Abstract
Background and Aims
Management of Crohn’s disease (CD) requires frequent monitoring for disease activity and response to therapy. In this study, we examined the clinical utility of a novel stool-derived eukaryotic RNA (seRNA)-based diagnostic in patients with CD.
Methods
Stool samples were collected from 68 individuals for up to 3 time points prior to, and after initiation of an advanced therapy. Stool samples underwent RNA extraction and sequencing using a custom capture panel (n = 1507 transcripts). seRNA signatures were compared to Crohn's Disease Activity Index scores and endoscopies, when available. Random forest models classified disease severity when compared to Crohn's Disease Activity Index scores. seRNA signatures were also used to assess expression of the therapy target and cell type abundance at various time points.
Results
Across all 102 samples collected from 68 individuals, the classifier successfully parsed individuals with active disease (n = 37) relative to those in remission (n = 65) with 87% sensitivity and 77% specificity, respectively. A second classifier, which was employed on subjects with active disease (n = 37), successfully parsed individuals with mild disease (n = 15) from those with moderate disease (n = 22) with 93% and 86% sensitivity, respectively. For the 16 subjects with longitudinal data, seRNA expression of the therapeutic target (eg, ITGA4/ITGB7 for vedolizumab or IL12/IL23 for ustekinumab) as well as lymphocyte burden was correlated with response.
Conclusion
A novel seRNA and informatic-based method reliably discriminates active disease from remission and stratifies mild from moderate CD activity. This demonstrates preliminary feasibility to predict therapeutic response and assess disease activity for patients with CD.
Keywords: Crohn’s Disease, Noninvasive Diagnostics, Stool Eukaryotic RNA, Disease Activity and Therapeutic Response
Introduction
Crohn’s disease (CD) is characterized by transmural inflammation of the gastrointestinal mucosa, affecting more than a million individuals in the United States.1 CD is a progressive disease where a majority of the patients ultimately develop complications of either stricturing or penetrating disease with intestinal surgery required for as many as 80% of patients, and a permanent stoma required in more than 10% of patients.2 Maintaining long-term remission is therefore the goal of treatment to effectively avoid complications, surgery, malignancy, and iatrogenic side effects.3, 4, 5 Currently, methods for assessing treatment effectiveness primarily include invasive tests such as colonoscopy and sigmoidoscopy, which require inconvenient and uncomfortable bowel preparation.6
Existing noninvasive diagnostics within the inflammatory bowel disease (IBD) space fall into 3 categories: blood-based protein biomarkers, stool-based protein biomarkers, or stool-based microbiome biomarkers. Serology markers typically include C-reactive protein (CRP).7,8 and fecal markers typically include calprotectin.9 Currently, there are no clinically used stool-based RNA biomarkers for diagnosing or monitoring CD. While noninvasive biomarkers are frequently employed for evaluation and monitoring of patients with CD, current guidelines only recommend noninvasive monitoring of disease activity in niche circumstances such as patients with established disease, with recently documented endoscopic remission. False negative and false positive rates of both fecal calprotectin and serum CRP prevent recommendations for empirical treatment for individuals who have discrepant clinical symptoms with noninvasive outcomes (eg, asymptomatic with elevated fecal calprotectin levels).10 These limitations include lack of specificity for CD activity, inability to quantify the extent and severity of inflammation, and inability to detect other conditions that would be identified via endoscopy, such as dysplasia, cytomegalovirus colitis, or stricturing disease.
Novel molecular tests that leverage blood-based biomarkers (eg, IBD sgi Diagnostic and Monitr) have been developed to improve sensitivity and specificity for predicting therapeutic response and monitoring mucosal healing during treatment.11,12 While these noninvasive options can be leveraged as alternatives to endoscopic procedures, they lack sufficient specificity for IBD and are not reliable for predicting therapeutic response or monitoring disease status during treatment.13,14
Numerous researchers and institutions have attempted to use RNA biomarkers in stool samples to evaluate gastrointestinal health with limited success. First, the human RNA biomarkers in stool samples are heavily degraded. Previous attempts to isolate eukaryotic RNA in stool have been largely unsuccessful with 25%–50% of extracted samples containing no viable RNA for analysis.15,16 Second, these degraded human signals are outnumbered by bacterial noise. Within the colon, there are approximately 1012 bacterial cells per gram of intestinal content and colonic microflora can include thousands of different species.17,18 Recent technology has provided insight into the preservation and stabilization of RNA biomarkers in stool samples.19,20 This research has leveraged RNA biomarkers in stool samples to develop diagnostics for gastrointestinal disease within the oncology space.21
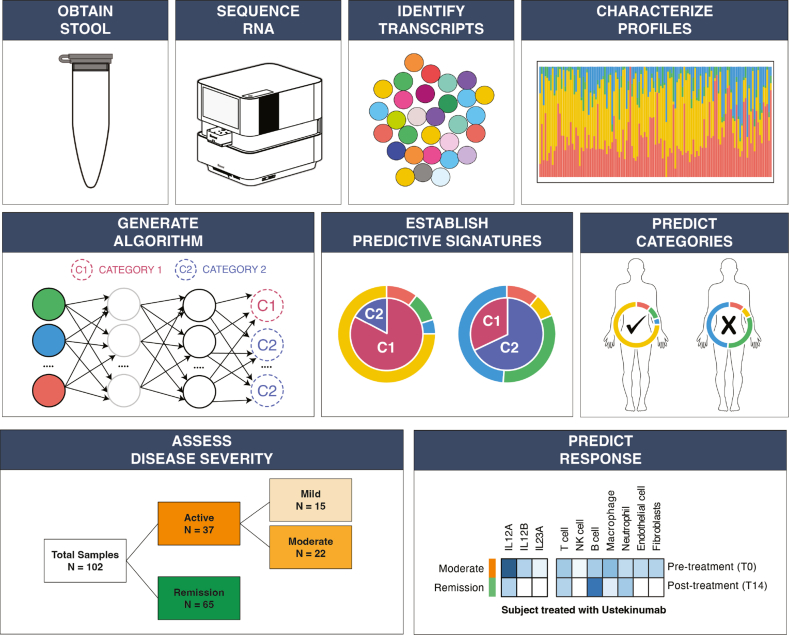
However, stool-based RNA biomarkers have never been assessed for the ability to improve disease monitoring within IBD. Here we describe a cohort of 68 subjects who provided stool samples as part of a prospective observational cohort. RNA sequencing data from these samples was assessed to monitor disease activity and predict therapeutic response during treatment. Results from this study provide evidence that RNA biomarkers in stool samples can be an effective method for noninvasive assessment of IBD (Figure 1).
Figure 1.
Overview of the analysis pipeline and the methods employed to understand if stool-derived eukaryotic RNA (seRNA) can be used to noninvasively assess disease severity and predict therapeutic response for subjects with Crohn’s disease (CD). Stool samples were collected and total seRNA extraction and sequencing was employed. Quantified seRNA transcripts were compared to clinical correlates to build algorithms, establish signatures, and predict outcomes. Model results were compared to CD Activity Index scores.
Methods
Study Overview and Sample Collection
Individuals with CD were identified for enrollment as part of a prospective observational cohort approved by the institutional review board (NCT03646708). Enrolled subjects were being evaluated at the Washington University School of Medicine IBD center, and stool samples were collected through the Digestive Disease Research Core Center. For all subjects, advanced therapies employed were based on treating physician’s recommendations. Crohn's Disease Activity Index (CDAI) was used to determine disease activity at all timepoints where stool samples were collected. Colonoscopy, fecal calprotectin, CRP, and other clinical correlates were completed at various time points based on physician recommendations (Figure A1). Enrolled subjects were asked to provided stool samples prior to treatment (T0), approximately 14 weeks (range = 6–22 weeks) after treatment (T14), and approximately 52 weeks after treatment (T52).
Stool-Derived Eukaryotic RNA (seRNA) Isolation
For all subjects, stool samples were collected fresh-frozen and stored at −80 degree celsius until extraction. All stool samples were shipped on dry ice to a centralized laboratory for processing. RNA was extracted as previously described.19,20 RNA integrity and mass were evaluated using an Agilent 2100 Bioanalyzer.22 RNA underwent reverse transcriptase, library preparation, and hybridization capture using a custom capture sequencing panel (human reference genome (hg38)).23 Sequencing was performed using an Illumina NextSeq 500.
Gene Panel for Targeted Sequencing
The gene panel for targeted sequencing was developed from literature reviews. One of the primary sources was the IBD Transcriptome and Metatranscriptome Meta-Analysis,24 from which differentially expressed (DE) gene sets were downloaded (https://ibd-meta-analysis.herokuapp.com). Each set was generated by contrasting IBD cohorts with control cohorts for a particular tissue type (colon, ileum, rectum, fibroblasts, epithelium, and stool-associated). Gene candidates were required to meet one of the following criteria: (1) DE in 3 or more tissues, (2) DE in 2 or more tissues with high expression, (3) DE in at least one tissue with extremely high expression. The gene panel was expanded by incorporating genes identified in the ClinGen database25 and genes marked with “elite” status under IBD in MalaCards database.26
Next, a probe set was developed according to the gene panel. For each gene, probes were deisgned to target regions on 2 disjoined exons. Glyceraldehyde-3-phosphate dehydrogenase, which served as the housekeeping transcriped, required full tiling of all exons. A target with max length of 120 basepairs was extracted by centering at each exon after excluding any untranslated region. The target candidates were ranked in descending order of the number of transcript isoforms that cover the locus. To enrich eukaryotic RNA signals, each candidate was compared against microbial genomes and removed if significant hits were detected. Furthermore, the candidate was compared against the human genome to ensure minimal effect of multiple hits.
Biomarker Panel Development
Biomarkers were derived from mining transcript interactions from each model. Selections for the top 3 performant biomarkers were based on the following: 1) interactions that had the most influence on making consistently accurate predictions, 2) if removing an interaction term (composite biomarker) from the model imposed an unsatisfactory error rate on performance, and 3) biomarker complexity (ie, the number of constituent transcripts that composed the biomarker).
Analysis and Statistical Considerations
Machine learning classifiers were constructed using custom code and the following open source R packages with associated dependencies: tidymodels (CRAN) (v1.1.0), caret (CRAN) (v6.0-94), caretEnsemble (CRAN) (v2.0.2), randomForest (CRAN) (v4.7-1.1), MLMetrics (CRAN) (v1.1.1). Models were constructed using 10-fold internal cross validation with 3 repeats. Models were assessed for performance using the receiver operating characteristic (ROC) area under the curve (AUC), sensitivity, and specificity. Gene ontologies were derived using custom code and the following open source R packages with associated dependencies: DESeq2 (Bioconductor) (v1.42.0), clusterProfiler (Bioconductor) (v4.10.0). Models were assessed using genome wide annotations for human (org.Hs.eg.db) (v3.19.1) and biological process sub ontology. Using a microenvironment cell populations-counter, the abundance of tissue-infiltrating immune and stromal cell populations were deconvoluted using the entire targeted transcript expression profile of the CD cohort.27
Results
Cohort Demographics and Clinical Information
In total 102 stool samples were obtained from 68 subjects (Table). Of the 68 subjects, 39 (57%) were female with 39 subjects (57%) being 17–40 year old and 28 (41%) being over 40 year old. Almost all participants (97%) had ileal or ileocolonic disease with 54% having stricturing or penetrating disease and 10% having concomitant perianal disease. Based on CDAI scores, 64% of subjects were in remission (score <150), 15% had mild disease (score ≥150 to <220), and 22% had moderate disease (score ≥220 to <450). The majority of participants were being treated with vedolizumab (26%) or ustekinumab (32%) with a small proportion of individuals being treated with infliximab (9%). Across all 68 subjects, 5 endoscopies were completed, 2 subjects had fecal calprotectin completed, 21 subjects had a CRP completed contemporaneously with stool sample (±14 days) (Figure A1).
Table.
Distribution of Demographic and Clinical Information for All Eligible Participants and Associated Stool Samples
| Characteristic | Eligible cohort |
|---|---|
| No. | 68 participants (102 stool samples) |
| Sex | |
| Female | 39 participants (58 stool samples) |
| Male | 29 participants (44 stool samples) |
| Age at diagnosis | |
| ≤16 y | 0 participants (0 stool samples) |
| 17–40 y | 39 participants (54 stool samples) |
| >40 y | 28 participants (46 stool samples) |
| Unknown | 1 participant (2 stool samples) |
| Location (not mutually exclusive) | |
| Ileal | 25 participants (39 stool samples) |
| Colonic | 0 participants (0 stool samples) |
| Ileocolonic | 41 participants (62 stool samples) |
| Upper gastrointestinal disease | 2 participants (3 stool samples) |
| Behavior (not mutually exclusive) | |
| Nonstricturing or nonpenetrating | 33 participants (48 stool samples) |
| Stricturing | 26 participants (39 stool samples) |
| Penetrating | 11 participants (18 stool samples) |
| Concomitant perianal disease | 7 participants (10 stool samples) |
| Stool sample collections | |
| Pre-treatment (T0) | 55% (56 stool samples) |
| 14 wk post-treatment (T14) | 32% (33 stool samples) |
| 52 wk post-treatment (T52) | 13% (13 stool samples) |
| Crohn’s disease activity index (CDAI) | |
| Remission (score <150) | 64% (65 stool samples) |
| Mild (score ≥150 to <220) | 15% (15 stool samples) |
| Moderate (score ≥220 to <450) | 22% (22 stool samples) |
| Treatment modalities (not mutually exclusive) | |
| Vedolizumab | 26% (18 participants) |
| Ustekinumab | 32% (22 participants) |
| Infliximab | 9% (6 participants) |
| Adalimumab | 1% (1 participants) |
| Concomitant anti-inflammatorya | 9% (6 participants) |
| Unknown | 6% (4 participants) |
Anti-inflammatory agents included prednisone, budesonide, azathioprine, or mesalamine.
Stool Sample Sequencing Quality and Integrity
Stool-derived eukaryotic RNA (seRNA) from all stool samples underwent custom capture sequencing (see Methods). All samples (n = 102) had a minimum of 0.75 million reads per sample with 82% having over 1 million reads per sample (Figure A2). There were 39 samples that had ≤25% alignment to the targeted regions of interest.
seRNA Classifier Models Stratify IBD Disease Activity
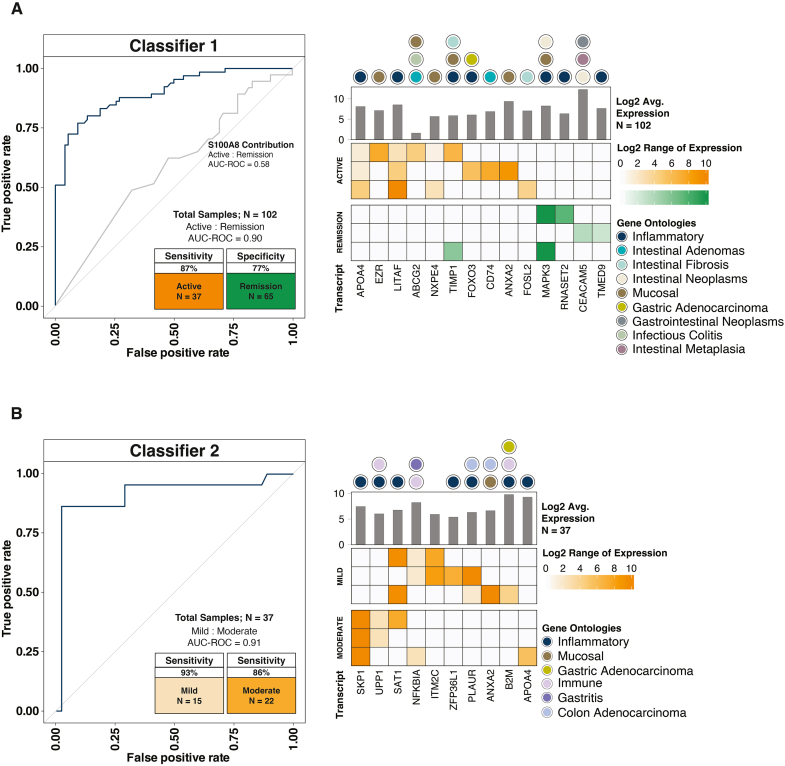
Using the concentrations for seRNA transcripts, 2 classifiers were constructed to predict disease activity (based on CDAI) for subjects with CD (n = 102 stool samples from 68 unique patients). For all individuals, Classifier 1 correctly predicted 32 of 37 subjects with active disease (sensitivity = 87%) and 50 of 65 subjects in remission (specificity = 77%). ROC AUC was 0.90 (Figure 2A). While the model identified a calprotectin subunit (S100A8) as a significant contributor to segregating subjects with various disease activities, ROC AUC for the surrogate fecal calprotectin protein marker (S100A8) in isolation was only 0.58 (Figure 2A). For the 37 subjects with active disease (ie, mild or moderate), Classifier 2 correctly classified 14 of 15 individuals with mild disease (sensitivity = 93%) and 19 of 22 individuals with moderate disease (sensitivity = 86%). ROC AUC was 0.91 (Figure 2B). Based on gene ontologies, the biomarker signatures leveraged to classify subjects were directly related to inflammation, mucosal disease, and intestinal neoplasia.
Figure 2.
Machine learning model classifiers show seRNA's ability to approximate disease activity. For each classifier, the top 3 biomarker signatures are shown. Each row in the heatmap indicates a composite biomarker, which is composed of highlighted transcripts in that row. The shade of the highlight indicates transcript expression. Gene ontologies for each transcript are also provided. (A) Classifier 1 predicted subjects with active disease (n = 37) relative to subjects in remission (n = 65) when compared to the Crohn’s Disease Activity Index (CDAI). The receiver operating characteristic (ROC) curve for the RNA calprotectin subunit (S100A8) is shown to provide a surrogate marker for fecal protein calprotectin (ROC area under the curve (AUC) = 0.58). (B) For subjects with active disease, Classifier 2 further parsed those with mild disease (n = 15) relative to those with moderate disease (n = 22) when compared to the CDAI.
Predicting Therapeutic Response Using seRNA Biomarkers
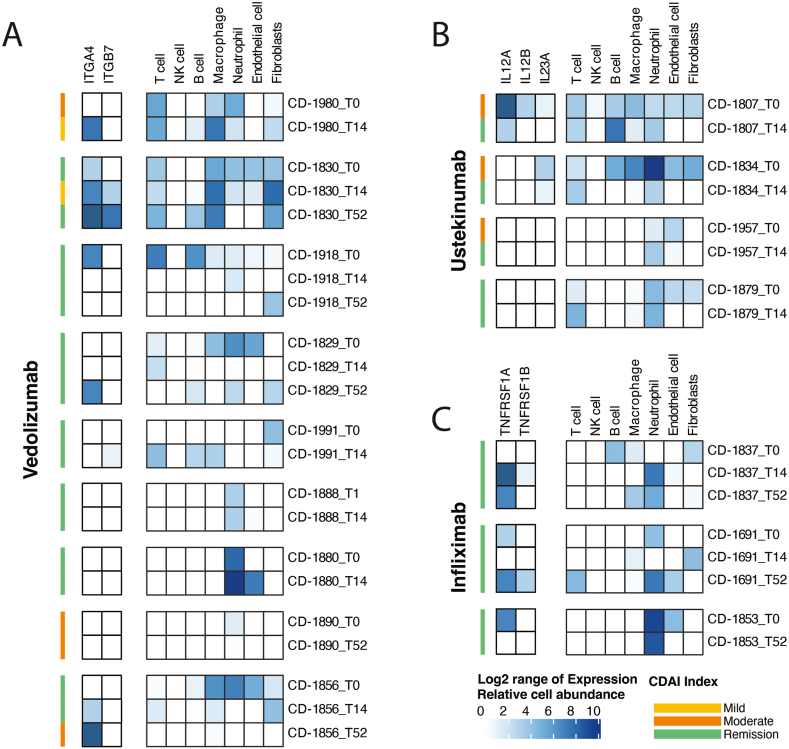
Across all subjects, 16 had a valid baseline stool sample with a valid subsequent time point (T14 or T52). Of these, 9 were being treated with vedolizumab (Figure 3A), 4 were being treated with ustekinumab (Figure 3B), and 3 were being treated with infliximab (Figure 3C). Of the 12 subjects being treated with vedolizumab and infliximab, only 2 had mild or moderate CDAI scores at baseline (10 had CDAI scores indicating remission). However, 75% of subjects being treated with ustekinumab had baseline disease activity (mild or moderate disease) with subsequent CDAI scores indicating remission after 14 weeks of treatment. For these subjects, 2 had high expression of the therapeutic target at T0 with depletion of the target transcript after treatment. Similarly, when looking at cell deconvolution of various resident lymphocytes over time, there was a high lymphocytic burden in the gastrointestinal tract at T0 with reduction in lymphocytic burden at T14 (Figure 3B).
Figure 3.
seRNA biomarkers demonstrated preliminary ability to predict response to targeted therapeutics. Each grouping represents a unique individual. Each stacked row represents a unique timepoints (eg, T0, T14, T52). For each timepoint, the Crohn’s disease activity index (CDAI) is provided. The first column of blue boxes indicates the expression of the transcript related to the therapy target, or its associated receptor. The second column of blue boxes indicates the abundance of various immune-related cell types for each timepoint. (A) Data for the 9 subjects on vedolizumab. (B) Data for the 4 subjects on ustekinumab. (C) Data for the 3 subjects on infliximab.
Discussion
This study provides preliminary evidence that RNA biomarkers in stool samples might be viable to predict therapeutic response prior to treatment and longitudinally dictate disease severity for patients with CD. Using stool samples to assess disease activity offers a range of substantial benefits. First, it provides a noninvasive and patient-friendly approach that does not require a physician appointment for sample collection. This mitigates the need for uncomfortable and sometimes risky invasive procedures, such as repeated endoscopies, and can improve patient compliance associated with disease monitoring. At-home stool sample collection also enable frequent and longitudinal assessments, allowing for more accurate tracking of disease progression. Furthermore, this method offers cost-effective and convenient disease monitoring, reducing health-care expenses and overall burden.
Stool-based RNA tests could address some of the limitations associated with fecal calprotectin, CRP, and other noninvasive approaches.10 Specifically, the output from the RNA test provides an ordinal value, which could be used to quantify disease activity, extent of disease, or potentially point to disease location. Research is ongoing to determine the bounds of RNA technology as it relates to advancing noninvasive diagnostics for patients with CD. Further, a future version of this test could be used in conjunction with other noninvasive RNA-based diagnostics that assess other disease states such as presence of dysplasia.21
With the introduction of new therapies that incldue novel mechanisms of action, the utilization of RNA biomarkers in stool samples to predict therapeutic response could represent an interesting approach to precision medicine. Although the data generated in this study was primarily intended to assess feasibility, and represents individual case studies that may not be generalizable, the results demonstrate a promising avenue for further research. By analyzing RNA signatures in stool, providers could gain valuable insights into the dynamic molecular changes occurring within the gut mucosa. This approach enables the identification of specific gene expression patterns associated with therapeutic response in the pretreatment stool sample, thereby allowing for the selection of a more tailored and effective treatment strategy. Such personalized medicine approaches have the potential to significantly improve patient outcomes by optimizing the selection of advanced therapies, minimizing the risk of adverse effects, and enhancing the overall management of CD. This innovative use of RNA biomarkers in stool samples may ultimately revolutionize the way we treat CD, paving the way for precision medicine in gastroenterology and leading to better outcomes and quality of life for patients.
Beyond querying the targeted transcript of interest, the evaluation of cell deconvolution using stool samples to assess changes in cell types for patients with CD represents a novel approach for noninvasive assessment. By using stool samples, this method alleviates the discomfort of invasive procedures, and provides a comprehensive and longitudinal view of changes in cell types, which previously required repeated endoscopic evaluations, mucosal biopsies, and additional complex context including cytometry by time-of-flight or single cell sequencing. This novel technique opens up new avenues for the study of CD, offering a deeper insight into the complex immune and epithelial interactions underlying the disease, potentially paving the way for more effective diagnostic and therapeutic strategies.
Several limitations need to be acknowledged in the present study. First, there was a lack of stool samples collected at all 3 time points for all subjects, which may have impacted the ability to fully capture the dynamics of gut mucosa over time. This was due to opportunistic collection of stool samples from willing participants at clinic visits managed by care coordinators and not part of a larger-scale clinical trial. Additionally, the absence of endoscopic assessments at all timepoints hindered the ability to correlate RNA signatures with disease severity, potentially limiting our understanding of the relationship between gene expression and disease progression. The use of a surrogate marker for clinical activity (ie, CDAI) has limitations; as such, results from this study are preliminary and demonstrate feasibility rather than clinical utility. Furthermore, the capture panel used in this study did not offer a comprehensive assessment of the transcriptome, and there is potential for further improvement by expanding its coverage to evaluate cellular pathways more comprehensively and enhance the accuracy of cell abundance deconvolution. These limitations should be taken into consideration when interpreting the findings of this research and may serve as opportunities for future studies.
Future studies should prioritize the validation and refinement of the stool-derived transcripts identified in this study, aiming to better understand the overall role in predicting therapeutic response and monitoring mucosal healing. Robust validation is crucial to establish the reliability and generalizability of these innovative approaches in clinical practice. Large-scale, multicenter trials involving diverse patient populations should be conducted to confirm the accuracy and reproducibility of RNA biomarker-based predictions of disease activity and therapeutic response in patients with CD. Additionally, exploring the longitudinal stability of these RNA biomarkers over extended periods of time will be essential to assess their suitability for monitoring disease progression. Comparative studies, analyzing the performance of RNA biomarkers alongside established diagnostic and monitoring methods, can help ascertain their added value in CD management. Furthermore, investigations into the potential for stool-based RNA biomarkers to predict disease flares and long-term outcomes are warranted. By conducting rigorous validation studies, we can solidify the foundation for the integration of RNA biomarkers in stool samples as a valuable and reliable tool in the clinical management of CD, offering patients more precise and personalized treatment strategies in the future.
Acknowledgments
We would like to thank the patients and their families for their generous donations to the advancement of sciences. This study would not have been possible without the dedicated efforts of Andrew Barnell (M.B.A., Geneoscopy), Aja Jacobs (B.S., Geneoscopy), Ben Wedeking (B.S., Geneoscopy), Brett Miller (B.S., Geneoscopy), Cath Morrison (B.S., Geneoscopy), Kaitlyn Maly (B.S., Geneoscopy), Kimberly Kruse (M.Sc., Geneoscopy), Maya Crowder (B.S., Geneoscopy), Jim Fitzgibbon (M.B.A., Geneoscopy). All individuals in the acknowledgement were compensated by Geneoscopy as part of their efforts in completing the study.
Authors' Contributions
Erica K. Barnell, Ryan B. Ghannam drafted the manuscript; Ryan B. Ghannam, Richard Roberts, Spencer King, Jack Land, Clayton Grass, Matthew A. Ciorba generated data for analysis; Erica K. Barnell provided analysis and interpretation of data; Erica K. Barnell, Ryan B. Ghannam, Richard Roberts, Spencer King, Jack Land, Clayton Grass, Matthew A. Ciorba, Patrick Donohue provided critical revision of the manuscript for important intellectual content; Erica K. Barnell, Matthew A. Ciorba, Patrick Donohue contributed to acquisition of data; Erica K. Barnell provided study supervision; Matthew A. Ciorba, Patrick Donohue advised on study development.
Footnotes
Conflicts of Interest: These authors disclose the following: Erica Barnell is an owner, employee, and member of Geneoscopy Inc. Erica Barnell is an inventor of the intellectual property owned by Geneoscopy Inc. Ryan B. Ghannam, Richard Roberts, Spencer King, Jack Land, Clayton Grass, are employees at Geneoscopy Inc. Parakkal Deepak has received research support under a sponsored research agreement unrelated to the data in the paper and/or served as a consultant with AbbVie, Pfizer, Boehringer Ingelheim, Bristol Myers Squibb, Janssen, Merck, Takeda Pharmaceuticals, CorEvitas, LLC, Alimentiv, Iterative Scopes, Roche Genentech, Sandoz, Landos Pharma, Teva Pharma, Agomab and Fresenius Kabi. The remaining authors disclose no conflicts.
Funding: Dr Deepak is supported by a Junior Faculty Development Award from the American College of Gastroenterology which funded the observational cohort in the study and is also supported by the IBD Plexus of the Crohn's & Colitis Foundation. The work in this manuscript was additionally supported by the Washington University Division of Gastroenterology Digestive Disease Research Center Core (NIDDK P30 DK052574). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Ethical Statement: IRB approval was obtained from the Washington University School of Medicine Institutional Review Board (IRB #20111107; Digestive Disease Reserch Core Center Biobank).
Data Transparency Statement: Data, analytic methods, and study materials can be made available to other researchers upon request of the corresponding author.
Reporting Guidelines: None.
Material associated with this article can be found in the online version at https://doi.org/10.1016/j.gastha.2024.07.012.
Supplementary Data
References
- 1.Lewis J.D., Parlett L.E., Jonsson Funk M.L., et al. Incidence, prevalence, and racial and ethnic distribution of inflammatory bowel disease in the United States. Gastroenterology. 2023;165:1197–1205.e2. doi: 10.1053/j.gastro.2023.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosnes J., Gower-Rousseau C., Seksik P., et al. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 3.Ashton J.J., Green Z., Kolimarala V., et al. Inflammatory bowel disease: long-term therapeutic challenges. Expert Rev Gastroenterol Hepatol. 2019;13:1049–1063. doi: 10.1080/17474124.2019.1685872. [DOI] [PubMed] [Google Scholar]
- 4.Gupta N., Bostrom A.G., Kirschner B.S., et al. Incidence of stricturing and penetrating complications of Crohn's disease diagnosed in pediatric patients. Inflamm Bowel Dis. 2010;16:638–644. doi: 10.1002/ibd.21099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubinsky M.C., Lin Y.C., Dutridge D., et al. Serum immune responses predict rapid disease progression among children with Crohn's disease: immune responses predict disease progression. Am J Gastroenterol. 2006;101:360–367. doi: 10.1111/j.1572-0241.2006.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiceland C.M., Lodhia N. Endoscopy in inflammatory bowel disease: role in diagnosis, management, and treatment. World J Gastroenterol. 2018;24:4014–4020. doi: 10.3748/wjg.v24.i35.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuna A.T. Serological markers of inflammatory bowel disease. Biochem Med. 2013;23:28–42. doi: 10.11613/BM.2013.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Schaik F.D.M., Oldenburg B., Hart A.R., et al. Serological markers predict inflammatory bowel disease years before the diagnosis. Gut. 2013;62:683–688. doi: 10.1136/gutjnl-2012-302717. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann F.S., Burri E., Beglinger C. The role and utility of faecal markers in inflammatory bowel disease. Therap Adv Gastroenterol. 2015;8:23–36. doi: 10.1177/1756283X14553384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ananthakrishnan A.N., Adler J., Chachu K.A., et al. AGA clinical practice guideline on the role of biomarkers for the management of Crohn's disease. Gastroenterology. 2023;165:1367–1399. doi: 10.1053/j.gastro.2023.09.029. [DOI] [PubMed] [Google Scholar]
- 11.Plevy S., Silverberg M.S., Lockton S., et al. Combined serological, genetic, and inflammatory markers differentiate non-IBD, Crohn's disease, and ulcerative colitis patients. Inflamm Bowel Dis. 2013;19:1139–1148. doi: 10.1097/MIB.0b013e318280b19e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Haens G., Kelly O., Battat R., et al. Development and validation of a test to monitor endoscopic activity in patients with Crohn's disease based on serum levels of proteins. Gastroenterology. 2020;158:515–526.e10. doi: 10.1053/j.gastro.2019.10.034. [DOI] [PubMed] [Google Scholar]
- 13.Chen P., Zhou G., Lin J., et al. Serum biomarkers for inflammatory bowel disease. Front Med (Lausanne) 2020;7:123. doi: 10.3389/fmed.2020.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iborra M., Beltrán B., Nos P. Noninvasive testing for mucosal inflammation in inflammatory bowel disease. Gastrointest Endosc Clin N Am. 2016;26:641–656. doi: 10.1016/j.giec.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Koga Y., Yasunaga M., Moriya Y., et al. Detection of colorectal cancer cells from feces using quantitative real-time RT-PCR for colorectal cancer diagnosis. Cancer Sci. 2008;99:1977–1983. doi: 10.1111/j.1349-7006.2008.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertucci F., Salas S., Eysteries S., et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 17.Mai V., Morris J.G., Jr. Colonic bacterial flora: changing understandings in the molecular age. J Nutr. 2004;134:459–464. doi: 10.1093/jn/134.2.459. [DOI] [PubMed] [Google Scholar]
- 18.Quigley E.M.M. Gut bacteria in health and disease. Gastroenterol Hepatol (N Y) 2013;9:560–569. [PMC free article] [PubMed] [Google Scholar]
- 19.Barnell E.K., Kang Y., Wurtzler E.M., et al. Noninvasive detection of high-risk adenomas using stool-derived eukaryotic RNA sequences as biomarkers. Gastroenterology. 2019;157:884–887.e3. doi: 10.1053/j.gastro.2019.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnell E.K., Kang Y., Barnell A.R., et al. Multitarget stool RNA test for noninvasive detection of colorectal neoplasia in a multicenter, prospective, and retrospective cohort. Clin Transl Gastroenterol. 2021;12 doi: 10.14309/ctg.0000000000000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnell E.K., Wurtzler E.M., La Rocca J., et al. Multitarget stool RNA test for colorectal cancer screening. JAMA. 2023;330:1760–1768. doi: 10.1001/jama.2023.22231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sodowich B.I., Fadl I., Burns C. Method validation of in vitro RNA transcript analysis on the agilent 2100 bioanalyzer. Electrophoresis. 2007;28:2368–2378. doi: 10.1002/elps.200600673. [DOI] [PubMed] [Google Scholar]
- 23.Boyle E.A., O'Roak B.J., Martin B.K., et al. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30:2670–2672. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massimino L., Lamparelli L.A., Houshyar Y., et al. The inflammatory bowel disease transcriptome and metatranscriptome meta-analysis (IBD TaMMA) framework. Nat Comput Sci. 2021;1:511–515. doi: 10.1038/s43588-021-00114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehm H.L., Berg J.S., Brooks L.D., et al. ClinGen — the clinical genome resource. N Engl J Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappaport N., Nativ N., Stelzer G., et al. MalaCards: an integrated compendium for diseases and their annotation. Database (Oxford) 2013;2013 doi: 10.1093/database/bat018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becht E., Giraldo N.A., Lacroix L., et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.