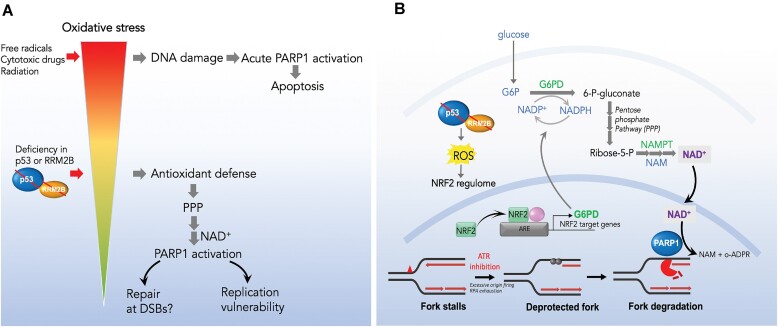
Abstract
Mechanisms underlying p53-mediated protection of the replicating genome remain elusive, despite the quintessential role of p53 in maintaining genomic stability. Here, we uncover an unexpected function of p53 in curbing replication stress by limiting PARP1 activity and preventing the unscheduled degradation of deprotected stalled forks. We searched for p53-dependent factors and elucidated RRM2B as a prime factor. Deficiency in p53/RRM2B results in the activation of an NRF2 antioxidant transcriptional program, with a concomitant elevation in basal PARylation in cells. Dissecting the consequences of p53/RRM2B loss revealed a crosstalk between redox metabolism and genome integrity that is negotiated through a hitherto undescribed NRF2-PARP1 axis, and pinpoint G6PD as a primary oxidative stress-induced NRF2 target and activator of basal PARylation. This study elucidates how loss of p53 could be destabilizing for the replicating genome and, importantly, describes an unanticipated crosstalk between redox metabolism, PARP1 and p53 tumor suppressor pathway that is broadly relevant in cancers and can be leveraged therapeutically.
Graphical Abstract
Graphical Abstract.
Introduction
The correct recovery of stalled replication forks has emerged as a critical process in the maintenance of genomic stability (1,2). Dissecting the events from fork stalling to recovery or breakdown is imperative in understanding how cells cope with stochastic fork impediments and further elucidates how replication stress drives genetic instability in cancers. The ATR checkpoint pathway is critical for maintaining the stability of disrupted replication forks which are otherwise susceptible to breakage (3–5). Fork remodeling and the restoration of DNA replication are precisely regulated in cells and is a point of convergence for multiple fork protection and repair pathways (1,6,7). The emerging interplay between a vast number of factors involved in homologous recombination (HR), chromatin remodelers and nucleolytic enzymes highlights the complexity of fork maintenance pathways that are only beginning to be unraveled.
Fork degradation is shown to be a prominent mechanism of genomic instability. Under conditions of BRCA deficiency or mutations in the Fanconi Anemia pathway, excessive nucleolytic resection of the nascent DNA occurs, in part due to the impairment of Rad51 protection, resulting in chromosomal abnormalities (8–10). A broader network of RAD51-dependent and -independent factors have been elucidated as determinants of nascent DNA protection that is linked to genomic stability and chemosensitivity (7,9,11–16). It is proposed that the transient reversal of replication fork precedes fork degradation and is mediated by annealing of nascent DNA strands to create a regressed 4-way junction. This may allow for the temporary stabilization of replication fork until the obstructing lesion is removed or repaired (17,18). Fork reversal may also facilitate fork restart through template switching and homologous recombination to bypass DNA lesions by using an alternative undamaged DNA template (19–21). Among the best characterized fork remodeling enzymes that catalyse fork reversal include the SNF2 family DNA translocases SMARCAL1, ZRANB3, and HTLF, and also the F-box DNA helicase, FBH1 (22–26). Regression of the stalled replication fork involves converting the three-armed replication fork into a Holiday Junction, with the nascent DNA strands forming the extruded arm of the regressed fork structure (24–27). Unscheduled nucleolytic degradation of the extruded arm of a reversed fork (RF) has emerged as a pathological consequence of defects in the BRCA2/RAD51 pathway(8,14). Deprotection of the nascent strands as a result of an impairment in RAD51 nucleofilaments formation leads to MRE11 and EXO1-dependent fork degradation and genomic instability in BRCA1/2-deficient cells upon replication stress (10). Further, CtIP depletion in BRCA1-deficient setting synergistically promotes fork degradation (28), highlighting multiple independent mechanisms that act complementarily in ensuring the integrity of stalled forks. These studies distinguish BRCA1 and BRCA2 roles in replication fork protection from their well-established roles in homologous recombination-mediated repair of DNA double-stranded breaks (10,29). RAD52, on the other hand, prevents SMARCAL1-catalysed fork reversal and therefore avoids an excessive degradation of reversed forks by MRE11 nuclease, playing non-complementary roles from BRCA2/RAD51 in the recovery of arrested forks (13). PARP1 stabilizes replication forks genome wide and at telomeres (30,31) and antagonizes the subsequent restoration of fork restart mediated by RecQ1-catalysed reverse branch migration repaired (30,32). PARP1 also recruits MRE11 to stalled replication forks for end processing and subsequent recombination repair and restart of stalled forks (33,34). However, activation of PARP1 appeared to be a double-edged sword (35,36), and the physiological context and extent to which PARP1 is regulated to effect proper control over fork dynamics is still unclear.
The tumor suppressor p53 plays a quintessential role in linking genomic stability and cancer susceptibility. p53 controls a plethora of cellular signals and processes (37) and exerts canonical functions in apoptosis or cell cycle arrest, largely acting post damage (38–41). Recent hints of the noncanonical actions of p53 distinguishes from its usual modus operandi and implicates p53 as an important regulator of DNA replication stress responses (42–44). Despite the vital role of p53 as the guardian of the genome, the specific mechanisms by which it safeguards DNA replication remained elusive. The observed synergism between p53 loss and ATR inhibition hints that the p53 pathway may be involved in replication fork maintenance although the underlying mechanisms are not fully elucidated (45). ATR plays diverse roles in DNA replication fork protection. ATR acts as a first responder to replication stress and is activated by Replication Protein A (RPA)-coated single-stranded DNA generated by the uncoupling of replicative polymerase (46). ATR acts globally to impact DNA replication origin firing (47,48) and its long-range effects on origin firing are mediated in part through its direct regulation of CHK1 kinase (49–51). In addition to its direct phosphorylation of RPA which promotes the recruitment of critical HR and DDR factors (52–54) and prevents excessive ssDNA accumulation at stalled forks (55), interestingly, global unscheduled firing of origins when ATR is inhibited in turn results in local fork deprotection that is in part mediated by the global exhaustion of RPA (56). Replication gaps have emerged as a potential vulnerability in cancer cells (57,58) particularly in cells with compromised BRCA1/2 functions, and are repaired by post-replicative gap repair mechanisms (59–62). These gaps are initiated by PrimPol repriming and extended by resection enzymes EXO1 and DNA2, and is critical for ATR activation upon replication stress (63). Loss of ATR results in fork destabilizing outcomes that are mediated in part by ATR-dependent inhibition of regressed forks (64,65), RNF4/PLK1 activation (66) or ATR/CHK1-dependent phosphorylation of EXO1 (67). Consequently, delayed or defective ATR signaling results in impaired fork stabilization (68), genomic and chromosomal instability (69), developmental defects and accelerated organismal aging (70). Not surprisingly, ATR signaling acts as an important barrier to cancer progression; impaired ATR signaling under chronic replication stress induced by oncogenic conditions promotes genomic instability (71–74). Therefore, elucidating oncogenic conditions which drives synergistic effects with ATR impairment remains an important goal for dissecting the multiple layers of replication vulnerability.
A plethora of evidence points towards uncontrolled fork degradation as a predictor of chemosensitivity to replication inhibitors, conferring genomic instability, particularly in cells with compromised fork protection. The best characterized mechanism ensuring protection against uncontrolled fork degradation involves the BRCA proteins and components of the Fanconi Anemia tumor suppressor pathway (7,14,16). Numerous additional factors and pathways influencing fork protection have also been recently elucidated, either by impacting RAD51 nucleofilament formation or through entirely distinct mechanisms (75–79). In this study, we uncover an unexpected role of p53 in limiting replication stress by preventing the nucleolytic attack of newly replicated DNA when forks are deprotected by ATR inhibition. We provide evidence that activation of an NRF2 antioxidant program underlies an unknown dysregulation in cellular PARylation in p53- or RRM2B-deficient cells which dictates the susceptibility of nascent DNA to unscheduled degradation catalysed by MRE11/EXO1. Altogether, this study highlights an important crosstalk between redox metabolism, PARP1 and the p53 tumor suppressor pathway in maintaining replication integrity that is negotiated through the NRF2-PARP1 axis identified herein. These findings have broad underpinnings for mechanisms of cancer development and potential therapeutic interventions.
Materials and methods
Cell culture
HCT116 wild-type (WT) and HCT116 p53-deficient (p53KO) cells were kind gifts from Dr Bert Vogelstein (John Hopkins University School of Medicine, Baltimore, MD). HCT116 parental wildtype (WT) and p53KO were cultured in McCoy's 5A medium (GE Healthcare Life Science, SH30200.01). A549 and U20S cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (GE Healthcare Life Science, SH30022.01). All media were supplemented with 10% (v/v) fetal bovine serum (FBS) (GE Healthcare Life Science, SH30071.03) and 1% penicillin–streptomycin (Pen–Strep) (Gibco, 15070–063). Cells were incubated at 37°C in a humidified 5% CO2 atmosphere. Cell lines were tested for mycoplasma contamination before use.
Drugs
Menadione (Sigma Aldrich, M5625), Trolox (Sigma Aldrich, 238813–1G), Tempo (Sigma Adrich, 176141-1G), NAC (Sigma Adrich, A9165-5G), PARGi (Tocris Bioscience, 00017273), Doxycycline (Clontech Laboratories, 631311), Doxorubicin (Selleck Chem, S1208), ML385 (Sigma Aldrich, SML 1833-5MG), Prexasertib HCL (Selleck Chem, S7178), Aphidicolin (Sigma Aldrich, 89458), PHA-767491 (CDC7i) (Selleck Chem, S2742), Roscovitine (Calbiochem, 557364), Hydroxyurea (Sigma Aldrich, H8627), 5-Chloro-2′-deoxyuridine (CIdU) (Sigma Aldrich, C6891), 5-Iodo-2′-deoxyuridine (IdU) (Sigma Aldrich, I7125), PJ34 (Calbiochem, 528150), VE-821 (Selleck Chemicals, S8007), Olaparib (AZD2281, Ku-0059436) (Selleck Chem, S1060), 5-ethynyl-2-deoxyuridine (Thermo Fisher Scientific, E10187). Dimethyl fumarate (Sigma Aldrich, 242926), G6PDi-1 (Sigma Aldrich, SML2980) and G6PD activator AG1 (MedchemExpress, HY-123962). Most compounds were reconstituted in DMSO unless otherwise suggested.
Antibodies
Primary antibodies used were: anti-γH2AX antibodies (Abcam, ab2893 and Millipore, JBW301), anti-NRF2 antibody (Santa Cruz, sc-365949), anti-Poly (ADP-Ribose) Polymer antibody [10H] (Abcam, ab14459), anti-PAR polyclonal antibody (Trevigen, 4336-BPC-100), anti-poly/mono-ADP ribose (E6F6A) antibody (CST, 83732), anti-GAPDH antibody (Abcam, ab9485), anti-Exonuclease 1 antibody (Thermo Fisher Scientific, A302-640A), anti-RRM2B antibody (Abcam, ab8105), anti-p53 antibody (DO-1) (Santa Cruz, sc-126), anti-HSP90 (AC88) (Abcam, ab13492), anti-BrdU antibody (BU1/75) (Abcam, ab6326), anti-RPA70 antibody (Abcam, ab79398), anti-RPA2(pS4/S8) (Novus Biologicals, NBP1-23017), anti-BrdU antibody (BD Biosciences, 347580), anti-β-Actin antibody (Sigma Aldrich, A5441), DYKDDDDK Tag antibody (Cell Signaling Technology, CST, #2368), anti-G6PD antibody (Santa Cruz, sc-373886), anti-CHK1 antibody (CST, 2345), anti-phosphoCHK1 (Ser317) antibody, (CST, 2344), anti-CHK2 antibody (CST, 2662), anti-phosphoCHK2 (Thr68) antibody (2197), anti-phosphoATM (Ser1981) antibody (CST, 4526), anti-ATR antibody (CST,2790) and anti-phosphoATR (Thr-1989) antibody (Abcam, ab223258). Secondary antibodies used are: anti-Mouse IgG (HL) Alexa Fluor 488 (Thermo Fisher Scientific, A-11001), Donkey anti-Rat IgG (HL) Cy3 (Jackson ImmunoResearch Labs, 712-166-153) and Goat polyclonal anti-Mouse IgG2a Alexa Fluor 350 (Thermo Fisher Scientific, A-21130).
Transfection
Transient transfections were carried out according to the manufacturer's instructions. RNAiMAX (Invitrogen, 13778075) was used for small interfering RNA (siRNA) transfection. ON-TARGETplus SMARTpool® siRNAs were obtained from GE Dharmacon: siNRF2 (Dharmacon, L-003755–00-0005), siRRM2B (Dharmacon, L-010575-00-0005), siRRM2 (Dharmacon, L-010379-00-0005), siMRE11 (Dharmacon, L-009271-00-0005), siEXO1 (Dharmacon, L-013120-00-0005), siPARP (Dharmacon, L-006656-03-0005), siCDC7 (Dharmacon, L-003234–00-0005), siP53 (Dharmacon, J-003329–16-0020), siP21 (Dharmacon, L-003471-00), siMDM2 (Dharmacon, L-003279-00), Non-targeting Pool (Dharmacon, D-001810-10-20), and siG6PD (Dharmacon, L-008181-02-0005). siRNAs against OGG1, MSH2, PMS2, XPC, RAD51, DDB2, POLH and PCNA were sourced from a custom Dharmacon siRNA library.
Viral transduction and generation of shp53 cell lines
HEK293 cells were transfected with 6 μg plasmid (pLKO.1 shRNA-p53 or pLKO.1 puro empty vector (EV) and Trans-Lentiviral Packaging Mix (Dharmacon, TLP4606). Media was replaced with fresh media containing 1 mM sodium pyruvate, 0.1 mM MEM non-essential amino acids, 10% FBS and 2 mM l-glut. 48 h later, viral supernatant was collected. 40 000 cells per well in 12-well plate was seeded and the supernatant was added to the cells and incubated for 48 h before selection in puromycin (1.5 μg/ml).
DNA Fiber labeling
Cells were pulse-labelled with nucleotide analogs 5-chloro-2′-deoxyuridine (CldU) (Sigma Aldrich, C6891-100 mg) and 5-Iodo-2′-deoxyuridine (IdU) (Sigma Aldrich, 17125–5G) using concentrations of 250 and 30 μM, respectively. For fork degradation experiments, the duration of drug treatment after CldU/IdU pulse-labelling was 5 h. The labelled cells were harvested, resuspended in PBS to 2.5 × 105 cells and the cell suspension mixed with lysis buffer (200 mM Tris–HCl pH 7.4, 50 mM EDTA (Promega, V4231), 0.5% SDS (Sigma Aldrich, L3771-1KG) on a glass slide. Slides were inclined at an angle of 45° for the suspension to spread on the slide. Upon drying, the DNA fiber spreads were fixed in 3:1 methanol–acetic acid solution overnight. DNA was denatured with 2.5 N HCl for 80 min. Slides were immersed into filtered blocking solution (2% BSA (Sigma Aldrich, A7906), 0.1% Tween 20 (Promega, H5151), 1× PBS; 0.22 μm (Gibco, 10010023)) for 40 min followed by primary antibody incubation. Anti-BrdU antibody (BU1/75 (ICR1) (Abcam, ab6326) and mouse monoclonal anti-BrdU antibody (BD Biosciences, 347580) used at 1:450 and 1:100 dilutions in the blocking buffer respectively for 2 h 30 min at room temperature in the dark. Secondary antibody incubation was done with the following secondary antibodies: anti-Mouse IgG Alexa 488 (ThermoFisher, A-11001) and anti-Rat Cy3, (Immuno Research, 712-166-1530) at a dilution of 1:300 in the blocking solution for 1 h at room temperature in the dark. Single-stranded DNA was stained with anti-DNA antibody, single stranded, clone 16–19 (Chemicon, MAB3034) and Isotype-specific Alexa Fluor 350 Goat anti-Mouse IgG2a (gamma2a) (Invitrogen, A21130), used at a dilution of 1:100 and 1:250 respectively for 30 min at 37°C in a humidified chamber. Slides were mounted using ProLong® Gold Antifade (Invitrogen, P36930). Images were acquired using the microscope (ZEISS Axio Imager.Z1) and at least 100 DNA fibers were quantified using ImageJ (RRID: SCR_003070). Nucleolytic degradation of nascent DNA is indicated by the ratio between the second label (IdU) and the first label (CIdU). Fork speed was measured by dividing the length of both tracts (kb) by the labeling time and converting the values from μm into kb by multiplying the length by 2.59 (80–82). Additionally, the frequency of fork stalling was determined by quantifying the percentages of stalled forks relative to the total number of ongoing forks.
Cloning of pTRIPZ plasmids and lentiviral transduction of cell lines
HCT116 WT and p53KO cells were stably transfected with doxycycline-inducible pTRIPZ plasmids: pTRIPZ-RRM2B(WT)DYK, pTRIPZ-RRM2B(Q127K) DYK, pTRIPZ-RRM2B(Y331F) DYK, pTRIPZ-G6PD(WT) DYK, pTRIPZ-G6PD(K171Q) DYK and pTRIPZ-EV (empty vector). RRM2B was cloned using forward primer, 5′-GAATGCACCGGTGATAGAACCATGGGCGACCCGGA AAGGC-3′ (AgeI restriction site in underlined) and reverse primer, 5′- GCATTCACGCGTTTATCACTTATCGTCGTCATCCTTGTAATCAAAATCTGCATCCAAGG-3′ (MluI restriction site underlined and DYK sequence in bold) to include a DYK tag at the C- terminus of RRM2B sequence. pTRIPZ plasmid and PCR-amplified RRM2B target was restriction digested BsrGI and MluI at 37°C for 2 h prior to ligation of digested plasmid and target RRM2B. pTRIPZ-RRM2B was transformed into TOP10 competent cells (Invitrogen, C404010) and single colonies selected for validation. G6PD gene (NM_001042351.3) was cloned into pTRIPZ vector at the AgeI (5′) and MluI (3′) restriction sites. The 5′ end of the cloned G6PD sequence would be: ATTCACCGGTGCCACCATGGCAGAGCAGGTGGCCC. The 3′ end of the cloned G6PD would be: GTGAACCCCCACAAGCTC GATTACAAGGATGACGACGATAAGTGATAAACGCGTAAT. The sites in bold represent the restriction cut sites of AgeI and MluI, while the underlined sequence corresponds to the complementary to G6PD gene. Additionally, the DYK sequence is inserted before the stop codon (TGATAA) at the C-terminus of G6PD.
6 μg of extracted plasmid pTRIPz-RRM2B (DYK) was then transfected into HEK 293T cells with Lipofectamine 2000 (Invitrogen, 11668019), Opti-MEM (Gibco, 31985070), and lentiviral packaging mix (Dharmacon, TLP5912). Media was changed the next day after transfection. Virus supernatant was collected at 72 h and filtered, then 500 ul of virus supernatant was added directly to HCT116 cells. Puromycin was added to select HCT116 cells stably transduced with pTRIPZ plasmids.
Patient-derived primary hepatocellular carcinoma cells
Hepatocellular Carcinoma (HCC) tissues were obtained from Singapore General Hospital (SGH) and National University (NUH) with patients’ consent and under approval from Central Institution Review Board of SingHealth (CIRB 2012/669/B). Tissue was digested with 1 mg/mL collagenase IV (Gibco) and 1 mg/mL dispase II (Thermo Fisher Scientific) in F12 media for 1 h while shaking in a 37°C incubator. Cell suspension was then put through a cell strainer and centrifuged for 5 min at 1500 rpm. Cells were then washed twice and resuspended in primary culture medium and seeded onto 6-well culture plates (83). The culture medium was then changed twice a week and cells were sub-passaged when they reached 70–80% confluency.
Whole-exome sequencing and identification of TP53 mutations in patient-derived HCC cell lines
Quantity of 500 ng to 1 μg of genomic DNA was sheared using Covaris to a size of 300–400 bp and subjected to library preparation using NEBNext® End repair, A-tailing and Ligation modules (New England Biolabs). 3–6 samples were pooled together and hybridized using the SeqCap EZ Human Exome Library v3.0 (Nimblegen, Roche) kit. Captured regions were washed, purified, amplified and subjected to 2 × 101 sequencing on the Hiseq 2000 to obtain a mean coverage of 114×. TP53 gene mutations for each sample were called using MuTect (84) (v.1.1.7).
Pulsed-field gel electrophoresis (PFGE)
Cells were seeded in 10 cm dishes at 1.0 × 106 cells per sample and treated with drugs accordingly. Cells were harvested and resuspended to a concentration of 106 per milliliter and mixed with an equal volume of molten 2% low melting point (LMP) agarose gel before adding to plug molds and left to solidify at 4°C. Plugs were then incubated in lysis buffer (100 mM EDTA pH 8.0, 0.2% sodium deoxycholate (w/v), 1% sodium lauryl sarcosine (w/v)) with proteinase K (0.5 mg/ml) for 36–72 h at 37°C. The plugs were washed three times in washing buffer (Tris–HCl 20 mM, EDTA 50 mM, pH 8.0), and embedded into 0.9% agarose gel (Pulsed-field certified agarose from Bio-Rad no. 162-0137). DNA breaks were separated on a contour-clamped homogeneous electric field (CHEF) III Mapper system (BIO-RAD) in 1× Tris-borate EDTA (TBE) buffer with the following conditions: 5.5 V/cm for 9 h at 14°C, with a 30–18 s switch time and pump of 90–100%; 4.5 V/cm for 6 h at 14°C, with 18 to 9 s, switch time; and 4.0 V/cm for 6 h at 14°C, with a 9 to 5 s switch time. Gel running conditions were optimized to detect the migration of broken DNA as a single band during electrophoresis. Gels were stained in TBE buffer containing SYBR® Gold nucleic acid gel stain and photographed under UV light. Lambda ladder PFG Marker was used as a marker.
Western blot analysis
Cells were lysed in RIPA buffer (50 mM Tris–HCl pH 8, 0.5% Na-deoxycholate, 150 mM NaCl, 1% IGEpal CA-630, 0.1% SDS, 0.1 mM PMSF, 0.1 mM Na2VO3, 10 μg/ml protease inhibitor mix, 1 mM NaF, 1 mM DTT) for 30 min on ice and then sonicated briefly. Pierce BCA protein assay kit (Thermo Fisher Scientific) was used to determine the concentration of protein. Cell lysates were mixed with 4× LDS Buffer (Invitrogen) in dH2O with 0.1 M DTT. The blotting was performed using standard methods. Blots were incubated with primary antibodies overnight followed by Horse radish peroxidase–linked secondary antibody incubation (1:5000). The blots were visualized using Amersham™ ECL™ western blotting detection reagent.
Comet assay
180 000 cells were seeded onto a 6-well plate and treated with HU and ATRi for 6, 12 or 24 h after 24 h of cell seeding. Comet Assay (Cell Biolab, Inc., USA, STA-351) was performed according to the manufacturer's protocol, with the following modification. Lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris base, 200 mM NaOH, 1% sodium lauroyl sarcosinate and 1% triton X-100. pH 10), alkaline solution (300 mM NaOH, 1 mM EDTA) was prepared and chilled before use. SuperFrost Gold (Thermo Fisher Scientific, K5800AMNZ72) was precoated with 1% low melting agarose (Promega, PR-V2111) and warmed at 37°C before use. Cells were harvested via trypsinization. It was counted and resuspended to obtain a final concentration of 1 × 106 cells/ml before diluting with comet agarose in 1:10. 75 μl of the cell agarose mixture was then added to the coated slides and left to solidify. The slides were first immersed into the pre-chilled lysis buffer for 50 min at 4°C followed by incubation in the alkaline solution for 30 min at 4°C. Prior to the electrophoresis, the slides were immersed in pre-chilled 1× TBE buffer twice for 5 min each. The electrophoresis was carried out at 24 V for 20 min at 4°C. After that, it was washed twice for 2 min each in chilled deionized water and then 70% ethanol for 5 min before air drying. 100 μl of vista green (diluted 1:10 000 TE buffer) was added to the gel spot and left to incubate for 15 min in the dark. Imaging of the comet was performed using fluorescent microscope (ZEISS Axio Imager.Z1) with FITC filter. Tail length and head length was measured to calculate the olive tail moment (OTM) according to the following equation: (tail length-head length) × % tail DNA. The average OTM for each sample was obtained through analysis and measurement of the cells per condition.
Immunofluorescence staining
For immunofluorescence staining, cells were seeded on glass coverslips, left untreated or treated accordingly the next day. Cells were washed with PBS and fixed with 4% paraformaldehyde at room temperature for 10 min and permeabilized with 0.5% Triton X-100 in PBS or Cytoskeleton (CSK) buffer containing 10 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose, 0.2% Triton X-100 and distilled water to the final volume. Cells were then blocked in 3% BSA in PBS for 30 min before incubating in primary antibodies (diluted in 3% BSA in PBS) overnight in 4°C followed by secondary antibody for 1 h in the dark. Nuclei were counterstained with DAPI and mounted using ProLong® Gold Antifade (Thermo Fisher Scientific, P36830). Immunostained cells were visualized using fluorescence microscopy (ZEISS Axio Imager.Z1). Quantification of immunofluorescence signals was performed using ImageJ.
Quantitative image-based cytometry
Quantitative image-based cytometry was adapted from Besse et al. (85). QIBC was performed with a minimum of 4000 cells acquired using the 20× objective (NA 0.9) (ZEISS Axio Imager.Z1). The images were captured with settings that ensured non-saturation, and consistent parameters were used for all coverslips in the experiment. Following acquisition, images were analysed using ImageJ. Cell nuclei were identified as individual objects through image segmentation using the DAPI signal. Nuclei that overlapped are excluded from quantification. Following image segmentation, automated individual cell multi-parameter extraction, including total nuclei DNA content measured by DAPI or mean fluorescence intensities of γH2AX or RPA2S4S8 within the nuclear masks per cell was performed. DAPI and γH2AX/RPA2S4S8 intensities for individual cells were plotted using Prism in scatterplot.
EdU incorporation and RPA1 immunofluorescence
150 000 cells were seeded on glass coverslips and incubated for 48 h. The cells were stained for 40 min with EDU 10 μM, the cells were treated with HU/ATRi for 6 h. the cells were washed by PBS and fixed with 3.7% paraformaldehyde in PBS for 10 min. After removing the fixation buffer and washing the cells with PBS, the cells were permeabilized with 0.5% triton X-100. The cells were washed three times with PBS. The Click-iT reaction cocktail prepared and added as mentioned in Click-iT EdU Imaging Kit, Invitrogen, Cat.No10338. Cells were incubated for 30 min with the reaction cocktail at room temperature, protected from light. The reaction cocktail was removed and the cells were washed two times with 3% BSA in PBS. The cells were incubated with RPA1 antibody (Abcam, ab2172) overnight. The cells were washed by PBS and incubated with anti-Rabbit Alexa Fluor 594 secondary antibody (Invitrogen, A11012). Nuclei were counterstained with Hoechst stain (Invitrogen, 33342) and mounted using ProLong® Gold Antifade (Thermo Fisher Scientific, P36830). The cells were immunostained and visualized using fluorescence microscopy (ZEISS Axio Imager.Z1).
Detection of EdU-positive cells and cell cycle phases by flow cytometry
150 000 cells were seeded in 6-well plates and incubated for 48 h. Cells were then incubated for 30 min with 5-Ethynyl-2′-deoxyuridine (EdU)(10 μM) and treated with drugs for different time points (6,12 or 24 h). Cells were harvested by trypsinization, fixed and permeabilized following as indicated in Click-iT®EdU Flow Cytometry Assay Kit (Invitrogen, 10418). The cells were incubated at room temperature with the Click-iT reaction buffer as indicated in the kit. DAPI (0.3 μg/ml) was used to detect the DNA content. The labeled cells were analysed by flow cytometry. For cell cycle analysis using propidium iodide, cells were harvested and fixed in 70% ethanol/PBS. Fixed cells were resuspended in PBS containing 0.05 mg/ml Propidium iodide and 0.04 mg/ml RNase A and analysed by flow cytometry. Total cell populations were gated by FSC/SSC, followed by singlet selection using DAPI-A/DAPI-W. Gated cell populations were then displayed in dot plots with 488-A (y-axis) and DAPI-A (x-axis).
Colony forming assay
100 000 cells were seeded in 6 well plate and incubated for 48 h at 37°C. Cells were then treated with the indicated drugs for 24–48 h. Following that, cells were recovered in fresh media for 10 days before staining with 1× crystal violet solution diluted from 10X (8% glutaraldehyde v/v for fixation and 0.5% crystal violet w/v for staining in 20% methanol) for 2 h.
Cellular ROS assay
150 000 cells were seeded on glass coverslips. The cells were stained with 2′,7′-dichlorofluorescin diacetate (H2DCFDA) (Invitrogen, D399) 20–50 μM for 30 min. The cells were washed with PBS. The cells were visualized using fluorescence microscopy (ZEISS Axio Imager.Z1).
Extraction of metabolites for LC–MS analysis
Solvents used for extraction of cells are Optima™ grade methanol from Fisher Chemical (Pennsylvania, USA), tricine and chloroform from Merck (Darmstadt, Germany). Cells were quenched and metabolites were extracted using a two-phase liquid-liquid extraction protocol as previously described (86). The polar fractions were collected and dried under vacuum at 4°C using a centrifugal vacuum concentrator (CentriVap, Labconco, USA). These dried extracts were then stored at –80°C prior LC–MS analysis.
Mobile phases prepared for LC analysis were laboratory grade water from a Satorius water purification system (Goettingen, Germany) and liquid chromatography gradient grade for acetonitrile from Merck (Darmstadt, Germany). In addition, ammonium bicarbonate from Sigma-Aldrich (St. Louis, Missouri, USA)) was used as an additive.
The dried extracts were reconstituted in 10mM ammonium bicarbonate and each sample was analysed in triplicate using an ultra-performance liquid chromatography system (UPLC, Acquity, Waters, USA) in tandem with a mass spectrometer (Q Exactive, Thermo Fisher Scientific, USA). For UPLC separation, a reversed phase column (Sunniest C18-HT column, 2.1 × 100 mm, ChromaNik Technologies Inc., Japan) was used with 10mM ammonium bicarbonate as solvent A and acetonitrile as solvent B. Solvent A was kept at 99.9% for 2.5 min before decreasing linearly to 50% in 6 min. This was followed by another 3 min wash with 2% A and 2 min equilibration back at 99.9% A. The flow rate used was 0.15 ul/min. For mass spectrometry parameters, analytes are detected at negative mode. Sheath gas flow rate was at 40 arbitrary units, auxiliary gas at 10 arbitrary units and sweep gas rate 2 arbitrary units. The capillary and auxiliary gas heater temperature were at 250°C. The spray voltage was at 2.5 kV. Raw LC–MS data obtained were pre-processed using the XCMS peak finding algorithm (87) and peak areas normalized based on the cell number in each sample. The identities of detected mass peaks were then confirmed by comparison of their MSMS spectra with commercially available metabolite standards.
Computational analysis
Analysis of transcriptomic data from clinical datasets
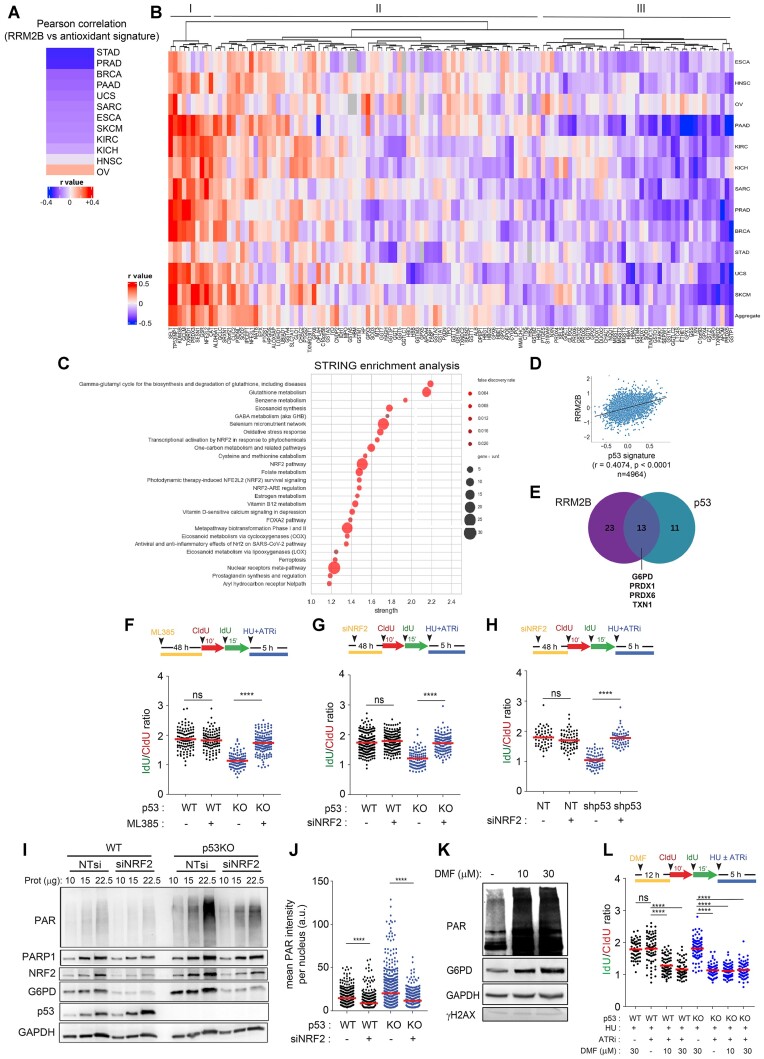
Batch effects normalized mRNA data and molecular subtype data was downloaded from the TCGA project (accessed via the XENA browser). Subsequent data pre-processing steps included imputing missing data with zeros and Z normalizing the data. An antioxidant gene signature (Supplementary Table S2) was obtained from previous work Ren et al. (88). Then, correlation and significance of the antioxidant gene signature against RRM2B expression was quantified in selected TCGA datasets using Pearson correlation coefficient (r value) and two tailed P value (Bonferroni corrected) respectively. This analysis was repeated using a p53 gene signature (13 gene set) to determine the functionality of the p53 pathway (89). The resulting r values were visualized in form of a heatmap generated using Complex Heatmap in R (90,91). Pearson correlation coefficient was calculated between the expression of each gene in the 125 gene antioxidant signature and RRM2B expression. The resulting r values are then visualized in R using the ComplexHeatmap package. Then, hierarchical clustering was applied in R using the same package (90,91). A negative r value means that the variables tested are inversely related. Genes from Cluster III (Figure 5B) was inputted into STRING where enriched pathways were determined using WikiPathways (92). To better understand the antioxidant gene(s) coregulated by RRM2B and p53, antioxidant genes which sufficiently negatively correlate with both p53 or RRM2B (r←0.10) (P< 0.001) (Figure 5E) were selected and overlapped. The resulting genes were then filtered for NRF2 targets. The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Figure 5.
A NRF2-PARP1 axis connects redox homeostasis to replication fork integrity. (A) Heatmap showing the Pearson correlation coefficient (r value) of RRM2B gene expression (log2 normalized) against an antioxidant gene signature for 12 TCGA transcriptome datasets (STAD: Stomach Adenocarcinoma; PRAD: Prostate Adenocarcinoma; BRCA: Breast Invasive Carcinoma; PAAD: Pancreatic Adenocarcinoma; UCS: Uterine Carcinosarcoma; SARC: Sarcoma; ESCA: Oesophageal Carcinoma; SKCM: Skin Cutaneous Melanoma; KIRC: Kidney Renal Clear Cell Carcinoma; KICH: Kidney Chromophobe; HNSC: Head and Neck Squamous Cell Carcinoma; OV: Ovarian Carcinoma). A negative r value indicates that the variables tested are inversely related. All results are significant at P< 0.0001. (B), The Pearson correlation coefficients of the expression of each gene in the antioxidant gene signature (log2 normalized, 125 genes) against RRM2B expression (log2 normalized) were calculated across 12 TCGA datasets (as described in A). Tumor samples n = 4964. A hierarchical heatmap was constructed to depict the Pearson correlation coefficient (r value). All results achieved significance at P< 0.001. (C) Genes are clustered in (D), according to their Pearson correlation coefficient (r value). STRING enrichment analysis (WikiPathways) of genes identified in cluster III in (B) sorted by enrichment strength, Log10(observed/expected), with false discovery rate and observed gene count indicated. Similar gene enrichment analysis was performed using a p53 gene signature instead of RRM2B gene expression (Supplementary Table S3). (D) Pearson correlation coefficient (r value) between RRM2B gene expression and p53 gene signature score that predicts for wildtype p53 functionality were calculated across 12 TCGA transcriptome datasets used in (A) and (B) (tumor samples n = 4964). (E) Venn diagram showing overlap of antioxidant genes that are enriched with reduced RRM2B gene expression or reduced p53 gene signature score, identified by their Pearson correlation coefficient (r value). Pearson correlation coefficient of r←0.1 (P< 0.001) was used as a cut-off. Four NRF2 target antioxidant genes are commonly enriched (G6PD, PRDX1, PRDX6 and TXN1). (F) Schematic of the fork degradation assay in HCT116 parental (WT) and p53KO cells pretreated with an NRF2 inhibitor, ML385 (30 μM, 48 h). Cells were subjected to HU (2 mM) in the presence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (G) Schematic of the fork degradation assay in HCT116 parental (WT) and p53KO cells transfected with NRF2-targeted siRNAs. Cells were subjected to HU (2 mM) in the presence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (H) Schematic of the fork degradation assay in HCT116 parental (WT) stably transfected with shp53 or empty vector (EV) control. As in (G), cells were transfected with NRF2-targeted siRNAs. Cells were subjected to HU (2 mM) in the presence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (I) Total PARylation in HCT116 (WT) and p53KO cells transfected with NRF2-targeted siRNAs was analysed by western blot using an anti-PAR antibody (CST, 83732). Total PARP1, NRF2, G6PD and p53 protein expression was also detected in immunoblots. GAPDH was used as a loading control. (J) HCT116 parental (WT) cells transfected with siNRF2 were immunostained for total PAR (anti-PAR [10H] antibody, Abcam ab14459). Mean PAR intensity per nucleus were quantified and plotted. n ≥ 200 in each condition (mean ± SD; n = 3; two-tailed t-test). (K) HCT116 parental (WT) cells were treated with dimethyl fumarate (DMF, NRF2 activator) at 10 or 30 μM for 12 h. Total PARylation, G6PD and γH2AX protein expression analysed by western blot. GAPDH used as a loading control. (L) Schematic of the fork degradation assay in HCT116 parental (WT) and p53KO cells pretreated with Dimethyl Fumarate (DMF) (10 or 30 μM, 12 h). Cells were subjected to HU (2 mM) in the presence or absence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. In (F), (G), (H) and (l) red horizontal bar represents mean of CldU/IdU ratios; P value was calculated from n ≥ 100 DNA fibers using Mann–Whitney test (P< 0.0001 ****; ns = not significant). Data Representative of n = 3 independent experiments. Western blots in (I) and (J) were repeated independently at least three times with similar results. GAPDH was used as loading control.
Gene expression profiling analysis
Microarray analysis of gene expression in isogenic HCT116 parental and p53KO cells was performed. Three biological and three technical replicates of each line were performed to yield a total of 9 readings in order to increase data consistency. The average signal strength was assembled into a count matrix which was recorded using the Illumina Human HT-12 v4.0 BeadChip Platform. The count matrix and metadata file were input into the DEseq2 package (93) and the Limma package (94) on R (https://www.R-project.org/) (95). A Differential Gene Expression Analysis in DEseq2 was conducted where signals were normalised, and shrinkage was performed using the ashr method (96). A p-value of 0.05 and an arbitrary log2 threshold of 0.32 was used to select for Differentially Expressed Genes which were then plotted as a Volcano Plot and Heatmap using ggplot2 and pheatmap, respectively. A network analysis of the identified DEGs related to the NRF2 pathway was performed using STRING through the web interface. The largest network cluster was then selected to be further analysed. A medium confidence network was then plotted in Cytoscape (97) with the genes linked with high confidence highlighted in yellow. A STRING Enrichment (98) was then conducted on the genes linked with high confidence, and the results were plotted using R. Highest significance gene-sets based on FDR scores were selected and sorted by gene ratio.
Statistical analysis
All data are reported as mean ± SD unless otherwise stated. Data normality were assumed. Statistical significance between two samples was determined using Student's unpaired t test or Mann–Whitney U test. P-values <0.05 were considered significant and represented in graphs as: P< 0.0001 is ****; P< 0.001 is ***; P< 0.01 is **; P< 0.05 is *. All analyses were performed using GraphPad Prism.
Results
Unscheduled degradation of nascent DNA at deprotected forks is dependent on p53
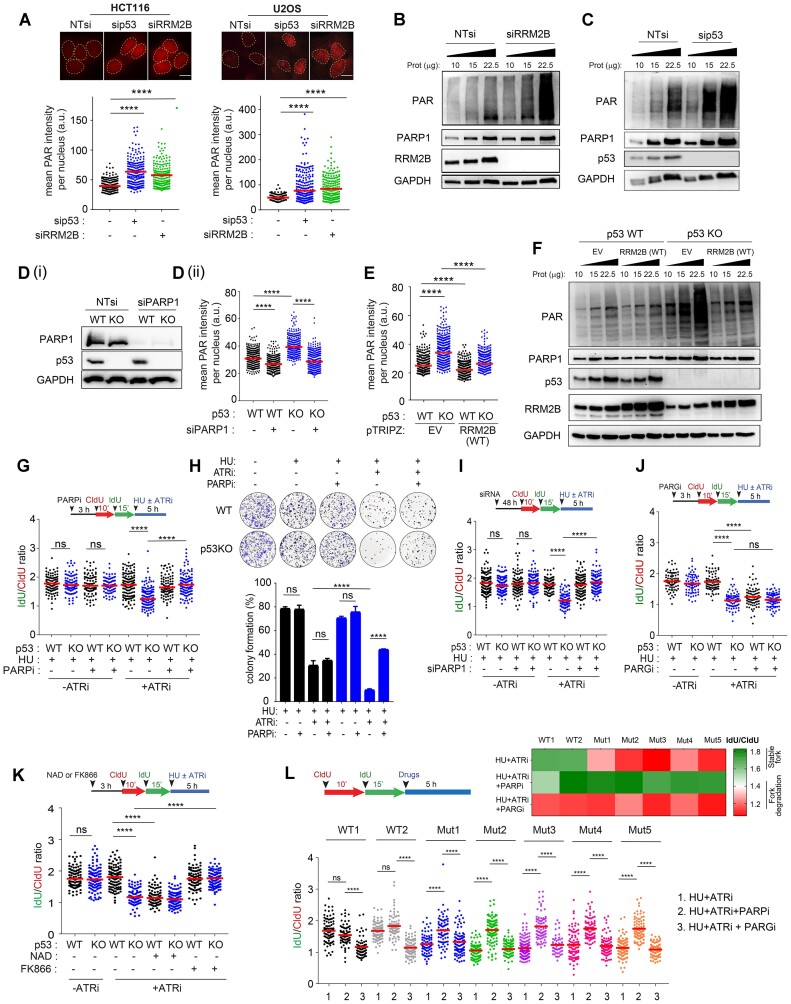
Replication stress (RS) activates many pathways to protect arrested replication forks from aberrant nucleolytic processing or collapse (99,100). The ATR pathway stabilizes stalled replication forks and is paramount for maintaining genomic integrity upon replication stress (3,101). Despite the critical role of ATR in fork protection, we unexpectedly observed that nascent DNA at hydroxyurea-arrested forks remains stable in response to ATR inhibition (Figure 1A(i)). Given the established but not fully elucidated synergism between deficiencies in p53 and ATR (45,102), we hypothesize that p53 plays a critical role in preserving fork integrity, particularly when forks are rendered vulnerable due to ATR inhibition.
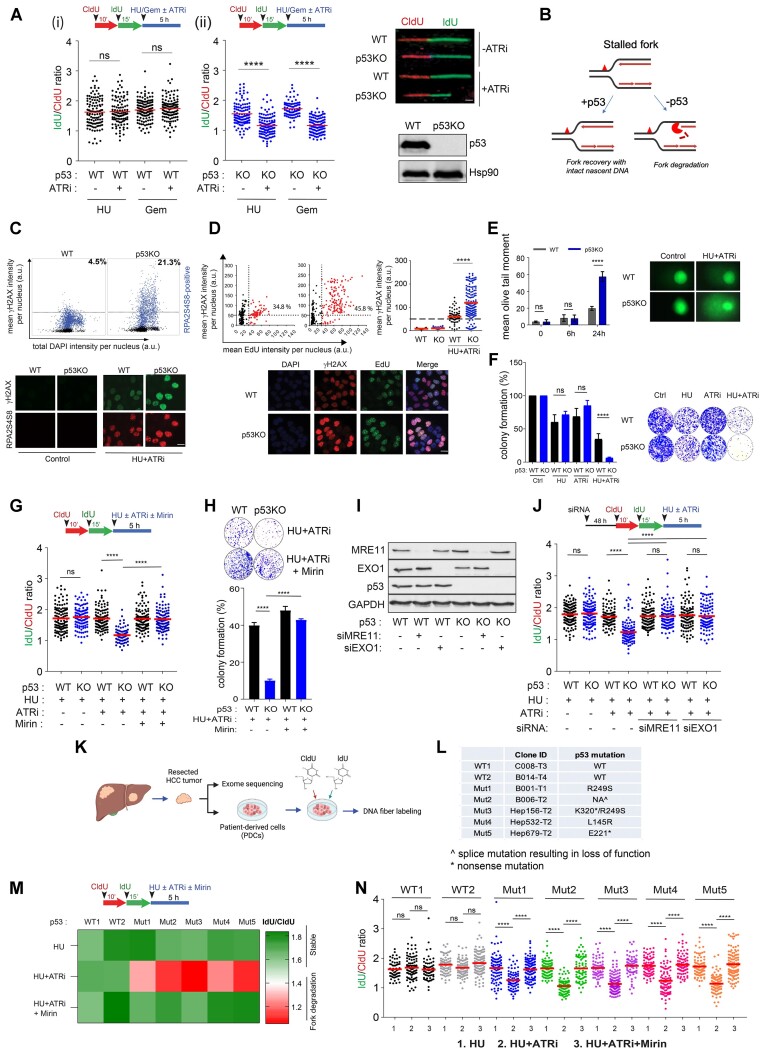
Figure 1.
Loss of p53 promotes MRE11/EXO1-directed fork degradation at deprotected forks. (A), Schematic of the fork degradation assay in (i) HCT116 parental (WT) and (ii) isogenic p53KO cells. Red and green indicate CldU and IdU labeling, respectively, prior to replication stress induced either by HU (2 mM) or gemcitabine (1 μM) and in the presence or absence of an ATR inhibitor (ATRi), VE-821 (2 μM). IdU/CIdU ratios of individual replication forks presented in a scatterplot. Representative of at least three independent experiments. Right: Representative images of single DNA fibers shown. Scale bar, 50 μm. n > 100. Immunoblots showing p53 protein levels in HCT116 parental (WT) and isogenic p53KO cells. Hsp90 included as loading control. (B) Schematic model illustrating the fate of deprotected stalled forks in cells, in the presence or absence of p53. (C) Quantitative image-based cytometry (QIBC) of HCT116 cells treated with HU (2 mM) and ATRi (2 μM) for 6 h. Cells were co-immunostained for γH2AX (Millipore, JBW301) and RPA2S4S8 (Novus Biologicals, NBP1-23017). Mean γH2AX, RPA2S4S8 and total DAPI intensity per nucleus were quantified and plotted. n > 4000. Representative IF images are shown (bottom). Scale bar, 20 μm. (D) Percentages of EdU/γH2AX co-immunostained cells are indicated in HCT116 parental (WT) and p53KO cells pulsed-labelled with EdU (10 μM) for 15 min followed by treatment with HU (2 mM) and ATRi (2 μM) for 6 h. Mean γH2AX intensity per nucleus also plotted (right). Representative IF images shown (bottom). Scale bar, 20 μm. (E) HCT116 parental (WT) and p53KO cells treated with HU (2mM) and ATRi (2 μM) at the indicated timepoints were subjected to a comet assay and mean olive tail moment plotted. Experiments in (C) to (E) are repeated three times with similar results. (F) Colony forming assay of HCT116 (WT) and p53KO cells treated with HU (2 mM) and in the presence or absence of an ATR inhibitor VE-821 (2 μM) for 24 h and recovered in fresh media for eight days. DMSO-treated control included (Ctrl). Colonies are quantified and expressed as percentage of DMSO-treated controls. Representative of n = 3 independent experiments. (G), Schematic of the fork degradation assay in HCT116 parental (WT) and isogenic p53KO cells with or without mirin (50 μM). HU (2 mM) in the presence or absence of ATRi (2 μM) used as indicated. Representative of at least three independent experiments. (H) Colony forming assay of HCT116 (WT) and p53KO cells treated with or without mirin (50 μM) in the presence of HU (2 mM) and ATRi (2 μM) (24 h). Representative of n = 3 experiments. Colonies are quantified and expressed as percentage of DMSO-treated controls. (I) Western blot to validate the knockdown efficiencies of MRE11- and EXO1-targeted siRNAs in HCT116 cells (WT and p53KO). Representative of n = 2 experiments. (J) Schematic of the fork degradation assay in HCT116 parental (WT) and isogenic p53KO cells in HCT116 parental (WT) and p53 KO cells transfected with MRE11- or EXO1-targeted siRNAs. HU (2 mM) in the presence or absence of ATRi (2 μM) were used as indicated. Representative of at least three independent experiments. (K) Hepatocellular Carcinoma (HCC) patient-derived cells (PDCs) were subjected to whole exome sequencing and their p53 genetic status determined, and indicated in (L, M). Schematic of the fork degradation assay in p53 wildtype and p53 mutant HCC PDCs shown, to assess effects of HU (2 mM), ATRi (2 μM) and mirin (50 μM) on nascent DNA stability. IdU/CldU ratios for individual replication forks were calculated and mean of IdU/CldU ratios are represented in a heatmap, as well as in a scatterplot in (N). Representative of n = 3 independent experiments. In (A), (G), (J) and (N), mean of IdU/CldU ratios indicated by a horizontal red bar and P value was calculated from n ≥ 100 DNA fibers using Mann–Whitney test (P< 0.0001 ****).
To directly visualize the stability of newly replicated DNA, nascent DNA was labelled with halogenated nucleotides (CldU and IdU) before replication stress (RS) was induced with hydroxyurea (HU) or gemcitabine. Progressive degradation of stalled forks was assessed on the basis of the shortening of preformed DNA tracks during fork blockade and quantitative reductions in the IdU/CldU ratio (Supplementary Figure S1A). Stalled forks appeared to be resilient to degradation even after exposure to an ATR inhibitor (IdU/CldUHU= 1.63; IdU/CldUHU+ATRi= 1.64; IdU/CldUGem= 1.70; IdU/CldUGem+ATRi= 1.74) (Figure 1A(i)). Similarly, the mean IdU/CldU ratio remained unchanged in HCT116 colorectal carcinoma cells under increased replication stress induced by a higher concentration of HU (Supplementary Figure S1B(i)), which was shown to promote fork degradation in BRCA-deficient cells (8,14), or various combinations with ATR inhibitor (2–10 μM) (Supplementary Figure S1B(i)). To investigate the involvement of p53 in fork stability, a fork degradation assay was performed using a well-characterized isogenic HCT116 p53KO cell line (103). Strikingly, the mean IdU/CldU ratio was significantly reduced in isogenic p53KO cells (IdU/CldUHU+ATRi= 1.16 and IdU/CldUGem+ATRi = 1.16) (Figure 1A(ii) and Supplementary Figure S1B(ii)) in contrast to HCT116 parental cells; thus, stalled and deprotected forks are more susceptible to nucleolytic degradation in the absence of p53 (Figure 1B). The same phenotype was observed in hTERT immortalized p53-null retinal pigmental epithelial cells (RPE1-hTERT p53−/−) (104) and in HCT116 or A549 cells transfected with siRNAs targeting p53 (Supplementary Figure S1C).The results are reproducible when aphidicolin, an inhibitor of replicative polymerases, is used instead of hydroxyurea (Supplementary Figure S1D). Consistent with the notion that fork degradation is a source of replication stress and DNA damage, quantitative image-based cytometry (QIBC) showed that p53 disruption induced DNA damage and DNA replication stress, as evidenced by a pan-nuclear γH2AX pattern and RPA2 phosphorylation at serine 4 and serine 8 (RPA2S4S8) in S/G2 cells (Figure 1C and Supplementary Figure S1E) and in EdU-positive S phase cells lacking p53 (Figure 1d). Western blot of γH2AX, phospho-ATM, phospho-CHK2, phospho-RPA2(S4S8) and phospho-RPA2(S33) corroborated these observations, indicating pronounced DNA damage and replication stress in cells devoid of p53 (Supplementary Figure S1F(i) and (ii)). Although the ATR-CHK1 pathway is promptly activated in both HCT116 WT and p53KO cells in response to the initial replication stress induced by hydroxyurea (Supplementary Figure S1F(ii)), replication stress at forks deprotected by ATR inhibition eventually caused extensive DNA breaks, detected by comet assay or pulsed-field gel electrophoresis (PFGE), that were significantly exacerbated in p53KO cells (Figure 1E and Supplementary Figure S1G). Accompanying these molecular changes was a significant loss in cell viability to a far greater extent in p53KO cells (Figure 1F). Interestingly, we found that ATR inhibition induces unscheduled origin firing rapidly (Supplementary Figure S1H), in concordance with the reported (105). To elucidate whether ATR’s function in regulating origin firing might be causative for the observed fork degradation in p53KO cells, we depleted Cdc7, a prototypical regulator of origin firing (106–108). Depletion of Cdc7 by gene-targeted siRNAs which will downregulate replication initiation (106–108), fully restored the stability of nascent DNA strand in p53KO cells (Supplementary Figure S1I), suggesting that excessive origin firing induced by ATR inhibition deprotects stalled replication forks, rendering them susceptible to fork degradation in the absence of p53.
Together, these results provide new evidence that the p53 pathway restrains pathological outcomes of replication stress at deprotected stalled forks by preventing fork degradation; this is mechanistically distinct from the role of p53 in negotiating transcription-replication conflict we had previously reported (42).
p53 prevents fork degradation mediated by MRE11/EXO1 exonucleases in established and patient-derived cancer cells
To identify the mechanism of fork degradation at stalled forks, we investigated whether specific nucleases are responsible for the observed nucleolytic processing of stalled forks in p53KO cells. Compelling evidence has shown that MRE11 and EXO1 are responsible for the extensive fork degradation at stalled forks, specifically when fork protection mechanisms are attenuated (8). To investigate whether MRE11 is responsible for the fork degradation observed in p53KO cells, we used mirin, a chemical inhibitor of MRE11 nuclease activity (109). After mirin treatment, the IdU/CldU ratio was restored to near wildtype levels in p53KO cells (IdU/CldUKO+mirin = 1.69; IdU/CldUWT+mirin = 1.70 (Figure 1G). Additionally, mirin suppressed γH2AX and RPA2S4S8 signals in p53KO cells (Supplementary Figure S1J) and partially restored the viability of p53KO cells (Figure 1H), indicating that extensive fork degradation contributes to replication catastrophe and cell lethality triggered by prolonged fork stalling and ATR inhibition in p53KO cells. To exclude any possible off-target effects of mirin, we knocked down MRE11 using short-interfering RNAs (Supplementary Figure S1K). Consistently, MRE11 depletion substantially protected the nascent DNA tracts in p53KO cells from degradation and suppressed replication stress signals (Figure 1J and Supplementary Figure S1L). Likewise, depleting EXO1 exonuclease, which was found to generate long tracts of ssDNA in both MMR and DSB repair (110) and to catalyse extensive fork degradation (111), fully restored nascent DNA tracts in p53KO cells (Figure 1J and Supplementary Figure S1M), linking nucleolytic degradation of deprotected stalled forks by MRE11/EXO1 to pronounced genomic instability in p53KO cells upon replication stress.
Our data are consistent with the theory that fork degradation precedes fork breakage in p53-deficient cells. Before DSBs became detectable (Supplementary Figure S1N), γH2AX and RPA2S4S8 signals had increased, accompanied by fork degradation in p53-deficient cells, as early as 6 h after drug treatment (Supplementary Figure S1O, P). To further exclude the possibility that the observed shortening of IdU tracts may be due to fork cleavage, we changed the dual fibre labelling scheme so that forks were stalled just before the second label (IdU). If fork cleavage and any subsequent DSB processing was the primary cause of the shortening of the nascent tracts during fork stalling, synthesis of the second label will not occur (Supplementary Figure S1Q(i)). The majority of fibers in p53KO cells treated with HU + ATRi showed continuous CldU-IdU labeling in p53KO cells. Importantly, preformed CldU tracts (1st label) significantly shortened in p53KO cells (CldU/IdUp53KO = 0.58) compared to WT cells (CldU/IdUWT = 0.92) (Supplementary Figure S1Q(ii)) indicating bona fide fork degradation. Together, the results support the notion that the p53 pathway is critical for preventing MRE11/EXO1-mediated degradation at deprotected forks.
While established cancer cell lines provide tractable models to interrogate molecular pathways, to determine whether our observations are clinically relevant, we interrogated a panel of patient-derived progenitor cells (PDCs) obtained from freshly resected hepatocellular carcinoma tissues. Whole-exome sequencing was performed, and p53 genetic status was evaluated (Figure 1K and L; Supplementary Table S1) prior to the fork degradation assay. Strikingly, compared to that in PDCs harbouring wild-type p53, nascent DNA at stalled forks in early-passage HCC PDCs bearing p53 mutations was severely compromised in response to ATRi (IdU/CldUWTavg_HU+ATRi= 1.68; IdU/CldUp53MUTavg_ HU+ATRi= 1.16) and was protected when MRE11 was inhibited (Figure 1M and N). These findings indicate that the same mechanisms governing fork degradation are relevant in clinically derived cancer cells. Together, these results provide substantial evidence that a functional p53 pathway limits excessive fork degradation at deprotected forks in established cancer cell lines as well as in patient-derived cells, confirming the universality of these observations.
Extensive degradation of nascent DNA is perpetuated by the loss of a critical p53-regulated factor, RRM2B
To further delineate the mechanism of p53 in fork maintenance, we next considered how the p53 pathway may act via partner proteins or by promoting the expression of genes involved in the DNA damage response. We reasoned that knockdown of a p53-regulated DDR gene would phenocopy the outcome of p53 deficiency and promote fork degradation. To investigate this, we assessed the effects of siRNAs targeted against candidate DDR genes under conditions of replication stress. Strikingly, among the DDR genes investigated, only knockdown of RRM2B mimicked p53 depletion, promoting significant fork degradation when ATR was concomitantly inhibited (Figure 2A and B, Table 1, Supplementary Figure S2A) and replication-associated DNA damage in EdU-positive cells (Supplementary Figure S2B). Moreover, simultaneous disruption of RRM2B and p53 did not further enhance fork degradation (IdU/CldUNT= 1.60; IdU/CldUsip53= 1.24; IdU/CldUsiRRM2B = 1.25; IdU/CldUsiRRM2B+sip53 = 1.27) (Figure 2C), indicating that RRM2B and p53 act in the same fork protection pathway. In contrast, downregulation of canonical p53 targets such as p21 and MDM2 had no impact on fork degradation (Figure 2A).
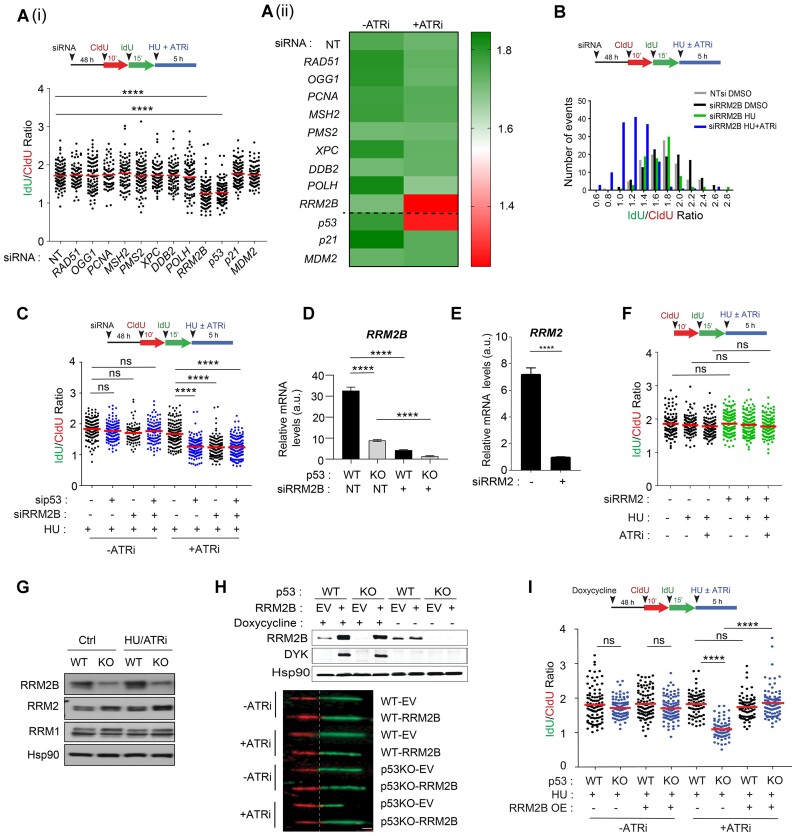
Figure 2.
RRM2B is a prime p53-dependent factor that determines nascent DNA resiliency. (A) (i) Schematic of the fork degradation assay in HCT116 parental (WT) cells transfected with siRNAs against the indicated DNA damage responsive genes and p53-regulated canonical target genes p21 and MDM2. Cells were treated with HU (2 mM) and ATR inhibitor (2 μM). Mean of IdU/CldU ratios represented in scatterplot and in (ii) heatmap. Controls (-ATRi) are included here and in Supplementary Figure S2A(ii) and A(iii). (B) Schematic of the fork degradation assay in HCT116 parental (WT) cells transfected with siRNAs against RRM2B (siRRM2B). Non-targeting (NT) siRNAs used as control. Histogram of IdU/CldU ratios of individual replication forks plotted in GraphPad Prism. Transfected cells were treated with HU (2 mM), and with or without ATRi (2 μM). Representative of three independent experiments. (C) Schematic of the fork degradation assay in HCT116 parental (WT) cells transfected with siRNAs targeting p53 or RRM2B. Cells were treated with HU (2 mM) and in the presence or absence of ATR inhibitor (2 μM). Representative of n = 3 independent experiments. (D) RRM2B transcript levels were quantified in HCT116 (WT) and p53KO cells transfected with siRNAs against RRM2B (siRRM2B) or NT siRNAs. (E) qPCR analysis of transcript levels of RRM2 in HCT116 parental (WT) cells transfected with siRNAs against RRM2 (siRRM2). (F) Schematic of the fork degradation assay in HCT116 parental (WT) cells transfected with siRNAs targeting RRM2. Treatment with HU (2 mM) in the presence or absence of ATR inhibitor (2 μM) as indicated. IdU/CldU ratios of individual replication forks were plotted. Experiment was repeated three times with similar results. (G) Immunoblots showing RRM2B, RRM2 and RRM1 protein levels in HCT116 parental (WT) and p53KO cells treated with HU (2 mM) and ATR inhibitor (2 μM) (6 h). DMSO-treated cells as control (Ctrl). Results are representative of n = 3. (H) Stable expression of pTRIPZ-RRM2B(DYK) in HCT116 parental (WT) and p53KO cells. EV represent empty pTRIPZ vector control. Induced expression of RRM2B-DYK achieved using doxycycline (1.5 μg/ml) treatment for 48 h. DYKDDDDK Tag antibody (CST, #2368) detects exogenous RRM2B(DYK) protein and anti-RRM2B antibody detects total RRM2B protein in immunoblots. Representative images of single DNA fibers. Scale bar, 50 μm. (I) Schematic of the fork degradation assay in HCT116 parental (WT) cells and isogenic p53KO cells overexpressing wildtype RRM2B (OE). Empty pTRIPZ vector used as control (EV). Treatment with HU (2 mM) in the presence or absence of ATR inhibitor (2 μM) as indicated. Scatterplot showing IdU/CldU ratios in individual experimental condition. In (A), (C), (F) and (I), mean of IdU/CldU ratios indicated by a red horizontal bar and P value was calculated from n ≥ 100 DNA fibers using Mann–Whitney test (P< 0.0001 ****; ns = not significant). qPCR or western blots in (G) and (H) were repeated independently at least three times with similar results.
Table 1.
Primer sequences used in qPCR
| Gene names | Primer sequence |
|---|---|
| XPC | 5′-GGATGAAGCCCTCAGCGATGG-3′ (F) |
| 5′-CTTGAGGTCACTTGGAAAG-3′ (R) | |
| RAD51 | 5′-GGCCCACAACCCATTTCACG-3′ (F) |
| 5′-GGCAACAGCCTCCACAGTATGG-3′ (R) | |
| POLH | 5′-GTGGGAAAGCTAACCTCACC-3′ (F) |
| 5′-CTTCAACACTGGCTTCCCGG-3′ (R) | |
| DDB2 | 5′-GAGCGAGATCCGAGTTTAC-3′ (F) |
| 5′-CTGGGTATCGGCCCACAAC-3′ (R) | |
| OGG1 | 5′-GCCTTCTGGACAATCTTTCCG-3′ (F) |
| 5′-GCAGGACTTTGCTCCCTCCAC-3′ (R) | |
| PCNA | 5′-CATGGGCGTGAACCTCACC-3′ (F) |
| 5′-CTTTACTACACAGCTGTACTC-3′ (R) | |
| PMS2 | 5′-CAGGGGCAGAGGCTCATAG-3′ (F) |
| 5′-CAGTTAAGTTGAGAGTCTGAGG-3′ (R) | |
| MSH2 | 5′-GGATAAGAACAGAATAGAGGAG-3′ (F) |
| 5′-CAAAAGCTTCCACTAAATTC-3′ (R) | |
| RRM2B | 5′-ACTCAGAGAT GTACAGTTTG CTG-3′ (F) |
| 5′-TTC TTA ACA TAG GGC ATG GTT TCA-3′ (R) | |
| EXO1 | 5′-GGGGAAAGTCTCGGAAGCTCGAGA-3′ (F) |
| 5′-GGCAATCTACCCCCTGAGACC-3′ (R) | |
| MRE11 | 5′-CCAACAAAGGAAGAGGCCGAGG-3′ (F) |
| 5′-CGGGTAGAAGTCTCCAGACC-3′ (R) | |
| p53 | 5′-TTCACCCTTCAGATCCGTGG-3′ (F) |
| 5′-CAGCTCTCGGAACATCTCGAA-3′ (R) | |
| NRF2 | 5′-CAGCGACGGAAAGAGTATGA-3′ (F) |
| 5′-TGGGCAACCTGGGAGTAG-3′ (R) | |
| NQO1 | 5′-GGTTTGGAGTCCCTGCCATT-3′ (F) |
| 5′-TTGCAGAGAGTACATGGAGCC-3′ (R) | |
| G6PD | 5′-GACGACGAAGCGCAGACA-3′ (F) |
| 5′-TCCGACTGATGGAAGGCATC-3′ (R) | |
| PGD | 5′-GGCGTACCCGTCACCCTC-3′ (F) |
| 5′-CACCGAGCAAAGACAGCTTC-3′ (R) | |
| GSTA4 | 5′-CTCCGAGTGGACTCCAGAAA-3′ (F) |
| 5′-TCATCAAACTCGACTCCGGC-3′ (R) | |
| GSR | 5′-CCCGAATACCAAGGACCTG-3′ (F) |
| 5′-CCCTTGTCATCGGTTTGAATCC-3′ (R) | |
| GCLM | 5′-GCCCGTCCACGCACAGCGAG-3′ (F) |
| 5′-CTGGGTTGATTTGGGAACTC-3′ (R) | |
| TXN | 5′-CAGACTCCAGCAGCCAAGAT-3′ (F) |
| 5′-AGCAACATCATGAAAGAAAGGCT-3′ (R) | |
| PRDX1 | 5′-TTGGTATCAGACCCGAAGCG-3′ (F) |
| 5′-AAAGGCCCCTGAACGAGATG-3′ (R) | |
| Catalase | 5′-GCGGAGATTCAACACTGCC-3′ (F) |
| 5′-CGTTCACATAGAATGCCCGC-3′ (R) | |
| IDH | 5′-ATCATCATAGGTCGTCATGC-3′ (F) |
| 5′-CAACAAAATCAGTTGCTC-3′ (R) | |
| PTGR1 | 5′-CACTGTTATCGGCCAGATGAAG-3′ (F) |
| 5′-CGAAGCTCCTGATAGATAAC-3′ (R) | |
| TXNRD1 | 5′-GATAGGCGGCCATGGTCCAACC-3′ (F) |
| 5′-GCCAGACCTCCTGAGCCACCTCC-3′ (R) | |
| TALDO1 | 5′-CCGACACGGGCGACTTCCAC-3′ (F) |
| 5′-CTGGGGCTTGTACTCGTCG-3′ (R) | |
| RRM2 | 5′-GCAGCAAGCGATGGCATAG-3′ (F) |
| 5′-CTTCTTGGCTAAATCGCTCC-3′ (R) | |
| CDC7 | 5′- CCA CAG CAC AGT TAC AAG TAG-3′ (F) |
| 5′- CTC CCA TGA CAT TAT CTT GCC-3′ (R) |
Further molecular analysis of the mRNA and protein levels of RRM2B and chromatin immunoprecipitation (ChIP) confirmed that RRM2B is a p53-dependent target gene under basal and replication stress conditions (Figure 2D, Supplementary Figure S2C and D). RRM2B is a p53-inducible small subunit (p53R2) of the ribonucleotide reductase (RNR) enzyme that catalyses the de novo synthesis of dNTPs to maintain the balanced dNTP pool needed for DNA replication and repair under hypoxic conditions (112) and is also required for the maintenance of mitochondrial DNA (113). RRM2B has also been shown to act following oxidative stress, to prevent the onset of oxidative-stress induced cellular senescence (114). Although RRM2B is known to function in mtDNA replication and maintenance, its role in regulating nuclear DNA replication and genomic integrity remains unclear. Given the role of RRM2B in counteracting fork degradation at deprotected forks, we next investigated whether another ribonucleotide reductase subunit, RRM2, was involved (115). Despite the crucial role of RRM2 in preserving the global nucleotide pool (116), as evidenced by widespread replication fork slowing in the absence or presence of stress-induced obstacles when RRM2 (but not RRM2B) was suppressed (Supplementary Figure S2E and F) (115), knockdown of RRM2 did not promote fork degradation in response to HU and ATRi (Figure 2E and F). Additionally, RRM2B, but not RRM2 or RRM1, is tightly regulated by p53 in untreated cells under basal conditions and in cells exposed to replication stress (Figure 2G). Altogether, this evidence confirms RRM2B as a key factor mediating the protective effects of p53 and suggests that the function of RRM2B in regulating fork degradation is mechanistically distinct from that of RRM2.
To confirm the role of RRM2B in preventing fork degradation, particularly in the absence of p53, we investigated whether restoring RRM2B levels in p53KO cells mitigates fork degradation. We genetically modified HCT116 parental and isogenic p53KO cells to enable ectopic expression of DYK-tagged RRM2B under a doxycycline-inducible promoter (Figure 2H). Overexpression of wildtype RRM2B abolished fork degradation in p53KO cells compared to empty vector control cells (IdU/CldURRM2B_OE = 1.84 versus IdU/CldUEV = 1.08). (Figure 2I) and significantly reduced DNA damage and replication stress signalling in p53KO cells (Supplementary Figure S2G). Collectively, these findings highlight RRM2B as a critical p53-regulated factor whose absence promotes fork degradation and exacerbates replication stress when stalled forks are deprotected by ATR inhibition.
The p53-RRM2B pathway prevents disproportionate activation of PARylation
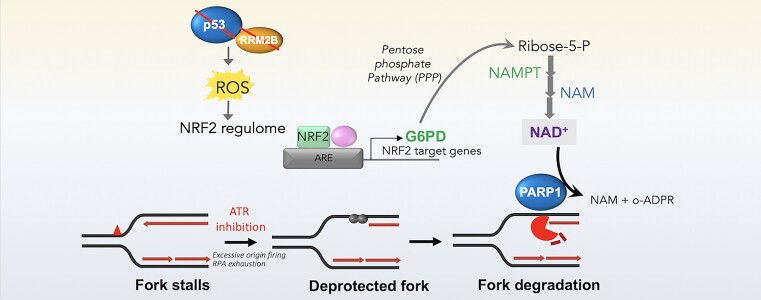
To further elucidate the mechanism by which fork stalling leads to degradation in p53/RRM2B-deficient cells, we searched for factors that guard replication fork stability. PARP1 stabilizes the replication fork and regulates fork progression (117,118) in part by controlling replication fork speed and stabilizing regressed forks (18,118). Although PARP1 is an important regulator of the balance between fork restoration and resection (33,119), the extent to which PARP1 is regulated to control replication fork dynamics remains unclear. Moreover, how p53 and RRM2B regulate PARP1 and the implications for the DNA replication stress response are poorly understood. To investigate the effects of p53 or RRM2B on PARylation in cells, immunodetection of PAR was performed using a commonly used anti-PAR antibody (10H) (120). Single-cell IF analyses of multiple panels of cells revealed a 1.8–2.0-fold increase in basal PAR signals in HCT116 and U2OS cells transfected with sip53 or siRRM2B compared to control cells (Figure 3A) and in isogenic HCT116 p53KO cells compared to HCT116 parental (WT) cells (Supplementary Figure S3A(i)). Western blot analyses confirmed that basal cellular PARylation was significantly elevated in HCT116 parental (WT) cells depleted of p53 or RRM2B (Figure 3B and C) and in isogenic HCT116 p53KO cells (Supplementary Figure S3A(ii)). The specificity of the detected PAR signals in western blot or IF analyses was confirmed either by chemical inhibition of PARP1 (Supplementary Figure S3B) or an siRNA targeting PARP1 (Figure 3D). Importantly, genetic complementation with wild-type RRM2B suppressed basal PARylation in p53KO cells to a level comparable to that in wild-type cells (Figure 3E and F). As most PARylation is executed by PARP1 (121), our results led to two important predictions: (i) the absence of p53 or RRM2B results in an imbalance or dysregulation of PARP1 activation, leading to excessive PARylation and (ii) excessive PARP1 activation may contribute to aberrant fork processing and replication stress at deprotected forks.
Figure 3.
p53/RRM2B loss results in hyperPARylation compromising nascent fork stability. (A) sip53 or siRRM2B-transfected HCT116 and U2OS parental cells were immunostained for total PAR (anti-PAR [10H] antibody, Abcam ab14459). Mean PAR intensity per nucleus was measured and plotted. Representative immunofluorescent images were shown (top). n ≥ 200 in each condition. Scale bar, 20 μm (mean ± SD; n = 3; two-tailed t-test). (B, C), Total cellular PARylation was analysed in whole cell lysates (WCL) harvested from sip53- or siRRM2B-transfected HCT116 cells. Total protein from WCL loaded on gel indicated in μg. (D)(i) HCT116 parental (WT) and p53KO cells were transfected with siPARP1 or non-targeting (NT) siRNAs. Western blot showing knockdown of PARP1 in HCT116 parental (WT) cells and p53KO cells. (ii) siRNA-transfected cells were immunostained using anti-PAR [10H] antibody (Abcam ab14459). Mean PAR intensity per nucleus was measured and plotted. n ≥ 200 in each condition. (mean ± SD; n = 3; two-tailed t-test). (E) Mean PAR intensity per nucleus was measured in HCT116 parental (WT) and p53KO cells with stable expression of pTRIPZ-RRM2B(WT). EV represent pTRIPZ empty vector control. Induced expression of RRM2B(WT) achieved using doxycycline (1.5 μg/ml, 48 h). n ≥ 200 in each condition (mean ± SD; n = 3; two-tailed t-test). (F) HCT116 parental (WT) and p53KO cells with stable expression of pTRIPZ-RRM2B(WT) or pTRIPZ-empty vector (EV) were subjected to western blot analysis of total PAR (anti-PAR [10H] antibody, Abcam ab14459). Overexpression of RRM2B was verified using anti-RRM2B antibody. Total protein from WCL loaded on gel indicated in μg. (G) Schematic of the fork degradation assay in HCT116 parental (WT) cells and p53KO cells pretreated with PARPi (50 μM). HU (2 mM) in the presence or absence of ATRi (2 μM) were used as indicated. IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (H) Colony forming assay was performed on HCT116 parental (WT) and p53KO cells following treatment with HU (2 mM)/ATRi (2 μM) ± PARPi (50 μM) for 24 h before recovery in fresh media for eight days. Experiment is repeated independently at least three times with similar results. (I) Schematic of the fork degradation assay in HCT116 parental (WT) cells and p53KO cells transfected with pooled targeted siRNAs against PARP1. Cells were subjected to 2 mM HU treatment in the presence or absence of ATRi (2 μM), as indicated. IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (J) Schematic of the fork degradation assay in HCT116 parental (WT) cells and p53KO cells pretreated with PARG inhibitor (0.6 μM). Cells were subjected to 2 mM HU treatment in the presence or absence of ATRi (2 μM), as indicated. IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (K) Schematic of the fork degradation assay in HCT116 parental (WT) cells and p53KO cells pretreated with NAD (1 mM) or FK866 (1 μM). Cells were then subjected to 2 mM HU treatment in the presence or absence of ATRi (2 μM), as indicated. IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (L) Schematic of the fork degradation assay in hepatocellular carcinoma (HCC) patient-derived cells. Cells were pretreated with PARPi (50 μM) or PARGi (0.6 μM) as indicated, followed by HU (2 mM) and ATR inhibitor (2 μM). Mean of IdU/CldU ratios shown in heatmap. IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. In (G), (I), (J), (K) and (L), results are representative of at least n = 3 independent experiments. Mean of IdU/CldU ratios indicated by a red horizontal bar and P value was calculated from n ≥ 100 DNA fibers using Mann–Whitney test (P< 0.0001 ****; ns = not significant). Western blots in (B), (C), (D) and (F), were repeated independently at least three times with similar results.
Hyper-PARylation compromises the stability of nascent DNA
Our results reveal a previously unknown dependency of basal PARylation on the p53-RRM2B pathway. We next considered whether hyper-PARylation at deprotected stalled forks could counteract the stability of nascent DNA. Because PARP1 stabilizes stalled forks by counteracting fork restart (32,33), we reasoned that hyper-PARylation may cause fork remodelling events permissive for fork degradation. Indeed, we observed that transient inhibition of PARP1 with olaparib abolished fork degradation in p53KO cells (IdU/CldUp53KO = 1.30; IdU/CldUp53KO+PARPi = 1.76; IdU/CldUWT = 1.73; IdU/CldUWT+PARPi = 1.69) (Figure 3G) and restored the viability of p53 KO cells (Figure 3H), linking fork degradation to chemosensitivity. To rule out possible side effects of the inhibitor, we genetically ablated PARP1 and confirmed that PARP1 activation is a key factor driving unscheduled fork degradation in p53KO cells (IdU/CldUp53KO = 1.19 to IdU/CldUp53KO+siPARP1 = 1.86) (Figure 3I). Finally, an alternative CldU/IdU labelling scheme that was used earlier to differentiate fork degradation and breakage (Supplementary Figure S1Q) also demonstrated that transient PARP1 inhibition suppresses bona fide fork degradation in p53KO cells (Supplementary Figure S3C).
The above data indicated that a hyper-PARylated state in p53KO cells predicts unscheduled fork degradation at deprotected forks. Indeed, by directly blocking the catabolism of PAR chains using a PARG inhibitor, endogenous PARylation (Supplementary Figure S3D) and fork degradation could be induced in ATRi-treated HCT116 parental (WT) cells (Figure 3J) which was previously resilient to fork degradation (Figure 1A(i)), but not in the absence of ATRi (Supplementary Figure S3E). This was followed by a concomitant increase in γH2AX signals in HU + ATRi-treated HCT116 parental (WT) cells (Supplementary Figure S3F). Second, by altering the availability of nicotinamide adenine dinucleotide (NAD), which is used in its oxidized form (NAD+) by PARP1 to ADP (ribosyl)ate itself and its target proteins, we could determine the fate of the nascent DNA strand. NAD supplementation increased basal PARylation (Supplementary Figure S3G) and effectively promoted fork degradation even in HCT116 (WT) cells (Figure 3K). Conversely, a potent small molecule inhibitor (FK866) that inhibits nicotinamide phosphoribosyl transferase (NAMPT) (122) reduced total cellular PARylation (Supplementary Figure S3H), abolished fork degradation (Figure 3K) and suppressed replication-associated DNA damage in p53KO cells (Supplementary Figure S3I). Notably, the importance of PARP1 in the regulation of fork stability was confirmed in patient-derived HCC cells (Figure 3L), confirming the physiological relevance of these mechanisms for fork and genomic integrity. While olaparib caused a marked reduction in γH2AX and RPA2S4S8 levels and replication-associated breaks in HU and ATRi-treated p53KO cells (Supplementary Figure S3J–L), it did not rescue genome-wide DNA damage and DSBs induced by doxorubicin (Supplementary Figure S3M and N). We conclude that hyper-PARylation exacerbates replication stress-induced damage at deprotected stalled forks and that dysregulation of PARP1 is the key factor underlying replication vulnerability in p53KO cells.
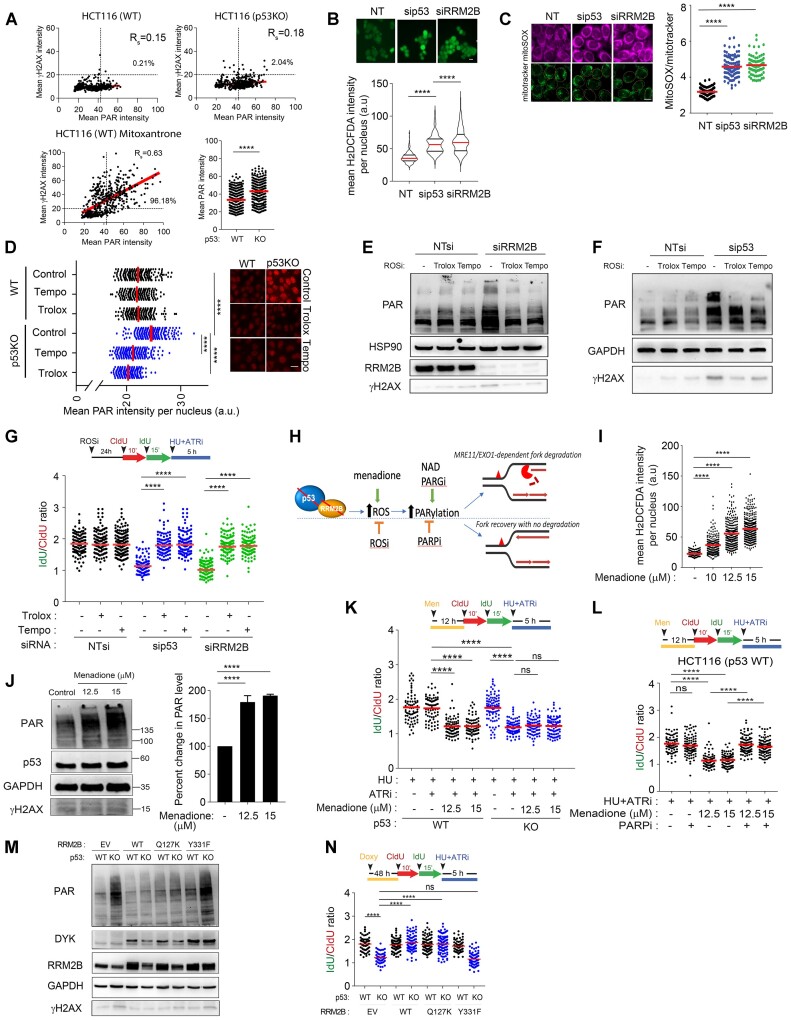
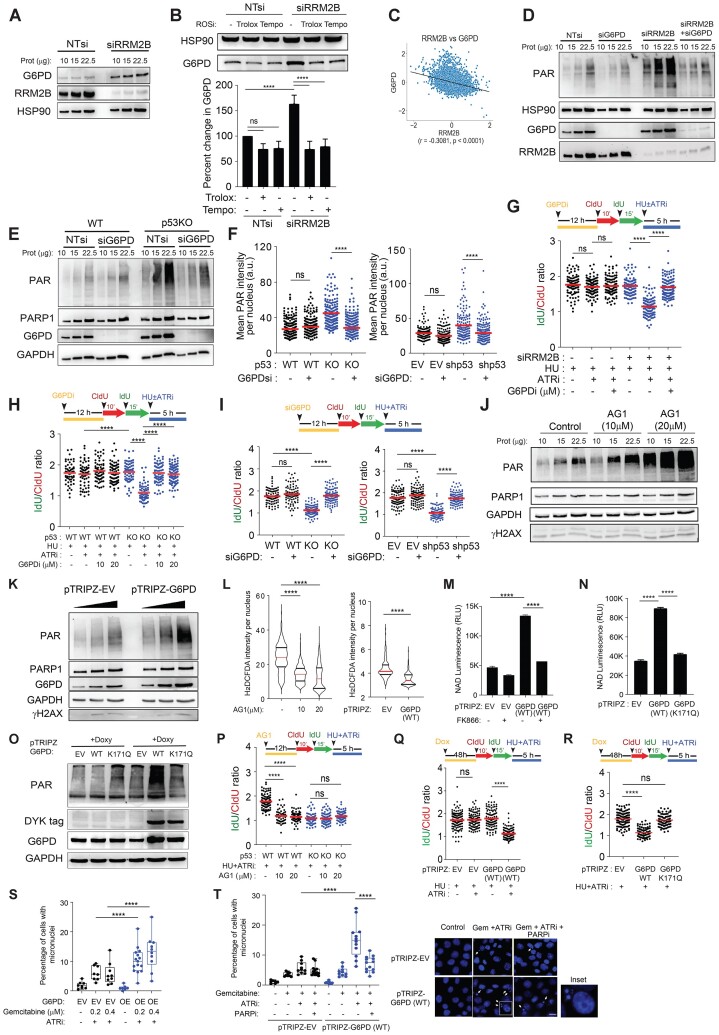
Crosstalk between the oxidative stress response and fork degradation mediated by the p53-RRM2B pathway
Notably, we observed that the increased PARylation in p53-deficient cells is not directly caused by acute drug-induced DNA replication stress, since it occurred in untreated cells (Supplementary Figure S4A). Next, to investigate whether the increased in basal PARylation in p53-/RRM2B-deficient cells correlated with any changes in DNA damage signaling, we established a dual immunostaining method relying on anti-γH2AX and anti-PAR (10H) antibodies. Treatment with mitoxantrone, an anthracycline that inhibits topoisomerase II and also intercalates the DNA resulting in DNA breaks (123), induced abundant DNA damage-induced PARylation, as reflected by the significant correlation between PAR and γH2AX signals (Spearman correlation coefficient, Rsmito = 0.63) (Figure 4A). In contrast, a marked increase in PAR signals was observed in p53KO cells; however, the lack of a significant correlation with γH2AX (Spearman correlation coefficient, RsWT = 0.15, Rsp53KO= 0.18), suggest that hyper-PARylation in p53KO cells did not induce a γH2AX-dependent DDR response.
Figure 4.
Oxidative stress induced hyper-PARylation underpins fork degradation in RRM2B/p53-deficient cells. (A), Single-cell dual IF analysis was performed using anti-PAR [10H] (Abcam, ab14459) and anti-γH2AX (Abcam, ab2893) antibodies. Mean γH2AX and PAR intensities per nucleus were quantified. Percentages of cells positively stained for γH2AX and PAR as indicated, in untreated HCT116 cells (WT) and p53KO cells, and in mitoxantrone-treated HCT116 cells (WT) cells (1.25 μM, 6 h). Bottom right panel: Mean PAR intensity per nucleus in HCT116 cells (WT) and p53KO were represented in a scatterplot. Representative of n = 3 independent experiments and n ≥ 200 in each condition. (B) Mean H2DCFDA (2′,7′-dichlorofluorescin diacetate) (Invitrogen, D399) intensity per nucleus of sip53 or siRRM2B-transfected HCT116 parental (WT) cells quantified and plotted. Representative images shown. Scale bar, 20 μm (mean ± SD; n = 3; two-tailed t-test). (C) MitoSOX/Mitotracker ratios were quantified in sip53 or siRRM2B-transfected HCT116 parental (WT) cells and plotted. Representative images on the left of the graph. Scale bar, 20 μm (mean ± SD; n = 3; two-tailed t-test). (D) Total PARylation was detected by immunofluorescence using anti-PAR antibody (Abcam, ab14459) in HCT116 WT and p53KOcells following treatment with Tempo (1 mM) or Trolox (0.2 mM). Mean PAR intensity per nucleus was measured and plotted. Representative images shown. Scale bar, 20 μm (mean ± SD; n = 3; two-tailed t-test). (E) Total cellular PARylation in siRRM2B- or (F) sip53-transfected HCT116 parental (WT) cells were treated with Trolox (0.2 mM) or Tempo (1 mM) and analysed by western blot. (G) Schematic of the fork degradation assay in HCT116 parental (WT) transfected with sip53 or siRRM2B and pretreated with Tempo (1 mM) or Trolox (0.2 mM). Cells were subjected to HU (2 mM) in the presence or absence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (H) Schematic diagram illustrating oxidative stress induced PARylation as key factors influencing the stability of nascent DNA at stalled deprotected forks. (I, J) HCT116 cells (WT) cells were treated with an oxidative stress inducer, menadione, at the indicated concentrations for 12 h. Mean H2DCFDA (Invitrogen, D399) intensity per nucleus quantified and plotted in (I), and total cellular PARylation was analysed in western blot using an anti-PAR antibody (CST, 83732). γH2AX (Abcam, ab2893) was also detected as a surrogate marker of DNA damage. PAR levels were quantified (mean ± SD; n = 3; two-tailed t-test). (K) Schematic of the fork degradation assay in HCT116 parental (WT) and p53KO cells pre-treated with menadione, and then subjected to HU (2 mM) in the presence or absence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (L) Schematic of the fork degradation assay in HCT116 parental (WT) cells pre-treated or not with menadione. Cells were then subjected to treatment with HU (2 mM) and ATRi (2 μM), and in the presence or absence of olaparib (50 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (M) pTRIPZ-RRM2B(Q127K) and pTRIPZ-RRM2B (Y331F) were stably expressed in HCT116 parental (WT) and p53KO cells, under doxycycline induction (1.5 μg/ml, 48 h). Total PARylation was analysed in western blot using an anti-PAR antibody (CST, 83732). (N) Schematic of the fork degradation assay in HCT116 parental (WT) and p53KO cells overexpressing WT RRM2B, RRM2B(Q127K) or RRM2B(Y331F). pTRIPZ-EV (empty vector) included as control. Cells were treated with doxycycline (Doxy) (1.5 μg/ml, 48 h) prior to the fork degradation experiment. IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. In (G), (K), (L) and (N), red horizontal bar represent mean of CldU/IdU ratios; P value was calculated from n ≥ 100 DNA fibers using Mann–Whitney test (P< 0.0001 ****; ns = not significant). Data representative of n = 3 independent experiments. Western blots in (E), (F), (J) and (M) were repeated independently at least three times with similar results. GAPDH or HSP90 were used as loading control.
To determine the specific mechanism of hyper-PARylation in p53KO cells, we considered whether RRM2B’s roles in mitochondrial metabolism and cellular redox might be pivotal. Two key observations indicated that dysregulated cellular redox in p53/RRM2B-deficient cells induced robust PARylation signalling in otherwise unstressed cells. First, heightened signals from 2′,7′-dichlorofluorescin diacetate (H2DCFDA), a probe sensitive to hydroxyl radicals, peroxyl radicals, and various other ROS, and mitoSOX, which measures the formation of mitochondrial superoxide, confirmed that oxidative stress responses are specifically induced in HCT116 parental (WT) cells transfected with siRNAs against p53 or RRM2B (Figure 4B and C), in isogenic p53KO cells, as well as in HCT116 parental (WT) cells with stable expression of shp53 (Supplementary Figure S4B). Second, the ROS scavengers Trolox (vitamin E analogue) and Tempo (nitroxide radical) markedly reduced H2DCFDA intensities (Supplementary Figure S4C) and PAR signals in p53KO cells (Figure 4D) and in cells transfected with siRRM2B or sip53 (Figure 4E and F), indicating the importance of an oxidative stress response in sustaining PARP1 activation. Although extreme oxidative stress induces DNA damage and subsequent damage-induced PARylation and apoptosis (124), our results suggest that a sublethal level of ROS in p53-/RRM2B-deficient cells sustains PARP1 activation through an alternative mechanism.
We then sought to determine whether a sublethal level of ROS in p53-/RRM2B-deficient cells is a critical determinant of fork stability. Significantly, pretreatment with Trolox or Tempo prior to replication stress completely suppressed fork degradation in sip53- or siRRM2B-transfected cells (IdU/CldUsip53= 1.11; IdU/CldUsip53+Trolox= 1.83; IdU/CldUsip53+Tempo= 1.78; IdU/CldUsiRRM2B= 1.03; IdU/CldUsiRRM2B+Trolox= 1.80; IdU/CldUsiRRM2B+Tempo= 1.78) (Figure 4G). These results were confirmed in p53KO cells (Supplementary Figure S4D), providing compelling evidence that a ROS-mediated mechanism sustained hyper-PARylation in p53KO cells and promoted fork degradation. Moreover, inhibiting ROS promoted p53KO cell resistance to replication stress and ATR inhibition (Supplementary Figure S4E) and suppressed replication-associated γH2AX (Supplementary Figure S4F and G). Finally, we made the reverse prediction that a forced increase in ROS levels might be sufficient to induce hyper-PARylation in cells and promote the degradation of deprotected forks, phenocopying p53 or RRM2B loss (Figure 4H). To achieve a mild and controlled increase in ROS levels, we titrated doses of a known pharmacologic inducer of oxidative stress, menadione (Figure 4I) (125). Low doses of menadione increased basal cellular PARylation (Figure 4J) and induced pronounced fork degradation in HU + ATRi-treated HCT116 parental (WT) cells (IdU/CldUWT = 1.74 to IdU/CldUWT+Men= 1.18) to the same extent as in p53KO cells (Figure 4K). Moreover, the effects of menadione on fork degradation were completely mitigated by olaparib, confirming that mild oxidative stress induced hyper-PARylation is a critical determinant of nascent DNA stability in p53KO cells (Figure 4L).
To ascertain that hyper-PARylation is due to the impairment of RRM2B function and its consequential mitigation of oxidative stress, rather than its involvement in replenishing deoxyribonucleotide triphosphates (dNTPs) in DNA repair, stable HCT116 cell lines (wild-type and p53 knockout) were engineered to express pTRIPZ-RRM2B variants (wild-type, Q127K, and Y331F). Q127K mutation in RRM2B is known to hinder its catalytic function in generating dNTPs (112). A mutagenesis study targeting Y331 in RRM2B revealed the abrogation of antioxidant activity in the Y331F mutant, emphasizing the critical role of Y331 in scavenging reactive oxygen species (ROS) (126).
RRM2B overexpression was validated using qPCR analysis (Supplementary Figure S4H). Concurrently, the assessment of antioxidant functionality of wildtype RRM2B and its mutant derivatives (Q127K and Y331F) was affirmed through measurements of 2′,7′-dichlorofluorescin diacetate (H2DCFDA). Our results show that overexpression of wildtype RRM2B and RRM2B(Q127K) effectively reduced ROS levels. Conversely, RRM2B(Y331F) failed to suppress ROS in p53KO cells, consistent with the reported role of Y331 in RRM2B’s antioxidant activity (126) (Supplementary Figure S4I). Western blot analysis demonstrated reduced PAR expression in p53KO cells overexpressing RRM2B(WT) and RRM2B(Q127K) (Figure 4M), indicating the capability of catalytically inactive RRM2B in mitigating PARylation levels in p53KO cells. Conversely, overexpression of RRM2B(Y331F) failed to suppress hyper-PARylation, highlighting the significance of RRM2B antioxidant activity in PAR regulation in p53KO cells (Figure 4M). Finally, overexpression of RRM2B(WT) and RRM2B(Q127K) but not RRM2B(Y331F) successfully rescued fork degradation in p53KO cells (Figure 4N). Collectively, these data support the notion that the antioxidant function ascribed to RRM2B is critical in counteracting endogenous ROS and hyperPARylation, and preventing unscheduled fork degradation at deprotected stalled forks.
The NRF2-PARP1 axis connects redox homeostasis to replication fork integrity
The above results prompted us to investigate the mechanism by which low ROS levels trigger increased PARylation. Our data suggest that this process operates independently from the conventional activation of PARP1 triggered by ROS-induced DNA damage, a phenomenon well documented in cells under severe oxidative stress (124,127). To precisely define the mechanisms that connect ROS, hyper-PARylation and impairments in p53/RRM2B, we hypothesized that specific antioxidant-driven transcriptional programs may be activated in the absence of p53, the cause of RRM2B deficiency. Hence, bioinformatics analysis was employed to elucidate clinically relevant pathways and evaluate the enrichment of antioxidant pathways associated with global alterations in RRM2B gene expression in patient cohorts. Patient transcriptome data across 12 cancer datasets (aggregated n = 4964) was first downloaded from TCGA in csv form. Python and R was utilized to determine the Pearson correlation between RRM2B expression and antioxidant gene response, quantified using an antioxidant gene signature comprising 125 genes derived from Ren et al. (2021) (88) (Figure 5A, Supplementary Table S2). To further dissect the relationship between each gene in the antioxidant gene signature and RRM2B expression, Pearson correlation was calculated between the expression of each gene and that of RRM2B. The result was visualized using R (Figure 5B). Negative r value (r←0.1) indicates an enrichment of the antioxidant gene with reduction in RRM2B expression. Hierarchical clustering was applied on the correlation coefficients (r values) to derive genes with similar relationships to RRM2B (Figure 5B) (Supplementary Table S3). The cluster with significant r value < -0.1 (identified as Cluster III in Figure 5B) from this analysis was analysed using STRING and this revealed that NRF2-related pathways were among the most significantly enriched pathways (Figure 5C). Instead of RRM2B expression, similar analysis was conducted using a p53 gene signature (89) on the same TCGA transcriptomic dataset (Supplementary Table S4). Antioxidant genes that correlate significantly with RRM2B and p53 were then overlapped, and additional filtering for NRF2 targets revealed a set of four putative antioxidant genes (Figure 5D and E, Supplementary Table S5) (128). To cross-validate the results from the gene expression analysis of the TCGA dataset, we screened for NRF2-regulated genes that were differentially upregulated (DEGs) in HCT116 p53KO cells using gene expression profiling (Supplementary Figure S5A–D and Supplementary Table S6). This revealed that NRF2 targets, G6PD, TXN1 and PRDX1, were commonly enriched in both analyses (68) (Supplementary Figure S5E–G). We next sought to validate the role of NRF2 in the context of replication integrity in p53-defective cells.
Constitutive activation of NRF2 occurs in various types of cancers, and its role in promoting chemo- or radioresistance and tumour aggressiveness has been ascribed mainly to the broad antioxidant functions of NRF2 (128,129). Our results raise the possibility that antioxidant responses induced by NRF2 may counteract fork and genomic stability, presenting a previously unknown vulnerability. To test this, we used a pharmacological inhibitor of NRF2, ML385 (130) or gene-targeted siRNAs. Remarkably, pretreatment with ML385 (Figure 5F) or genetic ablation of NRF2 completely suppressed fork degradation in p53-deficient cells, providing concrete evidence for the role of NRF2 in promoting fork degradation (Figure 5G and H, Supplementary Figure S5H and I). Further suggesting that PARP1 is the intermediate factor linking NRF2 redox homeostasis to fork degradation, transient inhibition of NRF2 by siRNA (Figure 5I and J) or ML385 (Supplementary Figure S5J) resulted in obvious suppression of PARylation in HCT116 p53KO cells, this is consistent with the notion that the observed hyperPARylation in p53KO cells is largely driven by NRF2. Finally, to investigate whether NRF2-dependent activation of PARP1 is critical to nascent DNA stability, small molecule-induced activation of NRF2 was achieved using dimethyl fumarate (DMF) (131,132). Pretreatment of HCT116 cells with DMF caused a pronounced increase in basal PARylation without inducing γH2AX (Figure 5K). As predicted by our previous result, activation of NRF2 by DMF caused pronounced fork degradation in HCT116 parental (WT) cells but did not further aggravate fork degradation in p53KO cells (Figure 5L), indicating that NRF2 activation and p53 loss compromise nascent DNA stability via the same mechanism. Collectively, these data revealed the role of the newly identified NRF2-PARP1 axis in counteracting fork integrity.
NRF2-dependent G6PD activation underlies replication vulnerability in p53/RRM2-deficient cells
The intriguing involvement of NRF2 in fork stability led us to question which NRF2 target gene might be involved. G6PD is a robust NRF2 target; expression of the G6PD protein was clearly induced upon NRF2 activation by DMF (Figure 5K), and G6PD mRNA levels were reduced by siRNA-mediated depletion of NRF2 (Supplementary Figure S5F). Consistent with our model, G6PD protein was enriched when RRM2B is knocked down (Figure 6A) and suppressed by Tempo or Trolox (Figure 6B), leading us to conclude that G6PD induction in RRM2B-compromised cells is specifically driven by redox-dependent NRF2 activation. An inversed correlation between RRM2B and G6PD gene expression was also significant in a TCGA transcriptome analysis (Figure 6C).
Figure 6.
NRF2-dependent G6PD activation underlies the replication vulnerability in p53/RRM2-deficient cells. (A) HCT116 (WT) cells transfected with siRRM2B. G6PD protein expression analysed in whole cell lysates by western blot. Representative of n = 3 experiments. (B) siRRM2B or non-targeting (NT) siRNA-transfected HCT116 (WT) cells were treated with antioxidants, Trolox (0.2 mM) or Tempo (1 mM) for 12 h. Total G6PD protein was detected using western blot. Ratios of G6PD to HSP90 (loading control) were calculated and plotted (mean ± SD; n = 3; two-tailed t-test). (C) Scatterplot showing normalized (log2) gene expression values of RRM2B and G6PD expression in aggregated 12 TCGA databases (tumor samples n = 4964) represented in Figure 5A. Pearson correlation coefficient indicated, r = –0.3081, P< 0.001. (D) Parental HCT116 (WT) cells were transfected with siRRM2B and/or siG6PD. Total PARylation detected using an anti-PAR antibody (CST, 83732). Specific knockdown of RRM2B and G6PD validated by western blot. (E) HCT116 (WT) and p53KO cells were transfected with siG6PD. Total PARylation detected by western blot using an anti-PAR antibody (CST, 83732). (F) Immunostaining for PAR (Abcam ab14459) was performed in HCT116 (WT) and isogenic p53KO cells transfected with siG6PD, or in HCT116 cells stably transfected with short hairpin RNA against p53 (shp53) or empty vector (EV) as control. Mean PAR intensity per nucleus was quantified and plotted (mean ± SD; n = 3; two-tailed t-test). (G) Schematic of the fork degradation assay in HCT116 parental (WT) cells transfected with siRRM2B and treated with G6PDi (20 μM, 12 h). Cells were subjected to HU (2 mM) in the presence or absence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (H) Schematic of the fork degradation assay in HCT116 parental (WT) and isogenic p53KO cells pre-treated with G6PDi (10 or 20 μM, 12 h). Cells were subjected to HU (2 mM) in the presence or absence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (I) Schematic of the fork degradation assay in HCT116 parental (WT) cells and isogenic p53KO cells transfected with G6PD-targeted siRNAs, or in HCT116 (WT) cells harboring shp53 or empty vector (EV) control transfected with G6PD-targeted siRNAs. IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (J) Total PARylation in HCT116 (WT) cells treated with a selective G6PD activator (AG1) (10 or 20 μM, 12 h) was detected using an anti-PAR [10H] antibody (Abcam ab14459). PARP1 and γH2AX were also detected in immunoblots. (K) Stable expression of pTRIPZ-G6PD and pTRIPZ-EV (empty vector) in HCT116 parental (WT) cells. Total PARylation detected using an anti-PAR antibody (CST, 83732) in western blot. Total PARP1, G6PD and γH2AX proteins were detected. (L) Mean H2DCFDA intensities per nucleus were quantified and plotted in HCT116 cells pre-treated with specific G6PD inducer AG1 (10 or 20 μM, 12 h), or in HCT116 (WT) cells stably expressing pTRIPZ-G6PD(WT)-DYK and pTRIPZ-EV (empty vector). (M) NAD levels were measured in HCT116 (WT) cells stably expressing pTRIPZ-G6PD(WT)-DYK or pTRIPZ-EV (control) and treated with FK866 (1 μM, 12 h) (mean ± SD; n = 3; two-tailed t-test). (N) NAD levels were measured in HCT116 (WT) cells stably expressing pTRIPZ-G6PD(WT)-DYK or pTRIPZ-G6PD(K171Q)-DYK. pTRIPZ-EV included as control (mean ± SD; n = 3; two-tailed t-test). (O) Total PARylation in HCT116 (WT) cells stably expressing pTRIPZ-G6PD(WT)-DYK, pTRIPZ-G6PD(K171Q)-DYK or pTRIPZ-EV detected using an anti-PAR antibody (CST, 83732). Induced expression of G6PD(WT)-DYK or G6PD(K171Q)-DYK was achieved by doxycycline (1.5 μg/ml, 48 h) and detected using an DYKDDDDK Tag antibody (CST, #2368) by western blot. Total G6PD was also detected using an anti-G6PD antibody. (P) Schematic of the fork degradation assay in HCT116 parental (WT) cells and isogenic p53KO cells pretreated with AG1 at the indicated concentrations. Cells were then exposed to HU (2 mM) and ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (Q) Schematic of the fork degradation assay in HCT116 parental (WT) cells stably expressing pTRIPZ-G6PD or pTRIPZ-EV (empty vector). G6PD(WT)-DYK was induced by doxycycline (1.5 μg/ml, 48 h). Cells were then exposed to HU (2 mM) in the presence or absence of ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (R) Schematic of the fork degradation assay in parental (WT) cells stably expressing pTRIPZ-G6PD(WT)-DYK, pTRIPZ-G6PD(K171Q)-DYK or pTRIPZ-EV (empty vector). Cells were treated with doxycycline (1.5 μg/ml, 48 h) and then exposed to HU (2 mM) and ATRi (2 μM). IdU/CIdU ratios for individual replication forks plotted. Representative of n = 3 experiments. (S) Percentages of cells with micronuclei were quantified in HCT116 parental (WT) cells stably expressing G6PD(WT)-DYK or pTRIPZ-EV (empty vector). Cells were treated with doxycycline (1.5 μg/ml, 48 h), followed by gemcitabine (0.2 or 0.4 μM, 24 h). Cells were recovered in fresh media for 48 h before micronuclei were assessed by DAPI staining and immunofluorescence. (T) As in s, Percentages of cells with micronuclei were quantified in HCT116 parental (WT) cells stably expressing G6PD(WT)-DYK or pTRIPZ-EV (empty vector) and drugged with gemcitabine (0.4 μM) in the presence or absence of olaparib (PARPi, 50 μM) for 24 h. Cells were recovered in fresh media for 48 h before micronuclei were assessed by DAPI staining. In (S) and (T), P value was calculated by Mann–Whitney test (P< 0.0001 ****; ns = not significant; n = 3). In (G), (H), (I), (P), (Q) and (R), red horizontal bar represent mean of CldU/IdU ratios; P value was calculated from n ≥ 100 DNA fibers using Mann–Whitney test (P< 0.0001 ****; ns = not significant). Western blots in (A), (B), (D), (E), (J), (K) and (O) were repeated independently at least three times with similar results. GAPDH or HSP90 were used as loading controls.
G6PD is a key rate-limiting enzyme in the pentose phosphate pathway (PPP) and a major regulator of the cellular redox state. In addition to its well-described function in NADPH regeneration and curbing ROS (133–135), the PPP also generates ribose-5-phosphate (R5P), which is not only a precursor of nucleotides but also an important substrate for the NAMPT-dependent synthesis of nicotinamide mononucleotide (NMN) via the salvage pathway, a key route for NAD biosynthesis (136). Remarkably, depletion of G6PD using siRNAs reversed the effects of RRM2B or p53 depletion, resulting in an obvious suppression of PARylation (Figure 6D-F). Furthermore, a small molecule inhibitor of G6PD that interferes with G6PD activity (137) effectively suppressed PAR signals in p53KO cells (Supplementary Figure S6A). Together, these results identified G6PD as an important driver of basal PARylation, particularly in the absence of p53 or RRM2B. Consistent with our prediction that G6PD-induced hyper-PARylation underlies the prominent fork degradation observed at deprotected forks, inhibiting G6PD was found to fully restore nascent DNA stability in siRRM2B-transfected cells (IdU/CldUsiRRM2B = 1.15; IdU/CldUsiRRM2B+G6PDi= 1.67) (Figure 6G) and in HCT116 p53KO cells (IdU/CldUp53KO = 1.10; IdU/CldUp53KO+G6PDi= 1.73) (Figure 6H). Similarly, siRNAs targeting G6PD rescued fork integrity in HCT116 p53KO and shp53-transfected cells (IdU/CldUsiG6PD-p53KO = 1.79; IdU/CldUsiG6PD-shp53= 1.71) (Figure 6I). Moreover, inhibiting G6PD significantly suppressed replication-associated damage in p53KO cells (Supplementary Figure S6B) and partially rescued colony growth, particularly in HCT116 p53KO cells (Supplementary Figure S6C). Our data demonstrates that G6PD activation is a major driver of unscheduled fork degradation and replication vulnerability in p53/RRM2B-deficient cells.
One potential caveat is that these results were obtained under conditions in which G6PD activation was induced by ROS when p53 or RRM2B was compromised. To confirm that these observations were independent of any direct ROS-induced effects, we revisited some key hypotheses by using independent means of altering G6PD activity. First, we reasoned that if our model is generally applicable, enforced activation of G6PD would allow us to determine whether G6PD activation alone is sufficient to impact overall cellular PARylation. To this end, we exploited a small molecule activator of G6PD, AG1, which promotes the oligomerization of G6PD to catalytically competent forms (138). As predicted by our model, AG1 induced a robust induction of cellular PARylation in HCT116 WT cells at concentrations previously shown to activate G6PD in cells (139) (Figure 6J and Supplementary Figure S6D), in the absence of any other exogenous damage. To corroborate this finding, we genetically modified HCT116 WT cells to allow the ectopic expression of wildtype G6PD using a pTRIPZ-G6PD(WT)-DYK construct. Similar to AG1 treatment, controlled overexpression of G6PD (Supplementary Figure S6E) resulted in hyperPARylation in HCT116 WT cells, verified by western blot (Figure 6K) or in IF analyses (Supplementary Figure S6F), confirming the role of G6PD in the regulation of cellular PARylation in a broader context. Importantly, G6PD-induced hyperPARylation is independent of any direct ROS-induced effects, as indicated by the reduction in ROS upon AG1 treatment or when G6PD is overexpressed (Figure 6L). Using liquid chromatography mass spectrometry -based metabolomics, we demonstrated that relative to control (pTRIPZ-EV) cells, G6PD overexpressed (pTRIPZ-G6PD) HCT116 cells exhibited increased levels of metabolites in the Pentose Phosphate Pathway (PPP) and nucleotide biosynthesis pathways (Supplementary Figure S6G), consistent with the established role of G6PD in promoting nucleotide biosynthesis and availability (140). Importantly, we also demonstrated that cellular NAD+ levels are increased in wild-type G6PD-overexpressed (pTRIPZ-G6PD) HCT116 cells but reduced by FK866, a NAMPT inhibitor, supporting the idea that heightened G6PD activity may amplify PARP1 activation by promoting NAD+ availability through increased flux in PPP and the NAD+ salvage pathway. The end product of the PPP, the nucleotide precursor ribose-5-phosphate (R5P), contributes to NAD+ synthesis via phosphoribosylpyrophosphate (PRPP) and NAMPT in the salvage pathway (141). Consistently, PRPP levels are elevated in G6PD-overexpressed (pTRIPZ-G6PD) HCT116 cells (Supplementary Figure S6G). Moreover, inhibition of NAMPT rescued fork degradation, as illustrated in Figure 3K. In contrast, a G6PD catalytic dead mutant, G6PD(K171Q) (134), failed to elevate cellular NAD+ (Figure 6N). As anticipated, G6PD(K171Q) also exhibited impaired induction of hyperPARylation: cellular PARylation levels in cells overexpressing G6PD(K171Q) were significantly lower than those in cells overexpressing wild-type G6PD (Figure 6O).
Accordingly, replication stress can induce lagging chromatin, anaphase bridges and micronuclei (142,143). Consistent with the notion that enhanced G6PD activity increases replication-induced genomic instability, we demonstrated that micronuclei formation is enriched in HCT116 (WT) cells overexpressing G6PD and challenged with gemcitabine and ATRi (Figure 6S) and suppressed when PARP1 is inhibited (Figure 6T). Together, these findings revealed G6PD-induced hyperPARylation as a primary mechanism that counteracts fork stability when p53 or RRM2B is compromised. Importantly, our results also highlight an unexpected role of G6PD in the regulation of replication fork stability in addition to its well-described function in NADPH regeneration and curbing ROS, thereby revealing a hitherto unknown crosstalk between the cellular redox regulatory network and replication fork stability that is broadly relevant.
Discussion
Replication stress is a major driver of genomic instability. Recent studies have revealed the diverse interaction among HR factors, chromatin and fork remodellers and nucleolytic enzymes to ensure the correct recovery of stalled forks. In this study, we report an unprecedented role of p53 in promoting genomic integrity by limiting extensive nascent DNA degradation at arrested replication forks, particularly upon inhibition of ATR. We further clarified an unanticipated interaction between redox homeostasis and genome integrity, facilitated by an NRF2-PARP1 axis that establishes a connection between p53 tumor suppressor function and the stability of replication forks. This highlights the susceptibility of p53-defective cancers to replication stress and underscores the implication for replication and therapeutic intervention.
p53/RRM2B Suppresses unscheduled fork degradation catalysed by MRE11/EXO1
A salient point of our model, outlined in Figure 7, is underpinned by the observation that the vulnerability of nascent DNA at deprotected stalled forks is critically dependent on hyper-PARylation induced de novo by the loss of p53 and RRM2B. Two important conclusions were drawn from our studies: first, p53 acts through its transcriptional target, RRM2B, to maintain normal endogenous PARylation. As most PARylation results from PARP1 activity, we concluded that impairment of p53, as a result of deficiency in RRM2B, causes dysregulation of PARP1 activation, resulting in hyper-PARylation. Second, excessive PARP1 activation is detrimental to fork stability. Our findings align with a model in which hyper-PARylation promotes the onset of irreversible fork degradation catalysed by MRE11/EXO1 at deprotected forks, leading to widespread replication catastrophe and cell lethality in the absence of p53 or RRM2B (Figure 7). At deprotected forks, RRM2B deficiency provokes over-resection of nascent DNA strands, whereas restoring wildtype RRM2B in p53-defective cells completely restored nascent DNA stability and suppressed γH2AX and RPA2S4/S8 phosphorylation, providing unequivocal evidence for a critical role of RRM2B in replication fork protection. These findings suggest that the function of p53 in stabilizing arrested replication fork is likely to entail a different mode of action, compared to a proposed role of p53 in binding directly to the replication fork and suppressing RAD52-dependent fork restart (144), and in preventing transcription-replication interference, as we had previously discussed (42). Our findings here are further recapitulated in patient-derived cells from freshly resected tumors, highlighting their significance in clinically relevant models (Figure 2).
Figure 7.
p53/RRM2B deficiency promotes the activation of NRF2-PARP1 axis and fork degradation. (A) The diagram illustrates a conceptual model wherein mild oxidative stress induced by the loss of p53/RRM2B triggers the activation of PARP1 through the Pentose Phosphate Pathway (PPP). Sustained PARP1 activation confers replication forks vulnerabilities that drive replication catastrophe and subsequent lethality in the absence of functional p53 or RRM2B. This distinguishes from acute and excessive oxidative stress generated by a variety of pathological conditions, driving DNA damage and PARP1 activation. (B) A proposed model providing a mechanistic insight into the activation of NRF2/G6PD in the absence of p53/RRM2B, specifically in response to mild reactive oxygen species (ROS). This activation leads to an enhanced production of NAD+ through the salvage pathway. The augmented NAD+ levels subsequently trigger PARP1 activation, which, in turn, instigates unscheduled degradation of replication forks at deprotected stalled sites induced by ATR inhibition. ATR inhibition induces unscheduled origin firing which potentially results in an exhaustion of RPA in cells and deprotection of stalled forks, rendering them susceptible to fork degradation induced by hyperPARylation.
Hyper-PARylation determines fork degradation at deprotected stalled forks
Our findings point to the importance of regulating PARP1 to ensure fork stability and highlight a point of convergence for multiple factors, including redox pathways, in fork and genomic stability. Several observations support this interpretation: (i) in addition to inhibiting PARP1 with a small molecule or targeted siRNAs, we show that fine-tuning the levels of cellular PARylation, either by toggling NAD availability through (ii) NAD supplementation, (iii) inhibiting NAMPT or (iv) by inhibiting PARG, which hydrolyses poly (ADP-ribose) (145), can determine the fate of the nascent DNA strand, that is marked by extensive fork degradation at deprotected forks and widespread replication catastrophe. Conversely, (v) suppressing ROS with ROS scavengers completely abolished fork degradation in p53/RRM2B-deficient cells. Since these phenomena are entirely PARP1-dependent, our results support a model in which PARylation is a critical biomarker of nascent DNA stability at deprotected forks.
PARP1 has well established roles in base excision and single-strand break repair processes and is reported to counteract fork resection at DNA double-strand breaks (DSBs) (117,119,146). Unexpectedly, here we show that hyper-PARylation promotes fork degradation at forks deprotected by ATR inhibition, indicating the diverse functions of PARP1 at DSBs and replication forks (119,147). Our data suggest that, unlike the situation at DSBs, hyper-PARP1 activation at deprotected forks causes extensive resection of DNA, mediated by MRE11 and EXO1. Although limited processing of nascent DNA is required for RAD51-mediated fork restart (33), however, in the absence of proper fork protection mechanisms, promiscuous nucleases such as MRE11 and EXO1 are reported to degrade stalled forks (8,14). Emerging data suggest that PARP1 stabilizes fork structures genome-wide and at telomeres (31,117). Fork remodelling promoted by PARP1 is thought to favour the stabilization of reverse forks (18,31), which are regressed 4-way junctions formed temporarily to allow fork repair and restart through alternative recombination and translesion bypass mechanisms (18,148). A possible interpretation of our results is that hyper-PARylation could promote fork remodelling events that are permissive for fork degradation. This view is reinforced by previous reports demonstrating that PARP1 stabilizes regressed forks by counteracting RECQ1-dependent fork restart activity (32) and that the extruded end of the regressed forks serves as an entry point for nucleolytic degradation of the nascent DNA, especially in the absence of fork protection factors (e.g. BRCA1/2, RAD51 and RAD51 paralogues, RAD52) (8,13,149). Although it is generally assumed that changes in the PARylation state of proteins at stalled forks, to either facilitate or destabilize reversed forks during DNA replication fork recovery, are typically short-lived and dynamic, our findings reveal that excessive PARylation or sustained activation of PARP1 in the absence of proper fork protection mechanism, can actually undermine fork protection. Our findings point towards the dynamic roles of PARP1 in the maintenance of genomic stability.
An unanticipated crosstalk between the NRF2 redox pathway and the regulatory dynamics of PARP1
Collectively, our results provide a basis for elucidating the replication vulnerability of cancers bearing p53 mutations and factors influencing fork protection including an unanticipated interaction with redox pathways. The interaction between p53 and PARP1 have been reported (150), but previous observations have mainly centered around p53 as a target for PARylation (151,152). A surprising finding from our study was that the loss of p53 or RRM2B promotes hyper-PARylation, orchestrated through an NRF2 antioxidant program, introducing a novel paradigm in the context of PARP1 modulation. We leveraged large-scale transcriptomics data from extensive clinical tumour datasets to identify and validate antioxidant pathways correlated with impaired p53 function or diminished RRM2B gene expression and identified G6PD as the dominant NRF2-regulated factor in the regulation of PARylation and fork stability. Other than its multifaceted cell-protective redox functions (129), we propose, for the first time, that NRF2 may not only challenge but also significantly compromise replication and genomic integrity. In contrast to the situation at DSBs, where NRF2 promotes DSB repair by regulating DNA repair genes, including 8-oxoguanine DNA glycosylase (OGG1) (153) and 53 binding protein 1 (53BP1) (154), our findings align with a model that oxidative stress-induced NRF2 activation counteracts fork integrity (Figure 7). Further dissection of the phenomenon revealed that NRF2-dependent activation of G6PD drives the degradation of nascent DNA at deprotected forks. Toggling G6PD activity directly (Figure 6) or indirectly by modulating ROS levels (Figure 4) convincingly demonstrated that ROS-induced activation of G6PD is tightly coupled to redox-mediated hyper-PARylation and fork degradation. This discovery further implies that the role of mild oxidative stress in perpetuating fork degradation and damage, as revealed here, entails a mechanism (Figure 7A) distinct from how prolonged acute oxidative stress results in extensive damage-induced PARylation (127,155) or directly activates MRE11 through ATM-dependent phosphorylation (156). To exclude the possibility that multiple factors promote fork instability in this scenario, we demonstrated that overexpression of the wild-type G6PD enzyme effectively promoted fork degradation while reducing overall cellular ROS levels, therefore indicating that fork degradation induced by G6PD is uncoupled from any direct ROS-induced effects. Rather, G6PD, a rate-limiting enzyme in the PPP, amplify PARP1 activation by promoting NAD+ levels through increased flux in PPP, thereby establishing a molecular mechanism that links cellular redox and fork integrity through the activation of PARP1.
We present a model illustrating how hyper-PARylation, triggered by the loss by p53/RRM2B and mild oxidative stress, confers vulnerability at replication forks (Figure 7). Beyond the potential implications for tumorigenicity linked to p53 mutations, our results identified PARP1 as an important sensor and transducer of redox stress, transducing antioxidant signals to impaired replication stress responses, thereby revealing a therapeutic vulnerability to replication inhibitors. In addition, our discovery of the ability of NRF2/G6PD to promote unscheduled fork degradation at deprotected stalled forks, achieved through the regulation of PARP1 activation, exposes a complex network of fork determinants. This calls for a more comprehensive interrogation of the myriad factors influencing oxidative stress and PARylation as the next step to unveil the multiple layers of vulnerabilities that drive replication catastrophe and lethality in cancers. The integration of these insights can provide a systematic framework for identifying new therapeutic strategies through the combination of diverse pharmacological approaches.
Supplementary Material
Acknowledgements
We thank Prof Luis Toledo and Prof Jiri Lukas (University of Copenhagen) for their invaluable advice on QIBC, Prof Ashok Venkitaraman for his critical comments and valuable insights, Prof Bert Vogelstein (John Hopkins) for the HCT116 isogenic cell lines, and all members of the CCF Lab, especially Lu Sixiao for reading and editing. We sincerely apologize to authors whose articles are not cited here due to space constraints.
Author contributions: Conceptualization: C.F.C.,. G.A., O.A.; Methodology: G.A., O.A., T.W.N., P.Y., B.K.P.G., J.Z.Z.Y., S.H.; Investigation: G.A., O.A., T.W.N., P.Y., L.Y., J.Y., C.Z.J.P., S.H.; Visualization: J.C., G.A., S.H.; Supervision: C.F.C., W.L.T., P.C.; Writing—original draft: C.F.C.; Writing—review & editing: C.F.C., G.A., J.C.
Contributor Information
Gamal Ahmed Elfar, NUS Department of Pathology, National University of Singapore, Yong Loo Lin School of Medicine, Singapore; Institute of Molecular and Cell Biology, Agency for Science, Technology and Research (A*STAR), Singapore.
Obed Aning, NUS Department of Pathology, National University of Singapore, Yong Loo Lin School of Medicine, Singapore.
Tsz Wai Ngai, NUS Department of Pathology, National University of Singapore, Yong Loo Lin School of Medicine, Singapore.
Pearlyn Yeo, NUS Department of Pathology, National University of Singapore, Yong Loo Lin School of Medicine, Singapore.
Joel Wai Kit Chan, Institute of Molecular and Cell Biology, Agency for Science, Technology and Research (A*STAR), Singapore.
Shang Hong Sim, Institute of Molecular and Cell Biology, Agency for Science, Technology and Research (A*STAR), Singapore.
Leonard Goh, NUS Department of Pathology, National University of Singapore, Yong Loo Lin School of Medicine, Singapore.
Ju Yuan, Genome Institute of Singapore, Agency for Science, Technology and Research (A*STAR), Singapore.
Cheryl Zi Jin Phua, Genome Institute of Singapore, Agency for Science, Technology and Research (A*STAR), Singapore.
Joanna Zhen Zhen Yeo, Genome Institute of Singapore, Agency for Science, Technology and Research (A*STAR), Singapore; School of Biological Sciences, Nanyang Technological University, Singapore.
Shi Ya Mak, Bioprocessing Technology Institute (BTI), Agency for Science, Technology and Research (A*STAR), Singapore.
Brian Kim Poh Goh, Department of Hepatopancreatobiliary and Transplant Surgery, Singapore General Hospital, Singapore and National Cancer Centre Singapore, Singapore.
Pierce Kah-Hoe Chow, Department of Hepatopancreatobiliary and Transplant Surgery, Singapore General Hospital, Singapore and National Cancer Centre Singapore, Singapore; Surgery Academic ClinicalProgramme, Duke-NUS Medical School, National University of Singapore, Singapore.
Wai Leong Tam, Genome Institute of Singapore, Agency for Science, Technology and Research (A*STAR), Singapore; School of Biological Sciences, Nanyang Technological University, Singapore; Cancer Science Institute of Singapore, National University of Singapore, Singapore; NUS Center for Cancer Research, Yong Loo Lin School of Medicine, National University Singapore, Singapore.
Ying Swan Ho, Bioprocessing Technology Institute (BTI), Agency for Science, Technology and Research (A*STAR), Singapore.
Chit Fang Cheok, NUS Department of Pathology, National University of Singapore, Yong Loo Lin School of Medicine, Singapore; Institute of Molecular and Cell Biology, Agency for Science, Technology and Research (A*STAR), Singapore; NUS Center for Cancer Research, Yong Loo Lin School of Medicine, National University Singapore, Singapore.
Data availability
The authors declare that all data supporting the results in this study are available within the paper and its Supplementary Material, or upon request from the authors. Microarray data have been deposited to the Gene Expression Omnibus (GEO), with the dataset identifier GSE207647. Scripts were written in R and Python (157) and were run on R Studio () and PyCharm. All scripts are made available on https://github.com/shanghongsim/HU-ATRi-Code-Repository and https://doi.org/10.5281/zenodo.13627574.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Funding for open access charge: NUS startup grant.
Conflict of interest statement. None declared.
References
- 1. Yeeles J.T., Poli J., Marians K.J., Pasero P.. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 2013; 5:a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berti M., Cortez D., Lopes M.. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 2020; 21:633–651. [DOI] [PubMed] [Google Scholar]
- 3. Cimprich K.A., Cortez D.. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008; 9:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casper A.M., Nghiem P., Arlt M.F., Glover T.W.. ATR regulates fragile site stability. Cell. 2002; 111:779–789. [DOI] [PubMed] [Google Scholar]
- 5. Cortez D., Guntuku S., Qin J., Elledge S.J.. ATR and ATRIP: partners in checkpoint signaling. Science. 2001; 294:1713–1716. [DOI] [PubMed] [Google Scholar]
- 6. Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J.. The importance of repairing stalled replication forks. Nature. 2000; 404:37–41. [DOI] [PubMed] [Google Scholar]
- 7. Berti M., Vindigni A.. Replication stress: getting back on track. Nat. Struct. Mol. Biol. 2016; 23:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P.. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 2017; 8:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vallerga M.B., Mansilla S.F., Federico M.B., Bertolin A.P., Gottifredi V.. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6624–E6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., Lopes M.. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015; 208:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhat K.P., Cortez D.. RPA and RAD51: fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018; 25:446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malacaria E., Pugliese G.M., Honda M., Marabitti V., Aiello F.A., Spies M., Franchitto A., Pichierri P.. Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation. Nat. Commun. 2019; 10:1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E.H., Hendrickson E.A.et al.. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 2015; 208:545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zadorozhny K., Sannino V., Belan O., Mlcouskova J., Spirek M., Costanzo V., Krejci L.. Fanconi-anemia-associated mutations destabilize RAD51 Filaments and impair replication fork protection. Cell Rep. 2017; 21:333–340. [DOI] [PubMed] [Google Scholar]
- 17. Marians K.J. Lesion bypass and the reactivation of stalled replication forks. Annu. Rev. Biochem. 2018; 87:217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012; 19:417–423. [DOI] [PubMed] [Google Scholar]
- 19. Marians K.J. Lesion bypass and the reactivation of stalled replication forks. Annu. Rev. Biochem. 2018; 87:217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., Haracska L.. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007; 28:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kile A.C., Chavez D.A., Bacal J., Eldirany S., Korzhnev D.M., Bezsonova I., Eichman B.F., Cimprich K.A.. HLTF’s ancient HIRAN domain binds 3' DNA ends to drive replication fork reversal. Mol. Cell. 2015; 58:1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poole L.A., Cortez D.. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 2017; 52:696–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fugger K., Mistrik M., Neelsen K.J., Yao Q., Zellweger R., Kousholt A.N., Haahr P., Chu W.K., Bartek J., Lopes M.. FBH1 catalyzes regression of stalled replication forks. Cell Rep. 2015; 10:1749–1757. [DOI] [PubMed] [Google Scholar]
- 24. Betous R., Mason A.C., Rambo R.P., Bansbach C.E., Badu-Nkansah A., Sirbu B.M., Eichman B.F., Cortez D.. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012; 26:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L.et al.. Smarcal1-Mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell. 2017; 67:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Achar Y.J., Balogh D., Haracska L.. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14073–14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vujanovic M., Krietsch J., Raso M.C., Terraneo N., Zellweger R., Schmid J.A., Taglialatela A., Huang J.W., Holland C.L., Zwicky K.et al.. Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol. Cell. 2017; 67:882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Przetocka S., Porro A., Bolck H.A., Walker C., Lezaja A., Trenner A., von Aesch C., Himmels S.-F., D’Andrea A.D., Ceccaldi R. CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol. Cell. 2018; 72:568–582. [DOI] [PubMed] [Google Scholar]
- 29. Roy R., Chun J., Powell S.N.. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012; 12:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012; 19:417–423. [DOI] [PubMed] [Google Scholar]
- 31. Margalef P., Kotsantis P., Borel V., Bellelli R., Panier S., Boulton S.J.. Stabilization of reversed replication forks by Telomerase drives Telomere catastrophe. Cell. 2018; 172:439–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R.. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 2013; 20:347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T.. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009; 28:2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haince J.-F., McDonald D., Rodrigue A., Déry U., Masson J.-Y., Hendzel M.J., Poirier G.G.. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008; 283:1197–1208. [DOI] [PubMed] [Google Scholar]
- 35. Pillay N., Tighe A., Nelson L., Littler S., Coulson-Gilmer C., Bah N., Golder A., Bakker B., Spierings D.C., James D.I.. DNA replication vulnerabilities render ovarian cancer cells sensitive to poly (ADP-ribose) glycohydrolase inhibitors. Cancer Cell. 2019; 35:519–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alano C.C., Garnier P., Ying W., Higashi Y., Kauppinen T.M., Swanson R.A.. NAD+ depletion is necessary and sufficient forPoly (ADP-ribose) polymerase-1-mediated neuronal death. J. Neurosci. 2010; 30:2967–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bieging K.T., Mello S.S., Attardi L.D.. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014; 14:359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vogelstein B., Lane D., Levine A.J.. Surfing the p53 network. Nature. 2000; 408:307–310. [DOI] [PubMed] [Google Scholar]
- 39. Bargonetti J., Friedman P.N., Kern S.E., Vogelstein B., Prives C.. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell. 1991; 65:1083–1091. [DOI] [PubMed] [Google Scholar]
- 40. Kern S.E., Kinzler K.W., Bruskin A., Jarosz D., Friedman P., Prives C., Vogelstein B.. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991; 252:1708–1711. [DOI] [PubMed] [Google Scholar]
- 41. Hassin O., Oren M.. Drugging p53 in cancer: one protein, many targets. Nat. Rev. Drug Discov. 2023; 22:127–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeo C.Q.X., Alexander I., Lin Z., Lim S., Aning O.A., Kumar R., Sangthongpitag K., Pendharkar V., Ho V.H., Cheok C.F.. p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 2016; 15:132–146. [DOI] [PubMed] [Google Scholar]
- 43. Klusmann I., Rodewald S., Müller L., Friedrich M., Wienken M., Li Y., Schulz-Heddergott R., Dobbelstein M.. p53 activity results in DNA replication fork processivity. Cell Rep. 2016; 17:1845–1857. [DOI] [PubMed] [Google Scholar]
- 44. Roy S., Tomaszowski K.H., Luzwick J.W., Park S., Li J., Murphy M., Schlacher K.. p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and POLtheta pathways. eLife. 2018; 7:e31723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reaper P.M., Griffiths M.R., Long J.M., Charrier J.-D., MacCormick S., Charlton P.A., Golec J.M., Pollard J.R.. Selective killing of ATM-or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 2011; 7:428–430. [DOI] [PubMed] [Google Scholar]
- 46. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 47. Böhly N., Schmidt A.-K., Zhang X., Slusarenko B.O., Hennecke M., Kschischo M., Bastians H.. Increased replication origin firing links replication stress to whole chromosomal instability in human cancer. Cell Rep. 2022; 41:111836. [DOI] [PubMed] [Google Scholar]
- 48. Moiseeva T., Hood B., Schamus S., O’Connor M.J., Conrads T.P., Bakkenist C.J.. ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and and-1. Nat. Commun. 2017; 8:1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petermann E., Woodcock M., Helleday T.. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:16090–16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao H., Piwnica-Worms H.. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 2001; 21:4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu Q., Guntuku S., Cui X.-S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A.. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 2000; 14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 52. Dubois J.-C., Yates M., Gaudreau-Lapierre A., Clément G., Cappadocia L., Gaudreau L., Zou L., Maréchal A.. A phosphorylation-and-ubiquitylation circuitry driving ATR activation and homologous recombination. Nucleic Acids Res. 2017; 45:8859–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu S., Opiyo S.O., Manthey K., Glanzer J.G., Ashley A.K., Amerin C., Troksa K., Shrivastav M., Nickoloff J.A., Oakley G.G.. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012; 40:10780–10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marechal A., Zou L.. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015; 25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vassin V.M., Anantha R.W., Sokolova E., Kanner S., Borowiec J.A.. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. Cell Sci. 2009; 122:4070–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Toledo L.I., Altmeyer M., Rask M.-B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J.. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013; 155:1088–1103. [DOI] [PubMed] [Google Scholar]
- 57. Nayak S., Calvo J.A., Cong K., Peng M., Berthiaume E., Jackson J., Zaino A.M., Vindigni A., Hadden M.K., Cantor S.B.. Inhibition of the translesion synthesis polymerase REV1 exploits replication gaps as a cancer vulnerability. Sci. Adv. 2020; 6:eaaz7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Panzarino N.J., Krais J.J., Cong K., Peng M., Mosqueda M., Nayak S.U., Bond S.M., Calvo J.A., Doshi M.B., Bere M.et al.. Replication gaps underlie BRCA deficiency and therapy response. Cancer Res. 2021; 81:1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Belan O., Sebald M., Adamowicz M., Anand R., Vancevska A., Neves J., Grinkevich V., Hewitt G., Segura-Bayona S., Bellelli R.et al.. POLQ seals post-replicative ssDNA gaps to maintain genome stability in BRCA-deficient cancer cells. Mol. Cell. 2022; 82:4664–4680. [DOI] [PubMed] [Google Scholar]
- 60. Mann A., Ramirez-Otero M.A., De Antoni A., Hanthi Y.W., Sannino V., Baldi G., Falbo L., Schrempf A., Bernardo S., Loizou J.et al.. POLtheta prevents MRE11-NBS1-CtIP-dependent fork breakage in the absence of BRCA2/RAD51 by filling lagging-strand gaps. Mol. Cell. 2022; 82:4218–4231. [DOI] [PubMed] [Google Scholar]
- 61. Schrempf A., Bernardo S., Arasa Verge E.A., Ramirez Otero M.A., Wilson J., Kirchhofer D., Timelthaler G., Ambros A.M., Kaya A., Wieder M.et al.. POLtheta processes ssDNA gaps and promotes replication fork progression in BRCA1-deficient cells. Cell Rep. 2022; 41:111716. [DOI] [PubMed] [Google Scholar]
- 62. Taglialatela A., Leuzzi G., Sannino V., Cuella-Martin R., Huang J.W., Wu-Baer F., Baer R., Costanzo V., Ciccia A.. REV1-Polzeta maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps. Mol. Cell. 2021; 81:4008–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garcia-Rodriguez N., Dominguez-Garcia I., Dominguez-Perez M.D.C., Huertas P.. EXO1 and DNA2-mediated ssDNA gap expansion is essential for ATR activation and to maintain viability in BRCA1-deficient cells. Nucleic Acids Res. 2024; 52:6376–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Leung W., Simoneau A., Saxena S., Jackson J., Patel P.S., Limbu M., Vindigni A., Zou L.. ATR protects ongoing and newly assembled DNA replication forks through distinct mechanisms. Cell Rep. 2023; 42:112792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Couch F.B., Bansbach C.E., Driscoll R., Luzwick J.W., Glick G.G., Betous R., Carroll C.M., Jung S.Y., Qin J., Cimprich K.A.et al.. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013; 27:1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ragland R.L., Patel S., Rivard R.S., Smith K., Peters A.A., Bielinsky A.-K., Brown E.J.. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013; 27:2259–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li S., Lavagnino Z., Lemacon D., Kong L., Ustione A., Ng X., Zhang Y., Wang Y., Zheng B., Piwnica-Worms H.. Ca2+-stimulated AMPK-dependent phosphorylation of Exo1 protects stressed replication forks from aberrant resection. Mol. Cell. 2019; 74:1123–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Friedel A.M., Pike B.L., Gasser S.M.. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 2009; 21:237–244. [DOI] [PubMed] [Google Scholar]
- 69. Brown E.J., Baltimore D.. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000; 14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 70. Murga M., Bunting S., Montaña M.F., Soria R., Mulero F., Cañamero M., Lee Y., McKinnon P.J., Nussenzweig A., Fernandez-Capetillo O.. A mouse model of the ATR-Seckel Syndrome reveals that replicative stress during embryogenesis limits mammalian lifespan. Nat. Genet. 2009; 41:891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gilad O., Nabet B.Y., Ragland R.L., Schoppy D.W., Smith K.D., Durham A.C., Brown E.J.. Combining ATR suppression with oncogenic ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010; 70:9693–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schoppy D.W., Ragland R.L., Gilad O., Shastri N., Peters A.A., Murga M., Fernandez-Capetillo O., Diehl J.A., Brown E.J.. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J. Clin. Invest. 2012; 122:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Murga M., Campaner S., Lopez-Contreras A.J., Toledo L.I., Soria R., Montaña M.F., D’artista L., Schleker T., Guerra C., Garcia E. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 2011; 18:1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Igarashi T., Mazevet M., Yasuhara T., Yano K., Mochizuki A., Nishino M., Yoshida T., Yoshida Y., Takamatsu N., Yoshimi A.. An ATR-PrimPol pathway confers tolerance to oncogenic KRAS-induced and heterochromatin-associated replication stress. Nat. Commun. 2023; 14:4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dibitetto D., Liptay M., Vivalda F., Dogan H., Gogola E., González Fernández M., Duarte A., Schmid J.A., Decollogny M., Francica P.. H2AX promotes replication fork degradation and chemosensitivity in BRCA-deficient tumours. Nat. Commun. 2024; 15:4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Moore C.E., Yalcindag S.E., Czeladko H., Ravindranathan R., Wijesekara Hanthi Y., Levy J.C., Sannino V., Schindler D., Ciccia A., Costanzo V.. RFWD3 promotes ZRANB3 recruitment to regulate the remodeling of stalled replication forks. J. Cell Biol. 2023; 222:e202106022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Okamoto Y., Abe M., Mu A., Tempaku Y., Rogers C.B., Mochizuki A.L., Katsuki Y., Kanemaki M.T., Takaori-Kondo A., Sobeck A.. SLFN11 promotes stalled fork degradation that underlies the phenotype in Fanconi anemia cells. Blood. 2021; 137:336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Batenburg N.L., Sowa D.J., Walker J.R., Andres S.N., Zhu X.-D.. CSB and SMARCAL1 compete for RPA32 at stalled forks and differentially control the fate of stalled forks in BRCA2-deficient cells. Nucleic Acids Res. 2024; 52:5067–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen J., Wu M., Yang Y., Ruan C., Luo Y., Song L., Wu T., Huang J., Yang B., Liu T.. TFIP11 promotes replication fork reversal to preserve genome stability. Nat. Commun. 2024; 15:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Daigaku Y., Davies A.A., Ulrich H.D.. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010; 465:951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Quinet A., Carvajal-Maldonado D., Lemacon D., Vindigni A.. Chapter three-DNA fiber analysis: mind the gap. Methods Enzymol. 2017; 591:55–82. [DOI] [PubMed] [Google Scholar]
- 82. Jackson D.A., Pombo A.. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998; 140:1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Qiu Z., Zou K., Zhuang L., Qin J., Li H., Li C., Zhang Z., Chen X., Cen J., Meng Z.et al.. Hepatocellular carcinoma cell lines retain the genomic and transcriptomic landscapes of primary human cancers. Sci. Rep. 2016; 6:27411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G.. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013; 31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Besse L., Rumiac T., Reynaud-Angelin A., Messaoudi C., Soler M.-N., Lambert S.A., Pennaneach V.. Protocol for automated multivariate quantitative-image-based cytometry analysis by fluorescence microscopy of asynchronous adherent cells. STAR Protocols. 2023; 4:102446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kumar R., Coronel L., Somalanka B., Raju A., Aning O.A., An O., Ho Y.S., Chen S., Mak S.Y., Hor P.Y.et al.. Mitochondrial uncoupling reveals a novel therapeutic opportunity for p53-defective cancers. Nat. Commun. 2018; 9:3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Smith C.A., Want E.J., O’Maille G., Abagyan R., Siuzdak G.. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006; 78:779–787. [DOI] [PubMed] [Google Scholar]
- 88. Ren X., Ma L., Wang N., Zhou R., Wu J., Xie X., Zhang H., Liu D., Ma X., Dang C.. Antioxidant gene signature impacts the immune infiltration and predicts the prognosis of kidney renal clear cell carcinoma. Front. Genet. 2021; 12:721252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jeay S., Gaulis S., Ferretti S., Bitter H., Ito M., Valat T., Murakami M., Ruetz S., Guthy D.A., Rynn C.. A distinct p53 target gene set predicts for response to the selective p53–HDM2 inhibitor NVP-CGM097. eLife. 2015; 4:e06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gu Z., Eils R., Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- 91. Gu Z. Complex heatmap visualization. Imeta. 2022; 1:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pico A.R., Kelder T., Van Iersel M.P., Hanspers K., Conklin B.R., Evelo C.. WikiPathways: pathway editing for the people. PLoS Biol. 2008; 6:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2023; Vienna, Austria: Available athttps://www.R-project.org/. [Google Scholar]
- 96. Stephens M. False discovery rates: a new deal. Biostatistics. 2017; 18:275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Doncheva N.T., Morris J.H., Gorodkin J., Jensen L.J.. Cytoscape StringApp: network analysis and visualization of proteomics data. J. Proteome Res. 2018; 18:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Saxena S., Zou L.. Hallmarks of DNA replication stress. Mol. Cell. 2022; 82:2298–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Saldivar J.C., Cortez D., Cimprich K.A.. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017; 18:622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cheok C.F., Verma C.S., Baselga J., Lane D.P.. Translating p53 into the clinic. Nat. Rev. Clin. Oncol. 2011; 8:25–37. [DOI] [PubMed] [Google Scholar]
- 103. Bunz F., Hwang P.M., Torrance C., Waldman T., Zhang Y., Dillehay L., Williams J., Lengauer C., Kinzler K.W., Vogelstein B.. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest. 1999; 104:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Crozier L., Foy R., Mouery B.L., Whitaker R.H., Corno A., Spanos C., Ly T., Gowen Cook J., Saurin A.T.. CDK4/6 inhibitors induce replication stress to cause long-term cell cycle withdrawal. EMBO J. 2022; 41:e108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Moiseeva T., Hood B., Schamus S., O’Connor M.J., Conrads T.P., Bakkenist C.J.. ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and and-1. Nat. Commun. 2017; 8:1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ito S., Ishii A., Kakusho N., Taniyama C., Yamazaki S., Fukatsu R., Sakaue-Sawano A., Miyawaki A., Masai H.. Mechanism of cancer cell death induced by depletion of an essential replication regulator. PLoS One. 2012; 7:e36372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Jones R., Mortusewicz O., Afzal I., Lorvellec M., Garcia P., Helleday T., Petermann E.. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013; 32:3744–3753. [DOI] [PubMed] [Google Scholar]
- 108. Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., Obuse C., Tsurimoto T., Yoshikawa H.. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998; 395:618–621. [DOI] [PubMed] [Google Scholar]
- 109. Dupré A., Boyer-Chatenet L., Sattler R.M., Modi A.P., Lee J.-H., Nicolette M.L., Kopelovich L., Jasin M., Baer R., Paull T.T.. A forward chemical genetic screen reveals an inhibitor of the Mre11–Rad50–Nbs1 complex. Nat. Chem. Biol. 2008; 4:119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Keijzers G., Bakula D., Petr M.A., Madsen N.G.K., Teklu A., Mkrtchyan G., Osborne B., Scheibye-Knudsen M.. Human exonuclease 1 (EXO1) regulatory functions in DNA replication with putative roles in cancer. Int. J. Mol. Sci. 2018; 20:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lemaçon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N.. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017; 8:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Foskolou I.P., Jorgensen C., Leszczynska K.B., Olcina M.M., Tarhonskaya H., Haisma B., D’Angiolella V., Myers W.K., Domene C., Flashman E.. Ribonucleotide reductase requires subunit switching in hypoxia to maintain DNA replication. Mol. Cell. 2017; 66:206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pontarin G., Ferraro P., Bee L., Reichard P., Bianchi V.. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:13302–13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kuo M.-L., Sy A.J., Xue L., Chi M., M.T. Y., T. C., M.-I. C., L. C., Yen Y. RRM2B suppresses activation of the oxidative stress pathway and is up-regulated by p53 during senescence. Sci. Rep. 2012; 2:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Saxena S., Somyajit K., Nagaraju G.. XRCC2 regulates replication fork progression during dNTP alterations. Cell Rep. 2018; 25:3273–3282. [DOI] [PubMed] [Google Scholar]
- 116. Reichard P. From RNA to DNA, why so many ribonucleotide reductases?. Science. 1993; 260:1773–1777. [DOI] [PubMed] [Google Scholar]
- 117. Ronson G.E., Piberger A.L., Higgs M.R., Olsen A.L., Stewart G.S., McHugh P.J., Petermann E., Lakin N.D.. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat. Commun. 2018; 9:746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sugimura K., Takebayashi S.-i., Taguchi H., Takeda S., Okumura K.. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol. 2008; 183:1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Caron M.-C., Sharma A.K., O’Sullivan J., Myler L.R., Ferreira M.T., Rodrigue A., Coulombe Y., Ethier C., Gagné J.-P., Langelier M.-F. Poly (ADP-ribose) polymerase-1 antagonizes DNA resection at double-strand breaks. Nat. Commun. 2019; 10:2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Santofimia-Castaño P., Huang C., Liu X., Xia Y., Audebert S., Camoin L., Peng L., Lomberk G., Urrutia R., Soubeyran P.. NUPR1 protects against hyperPARylation-dependent cell death. Commun. Biol. 2022; 5:732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Huang D., Kraus W.L.. The expanding universe of PARP1-mediated molecular and therapeutic mechanisms. Mol. Cell. 2022; 82:2315–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hasmann M., Schemainda I.. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosiss. Cancer Res. 2003; 63:7436–7442. [PubMed] [Google Scholar]
- 123. Evison B.J., Sleebs B.E., Watson K.G., Phillips D.R., Cutts S.M.. Mitoxantrone, more than just another topoisomerase II poison. Med. Res. Rev. 2016; 36:248–299. [DOI] [PubMed] [Google Scholar]
- 124. Fang E.F., Scheibye-Knudsen M., Brace L.E., Kassahun H., SenGupta T., Nilsen H., Mitchell J.R., Croteau D.L., Bohr V.A.. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell. 2014; 157:882–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Loor G., Kondapalli J., Schriewer J.M., Chandel N.S., Hoek T.L.V., Schumacker P.T.. Menadione triggers cell death through ROS-dependent mechanisms involving PARP activation without requiring apoptosis. Free Radic. Biol. Med. 2010; 49:1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Xue L., Zhou B., Liu X., Wang T., Shih J., Qi C., Heung Y., Yen Y.. Structurally dependent redox property of ribonucleotide reductase subunit p53R2. Cancer Res. 2006; 66:1900–1905. [DOI] [PubMed] [Google Scholar]
- 127. Hegedűs C., Virág L.. Inputs and outputs of poly (ADP-ribosyl) ation: relevance to oxidative stress. Redox. Biol. 2014; 2:978–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T.W., Wasserman W.W.. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-seq profiling and network analysis. Nucleic Acids Res. 2010; 38:5718–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. de la Vega M.R., Chapman E., Zhang D.D.. NRF2 and the hallmarks of cancer. Cancer Cell. 2018; 34:21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Singh A., Venkannagari S., Oh K., Zhang Y., Rohde J., Liu L., Nimmagadda S., Sudini K., Brimacombe K., Gajghate S.et al.. Small Molecule Inhibitor of NRF2 Selectively Intervenes Therapeutic Resistance in KEAP1-Deficient NSCLC Tumors. ACS Chem. Biol. 2016; 11:3214–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gopal S., Mikulskis A., Gold R., Fox R.J., Dawson K.T., Amaravadi L.. Evidence of activation of the Nrf2 pathway in multiple sclerosis patients treated with delayed-release dimethyl fumarate in the phase 3 DEFINE and CONFIRM studies. Multiple Sclerosis J. 2017; 23:1875–1883. [DOI] [PubMed] [Google Scholar]
- 132. Linker R.A., Lee D.-H., Ryan S., van Dam A.M., Conrad R., Bista P., Zeng W., Hronowsky X., Buko A., Chollate S.. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011; 134:678–692. [DOI] [PubMed] [Google Scholar]
- 133. Nóbrega-Pereira S., Fernandez-Marcos P.J., Brioche T., Gomez-Cabrera M.C., Salvador-Pascual A., Flores J.M., Vina J., Serrano M.. G6PD protects from oxidative damage and improves healthspan in mice. Nat. Commun. 2016; 7:10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhang Y., Xu Y., Lu W., Li J., Yu S., Brown E.J., Stanger B.Z., Rabinowitz J.D., Yang X.. G6PD-mediated increase in de novo NADP+ biosynthesis promotes antioxidant defense and tumor metastasis. Sci. Adv. 2022; 8:eabo0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Rao X., Duan X., Mao W., Li X., Li Z., Li Q., Zheng Z., Xu H., Chen M., Wang P.G.. O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nat. Commun. 2015; 6:8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Xie N., Zhang L., Gao W., Huang C., Huber P.E., Zhou X., Li C., Shen G., Zou B.. NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Targeted Ther. 2020; 5:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ghergurovich J.M., García-Cañaveras J.C., Wang J., Schmidt E., Zhang Z., TeSlaa T., Patel H., Chen L., Britt E.C., Piqueras-Nebot M.. A small molecule G6PD inhibitor reveals immune dependence on pentose phosphate pathway. Nat. Chem. Biol. 2020; 16:731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Raub A.G., Hwang S., Horikoshi N., Cunningham A.D., Rahighi S., Wakatsuki S., Mochly-Rosen D.. Small-molecule activators of glucose-6-phosphate dehydrogenase (G6PD) bridging the dimer interface. ChemMedChem. 2019; 14:1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hwang S., Mruk K., Rahighi S., Raub A.G., Chen C.-H., Dorn L.E., Horikoshi N., Wakatsuki S., Chen J.K., Mochly-Rosen D.. Correcting glucose-6-phosphate dehydrogenase deficiency with a small-molecule activator. Nat. Commun. 2018; 9:4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhang Y., Xu Y., Lu W., Ghergurovich J.M., Guo L., Blair I.A., Rabinowitz J.D., Yang X.. Upregulation of antioxidant capacity and nucleotide precursor availability suffices for oncogenic transformation. Cell Metab. 2021; 33:94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Navas L.E., Carnero A.. NAD+ metabolism, stemness, the immune response, and cancer. Signal Transduct. Targeted Ther. 2021; 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D.. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009; 11:753–760. [DOI] [PubMed] [Google Scholar]
- 143. Naim V., Rosselli F.. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 2009; 11:761–768. [DOI] [PubMed] [Google Scholar]
- 144. Roy S., Tomaszowski K.-H., Luzwick J.W., Park S., Li J., Murphy M., Schlacher K.. p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and polθ pathways. eLife. 2018; 7:e31723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Oka S., Kato J., Moss J.. Identification and characterization of a mammalian 39-kDa poly (ADP-ribose) glycohydrolase. J. Biol. Chem. 2006; 281:705–713. [DOI] [PubMed] [Google Scholar]
- 146. Sefer A., Kallis E., Eilert T., Röcker C., Kolesnikova O., Neuhaus D., Eustermann S., Michaelis J.. Structural dynamics of DNA strand break sensing by PARP-1 at a single-molecule level. Nat. Commun. 2022; 13:6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Beck C., Robert I., Reina-San-Martin B., Schreiber V., Dantzer F.. Poly (ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp. Cell. Res. 2014; 329:18–25. [DOI] [PubMed] [Google Scholar]
- 148. Liao H., Ji F., Helleday T., Ying S.. Mechanisms for stalled replication fork stabilization: new targets for synthetic lethality strategies in cancer treatments. EMBO Rep. 2018; 19:e46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Berti M., Teloni F., Mijic S., Ursich S., Fuchs J., Palumbieri M.D., Krietsch J., Schmid J.A., Garcin E.B., Gon S.. Sequential role of RAD51 paralog complexes in replication fork remodeling and restart. Nat. Commun. 2020; 11:3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Wiman K. p53 talks to PARP: the increasing complexity of p53-induced cell death. Cell Death Differ. 2013; 20:1438–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Fischbach A., Krüger A., Hampp S., Assmann G., Rank L., Hufnagel M., Stöckl M.T., Fischer J.M., Veith S., Rossatti P.. The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly (ADP-ribosyl) ation of p53 by PARP1. Nucleic Acids Res. 2018; 46:804–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Süsse S., Scholz C.J.r., Bürkle A., Wiesmüller L.. Poly (ADP-ribose) polymerase (PARP-1) and p53 independently function in regulating double-strand break repair in primate cells. Nucleic Acids Res. 2004; 32:669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Shang Q., Pan C., Zhang X., Yang T., Hu T., Zheng L., Cao S., Feng C., Hu X., Chai X.. Nuclear factor Nrf2 promotes glycosidase OGG1 expression by activating the AKT pathway to enhance leukemia cell resistance to cytarabine. J. Biol. Chem. 2023; 299:102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Kim S.B., Pandita R.K., Eskiocak U., Ly P., Kaisani A., Kumar R., Cornelius C., Wright W.E., Pandita T.K., Shay J.W.. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2949–E2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Luo X., Kraus W.L.. On PAR with PARP: cellular stress signaling through poly (ADP-ribose) and PARP-1. Genes Dev. 2012; 26:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Somyajit K., Spies J., Coscia F., Kirik U., Rask M.-B., Lee J.-H., Neelsen K.J., Mund A., Jensen L.J., Paull T.T.. Homology-directed repair protects the replicating genome from metabolic assaults. Dev. Cell. 2021; 56:461–477. [DOI] [PubMed] [Google Scholar]
- 157. Van Rossum G., Drake F.. Python 3 Reference Manual Createspace. 2009; CA: Scotts Valley. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the results in this study are available within the paper and its Supplementary Material, or upon request from the authors. Microarray data have been deposited to the Gene Expression Omnibus (GEO), with the dataset identifier GSE207647. Scripts were written in R and Python (157) and were run on R Studio () and PyCharm. All scripts are made available on https://github.com/shanghongsim/HU-ATRi-Code-Repository and https://doi.org/10.5281/zenodo.13627574.