The whole genome of P. nitroreducens was compared with all Pseudomonas sequences in the National Center for Biotechnology Information database. Conserved and interspecific genes from P. nitroreducens were selected for this design. Finally, the pTX1 gene was selected. For the degenerate primers of 3/17β-HSD enzymes, we compared the 3/17β-HSD gene sequence in the Basic Local Alignment Search Tool and selected all homologous genes for the design of primers targeting the conserved regions. Eub universal primers were used to calculate the total amount of bacteria in fecal DNA. qPCR assay was used to quantitate P. nitroreducens by targeting the pTX1 gene and total bacteria using universal Eub-primers targeting the 16S rRNA gene from the domain bacteria. Likewise, the quantification of 3/17β-HSD gene abundance was conducted through comparative analysis utilizing universal Eub-primers.
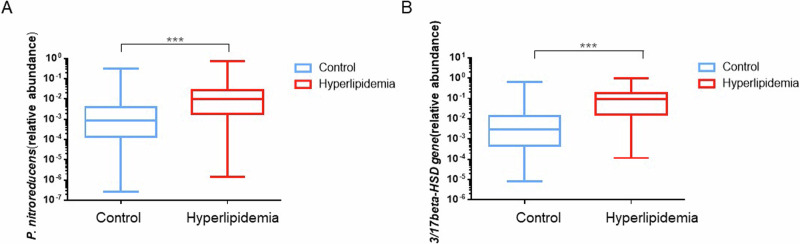
qPCR screening showed that the relative abundance of P. nitroreducens was log10[−1.99 (−2.74, −1.56)] and log10[−3.07 (−3.85, −2.39)] in patients with hyperlipidemia and controls, respectively. The relative abundance of 3/17β-HSD in patients with hyperlipidemia and controls was log10 [−1.02 (−1.80, −0.75)] and log10 [−1.54 (−3.35, −1.87)], respectively; this indicates that the relative abundance of intestinal P. nitroreducens and the 3/17β-HSD gene is higher in individuals with hyperlipidemia (Fig. 6). To further investigate the potential relationship between P. nitroreducens and the 3/17β-HSD gene with testosterone and lipid levels, we conducted a correlation analysis. The results indicated a correlation between the relative abundance of P. nitroreducens and the 3/17β-HSD gene with the levels of testosterone, TC, TG, HDL-C, and LDL-C (Supplementary Fig. 6).
Fig. 6. Abundance of P. nitroreducens and 3/17β-HSD in male patients with hyperlipidemia.
A Abundance of P. nitroreducens in the hyperlipidemia group (n = 158) and control group (n = 151); B Abundance of 3/17β-HSD in the hyperlipidemia group (n = 158) and control group (n = 151). Data are presented as interquartile range, and Mann–Whitney U-test was performed to calculate statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
The onset of testosterone deficiency-induced metabolic disorders is widely recognized, especially because of its promoting effect on hyperlipidemia and obesity25,26. Although the mechanism underlying testosterone deficiency is not fully understood, it can be caused by aging, Klinefelter’s syndrome, and other factors27. The gut microbiota reportedly regulates testosterone levels28. The colonization of filamentous bacteria in the intestine causes increased serum testosterone levels13, whereas the colonization of Mycobacterium neoaurum can cause decreased testosterone levels29. These results suggest that the gut microbiota can regulate serum testosterone levels through certain pathways. Over 50 years ago, certain bacteria were found to be able to degrade steroids; however, research on them was mostly limited to ecology and environmental protection14. Additionally, the effect of the gut microbiota on regulating blood lipid metabolism by testosterone degradation has not been confirmed.
Our study demonstrated that the presence of bacteria in the gut, which can metabolize testosterone, can affect serum testosterone levels. Previous studies have shown that the gut microbiota can regulate serum testosterone levels through certain pathways13,29. The regulation of androgens by the gut microbiota is currently not fully understood. Specific bacteria, such as Clostridium scindens, can synthesize androgens from glucocorticoids, and certain bacteria can produce 5α-reductase to convert testosterone into the more active metabolite, dihydrotestosterone30. This study found that P. nitroreducens and E. coli BL21 (DE3)/pJT06 containing the 3/17β-HSD gene in the intestine can metabolize testosterone, thereby reducing serum testosterone levels; this may be due to the degradation of the bound form of testosterone in the intestine by the 3/17β-HSD enzyme expressed by P. nitroreducens or E. coli BL21 (DE3)/pJT06 before reabsorption into the circulation.
This research was based on the fact that testosterone deficiency can cause hyperlipidemia. Testosterone alters the expression of genes involved in lipoprotein assembly and secretion. It upregulates the expression of scavenger receptor B1 (SR-B1) and hepatic lipase in hepatocytes. Hepatic lipase hydrolyzes phospholipids on the surface of HDL-C and induces SR-B1 to selectively absorb HDL-C, thereby promoting the reverse transport of cholesterol and playing an anti-atherosclerotic role31. Additionally, testosterone deficiency can decrease the expression of LDL receptor (LDLR) mRNA in the liver of pigs. LDLR is an important regulator of serum LDL-C levels in humans and animals, and decreased LDLR expression can increase serum LDL-C levels32. Moreover, testosterone can regulate the expression of microsomal TG transfer protein (MTP) in the liver of rats. Lack of testosterone can decrease MTP levels, block TG transport, and increase serum TG levels33,34. Furthermore, testosterone deficiency promotes increased liver adipogenesis and obesity. Testosterone therapy in patients with obesity leads to decreased TC, TG, and LDL-C levels35.
Our study has potential clinical applications. Antibiotic treatments targeting testosterone-degrading bacteria can improve testosterone levels and ameliorate dyslipidemia. In our study, treating P. nitroreducens with imipenem increased testosterone levels and decreased serum TC, TG, and LDL-C levels. However, adverse consequences of antibiotic treatment, such as dysbiosis and antibiotic resistance, require attention. Supplementation with exogenous testosterone can ameliorate testosterone-induced dyslipidemia. The findings of this study suggest that adding external testosterone can ameliorate blood lipid irregularities induced by E. coli BL21 (DE3)/pJT06. Nonetheless, more investigation is warranted to ascertain whether testosterone supplementation confers growth advantages to bacteria that metabolize testosterone. It has been noted that testosterone supplementation can disrupt the balance of gut microbiota36. Studies have revealed a link between exogenous testosterone supplementation and changes in the diversity and composition of gut microbiota, as well as the incidence of colorectal cancer37. Therefore, before testosterone supplementation, strict dosage control is necessary to prevent side effects. In addition to testosterone supplementation, inhibiting intestinal 3/17β-HSD activity is an alternative or combined regimen approach. Soy can inhibit 3/17β-HSD activity in the intestine. Isoflavones are naturally occurring phytoestrogens in soybeans that can competitively inhibit 3/17β-HSD expression by producing strong inhibitory effects (Ki, 0.01–0.02 μm)38. Over 80% of circulating estradiol in males originates from the aromatization process of testosterone39. Consequently, reductions in serum testosterone levels frequently coincide with declines in serum estradiol levels. Studies have shown that certain symptoms of testosterone deficiency may stem from decreased estrogen levels40,41. Estrogen can influence lipid metabolism in males42–44. Hence, supplementation with phytoestrogens can potentially mitigate lipid levels by binding to estrogen receptors. Epidemiological evidence indicates an inverse relationship between dietary isoflavone intake and hyperlipidemia45. Therefore, a diet rich in soy (or isoflavones) could serve as a lifestyle intervention to increase serum testosterone levels and ameliorate dyslipidemia caused by intestinal 3/17β-HSD.
However, some limitations need to be taken into account in future research. First, the intestine contains a large number of anaerobic bacteria that can be difficult to cultivate. Further investigations are warranted to elucidate their potential impact on testosterone metabolism. In addition, there is more than one type of bacteria expressing 3/17β-HSD in the intestine and the abundance of other bacteria with 3/17β-HSD expression ability should also be evaluated. Moreover, besides 3/17β-HSD, there are many other enzymes in the intestine that have the ability to metabolize testosterone, and this study did not consider their impact on testosterone levels. Furthermore, the gut environmental factors that affect enzyme activity, such as gut pH and enzyme inhibitors, also affect the ability of 3/17β-HSD to metabolize testosterone. In summary, this study identified bacteria expressing the 3/17β-HSD gene in the intestine, which may cause testosterone deficiency and subsequent onset and progression of hyperlipidemia. Interventions targeting intestinal 3/17β-HSD in patients with hyperlipidemia are the main focus of our future research.
Supplementary information
Acknowledgements
Funding from the National Natural Science Foundation of China (81772265) supported the analysis and interpretation of data. All authors have contributed significantly and agreed with the content of the manuscript. We are grateful to all the people who participated in our study.
Author contributions
Conception and design: J. Tao, W. Dai, Y. Lyu, Y. Li, Z. Zhao, X. Jiang; Administrative support: Y. Li; Provision of study materials or patients: Y. Lyu, H. Liu, J. Le, T. Sun, Q. Yao; Collection and assembly of data: J. Tao, W. Dai, H. Liu, J. Le; Data analysis and interpretation: J. Tao, Y. Li; Manuscript writing: All authors; Final approval of manuscript: All authors.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. The sequencing data from this study have been submitted to NCBI’s Sequence Read Archive under accession no. PRJNA1020053.
Competing interests
The authors declare no competing interests.
Ethical approval
All protocols were approved by the Renmin Hospital of Wuhan University Institutional Review Board (approval no. WDRY2023-K023). Informed consent was waived by the committee. Research records or samples intended for publication will adhere to strict confidentiality standards, excluding any names or other identifying information of the subjects. Subject information, encompassing medical records, clinical examination results, and related data, is uniquely identified by a designated code. Only research doctors and authorized personnel can associate this code with the names of subjects through a checklist, which is securely stored in the research center. The research data is kept at the Clinical Laboratory Center and can be viewed by researchers, research regulatory authorities, and ethics committees. Public reports on the results of this study will not disclose the personal identity of the subjects. Animal studies were conducted in strict accordance with protocols approved by the Animal Experiment Center Renmin Hospital of Wuhan University (approval no. S0272012259A).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jun Tao, Wen Dai, Yongnan Lyu.
Contributor Information
Zhiming Zhao, Email: heat.zhao@163.com.
Xuejun Jiang, Email: xjjiang@whu.edu.cn.
Yan Li, Email: yanlitf1120@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00599-1.
References
- 1.Cui, B. et al. Association between cooking patterns and the prevalence of hyperlipidemia in Eastern China. BMC Public Health24, 75 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yanai, H., Adachi, H., Hakoshima, M. & Katsuyama, H. Postprandial hyperlipidemia: its pathophysiology, diagnosis, atherogenesis, and treatments. Int. J. Mol. Sci.24, 13942 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ascaso, J. F. et al. Prevalence of metabolic syndrome and cardiovascular disease in a hypertriglyceridemic population. Eur. J. Intern. Med.22, 177–181 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Simon, D. et al. Association between plasma total testosterone and cardiovascular risk factors in healthy adult men: the Telecom Study. J. Clin. Endocrinol. Metab.82, 682–685 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Zhang, N. et al. The relationship between endogenous testosterone and lipid profile in middle-aged and elderly Chinese men. Eur. J. Endocrinol.170, 487–494 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Basaria, S. et al. Adverse events associated with testosterone administration. N. Engl. J. Med.363, 109–122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corona, G. et al. Testosterone and metabolic syndrome: a meta-analysis study. J. Sex. Med.8, 272–283 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Kelly, D. M. & Jones, T. H. Testosterone: a metabolic hormone in health and disease. J. Endocrinol.217, R25–R45 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Li, C. Y. et al. Major glucuronide metabolites of testosterone are primarily transported by MRP2 and MRP3 in human liver, intestine and kidney. J. Steroid Biochem. Mol. Biol.191, 105350 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auer, K. E. et al. Measurement of fecal testosterone metabolites in mice: replacement of invasive techniques. Animals10, 165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsiao, T. H. et al. Circulating androgen regulation by androgen-catabolizing gut bacteria in male mouse gut. Gut Microbes15, 2183685 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markle, J. G. et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science339, 1084–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Yurkovetskiy, L. et al. Gender bias in autoimmunity is influenced by microbiota. Immunity39, 400–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch, S. V. & Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med.375, 2369–2379 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Fu, J. et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res.117, 817–824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takagi, T. et al. Changes in the gut microbiota are associated with hypertension, hyperlipidemia, and type 2 diabetes mellitus in Japanese subjects. Nutrients12, 2996 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holert, J. et al. Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. mBio9, e02345–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy, S. M. et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: Executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation139, e1046–e1081 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Skye, S. M. et al. Microbial transplantation with human gut commensals containing cutC is sufficient to transmit enhanced platelet reactivity and thrombosis potential. Circ. Res.123, 1164–1176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan, J. et al. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metab.30, 675–688.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Chevalier, G. et al. Effect of gut microbiota on depressive-like behaviors in mice is mediated by the endocannabinoid system. Nat. Commun.11, 6363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bárcena, C. et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med.25, 1234–1242 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, D. et al. Gut-microbiome-expressed 3β-hydroxysteroid dehydrogenase degrades estradiol and is linked to depression in premenopausal females. Cell Metab.35, 685–694.e5 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Kelly, D. M. & Jones, T. H. Testosterone and obesity. Obes. Rev.16, 581–606 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Navarro, G., Allard, C., Xu, W. & Mauvais-Jarvis, F. The role of androgens in metabolism, obesity, and diabetes in males and females. Obesity23, 713–719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basaria, S. Male hypogonadism. Lancet383, 1250–1263 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Shin, J. H. et al. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res. Microbiol.170, 192–201 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Li, D. et al. 3β-Hydroxysteroid dehydrogenase expressed by gut microbes degrades testosterone and is linked to depression in males. Cell Host Microbe30, 329–339.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Ridlon, J. M. et al. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res.54, 2437–2449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langer, C. et al. Testosterone up-regulates scavenger receptor BI and stimulates cholesterol efflux from macrophages. Biochem. Biophys. Res. Commun.296, 1051–1057 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Cai, Z. et al. Effect of testosterone deficiency on cholesterol metabolism in pigs fed a high-fat and high-cholesterol diet. Lipids Health Dis.14, 18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han, C. C. et al. Effect of cholesterol on lipogenesis and VLDL-TG assembly and secretion in goose primary hepatocytes. Mol. Cell Biochem.374, 163–172 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Senmaru, T. et al. Testosterone deficiency induces markedly decreased serum triglycerides, increased small dense LDL, and hepatic steatosis mediated by dysregulation of lipid assembly and secretion in mice fed a high-fat diet. Metabolism62, 851–860 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Mårin, P., Odén, B. & Björntorp, P. Assimilation and mobilization of triglycerides in subcutaneous abdominal and femoral adipose tissue in vivo in men: effects of androgens. J. Clin. Endocrinol. Metab.80, 239–243 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Li, L. Y. et al. Alterations of gut microbiota diversity, composition and metabonomics in testosterone-induced benign prostatic hyperplasia rats. Mil. Med. Res.9, 12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song, C. H. et al. Changes in gut microbiome upon orchiectomy and testosterone administration in AOM/DSS-induced colon cancer mouse model. Cancer Res. Treat.55, 196–218 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keung, W. M. Dietary estrogenic isoflavones are potent inhibitors of beta-hydroxysteroid dehydrogenase of P. testosteronii. Biochem. Biophys. Res. Commun.215, 1137–1144 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Longcope, C., Kato, T. & Horton, R. Conversion of blood androgens to estrogens in normal adult men and women. J. Clin. Invest.48, 2191–2201 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khosla, S. et al. Relationship of serum sex steroid levels and bone turnover markers with bone mineral density in men and women: a key role for bioavailable estrogen. J. Clin. Endocrinol. Metab.83, 2266–2274 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Hammes, S. R. & Levin, E. R. Impact of estrogens in males and androgens in females. J. Clin. Invest.129, 1818–1826 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carani, C. et al. Effect of testosterone and estradiol in a man with aromatase deficiency. N. Engl. J. Med.337, 91–95 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Jones, M. E., Boon, W. C., Proietto, J. & Simpson, E. R. Of mice and men: the evolving phenotype of aromatase deficiency. Trends Endocrinol. Metab.17, 55–64 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Finkelstein, J. S. et al. Gonadal steroids and body composition, strength, and sexual function in men. N. Engl. J. Med.369, 1011–1022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, X. et al. Dietary isoflavones intake is inversely associated with non-alcoholic fatty liver disease, hyperlipidaemia and hypertension. Int. J. Food Sci. Nutr.73, 60–70 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The sequencing data from this study have been submitted to NCBI’s Sequence Read Archive under accession no. PRJNA1020053.