Abstract
Naturally occurring cell-derived extracellular vesicles (EVs) have emerged as attractive nanocarriers for drug delivery. However, production of large quantities of EVs for clinical applications in a scalable manner remains a significant challenge. This study investigated at the single cell level how sonoporation, or membrane poration produced by ultrasound-induced microbubble cavitation, impacts EV production using mouse macrophage RAW 264.7 cells stably expressing CD63-GFP as a model system. Real-time fluorescence videomicroscopy detected rapid changes in CD63-GFP, a tetraspanin family member highly enriched in intraluminal vesicles tagged with GFP, to track changes in multivesicular bodies (MVBs), which are the cellular compartments where exosomes originate within the cells. Our results revealed distinct dynamic changes in CD63-GFP intensity and distribution in RAW 264.7 cells in terms of response time and duration depending on whether the cells were directly or indirectly impacted by sonoporation, suggesting reorganization of MVBs in response to direct and indirect mechanisms resulted from the mechanical impact of ultrasound pulse on the cells. Analysis of the supernatant from sonoporation-treated RAW 264.7 cells expressing CD63-GFP demonstrated a delayed and sustained increase in the production of CD63-GFP-positive EVs. These results show the robust and detailed effect of sonoporation and reveal insights into sonoporation-induced EV release useful for guiding the application of sonoporation to enhance large-scale EV production.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79042-6.
Keywords: Extracellular vesicle, Exosome, Sonoporation, Ultrasound, Macrophage
Subject terms: Permeation and transport, Biological techniques, Biophysics, Cell biology
Introduction
Nanoparticle-based delivery platforms have garnered significant attention as a promising drug delivery strategy for treating various diseases1,2. However, their clinical efficacy is restricted due to dose-dependent toxicity, inefficient crossing through endothelial barriers, rapid clearance, and non-specific tissue accumulation3. In contrast, naturally occurring cell-derived extracellular vesicles (EVs) have the inherent ability to diffuse through tissues. They play vital roles in intercellular communications among non-adjacent cells by transporting various bioactive molecules, such as proteins, lipids, and nucleic acids, between cells and have emerged as attractive nanocarriers for drug delivery in EV-based therapeutic approaches4,5.
EVs encompass various subtypes distinguished by their size, biogenesis, cargo molecules, and density6,7. Two main categories of EVs have been broadly classified: small EVs (sEVs; <200 nm), which include exosomes, and large EVs (> 200 nm), such as plasma membrane-derived microvesicles8. Among these, exosomes (30–120 nm in diameter) are the most extensively studied subtype of sEVs and are known for their significant biological functionality4. Notably, exosomes undergo unique biogenesis: they form as intraluminal vesicles within multivesicular bodies (MVBs) in late endosomes, which then fuse with the plasma membrane to release exosomes into the extracellular spaces. This endosomal origin allows exosomes to encapsulate and protect therapeutic molecules, such as RNA, proteins, and even DNA, from degradation. Their small size facilitates efficient tissue penetration and uptake by recipient cells. Their biocompatibility, low immunogenicity, and inherent ability to target specific tissues and cells further enhance their potential as drug delivery vehicles for the treatment of complex diseases like cancer and neurodegenerative disorders9.
While the therapeutic macromolecules (e.g., DNA, mRNA, and miRNA) can be effectively protected and delivered to target cells and tissues10–12, many challenges still remain, including producing large quantities of EVs for clinical applications in a scalable manner. Overcoming these hurdles is crucial for realizing the full potential of EVs in therapeutic applications could profoundly impact the advancement of EV-based drug delivery approaches.
Several strategies have been pursued to increase therapeutic EV production, including multi-layer culture flasks, fixed-bed bioreactors, stirred tank bioreactors, or continuous production in perfusion reactors. These approaches focus on maximizing culture surface area compared to conventional planar cell culture13, but the yields often fall short of clinical needs. Scaffolds, spheroid cultures14, or microcarrier-based 3D cultures15 have also been explored. Despite advances in bioreactor designs and 3D culture systems, scaling up therapeutic EV production remains a major challenge. Mechanical stretching of cells in 2D cultures has been exploited to increase EV generation16, but implementation in a 3D environment poses significant challenges.
Ultrasound technologies have a long history in medicine, and recent developments include a diverse range of therapeutic applications based on their capability to induce desired cellular and physiological effects17, e.g., ultrasound neurostimulation by activating ion channels18–20 and intracellular macromolecule delivery via sonoporation21–23. Sonoporation is the formation of pores on cell membranes by ultrasound, often facilitated by attaching gaseous microbubbles to the cell membrane24–26. Sonoporation has been used to increase intracellular uptake of macromolecules for non-viral drug and gene delivery22,23. Mechanistic studies have revealed a number of downstream effects of sonoporation, including cytoskeletal reorganization27,28 and calcium influx in the sonoporated cells along with intercellular calcium waves24,29–33 Interestingly, ultrasound exposure has been shown to promote EV release from cells, and it is hypothesized that ultrasound-induced Ca2+ influx from the extracellular space34,35 may trigger Ca2+-dependent activation of ESCRT pathways and cytoskeletal remodeling, processes known to orchestrate EV production34,36,37. However, the biophysical mechanisms underlying ultrasound-enhanced EV production remain incompletely understood. The detailed process how sonoporation impacts the cells to enhance EV release at the single cell level has not been examined.
While previous mechanistic studies of sonoporation focused on pore formation and resealing, downstream effects such as cytoskeletal changes, apoptosis, and calcium signaling waves32,33, the cellular responses that impact EV generation by ultrasound have not been examined at the single-cell level. A recent study has demonstrated that MVB fusion with the plasma membrane and subsequent exosome release is triggered by Ca2+ influx during plasma membrane wounding38. This process is dependent on annexin A6, a well-known plasma membrane repair protein39. Such insights are important to guide the successful development of ultrasound-based EV production technology. Thus, in this study, we aim to elucidate the dynamic cellular responses to ultrasound-mediated membrane poration in the context of EV production. We utilized mouse macrophage RAW 264.7 cells as a model system, given the significant role of EVs in immune cell communication, their potential as therapeutic targets, and their promising applications in immunotherapy40,41. Single-cell sonoporation experiments examined the dynamic changes of CD63 in RAW 264.7 cells at the single-cell level. Here, CD63, a tetraspanin family member highly enriched in intraluminal vesicles within MVBs and a typical exosome marker, was tagged with GFP to enable real-time fluorescence videomicroscopy42 to assess exosome production in response to sonoporation. Ensemble cell sonoporation experiments examined whether sonoporoation could increase exosome production via bulk analysis of EV production from sonoporated cells in post-sonoporation supernatant collected at various time points. Together, the analysis of CD63-GFP changes at the single-cell level and bulk characterization of CD63-GFP-positive EVs in the conditioned media provide a complementary assessment of the temporal dynamics of exosome production from RAW 264.7 cells following sonoporation.
Materials and methods
Cell culture
Mouse macrophage RAW 264.7 cells stably expressing CD63-GFP were produced using Sleeping Beauty transposons43,44. The CD63-GFP gene was sub-cloned under the CAG promoter in the pKT2/CAGXSP vector through recombination cloning (In-Fusion HD Cloning Kit, Clontech), as described before44. The cells were cultured in Dulbecco’s-modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin streptomycin. For sonoporation experiments, RAW 264.7 cells were seeded as single cells on glass bottom dishes at a density of 8,000 cells/cm2. Cells were maintained at 37℃ in 100% humidified air containing 5% CO2, and the medium was changed every other day. Both single-cell and ensemble sonoporation experiments were conducted two days after cell seeding (day 2).
Attachment of microbubbles to cells for sonoporation
To attach microbubbles to cells for sonoporation, pre-formed microbubbles were functionalized with Arg-Gly-Asp (RGD) peptides to allow their attachment to cells via RGD-integrin binding. Here, biotinylated SIMB4-5 microbubbles (Advanced Microbubbles Laboratories LLC) were first incubated at room temperature with streptavidin (Fisher Scientific) at a volume ratio of 20:1 for one hour. After washing with PBS to remove excess streptavidin, the microbubbles were then conjugated to Arg-Gly-Asp-biotin (RGD-biotin) (Biosynth, 2 mg/mL) at a volume ratio of 10:1 for one hour. The microbubbles were then washed with PBS to remove excess biotin-RGD. RGD-microbubbles were diluted 10 times using a culture medium before use. Microbubble size distribution was measured using image analysis after bubble attachment and compared with the size distribution provided by the manufacturer.
For the single-cell sonoporation experiments, to attach the RGD-microbubbles to the cells, the culture medium in the cell culture dish was removed first, followed by an addition of 40 µL of the RGD-microbubble solution. For the supernatant experiments, 100 µL of the RGD-microbubble solution was added to each well immediately following the removal of the culture medium. The cell culture dish was then inverted, allowing the microbubbles to float upwards for 10 min at 37 °C to allow the microbubbles to be attached to the cells via RGD-integrin binding. The dish was flipped back, and gentle washing using PBS was performed to remove unbound microbubbles. The dish, including the cells with attached microbubbles, was then placed on a microscope stage, ready for single-cell sonoporation experiments. For the ensemble sonoporation experiments, RAW 264.7 cells were seeded in 6-well plates (Thermo Fisher Scientific) at a density of 20,000 cells/cm2 with the cell culture medium replenished daily. Sonoporation experiments were conducted two days after cell seeding. Supernatant was collected and analyzed at 0 h, 2 h, 4 h, 24 h, and 48 h.
Single-cell sonoporation experiments
The ultrasound system for single-cell sonoporation used in this study was described previously24,45. Briefly, the setup included a single element planar ultrasound transducer (Advanced Devices, Wakefield, MA, USA) with a center frequency of 1.25 MHz, 6-dB beam width of 3.54 mm, and Rayleigh distance of 9 mm. The transducer was positioned at 45° relative to the horizontal direction to minimize the artifacts of standing waves and for microscopic imaging of the RAW 264.7 cells on the bottom of a standard cell culture dish. The transducer was driven by a waveform generator (Agilent Technologies 33250 A) and a 75 W power amplifier (Amplifier Research 75A250) and was calibrated using a fiber optic hydrophone (Onda HFO 690) by measuring the acoustic pressure field in de-gassed, room temperature water in a free field condition. Prior to sonoporation experiment, an alignment procedure was performed using a small mental wire in an identical dish filled with medium but no cells. The metal wire served as a target to align the acoustic and optical field using a pulse/receiver (Panametrics) to maximize the acoustic field within the optical field of view. For experiments, the active surface of the transducer was submerged in the medium, pointing at the cells on the bottom of the dish to induce sonoporation. A single pulse containing 10 cycles (duration ~ 8 µs), with peak pressure of 0.26 MPa, was applied to generate sonoporation.
Fluorescence videomicroscopy
Imaging of RAW 264.7 cells was performed to visualize the cellular changes due to sonoporation using a fluorescence imaging system as described in our previous work24,45 that included a fluorescence illuminator (X-Cite Series 120PCQ) with a 120 W lamp. The excitation light was directed through a 20x Plan Fluor or 40x Plan Fluor objectives (Nikon) to the cells. The glass bottom dish with RAW 246.7 cells was placed on the stage of an inverted microscope (Nikon Eclipse Ti-U). Videos of the cells were recorded using a Photometrics QuantEM: 512SC camera.
To verify sonoporation of the cells, propidium iodide (PI) (Millipore Sigma) was used as an indicator of intracellular transport through the pores on the membrane due to sonoporation. PI became fluorescent (excitation wavelength at 538 nm and emission at 610 nm) when bound to the nucleic acids inside the cells. PI solution (50 µg/mL) was added to the medium of the RAW 264.7 cells before ultrasound application. Real-time fluorescence microscopy was then used to detect the increased intracellular fluorescence signals of PI entering the cells resulting from sonoporation.
For detecting the CD63-GFP signal changes in the cells subjected to sonoporation, real-time fluorescence imaging was performed with excitation and emission wavelengths of 488 nm and 509 nm, respectively. The exposure duration was set at 400 ms. Videos of the cells were recorded for various time durations ranging from 5 to 20 min, starting at 10 s before ultrasound applications. NIS-Elements Advanced Research (Nikon) and Image J (The National Institutes of Health, Bethesda, MD, USA) were used to acquire and analyze the images/videos. A custom Matlab script was developed to post-process the recorded videos to quantify changes in the RAW 264.7 cells due to sonoporation as a function of time.
Ensemble cell sonoporation experiments and collection of supernatant
Ensemble sonoporation experiments were designed to assess EV production from RAW 264.7 cells over a longer duration. For these experiments, ultrasound application was performed using a single-element planar transducer (Advanced Devices, Wakefield, MA, USA) with a center frequency of 1 MHz submerged in water for acoustic coupling. The transducer was calibrated using a fiber optic hydrophone (Onda HFO 690) by measuring the acoustic pressure field of the transducers in de-gassed, room-temperature water in a free field condition. The transducer was driven by a waveform generator (Agilent Technologies 33250 A) and a 75 W power amplifier (Amplifier Research 75A250) to generate an ultrasound pulse with an acoustic pressure of 0.3 MPa and 8 µs duration. In these experiments, cells were plated in a 6-well plate with only its bottom submerged in water for acoustic coupling and to ensure a sterile condition for the cells. The application of ultrasound pulse was from below and through the bottom of a 6-well plate to generate sonoporation of the cells. The transducer was fixed at an angle of 45° with respect to the vertical direction to avoid the artifacts of standing waves. A similar alignment procedure as the single-cell sonoporation was performed to place the ultrasound field on cells on dish bottom.
EV-depleted FBS was prepared by 18-h ultracentrifugation at 100,000×g, 4 °C46. EV-depleted cell culture medium was first created using DMEM supplemented with 10% EV-depleted FBS and 1% penicillin-streptomycin. Prior to the ultrasound application, each well was washed with 1 mL of PBS and replaced with 2.5 mL of the EV-depleted cell culture medium. The conditioned media of RAW 264.7 cells stably expressing CD63-GFP were collected immediately after sonoporation (0 h), followed by gentle washing with PBS, and replaced with 2.5 mL of EV-depleted cell culture medium in each well. The collected conditioned media were stored at -20 °C. The 6-well plate was placed in the incubator at 37 °C after supernatant collection. The supernatant was collected at 0 h, 2 h, 4 h, 24 h, and 48 h post-sonoporation using the same procedure. The same procedure was applied to a control group without ultrasound application.
Isolation, purification, and analysis of small EVs
To isolate the GFP-labeled EVs produced by RAW 264.7 cells stably expressing CD63-GFP from the collected supernatant, the supernatant was centrifuged at 600×g for 5 min to remove cells and debris, followed by a 0.22 μm filtration using PES membrane filters (Nalgene, 725–2520) to collect the small EVs (sEVs ∼ ≤ 200 μm). The sEVs were enriched using 50-nm porous membranes (Whatman, WHA110603) with holders (EMD Millipore, SX0002500) by vacuum pressure44. Next, the remaining sEV-enriched fraction in the supernatant was trapped on the membranes, followed by washing with 5 mL PBS. When 500 µL of the sample remained, the concentrated sEVs were carefully collected. To ensure sEV stability, a freezing solution (10%) containing 250 mM trehalose, 250 mM HEPES, and 2% bovine serum albumin (BSA) was used to protect them from freezing conditions47. sEVs were aliquoted into 200 µl portions and stored at -80 °C until analysis.
Nanoparticle tracking analysis
The size distribution and concentration of sEVs were determined using nanoparticle tracking analysis (NTA) (ZetaView, Particle Metrix). Prior to analysis, sEV samples were diluted in PBS to achieve an optimal particle concentration (1 × 109 – 1 × 1010 particles/mL). Automated measurements were taken at 11 distinct positions in the sample cell. The instrument’s outlier control feature selected high-quality videos for analysis. Particle size distribution (nm) and concentration (particles/mL) were calculated based on the principles of Brownian motion and light scattering.
Protein quantification using micro BCA assay
Protein concentration was determined using the micro bicinchoninic acid (BCA) assay (23235, Thermo Fisher Scientific), following the manufacturer’s protocol. Briefly, sEV samples (1 µL) were diluted in 99 µL PBS and incubated with the BCA reagent working solution at 37 °C for 2 h. Absorbance was measured at 562 nm using a microplate reader (Spark, Tecan). A standard curve was prepared using bovine serum albumin (BSA) standards ranging from 0.5 to 40 µg/mL. Protein concentrations were calculated by linear regression analysis of the standard curve.
Western blotting
Western blotting was performed to detect the presence of the sEV marker CD63. The isolated sEVs were lysed with 4x sample buffer (Bio-Rad) under non-reducing conditions (without β-mercaptoethanol). Equal amounts of protein (30 µg) were loaded onto a 4–20% Mini-PROTEIN TGX stain-free gel (Bio-Rad) and separated by electrophoresis. The proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane (IPFL00010, Millipore) using a semi-dry transfer system (Trans-Blot Turbo, Bio-Rad). After blocking with 5% non-fat milk in PBS with 0.05% Tween-20 (PBST) for 30 min at room temperature, the membrane was incubated overnight at 4 °C with anti-CD63 primary antibody (10628D, Ts63, Thermo Fisher, 1:1,000) diluted 1:1000 in PBST. Following incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (diluted 1:10,000) for 1 h at room temperature, the bands were visualized using an enhanced chemiluminescence (ECL) detection system (RPN2235, GE Healthcare) and imaged using a chemiluminescence imager (ChemiDoc MP Imaging System, Bio-Rad).
Bead-based flow cytometric analysis of GFP+ sEVs
The presence of GFP-positive sEVs (GFP+ sEVs) was analyzed using a bead-based flow cytometry assay, as previously described48. sEV samples (10 µg EV protein) were incubated with 5 µL of 4 μm aldehyde-activated latex beads (A37304, Invitrogen) in 500 µL HEPES (25 mM)/PBS overnight at 4 °C with. The beads were washed with HEPES/PBS at 5,000 × g for 5 min. The beads were resuspended in 100 µL of HEPES/PBS buffer. Finally, the samples were diluted with 400 µL of 2% paraformaldehyde (PFA) and incubated for 15 min at room temperature before flow cytometry.
Flow cytometric analysis was performed using a Cytek Aurora flow cytometer (Cytek), and data were analyzed with FlowJo software. Beads without sEVs and beads incubated with GFP-negative sEVs served as negative controls for setting gating thresholds. The percentage of GFP+ sEVs was calculated based on the population of bead-bound events displaying GFP fluorescence.
Single EV fluorescence microscopy analysis
To quantify GFP+ sEVs, a drop of the isolated sEVs was placed on hydrophobic PTFE printed slides (Electron Microscopy Sciences, 63429-04)49, as previously described. After 30 min of incubation at 4 °C, slides were washed twice with PBS and imaged using a Leica Thunder wide field fluorescence microscope (Leica Microsystems). Image analysis was conducted using ImageJ/Fiji software with the EVAnalyzer plugin50. The ‘EVCount’ function was used to quantify the number of GFP+ sEVs. Statistical analysis was performed on the data produced by EVAnalyzer, removing outliers using the automated Triangle option as the threshold setting.
Transmission electron microscopy
sEVs were visualized using transmission electron microscopy (TEM) to assess their morphology and size. Isolated sEV samples were fixed in 1% paraformaldehyde for 30 min at room temperature. A small volume (50 µL) of the sEV suspension was placed onto a Formvar/carbon-coated gold grid and allowed to adsorb for 20 min. Excess liquid was gently removed using filter paper. The grids were then stained with 1% uranyl acetate for contrast enhancement for 10 min, followed by drying at room temperature. Samples were observed using a JEOL 1400 Flash Transmission Electron Microscope equipped with an integrated Matataki Flash CMOS bottom-mounted camera. The 1400 Flash was operated at 100 kV.
Statistical analysis
All experiments were repeated for at least four independent batches. Unpaired two-tailed Student’s t-test was performed to compare results from an experiment and a control group. Differences between two groups are considered statistically significant if p < 0.05.
Results
Controlled sonoporation of single RAW 264.7 cells
In microbubble-augmented sonoporation, a short ultrasound pulse is typically applied to induce cavitation of cell-bound microbubbles, generating a concentrated mechanical impact that induces pores on the cell membrane. Due to the small sizes and short lifetime of the pores24,25, direct observation of their formation is challenging in practice, even with an ultrahigh-speed camera or other techniques such as patch clamping23–25.
In this study, functionalized microbubbles (Advanced Microbubbles Laboratories LLC) were attached to of RAW 264.7 cells via RGD-integrin binding. The bubbles exhibited a size distribution with a peak diameter of 4.0 μm after attachment with the cells (Fig. S1a), slightly smaller than the diameter of the free bubbles before attachment at 4.7 μm (Fig. S1b). To avoid multibubble interactions during ultrasound application, the number density of microbubble was chosen to ensure a bubble to cell ratio of 1:2 in our experiment.
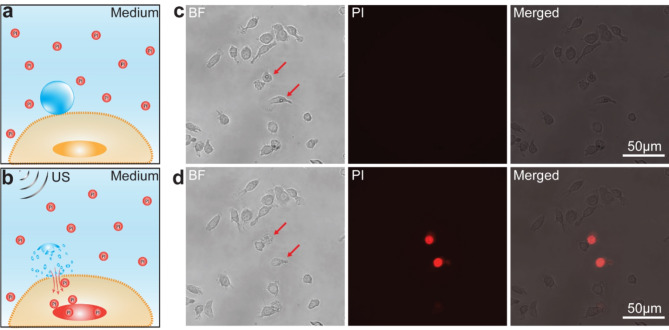
Sonoporation of RAW 264.7 cells was verified using propidium iodide (PI) as an indicator of intracellular transport through the pores on the membrane, as PI only becomes fluorescent when bound to the nucleic acids inside the cells (Fig. 1a,b). As shown in Fig. 1c,d and Supplemental video 1, application of an ultrasound pulse (duration 8 µs, 0.34 MPa) successfully generated sonoporation of single RAW 264.7 cells targeted by microbubbles, resulting in intracellular transport and uptake of PI into the cells. Other cells without attached microbubbles did not show PI uptake even though the ultrasound field was broadly applied to all cells in the field of view. Thus, sonoporation of individual RAW 264.7 cells relied on cavitation of cell-anchored microbubbles, which generated localized membrane poration.
Fig. 1.
Sonoporation of single RAW 264.7 cells. (a) Schematics of a cell-anchored microbubble (blue) in the presence of PI molecules (red circles) in the medium. (b) Schematics of microbubble-mediated sonoporation of a cell generated by an ultrasound pulse, resulting in intracellular transport of PI. (c) An example of RAW 264.7 cells before sonoporation. Left: a bright field (BF) image showing two cells (red arrows) with attached microbubbles; Middle: corresponding PI image of the cells; Right: overlaid BF and PI fluorescence images. (d) The RAW 264.7 cells in (c) after sonoporation. Left: a BF image of cells; Middle: PI image showing increased fluorescence signal in two sonoporated cells; Right: overlaid PI and BF images.
Sonoporation generated dynamic changes in the endosome network within RAW 264.7 cells
In the single-cell sonoporation experiments, real-time fluorescence videomicroscopy detected dynamic changes in the CD63-GFP signals in RAW 264.7 cells subjected to an ultrasound pulse (Supplemental video 2). Our results show that sonoporation of RAW 264.7 cells depended on the presence of microbubbles (Fig. S2), and the dynamic changes in subcellular CD63-GFP signals of the RAW 264.7 cells varied at the single-cell level. The example in Fig. 2 shows that sonoporation of a single RAW 264.7 cell (Fig. 2a) resulted in rapid radial expansion of CD63-GFP signals into the immediate vicinity of a sonoporated cell (Fig. 2b), suggesting a dynamic cellular response involving the reorganization of MVBs, where CD63 proteins are concentrated, due to sonoporation of the cell as indicated by PI uptake (Fig. 2c). The increase of CD63-GFP fluorescence signal intensity beyond the original cell area, defined as the region of interest (ROI) in this case, occurred immediately after ultrasound application and stabilized about 15 s afterward (Fig. 2d), suggesting rapid and sustained effects of sonoporation on the RAW 264.7 cell.
Fig. 2.
Sonoporation-induced dynamic changes in RAW 264.7 cells. (a) A bright field (BF) image (left) and a PI image overlaid on the BF image before sonoporation. (b) Fluorescence images of CD63-GFP of two RAW 264.7 cells at 10s, 15s, 25s, 30s, and 40 s. Sonoporation was generated by applying an ultrasound pulse at 10 + s. White lines in each panel represent the outline of the original cell area before sonoporation and red square is the selected region. The region of interest (ROI) is defined as the region within the red square and outside the white line in each panel. (c) BF and PI images of the cells in (a) 20 min after sonoporation. (d) The CD63-GFP fluorescence intensity within the ROI vs. time. “US” indicates the time point of the ultrasound application. The vertical red dash line indicates the time point when the CD63-GFP signal within the ROI increased by 5% of the background level.
Effects of sonoporation of RAW 264.7 cells on adjacent cells
In addition to the changes in sonoporated RAW 264.7 cells, our results show that sonoporation of RAW 264.7 cells also affected adjacent cells (Fig. S2, supplemental video 2). In contrast to the outward expansion of CD63-GFP signals in all directions observed in a sonoporated cell (Fig. 2), the example in Fig. 3 shows that cell A (Fig. 3a) adjacent to cell B (Fig. 3a) appeared to form a localized protrusion (yellow arrow, Fig. 3b) as indicated by the increased CD63-GFP fluorescence signal. PI staining confirmed that non-sonoporated cell A was adjacent to a sonoporated cell B (Fig. 3c). The CD63-GFP fluorescence signal in the ROI started to increase at 60 s–1 min, much slower than sonoporated cells shown in Fig. 2d, and recovered at 210 s–3.5 min after ultrasound application (Fig. 3d).
Fig. 3.
Cellular responses to sonoporation of an adjacent cell. (a) A bright field (BF) (left) and overly of BF and PI image (right) of two RAW 264.7 cells, cell A and cell B, before sonoporation. (b) Fluorescence images of CD63-GFP of the RAW 264.7 cells in (a) at different time points after application of an ultrasound pulse at 10 s. White lines outline the original area of cell A before ultrasound application at 10 s. Red line in each panel encloses the selected region of interest (ROI). The yellow arrow in each panel indicates a protrusion with increased CD63-GFP signals from cell A after ultrasound application. (c) BF and PI images of the two cells in (a) 20 min after ultrasound application. The PI signal (red) indicate a non-sonoporated cell (cell A) adjacent to a sonoporated cell (cell B). (d) The CD63-GFP fluorescence signal intensity in the ROI in (b) vs. time. “US” indicates the time point when an ultrasound pulse was applied at 10 s. The vertical red dash line indicates the time point when the CD63-GFP signal within the ROI increased by 5% of the background level. The shaded area indicates the time duration with elevated CD63-GFP signal intensity in the ROI.
These differences in the changes between a sonoporated cell and non-sonoporated cell that was adjacent to a sonoporated cells suggest that the indirect impacts of sonoporation on RAW 264.7 cells differ from the direct effects of sonoporation. The rapid time scale of the reorganization of MVBs in the sonoporated cells indicate the direct mechanical impact of microbubble cavitation/collapse (Fig. 2). In contrast, in the RAW 264.7 cells indirectly affected by sonoporation of adjacent cells, the cellular responses were much slower and more transient, likely facilitated by the close-range cell-cell interaction via chemical signals, including Ca2+, diffusing across gap junction between the attached cells24.
Effects of sonoporation of RAW 264.7 cells on nearby cells without direct contact
Further examination of the results from single-cell sonoporation experiments revealed that cells nearby but not in direct contact with a sonoporated cell exhibited different changes (Supplemental video 3, Fig. S3) from sonoporated cell (Fig. 2) and cell adjacent to a sonoporated cell (Fig. 3). As illustrated by the example in Fig. 4, in a non-sonoporated cell (cell A, Fig. 4a) near a sonoporated cell (cell B, Fig. 4a), a protrusion, indicated by increased CD63-GFP signal, was formed (arrows, Fig. 4b), and the CD63-GFP signal intensity within the ROI started to increase around 6 min and recovered around 10 min (Fig. 4d), much slower and lasting longer than that in the cell adjacent to a sonoporated cell (Fig. 3d). Without direct contact with the sonoporated cells, the responses in the nearby cells were likely due to longer-range cell-cell interaction via paracrine signaling rather than diffusion through gap junction between cells in contact with each other (Fig. 3), resulting in reorganization of MVBs in RAW 264.7 cells due to indirect impact of sonoporation.
Fig. 4.
Impact on a RAW 264.7 cell by sonoporation of a cell without direct contact. (a) A bright field (BF) and overlay of PI and BF images of two cells, cell A and cell B, before ultrasound application. (b) CD63-GFP fluorescence images of the two RAW 264.7 cells in (a) at different time points after ultrasound application at 10 s. White solid lines outline the original area of cell A, while red lines define the region of interest (ROI). (c) BF and overlay of PI and BF images of the two cells in (a) after ultrasound application, indicating a non-sonoporated cell (cell A) and sonoporated cell (cell B). (d) The CD63-GFP signal intensity within the ROI in (b). ‘US’ indicates the time point when the ultrasound pulse was applied at 10+ s. The vertical red dash line indicates the response time point when the CD63-GFP signal in the protrusion increased by 5% of the background level. The shaded area indicates the time duration with elevated CD63-GFP signal intensity within the ROI.
Different characteristics of cellular responses to sonoporation
A systematic analysis was performed to quantify the three types of changes in the CD63-GFP signal in RAW 264.7 cells in response to sonoporation, as illustrated in Figs. 2, 3 and 4. The relative changes of the maximum CD63-GFP signal intensity in the respective ROI in each case, against the CD63-GFP signal intensity in the original cell area before sonoporation (Fig. 5a) show that sonoporated RAW 264.7 cells (Sono) exhibited the largest changes, 27.3 ± 7.3% (n = 25), compared to the cells adjacent to a sonoporated cell (Adjacent), 14.0 ± 10.1% (n = 16), and cells near but not in direct contact with a sonoporated cell (Nearby), 8.3 ± 4.3% (n = 16). These results indicate that the direct mechanical impact of microbubble cavitation driven by an ultrasound pulse not only generated sonoporation of RAW 264.7 cells but also induced robust reorganization of MVBs in the cells that were more than the chemical factor-based effects of sonoporation of adjacent or nearby cells.
Fig. 5.
Quantification of the changes in the CD63-GFP intensity of RAW 264.7 cells induced by sonoporation. (a) Percentage change of the maximum CD63-GFP intensity in the region of interest (ROI) defined in Figs. 2, 3 and 4 for sonoporated RAW 264.7 cells (Sono), cells adjacent, and cells near but not in contact with a sonoporated cell (Nearby), respectively. (b) Response time of the CD63-GFP signal in the each of three ROI after ultrasound for the three groups of cells in (a). (c) Duration of elevated CD63-GFP signal in the ROI for cells adjacent and near sonoporated cells in (a), respectively. d. Increase of the CD63-GFP signal in the protruded region for the two groups of cells in (c). * denotes p < 0.05, ** p < 0.01, *** p < 0.001, and n.s. means not significant.
The response time of these changes in three groups of cells after ultrasound application was 0.1 ± 0.04 min (n = 25) for sonoporated RAW 264.7 cells, much faster than the response time due to indirect impact of sonoporation, i.e., 2.1 ± 1.2 min (n = 16) and 4.3 ± 2.6 min (n = 16) for the Adjacent and Nearby cells, respectively (Fig. 5b). The Adjacent cells responded significantly faster than the Nearby cells (Fig. 5b), although were both influenced by the indirect impact of sonoporation. The duration of elevated CD63-GFP signal intensity within the respective ROI in the cells indirectly affected by sonoporation was 4.2 ± 1.6 min (n = 17) in the Adjacent RAW 264.7 cells, slightly shorter but the difference was not statistically different when compared with the duration of 4.7 ± 2.2 min (n = 16) in the Nearby RAW 264.7 cells (Fig. 5c).
Lastly, the maximum CD63-GFP intensity in the protruded region of the cells indirectly impacted by sonoporation of the Adjacent cells was 5.1 ± 2.5 a.u. (n = 17), higher than the change of 3.4 ± 1.9 min (n = 16) in Nearby cells (Fig. 5d), suggesting that the total mass of reorganized MVBs may be higher in Adjacent cells than the Nearby cells, possibly due to the closer range of cell-cell interaction by the chemical factors from sonoporated cells.
Taken together, these results suggest that sonoporation may elicit reorganization of MVBs in RAW 264.7 cells through both mechanical and chemical factor-based mechanisms. These results also suggest that the direct, mechanical impact of sonoporation induced much faster reorganization of MVBs in RAW 264.7 cells than the indirect, chemical signals from sonoporation of Adjacent or Nearby cells. These different dynamic responses of RAW 264.7 cells suggest different mechanisms involved in MVB reorganization by sonoporation at the single-cell level.
EV production from RAW 264.7 cells subjected to sonoporation
A set of ensemble sonoporation experiments was performed to examine the production of small extracellular vesicles (sEVs) from RAW 264.7 cells. In these experiments, RAW 264.7 cells stably expressing CD63-GFP were subjected to sonoporation and continuously cultured. The supernatant of the RAW 264.7 cells was collected after 2 h for the isolation and characterization of sEVs, with non-sonoporated cells serving as controls. The sEVs derived from the sonoporated cells exhibited a peak size of 92 nm in diameter, a concentration of 1.4 × 10¹⁰ particles/mL, and a total protein concentration of 3,398 µg/mL (Fig. S4a). In contrast, sEVs derived from the non-sonoporated cells showed a slightly larger peak size of 102.2 nm, with a higher concentration of 2.4 × 1010 particles/mL and a similar protein concentration of 3,369 µg/mL (Fig. S4a). Both sonoporated and control sEVs expressed CD63, a characteristic marker protein for sEVs (Fig. S4b). Transmission Electron Microscopy (TEM) confirmed the typical morphology of the isolated sEVs, revealing spherical or cup-shaped vesicles with diameters ranging from approximately 70–150 nm (Fig. 6a). Further, GFP-positive sEVs (GFP⁺-sEVs) were quantitatively analyzed using bead-based bulk EV assays, indicating a two-fold increase in GFP⁺-sEV-coated beads in sonoporated samples (0.29%) compared to controls (0.14%) (Fig. S4c). Next, the temporal pattern of sEV release was investigated. Supernatants were collected at various time points, and the medium was replenished after each collection. The total particle count in purified sEV samples at different time points was quantified using nanoparticle tracking analysis (NTA), which showed no significant differences between sonoporated and control cells (Fig. S4d). Subsequently, the temporal pattern of GFP⁺-sEV release was specifically analyzed by single EV fluorescence microscopy, as previously described49,50. Our results (Fig. 6b and c) show that the number of GFP+-sEVs released from the cells immediately after sonoporation (0 h) did not change significantly compared to the control cells without sonoporation.
Fig. 6.
Morphology of sEVs and number of GFP-positive sEVs in the supernatant of RAW 264.7 cells expressing CD63-GFP at different time points after sonoporation. (a) TEM images of control and sonoporated sEVs (yellow arrows). (b) Representative fluorescence images of individual GFP+-sEVs isolated at different time points following sonoporation treatments. The purified sEVs were immobilized on a hydrophobic surface and visualized by fluorescence microscopy. (c) The horizontal axis includes the time point of the supernatant collection and the duration of the culture after the last supernatant collection. 0 h ( 0 h): immediately after sonoporation; 2 h (∆2 h): supernatant collected 2 h after sonoporation and cells were in culture for 2 h; 4 h (∆2 h): supernatant collected 4 h after sonoporation, but cells were in culture for 2 h after the last supernatant collection at 2 h; 24 h (∆20 h): supernatant collected 24 h after sonoporation and cells in culture for 20 h after the last supernatant collection at 4 h; 48 h (∆24 h): supernatant collected 48 h after sonoporation and cells in culture for 24 h after the last supernatant collection at 24 h. n = 12 for all groups, ** denotes p < 0.01; n.s. means not significant.
0 h): immediately after sonoporation; 2 h (∆2 h): supernatant collected 2 h after sonoporation and cells were in culture for 2 h; 4 h (∆2 h): supernatant collected 4 h after sonoporation, but cells were in culture for 2 h after the last supernatant collection at 2 h; 24 h (∆20 h): supernatant collected 24 h after sonoporation and cells in culture for 20 h after the last supernatant collection at 4 h; 48 h (∆24 h): supernatant collected 48 h after sonoporation and cells in culture for 24 h after the last supernatant collection at 24 h. n = 12 for all groups, ** denotes p < 0.01; n.s. means not significant.
On the other hand, a significant increase in GFP+-sEVs was detected in the supernatant 2 h after sonoporation compared to control cells without sonoporation, demonstrating the effect of sonoporation on EV production in RAW 264.7 cells. EV production measured after an additional 2–20 h of culture of these cells was similar to that from control cells. However, after another 24 h of culture, RAW 264.7 cells subjected to sonoporation produced a significantly higher number of EVs than control cells, suggesting a delayed and sustained effect of sonoporation on EV production activity. It is noted that EV production levels at 24 h and 48 h were much higher than those at 4 h for both sonoporation and control groups. This increase can be attributed to the longer culture duration (20 h vs. 2 h) prior to the final supernatant collection and analysis. Control cells showed similar EV production levels at 24 and 48 h post-sonoporation, indicating a constant rate of EV production in non-sonoporated cells.
Discussions
Sonoportion can be used to both facilitate intracellular uptake of desired molecules by increasing plasma membrane permeability and enhance EV production from cells. Thus, the technique could be used to engineer therapeutic EVs encapsulating desired therapeutic cargoes. Utilizing ultrasound waves, sonoproation offers the advantage of non-invasive applications in vivo. For in vitro applications, sonoporation EV production can use standard labware and achieve high efficiency or high throughput operation by readily implementing a multi-channel system with an array of cost-effective ultrasound transducers51.
This study examined sonoporation-induced dynamic changes in CD63-GFP signal intensity and distribution in response to microbubble-enabled sonoporation at the single-cell level in RAW 264.7 cells. Our results show, for the first time, different changes in the sonoporated cells and the RAW 264.7 cells subjected to indirect impacts of sonoporation, depending on their proximity to the sonoporated cells. These different dynamic changes in CD63 reflect different dynamic characteristics in MVB reorganization due to the direct mechanical impact of sonoporation and the indirect effect of sonoporation via chemical factors released from sonoporated cells through cell-cell communication.
As our current study focused solely on CD63-GFP proteins within subcellular compartments and released EVs, it remains unclear whether sonoporation impacts the reorganization of other cellular compartments and the biogenesis of different EV subtypes, such as plasma membrane-derived microvesicles. Further research addressing these aspects would provide a more comprehensive understanding of sonoporation’s effects on EV production and cellular processes. It is also unclear whether the direct and indirect mechanisms of sonoporation impacting MVB reorganization also affect the properties of EVs characteristics.
Unlike in soluble factors or molecular solutions where cells are generally uniformly exposed to the agents, cellular attachment of microbubbles, due to their finite size, is discrete and often not uniform in numbers and locations. This heterogeneity, as clearly seen in our experiments, can lead to multiple mechanisms of action and complicate the interpretation of population-based observations and results.
Measurements in the single-cell sonoproation experiments in this study were limited to 2D images of the changes in the CD63-GFP signal intensity and distribution in a monolayer of RAW 264.7 cells. Our measurements focused on the extension areas beyond the original cell boundaries due to their superior signal-to-noise ratio, as these areas had no signal before sonoporation. However, it is possible that the CD63-GFP signal intensity and distribution inside the cells in 3D also undergo dynamic changes. Future studies using imaging techniques with higher sensitivity and spatiotemporal resolution are needed to investigate these potentially interesting outcomes.
Identifying Nearby cells in our analysis was relatively easy based on their apparent physical distance from sonoporated cells. This approach likely captured most cells in this category accurately. However, since identifying the Adjacent and Nearby cells in this study was performed manually without directly verifying gap junctions between two Adjacent cells, some of the Adjacent cells may belong to the group of Nearby cells. Thus, the observed variations in the data and differences between these two groups (Fig. 5) may disappear or become more significant with more precise classification. Future work is needed to overcome this limitation and verify the presence of functional gap junctions to better delineate the impact of chemical factors after sonoporation via gap junction transport or paracrine diffusion.
The ultrasound parameters in this study were chosen based on prior sonoporation studies for intracellular delivery with high cell viability24,45 and, thus, may not be optimal for EV production per se. Our single-cell sonoporation experiments examined the acute responses of single-cells based on the CD63-GFP signal intensity in the RAW 264.7 cells. On the other hand, in order to assess EVs released from the cells over a long period of time and distributed throughout the entire volume of the medium, the ensemble sonoporation experiments provided complementary information from the analysis of the supernatants and assess the longer-term effects of sonoporation on EV production. To match the conditions used in single-cell sonoporation experiments, only one ultrasound pulse was applied in the ensemble sonoporation experiments. Yet the one-time ultrasound application already yielded enhanced EV production at 2 h, and the effects appeared to last up to 48 h. The initial 2 h-delay from sonoporation may be attributed to the time required for cellular processes to react to the physical perturbation caused by sonoporation. The prolonged effect on EV production may be attributed to the activation of persistent stress-response signaling pathways. Numerous studies have demonstrated that cells exposed to various stressors, including thermal, oxidative, photodynamic, radiative, and shear stress, exhibit enhanced EV production52,53. This suggests that increased EV secretion may be a conserved cellular stress response, potentially functioning as a mechanism for membrane repair or removal of damaged components to maintain homeostasis. Future studies are needed to elucidate the underlying mechanisms driving this delayed and sustained increase in EV production.
This study was not designed to maximize EV production from the RAW 264.7 cells using sonoporation. It is possible that application of multiple ultrasound pulses could further increase EV production. However, since microbubble-enabled sonoporation depends on the cavitation of cell-anchored microbubbles, strategies applying multiple ultrasound pulses need to consider the complex situation resulting from the potentially significant changes in microbubbles due to exposure to ultrasound pulses, including collapse, shrinkages, and coalescence45. These complex scenarios could result in less predictable and repeatable outcomes for ensemble cell sonoporation experiments. Future studies seeking to maximize EV production need to systematically investigate the impact of different ultrasound pulse schemes and interactions with microbubbles.
Conclusions
The single-cell sonoporation experiments in this study reveal new findings on the rapid reorganization of MVBs in RAW 264.7 cells with different dynamic characteristics, depending on whether the cells were directly or indirectly affected by sonoporation. These results suggest different mechanisms in sonoporation-induced reorganization of MVBs in RAW 264.7 cells mediated by mechanical and chemical factors. The ensemble sonoporation experiments revealed a sustained increase in CD63-GFP-positive EV production in the supernatant of sonoporation-treated RAW 264.7 cells. These results demonstrate sonoporation’s ability to enhance EV production and provide insights for future studies to optimize sonoporation parameters for large-scale EV production applications.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. M. Bernard at the MSU Flow Cytometry Core Facility; Dr. A. Withrow at the Center for Advanced Microscopy at MSU; Dr. S. Makaremi at the IQ Microscopy Core Facility. This study was supported, in part, by funding from the NIH/NIBIB: R21-EB033554; R01-EB030565; NIH/NEI: R01-EY016077; and start-up funds from Michigan State University.
Author contributions
W.L., C. G., and N.A.S. performed the experiments, data analysis, and made the figures. M.A.G. and X.V. participated in the experiments involving isolation and analysis of EVs. M.K. and C.X.D. designed and led the execution of the study. All authors contributed to writing and reviewed the manuscript.
Data availability
Data included in this manuscript is available and will be shared upon request. Contact Masamitsu Kanada (kanadama@msu.edu) and Cheri X. Deng (cxdeng@umich.edu) for data request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Masamitsu Kanada, Email: kanadama@msu.edu.
Cheri X. Deng, Email: cxdeng@umich.edu
References
- 1.Shi, J., Kantoff, P. W., Wooster, R. & Farokhzad, O. C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer. 17, 20–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfram, J. & Ferrari, M. Clinical cancer nanomedicine. Nano Today. 25, 85–98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ouyang, B. et al. The dose threshold for nanoparticle tumour delivery. Nat. Mater.19, 1362–1371 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Mager, S. E. L. A., Breakefield, I. & Wood, X. O. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov.. 12, 347–357 (2013). [DOI] [PubMed] [Google Scholar]
- 5.van der Meel, R. et al. Extracellular vesicles as drug delivery systems: Lessons from the liposome field. J. Control Release. 195, 72–85 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Welsh, J. A. et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles. 13, e12404 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witwer, K. W. Minimal information for studies of extracellular vesicles 2023: Relevance to cell and gene therapies. Cytotherapy, 26, 1119–1121 (2024). [DOI] [PubMed]
- 8.Jeppesen, D. K., Zhang, Q., Franklin, J. L. & Coffey, R. J. Extracellular vesicles and nanoparticles: Emerging complexities. Trends Cell. Biol.33, 667–681 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M HR, et al. Exosomes: From garbage bins to promising therapeutic targets. Int. J. Mol. Sci.18, (2017). [DOI] [PMC free article] [PubMed]
- 10.Alvarez-Erviti, L. et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol.29, 341–345 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Haney, M. J. et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control Release. 207, 18–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamerkar, S. et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 546, 498–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gowen, A., Shahjin, F., Chand, S., Odegaard, K. E. & Yelamanchili, S. V. Mesenchymal stem cell-derived extracellular vesicles: Challenges in clinical applications. Front. Cell. Dev. Biol.8, 149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cha, J. M. et al. Efficient scalable production of therapeutic microvesicles derived from human mesenchymal stem cells. Sci. Rep.8, 1171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haraszti, R. A. et al. Exosomes produced from 3D cultures of MSCs by tangential flow filtration show higher yield and improved activity. Mol. Ther.26, 2838–2847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, Z. et al. Cyclic Stretch Force induces Periodontal Ligament cells to Secrete exosomes that suppress IL-1beta production through the inhibition of the NF-kappaB signaling pathway in macrophages. Front. Immunol.10, 1310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, X. et al. A review of low-intensity pulsed ultrasound for therapeutic applications. IEEE Trans. Biomed. Eng.66, 2704–2718 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Kubanek, J. et al. Ultrasound modulates ion channel currents. Sci. Rep.6, 24170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorum, B., Rietmeijer, R. A., Gopakumar, K., Adesnik, H. & Brohawn, S. G. Ultrasound activates mechanosensitive TRAAK K(+) channels through the lipid membrane. Proc. Natl. Acad. Sci. U.S.A.118, e2006980118 (2021). [DOI] [PMC free article] [PubMed]
- 20.Yoo, S., Mittelstein, D. R., Hurt, R. C., Lacroix, J. & Shapiro, M. G. Focused ultrasound excites cortical neurons via mechanosensitive calcium accumulation and ion channel amplification. Nat. Commun.13, 493 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helfield, B., Chen, X., Watkins, S. C. & Villanueva, F. S. Biophysical insight into mechanisms of sonoporation. Proc. Natl. Acad. Sci.113, 9983–9988 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rich, J., Tian, Z. & Huang, T. J. Sonoporation: past, Present, and Future. Adv. Mater. Technol.7, 2100885 (2022). [DOI] [PMC free article] [PubMed]
- 23.Deng, C. X., Sieling, F., Pan, H. & Cui, J. Ultrasound-induced cell membrane porosity. Ultrasound Med. Biol.30, 519–526 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Fan, Z., Liu, H., Mayer, M. & Deng, C. X. Spatiotemporally controlled single cell sonoporation. Proc. Natl. Acad. Sci. U.S.A.. 109, 16486–16491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, Y., Kumon, R. E., Cui, J. & Deng, C. X. The size of sonoporation pores on the cell membrane. Ultrasound Med. Biol.35, 1756–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, Y., Cui, J. & Deng, C. X. Dynamics of sonoporation correlated with acoustic cavitation activities. Biophys. J.94, L51–L53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, X., Leow, R. S., Hu, Y., Wan, J. M. & Yu, A. C. Single-site sonoporation disrupts actin cytoskeleton organization. J. R Soc. Interface. 11, 20140071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan, X., Zhou, Q., Wan, J. M. F. & Yu, A. C. H. Sonoporation generates downstream cellular impact after membrane resealing. Sci. Rep.11, 5161 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan, Z., Kumon, R. E., Park, J. & Deng, C. X. Intracellular delivery and calcium transients generated in sonoporation facilitated by microbubbles. J. Control Release. 142, 31–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumon, R. E. et al. Spatiotemporal effects of sonoporation measured by real-time calcium imaging. Ultrasound Med. Biol.35, 494–506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, Y., Shi, J., Cui, J. & Deng, C. X. Effects of extracellular calcium on cell membrane resealing in sonoporation. J. Control Release. 126, 34–43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan, Z., Kumon, R. E. & Deng, C. X. Mechanisms of microbubble-facilitated sonoporation for drug and gene delivery. Ther. Deliv. 5, 467–486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan, Z., Chen, D. & Deng, C. X. Improving ultrasound gene transfection efficiency by controlling ultrasound excitation of microbubbles. J. Control Release. 170, 401–413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuana, Y., Balachandran, B., van der Wurff-Jacobs, K. M. G., Schiffelers, R. M. & Moonen, C. T. Potential use of extracellular vesicles generated by microbubble-assisted ultrasound as drug nanocarriers for cancer treatment. Int. J. Mol. Sci.21, 3024 (2020). [DOI] [PMC free article] [PubMed]
- 35.Sridharan, B. & Lim, H. G. Exosomes and ultrasound: The future of theranostic applications. Mater. Today Bio. 19, 100556 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuana, Y. et al. Microbubbles-assisted Ultrasound triggers the release of extracellular vesicles. Int. J. Mol. Sci.18, 1610 (2017). [DOI] [PMC free article] [PubMed]
- 37.Boulanger, C. M., Amabile, N. & Tedgui, A. Circulating microparticles: A potential prognostic marker for atherosclerotic vascular disease. Hypertension. 48, 180–186 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Williams, J. K., Ngo, J. M., Lehman, I. M. & Schekman, R. Annexin A6 mediates calcium-dependent exosome secretion during plasma membrane repair. Elife12, e86556 (2023). [DOI] [PMC free article] [PubMed]
- 39.Demonbreun, A. R. et al. An actin-dependent annexin complex mediates plasma membrane repair in muscle. J. Cell. Biol.213, 705–718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buzas, E. I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol.23, 236–250 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, Y. et al. Macrophage-derived extracellular vesicles: Diverse mediators of pathology and therapeutics in multiple diseases. Cell. Death Dis.11, 924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung, B. H. et al. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun.11, 2092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mates, L. et al. Molecular evolution of a novel hyperactive sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet.41, 753–761 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Perez, G. I. et al. In vitro and in vivo analysis of extracellular vesicle-mediated metastasis using a bright, red-shifted bioluminescent reporter protein. Adv. Genet. (Hoboken). 3, 2100055 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan, Z., Chen, D. & Deng, C. X. Characterization of the dynamic activities of a population of microbubbles driven by pulsed ultrasound exposure in sonoporation. Ultrasound Med. Biol.40, 1260–1272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shelke, G. V., Lasser, C., Gho, Y. S. & Lotvall, J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles3, 24783 (2014). [DOI] [PMC free article] [PubMed]
- 47.Gorgens, A. et al. Identification of storage conditions stabilizing extracellular vesicles preparations. J. Extracell. Vesicles. 11, e12238 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez, G. I. et al. Phosphatidylserine-exposing annexin A1-Positive extracellular vesicles: Potential Cancer biomarkers. Vaccines 11, 639 (2023). [DOI] [PMC free article] [PubMed]
- 49.Ferguson, S. et al. Single-EV analysis (sEVA) of mutated proteins allows detection of stage 1 pancreatic cancer. Sci. Adv.8, eabm3453 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schurz, M. et al. EVAnalyzer: High content imaging for rigorous characterisation of single extracellular vesicles using standard laboratory equipment and a new open-source ImageJ/Fiji Plugin. J. Extracell. Vesicles. 11, e12282 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hendren, C., Li, W., Stegemann, J. P., Hall, T. L. & Deng, C. X. Multichannel resonant acoustic rheometry system for quantification of coagulation of multiple human plasma samples. Sci. Rep.13, 19237 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hedlund, M., Nagaeva, O., Kargl, D., Baranov, V. & Mincheva-Nilsson, L. Thermal- and oxidative stress causes enhanced release of NKG2D ligand-bearing immunosuppressive exosomes in leukemia/lymphoma T and B cells. PLoS ONE. 6, e16899 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo, S. et al. Stimulating Extracellular vesicles production from Engineered tissues by Mechanical forces. Nano Lett.21, 2497–2504 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in this manuscript is available and will be shared upon request. Contact Masamitsu Kanada (kanadama@msu.edu) and Cheri X. Deng (cxdeng@umich.edu) for data request.