Abstract
Background
Metastatic urothelial carcinoma (mUC) has a poor prognosis, despite recent therapeutic advancements. Prostaglandin Reductase 1 (PTGR1) is essential for activating acylfulvens, a promising class of drugs for treating a subset of urothelial carcinoma (UC) patients. The efficacy of acylfulvens depends on PTGR1 activity and defects in the nucleotide excision repair (NER) pathway, notably ERCC2 mutations present in 10–15% of bladder tumors. This study identifies patients eligible to be included in acylfulven clinical trials based on the presence of PTGR-1 by immunohistochemistry (IHC) staining, RNA expression level and mutations in the NER pathway. Additionally, it evaluates PTGR-1 expression as a prognostic biomarker.
Methods
Three PTGR-1 antibodies were tested in a kidney cancer cell line A498 and in tissues with known PTGR1 expression (liver, tonsils, pancreas, small intestine, etc.). Patients with untreated mUC receiving 1st line platinum from Dec. 2019 to Dec. 2021 in the Capital Region, Denmark, were included retrospectively. FFPE tumor samples were collected, and tissue microarrays (TMAs) were constructed. TMAs were stained with the best-performing PTGR1 antibody, RNA expression was analyzed using Nanostring nCounter PanCancer panel and gene mutations were assessed using a targeted genomic panel (TSO-500). Kaplan-Meier and multivariate Cox regression assessed overall survival and covariate impacts.
Results
The AB181131 PTGR1 antibody was the most reliable in validation tissues. Tumors from 71 mUC patients were used to construct the TMA, and 40% of tumors scored positive for PTGR1 by IHC staining. A normalized PTGR1 RNA cutoff at 2,550 normalized counts achieved an AUC of 0.9, defining 35 samples as positive with a sensitivity of 96% and specificity of 85% in relation to IHC-positivity. Differential expression showed a significant upregulation of PTGR1 RNA in PTGR1 IHC-positive cases. NER-deficiency and PTGR1 positivity (mutations in ERCC1, ERCC2, ERCC3, ERCC4) was seen in 9 patients (13%). Median overall survival was 16 months in the cohort. Overall survival (OS) analysis indicates that overexpression of PTGR1 RNA was associated with a reduced median OS (12 months vs. 25 months, p = 0.039, log-rank).
Conclusion
PTGR1 IHC staining pattern using the Abcam AB181131 antibody is highly correlated with PTGR1 RNA expression. 13% in our cohort were identified as NER deficient and PTGR1 positive. Lower levels of PTGR1 indicates better outcome in this cohort.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79334-x.
Keywords: Bladder cancer, Urothelial cancer, Biomarker, Validation study, DNA repair deficiency, PTGR1
Subject terms: Cancer genomics, Bladder cancer
Introduction
Despite recent therapeutic advancements, metastatic urothelial carcinoma (mUC) continues to carry a poor prognosis, with a median overall survival rarely exceeding two years from diagnosis1–3. Emerging therapies, such as the combination of enfortumab vedotin and pembrolizumab4, hold promise, but the high rates of de novo or acquired resistance to this and other therapies, such as platinum combination regimens5, necessitate the development of additional therapeutic strategies. To address this unmet clinical need, research efforts are focused on identifying novel therapies and predictive biomarkers to optimize patient selection for existing and future treatment strategies6–8.
The enzyme Prostaglandin Reductase 1 (PTGR1) plays an important role in the intracellular activation of Acylfulvens, a shelved class of anti-cancer drugs that might have potential therapeutic applications in patients with urothelial carcinoma9–13. The activation process is contingent upon the metabolic conversion of acylfulvenes, including irofulven and LP-184, into their active drug forms14–16. The activation pathway involves the enzyme NADPH-dependent alkenyl/one oxidoreductase (AOR), for which PTGR1 is essential. Within this pathway, PTGR1 facilitates the reduction of the alkenyl or ketone moiety present in acylfulvenes, utilizing NADPH as a cofactor. This reduction changes the prodrug into its pharmacologically active form, capable of inducing DNA damage and interrupting DNA repair processes17. In addition, PTGR1 levels may also have prognostic implications independent of treatment, as upregulation of PTGR1 has been implicated with poorer outcomes in multiple cancer types14,18,19. It has been suggested that cancer cells’ ability to protect themselves against free radical oxides by upregulating AOR (and, by extension, PTGR1) can improve survival in oxygen-deprived tumor environments18.
When PTGR1 is absent within cells, the active drug (e.g., Irofulven or LP-184)20 cannot undergo the necessary biochemical conversion to its active state, resulting in therapeutic resistance and reduced efficacy15. Thus, the presence of PTGR1 is a key determinant of the cancer cells’ susceptibility to irofulven and similar compounds. However, the critical role of PTGR1 in Acylfulven activation has not been considered in previous clinical trials, perhaps explaining the modest activity reported10,12,21.
In addition to conversion by PTGR1 to its active form, tumor cell sensitivity to irofulven depends upon the loss of nucleotide excision repair (NER) pathway function. In urothelial carcinoma (UC), NER dysfunction is primarily driven by inactivating mutations in the NER gene ERCC2 and, less commonly, by mutations in other NER genes (e.g., ERCC3)11,22. NER deficiency is associated with increased responsiveness to Irofulven/LP-184.20,23–25 UC exhibits one of the highest frequencies of ERCC2 mutations across tumor types, ranging from 12 to 15%, so Irofulven/Lp-184 has emerged as an attractive therapeutic strategy9,24–28.
To identify patients who could benefit from Acylfulven treatment, the cytoplasmic presence of PTGR1 protein and NER deficiency must be established. Techniques for PTGR1 protein assessment, such as immunoblotting, RNA-sequencing, and proteomics, are available but can be cumbersome and have the potential for inter-laboratory discrepancies. Immunohistochemistry (IHC) is a practical, easily reproducible, cost-effective option poised for clinical implementation using commercially available antibodies. However, the optimal antibody for the clinical application of PTGR1 IHC has yet to be identified.
Here, we validated an IHC antibody for identifying PTGR1-positive patients with urothelial carcinoma. Additionally, we incorporated a targeted sequencing panel to evaluate mutations in the tumor tissue’s nucleotide excision repair (NER) pathway. This dual approach can potentially identify patients who may benefit from treatment with Acylfulven compounds.
Methods
Patient selection
This retrospective study included patients treated for mUC within The Capital Region of Denmark from January 2021 through December 2022. Inclusion criteria included patients who had not received previous platinum therapy, had available tissue from the point of diagnosis and completed at least one cycle of platinum-based chemotherapy following the biopsy. Metadata and clinical annotations were retrieved from electronic patient records. All tissues were from the primary tumor.
Antibody selection for PTGR1 IHC
Antibodies tested included Sigma HPA036724, Abcam AB181131, and Abcam AB222818. The antibodies were selected based on publications, availability, and performance in immunoblots20. These commercially available antibodies were used to characterize PTGR1 expression in tissues and cell lines where the presence of PTGR1 was known or expected, specifically in the liver, tonsils, pancreas, smooth muscle, and small intestines. The kidney cancer cell line A498 (cell cultivation protocol in Appendix 1, source of cell line A498: National Center for Cancer Immune Therapy (CCIT-DK)), is known to have sensitivity to Irofulven and immunoblot, and RNA-seq has established the presence of PTGR116,20. A498 is included in the NCI-60 panel. The staining protocol for PTGR1 is described in detail in Appendix 2, along with detailed information about the selected antibodies. The antibody staining intensity was confirmed through negative and positive controls using known expression patterns of PTGR1 in the assessed tissues and cell line. The best-performing antibody was selected for staining of tissue microarrays (TMAs).
TMA-preparation
Formalin-fixed, paraffin-embedded (FFPE) tumor samples from each patient were retrieved for evaluation. Samples deemed too sparse or of inadequate quality were excluded from further analysis. This assessment was conducted using Hematoxylin & Eosin (HE) staining of full-slide sections. For each qualifying FFPE block, punch core biopsies were extracted from areas marked as tumor tissue and re-embedded into new FFPE blocks to construct TMAs. Two punch core biopsies of 2 mm diameter were obtained from each patient’s tumor. These biopsies were randomly allocated within the TMAs. Liver, pancreas, tonsil, small intestine, lung, and A498 punch-cores were incorporated in the TMAs as control tissue. TMAs were cut into 2-micron thick sections and stained with H&E and PTGR1 antibody using DAKO Autostainer (staining protocol in Appendix 2). All slides were scanned using Hamamatsu C13220 at 40x magnification and 230 nm/pixel at the Department of Pathology, Copenhagen University Hospital Rigshospitalet.
PTGR1 IHC assessment
The cytoplasmatic PTGR1 staining intensity was scored semiquantitatively: 0 for negative cytoplasmic immunoreactivity, 1 for weak positive cytoplasmic immunoreactivity, 2 for moderate positive cytoplasmic immunoreactivity, and 3 for strong positive cytoplasmic immunoreactivity. Three medical doctors, including two urological pathologists, independently conducted this evaluation in a blinded fashion.
The inter-rater reliability between the three assessors’ scores was calculated using Fleiss’s kappa coefficient. Additionally, Cohen’s kappa was calculated between matched cores across patients to determine consistency between the two cores for each patient and assessor, estimating intra-rater reliability, assuming the two cores should stain similarly.
In instances where tissue samples in the TMAs were compromised due to missing sections or the presence of artifacts, only the unaffected cores were considered for analysis. The mean scores of the cores were calculated across assessors and individual cores. In cases with poor agreement between the cores or between the assessors, the individual cores were re-examined to minimize the risk of human error.
Targeted RNA-expression
DNA and RNA were extracted from tumor samples preserved in RNA later using the AllPrep DNA/RNA/protein Extraction Kit (Qiagen), according to the manufacturer’s instructions. Nanostring nCounter29 Quantification was performed using a customized PanCancer® panel of 778 genes on FFPE samples (gene list in Appendix 3). Based on the pathological evaluation of matched HE-stained sections with a minimum of 30% tumor tissue, RNA isolation from 10 μm FFPE sections was performed using High Pure FFPET RNA Isolation kit (Roche, Basel, Schweiz) and Nanodrop and qRT-PCR were used to assess RNA quality.
For quality control (QC) of RNA-expression data, we applied the Nanostring nSolver software and Bioconductor packages Nanotube v. 1.8.0 for normalization with background correction using background thresholding of two standard deviations above the mean of negative controls and 40 housekeeping genes which are known or suspected to show minimal variability in expression, six internal positive controls, and eight negative internal controls (Appendix 3). QC was performed according to Nanostring’s “Gene Expression Data Analysis Guidelines” before and after normalization30. After pre- and post-normalization QC, 729 genes (excluding housekeeping genes) were included for further analysis. Differential expression (DE) analysis between IHC positive and IHC negative cases was performed using the robust empirical Bayes hyperparameter estimation method (Limma)31.
Targeted genomic panel
We used the Illumina TruSight Oncology (TSO500) panel, where output criteria included an average sequencing depth of at least 300x, a variant allele frequency (VAF) threshold of at least 5%, and a minimum of 10 reads. Sequence alignment against the human reference genome (hg38/GRCh38) was performed using Sentieon-genomics (version 202112) BWA-mem counterpart. Somatic mutations were reported through tumor/panel of normals joint analysis, functionally subtracting suspected germline variants from tumor variants using Strelka2 (version 2.7). Variants with allelic frequencies above 1% in the general population (GNOMAD, Excac) were excluded. Annotation of variants was performed using VEP v11032 and OncoKB API service33; pathogenicity were also examined through computational prediction software CancerVar (OPAI)34, SIFT, MetaSVM, Polyphen2, MetaLR, (accessible at https://cancervar.wglab.org/). Any ERCC2 missense mutation within either helicase domain was considered pathogenic24.
Statistical analysis
Differences in clinical variables between PTGR1 IHC positive and negative were summarized using descriptive statistics. Continuous variables were compared using a Welch T-test, and discrete variables were compared using the Fisher exact test.
To identify the cutoff value of the RNA-expression data, a range of potential cutoff values was generated to determine the optimal cutoff for PTGR1. Sensitivity and specificity for each cutoff were calculated to assess the performance at each threshold in distinguishing between PTGR1 IHC positive and negative cases. This involved comparing the predicted classification based on the cutoff to actual classifications derived from IHC scores. The optimal cutoff of normalized RNA count was identified using Youden’s index, which maximizes sensitivity and specificity35,36.
The Receiver Operating Characteristic (ROC) curve was plotted, illustrating the trade-off between sensitivity and specificity across the range of possible cutoffs. The area under the curve (AUC) was calculated to quantify the overall ability of the PTGR1 expression levels to classify samples according to the optimal cutoff. Overall survival was assessed using the Kaplan-Meyer and log-rank test to discern the differences between selected strata, time period included was defined from start of first-line treatment until death or censoring. The Cox proportional hazards model was used to assess the effect of covariates on overall survival, ensuring that variable inclusion adhered to a minimum of 10 events per variable. The proportional hazards assumption was evaluated using the Schoenfeld residuals test, which confirmed that the assumption was satisfied.
For differential expression (DE) analysis, a p-value < 0.0001 was considered statistically significant; for all other analyses, a p-value < 0.05 was considered statistically significant. R v. 4.3.2 38, Nanotube v. 1.8.039, pracma v 2.4.440 packages were applied for statistical analysis.
Ethical considerations
The study was approved by the Danish Regional Data Protection Agency (j.no.: P-2022-33) and the Regional Ethics Committees (file number H-22000555), under which the need for informed consent could be waived. We have adhered to the declaration of Helsinki40.
Results
PTGR1 immunohistochemistry
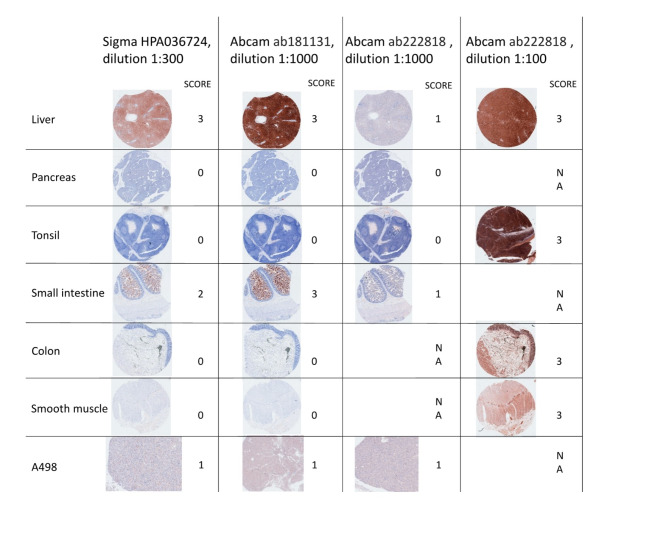
We first tested the performance of three commercially available PTGR1 antibodies using a panel of control tissues with known PTGR1 expression patterns. Each of the three antibodies was used at 1:1000 dilution. Abcam AB181131 performed best by the evaluation of two pathologists among the three antibodies (Fig. 1), with a clear staining pattern in validation tissues. The liver and small intestine stained strongly as expected (both tissues have high levels of PTGR1 RNA expression), and moderate staining was observed in the A498 cell line, which has also been shown to express PTGR1 at the RNA level robustly. Conversely, the pancreas, tonsil, and smooth muscle stained negative with all antibodies except AB222818 in dilution 1:100 (Fig. 1).
Fig. 1.
Antibody selection and validation in known tissues. Three antibodies were tested, ab222818 at two different concentrations, as the manufacturers recommended a dilution of 1:100, which stained all tissues positive. Scoring is done by cytoplasmatic coloring and intensity, from 0 (no staining) to 3 (full-house).
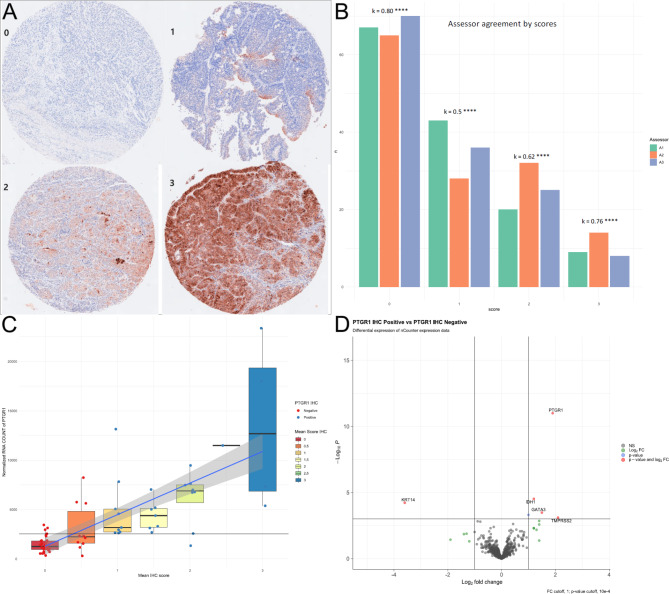
Given that it produced the most robust and reliable staining patterns across control tissues, the AB181131 antibody was used to stain the bladder tumor TMAs, which included 142 cores from 71 mUC patients. A positive score was defined as a mean score of one or above, showing the presence of PTGR1 protein in tumor tissue, and a negative was defined as a mean score below one. There was a strong correlation between the scores of the two cores from each patient, with Cohen’s kappa ranging between 0.68 and 0.8 for the three assessors (Table S1). Fleiss’ Kappa, which compares agreement across assessors, was approximately 0.7 overall for the three assessors, with the greatest agreement on negative (k = 0.8) and highly positive scores (k = 0.76) (Fig. 2A and B).
Fig. 2.
A: IHC scoring samples 0–3 of PTGR1 Abcam ab181131 from the UC cohort. B: Assessors scores and comparison of each core, and Fleiss Kappa across assessors, **** indicate p < 0.0001. C: Boxplot of IHC score, mean scores of assessors, plotted against normalized PTGR1 count. The blue line shows a linear regression line, with the grey area representing a 95% confidence interval. D: Differential expression between PTGR1 IHC positive (score > = 1) and negative (< 1).
Clinical characteristics and PTGR1 staining
Clinical characteristics of the mUC patient cohort are summarized in Table 1. Seventy-one patients were included in the study, 22 (31%) female and 49 (69%) male. The median age was 72.7 years (range 48.7 to 86.8). The primary tumor location was the bladder in 54 patients (76.1%), renal pelvis or ureter for 16 patients (22.5%), and urethra for 1 patient (1.4%). The majority of tumors had predominant (> 50%) urothelial carcinoma histology (68 patients, 95.8%), and 40 (56.3%) tumors were PTGR1 IHC negative, while 28 (39.4%) were PTGR1 IHC positive. No significant differences were observed in the demographic or clinical characteristics of PTGR1 IHC positive versus negative patients.
Table 1.
Showing clinical characteristics and PTGR1 IHC state. Ns = not significant, **** = p-value < 0.0001. #Two patients started immunotherapy as first-line treatment; one terminated treatment after 2 cycles due to toxicity, and the other after 3 cycles due to disease progression. Both patients received Carboplatin directly afterward and were therefore included in this cohort. Age is defined at baseline 1st line treatment.
| All | PTGR1.IHC.NEG | PTGR1.IHC.POS | p-value | |
|---|---|---|---|---|
| Gender | ||||
| Female | 22 (31%) | 14 (19.7%) | 8 (11.3%) | ns |
| Male | 49 (69%) | 28 (39.4%) | 21 (29.6%) | |
| Age in years – median (range) |
72.7 (48.7–86.8) |
72.6 (48.7–86.8) |
73.2 (49.6–82.2) |
ns |
| Primary tumor location | ||||
| Bladder | 54 (76.1%) | 30 (42.3%) | 24 (33.8%) | ns |
| Renal pelvis or ureter | 16 (22.5%) | 11 (15.5%) | 5 (7.0%) | |
| Urethra | 1 (1.4%) | 1 (1.4%) | 0 (0.0%) | |
| Histology | ||||
| Sarcomatoid differentiation | 1 (1.4%) | 0 (0.0%) | 1 (1.4%) | ns |
| Squamous cell differentiation | 2 (2.8%) | 2 (2.8%) | 0 (0.0%) | |
| Urothelial carcinoma (predominantly) | 68 (95.8%) | 40 (56.3%) | 28 (39.4%) | |
| Smoke | ||||
| Current Smoker | 25 (35.2%) | 13 (18.3%) | 12 (16.9%) | ns |
| Former Smoker (> 7 days cessation) | 34 (47.9%) | 21 (29.6%) | 13 (18.3%) | |
| Never Smoker | 12 (16.9%) | 8 (11.3%) | 4 (5.6%) | |
| ECOG PS | ||||
| 0 | 39 (54.9%) | 21 (29.6%) | 18 (25.4%) | ns |
| 1 | 28 (39.4%) | 18 (25.4%) | 10 (14.1%) | |
| 2 | 4 (5.6%) | 3 (4.2%) | 1 (1.4%) | |
| First line treatment after biopsy # | ||||
| Atezolizumab | 1 (1.4%) | 0 (0.0%) | 1 (1.4%) | ns |
| Carboplatin based | 34 (47.9%) | 19 (26.8%) | 15 (21.1%) | |
| Cisplatin based | 27 (38.0%) | 16 (22.5%) | 11 (15.5%) | |
| Cisplatin/Carboplatin based (platin shift) | 8 (11.3%) | 6 (8.5%) | 2 (2.8%) | |
| Pembrolizumab | 1 (1.4%) | 1 (1.4%) | 0 (0.0%) | |
|
PTGR-1 RNA count (optimized by IHC positivity) |
||||
| PTGR1 count high | 34 (47.9%) | 6 (8.5%) | 28 (39.4%) | **** |
| PTGR1 count low | 37 (52.1%) | 36 (50.7%) | 1 (1.4%) |
Correlation between PTGR1 staining and transcript levels
There was a strong association between PTGR1 IHC status and PTGR1 RNA count, with an optimal RNA cutoff of 2550 counts per million (CPM) in the normalized RNA count dataset to discriminate PTGR1 IHC positive vs. negative cases. This cutoff provided a sensitivity of 0.96, a specificity of 0.85, and an area under the curve (AUC) of 0.9 ( S1). An agreement was observed in 64 out of 71 cases (90%) (p = 0.0001, Fisher exact test) (Bottom of Table 1; Fig. 2C).
Differential expression analysis showed that PTGR1 IHC-positive cases exhibited nearly a 2 log-fold increase in PTGR1 RNA levels compared to PTGR1 IHC-negative cases (log2fc = 1.9, p = 10− 11). Additionally, KRT14, a basal marker, was significantly down-regulated (log2fc = -3.6, p = 6 × 10− 5), whereas GATA3, a luminal marker, was significantly up-regulated (log2fc = 1.5, p = 3.3 × 10− 4) in the PTGR1 IHC-positive cases (Fig. 2D).
PTGR1 IHC staining and NER gene mutations
Targeted DNA sequencing data (TSO-500) were available for 70 patients (one of the 71 patients in the cohort had insufficient tissue for DNA analysis). ERCC2 mutations were present in 10 tumors, including helicase domain missense mutations (n = 6), frameshift (n = 2), and splice-site (n = 2) (Fig. 3A and B). All variants were processed in OncoKB and CancerVar for computational pathogenicity prediction (Table S3). Most algorithms considered all the variants pathogenic. All of the missense mutations in ERCC2 were within the helicase domains and were therefore considered pathogenic based on previous functional work24 (Fig. 3B); however, the functional implications of the ERCC2 frameshift and splice-site mutations are less clear. The distribution of ERCC2 mutations among PTGR1 scores was even, with 5 ERCC2-mutant patients PTGR1-positive and 5 patients negative (Fig. 3B).
Fig. 3.
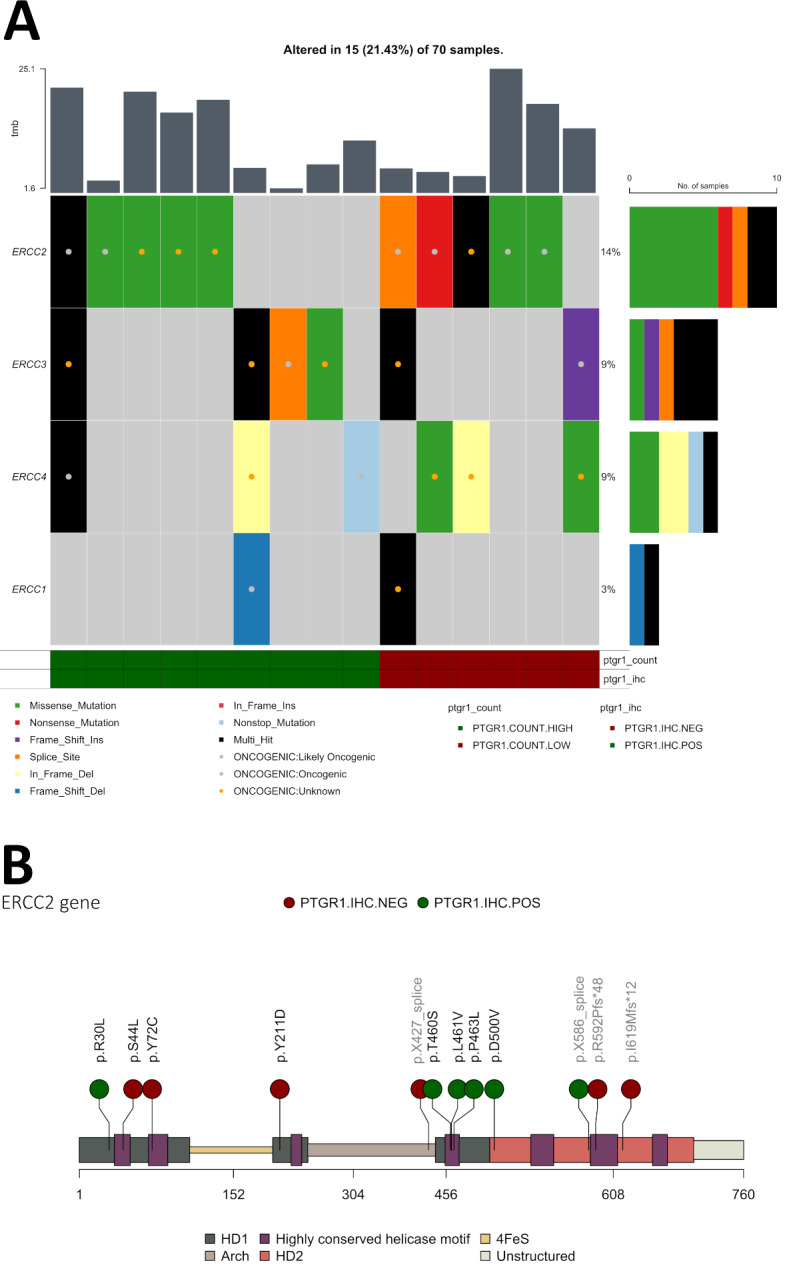
A: Oncoplot showing ERCC1-4, non-mutated cases are filtered out, of the 70 patients, sorted by PTGR1 positive and PTGR1 negative (by both IHC and RNA expression). Variants known pathogenicity by OncoKB is marked with a colored dot. B: mapping the mutations of ERCC2 along the gene, depicting the helicases and genomic structures. Missense mutations in the helicase domain are considered pathogenic, all missense mutations are within the helicase domains. Splice site and frameshift mutations are noted in light-grey text. Each dot is colored by PTGR1 IHC status.
The less common NER genes included in the TSO-500 panel with oncogenic or likely oncogenic mutations (according to OncoKB) included ERCC1 (1 patient), ERCC3 (2 patients), ERCC4 (2 patients), and ERCC5 (only variants of unknown significance, not considered pathogenic, therefore not included in the figure) (Fig. 3A). Four of these five cases were PTGR1 IHC positive (Fig. 3A). In total, nine patients (13%) have a pathogenic NER mutation and PTGR1 positivity. Additionally, we found no differentially mutated genes between the PTGR1 expression groups (data not shown).
Overall survival (OS) analysis showed a significant difference between the PTGR1 RNA count high and low groups in the cohort, with a median survival of 12.2 and 25.3 months (p = 0.038, log-rank), respectively (Figure S2), and we observed 51 events in the follow-up period. The median OS in the events group (n = 51) was 11.6 months (IQR 7.6–16.7 months), and the median follow-up for those without events (n = 20) was 28 months (IQR 23.3–39 months). In multivariate Cox regression analysis, ECOG PS, age, choice of first-line treatment, and PTGR1 status were significantly associated with survival, and PTGR1-high had an HR of 2.68 (95% CI 1.48.-4.85, Cox PH) (Table S4). A similar multivariate Cox regression analysis performed for PTGR1 IHC positive vs. negative cases did show a non-significant difference (HR 1.75 (95% CI 0.97–3.18, Cox PH) (Table S5).
Discussion
In this validation study, the Abcam AB181131 antibody demonstrated robust performance in identifying PTGR1 expression within urothelial carcinoma samples, identifying a potentially useful biomarker to identify mUC cases that may be more sensitive to acylfulvene treatment. For this cohort of mUC patients (n = 71), 41% exhibited PTGR1 positivity by IHC, aligning with RNA expression data.
Notably, the AB181131 antibody, validated for Western blot41, outperformed the two manufacturer-validated antibodies in IHC, which displayed poorer staining patterns. The Sigma HPA036724 and Abcam AB222818 antibodies yielded suboptimal staining in validation tissues, excluding them from application to the patient TMAs.
The IHC positivity cutoff was determined by the staining patterns observed in positive normal tissues (liver, small intestine epithelial lining) and the Irofulvene-sensitive A498 kidney cancer cell line, known for PTGR1 expression20. The inter-observer agreement among two genitourinary pathologists and a trained medical doctor demonstrated good to excellent Kappa values across all staining intensities, particularly distinguishing negative (score 0) from positive values (score 1–3). This aligns with published standards for antibody selection42–45.
The clinical characteristics of our cohort are similar to those reported in the literature for first-line settings3. Interestingly, PTGR1 positivity was evenly distributed, showing no association with the examined clinical parameters. Thus, PTGR1 testing is necessary for patient selection, as clinical features are inadequate for patient identification given its distribution. PTGR1 RNA expression strongly correlated with IHC staining intensity, substantiating the relationship between mRNA and protein expression. We compared the PTGR1 IHC stain of the TMAs and the A498 cell line, known to be Irofulven sensitive, thus more confidently setting the IHC cutoff at a clinically relevant threshold.
Recognizing that response to acylfulvens requires PTGR1 expression and NER deficiency, we examined our cohort for mutations in key NER genes. ERCC2, known for its frequent mutation in UC (10–15%)11, has been shown, along with ERCC3, to drive Irofulven and LP-184 sensitivity in kidney20, pancreatic16, and UC cell lines11. We found a prevalence of 14% ERCC2 mutations in our cohort, and we analyzed each variant type through multiple deleterious prediction software, reducing the risk of including or excluding important variants. However, the accuracy of prediction software is debated, and the functional impact of a genetic variant on the cellular phenotype is best assessed through cellular studies utilizing, for instance, CRISPR-Select46. Regardless, we included all variants deemed pathogenic by prediction software34 or OncoKB33 and if the missense mutation was located in either helicase domain of ERCC2 (Fig. 3B). The phenotypic impact of splice site or frameshift mutation is unclear24. ERCC2 mutations occurred with the same frequency in PTGR1-positive and PTGR1-negative cases, reducing the number of patients accessible for acylfulvene treatment. In our cohort, where mutational data was available for 70 patients, five had pathogenic ERCC2 mutation and PTGR1 presence (7%), underlining the high number of patients to be screened for acylfulvene trials. Incorporating a broader selection of NER genes, four additional patients are NER-deficient and PTGR1 positive, totaling 13% of the cohort.
In clinical practice, utilizing IHC for patient stratification offers a cost-effective and widely available alternative, which can facilitate further genomic analysis. This approach proves valuable for investigator-initiated trials with limited funding. Other biomarker studies, for instance, for FGFR3 mutations47, applied more advanced and expensive methods, such as polymerase chain reaction amplification (PCR), direct DNA sequencing, or next-generation sequencing (NGS)7,48–50. Due to NGS’s cost, data processing demands, and long turnaround time, which can delay treatment initiation, IHC is often preferred for initial patient screening, particularly when reliable IHC markers exist51.
PTGR1’s role in mitigating oxidative stress has been suggested as a tumorigenic protective mechanism, where cancer cells upregulate PTGR1 in hypoxic environments18,52. High PTGR1 RNA expression has been linked to decreased survival in some mUC studies14,19. To investigate this association, we examined the relationship between PTGR1 RNA expression, utilizing an IHC-based cutoff, and survival in our cohort. Our findings confirm previous reports, demonstrating that patients classified as PTGR1 RNA high exhibited poorer survival compared to those with PTGR1 RNA low (mOS 12 vs. 25 months). This significant difference was not found for PTGR1 IHC positive vs. negative, probably due to the discordance between RNA and IHC in seven patients. While PTGR1 as a prognostic clinical biomarker currently appears unfeasible, our results warrant further investigation and validation.
From the differential expression analysis, the PTGR1 IHC positive cases had, in addition to upregulation of PTGR1, significant upregulation of GATA3 and downregulation of KRT14, linked to the Luminal molecular subtype53–55. Interestingly, the luminal molecular subtype is characterized by a better prognosis in muscle-invasive UC, contrasting our finding of high PTGR1 being linked to worse survival. This association is still unclear and might be linked to the consensus subtyping system being constructed for muscle-invasive UC. Also, as we used a targeted RNA panel where only a few percent of genes in the RNA subtype panel overlapped, we could not confidently define the molecular subtypes from the consensus-defined centroids53.
While efforts were made to mitigate bias and limitations, we acknowledge discrepancies between IHC-based PTGR1 staining and RNA-based expression. These inconsistencies, though minor, likely arise from variations in tissue fixation, antigen retrieval, antibody-epitope interactions, or antibody limitations. Resource limitations necessitated a pathologist-led screening of commercially available PTGR1 antibodies. Hence, there might be superior, untested alternatives.
HE-stained TMAs were examined to minimize sampling bias, but the risk of reduced tumor representation on PTGR1 slides persists due to technical factors or inherent tumor heterogeneity. Intratumoral heterogeneity can further confound comparisons between RNA expression and IHC-based protein detection56,57. However, using the same FFPE sample for RNA, DNA, and TMAs likely limits this effect. The limited patient cohort may hinder the detection of statistically significant associations and potentially overestimate biomarker performance. In addition, our cohort is exclusively urothelial carcinoma, limiting interpretability for other histological tumor types.
Despite these limitations, this study supports the feasibility of Abcam AB181131 for immunohistochemical screening of patients with UC in Acylfulven trials without requiring RNA-expression validation. Future studies will explore the association between PTGR1, molecular subtypes, and survival.
Conclusion
PTGR1 IHC is a viable biomarker that may be useful for stratifying metastatic urothelial carcinoma patients for acylfulvene trials. Abcam AB181131 was the best-performing antibody. A strong association between PTGR1 IHC and RNA expression was observed. Lower PTGR1 levels were associated with prolonged survival, further confirmation of this is paramount.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Arianna Draghi and the National Center for Cancer Immunotherapy (CCIT-DK) for supplying cell line A498.
Author contributions
D.R.S., H.P., B.G.T, and S.L. conceived and designed the study. D.R.S. and L.C.M. acquired the data. D.R.S., S.L., Z.S., K.M., B.G.T., and J.B. analyzed and interpreted the data. B.G.T., L.C.M., and H.P. provided administrative, technical, and material support. D.R.S. drafted the manuscript. D.R.S. and H.P. acquired the funding. D.R.S., S.L., K.M., Z.S., L.H.D, L.C.M., J.B., M.R., B.G.T., and H.P. critically revised the manuscript for important intellectual content. All authors approved the submission of the final version of the manuscript.
Funding
This study received independent unrestricted research grants from Pfizer, Merck Serono, Lis Kluwer, and Sejer Persson funds. The funding sources were not involved in the design and conduct of the study, management, analysis, or interpretation of the data, preparation, writing, or review of this manuscript, or in the decision to submit this manuscript for publication.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). RNA gene list in Appendix 3. Analysis for differential expression in Appendix 4. Data on NER genes are in Appendix 5. Normalized RNA counts and IHC scores are in Appendix 6. Computational prediction software is accessible at https://cancervar.wglab.org.
Declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
D.R.S. received educational and research funding from Pfizer and Merck Serono and served on advisory boards for Merck Sharp and Dome (MSD), Bristol Meyers Squibb (BMS), and Johnson and Johnson, unrelated to this study. S.L. No COI to report. B.G.T. Speaker fee from BMS unrelated to this study. L.H.D: No COI to report. L.C.M.: No COI to report. Z.S: Research funding from Lantern Pharma Inc; M.R: received personal fees from AstraZeneca and MSD and served on advisory board of MSD outside the submitted work.; H.P: Research funding from Pfizer and Merck unrelated to this study. K.W.M: Advisory/consulting work for EMD Serono, Pfizer, UroGen, and Riva Therapeutics; has received research support from Pfizer and Novo Ventures; has equity in Riva Therapeutics; has received writing/editor fees from UpToDate; has received speaking fees from OncLive; and is a co-inventor listed on an institutional patent application for analysis of mutational signatures of DNA repair deficiency.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reesink, D. J. et al. Real-world outcomes of first-line chemotherapy for unresectable stage III and IV bladder cancer. World J. Urol.41, 1551–1562 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Galsky, M. D. et al. Real-world effectiveness of Chemotherapy in Elderly patients with metastatic bladder Cancer in the United States. Bladder Cancer. 4, 227–238 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niegisch, G. et al. A real-world data study to evaluate treatment patterns, clinical characteristics and survival outcomes for first- and second-line treatment in locally advanced and metastatic urothelial cancer patients in Germany. J. Cancer. 9, 1337–1348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powles, T. et al. Enfortumab Vedotin and Pembrolizumab in Untreated Advanced Urothelial Cancer. N. Engl. J. Med.390, 875–888 (2024). [DOI] [PubMed] [Google Scholar]
- 5.van der Heijden, M. S. et al. Nivolumab plus Gemcitabine–Cisplatin in Advanced Urothelial Carcinoma. N. Engl. J. Med.389, 1778–1789 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Scholtes, M. P. et al. Biomarker-oriented therapy in bladder and renal Cancer. Int. J. Mol. Sci.22, 1–18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun, T., Anker, J. & Galsky, M. D. Biomarkers for therapy selection in metastatic urothelial cancer. J. Cancer Metastasis Treat. vol. 10.20517/2394-4722.2021.171 (2022). 8 Preprint at. [Google Scholar]
- 8.Eturi, A., Bhasin, A., Zarrabi, K. K. & Tester, W. J. Predictive and prognostic biomarkers and Tumor antigens for targeted therapy in Urothelial Carcinoma. Molecules29, (2024). [DOI] [PMC free article] [PubMed]
- 9.Jiang, H. & Greenberg, R. A. Morning for irofulven, what could be fiNER? Clin. Cancer Res.27, 1833–1835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandre, J. et al. Phase I and pharmacokinetic study of irofulven administered weekly or biweekly in advanced solid tumor patients. Clin. Cancer Res.10, 3377–3385 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Börcsök, J. et al. Identification of a synthetic lethal relationship between nucleotide excision repair deficiency and irofulven sensitivity in urothelial cancer. Clin. Cancer Res.27, 2011–2022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaspers, N. G. J. et al. Anti-tumour compounds illudin S and Irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription- and replication-coupled repair pathways. DNA Repair. (Amst). 1, 1027–1038 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Dent, P., Grant, S. & Irofulven Resurgence for alkylating therapy in cancer? Cancer Biol. Ther.3, 1143–1144 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Wang, X. et al. Prostaglandin Reductase 1 as a Potential Therapeutic Target for Cancer Therapy. Frontiers in Pharmacology vol. 12 Preprint at (2021). 10.3389/fphar.2021.717730 [DOI] [PMC free article] [PubMed]
- 15.Yu, X. et al. Up-regulation of human prostaglandin reductase 1 improves the efficacy of hydroxymethylacylfulvene, an antitumor chemotherapeutic agent. J. Pharmacol. Exp. Ther.343, 426–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Restifo, D. et al. Conditional dependency of LP-184 on prostaglandin reductase 1 is synthetic lethal in pancreatic cancers with DNA damage repair deficiencies. Mol. Cancer Ther.10.1158/1535-7163.MCT-22-0818 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woynarowski, J. M. et al. Cell cycle effects and induction of premitotic apoptosis by irofulven in synchronized cancer cells. Cancer Biol. Ther.3, 1137–1142 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Sánchez-Rodríguez, R. et al. Ptgr1 expression is regulated by NRF2 in rat hepatocarcinogenesis and promotes cell proliferation and resistance to oxidative stress. Free Radic Biol. Med.102, 87–99 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Tapak, L., Saidijam, M., Sadeghifar, M., Poorolajal, J. & Mahjub, H. Competing Risks Data Analysis with High-dimensional covariates: an application in bladder Cancer. Genomics Proteom. Bioinf.13, 169–176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prosz, A. et al. Nucleotide excision repair deficiency is a targetable therapeutic vulnerability in clear cell renal cell carcinoma. Scientific Reports 2023 13:1 13, 1–10 (2023). [DOI] [PMC free article] [PubMed]
- 21.Seiden, M. V. et al. A phase II study of irofulven in women with recurrent and heavily pretreated ovarian cancer. Gynecol. Oncol.101, 55–61 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Koeppel, F. et al. Irofulven cytotoxicity depends on transcription-coupled nucleotide excision repair and is correlated with XPG expression in solid tumor cells. Clin. Cancer Res.10, 5604–5613 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Topka, S. et al. Targeting germline- and tumor-associated nucleotide excision repair defects in cancer. Clin. Cancer Res.27, 1997–2010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Q. et al. ERCC2 helicase domain mutations Confer Nucleotide Excision Repair Deficiency and Drive Cisplatin Sensitivity in muscle-invasive bladder Cancer. Clin. Cancer Res.25, 977–988 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J. et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet.48, 600–606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Allen, E. M. et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 4, 1140–1153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, D. et al. Clinical validation of chemotherapy response biomarker ERCC2 in muscle-invasive urothelial bladder carcinoma. JAMA Oncology vol. 2 1094–1096 Preprint at (2016). 10.1001/jamaoncol.2016.1056 [DOI] [PMC free article] [PubMed]
- 28.Robertson, A. G. et al. Comprehensive molecular characterization of muscle-invasive bladder Cancer. Cell. 171, 540–556e25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren, S. & Simultaneous multiplexed detection of RNA and protein on the NanoString® nCounter® platform. in Methods in Molecular Biology vol. 1783 105–120 (2018). (Methods Mol Biol. [DOI] [PubMed]
- 30.NanoString Technologies Inc. MAN-C0011-04 Gene Expression Data Analysis Guidelines. (2017).
- 31.Phipson, B., Lee, S., Majewski, I. J., Alexander, W. S. & Smyth, G. K. Robust Hyperparameter Estimation Protects Against Hypervariable Genes and Improves Power To Detect Differential Expression. Ann. Appl. Stat.10, 946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaren, W. et al. The Ensembl variant effect predictor. Genome Biol.17, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakravarty, D. et al. OncoKB: a Precision Oncology Knowledge Base. JCO Precis Oncol.1, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, Q. et al. CancerVar: an artificial intelligence–empowered platform for clinical interpretation of somatic mutations in cancer. Sci. Adv.8, 1624 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruopp, M. D., Perkins, N. J., Whitcomb, B. W. & Schisterman, E. F. Youden Index and Optimal Cut-Point estimated from observations affected by a lower limit of detection. Biom J.50, 419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youden, W. J. Index for rating diagnostic tests. Cancer. 3, 32–35 (1950). [DOI] [PubMed] [Google Scholar]
- 37.R Core Team & R Core Team. (2020). Preprint at (2020). https://www.r-project.org/
- 38.Class, C. A., Lukan, C. J., Bristow, C. A. & Do, K. A. Easy NanoString nCounter data analysis with the NanoTube. Bioinformatics39, (2023). [DOI] [PMC free article] [PubMed]
- 39.Hans Borchers, M. W. Title Practical Numerical Math Functions Depends R ( > = 3.1.0). (2023).
- 40.World Medical Association declaration of Helsinki. Ethical principles for medical research involving human subjects. JAMA vol. 310 2191–2194 Preprint at (2013). 10.1001/jama.2013.281053 [DOI] [PubMed]
- 41.Prosz, A. et al. Nucleotide excision repair deficiency is a targetable therapeutic vulnerability in clear cell renal cell carcinoma. bioRxiv 11, 02.07.527498 (2023). (2023). [DOI] [PMC free article] [PubMed]
- 42.MacNeil, T. et al. Antibody validation for protein expression on tissue slides: a protocol for immunohistochemistry. Biotechniques. 69, 461–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McShane, L. M. et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst.97, 1180–1184 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Walker, R. A. Quantification of immunohistochemistry–issues concerning methods, utility and semiquantitative assessment I. Histopathology. 49, 406–410 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Thomsen, N. O. B., Olsen, L. H. & Nielsen, S. T. Kappa statistics in the assessment of observer variation: the significance of multiple observers classifying ankle fractures. J. Orthop. Sci.7, 163–166 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Niu, Y. et al. Multiparametric and accurate functional analysis of genetic sequence variants using CRISPR-Select. Nat. Genet.54, 1983–1993 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grantzau, T. et al. PD-L1 expression and FGFR-mutations among Danish patients diagnosed with metastatic urothelial carcinoma: a retrospective and descriptive study. APMIS. 130, 498–506 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powles, T. et al. An adaptive, biomarker-directed platform study of durvalumab in combination with targeted therapies in advanced urothelial cancer. Nat. Med.27, 793–801 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Crabb, S. J. et al. Double-Blind, Biomarker-Selected, phase II clinical trial of maintenance poly ADP-Ribose polymerase inhibition with Rucaparib Following Chemotherapy for Metastatic Urothelial Carcinoma. J. Clin. Oncol.41, 54–64 (2023). A Randomized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fulton, B. et al. ATLANTIS: a randomised multi-arm phase II biomarker-directed umbrella screening trial of maintenance targeted therapy after chemotherapy in patients with advanced or metastatic urothelial cancer. Trials21, (2020). [DOI] [PMC free article] [PubMed]
- 51.Sjödahl, G. Molecular Subtype Profiling of Urothelial Carcinoma Using a subtype-specific Immunohistochemistry Panelvol. 165553–64 (Humana Press Inc., 2018). in Methods in Molecular Biology. [DOI] [PubMed]
- 52.Ye, J. et al. Metformin escape in prostate cancer by activating the PTGR1 transcriptional program through a novel super-enhancer. Signal Transduction and Targeted Therapy 2023 8:1 8, 1–15 (2023). [DOI] [PMC free article] [PubMed]
- 53.Kamoun, A. et al. A Consensus Molecular classification of muscle-invasive bladder Cancer. Eur. Urol.77, 420–433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi, W. et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell.25, 152–165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dadhania, V. et al. Meta-analysis of the luminal and basal subtypes of bladder Cancer and the identification of signature immunohistochemical markers for clinical use. EBioMedicine. 12, 105–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavallee, E., Sfakianos, J. P. & Mulholland, D. J. Tumor heterogeneity and consequences for bladder cancer treatment. Cancers (Basel) 13, (2021). [DOI] [PMC free article] [PubMed]
- 57.Meeks, J. J. et al. Genomic heterogeneity in bladder cancer: challenges and possible solutions to improve outcomes. Nat. Rev. Urol.17, 259–270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). RNA gene list in Appendix 3. Analysis for differential expression in Appendix 4. Data on NER genes are in Appendix 5. Normalized RNA counts and IHC scores are in Appendix 6. Computational prediction software is accessible at https://cancervar.wglab.org.