Abstract
RAD54L is a DNA motor protein with multiple roles in homologous recombination DNA repair. In vitro, RAD54L was shown to also catalyze the reversal and restoration of model replication forks. In cells, however, little is known about how RAD54L may regulate the dynamics of DNA replication. Here, we show that RAD54L restrains the progression of replication forks and functions as a fork remodeler in human cancer cell lines and non-transformed cells. Analogous to HLTF, SMARCAL1 and FBH1, and consistent with a role in fork reversal, RAD54L decelerates fork progression in response to replication stress and suppresses the formation of replication-associated ssDNA gaps. Interestingly, loss of RAD54L prevents nascent strand DNA degradation in both BRCA1/2- and 53BP1-deficient cells, suggesting that RAD54L functions in both pathways of RAD51-mediated replication fork reversal. In the HLTF/SMARCAL1 pathway, RAD54L is critical, but its ability to catalyze branch migration is dispensable, indicative of its function downstream of HLTF/SMARCAL1. Conversely, in the FBH1 pathway, branch migration activity of RAD54L is essential, and FBH1 engagement is dependent on its concerted action with RAD54L. Collectively, our results reveal disparate requirements for RAD54L in two distinct RAD51-mediated fork reversal pathways, positing its potential as a future therapeutic target.
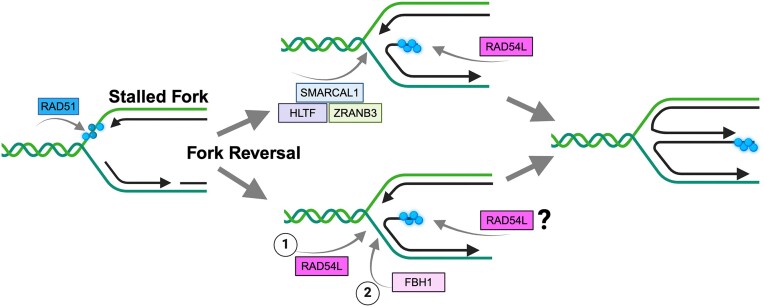
Graphical Abstract
Graphical Abstract.
Introduction
Faithful and complete DNA replication is critical for genome stability (1). Yet, faithful and complete DNA replication is challenged by genotoxic insult from endogenous and exogenous sources, leading to obstacles in replication fork progression and replication stress. To ameliorate replication stress, cells have acquired several mechanisms that promote the stabilization of stressed replication forks. One such mechanism is through fork reversal (2,3), a process that involves annealing of the nascent and parental DNA strands (4,5). Fork reversal creates a 4-way DNA structure and a double-stranded DNA end and is associated with a slowdown in fork progression, providing time for lesion bypass and repair (3,6). Efficient fork reversal requires the RAD51 recombinase (2,4,7–9), the key protein in homologous recombination DNA repair (HR), and several ATP-dependent DNA motor proteins of the SWI2/SNF2 family including SMARCAL1, ZRANB3, HLTF and the F-box DNA Helicase 1 (FBH1) (10–14). Stringent regulation of fork reversal is critical for genome stability and depends on a growing list of fork reversal and protection factors (2,14). If these factors are dysregulated, fork reversal can lead to unscheduled fork degradation and genome instability (3,14,15). Notably, several key proteins in HR, such as BRCA1 and BRCA2, have critical roles in protecting reversed forks from nucleolytic degradation (3,15,16).
HR is an essential DNA repair pathway that is also required for robust DNA replication (17). HR between sister chromatids ensures that ssDNA gaps are sealed correctly, and that collapsed replication forks are rescued through joint molecule formation with the undamaged sister. Joint molecule formation relies on the DNA strand exchange activity of RAD51 and its auxiliary proteins, including RAD54L (18).
RAD54L is a member of the SWI2/SNF2 family of DNA-dependent ATPases (19) and was shown to have multiple roles in HR (20). During strand invasion, RAD54L utilizes its ATPase activity to convert the synaptic complex into a displacement-loop (21,22). RAD54L also removes RAD51 from heteroduplex DNA to allow access of a DNA polymerase for repair synthesis (23,24). Independent of its ATPase activity, RAD54L functions at the pre-synaptic stage to stabilize the RAD51 filament (23,25–27). In vitro, purified RAD54L promotes the branch migration (BM) of Holliday junctions (HJs) and the reversal and restoration of model replication forks (28,29). In cells, however, the role of RAD54L in replication fork dynamics has remained enigmatic.
Previous studies in mouse embryonic stem and HeLa cells have shown that loss of RAD54L does not lead to the nucleolytic degradation of stalled replication forks (15,30,31). The results from these studies suggest that RAD54L plays no major role as a classical fork protection factor. However, in one of our studies (31) we noticed that RAD54L may function in replication fork restraint. As fork restraint is linked to fork reversal (3,32), we wondered if RAD54L may contribute to fork reversal in human cells.
Here, we show that RAD54L restrains the progression of replication forks in several human cell lines, including cancer and near-normal cells. In unperturbed cells, replication-associated ssDNA gaps are significantly more prevalent in the absence of RAD54L. Treatment of cells with a PARP inhibitor or hydroxyurea further enhances the creation of S1 nuclease-sensitive sites in RAD54L-deficient cells. We provide evidence that RAD54L functions in two distinct pathways of RAD51-mediated fork reversal. In the FBH1 pathway, RAD54L’s engagement largely depends on its ability to catalyze BM. In contrast, although RAD54L is also required in the HLTF/SMARCAL1 pathway, its ability to catalyze BM is dispensable here. Collectively, our results identify disparate requirements for RAD54L in two fork reversal pathways and provide new mechanistic insights on the cooperativity between RAD54L and FBH1 in driving fork reversal.
Materials and methods
Cell lines, transfections, siRNAs and western blots
HeLa and MCF7 cells were obtained from ATCC and maintained as recommended. Hs578T cells were a gift from Dr Joe Gray (OHSU) and maintained as described (33). hTERT RPE-1 cells were provided by Dr Tingting Yao (CSU) and maintained as described (34). HeLa and Hs578T cells that are knockout (KO) for RAD51AP1 or RAD54L, and HeLa RAD54L KO cells ectopically expressing RAD54L were maintained as described previously (27,31,35). DLD1 (BRCA2 KO) cells and DLD1 cells expressing the wild type BRCA2 protein were maintained as described (36).
The negative control siRNA (Ctrl) and siRNAs targeting RAD54L, BRCA1, or BRCA2 were described earlier (27,37–39) and obtained from IDT (Supplementary Table S2). For knockdown of HLTF, FBH1 or 53BP1, pools of three target-specific siRNAs were purchased from Santa Cruz Biotechnology or IDT (Supplementary Table S2). SiRNA forward transfections with Lipofectamine RNAiMAX (Thermo Fisher Scientific) were performed on two consecutive days. The concentration of siRNAs in transfections was 20 nM each. Cells were treated with drugs 96 h after the first transfection.
Western blot analyses were performed according to our standard protocols (40). The following primary antibodies were used: α-RAD51AP1 ((41); 1:6000), α-RAD54L (F-11; sc-374598; Santa Cruz Biotechnology; 1:500); α-RAD51 (Ab-1; EMD Millipore; 1:3000), α-PARP1 (ab6079; Abcam; 1:1000), α-α-Tubulin (DM1A; Santa Cruz Biotechnology; 1:1000), α-Histone H3 (ab1791; Abcam; 1:10 000), α-HLTF (E9H5I; 45965; Cell Signaling; 1:6000) α-FBH1 (sc-81563; 1:500), α-BRCA2 (OP95; EMD Millipore; 1:500), α-BRCA1 (MS110; ab16780; Abcam; 1:50), α-PCNA (sc-25280; Santa Cruz Biotechnology; 1:1000), α-NUCKS1 ((42); 1:10000), α-MSH2 (ab52266; Abcam; 1:5000), α-53BP1 (A300-272A; Bethyl Laboratories; 1:10 000). HRP-conjugated goat anti-rabbit or goat anti-mouse IgG (Jackson ImmunoResearch; 1:10 000) were used as secondaries. Western blot signals were acquired using a Chemidoc XRS + gel imaging system and ImageLab software version 5.2.1 (BioRad).
Generation of the MCF7 RAD51AP1 and RAD54L KO cells
To generate MCF7 RAD51AP1 KO cells, a cocktail of three different CRISPR/Cas9 knockout plasmids (Santa Cruz Biotechnology (sc-408187)) each encoding Cas9 nuclease and one of three different RAD51AP1-specific gRNAs targeting exons 2, 3 or 5/6 (Supplementary Table S3) was used to transfect parental MCF7 cells. To generate MCF7 RAD54L KO cells, a combination of two RAD54L CRISPR/Cas9-nic KO plasmids each containing one of two different sgRNAs (i.e. sgRNA (54L)-A and sgRNA (54L)-B; Santa Cruz Biotechnology (sc-401750-NIC); Supplementary Table S3) was used. Disruption of RAD51AP1 or RAD54L was validated by sequence analysis after genomic DNA was isolated from a selection of edited and non-edited clonal isolates using DNeasy Blood & Tissue Kit (Qiagen). RAD51AP1 and RAD54L genomic DNA sequences were amplified by PCR primers flanking the sgRNA target sites (Supplementary Table S4). PCR products were gel purified, cloned into pCR4-TOPO (Invitrogen) and transformed into TOP10 competent Escherichia coli. Plasmid DNA was prepared using ZR Plasmid Miniprep-Classic Kit (Zymo Research) and submitted for sequencing. For each cell line, 15–20 individually cloned amplicons were analyzed by Sanger sequencing.
iPOND assay
The iPOND assay was performed as described (43). Briefly, 3 × 106 HeLa cells were plated in six 145 mm plates per condition 72 h prior to treatment. Cells were pulse-labeled in 10 μM EdU (Thermo Fisher Scientific) for 20 min and washed twice in PBS. Cells were either fixed immediately or incubated for 2 h in medium containing 3 mM HU. Cells were fixed in 10 ml 1% paraformaldehyde for 20 min prior to quenching the reaction by addition of 1 ml 1.25 M glycine. The cells were washed three times in PBS and harvested using a cell scraper. Cells were spun at 900 × g and 4°C for 5 min, washed three times in ice-cold PBS twice, and then flash frozen. To begin protein extraction, 1 × 106 cells were permeabilized in 1 ml 0.25% Triton X-100/PBS at RT for 30 min. Cell were washed once in 0.5% BSA/PBS and once in PBS before performing the Click-iT reaction with Biotin-Azide (Thermo Fisher Scientific) at RT for 2 h. Then, the cells were pelleted, washed once in 0.5% BSA/PBS and once in PBS, and lysed in lysis buffer (50 mM Tris–HCl [pH 8.0], 1% SDS, and protease/phosphatase inhibitors (Thermo Fisher Scientific)). Cells were sonicated at high setting with 30 s on/off cycles for 45 min using a Bioruptor UCD-200 (Diagenode). Lysates were cleared by centrifugation at 16 100 × g. Cleared lysates were diluted with PBS containing protease and phosphatase inhibitors to reduce the final SDS concentration to 0.5% and then incubated with 50 μl/1 × 107 cells equilibrated streptavidin agarose beads (Novex) at 4°C for 16 h. Beads were washed once in chilled lysis buffer, once with 1M NaCl, and twice in lysis buffer before bound proteins were eluted in 2 × LDS (1:1 v/v of packed beads) and 95°C for 45 min. Captured proteins were fractionated on NuPAGE gels (Thermo Fisher Scientific) for immunoblot analyses.
Site-directed mutagenesis and lentiviral transduction
Mutations in RAD54L were generated in pENTR1A-RAD54L-HA (31) using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) (Supplementary Table S5). pENTR1A constructs were transferred into pLentiCMV/TO DEST#2 (44) using Gateway LR Clonase II (Thermo Fisher Scientific) for the production of lentiviral particles in HEK293FT cells (Thermo Fisher Scientific), as described (44). Lentivirus was used to transduce HeLa RAD54L KO cells in 6 μg/ml polybrene, as described (31,44).
DNA fiber assay
The DNA fiber assay was performed as described (31). In the modified fiber assay with S1 nuclease, cells were labeled for 25 min each with 25 μM 5-chloro-2′-deoxyuridine (CldU) and 250 μM 5-Iodo-2′-deoxyuridine (IdU) followed by 10 min in 200 μM thymidine, as described (45). Alternatively, cells were incubated in 25 μM CldU for 25 min and in 250 μM IdU for 30 or 50 min and with or without the thymidine chase, as indicated. Cells were harvested in cold PBS, mixed with unlabeled cells (1:1), and centrifuged at 1500 × g for 5 min at 4°C. Cells were resuspended in hypotonic buffer (10 mM HEPES [pH 7.5], 150 mM NaCl, 0.3 M sucrose and 0.5% Triton-X-100) and incubated on ice for 15 min before centrifugation as above. Cells were resuspended in S1-nuclease buffer (30 mM sodium acetate [pH 4.6], 10 mM zinc acetate, 5% glycerol and 50 mM NaCl) with or without 10U/ml S1 nuclease enzyme (Thermo Fisher Scientific) and incubated for 10 min at 37°C. Cells were centrifuged as above and resuspended in 50 μl lysis buffer (0.5% SDS, 200 mM Tris pH 7.5, 50 mM EDTA). The fibers were spread, imaged, and measured as described (10). Two slides per sample were prepared for each experimental repeat, and each pair of slides was blinded after immunodetection to avoid bias.
Fiber tract lengths in HeLa cell lines and hTERT RPE-1 cells were measured to evaluate the consequences of mild replication stress, as described (32). To do so, cells were pulse labeled with CldU for 20 min followed by IdU containing 25 μM hydroxyurea (Sigma) or 25 nM camptothecin (Sigma) for 20 min (HeLa cells), or 30 min (Hela and hTERT RPE-1 cells). Only IdU tracts following a CldU tract were analyzed.
Fiber tracts were converted to kb, as described (46), using a conversion factor of 2.59 kb/μm (47). Fork speeds (kb/min) were obtained by dividing the lengths of the tracts (in kb) by the labeling time (in min).
Comet assay
The Comet assay (Trevigen) was performed as recommended by the manufacturer. Cells seeded in 60-mm plates were incubated for 48 hours before 5-hour treatment with 4 mM HU. Cells were harvested in cold PBS and combined with molten low melting (LM) agarose at 1:10. Fifty μl of this mixture were spread on comet slides and incubated for 30 min at 4°C in the dark. The slides were then incubated overnight at 4°C in lysis buffer. Following a 30 min incubation in neutral comet electrophoresis buffer (10 mM Tris base, 250 mM sodium acetate) at 4°C, electrophoresis was performed at 25V and 4°C for 30 min in neutral electrophoresis buffer. The slides were then incubated in DNA precipitation solution (7.5 M NH4Ac and 95% ethanol) for 30 min followed by 30 min incubation in 70% ethanol. Slides were dried at 37°C for 1 hour and stained with 0.3 × SYBR Gold (Thermo Fisher Scientific). Images were acquired on a Zeiss Axio-Imager.Z2 microscope equipped with Zen Blue software (Carl Zeiss Microscopy) using a 20× objective, and 100 comets were measured per condition. The lengths of the comet tails were measured using ImageJ software (https://imagej.net).
Statistics and reproducibility
GraphPad Prism 9 software was used to perform statistical analyses on data obtained from three independent experiments, unless stated otherwise. To assess statistical significance ANOVA tests were performed as indicated, and P ≤ 0.05 was considered significant.
Results
RAD54L is recruited to stalled DNA replication forks
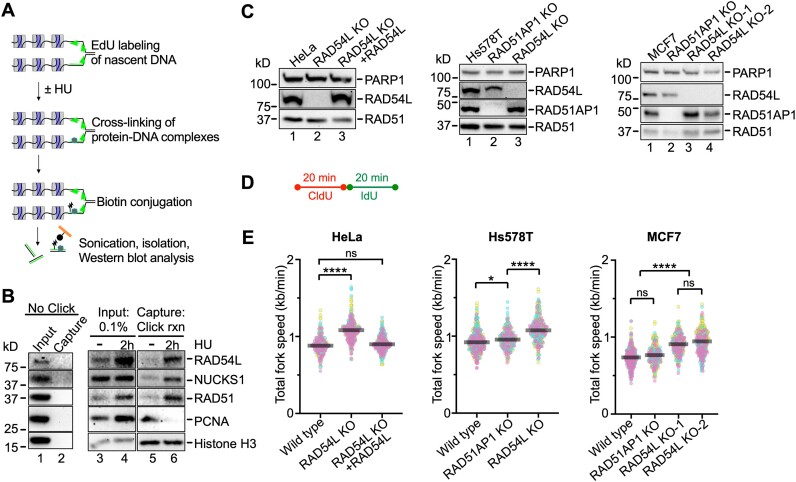
To begin exploring the involvement of RAD54L in replication fork progression in human cells, we applied the isolation of proteins on nascent DNA (iPOND) assay (43). We show that RAD54L is enriched at replication forks upon a 2 h-treatment of cells with 3 mM hydroxyurea (HU) (Figure 1A, B), in accord with results from others (30). Enrichment of RAD54L mirrored that of its binding partners RAD51 and NUCKS1. In contrast, enrichment of PCNA was downregulated after HU, as expected. These results demonstrate that RAD54L is recruited to stalled replication forks.
Figure 1.
RAD54L is enriched at stalled replication forks and restrains replication fork progression. (A) Schematic illustration of the iPOND assay. (B) Western blots of the input and iPOND samples probed for proteins, as indicated. NUCKS1 is a RAD54L interacting protein (27). Click rxn, Click reaction; the ‘no Click’ condition represents cells pulsed with EdU and processed without biotin-azide in the Click reaction step. (C) Western blots of HeLa, Hs578T and MCF7 cells and derivatives, as indicated. Loading control: PARP1. (D) Schematic of the protocol for the DNA fiber assay to determine replication speed. (E) SuperPlots (77) with medians of total fork speeds (i.e. CldU + IdU tracts) in unperturbed parental and RAD54L KO cells generated in HeLa, Hs578T, and MCF7 cells. RAD51AP1 KOs and the RAD54L KO ectopically expressing RAD54L-HA (31) are shown for comparison purposes (n = 3; 89–106 fiber tracts/experiment analyzed). Fork speeds were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗P< 0.05; ∗∗∗∗P< 0.0001).
RAD54L restrains replication fork progression in unperturbed cells
Previously, we generated HeLa RAD54L KO cells and RAD54L KO cells ectopically expressing HA-tagged RAD54L, which fully rescues the sensitivity of RAD54L KO cells to mitomycin C (MMC) and Olaparib (31). Using the single molecule DNA fiber assay and 20 min of consecutive pulse labeling each with two thymidine analogs, we noticed that RAD54L KO cells had longer replication tracts than the controls (31). These results were independently replicated here and plotted after conversion of the fiber tract lengths to replication speeds (Figure 1C–E; Supplementary Figure S1A). We also previously generated RAD54L KOs in Hs578T cells (31) and in this study in MCF7 cells (Figure 1C; Supplementary Table S6). We show that both Hs578T RAD54L KO and MCF7 RAD54L KO cells replicate faster than parental cells (Figure 1E; Supplementary Figure S1B, C). Moreover, as in the HeLa cell derivatives, fork speeds in the RAD54L KOs were significantly faster than those in Hs578T or MCF7 cells deficient in RAD51AP1 (Figure 1E; Supplementary Figure S1B, C). RAD51AP1, like RAD54L, is a RAD51-associated HR protein that enhances the activity of the RAD51 recombinase (48,49). These results show that loss of RAD54L accelerates DNA replication and that this effect is not cell-type specific and not shared with other RAD51-associated proteins.
RAD54L is dispensable for the recovery of cells from stalled DNA replication
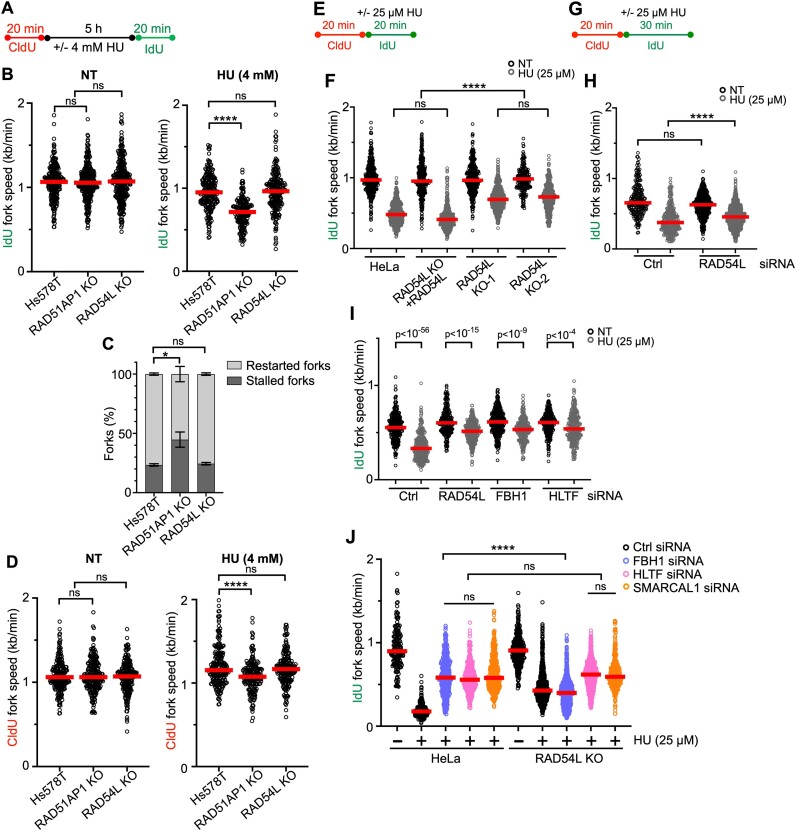
To understand the fate of stalled replication forks, we treated the Hs578T cells and derivatives with 4 mM HU for 5 h, which blocks DNA synthesis and stalls replication fork movement (14). Using the DNA fiber assay, we then monitored the recovery of cells from stalled replication (Figure 2A). We determined the ability of Hs578T cells and derivatives to restart DNA replication by measuring speeds of IdU tracts preceded by a CldU tract. The results show that RAD54L-deficient Hs578T cells recover as fast as parental cells from stalled DNA replication (Figure 2B, right panel; Supplementary Figure S2A). In contrast, RAD51AP1-deficient Hs578T cells replicate significantly slower after HU (Figure 2B, right panel; Supplementary Figure S2A), and, accordingly, contain significantly more stalled and fewer restarted forks than parental cells or the RAD54L KO (P< 0.05; Figure 2C). These results demonstrate that RAD54L is dispensable for the recovery from stalled DNA replication in human cells.
Figure 2.
Loss of RAD54L does not hinder replication restart, leads to faster replication during mild replication stress, and partially restores fork restraint in cells with FBH1 knockdown. (A) Schematic of the DNA fiber assay protocol used to assess replication restart. (B) Dot plot with medians of IdU fork speeds in untreated (NT) or HU-treated parental Hs578T, RAD51AP1 KO, and RAD54L KO cells (n = 3; 65–113 fiber tracts/experiment analyzed). (C) Fractions of stalled and restarted forks after 4 mM HU in the Hs578T cells and derivatives shown in (B). (D) Dot plot with medians of CldU fork speeds in untreated (NT) or HU-treated Hs578T cells and derivatives (n = 3; 65–80 fiber tracts/experiment analyzed). (E) Schematic of the DNA fiber assay protocol used to evaluate the progression of replication forks during mild replication stress in (F). (F) Dot plot with medians of IdU fork speeds in parental HeLa cells, RAD54L KO cells expressing RAD54L KO, and two independently isolated RAD54L KO cell lines treated with or without 25 μM HU during the IdU pulse (n = 3; 82–220 fiber tracts/experiment analyzed). (G) Schematic of the DNA fiber assay protocol used to evaluate the progression of replication forks during mild replication stress in (H), (I) and (J). (H) Dot plot with medians of IdU fork speeds in hTERT RPE-1 cells transfected with Ctrl or RAD54L siRNA and treated with or without 25 μM HU during the IdU pulse (n = 3; 76–293 fiber tracts/experiment analyzed). (I) Dot plot with medians of IdU fork speeds in HeLa cells transfected with control (Ctrl), RAD54L, FBH1 or HLTF siRNA and treated with or without 25 μM HU during the IdU pulse (n = 3; 92–139 fiber tracts/experiment analyzed). (J) Dot plot with medians of IdU fork speeds in HeLa and RAD54L KO cells transfected with Ctrl, FBH1, HLTF or SMARCAL1 siRNA (n = 3; 122–420 fiber tracts/experiment analyzed). All data were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗P< 0.05; ∗∗∗∗P< 0.0001; or as indicated).
RAD54L slows replication fork progression upon nucleotide depletion
In response to replication stress, replication forks reverse into four-way junctions through annealing of the nascent DNA strands (2,9). Reversed forks must be protected from nucleolytic attack to prevent fork attrition (10,13,15,50,51). To assess if RAD54L in Hs578T cells functions in the protection of replication forks from unprogrammed nuclease degradation, CldU fork speeds in cells exposed to HU were measured. The results show that CldU fork speeds in Hs578T RAD54L KO cells are not significantly different from CldU fork speeds in parental cells (Figure 2D, right panel; Supplementary Figure S2A). Hence, RAD54L appears to play no major role in protecting reversed forks from nuclease attrition in cells with otherwise unaltered fork protection pathways. These results are in accord with earlier studies by us and others using different cell types (15,30,31).
Previous studies have shown that a defect in replication fork restraint is linked to compromised fork reversal (2,32,52,53). It is also known that RAD54L has branch migration (BM) activity and can reverse model replication forks in vitro (28,29,54). Prompted by our findings that identified a defect in replication fork restraint in RAD54L-deficient cells, we used an established protocol of the DNA fiber assay to monitor replication fork progression (46) under a low concentration of HU (25 μM) given within the IdU pulse (Figure 2E). In these experiments, we measured IdU fork speeds in IdU tracts with a preceding CldU tract. The results show that IdU fork speeds in two independently isolated RAD54L KO lines (31) are significantly faster than IdU fork speeds in parental cells or RAD54L KO cells expressing RAD54L (P< 0.0001; Figure 2F; Supplementary Figure S2B). These results suggest that in response to HU, which induces fork reversal (50), accelerated fork progression is a consequence of RAD54L deficiency. These results also suggest that RAD54L may catalyze the reversal of replication forks in human cells.
Next, we tested if loss of RAD54L expression would lead to a defect in fork restraint in non-cancerous hTERT RPE-1 cells. To do so, we depleted RAD54L in hTERT RPE-1 cells (Supplementary Figure S2C) and measured fork speeds with the fiber assay using an extended (i.e. 30 min) IdU pulse (Figure 2G). In the presence of HU, IdU forks proceeded significantly faster in hTERT RPE-1 cells depleted for RAD54L than in hTERT RPE-1 cells transfected with a negative control (Ctrl) siRNA (P< 0.0001; Figure 2H; Supplementary Figure S2D).
Next, we compared the consequences of RAD54L loss to loss of HLTF or FBH1, two established fork remodelers that each function in a different RAD51-mediated fork reversal pathway (14,32,53,55,56). To do so, we depleted RAD54L, HLTF, or FBH1 in HeLa cells (Supplementary Figure S2E) and measured IdU fork speeds in the presence of 25 μM HU (Figure 2G). In the presence of HU, loss of HLTF or FBH1 led to significantly faster DNA replication (Figure 2I; Supplementary Figure S2F), in accord with prior studies in different cell types (14,32). Moreover, replication also proceeded faster in HU-treated RAD54L-depleted cells than in HU-treated HeLa cells transfected with Ctrl siRNA (Figure 2I; Supplementary Figure S2F). Collectively, our results show that RAD54L’s ability to restrain fork progression is not confined to cancer cell lines, occurs in both unperturbed cells and upon exposure of cells to mild replication stress, and simulates that of established fork remodelers (Figure 1E; Figure 2F–I).
FBH1 activity in replication fork reversal is dependent on RAD54L
To better understand how RAD54L is engaged in each of the two RAD51-mediated fork reversal pathways, we depleted FBH1, HLTF, or SMARCAL1 in both HeLa and RAD54L KO cells (Supplementary Figure S2G) and monitored IdU fork speeds in unperturbed cells and under mild HU-induced replication stress (Figure 2G). In unperturbed cells, IdU fork speeds in HeLa and RAD54L KO cells transfected with Ctrl, FBH1, or HLTF siRNA were not significantly different from each other (P> 0.999; Supplementary Figure S2H). SMARCAL1 knockdown, however, significantly accelerated IdU fork progression in unperturbed RAD54L KO cells compared to wild-type cells (P< 0.0001; Supplementary Figure S2H). Indicative of proficient fork reversal, IdU fork progression was significantly slower in HU-treated HeLa cells than in unperturbed HeLa cells (P< 0.0001; Figure 2J). Loss of either HLTF or SMARCAL1 in RAD54L KO cells had no effect on IdU fork speeds when compared to the equivalent knockdown condition in HeLa cells (Figure 2J; Supplementary Figure S2K). These results suggest that both HLTF and SMARCAL1 may function independently or upstream of RAD54L. In contrast, IdU fork speeds in RAD54L KO cells with FBH1 knockdown were significantly slower than those in HeLa cells with FBH1 knockdown (P< 0.0001; Figure 2J; Supplementary Figure S2K), suggesting that the activity of FBH1 in fork reversal may depend on RAD54L.
Accelerated replication fork progression in RAD54L-deficient cells is associated with ssDNA gap formation
To test if accelerated DNA replication under conditions of mild replication stress is associated with the presence of ssDNA formed at replication gaps, we used the DNA fiber assay followed by S1 nuclease, which cleaves replication intermediates that contain ssDNA (46). Here, we used an extended 50 min HU-containing IdU pulse followed by S1 nuclease (Supplementary Figure S2I). Indeed, RAD54L KO cells depleted for HLTF showed fastest IdU fork speed along with the highest sensitivity to S1 nuclease digest (Supplementary Figure S2J). In contrast, IdU forks in RAD54L KO cells transfected with Ctrl or FBH1 siRNA did not proceed as quickly and, consequently, were also less sensitive to digest with S1 nuclease (Supplementary Figure S2J). These results further point to a concerted action between FBH1 and RAD54L in fork reversal.
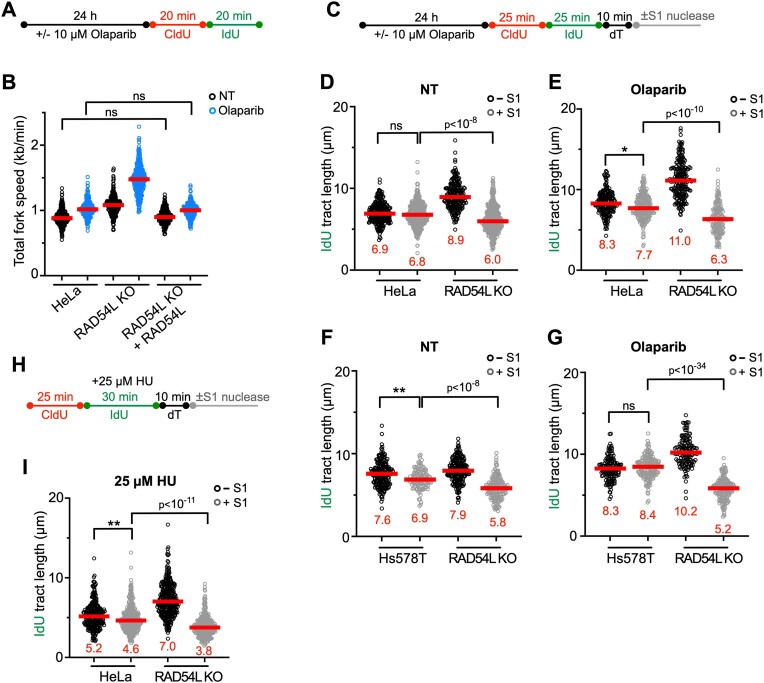
Treatment of cells with a PARP1 inhibitor (PARPi) also leads to accelerated replication fork progression, impediments in nascent DNA strand maturation, and fork collapse (57–60). RAD54L-deficient cells are sensitive to the PARPi Olaparib (31,34), and treatment of RAD54L KO cells with Olaparib further exacerbates fork progression (Figure 3A, B; Supplementary Figure S3A). Expression of ectopic RAD54L in RAD54L KO cells reverses accelerated fork progression (Figure 3B; Supplementary Figure S3A).
Figure 3.
Loss of RAD54L accelerates replication fork progression and ssDNA gap formation. (A) Schematic of the DNA fiber assay protocol to assess fork progression in (B). (B) Dot plot with medians of total fork speeds (i.e. in CldU + IdU tracts) in HeLa, RAD54L KO and RAD54L KO + RAD54L cells with or without Olaparib (n = 3; 86–100 fiber tracts/experiment analyzed). (C) Schematic of the DNA fiber assay protocol with S1 nuclease in unperturbed and Olaparib-treated cells used in (D)–(G). (D) Dot plot with medians of IdU fiber tract lengths in untreated (NT) HeLa and RAD54L KO cells with or without S1 nuclease (n = 3; 74–149 fiber tracts/experiment analyzed). (E) Dot plot with medians of IdU fiber tract lengths in HeLa and RAD54L KO cells treated with Olaparib and with or without S1 nuclease (n = 3; 60–80 fiber tracts/experiment analyzed). (F) Dot plot with medians of IdU fiber tract lengths in untreated (NT) Hs578T and RAD54L KO cells with or without S1 nuclease (n = 3; 42–70 fiber tracts/experiment analyzed). (G) Dot plot with medians of IdU fiber tract lengths in Hs578T and RAD54L KO cells treated with Olaparib and with or without S1 nuclease (n = 3; 47–63 fiber tracts/experiment analyzed). (H) Schematic of the DNA fiber assay protocol with S1 nuclease in HU-treated cells used in (I). (I) Dot plot with medians of IdU fiber tract lengths in HU-treated HeLa and RAD54L KO cells with or without S1 nuclease (n = 2; 136–169 fiber tracts/experiment analyzed). All data were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗P< 0.05; ∗∗P< 0.01; or as indicated).
Next, we incubated unperturbed cells and cells treated with Olaparib with and without S1 nuclease (Figure 3C). In these experiments, we integrated a 10-min incubation of cells with thymidine (45) to allow the formation of ssDNA gaps. Under unperturbed conditions in both HeLa and Hs578T cells, S1 treatment resulted in significantly more shortened replication tracts in the absence of RAD54L (Figure 3D, F; Supplementary Figure S3A, B). Treatment of cells with Olaparib and S1 nuclease exacerbated replication tract shortening in the absence of RAD54L (Figure 3E, G; Supplementary Figure S3A, B).
Last, we subjected HeLa and RAD54L KO cells to S1 nuclease following mild replication stress after incubation of cells in 25 μM HU (Figure 3H). Indicative of reduced fork reversal, IdU tracts were longer and significantly more sensitive to S1 nuclease in RAD54L KO than in HeLa cells (P< 10−11; Figure 3I; Supplementary Figure S3C).
Collectively, these results show that replication in the absence of RAD54L not only is accelerated (under unperturbed conditions, in the presence of HU, and after Olaparib), but also is proceeding with an elevated production of ssDNA gaps.
Loss of RAD54L restores replication fork stability in both BRCA1/2- and 53BP1-deficient cells
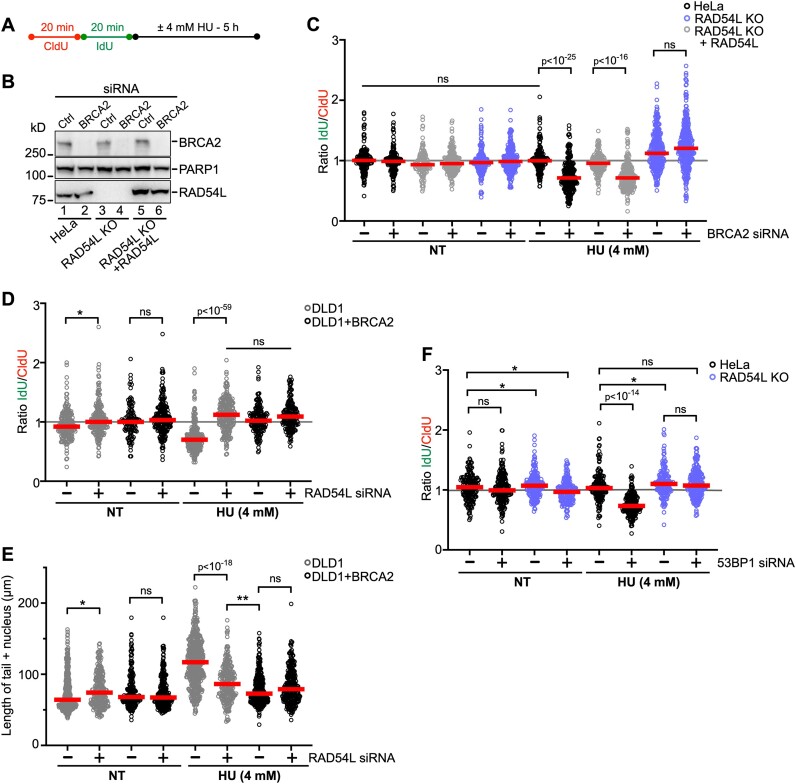
The fork reversal activities of SMARCAL1, ZRANB3, and HTLF lead to the degradation of nascent strand DNA in BRCA1/2-deficient cells upon treatment with HU (9,10). Given our results that suggest that RAD54L may function similarly to these established fork remodelers, we tested if RAD54L loss prevents nascent strand degradation in BRCA1/2-deficient cells (Figure 4A). We depleted BRCA2 in HeLa, RAD54L KO, and RAD54L-rescued cell lines (Figure 4B) and measured fork degradation by IdU/CldU ratios in response to a 5 h treatment of cells with 4 mM HU. As expected, BRCA2 knockdown in HeLa and RAD54L KO + RAD54L cells led to significantly reduced IdU/CldU tract ratios (P< 0.0001; Figure 4C; Supplementary Figure S4A). In contrast, forks in RAD54L KO cells with BRCA2 knockdown withstood fork attrition (Figure 4C; Supplementary Figure S4A). Similarly, IdU/CldU tract ratios in HeLa cells with BRCA1 knockdown were reduced, while no such effect was observed in RAD54L KO cells with BRCA1 knockdown (Supplementary Figure S4B-D). These results show that RAD54L loss in BRCA1/2-deficient HeLa cells prevents nascent strand DNA degradation in response to HU.
Figure 4.
Loss of RAD54L prevents fork degradation in both BRCA2- and 53BP1-deficient cells. (A) Schematic of the DNA fiber assay protocol. This experimental scheme was followed to assess fork degradation in (C), (D) and (F) and double-strand break formation in (E). (B) Representative Western blots to show extent of BRCA2 knockdown in HeLa cells and derivatives for the experiments shown in (C). Loading control: PARP1. (C) Dot plot with medians of IdU/CldU tract length ratios in HeLa, RAD54L KO, and RAD54L KO + RAD54L cells transfected with Ctrl (–) or BRCA2 siRNA and treated with or without HU (n = 3; 42–117 fiber tracts/experiment analyzed). (D) Dot plot with medians of IdU/CldU tract length ratios in DLD1 and DLD1 + BRCA2 cells transfected with Ctrl (–) or RAD54L siRNA and treated with or without HU (n = 3; 38–89 fiber tracts/experiment analyzed). (E) Dot plot with medians after neutral Comet assay in DLD1 and DLD1 + BRCA2 cells transfected with Ctrl (–) or RAD54L siRNA and treated with or without HU (n = 3; 74–206 Comet tails/experiment analyzed). (F) Dot plot with medians of IdU/CldU tract length ratios in HeLa and RAD54L KO cells transfected with Ctrl (–) or 53BP1 siRNA and treated with or without HU (n = 3; 60–82 fiber tracts/experiment analyzed). IdU/CldU ratios and lengths of Comet tails were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗P< 0.05; ∗∗P< 0.01; or as indicated). NT: not treated.
Next, we used DLD1 (BRCA2 KO) cells and a DLD1 cell line stably expressing the wild type BRCA2 protein (36) and depleted RAD54L in these two cell lines (Supplementary Figure S4E). IdU/CldU tract length ratios were significantly reduced in DLD1 cells after treatment with HU (Figure 4D; Supplementary Figure S4F), as shown previously (36). In contrast, knockdown of RAD54L in HU-treated DLD1 cells gave rise to IdU/CldU tract ratios with a distribution not significantly different to that from DLD1 + BRCA2 cells (Figure 4D; Supplementary Figure S4F). These results show that RAD54L loss in BRCA2-deficient DLD1 cells prevents nascent strand DNA degradation after treatment of cells with HU.
To assess the impact of RAD54L deficiency on DSB formation, we subjected DLD1 and DLD1 + BRCA2 cells (transfected with Ctrl or RAD54L siRNA) to DSB detection by neutral comet assay following a 5-h incubation of cells in 4 mM HU. Consistent with blocked fork degradation (Figure 4D), depletion of RAD54L in DLD1 + BRCA2 cells had no significant impact on DSB formation (Figure 4E; Supplementary Figure S4G). In contrast, depletion of RAD54L in DLD1 (BRCA2 KO) cells led to significantly reduced DSB formation (P< 10−18; Figure 4E; Supplementary Figure S4G). These results show that RAD54L activity contributes to the formation of DSBs in BRCA2-deficient DLD1 cells treated with HU.
Aside from the SMARCAL1/HLTF/ZRANB3 pathway, a second RAD51-dependent fork reversal pathway has been described. This pathway relies on the FBH1 DNA helicase, and the 53BP1 protein is one critical fork protection factor in this pathway (14,56). Using HeLa and RAD54L KO cells, we tested if RAD54L activity would also contribute to fork degradation in 53BP1-depleted cells (Supplementary Figure S4H). Upon treatment of 53BP1-depleted HeLa cells with HU, IdU/CldU tract length ratios were reduced (P< 0.0001; Figure 4F; Supplementary Figure S4I), indicative of fork attrition. However, IdU/CldU ratios in HU-treated 53BP1-depleted RAD54L KO cells were not different from those in RAD54L KO cells transfected with Ctrl siRNA (Figure 4F; Supplementary Figure S4I). Collectively, our results show that RAD54L activity contributes to nascent strand DNA degradation in both BRCA1/2- and 53BP1-deficient cells, suggesting that RAD54L functions in each of the two described RAD51-mediated fork reversal pathways.
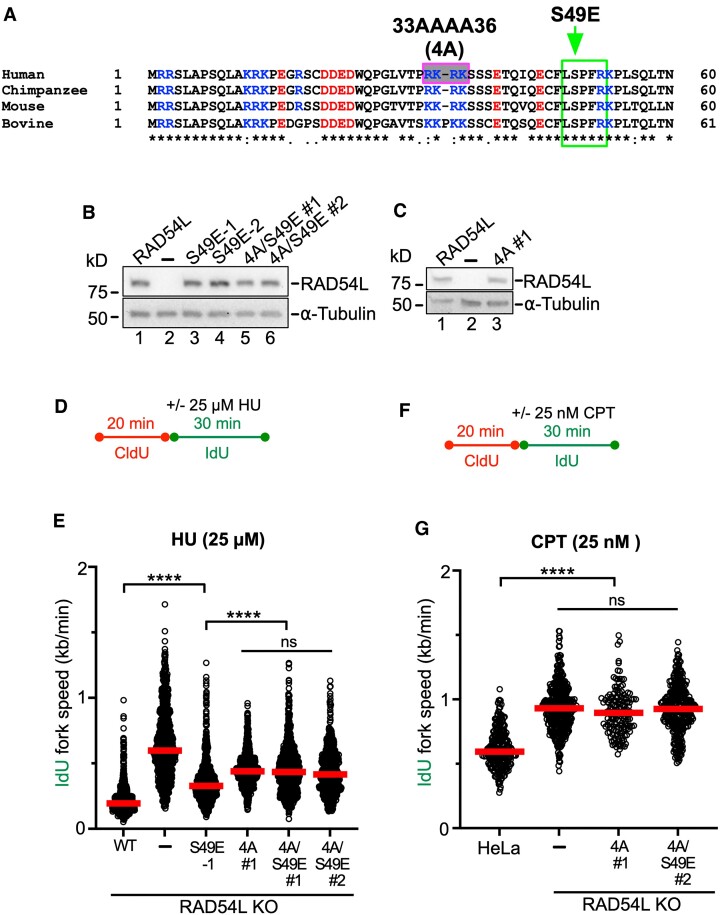
RAD54L’s branch migration activity contributes to its engagement in replication fork restraint
Fork reversal is a two-step process, in which the initiation of fork regression is followed by branch migration (BM) to drive extensive reversal (61). RAD54L catalyzes BM in vitro, and its N-terminal domain is essential for this activity (29). As N-terminal mutations in RAD54L that selectively inhibit its BM activity have been described (29), we asked if RAD54L BM mutants (Figure 5A) would show defects in fork restraint. To this end, we expressed mutant RAD54L-S49E (deficient in oligomerization (29)), RAD54L-4A (deficient in binding to HJ-like DNA structures (29)), and RAD54L-4A/S49E (deficient in both binding to HJs and oligomerization (29)) in RAD54L KO cells (Figure 5B, C). To assess defects in fork restraint, we chose the protocol as depicted in Figure 5D and compared the fork speeds of RAD54L KO cells expressing mutant RAD54L to that of RAD54L KO cells with and without wild type RAD54L. Under unperturbed conditions, RAD54L KO cells and RAD54L KO cells expressing RAD54L-S49E show a small but significant defect in fork restraint (P< 0.05; Supplementary Figure S5A, B). In the presence of low concentrations of HU, RAD54L KO cells exhibit a significant defect in fork restraint (P< 0.001; Figure 5E; Supplementary Figure S5B), as shown above (Figure 2F) and reproduced herein independently. Importantly, fork restraint also is significantly impaired in RAD54L KO cells expressing RAD54L-S49E (P< 0.0001) and further exacerbated in RAD54L KO cells expressing RAD54L-4A or the compound mutant RAD54L-4A/S49E (Figure 5E; Supplementary Figure S5B). These results show that RAD54L BM activity is required for fork restraint. As expected, ATPase-deficient RAD54L (K189R; (62)) is as defective in fork restraint as the RAD54L KO, as IdU fork speeds in RAD54L KO cells expressing RAD54L-K189R are not significantly different from those in RAD54L KO cells (P= 0.343 and P> 0.999 for K189R #1 and K189R #2, respectively; Supplementary Figure S5C–F).
Figure 5.
RAD54L’s branch migration activity contributes to its ability to restrain replication fork progression. (A) ClustalW sequence alignment of the N-terminal domains of RAD54L from human (Homo sapiens), chimpanzee (Pan troglodytes), mouse (Mus musculus) and bovine (Bos taurus). Basic residues are shown in blue, and acidic residues are shown in red. The pink/grey box indicates basic residues mutated to alanines, and the green box indicates CDK2 phosphorylation consensus sequence and mutated serine to glutamate, as previously described (29). (B) Representative western blots to show expression of wild type RAD54L (lane 1) and mutant RAD54L (lanes 3–6) in HeLa RAD54L KO cells. RAD54L-S49E expressing lines are clonal isolates; RAD54L-4A/S49E expressing cells are puromycin-resistant cell populations. Loading control: α-tubulin. (C) Representative western blots to show expression of wild type RAD54L (lane 1) and RAD54L-4A (a puromycin-resistant cell population; lane 3) in HeLa RAD54L KO cells. Loading control: α-tubulin. (D) Schematic of the DNA fiber assay protocol used in (E). (E) Dot plot with medians of IdU fork speeds in HU-treated HeLa RAD54L KO cells expressing wild type RAD54L (WT) or RAD54L BM mutants (n = 3; 238–375 fiber tracts/experiment analyzed). (F) Schematic of the DNA fiber assay protocol used in (G). (G) Dot plot with medians of IdU fork speeds in CPT-treated HeLa, RAD54L KO and RAD54L KO cells expressing RAD54L BM mutants (n = 3; 50–134 fiber tracts/experiment analyzed). Data were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗P< 0.05; ∗∗∗∗P< 0.0001).
Next, we wondered if the defect in fork restraint of the RAD54L BM mutants could also be observed after treatment of cells with another replication stress-inducing drug. To do so, we treated cells with sublethal concentrations (25 nM) of camptothecin (CPT; Figure 5F), a topoisomerase 1 inhibitor, shown to induce fork slowing and reversal. Of note, no DSB formation is expected from these CPT conditions (63). In CPT, RAD54L-4A and -4A/S49E expressing RAD54L KO cells showed accelerated IdU fork speeds (P< 0.0001) indicative of significant a defect in fork restraint, and this defect was not significantly different from that in RAD54L KO cells (P= 0.437; Figure 5G; Supplementary Figure S5G). Together, these results lead us to conclude that RAD54L BM activity is critical for fork restraint under conditions of mild replication stress.
The requirement for RAD54L branch migration activity is specific to the FBH1 pathway
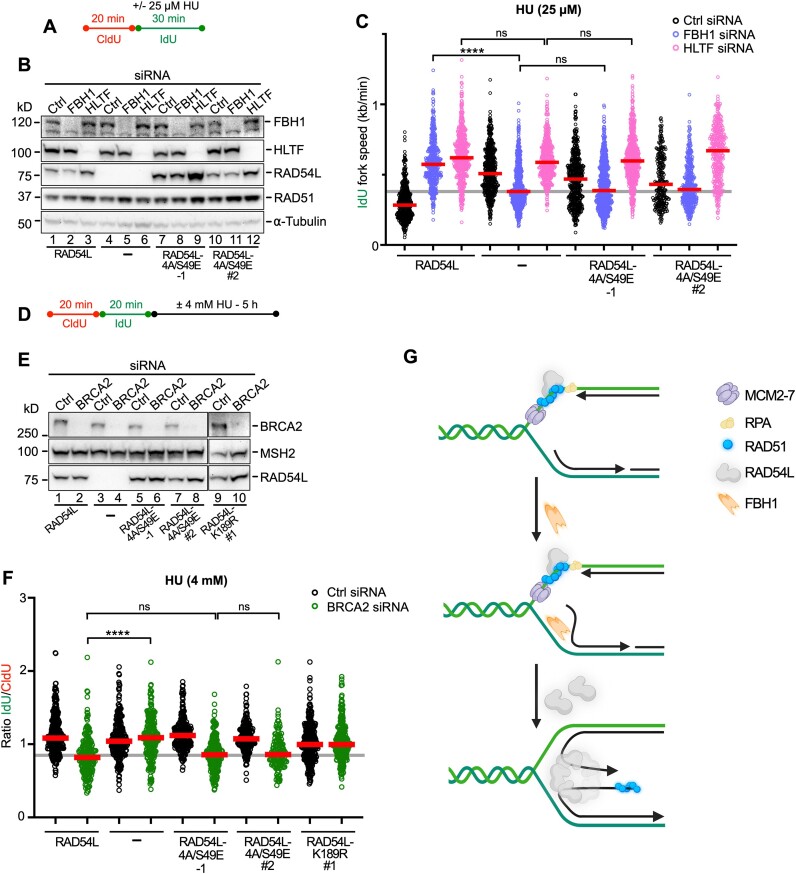
To dissect if RAD54L’s BM activity is required in both the FBH1 and the HLTF/SMARCAL1 pathways, we first analyzed replication speeds in the presence of 25 μM HU (Figure 6A) using RAD54L KO cells and RAD54L KO cells expressing wild type RAD54L or RAD54L-4A/S49E and with or without FBH1 or HLTF knockdown (Figure 6B). IdU fork speeds were similar in all cell types with HLTF knockdown (Figure 6C; Supplementary Figure S6A), suggesting that loss of fork reversal in the absence of HLTF is independent of RAD54L BM activity. In contrast, after FBH1 knockdown IdU fork speeds in RAD54L KO cells and RAD54L KO cells expressing the RAD54L-4A/S49E mutant were significantly slower than in RAD54L KO cells with wild type RAD54L (P< 0.0001; Figure 6C; Supplementary Figure S6A). This shows that the consequences of FBH1 loss in RAD54L-4A/S49E cells mirror those of FBH1 loss in RAD54L KO cells, suggesting that RAD54L BM activity is essential in the FBH1 pathway of RAD51-mediated fork reversal.
Figure 6.
RAD54L’s branch migration activity is specifically required for the FBH1 pathway of RAD51-mediated fork reversal. (A) Schematic of the DNA fiber assay protocol used in (C). (B) Representative Western blots to show extent of FBH1 and HLTF knockdown in RAD54L KO cells (lanes 4–6) and RAD54L KO cells expressing wild type RAD54L (lanes 1–3) or mutant RAD54L-4A/S49E (lanes 7–12). RAD54L-4A/S49E-1 dignifies a clonal isolate; RAD54L-4A/S49E #2 dignifies a puromycin-resistant cell population. Loading control: α-tubulin. (C) Dot plot with medians of IdU fork speeds in HU-treated RAD54L KO cells and RAD54L KO cells expressing wild type RAD54L or RAD54L-4A/S49E and transfected with Ctrl, FBH1 or HLTF siRNA (n = 3; 85–224 fiber tracts/experiment analyzed; n = 1 for RAD54L-4A/S49E #2). (D) Schematic of the DNA fiber assay protocol used in (F). (E) Representative western blots to show extent of BRCA2 knockdown in RAD54L KO cells (lanes 3–4) and RAD54L KO cells expressing wild type RAD54L (lanes 1–2) or mutant RAD54L-4A/S49E (lanes 5–8) and RAD54L-K189R (lanes 9–10). RAD54L-4A/S49E-1 dignifies a clonal isolate; RAD54L-4A/S49E #2 and RAD54L-K189R #1 dignifies puromycin-resistant cell populations. Loading control: MSH2. (F) Dot plot with medians of IdU/CldU tract length ratios in HU-treated RAD54L KO cells and RAD54L KO cells expressing wild type RAD54L or mutant RAD54L-4A/S49E and RAD54L-K189R transfected with Ctrl or BRCA2 siRNA (n = 3; 46–104 fiber tracts/experiment analyzed; n = 1 for RAD54L-K189R #1). Data were analyzed by Kruskal–Wallis test followed by Dunn's multiple comparisons test (ns, not significant; ∗∗∗∗P< 0.0001). (G) Model to explain the role of RAD54L in the FBH1 pathway of RAD51-mediated fork reversal. RAD54L may be recruited through its interaction with RAD51 on extended parental ssDNA. FBH1 is recruited to stalled replication forks by PCNA ((78); PCNA is not shown here). Unwinding of lagging strand DNA through FBH1 (72) may initiate nascent strand annealing. RAD54L may then oligomerize on a 4-way junction and drive reversal through BM. The dependency of FBH1 on RAD54L may be indicative of coordinated recruitment and/or post-translational modification orchestrating sequential coaction. The number of subunits in the RAD54L BM oligomer remain to be determined. Schematic created with Biorender.com.
We then tested RAD54L KO cells and RAD54L KO cells with wild type RAD54L, RAD54L-4A/S49E and RAD54L-K189R in the fork resection assay (Figure 6D) after BRCA2 knockdown (Figure 6E). In accord with our earlier results (Figure 4C), after a 5-h treatment of cells in 4 mM HU IdU/CldU ratios were >1 (indicative of fork protection) in RAD54L KO cells with BRCA2 knockdown (P< 0.0001; Figure 6F; Supplementary Figure S6B). In contrast, IdU/CldU ratios in RAD54L KO cells expressing RAD54L-4A/S49E and depleted for BRCA2 were comparable to those in cells with wild type RAD54L (P> 0.999) and significantly lower than in RAD54L KO cells with BRCA2 knockdown (P< 0.0001). These results show that the RAD54L BM mutant behaves like wild type RAD54L in the HLTF-mediated fork reversal pathway, in which the BRCA2 protein was shown to function as a major fork protection factor (10). These results suggest that loss of a RAD54L function other than its BM activity is required to prevent fork attrition in the absence of BRCA2 in the HLTF pathway. In support of this premise, we show that the ATPase-dead RAD54L-K189R fully rescues fork protection after HU (Figure 6F).
Discussion
Our work establishes that RAD54L activity decelerates the progression of replication forks in human cells. Our results corroborate biochemical evidence (28) and are in strong support of RAD54L’s engagement in replication fork reversal in human cells. In biochemical assays (28), purified RAD54L was shown to use its BM activity to catalyze both the regression and restoration of model replication forks. In the presence of RAD51, however, as would be the situation in cells, the reaction was shown to be shifted toward the accumulation of the chicken foot structure (28). Our results in cells are in accord with this in vitro investigation and now have added one more attribute to the multi-functional RAD54L protein, limiting replication stress through fork restraint in human cells.
We have dissected that RAD54L functions with disparate requirements in the two distinct RAD51-mediated fork reversal pathways that have been described earlier (14). We show that RAD54L’s ability to catalyze BM is critical for fork restraint when engaged in the FBH1 pathway. In the HLTF/SMARCAL1 pathway, however, RAD54L BM activity is dispensable, suggesting that RAD54L attributes other than its BM activity contribute to fork reversal here. Concurringly, while the RAD54L BM mutant fails block fork attrition in BRCA2-deficient cells, RAD54L defective in ATP hydrolysis can do so.
RAD54L BM activity relies on its ability to function as an ATPase and form high-order RAD54L oligomers on HJ-like DNA structures (64,65). Importantly, the 4A, S49E and S49E/4A mutants tested here interfere with RAD54L oligomerization and compromise BM activity but show no defect in ATP hydrolysis or in their ability to stimulate the RAD51-mediated strand exchange reaction (29). Consequently, we infer that RAD54L engagement in the HLTF pathway may rely on the stimulation of RAD51-mediated strand invasion, as suggested recently (9). This scenario could explain why fork degradation transpires in BRCA2-depleted RAD54L KO cells expressing RAD54L-4A/S49E in contrast to cells with BRCA2 knockdown and complete loss of RAD54L or expression of RAD54L-K189R. Accordingly, under mild replication stress and with HLTF/SMARCAL1 knockdown, RAD54L status has no effect on replication speed, as would be expected with HLTF/SMARCAL1 functioning prior to and largely independently of RAD54L. Nonetheless, RAD54L is required during later stages in the HLTF/SMARCAL1-mediated fork reversal reaction, and we have established that this RAD54L engagement is not related to its ability to drive BM.
Genetic and biochemical studies have delineated that fork reversal is catalyzed by three replication fork remodelers of the SNF2 family (SMARCAL1, ZRANB3 and HLTF) or by the FBH1 helicase (14,56,66,67). Loss of either SMARCAL1, ZRANB3 or HLTF was shown to block nascent strand degradation in BRCA1/2-deficent cells, results that suggest their cooperation and non-redundant activities in fork reversal (10). Further, current models posit that fork reversal is a dynamic reaction that involves orchestrated actions of several proteins with specialized substrate preference (10,67,68). For example, in biochemical reconstitution assays ZRANB3 and HLTF were shown to be highly efficient in BM activity, while SMARCAL1 was little efficient in this regard. Moreover, SMARCAL1 was efficient in annealing RPA-coated DNA, while ZRANB3 and HLTF had much reduced capacity (67).
To date, much less is known about the molecular details of the FBH1 pathway of RAD51-mediated fork reversal. However, it is known that FBH1’s functional helicase domain is needed for the catalysis of fork reversal, but its ubiquitin ligase activity is not (14,56). While BRCA1/2, FANCD2 (10,13,15,16) and other proteins (69–71) protect reversed forks generated by SMARCAL1, HLTF, and ZRANB3, forks reversed by FBH1 are protected by 53BP1, and loss of FBH1 rescues fork degradation in the absence of 53BP1 (14). Importantly, we show here that loss of RAD54L also rescues fork degradation in the absence of 53BP1. Moreover, under conditions of mild replication stress and in the absence of RAD54L, the reduction of fork restraint in cells with FBH1 knockdown is significantly ameliorated, in support of a concerted action between FBH1 and RAD54L in fork reversal. In contrast, RAD54L does not affect fork restraint in cells depleted for SMARCAL1 or HLTF, as their activities in fork reversal do not depend on RAD54L. Unlike in the SMARCAL1/HLTF pathway, in which RAD54L-4A/S49E behaves identical to wild type RAD54L, RAD54L KO cells expressing RAD54L-4A/S49E mirror RAD54L KO cells in the FBH1 pathway. These results are in strong support of FBH1 relying on RAD54L to drive BM to catalyze fork reversal. We suggest a model in which the combined activities of RAD54L and FBH1 lead to fork reversal (Figure 6E). In this model, FBH1 may help unwind the growing lagging strand to promote nascent strand annealing, as suggested previously (72), and RAD54L may regress the 4-way junction through its BM activity, as suggested by the results described here.
In human cells, a RAD54L paralog- RAD54B- was identified (73). RAD54B shares extensive homology with RAD54L, particularly in the central region that contains the seven helicase motifs (74). Within the N-terminal domains, however, there is little sequence conservation between RAD54L and RAD54B (74). Of note, RAD54B can partially compensate for RAD54L deficiency in cell survival assays (31), and it would be interesting to assess if RAD54B can similarly fulfil such a back-up role for RAD54L in fork reversal. Such role for RAD54B could potentially explain why the reduction in fork restraint in RAD54L-deficient cells is milder than that in the absence of FBH1.
A growing body of work has provided evidence that replication-associated ssDNA gaps affect the response of cancer cells to chemotherapy, suggesting that enzymes that limit ssDNA gap accumulation may represent useful targets in cancer therapy (58,75,76). We propose that RAD54L may be one such target, as ssDNA gap formation is enhanced in its absence and further exacerbated through treatment of cells with PARPi. This proposal may be particularly attractive, as RAD54L not only is a critical protein in at least two different mechanisms of fork reversal but also is a key player in the HR DNA repair pathway, a pathway frequently associated with resistance to cancer therapy.
Supplementary Material
Acknowledgements
The authors would like to thank Drs Tingting Yao and Joe Gray for providing hTERT RPE-1 and Hs578T cell lines, respectively. Graphical Abstract and Figure 6G were created with BioRender.com.
Author contributions: Mollie E. Uhrig: Investigation, methodology, validation, formal analysis, writing of the manuscript. Neelam Sharma: Investigation, methodology, validation. Petey Maxwell: Validation. Jordi Gomez: Validation. Platon Selemenakis: Critical reagents. Alexander V. Mazin: Conceptualization, funding acquisition. Claudia Wiese: Conceptualization, formal analysis, writing of the manuscript, funding acquisition.
Notes
Present address: Mollie E. Uhrig, Graduate Program in Cell and Molecular Biology, Colorado State University, Fort Collins, CO 80523, USA.
Present address: Platon Selemenakis, Virovek Inc., Houston, TX 77070, USA.
Contributor Information
Mollie E Uhrig, Department of Environmental and Radiological Health Sciences, Colorado State University, Fort Collins, CO 80523, USA.
Neelam Sharma, Department of Environmental and Radiological Health Sciences, Colorado State University, Fort Collins, CO 80523, USA.
Petey Maxwell, Department of Environmental and Radiological Health Sciences, Colorado State University, Fort Collins, CO 80523, USA.
Jordi Gomez, Department of Environmental and Radiological Health Sciences, Colorado State University, Fort Collins, CO 80523, USA.
Platon Selemenakis, Department of Environmental and Radiological Health Sciences, Colorado State University, Fort Collins, CO 80523, USA.
Alexander V Mazin, Department of Biochemistry and Structural Biology, UT Health San Antonio, San Antonio, TX 78229, USA.
Claudia Wiese, Department of Environmental and Radiological Health Sciences, Colorado State University, Fort Collins, CO 80523, USA.
Data availability
The source data underlying all figures are provided within this paper and in Supplementary Table S1.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health [R01GM144579, R01GM136717, R01CA237286]; Cancer Prevention and Research Institute of Texas (CPRIT) [REI RR210023]; Congressionally Directed Medical Research Programs [BC191160]; A.V.M. is the holder of the Joe R. and Teresa Lozano Long Chair in Cancer Research. Funding for open access charge: NIH [R01GM144579].
Conflict of interest statement. None declared.
References
- 1. Saxena S., Zou L.. Hallmarks of DNA replication stress. Mol. Cell. 2022; 82:2298–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., Lopes M.. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015; 208:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thakar T., Moldovan G.L.. The emerging determinants of replication fork stability. Nucleic Acids Res. 2021; 49:7224–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kavlashvili T., Liu W., Mohamed T.M., Cortez D., Dewar J.M.. Replication fork uncoupling causes nascent strand degradation and fork reversal. Nat. Struct. Mol. Biol. 2023; 30:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berti M., Cortez D., Lopes M.. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 2020; 21:633–651. [DOI] [PubMed] [Google Scholar]
- 6. Tye S., Ronson G.E., Morris J.R.. A fork in the road: where homologous recombination and stalled replication fork protection part ways. Semin. Cell Dev. Biol. 2020; 113:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mason J.M., Chan Y.L., Weichselbaum R.W., Bishop D.K.. Non-enzymatic roles of human RAD51 at stalled replication forks. Nat. Commun. 2019; 10:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hashimoto Y., Ray Chaudhuri A., Lopes M., Costanzo V.. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010; 17:1305–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu W., Saito Y., Jackson J., Bhowmick R., Kanemaki M.T., Vindigni A., Cortez D.. RAD51 bypasses the CMG helicase to promote replication fork reversal. Science. 2023; 380:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taglialatela A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R.et al.. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-Family fork remodelers. Mol. Cell. 2017; 68:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L.et al.. Smarcal1-mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell. 2017; 67:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P.et al.. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 2017; 8:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemacon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N.et al.. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017; 8:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu W., Krishnamoorthy A., Zhao R., Cortez D.. Two replication fork remodeling pathways generate nuclease substrates for distinct fork protection factors. Sci. Adv. 2020; 6:eabc3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costes A., Lambert S.A.. Homologous recombination as a replication fork escort: fork-protection and recovery. Biomolecules. 2012; 3:39–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao W., Wiese C., Kwon Y., Hromas R., Sung P.. The BRCA tumor suppressor network in chromosome damage repair by homologous recombination. Annu. Rev. Biochem. 2019; 88:221–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flaus A., Martin D.M., Barton G.J., Owen-Hughes T.. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006; 34:2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ceballos S.J., Heyer W.D.. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta. 2011; 1809:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sigurdsson S., Van Komen S., Petukhova G., Sung P.. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 2002; 277:42790–42794. [DOI] [PubMed] [Google Scholar]
- 22. Crickard J.B., Moevus C.J., Kwon Y., Sung P., Greene E.C.. Rad54 Drives ATP hydrolysis-dependent DNA sequence alignment during homologous recombination. Cell. 2020; 181:1380–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agarwal S., van Cappellen W.A., Guenole A., Eppink B., Linsen S.E., Meijering E., Houtsmuller A., Kanaar R., Essers J.. ATP-dependent and independent functions of Rad54 in genome maintenance. J. Cell Biol. 2011; 192:735–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wright W.D., Heyer W.D.. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol. Cell. 2014; 53:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mazin A.V., Alexeev A.A., Kowalczykowski S.C.. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003; 278:14029–14036. [DOI] [PubMed] [Google Scholar]
- 26. Sanchez H., Kertokalio A., van Rossum-Fikkert S., Kanaar R., Wyman C.. Combined optical and topographic imaging reveals different arrangements of human RAD54 with presynaptic and postsynaptic RAD51-DNA filaments. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11385–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maranon D.G., Sharma N., Huang Y., Selemenakis P., Wang M., Altina N., Zhao W., Wiese C.. NUCKS1 promotes RAD54 activity in homologous recombination DNA repair. J. Cell Biol. 2020; 219:e201911049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bugreev D.V., Rossi M.J., Mazin A.V.. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 2011; 39:2153–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goyal N., Rossi M.J., Mazina O.M., Chi Y., Moritz R.L., Clurman B.E., Mazin A.V.. RAD54 N-terminal domain is a DNA sensor that couples ATP hydrolysis with branch migration of Holliday junctions. Nat. Commun. 2018; 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spies J., Waizenegger A., Barton O., Surder M., Wright W.D., Heyer W.D., Lobrich M.. Nek1 regulates Rad54 to orchestrate homologous recombination and replication fork stability. Mol. Cell. 2016; 62:903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Selemenakis P., Sharma N., Uhrig M.E., Katz J., Kwon Y., Sung P., Wiese C.. RAD51AP1 and RAD54L can underpin two distinct RAD51-dependent routes of DNA damage repair via homologous recombination. Front. Cell Dev. Biol. 2022; 10:866601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bai G., Kermi C., Stoy H., Schiltz C.J., Bacal J., Zaino A.M., Hadden M.K., Eichman B.F., Lopes M., Cimprich K.A.. HLTF promotes fork reversal, limiting replication stress resistance and preventing multiple mechanisms of unrestrained DNA synthesis. Mol. Cell. 2020; 78:1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Colston K.W., Perks C.M., Xie S.P., Holly J.M.. Growth inhibition of both MCF-7 and Hs578T human breast cancer cell lines by vitamin D analogues is associated with increased expression of insulin-like growth factor binding protein-3. J. Mol. Endocrinol. 1998; 20:157–162. [DOI] [PubMed] [Google Scholar]
- 34. Olivieri M., Cho T., Alvarez-Quilon A., Li K., Schellenberg M.J., Zimmermann M., Hustedt N., Rossi S.E., Adam S., Melo H.et al.. A genetic map of the response to DNA damage in Human cells. Cell. 2020; 182:481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liang F., Miller A.S., Longerich S., Tang C., Maranon D., Williamson E.A., Hromas R., Wiese C., Kupfer G.M., Sung P.. DNA requirement in FANCD2 deubiquitination by USP1-UAF1-RAD51AP1 in the Fanconi anemia DNA damage response. Nat. Commun. 2019; 10:2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kwon Y., Rosner H., Zhao W., Selemenakis P., He Z., Kawale A.S., Katz J.N., Rogers C.M., Neal F.E., Badamchi Shabestari A.et al.. DNA binding and RAD51 engagement by the BRCA2 C-terminus orchestrate DNA repair and replication fork preservation. Nat. Commun. 2023; 14:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parplys A.C., Zhao W., Sharma N., Groesser T., Liang F., Maranon D.G., Leung S.G., Grundt K., Dray E., Idate R.et al.. NUCKS1 is a novel RAD51AP1 paralog important for homologous recombination and genome stability. Nucleic Acids Res. 2015; 43:9817–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao W., Steinfeld J.B., Liang F., Chen X., Maranon D.G., Jian Ma C., Kwon Y., Rao T., Wang W., Sheng C.et al.. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature. 2017; 550:360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao W., Vaithiyalingam S., San Filippo J., Maranon D.G., Jimenez-Sainz J., Fontenay G.V., Kwon Y., Leung S.G., Lu L., Jensen R.B.et al.. Promotion of BRCA2-dependent homologous recombination by DSS1 via RPA targeting and DNA mimicry. Mol. Cell. 2015; 59:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wiese C., Hinz J.M., Tebbs R.S., Nham P.B., Urbin S.S., Collins D.W., Thompson L.H., Schild D.. Disparate requirements for the Walker A and B ATPase motifs of human RAD51D in homologous recombination. Nucleic Acids Res. 2006; 34:2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dray E., Etchin J., Wiese C., Saro D., Williams G.J., Hammel M., Yu X., Galkin V.E., Liu D., Tsai M.S.et al.. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010; 17:1255–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ostvold A.C., Norum J.H., Mathiesen S., Wanvik B., Sefland I., Grundt K.. Molecular cloning of a mammalian nuclear phosphoprotein NUCKS, which serves as a substrate for Cdk1 in vivo. Eur. J. Biochem. 2001; 268:2430–2440. [DOI] [PubMed] [Google Scholar]
- 43. Sirbu B.M., Couch F.B., Cortez D.. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 2012; 7:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Campeau E., Ruhl V.E., Rodier F., Smith C.L., Rahmberg B.L., Fuss J.O., Campisi J., Yaswen P., Cooper P.K., Kaufman P.D.. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009; 4:e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Somyajit K., Spies J., Coscia F., Kirik U., Rask M.B., Lee J.H., Neelsen K.J., Mund A., Jensen L.J., Paull T.T.et al.. Homology-directed repair protects the replicating genome from metabolic assaults. Dev. Cell. 2021; 56:461–477. [DOI] [PubMed] [Google Scholar]
- 46. Quinet A., Carvajal-Maldonado D., Lemacon D., Vindigni A.. DNA Fiber analysis: mind the gap!. Methods Enzymol. 2017; 591:55–82. [DOI] [PubMed] [Google Scholar]
- 47. Jackson D.A., Pombo A.. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998; 140:1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wiese C., Dray E., Groesser T., San Filippo J., Shi I., Collins D.W., Tsai M.S., Williams G.J., Rydberg B., Sung P.et al.. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell. 2007; 28:482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Modesti M., Budzowska M., Baldeyron C., Demmers J.A., Ghirlando R., Kanaar R.. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell. 2007; 28:468–481. [DOI] [PubMed] [Google Scholar]
- 50. Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T.. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010; 37:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E.H., Hendrickson E.A.et al.. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 2015; 208:545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vujanovic M., Krietsch J., Raso M.C., Terraneo N., Zellweger R., Schmid J.A., Taglialatela A., Huang J.W., Holland C.L., Zwicky K.et al.. Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol. Cell. 2017; 67:882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kile A.C., Chavez D.A., Bacal J., Eldirany S., Korzhnev D.M., Bezsonova I., Eichman B.F., Cimprich K.A.. HLTF’s ancient HIRAN domain binds 3' DNA ends to drive replication fork reversal. Mol. Cell. 2015; 58:1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mazina O.M., Rossi M.J., Deakyne J.S., Huang F., Mazin A.V.. Polarity and bypass of DNA heterology during branch migration of Holliday junctions by human RAD54, BLM, and RECQ1 proteins. J. Biol. Chem. 2012; 287:11820–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Unk I., Hajdu I., Blastyak A., Haracska L.. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst.). 2010; 9:257–267. [DOI] [PubMed] [Google Scholar]
- 56. Fugger K., Mistrik M., Neelsen K.J., Yao Q., Zellweger R., Kousholt A.N., Haahr P., Chu W.K., Bartek J., Lopes M.et al.. FBH1 Catalyzes regression of stalled replication forks. Cell Rep. 2015; 10:1749–1757. [DOI] [PubMed] [Google Scholar]
- 57. Vaitsiankova A., Burdova K., Sobol M., Gautam A., Benada O., Hanzlikova H., Caldecott K.W.. PARP inhibition impedes the maturation of nascent DNA strands during DNA replication. Nat. Struct. Mol. Biol. 2022; 29:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hanzlikova H., Kalasova I., Demin A.A., Pennicott L.E., Cihlarova Z., Caldecott K.W.. The importance of poly(ADP-Ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol. Cell. 2018; 71:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maya-Mendoza A., Moudry P., Merchut-Maya J.M., Lee M., Strauss R., Bartek J.. High speed of fork progression induces DNA replication stress and genomic instability. Nature. 2018; 559:279–284. [DOI] [PubMed] [Google Scholar]
- 60. Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T.. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005; 434:913–917. [DOI] [PubMed] [Google Scholar]
- 61. Tian T., Bu M., Chen X., Ding L., Yang Y., Han J., Feng X.H., Xu P., Liu T., Ying S.et al.. The ZATT-TOP2A-PICH axis drives extensive replication fork reversal to promote genome stability. Mol. Cell. 2021; 81:198–211. [DOI] [PubMed] [Google Scholar]
- 62. Swagemakers S.M., Essers J., de Wit J., Hoeijmakers J.H., Kanaar R.. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem. 1998; 273:28292–28297. [DOI] [PubMed] [Google Scholar]
- 63. Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012; 19:417–423. [DOI] [PubMed] [Google Scholar]
- 64. Bugreev D.V., Mazina O.M., Mazin A.V.. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006; 442:590–593. [DOI] [PubMed] [Google Scholar]
- 65. Mazina O.M., Rossi M.J., Thomaa N.H., Mazin A.V.. Interactions of human rad54 protein with branched DNA molecules. J. Biol. Chem. 2007; 282:21068–21080. [DOI] [PubMed] [Google Scholar]
- 66. Joseph S.A., Taglialatela A., Leuzzi G., Huang J.W., Cuella-Martin R., Ciccia A.. Time for remodeling: sNF2-family DNA translocases in replication fork metabolism and human disease. DNA Repair (Amst.). 2020; 95:102943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Halder S., Ranjha L., Taglialatela A., Ciccia A., Cejka P.. Strand annealing and motor driven activities of SMARCAL1 and ZRANB3 are stimulated by RAD51 and the paralog complex. Nucleic Acids Res. 2022; 50:8008–8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Neelsen K.J., Lopes M.. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015; 16:207–220. [DOI] [PubMed] [Google Scholar]
- 69. Xu S., Wu X., Wu L., Castillo A., Liu J., Atkinson E., Paul A., Su D., Schlacher K., Komatsu Y.et al.. Abro1 maintains genome stability and limits replication stress by protecting replication fork stability. Genes Dev. 2017; 31:1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ye Z., Xu S., Shi Y., Cheng X., Zhang Y., Roy S., Namjoshi S., Longo M.A., Link T.M., Schlacher K.et al.. GRB2 stabilizes RAD51 at reversed replication forks suppressing genomic instability and innate immunity against cancer. Nat. Commun. 2024; 15:2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Higgs M.R., Reynolds J.J., Winczura A., Blackford A.N., Borel V., Miller E.S., Zlatanou A., Nieminuszczy J., Ryan E.L., Davies N.J.et al.. BOD1L Is required to suppress deleterious resection of stressed replication forks. Mol. Cell. 2015; 59:462–477. [DOI] [PubMed] [Google Scholar]
- 72. Masuda-Ozawa T., Hoang T., Seo Y.S., Chen L.F., Spies M.. Single-molecule sorting reveals how ubiquitylation affects substrate recognition and activities of FBH1 helicase. Nucleic Acids Res. 2013; 41:3576–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hiramoto T., Nakanishi T., Sumiyoshi T., Fukuda T., Matsuura S., Tauchi H., Komatsu K., Shibasaki Y., Inui H., Watatani M.et al.. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene. 1999; 18:3422–3426. [DOI] [PubMed] [Google Scholar]
- 74. Tanaka K., Kagawa W., Kinebuchi T., Kurumizaka H., Miyagawa K.. Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic Acids Res. 2002; 30:1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cybulla E., Vindigni A.. Leveraging the replication stress response to optimize cancer therapy. Nat. Rev. Cancer. 2023; 23:6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cong K., Peng M., Kousholt A.N., Lee W.T.C., Lee S., Nayak S., Krais J., VanderVere-Carozza P.S., Pawelczak K.S., Calvo J.et al.. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell. 2021; 81:3128–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lord S.J., Velle K.B., Mullins R.D., Fritz-Laylin L.K.. SuperPlots: communicating reproducibility and variability in cell biology. J. Cell Biol. 2020; 219:e202001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bacquin A., Pouvelle C., Siaud N., Perderiset M., Salome-Desnoulez S., Tellier-Lebegue C., Lopez B., Charbonnier J.B., Kannouche P.L.. The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells. Nucleic Acids Res. 2013; 41:6501–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data underlying all figures are provided within this paper and in Supplementary Table S1.