Abstract
Self-assembling DNA nanoparticles have the potential to significantly advance the targeted delivery of molecular cargo owing to their chemical and architectural flexibility. Recently, it has been demonstrated that the genetic code embedded in DNA nanoparticles produced by the method of DNA origami or related techniques can be recognized and copied by RNA polymerase in vitro. Further, sculpted DNA nanoparticles can serve as a substrate for Cas9-mediated gene modification and gene expression in cell culture. In the present study, we further investigate the ability of DNA origami nanoparticles to be expressed in a human cell line with emphasis on the impact of single-stranded DNA (ssDNA) domains and the contributions of the architectural disposition of genetic control elements, namely promoter and enhancer sequences. Our findings suggest that while cells possess the remarkable capability to express genes within highly folded architectures, the presence and relative density and location of ssDNA domains appears to influence overall levels of gene expression. These results suggest that it may be possible to nuance folded DNA nanoparticle architecture to regulate the rate and/or level of gene expression. Considering the highly malleable architecture and chemistry of self-assembling DNA nanoparticles, these findings motivate further exploration of their potential as an economic nanotechnology platform for targeted gene editing, nucleic acid-based vaccines, and related biotherapeutic applications.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78399-y.
Keywords: DNA nanotechnology, DNA origami, Human cell line, Flow cytometry, Green fluorescent protein (GFP), Gene expression
Subject terms: Biotechnology, Cell biology, Molecular biology, Gene delivery, Gene expression analysis, Genetics, Gene expression, Genetic association study
Introduction
Targeted delivery of biomaterials in vivo has significantly improved therapeutic agent efficacy and safety in biomedicine by increasing accumulation at specific target sites and reducing off-target effects1–5. Currently, various molecules, as well as application approaches, are being explored to target specific tissues, cells, and even subcellular compartments in vivo2,3,5–10. A promising strategy for the targeted delivery of nucleic acid biomedicines is the use of self-assembling DNA nanoparticles constructed by the method of DNA origami and related techniques11,12. This approach provides both chemical and architectural flexibility, thereby establishing a platform for precise control over the nanoscale molecular arrangement of targeting agents for nanoparticle drug delivery vehicles13–17. Thus, a wide range of site-specifying conjugates, attachment strategies, and active switching mechanisms are being explored to fully leverage the DNA nanoparticle delivery platform for biomedicine17–19. Nonetheless, to date, self-assembled DNA nanoparticles have primarily been envisioned as delivery vessels with internal or attached molecular cargo. However, an additional dimension has recently been added to the potential utility of sculpted DNA nanoparticles wherein the particle itself, constructed from biologically active genetic elements, may be both the carrier and the cargo20–22. Highly compact and crosslinked DNA origami nanoparticles are recognized and copied by RNA polymerase in vitro23. Further, a DNA origami nanoparticle has served as a CRISPR-mediated insertion element in cell culture21, and fully folded DNA nanostructures were shown to be expressed in cell culture, showcasing the cells’ versatile capability to express genes embedded in sculpted DNA nanoparticles20. Interestingly, in the latter case gene expression was achieved when utilizing either the sense or antisense strands as the nanoparticle scaffold, suggesting that DNA origami nanoparticles are capable of supporting both DNA replication and transcription in the cellular milieu20.
In the current report, we expand upon these studies, focusing on architectural details involved in DNA nanoparticle gene expression in cell culture. In general, folded DNA nanoparticles tend to be more resistant to nuclease degradation than their linear counterparts, presumably owing to the former’s compact architecture24. However, these nanoparticles necessarily include nicks in duplex domains at juxtaposed staples and, often, single-stranded DNA (ssDNA) domains to facilitate folding, making them susceptible to nuclease-mediated degradation. A single scission at a ssDNA domain could render a folded DNA nanoparticle genetically inactive while still retaining its overall architecture. Thus, it may be possible to modulate DNA nanoparticle efficacy in cell culture by modulating the relative abundance and locations of double-stranded DNA (dsDNA) and ssDNA domains.
Materials and methods
Primers for amplification of the Green Fluorescent Protein (GFP) gene were designed using SnapGene and staples for DNA nanoparticles were designed using caDNAno (http://www.cadnano.org). All oligonucleotides used in the study were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA) and their sequences are listed in Supplementary Tables 1 and 2. Phusion® DNA polymerase (Cat # M0530S) and LongAmp® Taq DNA polymerase (Cat # M0323L) were used for PCR asymmetric PCR (aPCR) respectively and they were purchased from New England Biolabs (NEB, Ipswich, MA, USA) along with their corresponding reaction components including buffers (5× Phusion® HF buffer (NEB, Cat # M0530S) and 5× LongAmp® Taq buffer (NEB, Cat # M0323L) and 20 mM dNTP mix (NEB, Cat # N0447S). pCMV-T7-EGFP was used as template for PCR and it was purchased from Addgene (Addgene, Watertown, MA, USA, Cat # 133962). For agarose gel electrophoresis, 6× loading dye was purchased from NEB (Cat # B7024S). SYBR safe and SYBR gold were used for staining and they were purchased from Invitrogen (Invitrogen, Waltham, MA, USA, Cat # S33102 and Cat # S11494 respectively). For DNA purification, a Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA, Cat # D4001) and a Zymoclean Gel RNA Recovery Kit from Zymo Research (Zymo Research, Cat # R1011) was used to extract dsDNA and ssDNA from agarose gels respectively. Ultrafiltration performed as the purification of DNA nanoparticles was performed using an Amicon ultracentrifugation filter with a 100 kDa molecular cutoff (Millipore Sigma, Burlington, MA, USA, Cat # UFC510096). The transfection of DNA nanoparticles was performed in HTR-8/SVneo (American Type Culture Collection (ATCC), Gaithersburg, MD, USA, Cat # CRL-3271) cells using jetPRIME transfection reagent (VWR, Batavia, Il, USA, Cat # 89129-924). The cells were processed for flow cytometry using 0.25% trypsin (Thermofisher Scientific, Madison, WI, USA, Cat # 25200056), and 4% Paraformaldehyde (PFA) (Fisher Scientific, Cat # AAJ61899AK). Gene-bearing DNA origami scaffolds were prepared as previously described25, summarized as follows.
Standard PCR
Each PCR reaction mixture was prepared in 50 µL final volume, composed of 1× Phusion® HF buffer (NEB), 200 nM dNTP mix (NEB), 500 nM sense primer (GFP sense = undesired sense strand), 500 nM antisense primer (GFP anti = desired antisense strand), 10 ng pCMV-T7-EGFP (Addgene), 0.5 µL Phusion® DNA polymerase (NEB), and nuclease-free water to volume. Each PCR was performed using the following thermocycler steps: 30 s at 98 °C, 30 s at 58 °C, and 1 min at 72 °C for 30 cycles.
The reaction product was mixed with 6× loading dye (1× buffer components: 2.5% Ficoll®-400, 10 mM EDTA, 3.3 mM Tris-HCl, 0.08% SDS, 0.02% Dye 1, 0.0008% Dye 2, pH 8 at 25 °C; NEB, ) and then loaded onto a 1% agarose gel pre-stained with SYBR safe DNA dye (Invitrogen). Electrophoresis was carried out at 8 V/cm for 1 h. The SYBR Safe-containing DNA was visualized using a 490 nm wavelength (blue) transilluminator and an amber filter.
Asymmetric PCR (aPCR)
Primers used in aPCR were identical to those used in standard PCR. Along with sense and antisense primers, a 3′ terminal modified primer (3′ GFP blocker) was used.
For the generation of a scaffold, each aPCR reaction was carried out in 50 µL total volume, composed of 1× LongAmp® Taq buffer (60 mM Tris-SO4, 20 mM (NH4)2SO4, 2 mM MgSO4, 3% glycerol, 0.06% IGEPAL® CA-630, 0.05% Tween® 20, pH 9.1 at 25 °C) from NEB, 500 nM GFP anti, 25 nM GFP sense, 475 nM 3′ GFP blocker, 300 nM dNTP mix (NEB), 10 ng double-stranded GFP gene (dsGFP; generated by standard PCR), 2 µL LongAmp® Taq DNA polymerase (NEB), and nuclease-free water to final volume. Each aPCR was performed using the following thermocycler steps: 30 s at 94 °C, 30 s at 59 °C, and 2 min at 65 °C for 25 cycles. The size of both PCR and aPCR amplicons is 1771 bp.
The reaction product was loaded onto a 1% agarose gel pre-stained with 1× SYBR Safe (Invitrogen), electrophoresed, and visualized as above.
Double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) purification
dsDNA and ssDNA in agarose gels were extracted using Zymoclean Gel DNA Recovery Kit and Zymoclean Gel RNA Recovery Kit from Zymo Research (Zymo Research) respectively. Gel bands containing target dsDNA and ssDNA were removed using a clean razor blade. Three times the gel slice volume of the provided agarose dissolving/binding buffer was added to each gel fragment and incubated at 55 °C on a heat block for 15 min. Each dissolved dsDNA gel solution was transferred to a provided silica-based spin column and centrifuged at 10,000 relative centrifugal force (rcf) for 60 s in a table-top centrifuge. 200 µL of ethanol-based DNA wash buffer was added to each spin column and centrifuged at 10,000 rcf for 30 s. A washing step was repeated before centrifuging at 10,000 rcf for 60 s for the complete removal of ethanol. Flow-through from all steps was discarded. After transferring each spin column to a clean microcentrifuge tube, 6–20 µL of the provided elution buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.5) was added directly to the matrix of each spin column followed by centrifugation at 10,000 rcf for 60 s for DNA collection. For ssDNA purification, overall purification steps are same with minor differences in centrifugation speed, time, and amount of buffer used. All centrifugation conducted for ssDNA purification except for the final elution step was done at 12,000 rcf. Initial centrifugation of dissolved ssDNA gel solution was done for 2 min followed by additional step of adding 400 µL RNA Prep buffer and centrifugation for 1 min. The washing step was conducted with 800 µL ethanol-based wash buffer and centrifugation of 30 s, and this step was repeated with 400 µL ethanol-based wash buffer and centrifugation for 2 min to fully remove residual ethanol. A fraction of each purified dsDNA and ssDNA was mixed with 6× loading dye (NEB) and loaded onto a 1% agarose gel pre-stained with 1× SYBR safe (Invitrogen). Electrophoresis and visualization were carried out as above. The yield of the purified dsDNA sample was evaluated by measuring band intensities relative to a known control (1 kb DNA ladder; NEB, Cat # N0468S) using GelAnalyzer 19.1 available at http://www.gelanalyzer.com (accessed on 19 August 2021).
DNA nanoparticle construction
DNA nanoparticles were designed using caDNAno (http://www.cadnano.org). DNA nanoparticles were prepared by mixing single-stranded GFP gene (ssGFP; generated by aPCR) to a final concentration of 91.4 nM (2.5 µg of scaffold) and each staple to a final concentration of 731.2 nM (1:8 scaffold: staple ratio) in 1× TAE buffer supplemented with 12.5 mM Mg(OAc)2 in a final volume of 50 µL (TAEM). The staple set used for each DNA nanoparticle is listed in Supplementary Table 3. The mixture was incubated at 90 °C for 10 min in a water bath followed by gradual cooling to room temperature. After construction, DNA nanoparticles were purified by ultrafiltration. The final volume of each DNA nanoparticle was scaled up to 500 µL by adding 1× Phosphate-buffered saline containing 12.5 mM Mg(OAc)2 (PBSM). For the purification of final products, excess staples were removed using an Amicon ultracentrifugation filter with a 100 kDa molecular cutoff (Millipore Sigma). 500 µL DNA nanoparticle product was applied to each filter unit followed by centrifugation at 14,000 rcf for 3 min. This step was repeated three times with the addition of 400 µL of 1× PBSM to ensure the full removal of staples. All the flowthroughs were discarded. For the elution of purified DNA nanoparticles, the centrifugation was done at 14,000 rcf for 5 min with the filter unit inverted. The purified products were verified by mixing with 6× loading dye (NEB) and then loading onto 1% agarose gel containing 12.5 mM Mg(OAc)2. Electrophoresis was carried out in TAEM buffer at 6 V/cm for 90 min. The gel was post-stained with 1× SYBR gold DNA dye (Invitrogen) and visualized as above.
The quantity of each purified DNA nanoparticle was determined by measuring the quantity of the scaffold. A fraction of each purified DNA nanoparticle (2 µL) was mixed with 18 µL nuclease-free water followed by heating the samples at 70 °C for 10 min to purposely disintegrate the nanoparticle (i.e., remove staples.) The heated mixtures were mixed with 6× loading dye (NEB, Cat # B7025S) and were gel electrophoresed within a 1% agarose gel pre-strained with 1× SYBR safe (Invitrogen, Cat # S33102) as described above. Quantification was carried out using GelAnalyzer 19.1 as described above.
DNA nanoparticle transfection
The transfection of DNA nanoparticles was performed for a total of three biological replicates using HTR-8/SVneo (ATCC) cells and jetPRIME transfection reagent (VWR). HTR-8/SVneo is an immortalized epithelial-like cell line that is often used as a model for trophoblast cells of the placenta. The cell line was originally derived from cells that grew out of chorionic explants of human first-trimester placenta. Cells were cultured as described previously26. Briefly, 50,000 cells were seeded per 24-well plate and transfections were performed after 24 h using 141.5 fmol (corresponding to 500 ng of GFP plasmid) of origami samples with a 1:2 ratio of the jetPRIME reagent. Media was replaced 24 h after transfection. Images were taken after 48 h of transfection (Olympus Fluorescent Microscope, AST-0008943) and the GFP images were processed using ImageJ software27. The cells were dissociated using 0.25% trypsin (Thermofisher Scientific), and then fixed with 4% PFA (Fisher Scientific) for 15 min at room temperature, followed by a 2× PBS wash and resuspension in 500ul of PBS. The PFA fixed cells were kept on ice and transported to The Iowa State University Flow Cytometry Facility for assessing percentage of GFP positive cells.
Flow cytometry
Data acquisition and analysis were conducted using an unmodified BD FACSCanto flow cytometer (BD Biosciences, San Jose, CA) with a 488 nm laser for GFP excitation and measurement of cell forward (FSC) and side scatter (SSC) properties. Green fluorescence was collected through a 525/50 nm band pass (BP) filter, while forward and side scatter data were acquired using 488/10 nm BP filters. Voltages for scatter and fluorescence detectors were adjusted based on untreated or mock-transfected cells. Data from these negative controls were used to differentiate GFP(-) and GFP(+) events. GFP fluorescence data were presented on four-decade log scales with bi-exponential scaling, while FSC and SSC were displayed using linear amplification. In order to differentiate intact cells from debris and other extraneous events, FSC and SSC properties of the cells were used for establishing the analysis standards. Data acquisition employed a medium flow rate of 60 µL/min, collecting a minimum of 10,000 gated cell events for each sample. The FACSDiva software (ver. 8.0.1, BD Biosciences) was used for data analysis, and percentages of GFP positive cells were adjusted relative to the positive control.
Results
DNA nanoparticle construction, purification, and verification
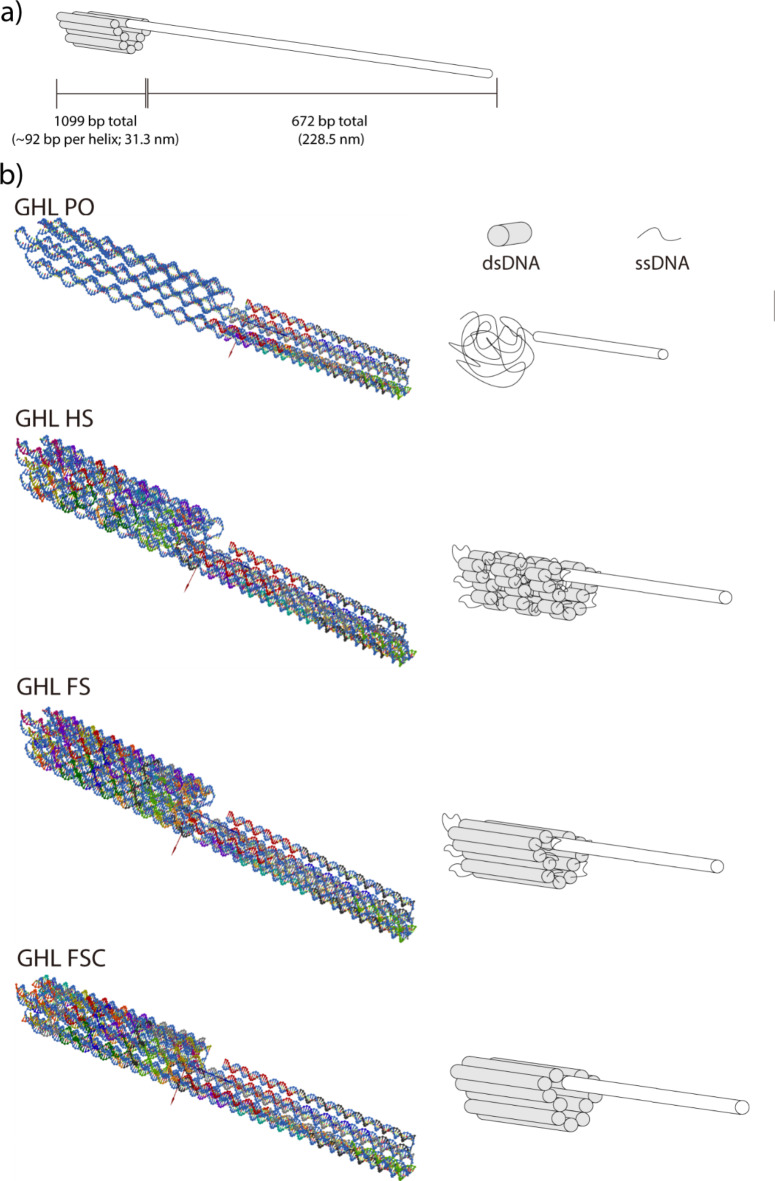
To assess the capability of DNA nanoparticles to be able to express embedded genes in cell culture, a custom scaffold was synthesized by asymmetric PCR (aPCR) as previously described25. The scaffold contained a human Cytomegalovirus (CMV) promoter and a CMV enhancer to support gene expression in human cells, and the Enhanced Green Fluorescent Protein (EGFP) gene for fluorescence-based visualization of gene expression. The design of DNA nanoparticles in this study was carried out using caDNAno28. The overall architecture was roughly cylindrical, featuring a consistent crossover pattern across various forms of the structure (Fig. 1). The DNA nanoparticles, referred to as GHL (GFP gene Honeycomb structure with a Linear promoter and enhancer), incorporated linear duplex promoter and enhancer domains (i.e., not folded using staple oligonucleotides) to investigate the gene expression capability of a folded architecture. We previously observed a positive correlation between the abundance of ssDNA domains within DNA nanoparticles and overall levels of transcription in a nuclease-free in vitro system23. We proposed that the increased structural flexibility conferred by ssDNA in that system facilitated polymerase activity. However, ssDNA is generally more susceptible to nuclease-mediated scission than dsDNA24,29. Therefore, to assess the influence of ssDNA domains on gene expression in a cellular milieu, multiple DNA nanoparticles with an identical overall design but with varying levels of ssDNA were prepared. These variants were labeled as GHL PO (DNA nanoparticle with linear duplex promoter/enhancer with single-stranded gene; Promoter/enhancer domains were Only duplex within the structure), GHL HS (DNA nanoparticle with linear duplex promoter/enhancer and a half complement of staples; Half-Set of staples used), and GHL FS (DNA nanoparticle with linear duplex promoter/enhancer and a mostly full complement of staples; Full-Set of staples used), in order of increasing number of staples (Table 1; Fig. 1, and Supplementary Fig. 1). Due to different levels of ssDNA domains, each sample has slightly varied molecular weight (Supplementary Table 4). Notably, although GHL FS had the greatest number of staples among the aforementioned DNA nanoparticles, it retained ssDNA domains at the ends of duplex DNA helices primarily to minimize end-to-end adhesion of DNA nanoparticles30 but also to enhance flexibility and potentially facilitate folding31. Finally, to fully eliminate ssDNA domains within the nanoparticle, the staples at the edges of GHL FS were replaced for the full coverage of ssDNA domains, resulting in the GHL FSC (DNA nanoparticle with linear duplex promoter/enhancer, a full complement of staples (Full-Set of staples used), and fully Covered single-stranded end of helices) variant (Table 1; Fig. 1, and Supplementary Fig. 1).
Fig. 1.
Schematic diagram and 3D rendered model of various DNA nanoparticles. DNA nanoparticles with linear promoter/enhancer (GFP gene Honeycomb structure with a Linear promoter and enhancer; GHL). GHL PO: DNA nanoparticle with linear promoter/enhancer and single-stranded gene; GHL HS: DNA nanoparticle with linear promoter/enhancer and a half complement of staples; GHL FS: DNA nanoparticle with linear promoter/enhancer and a mostly full complement of staples; GHL FSC: DNA nanoparticle with linear promoter/enhancer and a full complement of staples. (a) Overall architectural scheme of DNA nanoparticles and its size. (b) 3D rendered models and corresponding schematic diagrams of various DNA nanoparticles. 3D models were rendered using oxDNA. For the oxDNA images, regions that are relatively more compact and comprised of strands with various colors represent duplex domains. Regions that are relatively less compact and with single-colored strands represent ssDNA domains. For schematic diagrams, white represents long, linear duplex DNA, gray represents folded duplex DNA and thin black lines represent single-stranded DNA.
Table 1.
Names of DNA samples used in this study.
| Name | Contents |
|---|---|
| dsGFP | Fully linear double-stranded GFP gene |
| dsGFP-a | Fully linear double-stranded GFP gene, produced as a byproduct of aPCR |
| ssGFP | Single-stranded antisense strand for nanoparticle scaffold |
| GHL PO | DNA nanoparticle with a linear duplex promoter/enhancer and single-stranded GFP gene; variant of GFP gene Honeycomb structure with a Linear promoter and enhancer, Promoter/enhancer domains are Only duplexes within the structure |
| GHL HS | DNA nanoparticle with a linear duplex promoter/enhancer and a half complement of staples (i.e., loosely folded); variant of GFP gene Honeycomb structure with a Linear promoter and enhancer, the structure folded with a Half-Set of staples |
| GHL FS | DNA nanoparticle with a linear duplex promoter/enhancer and a mostly full complement of staples (i.e., tightly folded with single-stranded edges); variant of GFP gene Honeycomb structure with a Linear promoter and enhancer, the structure folded with a Full-Set of staples |
| GHL FSC | DNA nanoparticle with a linear duplex promoter/enhancer and a full complement of staples (i.e., tightly folded with no ssDNA domains); variant of GFP gene Honeycomb structure with a Linear promoter and enhancer, the structure folded with a Full-Set of staples and Covered single-stranded ends |
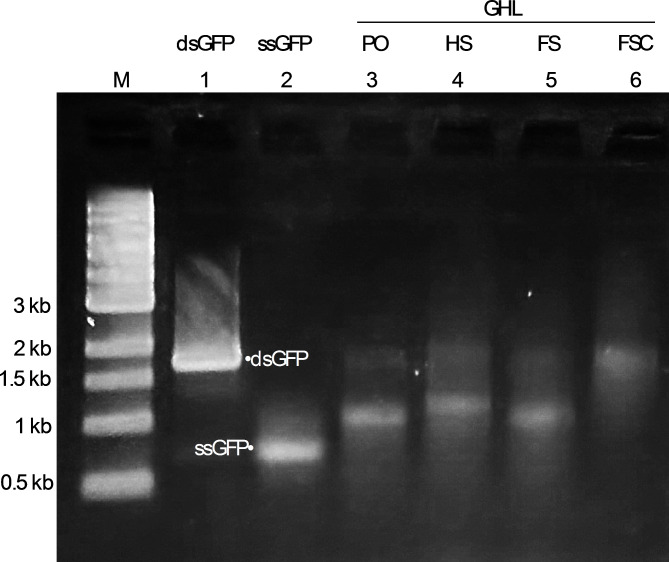
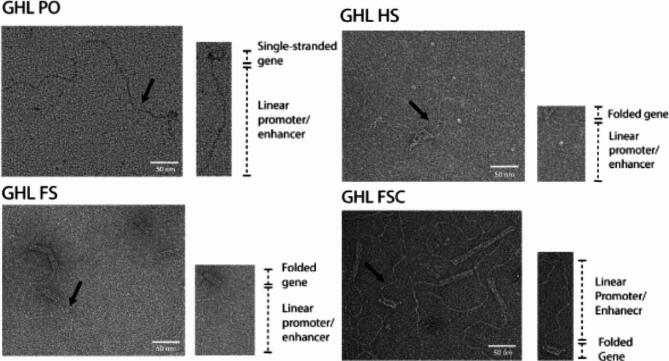
To mitigate any influence of the presence of excess staples, they were removed by ultrafiltration. Electrophoretic analysis confirmed effective staple elimination. However, some nanoparticle samples showed noticeable smearing, which often indicates sample aggregation (Fig. 2). This aggregation was likely attributable to end-to-end adhesion (pi bond stacking) in these samples, a phenomenon known to occur in DNA origami with blunt-ended helices (32,33; Fig. 3).
Fig. 2.
Generation and purification of DNA nanoparticles from GFP gene-containing scaffolds. Representative agarose gel electrophoresis display of the purified DNA nanoparticles used in this study. M: molecular marker; 1: dsGFP (fully duplex GFP gene); 2: ssGFP (single-stranded antisense strand (i.e., scaffolds for DNA nanoparticles)); GHL: DNA nanoparticles with linear promoter/enhancer; 3: PO (GHL PO; DNA nanoparticle with linear promoter/enhancer and single-stranded gene); 4: HS (GHL HS; DNA nanoparticle with linear promoter/enhancer and a half complement of staples); 5: FS (GHL FS; DNA nanoparticle with linear promoter/enhancer and a mostly full complement of staples); 6: FSC (GHL FSC; DNA nanoparticle with linear promoter/enhancer and a full complement of staples). For lanes 1 and 2, the band at approximately 1.7 kb represents dsGFP, and the band at approximately 0.7 kb represents ssGFP. The bands between 1.7 kb and 1 kb for lanes 3, 4, 5, and 6 represent successfully self-assembled DNA nanoparticles with linear duplex promoter/enhancer.
Fig. 3.
Verification of the structural integrity of ultracentrifugation-filtered DNA nanoparticles by transmission electron microscopy (TEM). A representative DNA nanoparticle with key structural elements is located to the lower right of each representative TEM field. The black arrows point to the representative DNA nanoparticles within the TEM field. ssDNA and loosely crosslinked domains appear as relatively amorphous consolidated networks whereas linear duplexes containing the promoter and enhancer appear as curved tails due to the enhanced persistence length of dsDNA relative to ssDNA31. Compact, highly crosslinked origami regions display the canonical multi-helix particle morphology. Scale bar: 50 nm.
Gene expression of DNA nanoparticles in cultured cells
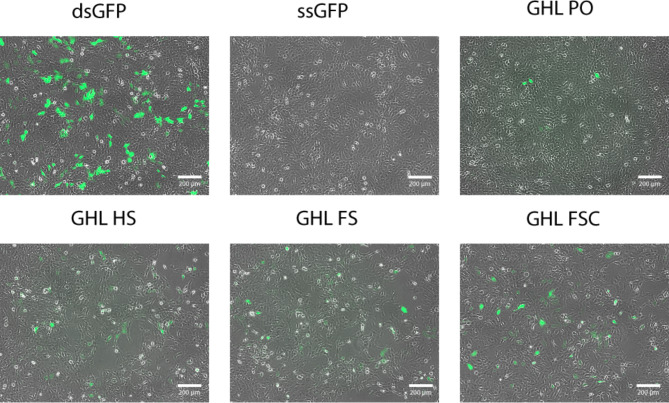
Expression of the genetic content of DNA nanoparticles was evaluated by transfection of a human trophoblast cell line in culture and subsequent image analysis of GFP production. Each sample was initially qualitatively analyzed for the presence of fluorescence indicative of successful GFP gene expression. All samples, except the ssDNA negative control, expressed GFP, with the linear duplex (control) sample clearly capable of the highest level of gene expression (Fig. 4).
Fig. 4.
Qualitative fluorescence analysis of GFP expression from various DNA nanoparticle architectures in mammalian cell culture. All architectures were capable of supporting GFP expression at a level greater than single stranded DNA (ssGFP) but not as efficiently as linear duplex DNA (dsGFP). Scale bar: 200 μm.
For quantitative analysis of gene expression, each sample was transfected in three biological replicates and analyzed by flow cytometry to quantify the number of GFP-positive cells per sample. As a positive control, a linear duplex GFP PCR product (dsGFP) made with a high-fidelity Taq-based system (Phusion) was used to establish the maximum expression potential for this gene ensemble under the transfection conditions used. To evaluate the impact of the potential for the introduction of coding errors during aPCR using the lower fidelity enzyme LongAmp Taq, the dsDNA product of the aPCR reaction (dsGFP-a) was included in these analyses. This dsDNA control exhibited a reduced level of gene expression in comparison to dsGFP produced by the higher fidelity Phusion PCR system as would be expected for a Taq-based system known to introduce sequence errors (i.e., LongAmp vs. Phusion).
A single-stranded antisense strand (ssGFP) comprised of the bare origami scaffold was employed as a negative control based on the previous reports that ssDNA is highly susceptible to nuclease degradation in cell culture and should, therefore, result in a minimum level of gene expression24,29. As expected, and consistent with initial qualitative analyses (Fig. 4), ssGFP exhibited a basal level of gene expression that was substantially lower than all of the experimental samples.
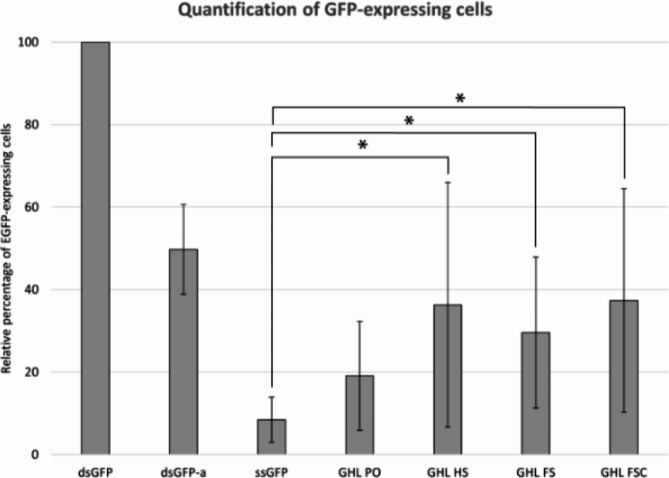
Expression levels were quantitatively assessed by flow cytometry analysis (Fig. 5 and Supplementary Fig. 2). All of the DNA nanoparticles in this study supported gene expression, exhibiting roughly equivalent, but not identical, levels of gene expression regardless of their folded states and in agreement with previous reports on the expression of DNA nanoparticles in mammalian cells20. P-values were calculated by a student t-test between DNA nanoparticle samples and ssGFP (negative control). Three out of four samples (GHL HS, GHL FS, and GHL FSC) showed differences with p-values less than 0.1 (Supplementary Table 5). GHL PO had a p-value greater than 0.1 presumably due to its structural similarity to ssGFP. Flow cytometry further revealed that GFP-expressing cells transfected with DNA nanoparticles exhibited a generally reduced level of fluorescence in comparison to positive controls. The details of the influence of DNA nanoparticle architecture as a mechanism to regulate gene expression levels remain to be determined.
Fig. 5.
Quantification of GFP-expressing cells from various DNA samples. GFP-expressing cells were identified by flow cytometry. The percentage of GFP-expressing cells for each DNA sample was calculated and adjusted relative to a positive control (dsGFP). The average relative percentage of each sample is represented as a bar. Error bars represent one standard deviation from the mean of three independent experiments (* represents p-value less than 0.1). P-values were calculated by student t-test between DNA nanoparticle samples and ssGFP (negative control). The raw data is presented in Supplementary Table 6.
Discussion
Self-assembling DNA nanoparticles represent a potentially powerful platform for exploring the uses of architecturally engineered genetic material in living systems20–22,34. In the present study, we explored the impact of various DNA origami nanoparticle design parameters on gene expression in mammalian cell culture. These parameters include the presence of ssDNA domains and the overall flexibility of the architectures mediated by the ratio of ssDNA to dsDNA domains.
Previous study has shown that sculpted DNA nanoparticles are somewhat immunogenic and the level of immunogenicity can be subtly influenced by architectural variation35. Although immunogenicity is not directly relevant to the current study (evaluating expression of DNA nanoparticles in cell culture) it will be of fundamental importance in ongoing work in which sculpted DNA nanoparticle expression is studied in living systems in which various factors including ss/dsDNA ratio, nanoparticle shape and dimensions, and, critically, the influence of readily modifiable surface moieties (e.g., CpG domains36) offer a suite of fine tuning opportunities to optimize performance.
Our results revealed that the presence of ssDNA domains in DNA nanoparticles has a minor adverse impact on gene expression. For example, the construct GHL PO, containing the maximum number of ssDNA domains amongst the nanoparticles examined in this study, supported the minimum level of gene expression (Fig. 5). Based on this observation one might anticipate a graded inverse relationship between levels of gene expression and relative abundance of ssDNA and concomitant single-strand specific nuclease sensitivity. However, a comparison between GHL PO and the other DNA nanoparticles studied here suggests that once the staple coverage reached a threshold level of dsDNA coverage, the susceptibility of gene expression to ssDNA nuclease plateaued (Fig. 5). It is noteworthy that the samples analyzed by flow cytometry were prepared in phosphate buffered saline supplemented with magnesium (PBSM), rather than the more conventional Tris-acetate EDTA supplemented with magnesium buffer (TAEM), the former of which has been shown to enhance DNA nanoparticle resistance to nuclease attack37. Thus, the preparation conditions may have influenced the observed levels of resistance to nuclease. Additional variables could have influenced the observed adverse effect on gene expression such as potential differences on transfection efficiencies between ssDNA and dsDNA. These considerations notwithstanding, it is remarkable that a recent study by Kretzmann et al.20 demonstrated the ability of cells to process complex structured genes. Particularly striking was the observation that DNA nanoparticles constructed using sense strand scaffolds, rather than the gene-encoding antisense strand, still expressed the encoded gene within cells. This would require that the sense strand first be replicated to create the template strand which may then be transcribed to create the gene product. Thus, it may be the case that self-assembling, sculpted DNA nanoparticles are both transcription- and replication-competent in cell culture20. It is noteworthy that in the Kretzmann et al. study the constructs employed included a eukaryotic origin of replication whereas the constructs used in the present study did not. Thus, it may be possible to not only modulate transcriptional activity as a function of DNA nanoparticle preparation method and architectural details but to also regulate expression longevity by modulation of the ability to self-replicate in the cellular milieu. Further studies are required to understand the limitations and flexibility of the myriad construct variations possible with the self-assembling DNA nanoparticle platform.
Finally, it is noteworthy that the chemical malleability of DNA nanoparticles enables the incorporation of diverse molecular and chemical species, offering advantages such as enhanced cellular uptake, improved stability, and intra-/intermolecular targeting38–44. For example, coating DNA nanoparticles with lipid moieties through electrostatic interactions enhances stability under physiological conditions and promotes increased cellular uptake22,45–47. Wu et al. demonstrated this approach by coating DNA nanoparticles with lipid fatty acids to enhance cell penetration22. Additionally, introducing nuclear localization signals (NLS) through intercalation or conjugation significantly enhances the nuclear import of transfected DNA, resulting in improved overall gene expression6,48,49. For example, Liedl et al. enhanced gene expression in DNA nanoparticles by incorporating DNA nuclear targeting sequences (DTS)34. In summary, self-assembling DNA nanoparticles hold great potential as a platform for targeted in vivo gene expression and gene modification.
Conclusion
This study demonstrates that self-assembling DNA nanoparticles are capable of gene expression in a human cell culture system. In contrast to our previous findings in a nuclease-free test tube system where increased single-stranded DNA (ssDNA) domains enhanced transcription efficiency, in cell culture we observed a moderate inverse relationship between gene expression and the degree of exposed ssDNA domains23. This may be attributed to the increased susceptibility of ssDNA to degradation by cellular nucleases in comparison to double-stranded DNA (dsDNA)24,29. Combined with previous research, these findings emphasize the potential of gene-bearing, self-assembling DNA nanoparticles containing nuanced architectural features to serve as a platform technology for biomedicine and related fields.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- GFP
Green fluorescent protein
- IDT
Integrated DNA Technologies
- NEB
New England Biolabs
- dsDNA
Double-stranded DNA
- ssDNA
Single-stranded DNA
- dsGFP
Double-stranded GFP gene
- dsGFP-a
Double-stranded GFP gene obtained from aPCR
- ssGFP
Single-stranded GFP gene
- rcf
Relative centrifugal force
- TAEM
Tris-Acetate-EDTA supplemented with 12.5 mM of Mg(OAc)2
- PBSM
Phosphate-buffered saline containing 12.5 mM Mg(OAc)2
- ATCC
American Type Culture Collection
- PFA
Paraformaldehyde
- TEM
Transmission electron microscopy
- NLS
Nuclear localization signals
- DTS
DNA nuclear targeting sequences
Author contributions
Eric Henderson contributed the concept of the project and editing of the manuscript. Chang Yong Oh perfomed majority of experiments and wrote the manuscriptHaninder Kaur performed transfection experiments and contributed to the editing of statistics.Geetu Tuteja contributed to the editing of the statistics.
Funding
Presidential Interdisciplinary Research Seed Grant (PIRS), Iowa State University, Ames IA 50011.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information file.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harada, H., Kizaka-Kondoh, S. & Hiraoka, M. Antitumor protein therapy; application of the protein transduction domain to the development of a protein drug for cancer treatment. Breast Cancer13, 16–26 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Marchetti, G. M., Burwell, T. J., Peterson, N. C. et al. Targeted drug delivery via caveolae-associated protein PV1 improves lung fibrosis. Commun. Biol. 2 (2019). [DOI] [PMC free article] [PubMed]
- 3.Yan, Y., Zhou, L., Sun, Z., Song, D. & Cheng, Y. Targeted and intracellular delivery of protein therapeutics by a boronated polymer for the treatment of bone tumors. Bioact. Mater.7, 333–340 (2022). [DOI] [PMC free article] [PubMed]
- 4.Rosenbaum, M. I., Clemmensen, L. S., Bredt, D. S., Bettler, B. & Strømgaard, K. Targeting receptor complexes: a new dimension in drug discovery. Nat. Rev. Drug Discov.19, 884–901 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Zhao, Z., Ukidve, A., Kim, J. & Mitragotri, S. Targeting strategies for tissue-specific drug delivery. Cell181, 151–167 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Brandén, L. J., Mohamed, A. J. & Smith, C. I. E. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol.17, 784–787 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Iwasaki, T., Murakami, N. & Kawano, T. A polylysine–polyhistidine fusion peptide for lysosome-targeted protein delivery. Biochem. Biophys. Res. Commun.533, 905–912 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Spencer, B. J. & Verma, I. M. Targeted delivery of proteins across the blood-brain barrier (2007). [DOI] [PMC free article] [PubMed]
- 9.Liang, K. et al. Targeted intracellular protein delivery based on hyaluronic acid-green tea catechin nanogels. Acta Biomater.33, 142–152 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Schneider, A. F. L., Wallabregue, A. L. D., Franz, L. & Hackenberger, C. P. R. Targeted subcellular protein delivery using cleavable cyclic cell-penetrating peptides. Bioconjug. Chem.30, 400–404 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Nieves, D., Gaus, K. & Baker, M. DNA-based super-resolution microscopy: DNA-PAINT. Genes (Basel)9, 621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Mela, I. et al. DNA nanostructures for targeted antimicrobial delivery. Angew. Chem. Int. Ed.59, 12698–12702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge, Z. et al. DNA origami-enabled engineering of ligand–drug conjugates for targeted drug delivery. Small16 (2020). [DOI] [PubMed]
- 15.Pal, S., Rakshit, T. & Folate-Functionalized, D. N. A. Origami for targeted delivery of doxorubicin to triple-negative breast cancer. Front. Chem. 9 (2021). [DOI] [PMC free article] [PubMed]
- 16.Zhang, Q. et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano8, 6633–6643 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Ma, W. et al. The biological applications of DNA nanomaterials: current challenges and future directions. Signal Transduct. Target. Therapy6 (2021). [DOI] [PMC free article] [PubMed]
- 18.Knappe, G. A., Wamhoff, E-C. & Bathe, M. Functionalizing DNA origami to investigate and interact with biological systems. Nat. Rev. Mater.8, 123–138 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daljit Singh, J. K., Luu, M. T., Abbas, A. & Wickham, S. F. J. Switchable DNA-origami nanostructures that respond to their environment and their applications. Biophys. Rev.10, 1283–1293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretzmann, J. A. et al. Gene-encoding DNA origami for mammalian cell expression. Nat. Commun.14, 1017 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin-Shiao, E. et al. CRISPR-Cas9-mediated nuclear transport and genomic integration of nanostructured genes in human primary cells. Nucleic Acids Res.50, 1256–1268 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, X. et al. Genetically encoded DNA origami for gene therapy in vivo. J. Am. Chem. Soc.145, 9343–9353 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Oh, C. Y. & Henderson, E. R. In vitro transcription of self-assembling DNA nanoparticles. Sci. Rep.13, 12961 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrasekaran, A. R. Nuclease resistance of DNA nanostructures. Nat. Rev. Chem.5, 225–239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh, C-Y. & Henderson, E. R. A comparison of methods for the production of kilobase-length single-stranded DNA. DNA2, 56–67 (2022). [Google Scholar]
- 26.Vu, H. T., Kaur, H., Kies, K. R., Starks, R. R. & Tuteja, G. Identifying novel regulators of placental development using time-series transcriptome data. Life Sci. Alliance6, e202201788 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res.37, 5001–5006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anindya, R. Single-stranded DNA damage: protecting the single-stranded DNA from chemical attack. DNA Repair (Amst). 87, 102804 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Berengut, J. F., Berg, W. R., Rizzuto, F. J. & Lee, L. K. Passivating blunt-ended helices to control monodispersity and multi‐subunit assembly of DNA origami structures. Small Struct.5, 2300441 (2024). [Google Scholar]
- 31.Yang, M., Bakker, D., Raghu, D. & Li, I. T. S. A single strand: a simplified approach to DNA origami. Front. Chem.11 (2023). [DOI] [PMC free article] [PubMed]
- 32.Baptist, A. V. & Heuer-Jungemann, A. Lyophilization reduces aggregation of three-dimensional DNA origami at high concentrations. ACS Omega8, 18225–18233 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagenbauer, K. F. et al. How we make DNA origami. ChemBioChem18, 1873–1885 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Liedl, A., Grießing, J., Kretzmann, J. A. & Dietz, H. Active nuclear import of mammalian cell-expressible DNA origami. J. Am. Chem. Soc.145, 4946–4950 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linko, V. & Keller, A. Stability of DNA Origami nanostructures in physiological media: the role of molecular interactions. Small19, 2301935 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Mathur, D., Galvan, A. R., Green, C. M., Liu, K. & Medintz, I. L. Uptake and stability of DNA nanostructures in cells: a cross-sectional overview of the current state of the art. Nanoscale15, 2516–2528 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephanopoulos, N. Strategies for stabilizing DNA nanostructures to Biological conditions. ChemBioChem20, 2191–2197 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Mikkilä, J. et al. Virus-encapsulated DNA origami nanostructures for cellular delivery. Nano Lett.14, 2196–2200 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Jiang, Q., Liu, S., Liu, J., Wang, Z. G. & Ding, B. Rationally designed DNA-Origami nanomaterials for drug delivery in vivo. Adv. Mater. 31 (2019). [DOI] [PubMed]
- 40.Xu, R., Li, Y., Zhu, C., Liu, D. & Yang, Y. R. Cellular ingestible DNA nanostructures for biomedical applications. Adv. Nanobiomed. Res.3, 2200119 (2023). [Google Scholar]
- 41.Udomprasert, A. & Kangsamaksin, T. DNA origami applications in cancer therapy. Cancer Sci.108, 1535–1543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas, C. R. et al. DNA Origami nanostructures elicit dose-dependent immunogenicity and are nontoxic up to high doses in vivo. Small 18(26), 2108063 (2022). [DOI] [PMC free article] [PubMed]
- 43.Agarwal, N. P., Matthies, M., Gür, F. N., Osada, K. & Schmidt, T. L. Block copolymer micellization as a protection strategy for DNA origami. Angew. Chem.129, 5552–5556 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Wang, S-T. et al. DNA origami protection and molecular interfacing through engineered sequence-defined peptoids. PNAS117, 6339–6348 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bastings, M. M. C. et al. Modulation of the cellular uptake of DNA origami through control over mass and shape. Nano Lett.18, 3557–3564 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Zanta, M. A., Belguise-Valladier, P. & Behr, J-P. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus (1999). [DOI] [PMC free article] [PubMed]
- 47.Bogacheva, M. et al. Arginine-rich cross-linking peptides with different SV40 nuclear localization signal content as vectors for intranuclear DNA delivery. Bioorg. Med. Chem. Lett.27, 4781–4785 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Vinogradov, S. V., Zhang, H., Mitin, A. & Warren, G. Intercalating conjugates of PEG with nuclear localization signal (NLS) peptide. Polym. Prepr. 49, 434–435 (2008). [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng, Y.C. et al. Fine tunning of CpG spatial distribution with DNA origami for improved cancer vaccination. Nature Nanotechnology 19, 1055-1065 (2024). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information file.