Abstract
Integrons are genetic elements that increase the evolvability of bacteria by capturing new genes and stockpiling them in arrays. Sedentary chromosomal integrons (SCIs) can be massive and highly stabilized structures encoding hundreds of genes, whose function remains generally unknown. SCIs have co-evolved with the host for aeons and are highly intertwined with their physiology from a mechanistic point of view. But, paradoxically, other aspects, like their variable content and location within the genome, suggest a high genetic and functional independence. In this work, we have explored the connection of SCIs to their host genome using as a model the Superintegron (SI), a 179-cassette long SCI in the genome of Vibrio cholerae N16961. We have relocated and deleted the SI using SeqDelTA, a novel method that allows to counteract the strong stabilization conferred by toxin–antitoxin systems within the array. We have characterized in depth the impact in V. cholerae’s physiology, measuring fitness, chromosome replication dynamics, persistence, transcriptomics, phenomics, natural competence, virulence and resistance against protist grazing. The deletion of the SI did not produce detectable effects in any condition, proving that—despite millions of years of co-evolution—SCIs are genetically and functionally isolated units of genomes.
Graphical Abstract
Graphical Abstract.
Introduction
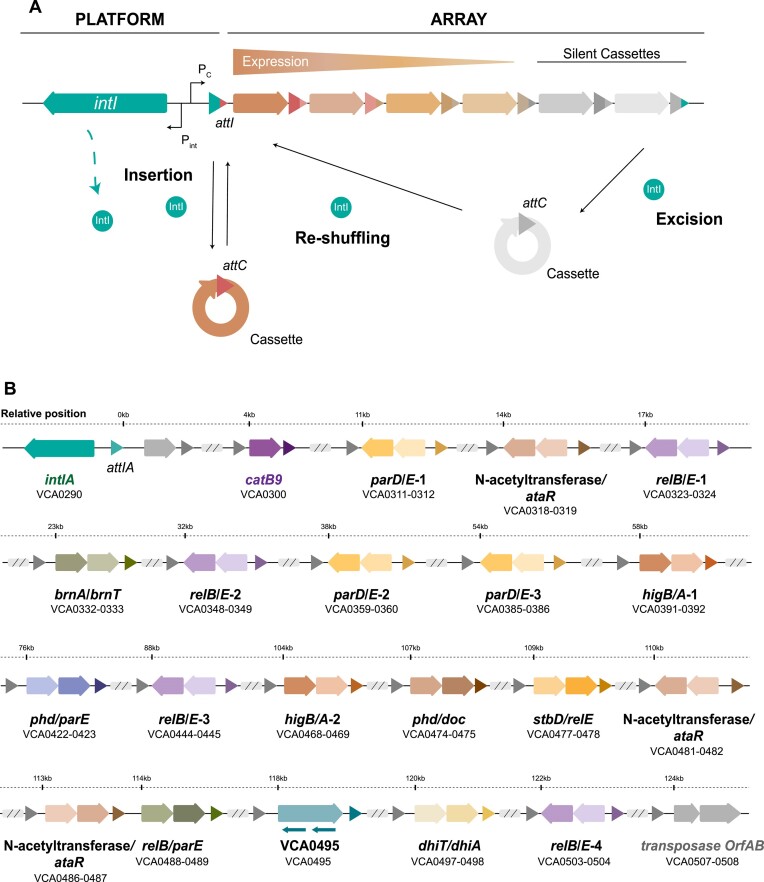
Integrons are genetic elements that allow bacteria to adapt swiftly to changing environments (1–3). They recruit new genes encoded in small mobile genetic elements called integron cassettes (IC) (or gene cassettes), stocking them in an array to form a genetic memory of adaptive functions. According to the needs of their hosts, integrons can also modulate cassette expression by reshuffling their order within the array (4,5). The integron platform comprises the gene encoding the integrase (intI)—the recombinase that governs all reactions—the integration site where cassettes are incorporated (attI site), and two promoters in opposite orientations that drive the expression of the integrase (Pint) and that of cassettes in the array (Pc), which are generally promoterless (Figure 1A). Integrons are well known for their role in the rise of multidrug resistance and the spread of resistance genes among clinically relevant Gram-negative species (6–8). Encoded on plasmids, these Mobile Integrons (MIs) act as vehicles for more than 170 resistance genes against the most relevant families of antibiotics (9,10). Yet, MIs are only a small subset of all integrons that have been mobilized from the chromosomes of environmental bacteria by transposons. The vast majority of integrons in nature are sedentary chromosomal integrons (SCIs), which are found in a variety of phyla and have an ancient origin (11–13). These SCIs are a highly variable region of the genome, representing a hotspot for genetic diversity (14). On the antipodes of what is observed for MIs, the functions encoded in SCIs are generally not related to antimicrobial resistance and are mostly unknown (15). The colossal repertoire of cassettes in the environment holds a high potential for biotechnology that is yet to be explored (16).
Figure 1.
(A) Schematic representation of an integron. On the left side, the functional platform includes the integrase gene (intI1), the cassette promoter (PC), the integrase promoters (Pint) and the integron insertion site (attI). On the right side, the variable array of ICs is shown. Hybrid attI and attC sites are marked with corresponding colors. The arrows within the cassettes indicate the orientation of the open reading frame, with the color intensity of each arrow representing the expression level. The diagram also illustrates the primary reactions mediated by the IntI recombinase: cassette insertion (attI × attC), excision (attC × attC) and reshuffling. (B) Diagram showing the organization and relative position of the relevant genes within the SI cassette array. Arrows represent individual genes contained within cassettes, while gray triangles denote the attC sites. TA systems are illustrated with colored arrows: darker arrows indicate toxins and lighter arrows indicate antitoxins. The orientation and position of each arrow reflect the transcriptional direction of the genes. Additionally, smaller arrows mark regions corresponding to non-coding RNAs.
The paradigm of SCI is the Superintegron (SI) (17), a massive structure encoded in the secondary chromosome of Vibrio cholerae, the causative agent of cholera disease. In strain N16961, where it was first described, the SI is 126 kb long, embodying 3% of the genome of this bacterium and containing 179 ICs. Although the model of integron predicts that the SI should be mostly silent, a recent RNA-seq analysis shows that many cassettes are expressed at biologically relevant levels (18). The functions of cassettes are mostly cryptic, except for the notable exception of 19 cassettes that encode toxin–antitoxin (TA) systems (19,20) and a chloramphenicol resistance gene (21) (Figure 1B). TA cassettes are scattered along the SI and serve to stabilize the array through a post-segregational killing mechanism: if the TA system is lost, the labile antitoxin is degraded before the more stable toxin, leading to cell death (22,23).
Despite being the best-studied SCI, the knowledge on the SI is limited for two reasons. First, the long cassette array interferes with the genetic tools commonly used in the field to deliver recombination experiments that would allow the characterization of the system. Second, its integrase cannot be studied in heterologous genetic backgrounds because it seems to require additional host factors (24–26).
SCIs have co-evolved with the genome of the host for millions of years (27,28). Indeed, large SCIs like the SI are ubiquitous in Vibrio species, and the phylogenetic signal of integrases mimics that of the species (1). This suggests that their acquisition predates the radiation of the genus, an event that occurred >300 million years ago (29). The timescale of this co-evolution process has allowed for the intertwining between host physiology and integron activity at many different levels. A blatant example is that integrase expression is under the control of the host’s SOS response. Yet, many other aspects are rather more subtle and represent better the depth of such connections (30). Examples of this are (i) the balanced interplay between the integrase and single-strand binding (SSB) proteins of the host on attC sites, that impede attC site folding to avoid its formation unless needed for recombination, i.e. when the integrase is expressed (31,32); or (ii) the fact that SCIs have a specific orientation related to the origin of replication, to maximize their carrying capacity (23,33). A striking observation is that some aspects of SCIs remain extremely plastic, such as their content and their location—despite their size, SCIs can be found in either chromosome depending on the species of Vibrio. Hence, it remains unclear whether the intertwining between host and integron is restricted to the functioning of the platform and the mechanistic of recombination (Pint and Pc promoters and attC sites), or if it is extensive to the array of functions encoded in the variable part. In other words, we ignore if cassettes are streamlined to act as plug-and-play add-ons to the pool of functions encoded in the genome without interacting with it, or whether they can modify or interfere with the host physiology in any other way.
In this work, we have addressed the question of whether chromosomal integrons are genetically and functionally isolated units of the genome by relocating and deleting the SI from V. cholerae. To overcome the high stabilization of the structure by the TAs, we have designed an approach called SeqDelTA (Sequential Deletion of Toxin–Antitoxin Systems) that exploits natural competence and homologous recombination to deliver a set of 16 consecutive allelic replacements. At each step, a toxin is inactivated without altering the cognate antitoxin, deleting a piece of the SI. After the last allelic replacement, we produce the scarless deletion of the SI through a counter-selectable suicide vector. The resulting strain has been sequenced, and unintended mutations located elsewhere in the genome have been corrected. Incidentally, this strain represents a milestone in the study of chromosomal integrons, since it provides, for the first time, the possibility of delivering experimental studies using common tools, without interference from the SI. To understand how the deletion of the SI affects V. cholerae, we have characterized in depth this strain from a variety of perspectives, including chromosome replication dynamics, fitness, natural competence, antibiotic persistence, transcriptomics, phenomics, virulence and protist grazing assays. We observe no significant variations in the physiology of the ΔSI strain compared to the wild-type. We conclude that SCIs are functionally isolated units of the genome, despite eons of co-evolution with their hosts. This has strong implications in the type of functions one can expect to find in ICs.
Materials and methods
SI relocation from Chr2 to Chr1
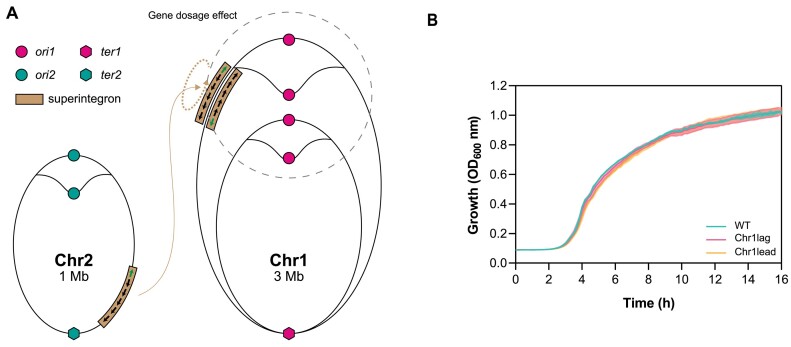
All strains, plasmids and primers used for the SI relocation are listed in Supplementary Tables S1, S4 and S5. To relocate the whole SI, we used a genetic tool based on the recombination of two bacteriophage attachment sites (34). Prophage excision from the host chromosome relies on site-specific recombination between two sequences flanking the phage, termed attachment sites attL and attR. This recombination is carried out by phage-specific recombinases ‘Int’ and their cognate RDF (recombination directionality factor), or excisionase, Xis. This tool uses the attachment sites from bacteriophages HK and λ (Supplementary Figure S1). The attL/attR pairs of each site were associated with parts of a genetic marker that is reconstituted when the sites recombine: HK sites were associated with parts of a bla gene (ampicillin resistance) and the λ sites with parts of lacZ. Co-expression of the HK and λ integrases/excisionases triggers the recombination of the partner attL/attR sites, which allows for the reconstitution of fully functional bla and lacZ markers and the selection of clones in which the SCI relocation has occurred. Blue colonies growing in LB supplemented with carbenicillin and X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) were polymerase chain reaction (PCR) verified and used for growth curves. The SI was relocated between genes VC0018 and VC0019 in both orientations (Figure 2A).
Figure 2.
(A) Scheme of the relocation of the SI from Chr2 to Chr1, where it is inserted in both orientations (modified from (76)). (B) Growth curves of WT and relocated-SI strains (Chr1lag and Chr1lead). No significant growth differences are observed.
SeqDelTA
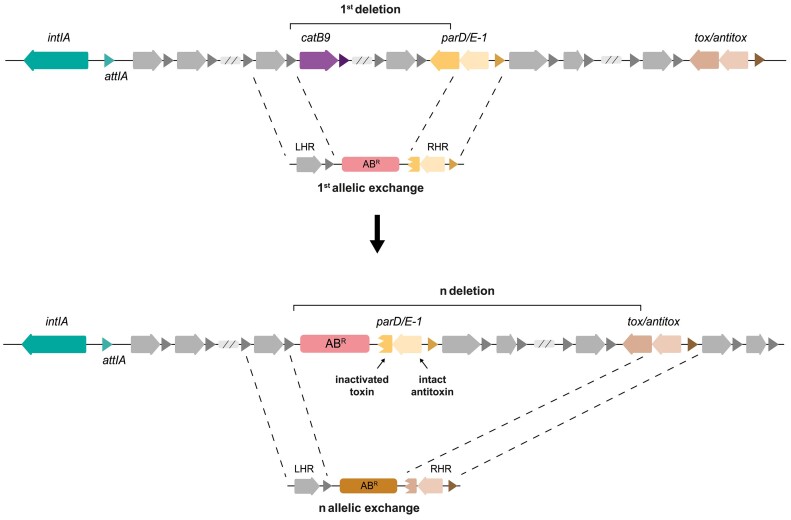
All strains, plasmids and primers for the deletion of the SI are listed in Supplementary Tables S2, S3, S4 and S5. To generate the ΔSI mutant of V. cholerae N16961, we developed SeqDelTA, as depicted in Figure 3. After identifying the 19 TA systems contained in the V. cholerae N16961 SI, we prepared successive constructs designed with homology regions (HR) and a selective marker. A left homology region (LHR) was designed and maintained throughout the procedure. This region corresponded to the 5′ region of the catB9 gene (VCA0300), and was amplified with primers VCA 299 F and LRHI VCA 300 R. For the successive deletions, different right homology regions (RHD) were designed to maintain antitoxin gene but to eliminate part of the corresponding toxin gene, ensuring that the event was not lethal to the bacteria (Figure 3 and Supplementary Table S2). As selective markers, three different antibiotic resistance genes (ZeoR, CmR and CarbR) were used and amplified with the same primers, facilitating the consequent design and assembly of the HRs. These three fragments (LHR, resistance marker and RHR) were then assembled by splicing by overlap extension PCR (SOE-PCR), placing the resistance marker between the LHR and the RHR. Each successive construct was introduced by natural transformation (described below) into the V. cholerae strain obtained from the previous TA system deletion. This strategy allowed the removal of the resistance marker introduced in the previous step while advancing the deletion of TA systems by changes in the RHR. When necessary, due to gene orientation and/or gene order in the TA system, alternative LHRs were designed, and two resistance markers were maintained in consecutive allelic replacements before the new step allowed deletion of both (Supplementary Table S2).
Figure 3.
Schematic representation of two successive allelic replacements using SeqDelTA. The LHR is conserved during the deletion process, allowing the recycling of the resistance markers at each step. The RHR is redesigned at each step to inactivate the following TA module and delete the cassette cargo in between. The inactivation of the TA system is performed by knocking the toxin but leaving the antitoxin intact. A total of 18 sequential replacements were performed. The last deletion step was carried out using a suicide plasmid.
The last deletion was performed using the R6K origin of replication-plasmid pMP7 as described in (25,35). pMP7 replication is dependent of the cell π protein and encodes the ccdB toxin under the control of the inducible promoter PBAD. Five hundred bp of each side of the SI were amplified with primers RHI link pMP7 F and LRHI + D R for the LHR, and RHD F and RHD link pMP7 R for the RHR. The two fragments were then joined by SOE-PCR and digested with enzymes NaeI y HindIII. Finally, the insert was ligated with the empty pMP7 backbone, giving rise to pMP7_ΔintIA attIA zeoR. We started the cloning steps by transforming the pMP7_ΔintIA attIA zeoR plasmid into ccdB-resistant Escherichia coli π3813 (thy::pyr +) competent cells. Transformants were selected in LB agar plates containing Cm (25 μg/ml), thymidine (dT; 0.3 mM) and glucose (1%). For conjugal transfer of pMP7_ΔintIA attIA zeoR into V. cholerae, the plasmid was first transformed into the donor strain E. coli β3914 (dap::pir +). Transformants were selected in LB agar plates supplemented with Cm (25 μg/ml), 2,6-diaminopimelic acid (DAP; 0,3 mM) and glucose (1%). Conjugation assay was performed by growing both donor (E. coli β3914) and recipient (V. cholerae A066) strains to a OD600 of 0.2. Then, cells were mixed in a ratio of 1/10 (donor/recipient) and transferred to a mating filter placed on a LB agar plate supplemented with DAP (0.3 mM) and glucose (1%) and conjugation was performed overnight at 37°C. Integration of the pMP7_ΔintIA attIA zeoR into the Vibrio genome was selected by washing the filter in 5 ml of LB and plating serial dilutions on Cm (2.5 μg/ml) and glucose (1%) plates, but lacking DAP. Afterward, Cm-resistant colonies were grown in liquid LB medium and plated in LB agar plates supplemented with L-arabinose (0.2%) to express ccdB and select for the second crossover that implies the excision of the pMP7 backbone. After this process, we Sanger-sequenced the SI region and selected a V. cholerae ΔSI strain (A101).
Natural transformation assays
Transformation assays were performed as previously described in (36) with some modifications. Briefly, strains were inoculated from frozen stocks into LB broth and grown rolling overnight at 30°C. Overnight cultures were diluted 1:100 and grown to an OD600 of 1.0. Cells were then washed and resuspended in DASW (defined artificial seawater) (Instant Ocean®) 0.5× and inoculated in a final volume of 1 ml DASW with 150 μl of chitin (Apollo Scientific®) slurry. Cells were incubated in static conditions at 30°C for ∼18 h. Then, 550 μl of supernatant was removed from the reactions, and the purified transforming DNA (tDNA) was added. DNA and cells were incubated for ∼18 h in static conditions at 30°C. When needed, reactions were outgrown by adding 1 ml LB broth and shaking at 30°C for 2 h. Cells were plated in LB agar with the corresponding antibiotic.
To measure natural competence frequencies, cells were incubated with 2 μg of a PCR product carrying a kanamycin resistance cassette flanked by 1 kb long homology regions targeting the lacZ gene. Primers and strains used for this assay are listed in Supplementary Tables S3 and S5. In these assays, cells were incubated for 8 h with tDNA before plating in selective medium (LB plates supplemented with kanamycin 75 μg/ml). Frequencies were calculated as the number of colony-forming units (CFUs) growing in kanamycin over the total CFUs.
DNA isolation, Whole Genome Sequencing (WGS) and data analysis
Genomic DNA of the obtained V. cholerae ΔSI strain (A101) was extracted using the DNeasy® Blood and Tissue Kit (QIAGEN) following the manufacturer’s protocol. The DNA quantity was determined using a BioSpectrometer (Eppendorf). WGS sequencing was performed at Institut Pasteur using Illumina paired-end reads, at MiGS (Pittsburgh, USA), now SeqCoast Genomics (Portsmouth, USA) and inhouse at MBA (Madrid) using MinIon. Data analysis was accomplished with Geneious Prime software and the genetic variants were identified by mapping the generated reads to the V. cholerae N16961 reference genome (Accession number: chromosome 1 CP028827.1; chromosome 2 CP028828.1).
Marker Frequency Analysis (MFA)
Marker frequency analysis (MFA) was performed as described in (37). Cells were grown in LB at 37°C with shaking and genomic DNA was isolated using the DNeasy® Tissue Kit (Qiagen) from exponential and stationary phase cultures. Genomic DNA was sequenced with Illumina technology. Short reads were aligned to the reference genome using Bowtie2 (38) and R2R. The number of reads starting per bp (N) were calculated for 1 and 10 kbp windows. The exponential phase data were normalized with a factor calculated as the deviation of observed N from calculated N for each window of the stationary phase culture data. Any window including repeated sequences was excluded and N was plotted as a function of the absolute position, centered on the replication origins.
Correction of V. cholerae ΔSI mutations
All the strains, plasmids and primers are listed in Supplementary Tables S3, S4 and S5. Three mutations were found in V. cholerae ΔSI strain (A101): rocS (VC0653), cry2 (VC01392) and rpoS (VC0534). While rocS and rpoS mutations led to frameshifts, cry2 contained a non-synonymous mutation. To revert the mutations to the WT variant, allelic exchanges were performed using the pMP7, as previously described. To provide homology for the allelic replacement, 1000 bp-fragments of the N16961 strain genes rocS, rpoS and cry2 were amplified with primers rocS_pMP7 F/R, rpoS_pMP7 F/R and cry2_pMP7 F/R, respectively, which contain 20 bp-homology fragments with the pMP7 cloning site; pMP7 backbone was amplified with primers pMP7_bb_Gibson F/R. Cloning of the amplified fragments was performed using Gibson assembly (39), giving rise to pMP7_rocS, pMP7_rpoS and pMP7_cry2. We performed the mating and allelic exchanges as previously mentioned, starting with pMP7_rocS, then pMP7_rpoS, and finally, pMP7_cry2. After each step, we Sanger-sequenced the targeted gene and selected a V. cholerae strain corrected for rocS+ (A677), rocS+, rpoS+ (A684), and finally rocS+, rpoS+, cry2+ (B522). We verified by Illumina whole-genome sequencing that V. cholerae ΔSI (B522) did not contain any other unintended mutations.
RNA extraction and preparation for RNA-seq
Total RNA was extracted from V. cholerae WT and ΔSI cultures grown in LB at 37°C in both exponential (OD600 0.8) and stationary (OD600 2.8) growth phases using the RNeasy® Mini Kit (QIAGEN), following the manufacturer’s protocol. To eliminate any residual DNA, RNA was treated using the TURBO DNA-free™ Kit (Invitrogen). RNA concentration was measured using a BioSpectrometer (Eppendorf), and RNA integrity was determined using a Qubit™ 4 fluorometer (Invitrogen) with the RNA IQ Assay Kit (Invitrogen). Ribodepletion RNA library and sequencing was performed at the Oxford Genomics Centre using a NovaSeq6000 sequencing system (Illumina). Three biological samples per condition were sequenced.
RNA-seq data analysis
Raw Illumina reads were trimmed and adapter-removed using Trim Galore v0.6.6 (https://github.com/FelixKrueger/TrimGalore) with a quality threshold of 20 and removing reads <50 bp. Trimmed paired reads were mapped to the V. cholerae N16961 reference genome (accession number: chromosome 1 CP028827.1; chromosome 2 CP028828.1) using BWA-MEM v0.7.17 (40). featureCounts from the Rsubread v2.10.2 package (41) was used to obtain read counts per annotated feature, including coding sequences (CDS), non-coding RNA, transfer-messenger RNA, ribonuclease P, tRNA, antisense RNA and signal recognition particle RNA. Differential expression analysis was performed from raw counts using DESeq2 v1.36.0 (42), by comparing the expression data of the deletion mutant against the WT strain.
Differentially expressed genes (DEGs) of CDSs were annotated with Gene Ontology (GO) terms retrieved from UniProt on 7 July 2023 (43). Gene set enrichment analysis (GSEA) was performed to identify sets of biological processes, molecular functions or cellular components with different expression profiles between the SI deletion mutant and WT strains, in both exponential and stationary phases. For this, a pre-ranked list of DEGs, ordered by log2 fold changes, was provided to the GSEA function of the clusterProfiler v4.8.1 (44). Genes belonging to the SI (positions 31 2057–43 8942 bp of chromosome 2) were not included to prevent biased enrichment results caused by the acute downregulation of the superintegron genes. GSEA was run with default parameters, correcting for multiple tests with the Benjamini–Hochberg procedure.
Reverse transcription-quantitative PCR (RT-qPCR)
Independent samples of total RNA from V. cholerae WT and ΔSI cultures grown in LB at 37°C in both exponential (OD600 0.8) and stationary (OD600 2.8) were isolated as described previously. To eliminate any residual DNA, RNA was treated using the TURBO DNA-free™ Kit (Invitrogen) and RNA concentration was measured using a BioSpectrometer (Eppendorf). Before cDNA synthesis, a previous DNA wipe-out step was performed using the QuantiTect® Reverse Transcription Kit (QIAGEN) following the manufacturer’s instructions; cDNA synthesis was carried out afterward using the same kit using the temperature steps: 42°C for 15 min; 95°C for 3 min.
Reverse transcription quantitative PCR (RT-qPCR) was performed in an Applied Biosystems QuantStudio 3 using the Fast SYBR™ Green Master Mix (Thermo Fischer Scientific). Primers used are listed in Supplementary Table S5, and gyrA was used as housekeeping gene. Relative changes in gene expression for the SI deletion mutant with respect to the WT were determined according to the threshold cycle method (2−ΔΔCT). Mean values were obtained from two independent biological replicates with three technical replicates each.
Note on the controls used for the search for phenotypes
The original V. cholerae N16961 isolate contains mutations in the hapR gene and is not naturally competent. The deletion of the SI has been performed in a well-known derivative strain in which natural competence is restored by the insertion of a transposon carrying a functional hapR gene (strain A001 in the Supplementary Table S3). It is known that V. cholerae cultures have a tendency toward the hapR− genotype. Through genome sequencing we observed this phenomenon in our stock of the parental strain, which seemed to rise through homologous recombination between both hapR alleles. Since hapR− strains have certain phenotypes, such as distinct growth curves or a decreased virulence, we performed most phenotypic experiments using as a control a strain of V. cholerae N16961, in which mutations in hapR have been corrected without the addition of a second allele (A096 in our strain list). In RNA-seq and MFA experiments were we could use as a control the parental strain (A001), by confirming through sequence analysis that all replicates contained a functional hapR allele.
Growth curves
Overnight cultures were prepared by inoculating single colonies from LB agar plates in LB liquid medium and incubating for 16–20 h at 37°C. Cultures were then diluted 1:1000 in fresh medium and 200 μl were transferred into 96-well plates (Nunc, Thermo Scientific). OD600 was measured every 15 min for 16 h (36 h for 20°C) using a Biotek Synergy HTX plate reader (Agilent) at different temperatures (20, 30, 37 and 42°C). Vmax and area under the curve (AUC) were determined using Gen5 software and MATLAB, version R2022a (Mathworks), respectively. At least 10 independent replicates were included in the experiment and significant differences were assessed by performing an unpaired t-test.
Growth curves in the presence of subinhibitory concentrations of norfloxacin, ceftriaxone and tannic acid were performed by diluting the compounds at the desired concentrations in MH medium. Overnight cultures of V. cholerae WT and ΔSI were diluted 1:1000 in the antimicrobial-containing MH media in 96-well plates (Nunc, Thermo Scientific). OD600 was measured every 20 min for 24 h using a Biotek Synergy HTX plate reader (Agilent). At least three biological replicates were included in the experiment.
Competition assays
Competition assays were performed by flow cytometry to measure the fitness values of V. cholerae WT, the ΔSI mutant and the corrected intermediate deletion strains, relative to an E. coli DH5α strain as a common competitor carrying a pSU38 plasmid with the gfp under the control of the constitutive promoter PCS (strain A370, Supplementary Table S3). The competition procedure was performed as described in (45) with some modifications. Briefly, pre-cultures were prepared by inoculating single colonies from a LB agar plate in 200 μl of liquid LB in a 96-well plate (Nunc, Thermo Scientific). After 22 h of growth at 37°C with 250 rpm shaking, cultures were mixed at 1:1 proportion and diluted 1:400 in fresh medium. In order to confirm the initial proportions of E. coli carrying gfp and the non-fluorescent V. cholerae strains, cells were diluted 1:400 in NaCl (0.9%) and 30.000 events per sample were recorded using a CytoFLEX S flow cytometer (Beckman Coulter). Bacterial mixtures were competed for 22 h at 37°C with 250 rpm shaking, and final proportions were determined as stated above. The fitness values of both V. cholerae WT and ΔSI, relative to E. coli PCS::gfp were calculated using the formula: w = ln(Nfinal,gfp-/Ninitial,gfp-)/ln(Nfinal,gfp+/Ninitial,gfp+), where w is the relative fitness of the non-GFP-tagged V. cholerae strains, Ninitial,gfp- and Nfinal,gfp- are the numbers of non-GFP-tagged V. cholerae at the beginning and end of the competition, and Ninital,gfp+ and Nfinal,gfp+ are the numbers of E. coli PCS::gfp cells at the beginning and end of the competition, respectively. Twelve/six biological replicates were performed for each competition experiment and significant differences were assessed by performing unpaired t-test.
Phenotypic microarray assay
Phenotype Microarrays (Biolog) PM1-PM20 were tested as previously described in (46) with some modifications. Briefly, single colonies of V. cholerae WT and ΔSI were inoculated in liquid LB medium and grown overnight at 37°C. For PM1 and PM2 plates inoculation, 120 μl of Redox Dye mix D (100×), 1 ml of sterile water and 880 μl of a 85% T-cell suspension in NaCl (0.9%) were added to 10 ml of IF-0 (1.2×), resulting in a final volume of 12 ml per plate. For PM3-PM8 plates inoculation, which requires an appropriate carbon source, 120 μl of sodium pyruvate (2 M) was added as an additive and mixed with 120 μl of Redox Dye mix D (100×), 880 μl of sterile water and 880 μl of a 85% T-cell suspension in NaCl (0.9%). Then, this solution was added to 10 ml of IF-0 (1.2×), resulting in a final volume of 12 ml per plate. For PM9-PM20 plates inoculation, 120 μl of Redox Dye mix D (100×), 1 ml of sterile water and 880 μl of 1:200 dilution of the 85% T-cell suspension in NaCl (0.9%) were added to 10 ml of IF-10 (1.2×), resulting in a final volume of 12 ml per plate. Each plate was inoculated with 100 μl per well of the respective prepared solution and then sealed using Breathe-Easy® sealing membranes (Sigma-Aldrich). Plates were then transferred to a Biostack Microplate Stacker (Agilent) to process several plates per day and incubated at 37°C using a Memmert IF750 (Memmert). OD590, which indicates the reduction of the tetrazolium dye, was measured every 20 min during 24 h using a Biotek Synergy HTX plate reader (Agilent). Experiments were conducted in duplicate and growth characteristics were determined by calculating the AUC using MATLAB, version R2022a (Mathworks).
Antibiotic persistence assay
Three colonies of V. cholerae WT and ΔSI were inoculated in LB liquid medium and grown overnight at 37°C. Overnight cultures were diluted 1:100 in 1 ml of fresh LB medium in 24-well plates and grown at 37°C on 150 rpm shaker for 3 h. After 3 h incubation, cultures were challenged individually with 10-fold minimum inhibitory concentration (MIC) of ciprofloxacin (MIC = 0008 μg/ml) or ampicillin (MIC = 16 μg/ml) and incubated for 6 h at 37°C on 150 rpm shaker. The CFUs/ml were enumerated at times 0 h (before adding the antibiotics), 2, 4 and 6 h by serially diluting the cells and plating on LB agar.
Resistance to protozoan predation
Tetrahymena elliotti strain 4EA was routinely cultured at 28°C in 2% proteose peptone supplemented with 10 μM FeCl3 and 250 μg/ml of both streptomycin sulfate and penicillin G. Log-phase V. cholerae bacteria (OD600= 0.6) and ciliates (3 × 105 cells/ml) were washed and suspended in 0.01 M Tris-HCl pH 7.5 buffer. Co-cultures were established by combining ciliates and bacteria (1:200 ratio) in a 96-well plate (Costar) for up to 48 h at 28°C. Control bacteria were incubated in 0.01 M Tris-HCl pH 7.5 buffer in the absence of ciliates for the same period. Every 24 h, intracellular bacteria were released by lysing protozoan cells with 1% Triton X-100. A number of viable bacteria were quantified by CFU counts on LB agar plates.
Acanthamoeba castellanii strain Neff was routinely propagated at 30°C in peptone-yeast-glucose (PYG) medium (Pronadisa) supplemented with 250 μg/ml of both streptomycin sulfate and penicillin G. For grazing resistance experiments, 24 h old amoebae were washed in AC buffer (4 mM MgSO4·7H2O, 0.4 mM CaCl2, 3.4 mM sodium citrate dihydrate, 0.05 mM Fe(NH4)2(SO4)2·6H2O, 0.05 mM Na2HPO4·7H2O, 2.5 mM KH2PO4, 0.05 mM NH4Cl, pH 6.5) and seeded at a concentration of 1 × 105 cell/well in 96-well tissue culture plate (Costar). Amoebae were allowed to adhere to the bottom of the wells for 1 h before addition of bacteria. Log-phase V. cholerae cells (OD600= 0.6) were washed in AC buffer an added to the amoebae at a multiplicity of infection (MOI) of 500. Plates were centrifuged at 500 × g for 10 min to synchronize bacterial uptake. Co-cultures were incubated statically at 30°C for up to 48 h. Control bacteria were incubated in AC buffer in the absence of amoebae for the same period. Every 24 h, intracellular bacteria were released by lysing protozoan cells with 1% Triton X-100. The number of viable bacteria were quantified by CFU counts on LB agar plates.
Caenorhabditis elegans virulence assay
Caenorhabditis elegans N2 Bristol individuals were routinely maintained at 20°C on potato dextrose agar (PDA; Sigma-Aldrich) plates seeded with E. coli OP50. Overnight LB cultures of the V. cholerae strains and E. coli OP50 were diluted 1:100 and grown at 37°C until an OD600 of 0.8 was reached. Then, 50 μl from each strain were spread on 6-cm-diameter plates containing PDA medium. Plates were incubated for 16–20 h at 37°C to form a bacterial lawn and 50 L4-stage hermaphrodite individuals were then placed on the PDA plates and incubated at 20°C. Live C. elegans were scored every 48 h for 8 days. An individual was considered dead when it no longer responded to touch. E. coli OP50 was used as a negative control of virulence. At least, three independent experiments were performed.
Biofilm formation assay
Overnight cultures were diluted 1:100 in fresh LB medium. Hundred microliters were transferred to into 96-well Serocluster™ plates (Costar) and incubated at 37°C for 20 h without shaking. After incubation, all the wells were washed with sterile distilled water for three times and dried. Biofilm mass was stained with 125 μl cristal violet (1%) per well and incubated for 15 min at room temperature. Cristal violet was removed by performing three washes with sterile distilled water. The stained biofilm mass was then detached from the wells using acetic acid (30%) and transferred to a new 96-well plate (Nunc, Thermo Scientific). OD570 was measured using a Biotek Synergy HTX plate reader (Agilent). At least 20 independent experiments were performed per strain. One-way analysis of variance (ANOVA) was used to determine significant differences among the three groups.
Swarming motility assay
Swarming motility assays were performed as described in (47). Briefly, 3 μl of overnight cultures were soaked on motility agar plates containing 10 g/l tryptone, 5 g/l NaCl and 0,25% bacto-agar. The swarming motility halos were measured and compared after growth at 37°C for 6 h. At least three independent experiments were performed per strain. One-way ANOVA was used to determine significant differences among the three groups.
Results
Genomic position of the SI does not alter V. cholerae growth
SCIs are present in all Vibrio species, suggesting their acquisition before the genus radiation (1). Despite their size, the genomic location of SCIs in Vibriospecies is remarkably plastic: they can be found in the same position as the V. cholerae SI but also in other unrelated locations, e.g. in the chromosome 1 (48). However, the orientation is usually conserved: the bottom strands of attC sites are usually carried by the leading strand template (33) to avoid the loss of cassettes (23).
We ignore whether SCI relocations in different species have been innocuous for the cell or have entailed complex changes that required subsequent adaptation. To test this, here we used the Φ HK recombination machinery to relocate the V. cholerae SI. We moved it from the right arm of chromosome 2—close to the replication terminus—to chromosome 1, near the origin of replication (between VC0018 and VC10019), where we placed it in both orientations (Figure 2A and Supplementary Figure S1). This relocation could have a potential impact on cell growth because of the higher gene dosage effect near the Ori of chromosome 1. Additionally, it could also generate conflicts between replication and transcription machineries depending on the orientation of the array. We measured growth of both mutants and could not observe differences with the WT (Figure 2B), suggesting that SCIs are genetically independent units of the genome.
SeqDelTA, a tool to erase chromosomal integrons
To further assess the level of genetic and functional independence, we sought to delete the SI. The deletion of the SI in V. cholerae is a longstanding milestone in the field that should allow to deliver the kind of experiments that have advanced the field in the last years using the Class 1 integron as a model. Previous efforts to delete the SI were unfruitful because a one-step deletion approach was used in the presence of—at that time—unknown TA systems. Post-segregational killing due to these uncharacterized TAs made the deletion impossible. The discovery and characterization of these TA systems (19,20), has enabled the development of SeqDelTA, a multi-step approach that allows the sequential inactivation of TA systems while deleting, at each step, the cassette cargo between two TAs (Figure 3).
The SeqDelTA approach exploits the natural competence of V. cholerae to generate a series of sequential allelic replacements. To do so, we introduce linear DNA fragments containing a resistance marker flanked by left and right homology regions (LHR and RHR). At each step, these homology regions correspond to specific sequences, allowing the deletion of SI fragments. The SI spans from gene VCA0291 (the integrase) to VCA0506 (nucleotides 309,750—435,034). As LHR, we generally targeted the 5′ region of catB9 chloramphenicol resistance cassette at the ninth position of the array (VCA0300). This choice was initially made to avoid interference with expression from the PC promoter, since the strain is susceptible to chloramphenicol, although we have recently shown that this gene is transcriptionally active (18). In contrast, RHRs vary at each step of the deletion process, allowing the deletion of an SI fragment spanning from the LHR until the next TA system in the array. They are specifically designed to inactivate the toxin in the TA while keeping the antitoxin gene intact, ensuring the viability of the replacement. Sequential deletions facilitate the removal of the resistance marker from the previous step, allowing for easy alternation of resistance markers. This scheme for SeqDelTA was generally kept constant. In the cases in which the orientation and/or the order of the genes in the TA system were not favorable (e.g. the phd/parE TA system where the toxin is positioned after the anti-toxin [see Figure 1B]), we used alternative LHRs and maintained two resistance markers in consecutive allelic replacements. Then, a third replacement allowed the removal of both markers. After each allelic exchange, several colonies growing on the appropriate antibiotic were verified through PCR and checked phenotypically for the loss of the previous marker where applicable. The growth curve of PCR-verified clones was then analyzed to avoid the hitchhiking of mutations with important deleterious effects. A clone without growth defects was chosen to continue the deletion process. After the last allelic replacement, we had erased all cassettes from VCA0300 (catB9) to VCA0503 (the last toxin). We then used a counter-selectable integrative vector (pMP7 (35)) to deliver the scarless deletion of the remainder of the SI, including the integrase, the first 8 and the last 2 cassettes from the array, as well as a transposase inserted immediately downstream the last cassette (VCA0291 to VCA0508, see Figure 1B). A PCR from the borders of the integron confirmed the deletion of the SI.
Whole genome sequencing (WGS) and marker frequency analysis (MFA)
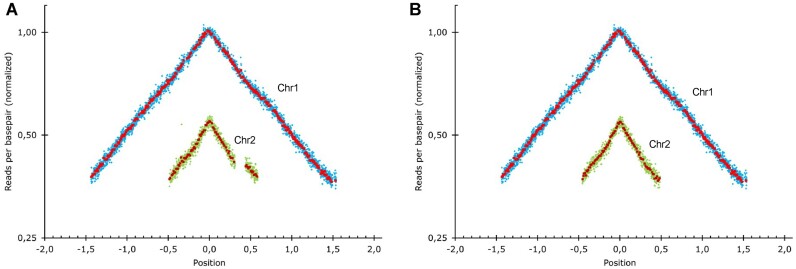
We performed WGS on the obtained V. cholerae ΔSI using Illumina short reads and confirmed the scarless deletion of the SI. We also used this approach to perform MFA, a technique that allows to assess the dynamics of chromosome replication. MFA compares the abundance of reads across the genome between exponential and stationary growth conditions. In fast growing bacteria as V. cholerae, this comparison reveals the gene dosage gradient from the origin to the terminus of replication as an inverted ‘V’. MFA allows to observe the relative timing of replication of both chromosomes, and the speed of replication across the genome (37). As shown in Figure 4, no major alterations in chromosome replication dynamics could be observed, compared to the WT strain.
Figure 4.
Marker frequency analysis of the two chromosomes of V. cholerae ΔSI mapped against V. cholerae WT genome (A) and against V. cholerae ΔSI genome (B). The gap on chromosome 2 in panel (A) reflects the SIs deletion. The genome position is represented relative to the oriC (set to 0). Blue and green rhombus represent the average of 1 kbp windows, and red and garnet squares the average of 10 kbp windows in Chr1 and Chr 2, respectively.
MFA can reveal the presence of large chromosomal rearrangements (such as inversions), through inconsistencies in the slope of the inverted V shape, but lacks resolution for smaller rearrangements like IS movements. We therefore sequenced V. cholerae ΔSI using MinION long reads to detect the presence of any rearrangement that could go undetected by the assembly of short reads. The combination of small and long reads allowed us to obtain a high-quality genome sequence in which we could rule out genomic rearrangements while detecting the presence of unintended single-nucleotide polymorphisms (SNPs). The V. cholerae ΔSI strain bore three unintended mutations in rocS (VC0653), cry2 (VC01392) and rpoS (VC0534) genes. RocS is a diguanylate cyclase involved in the smooth-to-rugose switch in our strain. The ΔSI strain had a single base deletion that altered the reading frame of rocS. We could observe the loss of motility and increase in biofilm formation described in rocS− mutants (49,50). We were able to map the emergence of this mutation to the third deletion step of SeqDelTA. Interestingly, 12 clones from two independent experiments had been kept from this deletion step, of which 11 had mutations in rocS. We found eight different types of mutations among them, pointing to an unusually high mutation rate. Indeed, the sequence of rocS is rich in homopolymeric tracts and is likely to act as a contingency locus, allowing for a frequent smooth-to-rugose switch (51). cry2 presented a non-synonymous (Pro129Lys) mutation that appeared in the 12th step. This gene encodes a protein annotated as a photolyase, for which no experimental evidence of its function could be found. RpoS is an alternative sigma factor related to stress response and entry in stationary phase. It is a major regulator of cell physiology and influences many relevant phenotypes, such as oxidative stress, natural competence, motility or colonization (52–54). In the last deletion step 16, V. cholerae ΔSI acquired a single-base insertion in rpoS giving rise to a frameshift.
To understand the impact of the deletion of the SI in the bacterium’s physiology we need to avoid the interference of the mutations in these three genes. To do so, we restored the WT sequence of the three alleles using the counter-selectable suicide vector pMP7. We then verified the restoration of the three genes, as well as the absence of other mutations arising during this process again through WGS using short and long reads. As a result, we could confirm that the ΔSI strain is genetically identical to the WT except for the absence of the SI and the loss of one of the two copies of satellite phage RS1.
Transcriptomics
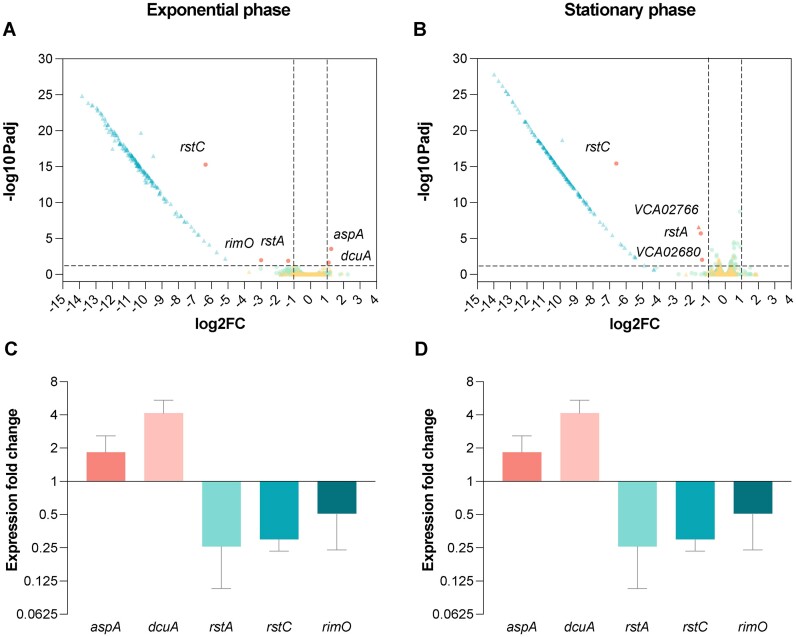
Regulatory networks are generally finely tuned and often affected by minor genetic changes. To address the level of functional isolation of the SI, we sought to assess the changes in expression patterns of the genome in the absence of the SI. Indeed, the deletion of the SI could have pleiotropic effects through direct and indirect links to regulatory networks within the cell. To unveil such effects, we have performed transcriptomic analysis of the ΔSI and the parental strain in exponential and stationary phase (Figure 5A and B). As expected, all genes contained within the SI gave a clear signal of decreased expression. Unexpectedly, the transcriptome of the rest of the genome remained fundamentally unaltered, with few exceptions that were confirmed through RT-qPCR using independent RNA extractions.
Figure 5.
(A) Volcano plots showing gene expression measured by RNA-seq in V. cholerae ΔSI in comparison to the WT strain in exponential and (B) stationary growth phase. Green dots represent chromosome 1 genes; yellow triangles represent chromosome 2 genes, while blue triangles represent those genes from the SI; red dots or triangles represent downregulated or upregulated genes from chromosome 1 or 2, respectively. The vertical dashed lines indicate the log2 fold change (FC) cutoffs, and the horizontal dashed line indicates the threshold of the Padj value (<0.05). (C) Validation of the DEGs by RT-qPCR during exponential and (D) stationary growth phase. Differential expression values represent the fold change in gene expression compared to the WT strain. Error bars indicate standard deviation of two biological replicates with three technical replicates each.
The loss of one of the tandem copies of Φ RS1 in the ΔSI strain (either due to homologous recombination between both copies or to the excision of one copy of the prophage) led to the downregulation in both exponential growth and stationary phase of rstA (VC01411) and rstC (VC01409) that was confirmed through RT-qPCR on independent samples. In the WT strain, rstA is found three times as part of the Φ CTX (VC01397) and as part of the two adjacent copies of the satellite phage Φ RS1 (VC01407, VC01411), while rstC is exclusively found in the latter two copies (VC01409, VC01413) (Supplementary Figure S2). This downregulation through the loss of a Φ RS1 copy, a common variable region in V. cholerae strains (55,56), serves as an additional validation of transcriptomic results.
Besides prophage-related genes, in exponential growth phase we also found a 2-fold upregulation of aspA (VC00205; aspartate ammonia-lyase) and dcuA (VC00204; C4-dicarboxilase transporter) and a 6-fold downregulation of rimO (VC00523; ribosomal protein S12 methylthiotransferase). aspA and dcuA colocalize in V. cholerae chromosome 1. While aspA catalyzes the reversible conversion of L-aspartate to fumarate (57), dcuA is involved in the L-aspartate/fumarate antiport (58). Additionally, RNA-seq data showed genes VC02680 and VCA02766 to be downregulated 2-fold approximately in stationary phase, but RT-qPCR failed to confirm such downregulation (Figure 5C and D).
A GSEA can reveal the collective up- or downregulation of related genes, even if individually they show no significant changes in expression patterns. GSEA was performed here on the group of DEGs, revealing that the ΔSI strain showed an enrichment of the iron ion transmembrane transport process (GO:0034755; NES = 2,126432; Padj = 0.00714409) and the arginine biosynthetic process (GO:0006526; NES = 1 959 273; Padj = 0.02539212) in exponential phase and of isoleucine biosynthetic process in stationary phase (GO: 0009097; NES = 2 020 381; Padj = 0.0129091). However, neither aspA, dcuA, nor rimO are annotated with GO terms belonging to these biological processes. Collectively, these data indicate that deleting the SI does not have important consequences at the transcriptome level in V. cholerae.
Phenotype search
The working model of integrons suggested that most of the array of the SI should be silent, and only the cassettes closest to the Pc are expressed (see Figure 1A). Hence, it would be unlikely to find many phenotypic changes after the deletion of the SI. Yet, this seems not to be true, with independent reports showing both the presence of transcription start sites scattered along the SI and a large proportion of cassettes expressed at biologically relevant levels (18), in accordance with our RNA-seq results (59). To address the potential emergence of phenotypic changes and, therefore, the discovery of cassette functions, we implemented a broad approach to characterize a maximum of phenotypes.
Growth and Fitness
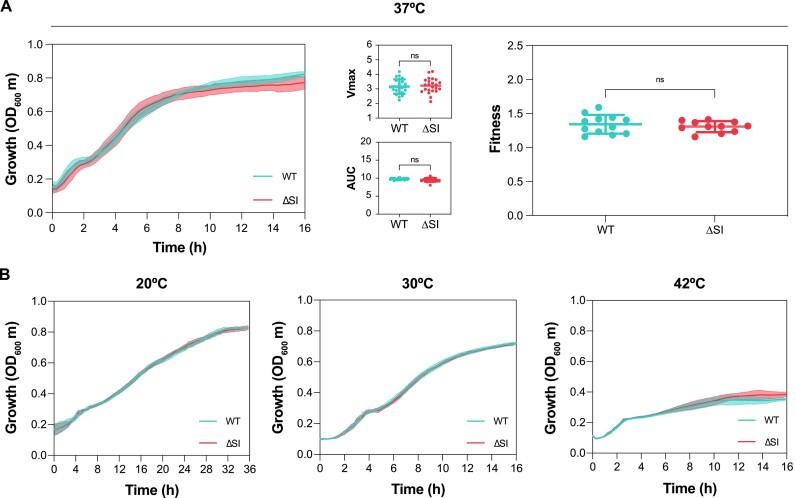
The deletion of V. cholerae SI implies the loss of 3% of the bacterial genome. Thus, it is expected that elimination of this genomic load would result in some growth and/or fitness consequences. To explore this possibility, we performed growth curves of 24 independent V. cholerae ΔSI colonies and compared with the WT in rich LB medium. As shown in Figure 6A, growth curves of both strains present similar shapes after 16 h of growth. We did not observe significant growth differences between strains measured as Vmax. We then measured the AUC, which combines more information about the growth curve, such as lag phase, Vmax, and the carrying capacity. The results showed similar AUC values for both strains. To confirm that the deletion of the SI does not influence bacterial fitness, we increased the sensitivity by performing competition assays using flow cytometry. Using a fluorescent E. coli strain as a competitor, we found no significant differences between the fitness values given by V. cholerae WT and ΔSI (Figure 6A). We hypothesized that this lack of cost could be the serendipitous net result of deleting costly and beneficial cassettes. To test this, we performed the same competitions using intermediate deletion strains (previously corrected for unintended point mutations). Again, we saw no significant differences between the fitness of the parental and these intermediate deletion strains (Supplementary Figure S3) supporting the lack of cost of the SI.
Figure 6.
(A) Growth curves of V. cholerae WT (blue) and ΔSI (red) strains in LB at 37°C. Growth parameters including Vmax and the AUC were extracted from growth curves in LB by measuring the optical density at 600 nm (OD600). Values correspond to the measurement of 24 independent colonies. Relative fitness of V. cholerae WT and ΔSI strains compared with E. coli DH5α PcS::gfp was performed in LB by inoculating cells at a ratio of 1:1. Fitness values were determined from 12 independent experiments by flow cytometry. The P-values were calculated by comparing each measure with that of the WT strain using an unpaired t-test. Ns: not significant. (B) Growth curves of V. cholerae WT (blue) and ΔSI (red) strains in LB at 20, 30 and 42°C, respectively. Values correspond to the measurement of 10 independent colonies.
V. cholerae is known to thrive in a variety of environments different from the human host. Its natural setting is brackish or marine water, where temperature is lower (12–30°C versus 37°C). To test the impact of deleting the SI in these conditions, we performed growth curves at 20 and 30°C. We also included 42°C as the heat-shock response could be induced, and the SI could pose an adaptive advantage in such conditions. We show that the growth of the WT and ΔSI strains is similar at all temperatures (Figure 6B). Altogether, these results indicate that deletion of the V. cholerae SI does not significantly affect bacterial growth.
Biolog Phenotype Microarrays
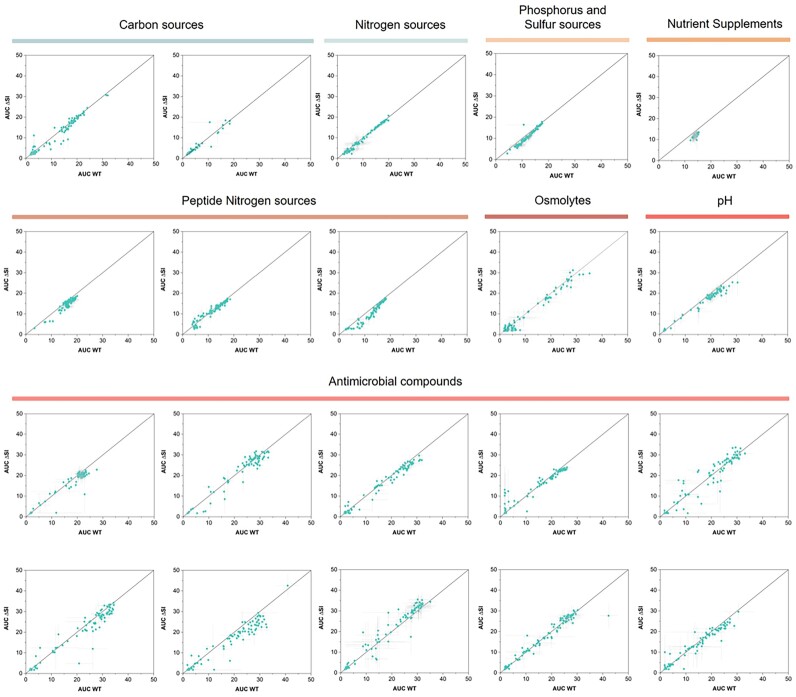
To address if we can observe phenotypic differences in the absence of the SI, we have used Biolog Phenotype Microarrays® (PMs), a phenomics technology that allows the testing of bacterial respiration and growth in nearly 2000 conditions (46). PMs test carbon utilization, nitrogen sources, phosphorus and sulfur compounds, biosynthetic pathways, osmotic, ionic and pH effects, as well as a broad set of chemicals and antibiotic compounds at four different concentrations. By using tetrazolium violet as a redox dye in the mixture, we monitor bacterial respiration for 24 h and compare the metabolism of two independent replicates of the WT and ΔSI strain in all the available conditions (Supplementary Figure S4).
To search for differences between the two strains, the mean AUCs given by both replicates of WT and ΔSI in a specific compound were graphically compared using Matlab software (R2022a). In Figure 7, we show the correlation of AUC values for WT and ΔSI strains in each condition. Distance to the origin of the axis represents metabolic activity, and deviations from the diagonal represent differences between strains. Generally, dots fall within the diagonal, showing that both strains behave similarly in the presence of the different compounds, which also supports the similarity in growth rates across a broad variety of conditions. It is of note, that the ΔSI shows a potential decrease in respiration when using dipeptides and tripeptides as nitrogen sources (PM6 and PM8) (see ‘Discussion’ section). Within antimicrobial-containing plates (PM11–20), we generally did not found differences between the WT and ΔSI. However, we have highlighted some isolated cases in Figure 7 where the AUC of one of the strains is at least the double of the other and the standard deviation does not cross the bisector. These compounds are ceftriaxone (G4, PM11), cadmium chloride (D4, PM14), methyl viologen (E12, PM15), norfloxacin (B3, PM16), tannic acid (F10, PM17), sodium metasilicate (E4, PM18), iodonitro tetrazolium violet (D8, PM19), thioridazine (C1, PM20) and benserazide (A10, PM20). Despite the lack of consistent results with other molecules of the same family, we believed these results deserved further verification. We selected three compounds (ceftriaxone, norfloxacin and tannic acid) and measured the MIC but found identical values for both strains: norfloxacin 50 ng/ml; ceftriaxone 25 ng/ml; tannic acid 250 ng/ml. We also performed growth curves using subinhibitory concentrations of the three compounds in MH but could not observe the same phenotype previously obtained using the phenotype microarrays (Supplementary Figure S5).
Figure 7.
Comparison of AUC values of V. cholerae WT and ΔSI in different substrates given by Biolog Phenotype Microarrays (PM1–PM20). The AUC values are the mean of two biological replicates for each strain. Error bars indicate the standard error. The points that are above the bisector indicate those conditions where the absence of the SI is beneficial for the bacteria, while those points that lay under the bisector indicate the conditions where the presence of the SI is beneficial for the bacteria.
Antibiotic persistence, grazing resistance, virulence, biofilm formation, natural competence and swarming motility
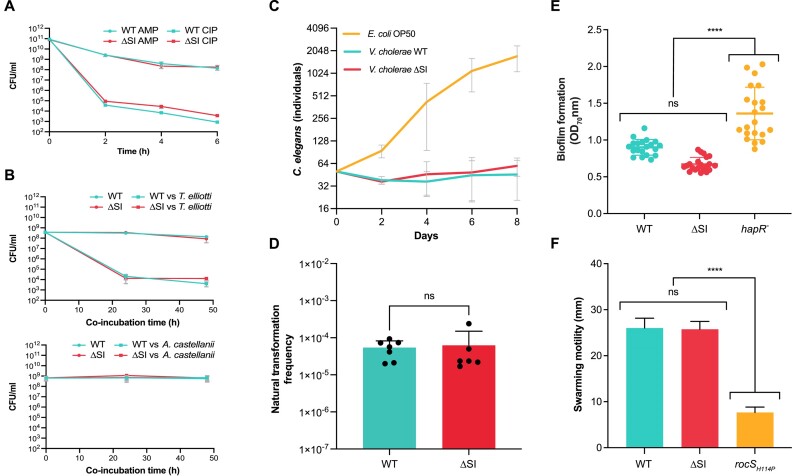
Antibiotic persistence is the ability of a bacterial population (or a part of it) to survive exposure to bactericidal drugs (60), without becoming resistant to the antibiotic. TA systems have been described to play a role in triggering persistence in bacteria by reversibly intoxicating the bacterium and producing a transient arrest of metabolic activity that renders antibiotics ineffective. This role is controversial, at least for some TAs, since several studies were later shown to be biased by the activity of prophages (61). Given the deletion of a variety of TA systems in the ΔSI mutant we decided to test antibiotic persistence upon exposure to high concentrations of antibiotics. We challenged growing cultures of V. cholerae WT and ΔSI to 10-fold the MIC of ampicillin and ciprofloxacin, and viability was measured along a 6 h-period (Figure 8A). Similar decreases in the number of viable cells were observed for both strains, of varying intensities depending on the antibiotic molecule, suggesting that TA systems encoded in the SI do not play a role in antibiotic persistence in V. cholerae.
Figure 8.
(A) Antibiotic persistence assay of V. cholerae WT and ΔSI mutant challenged with 10-fold the MIC of ampicillin (AMP) and ciprofloxacin (CIP), individually. CFUs/ml are plotted against time. Error bars indicate standard deviations from three independent biological replicates. (B) V. cholerae WT and ΔSI survival under predation by T. elliotti (MOI 1:10000) or A. castellanii (MOI 1:500). The number of surviving bacteria, incubated either in the absence (circles) or presence (squares) of ciliates/amoeba, was quantified by CFU counting after 0–48 h of co-incubation. Error bars indicate standard deviations from at least three independent biological replicates. (C) Virulence assay of V. cholerae WT and ΔSI mutant in a C. elegans model. E. coli OP50 was used as a negative control. Error bars indicate standard deviations from three independent biological replicates. (D) Natural competence assay of V. cholerae WT and ΔSI mutant. Transformation frequencies are given on the Y-axis and were calculated as the number of CFUs growing in kanamycin over the total CFUs. Bar charts show the mean ± s.d. from biological replicates (individual dots) from at least three independent experiments. (E) Biofilm formation of V. cholerae WT and ΔSI mutant. A V. cholerae hapR (frameshift) mutant was included as a positive control of biofilm formation. Data from at least 20 independent colonies is represented. (F) Swarming motility assay of V. cholerae WT and ΔSI mutant. A V. cholerae rocS (H114P) mutant was included as a non-motile control strain. Error bars indicate standard deviation of at least three biological replicates. P-values (****) were calculated by pairwise comparison using the one-way ANOVA test (P<0.0001). Ns: not significant.
V. cholerae interacts with various free-living protozoa in aquatic environments, as the model organisms Tetrahymena pyriformis and Acanthamoeba castellanii (62). These interactions can be complex and may impact the survival and infectivity of the pathogen. For instance, it has been reported that T. pyriformis and other ciliates can enhance V. cholerae’s environmental persistence by expelling hyper-infectious forms (63). Other studies indicate that predation by protozoa, particularly ciliates, can eliminate V. cholerae from the environment (64). Additionally, V. cholerae can survive intracellularly within A. castellanii, potentially providing a protected niche (65). Given the relevance of these interactions, we aimed to investigate whether the V. cholerae SI contains factors that may contribute to grazing resistance by protozoa. To assess this, survival assays were conducted by co-incubating V. cholerae WT and ΔSI with the protozoan predators T. elliotti and A. castellanii. Although we observed a different degree of grazing between the ciliates and the amoeba, there were no differences between V. cholerae WT and ΔSI in terms of intracellular survival within both protozoa under the tested conditions (Figure 8B).
V. cholerae expresses well-known virulence factors to colonize and cause infection in the mammalian host, as the cholera toxin or the toxin-coregulated pili (66,67). The nematode C. elegans has been widely used as an invertebrate infection model to screen and identify virulence factors of several human pathogens, including V. cholerae (68,69). TA systems in the SI are reported to be associated with virulence. For instance, the RelBE TA systems of the SI play a role in intestine colonization in a mouse model (70). However, evidence on the implication of other elements in the SI in the virulence of V. cholerae is lacking. To address this question, we performed a C. elegans killing assay. Hermaphrodites L4 worms raised on E. coli OP50 were transferred onto bacterial lawns of WT and ΔSI strains. The number of C. elegans individuals were counted every 48 h for 8 days. E. coli OP50 was used a control. As shown in Figure 8C, we did not observe any differences in lethality between V. cholerae WT and the ΔSI mutant.
Biofilm formation, natural competence, and motility are key behaviors in the lifestyle of V. cholerae, with strong implications for its survival in the environment, pathogenesis, dispersal and genetic variability. Motility is involved in regulating biofilm formation by participating in important processes, such as surface attachment or biofilm dispersal (71). Increased biofilm formation also entails a downregulation of natural competence through quorum sensing control (72,73). We investigated the role of the SI in these processes by measuring natural competence, biofilm formation, and swarming motility. As shown in Figure 8D–F, none of the processes was significantly affected in the ΔSI strain. As a control for biofilm formation, we included a V. cholerae strain with a frameshift mutation in hapR (a regulator of quorum sensing), leading to an increase in biofilm production. For motility, we tested a strain with an amino acid change mutation in rocS as a non-motile control.
Discussion
The working model of integrons is based on semiconservative recombination and the strong expression of cassettes upon integration, highlighting that cassettes must be adaptive to be maintained in the array. This, together with the intimate intertwining between integrons and their hosts (through the SOS response controlling the expression of the integrase (74), or through host factors (26,30)) put integrons at the service of the host to provide adaptation on demand (2,26,75). A clear example of this is their role in the rise and spread of multidrug resistance during the last decades. In Vibrionaceae, integrons provide genetic diversity upon which natural selection can act (48). It is therefore assumed that SCIs provide great adaptability too. Yet, a better understanding of how they do so remains out of reach because the functions encoded in the cassettes of chromosomal integrons are mostly unknown (15). Apart from antibiotic resistance genes and TA systems, only a handful of cassettes have been characterized (for a review, see (76) and (77)).
Chromosomal integrons have co-evolved with their hosts during aeons. It is therefore plausible that hosts have co-evolved with integrons and have adapted to the presence of such massive structures. Also, the functions encoded in cassettes could be intimately linked to the host’s physiology, somehow interfering or modulating housekeeping functions. Indeed, in silico analysis of cassettes in Vibrionaceae revealed potential functions like information storage, cellular processes and metabolism (15). This is certainly at odds with the variable nature of the array of cassettes, which would imply that cassettes provide exclusively ‘self-contained’ or independent functions that do not interfere with those encoded in the rest of the genome. Here, we have tested the hypothesis that SCIs are genetically and functionally independent of the host by relocating and deleting the SI of V. cholerae and testing a broad variety of phenotypes of importance in the lifestyle of this bacterium. The result is surprisingly clear: the ΔSI mutant behaves almost identically to the WT strain in most conditions tested, and differences (like those observed in the use of di-peptides) are, at best, very mild. This tilts the balance towards the ‘plug-and-play’ model of cassette function. Indeed, our data suggest that cassettes that are expressed have been selected to be as neutral as possible. In line with our results, a recent CRISPRi study in V. cholerae shows that guides against the SI are not deleterious, except when they silence the antitoxin genes (78). In this sense, it is possible that SCIs are biased to encode functions that are exclusively activated under specific cues and with little transcriptional and translational demand. A good example of this is the case of TAs. These systems often repress their own expression under normal conditions but can be triggered by infecting phage, delivering their function (abortive infection) without the need for additional RNA or protein synthesis (79). It is of note that coding such low-cost triggerable functions is not conditio sine qua non, since the shuffling ability of integrons can also serve to switch on and off the expression of cassettes. This has been shown for the catB9 cassette in the SI, which can move from ninth to second position, providing antibiotic resistance (80).
Focusing on the subtle differences that we have observed, we note that the deletion of the SI has a limited impact on gene expression, with only three genes (aspA, dcuA and rimO) being mildly affected. aspA and dcuA are involved in nitrogen metabolism. AspA carries out the reversible conversion of L-aspartate to fumarate, releasing ammonia for nitrogen assimilation (81). This enzyme has a specific interaction with DcuA, which functions as an L-aspartate/fumarate antiporter under both aerobic and anaerobic conditions (58). This interaction has been proposed to form a metabolon, where DcuA facilitates the uptake of aspartate, while AspA converts it to fumarate (82). The GSEA analysis revealed an enrichment in arginine and isoleucine biosynthetic processes in the ΔSI strain. Arginine synthesis is derived from glutamate, which, in turn, originates from L-aspartate. Additionally, the precursor molecule for isoleucine biosynthesis is pyruvate, a product of glycolysis, and an indirect byproduct of the aspartate metabolism (83). These findings, coupled with the observation that the ΔSI mutant exhibits impaired growth when utilizing certain dipeptides and tripeptides as nitrogen sources, may suggest a potential connection between these processes. However, it is important to note that these pathways are not directly linked, and further experimental data would be needed to ascertain whether a change in metabolic balance has occurred within the cell. Given the subtle signal from our data, we remain cautious about the biological significance of these observations.
The obtention of a ΔSI mutant of V. cholerae is a long-awaited milestone in the field, unblocking the study of SCIs. Getting rid of the interference of the cassettes within the SI allows to use this strain as a chassis to easily perform many of the experiments that have been delivered in E. coli with the class 1 integron, allowing to enrich the field with data from other integron models. It is also the perfect chassis to unveil cassette functions in a classical in trans approach, indeed one of the key questions that remains unanswered in the field and one with an important biotechnological potential.
Supplementary Material
Acknowledgements
Authors would like to thank Laurence Van Melderen, Nathalie Balaban and David Bikard for helpful discussion, and Claire Vit for technical assistance.
Author contributions: P.B.: Conceptualization, formal analysis, methodology, validation and writing-original draft; F.T.d.R.: Formal analysis, methodology, validation and writing-original draft; L.T.-C.: Methodology, analysis and validation; L.G.-P.: Methodology, analysis and validation; N.C.: Methodology, analysis and validation; F.O.: Methodology, analysis and validation; B.D.: Methodology, analysis and validation; O.S.: Methodology, analysis and validation; Á.S.M.: Formal analysis, writing—review and editing; D.M.: Conceptualization, writing—review and editing. C.L.: Conceptualization, analysis, validation, writing—review and editing; J.A.E.: Conceptualization, methodology, formal analysis, validation, writing—original draft, review and editing.
Contributor Information
Paula Blanco, Molecular Basis of Adaptation, Departamento de Sanidad Animal, Universidad Complutense de Madrid, Madrid 28040, Spain; VISAVET Health Surveillance Centre, Universidad Complutense de Madrid, Madrid 28040, Spain.
Filipa Trigo da Roza, Molecular Basis of Adaptation, Departamento de Sanidad Animal, Universidad Complutense de Madrid, Madrid 28040, Spain; VISAVET Health Surveillance Centre, Universidad Complutense de Madrid, Madrid 28040, Spain.
Laura Toribio-Celestino, Departamento de Microbiología Microbiana, Centro Nacional de Biotecnología–CSIC, Madrid 28049, Spain.
Lucía García-Pastor, Molecular Basis of Adaptation, Departamento de Sanidad Animal, Universidad Complutense de Madrid, Madrid 28040, Spain; VISAVET Health Surveillance Centre, Universidad Complutense de Madrid, Madrid 28040, Spain.
Niccolò Caselli, Departamento de Química Física, Universidad Complutense de Madrid, Madrid 28040, Spain.
Álvaro Morón, Departamento de Genética, Fisiología y Microbiología, Facultad de Ciencias Biológicas, Universidad Complutense de Madrid, Madrid 28040, Spain.
Francisco Ojeda, Molecular Basis of Adaptation, Departamento de Sanidad Animal, Universidad Complutense de Madrid, Madrid 28040, Spain; VISAVET Health Surveillance Centre, Universidad Complutense de Madrid, Madrid 28040, Spain.
Baptiste Darracq, Institut Pasteur, Université Paris Cité, CNRS UMR3525, Unité Plasticité du Génome Bactérien, 75015 Paris, France; Sorbonne Université, ED515, F-75005 Paris, France.
Ester Vergara, Molecular Basis of Adaptation, Departamento de Sanidad Animal, Universidad Complutense de Madrid, Madrid 28040, Spain; VISAVET Health Surveillance Centre, Universidad Complutense de Madrid, Madrid 28040, Spain.
Francisco Amaro, Departamento de Genética, Fisiología y Microbiología, Facultad de Ciencias Biológicas, Universidad Complutense de Madrid, Madrid 28040, Spain.
Álvaro San Millán, Departamento de Microbiología Microbiana, Centro Nacional de Biotecnología–CSIC, Madrid 28049, Spain.
Ole Skovgaard, Department of Science and Environment, Roskilde University, 4000 Roskilde, Denmark.
Didier Mazel, Institut Pasteur, Université Paris Cité, CNRS UMR3525, Unité Plasticité du Génome Bactérien, 75015 Paris, France.
Céline Loot, Institut Pasteur, Université Paris Cité, CNRS UMR3525, Unité Plasticité du Génome Bactérien, 75015 Paris, France.
José Antonio Escudero, Molecular Basis of Adaptation, Departamento de Sanidad Animal, Universidad Complutense de Madrid, Madrid 28040, Spain; VISAVET Health Surveillance Centre, Universidad Complutense de Madrid, Madrid 28040, Spain.
Data availability
The RNA-seq data included in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE247496.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
European Research Council (ERC) [803375-KRYPTONINT; 757440-PLASREVOLUTION] to J.A.E, and L.T.C. and A.S.M.; Ministerio de Ciencia, Innovación y Universidades [BIO2017-85056-P, PID2020-117499RB-100] to J.A.E.; Comunidad de Madrid [2016-T1/BIO-1105, 2020-5A/BIO-19726 to J.A.E.]; Ministerio de Ciencia, Innovación y Universidades [FJC 2020-043017-I] to P.B.; Fundação para Ciência e a Tecnologia [SFRH/BD/144108/2019] to F.T.R.; Centre National de la Recherche Scientifique [CNRS-UMR3525] to B.D., D.M., C.L.; Fondation pour la Recherche Médicale [EQU202103012569] to D.M.. Funding for open access charge: European Research Council Starting [803375].
Conflict of interest statement. Universidad Complutense de Madrid and Institut Pasteur have published a patent covering SeqDelTA and the V. cholerae ΔSI strain B522. Patent reference number: ES 2 970 040 A1. Inventors: Escudero, J.A.; Trigo da Roza, F.; Blanco, P.; Mazel, D. and Lopéz-Igual, R.
References
- 1. Mazel D. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 2006; 4:608–620. [DOI] [PubMed] [Google Scholar]
- 2. Escudero J., Loot C., Nivina A., Mazel D.. The Integron: adaptation on demand. Microbiol. Spectr. 2015; 13:MDNA3–A0019–2014. [DOI] [PubMed] [Google Scholar]
- 3. Stokes H.W., Hall R.M.. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 1989; 3:1669–1683. [DOI] [PubMed] [Google Scholar]
- 4. Souque C., Escudero J.A., Maclean R.C.. Integron activity accelerates the evolution of antibiotic resistance. eLife. 2021; 10:e62474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barraud O., Ploy M.C.. Diversity of class 1 integron gene cassette rearrangements selected under antibiotic pressure. J. Bacteriol. 2015; 197:2171–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitsuhashi S., Harada K., Hashimoto H., Egawa R.. On the drug-resistance of enteric bacteria. 4. Drug-resistance of Shigella prevalent in Japan. Jpn. J. Exp. Med. 1961; 31:47–52. [PubMed] [Google Scholar]
- 7. Gillings M., Boucher Y., Labbate M., Holmes A., Krishnan S., Holley M., Stokes H.W.. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 2008; 190:5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu Y.-G., Gillings M., Simonet P., Stekel D., Banwart S., Penuelas J.. Microbial mass movements. Science. 2017; 357:1099–1100. [DOI] [PubMed] [Google Scholar]
- 9. Hipólito A., García-Pastor L., Blanco P., Trigo da Roza F., Kieffer N., Vergara E., Jové T., Álvarez J., Escudero J.A.. The expression of aminoglycoside resistance genes in integron cassettes is not controlled by riboswitches. Nucleic Acids Res. 2022; 50:8566–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hipólito A., García-Pastor L., Vergara E., Jové T., Escudero J.A.. Profile and resistance levels of 136 integron resistance genes. Npj Antimicrob. Resist. 2023; 1:13. [Google Scholar]
- 11. Rowe-Magnus D.A., Guerout A.M., Ploncard P., Dychinco B., Davies J., Mazel D.. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl Acad. Sci. 2001; 98:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Néron B., Littner E., Haudiquet M., Perrin A., Cury J., Rocha E.P.C.. IntegronFinder 2.0: identification and analysis of integrons across bacteria, with a focus on antibiotic resistance in Klebsiella. Microorganisms. 2022; 10:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cury J., Jové T., Touchon M., Néron B., Rocha E.P.. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016; 44:4539–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clark C.A., Purins L., Kaewrakon P., Focareta T., Manning P.A.. The Vibrio cholerae 01 chromosomal integron. Microbiology. 2000; 146:2605–2612. [DOI] [PubMed] [Google Scholar]
- 15. Boucher Y., Labbate M., Koenig J.E., Stokes H.W.. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 2007; 15:301–309. [DOI] [PubMed] [Google Scholar]
- 16. Ghaly T.M., Penesyan A., Pritchard A., Qi Q., Rajabal V., Tetu S.G., Gillings M.R.. Methods for the targeted sequencing and analysis of integrons and their gene cassettes from complex microbial communities. Microb. Genomics. 2022; 8:000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mazel D., Dychinco B., Webb V.A., Davies J.. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998; 280:605–608. [DOI] [PubMed] [Google Scholar]
- 18. Blanco P., Hipólito A., García-Pastor L., Trigo da Roza F., Toribio-Celestino L., Ortega A.C., Vergara E., San Millán Á., Escudero J.A.. Identification of promoter activity in gene-less cassettes from Vibrionaceae superintegrons. Nucleic Acids Res. 2024; 52:2961–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iqbal N., Guérout A.M., Krin E., Le Roux F., Mazel D.. Comprehensive functional analysis of the 18Vibrio cholerae N16961 toxin-antitoxin systems substantiates their role in stabilizing the superintegron. J. Bacteriol. 2015; 197:2150–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krin E., Baharoglu Z., Sismeiro O., Varet H., Coppée J.-Y., Mazel D.. Systematic transcriptome analysis allows the identification of new type I and type II toxin/antitoxin systems located in the superintegron of Vibrio cholerae. Res. Microbiol. 2023; 174:103997. [DOI] [PubMed] [Google Scholar]
- 21. Rowe-Magnus D.A., Guerout A.M., Mazel D.. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 2002; 43:1657–1669. [DOI] [PubMed] [Google Scholar]
- 22. Szekeres S., Dauti M., Wilde C., Mazel D., Rowe-Magnus D.A.. Chromosomal toxin–antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 2007; 63:1588–1605. [DOI] [PubMed] [Google Scholar]
- 23. Richard E., Darracq B., Littner E., Vit C., Whiteway C., Bos J., Fournes F., Garriss G., Conte V., Lapaillerie D.et al.. Cassette recombination dynamics within chromosomal integrons are regulated by toxin-antitoxin systems. Sci. Adv. 2024; 10:eadj3498. [DOI] [PubMed] [Google Scholar]
- 24. Biskri L., Bouvier M., Guérout A.M., Boisnard S., Mazel D.. Comparative study of class 1 integron and Vibrio cholerae superintegron integrase activities. J. Bacteriol. 2005; 187:1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vit C., Richard E., Fournes F., Whiteway C., Eyer X., Lapaillerie D., Parissi V., Mazel D., Loot C.. Cassette recruitment in the chromosomal Integron of Vibrio cholerae. Nucleic Acids Res. 2021; 49:5654–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richard E., Darracq B., Loot C., Mazel D.. Unbridled integrons: a matter of host factors. Cells. 2022; 11:925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rowe-Magnus D.A., Mazel D.. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 2001; 4:565–569. [DOI] [PubMed] [Google Scholar]
- 28. Escudero J.A., Loot C., Parissi V., Nivina A., Bouchier C., Mazel D.. Unmasking the ancestral activity of integron integrases reveals a smooth evolutionary transition during functional innovation. Nat. Commun. 2016; 7:10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sawabe T., Kita-Tsukamoto K., Thompson F.L.. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 2007; 189:7932–7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loot C., Bikard D., Rachlin A., Mazel D.. Cellular pathways controlling integron cassette site folding. EMBO J. 2010; 29:2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loot C., Parissi V., Escudero J.A., Amarir-Bouhram J., Bikard D., Mazel D.. The integron integrase efficiently prevents the melting effect of Escherichia coli single-stranded DNA-binding protein on folded attC sites. J. Bacteriol. 2014; 196:762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grieb M.S., Nivina A., Cheeseman B.L., Hartmann A., Mazel D., Schlierf M.. Dynamic stepwise opening of integron attC DNA hairpins by SSB prevents toxicity and ensures functionality. Nucleic Acids Res. 2017; 45:10555–10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loot C., Nivina A., Cury J., Escudero J.A., Ducos-Galand M., Bikard D., Rocha E.P.C., Mazel D.. Differences in integron cassette excision dynamics shape a trade-off between evolvability and genetic capacitance. mBio. 2017; 8:e02296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bland M.J., Ducos-Galand M., Val M.-E., Mazel D.. An att site-based recombination reporter system for genome engineering and synthetic DNA assembly. BMC Biotechnol. 2017; 17:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Val M.-E., Skovgaard O., Ducos-Galand M., Bland M.J., Mazel D.. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet. 2012; 8:e1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marvig R.L., Blokesch M.. Natural transformation of Vibrio cholerae as a tool Optimizing the procedure. BMC Microbiol. 2010; 10:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skovgaard O., Bak M., Løbner-Olesen A., Tommerup N.. Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res. 2011; 21:1388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 40. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013; arXiv doi:16 March 2013, preprint: not peer reviewedhttps://arxiv.org/abs/1303.3997.
- 41. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 42. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bateman A., Martin M.-J., Orchard S., Magrane M., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., Bye-A-Jee H., Cukura A.et al.. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023; 51:D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L.et al.. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. The Innovation. 2021; 2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Herencias C., Rodríguez-Beltrán J., León-Sampedro R., Valle A.A.D., Palkovičová J., Cantón R., Millán Á.S.. Collateral sensitivity associated with antibiotic resistance plasmids. eLife. 2021; 10:e65130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bochner B.R., Gadzinski P., Panomitros E.. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001; 11:1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. García-Pastor L., Sánchez-Romero M.A., Gutiérrez G., Puerta-Fernández E., Casadesús J.. Formation of phenotypic lineages in Salmonella enterica by a pleiotropic fimbrial switch. PLoS Genet. 2018; 14:e1007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rowe-Magnus D.A., Guerout A.M., Biskri L., Bouige P., Mazel D.. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 2003; 13:428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rashid M.H., Rajanna C., Ali A., Karaolis D.K.R.. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 2003; 227:113–119. [DOI] [PubMed] [Google Scholar]
- 50. Lim B., Beyhan S., Meir J., Yildiz F.H.. Cyclic-diGMP signal transduction systems in Vibrio cholerae: Modulation of rugosity and biofilm formation. Mol. Microbiol. 2006; 60:331–348. [DOI] [PubMed] [Google Scholar]
- 51. Moxon R., Bayliss C., Hood D.. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 2006; 40:307–333. [DOI] [PubMed] [Google Scholar]
- 52. Wölflingseder M., Tutz S., Fengler V.H., Schild S., Reidl J.. Regulatory interplay of RpoS and RssB controls motility and colonization in Vibrio cholerae. Int. J. Med. Microbiol. 2022; 312:151555. [DOI] [PubMed] [Google Scholar]
- 53. Dalia A.B. RpoS is required for natural transformation of Vibrio cholerae through regulation of chitinases. Environ. Microbiol. 2016; 18:3758–3767. [DOI] [PubMed] [Google Scholar]
- 54. Baharoglu Z., Krin E., Mazel D.. RpoS plays a central role in the SOS induction by sub-lethal aminoglycoside concentrations in Vibrio cholerae. PLoS Genet. 2013; 9:e1003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee J.H., Han K.H., Choi S.Y., Lucas M.E.S., Mondlane C., Ansaruzzaman M., Nair G.B., Sack D.A., Seidlein L.V., Clemens J.D.et al.. Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J. Med. Microbiol. 2006; 55:165–170. [DOI] [PubMed] [Google Scholar]
- 56. Kamruzzaman M., Robins W.P., Bari S.M.N., Nahar S., Mekalanos J.J., Faruque S.M.. RS1 satellite phage promotes diversity of toxigenic Vibrio cholerae by driving CTX prophage loss and elimination of lysogenic immunity. Infect. Immun. 2014; 82:3636–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Falzone C.J., Karsten W.E., Conley J.D., Viola R.E.. L-Aspartase from Escherichia coli: substrate specificity and role of divalent metal ions. Biochemistry. 1988; 27:9089–9093. [DOI] [PubMed] [Google Scholar]
- 58. Strecker A., Schubert C., Zedler S., Steinmetz P., Unden G.. DcuA of aerobically grown Escherichia coli serves as a nitrogen shuttle (L-aspartate/fumarate) for nitrogen uptake. Mol. Microbiol. 2018; 109:801–811. [DOI] [PubMed] [Google Scholar]
- 59. Krin E., Pierlé S.A., Sismeiro O., Jagla B., Dillies M.A., Varet H., Irazoki O., Campoy S., Rouy Z., Cruveiller S.et al.. Expansion of the SOS regulon of Vibrio cholerae through extensive transcriptome analysis and experimental validation. BMC Genomics. 2018; 19:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Balaban N.Q., Helaine S., Lewis K., Ackermann M., Aldridge B., Andersson D.I., Brynildsen M.P., Bumann D., Camilli A., Collins J.J.et al.. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019; 17:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Harms A., Fino C., Sørensen M.A., Semsey S., Gerdes K.. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio. 2017; 8:e01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Noorian P., Hu J., Chen Z., Kjelleberg S., Wilkins M.R., Sun S., McDougald D.. Pyomelanin produced by Vibrio cholerae confers resistance to predation by Acanthamoeba castellanii. FEMS Microbiol. Ecol. 2017; 93:fix147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Espinoza-Vergara G., Noorian P., Silva-Valenzuela C.A., Raymond B.B.A., Allen C., Hoque M.M., Sun S., Johnson M.S., Pernice M., Kjelleberg S.et al.. Vibrio cholerae residing in food vacuoles expelled by protozoa are more infectious in vivo. Nat. Microbiol. 2019; 4:2466–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pérez M.E.M., Macek M., Galván M.T.C.. Do protozoa control the elimination of Vibrio cholerae in brackish water?. Int. Rev. Hydrobiol. 2004; 89:215–227. [Google Scholar]
- 65. Henst C.V.D., Scrignari T., Maclachlan C., Blokesch M.. An intracellular replication niche for Vibrio cholerae in the amoeba Acanthamoeba castellanii. ISME J. 2016; 10:897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhu J., Miller M.B., Vance R.E., Dziejman M., Bassler B.L., Mekalanos J.J.. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl Acad. Sci. USA. 2002; 99:3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ho L.S., Hava D.L., Waldor M.K., Camilli A.. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999; 99:625–634. [DOI] [PubMed] [Google Scholar]
- 68. Vaitkevicius K., Lindmark B., Ou G., Song T., Toma C., Iwanaga M., Zhu J., Andersson A., Hammarström M.L., Tuck S.et al.. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl Acad. Sci. USA. 2006; 103:9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sahu S.N., Lewis J., Patel I., Bozdag S., Lee J.H., LeClerc J.E., Cinar H.N.. Genomic analysis of immune response against Vibrio cholerae hemolysin in Caenorhabditis elegans. PLoS One. 2012; 7:e38200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang Y., Wang H., Hay A.J., Zhong Z., Zhu J., Kan B.. Functional RelBE-family toxin-antitoxin pairs affect biofilm maturation and intestine colonization in Vibrio cholerae. PLoS One. 2015; 10:e0135696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Teschler J.K., Nadell C.D., Drescher K., Yildiz F.H.. Mechanisms underlying Vibrio choleraebiofilm formation and dispersion. Annu. Rev. Microbiol. 2022; 76:503–532. [DOI] [PubMed] [Google Scholar]
- 72. Hammer B.K., Bassler B.L.. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 2003; 50:101–104. [DOI] [PubMed] [Google Scholar]
- 73. Suckow G., Seitz P., Blokesch M.. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J. Bacteriol. 2011; 193:4914–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guerin É., Cambray G., Sanchez-Alberola N., Campoy S., Erill I., Da Re S., Gonzalez-Zorn B., Barbé J., Ploy M.C., Mazel D.. The SOS response controls integron recombination. Science. 2009; 324:1034. [DOI] [PubMed] [Google Scholar]
- 75. Fonseca É.L., Vicente A.C.. Integron functionality and genome innovation: an update on the subtle and smart strategy of integrase and gene cassette expression regulation. Microorganisms. 2022; 10:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Escudero J.A., Mazel D.. Genomic plasticity of Vibrio cholerae. Int. Microbiol. 2017; 20:138–148. [DOI] [PubMed] [Google Scholar]
- 77. Rapa R.A., Labbate M.. The function of integron-associated gene cassettes in Vibrio species: The tip of the iceberg. Front. Microbiol. 2013; 4:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Debatisse K., Niault T., Peeters S., Maire A., Darracq B., Baharoglu Z., Bikard D., Mazel D., Loot C.. Fine-tuning of a CRISPRi screen in the seventh pandemic Vibrio cholerae. 2024; bioRxiv doi:03 July 2024, preprint: not peer reviewed 10.1101/2024.07.03.601881. [DOI] [PMC free article] [PubMed]
- 79. Laub M.T., Typas A.. Principles of bacterial innate immunity against viruses. Curr. Opin. Immunol. 2024; 89:102445. [DOI] [PubMed] [Google Scholar]
- 80. Baharoglu Z., Bikard D., Mazel D.. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 2010; 6:e1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rudolph F.B., Fromm H.J.. The purification and properties of aspartase from Escherichia coli. Arch. Biochem. Biophys. 1971; 147:92–98. [DOI] [PubMed] [Google Scholar]
- 82. Schubert C., Kim N.Y., Unden G., Kim O.B.. C4-dicarboxylate metabolons: interaction of C4-dicarboxylate transporters of Escherichia coli with cytosolic enzymes. FEMS Microbiol. Lett. 2022; 369:fnac078. [DOI] [PubMed] [Google Scholar]
- 83. Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data included in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE247496.