Abstract
The tumour suppressor factor p53 plays an essential role in regulating numerous cellular processes, including the cell cycle, DNA repair, apoptosis, autophagy, cell metabolism and immune response. TP53 is the most commonly mutated gene in human cancers. These mutations are primarily non-synonymous changes that produce mutant p53 proteins characterized by loss of function, a dominant negative effect on p53 tetramerisation and gain of function (GOF). GOF mutations not only disrupt the tumour-suppressive activities of p53 but also endow the mutant proteins with new oncogenic properties. Recent studies analysing different pathogenic features of mutant p53 in cancer-derived cell lines have demonstrated that restoring wild-type p53, rather than removing GOF mutations, reduces cancer cell growth. These findings suggest that therapeutic strategies for reactivating wild-type p53 function in cancer cells may bring a greater benefit than approaches halting mutant p53. This approach could involve the use of small molecules, gene therapy and other methods to re-establish wild-type p53 activity. This review describes the complexity of the biological activities of different p53 mutants and summarizes the current therapeutic approaches to restore p53 function.
Key words: dominant negative effect, gain-of-function, loss-of-function, missense mutations, therapy, TP53, tumour suppressor
1. Introduction
The p53 protein was first described in 1979 by several research groups as a cellular factor forming a complex with the simian virus 40 large tumour antigen (1,2). Subsequent studies demonstrated excessive accumulation of p53 both in cells expressing viral tumour antigens and in cancer cells negative for viral infections, whereas p53 levels were low in normal, uninfected cells (3,4). In the early 1980s, p53 was recognised as an oncoprotein whose upregulation by tumour viruses or other mechanisms could contribute to cellular transformation (5,6). Indeed, the TP53 cDNA cloned from various cancer cell lines was shown to immortalise primary cells, induce multilayer cell growth and promote tumourigenicity in animal models, thereby experimentally substantiating the oncogenic role of p53 in tumour development (7,8). In the late 1980s, DNA sequencing of the TP53 gene isolated from tumour cells revealed frequent missense mutations conferring oncogenic features to the mutant p53 proteins (9,10). Conversely, the expression of wild-type p53 in transformed cells was shown to suppress the transformed phenotype without inducing damaging effects in non-transformed cells (11,12). The critical role of p53 as a tumour suppressor was further demonstrated in patients with Li-Fraumeni syndrome, associated with monoallelic germline TP53 mutations and characterized by a high predisposition to various cancer types, including breast cancer, sarcomas, brain tumours, leukaemia and adrenal gland cancers (13,14). The observation that tumours developing in patients with Li-Fraumeni syndrome have lost the wild-type TP53 allele definitively established p53 as a tumour suppressor factor (15).
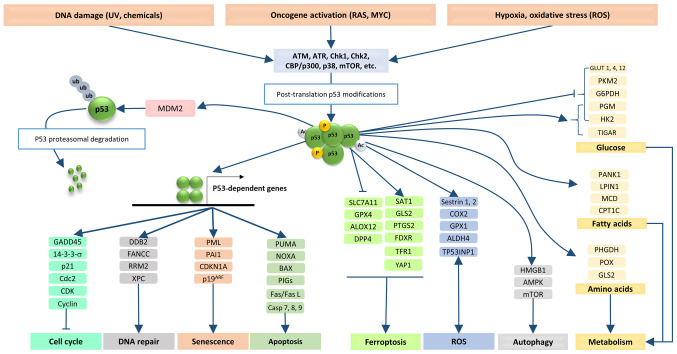
The scientific evidence accumulated over the past 40 years since the discovery of p53 has shown that this oncosuppressor is involved in regulating a broad range of cellular processes (16,17). These include cell cycle control, activation of DNA repair mechanisms in response to genetic damages, programmed cell death in response to severe cellular stress, induction of cell senescence and regulation of metabolic pathways (Fig. 1) (18,19). Therefore, mutations within the TP53 gene and expression of mutant p53 proteins can be considered to be involved in virtually all hallmarks of cancer (20).
Figure 1.
Under physiological conditions, cell stress promotes the activation and stabilization of p53 by post-translational modifications (phosphorylation, acetylation, etc.). The stabilized p53 forms tetramers, which bind the p53-dependent promoters and activate the expression of genes involved in different biological functions, such as cell cycle control, DNA repair, senescence and apoptosis. The p53 protein directly interacts with numerous other proteins and regulates cellular pathways such as ferroptosis, ROS, autophagy and metabolism. The level of p53 protein is regulated by a p53-MDM2/MDMX feedback loop via proteasomal degradation of p53. 14-3-3-s, 14-3-3 protein sigma; ALDH4, aldehyde dehydrogenase 9 family member A1; ALOX12, arachidonate 12-lipoxygenase, 12S type; AMPK, protein kinase AMP-activated catalytic subunit alpha 2; ATM, ataxia telangiectasia mutant; ATR, ataxia telangiectasia related; BAX, BCL2 associated X, apoptosis regulator; Casp 7, caspase 7; CBP/p300, CREB-binding protein/p300; Cdc2, cyclin-dependent kinase 1; CDK, cyclin-dependent kinase; CDKN1A, CDK inhibitor 1; CHK1, checkpoint kinase 1; COX2, prostaglandin-endoperoxide synthase 2; CPT1C, carnitine palmitoyltransferase 1C; DDB2, damage specific DNA binding protein 2; DPP4, dipeptidyl peptidase 4; FANCC, Fanconi anemia complementation group C; Fas/Fas L, Fas cell surface death receptor/Fas cell surface death receptor ligand; FDXR, ferredoxin reductase; G6PDH, glucose-6-phosphate dehydrogenase; GADD45, growth arrest and DNA damage-inducible 45; GLS2, glutaminase 2; GLUT 1, solute carrier family 2 member 1; GPX1, glutathione peroxidase 1; HK2, hexokinase 2; HMGB1, high mobility group box 1; LPIN1, lipin 1; MCD, malonyl-CoA decarboxylase; mTOR, mechanistic target of rapamycin kinase; NOXA, NADPH oxidase activator; P19ARF, CDK inhibitor 2A; p38, p38 kinase; PAI1, serpin family E member 1; PANK1, pantothenate kinase 1; PGM, phosphoglucomutase 1; PHGDH, phosphoglycerate dehydrogenase; PIGs, phosphatidylinositol glycan anchor biosynthesis class S; PKM2, pyruvate kinase M1/2; PML, promyelocytic leukemia protein; POX, proline dehydrogenase 1; PTGS2, prostaglandin-endoperoxide synthase 2; PUMA, BCL2 binding component 3; RRM2, ribonucleotide reductase regulatory subunit M2; ROS, reactive oxygen species; SAT1, spermidine/spermine N1-acetyltransferase 1; SLC7A11, solute carrier family 7 member 11; TFR1, transferrin receptor; TIGAR, TP53 induced glycolysis regulatory phosphatase; TP53INP1, tumor protein p53 inducible nuclear protein 1; XPC, xeroderma pigmentosum, complementation group C; YAP1, Yes1-associated transcriptional regulator.
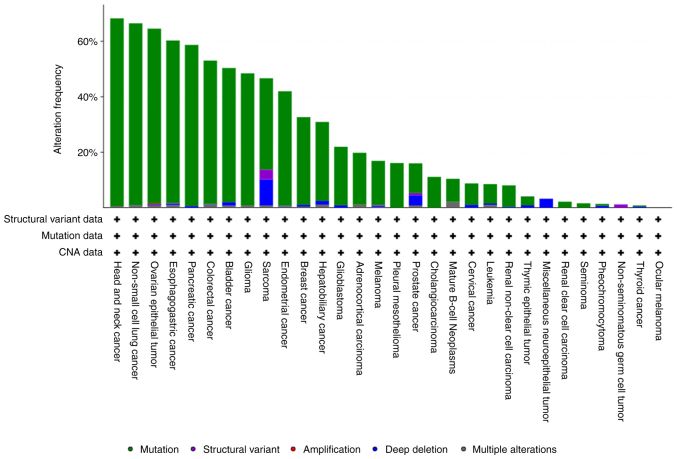
TP53 is the most commonly mutated gene in human cancers, with mutation frequencies exceeding 50% in at least 20 tumour types, including colorectal, ovarian and oesophageal carcinoma and lung cancer (Fig. 2). Certain nucleotide changes in the TP53 gene have been recognized as molecular signatures of carcinogen exposure in tumours developing in specific organs, such as aflatoxin B1 and G to T transversion at codon 249 in hepatocellular carcinoma, tobacco smoking and G to T transversion at specific G:C base pairs in lung cancer, and ultraviolet light irradiation leading to CC to TT tandem mutations in skin cancers (21,22). In addition, a significant proportion of TP53 mutations consist of G:C to A:T transitions at CpG sites, possibly originating from spontaneous deamination of DNA bases (23). However, numerous aspects regarding the cause of TP53 mutations, their diverse activities in cancer cells and therapeutic opportunities in tumours harbouring p53 mutations remain mostly unknown.
Figure 2.
TP53 gene mutations in the curated set of non-redundant studies including TCGA and non-TCGA datasets (n=10,953 patients from 32 studies) that do not include overlapping samples (www.cbioportal.org/). TCGA, The Cancer Genome Atlas; CNA, copy number alterations.
2. Mutations in TP53 gene
The human TP53 gene is ~20 kilobases long, contains 11 exons and encodes a zinc-coordinated protein composed of 393 amino acids (24). In normal cells, p53 protein is present at low levels, with an unstable conformation and short half-life due to continuous degradation mediated by its negative regulator mouse double minute 2 (MDM2) (25,26).
The p53 protein contains two transcriptional activation domains (TAD), namely TAD1 comprising amino acids (aa)21-28 and TAD2 comprising aa47-55, and a proline-rich sequence (aa62-94) at the N-terminal region, a DNA-binding domain (DBD, aa94-292) and a tetramerisation domain (TET, aa318-355) in the central region, as well as a nuclear localization signal and sites of post-translational modifications at the C-terminal domain (RD, aa363-393) (Fig. 3) (27-29). The p53 TAD1 induces the transcriptional activation of genes involved in cell cycle arrest, such as cyclin dependent kinase inhibitor 1A CDKN1A), and regulates apoptosis by activating the BCL2 associated X, apoptosis regulator (BAX), BCL2 binding component 3 (PUMA), NADPH oxidase activator (NOXA) and BCL2 associated agonist of cell death genes. Both TAD1 and TAD2 participate in tumour suppression and binding to MDM2 (Fig. 1) (30,31). The p53 DBD domain interacts with specific p53-responsive elements consisting of two copies of 5′-RRRCWWGYYY-3′ DNA sequences that are located in the regulatory regions (enhancers and promoters) of genes regulated by the p53 protein (32). The TET domain, involved in p53 tetramerisation, allows the appropriate conformation of p53 and interaction with target DNA sequences (33). The RD region regulates the binding of DBD to p53-responsive elements in the p53-regulated genes, depending on post-translational modifications such as acetylation, phosphorylation and sumoylation (34-40). The p53 C-terminal domain also contains nuclear export and localisation signals, which are important for its vehiculation to the nucleus as well as for transferring p53 into the cytoplasm for degradation (41,42).
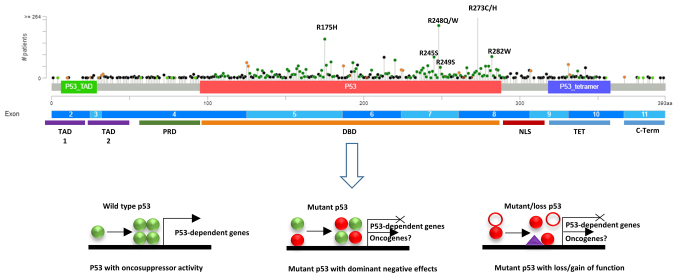
Figure 3.
Lollipop plot produced by the MutationMapper tool of cBioPortal shows the frequency and position of TP53 mutations in 10953 tumours from 32 studies. Domain organization of p53 is also described, followed by the C-terminus containing the nuclear export signal. TAD, transactivation domain; PRD, proline rich domain; DBD, DNA binding domain; NLS, nuclear localization signal; NLS, nuclear localization signal; TET, tetramerisation domain.
Mutations in the TP53 gene have been shown to confer to cells new biological features, such as increased proliferation, deregulated metabolism, metastatic potential and an altered tumour microenvironment, as well as resistance to chemotherapy and radiotherapy (43). Importantly, >90% of tumours carrying a mutant TP53 gene show loss of the second allele by mutation, chromosomal deletion or loss of heterozygosity, strongly supporting the tumour suppressor model (44).
The main genetic alterations of the TP53 gene include truncating and splice sites mutations, in-frame insertions/deletions (indels), frameshift indels and missense mutations arising from diverse mechanisms, playing specific roles in tumour development (45,46). Missense mutations, resulting in single amino acid changes, represent the vast majority of mutations occurring in >190 different codons of the TP53 gene (>80%), with the highest frequency in the DNA-binding domain (Fig. 2). Post-translational modifications of mutant p53 proteins, such as phosphorylation and acetylation, contribute to the stabilization of mutant proteins, leading to their accumulation in cancer cells (44,47). For instance, phosphorylation on Ser15 and Ser37 in mutant p53 has been shown to stabilise the protein and to enhance the oncogenic activity in ovarian cancer (48).
Different missense mutations in p53 may confer new biological activities to the mutant oncosuppressor, which can be grouped into three possible mechanisms: i) Loss of function (LOF) in transcriptional regulation of p53-dependent genes; ii) dominant negative effect (DNE) on the activity of wild-type p53 via the formation of mixed heterotetramers; and iii) gain of function (GOF) in terms of oncogenic activity (Fig. 3) (49-52). While LOF and DNE of mutant p53 have been proven crucial for cell proliferation and malignant transformation, the processes activated by mutant p53 characterised by GOF remain to be fully elucidated (52-56).
Approximately 30% of missense mutations in the TP53 gene occur at six hot spots and produce eight diverse p53 mutants, such as R175H, G245S, R248Q, R248W, R249S, R273C, R273H and R282W (57-59). These have been reported to possess GOF activity, although the molecular mechanisms behind their novel oncogenic functions have not been fully elucidated. The hot spot mutations determine amino acid changes in p53 that have been classified as contact mutants, which occur in the DNA-binding domain (R248Q, R248W, R273H and R273C), and conformational mutants, which cause abnormal protein folding (R175H, G245S, R249S and R282H) (60,61). Contact mutants directly affect the ability of mutant p53 to control the transcription of target genes, while conformational mutants cause the loss of zinc coordination and DNA-binding activity (62).
Mutant p53 has been shown to bind nuclear transcription factors, such as nuclear transcription factor Y (NF-Y), tumour protein p73, nuclear factor erythroid 2-related factor 2 and protein C-ets-1, to activate the transcription of their target genes and promote malignant transformation by inducing overexpression of cell cycle genes (63,64). Complexes formed by mutant p53 and the transcriptional co-regulator tyrosine-protein kinase Yes-associated protein and the nuclear transcription factor NF-Y were shown to promote the aberrant expression of cell cycle-related genes, such as cyclin A, cyclin B and cyclin-dependent kinase 1 (65). In addition, mutant p53 has been shown to antagonise the tumour suppression activity mediated by p63/p73 via the Notch1 signalling pathway in colorectal and pancreatic cancers (66). The p53 mutations causing nuclear delocalization are also involved in regulating oncogenic activity. Indeed, in colon cancer cells, the p53 P151H and R282W mutants located in the nucleus were shown to hinder autophagy, while p53 E258K, R273H and R273L mutants located in the cytoplasm were unable to inhibit autophagy (67).
A recent study by Wang et al (68) addressed the importance of mutant p53 GOF in neoplastic transformation by inactivating 12 TP53 mutations in a panel of 15 human cancer cell lines derived from breast cancer, colorectal cancer, lymphoma, hepatocellular carcinoma, leukaemia, osteosarcoma and lung cancer. They found that removing mutant TP53 using an inducible clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) platform neither reduced in vitro cell survival and proliferation, nor did it affect the killing of cancer cells in response to drug treatment. Furthermore, removing mutant p53 did not impact mitochondrial activity and intracellular reactive oxygen species (ROS) in the tested cell lines. In addition, knocking out the mutant TP53 did not affect the local growth of tumours derived from cancer cell lines or the number of lung metastasis in NOD scid gamma mice (68). Conversely, changing mutant TP53 to wild-type TP53 induced a reduction in tumour cell proliferation in vitro, suggesting that drugs capable of reshaping the structure of mutant p53 to that of wild-type p53 could be more effective as anticancer therapies.
The clinical outcome of p53 mutations has been extensively studied, yielding sometimes divergent results (53,69). Early studies in patients with Li-Fraumeni syndrome showed that the germline mutation p53 R248Q was associated with the development of cancer at a younger age compared with those with non-mutated TP53 (70). Conversely, somatic mutations in p53, commonly associated with a wide range of cancers, are more frequent in older populations (71). Overall, the majority of studies have demonstrated an association between TP53 mutations and more aggressive tumours, resistance to treatments and poor overall outcomes in several cancer types (17,72).
The chemoresistance of cancer cells to various drugs is strongly dependent on TP53 mutational status in numerous cancer types (50). Several studies have shown that mutant p53 causes an increased expression of the multidrug resistance 1 gene, an important regulator of the membrane pump, which induces chemoresistance by increasing the efflux of drugs out of cells (73). Conversely, depletion of mutant p53 reduces cell resistance to cisplatin, adriamycin and etoposide in multiple cancer cell lines (74). Mutations in the TP53 gene identified in colorectal cancer have been associated with resistance to insulin-like growth factor-1 receptor inhibitors (75). In osteosarcoma, the p53 R273H mutant is associated with reduced expression of procaspase-3 and failure of methotrexate and doxorubicin to induce apoptosis (76). In colon cancer, mutant p53 that fails to activate the PUMA promoter causes evasion from apoptosis and reduces the sensitivity of tumour cells to 5-fluorouracil (77). Considering all these findings, it is highly plausible that mutant p53 plays a critical role in the chemoresistance of several tumour types.
The response to radiation therapy is also influenced by the p53 status. A study of 60 different cancer cell lines carrying mutant p53 revealed an inability to induce key regulatory genes such as CDKN1A, MDM2 and growth arrest and DNA damage inducible alpha (GADD45), leading to a failure in G1 phase arrest after gamma-irradiation (78). This lack of response resulted in increased radioresistance. Similarly, ovarian cancer, head and neck cancer, hepatocellular carcinoma, cervical cancer and endometrial cancer cells carrying mutant p53 are more resistant to radiation therapy compared to cells carrying wild-type p53 (79). Therefore, additional research is necessary to fully characterise the relationship between mutant p53 and response to radiotherapy, which is crucial for improving patient treatment.
3. Therapeutic strategies to target mutant p53
The primary effect of p53 mutations in cancer is the loss of tumour suppressor function, making therapeutic reactivation of p53 a priority. Several therapeutic approaches include refining earlier strategies with improved understanding and delivery methods or utilising novel drug design techniques. New strategies encompass gene therapy to restore normal p53 function, inhibition of MDM2-p53 interaction, p53-based vaccines and the use of small molecules capable of reinstating a wild-type-like status of mutant p53. Several excellent reviews have reported state-of-the-art therapeutic strategies to enhance wild-type p53 activity as well as p53-based immunotherapy (80,81). The present review reassesses the progress in promising therapeutic strategies targeting mutant p53 for degradation or reinstating its wild-type-like conformation to restore its oncosuppressor activities.
Degradation of mutant p53 has emerged as a promising antitumour strategy. Various compounds, including gambogic acid (GA), capsaicin, MCB-613 and NSC59984, have demonstrated the ability to effectively degrade mutant p53 (82). GA was reported to reduce the expression of MDM2, thereby increasing the levels of wild-type p53 and inhibiting tumour growth (83). Later studies revealed that GA also reduces mutant p53 levels by hindering the formation of heat shock protein (Hsp)90/mutant p53 complexes, leading to the ubiquitin/proteasome-mediated degradation of mutant p53 (84). However, GA was observed to drive other mechanisms critical for malignant cell death, such as the activation of the intrinsic apoptotic pathway in TP53-null prostate cancer cells (85). Thus, GA inhibits tumour growth by activating several cell growth inhibitory pathways, with mutant p53 degradation being one possible contributing mechanism.
Capsaicin, the main bioactive component of chili peppers, has been shown to induce autophagy and mutant p53 protein degradation in cells carrying mutant p53 and in H1299 (p53 null) overexpressing p53 R175H and p53 R273H mutant proteins (86). The small molecule MCB-613 has been demonstrated to cause fast and selective degradation of p53 R175H protein in ovarian cancer cells by inhibiting ubiquitin carboxyl-terminal hydrolase 15, enabling selective depletion of oncogenic p53 R175H levels (87). In addition, the small molecule NSC59984 has been shown to specifically restore the p53 pathway through p73 and deplete mutant p53 proteins in colorectal cancer cells (88).
Several drugs effective in degrading mutant p53, such as ganetespib, statins and suberoylanilide hydroxamic acid (SAHA), are currently under evaluation in clinical trials. Ganetespib is a potent Hsp90 inhibitor that has shown remarkable effectiveness in degrading mutant p53 and killing cancer cells. It has exhibited strong cytotoxic effects across various haematological and solid tumours and has been successful in inhibiting tumour growth and extending survival in mouse models with specific mutant p53 expressions (89). Combining ganetespib with chemotherapy agents such as cyclophosphamide has been shown to further enhance its tumour-suppressing effects (90).
It has been shown that statins, which target the rate-limiting enzyme in cholesterol biosynthesis, inhibit the growth of cancer cells expressing mutant TP53 and increase their sensitivity to chemotherapeutics (91,92). Statins induce degradation of certain forms of mutant p53 through a mechanism involving the inhibition of mutant p53 interaction with DnaJ homolog subfamily A member 1, leading to E3 ubiquitin-protein ligase CHIP-mediated degradation while sparing wild-type p53 and other mutants (93). However, there are conflicting results regarding the ability of statins to selectively kill tumour cells depending on the TP53 mutation status (94). Hence, the mechanisms by which statins kill tumour cells require further investigation.
Similarly to statins, SAHA targets mutant p53 by disrupting its association with Hsp90 via histone deacetylase 6 (HDAC6) inhibition, resulting in the reactivation of degradation pathways and exhibiting selective toxicity towards cancer cells harbouring mutant p53 (95). These findings underscore the potential of mutant p53 degradation as an effective therapeutic approach against cancer. However, clinical trials are necessary to fully establish the safety and efficacy of these treatments for widespread clinical use.
Restoration of wild-type activity by using small molecules capable of restoring a wild-type-like structure and the ability to promote the transcription of p53-dependent genes or to inhibit the oncogenic function of GOF mutant p53 is gaining attention (96,97). Several compounds have been developed to reinstate the wild-type-like activity of mutant p53 (Table I). These molecules can target mutant p53 and activate the transcription of p53-dependent genes by restoring the structural conformation of wild-type p53. Such compounds exert antitumour activity, inhibiting cell proliferation and tumour growth (98).
Table I.
Therapeutic compounds changing mutant p53 conformation to wild-type-like structure that are under evaluation in clinical trials.
| Agent | Target | Class | Mechanism | Phases of clinical trials | Clinical trial nos. |
|---|---|---|---|---|---|
| APR-246 | p53 R175H p53 R273H |
Small molecule | Wild-type-like conformation of p53 by binding to thiol groups | I-III | NCT03072043, NCT03588078, NCT03745716, NCT04383938, NCT04419389, NCT03931291, NCT04214860 |
| PEITC | R175H P223L |
Phytochemical | Wild-type-like conformation of p53, oxidative stress | I-II | NCT01790204, NCT03034603 |
| PC14586 | Y220C | Small molecule | Selectively restores wild-type p53 conformation | I | NCT04585750 |
| ATO | R175H V272M R282W E285K Y234C |
Small molecule | Restores wild-type p53 conformation | I-II | NCT03855371, NCT04489706, NCT04695223, NCT04869475 |
| Sodium stibogluconate | 65 mutants | Pentavalent antimony compound | Restores wild-type p53 conformation | II | NCT04906031 |
| COTI2 | R175H R273H R273C R282W |
Zn2+ chelator | Inhibition of mutant p53 misfolding | I | NCT02433626 |
Among these, p53 reactivation with induction of massive apoptosis 1 (PRIMA-1) is a low-molecular-weight compound that was identified in 2002 through chemical screening and was observed to restore the wild-type p53 conformation to several p53 mutants (99). PRIMA-1 and its methylated form, namely APR-246, were found to delay tumour growth and increase apoptosis in tumour cells with mutant TP53 but not in those with wild-type TP53 via reactivation of p53-dependent target genes and induction of the pro-apoptotic protein NOXA (100-102). APR-246, alone or in combination with other drugs, is currently being tested in phase I-III clinical trials, including TP53-mutant myeloid malignancies, high-grade serous ovarian cancer, oesophageal cancer and melanoma (103-105). Two phase I/II trials showed significant effects when APR-246 was combined with azacitidine in patients with myelodysplastic syndrome (MDS) or acute myeloid leukaemia (AML) carrying p53 mutations. Azacitidine is the first Food and Drug Administration (FDA)-approved drug for treating MDS and the combination with APR-246 demonstrated substantial therapeutic potential in these patients (104,105). Despite their initial specificity for mutant TP53, both PRIMA-1 and APR-246 could also induce autophagy in certain cancer cells independently of their TP53 status (106). This highlights the complexity of the molecular mechanisms implicated in APR-246 activity, which remain to be fully elucidated.
Phenethyl isothiocyanate (PEITC), a natural compound present at high levels in cruciferous vegetables, has been shown to possess chemotherapeutic activity by targeting mutant TP53. Unlike APR-246, which can restore wild-type p53 function in several mutant p53 proteins, PEITC specifically targets the mutant R175 p53, restoring wild-type p53 conformation and transactivation functions (107). In addition, PEITC exerts significant anti-cancer activity in cells with structural TP53 mutants, such as P223L, but not in those with contact TP53 mutants (108). Importantly, PEITC is able to reduce the quantity of mutant p53 protein post-transcriptionally, with minimal impact on wild-type p53 levels. This dual action suggests that PEITC reactivates wild-type p53 functions while decreasing mutant p53 protein levels (107). However, further research is required to validate the efficacy of PEITC and clarify the critical mechanisms for killing cancer cells.
The small molecule PC14586 specifically targets the Y220C mutant. This orally bioavailable drug has achieved clinical responses in patients with various tumour types, including ovarian, breast, prostate, lung and endometrial cancers. In patients with solid tumours carrying the p53 Y220C mutation and wild-type KRAS, the overall response rate was 38% (6 of 16 evaluable patients) (109). Due to these promising results, the FDA granted fast-track designation of PC14586 for treating locally advanced or metastatic solid tumours with the p53 Y220C mutation. However, considering that the Y220C mutation is relatively rare, applying similar structure-based approaches to other p53 mutants may prove significantly challenging for several tumour types.
Arsenic trioxide (ATO) is a small molecule that can restore wild-type p53 function in tumour cells expressing structural p53 mutants by binding to their DNA-binding domain. This binding induces transcriptional activities characteristic of wild-type p53, suppressing tumour growth both in vitro and in vivo (110). Although ATO shows promise as a therapy for cancers expressing mutant p53, further studies are required to validate its specificity and efficiency.
Sodium stibogluconate (SSG), originally an antiparasitic drug, has recently been identified as a potent reactivator of temperature-sensitive p53 mutants in a high-throughput screen. SSG works by releasing antimony ions that bind to a specific pocket in the mutant p53 protein, increasing its stability and restoring its tumour-suppressing functions (111). It has shown effectiveness in rescuing 65 different p53 mutations, though none were hotspot mutations. Given its existing approval as an antiparasitic, repurposing SSG for cancer treatment is a cost-effective strategy. A phase II clinical trial is underway for patients with specific p53 mutations in MDS or AML cells.
A thiosemicarbazone, namely COTI-2, shows preferential activity against p53-mutated cancer cells but also affects p53-wild-type cells to a certain extent. In head and neck squamous cell carcinoma, COTI-2 demonstrated both p53-dependent and p53-independent effects, inducing cell death by causing DNA damage, replication stress and activating p53 target genes through the p53 family member p63 (112). COTI-2 has progressed to phase I clinical trials, while other related thiosemicarbazones targeting p53 R175H have not yet entered clinical trials despite promising preclinical results.
Numerous other compounds targeting mutant p53 are showing promising results as anticancer agents. However, they have yet to be evaluated in clinical trials. These include the small molecule reactivation of TP53 and induction of tumour cell apoptosis (RITA), which was identified by chemical screening and initially reported to inhibit the p53-MDM2 interaction, thereby activating the anti-tumour effects of wild-type p53 (113). A subsequent study revealed that RITA could also suppress proliferation and induce apoptosis in tumour cells expressing mutant TP53 by restoring wild-type p53 transcriptional activities in several hotspot p53 mutants (114). This was demonstrated by the expression of wild-type p53 target genes such as GADD45, BCL2 binding component 3, BAX and CDKN1A in these cells. Furthermore, RITA was also shown to induce apoptosis in tumour cells with wild-type p53 and also in TP53-deficient cancer cells, suggesting that its anticancer effects may not be specifically dependent on mutant p53 (115).
Similarly to the above-described small molecules, CP-31398 has been shown to restore functional activity to mutant p53 proteins, enabling them to exert wild-type p53 transcriptional activity (116). This synthetic compound induces cell death in a p53-dependent manner and is effective only in tumour cells expressing wild-type or mutant p53 but not in p53-deficient cells (117). CP-31398 increases levels of wild-type p53 by preventing its ubiquitination and proteasomal degradation, thus promoting cell cycle arrest and apoptosis (118). In addition, CP-31398 has been shown to restore wild-type p53 function in various p53 mutants, delaying growth in hepatocellular and colorectal cancer cells expressing different p53 mutations both in vitro and in vivo (119,120). The inhibitory effects on cell growth are consistent across different cancer types carrying diverse p53 mutations. Furthermore, CP-31398 has been shown to increase ROS production, triggering apoptosis in multiple myeloma cells regardless of p53 status (121). Further research is needed to fully understand the mechanisms behind the restoration of p53 activity.
Several compounds have been identified that target uncommon p53 mutants or distinct groups of mutants. Among these, PK7088 is a small molecule that specifically binds the unique surface crevice created by the amino acid change Y220C in mutant p53, converting the mutant structure to the wild-type conformation (122). PK7088 restores p53-dependent cell cycle arrest and apoptosis by activating p21 and NOXA expression, respectively.
Other mutants of p53 targeted by specific compounds include the amino acid change R175H, which causes structural modification of the p53 DNA-binding domain (123). The p53 R175H is specifically targeted by the ZMC1 (NSC319726) molecule, which is a metal ion chelator promoting p53-dependent apoptosis in vitro and inducing tumour regression in vivo (124). NSC319726 selectively kills cells carrying the p53 R175H mutation without impacting non-transformed cells or those with wild-type p53. Related compounds like ZMC2 and ZMC3 from the thiosemicarbazone family also induced a wild-type-like conformation of p53 R175H in vitro (125). Mutant p53 can interact with various oncogenic factors, forming complexes that promote cancer cell growth. Compounds that disrupt such interactions can inhibit cancer cell proliferation and induce apoptosis. For instance, molecules that inhibit the interaction between mutant p53 and its chaperone HSP90 cause enhanced ubiquitination and proteasomal degradation of mutant p53 while promoting apoptotic pathways and the death of cancer cells. The HDAC inhibitor SAHA has been reported to synergise with the HSP90 inhibitor 17AAG to degrade mutant TP53, inducing apoptosis and decreasing tumour growth in xenografts (126). Another HDAC inhibitor, FK228, also inhibits growth and induces apoptosis in tumour cells. Unlike SAHA, which specifically kills tumours carrying mutant TP53, FK228 induces cell death in both mutant and wild-type TP53-expressing tumour cells (127-129).
Other attractive therapeutic approaches include gene therapy to repair or replace the mutant TP53 gene, for example using the CRISPR/Cas9 technique, as well as RNA interference to silence critical genes, including the mutant TP53 gene that is required for tumour cell proliferation (130,131). Immunotherapeutic strategies, such as vaccines targeting mutant p53 neoantigens, the use of bispecific antibodies and adoptive T-cell therapies using engineered T-cells that recognise mutant p53-expressing cells also show promise for the treatment of p53-mutant tumours (58,80,132,133).
Small molecules that interfere with the binding of p53 to its negative regulator MDM2 may be effective in increasing the levels of p53 in tumours carrying the wild-type TP53 gene (80,134). Despite the promising results of certain clinical trials of MDM2 inhibitors and mutant p53-restoring compounds, none of these agents have been approved by the FDA.
In conclusion, targeting mutant p53 in cancer therapy is an advancing field with substantial therapeutic potential. Understanding the diverse roles of mutant p53 in tumour progression and utilising emerging strategies to counteract its oncogenic functions are essential for developing effective cancer treatments.
4. Conclusions
Mutations in the TP53 gene or functional inactivation of the p53 protein play a crucial role in cell transformation and the development of cancers. Missense nucleotide changes in the TP53 gene are the most frequent mutations observed in human cancers, making mutant p53 a key target for cancer research and development of therapies. Most of these genetic alterations are specific to cancer histotypes (135). Targeting mutant p53 to restore the activity of wild-type p53, by using specific drugs and immunotherapies, may be effective in all these cancers presenting a high frequency of mutations in the TP53 gene.
The screening of chemical libraries has allowed the identification of small molecules capable of specifically binding to mutant p53, guiding its folding to a wild-type-like p53 and restoring tumour suppressor activity. Such therapeutic strategies are likely to have a significant clinical impact in the treatment of numerous types of human cancers characterised by these mutations.
Nonetheless, important considerations remain to be resolved. These include questions related to the functional inactivation of the p53 protein mediated by viral oncoproteins that can bind to and degrade wild-type p53 in tumours caused by infectious agents (4). The BamHI Z fragment leftward open reading frame 1 encoded by Epstein Barr virus, the Early 6 protein of high-risk human papillomavirus and the non-structural protein 5 of hepatitis C virus have been shown to directly bind to and degrade p53 (136-138). Other viruses have been shown to inhibit the activity of wild-type p53 through the modulation of p300/CREB-binding protein nuclear factors, causing a decrease in its levels in infected cells. No drugs have been developed to disrupt the complex formed by viral oncoproteins and p53.
In conclusion, therapies targeting mutant p53 offer several potential advantages, such as specificity for cancer cells, thereby minimizing damage to normal cells, wide applicability given the high prevalence of p53 mutations in numerous tumour types, and the potential to reverse cancer progression, as these drugs can reinstate the normal tumour-suppressive function of p53. However, only a small number of these drugs have reached late-stage clinical trials, commonly due to off-target effects and nonspecific toxicity, which hinder the evaluation of their efficacy in clinical trials. Therefore, further efforts are needed to achieve the goal of targeting all forms of p53 dysfunction with specific molecules or drugs to address them effectively.
Acknowledgements
Not applicable.
Funding Statement
This work was partly funded by grants from the Italian Ministry of Health Ricerca Corrente L1/10; Italian Ministry of Health Progetto Finalizzato (grant no. RF-2018-12366163) and the Italian Association for Cancer Research (grant no. AIRC-IG-2021-ID-26111).
Availability of data and materials
Not applicable.
Author's contributions
Conceptualization, MLT; data curation, MLT; writing, review and editing, MLT. The author has read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The author declares that she has no competing interests.
References
- 1.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 2.Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 3.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tornesello ML, Annunziata C, Tornesello AL, Buonaguro L, Buonaguro FM. Human Oncoviruses and p53 tumor suppressor pathway deregulation at the origin of human cancers. Cancers (Basel) 2018;10:213. doi: 10.3390/cancers10070213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotter V. p53, a transformation-related cellular-encoded protein, can be used as a biochemical marker for the detection of primary mouse tumor cells. Proc Natl Acad Sci USA. 1983;80:2613–2617. doi: 10.1073/pnas.80.9.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oren M, Levine AJ. Molecular cloning of a cDNA specific for the murine p53 cellular tumor antigen. Proc Natl Acad Sci USA. 1983;80:56–59. doi: 10.1073/pnas.80.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins JR, Rudge K, Currie GA. Cellular immortalization by a cDNA clone encoding the transformation-associated phosphoprotein p53. Nature. 1984;312:651–654. doi: 10.1038/312651a0. [DOI] [PubMed] [Google Scholar]
- 8.Eliyahu D, Michalovitz D, Oren M. Overproduction of p53 antigen makes established cells highly tumorigenic. Nature. 1985;316:158–160. doi: 10.1038/316158a0. [DOI] [PubMed] [Google Scholar]
- 9.Finlay CA, Hinds PW, Tan TH, Eliyahu D, Oren M, Levine AJ. Activating mutations for transformation by p53 produce a gene product that forms an hsc70-p53 complex with an altered half-life. Mol Cell Biol. 1988;8:531–539. doi: 10.1128/mcb.8.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 11.Eliyahu D, Michalovitz D, Eliyahu S, Pinhasi-Kimhi O, Oren M. Wild-type p53 can inhibit oncogene-mediated focus formation. Proc Natl Acad Sci USA. 1989;86:8763–8767. doi: 10.1073/pnas.86.22.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 13.Schneider K, Zelley K, Nichols KE, Garber J. Li-Fraumeni Syndrome. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, Amemiya A, editors. GeneReviews(®) University of Washington; Seattle, WA: 1993. [PubMed] [Google Scholar]
- 14.Fortuno C, Feng BJ, Carroll C, Innella G, Kohlmann W, Lázaro C, Br unet J, Feliubadaló L, Iglesias S, Menéndez M, et al. Cancer risks associated with TP53 pathogenic variants: Maximum likelihood analysis of extended pedigrees for diagnosis of first cancers beyond the Li-Fraumeni Syndrome Spectrum. JCO Precis Oncol. 2024;8:e2300453. doi: 10.1200/PO.23.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2:475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20:471–480. doi: 10.1038/s41568-020-0262-1. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Su Z, Tavana O, Gu W. Understanding the complexity of p53 in a new era of tumor suppression. Cancer Cell. 2024;42:946–967. doi: 10.1016/j.ccell.2024.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Guo M, Wei H, Chen Y. Targeting p53 pathways: Mechanisms, structures, and advances in therapy. Signal Transduct Target Ther. 2023;8:92. doi: 10.1038/s41392-023-01347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Indeglia A, Murphy ME. Elucidating the chain of command: Our current understanding of critical target genes for p53-mediated tumor suppression. Crit Rev Biochem Mol Biol. 2024;59:128–138. doi: 10.1080/10409238.2024.2344465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: A tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004;(157):247–270. [PubMed] [Google Scholar]
- 22.Tornesello ML, Buonaguro L, Tatangelo F, Botti G, Izzo F, Buonaguro FM. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102:74–83. doi: 10.1016/j.ygeno.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Hainaut P, Pfeifer GP. Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb Perspect Med. 2016;6:a026179. doi: 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sammons MA, Nguyen TT, McDade SS, Fischer M. Tumor suppressor p53: From engaging DNA to target gene regulation. Nucleic Acids Res. 2020;48:8848–8869. doi: 10.1093/nar/gkaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 26.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 27.Raj N, Attardi LD. The Transactivation Domains of the p53 Protein. Cold Spring Harb Perspect Med. 2017;7:a026047. doi: 10.1101/cshperspect.a026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krois AS, Park S, Martinez-Yamout MA, Dyson HJ, Wright PE. Mapping Interactions of the Intrinsically Disordered C-Terminal Regions of Tetrameric p53 by Segmental Isotope Labeling and NMR. Biochemistry. 2022;61:2709–2719. doi: 10.1021/acs.biochem.2c00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruse JP, Gu W. SnapShot: p53 posttranslational modifications. Cell. 2008;133:930–930.e1. doi: 10.1016/j.cell.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 32.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 33.Halazonetis TD, Kandil AN. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993;12:5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/S0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 35.Hernández Borrero LJ, El-Deiry WS. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim Biophys Acta Rev Cancer. 2021;1876:188556. doi: 10.1016/j.bbcan.2021.188556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins LM, Durell SR, Mazur SJ, Appella E. p53 N-terminal phosphorylation: A defining layer of complex regulation. Carcinogenesis. 2012;33:1441–1449. doi: 10.1093/carcin/bgs145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 38.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stindt MH, Carter S, Vigneron AM, Ryan KM, Vousden KH. MDM2 promotes SUMO-2/3 modification of p53 to modulate transcriptional activity. Cell Cycle. 2011;10:3176–3188. doi: 10.4161/cc.10.18.17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West LE, Gozani O. Regulation of p53 function by lysine methylation. Epigenomics. 2011;3:361–369. doi: 10.2217/epi.11.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laptenko O, Shiff I, Freed-Pastor W, Zupnick A, Mattia M, Freulich E, Shamir I, Kadouri N, Kahan T, Manfredi J, et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell. 2015;57:1034–1046. doi: 10.1016/j.molcel.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirangi TR, Zaika A, Moll UM. Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J. 2002;16:420–422. doi: 10.1096/fj.01-0617fje. [DOI] [PubMed] [Google Scholar]
- 43.Zhu G, Pan C, Bei JX, Li B, Liang C, Xu Y, Fu X. Mutant p53 in cancer progression and targeted therapies. Front Oncol. 2020;10:595187. doi: 10.3389/fonc.2020.595187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, Li X, Babur O, Hsu TK, Lichtarge O, et al. Integrated Analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 2019;28:1370–1384 e5. doi: 10.1016/j.celrep.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Zhang T, Su W, Dou Z, Zhao D, Jin X, Lei H, Wang J, Xie X, Cheng B, et al. Mutant p53 in cancer: From molecular mechanism to therapeutic modulation. Cell Death Dis. 2022;13:974. doi: 10.1038/s41419-022-05408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shirole NH, Pal D, Kastenhuber ER, Senturk S, Boroda J, Pisterzi P, Miller M, Munoz G, Anderluh M, Ladanyi M, et al. TP53 exon-6 truncating mutations produce separation of function isoforms with pro-tumorigenic functions. Elife. 2016;5:e17929. doi: 10.7554/eLife.17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castrogiovanni C, Waterschoot B, De Backer O, Dumont P. Serine 392 phosphorylation modulates p53 mitochondrial translocation and transcription-independent apoptosis. Cell Death Differ. 2018;25:190–203. doi: 10.1038/cdd.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonego M, Schiappacassi M, Lovisa S, Dall'Acqua A, Bagnoli M, Lovat F, Libra M, D'Andrea S, Canzonieri V, Militello L, et al. Stathmin regulates mutant p53 stability and transcriptional activity in ovarian cancer. EMBO Mol Med. 2013;5:707–722. doi: 10.1002/emmm.201201504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Nakayama M, Hong CP, Oshima H, Oshima M. Gain-of-Function p53 mutation acts as a genetic switch for TGFβ signaling-induced epithelial-to-mesenchymal transition in intestinal tumors. Cancer Res. 2024;84:56–68. doi: 10.1158/0008-5472.CAN-23-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarado-Ortiz E, de la Cruz-López KG, Becerril-Rico J, Sarabia-Sánchez MA, Ortiz-Sánchez E, García-Carrancá A. Mutant p53 Gain-of-Function: Role in cancer development, progression, and therapeutic approaches. Front Cell Dev Biol. 2021;8:607670. doi: 10.3389/fcell.2020.607670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, Giacomelli AO, Wong W, Kim J, Chao S, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science. 2019;365:599–604. doi: 10.1126/science.aax3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aubrey BJ, Janic A, Chen Y, Chang C, Lieschke EC, Diepstraten ST, Kueh AJ, Bernardini JP, Dewson G, O'Reilly LA, et al. Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes Dev. 2018;32:1420–1429. doi: 10.1101/gad.314286.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Shchors K, Persson AI, Rostker F, Tihan T, Lyubynska N, Li N, Swigart LB, Berger MS, Hanahan D, Weiss WA, Evan GI. Using a preclinical mouse model of high-grade astrocytoma to optimize p53 restoration therapy. Proc Natl Acad Sci USA. 2013;110:E1480–E1489. doi: 10.1073/pnas.1219142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 57.Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018;25:154–160. doi: 10.1038/cdd.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang C, Lou G, Jin WL. The arsenal of TP53 mutants therapies: Neoantigens and bispecific antibodies. Signal Transduct Target Ther. 2021;6:219. doi: 10.1038/s41392-021-00635-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCann JJ, Vasilevskaya IA, McNair C, Gallagher P, Neupane NP, de Leeuw R, Shafi AA, Dylgjeri E, Mandigo AC, Schiewer MJ, Knudsen KE. Mutant p53 elicits context-dependent pro-tumorigenic phenotypes. Oncogene. 2022;41:444–458. doi: 10.1038/s41388-021-01903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiong S, Chachad D, Zhang Y, Gencel-Augusto J, Sirito M, Pant V, Yang P, Sun C, Chau G, Qi Y, et al. Differential Gain-of-Function Activity of Three p53 Hotspot mutants in vivo. Cancer Res. 2022;82:1926–1936. doi: 10.1158/0008-5472.CAN-21-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rasquinha JA, Bej A, Dutta S, Mukherjee S. Intrinsic differences in backbone dynamics between wild type and DNA-Contact Mutants of the p53 DNA binding domain revealed by nuclear magnetic resonance spectroscopy. Biochemistry. 2017;56:4962–4971. doi: 10.1021/acs.biochem.7b00514. [DOI] [PubMed] [Google Scholar]
- 62.Salari A, Thomay K, Lentes J, Ebersold J, Hagedorn M, Skawran B, Davenport C, Schambach A, Schlegelberger B, Göhring G. Effect of TP53 contact and conformational mutations on cell survival and erythropoiesis of human hematopoietic stem cells in a long term culture model. Oncotarget. 2018;9:29869–29876. doi: 10.18632/oncotarget.25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfister NT, Prives C. Transcriptional regulation by wild-type and cancer-related mutant forms of p53. Cold Spring Harb Perspect Med. 2017;7:a026054. doi: 10.1101/cshperspect.a026054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 65.Di Agostino S, Sorrentino G, Ingallina E, Valenti F, Ferraiuolo M, Bicciato S, Piazza S, Strano S, Del Sal G, Blandino G. YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep. 2016;17:188–201. doi: 10.15252/embr.201540488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Sun W, Kong X, Zhang Y, Yang HJ, Ren C, Jiang Y, Chen M, Chen X. Mutant p53 antagonizes p63/p73-mediated tumor suppression via Notch1. Proc Natl Acad Sci USA. 2019;116:24259–24267. doi: 10.1073/pnas.1913919116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, Soussi T, Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 68.Wang Z, Burigotto M, Ghetti S, Vaillant F, Tan T, Capaldo BD, Palmieri M, Hirokawa Y, Tai L, Simpson DS, et al. Loss-of-Function but not gain-of-function properties of mutant TP53 are critical for the proliferation, survival, and metastasis of a broad range of cancer cells. Cancer Discov. 2024;14:362–379. doi: 10.1158/2159-8290.CD-23-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roszkowska KA, Piecuch A, Sady M, Gajewski Z, Flis S. Gain of Function (GOF) Mutant p53 in cancer-current therapeutic approaches. Int J Mol Sci. 2022;23:13287. doi: 10.3390/ijms232113287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bougeard G, Sesboue R, Baert-Desurmont S, Martin C, Tinat J, Brugières L, Chompret A, de Paillerets BB, Stoppa-Lyonnet D, Bonaïti-Pellié C, et al. Molecular basis of the Li-Fraumeni syndrome: An update from the French LFS families. J Med Genet. 2008;45:535–538. doi: 10.1136/jmg.2008.057570. [DOI] [PubMed] [Google Scholar]
- 71.Levine AJ. Spontaneous and inherited TP53 genetic alterations. Oncogene. 2021;40:5975–5983. doi: 10.1038/s41388-021-01991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robles AI, Jen J, Harris CC. Clinical Outcomes of TP53 mutations in cancers. Cold Spring Harb Perspect Med. 2016;6:a026294. doi: 10.1101/cshperspect.a026294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sampath J, Sun D, Kidd VJ, Grenet J, Gandhi A, Shapiro LH, Wang Q, Zambetti GP, Schuetz JD. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J Biol Chem. 2001;276:39359–39367. doi: 10.1074/jbc.M103429200. [DOI] [PubMed] [Google Scholar]
- 74.Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–309. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- 75.Wang Q, Wei F, Lv G, Li C, Liu T, Hadjipanayis CG, Zhang G, Hao C, Bellail AC. The association of TP53 mutations with the resistance of colorectal carcinoma to the insulin-like growth factor-1 receptor inhibitor picropodophyllin. BMC Cancer. 2013;13:521. doi: 10.1186/1471-2407-13-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong RP, Tsang WP, Chau PY, Co NN, Tsang TY, Kwok TT. p53-R273H gains new function in induction of drug resistance through down-regulation of procaspase-3. Mol Cancer Ther. 2007;6:1054–1061. doi: 10.1158/1535-7163.MCT-06-0336. [DOI] [PubMed] [Google Scholar]
- 77.Huang Y, Liu N, Liu J, Liu Y, Zhang C, Long S, Luo G, Zhang L, Zhang Y. Mutant p53 drives cancer chemotherapy resistance due to loss of function on activating transcription of PUMA. Cell Cycle. 2019;18:3442–3455. doi: 10.1080/15384101.2019.1688951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O'Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, Scudiero DA, Monks A, Sausville EA. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–4300. [PubMed] [Google Scholar]
- 79.Kong X, Yu D, Wang Z, Li S. Relationship between p53 status and the bioeffect of ionizing radiation. Oncol Lett. 2021;22:661. doi: 10.3892/ol.2021.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hassin O, Oren M. Drugging p53 in cancer: One protein, many targets. Nat Rev Drug Discov. 2023;22:127–144. doi: 10.1038/s41573-022-00571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peuget S, Zhou X, Selivanova G. Translating p53-based therapies for cancer into the clinic. Nat Rev Cancer. 2024;24:192–215. doi: 10.1038/s41568-023-00658-3. [DOI] [PubMed] [Google Scholar]
- 82.Hu J, Cao J, Topatana W, Juengpanich S, Li S, Zhang B, Shen J, Cai L, Cai X, Chen M. Targeting mutant p53 for cancer therapy: Direct and indirect strategies. J Hematol Oncol. 2021;14:157. doi: 10.1186/s13045-021-01169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gu H, Wang X, Rao S, Wang J, Zhao J, Ren FL, Mu R, Yang Y, Qi Q, Liu W, et al. Gambogic acid mediates apoptosis as a p53 inducer through down-regulation of mdm2 in wild-type p53-expressing cancer cells. Mol Cancer Ther. 2008;7:3298–3305. doi: 10.1158/1535-7163.MCT-08-0212. [DOI] [PubMed] [Google Scholar]
- 84.Wang J, Zhao Q, Qi Q, Gu HY, Rong JJ, Mu R, Zou MJ, Tao L, You QD, Guo QL. Gambogic acid-induced degradation of mutant p53 is mediated by proteasome and related to CHIP. J Cell Biochem. 2011;112:509–519. doi: 10.1002/jcb.22941. [DOI] [PubMed] [Google Scholar]
- 85.Pan H, Lu LY, Wang XQ, Li BX, Kelly K, Lin HS. Gambogic acid induces cell apoptosis and inhibits MAPK Pathway in PTEN(-/-)/p53(-/-) prostate cancer cells in vitro and ex vivo. Chin J Integr Med. 2018;24:109–116. doi: 10.1007/s11655-017-2410-3. [DOI] [PubMed] [Google Scholar]
- 86.Garufi A, Pistritto G, Cirone M, D'Orazi G. Reactivation of mutant p53 by capsaicin, the major constituent of peppers. J Exp Clin Cancer Res. 2016;35:136. doi: 10.1186/s13046-016-0417-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Padmanabhan A, Candelaria N, Wong KK, Nikolai BC, Lonard DM, O'Malley BW, Richards JS. USP15-dependent lysosomal pathway controls p53-R175H turnover in ovarian cancer cells. Nat Commun. 2018;9:1270. doi: 10.1038/s41467-018-03599-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang S, Zhou L, Hong B, van den Heuvel AP, Prabhu VV, Warfel NA, Kline CL, Dicker DT, Kopelovich L, El-Deiry WS. Small-Molecule NSC59984 Restores p53 pathway signaling and antitumor effects against colorectal cancer via p73 activation and degradation of mutant p53. Cancer Res. 2015;75:3842–3852. doi: 10.1158/0008-5472.CAN-13-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jhaveri K, Modi S. Ganetespib: Research and clinical development. Onco Targets Ther. 2015;8:1849–1858. doi: 10.2147/OTT.S65804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alexandrova EM, Xu S, Moll UM. Ganetespib synergizes with cyclophosphamide to improve survival of mice with autochthonous tumors in a mutant p53-dependent manner. Cell Death Dis. 2017;8:e2683. doi: 10.1038/cddis.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong WW, Dimitroulakos J, Minden MD, Penn LZ. HMG-CoA reductase inhibitors and the malignant cell: the statin family of drugs as triggers of tumor-specific apoptosis. Leukemia. 2002;16:508–519. doi: 10.1038/sj.leu.2402476. [DOI] [PubMed] [Google Scholar]
- 92.Kornblau SM, Banker DE, Stirewalt D, Shen D, Lemker E, Verstovsek S, Estrov Z, Faderl S, Cortes J, Beran M, et al. Blockade of adaptive defensive changes in cholesterol uptake and synthesis in AML by the addition of pravastatin to idarubicin + high-dose Ara-C: A phase 1 study. Blood. 2007;109:2999–3006. doi: 10.1182/blood-2006-08-044446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parrales A, Ranjan A, Iyer SV, Padhye S, Weir SJ, Roy A, Iwakuma T. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat Cell Biol. 2016;18:1233–1243. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martirosyan A, Clendening JW, Goard CA, Penn LZ. Lovastatin induces apoptosis of ovarian cancer cells and synergizes with doxorubicin: Potential therapeutic relevance. BMC Cancer. 2010;10:103. doi: 10.1186/1471-2407-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18:1904–1913. doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Durairaj G, Demir O, Lim B, Baronio R, Tifrea D, Hall LV, DeForest JC, Lauinger L, Jebril Fallatah MM, Yu C, et al. Discovery of compounds that reactivate p53 mutants in vitro and in vivo. Cell Chem Biol. 2022;29:1381–1395 e13. doi: 10.1016/j.chembiol.2022.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Di Agostino S, Fontemaggi G, Strano S, Blandino G, D'Orazi G. Targeting mutant p53 in cancer: The latest insights. J Exp Clin Cancer Res. 2019;38:290. doi: 10.1186/s13046-019-1302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang Y, Jiao Z, Fu Y, Hou Y, Sun J, Hu F, Yu S, Gong K, Liu Y, Zhao G. An overview of the functions of p53 and drugs acting either on wild- or mutant-type p53. Eur J Med Chem. 2024;265:116121. doi: 10.1016/j.ejmech.2024.116121. [DOI] [PubMed] [Google Scholar]
- 99.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 100.Saha MN, Jiang H, Yang Y, Reece D, Chang H. PRIMA-1Met/APR-246 displays high antitumor activity in multiple myeloma by induction of p73 and Noxa. Mol Cancer Ther. 2013;12:2331–2341. doi: 10.1158/1535-7163.MCT-12-1166. [DOI] [PubMed] [Google Scholar]
- 101.Furukawa H, Makino T, Yamasaki M, Tanaka K, Miyazaki Y, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mori M, Doki Y. PRIMA-1 induces p53-mediated apoptosis by upregulating Noxa in esophageal squamous cell carcinoma with TP53 missense mutation. Cancer Sci. 2018;109:412–421. doi: 10.1111/cas.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zandi R, Selivanova G, Christensen CL, Gerds TA, Willumsen BM, Poulsen HS. PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin Cancer Res. 2011;17:2830–2841. doi: 10.1158/1078-0432.CCR-10-3168. [DOI] [PubMed] [Google Scholar]
- 103.Li XL, Zhou J, Chan ZL, Chooi JY, Chen ZR, Chng WJ. PRIMA-1met (APR-246) inhibits growth of colorectal cancer cells with different p53 status through distinct mechanisms. Oncotarget. 2015;6:36689–36699. doi: 10.18632/oncotarget.5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sallman DA, DeZern AE, Garcia-Manero G, Steensma DP, Roboz GJ, Sekeres MA, Cluzeau T, Sweet KL, McLemore A, McGraw KL, et al. Eprenetapopt (APR-246) and Azacitidine in TP53-Mutant myelodysplastic syndromes. J Clin Oncol. 2021;39:1584–1594. doi: 10.1200/JCO.20.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cluzeau T, Sebert M, Rahmé R, Cuzzubbo S, Lehmann-Che J, Madelaine I, Peterlin P, Bève B, Attalah H, Chermat F, et al. Eprenetapopt Plus Azacitidine in TP53-Mutated myelodysplastic syndromes and acute myeloid leukemia: A phase II study by the groupe francophone des myélodysplasies (GFM) J Clin Oncol. 2021;39:1575–1583. doi: 10.1200/JCO.20.02342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grellety T, Laroche-Clary A, Chaire V, Lagarde P, Chibon F, Neuville A, Italiano A. PRIMA-1(MET) induces death in soft-tissue sarcomas cell independent of p53. BMC Cancer. 2015;15:684. doi: 10.1186/s12885-015-1667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aggarwal M, Saxena R, Sinclair E, Fu Y, Jacobs A, Dyba M, Wang X, Cruz I, Berry D, Kallakury B, et al. Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth. Cell Death Differ. 2016;23:1615–1627. doi: 10.1038/cdd.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aggarwal M, Saxena R, Asif N, Sinclair E, Tan J, Cruz I, Berry D, Kallakury B, Pham Q, Wang TTY, Chung FL. p53 mutant-type in human prostate cancer cells determines the sensitivity to phenethyl isothiocyanate induced growth inhibition. J Exp Clin Cancer Res. 2019;38:307. doi: 10.1186/s13046-019-1267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dumbrava ECJ, Tolcher ML, Shapiro AW, Thompson G, El-Khoueiry JA, Vandross AB, Kummar AL, Parikh S, Munster AR, Daly PM, et al. First-in-human study of PC14586, a small molecule structural corrector of Y220C mutant p53, in patients with advanced solid tumors harboring a TP53 Y220C mutation. J Clin Oncol. 2022;40:1. doi: 10.1200/JCO.2022.40.16_suppl.3003. [DOI] [Google Scholar]
- 110.Chen S, Wu JL, Liang Y, Tang YG, Song HX, Wu LL, Xing YF, Yan N, Li YT, Wang ZY, et al. Arsenic trioxide rescues structural p53 mutations through a cryptic allosteric site. Cancer Cell. 2021;39:225–239.e8. doi: 10.1016/j.ccell.2020.11.013. [DOI] [PubMed] [Google Scholar]
- 111.Tang Y, Song H, Wang Z, Xiao S, Xiang X, Zhan H, Wu L, Wu J, Xing Y, Tan Y, et al. Repurposing antiparasitic antimonials to noncovalently rescue temperature-sensitive p53 mutations. Cell Rep. 2022;39:110622. doi: 10.1016/j.celrep.2022.110622. [DOI] [PubMed] [Google Scholar]
- 112.Lindemann A, Patel AA, Silver NL, Tang L, Liu Z, Wang L, Tanaka N, Rao X, Takahashi H, Maduka NK, et al. COTI-2, A novel thiosemicarbazone derivative, exhibits antitumor activity in HNSCC through p53-dependent and -independent Mechanisms. Clin Cancer Res. 2019;25:5650–5662. doi: 10.1158/1078-0432.CCR-19-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 114.Zhao CY, Grinkevich VV, Nikulenkov F, Bao W, Selivanova G. Rescue of the apoptotic-inducing function of mutant p53 by small molecule RITA. Cell Cycle. 2010;9:1847–1855. doi: 10.4161/cc.9.9.11545. [DOI] [PubMed] [Google Scholar]
- 115.Burmakin M, Shi Y, Hedström E, Kogner P, Selivanova G. Dual targeting of wild-type and mutant p53 by small molecule RITA results in the inhibition of N-Myc and key survival oncogenes and kills neuroblastoma cells in vivo and in vitro. Clin Cancer Res. 2013;19:5092–5103. doi: 10.1158/1078-0432.CCR-12-2211. [DOI] [PubMed] [Google Scholar]
- 116.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 117.Takimoto R, Wang W, Dicker DT, Rastinejad F, Lyssikatos J, el-Deiry WS. The mutant p53-conformation modifying drug, CP-31398, can induce apoptosis of human cancer cells and can stabilize wild-type p53 protein. Cancer Biol Ther. 2002;1:47–55. doi: 10.4161/cbt.1.1.41. [DOI] [PubMed] [Google Scholar]
- 118.Wang W, Takimoto R, Rastinejad F, El-Deiry WS. Stabilization of p53 by CP-31398 inhibits ubiquitination without altering phosphorylation at serine 15 or 20 or MDM2 binding. Mol Cell Biol. 2003;23:2171–2181. doi: 10.1128/MCB.23.6.2171-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.He XX, Zhang YN, Yan JW, Yan JJ, Wu Q, Song YH. CP-31398 inhibits the growth of p53-mutated liver cancer cells in vitro and in vivo. Tumour Biol. 2016;37:807–815. doi: 10.1007/s13277-015-3857-5. [DOI] [PubMed] [Google Scholar]
- 120.He X, Kong X, Yan J, Zhang Y, Wu Q, Chang Y, Shang H, Dou Q, Song Y, Liu F. CP-31398 prevents the growth of p53-mutated colorectal cancer cells in vitro and in vivo. Tumour Biol. 2015;36:1437–1444. doi: 10.1007/s13277-014-2389-8. [DOI] [PubMed] [Google Scholar]
- 121.Arihara Y, Takada K, Kamihara Y, Hayasaka N, Nakamura H, Murase K, Ikeda H, Iyama S, Sato T, Miyanishi K, et al. Small molecule CP-31398 induces reactive oxygen species-dependent apoptosis in human multiple myeloma. Oncotarget. 2017;8:65889–65899. doi: 10.18632/oncotarget.19508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu X, Wilcken R, Joerger AC, Chuckowree IS, Amin J, Spencer J, Fersht AR. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 2013;41:6034–6044. doi: 10.1093/nar/gkt305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Blanden AR, Yu X, Blayney AJ, Demas C, Ha JH, Liu Y, Withers T, Carpizo DR, Loh SN. Zinc shapes the folding landscape of p53 and establishes a pathway for reactivating structurally diverse cancer mutants. Elife. 2020;9:e61487. doi: 10.7554/eLife.61487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yu X, Blanden A, Tsang AT, Zaman S, Liu Y, Gilleran J, Bencivenga AF, Kimball SD, Loh SN, Carpizo DR. Thiosemicarbazones functioning as zinc metallochaperones to reactivate mutant p53. Mol Pharmacol. 2017;91:567–575. doi: 10.1124/mol.116.107409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA, Lozano G, Dobbelstein M, Moll UM. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature. 2015;523:352–356. doi: 10.1038/nature14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shi XY, Ding W, Li TQ, Zhang YX, Zhao SC. Histone Deacetylase (HDAC) inhibitor, suberoylanilide hydroxamic acid (SAHA), induces apoptosis in prostate cancer cell lines via the Akt/FOXO3a signaling pathway. Med Sci Monit. 2017;23:5793–5802. doi: 10.12659/MSM.904597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Panicker J, Li Z, McMahon C, Sizer C, Steadman K, Piekarz R, Bates SE, Thiele CJ. Romidepsin (FK228/depsipeptide) controls growth and induces apoptosis in neuroblastoma tumor cells. Cell Cycle. 2010;9:1830–1838. doi: 10.4161/cc.9.9.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- 130.Lo YH, Kolahi KS, Du Y, Chang CY, Krokhotin A, Nair A, Sobba WD, Karlsson K, Jones SJ, Longacre TA, et al. A CRISPR/Cas9-Engineered ARID1A-Deficient human gastric cancer organoid model reveals essential and nonessential modes of oncogenic transformation. Cancer Discov. 2021;11:1562–1581. doi: 10.1158/2159-8290.CD-20-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Geisinger JM, Stearns T. CRISPR/Cas9 treatment causes extended TP53-dependent cell cycle arrest in human cells. Nucleic Acids Res. 2020;48:9067–9081. doi: 10.1093/nar/gkaa603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alvarez MM, Biayna J, Supek F. TP53-dependent toxicity of CRISPR/Cas9 cuts is differential across genomic loci and can confound genetic screening. Nat Commun. 2022;13:4520. doi: 10.1038/s41467-022-32285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hsiue EH, Wright KM, Douglass J, Hwang MS, Mog BJ, Pearlman AH, Paul S, DiNapoli SR, Konig MF, Wang Q, et al. Targeting a neoantigen derived from a common TP53 mutation. Science. 2021;371:eabc8697. doi: 10.1126/science.abc8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Z, Strasser A, Kelly GL. Should mutant TP53 be targeted for cancer therapy? Cell Death Differ. 2022;29:911–920. doi: 10.1038/s41418-022-00962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chatterjee K, Das P, Chattopadhyay NR, Mal S, Choudhuri T. The interplay between Epstein-Bar virus (EBV) with the p53 and its homologs during EBV associated malignancies. Heliyon. 2019;5:e02624. doi: 10.1016/j.heliyon.2019.e02624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martinez-Zapien D, Ruiz FX, Poirson J, Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Vande Pol S, Podjarny A, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC, Wang YJ, Kato N, Omata M, Chang FY, Lee SD. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21:4801–4811. doi: 10.1038/sj.onc.1205589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.