Abstract
Assignment of patients diagnosed with acute myeloid leukemia (AML) to the 2022 European LeukemiaNet (ELN) favorable genetic risk group has important clinical implications, as allogeneic stem cell transplantation in first complete remission (CR) is not advised due to a relatively good outcome of patients receiving chemotherapy alone and transplant-associated mortality. However, not all favorable genetic risk patients experience long-term relapse-free survival (RFS), making recognition of patients who would most likely be cured of high importance. We analyzed 297 patients aged <60 years with de novo AML classified as 2022 ELN favorable genetic risk who achieved a CR and had RNA sequencing (RNA-seq) and gene mutation data from diagnostic samples available (Alliance trial A152010). To identify prognostically relevant transcripts that can distinguish patients cured from patients susceptible to lower or higher risk of relapse or death, we fit a regularized mixture cure model (MCM) where RNA-seq expression values were our candidate covariates. To validate the identified transcripts, we analyzed 75 patients with de novo AML aged <60 years included in the 2022 ELN favorable genetic risk group who achieved a CR in an independent test set from Gene Expression Omnibus (GSE37642). Our MCM identified 145 transcripts associated with cure or long-term RFS and 149 transcripts associated with latency or shorter-term time to relapse. The area under the curve and C-statistic were, respectively, 0.946 and 0.856 for our training set and 0.877 and 0.857 for our test set. Our results suggest that the favorable risk group includes distinct transcriptionally defined subgroups with different biological properties, which may be useful for refining this genetic risk category.
Graphical abstract

Public summary
-
•
Assignment of AML patients to the 2022 ELN Favorable genetic-risk group has important clinical implications.
-
•
Using our training set our model identified three subgroups that differed significantly by clinical outcome.
-
•
We validated our model using an independent test set which demonstrated excellent performance.
-
•
The identified transcripts may usefully differentiate patient subgroups and serve as a prognostic signature.
-
•
Our novel regularized mixture cure model can be used to fit models to high-dimensional covariate spaces.
Introduction
The European LeukemiaNet (ELN) genetic risk classification was updated in 2022 by an international expert panel and stratifies patients with acute myeloid leukemia (AML) into three genetic risk groups: favorable, intermediate, and adverse.1 Evaluation of these genetic risk groups using large cohorts of patients with AML demonstrated that, generally, they are associated with achievement of complete remission (CR), relapse rates, duration of disease-free survival, and overall survival (OS).2,3 However, for some endpoints, clinical outcome did not differ between genetic risk groups in selected subsets of patients; for example, African Americans, Hispanics, and patients aged 60 years and older.2 Moreover, the 2022 ELN classification conferred slightly worse accuracy in predicting OS than the 2017 ELN one.3 Therefore, performance of the 2022 ELN genetic risk groups in predicting clinical outcomes is still suboptimal and may benefit from inclusion of thus far unrecognized, prognostically significant features.
The Cox proportional hazards (PH) model is the most frequently used method for assessing the effect of a covariate on a time-to-event outcome, such as OS or relapse-free survival (RFS), and assumes independent time-to-event, non-informative censoring, a constant hazard ratio over time, and that all subjects are at risk of the event of interest throughout the observation period. Various groups have shown that advances in therapy for leukemia and myelodysplastic syndrome have increased OS rates.4,5 Thus, observed increases in long-term survival of patients with AML, especially those in the ELN favorable genetic risk group, indicate that a non-negligible subset of treated patients are cured; in other words, experience long-term RFS. Therefore, when a cured subset comprises the dataset, the Cox PH assumption that all subjects are at risk of the event of interest throughout the observation period and the assumption that there is a constant hazard ratio are violated.6 Additionally, estimating the effects of covariates on both the probability of cure and the time to event for patients susceptible to the event (i.e., relapse or death; these patients are hereafter referred to as susceptible) could be informative7 but cannot be estimated using a Cox PH model.8 When a cure fraction exists, mixture cure models should be used instead.
We posit that, due to substantial differences between ELN genetic risk and age groups, development of signatures within homogeneous subgroups of AML is warranted. We recently developed a high-dimensional mixture cure model (MCM) that can be fit when the number of predictors (transcripts) exceeds the number of samples.9 Because of genetic heterogeneity of the 2022 ELN favorable genetic risk group, here we used RNA sequencing (RNA-seq) on a well-characterized cohort of 2022 ELN favorable genetic risk patients with long-term follow-up and fit our novel high-dimensional MCM to identify prognostically relevant transcripts that could potentially better distinguish subgroups of these patients with disparate outcomes. Subsequently, we assessed whether the identified genes were prognostically relevant using an independent test set.
Results
Baseline demographic and clinical characteristics of the 297 patients in our training set and 75 patients in our test set appear in Table 1. For our training set, patients alive at their last follow-up were followed an average of 9.3 years (range, 7.3 months to 21.5 years), whereas the estimated time at which 95% of patients should experience relapse or death was 5.2 years. These estimates, together with the long plateau in the RFS curve (Figure 1A) and the rejected null hypothesis of insufficient follow-up (p < 0.0001), indicate that the training set had sufficient follow-up. The estimated proportion of cure was 40.3%, which is significantly larger than 0 when using either the non-parametric (p = 0.0004) or parametric test (p < 0.0001). The training and test sets differed only by percentage of blasts in the bone marrow (p < 0.001) and frequency of WT1 mutations (p = 0.006).
Table 1.
Demographic, clinical, and gene mutation data in the training (Alliance) and test (AMLCG) sets
| Characteristica | Training set (n = 297) | Test set (n = 75) | p Value |
|---|---|---|---|
| Age, years | 44 (17–59) | 42 (18–59) | 0.570 |
| Sex | 0.368 | ||
| Female | 133 (44.8) | 38 (50.7) | |
| Male | 164 (55.2) | 37 (49.3) | |
| Hemoglobin, g/dL | 9.2 (2.3–25.1) | 8.85 (3.5–13.4) | 0.365 |
| Platelet count, ×109/L | 47 (7–433) | 40 (4–301) | 0.072 |
| White blood cell count, ×109/L | 26.75 (0.40–303.60) | 24.05 (1.10–316.0) | 0.487 |
| Bone marrow blasts, % | 63 (2–97) | 75 (20–100) | <0.001 |
| Blood blasts, % | 54 (0–97) | N/A | N/A |
| DNMT3A | 0.524 | ||
| Mutated | 63 (21.2) | 13 (17.3) | |
| Wild-type | 234 (78.8) | 62 (82.7) | |
| NRAS | 0.092 | ||
| Mutated | 63 (21.2) | 23 (30.7) | |
| Wild-type | 234 (78.8) | 52 (69.3) | |
| SF3B1 | 0.588 | ||
| Mutated | 5 (1.7) | 0 | |
| Wild-type | 292 (98.3) | 75 (100.0) | |
| IDH1 | 0.598 | ||
| Mutated | 18 (6.1) | 6 (8.0) | |
| Wild-type | 279 (93.9) | 69 (92.0) | |
| CEBPAbzip | 1.00 | ||
| Mutated | 58 (19.5) | 14 (18.7) | |
| Wild-type | 239 (80.5) | 61 (81.3) | |
| GATA2 | 0.319 | ||
| Mutated | 23 (7.7) | 3 (4.0) | |
| Wild-type | 274 (92.3) | 72 (96.0) | |
| TET2 | 0.482 | ||
| Mutated | 23 (7.7) | 8 (10.7) | |
| Wild-type | 274 (92.3) | 67 (89.3) | |
| NPM1 | 0.150 | ||
| Mutated | 132 (44.4) | 26 (34.7) | |
| Wild-type | 165 (55.6) | 49 (65.3) | |
| WT1 | 0.006 | ||
| Mutated | 17 (5.7) | 12 (16.0) | – |
| Wild-type | 280 (94.3) | 63 (84.0) | |
| PTPN11 | 0.835 | ||
| Mutated | 32 (10.8) | 7 (9.3) | |
| Wild-type | 265 (89.2) | 68 (90.7) | |
| FLT3-TKD | 0.589 | ||
| Present | 43 (14.6) | 13 (17.3) | |
| Absent | 252 (85.4) | 62 (82.7) | |
| FLT3-ITD | 0.107 | ||
| Present | 10 (3.4) | 6 (8.0) | |
| Absent | 284 (96.6) | 69 (92.0) | |
| IDH2 | 0.556 | ||
| Mutated | 14 (4.7) | 5 (6.7) | |
| Wild-type | 283 (95.3) | 70 (93.3) | |
| MLL-PTD | 0.097 | ||
| Present | 3 (4.8) | 0 | |
| Absent | 60 (95.2) | 73 (100.0) | |
| TP53 | 1.00 | ||
| Mutated | 1 (0.3) | 0 | |
| Wild-type | 296 (99.7) | 75 (100.0) | |
| SRSF2 | 1.00 | ||
| Mutated | 3 (1.0) | 1 (1.3) | |
| Wild-type | 292 (99.0) | 74 (98.7) | |
| ASXL1 | 0.265 | ||
| Mutated | 3 (1.0) | 2 (2.7) | |
| Wild-type | 294 (99.0) | 73 (97.3) | |
| RUNX1 | 1.00 | ||
| Mutated | 2 (0.7) | 0 | |
| Wild-type | 295 (99.3) | 75 (100.0) | |
| BCOR | 1.00 | ||
| Mutated | 3 (1.0) | 1 (1.3) | |
| Wild-type | 294 (99.0) | 74 (98.7) |
Continuous variables were summarized by reporting the median (range), while categorical variables were summarized by reporting frequency (percentage).
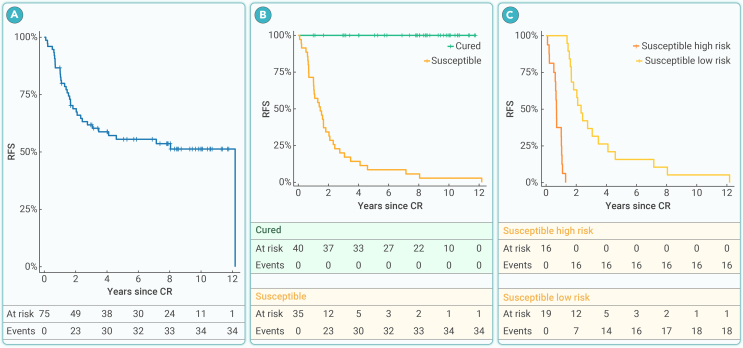
Figure 1.
Outcomes of patients with AML in the training set
(A) Kaplan-Meier curve for RFS.
(B) Kaplan-Meier curves for RFS of patients predicted to be cured and those susceptible to relapse or death using our MCM signature (p < 0.0001, log rank test).
(C) Kaplan-Meier curves for RFS of patients predicted to be susceptible to relapse or death, stratified by high versus low risk of relapse or death using our MCM signature (p < 0.0001, log rank test).
When analyzing the RNA-seq data for our training set, there were originally 23,549 transcripts. After filtering to remove transcripts with lower and less variable expression values, 7,993 transcripts remained for fitting a penalized Weibull MCM. The MCM included 145 transcripts in the incidence portion of the model, which are associated with long-term cure (Table S1), while 149 transcripts were included in the latency portion of the model, which are associated with short-term time to relapse (Table S2), with seven transcripts appearing in both portions of the model (SPRYD7, ZNF714, KLF11, NR1D2, EOLA1, RNU4-1, and CFAP45). As expected for the training set, the Kaplan-Meier curves for patients predicted to be cured versus those predicted to be susceptible (Figure 1B) and for patients predicted to be susceptible with lower versus higher risk of relapse or death (Figure 1C) were well separated. At 5 years, the MCM signature yielded an area under the receiver operating characteristic curve (AUC) of 0.946 and a C-statistic of 0.856, indicating good predictive capability of the selected transcripts. Interestingly, when examining baseline demographics and clinical characteristics among the three resulting MCM risk groups in the training set (namely, cured, susceptible with lower risk, and susceptible with higher risk), only frequency of NPM1 mutation (p = 0.014) differed significantly (Table 2).
Table 2.
Demographic, clinical, and gene mutation data in the training set, categorized by the penalized MCM as cured or susceptible with low risk or high risk
| Characteristica | Cured (n = 129) | Low risk (n = 79) | High risk (n = 89) | p Value |
|---|---|---|---|---|
| Age, years | 44 (18–59) | 43 (19–59) | 44 (17–59) | 0.495 |
| Sex | 0.785 | |||
| Female | 56 (43.4) | 38 (48.1) | 39 (43.8) | |
| Male | 73 (56.6) | 41 (51.9) | 50 (56.2) | |
| Hemoglobin, g/dL | 9.2 (3.1–25.1) | 9.1 (4.9–14.0) | 9.1 (2.3–14.2) | 0.680 |
| Platelet count, ×109/L | 50 (8–369) | 47 (7–433) | 43 (7–271) | 0.106 |
| White blood cell count, ×109/L | 23.5 (1.8–298.4) | 29.9 (0.6–158.5) | 27.8 (0.4–303.6) | 0.419 |
| Bone marrow blasts, % | 61.5 (2–97) | 65.0 (18–95) | 63.0 (23–95) | 0.068 |
| Blood blasts, % | 49 (0–97) | 57 (0–97) | 56 (1–97) | 0.198 |
| Cytogenetic groups, % | ||||
| Core-binding factor AML | 52 (40.3) | 24 (30.4) | 36 (40.4) | 0.292 |
| inv(16)(p13.1q22) | 30 (23.3) | 20 (25.3) | 23 (25.8) | 0.898 |
| t(8;21)(q22;q22) | 22 (17.1) | 4 (5.1) | 13 (14.6) | 0.029 |
| Sole +8 | 0 | 2 (2.5) | 1 (1.1) | 0.108 |
| DNMT3A | 0.762 | |||
| Mutated | 30 (23.3) | 15 (19.0) | 18 (20.2) | |
| Wild-type | 99 (76.7) | 64 (81.0) | 71 (79.8) | |
| NRAS | 0.232 | |||
| Mutated | 26 (20.2) | 13 (16.5) | 24 (27.0) | |
| Wild-type | 103 (79.8) | 66 (83.5) | 65 (73.0) | |
| SF3B1 | 0.279 | |||
| Mutated | 2 (1.6) | 0 | 3 (3.4) | |
| Wild-type | 127 (98.4) | 79 (100.0) | 86 (96.6) | |
| IDH1 | 0.318 | |||
| Mutated | 5 (3.9) | 7 (8.9) | 6 (6.7) | |
| Wild-type | 124 (96.1) | 72 (91.1) | 83 (93.3) | |
| CEBPAbzip | 0.612 | |||
| Mutated | 25 (19.4) | 13 (16.5) | 20 (22.5) | |
| Wild-type | 104 (80.6) | 66 (83.5) | 69 (77.5) | |
| GATA2 | 0.488 | |||
| Mutated | 10 (7.8) | 4 (5.1) | 9 (10.1) | |
| Wild-type | 119 (92.2) | 75 (94.9) | 80 (89.9) | |
| TET2 | 0.138 | |||
| Mutated | 9 (7.0) | 10 (12.7) | 4 (4.5) | |
| Wild-type | 120 (93.0) | 69 (87.3) | 85 (95.5) | |
| NPM1 | 0.014 | |||
| Mutated | 53 (41.1) | 46 (58.2) | 33 (37.1) | |
| Wild-type | 76 (58.9) | 33 (41.8) | 56 (62.9) | |
| WT1 | 0.182 | |||
| Mutated | 4 (3.1) | 7 (8.9) | 6 (6.7) | |
| Wild-type | 125 (96.9) | 72 (91.1) | 83 (93.3) | |
| PTPN11 | 0.158 | |||
| Mutated | 10 (7.8) | 13 (16.5) | 9 (10.1) | |
| Wild-type | 119 (92.2) | 66 (83.5) | 80 (89.9) | |
| FLT3 TKD | 0.076 | |||
| Present | 21 (16.4) | 15 (19.2) | 7 (7.9) | |
| Absent | 107 (83.6) | 63 (80.8) | 82 (92.1) | |
| FLT3 ITD | 1.00 | |||
| Present | 4 (3.1) | 3 (3.8) | 3 (3.4) | |
| Absent | 124 (96.9) | 75 (96.2) | 85 (96.6) | |
| IDH2 | 0.342 | |||
| Mutated | 4 (3.1) | 6 (7.6) | 4 (4.5) | |
| Wild-type | 125 (96.9) | 73 (92.4) | 85 (95.5) | |
| MLL-PTD | 0.274 | |||
| Present | 3 (10.0) | 0 | 0 | |
| Absent | 27 (90.0) | 23 (100.0) | 10 (100.0) | |
| TP53 | 0.566 | |||
| Mutated | 0 | 0 | 1 (1.1) | |
| Wild-type | 129 (100.0) | 79 (100.0) | 88 (98.9) | |
| SRSF2 | 0.180 | |||
| Mutated | 0 (0.0) | 1 (1.3) | 2 (2.3) | |
| Wild-type | 128 (100.0) | 78 (98.7) | 86 (97.7) | |
| ASXL1 | 0.790 | |||
| Mutated | 2 (1.6) | 0 | 1 (1.1) | |
| Wild-type | 127 (98.4) | 79 (100.0) | 88 (98.9) | |
| RUNX1 | 0.319 | |||
| Mutated | 0 | 1 (1.3) | 1 (1.1) | |
| Wild-type | 129 (100.0) | 78 (98.7) | 88 (98.9) | |
| BCOR | 0.469 | |||
| Mutated | 1 (0.8) | 0 | 2 (2.2) | |
| Wild-type | 128 (99.2) | 79 (100.0) | 87 (97.8) |
Continuous variables were summarized by reporting the median (range), while categorical variables were summarized by reporting frequency (percentage).
Patients in the test set were diagnosed with AML more recently, and their median follow-up among patients alive at their last follow-up was 8.6 years (range, 1.0–12.6 years). While the Kaplan-Meier curve does not necessarily show that a cure fraction is present, because this patient cohort consists of younger ELN favorable genetic risk patients with gene expression data available, we considered it a useful proxy test set. When mapping the 287 unique transcripts from our training set MCM signature to the Affymetrix GeneChip test set for validation, there were 50 incidence probe sets (Table S3) and 44 latency probe sets (Table S4) that mapped to 41 and 37 unique genes in the test set MCM, respectively. At 5 years, the test set MCM produced an AUC of 0.877 and a C-statistic of 0.857, indicating good predictive capability of the selected genes. Figure 2B presents the Kaplan-Meier estimates for patients predicted to be cured versus patients predicted to be susceptible in the test set. As desired, those predicted to be cured had a survival probability of 1 throughout the observation period, while those predicted to be susceptible group had an estimated survival curve descending toward 0. Figure 2C presents the Kaplan-Meier estimates for patients predicted to be susceptible with higher or lower risk of relapse in the test set. These two risk groups among patients predicted to be susceptible were well separated (p < 0.0001). Baseline demographic and clinical characteristics of patients in the test set who were classified as cured, susceptible with lower risk, and susceptible with higher risk by the MCM signature are summarized in Table 3. There was a significant difference between the three MCM risk groups with respect to frequencies of DNMT3A (p = 0.018) and IDH1 (p = 0.011) mutations.
Figure 2.
Outcomes of patients with AML in the test set
(A) Kaplan-Meier curve for RFS.
(B) Kaplan-Meier curves for RFS of patients predicted to be cured and those susceptible to relapse or death using our MCM signature (p < 0.0001, log rank test).
(C) Kaplan-Meier curves for RFS of patients predicted to be susceptible to relapse or death, stratified by high versus low risk of relapse or death using our MCM signature (p < 0.0001, log rank test).
Table 3.
Demographic, clinical, and gene mutation data in the test set, categorized by the MCM as cured or susceptible with low or high risk of relapse or death
| Characteristica | Cured (n = 40) | Low risk (n = 19) | High risk (n = 16) | p Value |
|---|---|---|---|---|
| Age, years | 40.5 (18–59) | 43 (20–57) | 47.5 (22–58) | 0.319 |
| Sex | 0.069 | |||
| Female | 17 (42.5) | 14 (73.7) | 7 (43.8) | |
| Male | 23 (57.5) | 5 (26.3) | 9 (56.2) | |
| Hemoglobin, g/dL | 8.7 (3.5–13.4) | 9.2 (6.0–12.4) | 8.95 (4.2–13.3) | 0.944 |
| Platelet count, ×109/L | 35 (4–273) | 42 (11–268) | 47 (14–301) | 0.547 |
| White blood cell count, ×109/L | 20.35 (3.0–262.5) | 26 (1.1–206.0) | 43.48 (1.33–316.0) | 0.758 |
| Bone marrow blasts, % | 70 (20–97) | 70 (30–100) | 86.5 (30–95) | 0.549 |
| DNMT3A | 0.018 | |||
| Mutated | 3 (7.5) | 7 (36.8) | 3 (18.8) | |
| Wild-type | 37 (92.5) | 12 (63.2) | 13 (81.2) | |
| NRAS | 0.630 | |||
| Mutated | 14 (35.0) | 4 (21.1) | 5 (31.2) | |
| Wild-type | 26 (65.0) | 15 (78.9) | 11 (68.8) | |
| SF3B1 | N/A | |||
| Mutated | 0 | 0 | 0 | |
| Wild-type | 40 (100.0) | 19 (100.0) | 16 (100.0) | |
| IDH1 | 0.011 | |||
| Mutated | 0 | 3 (15.8) | 3 (18.8) | |
| Wild-type | 40 (100.0) | 16 (84.2) | 13 (81.2) | |
| CEBPAbzip | 0.117 | |||
| Mutated | 6 (15.0) | 2 (10.5) | 6 (37.5) | |
| Wild-type | 34 (85.0) | 17 (89.5) | 10 (62.5) | |
| GATA2 | 1.00 | |||
| Mutated | 2 (5.0) | 1 (5.3) | 0 | |
| Wild-type | 38 (95.0) | 18 (94.7) | 16 (100.0) | |
| TET2 | 0.414 | |||
| Mutated | 3 (7.5) | 2 (10.5) | 3 (18.8) | |
| Wild-type | 37 (92.5) | 17 (89.5) | 13 (81.2) | |
| NPM1 | 0.184 | |||
| Mutated | 11 (27.5) | 10 (52.6) | 5 (31.2) | |
| Wild-type | 29 (72.5) | 9 (47.4) | 11 (68.8) | |
| WT1 | 1.00 | |||
| Mutated | 7 (17.5) | 3 (15.8) | 2 (12.5) | |
| Wild-type | 33 (82.5) | 16 (84.2) | 14 (87.5) | |
| PTPN11 | 0.334 | |||
| Mutated | 4 (10.0) | 3 (15.8) | 0 | |
| Wild-type | 36 (90.0) | 16 (84.2) | 16 (100.0) | |
| FLT3 TKD | 0.329 | |||
| Present | 7 (17.5) | 5 (26.3) | 1 (6.2) | |
| Absent | 33 (82.5) | 14 (73.7) | 15 (93.8) | |
| FLT3 ITD | 0.862 | |||
| Present | 3 (7.5) | 2 (10.5) | 1 (6.2) | |
| Absent | 37 (92.5) | 17 (89.5) | 15 (93.8) | |
| IDH2 | 0.826 | |||
| Mutated | 2 (5.0) | 2 (10.5) | 1 (6.2) | |
| Wild-type | 38 (95.0) | 17 (89.5) | 15 (93.8) | |
| MLL-PTD | N/A | |||
| Present | 0 | 0 | 0 | |
| Absent | 39 (100.0) | 19 (100.0) | 15 (100.0) | |
| TP53 | N/A | |||
| Mutated | 0 | 0 | 0 | |
| Wild-type | 40 (100.0) | 19 (100.0) | 16 (100.0) | |
| SRSF2 | 0.467 | |||
| Mutated | 0 | 1 (5.3) | 0 | |
| Wild-type | 40 (100.0) | 18 (94.7) | 16 (100.0) | |
| ASXL1 | 1.000 | |||
| Mutated | 2 (5.0) | 0 | 0 | |
| Wild-type | 38 (95.0) | 19 (100.0) | 16 (100.0) | |
| RUNX1 | N/A | |||
| Mutated | 0 | 0 | 0 | |
| Wild-type | 40 (100.0) | 19 (100.0) | 16 (100.0) | |
| BCOR | 0.467 | |||
| Mutated | 0 | 1 (5.3) | 0 | |
| Wild-type | 40 (100.0) | 18 (94.7) | 16 (100.0) |
Continuous variables were summarized by reporting the median (range), while categorical variables were summarized by reporting frequency (percentage).
Several genes included in our MCM signature were identified in previously published studies as having relevance in AML. Notably, the signature was enriched for genes previously reported in AML patients with favorable-risk chromosome abnormalities; namely, inv(16)(p13.1q22) and t(15;17)(q24;q21). Seventeen genes in our MCM signature (DOCK4, NUP50, SPRY2, HLX, NR4A1, MEST, MELTF, HSPB1, PML, DUSP2, ARHGEF12, IDS, SPRYD7, TRIM16, PPP1R16B, MAPKBP1, and RAB11FIP3) were identified as differentially expressed in a comparison of pediatric patients with AML harboring inv(16) with those who did not have inv(16).10 However, in our analyses, these genes were differentially expressed irrespective of favorable-risk-defining genetic lesions. Eight genes in our MCM signature (CDKN1C, STAB1, ST3GAL6, CST7, IGFBP2, TLE1, LTK, and CTSW) were among the top 100 genes in pediatric patients with AML that were specific to t(15;17), which encodes the PML-RARA protein.11 Relatedly, ZNF506 regulates transcription and is regulated by the PML protein nuclear bodies12; both ZNF506 and PML are in our MCM signature, whereas NR4A1, which plays a role in apoptosis of T lymphocytes, is transcriptionally repressed by PML.13 Subsequent pathway analyses revealed an enrichment of genes (10 of 17) involved in blood vessel morphogenesis (angiogenesis), which represents a previously established important biologic process in AML and other hematologic malignancies given the essential contribution of vascular endothelial cells to the vascular niches of the bone marrow microenvironment.14
Other genes in our MCM signature are related to other blood cancers. In a study of chronic lymphocytic leukemia, 18 genes from our MCM signature (TNFSF9, MFHAS1, NAGLU, FXYD5, HMGCS1, HMG20B, TSTD1, TMC8, CDT1, RALA, PIM2, DUSP5, AGPAT1, RGS1, GPR160, INTS6, TIFA, and LSP1) were among those upregulated at least 2-fold by CD5 when comparing empty vector and CD5-transfected B cells. In a study of T cell prolymphocytic leukemia (T-PLL), among genes downregulated in purified inv(14)/t(14;14)-positive T-PLL blood samples compared to purified CD3+ peripheral blood samples from healthy donors, 14 were in our MCM signature, namely, F2R, ACSL3, DUSP5, RDX, CST7, FAM98A, IDS, FAS, ENTPD4, INTS6, PPP1R16B, PRKCB, H3-3B, and CTSW.15 GAR1, which contributes to telomerase activity, was downregulated in mononuclear cells from chronic lymphocytic leukemia patients compared to normal controls.16
Topp Gene (accessed on October 2, 2023) was used to provide biological insights with respect to genes included in the training set MCM. Several genes in our MCM signature were over-represented with respect to specific molecular functions and pathways. The molecular function of four genes in our MCM signature (DUSP2, DUSP5, DUSP7, and DUSP14; false discovery rate [FDR] = 0.011) is mitogen-activated protein kinase phosphatase activity, whereas 40 genes are involved in transcription regulator activity (HIC1, ERF, SERTAD2, HLX, ETV5, NR4A1, ZNF440, KLF2, TSTD1, JUN, PRDM8, ZNF506, ZBTB18, MYRF, ZNF14, NR1D2, PML, ZNF487, KLF11, ZBTB2, MAGED1, ZNF555, MAFK, MED31, ZNF507, NR4A2, RUNX2, ZBED3, ZNF865, HIVEP3, ZNF865, CITED4, GZF1, TLE1, ZNF335, TADA2B, SMARCA2, CDYL2, PRKCB, and MXD1; FDR = 0.026). Three genes in our MCM signature are involved in fatty acid synthase activity (ELOVL5, ELOVL6, and MCAT; FDR = 0.038). Among genes in our MCM signature, eight (CDKN1C, SERTAD2, JUN, MFGE8, FGD4, PRRG4, SEC14L1, and DNAJC15) belong to the glucocorticoid receptor pathway (FDR = 0.003), 10 (NAGLU, NEU1, AP3S1, IDS, ENTPD4, LIPA, ARSA, PPT1, GGA1, and CTSW) belong to the lysosome pathway (FDR = 0.003), and four (ELOVL5, ACSL3, ELOVL6, and FADS1) belong to the omega 9 fatty acid synthesis pathway (FDR = 0.009).
Discussion
The 2022 ELN genetic risk classification and its 2017 predecessor differentiate prognosis between three risk groups: favorable, intermediate, and adverse. Patients aged <60 years included in the 2017 ELN favorable group are generally expected to do well, with a 3-year OS rate reported as 64% in one large cohort17 and a 5-year OS rate reported as 64.2% in another.18 Although a sizable proportion of patients classified in the 2022 ELN favorable genetic risk group experience long-term RFS, others are susceptible to relapse. In this study, we applied an MCM to high-dimensional RNA-seq data and identified important subsets of genes associated with long-term cure and time to relapse or death. The identified subgroups (cured and susceptible with lower risk or higher risk of relapse or death) differed significantly by clinical outcome, suggesting that the 2022 ELN favorable genetic-risk group is heterogeneous and includes distinct subgroups having different expression profiles and biological properties, which may include different sensitivities to therapeutic agents. While we could not directly apply the coefficients from our training set to our test set due to the differences between the RNA-seq and Affymetrix platforms, the estimated 5-year AUC of 0.877 and C-statistic of 0.857 for the more limited set of genes in the Affymetrix GeneChip test set suggests good predictive capability of the selected genes. Thus, the identified transcripts may be useful in differentiating patient subgroups and may serve as a prognostic signature to better inform treatment decisions for 2022 ELN favorable AML patients with different expression characteristics.
In our previous article by Fu et al.,9 we compared our MCM with two competing mixture cure modeling methods that can handle a high-dimensional covariate space: C-mix by Bussy et al.19 and sign consistency in cure rate models (SCinCRM) by Shi et al.20 The C-Mix model includes the MCM as a special case and applies to high-dimensional data, but it only allows covariates into the incidence portion of the model so that covariates are not included in the latency portion of the model. The SCinCRM model can be used to fit a high-dimensional MCM but imposes a sign-based penalty to promote the same coefficient sign in the incidence and latency portions of the model. Our MCM does not suffer from either restriction. Our extensive simulation studies described in Fu et al.9 demonstrated that our method outperformed both of these high-dimensional mixture cure methods and additionally outperformed a penalized Weibull model that does not take cure into account. While we did not reiterate our previous findings, we did examine results when fitting a penalized Cox PH model, as that model would be the most likely alternative method used in a high-dimensional time-to-event data analysis (supplemental information). Our MCM outperformed the penalized Cox PH model and allows for the identification of three groups; namely, cured, susceptible with lower risk, and susceptible with higher risk.
Several gene mutations as well as differences in expression of specific genes, such as BAALC, ERG, EVI1, and MN1,21 have been identified as having prognostic relevance, though they have not yet entered standards for clinical decision-making.22 Recently, a genome-wide association study of 104 patients with core-binding factor AML identified prognostic models for OS and event-free survival (EFS) where each model included six SNPs along with age group (<55 vs. ≥ 55 years), the presence versus absence of exon 17 mutations in the KIT gene, and lactate dehydrogenase level.23 Others found that inclusion of gene expression in addition to cytogenetic and gene mutation data could enhance the prognostic performance of the 2010 ELN genetic risk classification.24 Research to elucidate mutations and transcriptional expression associated with ex vivo drug activity is underway.25 In a review article describing the role of cytogenetics and the addition of mutation data in AML diagnosis and treatment decision-making, the authors predicted that gene expression profiling and next-generation sequencing will be at the center of future AML diagnostics.26 As next-generation sequencing turnaround times and costs decrease, global genomics and transcriptomics could be included in genetic risk stratification systems.27
Previous studies applied different methods to high-dimensional gene expression data for exploring prognostic subgroups in AML. For example, unsupervised clustering of gene expression data was applied to identify novel subgroups of AML patients.28 Different genes discriminated between the identified subgroups, suggesting there is heterogeneity among functional pathways that lead to AML. Unsupervised clustering of gene expression profiles in core-binding factor AML, which constitutes a large proportion of ELN favorable patients, identified a subgroup having shorter OS.29 These previous studies used unsupervised methods to identify subgroups followed by class comparisons to identify differentially expressed genes and then applied survival analytic methods to determine whether the identified subgroups differed by outcome. This is in contrast to others, who used gene expression profiling to classify favorable-risk AML patients with balanced rearrangements30 or to identify genes associated with resistance to induction chemotherapy in AML31 or resistant disease.32 The latter study32 combined gene expression with cytogenetics to develop a risk score that outperformed existing classifiers, including those based on age, sex, performance status, white blood cell count, platelet count, bone marrow blasts, type of AML, mutation status of NPM1 and FLT3-ITD, and cytogenetics,33 a modified classifier that additionally included mutations,34 or LSC17 stemness.35,36 However, due to the low proportion of ELN favorable AML patients with resistant disease, this classifier’s prognostic performance for OS was limited to only the intermediate and adverse ELN prognostic groups. A meta-analysis of gene expression across four microarray studies further identified a compound covariate 24-gene expression signature that significantly enhanced ELN in predicting OS and EFS, indicating that molecular signatures can improve the prognostic classification of AML beyond ELN.37 However, their goal was to identify genes that are consistently associated with AML prognosis, so that no age restriction was imposed, and patients with various cytogenetic and molecular abnormalities were included; thus, training and validation data spanned all ELN genetic risk groups. Recently, transcriptomic data had better prognostic performance than clinical, cytogenetic, and somatic mutation data in predicting OS, although the median follow-up in the training data was only 263 days38
In recognition that there is heterogeneity in patient outcomes even within well-defined molecular subgroups, researchers used various regularized regression methods predicting OS using 231 predictors that included fusion genes, copy number alterations, point mutations, gene-gene interactions, demographic features, clinical risk factors, and treatment received to identify which patients should be offered allogeneic hematopoietic cell transplant in the first CR in a large diverse cohort of 1,540 AML patients.39 In contrast, we focused our analyses on younger 2022 ELN favorable genetic risk AML patients. While we examined the association between demographic and select mutations at diagnosis, we derived a penalized MCM to predict RFS using only transcript expression assessed in diagnostic samples as our predictors. Our model was effective in separating 2022 ELN favorable genetic risk AML patients into those cured or having a long-term durable CR and those susceptible to relapse or death with either lower or higher risk. We also validated our MCM signature by assessing its performance using a test set that used a different assay to measure gene expression, demonstrating its independence.
With respect to signature-defining genes, we noted an inclusion of genes that have been reported previously in studies on different genetic subsets of ELN favorable risk AML; namely, inv(16) and t(15;17). However, as the genes reported previously in an inv(16)-associated signature were found to be deregulated in our cohort irrespective of cytogenetic subgroup, and as our patient cohort did not include any t(15;17) AML, it is tempting to speculate that these genes indeed represent a favorable risk signature rather than genomic responses associated with the presence or absence of recurrent genetic lesions.
Materials and methods
Patients and treatment
Based on previous research showing that outcome analyses should be stratified by age group,40 our training set included 297 patients aged <60 years (range, 17–59) diagnosed with de novo AML (excluding acute promyelocytic leukemia) between 1986 and 2016 and enrolled on frontline Cancer and Leukemia Group B (CALGB) clinical trials and companion cytogenetic (CALGB 8461), leukemia tissue bank (CALGB 9665), and molecular (CALGB 20202) studies. CALGB is now part of Alliance for Clinical Trials in Oncology (Alliance). All patients achieved CR and were classified as favorable genetic risk according to the 2022 ELN.1 Generally, patients received intensive cytarabine and daunorubicin or idarubicin-based induction treatment. No patient received allogeneic hematopoietic stem cell transplantation (HSCT) in the first CR on study protocols, and off-study patients who received an HSCT were excluded because of missing or incomplete follow-up data. All patients’ karyotypes underwent central review,41 and, additionally, all patients had RNA-seq and selected gene mutation data from diagnostic samples available. To validate the performance of our model, we employed an independent test set from the Acute Myeloid Leukemia German Cooperative Group (AMLCG) available under GEO: GSE37642,32,37,42,43 where diagnostic samples were hybridized to Affymetrix GeneChips. Patients were initially enrolled in the AMLCG 1999 study (ClinicalTrials.gov: NCT00266136), which actively recruited between 1999 and 2005. We restricted our analysis of the test set to the 75 patients with de novo AML aged <60 years who were treated on AMLCG trials, achieved a CR, were classified as favorable genetic risk according to the 2022 ELN,1 and had gene expression data from pre-treatment samples and relapse data available. In accordance with the Declaration of Helsinki, patients provided study-specific written informed consent for participation in the treatment studies. Institutional review board approval of all CALGB/Alliance and AMLCG protocols was obtained before any research was performed.
Clinical endpoints and statistical analysis
We estimated RFS as the time from achievement of a CR to relapse, death, or last follow-up, censoring for patients alive without relapse.1 The Kaplan-Meier method was used to estimate the proportion of cured patients, or cured fraction, as the survival estimate at the maximum follow-up time.6,44 The term “cured” typically implies that the patient is immune to the event of interest. However, because we considered both relapse and death as events, here, “cured” is synonymous with long-term RFS; in other words, patency of CR. To test whether there was a significant cured fraction, the rate parameter for an exponential survival model was estimated45 and then used to simulate time-to-event data from an exponential distribution under the condition of the null hypothesis, where 10,000 simulations were used.44 For each simulated dataset, the proportion of patients susceptible to the event (uncured) was estimated. The p value was calculated as the proportion of times the observed susceptible fraction estimate exceeded the simulated estimate.44 We additionally tested for the presence of a cured fraction by fitting a parametric MCM and the parametric survival model and performing a boundary likelihood ratio test.44
We also need to have decisive evidence that there is sufficient follow-up44,46 to ensure consistency of Kaplan-Meier estimates and to ensure that we can reliably detect whether a cured fraction exists. In other words, we need to test that we have followed all patients long enough so that we would have observed the event times for all susceptible subjects. We estimated the time when 95% of subjects should experience the event47 and compared that estimate with our median follow-up among patients relapse free and alive. We also performed an inferential test examining whether there is sufficient follow-up.48
Pre-processing methods applied to the RNA-seq data are described in the supplemental information. The dimensionality of the RNA-seq dataset was reduced by removing transcripts with lower and less variable expression values and by retaining transcripts having a mean log expression greater than 4 and standard deviation of logged expression values greater than 0.4. The filtered expression values were then used as candidate covariates in our penalized Weibull MCM.9 The final model was selected as that attaining the minimum corrected Akaike information criterion. Thereafter, the linear predictor for the incidence portion of the model was used to predict cured versus susceptible using zero as the cutpoint. Likewise, the linear predictor for the latency portion of the model was used to predict higher versus lower risk of relapse or death using zero as the cutpoint. When comparing whether there were significant differences with respect to baseline variables among the three subgroups in the training set (cured, susceptible with lower risk, and susceptible with higher risk), we used a Kruskal-Wallis test for continuous variables and Fisher’s exact test for categorical variables.
To validate the relevance and importance of our identified RNA transcripts, we employed an independent test set from the AMLCG (derived from GEO: GSE37642).32,37,42,43 The GEO: GSE37642 dataset includes clinical and gene expression data for 562 patients with AML. We restricted our analysis to the 75 patients with de novo AML aged <60 years who achieved a CR and were classified as having favorable genetic risk according to the 2022 ELN with RFS recorded. Because the gene expression levels were measured using the Affymetrix HG-U133Plus2, HG-U133A, and HG-U133B GeneChips, we could not normalize these data with our training set RNA-seq data. That is, differences between RNA-seq and Affymetrix with respect to the dynamic range, the structure of the expression data (count data for RNA-seq versus relative abundance for Affymetrix), sequencing versus fixed hybridized probe design, and lack of a one-to-one transcript to probe set correspondence precluded a direct comparison. Instead, we matched these GeneChips on probe set ID to form an integrated dataset. We then mapped the transcripts included in our training set MCM signature to their corresponding probe sets via gene symbols. Due to differences in the scale of gene expression data between the training and test sets and the inability to completely match all RNA-seq MCM signature transcripts to probe sets, we fit a penalized Weibull MCM to the test set using available probe sets that matched transcripts in our training set MCM signature. Again, the linear predictor for the incidence portion of the model was used to predict cured versus susceptible using zero as the cutpoint. Likewise, the linear predictor for the latency portion of the model was used to predict higher versus lower risk of relapse/death using zero as the cutpoint. The AUC and C-statistic designed for MCMs were calculated to assess predictive performance.49 When comparing whether there were significant differences with respect to baseline variables among the three subgroups in the test set (cured, susceptible with lower risk, and susceptible with higher risk), we used a Kruskal-Wallis test for continuous variables and Fisher’s exact test for categorical variables.
Data and code availability
The filtered gene expression data used for the training and test sets along with the corresponding R code are provided at https://github.com/kelliejarcher/AML_2022_ELN_Favorable. Data are further summarized in the supplemental information files. The full dataset from which the test set was derived is available from GEO: GSE37642.
Acknowledgments
Research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under award R01LM013879. Research reported in this publication was also supported in part by the National Cancer Institute at the National Institutes of Health under award R01CA262496, R01CA284595, R01CA283574, U10CA180821, U10CA180882, U24CA196171, UG1CA233327, UG1CA233331, UG1CA233338, UG1CA233339, R35CA197734, and P30CA016058; the Coleman Leukemia Research Foundation; an ASH Junior Faculty Scholar Award and ASH Bridge Grant (to A.-K.E.); the Leukemia Research Foundation (to A.-K.E.); the Leukemia & Lymphoma Society (to A.-K.E.); The D. Warren Brown Foundation; and by an allocation of computing resources from The Ohio Supercomputer Center and Shared Resources (Leukemia Tissue Bank). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Support to Alliance for Clinical Trials in Oncology and Alliance Foundation Trials, LLC programs is listed at https://acknowledgments.alliancefound.org. The authors are grateful to the patients who consented to participate in clinical trials and the families who supported them; to Christopher Manning and the Alliance Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for sample processing and storage services; and to Lisa J. Sterling for data management.
Author contributions
K.J.A. and A.-K.E. conceived and designed the study; H.F. and K.J.A. developed the statistical method and computational code; K.J.A. performed the statistical analyses; K.J.A., H.F., K.M., A.S.M., D.N., T.H., and A.-K.E. wrote the manuscript; K.J.A., H.F., K.M., D.N., A.S.M., G.L.U., W.S., J.C.B., C.R., U.K., C.S., D.G., W.E.B., B.J.W., J.B., K.S., T.H., K.H.M., W.H., and A.-K.E. edited the manuscript; and J.C.B., T.H., K.H.M., W.H., C.R., U.K., C.S., D.G., W.E.B., B.J.W., J.B., K.S., and A.-K.E. provided study materials or patients. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published Online: October 21, 2024
Footnotes
It can be found online at https://doi.org/10.1016/j.xinn.2024.100719.
Lead contact website
Supplemental information
References
- 1.Döhner H., Wei A.H., Appelbaum F.R., et al. Diagnosis and management of AML in adults: 2022 ELN recommendations from an international expert panel. Blood. 2022;140:1345–1377. doi: 10.1182/blood.2022016867. [DOI] [PubMed] [Google Scholar]
- 2.Mrózek K., Kohlschmidt J., Blachly J.S., et al. Outcome prediction by the 2022 European LeukemiaNet genetic-risk classification for adults with acute myeloid leukemia: an Alliance study. Leukemia. 2023;37:788–798. doi: 10.1038/s41375-023-01846-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rausch C., Rothenberg-Thurley M., Dufour A., et al. Validation and refinement of the 2022 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2023;37:1234–1244. doi: 10.1038/s41375-023-01884-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kantarjian H., O’Brien S., Cortes J., et al. Therapeutic advances in leukemia and myelodysplastic syndrome over the past 40 years. Cancer. 2008;113:1933–1952. doi: 10.1002/cncr.23655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derolf A.R., Kristinsson S.Y., Andersson T.M.-L., et al. Improved patient survival for acute myeloid leukemia: a population-based study of 9729 patients diagnosed in Sweden between 1973 and 2005. Blood. 2009;113:3666–3672. doi: 10.1182/blood-2008-09-179341. [DOI] [PubMed] [Google Scholar]
- 6.Goldman A.I. The cure model and time confounded risk in the analysis of survival and other timed events. J. Clin. Epidemiol. 1991;44:1327–1340. doi: 10.1016/0895-4356(91)90094-p. [DOI] [PubMed] [Google Scholar]
- 7.Andersson T.M.-L., Lambert P.C., Derolf A.R., et al. Temporal trends in the proportion cured among adults diagnosed with acute myeloid leukaemia in Sweden 1973-2001, a population-based study. Br. J. Haematol. 2010;148:918–924. doi: 10.1111/j.1365-2141.2009.08026.x. [DOI] [PubMed] [Google Scholar]
- 8.Sposto R. Cure model analysis in cancer: an application to data from the Children’s Cancer Group. Stat. Med. 2002;21:293–312. doi: 10.1002/sim.987. [DOI] [PubMed] [Google Scholar]
- 9.Fu H., Nicolet D., Mrózek K., et al. Controlled variable selection in Weibull mixture cure models for high-dimensional data. Stat. Med. 2022;41:4340–4366. doi: 10.1002/sim.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yagi T., Morimoto A., Eguchi M., et al. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849–1856. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
- 11.Ross M.E., Mahfouz R., Onciu M., et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 12.Fleischer S., Wiemann S., Will H., et al. PML-associated repressor of transcription (PAROT), a novel KRAB-zinc finger repressor, is regulated through association with PML nuclear bodies. Exp. Cell Res. 2006;312:901–912. doi: 10.1016/j.yexcr.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Wu W.-S., Xu Z.-X., Ran R., et al. Promyelocytic leukemia protein PML inhibits Nur77-mediated transcription through specific functional interactions. Oncogene. 2002;21:3925–3933. doi: 10.1038/sj.onc.1205491. [DOI] [PubMed] [Google Scholar]
- 14.Testa U., Castelli G., Pelosi E. Angiogenesis in acute myeloid leukemia. J. Cancer Metastasis Treat. 2020;2020 doi: 10.20517/2394-4722.2020.111. [DOI] [Google Scholar]
- 15.Dürig J., Bug S., Klein-Hitpass L., et al. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32) Leukemia. 2007;21:2153–2163. doi: 10.1038/sj.leu.2404877. [DOI] [PubMed] [Google Scholar]
- 16.Dos Santos P.C., Panero J., Stanganelli C., et al. Dysregulation of H/ACA ribonucleoprotein components in chronic lymphocytic leukemia. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisfeld A.-K., Kohlschmidt J., Mims A., et al. Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years. Leukemia. 2020;34:3215–3227. doi: 10.1038/s41375-020-0872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herold T., Rothenberg-Thurley M., Grunwald V.V., et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2020;34:3161–3172. doi: 10.1038/s41375-020-0806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bussy S., Guilloux A., Gaïffas S., et al. C-mix: A high-dimensional mixture model for censored durations, with applications to genetic data. Stat. Methods Med. Res. 2019;28:1523–1539. doi: 10.1177/0962280218766389. [DOI] [PubMed] [Google Scholar]
- 20.Shi X., Ma S., Huang Y. Promoting sign consistency in the cure model estimation and selection. Stat. Methods Med. Res. 2020;29:15–28. doi: 10.1177/0962280218820356. [DOI] [PubMed] [Google Scholar]
- 21.Baldus C.D., Mrózek K., Marcucci G., et al. Clinical outcome of de novo acute myeloid leukaemia patients with normal cytogenetics is affected by molecular genetic alterations: a concise review. Br. J. Haematol. 2007;137:387–400. doi: 10.1111/j.1365-2141.2007.06566.x. [DOI] [PubMed] [Google Scholar]
- 22.Marcucci G., Haferlach T., Döhner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J. Clin. Oncol. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 23.Park S., Choi H., Kim H.J., et al. Genome-wide genotype-based risk model for survival in core binding factor acute myeloid leukemia patients. Ann. Hematol. 2018;97:955–965. doi: 10.1007/s00277-018-3260-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang M., Lindberg J., Klevebring D., et al. Validation of risk stratification models in acute myeloid leukemia using sequencing-based molecular profiling. Leukemia. 2017;31:2029–2036. doi: 10.1038/leu.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyner J.W., Tognon C.E., Bottomly D., et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visani G., Loscocco F., Isidori A., et al. Genetic profiling in acute myeloid leukemia: a path to predicting treatment outcome. Expert Rev. Hematol. 2018;11:455–461. doi: 10.1080/17474086.2018.1475225. [DOI] [PubMed] [Google Scholar]
- 27.Haferlach T., Schmidts I. The power and potential of integrated diagnostics in acute myeloid leukaemia. Br. J. Haematol. 2020;188:36–48. doi: 10.1111/bjh.16360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valk P.J.M., Verhaak R.G.W., Beijen M.A., et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 29.Bullinger L., Rücker F.G., Kurz S., et al. Gene-expression profiling identifies distinct subclasses of core binding factor acute myeloid leukemia. Blood. 2007;110:1291–1300. doi: 10.1182/blood-2006-10-049783. [DOI] [PubMed] [Google Scholar]
- 30.Schoch C., Kohlmann A., Schnittger S., et al. Acute myeloid leukemias with reciprocal rearrangements can be distinguished by specific gene expression profiles. Proc. Natl. Acad. Sci. USA. 2002;99:10008–10013. doi: 10.1073/pnas.142103599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heuser M., Wingen L.U., Steinemann D., et al. Gene-expression profiles and their association with drug resistance in adult acute myeloid leukemia. Haematologica. 2005;90:1484–1492. doi: 10.3324/%x. [DOI] [PubMed] [Google Scholar]
- 32.Herold T., Jurinovic V., Batcha A.M.N., et al. A 29-gene and cytogenetic score for the prediction of resistance to induction treatment in acute myeloid leukemia. Haematologica. 2018;103:456–465. doi: 10.3324/haematol.2017.178442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter R.B., Othus M., Burnett A.K., et al. Resistance prediction in AML: analysis of 4601 patients from MRC/NCRI, HOVON/SAKK, SWOG and MD Anderson Cancer Center. Leukemia. 2015;29:312–320. doi: 10.1038/leu.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter R.B., Othus M., Paietta E.M., et al. Effect of genetic profiling on prediction of therapeutic resistance and survival in adult acute myeloid leukemia. Leukemia. 2015;29:2104–2107. doi: 10.1038/leu.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng S.W.K., Mitchell A., Kennedy J.A., et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 36.Bill M., Nicolet D., Kohlschmidt J., et al. Mutations associated with a 17-gene leukemia stem cell score and the score’s prognostic relevance in the context of the European LeukemiaNet classification of acute myeloid leukemia. Haematologica. 2020;105:721–729. doi: 10.3324/haematol.2019.225003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Herold T., He C., et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J. Clin. Oncol. 2013;31:1172–1181. doi: 10.1200/JCO.2012.44.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang M., Lindberg J., Klevebring D., et al. Development and validation of a novel RNA sequencing–based prognostic score for acute myeloid leukemia. J. Natl. Cancer Inst. 2018;110:1094–1101. doi: 10.1093/jnci/djy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerstung M., Papaemmanuil E., Martincorena I., et al. Precision oncology for acute myeloid leukemia using a knowledge bank approach. Nat. Genet. 2017;49:332–340. doi: 10.1038/ng.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mrózek K., Marcucci G., Nicolet D., et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J. Clin. Oncol. 2012;30:4515–4523. doi: 10.1200/JCO.2012.43.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mrózek K., Carroll A.J., Maharry K., et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: the Cancer and Leukemia Group B experience. Int. J. Oncol. 2008;33:239–244. doi: 10.3892/ijo_00000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herold T., Metzeler K.H., Vosberg S., et al. Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood. 2014;124:1304–1311. doi: 10.1182/blood-2013-12-540716. [DOI] [PubMed] [Google Scholar]
- 43.Kuett A., Rieger C., Perathoner D., et al. IL-8 as mediator in the microenvironment-leukaemia network in acute myeloid leukaemia. Sci. Rep. 2015;5 doi: 10.1038/srep18411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maller R.A., Zhou X. Wiley; 1996. Survival Analysis with Long-Term Survivors. [Google Scholar]
- 45.Jackson C.H. flexsurv: a platform for parametric survival modeling in R. J. Stat. Software. 2016;70:i08–i33. doi: 10.18637/jss.v070.i08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Othus M., Bansal A., Erba H., et al. Bias in mean survival from fitting cure models with limited follow-up. Value Health. 2020;23:1034–1039. doi: 10.1016/j.jval.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldman A.I. Survivorship analysis when cure is a possibility: A Monte Carlo study. Stat. Med. 1984;3:153–163. doi: 10.1002/sim.4780030208. [DOI] [PubMed] [Google Scholar]
- 48.Maller R.A., Zhou S. Testing for sufficient follow-up and outliers in survival data. J. Am. Stat. Assoc. 1994;89:1499–1506. doi: 10.2307/2291012. [DOI] [Google Scholar]
- 49.Asano J., Hirakawa A. Assessing the prediction accuracy of a cure model for censored survival data with long-term survivors: application to breast cancer data. J. Biopharm. Stat. 2017;27:918–932. doi: 10.1080/10543406.2017.1293082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The filtered gene expression data used for the training and test sets along with the corresponding R code are provided at https://github.com/kelliejarcher/AML_2022_ELN_Favorable. Data are further summarized in the supplemental information files. The full dataset from which the test set was derived is available from GEO: GSE37642.