Significance
Nuclear receptors (NRs) are important transcriptional regulators of homeostasis and metabolism. This study explores the effects of six activated NR in detail, specifically in the context of the human small intestine. By using organoids and pharmacological small molecules, NR were activated and characterized individually. By studying the consequences downstream of an activated nuclear receptor one by one, this work contributes to the understanding of NR in intestinal homeostasis in humans.
Keywords: nuclear receptors, small intestine, lipid absorption, organoids
Abstract
Nuclear receptors (NRs) are widely expressed transcription factors that bind small, lipophilic compounds and regulate diverse biological processes. In the small intestine, NRs are known to act as sensors that control transcriptional responses to endogenous and exogenous signals, yet their downstream effects have not been characterized extensively. Here, we investigate the activation of six different NRs individually in human intestinal organoids using small molecules agonists. We observe changes in key enterocyte functions such as lipid, glucose, and amino acid absorption, the regulation of electrolyte balance, and drug metabolism. Our findings reinforce PXR, LXR, FXR, and PPARα as regulators of lipid absorption. Furthermore, known hepatic effects of AHR and VDR activation were recapitulated in the human small intestine. Finally, we identify unique target genes for intestinal PXR activation (ERG28, TMEM97, and TM7SF2), LXR activation (RAB6B), and VDR activation (CA12). This study provides an unbiased and comprehensive transcriptomic description of individual NR pathways in the human small intestine. By gaining a deeper understanding of the effects of individual NRs, we might better harness their pharmacological and therapeutic potential.
Nuclear receptors (NRs) act as transcription factors, regulating the expression of a large number of genes in response to various intracellular and extracellular signals. The activity of NRs is regulated by binding of lipophilic ligands such as steroids, retinoids, lipids, bile acids, hormones, and xenobiotics. In the small intestine, such ligands derive from the body proper (e.g., endocrine signals) as well as from the intestinal lumen. Luminal ligands include dietary nutrients and their derivatives, bacterial metabolites, and drugs (1). NRs respond to these exogenous or endogenous signals by regulating genes required for the absorption and metabolism of their ligands. As such, the liver X receptor [LXRα and β (NR1H3/2, respectively)] and the peroxisome proliferator-activated receptor α [PPARα (NR1C1)] bind cholesterol and dietary lipid derivatives and are implicated in the handling of these molecules (2, 3). Farnesoid X receptor [FXR (NR1H4)] is involved in regulating bile acids through the enterohepatic cycle (4). The pregnane X receptor [PXR (NR1I2)] and Aryl hydrocarbon receptor (AHR) are activated by xenobiotics and bacterial metabolites and induce enzymes required for their metabolism (5, 6). The Vitamin D receptor [VDR (NR1I1)] is activated by calcitriol, the active form of vitamin D, and is an important regulator of calcium and phosphate homeostasis (7).
NRs have been extensively studied in the kidney and liver but are increasingly appreciated for their role in intestinal physiology and disease. The intestinal epithelium is maintained by highly proliferative LRG5+ stem cells, located at the bottom of the crypts of Lieberkühn. Their daughter cells migrate along the crypt–villus axis while differentiating into functional cell types that fall into two major lineages (8): the secretory lineage, which includes Paneth, tuft, goblet, and enteroendocrine cells, and the absorptive lineage, which consists solely of enterocytes and constitutes 80 to 90% of the villus epithelium. Since enterocytes are the most abundant cell type of the intestinal epithelium and play a crucial role in overall intestinal function, the current study focused on the effects of intestinal NR activation in this cell type. Within the intestinal epithelium, NRs are known to regulate the absorption and metabolism of lipids and bile acids (9, 10). Furthermore, NRs are thought to play an important role in xenobiotic metabolism, mucosal immunity, tolerance to the microbiota, and in maintaining the intestinal barrier (5, 11, 12). Aberrant NR signaling is implicated in many digestive tract diseases such as inflammatory bowel disease, metabolic dysfunction-associated steatotic liver disease and hepatocellular carcinoma (FXR), colorectal cancer (FXR, AHR), and metabolic syndrome (PPARα, LXR, and FXR) (13–17). While NRs have been extensively studied in specific disease settings and especially in the kidney and liver, their role in the small intestinal epithelium is less well understood.
Direct analysis of the effects of individual NR in the human intestine has proven challenging for several reasons. NRs are expressed in almost every tissue and are known to regulate interactions between tissues (e.g., regulation of the enterohepatic circulation by FXR) (18, 19). Studying the direct effects of NRs on the intestinal epithelium is confounded in vivo by NR-mediated effects from other organs and tissues. Furthermore, transcriptional changes downstream of NRs can be tissue-specific and are thus poorly generalizable from a single in vitro model (20, 21). Therefore, suitable reductionist intestinal models are needed to better understand intestinal-specific NR signaling in humans. Recently, tissue-specific knockouts have yielded more insights into NR biology, but these remain restricted to animal models. Furthermore, a large part of our understanding of NRs in human intestine comes from the use of malignant cells not originating from the small intestine, such as the colon cancer cell line Caco-2. Cell lines are also limited in their differentiation and often only poorly express NRs and their target genes (22). Studies with human subjects are limited by the fact that—when applying dietary intervention—diet components or their metabolites are recognized by multiple different NRs. This simultaneous activation could obscure effects and prevent their association with an individual NR pathway. Furthermore, some NR ligands engage membrane receptors, such as the free fatty acids receptor and the bile acid receptor GPBAR1, leading to further complexity (23). In summary, investigating the effects of a specific NR pathway in isolation has proven to be challenging in humans and has prevented an in-depth understanding of distinct NR signaling events in the human intestine.
Human intestinal organoids provide a reductionist model that enables in-depth investigation of individual NR without systemic interference. Organoids recapitulate functional characteristics of the in vivo organ and allow for indefinite expansion of healthy human cells in vitro. Previously, mouse-derived organoids have been applied to study NR signaling and were shown to more closely resemble NRs expression of the tissue as opposed to cell lines (24). Thus, organoids enable the evaluation of NR effects directly in healthy human intestinal cells. NRs are prominent drug targets—accounting for roughly 10 to 20% of all approved FDA drugs—because they are important transcription factors that can be readily modulated with lipophilic small molecules (25). This has resulted in the development of pharmacological small molecules that bind NRs with high specificity and potency, making it possible to potently modulate individual NRs (26–31). Furthermore, these small molecules can readily enter cells, irrespective of cell polarity, as they are taken up mainly through passive diffusion (32). Here, we specifically activate individual NRs with small molecules in adult stem cell-derived human small intestinal organoids, to characterize the role of six NRs within enterocytes of the healthy human small intestine.
Results
Enterocyte Differentiation Is not Affected by Nuclear Receptor Agonists.
Human intestinal organoids allow for near-indefinite expansion of healthy human cells in vitro and—combined with small molecule NR agonists—enable the evaluation of individual NRs in healthy human intestinal cells. A switch from organoid expansion medium—mimicking the niche signals in the crypt bottom—to differentiation medium, results in organoids containing the various differentiated cell types within a week. Absorptive enterocytes represent the most abundant cell type in these conditions (33, 34). We differentiated organoids in the presence of BMP to deplete stem and progenitor cells and to enrich for terminally differentiated, villus tip enterocytes (35). To study the impact of activation of individual NRs, we added small molecules to activate one of six NRs at the initiation of enterocyte differentiation (Fig. 1A). Addition of an NR agonist did not greatly affect organoid morphology (Fig. 1B). Differentiation reduced expression of the stem cell marker OLFM4 (36), while inducing BMP target genes ID1, ID2, and ID3 and generic enterocyte markers FABP1, VIL1, ALPI, and SI (Fig. 1C). These differentiation effects were not altered by addition of NR agonist, demonstrating that the modulation of nuclear receptor pathways did not affect enterocyte lineage allocation and differentiation.
Fig. 1.
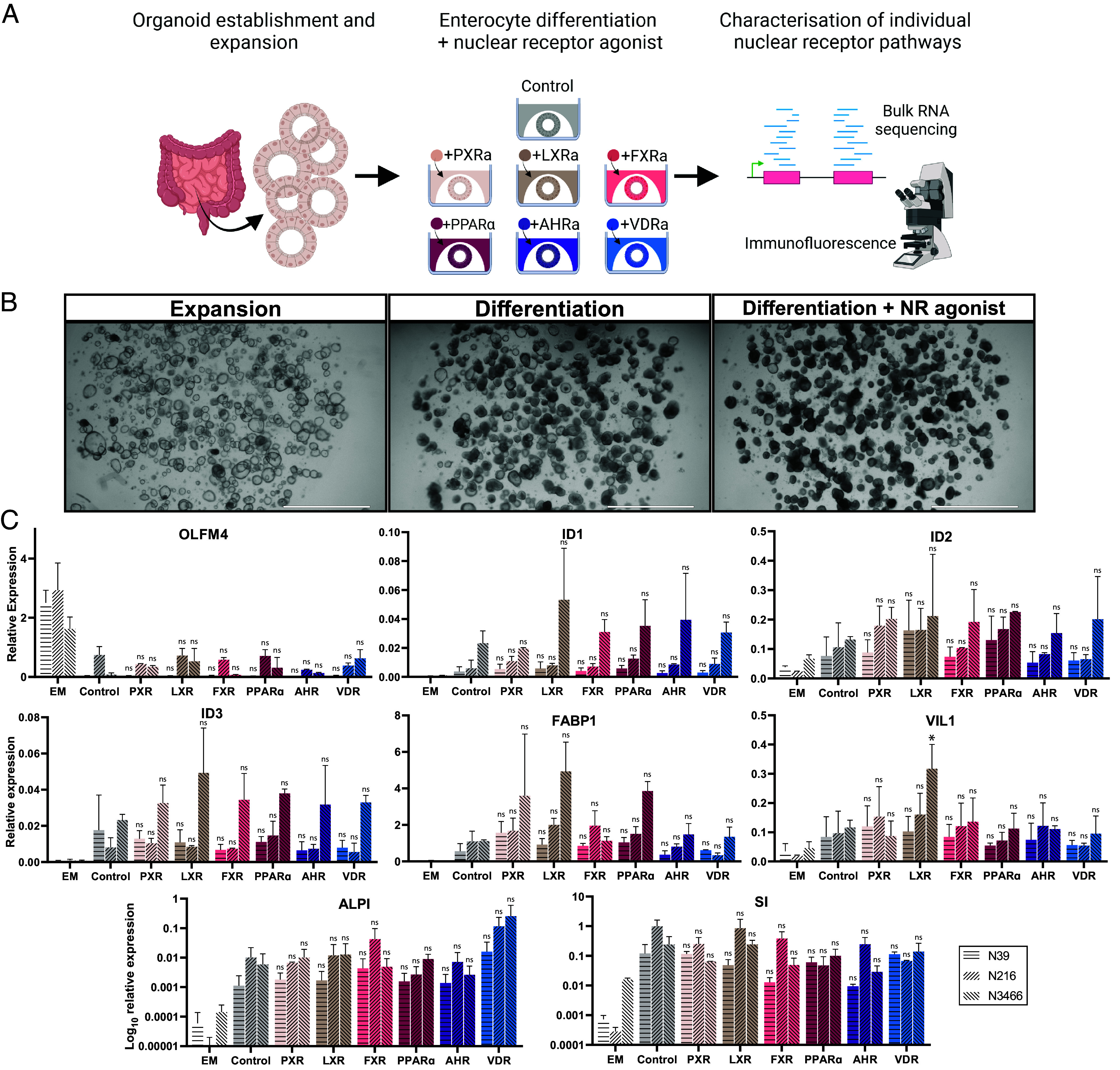
Addition of NRs does not affect enterocyte differentiation. (A) Overview of experimental setup. Human small intestinal organoids were differentiated without or with one of the shown nuclear receptor agonists, followed by transcriptomic characterization and immunofluorescence. (B) Representative bright-field images of intestinal organoids before differentiation and after 5 d of differentiation without or with the addition of a nuclear receptor agonist. (Scale bar, 2,000 μm.) (C) Quantitative PCR analysis of selected markers in human small intestinal organoids in expansion (EM) or differentiated in the presence of BMP without (Control) and with specified nuclear receptor agonists for 7 d. Shown is n = 2 with three donors, tested with one-way ANOVA compared to the control for each donor. Error bars = SD, *P < 0.05.
Small-Molecule Activation of NRs Reveals Unique Transcriptional Profiles.
We performed bulk RNA sequencing on differentiated organoids exposed to each of the six nuclear receptor agonists with appropriate controls. This allowed for unbiased characterization of the transcriptional profiles of individual NRs through differential gene expression and gene ontology analysis (Fig. 2 and SI Appendix, Fig. S1). On average, a given NR agonist led to the differential expression of ~600 genes. This included the induction of previously established target genes for each NR, validating our experimental setup (7, 19, 37–53). The vast majority of differentially expressed genes were upregulated, which is in line with the role of NRs as transcriptional activators. The outcome of the bulk RNA sequencing experiments was validated in three independent ileum organoid lines by quantitative PCR analysis of the three strongest induced genes for each condition (SI Appendix, Fig. S2). Taken together, these results indicated that human small intestine organoids are a suitable model to characterize transcriptional consequences of nuclear receptor activation. We list the most important changes for each nuclear receptor activation below.
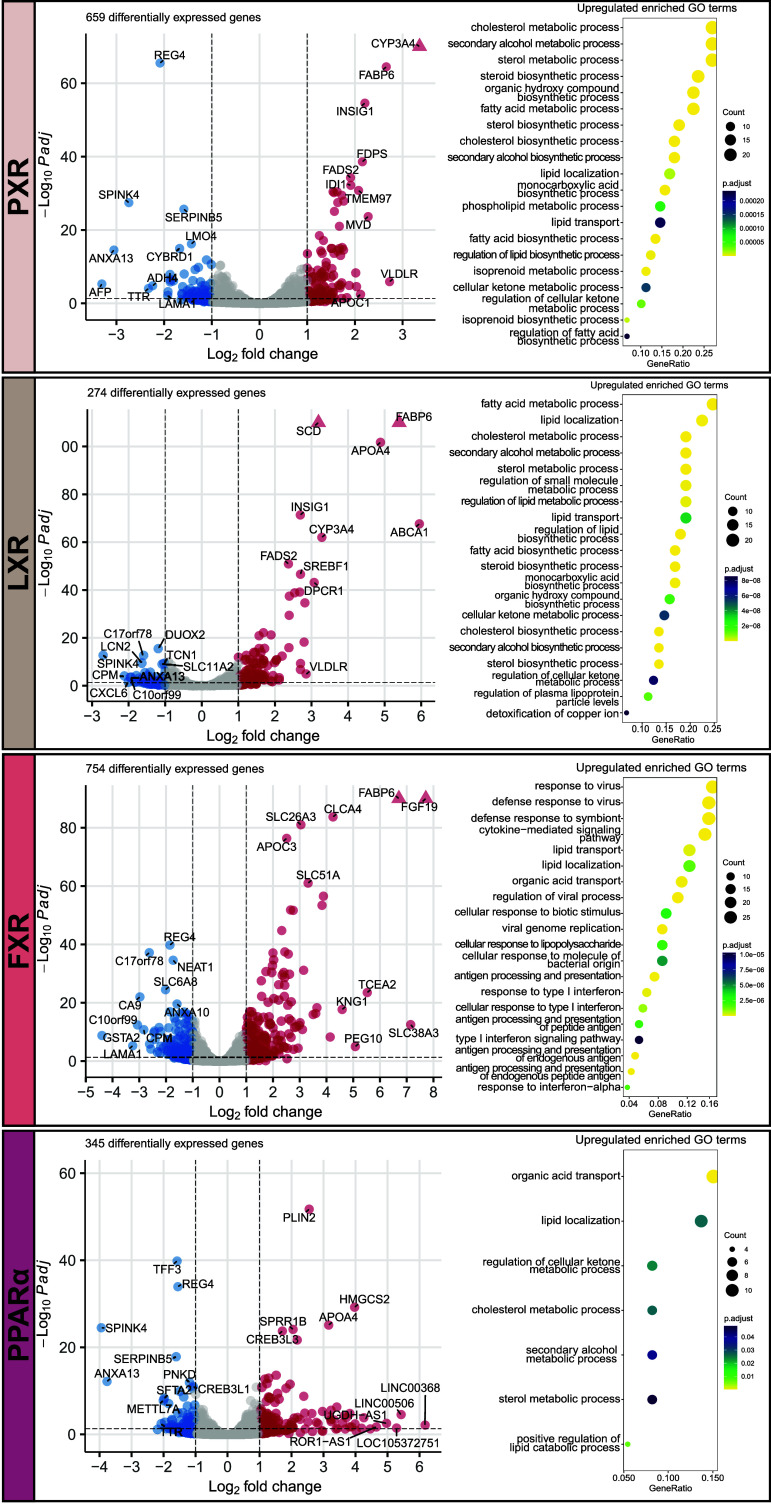
Fig. 2.
NRs induce characteristic transcriptome profiles in the human intestine. Volcano plots and enriched upregulated GO processes showing transcriptional profiles induced by PXR agonist, LXR agonist, FXR agonist, and PPARα agonist. Labeled genes are the top 10 differentially up- or down-regulated genes with the highest log2 fold change. Triangles indicate –log10 P-adjusted value higher than y-axis limit.
PXR activation upregulated the xenobiotic metabolism gene CYP3A4 most potently, but other genes of this family, such as CYP51A1, CYP2C9, CYP2C19, and CYP4F2, were upregulated as well (Fig. 2 and Dataset S1). Furthermore, activation of PXR strongly upregulated genes related to fatty acid transport and metabolism (FABP6 and FADS2), regulation of cellular cholesterol levels (TMEM97), and lipoproteins (APOC1 and VLDLR). GO term analysis further substantiated PXR as important positive regulator of lipid and cholesterol handling since many lipid-, cholesterol-, and sterol-related GO processes were enriched after PXR activation (Fig. 2).
LXR activation broadly upregulated genes related to the basolateral export of cholesterol (ABCA1), fatty acid transport and metabolism (FABP6, SCD, and FADS2), and lipoproteins (APOA4, SREBF1, was VLDLR). Again, the majority of enriched GO processes were processes related to lipid and cholesterol (Fig. 2).
FXR activation strongly induced the expression of FGF19 in intestinal organoids, which is a well-established signaling molecule in the enterohepatic bile acid feedback loop (4). Furthermore, lipid absorption-related genes FABP6 and APOC3 were strongly upregulated and the GO terms “lipid transport” and “lipid localization” were enriched (Fig. 2). These enriched GO terms highlighted the upregulation of genes such as FABP2, APOA4, APOA1, and APOL2 (Dataset S2). Finally, FXR activation led to upregulation of genes of the interferon I response and viral immunity, as shown by GO term analysis, which included antigen processing-related genes TAP1, TAP2 (transportation of cytosolic antigens), HLA-B, HLA-C, HLA-E, HLA-DRA, HLA-F, and HLA-DRB1 (MHC complex) and many chemokines and interferon-inducible genes.
PPARα activation most notably induced the expression of PLIN2, a gene which promotes the formation of intracellular lipid droplets. For PPARα, the most striking enriched GO terms were “organic acid transport” and “lipid localization” (Fig. 2). The latter included upregulated genes FABP1, FABP2, APOA4, VLDLR, CPT1A, and HILPDA (Dataset S2).
AHR activation induced GO processes related to xenobiotic response and responses to metals and upregulated xenobiotic metabolism genes CYP1A1, CYP1B1, and CYP1A2 and multiple metallothioneins including MT2A, MT1G, MT1E, MT1H, and MT1F (SI Appendix, Fig. S1 and Dataset S2).
VDR activation led to the expected induction of genes involved in calcium- (TRPV6, S100G) and phosphate (SLC34A3) uptake. Furthermore, VDR activation upregulated the CYP genes CYP24A1, CYP3A4, CYP2C8, CYP2C9, and CYP2C19 and downregulated GO processes of hormone metabolism and ROS biosynthesis (SI Appendix, Fig. S1 and Dataset S2).
NRs Regulate Key Enterocyte Functions.
Our unbiased approach revealed specific transcriptional profiles for individual NRs but also indicated that multiple NRs appeared to regulate overlapping gene programs, such as the lipid absorption program induced by LXR, FXR, PPARα, and PXR activation. We then compared the effects of each nuclear receptor on key enterocyte functions (Fig. 3). We included the following five functions of the small intestinal enterocyte: lipid absorption, xenobiotic metabolism, the maintenance of electrolyte balance, amino acid absorption, and glucose absorption.
Fig. 3.
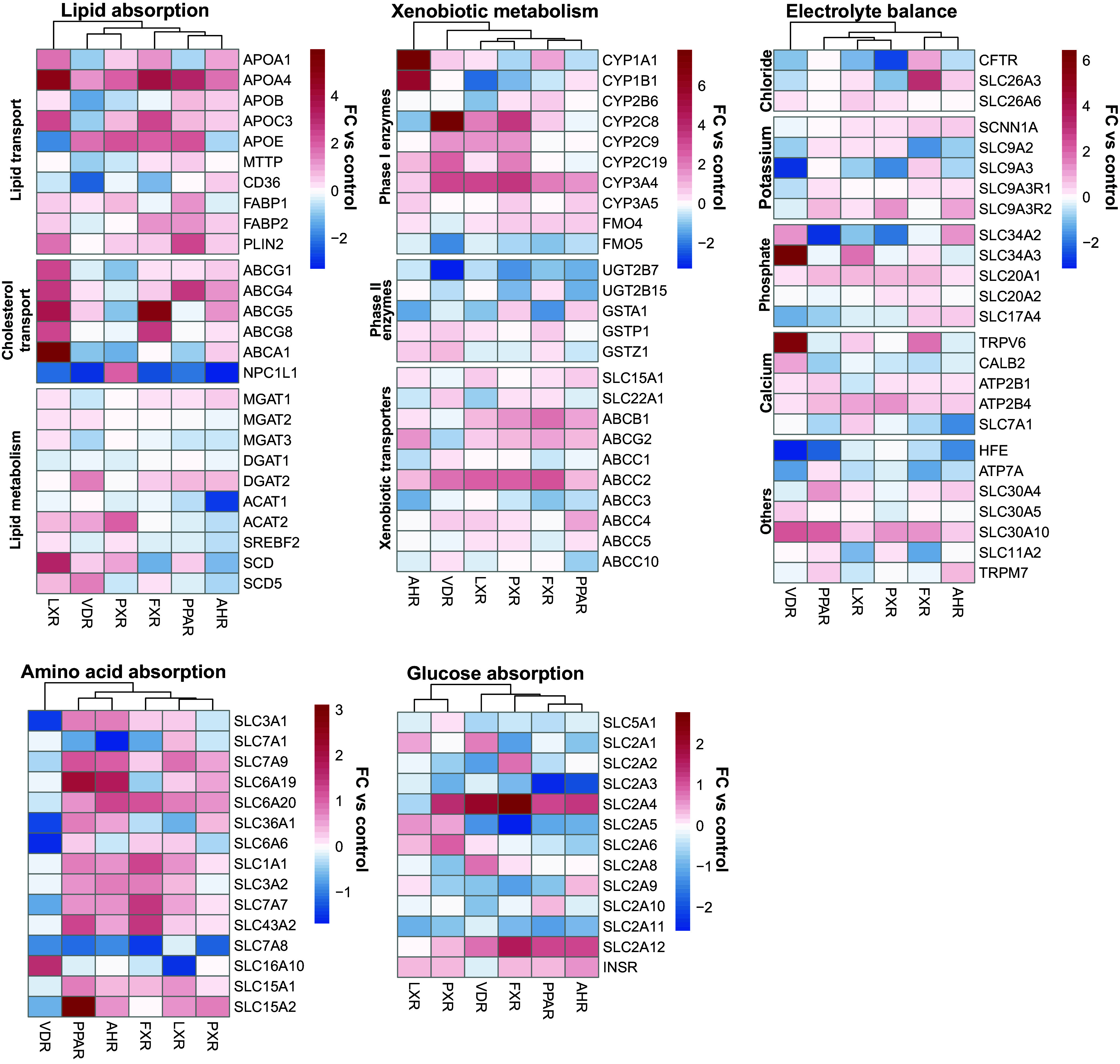
NRs modulate key enterocyte functions. Shown are log2 normalized counts as fold changes compared to the control (no addition of nuclear receptor agonists) for lists of genes known to be involved in their respective enterocyte functions.
Lipid absorption-related metabolic genes appeared to be less strongly regulated by NRs as compared transporters of fatty acid and cholesterol. Interestingly, LXR activation—and to a lesser extent FXR and AHR activation—induced all enterocyte cholesterol exporters and downregulated NPC1L1, which imports cholesterol into enterocytes. For PXR activation, the inverse was true.
Xenobiotic metabolism consists of phase I and phase II reactions, as well as excretion through transporters. Several important phase I enzymes, genes that introduce reactive groups into their substrates during the first step of xenobiotic metabolism, were strongly induced by AHR and VDR activation. However, there was no clearly defined pattern in the regulation of phase I, phase II enzymes and xenobiotic transporters.
Electrolyte balance maintenance is an important part of intestinal homeostasis. Active, transcellular calcium uptake in the small intestine is known to be regulated by vitamin D (54). Indeed, VDR activation led to an upregulation of multiple genes associated with calcium transport. Multiple chloride transporters were induced by FXR activation. Potassium transporters appeared to be predominantly induced by PPARα activation, albeit weakly. Genes falling into other electrolyte groups did not appear to be clearly regulated by any one NR.
Amino acid absorption-related genes were in general induced by all NRs except VDR, which generally downregulated amino acid transporters. As exceptions, amino acid transporters SLC7A8 (LAT2) and SLC16A10 (TAT1) were mostly downregulated after activation of any NR.
Glucose absorption-related genes did not show a clear regulatory pattern by any single NR. Of note, the insulin receptor was induced by activation of all NRs except for VDR.
In conclusion, the NRs PXR, LXR, FXR, PPARα, AHR, and VDR appear to regulate well-known genes involved in key intestinal functions. Furthermore, different NRs showed similar effects on the investigated genes, suggesting the possibility that these NR pathways might show partial regulatory overlap.
Comparative Analysis Reveals Partial Redundancy and Unique Marker Genes.
Next, to better identify similarities in gene regulation by NR pathways in the small intestine, we performed an unbiased comparison of NR conditions. We performed principal component analysis and k-means clustering on the top 750 variable genes, which defined five clusters which indicated that activation of PXR and LXR led to similar transcriptional changes. AHR and PPARα activation also showed similarities in transcriptional changes (Fig. 4A). Expression patterns of the top 750 variable genes revealed groups of upregulated genes that were shared by different NRs within the aforementioned clusters (Fig. 4B). GO term enrichment analysis on these groups of upregulated genes showed that responses to copper, detoxification, and carboxylic acid transport was shared between AHR and PPARα activation (Fig. 4C). For PXR and LXR activation, shared upregulated genes were involved in secondary alcohol and sterol biosynthesis, purine ribonucleotide metabolism and cholesterol and secondary alcohol metabolism.
Fig. 4.
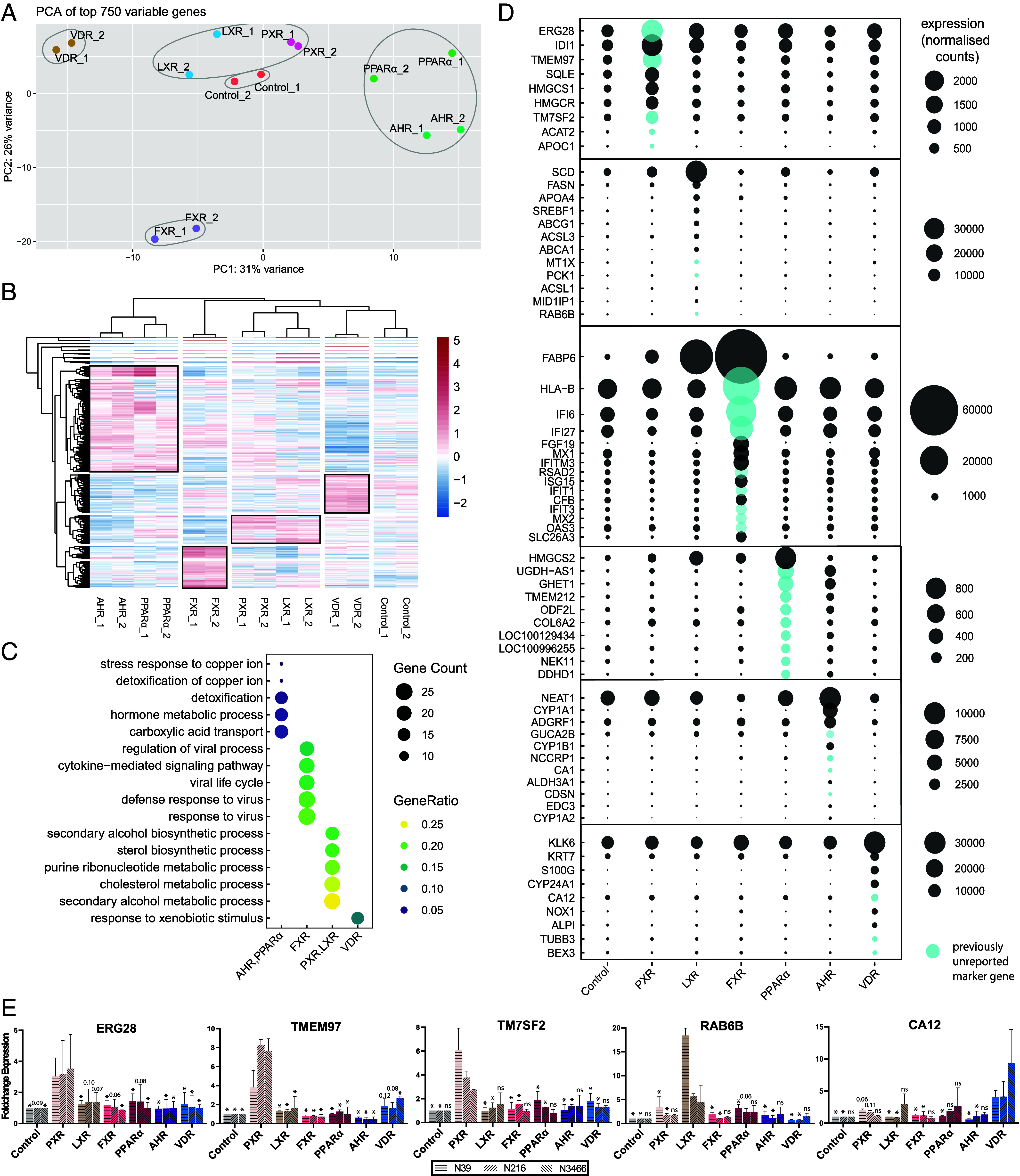
Comparison of individual nuclear receptor characterizations reveal similarities and= unique marker genes. (A) PCA of the top 750 variable genes. Number of k-means clusters were determined with the elbow method. (B) Clustered heatmap of the top 750 variable genes. Highlighted groups by row show clusters of upregulated genes that characterize the groups of NRs. (C) Enriched GO processes for each highlighted group of gene in B. (D) Putative marker genes for activation of NRs were determined among differentially expressed genes from all NR conditions. Genes that were expressed at least twofold higher in a specific NR compared to all others were considered putative marker genes. Previously unreported marker genes are marked in blue. Expression in normalized counts. (E) Unique NR marker genes resulting from qPCR validation of the three highest expressed putative marker genes for each NR condition. n = 2 with three donors. Tested with one-way ANOVA compared to the condition of the marker gene for each donor. Error bars = SD, *P < 0.05.
To further highlight similarities in transcriptional responses after activation of a single NR, we compared the GO terms enriched after separate activation of each individual NR (SI Appendix, Fig. S3). Activation of PXR and LXR shared multiple enriched GO terms related to lipid and cholesterol metabolism and lipid localization. Furthermore, FXR and PPARα activation shared a clear subset of enriched GO terms with the cluster containing PXR and LXR activation: lipid localization for FXR activation; and lipid localization and ketone, cholesterol, secondary alcohol, and sterol metabolism for PPARα activation. Through this unbiased comparison, NRs again appeared to drive partially overlapping lipid-related processes within the small intestine.
We next attempted to deduce unique target genes specific to single NRs. Genes that were induced at least twofold by a given NR (compared to all other conditions) were considered putative marker genes. Results of this analysis revealed previously reported marker genes for all NRs, validating their expression within the human small intestine and our approach for target gene identification (Fig. 4D). The three highest expressed putative marker genes, for each NR condition, were validated by qPCR in organoids of three independent donors (Fig. 4E). This led to the identification of the unique marker genes for intestinal activation of PXR (ERG28, TMEM97, and TM7SF2), LXR (RAB6B), and VDR (CA12).
Nuclear Receptor Modulation Leads to Phenotypic Changes in Intestinal Organoids.
To validate results obtained from the transcriptomic analysis, we studied how individual activation of PXR, LXR, FXR, or PPARα impacted lipid absorption in human intestinal organoids. Activation of VDR and AHR did not strongly induce genes involved in lipid absorption and were thus excluded from these experiments. First, we validated the transcriptomic results by studying protein expression of key lipid absorption genes. PLIN2 is a gene involved in the storage of absorbed lipids within enterocytes. Gene expression of PLIN2 was strongly induced by activation of PPARα (Fig. 5A). With PPARα activation, large areas of organoids stained for PLIN2 by immunofluorescence (Fig. 5B). Quantification showed a clear increase in PLIN2 protein in three donors after PPARα activation (Fig. 5C). Next, we investigated apolipoproteins, major components of chylomicrons, which are important for basolateral trafficking of absorbed dietary lipids. APOA4 RNA expression was induced by LXR activation in all donors and to some extent by FXR activation in one donor (Fig. 5D). Measuring the secreted level of APOA4 in this organoid culture confirmed that LXR, but not FXR, led to APOA4 induction at the protein level (Fig. 5E). Another apolipoprotein, APOA1, was similarly induced by LXR activation (Fig. 5F). LXR activation resulted in a clear increase in APOA1 protein across all donors (Fig. 5 G and H). Finally, we investigated the presence of intracellular lipids in organoids after NR exposure by staining with a lipid dye (Fig. 5 I and J). While intracellular lipid levels were low in control organoids, activation of PXR, LXR, FXR, or PPARα increased cellular lipid contents, albeit to different extents. LXR activation yielded the biggest and most homogeneous increase in cellular lipid contents. PXR and FXR activation led instead to a heterogeneous and smaller increase in lipids, whereas PPARα activation indicated a slight increase in cellular lipid levels. In conclusion, NR activation in the intestine drives induction of lipid-related genes at the transcriptomic and protein level and increased intracellular lipid contents.
Fig. 5.
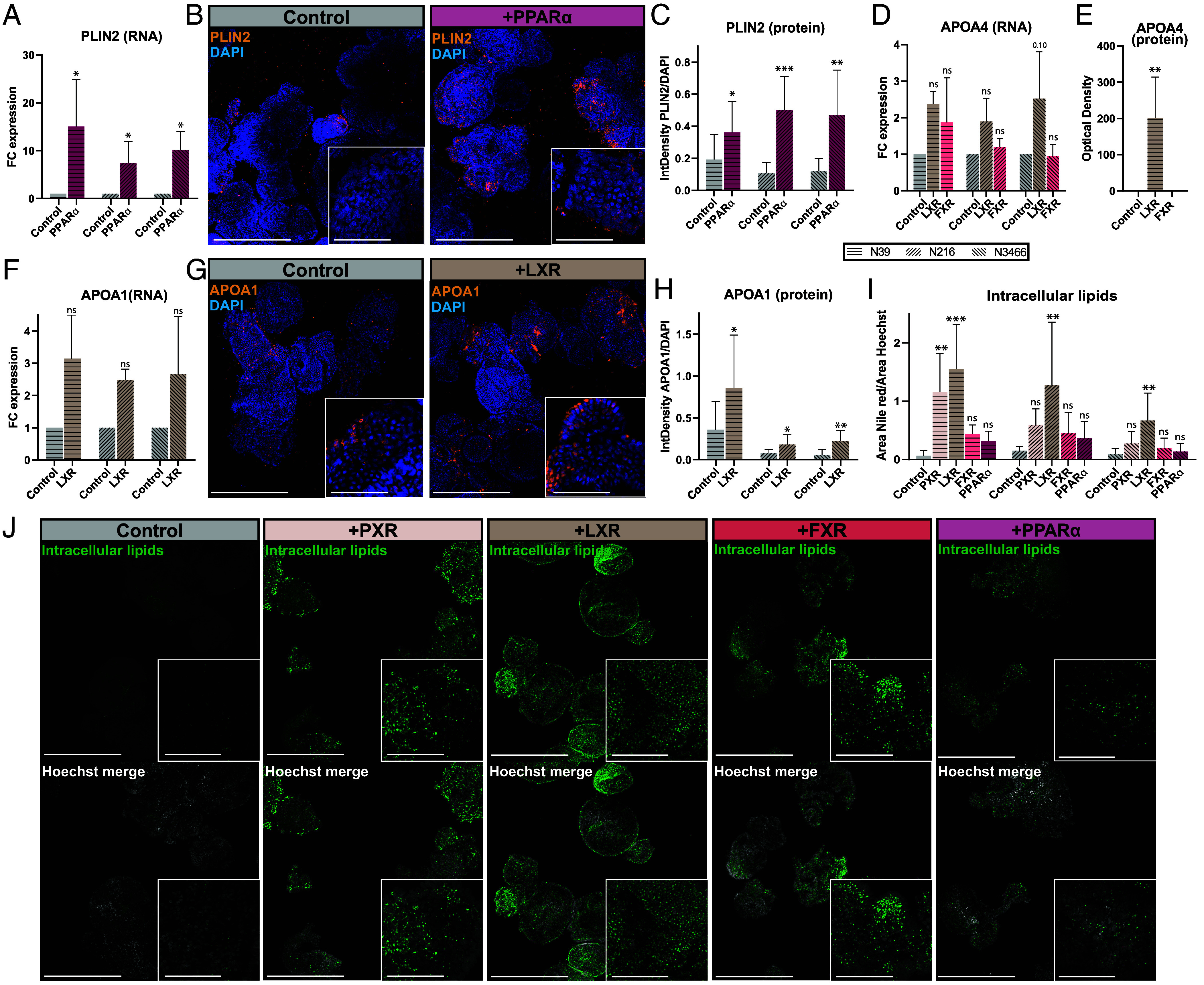
NRs induce lipid absorption-related genes at both the mRNA and protein level in enterocytes. (A) Expression of PLIN2 across different nuclear receptor agonists. n = 2 with 3 donors. (B) Representative max projection images of PLIN2 stainings at 10× (Scale bar, 400 μm) and at 63x in the insert (Scale bar, 100 μm). n = 2 with 3 donors (C) Quantification of PLIN2 stainings, based on approximately 80 organoids per condition, imaged at 10× magnification. n = 2 with 3 donors. (D) Expression of APOA4 across different nuclear receptor agonists. n = 2 with 3 donors. (E) APOA4 ELISA for donor N39. (F) Expression of APOA1 across different nuclear receptor agonists. n = 2 with 3 donors. (G) Representative max projection images of APOA1 stainings at 10× (Scale bar, 400 μm) and at 63× in the insert (Scale bar, 100 μm). n = 2 with 3 donors (H) Quantification of APOA1 stainings, based on approximately 40 organoids per condition, imaged at 10× magnification. n = 2 with 3 donors. (I) Quantification of intracellular lipid contents, based on approximately 60 organoids per condition, imaged at 10× magnification. n = 2 with 3 donors. (J) Representative confocal images of intracellular lipid contents, stained with Nile red. Organoids were imaged at 10× (Scale bar, 400 μm) and 63x in the inserts (Scale bar, 100 μm) (inserts). n = 2 with 3 donors. Error bars =SD, *P < 0.05, **P < 0.005, ***P < 0.0005 by one-way ANOVA of each NR condition compared to the control or t test compared to the control.
Discussion
While NRs have been extensively studied in the liver and kidney, their role in intestinal biology is less well understood. To our knowledge, this study is the first to characterize the genome-wide effects of activation of individual nuclear receptor pathways in human small intestinal cells. Here, we characterized PXR, LXR, FXR, PPARα, AHR, and VDR. These NRs were selected based on a combination of the following factors: They are expressed in the intestine in vivo, their ligands are known and agonists are available, and they are known to play a role in absorption and metabolism (6, 18, 21, 55–58). Genome-wide approaches have been used previously to characterize these NRs in various cell lines or primary cells, with a majority of studies focusing on immune cells or the liver. However, only PPARα, VDR, and FXR activation has been studied in the small intestine of mice ( 21, 57–63). With intestinal organoids and selective small molecules to activate specific NRs, we characterized the transcriptome consequences of activation of individual NRs in the small intestine in humans. We found that NRs regulate enterocyte gene expression as well as key intestinal functions including lipid absorption, amino acid absorption, glucose absorption, maintenance of electrolyte balance, and xenobiotic metabolism.
Lipid absorption appeared to be particularly amenable to regulation by NRs. Genes associated with lipid transport, localization, and metabolism such as FABP6, FADS2, APOA4, and, APOC3 were positively regulated by PXR, LXR, FXR, and PPARα activation. Furthermore, LXR induced multiple apolipoproteins at the protein level. LXR also upregulated cholesterol exporters ABCG5 and ABCA1 while downregulating cholesterol importer NPC1L1. Intracellular lipid levels were altered by activation of NRs. Activation of PXR, FXR, and PPARα caused a moderate increase of intracellular lipids while activation of LXR led to the largest increase of intracellular lipids in organoids. LXR has been previously described to be involved in intestinal cholesterol transport and lipid metabolism (56). While activation of PPARα and PXR is well known to increase the uptake and accumulation of lipids within the liver, our data support a similar function for these NRs within the intestine (55). Furthermore, we found that activation of PPARα in the intestine increased PLIN2 at the RNA and protein level and upregulated other lipid absorption-related genes. Others have recently shown that PPARα antagonism leads to a reduction of lipid absorption, lipid droplets, and PLIN2 levels in human biopsies (64). Our data thus further substantiate PPARα as a positive regulator of lipid absorption in the human small intestine. Overall, our findings support the role of PXR, LXR, FXR, and PPARα as key lipid sensors and regulators of lipid absorption in the human small intestine.
Besides lipid absorption, our work highlights that NRs regulating several other processes in the human intestine, including xenobiotic metabolism, electrolyte transport, and immunomodulation. Activation of AHR and PXR induced certain CYP enzyme family members, in line with their established role in xenobiotic metabolism and detoxification (6). Electrolyte and heavy metal transport and handling were affected as well. While AHR activation is known to induce MT2A, a metallothionein that protects against metal toxicity, we found AHR activation to also induce other members of this family including MT1G, MT1E, MT1H, and MT1F (65). This further implicates AHR as a regulator of xenobiotic metabolism and metal toxicity in the human small intestine. VDR activation has previously been established to regulate intestinal calcium absorption and barrier integrity (12, 54). In organoids, the calcium-binding protein S100G was strongly upregulated as well. We additionally find that VDR signaling controls expression of enzymes involved in vitamin D metabolism, including CYP24A1 and CYP3A4—as reported previously—but also CYP2C8 (66, 67). After FXR activation, we find a majority of the enriched GO processes to be related to the type I immune response and multiple immune response-related genes to be upregulated. The immunomodulatory function of FXR is well appreciated in cells of the innate immune system (68, 69). In intestinal epithelium cells, FXR has been reported to have an anti-inflammatory effect, by reducing the expression of cytokines IL-1b, IL-6, and CCL2 (69). We did not observe a differential expression of IL-1b, IL-6, but CCL2 was slightly upregulated. Furthermore, the upregulation of multiple MHC genes, antigen processing-related genes, and interferon-inducible genes by FXR activation in our intestinal organoids suggests that FXR agonists might have unappreciated immunomodulatory functions in epithelial cells.
The comprehensive understanding of individual nuclear receptor signaling and their impact on enterocyte functions may hold clinical relevance. The small intestine plays a pivotal role in the pathogenesis of many diseases and serves as the first site of xenobiotic metabolism for therapeutic drugs. Dysregulation of nuclear receptor pathways are contributing factors to various metabolic diseases, inflammation-related disorders and gastrointestinal diseases (14, 16). Our insights into the unique transcriptome profiles of different NR pathways may aid to better understand the pathophysiology of gastrointestinal diseases that result from aberrant NR signaling. One such contribution is the identification of unique marker genes for activation of individual NR pathways in the human small intestine. Our analyses confirm previously established marker genes for activation of multiple NR pathways and identify unique marker genes for intestinal activation of PXR (ERG28, TMEM97, and TM7SF2), LXR (RAB6B), and VDR (CA12). Furthermore, NR have been thoroughly investigated as drug targets and there is a large interest in their therapeutic application. In the liver, NR agonists are currently investigated for treatment of steatosis, fibrosis, cholestatic liver disease, and more. Some of these agonists already showed promising results in phase II clinical trails (70). Similarly, since NRs regulate metabolism and absorption, there has been increasing focus on their potential applications in treating metabolic diseases, including metabolic syndrome (15). The effects that these NR targeting drugs could have on the intestine are twofold. First, we show that drug-metabolising CYP genes are upregulated in the intestine in response to NR small molecules. Considering that the intestine is a site of first-pass metabolism for orally administrated drugs, NR small molecule drugs targeting other tissues will likely affect their own metabolism also in the intestine. Second, modulating NR signaling in the intestine might be leveraged to prevent excessive uptake or aberrant metabolism of lipids or cholesterol in the intestine, for example in the context of metabolic syndrome or congenital diseases of aberrant lipid or cholesterol absorption. To summarize, a better understanding NR biology in the intestine could facilitate more extensive preclinical research into the therapeutical application of NR small molecules.
While NRs have been extensively studied in the liver, kidney, and other tissues, their roles in the small intestine are less well appreciated. Although nuclear receptor signaling has previously been studied in the small intestine, these studies on animal models or human cell lines are limited—respectively—by confounding systemic effects or low expression of NRs. By utilizing human primary healthy tissue-derived intestinal organoids, we were able to specifically activate and study NR pathways in the human small intestine in vitro. Activation of NR in intestinal organoids resulted in gene expression profiles that are consistent with the current understanding of NR pathways in other organ systems, while also expanding upon this knowledge by characterizing their intestinal-specific effects. While our study focuses on the effects of NRs on enterocytes, other, rarer small intestinal cell types—such as goblet cells, Paneth cells, tuft cells, and enteroendocrine cells—also express NRs. Future studies may focus on investigating NR effects in these cell types to gain a broader understanding of NR signaling in intestinal function. Our study provides an unbiased characterization of the transcriptome profile for individual nuclear receptor pathways in human small intestinal enterocytes and broadens our understanding of the critical roles that NRs have in intestinal homeostasis.
Materials and Methods
Experimental Model and Subject Details.
Human small intestinal organoids were derived from ileum tissues of three patients with their informed consent. Donor N39 was previously established from an intestinal endoscopic biopsy from the ileum (71). The study was approved by the ethics committee of the University Medical Center Utrecht. The other human organoid lines (N216 and N3466) were obtained from healthy adjacent tissue from tumor resections. The study was approved by the ethics committee of the NKI Institutional Review Board (IRB) with code CFMPB582. This study is compliant with all relevant ethical regulations regarding research involving human participants.
Culturing of Intestinal Organoids.
rganoids were cultured in human small intestine expansion medium (EM): Advanced Dulbecco’s Modified Eagle/F12 medium (DMEM) (Gibco), supplemented with 1% GlutaMAX (Thermofisher), 1% HEPES (Thermofisher), and 1% penicillin-streptomycin (Thermofisher) henceforth referred to as ADF+++. EM was completed with additional 20% R-spondin1-conditioned medium (in-house), 2% Noggin surrogate (U-protein Express), 2% B27 (with vitamin A) (Thermofisher), 10 mM nicotinamide (Sigma-Aldrich), 1mM N-acetyl cysteine (Sigma-Aldrich), 3 μM p38 inhibitor SB202190 (Sigma-Aldrich), 0.15 nM Wnt3a surrogate (U-protein express), 50 ng/mL EGF (Peprotech), 0.5 μM TGF-β inhibitor (A83-01) (Tocris), 1 μM prostaglandin E2 (Tocris), and 50 μg/mL primocin (Invivogen). Organoids were passaged by disruption every 7 to 10 d and cultured in Cultrex® Basement Membrane Extract (R&Dsystems).
Differentiation and Nuclear Receptor Agonist Exposure.
Differentiation experiments were performed on organoids five days after passaging. Before the addition of the differentiation medium, the EM was replaced with ADF+++ for 2 h to wash out EM components. Differentiation lasted seven days with the medium refreshed every 2–3 d. The base differentiation medium (ER+BMP) consisted of ADF+++, 10% R-spondin1-conditioned medium (produced in-house), 2% B27 (with vitamin A) (Thermofisher), 1.25mM N-acetyl cysteine (Sigma-Aldrich), 50 ng/mL EGF (Peprotech), 50 μg/mL primocin (Invivogen), 50 ng/mL BMP2 (Peprotech), and 50 ng/mL BMP4 (Peprotech). NRs agonists were added during the ER+BMP differentiation and included 5 µM SR12813 (PXR activation), 5 µM GW3965 (LXR activation), 5 µM GW4064 (FXR activation), 500 µM Clofibric acid (PPARα activation), 1 µM FICZ (AHR activation), and 5 μM Calcitriol (VDR activation).
Microscopy and Quantification.
To prepare samples for imaging, the cultures were incubated with 1 mg/mL Dispase for 30 min prior to harvesting. Samples were then washed in ADF+++ to remove the remaining BME. Organoids were fixed in 4% PFA for 30 min at RT, then permeabilized and blocked with 0.5% BSA-PBS containing 0.5% triton for 1 h at RT. Primary stainings (PLIN2: Proteintech #15294-1-AP, APOA1: Invitrogen #PA5-88109) were done at 1:50 at 4 °C overnight while secondary stainings (AF488 anti-rabbit Life technologies #A21206) were done at 1:400 at 4 °C overnight. Dapi was added 1:1,000 during secondary stainings. Organoids stained for intracellular lipids were not permeabilized but incubated directly after fixation with 1:2000 Nile red and 1:1,000 Hoechst for 20 min. Images were acquired on a SP8 Confocal Microscope (Leica) as Z-stacks covering the whole depth of the organoids, with 499 nm/510 to 535 nm for protein stainings and 515 nm/525 to 600 nm for Nile red. To avoid bleed-through, channels were imaged sequentially. To quantify intracellular lipids and protein stainings, images were max projected and analyzed with ImageJ. To separate signal from background, selections were created by thresholding nuclear stainings at 40 to 255, lipid stainings at 35 to 255, and protein stainings at 60 to 255. The integrated density was then measured from these selections. To correct for background signal, the integrated density was also measured from the inversed of the selections and subtracted from the true signal. The amount of protein or lipids was calculated as the background-corrected integrated density of the staining over the background-corrected integrated density of the nuclear staining.
Real-Time qPCR.
RNA was isolated from cells using the Nucleospin RNA kit (BIOKÉ) and cDNA was synthesized using MMLV Reverse Transcriptase (Promega). Real-time qPCR was performed on 12 ng cDNA per well with iQ SYBR Green (BIORAD) on a CFX384 Realtime system (BIORAD) using the primers indicated in Dataset S3. The expression of each gene was calculated relative to ACTB or as foldchange relative expression to ACTB compared to the control condition (differentiation without nuclear receptor agonist).
Measuring APOA4 Protein.
APOA4 was measured with an ELISA kit (Abcam # ab214567), according to the manufacturer’s instructions. Two-day-old culturing medium was collected from organoids differentiated for 13 d. These were spun down at 1,000 g for 10 min to remove cellular debris. Supernatants were snap-frozen in liquid nitrogen and stored at -80 °C until use. Optical density (OD) was measured on an Asys expert plus microplate reader (Biochrom).
Bulk Sequencing.
For bulk RNA sequencing, two independent samples of N39 organoids were differentiated for 7 d and collected in Eppendorf tubes containing 350 μL RLT buffer (RNeasy kit, QIAGEN). RNA was extracted using the RNeasy mini kit (QIAGEN) following the manufacturer’s instructions. Sequencing libraries were generated using a modified CELseq2 protocol (72). 75 bp paired-end sequencing of libraries was performed on an Illumina NextSeq platform. Reads were mapped to the GRCh37 genome assembly and counted. The readcounts were filtered to exclude reads with identical library- and molecule barcodes.
Individual Nuclear Receptor Characterization by Differential Expression and GO Term Analyses.
Differential gene expression was performed with DESeq 2 (package version 3.13) in R, for each nuclear receptor condition compared to the control differentiated organoids (73). Differential expression was considered significant with a p-adjusted value below 0.05 and a basemean of 50 transcripts across all conditions. Volcano plots were generated with the R package EnhancedVolcano and GO term analysis was performed using R package clusterProfiler (74, 75). For GO term analysis, lists of differentially expressed genes were separated into up- and down-regulated genes. Genes with a logFC of at least 1 and a PADJ of 0.05 were compared against GO biological pathways. A semantic similarity cutoff of 0.8 was used, to reduce redundancy in GO terms (76).
Nuclear Receptor Comparisons and Marker Identification Analyses.
Normalization was performed with DESeq2’s median of ratios normalization. Heatmaps were generated with pheatmap package version 1.0.12. Heatmaps depicting key enterocyte functions use a list of genes based on literature and show the log2 FC compared to the control, as average over the replicates. PCA was performed on the top 750 variable genes across all NR conditions and the control. The conditions were clustered based on k-means clustering using pheatmap. The amount of clusters for k-means clustering was determined with the elbow method. GO pathway analysis was performed on clusters containing more than 2 genes with a semantic similarity cutoff of 0.7. Marker genes were identified from a list of genes consisting all differentially expressed genes from all NR conditions. A gene with a normalized count at least 2 fold higher in one condition compared to all others was considered a putative marker gene for this condition.
Statistical Analysis.
Statistical analysis was performed with GraphPad Prism 9.2.0. In comparisons between one NR condition and the control, t tests were performed with the Holm–Sidak method for multiple comparisons. In cases of multiple nuclear receptor conditions, each NR condition was compared to the control using a one-way ANOVA with Dunnett’s test for multiple comparisons. For marker gene validation (Fig. 4), each condition was compared to the NR condition of the marker gene instead. For data plotted as fold change, statistics were performed on the data prior to calculating the fold change.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Acknowledgments
We thank Stieneke van den Brink for the in-house production of R-spondin1-conditioned medium. We thank Gijs van Son for preparing the bulk RNA sequencing data for the GEO depository. We thank Single Cell Discoveries for their bulk RNA-sequencing services. Part of Fig. 1 was created with the use of Biorender.com. This work was supported by the NWO Gravitation project Material Driven Regeneration, funded by the Ministry of Education, Culture and Science of the government of the Netherlands.
Author contributions
S.W., F.A.Y.Y., J.v.E., J.B., and H.C. designed research; S.W. and F.A.Y.Y. performed research; S.W., J.P., M.B.R., and M.C.V. analyzed data; and S.W., F.A.Y.Y., J.B., and H.C. wrote the paper.
Competing interests
H.C. is the head of Pharma Research and Early Development at Roche, Basel, and holds several patents related to organoid technology. The full disclosure is given at https://www.uu.nl/staff/JCClevers/.
Footnotes
Reviewers: V.D.-M., Amsterdam University Medical Center; and P.J., Universite de Montpellier.
Contributor Information
Joep Beumer, Email: joep.beumer@roche.com.
Hans Clevers, Email: h.clevers@hubrecht.eu.
Data, Materials, and Software Availability
The bulk RNA sequencing data have been deposited Gene Expression Omnibus (GEO) under accession number GSE273646 (77). Scripts of all bulk RNA sequencing analyses are available at https://github.com/Sam-Willemsen/Willemsen_et_al_Nuclear_receptors_PNAS (78).
Supporting Information
References
- 1.Wang Z., Chen W.-D., Wang Y.-D., Nuclear receptors: A bridge linking the gut microbiome and the host. Mol. Med. 27, 144 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo Sasso G., et al. , Intestinal specific LXR activation stimulates reverse cholesterol transport and protects from atherosclerosis. Cell Metab. 12, 187–193 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Hong F., Pan S., Guo Y., Xu P., Zhai Y., PPARs as nuclear receptors for nutrient and energy metabolism. Molecules 24, 2545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsubara T., Li F., Gonzalez F. J., FXR signaling in the enterohepatic system. Mol. Cell. Endocrinol. 368, 17–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatesh M., et al. , Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity 41, 296–310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradshaw T. D., Bell D. R., Relevance of the aryl hydrocarbon receptor (AhR) for clinical toxicology. Clin. Toxicol. 47, 632–642 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Christakos S., Vitamin D: A critical regulator of intestinal physiology. JBMR Plus 5, e10554 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker N., et al. , Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Alaynick W. A., Nuclear receptors, mitochondria and lipid metabolism. Mitochondrion 8, 329–337 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T., Chiang J. Y. L., Nuclear receptors in bile acid metabolism. Drug Metab. Rev. 45, 145–155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabacka M., Płonka P. M., Pierzchalska M., The PPARα regulation of the gut physiology in regard to interaction with microbiota, intestinal immunity, metabolism, and permeability. IJMS 23, 14156 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J., Zhang Y.-G., Vitamin D receptor influences intestinal barriers in health and disease. Cells 11, 1129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klepsch V., Moschen A. R., Tilg H., Baier G., Hermann-Kleiter N., Nuclear receptors regulate intestinal inflammation in the context of ibd. Front. Immunol. 10, 1070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L., Hu H., Gustafsson J. -Å., Zhou S., Nuclear receptors in cancer inflammation and immunity. Trends Immunol. 41, 172–185 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Sonoda J., Pei L., Evans R. M., Nuclear receptors: Decoding metabolic disease. FEBS Lett. 582, 2–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chávez-Talavera O., Tailleux A., Lefebvre P., Staels B., Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology 152, 1679–1694.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Saeed A., Dullaart R., Schreuder T., Blokzijl H., Faber K., Disturbed vitamin A metabolism in non-alcoholic fatty liver disease (NAFLD). Nutrients 10, 29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bookout A. L., et al. , Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126, 789–799 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grober J., et al. , Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. J. Biol. Chem. 274, 29749–29754 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Wolf I. M., Heitzer M. D., Grubisha M., DeFranco D. B., Coactivators and nuclear receptor transactivation. J. Cell. Biochem. 104, 1580–1586 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Thomas A. M., et al. , Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology 51, 1410–1419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmsen S., Koster A. S., Beijnen J. H., Schellens J. H. M., Meijerman I., Comparison of two immortalized human cell lines to study nuclear receptor-mediated CYP3A4 induction. Drug Metab. Dispos 36, 1166–1171 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Portincasa P., et al. , Bile acids and GPBAR-1: Dynamic interaction involving genes, environment and gut microbiome. Nutrients 12, 3709 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bijsmans I. T. G. W., et al. , Characterization of stem cell-derived liver and intestinal organoids as a model system to study nuclear receptor biology. Biochimica et Biophysica Acta (BBA) – Mol. Basis Dis. 1863, 687–700 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Santos R., et al. , A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donati S., et al. , Rapid nontranscriptional effects of calcifediol and calcitriol. Nutrients 14, 1291 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerdes D., et al. , Nongenomic actions of steroids—From the laboratory to clinical implications. J. Pediatric Endocrinol. Metab. 13, 853–878 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Lalwani N. D., et al. , Peroxisome proliferator-binding protein: Identification and partial characterization of nafenopin-, clofibric acid-, and ciprofibrate-binding proteins from rat liver. Proc. Natl. Acad. Sci. U.S.A. 84, 5242–5246 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toporova L., Grimaldi M., Boulahtouf A., Balaguer P., Assessing the selectivity of FXR, LXRs, CAR, and RORγ pharmaceutical ligands with reporter cell lines. Front. Pharmacol. 11, 1122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins J. L., et al. , Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J. Med. Chem. 45, 1963–1966 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Jönsson M. E., et al. , The tryptophan photoproduct 6-formylindolo[3,2-b]carbazole (FICZ) binds multiple AHRs and induces multiple CYP1 genes via AHR2 in zebrafish. Chem. Biol. Interact. 181, 447–454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosquera J., García I., Liz-Marzán L. M., Cellular uptake of nanoparticles versus small molecules: A matter of size. Acc. Chem. Res. 51, 2305–2313 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Sato T., et al. , Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Beumer J., Clevers H., Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 22, 39–53 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Beumer J., et al. , BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep. 38, 110438 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Van Der Flier L. G., Haegebarth A., Stange D. E., Van De Wetering M., Clevers H., OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137, 15–17 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Asman D. C., Takimoto K., Pitot H. C., Dunn T. J., Lindahl R., Organization and characterization of the rat class 3 aldehyde dehydrogenase gene. J. Biol. Chem. 268, 12530–12536 (1993). [PubMed] [Google Scholar]
- 38.Eltom S. E., Zhang L., Jefcoate C. R., Regulation of cytochrome P-450 (CYP) 1B1 in mouse Hepa-1 variant cell lines: A possible role for aryl hydrocarbon receptor nuclear translocator (ARNT) as a suppressor of CYP1B1 gene expression. Mol. Pharmacol. 55, 594–604 (1999). [PubMed] [Google Scholar]
- 39.Watson A. J., Hankinson O., Dioxin- and Ah receptor-dependent protein binding to xenobiotic responsive elements and G-rich DNA studied by in vivo footprinting. J. Biol. Chem. 267, 6874–6878 (1992). [PubMed] [Google Scholar]
- 40.Goodwin B., et al. , A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Zhu Y., Li F., Guo G. L., Tissue-specific function of farnesoid X receptor in liver and intestine. Pharmacol. Res. 63, 259–265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu K., Miyazaki M., Man W. C., Ntambi J. M., Stearoyl-coenzyme A desaturase 1 deficiency protects against hypertriglyceridemia and increases plasma high-density lipoprotein cholesterol induced by liver X receptor activation. Mol. Cell. Biol. 26, 6786–6798 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menke J. G., et al. , A novel liver X receptor agonist establishes species differences in the regulation of cholesterol 7α-hydroxylase (CYP7a). Endocrinology 143, 2548–2558 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Sabol S. L., Brewer H. B., Santamarina-Fojo S., The human ABCG1 gene: Identification of LXR response elements that modulate expression in macrophages and liver. J. Lipid Res. 46, 2151–2167 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Venkateswaran A., et al. , Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXRα. Proc. Natl. Acad. Sci. U.S.A. 97, 12097–12102 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janssen A. W. F., et al. , The impact of PPARα activation on whole genome gene expression in human precision cut liver slices. BMC Genomics 16, 760 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geick A., Eichelbaum M., Burk O., Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J. Biol. Chem. 276, 14581–14587 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Hariparsad N., et al. , Identification of pregnane-X receptor target genes and coactivator and corepressor binding to promoter elements in human hepatocytes. Nucleic Acids Res. 37, 1160–1173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehmann J. M., et al. , The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J. Clin. Invest. 102, 1016–1023 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C., et al. , All-trans-retinoic acid modulates ICAM-1 N-glycan composition by influencing GnT-III levels and inhibits cell adhesion and trans-endothelial migration. PLoS ONE 7, e52975 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isoherranen N., Zhong G., Biochemical and physiological importance of the CYP26 retinoic acid hydroxylases. Pharmacol. Therapeutics 204, 107400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada S., Kanda Y., Retinoic acid promotes barrier functions in human iPSC-derived intestinal epithelial monolayers. J. Pharmacol. Sci. 140, 337–344 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Dimitrov V., et al. , Vitamin D-regulated gene expression profiles: species-specificity and cell-specific effects on metabolism and immunity. Endocrinology 162, bqaa218 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fleet J. C., Vitamin D-mediated regulation of intestinal calcium absorption. Nutrients 14, 3351 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J., et al. , A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J. Biol. Chem. 281, 15013–15020 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zelcer N., Hong C., Boyadjian R., Tontonoz P., LXR regulates cholesterol uptake through idol-dependent ubiquitination of the LDL receptor. Science 325, 100–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bünger M., et al. , Genome-wide analysis of PPARα activation in murine small intestine. Physiol. Genomics 30, 192–204 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Li S., et al. , Analysis of 1,25-dihydroxyvitamin D3 genomic action reveals calcium-regulating and calcium-independent effects in mouse intestine and human enteroids. Mol. Cell. Biol. 41, e00372-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kandel B. A., et al. , Genomewide comparison of the inducible transcriptomes of nuclear receptors CAR, PXR and PPARα in primary human hepatocytes. Biochim. Biophys. Acta, Gene Regul Mech. 1859, 1218–1227 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Wiggins B. G., et al. , Endothelial sensing of AHR ligands regulates intestinal homeostasis. Nature 621, 821–829 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pehkonen P., et al. , Genome-wide landscape of liver X receptor chromatin binding and gene regulation in human macrophages. BMC Genomics 13, 50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trikha P., et al. , Defining the AHR-regulated transcriptome in NK cells reveals gene expression programs relevant to development and function. Blood Advances 5, 4605–4618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuoresmäki P., Väisänen S., Neme A., Heikkinen S., Carlberg C., Patterns of Genome-Wide VDR Locations. PLoS ONE 9, e96105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stojanović O., et al. , Dietary excess regulates absorption and surface of gut epithelium through intestinal PPARα. Nat. Commun. 12, 7031 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sato S., et al. , The aryl hydrocarbon receptor and glucocorticoid receptor interact to activate human metallothionein 2A. Toxicol. Appl. Pharmacol. 273, 90–99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouillon R., Bikle D., Vitamin D metabolism revised: Fall of dogmas. J. Bone Miner. Res. 34, 1985–1992 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thummel K. E., et al. , Transcriptional control of intestinal cytochrome P-4503A by 1α,25-dihydroxy Vitamin D3. Mol. Pharmacol. 60, 1399–1406 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Fiorucci S., Biagioli M., Zampella A., Distrutti E., Bile acids activated receptors regulate innate immunity. Front. Immunol. 9, 1853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fiorucci S., Zampella A., Ricci P., Distrutti E., Biagioli M., Immunomodulatory functions of FXR. Mol. Cell. Endocrinol. 551, 111650 (2022). [DOI] [PubMed] [Google Scholar]
- 70.Rudraiah S., Zhang X., Wang L., Nuclear receptors as therapeutic targets in liver disease: Are we there yet? Annu. Rev. Pharmacol. Toxicol. 56, 605–626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beumer J., et al. , High-resolution mRNA and secretome atlas of human enteroendocrine cells. Cell 182, 1062–1064 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Hashimshony T., et al. , CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blighe K. S. R., Lewis M., EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling (2024). https://github.com/kevinblighe/EnhancedVolcano. Accessed 6 September 2023.
- 75.Yu G., Wang L.-G., Han Y., Q.-Y., He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: A J. Integrative Biol. 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu G., et al. , GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978 (2010). [DOI] [PubMed] [Google Scholar]
- 77.Puschhof J., Beumer J., Willemsen S., Clevers H., Series GSE 273646; N39 APOA1-tdTomato drug screen. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE273646. Deposited 31 July 2024.
- 78.Willemsen S., Willemsen_et_al_Nuclear_receptors_PNAS_2024. GitHub. https://github.com/Sam-Willemsen/Willemsen_et_al_Nuclear_receptors_PNAS. Deposited 8 September 2024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Data Availability Statement
The bulk RNA sequencing data have been deposited Gene Expression Omnibus (GEO) under accession number GSE273646 (77). Scripts of all bulk RNA sequencing analyses are available at https://github.com/Sam-Willemsen/Willemsen_et_al_Nuclear_receptors_PNAS (78).