Abstract
Cyclophilin (CyP) D is a regulator of the mitochondrial F-ATP synthase. Here we report the discovery of a form of CyPD lacking the first 10 (mouse) or 13 (human) N-terminal residues (ΔN-CyPD), a protein region with species-specific features. NMR studies on recombinant human full-length CyPD (FL-CyPD) and ΔN-CyPD form revealed that the N-terminus is highly flexible, in contrast with the rigid globular part. We have studied the interactions of FL and ΔN-CyPD with F-ATP synthase at the OSCP subunit, a site where CyPD binding inhibits catalysis and favors the transition of the enzyme complex to the permeability transition pore. At variance from FL-CyPD, ΔN-CyPD binds OSCP in saline media, indicating that the N-terminus substantially decreases the binding affinity for OSCP. We also provide evidence that calpain 1 is responsible for generation of ΔN-CyPD in cells. Altogether, our work suggests the existence of a novel mechanism of modulation of CyPD through cleavage of its N-terminus that may have significant pathophysiological implications.
Subject terms: Biochemistry, Biophysics
Mitochondrial Cyclophilin D can be cleaved at the N-terminus, resulting in a truncated form with enhanced binding to the OSCP subunit of F-ATP Synthase.
Introduction
Cyclophilin (CyP) D is the only member of the CyP family (17 members in humans) localized to mitochondria. CyPs are highly conserved peptidyl-prolyl cis-trans isomerases (PPIase) whose activity is inhibited by Cyclosporin A (CsA) in the nanomolar range1. The conserved catalytic domain (cyclophilin-like domain, CLD)2 consists of a β-barrel arranged by 8 antiparallel β-sheets that form a compact hydrophobic core3 connected by several loops and 3 helical elements, i.e., the two α-helices H1 and H3, which pack against the barrel at opposite sides, and the small H2 between strands S6 and S7, also reported as a 310 helix4. The surface of the active site presents two distinct pockets, the S1’ pocket, which includes the highly conserved proline-binding residues, and the S2 pocket, which is responsible for interacting with residues P2 and P3 (relative to the substrate proline, P1)5. Access to this pocket is guarded by a set of gatekeeper residues, which dictate substrate specificity and include Thr86, Ser94, Arg95, Ala116, Thr120, Ser123, and Gln124 for CyPD5. To date, all the >60 structures of human CyPD derived from crystallographic studies in the PDB database6 are based on truncated forms of CyPD lacking 14 N-terminal residues, whose role in protein structure and function remains therefore to be clarified. PPIases function as protein folding chaperones, but the actual importance of this activity in vivo has been questioned, as CyPs could rather provide scaffolds or interaction hubs7 regardless of their enzymatic activity8.
One of the best-characterised functions of CyPD is regulation of the mitochondrial permeability transition pore (PTP)9–11, an unselective inner membrane channel whose opening requires matrix Ca2+ and is favored by oxidative stress12,13. Short openings provide a fast Ca2+ release channel while prolonged openings result in stable depolarization, thus preventing ATP synthesis, and lead to mitochondrial swelling and eventually to cell death by necrosis14 or through the release of proapoptotic proteins15. CyPD modulates the PTP by decreasing the Ca2+ load required for pore opening, an effect that is prevented by its binding of CsA9,10. The discovery that CyPD interacts with F-ATP synthase at subunit OSCP in a CsA-sensitive way16,17 put forward the suggestion that F-ATP synthase is involved in PTP formation. The demonstration that F-ATP synthase preparations form Ca2+-activated channels provided solid evidence for the F-ATP synthase role to generate the PTP17–20, which is also supported by genetic manipulation of the enzyme18,21–26 and by structural studies27.
CyPD interaction with F-ATP synthase influences cell bioenergetics as its binding to OSCP leads to a 30% inhibition of both ATP hydrolysis and synthesis, which are reactivated by CsA displacement of CyPD16. CyPD inhibition is counteracted also by the mitochondrial chaperone TRAP1, which competes for the binding at OSCP increasing the F-ATP synthase catalytic activity28. Based on the disruption of CyPD-OSCP interactions by CsA, a binding site overlapping with the CsA binding site on the catalytic region of CyPD appears likely. The binding site on OSCP overlaps with that of the F-ATP synthase inhibitor Benzodiazepine (Bz)-423, which also induces channel activity within F-ATP synthase17. A survey of electrostatic surface potentials of CyPD and OSCP suggested that the CyPD-OSCP interaction, which is disrupted by increased ionic strength and favoured by Pi, is electrostatic17. Recent work has revealed that CyPD undergoes several post-translational modifications including phosphorylation, acetylation and oxidation that mainly occur near the active site29,30 but also affect the N-terminus29,31,32 and the so-called “backface” located on the opposite side of the catalytic domain33. The impact of these modifications on mitochondrial pathophysiology has been well described34–36, while their consequences on CyPD structure are still to be defined.
Here we describe the structural properties and functional role of the N-terminal region of mammalian CyPD and report the presence of a novel form of CyPD truncated at the N-terminus within mammalian mitochondrial matrix, whose cleavage could be catalysed by calpain 1. We also show that the truncated form of CyPD has specific properties of interaction and modulation of F-ATP synthase, to which it binds more avidly in vitro, suggesting the existence of a previously unrecognized layer of PTP regulation.
Results
CyPD N-terminus is poorly conserved and does not regulate the PPIase activity
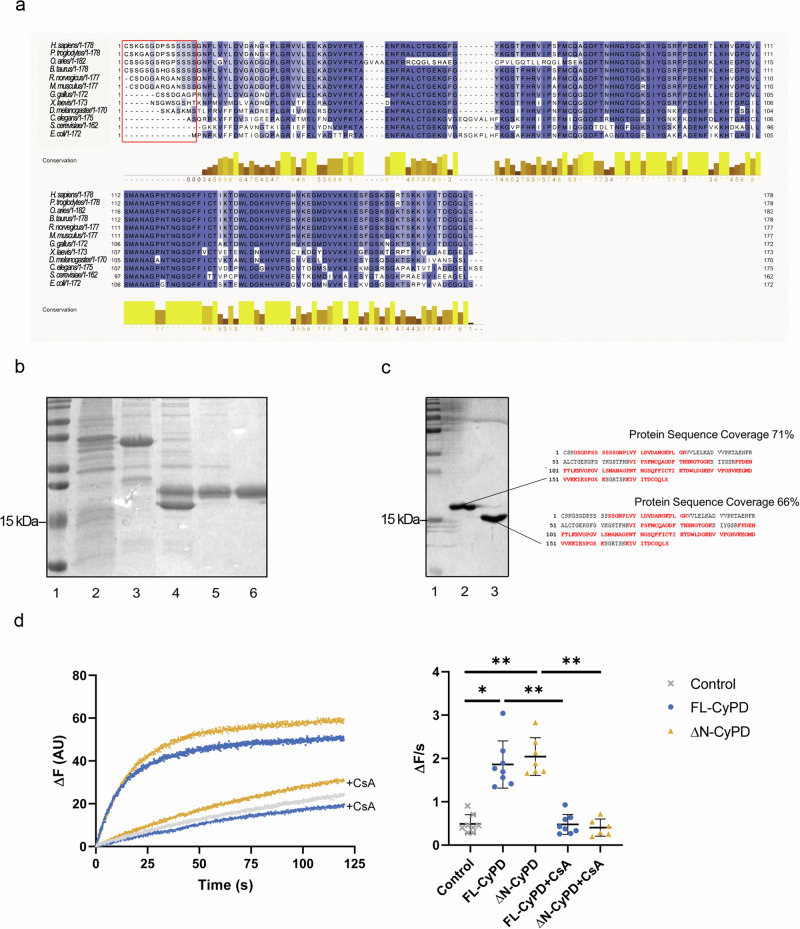
Sequence comparison of mature CyPD (excluding the mitochondrial-targeting sequence) revealed substantial variation in the N-terminus and a high degree of conservation across the rest of the sequence (Fig. 1a), which reached 100% for residues involved in the catalytic activity of CyPD such as Arg68 and Trp134. Mammalian CyPD harbors approximately 13-14 distinct N-terminal residues, whereas simpler organisms such as D. melanogaster and S. cerevisiae possess fewer residues or lack them altogether. This observation suggests a potential evolutionary divergence in the functional role of CyPD and prompted us to investigate whether this short sequence in the N-terminus (N-terminal tail, NTT) could contribute to shape the structural and functional features of CyPD. Thus, we produced recombinant human full-length CyPD (FL-CyPD) and a form devoid of N-terminal residues 1-13, which we will refer to as ΔN-CyPD (Fig. 1b and Supplementary Fig. 1). We could resolve FL-CyPD and ΔN-CyPD in 15% SDS-PAGE gels and confirmed their identity by mass spectrometry (Fig. 1c). Recombinant FL-CyPD and ΔN-CyPD displayed almost identical CsA-sensitive PPIase activity as determined by refolding of RNase T1 (Fig. 1d), clearly indicating that the NTT is not necessary for the catalytic activity.
Fig. 1. CyPD sequences and generation of full-length and truncated variants.
a Alignment of CyPD in different species. Residues are color-coded on a blue scale, from lightest to darkest, based on the percentage of conservation, while the red box highlights the N-terminal residues. b SDS-PAGE followed by Coomassie staining showing the principal steps of FL-CyPD production in E. coli strain BL21(DE3)pLysS. Lane 1, molecular weight markers; Lane 2, bacterial pellet after sonication; Lane 3, SUMO-CyPD eluted from the nickel-resin; Lane 4, SUMO and CyPD after the enzymatic reaction with Ulp1; Lane 5, flow through from the second incubation with nickel resin containing free CyPD and contaminants; Lane 6, purified CyPD after cationic exchange chromatography. c Mass spectrometry analysis of recombinant FL-CyPD (Lane 2) and ΔN-CyPD (Lane 3) separated on 15% SDS-PAGE (Lane 1, molecular weight markers) stained with colloidal Coomassie (left panel). Peptides from the bands of two recombinant proteins were sequenced and matched against human mature CyPD sequence (right panel). The univocally sequenced and matched peptides are shown in red, while residues in black represent non-detected ones (representative experiment out of five). Sequence coverages are indicated. d PPIase activity of recombinant FL-CyPD and ΔN-CyPD. PPIase activity was assessed monitoring the intrinsic fluorescence of RNase T1 during its refolding. Left panel: ΔF during time of the spontaneous reaction (grey line), compared with the refolding catalysed by FL-CyPD (blue) or ΔN-CyPD (orange), with or without 1:10 CsA. Representative traces of 4 different experiments. Right panel: slopes of the linear part of the traces were interpolated and used as a probe of FL-CyPD or ΔN-CyPD catalytic activity. Data were analysed according to the Kruskal-Wallis test followed by Dunn’s multiple comparisons test (**p < 0.01; *p < 0.05).
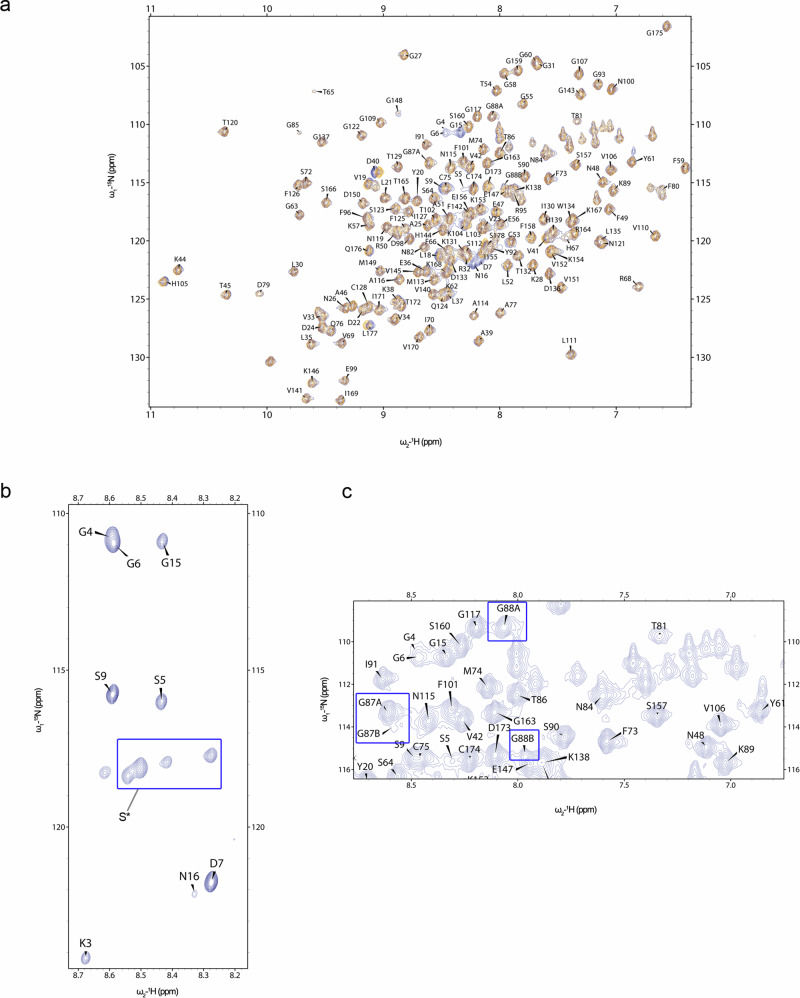
All CyPD structures deposited in the Protein Data Bank have been determined by X-ray crystallography and are all truncated forms at the N-terminus carrying a Lys-to-Ile substitution at position 146 (133 in the crystal, see e.g. PDB ID 4O8H37), thus closely resembling ΔN-CyPD. Since the structure of FL-CyPD has never been reported before, we have compared previously reported structures with the structural and dynamic features of FL-CyPD and ΔN-CyPD as derived from NMR experiments on uniformly [13C,15N]-labelled samples. 2D [1H,15N] HSQC spectra of FL-CyPD and ΔN-CyPD in CyPD buffer (20 mM NaPi, 2 mM DTE, pH 7.0) at 298 K show very well-dispersed peaks, typical for globular proteins, and superposition reveals a high similarity between the two protein forms (Fig. 2a). The assignment of the NMR chemical shifts in human ΔN-CyPD has already been described in the literature (BMRB ID: 25340) under similar experimental conditions38, while the chemical shifts assignment of human mature CyPD has never been reported. The chemical shift values of the backbone and Cβ atoms of both FL-CyPD and ΔN-CyPD were determined according to the standard triple resonance assignment procedure. Of note, although at 298 K the assignment completeness of the protein was over 90%, the chemical shifts of the FL-CyPD N-terminal residues were almost not observed. To improve the detection of the N-terminal region, 2D HSQC spectra were recorded in the temperature range between 283 K and 316 K. Signals from the N-terminal residues became visible only at low temperature between 283 K and 289 K, and they were clearly stronger than those of the other peaks (Fig. 2b). Analysis of 3D spectra at 283 K made possible to reliably assign the N-terminal segment, although the Ser10-14 stretch gave distinct but non-assignable signals that were therefore collectively assigned as S*. Signals from Gly78, Asp79 and Phe80 could only be assigned at 310K-316K. Moreover, signals from Lys161 and Ser162 were visible only when CyPD buffer contained 150 mM KCl. Last, a signal attributable to His83 was visible only when CsA was added at a 1:1 ratio. Except in the case of N-terminal residue signals, these observations were made for both FL-CyPD and ΔN-CyPD. Interestingly, both Gly87 and Gly88, adjacent to the gatekeeper Thr86, gave distinct signals in the [1H,15N] HSQC spectrum (Fig. 2c), indicating that these residues are in two distinct conformations under a slow chemical exchange regime in the NMR time scale. Indeed, from the chemical shift difference between the two forms, we could estimate the exchange constant to be larger than 1.8 ms. The assignment of the less populated forms, specified as Gly87B and Gly88B, was possible due to the signal propagation in the HNCA experiment, which indicated the sequential connectivity of two Gly residues, a unique feature in CyPD sequence.
Fig. 2. NMR chemical shifts assignment of human FL-CyPD and ΔN-CyPD.
a Overlay of the 1H-15N HSQC spectra (protein fingerprints) of FL-CyPD (blue) and ΔN-CyPD (orange) acquired at 298 K. b 1H-15N HSQC spectrum of FL-CyPD at 283 K. Counter levels were reduced to show the higher intensity of the signals from the N-terminal tail compared to the other residues. c Representative image from the 1H-15N HSQC spectrum acquired at 298 K of FL-CyPD showing the double forms of G87 and G88. Samples consisted of 500 μM CyPD in 20 mM NaPi pH 7.0. Spectra were acquired in a 700 MHz Bruker Spectrometer.
NMR highlights the pronounced flexibility of the N-terminal tail
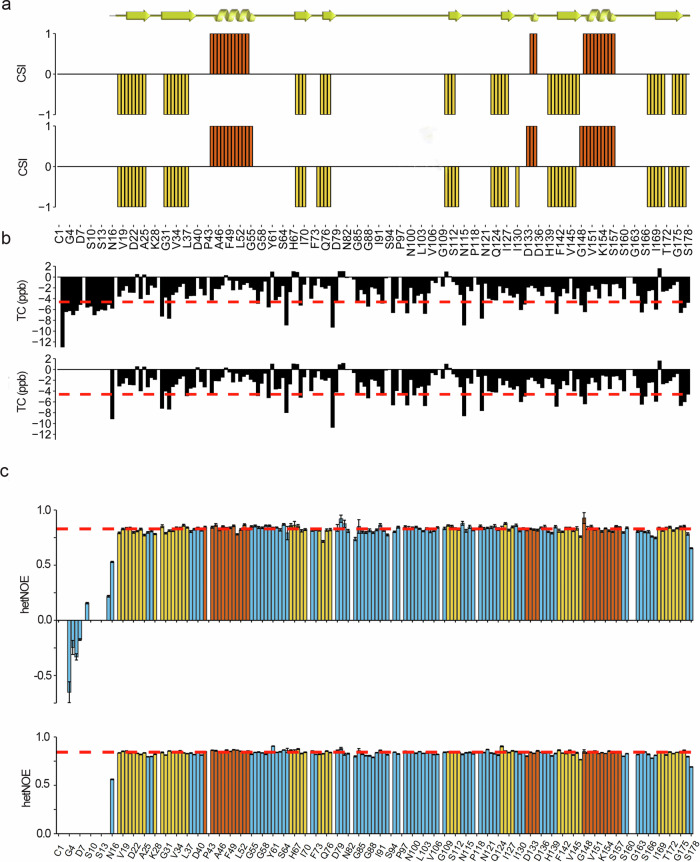
The secondary structure of FL-CyPD and ΔN-CyPD was determined using the software TALOS-N39 on the basis of the backbone assigned chemical shifts (13 Cα,13C’,15NH and 1HN) and 13Cβ of both proteins and their sequence (Fig. 3a). According to TALOS-N output, the secondary structures of FL-CyPD and ΔN-CyPD are highly similar and in good agreement with the crystallographic ones, comprising 3 α-helices and 8 β strands6. The only notable difference was in the last β-strand. Here, while the crystallographic structure comprised a continuous strand from residue 169 to residue 176, our analysis revealed two shorter strands running from residue 169 to residue 177, interrupted by what could be interpreted as a β-bulge at positions 172 and 173 (only 172 for ΔN-CyPD). Moreover, for FL-CyPD a two-residue helical element was found at position 136-137, corresponding to the 310 helix reported in the crystallographic structures at positions 135-137. Importantly, according to our analysis the NTT of FL-CyPD is unstructured, and the first secondary structural element (Strand 1) starts at residue Leu18.
Fig. 3. Structural and dynamical features of human FL-CyPD and ΔN-CyPD.
a Secondary structure of human FL-CyPD (upper barplot) and ΔN-CyPD (lower barplot) derived from the assigned main chain NMR chemical shifts according to the chemical shift indexing performed by TALOS-N (+ 1=helix, red; −1=strand, yellow; 0=random coil). The comparison with the crystallographic secondary structure, graphically represented above (ribbon=helix, arrow=strand), is also shown. b Temperature coefficients for each main chain amide group from FL-CyPD (upper barplot) and ΔN-CyPD (lower barplot). The plot is aligned with the one presented in (a), so the x-axis (representing the primary sequence) is the same. Red dashed line represents the limit above which a given amide group is considered involved in a H-bond (−4.6 ppb). c Barplots representing the hetNOE for each residue within FL-CyPD (upper barplot) and ΔN-CyPD (lower barplot). Bars are coloured according to the crystallographic secondary structure (blue=random coil; red=helix; yellow=strand). The red dotted lines represent the mean hetNOE of the secondary structure elements (0.828 for FL-CyPD, 0.842 for ΔN-CyPD). Error bars have been defined in the methods section.
A temperature titration from 283 K to 316 K in 3 K steps was performed to derive the temperature coefficients (TC) of main chain amide groups, which are diagnostic of their involvement in hydrogen bonding (Fig. 3b, Supplementary Data 1). Both FL-CyPD and ΔN-CyPD were found highly enriched in hydrogen bonds (i.e., TC > −4.6 ppb). Specifically, 124 backbone amide groups (~ 2/3 of the total amide groups) were found to be involved in hydrogen bonding, at the same positions for both CyPD forms and with comparable TCs. A close comparison with the crystal structure revealed no dramatic differences in terms of hydrogen bonding of the amide groups, most of which occur at the termini of secondary structure elements. Conversely, the 14 N-terminal residues of FL-CyPD were not observed to participate in hydrogen bond, coherently with the unstructured nature observed in the secondary structure computations.
The structural characterization of both FL-CyPD and ΔN-CyPD also involved the definition of the CsA binding site, again via solution NMR. To this end CsA was added at a 1:1 ratio, which should be sufficient for binding saturation given the low dissociation constant of the CsA:CyPD complex (13.4 nM)6. The resulting induced chemical shifts changes were analysed to identify the binding site positions (Supplementary Fig. 2) and clearly the same residues were affected to the same extent by CsA addition in both FL-CyPD and ΔN-CyPD. These residues were the only ones directly involved in the binding site defined by the crystal structure4, and no involvement of the N-terminal residues of FL-CyPD was observed, suggesting that CsA follows a “lock and key” kind of binding. The sidechain of the single Trp134 of CyPD was strongly perturbed (Δδ = 0.93 for both proteins), indicating its involvement in the binding as also reported by other groups5. Similarly, the side chain amide group of a His residue was strongly affected, most likely His139, whose NH side chain group has a < 4 Å distance from CsA in the crystal structure4.
We next investigated protein dynamics of backbone NHs in the ps-ns timescale by measuring the heteronuclear steady-state 15N{1H} nuclear Overhauser effect (hetNOE). Indeed, hetNOE correlates with local dynamics, with low hetNOE diagnostic for fast motions and typical values for secondary structured regions around 0.9. Figure 3c shows the hetNOEs measured at 298 K plotted against the residue number for FL-CyPD (upper panel) and ΔN-CyPD (lower panel). The hetNOE values span from –0.651 to 0.8 for FL-CyPD and from 0.746 to 0.906 for ΔN-CyPD with a mean value calculated for the residues in secondary structure elements of 0.828 for FL-CyPD, 0.842 for ΔN-CyPD. Overall, the hetNOEs of ΔN-CyPD are all around the mean value of the structured region, indicating low flexibility of the corresponding residues in the ps-ns time scale. The corresponding residues in FL-CyPD follow the same identical pattern, and range between 0.714 and 0.926. The N-terminal residues of FL-CyPD, instead show very low hetNOEs (from −0.651 to 0.219), notably with negative values in the stretch K3-G6, indicating fast motion dynamics of this region, in sharp contrast to the structured globular domain (Fig. 3c).
As mentioned above, Gly87 and Gly88 present double forms, named A and B. The TC values indicate that the Gly88 amide group is not involved in H-bonding in both conformations and in both FL-CyPD and ΔN-CyPD, whereas Gly87 amide group is indeed H-bonded in the “A” form while loses the H-bond in the “B” form, again in both variants (Supplementary Table 1). The hetNOE of Gly88B and Gly87B was found to be lower in both FL-CyPD and ΔN-CyPD than in the more populated counterpart (Supplementary Fig. 3), with a less pronounced hetNOE decrease in ΔN-CyPD. This suggests that Gly88B and Gly87B are more flexible than their respective “A” forms. Taken together, NMR analyses revealed that human FL-CyPD and ΔN-CyPD are structurally identical, except for the NTT of FL-CyPD, which is highly flexible, in sharp contrast with the remaining globular rigid part.
NTT influences CyPD binding and modulation of F-ATP synthase at physiological ionic strength
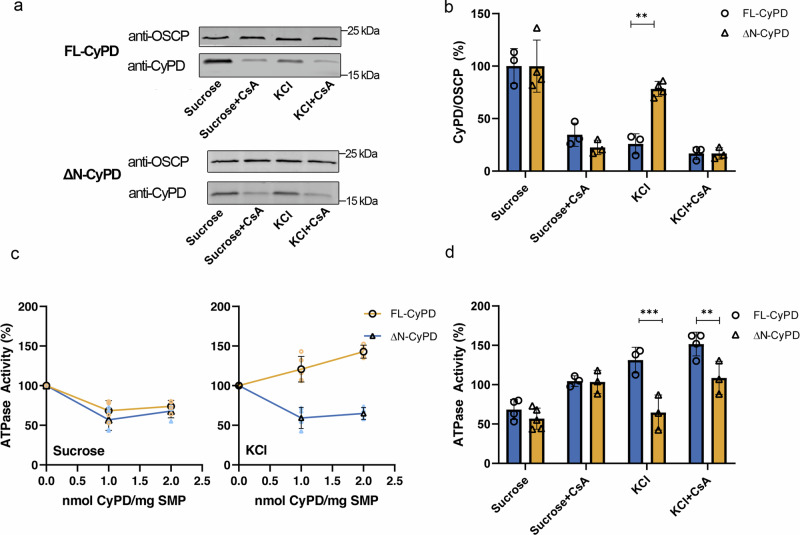
It is known that endogenous CyPD interacts with the OSCP subunit of F-ATP synthase and inhibits enzyme activity by about 30%, which is reactivated by CsA displacement of CyPD16,17. To assess whether the NTT of FL-CyPD plays a role in modulating the CyPD/OSCP interaction, we investigated whether FL-CyPD and ΔN-CyPD interact with and modulate F-ATP synthase to the same extent. For this purpose, we used submitochondrial particles (SMP), which retain most of the enzymatic machinery of oxidative phosphorylation including F-ATP synthase, whose catalytic sector is exposed to the solvent40 and therefore accessible to added exogenous recombinant CyPD. SMP were almost completely depleted of endogenous CyPD (Supplementary Fig. 4), the mitochondrial content of which is estimated to be between 0.1 and 0.4 nmol/mg protein41, and then incubated with 1 nmol of FL-CyPD or ΔN-CyPD/mg SMP. As reported for endogenous CyPD, in a sucrose-based buffer both recombinant FL-CyPD and ΔN-CyPD could be immunoprecipitated with OSCP, while in the KCl-based buffer only ΔN-CyPD retained its interaction (Fig. 4a, b) suggesting that at physiological ionic strength the NTT negatively affects the CyPD/OSCP interaction. In support of these findings, the above differences were found to be much more prominent in KCl-based buffer not supplemented with phosphate, which is required for the binding of endogenous CyPD in sucrose-based media16. Indeed, ΔN-CyPD still bound OSCP in a CsA-sensitive manner, while FL-CyPD binding was almost completely abrogated (Supplementary Fig. 5). Whenever CyPD was bound to OSCP, it could be displaced by CsA irrespective of the presence of the N-terminal domain (Fig. 4a, b), consistently with the NMR data (Supplementary Fig. 2). The functional relevance of these interactions is supported by the inhibitory effect on ATPase activity of SMP, which was always observed when they were treated with ΔN-CyPD but not with FL-CyPD in KCl, where we rather observed a slight stimulatory effect (Fig. 4c, d). It should be stressed that the ratio between 0.05 and 2 nmol of CyP/mg SMPs is within the 0.2-6 CyPD/F-ATP synthase molar ratio, based on the F-ATP synthase content of mitochondria40,42. The lower affinity between FL-CyPD and OSCP in KCl was also confirmed by surface plasmon resonance (SPR) experiments on a simplified binding system (Supplementary Fig. 6). Here recombinant OSCP with a Myc-tag was noncovalently captured onto a CM5 chip covalently derivatized with a monoclonal anti-Myc antibody, while increasing concentrations of FL-CyPD and ΔN-CyPD where injected into the mobile phase. Indeed, from the estimated Kd values, calculated as the ratio koff/kon, a > 90-fold increased affinity of ΔN-CyPD for OSCP was determined. Kinetic analysis of SPR data revealed that this remarkable increased affinity of ΔN-CyPD for OSCP is mainly due to an increase in the association rate constant by approximately 30-fold compared to FL-CyPD-OSCP, whereas the dissociation rate constant was only reduced 3-fold. Remarkably, both FL-CyPD and ΔN-CyPD retained comparable PPIase activity in KCl-containing medium (Supplementary Fig. 7), confirming that the NTT does not affect this enzyme activity.
Fig. 4. Binding and modulation of F-ATP synthase by FL-CyPD and ΔN-CyPD.
a Pig heart SMP depleted of endogenous CyPD (1 mg/ml) were incubated at 25°C for 15 min with 1 nmol of either FL-CyPD or ΔN-CyPD/mg SMP under two different conditions, namely in the sucrose-based buffer (250 mM sucrose) or the KCl-based buffer (125 mM KCl) supplemented with 10 mM KH2PO4, in the presence or absence of 2 μM CsA. Immunoprecipitation of OSCP was performed, followed by Western blotting with anti-OSCP or anti-CyPD antibodies (representative experiment out of three). b Each immunodetected band was analyzed by densitometry, and the ratio between the peak area of CyPD (FL-CyPD or ΔN-CyPD) and that of the corresponding OSCP subunit was measured and expressed relative to the ratio obtained in the sucrose buffer in the absence of CsA, which was taken as 100% (values are mean ± S.D. of at least three independent experiments). c ATPase activity of pig heart SMP depleted of endogenous CyPD and exposed to different concentrations of either FL-CyPD (◯) or ΔN-CyPD (△) in the sucrose-based buffer (left panel) or the KCl-based buffer (right panel). The oligomycin-sensitive ATP hydrolysis rate was determined spectrophotometrically. Mean ± S.D. values of at least four independent experiments are shown in black. d ATPase activity of pig heart SMP exposed to 1 nmol/mg SMP of either FL-CyPD or ΔN-CyPD in either sucrose-based buffer or KCl-based buffer in the presence or absence of 2 μM CsA. The ATPase activity in the absence of FL-CyPD or ΔN-CyPD was taken as 100%, and reported values are mean ± S.D. of at least 3 independent experiments. Data were analysed according to the two-way ANOVA analysis followed by Bonferroni’s multiple comparisons test (**p < 0.01; ***p < 0.005).
Overall, these results indicate that FL-CyPD and ΔN-CyPD differ in binding and modulation of F-ATP synthase at physiological ionic strength.
Ionic strength affects the conformation and dynamics of specific regions within CyPD
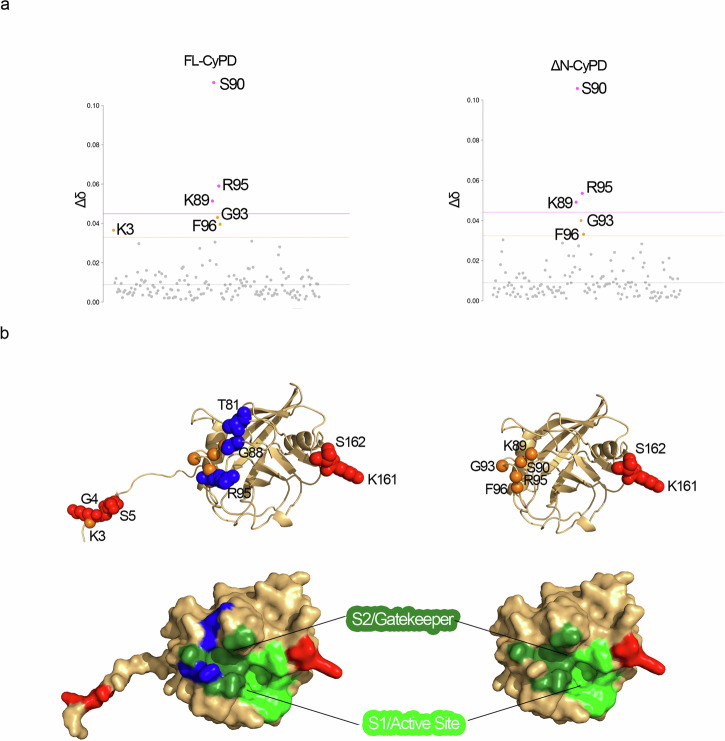
Given the prominent effects of KCl on the interaction between CyPD isoforms and F-ATP synthase, we used NMR to investigate possible structural effects. We compared CSP and lineshape of 1H-15N HSQC spectra of FL- and ΔN-CyPD in the absence and presence of KCl. Previous studies show that the addition of NaCl influences the chemical shift of the 1H and 15N nuclei of proteins; in particular, an overall negative deviation of 0.01 ppm/100 mM NaCl is observed43. We observed a similar behaviour with the addition of 6 residues of FL-CyPD and 5 residues of ΔN-CyPD, which were significantly affected by the addition of 150 mM KCl with a positive deviation of more than 0.01 ppm (Fig. 5a). Five of the six residues with changes of chemical shift were identical for ΔN-CyPD and FL-CyPD. These include Ser90 (the most perturbed one), Arg95, Lys89, Gly93 and Phe96, all clustering in the S2/gatekeeper region. The only residue specifically affected in FL-CyPD is Lys3, absent in the truncated form. Remarkably, one amide sidechain was also significantly affected in both proteins (Δδ = 0.087 for FL-CyPD, Δδ = 0.083 for ΔN-CyPD). The position in the HSQC spectrum suggests that it is a Gln or Asn sidechain amide group. Although, due to the lack of sidechain assignment, it is difficult to determine the residue identity, an educated guess can be made and points to Asn48, as its sidechain is 5.5 Å away from Ser90 amide group. The five common CSP are clustered in the long loop H1-S3 and may be interpreted in term of local conformational variations.
Fig. 5. Effect of KCl on structure and dynamics of FL-CyPD and ΔN-CyPD.
a CSP analysis of FL-CyPD (left panel) and ΔN-CyPD (right panel) after the addition of 150 mM KCl. Data points are coloured according to the chemical shift perturbation: pink points have a Δδ higher than S.D. (pink dotted line), orange points have a Δδ higher than S.D. (orange dotted line), grey points are non-perturbed residues. b KCl-induced variations plotted on the structures of FL-CyPD (left, Alphafold2 prediction) and ΔN-CyPD (right, PDB 2Z6W). Variations of chemical shifts shown in (a) are reported as orange spheres of the amide nitrogen of the corresponding residues. Lineshape variations were determined either considering the signal intensity or the integral at 0 mM KCl and at 150 mM KCl, only for those peaks that did not coalesce throughout the titration. Residues whose lineshape was significantly affected by salt addition are represented as spheres coloured in blue (signal intensity decrease) or red (signal intensity increase). The lower panel shows the surface representations of the above structures, showing the localization of the S2/gatekeeper and S1/active site regions and residues affected by KCl colored as above.
Measurement of the signal intensity, limited to non-superimposed residues, indicated a specific change in the lineshape, for a number of residues, due to KCl addition (Fig. 5b). The signals of Lys161 and Ser162, in the S1 region of CyPD, became detectable and assignable for both FL-CyPD and ΔN-CyPD, indicating an unquantifiable increase in their intensity, and thus showing a strong dependence in their dynamics on ionic strength (Fig. 5b). Additionally, a reduction in signal intensity was observed only for FL-CyPD, particularly at residues Thr81, Arg95 and Gly88B. These FL-CyPD-specific variations clustered at the S2/gatekeeper region. The NTT of FL-CyPD was also affected by KCl addition, as the signals from Lys3, Gly4 and Ser5 showed increased signal intensity. In particular, residue Lys3 also showed significant chemical shift perturbation, indicating salt-induced conformational variations as described above. Contribution of NTT and S2/gatekeeper region influencing CyPD binding to OSCP in the presence of KCl appears likely.
Truncation of NTT occurs in vivo and is impaired by Calpain inhibition
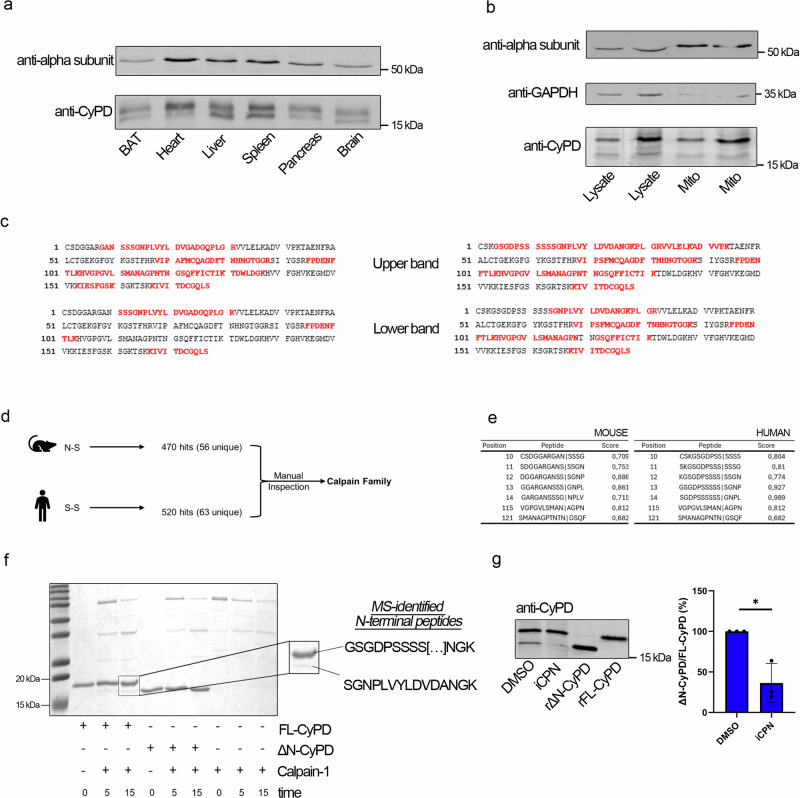
We noticed that two forms of CyPD differing at the N-termini were originally isolated from murine liver mitochondria41, while most but not all Western blot analyses reported in the literature showed only one prominent band immunoreactive with anti-CyPD antibodies14,44,45. Prompted by these observations, we performed Western blot analyses to detect CyPD in different tissues and cell types (Fig. 6a, b). In several murine tissue lysates, two distinct bands immunoreactive with anti-CyPD antibodies were clearly visible when an in-house made 15% SDS-PAGE gel was used (Fig. 6a) while commercial gradient gels were unable to resolve the two bands (Supplementary Fig. 8), thus suggesting a possible explanation for the rare detection of the truncated form in the literature. One band was detected at the expected molecular mass of about 19 kDa corresponding to FL-CyPD, and a second band was found at about 18 kDa, suggesting the presence of two forms of CyPD within different tissues. Intriguingly, the ratio between the lower and upper bands appeared to be tissue-specific, being consistently higher in liver and spleen than in other tissues (Supplementary Fig. 9). The presence of two distinct bands was also confirmed in total lysates and crude mitochondria from human skin-derived fibroblasts (Fig. 6b). Protein identification was performed by mass spectrometry (Fig. 6c). In both human and murine samples, the upper and lower bands corresponded to FL-CyPD and ΔN-CyPD, respectively. Indeed, the upper band N-terminal tryptic fragment was R | GANSSGNPLVYLDVGADGQPLGR and K | GSGDPSSSSSSGNPLVYLDVDANGK for the murine and human protein, respectively. Both the semitryptic fragments CSDGGAR (murine) and CSK (human) are not detected for technical limitations, therefore we can assume that the tryptic ones detected are diagnostic of the N-terminal sequence of the full-length form of CyPD. On the other hand, the N-terminal fragment of the lower band was semi-tryptic (SSSGNPLVYLDVGADGQPLGR and SGNPLVYLDVDANGK for the murine and human protein, respectively) indicating that it corresponds to the actual N-terminus of a protein lacking the first 10 and 13 residues, respectively. Overall, these experiments demonstrate the presence of a previously unreported, N-terminal cleaved form of mature CyPD in mammalian mitochondria, which appears to result from the cleavage of FL-CyPD.
Fig. 6. Detection of a truncated form of CyPD in cells and role of calpain-1 in CyPD cleavage.
a Western blot analysis of CyPD and F-ATP synthase α-subunit in lysates from different murine tissues. For each well, 30 μg of proteins were loaded. b Western blot analysis of CyPD and F-ATP synthase α-subunit in total lysates (Lysate, 10 μg and 20 μg in the first and second lane, respectively) or crude mitochondrial preparations (Mito, 5 μg and 10 μg in the third and fourth lane, respectively) from primary human skin fibroblasts. The cytosolic marker glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was almost undetectable in the mitochondrial fraction. c Mass spectrometry analysis of the upper band at ~19 kDa and of the lower band at ~18 kDa from murine heart lysates detected by anti-CyPD antibodies (left panel) or human skin fibroblasts (right panel). Peptides were sequenced and matched against murine (left panel) or human (right panel) mature CyPD sequence. The univocally sequenced and matched peptides are shown in red, while residues in black represent non-detected ones (representative experiment out of five). For murine samples, sequence coverages were 51% ± 9% for the 19-kDa band and 37% ± 21% for the 18-kDa band. For human samples, sequence coverages were 39% ± 16% for the 19-kDa band and 30% ± 15% for the 18-kDa band. d Workflow for the interrogation of the MEROPS database and its analysis. The murine (NS) and human (SS) cleavage sites were used as a query in the search for proteases. Then, among the different hits revealed by the database, a manual inspection was performed. Proteins of the calpain family were found as the most interesting hits. e Results for the GPS-CCD 1.0 analysis for mouse and human mature CyPD sequence. Each table shows the most probable target sites for calpain 1, assigning to each putative cleavage site a score. Letters in bold-red show the actual cleavage sites. f SDS-PAGE/Coomassie staining following the incubation of FL-CyPD or ΔN-CyPD with Calpain 1 (CPN1) at different times (0 min, 5 min, 15 min). On the right, a magnification of the enzymatic products of FL-CyPD+CPN1 after 15 minutes is shown. The two bands were manually excised for mass spectrometry analysis. The N-terminal peptide detected in each band is shown. g Western blot analysis of CyPD in total lysates from HEK293 cells treated for 30 min with 50 μM of the broad-spectrum calpain inhibitor PD150606 or with the DMSO concentration as vehicle control. The ratio between the lower and the higher band was quantified by densitometry and is reported as mean ± S.D. of three independent experiments. Data were analysed according to the one-sample t-test (*p = 0.044).
Given that mass spectrometry analysis revealed the cleavage site both in human (S-S) and mouse (N-S), we used them as queries in the MEROPS database (Fig. 6d). The search gave 470 hits (56 unique) when the murine cleavage site was used as query and 520 hits (63 unique) when the human one was adopted. We then manually inspected the results, searching for mitochondrial mammalian proteases. Ultimately, proteases of the calpain family were identified as the most interesting hits. The GPS-CCD 1.0 software46, which predicts the putative calpain cleavage sites on a given aminoacidic sequence, indicated that both human and murine FL-CyPD could be cleaved at the site we experimentally determined with a good score (0.927 and 0.709, respectively) (Fig. 6e). To validate the in silico results, we performed an incubation of commercial recombinant human calpain 1 with our recombinant FL-CyPD or ΔN-CyPD at a 1:5 ratio for 15 minutes. SDS-PAGE analysis revealed that already after 5 minutes of incubation a band with the same electrophoretic mobility of ΔN-CyPD appeared (Fig. 6f). Protein identity was confirmed by mass spectrometry (Fig. 6f) and Western blot analysis (Supplementary Fig. 10). Consistent with cleavage of FL-CyP D by calpain 1 in situ, treatment of HEK293 with calpain inhibitor markedly reduced the presence of ΔN-CyPD in total lysates (Fig. 6g).
Discussion
In this work, we have identified the N-terminus of mammalian FL-CyPD as a negative regulator of its binding to OSCP subunit of F-ATP synthase in a KCl-based buffer, which best resembles the physiological environment of the mitochondrial matrix, and demonstrated that cleavage of the N-terminus occurs in vivo. Inspection of mammalian CyPD structures in the Protein Data Bank derived from X-ray crystallography revealed that they all lack the NTT, suggesting NTT as a highly disordered region that likely hinders protein crystallization6. Therefore, our solution NMR study reports the first structural characterization of human FL-CyPD. Here we find that the NTT does not affect the overall structure and function of the protein, as FL-CyPD and ΔN-CyPD presented highly similar structural features and the same PPIase activity. Remarkably, we observed a difference from the crystallographic structure in the last β-strand, which is interrupted at positions 172 and 173. Considering that the nearby Cys174 (Cys203 with the MTS) is a site of multiple post-translational modifications45, the disruption of the ordered secondary structure suggests a local flexibility that could not be inferred from crystallographic data.
The NTT presents peculiar features compared to the globular region, with disorder-like characteristics, as it forms an unstructured and high flexible random coil, as evidenced by very low or even negative hetNOEs. In addition, other observations revealed that both proteins have a certain degree of conformational heterogeneity and flexibility. In particular, the double resonance of Gly87 and Gly88 (loop S7-S8), located in the S2/gatekeeper region, indicates that these residues can adopt two different conformations. The estimated half-life for each conformation is longer than 1.8 ms, presenting a conformational heterogeneity which was also captured in the crystal structure of CyPD in complex with CsA (PDB ID: 2Z6W)4, where Gly87, Gly88 and Lys89 are present in both conformations. Our findings, together with previous NMR relaxation data reporting microsecond exchange dynamics for Gly87 in CyPA47, indicate that the conformational dynamics of CyPD is not restricted to the catalytic residues, consistent with the plasticity required for catalysis.
FL-CyPD does not bind F-ATP synthase unless millimolar concentrations of Pi are present16, and it is easily removed by incubation of SMPs in isotonic KCl media48. Here, we showed that truncation of the NTT increases the affinity between CyPD and OSCP, making the binding possible at 125 mM KCl, an ionic strength closer to the physiological one at which FL-CyPD does not bind OSCP. Our NMR based observations highlighted that KCl has an effect on the structure and dynamics of CyPD. Specifically, KCl induces significant CSP (i.e., conformational variations) in the S2/gatekeeper region (residues Lys89, Ser90, Gly93, Arg95, Phe96) on both ΔN-CyPD and FL-CyPD. Furthermore, the dynamics of the two proteins is affected to the same residues in the S1/active site (Lys161, Ser162), as revealed by lineshape analysis. The observed common variations are consistent with the observed common impairment of the PPIase activity under 150 mM KCl (Supplementary Fig. 7). Lineshape analysis showed that the dynamics of FL-CyPD is specifically affected at residues residing in/surrounding the S2/gatekeeper region (Thr81, Arg95 and Gly88B), in addition to residues in the NTT (Gly4, Ser5, Lys3). Considering that the interaction between CyPD and OSCP is CsA-sensitive, and given the role of the S2/gatekeeper region in dictating the substrate specificity of different CyPs5, the contribution of the S2/gatekeeper region to OSCP binding appears likely. The observed decrease in signal intensity in KCl could indicate a slower motion of this region under physiological conditions for FL-CyPD, which in turn might contribute to minimize the interactions with OSCP. Indeed, it was recently shown in silico that phosphorylation of Ser162 (Ser191 considering the MTS), which is located near the S1/active site and is known to favour CyPD binding to OSCP29, increases protein dynamics in the S2/gatekeeper region49. Therefore, the dynamics of the S2/gatekeeper region appears a key factor in the interaction between CyPD and OSCP, and we have shown to be sensitive to the NTT truncation. The contribution of NTT itself in the FL-CyPD/OSCP interaction cannot be excluded. The increased dynamics of NTT in KCl may generate an overall unfavourable environment for the FL-CyPD/OSCP interaction. Removal of the NTT would then also release these constraints and increases the affinity of the interaction allowing binding of ΔN-CyPD in saline media in the absence of Pi.
The mechanistic description of how the long-range allosteric communication between the NTT and S2/gatekeeper region takes place requires further studies. However, other previously published evidence showed that the human CyPD contains a Ser residue at position 2 of the NTT and a stretch of Ser residues at positions 10-14 that are potentially phosphorylated, both negatively affecting propensity to cell death29,50. Phosphorylation of Ser2 by mitochondrial Akt2 in patient-derived tumor organotypic cultures treated with inhibitors of phosphatidylinositol-3 kinase opposes tumor cell death, conferring resistance to PI3K therapy50. In human cell lines mutation of the Ser stretch into Ala led to an increase of the Ser phosphorylation level of CyPD at Ser162 and increased PTP opening, suggesting that phosphorylation of the Ser stretch may act as a negative regulator of the PTP by inhibiting the further phosphorylation of the regulatory sites of CyPD29. These otherwise puzzling pieces of evidence can be explained in terms of allosteric communication between the (dynamics of the) NTT and the (dynamics of the) S1/S2 sites of CyPD. Taken together, these findings suggest that the S2/gatekeeper region is involved in the interactions between CyPD and OSCP and that its dynamics is sensitive to NTT truncation. We think that the N-terminal truncation of CyPD should not perturb the basic effects of the protein on PTP formation51, given that both FL-CyPD and ΔN-CyPD bind OSCP in a CsA-sensitive manner, although the affinity between FL-CyPD and OSCP is much lower in physiological-like conditions.
Importantly, in this study we have identified a truncated form of CyPD in a variety of mouse tissues and in human cells, indicating that the protein undergoes cleavage in situ. We have also shown that calpain 1 can cleave FL-CyPD in vitro and that HEK293 cells pre-treated with calpain inhibitor displayed a decrease in ΔN-CyPD levels. Based on these findings we propose a mechanism where calpain 1 is responsible for the proteolytic cleavage of FL-CyPD within the mitochondrial matrix. It is remarkable that the localization of calpain 1 and 2 within mitochondria has been well documented and their contribution to PTP formation has been demonstrated under various conditions52,53 including (i) cancer cells, where Ca2+-dependent mitochondrial depolarization and cell death triggered by hexokinase II displacement at mitochondria-endoplasmic reticulum contact sites was prevented by calpain inhibition54; (ii) mouse cardiomyocytes, where increased mitochondria-targeted calpain 1 increased the PTP propensity to open55; and (iii) rat hearts subjected to ischemia-reperfusion injury56.
The present data represent a necessary first step in the identification of the N-terminus of CyPD as an essential regulatory element of its binding to OSCP and potentially of the transition of F-ATP synthase from an energy-conserving to an energy-dissipating device. From an evolutionary perspective, this region, which is poorly conserved among CyPD orthologues, has been an active part of the Ppif gene, as it is shorter or absent in lower species such as yeast, where CyPD is not involved in PTP regulation57. Our findings suggest that CyPD has undergone a gain-of-function through acquisition of the NTT, which may determine how a highly conserved protein regulates the PTP in a species-specific manner. It is certainly worth noting that the NTT has evolved only in higher eukaryotes, and that it must serve function(s) that are independent of the PPIase activity, which is identical in FL- and ΔN-CyPD. Thus, cleavage of NTT is a novel post-translational modification of CyPD and a previously unrecognized event in F-ATP synthase regulation. Considering the existence of these two CyPD forms, future research must re-evaluate previous conclusions, recognizing the distinct functions and regulatory mechanisms imparted by the presence or absence of the NTT.
Methods
CyPD numbering
CyPD residues numbering can be quite confusing, and indeed the literature has not been consistent so far. This is due to the fact that human CyPD has a 29-residue mitochondrial targeting sequence absent in the mature form, which therefore affects the numbering of the latter (the first residue of the mature form can be numbered as “30”). Moreover, given that all the deposited structures of CyPD are truncated at the N-terminus, an additional numbering system considers as the first residue of mature CyPD what is actually the fifteenth. Here, we will consider the actual numbering of the mature form of CyPD (first residue will be “1”), reporting in brackets the corresponding numbering which considers the mitochondrial targeting sequence (MTS) (first residue will be “30”) whenever needed.
Cell and tissue sample preparation
Human dermal fibroblasts obtained from juvenile foreskin (purchased by PromoCell, Germany) were grown in Dulbecco’s modified Eagle’s medium (Gibco - ThermoFisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (Gibco - ThermoFisher Scientific) at 37 °C and with 5% CO2. Total lysate extract and crude mitochondria were prepared as described58 with minor modifications. Briefly, 2 × 107 cells/ml were suspended in grinding solution (250 mM sucrose, 1 mg/ml BSA, 2 mM EDTA pH 7.4) plus 1:50 v/v protease inhibitors cocktail P8340 (Sigma-Aldrich), and sonicated 3 times every 5 sec, with 30 sec intervals on ice. An aliquot was kept as total lysate extract. The remaining homogenate was used to isolate a crude mitochondrial fraction. Wild-type C57BL/6 adult male mice were purchased by the University of Padua’s Animal Care and Use Committee (authorization number: 43/2021, D2784.N.WUE). Brown adipose, brain, heart, liver, pancreas, and spleen tissues were harvested and immediately snap-frozen in liquid nitrogen. To prepare the total lysate extract frozen tissue samples were cut into small pieces and added with RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with protease inhibitors, as above. After incubation for 30 min on ice, the lysates were cleared by centrifugation at 10,000 × g for 10 min at 4°C. We have complied with all relevant ethical regulations for animal use.
Bioinformatics
Sequence Alignment was performed by retrieving sequences from the Uniprot database using Ppif as a query and filtering according to the Taxonomy. With the exception of E. coli, only mitochondrial proteins were considered. The sequences corresponding to H. sapiens, M. musculus, R. norvegicus, B. taurus and S. cerevisiae CyPD had already information about their localization and MTS within the Uniprot database. The sequences corresponding to P. troglodytes, O. aries, G. gallus, X. laevis, C. elegans and D. melanogaster CyPD were first predicted to be mitochondrial with the Deeploc 2.0 server59 and then analysed with TargetP 2.060 and MitoFates61 to identify the MTS. The alignment was performed with the online server CLUSTAW62 and visualized with Jalview v. 2.11.2.763.
To identify the protease responsible for the N-terminal cleavage of FL-CyPD, the MEROPS database was first used (https://www.ebi.ac.uk/merops/). The known cleavage sites in humans (S-S) and mice (N-S) were used as queries and a manual search focusing on mitochondrial mammalian proteases was then adopted. Subsequently, the GPS-CCD 1.0 software (http://ccd.biocuckoo.org/)46, designed to predict calpain 1 cleavage sites, was used with the strictest threshold to determine if FL-CyPD could be a potential target for calpain 1.
Expression and purification of recombinant FL-CyPD and ΔN-CyPD
The recombinant human full-length CyPD (FL-CyPD) and the N-terminal truncated form missing the first 13 residues (ΔN-CyPD) were expressed and purified following the same procedure. The nucleotide sequence encoding for the human FL-CyPD or the ΔN-CyPD protein was joined with that encoding the yeast SUMO protein tag bearing 6 histidine residues at their N-terminus64 into the pRSETA plasmid (purchased from Invitrogen) (Supplementary Fig. 1) and used to transform E. coli strain BL21(DE3)pLysS (purchased from Sigma-Aldrich). Briefly, the SUMO nucleotide sequence was obtained from yeast genomic DNA of Saccharomyces cerevisiae by PCR, using the primers 5’-ACA GCT AGC ATG TCG GAC TCA GAA GTC AAT CAA-3’ (Sumo-NheI) and 5’-GCA GGA TCC ACC AAT CTG TTC TCT GTG AGC CTC AAT AAT-3’ (Sumo-BamHI). The amplified sequences were cloned into the pRSETA plasmid between NheI and BamHI restriction sites in order to obtain the pSUMOB plasmid. The nucleotide sequence of FL-CyPD and ΔN-CyPD was obtained from a human cDNA placenta library (Clontech) by PCR with the primers 5’-GCA AAG CTT AGC TCA ACT GGC CAC AGT CTG TGA T-3’ (CID5) and 5’-GGT GGA TTC TGC AGC AAG GGC TCC GGC GAC CCG TCC-3’ (CID6) for FL-CyPD and 5’-GGT GGA TCC GGG AAC CCG CTC GTG TAC 3’ for the ΔN-CyPD, respectively. The amplified sequences were separately cloned into pSUMOB plasmids between the restriction sites BamHI and HindIII. Their correct sequences were confirmed by sequencing. The resulting vectors were used to transform E. coli strain BL21(DE3)pLysS, below indicated as E. coli cells. Bacteria were grown at 37°C in Luria-Bertani (LB) broth or its modified version65, which contained 0.3% KH2PO4, 0.05% NH4Cl, 0.024% MgSO4, or in modified LB containing 15N labelled nitrogen source and/or 13C labelled carbon source. Growth medium was always added with 100 mg/L of ampicillin. When 500 ml of transformed E. coli cells reached an O.D. of 0.6–0.8 at 600 nm, SUMO-FL-CyPD and SUMO-ΔN-CyPD expression was induced by adding 100 mg/L isopropyl thiogalactopyranoside (IPTG) for 6–7 hours at 25°C. Cells were then harvested via centrifugation at 2000 xg for 30 min at 4°C and stored at −20°C. The bacterial pellet containing SUMO-FL-CyPD or SUMO-ΔN-CyPD was resuspended in 40 ml of lysis solution (200 mM NaCl, 50 mM Tris/HCl pH 8.0, 1 mM DTE, 0.5% Triton, 1 tablet of Protease Inhibitor Cocktail EDTA-free cOmplete™) and sonicated on ice 20 times for 45 sec with 1 min intervals, 2 min every 5 cycles, using a Sonopuls UW 2070 (Bandelin) at 80% power. After centrifugation at 20,000 xg for 30 min at 4°C, the supernatants were incubated with 3 ml of the nickel-containing resin His-Trap FF® (Cytivia) and packed into a column. Bound proteins were eluted with 6 ml of 200 mM NaCl, 50 mM Tris/HCl pH 8.0, 1 mM DTE, 500 mM imidazole. Buffer exchange was performed by dialysis using Pur-A-Lyzer™ tubes (Sigma-Aldrich) with a 10-kDa cut-off and the external solution consisting of 200 mM NaCl, 20 mM NaPi pH 7.0, 1 mM DTE at room temperature (RT) overnight. The enzyme ULP1 (Sigma-Aldrich) was then added at a ratio of 1 U/mg protein and allowed to incubate for 24 h at RT. The enzymatic products were then incubated with the nickel-containing resin His-Trap FF® (Cytivia), as above. The flow-through, containing FL-CyPD or ΔN-CyPD freed of the SUMO moiety, was collected, diluted 8–10 times with FPLC buffer A (20 mM NaPi pH 7.0, 2 mM DTE) to reduce the NaCl concentration and loaded into a primed 1 ml HiTRAP SP-HP column (Cytivia) connected to an FPLC system (ÄKTA-Start, Cytivia). After sample loading, the column was washed with 5 ml of FPLC buffer A and eluted with a 0–20% gradient of FPLC buffer B (2 M NaCl, 20 mM NaPi pH 7.0, 2 mM DTE) in 1 ml-fractions. The CyPD-containing fractions were dialysed 6 times for 30 min against 500 ml of milliQ water and then against 1 L of 0.1 mM DTE overnight at RT. Then, protein was aliquoted and lyophilized in Lio 5 P (5 Pascal). Up to 10 mg of recombinant CyPDs could be obtained from 500 ml of bacterial culture.
Preparation of submitochondrial particles and incubation with FL-CyPD and ΔN-CyPD
Pig heart mitochondria were isolated in a buffer containing 250 mM sucrose, 10 mM Tris, 0.1 mM EGTA, pH 7.4 by standard differential centrifugation16. Submitochondrial particles (SMP) were prepared by exposure of heart mitochondria to ultrasonic energy in the presence of 15 mM MgCl2 and 1 mM ATP followed by a first centrifugation for 20 min at 18,000 x g and a second centrifugation of the supernatant for 30 min at 100,000 x g, as in ref. 16. The pellet containing SMP was washed once in KCl-based buffer (125 mM KCl, 10 mM Tris/HCl, 20 μM EGTA, pH 7.4) to minimize the amount of endogenous CyPD. The final pellet was suspended in a small volume of KCl-based buffer or sucrose-based buffer (250 mM sucrose, 10 mM Tris/HCl, 20 μM EGTA, pH 7.4) at 1 mg/ml and SMP were incubated with 0.05 - 2 nmol/mg SMP of either recombinant FL-CyPD or ΔN-CyPD. When needed, 2 μM cyclosporin A (CsA) and/or 10 mM KH2PO4 (Pi) was added. Afterwards, SMP were subjected to immunoprecipitation to isolate the OSCP subunit of F-ATP synthase or were used to determine the ATP hydrolysis rate. To immunoprecipitate OSCP, 250 μg SMP were centrifuged 20 min at 100,000 x g to release unbound CyPD. Then, SMP were suspended in IP buffer (150 mM NaCl, 50 mM Tris/HCl pH 8.0, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate) and incubated at 4°C overnight with 20 μl magnetic beads previously conjugated with 1 μg of mouse monoclonal anti-OSCP antibody (ab110276, Abcam). After two washings and protein elution from magnetic beads, Western blot analysis was employed to assess CyPD binding to OSCP.
Incubation of FL-CyPD and ΔN-CyPD with calpain 1
Recombinant FL-CyPD or ΔN-CyPD were incubated with human calpain 1 (ab91019, Abcam) at a 1:5 ratio for up to 15 minutes at 32°C in 50 mM Tris/HCl, 30 mM KCl, 1 mM DTE, 500 µM CaCl2 and then analysed by SDS-PAGE stained by Coomassie or Western blot. The two bands at 19 kDa and 18 kDa were manually excised from SDS-PAGE for mass spectrometry analysis.
Western blots
Cell and mouse tissue extracts and SMP were solubilized in Laemmli gel sample buffer containing 1% β-mercaptoethanol and separated on 15% SDS–PAGE gels. Separated proteins were transferred electrophoretically to PVDF membranes, which were probed in TBS (150 mM NaCl, 100 mM Tris pH 7.6 containing 2.5% Bovine Serum Albumin (BSA) and 0.1% Tween-20 with the following antibodies: rabbit Citrate Synthase (CS) 1:5000 (3H8L26, Invitrogen), mouse OSCP 1:2000 (ab110276, Abcam), mouse Cyclophilin D (CyPD) 1:2000 (ab110324, Abcam), mouse subunit α (ATP5A1) 1:500 (ab14748, Abcam). Immunoreactive bands were detected by enhanced chemiluminescence (Pierce) and analyzed with Quantity One software (Biorad). Alternatively, the signals were detected with an Odyssey CLx scanner (Li-Cor Biosciences) and quantified using ImageStudio software (Li-Cor Biosciences). Normalization was performed using Revert™ 700 Total Protein Stain (LI-COR Biosciences). All original images of Coomassie stained gels and immunoblots are shown in Supplementary Fig. 11.
Mass spectroscopy
Cell, mouse tissue extracts and recombinant proteins were analysed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Protein identification via MS was based on the following steps: (i) protein trypsin digestion, which specifically cleaves at the carboxyl side of arginine and lysine residues; (ii) LC-MS/MS separation and sequencing; (iii) Mascot software analysis for the protein identification via alignment of sequenced peptides against a database of known protein sequences. After SDS-PAGE separation and staining by colloidal Coomassie, the gel was incubated with milliQ water overnight under constant shaking at room temperature. The bands of interest were excised, destained by 3 min incubation with acetonitrile for three times and then left to dry on air. Protein bands were treated with 10 mM dithiothreitol (DTT) in 50 mM NH4HCO3 (AMBIC) pH 8.0, for 20 min at 37°C to reduce cysteine residues, then with 55 mM iodoacetamide (IAM) for the alkylation reaction. This latter was carried out in the dark at 37°C for 20 minutes. Excess reagent was finally removed by adding and discarding acetonitrile (ACN)/50 mM AMBIC sequentially for 3 times, and one more time with ACN only. The dehydrated gel bands were treated with 100 ng trypsin solution in 50 mM AMBIC. Finally, 50 mM AMBIC was added to cover the gel bands and samples were placed overnight at 37°C in a Thermoshaker. At the end of hydrolysis, peptide mixtures were collected in fresh tubes. The remaining gel pieces were treated with acetonitrile twice, in order to extract any peptides still present in the gel. The obtained mixtures were dried by a Speed-Vac system. Each peptide mixture was resuspended in 0.1% TFA in water and analyzed on a Thermo Scientific™ Q-Exactive Plus Orbitrap mass spectrometer connected to an Ultimate 3000 nanoLC system. Data were processed by Mascot software (Matrix Science, London, UK) using Swiss-Prot database.
NMR spectroscopy
NMR spectra were recorded at 700 MHz on a Bruker AVANCE NEO and at 500 MHz on a Bruker Advance, processed on Topspin v. 4.0.8 (Bruker) and analysed on NMRFAM-Sparky66 and/or with home-made scripts in R Statistical Software (v.3.4.4, R Core Team 2021). NMR Protein samples of about 350 or 550 μM 15N-13C FL-CyPD or 15N-13C ΔN-CyPD, respectively, were suspended in 20 mM NaPi pH 7.0 10% D2O, except were otherwise specified. TSP 50 μM was added as reference67. Protein backbone chemical shift assignment was performed with a set of 3D [1H, 13C, 15N] NMR experiments, i.e., HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB. All spectra were collected at 298 K except for additional 2D [1H, 15N] HSQC and 3D [1H, 13C, 15N] HNCA experiment acquired at 283 K and 310 K. The backbone resonance assignments together with Cβ chemical shifts were then used to predict the secondary structure elements of FL-CyPD and ΔN-CyPD employing the software TALOS-N39. Temperature coefficients of FL-CyPD and ΔN-CyPD have been derived by collecting 2D [1H, 15N] HSQC spectra in a temperature ramp from 283 K to 316 K with 3 K steps. 3D [1H, 13C, 15N] HNCA spectra were collected at 298 K, 283 K and 310 K to confidently assign peaks overlapping across the temperature ramp. To quantify dynamics in the ps-ns timescale, steady-state 15N {1H} NOE experiments were conducted for both FL-CyPD and ΔN-CyPD. The heteronuclear NOE of each residue was calculated as the ratio between the signal intensity of “NOE” (saturated) and the “noNOE” (non-saturated) spectra. The error on the hetNOE was computed as , were is the signal-to-noise ratio, as described in ref. 68. CsA binding to FL-CyPD and ΔN-CyPD was determined by chemical shift perturbation. 15N-13C FL-CyPD or 15N-13C ΔN-CyPD were suspended in 20 mM NaPi pH 7.0 10% D2O, 150 mM KCl, with 50 μM TSP added as a reference, in the presence or absence of CsA 1:1. 2D [1H, 15N] HSQC and 3D [1H, 13C, 15N] HNCA spectra were recorded. Chemical shift perturbation Δδ, was calculated for each residue as follows69:
where δX is the resonance of nucleus, while “apo” and “olo” refer to the chemical shift of nucleus X in the absence and presence of CsA, respectively. The criterium adopted to identify the most perturbed residues is to consider only those with , where and indicate the mean and the standard deviation, respectively. Specifically, is computed iteratively by excluding at each iteration the outliers, i.e., the most affected residues. The effect of the ionic strength on FL-CyPD and ΔN-CyPD was studied by collecting 2D [1H, 15N] HSQC spectra at 290 K of either 15N-13C FL-CyPD or 15N-13C ΔN-CyPD, respectively, suspended in 20 mM NaPi pH 7.0 10% D2O, 0–150 mM KCl, with 50 μM TSP added as a reference. Chemical shift perturbation between 0 mM and 150 mM KCl spectra was calculated as described above.
Surface plasmon resonance (SPR)
Mature human OSCP was fused with Myc-tagged SUMO at N -term and with β10 GFP at the C-term. The chimeric protein was produced in E. coli and purified as described28. SPR analyses were performed on a Biacore X-100 dual flow-cell instrument from GE Healthcare (Chicago, IL, USA) in 10 mM Tris-HCl, pH 7.4, containing 125 mM KCl and 10 mM potassium phosphate. Anti-Myc antibody was covalently immobilized via amine-coupling reaction on a CM5 sensor chip. Capture reaction was carried out by injecting a solution of OSCP (12 μM) onto the sensor chip for 180 s at a flow rate of 30 μl/min. Saturation of the antibody-derivatised sensor with OSCP chip was established by a marked increase of the SPR signal, expressed as response units (RU), which remained constant in the time-scale of SPR analysis. Binding of CypD to “captured” OSCP was determined in the single cycle mode, by injecting increasing CyPD concentrations (0-20 μM) in the mobile phase, with a contact time of 120 s at a flow rate of 30 μl/min, followed by passive dissociation of CyPD for 180 s. The data were subtracted for the corresponding base-line curves, obtained in a blank experiment on the reference flow cell and accounting for nonspecific binding, i.e. typically less than 2% of RUmax. After each set of SPR measurements, antibody-immobilized sensor chip was regenerated by injecting a 0.5% (w/v) solution of SDS for 30 s, to wash away OSCP-CyPD complex, and then loaded with OSCP for the capture reaction. The kinetic data were analyzed using the BIAevaluation software, using a single-site binding model to extract kon, koff and Kd values.
Biochemical assays
Protein concentration was determined with the Lowry70 or Bradford71 method for the particulate or soluble enzymes, respectively. Protein concentration of RIPA extracts was determined using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). The absorbance at 280 nm was used to quantify purified CyPD. ATP hydrolysis was followed spectrophotometrically by adding 50 μg SMP to the cuvette and recording NADH oxidation at 340 nm in the presence of 2 mM ATP and an ATP-regenerating system as described72. PPIase activity was determined according to the unfolding/refolding of RNase T173. Specifically, 50 μM RNase T1 (ThermoFisher) was unfolded in 20 mM Tris/HCl pH 8.0 and 8 M urea for 2 h at room temperature. Unfolded RNase T1 at a final concentration of 1.25 μM was then incubated with 25 nM FL-CyPD or ΔN-CyPD in 20 mM Tris/HCl pH 8.0 to start the refolding of RNase T1, or in 20 mM Tris/HCl pH 8.0 to monitor the non-enzymatic refolding. CsA sensitivity was assessed after pre-incubation of CyPD with 10-fold molar excess of CsA on ice. RNase T1 refolding was followed by the intrinsic fluorescence of tryptophan (excitation wavelength of 268 nm, slit 10 nm; emission wavelength of 325 nm, slit 5 nm) using a Cary Eclipse Fluorescence Spectrometer (Agilent). Temperature was kept at 10°C. PPIase activity was expressed as a ΔF/min from the linear part of the obtained curves.
Statistics and reproducibility
Data are presented as mean ± standard deviation (SD). Data groups were analysed with a two-way analysis of variance (ANOVA) followed by Tukey post hoc test or Kruskal-Wallis test followed by Dunn post hoc analysis. The decision on which test to use was made after checking the normality and homogeneity of variance of the samples. One sample t-test was performed when comparing the mean of a single sample to a known value or control. Results with a p value lower than 0.05 were considered significant. Each experiment was repeated at least three times. Analyses were performed with either R statistical software v.3.4.4 or GraphPad v.8.0.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
This work is in partial fulfillment of the requirements for a Ph.D. to Gabriele Coluccino. Supported by PRIN (2017LHFW42 to P.B. and G.L.) and Progetto ricerca collaborativa DM737, Università di Udine (G.L.). The authors thank Dr. Catia Mio (University of Udine) for sequencing, Dr. Giuseppe Cannino (University of Padova) and Dr. Marco Zancani (University of Udine) for their helpful discussion. The authors also thank “Centro Grandi Strumenti – Università di Pavia” for providing the access to the NMR facility. All of us are dismayed that our colleague Dr. Alessandro Negro passed away during the writing of the manuscript.
Author contributions
A.C., G.L., P.B. conceived the work. A.F., A.N., C.B., Cl.G., D.C., E.Z, G.C., L.A. M.Co., M.C.M., V.P.M. carried out experiments. G.L. and P.B., acquired funds. A.C., G.L. supervised the work. M.Ca., F.C., A.R., C.G., V. de F. contributed to study design and implementation. A.C., G.C., G.L., C.B., P.B. wrote the initial draft. All authors have reviewed and edited the manuscript.
Peer review
Peer review information
Communications Biology thanks George Porter and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Janesh Kumar and Kaliya Georgieva.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The source data underlying all graphs and charts present in the manuscript are provided in Supplementary Data 2. Chemical shift assignments of FL-CyPD and ΔN-CyPD have been deposited in the Biological Magnetic Resonance Bank (BMRB, https://bmrb.io/) with accession codes 52665 and 52661, respectively. The raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD057203. All data are also available upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alessandra Corazza, Email: alessandra.corazza@uniud.it.
Giovanna Lippe, Email: giovanna.lippe@uniud.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07172-8.
References
- 1.Daum, S. et al. Isoform-specific inhibition of cyclophilins. Biochemistry48, 6268–6277 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang, P. & Heitman, J. The cyclophilins. Genome Biol.6, 226 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ke, H. M., Zydowsky, L. D., Liu, J. & Walsh, C. T. Crystal structure of recombinant human T-cell cyclophilin A at 2.5 A resolution. Proc. Natl Acad. Sci. USA.88, 9483–9487 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kajitani, K. et al. Crystal structure of human cyclophilin D in complex with its inhibitor, cyclosporin A at 0.96-Å resolution. Proteins70, 1635–1639 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Davis, T. L. et al. Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol.8, e1000439 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlatter, D. et al. Crystal engineering yields crystals of cyclophilin D diffracting to 1.7 Å resolution. Acta Crystallogr D. Biol. Crystallogr61, 513–519 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Porter, G. A. & Beutner, G. Cyclophilin D, somehow a master regulator of mitochondrial function. Biomolecules8, 176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rein, T. Peptidylprolylisomerases, protein folders, or scaffolders? The Example of FKBP51 and FKBP52. Bioessays42, e1900250 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Baines, C. P. et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature434, 658–662 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Basso, E. et al. Properties of the permeability transition pore in mitochondria devoid of cyclophilin D. J. Biol. Chem.280, 18558–18561 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Beutner, G. et al. Coordinated metabolic responses to cyclophilin D deletion in the developing heart. iScience27, 109157 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernardi, P., Rasola, A., Forte, M. & Lippe, G. The mitochondrial permeability transition pore: channel formation by F-ATP synthase, integration in signal transduction, and role in pathophysiology. Physiol. Rev.95, 1111–1155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernardi, P., Carraro, M. & Lippe, G. The mitochondrial permeability transition: Recent progress and open questions. FEBS J.289, 7051–7074 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa, T. et al. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature434, 652–658 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Rasola, A. & Bernardi, P. Mitochondrial permeability transition in Ca(2+)-dependent apoptosis and necrosis. Cell Calcium50, 222–233 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Giorgio, V. et al. Cyclophilin D modulates mitochondrial F0F1-ATP synthase by interacting with the lateral stalk of the complex. J. Biol. Chem.284, 33982–33988 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giorgio, V. et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc. Natl Acad. Sci. USA110, 5887–5892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrer, A. et al. Defining the molecular mechanisms of the mitochondrial permeability transition through genetic manipulation of F-ATP synthase. Nat. Commun.12, 4835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urbani, A. et al. Purified F-ATP synthase forms a Ca2+-dependent high-conductance channel matching the mitochondrial permeability transition pore. Nat. Commun.10, 4341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mnatsakanyan, N. et al. A mitochondrial megachannel resides in monomeric F1FO ATP synthase. Nat. Commun.10, 5823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giorgio, V. et al. Ca2+ binding to F-ATP synthase β subunit triggers the mitochondrial permeability transition. EMBO Rep.18, 1065–1076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antoniel, M. et al. The unique histidine in OSCP subunit of F‐ATP synthase mediates inhibition of the permeability transition pore by acidic pH. EMBO Rep.19, 257–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo, L. et al. Arg-8 of yeast subunit e contributes to the stability of F-ATP synthase dimers and to the generation of the full-conductance mitochondrial megachannel. J. Biol. Chem.294, 10987–10997 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, L. et al. Arginine 107 of yeast ATP synthase subunit g mediates sensitivity of the mitochondrial permeability transition to phenylglyoxal. J. Biol. Chem.293, 14632–14645 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carraro, M. et al. The unique cysteine of F-ATP synthase OSCP subunit participates in modulation of the permeability transition pore. Cell Rep.32, 108095 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Morciano, G. et al. A naturally occurring mutation in ATP synthase subunit c is associated with increased damage following hypoxia/reoxygenation in STEMI patients. Cell Rep.35, 108983 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Pinke, G., Zhou, L. & Sazanov, L. A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol.27, 1077–1085 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Cannino, G. et al. The mitochondrial chaperone TRAP1 regulates F-ATP synthase channel formation. Cell Death Differ.29, 2335–2346 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurst, S. et al. Phosphorylation of cyclophilin D at serine 191 regulates mitochondrial permeability transition pore opening and cell death after ischemia-reperfusion. Cell Death Dis.11, 661 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hafner, A. V. et al. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging2, 914–923 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks, R. J. et al. Cyclophilin D-mediated regulation of the permeability transition pore is altered in mice lacking the mitochondrial calcium uniporter. Cardiovasc. Res.115, 385–394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jara, C., Cerpa, W., Tapia-Rojas, C. & Quintanilla, R. A. Tau deletion prevents cognitive impairment and mitochondrial dysfunction age associated by a mechanism dependent on cyclophilin-D. Front Neurosci.14, 586710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coluccino, G., Muraca, V. P., Corazza, A. & Lippe, G. Cyclophilin D in mitochondrial dysfunction: a key player in neurodegeneration? Biomolecules13, 1265 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasola, A. et al. Activation of mitochondrial ERK protects cancer cells from death through inhibition of the permeability transition. Proc. Natl Acad. Sci. USA.107, 726–731 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bochaton, T. et al. Inhibition of myocardial reperfusion injury by ischemic postconditioning requires sirtuin 3-mediated deacetylation of cyclophilin D. J. Mol. Cell Cardiol.84, 61–69 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Castillo, E. C. et al. Mitochondrial Hyperacetylation in the Failing Hearts of Obese Patients Mediated Partly by a Reduction in SIRT3: The Involvement of the Mitochondrial Permeability Transition Pore. Cell Physiol. Biochem53, 465–479 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Valasani, K. R. et al. High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules. Acta Crystallogr F. Struct. Biol. Commun.70, 717–722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holliday, M. J. et al. Structure and dynamics of GeoCyp: a thermophilic cyclophilin with a novel substrate binding mechanism that functions efficiently at low temperatures. Biochemistry54, 3207–3217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, Y. & Bax, A. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol.1260, 17–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomasetig, L., Di Pancrazio, F., Harris, D. A., Mavelli, I. & Lippe, G. Dimerization of F0F1ATP synthase from bovine heart is independent from the binding of the inhibitor protein IF1. Biochim Biophys. Acta1556, 133–141 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Connern, C. P. & Halestrap, A. P. Purification and N -terminal sequencing of peptidyl-prolyl cis-trans -isomerase from rat liver mitochondrial matrix reveals the existence of a distinct mitochondrial cyclophilin. Biochemical J.284, 381–385 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Pancrazio, F. et al. In vitro and in vivo studies of F(0)F(1)ATP synthase regulation by inhibitor protein IF(1) in goat heart. Biochim Biophys. Acta1659, 52–62 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Kukic, P., O’Meara, F., Hewage, C. & Erik Nielsen, J. Coupled effect of salt and pH on proteins probed with NMR spectroscopy. Chem. Phys. Lett.579, 114–121 (2013). [Google Scholar]
- 44.Beutner, G., Alanzalon, R. E. & Porter, G. A. Cyclophilin D regulates the dynamic assembly of mitochondrial ATP synthase into synthasomes. Sci. Rep.7, 14488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amanakis, G. et al. Cysteine 202 of cyclophilin D is a site of multiple post-translational modifications and plays a role in cardioprotection. Cardiovasc. Res.117, 212–223 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, Z. et al. GPS-CCD: a novel computational program for the prediction of calpain cleavage sites. PLOS ONE6, e19001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eisenmesser, E. Z., Bosco, D. A., Akke, M. & Kern, D. Enzyme dynamics during catalysis. Science295, 1520–1523 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Nicolli, A., Basso, E., Petronilli, V., Wenger, R. M. & Bernardi, P. Interactions of cyclophilin with the mitochondrial inner membrane and regulation of the permeability transition pore, a cyclosporin A-sensitive channel. J. Biol. Chem.271, 2185–2192 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Kumutima, J., Yao, X.-Q. & Hamelberg, D. Post-translational modifications of cyclophilin D fine-tune its conformational dynamics and activity: implications for its mitochondrial function. J. Phys. Chem. B126, 10844–10853 (2022). [DOI] [PubMed] [Google Scholar]
- 50.J. C. Ghosh, et al. Adaptive mitochondrial reprogramming and resistance to PI3K Therapy. JNCI.107 (2015). [DOI] [PMC free article] [PubMed]
- 51.Bernardi, P. et al. Identity, structure, and function of the mitochondrial permeability transition pore: controversies, consensus, recent advances, and future directions. Cell Death Differ.30, 1869–1885 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen, Q. & Lesnefsky, E. J. Heart mitochondria and calpain 1: Location, function, and targets. Biochim Biophys. Acta1852, 2372–2378 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Zhang, M., Wang, G. & Peng, T. Calpain-mediated mitochondrial damage: an emerging mechanism contributing to cardiac disease. Cells10, 2024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ciscato, F. et al. Hexokinase 2 displacement from mitochondria-associated membranes prompts Ca2+ -dependent death of cancer cells. EMBO Rep.21, e49117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao, T. et al. Increased calpain-1 in mitochondria induces dilated heart failure in mice: role of mitochondrial superoxide anion. Basic Res Cardiol.114, 17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shintani-Ishida, K. & Yoshida, K.-I. Mitochondrial m-calpain opens the mitochondrial permeability transition pore in ischemia-reperfusion. Int J. Cardiol.197, 26–32 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Frigo, E., Tommasin, L., Lippe, G., Carraro, M. & Bernardi, P. The haves and have-nots: the mitochondrial permeability transition pore across species. Cells12, 1409 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Domenis, R., Bisetto, E., Rossi, D., Comelli, M. & Mavelli, I. Glucose-modulated mitochondria adaptation in tumor cells: a focus on ATP synthase and inhibitor Factor 1. Int J. Mol. Sci.13, 1933–1950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thumuluri, V., Almagro Armenteros, J. J., Johansen, A. R., Nielsen, H. & Winther, O. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res50, W228–W234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almagro Armenteros, J. J. et al. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance2, e201900429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukasawa, Y. et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell Proteom.14, 1113–1126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butt, T. R., Edavettal, S. C., Hall, J. P. & Mattern, M. R. SUMO fusion technology for difficult-to-express proteins. Protein Expr. Purif.43, 1–9 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danquah, M. K. & Forde, G. M. Growth medium selection and its economic impact on plasmid DNA production. J. Biosci. Bioeng.104, 490–497 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wishart, D. S. et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR6, 135–140 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Kharchenko, V., Nowakowski, M., Jaremko, M., Ejchart, A. & Jaremko Dynamic 15N{1H} NOE measurements: a tool for studying protein dynamics. J. Biomol. NMR74, 707–716 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mulder, F. A., Schipper, D., Bott, R. & Boelens, R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol.292, 111–123 (1999). [DOI] [PubMed] [Google Scholar]
- 70.Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem.193, 265–275 (1951). [PubMed] [Google Scholar]
- 71.Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- 72.Das, A. M. & Harris, D. A. Regulation of the mitochondrial ATP synthase in intact rat cardiomyocytes. Biochem J.266, 355–361 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zemanova, L. et al. RNase T1 refolding assay for determining mitochondrial cyclophilin d activity: a novel in vitro method applicable in drug research and discovery. Biochemistry59, 1680–1687 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The source data underlying all graphs and charts present in the manuscript are provided in Supplementary Data 2. Chemical shift assignments of FL-CyPD and ΔN-CyPD have been deposited in the Biological Magnetic Resonance Bank (BMRB, https://bmrb.io/) with accession codes 52665 and 52661, respectively. The raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD057203. All data are also available upon request.