Abstract
ABSTRACT
Introduction
There are no established methods to identify children with atypical diabetes for further study. We aimed to develop strategies to systematically ascertain cases of atypical pediatric diabetes using electronic medical records (EMR).
Research design and methods
We tested two strategies in a large pediatric hospital in the USA. Strategy 1: we designed a questionnaire to rule out typical diabetes and applied it to the EMR of 100 youth with diabetes. Strategy 2: we built three electronic queries to generate reports of three atypical pediatric diabetes phenotypes: unknown type, type 2 diabetes (T2D) diagnosed <10 years old and autoantibody-negative type 1 diabetes (AbNegT1D).
Results
Strategy 1 identified six cases (6%) of atypical diabetes (mean diagnosis age=11±2.6 years, 16.6% men, 33% non-Hispanic white (NHW) and 66.6% Hispanic). Strategy 2: unknown diabetes type: n=68 (1%) out of 6676 patients with diabetes; mean diagnosis age=12.6±3.3 years, 32.8% men, 23.8% NHW, 47.6% Hispanic, 25.4% African American (AA), 3.2% other. T2D <10 years old: n=64 (6.6%) out of 1142 patients with T2D; mean diagnosis age=8.6±1.6 years, 20.3% men, 4.7% NHW, 65.6% Hispanic, 28.1% AA, 1.6% other. AbNegT1D: n=38 (5.6%) out of 680 patients with new onset T1D; mean diagnosis age=11.3±3.8 years; 57.9% men, 50% NHW, 19.4% Hispanic, 22.3% AA, 8.3% other.
Conclusions
In sum, we identified 1%–6.6% of atypical diabetes cases in a pediatric diabetes population with high racial and ethnic diversity using systematic review of the EMR. Better identification of these cases using unbiased approaches may advance precision diabetes.
Keywords: MODY, Children, Pediatric Diabetes
WHAT IS ALREADY KNOWN ON THIS TOPIC
This is a novel study, which introduces two strategies to systematically identify pediatric atypical diabetes mellitus cases using electronic medical records (EMR).
WHAT THIS STUDY ADDS
By implementing a specialized questionnaire and designing EMR queries, we were able to successfully identify 1%–6.6% of atypical diabetes cases, highlighting the potential for accurate identification with these strategies.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
The approach used in this study can significantly impact research practices by promoting precision diabetes care and improving understanding of diabetes heterogeneity in children.
Introduction
Type 1 diabetes (T1D) and type 2 diabetes (T2D) mellitus are heterogeneous diseases in which clinical presentation and disease progression may vary significantly within and between diabetes types.1 Furthermore, data obtained using molecular diagnostic methods and deep phenotyping techniques indicate the presence of atypical forms of diabetes mellitus with varied pathophysiology.2 Atypical diabetes mellitus comprises both rare genetic syndromes and clusters of phenotypically distinct forms of diabetes mellitus within a spectrum between classically presenting T1D and T2D.3 4
Atypical forms of diabetes mellitus are suspected clinically when diagnostic criteria and clinical phenotype do not meet usually accepted definitions of ‘classic’ T1D or T2D. T1D is characterized by evidence of islet autoimmunity (positivity for islet autoantibodies), rapid progression to near-complete insulin deficiency, insulin dependence, development of diabetic ketoacidosis if insulin therapy is interrupted, younger age of presentation and usually lack of obesity or evidence of insulin resistance. T2D is characterized by overweight/obesity and insulin resistance, older age at diagnosis, family history of diabetes mellitus, lack of insulin dependence (at least initially) and slowly progressing beta cell dysfunction. However, there is clearly heterogeneity within each of these types of diabetes mellitus, and investigations of these heterogenous forms have led to the identification of some well-characterized atypical diabetes syndromes such as latent autoimmune diabetes of adults and subtypes of ketosis-prone diabetes (KPD).5,7
Identification and characterization of atypical forms of diabetes mellitus that do not fit the classic definitions of T1D and T2D could improve the clinical classification of the condition and provide a foundation for ‘precision diabetes’ and targeted treatment. The value of this approach is evident from the considerable impact of identifying and characterizing monogenic forms of diabetes (maturity-onset diabetes of the young (MODY) and neonatal diabetes), which has dramatically improved the management of these patients with targeted therapies and family screening.7 8
Careful study of atypical forms of diabetes mellitus could also elucidate the complex pathophysiology underlying the common types of diabetes mellitus, by providing insight into new mechanisms that explain the heterogeneity of phenotypes in both ‘T1D’ and ‘T2D’.9 The Rare and Atypical Diabetes Network (RADIANT) is a group of universities, hospitals and clinics across the USA that aims to discover and define rare and atypical forms of diabetes mellitus through detailed phenotyping and genotyping of the participants and families referred for study.4
Because of a lack of established criteria to define ‘atypical diabetes’, assembling a cohort of pediatric patients with atypical diabetes mellitus to allow meaningful studies remains challenging. In addition, participants ascertained by self-referral or referral by healthcare providers have been shown to lack racial and ethnic diversity.10 On the other hand, the electronic medical records (EMR) provides a unique opportunity to identify individuals with atypical forms of diabetes mellitus in a more inclusive manner, utilizing data collected as part of routine clinical care. However, strategies to ‘query’ EMR databases need to be developed, as most such databases lack specific codes for atypical forms of diabetes mellitus. In this study, we aimed to identify diverse cases of rare or atypical pediatric diabetes mellitus in an extensive EMR database, as a foundation for future recruitment of participants in research studies such as RADIANT to understand their etiologies and pathogenesis. We hypothesized that systematic review of large EMR systems with prespecified criteria may facilitate the identification of diverse children with atypical forms of diabetes mellitus.
Research design and methods
We tested two strategies to query an EMR database (ie, Epic) for atypical diabetes mellitus cases at Texas Children’s Hospital (TCH), Houston, Texas. In strategy 1, we designed a questionnaire to rule out patients with typical diabetes mellitus, and this questionnaire was applied by a manual review of EMR. The questionnaire was designed to be used either by the patient (as self-referred) or by a physician. For this study, a physician (MFA) reviewed the EMR of 50 youth (0–21 years) with diabetes mellitus seen consecutively in the TCH Diabetes Outpatient Clinic in April 2019 and responded the questionnaire with the information available from the EMR of each patient. After revisions for clarity, a second version of the questionnaire was applied by the same physician (MFA) on 50 additional patients seen during the same month. Sex, race and ethnicity data were self-reported by parents and/or patients and available as documentation on the EMR.
The revised questionnaire included 19 questions (online supplemental material S1), many focused on excluding patients with gestational diabetes and diabetes associated with chronic corticosteroid use, cystic fibrosis, pancreatectomy, chronic pancreatitis, HIV infection or HIV medications, hemochromatosis, Cushing syndrome, acromegaly and lipodystrophy (questions 1–11). Other questions aimed to rule out classic T1D based on clinical presentation and presence of islet autoantibodies (questions 12–13) and to rule out T2D based on the clinical diagnosis documented in the EMR, presence of obesity or hypercholesterolemia at time of diagnosis (question 14). Subsections of question 14 aimed to identify patients with KPD and children diagnosed with T2D under the age of 10 years old for inclusion. Since the goal was to identify previously unknown forms of rare and atypical diabetes mellitus, subsequent questions aimed to exclude known forms of atypical diabetes mellitus including monogenic diabetes (question 15–16) and Wolfram syndrome, Mitochondrial Encephalopathy, Lactic Acidosis, and Stroke-like episodes and Maternally Inherited Diabetes and Deafness syndromes (question 17). At the end of the questionnaire, two free-text questions (questions 18 and 19) were included that could potentially be used for descriptions of suspected atypical features noted by individuals or their physicians.
The application of this questionnaire to the EMR of each patient took approximately 5–10 min, depending on how readily available the information was in their medical record.
In strategy 2, we aimed to identify three categories of atypical forms of diabetes mellitus using data from an available Diabetes Flowsheet embedded in the clinical encounter that must be completed by the treating pediatric endocrinologist or nurse practitioner as a mandatory part of the medical documentation for each outpatient visit. The Diabetes Flowsheet includes a question on diabetes type with a set of options including T1D, T2D, monogenic diabetes, steroid-induced diabetes, drug-induced diabetes, cystic fibrosis-related diabetes and ‘unknown’ diabetes type. In collaboration with EMR analysts, we built three electronic queries to generate reports of patients up to 21 years of age who attended the TCH Diabetes Outpatient Clinic from April 2019 to March 2020 and had diabetes mellitus of unknown type, T2D diagnosed before 10 years of age or autoantibody-negative T1D. The queries used data from Diabetes Flowsheet and demographic information including age, sex, race and ethnicity. We performed manual review of the participants under the category of unknown diabetes mellitus to better determine their characteristics.
Characteristics of both strategies were summarized using descriptive statistics.
Results
Strategy 1
The first questionnaire, done by a physician on initial 50 patient’s EMR, identified eight cases (16%) with suspected atypical diabetes mellitus. Following further manual review of the patients’ EMR, five cases were considered atypical (10%). The three excluded cases were two patients who had strong clinical characteristics of T2D and multiple family members with T2D, and one who had strong features of T2D treated with insulin. Consequently, the questionnaire was revised to avoid selecting patients as atypical in the setting of insulin treatment in patients with a phenotype of T2D because many pediatric patients with T2D are treated with insulin. The final revised questionnaire was applied to another 50 patients and identified 3 patients (6%) with possible atypical diabetes mellitus; of these, two were excluded at the stage of manual review because they lacked information regarding islet antibodies, but the clinical course suggested T1D (figure 1).
Figure 1. Flow diagram of the process of identification of atypical diabetes mellitus cases using a questionnaire (strategy 1). EMR, electronic medical record.
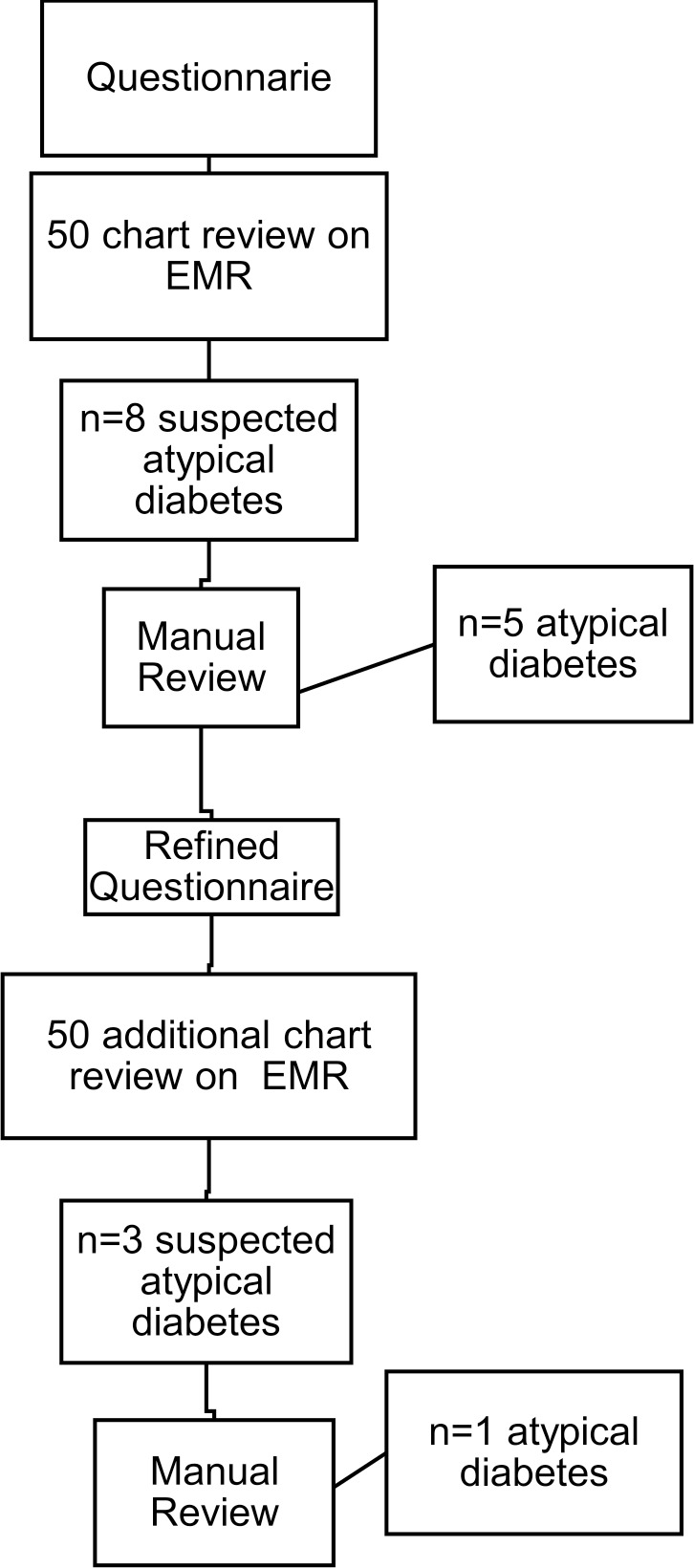
Ultimately, six cases of atypical diabetes mellitus (6%) were identified out of the 100 patients screened by the questionnaire (table 1). The mean age at diagnosis was 11±2.6 years, 16.6% were men, 33% non-Hispanic white (NHW) and 66.6% Hispanic (Hisp). Of the six cases, three had a clinical phenotype of T2D with onset before 10 years of age. All three had negative islet autoantibodies (including glutamic acid decarboxylase-65, insulinoma-associated antigen-2 [IA-2] and insulin autoantibodies (IAA)) and had been insulin-dependent since diagnosis. The fourth patient had a clinical diagnosis of T1D in the EMR, did not have obesity at diagnosis and had negative islet autoantibodies and a variant of unknown significance (VUS) in the PDX1 gene (heterozygous variant c.820G>T, pVal274Phe) on MODY genetic testing. The fifth patient had a clinical diagnosis of T2D in the EMR (although lacked obesity and dyslipidemia), with negative islet autoantibodies, multiple family members with ‘T2D’ and a negative MODY genetic test panel. The sixth patient had a clinical diagnosis of T1D, normal body mass index, required insulin from the time of diagnosis, had four islet autoantibodies negative (GAD65, IA-2, IAA and zinc transporter-8 (ZnT8) antibodies) and had a negative MODY genetic testing panel.
Table 1. Atypical diabetes mellitus cases identified with strategy 1 (questionnaire applied to EMR).
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | |
| Age (years)* | 9.1 | 9.5 | 8.8 | 10.8 | 15.7 | 12.5 |
| Sex | Male | Female | Female | Female | Female | Female |
| Race/ethnicity | Hispanic | Hispanic | NHW | Hispanic | Hispanic | NHW |
| BMI (%-ile)* | 99.14 | 99.48 | – | 66.28 | 81.07 | 5.72 |
| Diagnosis on EMR | T2D | T2D | T1D | T1D | T2D | T1D |
| Diabetes in parents | Yes | Yes | Yes | Yes | Yes | Yes |
| Acanthosis nigricans | Yes - minimal | Yes | No | No | No | No |
| C-peptide* (ng/mL) | – | 3.66 | – | 1.44 | 1.04 | 0.46 |
| Islet antibodies (GAD65, ICA512 and IAA) | Negative | Negative | Negative | Negative | Negative | Negative |
| ZnT8 antibody | Negative | – | Negative | Negative | Negative | Negative |
| Lipid profile*(mg/dL) | TG 114LDL 135HDL 52TChol 209 | TG 277LDL 79HDL 31TChol 166Non-HDL 135 | – | – | TG 150LDL 90HDL 43TChol 150Non-HDL 107 | TG 102LDL 84HDL 73TChol 177Non-HDL 104 |
| HbA1C (%)* | 5.9 | 9.6 | Not available | 11.7 | 12.9 | >14 |
| Recent HbA1C (%) | 10.8 | >14 | 8.7 | 8.5 | >14 | 8.2 |
| Treatment* | Metformin | Insulin | Insulin | Insulin and metformin | Insulin | Insulin |
| MODY genetic panel | – | – | Negative | VUS on PDX1[heterozygous variant,c.820G>T (p.Val274Phe)] | Negative | Negative |
At diagnosis.
BMIbody mass indexEMR, electronic medical record; HDL, high-density lipoprotein; %-ile, percentile; LDL, low-density lipoprotein; MODYmaturity-onset diabetes of the young NHW, non-Hispanic white; Non-HDL, non-HDL cholesterolTChol, total cholesterol; T1D, type 1 diabetes; T2D, type 2 diabetes; TG, triglycerides
A manual review was conducted on the 89 patients’ EMR identified as typical diabetes mellitus by the questionnaire and results were similar, no further cases of atypical diabetes mellitus were identified.
The six patients with atypical diabetes mellitus identified by the questionnaire and confirmed by manual review were referred to RADIANT for further investigation. Of those patients, only four patients started enrollment in the study. Participant 1 was later found to have a pathogenic VUS for MODY and was not further enrolled. Participant 2 did not enroll. Participant 3 enrolled, on wait list for future adjudication due to mainly suspected autoantibody-negative T1D. Participant 4 enrolled and currently completing stage 2 of the study. Participant 5 enrolled but had lost contact. Participant 6 enrolled and is currently completing stage 3 of the study.
Strategy 2
We identified 68 cases of unknown diabetes mellitus type (1%, out of 6676 total patients with diabetes mellitus <21 years of age). The mean age at diagnosis was 12.6±3.3 years, 32.8% were men, 23.8% NHW, 47.6% Hisp, 25.4% African American (AA) and 3.2% other. Out of 1142 children with a diagnosis of T2D in the EMR, 64 patients (6.6%) were diagnosed before age 10 years, mean age at diagnosis of 8.6±1.6 years, 20.3% men, 4.7% NHW, 65.6% Hisp, 28.1% AA, 1.6% other. Out of 680 patients with new-onset T1D, 38 (5.6%) were negative for islet autoantibodies (GAD, IA-2 and IAA), mean age at diagnosis 11.3±3.8 years; 57.9% men, 50% NHW, 19.4% Hisp, 22.3% AA, 8.3% other (table 2).
Table 2. Characteristics of patients identified with strategy 1 and strategy 2 (EMR generated queries to select pre-specified phenotypes of atypical pediatric diabetes).
| Strategy 1. Questionnaire | Strategy 2. EMR queries | |||
| Atypical diabetes (n=6/100) | Unknown type (n=68/6676) | T2D<10 y/o (n=64/1142) | Autoantibody negative T1D (n=38/680) | |
| Age at diagnosis, mean±SD (y/o) | 11±2.6 | 12.6±3.3 | 8.6±1.6 | 11.3±3.8 |
| Sex, n (%) | ||||
| Male | 1 (17%) | 22 (33%) | 13 (20%) | 22 (58%) |
| Female | 5 (83%) | 46 (67%) | 51 (80%) | 16 (42%) |
| Race/ethnicity, n (%) | ||||
| Non-Hispanic white | 2 (33%) | 16 (24%) | 3 (5%) | 19 (50%) |
| Hispanic | 33 (48%) | 42 (66%) | 7 (19%) | |
| African American | 4 (67%) | 17 (25%) | 18 (28%) | 9 (22%) |
| Other | – | 2 (3%) | 1 (1%) | 3 (8%) |
Unknown type: n=68/out of 6676 total of patients with diabetes under 21y/o, T2D<10y/o: n=64/out of 1142 patients with a diagnosis of T2D in the EMR, autoantibody negative T1D: n=38/out of 680 patients with new onset diagnosis of T1D.
EMR, electronic medical record; T1D, type 1 diabetes; T2D, type 2 diabetes
We conducted a manual review of the 68 cases denoted ‘unknown diabetes mellitus type’ to better delineate the forms of atypical diabetes mellitus subsumed under this category. Monogenic diabetes was suspected by the primary clinician in 41% but the majority (>80%) did not have a MODY genetic test. Of those patients who had a MODY genetic test report (n=5), four did not have any pathogenic variant in the genes included in their MODY panel and one had a variant of unknown clinical significance in one of the MODY-associated genes. 19% were classified as ‘likely T2D’ by their primary endocrinologist and 16% had a diagnosis of gestational diabetes. The remaining suspected diagnoses are summarized in table 3.
Table 3. Characteristics of patients with ‘unknown diabetes mellitus’ identified by EMR query.
| Cluster | N (%) |
| Suspected monogenic, negative MODY panel | 4 (6) |
| Suspected monogenic, VUS | 1 (1) |
| Suspected monogenic, MODY panel not done | 23 (34) |
| T1D vs T2D vs monogenic, MODY panel not done | 1 (1) |
| Syndromic | 2 (3) |
| T1D vs T2D | 3 (4) |
| Gestational diabetes | 11 (16) |
| Likely T2D | 13 (19) |
| Not determined (missing tests) | 3 (4) |
| Known MODY diagnosis | 2 (3) |
| Drug-induced or post-transplant vs T2D | 4 (6) |
| Stress induced hyperglycemia | 1 (1) |
EMRelectronic medical recordMODYmaturity-onset diabetes of the young T1Dtype 1 diabetesT2Dtype 2 diabetesVUSvariant of unknown significance
Conclusions
We developed two strategies to identify patients with atypical diabetes mellitus using EMR data. Using a clinical questionnaire and EMR queries we found that 1%–6.6% of pediatric patients with diabetes mellitus have atypical forms of diabetes. T1D and T2D compromise the vast majority of diabetes in the USA,1 and the current literature suggests 1%–6.5% of children with diabetes mellitus are estimated to have atypical forms of diabetes mellitus, consistent with our observation.11 12
In a prior RADIANT study, Parikh et al recently developed a data mining framework using phenotypical characteristics of two diabetes mellitus cohorts: one an adult patient cohort in a predominantly Hispanic population in South Texas, and the other a multiethnic pediatric cohort at TCH, Houston. The frequency of atypical diabetes mellitus in the adult diabetes cohort was 11.5%, whereas it was 5.3% in the pediatric diabetes mellitus cohort, very similar to the present study. In addition, the Parikh et al study demonstrated the existence of two distinct clusters of atypical diabetes mellitus phenotypes in the pediatric cohort.13
Of the atypical cases identified in our study, half were children with T2D diagnosed before 10 years of age. All had negative islet autoantibodies. This group exemplifies an atypical form of diabetes mellitus, since it is rare for children to be diagnosed with T2D before 10 years of age, especially if they are also pre-pubertal.14,16 The incidence of T2D increases with age17 and several studies have shown an association between puberty timing and the development of T2D.18 Information about the stage of pubertal development was not available in all patients, therefore we cannot rule out the presence of precocious puberty that could have contributed to the earlier presentation of diabetes mellitus.
The other half of the suspected atypical cases were identified by their physicians due to a phenotype of MODY; of these, all had negative MODY gene panel testing (with one VUS). Among the atypical cases identified using strategy 2, the majority of the ‘unknown diabetes mellitus type’ group also comprised suspected MODY cases.
There are no well-defined clinical practice guidelines once a patient with atypical forms of diabetes mellitus has been identified. Most of the patients with clinically suspected MODY had received a clinical diagnosis of either T1D or T2D as documented in the EMR. Subtle clinical phenotypes such as overlapping T1D versus T2D are less likely to be studied further. Leveraging the EMR to identify patients with atypical diabetes mellitus can have direct impact both on clinical and research efforts.
From the clinical care perspective, system-wide identification and monitoring of these cases may improve clinical outcomes. From the research perspective, better identification and characterization of atypical forms of diabetes mellitus may improve enrollment in clinical trials such as RADIANT and accelerate discovery of new pathophysiologic pathways to diabetes mellitus and the possibility of targeted treatment. The questionnaire used in strategy 1 aimed to exclude the more described forms of diabetes mellitus as categorized by the American Diabetes Association: T1D, T2D, gestational diabetes mellitus and specific diabetes types due to other known causes (eg, monogenic diabetes, diseases of the exocrine pancreas and drug or chemical-induced diabetes).1 The use of questionnaires as screening tools has proven helpful in clinical practice to identify rare forms of diabetes mellitus. For example, Shields et al developed and validated a calculator based on clinical criteria to determine an individual’s probability of having MODY as a tool for rational genetic testing with improved sensitivity and specificity for identifying MODY patients.19
In strategy 1, our questionnaire was effective in identifying atypical diabetes mellitus cases. Out of the 100 patients, only 11 required a more detailed manual review. In the clinical practice, identification of atypical diabetes mellitus may be easier and also non-biased using such a questionnaire. A questionnaire with 19 questions might seem time consuming for a busy clinical practice; however, the first 12 questions were close-ended questions with only the remaining seven questions requiring more detailed clinical information and some of those questions were not applicable to all patients. If validated, this questionnaire could potentially become a screening tool to be applied by clinicians as a guide in patients that have mixed phenotype of diabetes mellitus and may help to determine if they need further testing or referral for investigation of atypical forms of diabetes mellitus. The questionnaire featured a straightforward design primarily comprising binary questions. It could potentially serve as a self-referral tool accessible to patients or their families online. Alternatively, it could be made available to patients and families in the waiting room through an EMR-connected tablet. The latter strategy has been deemed a valuable tool in primary and specialty care clinics.20 21
In strategy 2, obtaining queries of patients with diabetes mellitus categorized as ‘unknown diabetes mellitus type’ was possible mainly due to the Diabetes Flowsheet included in our EMR system that is filled out at the clinic visit only by the pediatric endocrinologist or the diabetes nurse practitioner taking care of the patient. This approach streamlined the identification of atypical forms of diabetes mellitus from a vast amount of data that would otherwise require a significant manual chart review effort to complete. Institutions that have not yet integrated diabetes mellitus flowsheets into their EMR systems may consider incorporating these tools.
Our study reported a prevalence of 5.6% of autoantibody-negative T1D cases, however the prevalence reported in children has been as high as 19%.22 The race/ethnicity proportions of patients identified by the two strategies were similar to those reported in other studies, which assessed populations composed mainly of under-represented racial and ethnic groups (primarily Hisp and AA).23 24 However, in the subset of autoantibody-negative T1D cases identified using strategy 2, NHW (50%) and AA (22%) children comprised the majority of cases. Wang et al reported no significant racial/ethnic difference in prevalence of autoantibody-negative diabetes in younger children with newly diagnosed diabetes mellitus. However, in children older than 14 years, autoantibody-negative diabetes was much more prevalent in under-represented racial/ethnic groups compared with NHW (57.1% in Hisp, 70% in AA and 25.9% in NHW), due in part to a higher rate of T2D in Hisp and AA.22 Individuals from under-represented racial/ethnic groups with T1D and T2D have increased morbidity and poor long-term control of diabetes mellitus overall,25,27 yet very little is known on the prognosis of the atypical forms of diabetes mellitus that appear to be highly prevalent in these groups. Our approach allowed us to identify a significant number of children from under-represented racial/ethnic groups who are less likely to be referred for further evaluation due to multiple barriers to participation in biomedical research.10 Hence, it is important for physicians to be familiar with the concept and forms of atypical diabetes mellitus in persons belonging to underrepresented racial/ethnic groups.
The strength of our study is the use of an unbiased approach by leveraging EMR to identify pediatric patients with atypical diabetes mellitus. The limitations include that determination of atypical nature of a particular case was entirely limited to medical documentation in EMR. Also, EMR systems are designed differently, and the diabetes flowsheet used in this study might not be adaptable across organizations, limiting the use of our current approach. However, EMR usually has a significant amount of data for each patient enabling it as a valuable tool for screening purposes.
In conclusion, EMR-based approach revealed that approximately 1%–6.6% of pediatric diabetes mellitus cases are atypical. More studies are required to validate our clinical questionnaire and to streamline the process of EMR-based identification of atypical cases.
supplementary material
Footnotes
Funding: The RADIANT Study is funded by U54 DK118638 and U54 DK118612 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The work in this manuscript was also supported by the National Institutes of Health grant K23-DK129821 (MT).
Provenance and peer review: Not commissioned; externally peer-reviewed.
Patient consent for publication: Not applicable.
Collaborators: The RADIANT Study Group: Baylor College of Medicine: Ashok Balasubramanyam, M.D., PI1,2,3,4,5,6,7,8,9, Maria J. Redondo, M.D., Ph.D., M.P.H., PI1,3,6,7,8, Maaz Ahmed, William Craigen, M.D., Ansley Davis, Monica Dussan, M.D., Jordana Faruqi, M.D., M.P.H., Ruchi Gaba, M.D., Mark Herman, M.D., Shalini Jhangiani, Elizabeth Kubota-Mishra, M.S., D.O., Pengfei Liu, Ph.D.3, Iliana Migacz, M.A., Nkechinyere Osuji, Jennifer Posey, M.D., Ph.D. 1,3,7,9, Nalini Ram, M.B.B.S, Aniko Sabo, Ph.D.3,4,7, Alejandro Siller, M.D., Stephanie Sisley, M.D.3, Mustafa Tosur, M.D.1, Eric Venner, Ph.D.3 Past Staff: Marcela Astudillo, M.D., Adriana Cardenas, Hongzheng Dai, Ph.D., Dimpi Desai, M.D., Mary Fang, Erica Hattery, M.D., M.H.A.5,8, Adrienne Ideouzu, Julizza Jimenez, Nupur Kikani, M.D., Graciela Montes, David Murdock, M.D., Nikalina G. O’Brien, Lee-Jun Wong, Ph.D.3. Columbia University: Robin Goland, M.D., PI1,3,4,8, Wendy K. Chung, M.D., Ph.D.7, Anabel Evans, Rachelle Gandica, M.D., Rudolph Leibel, M.D.3, Jacqueline Lonier, M.D., Kaisha Mofford, James Pring. Indiana University: Carmella Evans-Molina, Ph.D., M.D., PI1,3,7, Farrah Anwar, M.D., Hannah Lease, RN, Angelica Mckibben, RN, Gabriela Monaco, M.D., Zeb Saeed, M.D., Emily Sims, M.D., Maria Spall. Past Staff: Marimar Hernandez-Perez, Ph.D., Kieren Mather, M.D., Kelly Moors, Anna Neyman, M.D. Massachusetts General Hospital: Miriam S. Udler, M.D., Ph.D., PI1,3,7,8, Jose C. Florez, M.D., Ph.D., PI1,2,3,4,5,6,7,8,9, Victoria Chen, Sara Cromer, M.D., Aaron Deutsch, M.D., Julia Douvas, Melton Fan, Cristinia Fernandez Hernandez, Evelyn Greaux6, Saadhvi Kartik, Raymond Kreienkamp, M.D., Ph.D., Mary Larkin, R.N., M.S., CDCES, Pam Ricevuto, Nopporn Thangthaeng R.N., Ph.D., CDCES, Armen I. Yerevanian, MD. Past Staff: Melissa Calverley, Kathy Chu, Mariella Facibene, Christopher Han, Dorit Koren, M.D., Micah Koss, William Marshall, Amy Sabean, R.N., Jordan Sherwood, M.D. NorthShore University HealthSystem/Endeavor Health: Liana K. Billings, M.D., M.M.Sc., PI1,3. SUNY Downstate Health Sciences University: Mary Ann Banerji, M.D., PI1,3, Necole Brown, Lina Soni, Lorraine Thomas. Past Staff: Jennifer Abrams, D.O., Kylnt Bally, M.D., Beisi Ji, M.D., Samara Skiwiersky. University of Chicago: Louis H. Philipson, M.D., Ph.D., PI1,2,3,4,5,6,7,8,9, Siri Atma W. Greeley, M.D., Ph.D., PI1,3, Graeme Bell, Ph.D.3,7, Shanna Banogon, Jui Desai, Anisa Dye, David Ehrmann, M.D.3,5,6,7, Lisa R. Letourneau-Freiberg, M.P.H.5,6,8, Carlin Lockwood, Kynnedie Maloz, Rochelle N. Naylor, M.D.1,3,7,8, Kaylee Oppenheimer, Erin Papciak, Karen Rodriguez, Rachel Son, Manu Sundaresan, Chelsea Wu. Past Staff: Colleen Bender, Lainie Friedman Ross, M.D., Ph.D., Persephone Tian. University of Colorado – Denver: Neda Rasouli, M.D., PI1,3,7,8, Chelsea Baker PA-C, Megan riff, MD, Courtney King, Wyatt Pfau, Avinash Pyreddy, Marjan Rezaei, Katlyn Sawyer, MD, Vatsala Singh. Past Staff: Mohsen Bahmani Kashkouli, Jules Barklow, Noosha Farhat, Andrew Her, Rebecca Lorch5, Carter Odean, M.S., Gregory Schleis, M.D., Chantal Underkofler. University of Maryland: Toni I. Pollin, Ph.D., M.S., PI1,3,8, Hadley Bryan, Ryan Jollie, M.P.H., Kristin Maloney, M.S., M.G.C.3, Jennifer Marron, RN, Ryan Miller, M.D., Paula Newton, M.D., Maria Eleni Nikita, M.D., Knightess Oyibo, MBChB, Kristi Silver, M.D., Hilary Whitlatch, M.D.1 Cindi Young. Past Staff: Kathleen Palmer, Stephanie Riley, M.S., Devon Nwaba, M.P.H.5,6, Elizabeth Streeten, M.D.1,3, Jessica Tiner, M.P.H. University of Michigan: Elif A. Oral, M.D., PI1,3,5,7,8, David Broome, M.D., Merve Celik-Gular, Tae-Hwa Chun, M.D., Ph.D., Anabela Dill Gomes, Maria Foss de Freitas, M.D., Brigid Gregg, M.D., Donatella Gilio, Seda Grigoryan, M.D., Diarratou Kaba, Melda Sonmez Ince, M.D., Adam Neidert, M.S., Carman Richison. Past Staff: Baris Akinci, M.D., Rita Hench, Salman Imam. University of North Carolina: John Buse, M.D., Ph.D., PI8, Jamie Diner, M.S.N., FNP-C, RN, CDCES3,8, Cassandra Donahue, RDN, LDN, CDCES, Rachael Fraser, Karla Fulghum RN, Faryal Gilani, Tahereh Ghorbani, M.D., Alex Kass, MSN, MBA, RN, CDCES5,6, Nina Jain, M.D., Klara Klein, M.D., Ph.D.1, Lauren Larison, BSN, RN, Brooke Matson, M.D., Ph.D, Catherine Morba. Past Staff: Sarah Adams, Chase Armstrong, Sue Kirkman, M.D.3,6,7,8. University of Rochester: Lainie Friedman Ross, M.D., Ph.D.3,5,6. University of Washington: Irl B. Hirsch, M.D., PI1,3, Jesica Baran, M.D., Rosanna Holod, Steven E. Kahn, M.B., Ch.B.1,3, Dori Khakpour6, Patali Mandava, Lori Sameshima. Past Staff: Xiaofu Dong, Thanmai Kalerus. Seattle Children’s: Catherine Pihoker, M.D., PI1,3,5,6,7, Beth Loots, M.P.H., M.S.W.6, Kathleen Santarelli. Past Staff: Cisco Pascual. Vanderbilt University: Kevin Niswender, M.D., Ph.D., PI, Norma Edwards, APRN, Justin Gregory, M.D., M.S.C.I., Alvin Powers, M.D.1,3, Andrea Ramirez, M.D., M.S.C.I.1,3. Past Staff: Jennifer Scott, Jordan Smith6. Washington University: Fumihiko Urano, M.D., Ph.D., PI1,3,7, Samantha Adamson, MD, PhD, Cris Brown, Joel Brune, RN, Mary Jane Clifton, Jing Hughes, M.D., Ph.D.1,3,7, Stacy Hurst, BSN, RN, CDE, Janet McGill, M.D., M.A.1,3,5,6,8, Isabella Paolicelli, R.N., Stephen Stone, M.D.1,3,7, Brittany Zwijack, RN. Past Staff: Toko Campbell, M.D., Jennifer May, M.D. Data Coordinating Center – University of South Florida: Jeffrey P. Krischer, Ph.D., PI1,2,3,4,5,6,7,8,9, Rajesh Adusumalli, M.S., Bruce Albritton, Analia Aquino, Paul Bransford, Nicholas Cadigan, Laura Gandolfo, Jennifer Garmeson, Joseph Gomes, M.S.Cp.E, Robert Gowing, Juan Herrera, Christina Karges, M.P.H., Callyn Kirk, M.S.P.H., Sarah Muller4, Jean Morissette, Hemang M. Parikh, Ph.D.8, Francisco Perez-Laras, Cassandra L. Remedios, M.S.7,8, Pablo Ruiz, Noah Sulman, Ph.D., Michael Toth, M.S.H.I., Lili Wurmser. Past Staff: Christopher Eberhard, M.S.4, Steven Fiske, Brandy Hutchinson, C.I.P., Sidhvi Nekkanti, Rebecca Wood, M.S. Genetics Core – Broad Institute: Jose C. Florez, M.D., Ph.D., PI1,2,3,4,5,6,7,8,9, MacKenzie Brandes, Noël Burtt, Jason Flannick, Ph.D.3, Ryan Koesterer, M.A., Phebe Olorunfemi, Miriam S. Udler, M.D., Ph.D.1,3,7,8 Past Staff: Ahmed Alkanaq, Ph.D.3, Lizz Caulkins. Central Laboratory – University of Florida: Clive Wasserfall, Ph.D.4, David Pittman4. Past Staff: William Winter, M.D.4. National Institutes of Diabetes and Digestive and Kidney Diseases: Beena Akolkar, Ph.D. 1,2,3,4,5,6,7,8,9. Past Staff: Christine Lee, M.D., M.S.1,2,3,4,5,6,7,8,9. Other contributors: David J. Carey, Ph.D.8, Geisinger Health System. Daniel Hood8, Regenstrief Institute. Santica M. Marcovina, Ph.D., Sc.D.4,5,6,7, Medspace Reference Laboratories. Christopher B. Newgard, Ph.D.3,4, Duke University Medical Center. Committees: 1 Adjudication, 2 Ancillary Studies/Data Access, 3Discovery, 4 Laboratory Implementation, 5 Protocol Implementation, 6 Protocol Oversight, 7 Publications and Presentations, 8 Recruitment and Retention, 9 Steering.
Presented at: Part of this study was presented at the 80th Virtual Scientific Sessions of the American Diabetes Association, June 12–16, 2020.
Ethics approval: This study was approved and the requirement for informed consent was waived by the Baylor College of Medicine Institutional Review Board (IRB). Research protocol H-45325.
Contributor Information
Marcela F Astudillo, Email: marc_astudillo1@hotmail.com.
William E Winter, Email: winter@pathology.ufl.edu.
Liana K Billings, Email: lbillings@northshore.org.
Raymond Kreienkamp, Email: raymond.kreienkamp@childrens.harvard.edu.
Ashok Balasubramanyam, Email: ashokb@bcm.edu.
Maria J Redondo, Email: redondo@bcm.edu.
Mustafa Tosur, Email: mustafa.tosur@bcm.edu.
RADIANT Study Group, Email: radiantpublications@epi.usf.edu.
RADIANT Study Group:
Ashok Balasubramanyam, Maria J Redondo, Maaz Ahmed, William Craigen, Ansley Davis, Monica Dussan, Jordana Faruqi, Ruchi Gaba, Mark Herman, Shalini Jhangiani, Elizabeth Kubota-Mishra, Pengfei Liu, Iliana Migacz, Nkechinyere Osuji, Jennifer Posey, Nalini Ram, Aniko Sabo, Alejandro Siller, Stephanie Sisley, Mustafa Tosur, Eric Venner, Marcela Astudillo, Adriana Cardenas, Hongzheng Dai, Dimpi Desai, Mary Fang, Erica Hattery, Adrienne Ideouzu, Julizza Jimenez, Nupur Kikani, Graciela Montes, David Murdock, Nikalina G O’Brien, Lee-Jun Wong, Robin Goland, Wendy K Chung, Anabel Evans, Rachelle Gandica, Rudolph Leibel, Jacqueline Lonier, Kaisha Mofford, James Pring, Carmella Evans-Molina, Farrah Anwar, Hannah Lease, Angelica Mckibben, Gabriela Monaco, Zeb Saeed, Emily Sims, Maria Spall, Marimar Hernandez-Perez, Kieren Mather, Kelly Moors, Anna Neyman, Miriam S Udler, Jose C Florez, Victoria Chen, Sara Cromer, Aaron Deutsch, Julia Douvas, Melton Fan, Cristinia Fernandez Hernandez, Evelyn Greaux, Saadhvi Kartik, Raymond Kreienkamp, Mary Larkin, Pam Ricevuto, Nopporn Thangthaeng, Armen I Yerevanian, Melissa Calverley, Kathy Chu, Mariella Facibene, Christopher Han, Dorit Koren, Micah Koss, William Marshall, Amy Sabean, Jordan Sherwood, Liana K Billings, Mary Ann Banerji, Necole Brown, Lina Soni, Lorraine Thomas, Jennifer Abrams, Kylnt Bally, Beisi Ji, Samara Skiwiersky, Louis H Philipson, SiriAtma W Greeley, Graeme Bell, Shanna Banogon, Jui Desai, Anisa Dye, David Ehrmann, Lisa R Letourneau-Freiberg, Carlin Lockwood, Kynnedie Maloz, Rochelle N Naylor, Kaylee Oppenheimer, Erin Papciak, Karen Rodriguez, Rachel Son, Manu Sundaresan, Chelsea Wu, Colleen Bender, Lainie Friedman Ross, Persephone Tian, Neda Rasouli, PA-C Chelsea Baker, Megan Riff, Courtney King, Wyatt Pfau, Avinash Pyreddy, Marjan Rezaei, Katlyn Sawyer, Vatsala Singh, Mohsen Bahmani Kashkouli, Jules Barklow, Noosha Farhat, Andrew Her, Rebecca Lorch, Carter Odean, Gregory Schleis, Chantal Underkofler, Toni I Pollin, Hadley Bryan, Ryan Jollie, Kristin Maloney, Jennifer Marron, Ryan Miller, Paula Newton, Maria Eleni Nikita, Knightess Oyibo, Kristi Silver, Hilary Whitlatch, Cindi Young, Kathleen Palmer, Stephanie Riley, Devon Nwaba, Elizabeth Streeten, Jessica Tiner, Elif A Oral, David Broome, Merve Celik-Gular, Tae-Hwa Chun, Anabela Dill Gomes, Maria Foss de Freitas, Brigid Gregg, Donatella Gilio, Seda Grigoryan, Diarratou Kaba, Melda Sonmez Ince, Adam Neidert, Carman Richison, Baris Akinci, Rita Hench, Salman Imam, John Buse, Jamie Diner, Cassandra Donahue, Rachael Fraser, Karla Fulghum, Faryal Gilani, Tahereh Ghorbani, Alex Kass, Nina Jain, Klara Klein, Lauren Larison, Brooke Matson, Catherine Morba, Sarah Adams, Chase Armstrong, Sue Kirkman, Lainie Friedman Ross, Irl B Hirsch, Jesica Baran, Rosanna Holod, Steven E Kahn, Dori Khakpour, Patali Mandava, Lori Sameshima, Xiaofu Dong, Thanmai Kalerus, Catherine Pihoker, Beth Loots, Kathleen Santarelli, Cisco Pascual, Kevin Niswender, Norma Edwards, Justin Gregory, Alvin Powers, Andrea Ramirez, Jennifer Scott, Jordan Smith, Fumihiko Urano, Samantha Adamson, Cris Brown, Joel Brune, Mary Jane Clifton, Jing Hughes, Stacy Hurst, Janet McGill, Isabella Paolicelli, Stephen Stone, Brittany Zwijack, Toko Campbell, Jennifer May, Jeffrey P Krischer, Rajesh Adusumalli, Bruce Albritton, Analia Aquino, Paul Bransford, Nicholas Cadigan, Laura Gandolfo, Jennifer Garmeson, Joseph Gomes, Robert Gowing, Juan Herrera, Christina Karges, Callyn Kirk, Sarah Muller, Jean Morissette, Hemang M Parikh, Francisco Perez-Laras, Cassandra L Remedios, Pablo Ruiz, Noah Sulman, Michael Toth, Lili Wurmser, Christopher Eberhard, Steven Fiske, Brandy Hutchinson, Sidhvi Nekkanti, Rebecca Wood, Jose C Florez, MacKenzie Brandes, Noël Burtt, Jason Flannick, Ryan Koesterer, Phebe Olorunfemi, Miriam S Udler, Ahmed Alkanaq, Lizz Caulkins, Clive Wasserfall, David Pittman, William Winter, Beena Akolkar, Christine Lee, David J Carey, Daniel Hood, Santica M Marcovina, and Christopher B Newgard
Data availability statement
Data are available upon reasonable request.
References
- 1.Care D, Suppl SS. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2020. Diabetes Care. 2020;43:S14–31. doi: 10.2337/dc20-S002. [DOI] [PubMed] [Google Scholar]
- 2.Steenkamp DW, Alexanian SM, Sternthal E. Approach to the patient with atypical diabetes. CMAJ. 2014;186:678–84. doi: 10.1503/cmaj.130185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasubramanyam A. Defining and Classifying New Subgroups of Diabetes. Annu Rev Med. 2021;72:63–74. doi: 10.1146/annurev-med-050219-034524. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanyam A, Redondo MJ, Craigen W, et al. The Rare and Atypical Diabetes Network (RADIANT) Study: Design and Early Results. Diabetes Care. 2023;46:1265–70. doi: 10.2337/dc22-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redondo MJ, Hagopian WA, Oram R, et al. The clinical consequences of heterogeneity within and between different diabetes types. Diabetologia. 2020;63:2040–8. doi: 10.1007/s00125-020-05211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balasubramanyam A, Nalini R, Hampe CS, et al. Syndromes of ketosis-prone diabetes mellitus. Endocr Rev. 2008;29:292–302. doi: 10.1210/er.2007-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hattersley AT, Patel KA. Precision diabetes: learning from monogenic diabetes. Diabetologia. 2017;60:769–77. doi: 10.1007/s00125-017-4226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tosur M, Philipson LH. Precision diabetes: Lessons learned from maturity-onset diabetes of the young (MODY) J Diabetes Investig. 2022;13:1465–71. doi: 10.1111/jdi.13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francis Y, Tiercelin C, Alexandre-Heyman L, et al. HNF1B-MODY Masquerading as Type 1 Diabetes: A Pitfall in the Etiological Diagnosis of Diabetes. J Endocr Soc. 2022;6:bvac087. doi: 10.1210/jendso/bvac087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tosur M, Gandolfo L, Balasubramanyam A, et al. Enrollment of underrepresented racial and ethnic groups in the Rare and Atypical Diabetes Network (RADIANT) J Clin Transl Sci. 2023;7:e47. doi: 10.1017/cts.2022.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacaud D, Schwandt A, de Beaufort C, et al. A description of clinician reported diagnosis of type 2 diabetes and other non-type 1 diabetes included in a large international multicentered pediatric diabetes registry (SWEET) Pediatr Diabetes. 2016;17 Suppl 23:24–31. doi: 10.1111/pedi.12426. [DOI] [PubMed] [Google Scholar]
- 12.Johansson BB, Irgens HU, Molnes J, et al. Targeted next-generation sequencing reveals MODY in up to 6.5% of antibody-negative diabetes cases listed in the Norwegian Childhood Diabetes Registry. Diabetologia. 2017;60:625–35. doi: 10.1007/s00125-016-4167-1. [DOI] [PubMed] [Google Scholar]
- 13.Parikh HM, Remedios CL, Hampe CS, et al. Data Mining Framework for Discovering and Clustering Phenotypes of Atypical Diabetes. J Clin Endocrinol Metab. 2023;108:834–46. doi: 10.1210/clinem/dgac632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astudillo M, Tosur M, Castillo B, et al. Type 2 diabetes in prepubertal children. Pediatr Diabetes. 2021;22:946–50. doi: 10.1111/pedi.13254. [DOI] [PubMed] [Google Scholar]
- 15.Pettitt DJ, Talton J, Dabelea D, et al. Prevalence of diabetes in U.S. youth in 2009: the SEARCH for diabetes in youth study. Diabetes Care. 2014;37:402–8. doi: 10.2337/dc13-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klingensmith GJ, Connor CG, Ruedy KJ, et al. Presentation of youth with type 2 diabetes in the Pediatric Diabetes Consortium. Pediatr Diabetes. 2016;17:266–73. doi: 10.1111/pedi.12281. [DOI] [PubMed] [Google Scholar]
- 17.Perng W, Conway R, Mayer-Davis E, et al. Youth-Onset Type 2 Diabetes: The Epidemiology of an Awakening Epidemic. Diabetes Care. 2023;46:490–9. doi: 10.2337/dci22-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng TS, Day FR, Lakshman R, et al. Association of puberty timing with type 2 diabetes: A systematic review and meta-analysis. PLoS Med. 2020;17:e1003017. doi: 10.1371/journal.pmed.1003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shields BM, McDonald TJ, Ellard S, et al. The development and validation of a clinical prediction model to determine the probability of MODY in patients with young-onset diabetes. Diabetologia. 2012;55:1265–72. doi: 10.1007/s00125-011-2418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogg W, Johnston S, Russell G, et al. Conducting waiting room surveys in practice-based primary care research: a user’s guide. Can Fam Physician. 2010;56:1375–6. [PMC free article] [PubMed] [Google Scholar]
- 21.Horn ME, Reinke EK, Mather RC, et al. Electronic health record-integrated approach for collection of patient-reported outcome measures: a retrospective evaluation. BMC Health Serv Res. 2021;21:626. doi: 10.1186/s12913-021-06626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Miao D, Babu S, et al. Prevalence of autoantibody-negative diabetes is not rare at all ages and increases with older age and obesity. J Clin Endocrinol Metab. 2007;92:88–92. doi: 10.1210/jc.2006-1494. [DOI] [PubMed] [Google Scholar]
- 23.Pihoker C, Gilliam LK, Ellard S, et al. Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. J Clin Endocrinol Metab. 2013;98:4055–62. doi: 10.1210/jc.2013-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haaland WC, Scaduto DI, Maldonado MR, et al. A-beta-subtype of ketosis-prone diabetes is not predominantly a monogenic diabetic syndrome. Diabetes Care. 2009;32:873–7. doi: 10.2337/dc08-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redondo MJ, Libman I, Cheng P, et al. Racial/Ethnic Minority Youth With Recent-Onset Type 1 Diabetes Have Poor Prognostic Factors. Diabetes Care. 2018;41:1017–24. doi: 10.2337/dc17-2335. [DOI] [PubMed] [Google Scholar]
- 26.TODAY Study Group Long-Term Complications in Youth-Onset Type 2 Diabetes. N Engl J Med. 2021;385:416–26. doi: 10.1056/NEJMoa2100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ezzatvar Y, Ramírez-Vélez R, Izquierdo M, et al. Racial differences in all-cause mortality and future complications among people with diabetes: a systematic review and meta-analysis of data from more than 2.4 million individuals. Diabetologia. 2021;64:2389–401. doi: 10.1007/s00125-021-05554-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request.


