Abstract
Purpose
In a nuclear or radiological event, an early diagnostic or prognostic tool is needed to distinguish unexposed from low- and highly exposed individuals with the latter requiring early and intensive medical care. Radiation-induced gene expression (GE) changes observed within hours and days after irradiation have shown potential to serve as biomarkers for either dose reconstruction (retrospective dosimetry) or the prediction of consecutively occurring acute or chronic health effects. The advantage of GE markers lies in their capability for early (1–3 days after irradiation), high-throughput, and point-of-care (POC) diagnosis required for the prediction of the acute radiation syndrome (ARS).
Conclusions
As a key session of the ConRad conference in 2021, experts from different institutions were invited to provide state-of-the-art information on a range of topics including: (1) Biodosimetry: What are the current efforts to enhance the applicability of this method to perform retrospective biodosimetry? (2) Effect prediction: Can we apply radiation-induced GE changes for prediction of acute health effects as an approach, complementary to and integrating retrospective dose estimation? (3) High-throughput and point-of-care diagnostics: What are the current developments to make the GE approach applicable as a high-throughput as well as a POC diagnostic platform? (4) Low level radiation: What is the lowest dose range where GE can be used for biodosimetry purposes? (5) Methodological considerations: Different aspects of radiation-induced GE related to more detailed analysis of exons, transcripts and next-generation sequencing (NGS) were reported.
Keywords: Gene expression, biodosimetry, effect prediction, radiation exposure, high dose, low dose
1. Introduction
Radiation exposure such as an accidental irradiation, the potential radioactive release after a nuclear accident or a potential terrorist attack on the civilian population using a nuclear weapon or a radioactive device may result in significant human exposures to ionizing radiation. In such a large-scale radiological emergency, early diagnosis of exposed individuals is clearly needed in order to evaluate the extent of radiation injuries and assign appropriate treatment (Kabacik et al. 2011; Boldt et al. 2012; Manning et al. 2013; Jacobs et al. 2020). In the absence of physical dosimetry devices, a combination of history of an individual’s location, clinical signs and symptoms, and individual hematology assessment, along with other methods such as biological changes occurring within hours and days after irradiation, can be used for either individual dose estimates (retrospective dosimetry) (Sullivan et al. 2013) or the prediction of consecutively occurring acute health effects (Port et al. 2016, 2021). Hereby, different assays emerged over the last decades including the dicentric chromosome assay (DCA), gamma-H2AX foci assay, cytokinesis block micronucleus assay or the ‘-omic’ assays (Sullivan et al. 2013). The DCA is currently considered the ‘gold-standard’, being very specific to ionizing radiation and showing a high sensitivity due to low background levels (IAEA 2011). However, like all cytogenetics-based assays, the DCA is labor intensive and time consuming (Rothkamm et al. 2013).
An alternative approach for retrospective assessment of radiation injury patients is the examination of radiation-induced GE changes. Application of GE in two radiation victims for late exposure validation generated promising results (Scherthan et al. 2007). However, authors emphasized the experimental approach and the requirement for validation of their results in bigger cohorts of radiation accident victims.
Gene expression (GE) analysis has been shown as an alternative tool for high throughput biodosimetry and prediction of clinical outcomes (Amundson et al. 2000, 2001; Meadows et al. 2008; Paul and Amundson 2008; Meadows et al. 2010; Paul et al. 2011; Port et al. 2016, 2018, 2019). The field of transcriptomics in this context identified promising biomarker panels, which are now to be validated to facilitate high-throughput automation and the development of field deployable devices (Homer et al. 2016; Larsen and Disbrow 2017; Lacombe et al. 2018).
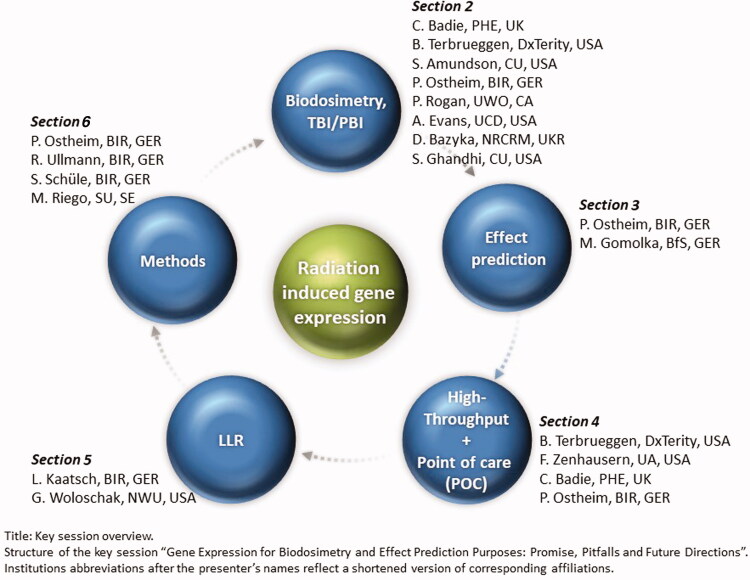
This article summarizes presentations from the key session at ConRad 2021 (Global Conference on Radiation Topics) related to five areas on GE for biodosimetry and effect prediction purposes pointing at promises, pitfalls and future directions (Figure 1). The key messages per chapter and presenters are also provided in Table 1.
Figure 1.
Key session overview. Structure of the key session ‘Gene expression for biodosimetry and effect prediction purposes: promise, pitfalls and future directions’. Institutions abbreviations after the presenter’s names reflect a shortened version of corresponding affiliations.
Table 1.
Summary of key messages per section and presentation.
| Section | Presenter | Key messages | |
|---|---|---|---|
| Biodosimetry | |||
| C. Badie | 1. In a large scale event, high-throughput measurements are required for individual dose estimation for radiobiological triage | ||
| 2. New sequencing technology: nanopore sequencing to detect radiation-responsive genes in human peripheral blood: portable, rapid, real-time biodosimetry platform | |||
| 3. Identification of specific FDXR transcript variants responsive to ionizing radiation (IR) –>in vivo validation in blood of radiotherapy patients | |||
| 4. High responsiveness of FDXR-201 and FDXR-208 | |||
| 5. New class of gene based radiation exposure markers: FDXR-218 and FDXR-219 without endogenous expression, but a clear detection after IR | |||
| B. Terbrueggen | see below | ||
| S. Amundson | 1. Development of algorithms to allow translation between animal and human responses, and between in vitro and in vivo responses | ||
| 2. Exposure modalities relevant for anticipating exposure scenarios: impact of relevant exposure factors such as dose rate, presence of neutrons, or exposure to internal emitters | |||
| 3. Gene expression (GE) approach shows great promise for dose reconstruction and injury prediction | |||
| 4. Better understanding and integration of factors that may impact both an individual’s physiological response to radiation and their GE response still needed | |||
| P. Ostheim | 1. GE changes in the peripheral blood (mRNA and miRNA) provide indications of the exposure pattern and a suggestion of the percentage of the exposed body area | ||
| 2. Dose-dependent GE responses to incorporated radionuclides (223Ra) | |||
| P. Rogan | 1. GE signatures for radiation exposure are susceptible to nonspecific changes caused by common, confounding hematological disorders | ||
| 2. Strategy: necessity to identify and mitigate confounder up front, include confounders as controls, exclude genes responsible for false positive calls | |||
| A. Evans | 1. Transcriptional biomarker can be used for assessing internalized 131I –>differentiate exposed from unexposed | ||
| 2. Dose and time dependent GE signature for internalized 131I over a period of 15 days after exposure | |||
| D. Bazyka | 1. Examination of 314 Chernobyl cleanup workers, and staff of the ‘Shelter’ and exclusion zone with various doses of external irradiation | ||
| 2. Demonstration of a dose–response relationship on a molecular level with promising results for biological dosimetry | |||
| S. Ghandhi | Examination of molecular response to internal radiation using mice injected with 137Cs –>sustained impact of dose and low dose-rate on immune response | ||
| Effect prediction | |||
| P. Ostheim | 1. Dose estimates are a ‘surrogate’ for prediction of acute health effects | ||
| 2. Concept of radiation-related biomarkers for effect prediction: integration of multiple radiation exposure characteristics as well as cell- and molecular-based processes like individual radiosensitivity via molecular changes lying downstream of the exposure and upstream of the effect | |||
| 3. Integrative approach for more robust and simplified prediction of later occurring acute health effects | |||
| M. Gomolka | 1. A radiation specific gene signature was identified in uranium workers 20–30 years after exposure | ||
| 2. Low and high dose exposure groups of uranium workers can by distinguished by gene expression analysis | |||
| 3. Deregulated genes are involved in immune response pathways, including interferon and pro-inflammatory responses | |||
| High-throughput and point-of-care (POC) diagnostics | |||
| B. Terbrueggen | 1. REDI-Dx: high throughput and blood-based biodosimetry test system | ||
| 2. Measurement of radiation responsive mRNA transcripts for estimation of absorbed radiation dose, extremely low false positive rate | |||
| 3. Up to 1200 samples within 24 hours and first results are available after 6.5 hours | |||
| 4. Only biodosimetry test for which the performance has been validated | |||
| F. Zenhausern | 1. Advances of paper-based vertical flow multiplex assay system (VeriFAST) for rapid biodosimetry in a mobile environment | ||
| 2. Simple user interface for self-collection and testing on a low-cost basis | |||
| 3. High sensitivity for a variety of nucleic acids to proteins multiplex panels of biosignatures for different bioeffects | |||
| 4. Future work and validation needed | |||
| C. Badie | Nanopore sequencing: portable, real-time and high-throughput biodosimetry platform for assessing radiation exposure (see above) | ||
| P. Ostheim | 1. Current progress in the development of point-of-care device based on microfluidic technology as a stand-alone platform | ||
| 2. Limitations in the linearity of GE values miniaturization restrictions have to be overcome | |||
| Low-level radiation | |||
| L. Kaatsch | 1. Diagnostic and interventional radiology provides alternative exposure model for LLR experiments, here computed tomography | ||
| 2. Modern CT diagnostics evoke genotoxic alterations with deregulation of well-known genomic biomarkers and DNA damage | |||
| 3. No increased biological effectiveness of varying X-ray spectra in dual-energy CT | |||
| G. Woloschak | 1. Irradiated animals tissue archive hosted at Northwestern University: repository of samples and data collected in the course of large-scale animal studies –>samples and existing datasets available upon request (janus.northwestern.edu) | ||
| 2. Difficult to do an analysis for the animals with very low dose rates 3. Mostly paraffin embedded tissue of differing quality, mRNA measurements challenging and miRNA measurements promising |
|||
| Methods | |||
| P. Ostheim | 1. Saliva, as a noninvasive easily accessible biofluid, contains presumably RNA biomarkers for prediction and diagnosis of several diseases and is poorly characterized for radiation biodosimetry | ||
| 2. Identification of challenges (high yield of bacterial RNA and low yield of human mRNA) and development of a robust methodology to process human whole saliva for GE analysis | |||
| 3. Next step: saliva samples from radiotherapy patients used to evaluate the applicability of this workflow for GE analysis for its use in a radiobiological context | |||
| R. Ullmann | Introduction of a novel RNASeq analysis strategy for whole genome exon- and not gene-based GE screens: rapid, budget-friendly and reproducible | ||
| S. Schüle | 1. Discrepancies in baseline and height of radiation-induced differential GE of commonly used radiation-induced genes have been observed among laboratories and in the validation of GE NGS data (gene-based examination) using qRT-PCR (exon-based examination). | ||
| 2. Identifying radiation responsive exons and primer-probe designs of genes widely used for biodosimetry and ARS prediction | |||
| 3. More meaningful comparison of NGS and qRT-PCR measurements (both exon-based) | |||
| M. Riego | Alternative transcript or splicing variants of known radiation-responsive genes can be used as a potential source of individually variable response to certain radiation qualities at an exon-level | ||
The table is ordered per main section of the key session ‘Gene expression for biodosimetry and effect prediction purposes: promise, pitfalls and future directions’ at ConRad 2021 as well as presenters. It summarizes the key contents of each presentation. Details are provided in the text of the manuscript.
In Section 2, different groups present their views on applicability of GE analysis for biodosimetry purposes. How to translate GE data from ex vivo experiments and animal studies for robust diagnostic use in humans and what exposure modalities are relevant for anticipating different exposure scenarios? What are potential confounders?
In Section 3, we describe the underlying concept and the applicability of radiation-related biomarkers of effect prediction, which can help to integrate multiple radiation exposure characteristics as well as cell- and molecular-based biological processes. This novel idea will be compared to the more established concept of dose estimation, i.e. GE analysis for biodosimetry purposes.
In Section 4, we will outline some of the current developments in this fast-moving field of radiation research to develop high-throughput, cost-effective and portable devices (point-of-care (POC) diagnostic).
In Section 5, we present work on transcriptional changes caused by low dose and different energy levels of computed tomography (CT) as an alternative exposure modality in this field.
In Section 6, we will give an insight on current developments for processing GE data, introducing novel analysis strategies for whole genome expression screening and identifying radiation responsive exon regions within genes. Furthermore, the capability of GE analysis in human whole saliva as an alternative body fluid will be discussed.
2. Biodosimetry
2.1. Gene expression biomarker for retrospective biodosimetry
Considering the causal pathway where biological processes in response to an exposure occur, the blood-based GE biomarkers can be used for upstream reconstruction of the exposure (Amundson et al. 2000, 2001). Examinations of the association of GE with dose represent the topic of the majority of molecular radiobiological publications. Herein, quantitative real-time PCR (qRT-PCR) is still the gold-standard methodology for providing robust and sensitive results. The technological developments in this field, especially microarray and RNA sequencing techniques have allowed an agnostic approach. This has resulted in the identification of complex responses comprising hundreds of genes for robustly discriminations, e.g. exposed from unexposed groups or different exposure levels (Abend et al. 2021; Amundson 2021).
FDXR is a gene known to show transcriptional responsiveness to IR over a large range of doses (O’Brien et al. 2018). Most recent results from Badie et al. even showed a new class of gene based radiation exposure markers: nanopore sequencing was performed to identify new promising genes as well as splicing variants of FDXR (Cruz-Garcia, O’Brien, Sipos, Mayes, Tichý, et al. 2020). In further work, a dose-dependent up-regulation for specific FDXR transcript variants using qRT-PCR was reported (Cruz-Garcia, O'Brien, Sipos, Mayes, Love, et al. 2020). Hereby, eight alternative transcripts could be validated in human whole blood from radiotherapy patients in response to IR.
2.2. Useful study design
The ultimate goal of the development of GE biomarkers is to produce biodosimetry methods that are useful for humans exposed to IR in vivo. Assessment of these biomarkers in human studies is limited because of a scarcity of human population radiation exposures, though in some cases clinical therapeutic radiation exposures have been used. Most groups rely on human blood irradiated ex vivo or on animal models like mice or ideally non-human primates (NHPs) (Port et al. 2018; Abend et al. 2021). However, there is a need to combine in vivo with ex vivo studies and to bridge the inter-species gap by assessing several species and to compare results with findings in irradiated patient populations and healthy humans (Amundson 2021). The idea is, that by learning how to translate between ex vivo and in vivo studies and between animals and humans, the community will be able to develop a novel radiation biodosimetry approach that is robust enough to be officially approved for its diagnostic use in humans.
Efforts were and are made to translate the ex vivo to in vivo models. Hereby, a previous study has shown that ex vivo irradiation is an appropriate model that can provide meaningful prediction of in vivo exposure levels, in this case using blood from hematopoietic stem cell transplant patients undergoing total body irradiation (TBI) (Paul et al. 2011). While this study only comprises a limited dose range, it exemplifies an early demonstration of transfer from ex vivo signatures to in vivo samples.
As mentioned above, a majority of the numerous proof-of-concept studies to build gene sets and signatures for biodosimetry purposes heavily rely on animal models, like mice, knowing that they show significant differences from human responses. Even NHPs do not correspond perfectly to human radiation responses, even when candidates from animal models have striking gene sequence homology to humans. For example, in a baboon model, FDXR was down-regulated (Port et al. 2016), whereas it appeared up-regulated in humans (Port et al. 2018) after radiation exposure. The question arises of how to bridge this inter-species gap. In a study looking at the interspecies translation of GE findings using an ex vivo irradiation model, it was possible to build a model for using NHP data that has similar accuracy of dose reconstruction in NHP and humans when selecting interspecies correlated genes and applying multiple regression-based cross-species conversion of expression values (Park et al. 2017). A successful inter-species validation using in vivo and ex vivo blood models in baboons and humans (leukemia patients) was recently reported (Port et al. 2018). It provided strong evidence for the applicability of a subset but not all candidate genes examined in both species in vivo as well as ex vivo.
As radiation accidents are known to be rare events, availability of human biological samples is even rarer. In this context, Bazyka et al. reported on promising GE dose–response relationships after irradiation in Chernobyl clean-up workers (Ilienko et al. 2018, 2020), and staff of the ‘Shelter’ and exclusion zone (Bazyka et al. 2018, 2020). The term ‘biological dosimeter’ was frequently used in this context, emphasizing the underlying association of dose to relative GE in this context.
Besides setting up new studies and exposure models, another way for performing GE studies is to use archived samples that already exist. Therefore, the irradiated animals tissue archive hosted at Northwestern University is a repository of samples and data collected in the course of large-scale animal studies performed between the 1950s and 1990s in several different national laboratories and institutes in the USA. These archival samples and existing datasets (including dogs, mice and rats) are available upon request (janus.northwestern.edu) and can be used for determination of low-dose rate effects or examination of incorporated radionuclides vs. external beam exposures. Investigations comprise the analysis of expression changes of proteins, miRNAs (mRNA are often degraded) and DNA damage.
2.3. Exposure modalities relevant for anticipated exposure scenarios
As mentioned above, the association of GE with dose represents the topic of the majority of molecular radiobiological publications (Abend et al. 2021). In comparison, the impact on GE in peripheral blood cells of dose rate, incorporated radionuclides, or neutrons, likely to be present in an improvised nuclear device (IND) detonation, have been less extensively examined.
Amundson et al. contributed to closing these gaps and examined the impact on GE of dose rate (Ghandhi et al. 2015, 2019), neutron exposure (Broustas, Xu, Harken, Chowdhury, et al. 2017; Broustas, Xu, Harken, Garty, et al. 2017; Broustas et al. 2018; Mukherjee et al. 2019) and exposure to incorporated emitters (Paul et al. 2014; Ghandhi et al. 2020; Shuryak et al. 2020). Their neutron studies used an accelerator-based biological irradiation facility simulating neutron exposure from an IND for irradiation of blood or small animals (mice) (Xu et al. 2015). Authors reported that the relative biological effectiveness (RBE) for neutrons differed for certain genes compared to X-ray exposure, suggesting that GE may provide information to differentiate exposures of different radiation quality (Broustas, Xu, Harken, Chowdhury, et al. 2017).
Considering a scenario with a radiologic dispersal device (‘dirty bomb’) (Rump et al. 2018), changes in GE are sought to be an easy method to detect incorporated radionuclides in human peripheral blood for improved internalized radionuclide diagnostic with the potential of high throughput diagnosis. Therefore, internalized radionuclide studies were set up, using murine models for examination of 137Cs incorporation (Ghandhi et al. 2020) as well as patients for examination of 223Ra incorporation (Ostheim et al. 2021). Dose-dependent GE responses to incorporated radionuclides could be found in these studies although the GE patterns appeared very complex in all incorporation studies, most likely resulting from continued irradiation at a decreasing dose rate. The human in vivo study for 223Ra incorporation was further challenged by the severe health conditions of the patients, potentially masking the underlying association between incorporated alpha-emitting radionuclides and GE changes.
In an effort to further study such variable dose-rates, an external 137Cs irradiator was used for long-term low dose rate studies (Garty et al. 2020). Interestingly, sex-specific differences were found in the applied murine model when looking at the relative response of different blood cell subtypes. Protracted exposures showed almost complete decline of T-cells in male but not female mice. In contrast, B-cells showed similar initial declines in both sexes, but a much more rapid recovery in female mice.
In vivo GE responses following 131I, another nuclide of interest following a ‘dirty bomb’ incident, were studied in relapsed and refractory neuroblastoma patients receiving 131I-tagged metaiodobenzylguanidine (131I-mIBG) as part of their radiotherapy regimens (Edmondson et al. 2016; Campbell et al. 2017). These studies utilized qRT-PCR to determine GE changes in human peripheral blood lymphocytes 3–4 days following 131I-mIBG treatment, providing both a GE-based biodosimetry model for 131I exposures (Edmondson et al. 2016) and identifying biomarkers of 131I toxicity in children (Campbell et al. 2017). Recently, Coleman et al. also found that transcript signatures in the peripheral blood may differentiate exposed from unexposed samples up to 15 days after 131I-mIBG treatment (Evans et al. 2021).
2.4. Total and partial body irradiation
For any type of radiological event, the exposure is expected to be heterogeneous and partial exposure more likely than whole body exposure. In an IND event, people may be partially shielded to prompt radiation by the structures surrounding them, and exposure to radionuclides from fallout will be non-uniform (Preston and Pierce 1988; Cullings et al. 2006; Sakata et al. 2012; Coleman and Koerner 2016). Although many of the early radiation dose reconstruction models were based on uniform total body exposure, new research in the field is directed at developing dose reconstruction models for partial exposures (Blakely et al. 2014; Sproull et al. 2017). The knowledge of the exposure pattern is an important characteristic when thinking about acute radiation health effects in a radiological scenario. In an NHP study, where animals were treated with different exposure patterns, 55 miRNAs (Ostheim et al. 2019) and about 20-times more mRNA species (Ostheim et al. 2020) associated with the exposed body area could be identified. Interestingly, 21 miRNAs revealed significant linear associations of GE changes with the percentage of the exposed body area. These results might provide indications of the exposure pattern and a suggestion of the percentage of the exposed body area.
2.5. Confounders of gene expression
There is an ongoing debate on the impact of confounders such as diseases but also demographic parameters (ethnicity, age, gender) on certain GE markers and e.g. the associated detection of unexposed or exposed individuals. Though few studies reported on a negligible effect of these factors, at least for discrimination of unexposed healthy donors from heavily exposed individuals (Agbenyegah et al. 2018), there are also hints that metabolomics changes (such as smoking (Paul and Amundson 2011), simulated bacterial infection and curcumin inflammation (Cruz-Garcia et al. 2018)), viral infections and blood-borne diseases can modify the normal baseline values of biomarkers used for diagnostic analysis of radiation exposure (Zhao et al. 2018). Most recent results, examining the impact of aging on GE response to X-ray irradiation using mouse blood, showed that age-dependent GE differences should be considered when developing gene signatures for use in radiation biodosimetry (Broustas et al. 2021). All these confounding conditions may lead to misclassification or high false positive rates for diagnosis of radiation exposure. In order to overcome these confounding issues and to mitigate confounder effects, Rogan et al. suggested a strategy for identification of gene signatures which (1) identifies and eliminates confounders up front, (2) validates designs by including patients with these conditions before they are applied clinically and (3) to exclude genes responsible for false positive calls, i.e. related to biological processes other than DNA damage response or apoptosis. While many highly performing radiation signatures are currently available, only those with performance that is not confounded by underlying pathologies may be useful for assessment, they conclude (Mucaki et al. 2021).
2.6. Challenges and promises
As outlined above there is still need for improvement in the translation of ex vivo to in vivo and from animal models to humans and to examine GE response to various radiation qualities, dose rates and exposures to internalized radionuclides as well as understanding the impact of potential confounders like sex, age, co-morbidities, and combined injuries.
Confounding factors might reduce the specificity of radiation-induced GE changes. This highlights the requirement for considering GE changes in context. In a radiological or nuclear scenario, clinical parameters (e.g. vomiting, diarrhea, and erythema), physical measurements, and different methods to detect biological changes should be considered holistically in order to minimize misclassifications (Blakely et al. 2010; Shuryak et al. 2020).
3. Effect prediction
3.1. The concept of effect prediction compared to retrospective dosimetry
As mentioned in Section 2.6, GE changes have been widely used as a tool for biodosimetry, but have limitations regarding diagnosis and triage of radiation injury patients suffering from acute radiation health effects considered here only (Port et al. 2019).
It is not uncommon assuming an increased hematological ARS (H-ARS) severity with increasing dose (Khvostunov et al. 2011). However, recently a non-linear relationship of dose and H-ARS severity degree was reported based on real case histories from radiation accidents (Abend et al. 2021). Exposures below 1 Gy roughly corresponded with no or a low-grade H-ARS (no hospitalization and intensive care are required) and doses above 5 Gy mainly correspond with severe H-ARS severity degrees (early hospitalization and intensive care are required). Knowing the dose in these situations provides a good estimate for acute health effects. However, whole body doses between 1 and 5 Gy poorly corresponded to different H-ARS degrees of severity, making an individual recommendation based only on dose almost impossible. That does not render absorbed dose uninformative, but reflects the complexity of the dose concept for acute health effect predictions.
Acute radiation syndrome (ARS) is caused by massive and sudden cell death and multiple radiation exposure characteristics (e.g. radiation quality, dose fractionation, dose rate, and partial/TBI) as well as biological processes (e.g. radiosensitivity, cell cycle dependency, and oxygenation) are known to contribute (Hall and Giaccia 2012). Examination of radiation-induced GE changes downstream of radiation exposure and upstream of the ARS event provide the basis of introducing radiation-related biomarkers for effect prediction (Abend and Port 2016; Port et al. 2018, 2021). Compared to dose estimation, this integrative approach appears more robust and easier to follow for clinicians, since GE changes are allocated to clinically defined ARS severity categories (see below) associated with certain treatment options.
In order to investigate long-term health effects of radiation, transcriptional profiles of 200 former uranium workers (‘Wismut cohort’) were examined by using a microarray platform (Kreuzer et al. 2011). Hereby, decades after irradiation, GE alterations of genes related to the immune system could be identified, which allowed discrimination between high and low radiation exposure groups (Maria Gomolka, data not published).
3.2. Predicting hematological ARS based on radiation-induced gene expression changes
METREPOL (MEdical TREatment ProtocOLs for Radiation Accident Victims) categorizes H-ARS into four severity degrees based on blood cell count (BCC) changes in the weeks after exposure: low (H1), medium (H2), severe (H3) and fatal (H4) H-ARS. Normal BCCs in METREPOL are not considered and are introduced as H0 herein. These severity degrees are associated with treatment decisions (Friesecke et al. 2001). Using a baboon model (Port et al. 2016) combined with ex vivo experiments (Ostheim et al. 2021) as well as measurements on healthy donors (Agbenyegah et al. 2018) and radiotherapy patients (Port et al. 2018) for validation purposes, a GE signature was developed that consists of only four genes in the most basic version (FDXR/DDB2 and WNT3/POU2AF1) for prediction of H-ARS. These genes allow triaging radiation exposed persons within the first days after radiation exposure: (1) unexposed individuals (H-ARS 0) to conserve clinical resources for those requiring it, (2) low-exposed individuals (H-ARS 1) requiring surveillance (late health effects) but no hospitalization or early treatment and (3) highly exposed individuals who will develop acute health effects, e.g. H-ARS 2–4 severity degree. The latter are in need of early intensive care. Merging existing H-ARS categories in those three categories appears appropriate to address urgent clinical questions such as prioritizing hospitalization as well as considering restricted medical resources. GE onset can be detected as early as 2–6 hours after irradiation, thus, underlining the potential as an early and high-throughput diagnostic tool (Ostheim et al. 2021).
Although this gene set has been validated in human patient models and using extensive ex vivo analysis, additional analysis for confirmation is required. Therefore, blood samples from a different species of NHPs (rhesus macaques) as well as blood samples from 92 chemotherapy-related female breast cancer patients will be analyzed for a further validation step of the diagnostic tool.
3.3. Challenges and promises
Although this narrowed gene set holds promise for applicability as a diagnostic tool for prediction of clinical outcomes and its use as a biomarker of acute health effects, the described approach is developed for H-ARS prediction. Although this is an important step in diagnosis of the ARS considering the bone marrow to be most radiosensitive, other organ systems are also affected by ARS (e.g. gastrointestinal or the neurovascular system). Our diagnostic tool does not directly cover those yet.
Again, a holistic view and the combination of clinical parameters, physical measurements and different methods to detect biological changes seems to be the most robust approach in order to minimize misclassifications (Shuryak et al. 2020). This is also important considering numerous confounding conditions that may affect the expression of certain genes of interest as outlined in Sections 2.5 and 2.6.
4. High-throughput and point-of-care diagnostics
In a large-scale radiological or nuclear event, high-throughput measurements and POC diagnostics are required for individual dose estimation and/or effect prediction with regard to an early radiobiological triage. It is required to determine the unexposed, low- and high-exposed individuals in order to initiate appropriate diagnostic and therapeutic interventions. Due to the early and high-throughput characteristics, blood-based GE markers seem to be attractive for this purpose. However, blood sampling and laboratory-based analysis are limiting factors to provide these results just in time. In order to overcome this ‘bottleneck’, several developments are currently under investigation, hereby striving to be as cost-effective and portable as possible.
Besides qRT-PCR using low-density arrays for measurement of up to 384 genes (Thermo Fisher Scientific, Waltham, MA) or the 12k open array format (OA, Thermo Fisher Scientific, Waltham, MA), next-generation sequencing (NGS) and nanopore sequencing based technologies with multiplexing of hundreds of samples in one reaction provide high-throughput capabilities.
4.1. High-throughput
In order to address the high-throughput applicability of GE measurements for dose estimation and early prediction of clinical outcomes, several experiments have proven that results can be provided from 1000 samples within 30 hours under optimal conditions (Port et al. 2019). Hereby, samples were processed automatically using the QIASymphony (Qiagen, Hilden, Germany), followed by targeted NGS. In total, 100 times more samples could be processed three times faster compared to established cytogenetic assays.
As part of an overall strategy for effective medical management of large-scale radiological or nuclear emergencies in the US, a high-throughput and blood-based biodosimetry test system called REDI-Dx was developed (Jacobs et al. 2020). The system consists of 18-plex GE assays (including housekeeping genes), which measure radiation responsive mRNA transcripts for estimation of absorbed radiation dose (range: 0–10 Gy). A single platform can process up to 1200 samples within 24 hours and first results are available after 6.5 hours. A validation study in human cancer patients TBI, irradiated NHPs (TBI) and non-irradiated human samples revealed sufficient sensitivity and specificity considering several potential confounders. The accuracy of the test would aid in the assignment of patients to the correct treatment category deduced from certain dose categories (>93%). Moreover, if the rate of false positives was very low, this would be especially effective in calming the non-irradiated concerned public. Until now, exact dose estimation is challenging, but dose categories can be discriminated sufficiently. Further work including, e.g. dose fractionation will be needed to ensure that this system will be robust and appropriate for its intended use.
4.2. POC
For a long time, the only POC capabilities for biodosimetry assessment were lymphocyte depletion kinetics and basic clinical examinations (Sullivan et al. 2013). Modern molecular technology in this field opened up new doors.
With the nanopore sequencing method mentioned in Section 2.1, the basis for the future development of a portable, real-time and high-throughput biodosimetry platform has been already introduced (Cruz-Garcia, O’Brien, Sipos, Mayes, Tichý, et al. 2020; Cruz-Garcia, O'Brien, Sipos, Mayes, Love, et al. 2020). The technology for sequencing of single-stranded DNA, respectively cDNA from RNA molecules, relies on the use of transmembrane proteins that are recording electrical changes characteristically for each of the DNA bases. Hereby, the minimum sequencing time required for dose estimation of irradiated samples using specific transcripts could be less than three minutes for a total of 50,000 reads in up to 50 samples. For gaining the full potential of this portable and rapid technology, some future improvements regarding sample processing and the bioinformatic pipeline for specific radiation-responsive transcript identification are required. For example, in order to overcome the bottleneck of RNA extraction prior to nanopore sequencing, a further module for in-field RNA extraction has to be slotted ahead.
The paper-based vertical flow multiplex assay system (VeriFAST) is a further development for rapid biodosimetry in a mobile environment. This versatile and low-cost platform for simultaneous testing of genes and proteins demonstrated a high sensitivity for a variety of nucleic acids and proteins (Chen et al. 2019; Devadhasan et al. 2021). One of the main advantages of this system is the kit configuration with a simple user interface for self-collection and testing in various mobile or remote environmental conditions. Future work will have to focus on optimization of assay chemistries and packaging, kit mass production, as well as regulatory development for specific applications. Further validation studies for robust and appropriate results with regard to biodosimetry assessment are lacking so far.
The microfluidic technology, that can handle small amounts of fluids in a confined and controlled environment, has been an emerging field for several years (Lacombe et al. 2016). Due to the potential of these stand-alone systems for automated user-friendly, reproducible and sensitive analyses, these devices may also be used in clinical biodosimetry. The idea is to miniaturize the established laboratory workflow for performing GE analysis as robustly as usual. Such a POC device would enable first responders worldwide to immediately triage radiation accident patients without a highly specialized laboratory, e.g. in hospitals, where potentially radiation exposed individuals will arrive in a radiological or nuclear event. Although expenses per cartridge have to be considered, the ‘lab on a chip’ technique would allow high mobility compared to the time consuming and often difficult shipment of human samples. Whereas several reports deal with the microfluidic technology used for GE analysis (Shinde et al. 2010), demonstrating some movement in this direction, no established qRT-PCR based microfluidic cartridge exists so far. Therefore, such a stand-alone device for RNA isolation and simultaneous detection of up to eight radiation responsive genes for prediction of individual radiological injury is currently under development at the Bundeswehr Institute of Radiobiology. At the moment, challenges like limitations in the linearity of GE values using a one-step or two-step qRT-PCR and miniaturization restrictions have to be overcome.
5. Low level radiation (LLR)
Investigation of health effects of low doses of radiation (mainly 100 mGy as cutoff) as a field of study has been riddled by difficulties since its inception (Paunesku and Woloschak 2018). This is most often ascribed to the fact that effects of low doses of radiation are subtle and difficult to distinguish from the glut of other ‘low grade’ stresses. Epidemiological LLR studies have to include hundreds of thousands of samples in order to provide statistically meaningful results concerning risk assessment. Even after the investigation of well controlled animal studies, the understanding of biological basis for risk from low dose radiation exposure is still not conclusive. As described in Section 2.3, a study was conducted using a repository of samples and data collected in the course of large-scale animal studies between the 1950s and 1990s in the USA. Hereby, it was hard to do an analysis for the animals with very low dose rates, because control animals developed cancer almost as often as LLR animals, which might be caused by the experimental stress conditions (Zander et al. 2021).
In the light of these limitations, there is a need of further insights into the biological processes induced by LLR to complement the existing knowledge base of risk assessment. An alternative exposure model for LLR experiments is provided by the field of diagnostic and interventional radiology (Kaatsch et al. 2020). Due to the increased usage of CT nowadays, this accounts for the majority of civilian radiation exposure. With regard to risk assessment and biological effects caused by varying energy levels in modern CT scanning techniques, a recent study investigated the biological effects of single- and dual-energy CT (DECT) with a special focus on the early transcriptomic response of peripheral blood cells. Using an ex vivo model, genotoxic alterations with a deregulation of well-known genomic biomarkers and DNA damage could be found, but no increased biological effectiveness was observed with regard to various X-ray spectra (80/150 kV) in DECT (Kaatsch et al. 2021). The results from this exploratory study are just the beginning of LLR experiments in a clinical environment and further, especially in vivo validation studies have to be performed. Further, the question whether there is a trend toward lower biological effectiveness caused by DECT has not been comprehensively answered yet.
6. Methodological considerations
6.1. New ways of data analysis
In previous inter-laboratory comparison exercises, differences in baseline and magnitude of radiation-induced differential GE of commonly used radiation-induced genes (e.g. FDXR) have been observed among laboratories (Port et al. 2018). These discrepancies could be caused by exon regions of the same gene with inherent differences in radiation-responsiveness, examined by different teams. In an effort to examine this issue, qRT-PCR based GE analysis using many different TaqMan® assays covering all exon regions of four genes (FDXR, DDB2, WNT3 and POU2AF1) was performed. Hereby, most promising exon regions and primer-probe designs were identified for these four genes by e.g. considering sufficiently detectable baseline gene copy numbers, height of radiation-induced fold-changes and inter-individual variance. These results have further implications when it comes to the validation of NGS results using qRT-PCR. In a conventional NGS workflow reads are allocated to genes, but with qRT-PCR exon-specific primer probe designs recognize only one specific exon of the whole gene, thus, discrepancies between both methods can occur. This analysis based on ‘radiation-responsive exons’ has the advantage that it comprises information of all splicing variants of a gene compared to the analysis of radiation-responsive transcripts which can be processed with nanopore sequencing only due to the large size of reads (see Section 2.1).
A RNASeq analysis strategy for whole genome GE screens was employed at the Bundeswehr Institute of Radiobiology enabling a rapid, budget-friendly and reproducible exon based NGS analysis. After providing the sequence files and the sample information, RNASeq quality check, sequence alignment, determination of expression levels and identification of both GE changes and differential exon usage are automatically performed with default settings. Finally, a comprehensive report is generated that summarizes the results and various data quality characteristics.
A further study from Sweden examined whether distinct alternative transcript or splice variants of known radiation-responsive genes in ex vivo irradiated human blood samples can be used as a potential source of individually variable response after mixed beam exposures (X-rays, alpha exposure or mixed beams of both). In summary, the preliminary results show different responses of the donors to certain radiation qualities at the exon-level (manuscript in preparation).
6.2. Saliva as an alternative biofluid used for GE related biodosimetry
As the great majority of GE biodosimetry studies have been performed using blood as the preferred source of tissue, the search for simple, less-invasive sampling methods plays an important role when considering sample collection for GE analysis in field conditions. Saliva as a blood plasma ultra-filtrate is thought as a ‘mirror of the body’ (Segal and Wong 2008; Yoshizawa et al. 2013). The collection of saliva samples represents an ideal noninvasive and easily-accessible alternative to blood. It has been shown to contain RNA biomarkers for prediction and diagnosis of several diseases (Li et al. 2004; Kaczor-Urbanowicz et al. 2017; Ghizoni et al. 2020), but is poorly characterized for radiation biodosimetry (Pernot et al. 2014; Lacombe et al. 2017).
In previous work, two problematic issues were identified, not coherently described before: (1) most of the isolated RNA originates from the oral microbiome and (2) the amount of isolated human RNA is comparatively low. Therefore, a robust workflow was developed to process human whole saliva for GE analysis, introducing a modified cDNA synthesis aiming at the poly(A)+-tail and a pre-amplification step prior to qRT-PCR (Ostheim et al. 2020). Recently, further efforts were made to advance this workflow and to examine the influence of sociodemographic and epidemiologic characteristics that could potentially influence salivary isolates (Ostheim et al. 2021).
7. Conclusions
Emergency response in the scenario of a radiological incident of mass population requires multiple biodosimetry technology platforms for early, high-throughput and rapid POC diagnostics regarding both dose estimation and acute health effect prediction. Employing radiation-induced GE changes provides great promise, but limitations inherent to this approach have to be further analyzed as well as specified and may best be addressed by combining other sources of information into a holistic approach including, e.g. clinical signs and symptoms or physical dosimetry.
Biographies
Patrick Ostheim, MD, is a Post-Doctoral Researcher of Radiobiology and a resident in Radiology at the Bundeswehr Institute of Radiobiology, Munich, Germany.
Sally A. Amundson, ScD, is an Associate Professor of Radiation Oncology and a faculty member of the Center for Radiological Research, Columbia University Irving Medical Center (CUIMC), New York, USA.
Christophe Badie, PhD, is a Researcher and leader of the Cancer Genetics and Cytogenetics group in the research department of Biological Effects at Public Health England’s Centre for Radiation, Chemical and Environmental Hazards (PHE CRCE), Faculty of Medicine at Imperial College of Science, Technology and Medicine, London, UK.
Dimitry Bazyka, MD, is a Professor of radiobiology and Director-General of the National Research Centre for Radiation Medicine of the Academy of Medical Sciences of Ukraine (RCRM), Kyiv, Ukraine.
Angela C. Evans is a Doctoral Student in the laboratory of Matthew A. Coleman at the University of California Davis, Sacramento, CA, USA and the Lawrence Livermore National Laboratory, Livermore, CA, USA.
Shanaz A. Ghandhi, PhD, is an Assistant Professor of Radiation Oncology at the Center for Radiological Research, Columbia University Irving Medical Center (CUIMC), New York, USA.
Maria Gomolka, PhD, is a biologist and senior scientist in the working group biological radiation effects, biological dosimetry, Federal Office for Radiation Protection, Oberschleissheim, Germany.
Milagrosa López Riego is a PhD Student at the Department of Molecular Biosciences, The Wenner-Gren Institute, Stockholm University, Sweden.
Peter K. Rogan, PhD, is a Professor in Biochemistry at the University of Western Ontario, London, Ontario, Canada, and cofounder of CytoGnomix Inc, London, Ontario, Canada.
Robert Terbrueggen, PhD, is a Researcher and the Founder/CEO of DxTerity Diagnostics, Los Angeles, CA, USA.
Gayle E. Woloschak, PhD, is a Professor of Radiation Oncology and Radiology at the Feinberg School of Medicine, Radiation Oncology, Northwestern University, Chicago, IL, USA.
Frederic Zenhausern, PhD, MBA, is a Professor of Radiation Oncology at the Department of Basic Medical Sciences, College of Medicine, The University of Arizona, Phoenix, AZ, USA, and Director of the Center for Applied Nanobioscience and Medicine, University of Arizona, Phoenix, AZ, USA.
Hanns L. Kaatsch, MD, is a Post-Doctoral Researcher of Radiobiology and a resident in Radiology at the Bundeswehr Institute of Radiobiology, Munich, Germany.
Simone Schüle, MD, is a Post-Doctoral Researcher of Radiobiology and a resident in Radiology at the Bundeswehr Institute of Radiobiology, Munich, Germany.
Reinhard Ullmann, PhD, is a biologist and senior scientist at the Bundeswehr Institute of Radiobiology, Munich, Germany.
Matthias Port, MD, is a Professor of Radiobiology and Internal Medicine and Head of the Bundeswehr Institute of Radiobiology, Munich, Germany.
Michael Abend, MD, is a Professor of Radiobiology and Deputy Head of the Bundeswehr Institute of Radiobiology, Munich, Germany.
Funding Statement
This work was supported by the German Ministry of Defense. SAA and SAG were supported through the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases Grant Number U19 AI067773. AE was supported by the National Cancer Institute Grant Number R01 CA172067. Work was also performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344.
Disclosure statement
The authors declare that they have no conflict of interest.
References
- Abend M, Blakely WF, Ostheim P, Schüle S, Port M.. 2021. Early molecular markers for retrospective biodosimetry and prediction of acute health effects. J Radiol Prot. [DOI] [PubMed] [Google Scholar]
- Abend M, Port M.. 2016. Combining radiation epidemiology with molecular biology-changing from health risk estimates to therapeutic intervention. Health Phys. 111(2):183–185. [DOI] [PubMed] [Google Scholar]
- Agbenyegah S, Abend M, Atkinson MJ, Combs SE, Trott KR, Port M, Majewski M.. 2018. Impact of inter-individual variance in the expression of a radiation-responsive gene panel used for triage. Radiat Res. 190(3):226–235. [DOI] [PubMed] [Google Scholar]
- Amundson SA. 2021. Transcriptomics for radiation biodosimetry: progress and challenges. Int J Radiat Biol. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundson SA, Bittner M, Meltzer P, Trent J, Fornace J.. 2001. Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat Res. 156(5 Pt 2):657–661.2.0.CO;2] [DOI] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, Fornace J.. 2000. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 154(3):342–346. [DOI] [PubMed] [Google Scholar]
- Bazyka D, Ilienko I, Golyarnik N, Belyaev O, Lyaskivska O.. 2020. Gene expression and cellular markers of occupational radiation exposure in Chernobyl shelter construction workers. Health Phys. 119(1):37–43. [DOI] [PubMed] [Google Scholar]
- Bazyka D, Ilienko I, Sushko V, Loganovsky K, Lyashenko L, Golyarnik N, Lyaskivska O, Nechaev S, Shvayko L, Bazyka K, et al. 2018. Biological markers of external and internal exposure in shelter construction workers: a 13-year experience. Radiat Prot Dosimetry. 182(1):146–153. [DOI] [PubMed] [Google Scholar]
- Blakely WF, Ossetrova NI, Whitnall MH, Sandgren DJ, Krivokrysenko VI, Shakhov A, Feinstein E.. 2010. Multiple parameter radiation injury assessment using a nonhuman primate radiation model-biodosimetry applications. Health Phys. 98(2):153–159. [DOI] [PubMed] [Google Scholar]
- Blakely WF, Sandgren DJ, Nagy V, Kim SY, Sigal GB, Ossetrova NI.. 2014. Further biodosimetry investigations using murine partial-body irradiation model. Radiat Prot Dosimetry. 159(1–4):46–51. [DOI] [PubMed] [Google Scholar]
- Boldt S, Knops K, Kriehuber R, Wolkenhauer O.. 2012. A frequency-based gene selection method to identify robust biomarkers for radiation dose prediction. Int J Radiat Biol. 88(3):267–276. [DOI] [PubMed] [Google Scholar]
- Broustas CG, Duval AJ, Amundson SA.. 2021. Impact of aging on gene expression response to X-ray irradiation using mouse blood. Sci Rep. 11(1):10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustas CG, Harken AD, Garty G, Amundson SA.. 2018. Identification of differentially expressed genes and pathways in mice exposed to mixed field neutron/photon radiation. BMC Genomics. 19(1):504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustas CG, Xu Y, Harken AD, Chowdhury M, Garty G, Amundson SA.. 2017. Impact of neutron exposure on global gene expression in a human peripheral blood model. Radiat Res. 187(4):433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustas CG, Xu Y, Harken AD, Garty G, Amundson SA.. 2017. Comparison of gene expression response to neutron and X-ray irradiation using mouse blood. BMC Genomics. 18(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K, Karski EE, Olow A, Edmondson DA, Kohlgruber AC, Coleman M, Haas-Kogan DA, Matthay KK, DuBois SG.. 2017. Peripheral blood biomarkers associated with toxicity and treatment characteristics after 131I-metaiodobenzylguanidine therapy in patients with neuroblastoma. Int J Radiat Oncol Biol Phys. 99(2):468–475. [DOI] [PubMed] [Google Scholar]
- Chen P, Gates-Hollingsworth M, Pandit S, Park A, Montgomery D, AuCoin D, Gu J, Zenhausern F.. 2019. Paper-based vertical flow immunoassay (VFI) for detection of bio-threat pathogens. Talanta. 191:81–88. [DOI] [PubMed] [Google Scholar]
- Coleman CN, Koerner JF.. 2016. Biodosimetry: medicine, science, and systems to support the medical decision-maker following a large scale nuclear or radiation incident. Radiat Prot Dosimetry. 172(1–3):38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Garcia L, O’Brien G, Sipos B, Mayes S, Tichý A, Sirák I, Davídková M, Marková M, Turner DJ, Badie C.. 2020. In vivo validation of alternative FDXR transcripts in human blood in response to ionizing radiation. Int J Mol Sci. 21(21):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Garcia L, O'Brien G, Donovan E, Gothard L, Boyle S, Laval A, Testard I, Ponge L, Woźniak G, Miszczyk L, et al. 2018. Influence of confounding factors on radiation dose estimation using in vivo validated transcriptional biomarkers. Health Phys. 115(1):90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Garcia L, O'Brien G, Sipos B, Mayes S, Love MI, Turner DJ, Badie C.. 2020. Generation of a transcriptional radiation exposure signature in human blood using long-read nanopore sequencing. Radiat Res. 193(2):143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullings HM, Fujita S, Funamoto S, Grant EJ, Kerr GD, Preston DL.. 2006. Dose estimation for atomic bomb survivor studies: its evolution and present status. Radiat Res. 166(1 Pt 2):219–254. [DOI] [PubMed] [Google Scholar]
- Devadhasan JP, Gu J, Chen P, Smith S, Thomas B, Gates-Hollingsworth M, Hau D, Pandit S, AuCoin D, Zenhausern F.. 2021. Critical comparison between large and mini vertical flow immunoassay platforms for Yersinia pestis detection. Anal Chem. 93(27):9337–9344. [DOI] [PubMed] [Google Scholar]
- Edmondson DA, Karski EE, Kohlgruber A, Koneru H, Matthay KK, Allen S, Hartmann CL, Peterson LE, DuBois SG, Coleman MA.. 2016. Transcript analysis for internal biodosimetry using peripheral blood from neuroblastoma patients treated with (131)I-mIBG, a targeted radionuclide. Radiat Res. 186(3):235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A, Setzkorn T, Edmondson D, Segelke H, Wilson P, Matthay K, Granger M, Marachelian A, Haas-Kogan D, DuBois S, et al. 2021. Peripheral blood transcript signatures after internal 131I-mIBG therapy in relapsed and refractory neuroblastoma patients identifies early and late biomarkers of internal 131I exposures. Rad Res. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesecke I, Beyrer K, Fliedner TM, METREPOL team. Medical treatment protocols for radiation accident victims as a basis for a computerised guidance system . 2001. How to cope with radiation accidents: the medical management. Br J Radiol. 74(878):121–122. [DOI] [PubMed] [Google Scholar]
- Garty G, Xu Y, Johnson GW, Smilenov LB, Joseph SK, Pujol-Canadell M, Turner HC, Ghandhi SA, Wang Q, Shih R, et al. 2020. VADER: a variable dose-rate external 137Cs irradiator for internal emitter and low dose rate studies. Sci Rep. 10(1):19899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Sima C, Weber WM, Melo DR, Rudqvist N, Morton SR, Turner HC, Amundson SA.. 2020. Dose and dose-rate effects in a mouse model of internal exposure to 137Cs. Part 1: global transcriptomic responses in blood. Radiat Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Smilenov LB, Elliston CD, Chowdhury M, Amundson SA.. 2015. Radiation dose-rate effects on gene expression for human biodosimetry functional and structural genomics. BMC Med Genomics. 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Smilenov L, Shuryak I, Pujol-Canadell M, Amundson SA.. 2019. Discordant gene responses to radiation in humans and mice and the role of hematopoietically humanized mice in the search for radiation biomarkers. Sci Rep. 9(1):19434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghizoni JS, Nichele R, de Oliveira MT, Pamato S, Pereira JR.. 2020. The utilization of saliva as an early diagnostic tool for oral cancer: microRNA as a biomarker. Clin Transl Oncol. 22(6):804–812. [DOI] [PubMed] [Google Scholar]
- Hall EJ, Giaccia AJ.. 2012. Radiobiology for the radiologist. 7th ed. Philadelphia: Wolters Kluwer. [Google Scholar]
- Homer MJ, Raulli R, DiCarlo-Cohen AL, Esker J, Hrdina C, Maidment BW, Moyer B, Rios C, Macchiarini F, Prasanna PG, et al. 2016. United States Department of Health and Human Services Biodosimetry and Radiological/Nuclear Medical Countermeasure Programs. Radiat Prot Dosimetry. 171(1):85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IAEA . 2011. Cytogenetic dosimetry: applications in preparedness for and response to radiation emergencies. Man Ser.; p. 247. [Google Scholar]
- Ilienko IM, Bazyka DA, Golyarnyk NA, Zvarych LM, Shvayko LI, Bazyka KD.. 2020. Changes in gene expression associated with non-cancer effects of the Chernobyl clean-up workers in the remote period after exposure. Probl Radiac Med Radiobiol. 25:456–477. [DOI] [PubMed] [Google Scholar]
- Ilienko IM, Golyarnik NA, Lyaskivska OV, Belayev OA, Bazyka DA.. 2018. Expression of biological markers induced by ionizing radiation at the late period after exposure in a wide range of doses. Probl Radiatsiinoi Medytsyny Ta Radiobiolohii. 2018(23):331–350. [DOI] [PubMed] [Google Scholar]
- Jacobs AR, Guyon T, Headley V, Nair M, Ricketts W, Gray G, Wong JYC, Chao N, Terbrueggen R.. 2020. Role of a high throughput biodosimetry test in treatment prioritization after a nuclear incident. Int J Radiat Biol. 96(1):57–66. [DOI] [PubMed] [Google Scholar]
- Kaatsch HL, Becker BV, Schüle S, Ostheim P, Nestler K, Jakobi J, Schäfer B, Hantke T, Brockmann MA, Abend M, et al. 2021. Gene expression changes and DNA damage after ex vivo exposure of peripheral blood cells to various CT photon spectra. Sci Rep. 11(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaatsch HL, Majewski M, Schrock G, Obermair R, Seidel J, Nestler K, Abend M, Waldeck S, Port M, Ullmann R, et al. 2020. CT irradiation-induced changes of gene expression within peripheral blood cells. Health Phys. 119(1):44–51. [DOI] [PubMed] [Google Scholar]
- Kabacik S, MacKay A, Tamber N, Manning G, Finnon P, Paillier F, Ashworth A, Bouffler S, Badie C.. 2011. Gene expression following ionising radiation: identification of biomarkers for dose estimation and prediction of individual response. Int J Radiat Biol. 87(2):115–129. [DOI] [PubMed] [Google Scholar]
- Kaczor-Urbanowicz KE, Carreras-Presas CM, Aro K, Tu M, Garcia-Godoy F, Wong DTW.. 2017. Saliva diagnostics – current views and directions. Exp Biol Med. 242(5):459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvostunov IK, Sevan’Kaev AV, Lloyd DC, Nugis VY, Voisin P.. 2011. Recent experience in applying the cytogenetic dosimetry assay. Radiat Meas. 46(9):832–836. [Google Scholar]
- Kreuzer M, Grosche B, Dufey F, Schnelzer M, Tschense A, Walsh L.. 2011. The German Uranium Miners Cohort Study (Wismut cohort), 1946–2003. Tech Rep [Internet]. [Google Scholar]
- Lacombe J, Brooks C, Hu C, Menashi E, Korn R, Yang F, Zenhausern F.. 2017. Analysis of saliva gene expression during head and neck cancer radiotherapy: a pilot study. Radiat Res. 188(1):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe J, Phillips SL, Zenhausern F.. 2016. Microfluidics as a new tool in radiation biology. Cancer Lett. 371(2):292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe J, Sima C, Amundson SA, Zenhausern F.. 2018. Candidate gene biodosimetry markers of exposure to external ionizing radiation in human blood: a systematic review. PLOS One. 13(6):e0198851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JC, Disbrow GL.. 2017. Project BioShield and the biomedical advanced research development authority: a 10-year progress report on meeting US preparedness objectives for threat agents. Clin Infect Dis. 64(10):1430–1434. [DOI] [PubMed] [Google Scholar]
- Li Y, St. John MAR, Zhou X, Kim Y, Sinha U, Jordan RCK, Eisele D, Abemayor E, Elashoff D, Park NH, et al. 2004. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 10(24):8442–8450. [DOI] [PubMed] [Google Scholar]
- Manning G, Kabacik S, Finnon P, Bouffler S, Badie C.. 2013. High and low dose responses of transcriptional biomarkers in ex vivo X-irradiated human blood. Int J Radiat Biol. 89(7):512–522. [DOI] [PubMed] [Google Scholar]
- Meadows SK, Dressman HK, Daher P, Himburg H, Russell JL, Doan P, Chao NJ, Lucas J, Nevins JR, Chute JP.. 2010. Diagnosis of partial body radiation exposure in mice using peripheral blood gene expression profiles. PLOS One. 5(7):e11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows SK, Dressman HK, Muramoto GG, Himburg H, Salter A, Wei Z, Ginsburg GS, Ginsburg G, Chao NJ, Nevins JR, et al. 2008. Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PLOS One. 3(4):e1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucaki E, Shirley B, Rogan PK. Improved radiation expression profiling by sequential application of sensitive and specific gene signatures. bioRxiv; 2021. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Grilj V, Broustas CG, Ghandhi SA, Harken AD, Garty G, Amundson SA.. 2019. Human transcriptomic response to mixed neutron–photon exposures relevant to an improvised nuclear device. Radiat Res. 192(2):189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien G, Cruz-Garcia L, Majewski M, Grepl J, Abend M, Port M, Tichý A, Sirak I, Malkova A, Donovan E, et al. 2018. FDXR is a biomarker of radiation exposure in vivo. Sci Rep. 8(1):684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheim P, Coker O, Schüle S, Hermann C, Combs SE, Trott KR, Atkinson M, Port M, Abend M.. 2021. Identifying a diagnostic window for the use of gene expression profiling to predict acute radiation syndrome. Radiat Res. 195(1):38–46. [DOI] [PubMed] [Google Scholar]
- Ostheim P, Haupt J, Herodin F, Valente M, Drouet M, Majewski M, Port M, Abend M.. 2019. MiRNA expression patterns differ by total- or partial-body radiation exposure in baboons. Radiat Res. 192(6):579–588. [DOI] [PubMed] [Google Scholar]
- Ostheim P, Haupt J, Schüle S, Herodin F, Valente M, Drouet M, Majewski M, Port M, Abend M.. 2020. Differentiating total- or partial-body irradiation in baboons using mRNA expression patterns: a proof of concept. Radiat Res. 194(5):476–484. [DOI] [PubMed] [Google Scholar]
- Ostheim P, Miederer M, Schreckenberger M, Nestler T, Hoffmann MA, Lassmann M, Eberlein U, Barsegian V, Rump A, Majewski M, et al. 2021. mRNA and small RNA gene expression changes in peripheral blood to detect internal Ra-223 exposure. Int J Radiat Biol. Under review. [DOI] [PubMed] [Google Scholar]
- Ostheim P, Tichý A, Sirak I, Davidkova M, Stastna MM, Kultova G, Paunesku T, Woloschak G, Majewski M, Port M, et al. 2020. Overcoming challenges in human saliva gene expression measurements. Sci Rep. 10(1):11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheim P, Worku S, Tichý A, Sirak I, Davidkova M, Markova Stastna M, Kultova G, Schüle S, Paunesku T, Woloschak G, et al. 2021. Examining potential confounding factors in gene expression analysis of human saliva and identifying potential housekeeping genes. Sci Rep. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JG, Paul S, Briones N, Zeng J, Gillis K, Wallstrom G, Labaer J, Amundson SA.. 2017. Developing human radiation biodosimetry models: testing cross-species conversion approaches using an ex vivo model system. Radiat Res. 187(6):708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Amundson SA.. 2008. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 71(4):1236–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Amundson SA.. 2011. Gene expression signatures of radiation exposure in peripheral white blood cells of smokers and non-smokers. Int J Radiat Biol. 87(8):791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Barker CA, Turner HC, McLane A, Wolden SL, Amundson SA.. 2011. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res. 175(3):257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Ghandhi SA, Weber W, Doyle-Eisele M, Melo D, Guilmette R, Amundson SA.. 2014. Gene expression response of mice after a single dose of 137Cs as an internal emitter. Radiat Res. 182(4):380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunesku T, Woloschak G.. 2018. Reflections on basic science studies involving low doses of ionizing radiation. Health Phys. 115(5):623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernot E, Cardis E, Badie C.. 2014. Usefulness of saliva samples for biomarker studies in radiation research. Cancer Epidemiol Biomarkers Prev. 23(12):2673–2680. [DOI] [PubMed] [Google Scholar]
- Port M, Hérodin F, Drouet M, Valente M, Majewski M, Ostheim P, Lamkowski A, Schüle S, Forcheron F, Tichy A, et al. 2021. Gene expression changes in irradiated baboons: a summary and interpretation of a decade of findings. Radiat Res. 195(6):501–521. [DOI] [PubMed] [Google Scholar]
- Port M, Herodin F, Valente M, Drouet M, Lamkowski A, Majewski M, Abend M.. 2016. First generation gene expression signature for early prediction of late occurring hematological acute radiation syndrome in baboons. Radiat Res. 186(1):39–54. [DOI] [PubMed] [Google Scholar]
- Port M, Majewski M, Abend M.. 2019. Radiation dose is of limited clinical usefulness in persons with acute radiation syndrome. Radiat Prot Dosimetry. 186(1):126–129. [DOI] [PubMed] [Google Scholar]
- Port M, Majewski M, Herodin F, Valente M, Drouet M, Forcheron F, Tichy A, Sirak I, Zavrelova A, Malkova A, et al. 2018. Validating baboon ex vivo and in vivo radiation-related gene expression with corresponding human data. Radiat Res. 189(4):389–398. [DOI] [PubMed] [Google Scholar]
- Port M, Ostheim P, Majewski M, Voss T, Haupt J, Lamkowski A, Abend M.. 2019. Rapid high-throughput diagnostic triage after a mass radiation exposure event using early gene expression changes. Radiat Res. 192(2):208–218. [DOI] [PubMed] [Google Scholar]
- Port M, Pieper B, Dörr HD, Hübsch A, Majewski M, Abend M.. 2018. Correlation of radiation dose estimates by DIC with the METREPOL hematological classes of disease severity. Radiat Res. 189(5):449–455. [DOI] [PubMed] [Google Scholar]
- Preston DL, Pierce DA.. 1988. The effect of changes in dosimetry on cancer mortality risk estimates in the atomic bomb survivors. Radiat Res. 114(3):437–466. [PubMed] [Google Scholar]
- Rothkamm K, Beinke C, Romm H, Badie C, Balagurunathan Y, Barnard S, Bernard N, Boulay-Greene H, Brengues M, De Amicis A, et al. 2013. Comparison of established and emerging biodosimetry assays. Radiat Res. 180(2):111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rump A, Becker B, Eder S, Lamkowski A, Abend M, Port M.. 2018. Medical management of victims contaminated with radionuclides after a “dirty bomb” attack. Mil Med Res. 5(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata R, Grant EJ, Ozasa K.. 2012. Long-term follow-up of atomic bomb survivors. Maturitas. 72(2):99–103. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Abend M, Müller K, Beinke C, Braselmann H, Zitzelsberger H, Köhn FM, Pillekamp H, Schiener R, Das O, et al. 2007. Radiation-induced late effects in two affected individuals of the Lilo radiation accident. Radiat Res. 167(5):615–623. [DOI] [PubMed] [Google Scholar]
- Shinde SM, Orozco C, Brengues M, Lenigk R, Montgomery DC, Zenhausern F.. 2010. Optimization of a microfluidic mixing process for gene expression-based bio-dosimetry. Qual Eng. 23(1):59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuryak I, Ghandhi SA, Turner HC, Weber W, Melo D, Amundson SA, Brenner DJ.. 2020. Dose and dose-rate effects in a mouse model of internal exposure from 137Cs. Part 2: integration of gamma-H2AX and gene expression biomarkers for retrospective radiation biodosimetry. Radiat Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproull M, Kramp T, Tandle A, Shankavaram U, Camphausen K.. 2017. Multivariate analysis of radiation responsive proteins to predict radiation exposure in total-body irradiation and partial-body irradiation models. Radiat Res. 187(2):251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JM, Prasanna PGS, Grace MB, Wathen LK, Wallace RL, Koerner JF, Coleman CN.. 2013. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 105(6):540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DT, Segal A, Wong DT.. 2008. Salivary diagnostics: enhancing disease detection and making medicine better. Eur J Dent Educ. 12(S1):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Randers-Pehrson G, Turner HC, Marino SA, Geard CR, Brenner DJ, Garty G.. 2015. Accelerator-based biological irradiation facility simulating neutron exposure from an improvised nuclear device. Radiat Res. 184(4):404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa JM, Schafer CA, Schafer JJ, Farrell JJ, Paster BJ, Wong DTW.. 2013. Salivary biomarkers: toward future clinical and diagnostic utilities. Clin Microbiol Rev. 26(4):781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander A, Paunesku T, Woloschak GE.. 2021. Analyses of cancer incidence and other morbidities in gamma irradiated B6CF1 mice. PLOS One. 15(8):e0231510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JZL, Mucaki EJ, Rogan PK.. 2018. Predicting ionizing radiation exposure using biochemically-inspired genomic machine learning. F1000Res. 7:233. [DOI] [PMC free article] [PubMed] [Google Scholar]