ABSTRACT
This Review delves into the mechanisms behind drug resistance in colorectal cancer (CRC), particularly examining the role of nutrient depletion and its contribution to multidrug resistance (MDR). The study highlights metabolic adaptations of cancer cells as well as metabolic adaptations of cancer cells under low nutrient availability, including shifts in glycolysis and lipid metabolism. It emphasizes the significance of MDR1 and its encoded efflux transporter, P-glycoprotein (P-gp/B1), in mediating drug resistance and how pathways such as HIF1α, AKT, and mTOR influence the expression of P-gp/B1 under limited nutrient availability. Additionally, the Review explores the dual roles of autophagy in drug sensitivity and resistance under nutrient limited conditions. It further investigates the involvement of lysosomes and mitochondria, focusing on their roles in drug sequestration and the challenges posed by lysosomal entrapment facilitated by non-enzymatic processes and ABC transporters like P-gp/B1. Finally, the Review underscores the importance of understanding the interplay between drug sequestration, lysosomal functions, nutrient depletion, and MDR1 gene modulation. It suggests innovative strategies, including structural modifications and nanotechnology, as promising approaches to overcoming drug resistance in cancer therapy.
Keywords: P-glycoprotein (P-gp/B1), Lysosomal drug sequestration, Mitochondrial drug sequestration, Colorectal cancer (CRC), Autophagy
Summary: This Review examines drug resistance in colorectal cancer, focusing on the role of nutrient depletion in multidrug resistance. It highlights metabolic adaptations, and the role of MDR1/P-glycoprotein, and suggests innovative strategies to counteract resistance.
Introduction
In 2020, more than 1.9 million incidences of colorectal cancer (CRC) were observed worldwide, ranking CRC as the third most common cancer. Additionally, 0.9 million deaths from CRC worldwide were recorded, ranking CRC as the second most common cause of cancer mortality (Morgan et al., 2023). For the past three decades, the first-line treatment of patients with CRC comprises of systemic chemotherapy involving 5-fluorouracil (5-FU), leucovorin, and oxaliplatin (Akhtar et al., 2014; Chionh et al., 2017). Despite new screening strategies and ongoing therapeutic developments, CRC is still one of the leading causes of cancer-related deaths (Sung et al., 2021). Nevertheless, the success of chemotherapy is restricted by drug resistance or low sensitivity (Silva and Gatenby, 2010). Therefore, it is particularly important to improve our understanding of basic processes such as the metabolism of cancer cells to gain insights into the divergent mechanisms they employ to provide survival and resistance mechanisms.
Several CRC cell lines are known for their ability to express characteristics of mature intestinal cells, such as enterocytes or mucus-producing cells (Simon-Assmann et al., 2007). This aspect adds a layer of complexity and relevance to their utilization. Hence, the CRC cell lines with the capacity to spontaneously differentiate, such as Caco-2 and T84 cell lines, are commonly employed as in vitro model systems to explore the mechanisms that control the differentiation-dedifferentiation processes of intestinal epithelial layer, to understand the causes of malignant growth, and find new therapeutic targets (Devriese et al., 2017). T84 cells in particular gain importance since they have the capacity to spontaneously differentiate into colonocytes, which are the epithelial cells of the colon and therefore provide important aspects of colon differentiation processes (Devriese et al., 2017).
CRC cell lines are integral to the field of cellular biology, serving not only as in vitro models that elucidate the processes of intestinal differentiation and malignant transformation, but also as unique systems that facilitate comprehensive investigations into the mechanisms underlying drug resistance (Cao et al., 2019). Beyond their primary application, CRC cell lines have contributed to understanding multidrug resistance (MDR), a phenomenon that poses challenges to the effectiveness of chemotherapeutic agents (Zhang et al., 2019).
The concept of MDR originates from the observation of cancer cells displaying resistance not only to a specific group of drugs, but also to a range of structurally and functionally unrelated drugs (Zhang et al., 2019). This fascinating phenomenon, observed across various cancer types, has prompted researchers to embark on an in-depth exploration of the intricate mechanisms contributing to this formidable obstacle in cancer treatment (Zhang et al., 2019).
The elucidation of drug sequestration within critical cellular organelles, including lysosomes and mitochondria, represents a landmark progression in our comprehension of the mechanisms underlying drug resistance. The study of Gotink et al. has revealed that CRC cell line HT-29 employes a strategic confinement tactic, entrapping drugs within such organelles (Gotink et al., 2011). This orchestrated drug entrapment acts to shield the cells from the detrimental effects of therapeutic agents (Gotink et al., 2011). An in vitro exploration into cellular dynamics elucidates that macroautophagy, a prominent cellular degradation process, plays a central role in this mechanism (Aguilera et al., 2012).
Metabolic adaptations in nutrient-depleted CRC cells
Cancer cells are highly proliferating cells requiring the activation of both anabolic and catabolic pathways to generate macromolecules – such as proteins, lipids, and nucleic acids – necessary for cell proliferation and meet cellular energy requirements at the same time (Schmitt et al., 2007). Most of the cancer cells within a tumor have a limited availability of nutrients, which drives them to rewire their metabolism, therefore altering several aspects of cancer cell growth, survival, and response of cells to chemotherapy agents (Scumaci et al., 2020). These adaptations involve intricate mechanisms to acquire and utilize essential nutrients, illustrating the complex interplay between cellular metabolism and cancer progression (Eng et al., 2010). Although alterations in metabolic pathways in cancer cells were reported nearly a century ago, the rewiring of metabolic mechanisms has been only recently recognized as an emerging hallmark of cancer (Zielinski et al., 2017).
Warburg effect and glycolytic shift
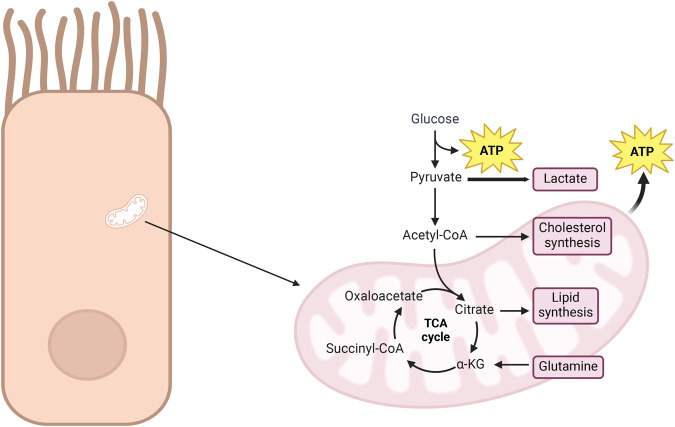
Warburg observed a unique trait in cancer cells compared to healthy ones. In the controlled in vitro environment of experiments, cancer cells demonstrate an increased utilization of glucose, transforming it into lactate, even in the presence of oxygen. This phenomenon is called the Warburg effect (Sun et al., 2019). According to Warburg, this metabolic shift is crucial for the transformation of normal cells into fast-growing cancer cells by affecting their usual method of respiration (Fig. 1). Unlike normal cells, cancer cells prefer the usage of glucose in the generation of building blocks to meet their proliferative demands (Icard et al., 2018). Arrest of glycolysis by over production of ATP and citrate is prevented via the downregulation of tricarboxylic acid (TCA) cycle. Citrate is mainly consumed to produce acetyl-CoA, which in turn is used in lipid biosynthesis and/or protein acetylation (Icard et al., 2018). Decreased mitochondrial activity also modulates reactive oxygen species levels to be in a concentration range compatible with cell proliferation (Icard et al., 2018). While most cancer cells use respiration for tumor growth, some tumors can grow using the TCA cycle without actively respiring. This flexibility highlights how cancer cells can adapt to different energy strategies (Sun et al., 2019).
Fig. 1.
The metabolic flexibility of cancer cells highlighting the Warburg effect, where glucose is preferentially converted into lactate despite the presence of oxygen, is emphasized in the diagram. Such a metabolic shift allows for rapid tumor expansion by supplying energy and essential biosynthetic precursors. Key intermediates of the TCA cycle are detailed, including the conversion of pyruvate into acetyl-CoA and the role of citrate in lipid and cholesterol synthesis. Cancer cells exploit this flexibility, relying on either glycolysis or the TCA cycle for energy production and biosynthesis to sustain their rapid growth, as demonstrated in the figure. The figure was generated by using BioRender.com (“Stimulated metabolic activity”).
Glycolysis and lipid metabolism
To sustain rapid proliferation, cancer cells, especially in late-stage CRC, demonstrate a remarkable shift towards high glycolytic activity, engaging in anaerobic fermentation of converting glucose to fructose. This metabolic reprogramming is pivotal, given the estimated substantial energy requirement of approximately 17,700 kcal over 3 months to support metastatic CRC patients (Lieffers et al., 2009). The preference for glycolysis not only serves as a prominent energy source but also facilitates the generation of metabolic intermediates crucial for anabolic processes, including nucleotide synthesis and fatty acid production, as demonstrated by in silico analyses (Peng et al., 2018).
Fatty acids and lipogenesis
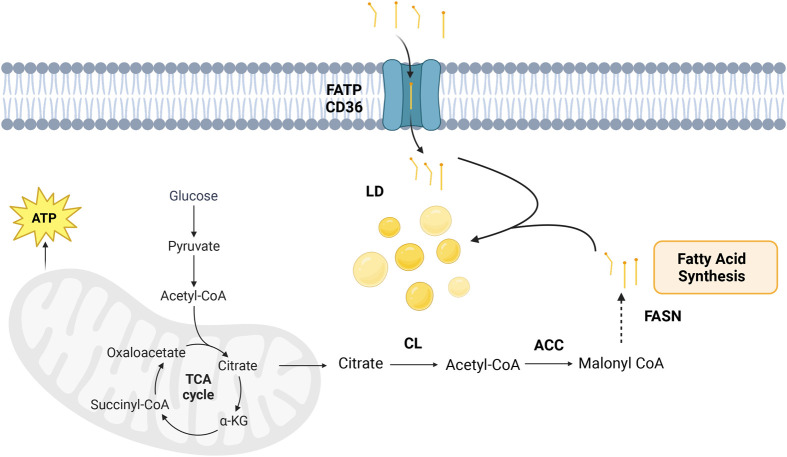
Dysregulation of fatty acid metabolism in CRC cells serves as a significant energy source and, therefore, can contribute to various anabolic processes essential for cancer progression (Ni et al., 2017). Transforming cells (in vitro and in vivo) exhibit elevated de novo fatty acid biosynthesis, a hallmark characterized by increased activity of lipogenic enzymes, such as ATP citrate lyase (CL) and fatty acid synthase (FASN) (Bueno et al., 2019). Elevated free fatty acids can be stored in lipid droplets to be used as an energy source or in membrane synthesis (Cruz et al., 2020). Additionally, increases in fatty acid uptake through elevated expression of fatty acid transporters such as FATP/CD36 provides free fatty acid supplies for the cells in vitro (Schneider et al., 2014). The process of TCA cycle leading to de novo fatty acid synthesis is explained in Fig. 2. Elevated lipogenesis through such a mechanism is fundamental for synthesizing new membranes, supporting the formation of lipid rafts for enhanced cell growth, receptor signaling, and generating signaling molecules crucial for activating proliferative pathways, notably the protein kinase B (AKT) pathway, as observed in vitro transforming cell lines and in vivo mouse models (Ni et al., 2017). Concurrently, fatty acids are catabolized through β-oxidation, revealing the intricate balance in lipid metabolism within cancer cell lines (Ni et al., 2017). Additionally, a high level of fatty acid availability has been shown to promote metastatic processes by providing an energy supply to support invasion and colonization, prevention of immune surveillance from detecting tumor cells, and promotion of cancer cell survival via enhanced antioxidative mechanisms (Corbet et al., 2020; Qiao et al., 2020; Wu et al., 2019). SW480 and SW620 cell lines were well excepted as matched primary and metastatic CRC cell lines, since they were isolated from the primary colorectal tumor and its lymph node metastasis from the same patient, respectively. Our previous work indicated that L-glutamine starvation leads to enhanced free fatty acid levels in SW620 cells compared to SW480 cells. In contrast, the levels of free fatty acids were comparable when cells were grown with L-glutamine. The primary source of the enhanced fatty acid levels was observed to be fatty acid uptake rather than enhanced synthesis. Moreover, enhanced fatty acid levels contributed towards a higher motility accompanied by loss of cell-cell contact only in SW620 cells. These results point out the possible role of fatty acids in the metastatic behavior of CRC cell lines, as well as the metabolic plasticity of CRC cell lines compensating for the retrieval of specific groups of nutrients (Güleç Taşkıran et al., 2024b).
Fig. 2.
The dysregulated process of de novo fatty acid synthesis in CRC cells. Enhanced glucose metabolism drives the production of acetyl-CoA, which feeds into the TCA cycle. Citrate is then exported to the cytoplasm, where CL catalyzes its conversion back into acetyl-CoA, a precursor for fatty acid biosynthesis. Key enzymes such as ACC and FASN drive this process, providing essential lipid components for cell membrane formation and energy storage, thereby supporting cancer cell proliferation and progression. The figure was generated by using BioRender.com (“Metabolic pathway in mitochondria”). LD, lipid droplets; CL, citrate lyase; ACC, acetyl-CoA carboxylase; FASN, fatty acid synthase; FATP/CD36, fatty acid transporters.
Nutrient depletion protocols in CRC cell lines
In the tumor microenvironment, nutrient limitation is a common phenomenon due to the irregular and often insufficient blood supply to the tumor core (Pan et al., 2022). This results in a heterogeneous distribution of nutrients and oxygen, creating regions within the tumor where cells experience significant nutrient deprivation (Pan et al., 2022). A variety of nutrient starvation or limitation protocols are applied in vitro to mimic the tumor microenvironment. These protocols help researchers study how cancer cells, including CRC cell lines, adapt to the nutrient scarce conditions typical of the tumor core.
Treatment of CRC cell lines with HBSS or EBBS, which depletes all sources of nutrients, is one of the most applied protocols (Alves et al., 2015; Lauzier et al., 2019). It has been shown that treatment of CRC cell lines with EBSS/HBSS can alter the autophagy flux, and different CRC cell lines exhibit differential metabolic adaptations due to their high heterogeneity (Alves et al., 2015; Lauzier et al., 2019). Complete depletion of serum, glucose, and all or some of the specific amino acids is one of the commonly applied types of nutrient limitation protocol used as well (Ji et al., 2021; Li et al., 2021; Zhao et al., 2021). It has been shown that serum starvation results in synergistic effects on CRC cell lines when combined with chemotherapy agents, and this synergistic behavior was attributed to changes in the transcription of genes responsible for cell metabolism and cancer's stress pathways (Cherkasova et al., 2023). Another study has shown that long-term serine deficiency affects cell proliferation by regulating transcriptional coactivator YAP, showing the importance of serine/glycine metabolism in cell proliferation regulation (Zhao et al., 2021).
The cells at the core of solid tumors have lowered access to nutrients and oxygen compared to cells at the periphery (Pan et al., 2022). While many cells undergo cell death, some are likely to survive and possess metabolic adaptations to nutrient-deficient conditions (Pan et al., 2022). Considering the low (but not non-) availability of nutrients, another approach is to apply a restricted amount of nutrients to the cells rather than complete depletion. Several CRC cell lines have been shown to enhance the expression of one or more epithelial and mesenchymal markers, suggesting the activation of hybrid/partial EMT upon incubation in a medium containing 10% of glucose, glutamine, and serum found in a complete medium (Hüsnügil et al., 2024). This study shows how CRC cell lines can activate adaptive mechanisms that help them survive and even become more motile to escape challenging metabolic conditions (Hüsnügil et al., 2024).
Cell-cycle regulators and oncogenic signaling
In the context of cancer, cell-cycle regulators play a pivotal role in maintaining a high rate of cellular growth (Lynch et al., 2012). Oncogenic signaling pathways, including AKT and mammalian target of rapamycin (mTOR), play a direct role in shaping glucose metabolism, boosting nutrient uptake, and promoting macromolecular biosynthesis to sustain uncontrolled cellular proliferation (Johnson et al., 2010). These pathways coordinate a metabolic shift marked by heightened glycolysis and suppressed oxidative metabolism, aligning with the energy requirements of cancer cells.
Autophagy and mitophagy
Devenport et al. discovered that even under nutrient-rich conditions, CRC cell lines demonstrate a substantial reliance on autophagy for growth in vitro, as evidenced by a marked reduction in growth upon inhibition of autophagy (Devenport et al., 2021). This underscores the adaptability of cancer cells to employ alternative mechanisms for nutrient acquisition, a critical aspect for proliferation even in the presence of abundant nutrients.
Mitophagy plays a vital role in nutrient replenishment and cell maintenance in tumors, as demonstrated by research conducted both in vitro and in vivo animal models (Devenport et al., 2021). Several studies have explored how mitophagy contributes to drug resistance. Yin et al. conducted a study with 90 CRC patients and reported altered PINK1 immunoexpression in both primary and liver metastasis lesions resected (Yin et al., 2021). Additionally, lesions of liver metastasis predict a worse prognosis with abnormal expression, possibly due to the abnormal function of mitophagy (Yin et al., 2021). Ke et al. conducted further investigations on more mitophagy related proteins and reported abnormal expression of PINK1, TOMM22, and TOMM40 in CRC using 51 normal samples versus 616 CRC patient samples (Ke et al., 2024). Tang et al. highlighted that multiple proteins engage various pathways to activate mitophagy, promoting proliferation and survival in both CRC patients and in vivo animal models (Tang et al., 2023). Since mitophagy has gathered attention in recent years, it is still a complex mechanism that needs further studies to fully explain its involvement to cancer progression.
Role of HIF, AKT, and mTOR in CRC metabolic plasticity
The interaction between hypoxia-inducible factor 1-alpha (HIF1α), AKT, and mTOR pathways is essential for regulating glycolysis, involving both the transcriptional upregulation and phosphorylation of key enzymes and transporters. Oxygen availability is a critical factor for cellular survival, and the tumor microenvironment's hypoxia-induced changes profoundly influence glycolytic enzymes and nutrient transporters (Ho et al., 2015). Dysregulation of hypoxia response mechanisms resonates across various cancers, impacting crucial aspects such as oxygen transport, angiogenesis, and immune cell function (Su et al., 2024). Under hypoxic conditions, cells undergo a metabolic shift from O2-dependent mitochondrial ATP production to O2-independent glycolysis (Ho et al., 2015). This transition is accompanied by heightened oxidative stress, nutrient deprivation, and endoplasmic reticulum stress (Zhang et al., 2020). Certain metabolites derived from the TCA cycle, including succinate, fumarate, and α-ketoglutarate, function as oncometabolites (William et al., 2021). These substances contribute to tumor growth by promoting oncogenic signaling, particularly through the upregulation and stabilization of HIF-1α, despite glycolysis being the favored metabolic pathway (Ho et al., 2015).
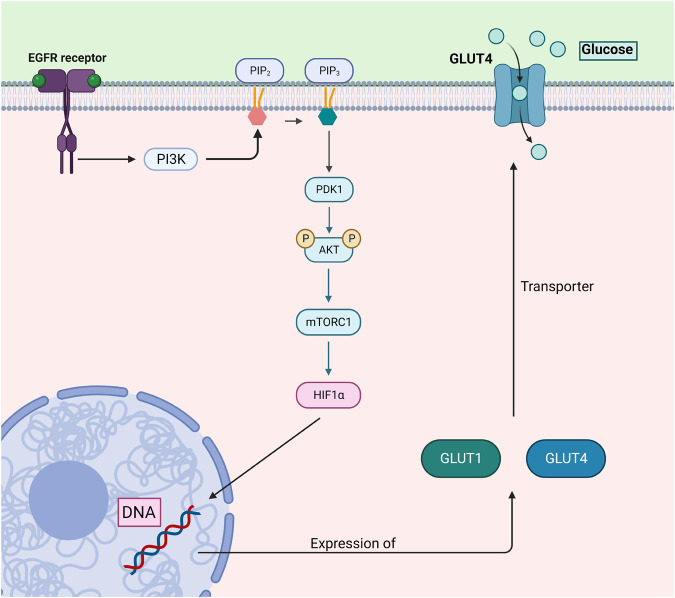
Furthermore, mTOR signaling is often disrupted in tumors, significantly impacting cellular growth and metastasis through complex pathways, such as AKT-dependent cytoskeletal remodeling (Hervieu et al., 2020; Qian et al., 2004). Aberrant PI3K/AKT/mTORC1 signaling can be seen in a variety of excised tumors of CRC patients (Lai et al., 2014). AKT promotes glucose uptake through glucose transporter protein type 1 (GLUT-1) and glucose transporter protein type 4 (GLUT-4) (Wieman et al., 2007). It exerts influence on metabolic processes by impacting downstream transcription factors responsible for metabolic control, including HIF1α (Guru et al., 2015) (Fig. 3). Additionally, AKT triggers lipogenesis by directly phosphorylating and activating ATP-citrate lyase, resulting in elevated cytoplasmic acetyl-CoA levels (Bauer et al., 2005). Furthermore, AKT enhances the uptake of L-glutamine through SLC5A1, a process mediated by Myc. AKT also stimulates the flow of glucose into the oxidative pentose phosphate pathway through the downstream effector mTORC1 (Düvel et al., 2010). mTORC1 further activates this pathway by triggering the activation of SREBP and the upregulation of glucose-6-phosphate dehydrogenase (Yecies, 2012).
Fig. 3.
This illustration details the PI3K/AKT/mTORC1/HIF1α pathway, focusing on its regulatory role in glucose uptake through GLUT1 and GLUT4 transporters. Activation of the EGFR receptor triggers PI3K signaling, leading to the phosphorylation of AKT via PDK1. AKT activation promotes downstream signaling such as mTORC1 and the stabilization of HIF1α, which regulates the expression of GLUT1 and GLUT4 transporters. These transporters facilitate glucose uptake to meet the energy demands of cancer cells, further enhancing glycolytic metabolism. Dysregulation of this pathway is associated with CRC, where aberrant mTORC1 signaling and HIF1α stabilization contribute to tumor growth and metastasis through enhanced glucose uptake and metabolic reprogramming. The figure was generated by using BioRender.com.
In brief, the above-mentioned pathways equip CRC cells with metabolic plasticity, which can confer survival advantages under nutrient-scarce conditions and, therefore, are highly actively modulated based on cellular needs.
MDR1 encodes P-glycoprotein, a key contributor to developing MDR
MDR in cancer cells, often mediated by the overexpression of P-gp/B1, encoded by the MDR1 gene, poses a significant challenge in chemotherapy (Halder et al., 2022). P-gp/B1, a member of the ATP-binding cassette (ABC) superfamily, is expressed on various cellular membranes, including the plasma membrane and Golgi membrane (Gericke et al., 2022). It is widely present in normal human tissues, including the liver, kidney, colon, adrenal gland, intestine, placenta, hematopoietic precursor cells, and endothelial cells at various barriers (Gericke et al., 2022).
P-gp/B1 is a key efflux pump that protects tissues from harmful agents and foreign substances such as xenobiotics (Zhou, 2008). The upregulation of P-gp/B1 in drug-resistant tumors significantly impacts the efficacy of anticancer drugs, leading to treatment failures, as it pumps out a range of anticancer drugs (Liu-Kreyche et al., 2019). This heightened expression in cancer cells confers MDR, making these cells resilient against the therapeutic effects of various drugs (Liu-Kreyche et al., 2019). The molecular mechanisms regulating MDR are multifaceted, involving the human ABC transporters, breast cancer resistance protein (BCRP/ABCG2), and multidrug resistance-associated proteins (MRP1-2/ABCC1-2) (Zaja et al., 2008). These efflux transporters interact with various anticancer agents, minimizing intracellular exposure to cytotoxic drugs and contributing to MDR (Liu-Kreyche et al., 2019).
Not only many efflux proteins are involved in MDR, but also various regulatory mechanisms are involved in the expression of P-gp/B1. In vitro research findings on K562 leukemia cells reveal that the upregulation of P-gp/B1 expression involves a two-step process: mRNA stabilization and relief from translational block (Yagüe et al., 2003). Unlike transcriptional activation, this upregulation in K562 cells is attributed to an increase in mRNA stability, a specific phenomenon for MDR1 mRNA (Yagüe et al., 2003). The complexities of how long mRNA lasts, especially its short lifespan in unmodified K562 cells, highlight the complicated system that controls how the MDR1 gene is turned on or off (Yagüe et al., 2003). Incubation of T84 CRC cells in a nutrient limited condition in vitro shown to be resulted in the survival of a population of cells with high viability, accompanied by reduced sensitivity to chemotherapeutic agents such as 5-FU or doxorubicin (Güleç Taşkıran et al., 2024a). The mechanism by which the cells lose chemosensitivity is attributed to lysosomal trapping of chemotherapy agents, but an increased mRNA level expression of MDR1 gene or mRNA stabilization was also observed in the nutrient depleted cells, suggesting the presence of multiple mechanisms for drug resistance (Güleç Taşkıran et al., 2024a). Considering the existence of starvation stress responsive element in MDR1 gene promoter, elevated MDR1 gene expression is a plausible observation (Tanimura et al., 1992). Understanding these molecular nuances provides valuable insights for potential therapeutic interventions targeting P-gp/B1 mediated MDR in CRCs.
Autophagy's impact on drug sensitivity: enhancing and inhibiting dynamics
Autophagy, a cellular process crucial for maintaining cellular homeostasis, exhibits dual roles in the context of cancer progression (Lock et al., 2011). In the early stages of tumorigenesis, autophagy acts as a tumor suppressor by degrading potentially harmful agents, damaged organelles, and misfolded proteins. It also maintains genomic stability by slowing damage repair, reduces chronic inflammation in the microenvironment, and limits the accumulation of reactive oxygen species (Degenhardt et al., 2006; Mathew et al., 2007). Under nutrient depletion conditions, autophagy plays a pivotal role, and induction of autophagy is regulated by factors such as AMP-activated protein kinase (AMPK) activation and mTORC1 inhibition (Yuan et al., 2013). In response to nutrient restriction (via the activation of AMPK and/or inhibition of mTORC1), autophagy is induced and serves pivotal roles for cell survival (Priault et al., 2010; Yang et al., 2014).
Several studies exhibited the role of autophagy in the proliferation of CRC cell lines under no and/or low nutrient availability. Zhang et al. exhibited that serum starvation-induced autophagy leads to the increased stability of LINC01615 (long non-coding RNA), which in turn promote the survival of CRC cell lines under serum-deprived conditions via pentose phosphate pathway (PPP) activation (Zhang et al., 2023). Tan et al. reported that Jumonji domain-containing protein 2B (JMJD2B) is involved in the promotion of CRC cell lines' survival upon glucose deprivation via enhancing the intracellular amino acid levels (Tan et al., 2020).
Autophagy serves as the primary mechanism for housekeeping, involving the recycling of redundant proteins and damaged organelles in normal cells. This process provides a survival advantage to tumor cells during tumorigenesis as exemplified by several studies mentioned (Gil et al., 2020; Lauzier et al., 2019). Nevertheless, if autophagy persists in a prolonged and excessive manner, it can unexpectedly lead to autophagic cell death. This contributes to the elimination of damaged or stressed cells, thereby enhancing the effectiveness of anticancer drugs (Su et al., 2024).
mTOR emerges as one of the main modulators of autophagy, influencing its dynamics and outcomes. Autophagy, acting as a guardian in drug resistance in cancer cell lines, supports cancer cell metabolism by recycling damaged components, preventing DNA damage, and inducing cancer drug resistance (Jung et al., 2009). This complex relationship is evident in various mechanisms, such as autophagy-regulated DNA damage response, autophagy-induced drug efflux (Liu et al., 2019; Orlotti et al., 2012).
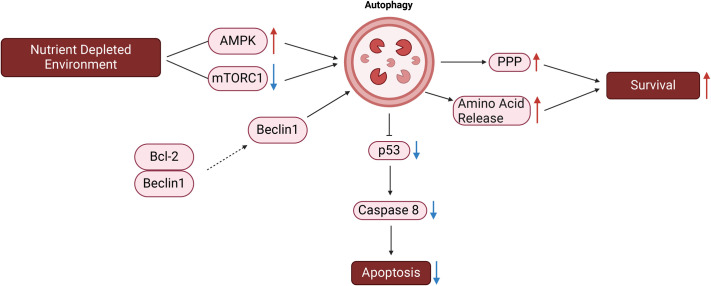
Furthermore, autophagy's involvement in drug resistance exceeds the nutrient stress, it is involved in the changes in apoptotic and survival signals regardless of the nutrient availability as well. Autophagy can degrade active caspase 8, a mitochondrial apoptosis pathway component, and exhibit a complex relationship with the tumor suppressor the tumor protein P53 (p53) (Hou et al., 2010). The disruption of the Beclin1/Bcl-2 complex and alterations in proapoptotic factors, antiapoptotic effectors, and survival signals contribute to autophagy-mediated drug resistance (Yang et al., 2020). Fig. 4 summarizes the involvement of autophagy induction in cancer cell survival and death.
Fig. 4.
This figure depicts how autophagy is activated in response to nutrient deprivation, playing a pivotal role in promoting cell survival and inhibiting apoptosis. Under nutrient stress, autophagy is initiated through AMPK activation and mTORC1 inhibition. Once autophagy is induced, it supports cell survival by enhancing amino acid release and stimulating the pentose phosphate pathway, which sustains metabolic processes. At the same time, autophagy suppresses apoptosis by reducing p53 activity and degrading active caspase 8, a key component of the mitochondrial apoptosis pathway. Beclin1, a core autophagy protein, is essential in initiating and regulating the autophagic process by interacting with various proteins to promote autophagosome formation. The disruption of the Beclin1-Bcl-2 complex further enhances autophagy, contributing to cell survival, particularly in CRC cells. This regulation allows cancer cells to evade apoptosis, leading to tumor progression and increased resistance to therapies. The figure was generated by using BioRender.com. PPP, pentose phosphate pathway.
Apart from its role as a guardian, autophagy can act as an executioner in tumor drug resistance (Gao et al., 2010). Autophagic cell death, induced by sustained autophagy, represents an alternative cell death approach when apoptosis fails, especially in drug-resistant tumors (Zhu et al., 2018). The intensity of oncogenic Ras signaling, and multiple activated signal pathways play crucial roles in determining whether autophagy acts as pro-survival or pro-death mediator (Elgendy et al., 2011).
Understanding the intricate interplay between autophagy, drug sensitivity, and nutrient availability provides valuable insights into developing targeted therapeutic strategies for overcoming drug resistance in various cancers.
Lysosomes and drug sequestration leads to drug resistance
Lysosomes are highly acidic cellular organelles and the lysosomal reservoir of the cells serves pivotal roles along with autophagy in cell survival when nutrients are scarce (Paquette et al., 2021). Lysosomes, integral components of the endolysosomal system (ES), have conventionally been perceived as the cell's hub for recycling and waste management, which in turn are involved regeneration of building blocks (Nguyen et al., 2017). Recently, lysosomes have been also recognized as a signaling hub of the cells as they integrate both external and internal nutritional information to sustain cellular homeostasis (Mony et al., 2016). The orchestration of ES biogenesis and signaling through lysosomes primarily hinges on the activities of the transcription factor EB (TFEB) and mTORC1 signaling – a process frequently disrupted in the throes of oncogenic transformations (Li et al., 2016). The connection between mTORC1 and lysosomes is significant, with mTORC1 regulating V-ATPase expression among other functions (Deleyto-Seldas and Efeyan, 2021).
Lysosomes are present in all eukaryotic cells, and their morphology, size, and abundance are precisely regulated by the coordinated lysosomal expression and regulation (CLEAR) gene network (Palmieri et al., 2011). Key regulators, including TFEB, transcription factor E3 (TFE3), and mitochondrial translational initiation factor (MTIF), play crucial roles in activating this network (Palmieri et al., 2011). The regulation of these key regulators relies highly on mTORC1 activity. Under low nutrient availability, mTORC1 is inactive and released from the lysosomal surface which frees TFEB and TFE3 (Raiborg, 2018). These two TF promote the expression of CLEAR genes, which are involved in the lysosomal activity and autophagosomal degradation (Raben and Puertollano, 2016).
Lysosomes play diverse roles, including biomolecule discard, endocytosis, autophagy, exocytosis, and plasma membrane repair (Huynh et al., 2004; Machado et al., 2015; Marszalowicz et al., 2014). They are crucial for cell signaling pathways such as mTOR and AMPK (Wang et al., 2019). In cancer cells, lysosomes detoxify acidic metabolites generated by the Warburg effect, regulating abnormal pH gradients that impact proliferation, tumorigenesis, and drug resistance (Lozupone et al., 2015). Involvement of lysosomes in cancer survival is not limited to elimination of detrimental effects of the Warburg effect, but they also provide energy and building blocks under low nutrient availability conditions. Bandyopadhyay et al. identified a starvation response that allows cells to store the essential amino acid leucine within lysosomes to sustain protein synthesis in vitro (Bandyopadhyay et al., 2022). Degradation of extracellular proteins by lysosomes also provides cell cancer cells with survival advantages when nutrients are scarce (Pechincha et al., 2022).
In addition to providing building block and energy, lysosomes exhibit a unique ability to sequester lipophilic, weakly basic chemotherapeutic drugs through a non-enzymatic, non-transporter mechanism (Kazmi et al., 2013). Among these drugs, including doxorubicin, daunorubicin, vincristine, lapatinib, and nintedanib, many become trapped in lysosomes upon crossing the lysosomal membrane (Hurwitz et al., 1997; Kallus et al., 2018). Lysosomes act as reservoirs, pulling drugs from their target sites, but this trapping is reversible (Angus et al., 2008). The ionized drug can cross the lysosomal membrane by passive diffusion when cytosolic concentrations decrease (Kazmi et al., 2013). Lysosomal inhibitors such as chloroquine and bafilomycin A1 can also lead to release of entrapped drugs from lysosomes into the cytoplasm as well (Guo et al., 2016).
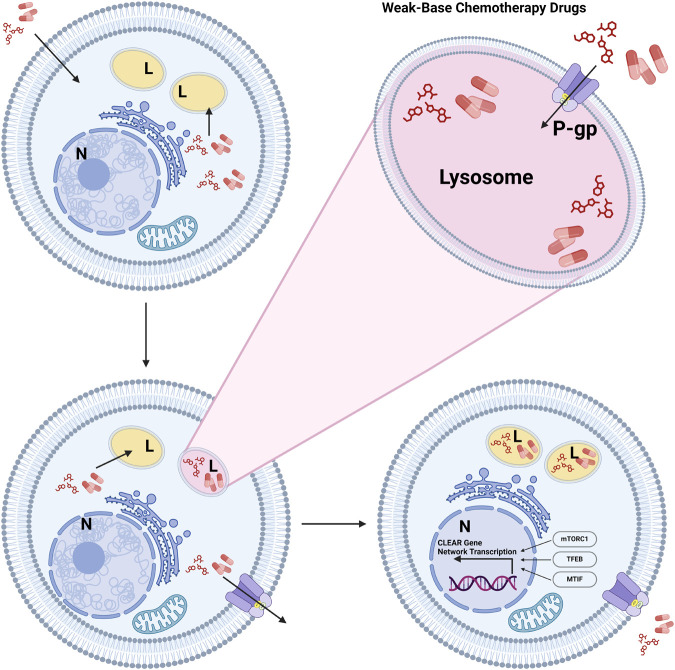
TFEB, regulated by mTORC1 phosphorylation, influences the distribution of chemotherapeutic drugs between cytosolic and nuclear compartments (Cotter et al., 2015). Lysosomal entrapment reduces the therapeutic efficacy of these drugs at intended targets, such as nuclear DNA (Yaghini et al., 2017). Lysosomes also contribute to drug sequestration through ATP-binding cassette transporters (Merlos Rodrigo et al., 2019). P-gp/B1, a representative ABC transporter, is present on cell and lysosomal membranes (Colombo et al., 2014). Drugs, despite being distributed between cytosolic and nuclear compartments, unintentionally become entrapped in lysosomes, diminishing their therapeutic effects at intended target sites (Yaghini et al., 2017). Nutrient-limited T84 CRC cells showed increased MDR1 mRNA expression and reduced sensitivity to doxorubicin and 5-FU in vitro. Furthermore, these cells also exhibit decreased nuclear localization and increased lysosomal localization of doxorubicin, indicating that MDR1 may contribute to the drug's sequestration in lysosomes (Fig. 5) (Güleç Taşkıran et al., 2024a).
Fig. 5.
This figure illustrates the process by which weak-base chemotherapy drugs become trapped in lysosomes, limiting their availability to cytosolic and nuclear compartments. Once inside the cell, drugs enter lysosomes through passive diffusion and are further concentrated by the P-gp/B1 transporter located on the lysosomal membrane. This sequestration reduces the drugs' therapeutic efficacy at their target sites, such as the nucleus. Lysosomal biogenesis and function are regulated by the CLEAR gene network, controlled by transcription factors TFEB, TFE3, and MITF is also depicted in the figure. The CLEAR network governs lysosomal size, abundance, and enzymatic activity, particularly under the influence of mTORC1 signaling. In cancer cells, this system can become dysregulated, enhancing the lysosome's capacity to sequester drugs, which can contribute to drug resistance. The figure was generated by using BioRender.com. N, nucleus; L, lysosome.
Mitochondria and drug sequestration leads to drug resistance
As the powerhouse of the cells, mitochondria support metabolic flexibility and affect cancer progression by regulating proliferation, autophagy, and apoptosis (Liu et al., 2023). Mitochondria is a double membraned organelle with its own DNA (mtDNA) (Meeusen and Nunnari, 2003). Emerging from the mitochondrial matrix, these organelles have four main parts: the mitochondrial matrix, inner mitochondrial membrane (IMM), intermembrane space, and outer mitochondrial membrane (OMM) (Sesso et al., 2012). They are crucial for processes like energy generation, calcium signaling, lipid metabolism, ROS production, and apoptosis, and they are linked to conditions such as neurodegenerative disorders, diabetes, drug toxicity, and aging (Chen et al., 2003; Denecker et al., 2001; Juárez-Flores et al., 2020; Robb-Gaspers, 1998; Sala-Vila et al., 2016). As well as the mentioned pathologies, mitochondria and mitochondrial plasticity play important roles in cancer progression as well. Even though cancers cells have a profound reliance on glycolytic flux and lactic acid release (Warburg effect), mitochondrial plasticity is commonly observed in metastatic cancer cells (Parida et al., 2023). Thanks to mitochondrial plasticity, disseminating cells can shift from glycolysis to TCA cycle and fatty acid utilization in mitochondria to provide survival advantages in the host tissue (Parida et al., 2023).
Importance of mitochondrial plasticity has been exemplified in many studies. Since cancer cells are reliant on glucose metabolism for their high energy demand, the use of 2-deoxy-dglucose (2DG) is commonly applied to restrict glucose availability (Wu et al., 2024). Even though the use of 2DG is a logical way of cutting glucose supply, the metabolic plasticity of cancer cells enables alternative energy source usage. Pyruvate kinase M2 (PKM2) has been shown to localize to mitochondria under glucose starvation, with this localization mitochondrial membrane permeability was shown to be increased keeping mitochondrial functions active and enhance survival of HCT-116 CRC cells (Qi et al., 2019). Another study exhibits the importance of mitochondrial compensation under glucose starvation via usage of 2DG loaded nanoparticles and Ce6 (for mitochondrial activity inhibition) in vitro (Wu et al., 2024). LnNP@mSiO2-GC loaded with 2DG and Ce6 has been shown to both inhibit glycolysis (2DG) and impair mitochondrial activity (Ce6), in this way bypasses the compensatory pathway of the TCA cycle and enhanced the efficacy of 2DG single agent treatment (Wu et al., 2024).
In addition to the role of mitochondrial function under nutrient stress, mitochondrial vesicular formation of multivesicular bodies associated proteins (MAPS) appears to be a conserved process across all cell lines, irrespective of drug sensitivity (Abunimer et al., 2018). Notably, the efficiency of this process correlates with P-gp/B1 expression, conferring resistance and heightened survival rates in P-gp/B1 overexpressing cell lines (Abunimer et al., 2018). MAPS function as both drug sinks and extracellular transport vectors, indicating that enhanced MAPS production and drug sequestration correlate with heightened resistance in cancerous cell lines (Abunimer et al., 2018). Yet, directing interventions toward MAPS to counteract chemo-resistance in humans may present hurdles, potentially leading to lethal toxicity in normal cells (Abunimer et al., 2018).
Mitochondria, central to cellular metabolism and energy production, serves as the specific target of many chemotherapeutic agents, particularly through the mitochondrial apoptosis pathway (Guo et al., 2019). The presence and role of P-gp/B1 in the mitochondrial membrane have been subject to controversy. Recent studies have indicated that P-gp/B1 is expressed in mitochondria and functions in pumping drugs into the mitochondria (Guo et al., 2019). Additionally, the exchange of mitochondria between cancer cells and endothelial cells has been observed, contributing to chemoresistance in cancer cells (Guo et al., 2019). Pasquier et al. presented that ovarian and breast cancer cell lines were able to form tunneling nanotubes with stromal cells under same culture conditions, and transfer of mitochondria from stromal cells to cancer cell lines has been observed with mitochondrial specific dye MitoTracker (Pasquier et al., 2013). Moreover, sorting of mitochondria receiving and non-receiving cancer cells showed that acquisition of mitochondria enables cancer cells with chemoresistance (Pasquier et al., 2013). This observation implies that cancer cells have the capacity to communicate resistant traits via mitochondrial transport. Consequently, delving into the functional status and proteomics of mitochondria in cells exhibiting drug resistance is pivotal for unraveling the intricate mechanisms underpinning multidrug resistance.
Nutrient sensing pathways and P-gp/B1 expression: insights into the PI3K/Akt/mTOR network's role in drug resistance
Navigating the intricate landscape of MDR reveals the paramount role of signaling pathways controlling P-gp/B1 expression (Yuan et al., 2020). Within this intricate web, the PI3K/Akt cascade stands out as a key regulator, acknowledged for its profound influence on P-gp/B1 expression (Yuan et al., 2020). Activated by phosphorylation, Akt initiates a sequence of events by phosphorylating inhibitor of nuclear factor kappa B (IκBα), which then causes IκBα to dissociate from NFκB. Subsequently, NFκB translocate to the nucleus, amplifying the expression of the MDR1 gene, the genetic blueprint for P-gp/B1 (Yuan et al., 2020).
Adding another layer of complexity, the downstream effector of Akt, mTORC1, plays a crucial role in nutrient sensing and MDR1 gene expression, as demonstrated in both in vivo and in vitro studies. Ma et al. demonstrated that inhibition of mTORC1 activity via Adriamycin treatment, reverses multidrug resistance in CRC cells, which is associated with increased autophagy, apoptosis and reduced MDR1 gene expression (Ma et al., 2015). Another study has been demonstrated the role of proton pump inhibitors (PPIs) in the expression of P-gp/B1 in vitro and in vivo (Chen et al., 2018). Treatment of the multidrug-resistant SGC7901 gastric adenocarcinoma cell line with PPIs resulted in diminished levels of P-gp/B1 through the PI3K/AKT/mTOR/HIF-1α signaling pathway (Chen et al., 2018). These studies accentuate the nuanced interplay between PI3K/Akt, mTOR, HIF-1α, and P-gp/B1 in orchestrating drug resistance.
This intricate regulatory network extends its reach into glucose metabolism, where aberrant PI3K/Akt activation emerges as a hallmark of cancer aggressiveness and drug resistance. HIF-1α, a downstream target of PI3K/Akt, takes command over genes encoding crucial glucose metabolism mediators, including GLUTs (Zhang et al., 2014b). Further elucidating this network, conducted both in vivo and in vitro experiments with Nuciferine, a bioactive compound that inhibits the AKT/PI3K/ERK pathway (Qi et al., 2016). This inhibition suppresses the activation of nuclear factor erythroid 2-related factor 2 (Nrf2) and HIF-1α, subsequently reducing the expression of P-gp/B1 and BCRP in HCT-8/T and A549/T cell lines (Liu et al., 2020). The association between PI3K/Akt, mTOR, HIF-1α, GLUT, and P-gp/B1 reveals a sophisticated regulatory network that influences drug resistance mechanisms, offering potential targets for therapeutic intervention.
Understanding these pathways is essential for developing targeted strategies to overcome multidrug resistance in cancer cells.
Strategies to overcome resistance mechanisms
In confronting the complexities of cancer, the interplay between drug resistance mechanisms and the metabolic reprogramming of cancer cells underscores a critical aspect of therapeutic challenges (Sun et al., 2020). Traditional modalities such as surgery, chemotherapy, radiotherapy, immunotherapy, and targeted therapy represent significant advances, yet the shadow of resistance looms large, diluting their long-term efficacy (Rulli et al., 2019). Central to addressing this issue are P-gp/B1 inhibitors, which aim to counteract cancer's defense mechanisms. Various strategies were proven effective in vitro, such as competitive and non-competitive inhibition, disruption of ATP hydrolysis, and altering cell membrane lipid composition (Al-Akra et al., 2018). This effort to sensitize cancer cells to therapy is further nuanced by the recognition that metabolic alterations in cancer cells, including those mediating P-gp/B1 activities to towards resilience against conventional treatments (Meschini et al., 2000). Notably, agents such as verapamil, cyclosporine A, dexverapamil, valspodar, and tariquidar have shown promise in vitro and in clinical trials as chemosensitizers. However, their clinical application has been hindered by issues like poor selectivity, low potency, high toxicity, and unpredictable pharmacokinetic interactions (Yahanda et al., 1992). Research efforts are increasingly directed towards exploring natural products and their structural alterations to develop novel P-gp/B1 inhibitors, potentially enhancing their safety and effectiveness (Li et al., 2014; Yoshida et al., 2006). Despite these efforts, the in vivo efficacy and safety of these inhibitors remains to be fully established, underscoring the need for further investigation (Li et al., 2014).
Interestingly, P-gp/B1 also plays a role in cancer metabolism or vice versa. Elevated expression of MDR1 has been shown in variety of tumors as opposed to normal tissue samples and in cancer cell lines as opposed to healthy cell (Noonan et al., 1990). Evidence from in vivo study shows that tumors with altered metabolic profiles, compared to treatment-naive tumors, have increased P-gp/B1 activity (Viale et al., 2014). Additionally, MDR1 expression profiles have been shown to be different between surgical samples from primary and metastatic tumors; metastatic CRC samples having the higher expression of MDR1 (Micsik et al., 2008). Considering the changing MDR1 expression throughout progression of cancerous cells – each having unique metabolic adaptation – metabolic adaptation has a great impact of MDR1 expression. This suggests a complex interaction between drug efflux mechanisms and metabolic reprogramming. This synergy between metabolic changes and P-gp/B1 activity illustrates the need for a multifaceted approach in cancer therapy, one that encompasses not just the inhibition of efflux pumps but also the targeting of metabolic vulnerabilities.
The metabolic reprogramming of cancer cells offers fertile ground for therapeutic intervention, with in silico research revealing how shifts in metabolism are integral to cancer progression and resistance (Sun et al., 2020). Therapy-resistant tumors exhibit enhanced reliance on mitochondrial metabolism compared to treatment-naive counterparts in vivo (Viale et al., 2014). This observation has led to strategies aiming to inhibit specific metabolic pathways to stymie tumor growth. For instance, research on in vivo tumor models has shown that anti-angiogenic kinase inhibitors have been shown to amplify AMPK signaling in breast and lung cancer models, shifting metabolism towards an oxidative phenotype (Navarro et al., 2016). This metabolic shift renders cells more susceptible to oxidative metabolism inhibitors, highlighting the potential of metabolic intervention in conjunction with traditional therapies (Navarro et al., 2016).
Additionally, inhibiting metabolic enzymes and pathways, such as lactate dehydrogenase (LDH) and pyruvate dehydrogenase kinase (PDHK) is another strategy in cancer treatment (Boudreau et al., 2016; Michelakis et al., 2010). Drugs targeting these pathways, like metformin, have shown the ability to kill chemotherapy-resistant breast tumor stem cells in vitro, highlighting the therapeutic potential of targeting cancer metabolism (Janzer et al., 2014). In the broader context, effective cancer treatment strategies are increasingly focusing on understanding the molecular and metabolic mechanisms behind resistance. Various approaches are being explored, from using miRNAs and siRNAs to target autophagy-related genes, to developing BH3 mimetics to induce apoptosis in cancer cells (Selvakumaran et al., 2013; Sümbül et al., 2014; Zhang et al., 2021). However, integrating metabolic interventions, either by directly targeting metabolic enzymes or by modulating pathways influenced by P-gp/B1, is becoming a crucial part of this evolving strategy. As research advances, the potential to disrupt the metabolic adaptations of cancer cells, thereby sensitizing them to both existing and novel therapies, offers a promising path forward in the relentless fight against cancer.
Concluding remarks
As we conclude this Review, we must explore the complex relationships between drug sequestration and MDR emergence. The MDR1 gene, which encodes P-gp/B1, a membrane-bound efflux transporter with the remarkable capacity to actively expel a wide variety of medicines out of the cells, is at the center of this link (Gericke et al., 2022). As essential parts of the ES, lysosomes are essential for drug sequestration because of their acidic environment and ability to accumulate weakly basic medicines (Colombo et al., 2014). Drugs, particularly chemotherapeutics, are sequestered within lysosomes by means unrelated to transporters or enzymes (Colombo et al., 2014). When the MDR1 gene is overexpressed in cancer cells, P-gp/B1 activity increases, which amplifies the drug's outflow from the intracellular environment. Because of this increasing efflux, prescription efficacy is waning, and multidrug resistance is starting to appear. Developing strategic approaches for overcoming the formidable challenge of multidrug resistance in cancer treatment requires a deep comprehension of the complicated interplay between drug sequestration, lysosomal complexity, and MDR1 gene regulation.
CRC cell lines are not only capable of metabolic plasticity to activate alternative energy pathways under nutrient stress to gain survival advantages. But also, they can enhance the P-gp/B1 expression as another survival mechanism when nutrients are scarce. In CRC cell lines, nutrient depletion can increase MDR1 expression and enhance drug sequestration within lysosomes, further increasing resistance (Güleç Taşkıran et al., 2024a). Addressing these nutrient-related factors is crucial for developing effective therapeutic strategies.
Innovative approaches have been prompted by the potential to mitigate drug resistance, especially regarding anthracyclines. Gong et al. aimed to reduce the intrinsic basicity of daunorubicin by making structural changes to the drug. The resulting daunorubicin derivatives significantly reduced lysosomal sequestration in vitro, as observed in two resistant cell lines (Gong et al., 2003). Remarkably, however, these changed compounds' durability and efficacy beyond in vitro evaluations are still unknown, providing a path for future research and possible therapeutic uses.
Our focus is on promising approaches to drug resistance that are derived from recent advancements in nanotechnology to combat drug resistance and advance cancer therapy. Liposomes, polymers, metals, and metal-oxide nanoparticles are examples of how nanomaterials are influencing new therapeutic paradigms (Doshi et al., 2015; Ghafari et al., 2020; Pandey et al., 2016; Yang et al., 2021). These nanocarriers function as vehicles for precisely delivering therapeutic drugs to specific areas while limiting effects on healthy tissues by utilizing the enhanced permeability and retention (EPR) effect in the tumor vasculature (Matsumura and Maeda, 1986). Some nanoparticles possess intrinsic characteristics that enable them to modify signaling pathways associated with autophagy regulation (Zhang et al., 2014a). Because of its dual purpose, nanotechnology is positioned as a treatment approach against a range of human cancers. These strategies are a significant advance in the fight against drug resistance and offer hope for more effective, targeted, and minimally invasive therapeutic approaches.
In conclusion, our Review highlights the MDR1 gene's role in drug resistance, the role of metabolic adaptation on MDR1 expression patterns by emphasizing lysosomal involvement in sequestration. Innovative strategies, such as structural modifications and nanotechnology interventions, offer promising avenues for combating multidrug resistance and advancing more effective cancer therapies.
Acknowledgements
The authors would like to acknowledge Prof Dr Özlem Darcansoy İşeri, Baskent University, for useful discussions.
Footnotes
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
References
- Abunimer, A. N., Mohammed, H., Cook, K. L., Soto-Pantoja, D. R., Campos, M. M. and Abu-Asab, M. S. (2018). Mitochondrial autophagosomes as a mechanism of drug resistance in breast carcinoma. Ultrastruct. Pathol. 42, 170-180. 10.1080/01913123.2017.1419328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera, M. O., Berón, W. and Colombo, M. I. (2012). The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy 8, 1590-1603. 10.4161/auto.21459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar, R., Chandel, S., Sarotra, P. and Medhi, B. (2014). Current status of pharmacological treatment of colorectal cancer. World J. Gastrointest. Oncol. 6, 177. 10.4251/wjgo.v6.i6.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Akra, L., Bae, D.-H., Sahni, S., Huang, M. L. H., Park, K. C., Lane, D. J. R., Jansson, P. J. and Richardson, D. R. (2018). Tumor stressors induce two mechanisms of intracellular P-glycoprotein–mediated resistance that are overcome by lysosomal-targeted thiosemicarbazones. J. Biol. Chem. 293, 3562-3587. 10.1074/jbc.M116.772699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves, S., Castro, L., Fernandes, M. S., Francisco, R., Castro, P., Priault, M., Chaves, S. R., Moyer, M. P., Oliveira, C., Seruca, R.et al. (2015). Colorectal cancer-related mutant KRAS alleles function as positive regulators of autophagy. Oncotarget 6, 30787-30802. 10.18632/oncotarget.5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus, D. W., Baker, J. A., Mason, R. and Martin, I. J. (2008). The potential influence of CO2, as an agent for euthanasia, on the pharmacokinetics of basic compounds in rodents. Drug Metab. Dispos. 36, 375-379. 10.1124/dmd.107.018879 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay, U., Todorova, P., Pavlova, N. N., Tada, Y., Thompson, C. B., Finley, L. W. S. and Overholtzer, M. (2022). Leucine retention in lysosomes is regulated by starvation. Proc. Natl. Acad. Sci. USA 119, e2114912119. 10.1073/pnas.2114912119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, D. E., Hatzivassiliou, G., Zhao, F., Andreadis, C. and Thompson, C. B. (2005). ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24, 6314-6322. 10.1038/sj.onc.1208773 [DOI] [PubMed] [Google Scholar]
- Boudreau, A., Purkey, H. E., Hitz, A., Robarge, K., Peterson, D., Labadie, S., Kwong, M., Hong, R., Gao, M., Del Nagro, C.et al. (2016). Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition. Nat. Chem. Biol. 12, 779-786. 10.1038/nchembio.2143 [DOI] [PubMed] [Google Scholar]
- Bueno, M. J., Jimenez-Renard, V., Samino, S., Capellades, J., Junza, A., López-Rodríguez, M. L., Garcia-Carceles, J., Lopez-Fabuel, I., Bolaños, J. P., Chandel, N. S.et al. (2019). Essentiality of fatty acid synthase in the 2D to anchorage-independent growth transition in transforming cells. Nat. Commun. 10, 5011. 10.1038/s41467-019-13028-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y., Chen, J., Huang, P., Ge, W., Hou, D. and Zhang, G. (2019). Inspecting human colon adenocarcinoma cell lines by using terahertz time-domain reflection spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 211, 356-362. 10.1016/j.saa.2018.12.023 [DOI] [PubMed] [Google Scholar]
- Chen, Q., Vazquez, E. J., Moghaddas, S., Hoppel, C. L. and Lesnefsky, E. J. (2003). Production of reactive oxygen species by mitochondria. J. Biol. Chem. 278, 36027-36031. 10.1074/jbc.M304854200 [DOI] [PubMed] [Google Scholar]
- Chen, M., Lu, J., Wei, W., Lv, Y., Zhang, X., Yao, Y., Wang, L., Ling, T. and Zou, X. (2018). Effects of proton pump inhibitors on reversing multidrug resistance via downregulating V-ATPases/PI3K/Akt/mTOR/HIF-1α signaling pathway through TSC1/2 complex and Rheb in human gastric adenocarcinoma cells in vitro and in vivo. Onco. Targets. Ther. 11, 6705-6722. 10.2147/OTT.S161198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova, V., Ilnytskyy, Y., Kovalchuk, O. and Kovalchuk, I. (2023). Transcriptome analysis of cisplatin, cannabidiol, and intermittent serum starvation alone and in various combinations on colorectal cancer cells. Int. J. Mol. Sci. 24, 14743. 10.3390/ijms241914743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chionh, F., Lau, D., Yeung, Y., Price, T. and Tebbutt, N. (2017). Oral versus intravenous fluoropyrimidines for colorectal cancer. Cochrane Database Syst. Rev. 7, CD008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, F., Trombetta, E., Cetrangolo, P., Maggioni, M., Razini, P., De Santis, F., Torrente, Y., Prati, D., Torresani, E. and Porretti, L. (2014). Giant lysosomes as a chemotherapy resistance mechanism in hepatocellular carcinoma cells. PLoS One 9, e114787. 10.1371/journal.pone.0114787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbet, C., Bastien, E., Santiago De Jesus, J. P., Dierge, E., Martherus, R., Vander Linden, C., Doix, B., Degavre, C., Guilbaud, C., Petit, L.et al. (2020). TGFβ2-induced formation of lipid droplets supports acidosis-driven EMT and the metastatic spreading of cancer cells. Nat. Commun. 11, 454. 10.1038/s41467-019-14262-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, K., Capecci, J., Sennoune, S., Huss, M., Maier, M., Martinez-Zaguilan, R. and Forgac, M. (2015). Activity of plasma membrane V-ATPases is critical for the invasion of MDA-MB231 breast cancer cells. J. Biol. Chem. 290, 3680-3692. 10.1074/jbc.M114.611210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, A. L. S., Barreto, E. A., Fazolini, N. P. B., Viola, J. P. B. and Bozza, P. T. (2020). Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death Dis. 11, 105. 10.1038/s41419-020-2297-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt, K., Mathew, R., Beaudoin, B., Bray, K., Anderson, D., Chen, G., Mukherjee, C., Shi, Y., Gélinas, C., Fan, Y.et al. (2006). Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleyto-Seldas, N. and Efeyan, A. (2021). The mTOR–autophagy axis and the control of metabolism. Front. Cell Dev. Biol. 9, 655731. 10.3389/fcell.2021.655731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecker, G., Vercammen, D., Steemans, M., Vanden Berghe, T., Brouckaert, G., Van Loo, G., Zhivotovsky, B., Fiers, W., Grooten, J., Declercq, W.et al. (2001). Death receptor-induced apoptotic and necrotic cell death: differential role of caspases and mitochondria. Cell Death Differ. 8, 829-840. 10.1038/sj.cdd.4400883 [DOI] [PubMed] [Google Scholar]
- Devenport, S. N., Singhal, R., Radyk, M. D., Taranto, J. G., Kerk, S. A., Chen, B., Goyert, J. W., Jain, C., Das, N. K., Oravecz-Wilson, K.et al. (2021). Colorectal cancer cells utilize autophagy to maintain mitochondrial metabolism for cell proliferation under nutrient stress. JCI Insight 6, e138835. 10.1172/jci.insight.138835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriese, S., Van Den Bossche, L., Van Welden, S., Holvoet, T., Pinheiro, I., Hindryckx, P., De Vos, M. and Laukens, D. (2017). T84 monolayers are superior to Caco-2 as a model system of colonocytes. Histochem. Cell Biol. 148, 85-93. 10.1007/s00418-017-1539-7 [DOI] [PubMed] [Google Scholar]
- Doshi, M., Krienke, M., Khederzadeh, S., Sanchez, H., Copik, A., Oyer, J. and Gesquiere, A. J. (2015). Conducting polymer nanoparticles for targeted cancer therapy. RSC Adv. 5, 37943-37956. 10.1039/C5RA05125H [DOI] [Google Scholar]
- Düvel, K., Yecies, J. L., Menon, S., Raman, P., Lipovsky, A. I., Souza, A. L., Triantafellow, E., Ma, Q., Gorski, R., Cleaver, S.et al. (2010). Activation of a metabolic gene regulatory network downstream of mTOR Complex 1. Mol. Cell 39, 171-183. 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendy, M., Sheridan, C., Brumatti, G. and Martin, S. J. (2011). Oncogenic ras-induced expression of noxa and beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol. Cell 42, 23-35. 10.1016/j.molcel.2011.02.009 [DOI] [PubMed] [Google Scholar]
- Eng, C. H., Yu, K., Lucas, J., White, E. and Abraham, R. T. (2010). Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci. Signal. 3, ra31. [DOI] [PubMed] [Google Scholar]
- Gao, P., Bauvy, C., Souquère, S., Tonelli, G., Liu, L., Zhu, Y., Qiao, Z., Bakula, D., Proikas-Cezanne, T., Pierron, G.et al. (2010). The Bcl-2 homology domain 3 mimetic gossypol induces both beclin 1-dependent and beclin 1-independent cytoprotective autophagy in cancer cells. J. Biol. Chem. 285, 25570-25581. 10.1074/jbc.M110.118125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gericke, B., Wienböker, I., Brandes, G. and Löscher, W. (2022). Is P-glycoprotein functionally expressed in the limiting membrane of endolysosomes? A biochemical and ultrastructural study in the rat liver. Cells 11, 1556. 10.3390/cells11091556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafari, M., Haghiralsadat, F., Khanamani Falahati–Pour, S. and Zavar Reza, J. (2020). Development of a novel liposomal nanoparticle formulation of cisplatin to breast cancer therapy. J. Cell. Biochem. 121, 3584-3592. 10.1002/jcb.29651 [DOI] [PubMed] [Google Scholar]
- Gil, J., Karpiński, P. and Sąsiadek, M. M. (2020). Transcriptomic profiling for the autophagy pathway in colorectal cancer. Int. J. Mol. Sci. 21, 7101. 10.3390/ijms21197101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Y., Duvvuri, M. and Krise, J. P. (2003). Separate roles for the Golgi apparatus and lysosomes in the sequestration of drugs in the multidrug-resistant human leukemic cell line HL-60. J. Biol. Chem. 278, 50234-50239. 10.1074/jbc.M306606200 [DOI] [PubMed] [Google Scholar]
- Gotink, K. J., Broxterman, H. J., Labots, M., De Haas, R. R., Dekker, H., Honeywell, R. J., Rudek, M. A., Beerepoot, L. V., Musters, R. J., Jansen, G.et al. (2011). Lysosomal sequestration of sunitinib: a novel mechanism of drug resistance. Clin. Cancer Res. 17, 7337-7346. 10.1158/1078-0432.CCR-11-1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güleç Taşkıran, A. E., Hüsnügil, H. H., Soltani, Z. E., Oral, G., Menemenli, N. S., Hampel, C., Huebner, K., Erlenbach-Wuensch, K., Sheraj, I., Schneider-Stock, R.et al. (2024a). Post-transcriptional regulation of Rab7a in lysosomal positioning and drug resistance in nutrient limited cancer cells. Traffic 25, e12956. 10.1111/tra.12956 [DOI] [PubMed] [Google Scholar]
- Güleç Taşkıran, A. E., Karaoğlu, D. A., Eylem, C. C., Ermiş, Ç., Güderer, İ., Nemutlu, E., Demirkol Canlı, S. and Banerjee, S. (2024b). Glutamine withdrawal leads to the preferential activation of lipid metabolism in metastatic colorectal cancer. Transl. Oncol. 48, 102078. 10.1016/j.tranon.2024.102078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B., Tam, A., Santi, S. A. and Parissenti, A. M. (2016). Role of autophagy and lysosomal drug sequestration in acquired resistance to doxorubicin in MCF-7 cells. BMC Cancer 16, 762. 10.1186/s12885-015-2026-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W., Dong, W., Li, M. and Shen, Y. (2019). Mitochondria P-glycoprotein confers paclitaxel resistance on ovarian cancer cells. Onco. Targets. Ther. 12, 3881-3891. 10.2147/OTT.S193433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guru, S. K., Pathania, A. S., Kumar, S., Ramesh, D., Kumar, M., Rana, S., Kumar, A., Malik, F., Sharma, P. R., Chandan, B. K.et al. (2015). Secalonic acid-D represses HIF1α/VEGF-mediated angiogenesis by regulating the Akt/mTOR/p70S6K signaling cascade. Cancer Res. 75, 2886-2896. 10.1158/0008-5472.CAN-14-2312 [DOI] [PubMed] [Google Scholar]
- Halder, J., Pradhan, D., Kar, B., Ghosh, G. and Rath, G. (2022). Nanotherapeutics approaches to overcome P-glycoprotein-mediated multi-drug resistance in cancer. Nanomedicine 40, 102494. 10.1016/j.nano.2021.102494 [DOI] [PubMed] [Google Scholar]
- Hervieu, A., Heuss, S. F., Zhang, C., Barrow-Mcgee, R., Joffre, C., Ménard, L., Clarke, P. A. and Kermorgant, S. (2020). A PI3K- and GTPase-independent Rac1-mTOR mechanism mediates MET-driven anchorage-independent cell growth but not migration. Sci. Signal. 13, eaba8627. 10.1126/scisignal.aba8627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, P.-C., Bihuniak, J. D., Macintyre, A. N., Staron, M., Liu, X., Amezquita, R., Tsui, Y.-C., Cui, G., Micevic, G., Perales, J. C.et al. (2015). Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217-1228. 10.1016/j.cell.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, W., Han, J., Lu, C., Goldstein, L. A. and Rabinowich, H. (2010). Autophagic degradation of active caspase-8. Autophagy 6, 891-900. 10.4161/auto.6.7.13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz, S. J., Terashima, M., Mizunuma, N. and Slapak, C. A. (1997). Vesicular anthracycline accumulation in doxorubicin-selected U-937 cells: participation of lysosomes. Blood 89, 3745-3754. 10.1182/blood.V89.10.3745 [DOI] [PubMed] [Google Scholar]
- Hüsnügil, H. H., Güleç Taşkıran, A. E., Güderer, I., Nehri, L. N., Oral, G., Menemenli, N. Ş., Özcan, Ö., Noghreh, A., Akyol, A. and Banerjee, S. (2024). Lysosomal alkalinization in nutrient restricted cancer cells activates cytoskeletal rearrangement to enhance partial epithelial to mesenchymal transition. Transl. Oncol. 41, 101860. 10.1016/j.tranon.2023.101860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, C., Roth, D., Ward, D. M., Kaplan, J. and Andrews, N. W. (2004). Defective lysosomal exocytosis and plasma membrane repair in Chediak–Higashi/beige cells. Proc. Natl. Acad. Sci. USA 101, 16795-16800. 10.1073/pnas.0405905101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icard, P., Shulman, S., Farhat, D., Steyaert, J.-M., Alifano, M. and Lincet, H. (2018). How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updat. 38, 1-11. 10.1016/j.drup.2018.03.001 [DOI] [PubMed] [Google Scholar]
- Janzer, A., German, N. J., Gonzalez-Herrera, K. N., Asara, J. M., Haigis, M. C. and Struhl, K. (2014). Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl. Acad. Sci. USA 111, 10574-10579. 10.1073/pnas.1409844111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, K. Y., Kim, K. M., Kim, Y. H., Shim, K. S., Lee, J. Y., Kim, T. and Chae, S. (2021). Serum starvation sensitizes anticancer effect of Anemarrhena asphodeloides via p38/JNK-induced cell cycle arrest and apoptosis in colorectal cancer cells. Am. J. Chin. Med. 49, 1001-1016. 10.1142/S0192415X21500488 [DOI] [PubMed] [Google Scholar]
- Johnson, S. M., Gulhati, P., Rampy, B. A., Han, Y., Rychahou, P. G., Doan, H. Q., Weiss, H. L. and Evers, M. B. (2010). Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J. Am. Coll. Surg. 210, 767-776. 10.1016/j.jamcollsurg.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Flores, D. L., Ezquerra, M., Gonzàlez-Casacuberta, Ï., Ormazabal, A., Morén, C., Tolosa, E., Fucho, R., Guitart-Mampel, M., Casado, M., Valldeoriola, F.et al. (2020). Disrupted mitochondrial and metabolic plasticity underlie comorbidity between age-related and degenerative disorders as Parkinson disease and type 2 diabetes mellitus. Antioxidants 9, 1063. 10.3390/antiox9111063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, C. H., Jun, C. B., Ro, S.-H., Kim, Y.-M., Otto, N. M., Cao, J., Kundu, M. and Kim, D.-H. (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992-2003. 10.1091/mbc.e08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallus, S., Englinger, B., Senkiv, J., Laemmerer, A., Heffeter, P., Berger, W., Kowol, C. R. and Keppler, B. K. (2018). Nanoformulations of anticancer FGFR inhibitors with improved therapeutic index. Nanomedicine. 14, 2632-2643. 10.1016/j.nano.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmi, F., Hensley, T., Pope, C., Funk, R. S., Loewen, G. J., Buckley, D. B. and Parkinson, A. (2013). Lysosomal sequestration (Trapping) of lipophilic amine (Cationic Amphiphilic) drugs in immortalized human hepatocytes (Fa2N-4 Cells). Drug Metab. Dispos. 41, 897-905. 10.1124/dmd.112.050054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, Q., Wang, Y., Deng, Y., Wang, J., Yuan, M., Liang, A., Wang, J. and Gong, Q. (2024). Identification and validation of mitophagy-related signatures as a novel prognostic model for colorectal cancer. Transl. Cancer Res. 13, 782-797. 10.21037/tcr-23-785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.-Y., Chen, Z.-G., Zhou, X., Fan, X.-R., Wang, H., Lai, P.-L., Su, Y.-C., Zhang, B.-Y., Bai, X.-C. and Li, Y.-F. (2014). DEPTOR expression negatively correlates with mTORC1 activity and tumor progression in colorectal cancer. Asian Pac. J. Cancer Prev. 15, 4589-4594. 10.7314/APJCP.2014.15.11.4589 [DOI] [PubMed] [Google Scholar]
- Lauzier, A., Normandeau-Guimond, J., Vaillancourt-Lavigueur, V., Boivin, V., Charbonneau, M., Rivard, N., Scott, M. S., Dubois, C. M. and Jean, S. (2019). Colorectal cancer cells respond differentially to autophagy inhibition in vivo. Sci. Rep. 9, 11316. 10.1038/s41598-019-47659-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Hu, J., Wang, B., Sheng, L., Liu, Z., Yang, S. and Li, Y. (2014). Inhibitory effects of herbal constituents on P-glycoprotein in vitro and in vivo: Herb–drug interactions mediated via P-gp. Toxicol. Appl. Pharmacol. 275, 163-175. 10.1016/j.taap.2013.12.015 [DOI] [PubMed] [Google Scholar]
- Li, R.-J., Xu, J., Fu, C., Zhang, J., Zheng, Y. G., Jia, H. and Liu, J. O. (2016). Regulation of mTORC1 by lysosomal calcium and calmodulin. Elife 5, e19360. 10.7554/eLife.19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Liu, C., Huang, Z., Shi, L., Zhong, C., Zhou, W., Meng, P., Li, Z., Wang, S., Luo, F.et al. (2021). AKR1B10 negatively regulates autophagy through reducing GAPDH upon glucose starvation in colon cancer. J. Cell Sci. 134, jcs255273. 10.1242/jcs.255273 [DOI] [PubMed] [Google Scholar]
- Lieffers, J. R., Mourtzakis, M., Hall, K. D., Mccargar, L. J., Prado, C. M. and Baracos, V. E. (2009). A viscerally driven cachexia syndrome in patients with advanced colorectal cancer: contributions of organ and tumor mass to whole-body energy demands. Am. J. Clin. Nutr. 89, 1173-1179. 10.3945/ajcn.2008.27273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Ren, Y., Hou, Y., Zhang, C., Wang, B., Li, X., Sun, R. and Liu, J. (2019). Dihydroartemisinin induces endothelial cell autophagy through suppression of the Akt/mTOR pathway. J. Cancer 10, 6057-6064. 10.7150/jca.33704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R.-M., Xu, P., Chen, Q., Feng, S. and Xie, Y. (2020). A multiple-targets alkaloid nuciferine overcomes paclitaxel-induced drug resistance in vitro and in vivo. Phytomedicine 79, 153342. 10.1016/j.phymed.2020.153342 [DOI] [PubMed] [Google Scholar]
- Liu, Y., Chen, Y., Wang, F., Lin, J., Tan, X., Chen, C., Wu, L., Zhang, X., Wang, Y., Shi, Y.et al. (2023). Caveolin-1 promotes glioma progression and maintains its mitochondrial inhibition resistance. Discov. Oncol. 14, 161. 10.1007/s12672-023-00765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Kreyche, P., Shen, H., Marino, A. M., Iyer, R. A., Humphreys, W. G. and Lai, Y. (2019). Lysosomal P-gp-MDR1 confers drug resistance of brentuximab vedotin and its cytotoxic payload monomethyl Auristatin E in tumor cells. Front. Pharmacol. 10, 749. 10.3389/fphar.2019.00749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock, R., Roy, S., Kenific, C. M., Su, J. S., Salas, E., Ronen, S. M. and Debnath, J. (2011). Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell 22, 165-178. 10.1091/mbc.e10-06-0500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, F., Borghi, M., Marzoli, F., Azzarito, T., Matarrese, P., Iessi, E., Venturi, G., Meschini, S., Canitano, A., Bona, R.et al. (2015). TM9SF4 is a novel V-ATPase-interacting protein that modulates tumor pH alterations associated with drug resistance and invasiveness of colon cancer cells. Oncogene 34, 5163-5174. 10.1038/onc.2014.437 [DOI] [PubMed] [Google Scholar]
- Lynch, T. P., Ferrer, C. M., Jackson, S. R., Shahriari, K. S., Vosseller, K. and Reginato, M. J. (2012). Critical role of O-linked β-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J. Biol. Chem. 287, 11070-11081. 10.1074/jbc.M111.302547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Q., Chang, Z., Wang, W. and Wang, B. (2015). Rapamycin-mediated mTOR inhibition reverses drug resistance to adriamycin in colon cancer cells. Hepatogastroenterology 62, 880-886. [PubMed] [Google Scholar]
- Machado, E., White-Gilbertson, S., Van De Vlekkert, D., Janke, L., Moshiach, S., Campos, Y., Finkelstein, D., Gomero, E., Mosca, R., Qiu, X.et al. (2015). Regulated lysosomal exocytosis mediates cancer progression. Sci. Adv. 1, e1500603. 10.1126/sciadv.1500603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalowicz, G. P., Snook, A. E., Magee, M. S., Merlino, D., Berman-Booty, L. D. and Waldman, S. A. (2014). GUCY2C lysosomotropic endocytosis delivers immunotoxin therapy to metastatic colorectal cancer. Oncotarget 5, 9460-9471. 10.18632/oncotarget.2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew, R., Kongara, S., Beaudoin, B., Karp, C. M., Bray, K., Degenhardt, K., Chen, G., Jin, S. and White, E. (2007). Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 21, 1367-1381. 10.1101/gad.1545107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura, Y. and Maeda, H. (1986). A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 46, 6387-6392. [PubMed] [Google Scholar]
- Meeusen, S. and Nunnari, J. (2003). Evidence for a two membrane–spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163, 503-510. 10.1083/jcb.200304040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos Rodrigo, M. A., Buchtelova, H., De Los Rios, V., Casal, J. I., Eckschlager, T., Hrabeta, J., Belhajova, M., Heger, Z. and Adam, V. (2019). Proteomic signature of neuroblastoma cells UKF-NB-4 reveals key role of lysosomal sequestration and the proteasome complex in acquiring chemoresistance to cisplatin. J. Proteome Res. 18, 1255-1263. 10.1021/acs.jproteome.8b00867 [DOI] [PubMed] [Google Scholar]
- Meschini, S., Calcabrini, A., Monti, E., Del Bufalo, D., Stringaro, A., Dolfini, E. and Arancia, G. (2000). Intracellular P-glycoprotein expression is associated with the intrinsic multidrug resistance phenotype in human colon adenocarcinoma cells. Int. J. Cancer 87, 615-628. [DOI] [PubMed] [Google Scholar]
- Michelakis, E. D., Sutendra, G., Dromparis, P., Webster, L., Haromy, A., Niven, E., Maguire, C., Gammer, T.-L., Mackey, J. R., Fulton, D.et al. (2010). Metabolic modulation of glioblastoma with dichloroacetate. Sci. Transl. Med. 2, 31ra34. 10.1126/scitranslmed.3000677 [DOI] [PubMed] [Google Scholar]
- Micsik, T., Mersich, T., Baranyai, Z., Besznyák, I., Zaránd, A., Dede, K., Nagy, P., Atkári, B., Jakab, F., Lőrincz, A.et al. (2008). Alterations of MDR1 and MRP1 functional activity of colorectal malignancies in a prospective clinical trial. Z. Gastroenterol. 46, a63. 10.1055/s-2008-1079667 [DOI] [Google Scholar]
- Mony, V. K., Benjamin, S. and O'rourke, E. J. (2016). A lysosome-centered view of nutrient homeostasis. Autophagy 12, 619-631. 10.1080/15548627.2016.1147671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, E., Arnold, M., Gini, A., Lorenzoni, V., Cabasag, C. J., Laversanne, M., Vignat, J., Ferlay, J., Murphy, N. and Bray, F. (2023). Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut 72, 338-344. 10.1136/gutjnl-2022-327736 [DOI] [PubMed] [Google Scholar]
- Navarro, P., Bueno, M. J., Zagorac, I., Mondejar, T., Sanchez, J., Mourón, S., Muñoz, J., Gómez-López, G., Jimenez-Renard, V., Mulero, F.et al. (2016). Targeting tumor mitochondrial metabolism overcomes resistance to antiangiogenics. Cell Rep. 15, 2705-2718. 10.1016/j.celrep.2016.05.052 [DOI] [PubMed] [Google Scholar]
- Nguyen, O. N. P., Grimm, C., Schneider, L. S., Chao, Y.-K., Atzberger, C., Bartel, K., Watermann, A., Ulrich, M., Mayr, D., Wahl-Schott, C.et al. (2017). Two-pore channel function is crucial for the migration of invasive cancer cells. Cancer Res. 77, 1427-1438. 10.1158/0008-5472.CAN-16-0852 [DOI] [PubMed] [Google Scholar]
- Ni, T., He, Z., Dai, Y., Yao, J., Guo, Q. and Wei, L. (2017). Oroxylin A suppresses the development and growth of colorectal cancer through reprogram of HIF1α-modulated fatty acid metabolism. Cell Death Dis. 8, e2865-e2865. 10.1038/cddis.2017.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan, K. E., Beck, C., Holzmayer, T. A., Chin, J. E., Wunder, J. S., Andrulis, I. L., Gazdar, A. F., Willman, C. L., Griffith, B., Von Hoff, D. D.et al. (1990). Quantitative analysis of MDR1 (multidrug resistance) gene expression in human tumors by polymerase chain reaction. Proc. Natl. Acad. Sci. USA 87, 7160-7164. 10.1073/pnas.87.18.7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlotti, N. I., Cimino-Reale, G., Borghini, E., Pennati, M., Sissi, C., Perrone, F., Palumbo, M., Daidone, M. G., Folini, M. and Zaffaroni, N. (2012). Autophagy acts as a safeguard mechanism against G-quadruplex ligand-mediated DNA damage. Autophagy 8, 1185-1196. 10.4161/auto.20519 [DOI] [PubMed] [Google Scholar]
- Palmieri, M., Impey, S., Kang, H., Di Ronza, A., Pelz, C., Sardiello, M. and Ballabio, A. (2011). Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852-3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Pan, M., Zorbas, C., Sugaya, M., Ishiguro, K., Kato, M., Nishida, M., Zhang, H.-F., Candeias, M. M., Okamoto, A., Ishikawa, T.et al. (2022). Glutamine deficiency in solid tumor cells confers resistance to ribosomal RNA synthesis inhibitors. Nat. Commun. 13, 3706. 10.1038/s41467-022-31418-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, N., Dhiman, S., Srivastava, T. and Majumder, S. (2016). Transition metal oxide nanoparticles are effective in inhibiting lung cancer cell survival in the hypoxic tumor microenvironment. Chem. Biol. Interact. 254, 221-230. 10.1016/j.cbi.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Paquette, M., El-Houjeiri, L., C., Zirden, L., Puustinen, P., Blanchette, P., Jeong, H., Dejgaard, K., Siegel, P. M. and Pause, A. (2021). AMPK-dependent phosphorylation is required for transcriptional activation of TFEB and TFE3. Autophagy 17, 3957-3975. 10.1080/15548627.2021.1898748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida, P. K., Marquez-Palencia, M., Ghosh, S., Khandelwal, N., Kim, K., Nair, V., Liu, X.-Z., Vu, H. S., Zacharias, L. G., Gonzalez-Ericsson, P. I.et al. (2023). Limiting mitochondrial plasticity by targeting DRP1 induces metabolic reprogramming and reduces breast cancer brain metastases. Nat. Cancer 4, 893-907. 10.1038/s43018-023-00563-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier, J., Guerrouahen, B. S., Al Thawadi, H., Ghiabi, P., Maleki, M., Abu-Kaoud, N., Jacob, A., Mirshahi, M., Galas, L., Rafii, S.et al. (2013). Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J. Transl. Med. 11, 94. 10.1186/1479-5876-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechincha, C., Groessl, S., Kalis, R., De Almeida, M., Zanotti, A., Wittmann, M., Schneider, M., De Campos, R. P., Rieser, S., Brandstetter, M.et al. (2022). Lysosomal enzyme trafficking factor LYSET enables nutritional usage of extracellular proteins. Science (80-.) 378, eabn5637. 10.1126/science.abn5637 [DOI] [PubMed] [Google Scholar]
- Peng, X., Chen, Z., Farshidfar, F., Xu, X., Lorenzi, P. L., Wang, Y., Cheng, F., Tan, L., Mojumdar, K., Du, D.et al. (2018). Molecular characterization and clinical relevance of metabolic expression subtypes in human cancers. Cell Rep. 23, 255-269.e4. 10.1016/j.celrep.2018.03.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault, M., Hue, E., Marhuenda, F., Pilet, P., Oliver, L. and Vallette, F. M. (2010). Differential dependence on beclin 1 for the regulation of pro-survival autophagy by Bcl-2 and Bcl-xL in HCT116 colorectal cancer cells. PLoS One 5, e8755. 10.1371/journal.pone.0008755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, H., Ning, X., Yu, C., Ji, X., Jin, Y., Mcnutt, M. A. and Yin, Y. (2019). Succinylation-dependent mitochondrial translocation of PKM2 promotes cell survival in response to nutritional stress. Cell Death Dis. 10, 170. 10.1038/s41419-018-1271-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Q., Li, R., Li, H., Cao, Y., Bai, M., Fan, X., Wang, S., Zhang, B. and Li, S. (2016). Identification of the anti-tumor activity and mechanisms of nuciferine through a network pharmacology approach. Acta Pharmacol. Sin. 37, 963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Y., Corum, L., Meng, Q., Blenis, J., Zheng, J. Z., Shi, X., Flynn, D. C. and Jiang, B.-H. (2004). PI3K induced actin filament remodeling through Akt and p70S6K1: implication of essential role in cell migration. Am. J. Physiol. Physiol. 286, C153-C163. 10.1152/ajpcell.00142.2003 [DOI] [PubMed] [Google Scholar]
- Qiao, S., Koh, S.-B., Vivekanandan, V., Salunke, D., Patra, K. C., Zaganjor, E., Ross, K., Mizukami, Y., Jeanfavre, S., Chen, A.et al. (2020). REDD1 loss reprograms lipid metabolism to drive progression of RAS mutant tumors. Genes Dev. 34, 751-766. 10.1101/gad.335166.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben, N. and Puertollano, R. (2016). TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu. Rev. Cell Dev. Biol. 32, 255-278. 10.1146/annurev-cellbio-111315-125407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C. (2018). How nutrients orchestrate lysosome positioning. Contact 1, 251525641875611. 10.1177/2515256418756111 [DOI] [Google Scholar]
- Robb-Gaspers, L. D. (1998). Integrating cytosolic calcium signals into mitochondrial metabolic responses. EMBO J. 17, 4987-5000. 10.1093/emboj/17.17.4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulli, E., Legramandi, L., Salvati, L. and Mandala, M. (2019). The impact of targeted therapies and immunotherapy in melanoma brain metastases: a systematic review and meta–analysis. Cancer 125, 3776-3789. 10.1002/cncr.32375 [DOI] [PubMed] [Google Scholar]
- Sala-Vila, A., Navarro-Lérida, I., Sánchez-Alvarez, M., Bosch, M., Calvo, C., López, J. A., Calvo, E., Ferguson, C., Giacomello, M., Serafini, A.et al. (2016). Interplay between hepatic mitochondria-associated membranes, lipid metabolism and caveolin-1 in mice. Sci. Rep. 6, 27351. 10.1038/srep27351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, T. L., Martignoni, M. E., Bachmann, J., Fechtner, K., Friess, H., Kinscherf, R. and Hildebrandt, W. (2007). Activity of the Akt-dependent anabolic and catabolic pathways in muscle and liver samples in cancer-related cachexia. J. Mol. Med. 85, 647-654. 10.1007/s00109-007-0177-2 [DOI] [PubMed] [Google Scholar]
- Schneider, H., Staudacher, S., Poppelreuther, M., Stremmel, W., Ehehalt, R. and Füllekrug, J. (2014). Protein mediated fatty acid uptake: synergy between CD36/FAT-facilitated transport and acyl-CoA synthetase-driven metabolism. Arch. Biochem. Biophys. 546, 8-18. 10.1016/j.abb.2014.01.025 [DOI] [PubMed] [Google Scholar]
- Scumaci, D., Olivo, E., Fiumara, C. V., La Chimia, M., De Angelis, M. T., Mauro, S., Costa, G., Ambrosio, F. A., Alcaro, S., Agosti, V.et al. (2020). DJ-1 proteoforms in breast cancer cells: the escape of metabolic epigenetic misregulation. Cells 9, 1968. 10.3390/cells9091968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumaran, M., Amaravadi, R. K., Vasilevskaya, I. A. and O'dwyer, P. J. (2013). Autophagy inhibition sensitizes colon cancer cells to antiangiogenic and cytotoxic therapy. Clin. Cancer Res. 19, 2995-3007. 10.1158/1078-0432.CCR-12-1542 [DOI] [PubMed] [Google Scholar]
- Sesso, A., Belizário, J. E., Marques, M. M., Higuchi, M. L., Schumacher, R. I., Colquhoun, A., Ito, E. and Kawakami, J. (2012). Mitochondrial swelling and incipient outer membrane rupture in preapoptotic and apoptotic cells. Anat. Rec. 295, 1647-1659. 10.1002/ar.22553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, A. S. and Gatenby, R. A. (2010). A theoretical quantitative model for evolution of cancer chemotherapy resistance. Biol. Direct 5, 25. 10.1186/1745-6150-5-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Assmann, P., Turck, N., Sidhoum-Jenny, M., Gradwohl, G. and Kedinger, M. (2007). In vitro models of intestinal epithelial cell differentiation. Cell Biol. Toxicol. 23, 241-256. 10.1007/s10565-006-0175-0 [DOI] [PubMed] [Google Scholar]
- Su, Y., Liu, J., Tian, Y., Dong, H., Shi, M., Zhang, J., Li, W., Huang, Q., Xiang, N., Wang, C.et al. (2024). HIF-1α mediates immunosuppression and chemoresistance in colorectal cancer by inhibiting CXCL9, −10 and −11. Biomed. Pharmacother. 173, 116427. 10.1016/j.biopha.2024.116427 [DOI] [PubMed] [Google Scholar]
- Sümbül, A. T., Göğebakan, B., Ergün, S., Yengil, E., Batmacı, C. Y., Tonyalı, Ö. and Yaldız, M. (2014). miR-204-5p expression in colorectal cancer: an autophagy-associated gene. Tumor Biol. 35, 12713-12719. 10.1007/s13277-014-2596-3 [DOI] [PubMed] [Google Scholar]
- Sun, H., Chen, L., Cao, S., Liang, Y. and Xu, Y. (2019). Warburg effects in cancer and normal proliferating cells: two tales of the same name. Genomics. Proteomics Bioinformatics 17, 273-286. 10.1016/j.gpb.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Zhou, Y., Skaro, M. F., Wu, Y., Qu, Z., Mao, F., Zhao, S. and Xu, Y. (2020). metabolic reprogramming in cancer is induced to increase proton production. Cancer Res. 80, 1143-1155. 10.1158/0008-5472.CAN-19-3392 [DOI] [PubMed] [Google Scholar]
- Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A. and Bray, F. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 71, 209-249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Tan, J., Wang, H.-L., Yang, J., Liu, Q.-Q., Li, C.-M., Wang, Y.-Q., Fu, L.-N., Gao, Q.-Y., Chen, Y.-X. and Fang, J.-Y. (2020). JMJD2B-induced amino acid alterations enhance the survival of colorectal cancer cells under glucose-deprivation via autophagy. Theranostics 10, 5763-5777. 10.7150/thno.38087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, J., Peng, W., Ji, J., Peng, C., Wang, T., Yang, P., Gu, J., Feng, Y., Jin, K., Wang, X.et al. (2023). GPR176 promotes cancer progression by interacting with G protein GNAS to restrain cell mitophagy in colorectal cancer. Adv. Sci. 10, 2205627. 10.1002/advs.202205627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura, H., Kohno, K., Sato, S., Uchiumi, T., Miyazaki, M., Kobayashi, M. and Kuwano, M. (1992). The human multidrug resistance 1 promoter has an element that responds to serum starvation. Biochem. Biophys. Res. Commun. 183, 917-924. 10.1016/0006-291X(92)90571-2 [DOI] [PubMed] [Google Scholar]
- Viale, A., Pettazzoni, P., Lyssiotis, C. A., Ying, H., Sánchez, N., Marchesini, M., Carugo, A., Green, T., Seth, S., Giuliani, V.et al. (2014). Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628-632. 10.1038/nature13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Shen, B., Qin, X., Liu, S. and Feng, J. (2019). Akt/mTOR and AMPK signaling pathways are responsible for liver X receptor agonist GW3965-enhanced gefitinib sensitivity in non-small cell lung cancer cell lines. Transl. Cancer Res. 8, 66-76. 10.21037/tcr.2018.12.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieman, H. L., Wofford, J. A. and Rathmell, J. C. (2007). Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of glut1 activity and trafficking. Mol. Biol. Cell 18, 1437-1446. 10.1091/mbc.e06-07-0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William, D., Erdmann, K., Ottemöller, J., Mangelis, A., Conrad, C., Peitzsch, M., Schröck, E., Eisenhofer, G., Zacharis, A., Füssel, S.et al. (2021). Targeted quantification of carbon metabolites identifies metabolic progression markers and an undiagnosed case of SDH-deficient clear cell renal cell carcinoma in a German Cohort. Metabolites 11, 764. 10.3390/metabo11110764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Han, Y., Rodriguez Sillke, Y., Deng, H., Siddiqui, S., Treese, C., Schmidt, F., Friedrich, M., Keye, J., Wan, J.et al. (2019). Lipid droplet–dependent fatty acid metabolism controls the immune suppressive phenotype of tumor–associated macrophages. EMBO Mol. Med. 11, e10698. 10.15252/emmm.201910698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Fan, Y., Wang, K., Miao, Y., Chang, Y., Ming, J., Wang, X., Lu, S., Liu, R., Zhang, F.et al. (2024). NIR-II imaging-guided precise photodynamic therapy for augmenting tumor-starvation therapy by glucose metabolism reprogramming interference. Sci. Bull. 69, 1263-1274. 10.1016/j.scib.2024.02.008 [DOI] [PubMed] [Google Scholar]
- Yaghini, E., Dondi, R., Tewari, K. M., Loizidou, M., Eggleston, I. M. and Macrobert, A. J. (2017). Endolysosomal targeting of a clinical chlorin photosensitiser for light-triggered delivery of nano-sized medicines. Sci. Rep. 7, 6059. 10.1038/s41598-017-06109-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagüe, E., Armesilla, A. L., Harrison, G., Elliott, J., Sardini, A., Higgins, C. F. and Raguz, S. (2003). P-glycoprotein (MDR1) expression in leukemic cells is regulated at two distinct steps, mRNA stabilization and translational initiation. J. Biol. Chem. 278, 10344-10352. 10.1074/jbc.M211093200 [DOI] [PubMed] [Google Scholar]
- Yahanda, A. M., Alder, K. M., Fisher, G. A., Brophy, N. A., Halsey, J., Hardy, R. I., Gosland, M. P., Lum, B. L. and Sikic, B. I. (1992). Phase I trial of etoposide with cyclosporine as a modulator of multidrug resistance. J. Clin. Oncol. 10, 1624-1634. 10.1200/JCO.1992.10.10.1624 [DOI] [PubMed] [Google Scholar]
- Yang, A., Rajeshkumar, N. V., Wang, X., Yabuuchi, S., Alexander, B. M., Chu, G. C., Von Hoff, D. D., Maitra, A. and Kimmelman, A. C. (2014). Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 4, 905-913. 10.1158/2159-8290.CD-14-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Song, R., Li, N., Sun, K., Shi, F., Liu, H., Shen, F., Jiang, S., Zhang, L. and Jin, Y. (2020). Silica dust exposure induces autophagy in alveolar macrophages through switching Beclin1 affinity from Bcl–2 to PIK3C3. Environ. Toxicol. 35, 758-767. 10.1002/tox.22910 [DOI] [PubMed] [Google Scholar]
- Yang, N., Gong, F., Zhou, Y., Hao, Y., Dong, Z., Lei, H., Zhong, L., Yang, X., Wang, X., Zhao, Y.et al. (2021). A general in-situ reduction method to prepare core-shell liquid-metal / metal nanoparticles for photothermally enhanced catalytic cancer therapy. Biomaterials 277, 121125. 10.1016/j.biomaterials.2021.121125 [DOI] [PubMed] [Google Scholar]
- Yecies, J. L. (2012). SREBP: A Key Effector of mTORC1 Signaling in Metabolism and Cancer. PhD Thesis, Harvard University, Cambridge, Massachusetts.
- Yin, K., Lee, J., Liu, Z., Kim, H., Martin, D. R., Wu, D., Liu, M. and Xue, X. (2021). Mitophagy protein PINK1 suppresses colon tumor growth by metabolic reprogramming via p53 activation and reducing acetyl-CoA production. Cell Death Differ. 28, 2421-2435. 10.1038/s41418-021-00760-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, N., Koizumi, M., Adachi, I. and Kawakami, J. (2006). Inhibition of P-glycoprotein-mediated transport by terpenoids contained in herbal medicines and natural products. Food Chem. Toxicol. 44, 2033-2039. 10.1016/j.fct.2006.07.003 [DOI] [PubMed] [Google Scholar]
- Yuan, H.-X., Russell, R. C. and Guan, K.-L. (2013). Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 9, 1983-1995. 10.4161/auto.26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Z., Liang, X., Zhan, Y., Wang, Z., Xu, J., Qiu, Y., Wang, J., Cao, Y., Le, V.-M., Ly, H.-T.et al. (2020). Targeting CD133 reverses drug-resistance via the AKT/NF-κB/MDR1 pathway in colorectal cancer. Br. J. Cancer 122, 1342-1353. 10.1038/s41416-020-0783-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaja, R., Caminada, D., Lončar, J., Fent, K. and Smital, T. (2008). Development and characterization of P-glycoprotein 1 (Pgp1, ABCB1)-mediated doxorubicin-resistant PLHC-1 hepatoma fish cell line. Toxicol. Appl. Pharmacol. 227, 207-218. 10.1016/j.taap.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Dong, Y., Zeng, X., Liang, X., Li, X., Tao, W., Chen, H., Jiang, Y., Mei, L. and Feng, S.-S. (2014a). The effect of autophagy inhibitors on drug delivery using biodegradable polymer nanoparticles in cancer treatment. Biomaterials 35, 1932-1943. 10.1016/j.biomaterials.2013.10.034 [DOI] [PubMed] [Google Scholar]
- Zhang, D., Li, J., Wang, F., Hu, J., Wang, S. and Sun, Y. (2014b). 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett. 355, 176-183. 10.1016/j.canlet.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Zhang, Y.-K., Wang, Y.-J., Lei, Z.-N., Zhang, G.-N., Zhang, X.-Y., Wang, D., Al-Rihani, S. B., Shukla, S., Ambudkar, S. V., Kaddoumi, A.et al. (2019). Regorafenib antagonizes BCRP-mediated multidrug resistance in colon cancer. Cancer Lett. 442, 104-112. 10.1016/j.canlet.2018.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Yang, J., Lin, C., Liu, W., Huo, Y., Yang, M., Jiang, S.-H., Sun, Y. and Hua, R. (2020). Endoplasmic Reticulum stress-dependent expression of ERO1L promotes aerobic glycolysis in Pancreatic Cancer. Theranostics 10, 8400-8414. 10.7150/thno.45124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Yue, P., Lu, T., Wang, Y., Wei, Y. and Wei, X. (2021). Role of lysosomes in physiological activities, diseases, and therapy. J. Hematol. Oncol. 14, 1-39. 10.1186/s13045-020-01025-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Xu, L., Ren, Z., Liu, X., Song, J., Zhang, P., Zhang, C., Gong, S., Wu, N., Zhang, X.et al. (2023). LINC01615 maintains cell survival in adaptation to nutrient starvation through the pentose phosphate pathway and modulates chemosensitivity in colorectal cancer. Cell. Mol. Life Sci. 80, 20. 10.1007/s00018-022-04675-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Fu, J., Hu, B., Chen, L., Wang, J., Fang, J., Ge, C., Lin, H., Pan, K., Fu, L.et al. (2021). Serine metabolism regulates YAP activity through USP7 in colon cancer. Front. Cell Dev. Biol. 9, 639111. 10.3389/fcell.2021.639111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S.-F. (2008). Structure, function and regulation of P-glycoprotein and its clinical relevance in drug disposition. Xenobiotica 38, 802-832. 10.1080/00498250701867889 [DOI] [PubMed] [Google Scholar]
- Zhu, J., Cai, Y., Xu, K., Ren, X., Sun, J., Lu, S., Chen, J. and Xu, P. (2018). Beclin1 overexpression suppresses tumor cell proliferation and survival via an autophagy-dependent pathway in human synovial sarcoma cells. Oncol. Rep. 40, 1927-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski, D. C., Jamshidi, N., Corbett, A. J., Bordbar, A., Thomas, A. and Palsson, B. O. (2017). Systems biology analysis of drivers underlying hallmarks of cancer cell metabolism. Sci. Rep. 7, 41241. 10.1038/srep41241 [DOI] [PMC free article] [PubMed] [Google Scholar]