Abstract
Exposure to saline environments significantly hampers the growth and productivity of oil crops, harmfully affecting their nutritional quality and suitability for biofuel production. This presents a critical challenge, as understanding salt tolerance mechanisms in crops is key to improving their performance in coastal and high-salinity regions. Our content might be read more properly: This review assembles current knowledge on protein-level changes related to salinity resistance in oil crops. From an extensive analysis of proteomic research, featured here are key genes and cellular pathways which react to salt stress. The literature evinces that cutting-edge proteomic approaches − such as 2D-DIGE, IF-MS/MS, and iTRAQ − have been required to reveal protein expression patterns in oil crops under salt conditions. These studies consistently uncover dramatic shifts in protein abundance associated with important physiological activities including antioxidant defence, stress-related signalling pathways, ion homeostasis, and osmotic regulation. Notably, proteins like ion channels (SOS1, NHX), osmolytes (proline, glycine betaine), antioxidant enzymes (SOD, CAT), and stress-related proteins (HSPs, LEA) play central roles in maintaining cellular balance and reducing oxidative stress. These findings underline the complex regulatory networks that govern oil crop salt tolerance. The application of this proteomic information can inform breeding and genetic engineering strategies to enhance salt resistance. Future research should aim to integrate multiple omics data to gain a comprehensive view of salinity responses and identify potential markers for crop improvement.
Keywords: Oil Crops, Proteomics, Salinity Stress Tolerance, Stress-Responsive Proteins
1. Introduction
Oil crops, such as soybeans, palm, and rapeseed, are invaluable not only for biodiesel production but also for their nutritional contributions, as they provide essential proteins and minerals crucial for both human and animal diets. They are a cornerstone of the global biofuel and food industries. The current global market value of these crops is estimated at approximately USD 265 billion.1 This reflects their immense economic significance, with oilseeds processed into vegetable oils and oil meals, the latter being a critical protein source for livestock. Furthermore, oilseeds' role in biodiesel production highlights their importance in the renewable energy sector, helping meet the rising global demand for sustainable energy sources. Oil crops also contain vitamins that help maintain good health, and they contain high amounts of dietary fibre beneficial to the digestive system. After cereals, they represent the second most important component of the agricultural economy within the field crop sector. However, these crops face threats from various biotic and abiotic stresses that limit photosynthetic rates and overall productivity. Among the most harmful are Fusarium spp., fungi that cause a range of diseases, leading to substantial economic losses globally.2
Abiotic stresses, particularly salinity are one the most significant problems for plant production as global food demand increases. At the whole plant level, these stressors result in slow early growth and reduced total productivity of a plant. A significant reduction in growth, oxidative damage and decreased photosynthesis at the plant level are major phenotypes for yet unchartered mechanisms of silencing.3 As it is, the diet of an ever-growing world population faces challenges made worse by global effects such as salinization that are degrading arable land.4 Traditional breeding methods have struggled to improve crop yield under saline conditions because salinity tolerance is controlled by multiple genes, each contributing to different physiological, biochemical, and molecular responses. Salinity impacts various processes, such as water uptake, ion homeostasis, and metabolic pathways, which makes it difficult to isolate and breed for just one trait. This complexity means that simply selecting crops based on one or two genes does not significantly improve overall salt tolerance. As a result, researchers are turning to more advanced techniques, such as molecular breeding, gene editing (e.g., CRISPR), and biotechnological approaches, to target multiple genes and pathways simultaneously for better results.5 These three salinity effects reduce water potential, cause ion imbalance or disrupt ion homeostasis, and cause toxicity. Early growth is slowed and plant productivity is limited as a result of the altered water status. The negative effect manifests itself at the plant level as plant mortality or a decrease in productivity. Salt stress disturbs all critical processes, such as plant yield, oxidative damage, photosynthesis, growth, water balance, and nutritional imbalance. Providing enough food for the expanding population is directly hampered by the shortage of arable land caused by salinization. Growing species that cannot just tolerate high salinity levels but also sustain optimal production levels in the presence of salt is thus necessary. However, attempts to raise crop yield under the impact of salt have typically been unsuccessful due to the multigenic and quantitative character of halotolerance. This has motivated researchers to combine both conventional breeding techniques and cutting-edge biotechnological strategies to increase crop adaptability to salt stress. Conventional methods include selective breeding and cross-breeding, where salt-tolerant varieties are developed by gradually selecting plants with better traits. On the other hand, cutting-edge approaches involve molecular breeding, genetic engineering (e.g., CRISPR/Cas9), and the use of omics technologies (such as genomics and proteomics) to identify and manipulate genes involved in stress tolerance. These combined methods enable a more comprehensive approach to improving crop resilience to saline conditions. The observable changes in plants under stress are closely linked to shifts in protein accumulation. Salt-tolerant canola breeds, for instance, do not experience significant reductions in leaf weight and plant height, which are typically associated with decreased chlorophyll concentrations under increasing salinity. These cultivars accumulate proline in their roots, a response linked to improved salt tolerance.6 We can learn a lot about the mechanisms behind plant tolerance or resistance from studies of in what form plants react to protein-level stress. Salt-tolerant canola breeds do not experience this loss in leaf weight and plant height, which is caused by the decrease in chlorophyll concentration with increasing salinity. Canola cultivars that are resistant to salt accumulate proline in their roots, whereas salt-sensitive cultivars accumulate it in their shoots.7 Improving salinity and temperature tolerance is one of the main targets for future plant breeding. For example, organisms that can survive well under the extreme environment of high temperatures have evolved a special mechanism for protection and stress adaptation by producing heat shock proteins (HSPs). Previous research on HSPs has also found that the presence of specific proteins, such as sti1 in response to salinity was associated with a tougher plant.8 Salt-stressed plants change their gene expression drastically in order to be able to live under challenging conditions. Plants experiencing salt stress have changes in gene expression on a large scale (transcriptome, proteome, and/or metabolome) later on. Stress proteome research has shown that stress conditions induced aggregation patterns of proteins differ drastically, which provided new information on stress-responsive mechanisms in plants.9 Of all environmental threat variables, the most dangerous element limiting agricultural output is salinity. Salinity tolerance is one of the measurable traits among species, and this trait is controlled by a number of genes in plants.10 Numerous genes involved in transcription control, signal transduction, ion transport, and metabolic balance have been found and characterized in the last ten years that affect salinity resistance in plants, including rice.11 Importantly, salinity stress not only induces the expression of certain genes but also represses others, depending on the plant's adaptive responses.
Studying molecular processes including plant development, the framework of its genome, and interactions with its environment are the goals of plant molecular biology.12 These in-depth multidimensional studies call for extensive testing tying together the entire genetic, functional, and structural elements. 'Omics' techniques are frequently employed in several fields of crop plant study, including rice. As technology developed over the past ten years, these methods quickly improved. The next part explains how 'omics-focused' methods have aided in identifying and analysing the mechanism underlying rice's tolerance to salinity as well as in creating some salt-tolerant germplasms.13 The most concise and straightforward method of explaining the function of the gene related to a specific protein is through its study. However, it should be noted that an organism's proteome and genome do not necessarily interact directly.14 Thus, the study of genomes is just as important as research conducted at the proteome levels. The study of proteins provides a framework for analysing complicated biological processes that involve a large number of proteins along with a matrix of interactions between those proteins. The principal method for revealing the molecular mechanisms involved in interactions between plants and various stressors, including salt stress, is proteomics.15 Stress from salt causes the expression of certain genes, which is ultimately reflected in the protein profile. Proteomic research in several rice tissue components has expanded the role of salinity. It has been demonstrated that the focus of the current rice proteome investigations has been on the identification of polypeptides based on their availability when subjected to various stressors.16 It was discovered that the complicated physiological reaction data from the proteomics study changed depending on how severe the stress was, which complicated the data evaluation and analysis integration. Post-translational modification (PTM) may now be used as a replacement to study the activities of stress signalling.17 It has been possible to recognize PTM in plants using a variety of methods. Several gel-free techniques for proteome differential analysis have also been discovered. Examples include: Using a multidimensional protein identification method without band broadening for chromatographic recognition, it is possible to successfully identify individual protein components,18 and affinity tags with isotope-coded.19 These strategies are seen as targeted tools for identifying changes in proteins caused by mass difference means across various proteomes.
Proteome analysis is among the methods most frequently employed for investigating how plants react to environmental stresses is important because it allows for easy protein extraction, generates highly reproducible two-dimensional electrophoresis gels, and provides high sensitivity in protein sequencing using mass spectrometry (MS).20 Proteomic studies on the expression of proteins under salt stress have been performed in crops such as rice,21 potato,22 soybean,23 and Chinese foxtail millet,24 this may give a more accurate assessment of cellular activities when salt stress is present. Early studies have shown that these proteins,25 oxidative stress-scavenging enzymes [for example, reactive oxygen species (ROS) up-feathered enzyme,26 and defence proteins27; may be essential molecular markers of increased salt tolerance. By employing the keywords “proteomics,” “salinity stress,” “oil crops,” and “tolerance” in databases such as PUBMED, Scopus, and Web of Science (WoS), and publishers like Taylor and Francis Online, Wiley, and Elsevier recent literature from the past decade was reviewed to advance the development of strategies for enhancing salinity tolerance in oil crops.
1.1. Salinity stress: Effects and mechanisms
Alterations-related salinity is induced in signalling pathways; signalling of growth hormone, biosensors and ion channels, besides the expression of ion transporter genes in crop plants.28, 29, 30, 31 Photosynthesis plays a crucial function in producing adenosine triphosphate (ATP), essential for CO2 fixation into sugars. However, various abiotic stresses disrupt photosynthetic processes.32, 33 For example, the stresses can induce lumen membrane damage and interfere with electron transport chain (ETC) / respiratory complexes; they disrupt enzymatic activity and protein synthesis pathways as well as breakdown of the Calvin cycle. Such perturbations are considered to lead to a disruption in ATP synthesis34 and then shift towards hydrolysis, a response that might eventually lead to ion deficiency because of both suppression and synthesis.35
Recent studies have observed a decrease in cucumber leaf water content following exposure to saline solutions. This decline was attributed to elevated Na levels and reduced K, which hinder photosynthesis due to Na's competitive uptake for ions. Gas exchange decreases with the increase in the concentration of Na and Chloride ions, including transpiration, light-dependent sugar photosynthesis output, and stomatal conductivity due to highly saturated conditions.36 Similarly, pepper plants exhibit reduced photosynthesis when subjected to a complete root system to partial exposure.37 Stomatal conductance, crucial for photosynthetic efficiency and biomass production, is negatively affected by the salt challenge.38 Salinity-induced reductions in the maximum quantum efficiency of photosystem II (PSII) have been documented.39 Besides, the high salt concentration also disrupts photosynthesis by non-stomatal restrictions involving inhibition of various enzymes present in the photosynthetic pathway and decreased chlorophyll a, b, or total carotenoids.40 Chlorophyll degradation under salt stress is linked to chlorophyllase activity and the absolute concentrations of chloride and Na.41 Additionally, carotenoid content decreases under salinity, while anthocyanin levels typically rise. Plant-derived antioxidants, including secondary metabolites like phenolic compounds, play crucial roles in stress adaptation. Tocopherol stabilizes membrane integrity and acts as a signalling molecule,42 while ascorbic acid scavenges ROS.43 It was shown that the abundance of carotenoids is decreased by ROS, and this carotenoid-rich bee pollen can be used as a shield against them.44 Supplementation of Putrescine improves salt tolerance in Indian mustard by enhancing carotenoid and glutathione contents while maintaining higher membrane stability.45 Paramount physiological responses indirectly associated with salinity stress include evaporation and plant transportation, which can be ameliorated by beneficial bacteria.46 In addition, the increased salinity induced by NaCl treatment promotes spermine and spermidine accumulation to favour salt tolerance induction but also enables better fruit quality.47, 48
1.2. Ionic toxicity: The salt-specific effect on plant cells
Under saline conditions, salinity stress induces ionic imbalances leading to disruptions in cellular respiration, carbon assimilation, and root system organization due to the inhibition of the uptake of essential minerals by plants. Salinity strain produces an imbalance of electrolytes fundamentally affecting different plant aspects due to disruption in various cellular metabolic pathways, light-dependent reactions, and root architecture by reducing the uptake of essential mineral elements under saline conditions.49 To cope with salinity stress, plants have developed elaborate strategies to regulate cellular ion distribution, which involves controlling the uptake and compartmentalization of ions, but these efforts are thwarted by a variety of factors. Plants have evolved mechanisms for maintaining cytoplasmic ion homeostasis by controlling both ionic uptake and compartmentalization to survive salinity stress.50 For this purpose, ion homeostasis is an important and very dynamic process, which has to establish a gradient between the milieu of the organism in order to allow the uptake of essential ions while enabling detoxification by excreting toxic ones. Maintaining homeostasis is a basic and active process in plants where they absorb needed ions but excrete toxic ones at a high energetic cost.51, 52, 53, 54 In roots, Na movement from soil to shoots occurs first following an apoplastic (space between cells) pathway, later using the symplastic mechanism throughout an epidermal layer, and is loaded into xylem tracheids. Transport of sodium from the plant base to the apex follows an extracellular route, changing to the symplast in the root epidermis, and finally xylem tracheids before reaching shoots and leaves where symptoms are most visible.55 In the cytoplasm, plants must deal with sodium and its toxic effects by keeping Na+ out of their living tissues, as well as exporting Na+ or storing it in vacuoles. Plants have developed strategies to protect the cytoplasm from the toxic effects of Na+ by (i) decreasing net influx and increasing efflux out of cells; or (ii) compartmentalization in vacuoles.56
1.3. Strategies plants employ to combat salinity stress
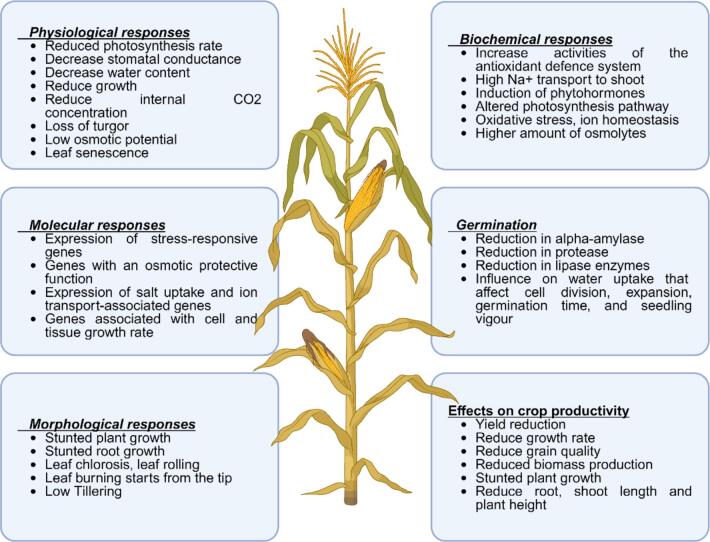
Achieving salt tolerance involves controlling cytoplasmic ion concentrations, osmotic adjustment, and enhanced antioxidant metabolism, such as increased ROS scavenging capability Plants have developed adaptable mechanisms to address salinity stress through alterations at the molecular physiological, biochemical and morphological levels.9, 57 Moreover, many genes and transcription factors (TFs) specific to salinity stress are upregulated upon exposure, facilitating plant adaptation to saline environments (Fig. 1). Many genes and TFs specific to salinity stress are upregulated upon exposure, facilitating plant adaptation to saline environments.
Fig. 1.
Illustrates plant reaction to salt-related stress, covering physiological, biochemical, molecular, and morphological aspects, along with effects on germination and crop productivity. Physiological responses include reduced photosynthesis, water content, growth, and leaf senescence. Biochemical responses involve increased antioxidants, Na+ transport, phytohormone induction, and oxidative stress. Molecular responses include stress-responsive genes, salt uptake, ion transport, and cell/tissue growth. Germination is affected by reduced alpha-amylase, protease, and water uptake. Morphological responses include stunted growth, leaf chlorosis, and low tillering. Crop productivity suffers from yield reduction, lower growth rate, grain quality, biomass, and root/shoot length. These complex adaptations affect agricultural productivity.
Plants have developed adaptable strategies to address salinity distress through molecular, biochemical, structural, and physiological alterations. When exposed to salt stress, they can mitigate damage either by reducing the concentration of salt ions to a tolerable level or by enhancing their resistance to salt stress.58 Plants employ four primary strategies to avoid salt-induced injury: excess salinity, plants use techniques including sodium expulsion, saline diffusion, ion accumulation, and salt blocking. Salt secretion, typically seen in halophytes, involves maintaining ion balance by excreting excess salt through specialized salt glands. Plants dilute internal salt levels with abundant water uptake or cell expansion. Salt accumulation is characterized by the sequestration of excess salt into the vacuoles to protect other cellular components. Salt exclusion involves plants using specialized structures to keep salt out of tissues. To avoid salt-induced injury, plants developed four major strategies: 1) salt excretion, 2) salt reduction, 3) salt accumulation, and 4) salt exclusion. For example, the excretion of salt in halophytes is a case where osmoregulatory requirements are met by maintaining ionic homeostasis through secreting excess salts via specialized glands for Na+. Salt dilution: plants absorb a large quantity of water, or cell size expands to dilute the internal salt levels. Salt accumulation defends the other cellular structures from damage by archiving extra salt in vacuoles. Salt exclusion is where plants use special structures that help stop the salt from entering their tissues.59 The genetic makeup of an organism remains largely consistent over time, with transcription being subject to various regulatory mechanisms. However, information regarding regulatory processes or alterations occurring after translation cannot be directly inferred from transcript expression alone. To address this, proteomic analysis, which involves the identification quantification reveals the molecular processes that drive plant answers to environmental constraints and elucidates the molecular mechanisms to recognize, respond, and adapt to these stresses.58, 60 Following translation, proteins undergo diverse modifications that influence their functionality, and some proteins are enzymatically degraded upon completing their tasks. Consequently, comprehensive insights into functional gene expression patterns can only be obtained through proteomic investigations.61 Quantitative assessment of protein levels is crucial for understanding how plants react to specific environmental challenges, including salt stress stressors such as salt stress.62
1.4. Introduction to proteomics and its significance in stress research
Technological advancements in conventional plant-breeding methods have revolutionized approaches to crop improvement. Abiotic and biotic stresses restrict plant growth, leading to reduced crop yields.63 Genomically-encoded proteins are crucial for plant endurance and adaptation to varying environmental conditions.64 Biotechnological applications in plant breeding depend significantly on insights from proteomic research. Proteomics offers unique advantages in assessing PTMs, revealing their functional impact on plant production. Cytoplasmic proteomic findings help elucidate precise cellular responses and interconnections among organelles throughout the plant life cycle and responses to biotic/abiotic stresses. Significant advancements in MS, sample extraction techniques, systematic software, and genome availability for various plant species have enabled large-scale plant proteomic studies, providing an inclusive understanding.65
Examining drought and its implications for global food security is crucial.66 Drought induces oxidative stress by generating excessive ROS, which causes plasma membrane damage and activates multiple stress-signalling pathways, including those mediated by ROS, Ca2+, and hormones. Mian plant action against drought stress involves root growth, turgor loss, and photosynthesis.67 Viable genetic improvement can be accomplished via effective genomic methods and gene-editing techniques like TALENs facilitating the characterisation of stress-related genes identified from potential gene candidates.68 This sophisticated data integration has the potential to reveal regulatory networks and pinpoint key master regulators.
Integrative data analysis is being robotically reinvented alongside advances in technology, even if slowly advancing the biological mindscape of all plant species. In recent years, proteomics has been widely applied in crop plants to better understand their responses and performances in an ever-changing global environment.69 The proteome is highly dynamic in any given cell, tissue, or organ with respect to temporal and spatial changes. In summary, plant protein modifications and interactions/network topology alterations are dramatically different among developmental stages or physiological conditions.70 Proteome analysis has developed substantially, and according to tools, techniques protocols, instrumentations as well bioinformatics applications have been modified in case of continuously emerging new trends.71
The complexity of plant cell proteins, which number in the thousands with vastly differing abundances, poses challenges for comprehensive coverage.72 Detailing the configurational organization73 of a variety of subcellular compartments with proteomics, circumventing traditional organelle purification. This event of subcellular proteomics has the ability to identify compartment-specific responses and cross-compartmental crosstalk observed in plant development, and abiotic stress pathways.74, 75 A comprehensive understanding of molecular-level mechanisms is crucial to grasping stress-related protein regulation in crops holistically. PTMs play central roles in cellular signalling events. PTM crosstalk finely tunes cellular responses to environmental changes,76 regulating protein activity, localization, and interactions across various cellular processes, intricately modulating plant responses to external stimuli.77 PTMs like dephosphorylation, phosphorylation, glycosylation, and ubiquitination regulate abscisic acid (ABA) signalling, aiding crop adaptation to stressful environments.78
1.5. Key proteins and pathways related to salinity tolerance
In response to salinity stress, plants accumulate various proteins. Typically, plant tissues react to salt stress by breaking down proteins related to salt stress.79 It was also found that salt-tolerant cultivars of many plants, including sunflowers express a larger number of proteins than salt-sensitive ones. This indicates that even a small amount of salt removes more proteins from the soluble fraction in cholera seedlings with regard to sunflower plants. Comparing plant species like sunflower and coriander further supports this fact; the level and type, not quantity, are fully responsible for the overall solubility of protein against NaCl stress.80 In the modern days, different types of cutting-edge proteomic techniques such as Tandem Mass Tagging (TMT) and Isobaric Tags for Relative and Absolute Quantitation (iTRAQ), Data-Independent Acquisition (DIA) for total proteome coverage have been adopted in plant research alongside specialized branches within proteomics including phosphoproteomics and glycoproteomics. For instance, single-cell proteomics, spatial proteomics as well degradome profiling. Interactome analysis is useful for mapping out protein–protein interactions, while targeted proteomics methods like selected reaction monitoring (SRM) and parallel reaction monitoring focus specifically on certain proteins. Meanwhile, Stable Isotope Labeling (SILAC) is finding its way into the field allowing for accurate quantification in plants. These strategies are useful for studying plant biology in combination with responses to different environmental stimuli. These methods, combined with bioinformatics tools, assist in identifying proteins specifically associated with salt stress. Proteins that react to salt stress are vital for various functions, including salt-responsive proteins which play crucial roles in various biological processes, including stress signal transduction, transcription regulation, protein metabolism, osmotic balance, ion regulation, ROS management, photosynthesis, carbohydrate and energy metabolism, and the maintenance of the cytoskeleton and cell wall.25 These proteins are found in various plant tissues such as roots, shoots, seedlings and mature leaves (vegetative parts), flowers and seeds (reproductive organs) or other detectable metabolites from plant callus tissue suspension cultures. It also covers the analysis of other subcellular organelles and membrane systems, which will be a future topic to cover in another article as this is outside the scope of current reviews focused on plasma membranes (Fig. 2)..81 The differential expression of salt stress-responsive proteins includes multiple biological functions with different post-transcriptional and post-translational regulation.82 These proteins also act directly to reshape new phenotypic traits that are better able to acclimate against future salt-exposed conditions via their involvement in major metabolic pathways. Therefore, the importance of individual proteins in salt tolerance mechanisms is more than protein abundance alone. Therefore, the importance of particular proteins in salt tolerance mechanisms is more than just protein concentration. Functionally, these proteins can be separated into salt-stress proteins that specifically accumulate under high salinity but not other stress conditions, and stress-associated proteins induced by oxidative or nitrosative stresses. Proteins like osmotin in soybean,83 and osmotic-like proteins in sesame,84 contribute to salt stress osmoregulation. In addition, several other enzyme complexes are involved in osmoregulation, such as those that regulate the biosynthesis of soluble sugars.85 Additionally, proteins like HSPs and late embryogenesis abundant (LEA) proteins protect cell membranes along with structural and enzymatic proteins from sodium toxicity related to dehydration due to salinity-induced osmotic stress.86 In this sense, new works have pointed out that PTMs are important for salt tolerance due to changes in phosphorylation and lysine acetylation states of many proteins acting on some metabolic pathways related to osmoregulation.87 Furthermore, ionic homeostasis is maintained as a crucial branch of salt tolerance through proteins located within the membrane playing key roles in ion transport and at the cytosolic level participating in intra-cellular communication.88 These proteins, including receptor-like protein kinases (RLKs) and primary cell wall sensors, regulate salt tolerance dynamically. Moreover, the involvement of specific proteins in different metabolic routes is also suggested by organelles as cellular compartments.89 Nevertheless, a protein encoded and isolated from one species may interact differently in other plant systems because of genetic diversity.90 Therefore, a broader physiological and biochemical approach to the identification of candidate proteins responsible for contributing towards salt tolerance is very desirable because each protein may have its individual function required under certain conditions of stress.
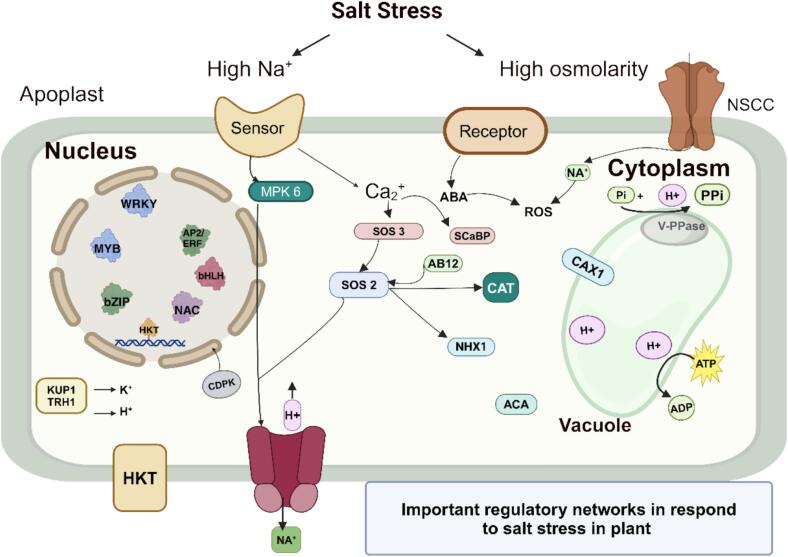
Fig. 2.
Salt Stress-responsive Gene/protein Regulatory Network in Plants. The figure illustrates various molecular components and pathways activated under salt stress conditions. Key elements include HKT, non-selective cation channels (NSCC), and Na+/H+ Exchanger (NHX1). The SOS pathway is highlighted with components such as SOS1, SOS2, and SOS3, along with SOS3-like calcium-binding protein (SCaBP). Calcium-dependent protein kinases (CDPK) and calcium-binding proteins modulate cellular responses. The role of ROS is mediated by enzymes like peroxidase (POD) and CAT. ABA and phosphatidic acid (PA) are also shown as important signalling molecules. Calcium transport and signalling are facilitated by autoinhibited Ca2+-ATPase (ACA) and cation/H+ exchanger 1 (CAX1). Additionally, TFs such as WRKY, MYB, AP2/ERF, bHLH, NAC, and bZIP are involved in stress response regulation. Other important components include potassium uptake permease (KUP1), transporter protein (TRH1), mitogen-activated protein kinase (MPK6), abscisic acid insensitive (ABI2), vacuolar H+-pyrophosphatase (V-PPase), ATP, adenosine diphosphate (ADP), and inorganic phosphate/inorganic pyrophosphate (Pi/PPi).
1.6. Proteomic findings in canola, soybean, and other oil crops
With the help of proteomics techniques, researchers have discovered the molecular mechanisms related to stress responses, lipid biosynthesis, and physiological responses in oil crops. One of the most commonly employed methods is Isobaric Tags for Relative and Absolute Quantitation (iTRAQ), which allows the quantification of multiple samples at the same time, making it perfect for stress response comparisons under different conditions. Two-Dimensional Gel Electrophoresis (2-DE) is another widely used approach to separate complex protein mixtures and has been frequently employed to identify stress-induced proteins that are differentially expressed. Another important technique is Liquid Chromatography coupled with Mass Spectrometry (LC-MS/MS), offering high sensitivity in the identification and quantification of proteins and their modifications. MALDI-TOF/TOF Mass Spectrometry (Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry) has emerged as the most powerful tool for precise protein identification and characterization, particularly in extreme environmental conditions. The proteomic techniques have provided useful insights into stress-responsive proteins in oil crops such as canola, soybean, and sesame, enhancing drought or salinity resistance and increasing crop production. Table 1 shows the applications of these proteomic tools in various studies to unravel genes implicated in stress tolerance, oil biosynthesis, and crop enhancement.
Table 1.
Summary of Plant Proteomic Studies on Salt Stress Responses.
| No | Plant | Proteomic technique | Major findings | References |
|---|---|---|---|---|
| 1 | Sesamum indicum L. |
|
The iTRAQ-based proteomics strategy yielded excellent data quality, demonstrated by uniform peptide length distribution and precise precursor ion tolerance factors. It also showed consistent protein quantitation across multiple experiments and repeats. | 97 |
| 2 | Salicornia europaea |
|
The finding outlines the key steps in calcium signalling that regulate nitrate uptake in S. europaea responding to salt stress | 98 |
| 3 | Brassica napus |
|
At the proteomic level, there are some specific proteins and essential signalling pathways that act as key players in recognizing as well as crosstalk between salt and drought stress response of plants. | 99 |
| 4 | Sesamum indicum L. |
|
The dynamic molecular responses of sesame impose feasibility and underscore the activation of fundamental pathways for adaptation and viability in saline environments. | 100 |
| 5 | Brassica napus |
|
Proteins were encoded by various genes involved in glycolysis, stress response, redox regulation, energy metabolism, and transport that contrasted in tolerant versus susceptible genotypes under salt stress conditions indicating several aspects of molecular mechanisms receiving a salt tolerance. | 101 |
| 6 | Carthamus tinctorius L. |
|
Salt-responsive proteins in safflower are primarily associated with key physiological processes essential for adaptation to salinity stress. | 102 |
| 7 | Ricinus communis |
|
This research provided insights into the specialized functions of cotyledons and true leaves responding to salt stress, emphasizing their unique roles in the overall salt tolerance mechanism that was activated during the seedling stage of Ricinus communis growth. | 103 |
| 8 | Elaeis guineensis Jacq. |
|
Salinity stress can induce complex regulatory mechanisms that affect biological networks at the levels of transcriptomic, proteomic and metabolomic responses. | 104 |
| 9 | Olea europaea L. |
|
The vulnerability of aboveground plant structures to both salinity and drought stressors highlights the critical role of photosynthesis and associated metabolic pathways in responding to these environmental challenges. | 105 |
| 10 | Brassica napus |
|
Under stress, the enzyme activity in the production of energy decreases while it increases the abundance of Phosphoribulokinase (PRK) | 7 |
| 11 | Brassica napus |
|
The bacterial infection of canola roots enhanced salt stress tolerance by upregulating proteins involved in energy metabolism and cell division. | 106 |
| 12 | Brassica napus |
|
The differential responses of antioxidant enzymes, osmoprotectant synthesis, and mitochondrial function in Hyola308 under varying levels of salt stress, were influenced by inoculation status. | 107 |
| 13 | Brassica napus |
|
The antioxidative defence system and photosynthesis are vital components contributing to salt tolerance in canola. | 108 |
| 14 | Carthamus tinctorius L. |
|
Salinity stress decreased fresh and dry weights and plant height, whereas proline content was increased. | 109 |
| 15 | Brassica napus |
|
Both SSA analysis and IPA suggest that the down-regulated brake proteins and JRTPDE3-metabolism of protein would inhibit protein synthesis and nitrogen transport to sink organs under salt stress, which agree with conceptions of molecular regulation of post-transcriptional process in soybean response to high salinity environment. | 110 |
| 16 | Glycine max (L.) |
|
Verified against published C08 RNA-seq data, the findings highlight the crucial role of solute transport and detection in regulating salt stress | 23 |
Proteomics studies of canola (Brassica napus), soybean (Glycine max), and other oil crops have provided essential information regarding the molecular basis underlying numerous physiological processes, stress responses, and lipid biosynthesis in these agriculturally important species. The use of proteomics techniques has provided insight into the extensive behaviour of proteins that act during growth, development, and stress tolerance, hence benefiting efforts to improve crop productivity.91 One important research area in canola, soybean, and other oil crops is the regulation of oil biosynthesis and the potential for more lipid accumulation. Nearly 500 differentially regulated proteins have been linked to lipid metabolism pathways, including fatty acid biosynthesis, lipid droplet formation, and oil body biogenesis. Insights into the functional roles of these proteins have been used to develop genetic manipulations or breeding strategies for increased oil content and improved quality in their respective crops.92 Proteomic analysis of canola, soybean, and other oil crops under abiotic and biotic stress has been discussed. These crops are usually exposed to stresses like drought, salinity, heat, or pathogen attacks that can reduce yield and oil production. Stress-responsive proteins belonging to signalling pathways, stress perception, and tolerance mechanisms have been identified in proteomic studies. One of the key steps is to develop stress-tolerant varieties that show enhanced resistance to biotic and abiotic environmental adversities.93, 94 Proteomic tools were used to survey agronomic and physiological traits in canola, soybean, and other oil crops. Protein comparisons in studies between cultivars within the same growth conditions have revealed proteins that are potentially involved with desired traits such as yield, seed quality, and nutritional properties.95 Proteomic research has also helped identify potential biomarkers for crop improvement and quality assessment.96 Proteomic findings have indicated that understanding specific proteins involved in various physiological processes has paved the way for improving stress-tolerance breeding, enhancing oil content and quality, and achieving higher agronomic recovery in biofuel crop productivity. Since oil crops provide essential food and energy resources, further research in this area is crucial for better understanding crop biology and achieving sustainable oil crop production.
1.7. Stress-induced proteins and their roles in salinity tolerance
Stress-induced proteins play pivotal activities in aiding plant growth roles in allowing plants to cope with environmental challenges, particularly salinity. On the other hand, plants have evolved different strategies to cope with this deleterious condition, involving stress-induced proteins as key factors in plant adaptive responses against abiotic stresses, including soil salinity, one of the major threats to global agricultural productivity. Nonetheless, plants have evolved several mechanisms to counteract the deleterious effects of salinity stress, and salt-responsive proteins play important roles in these adaptive responses.111 Osmotic adjustment is one of the main tasks of stress-induced proteins, providing a foundation that permits optimal expansion in saline conditions.112 In reaction to saline conditions under salt stress conditions undergo osmotic stress because of the elevated level of salt ions in the soil solution plants experience osmotic stress due to the high concentration of salt ions in the soil solution, which reduces water availability to the roots. Stress-induced proteins involved in osmoregulation help plants maintain cellular water balance by regulating the osmotic potential. Examples of such proteins include osmotin, salt shock proteins, and osmotin-like proteins, which accumulate in plant tissues under salinity stress pressure.113 Stress-induced proteins such as ion transporters, including K+ and Na+ channels and pumps, help regulate ion fluxes across cellular membranes, thereby maintaining optimal ion concentrations within the cell. This prevents the toxic buildup of sodium ions and helps the detrimental on plant physiology.114 Stress-induced proteins are essential in shielding cellular components from the negative effects of high salt levels. Elevated salt concentrations lead to oxidative stress, generating ROS that can harm proteins, lipids, and nucleic acids. Proteins with antioxidant functions, such as HSPs and LEA proteins, help neutralise ROS and protect cellular structures, thereby improving plant resilience to salinity stress Furthermore, stress-induced proteins play essential roles in protecting cellular components from salt-induced damage. High salt concentrations can induce oxidative stress in plant cells, leading to the production of ROS and subsequent oxidative damage to proteins, lipids, and nucleic acids. Stress-induced proteins with antioxidant properties, such as HSPs and LEA proteins, help scavenge ROS and protect cellular structures from oxidative damage, thus improving phyto-tolerance to salt-related stress.115, 116
1.8. Genes and transcription factors associated with salinity tolerance
Plants have evolved elaborate mechanisms to sense and counter salinity stress, which include myriad genetic and molecular entities.117 Within these, genes and TFs are key players in the fine regulation of molecular responses to salt stress, mediating plant tolerance and adaptability towards salinity conditions. Identification of the individual gene and TF responsible for salinity tolerance is essential to resolve the molecular complexity behind this trait, which ultimately leads to the breeding of salt-tolerant varieties.118 Many gene families have been recognized to play a vital role in the salinity adaptation process of plants. These proteins relate to ion channels, transporters, homeostasis, osmoticum balance, antioxidant defence as well as regulation pathways.
When salinity stress targets plants, the adaptive mechanisms become complex and consist of various molecular, biochemical, and physiological responses. For instance, osmotic stress triggers the trafficking of H(+)-ATPases to the plasma membrane, leading to the extrusion of toxic Na(+) through transporters such as SOS1 or sequestration into vacuoles by NHX transporters.119 Additionally, potassium transporters (HKT) play important roles in maintaining the balance of K+ absorption and Na+ exclusion120. Plants maintain osmotic balance by synthesizing osmoprotectants like proline,121 glycine betaine, and trehalose along with ion regulation. Antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) are activated to clear reactive oxygen species (ROS) and alleviate oxidative stress.122 Stress-responsive genes, including LEA proteins and HSPs, stabilize cellular structures under stress conditions.
Transcription factors (TFs) act as master regulators in these processes. DREB, AP2/ERF123, bZIP124, NAC125, and MYB126 families adjust the expression of stress-tolerant genes by regulating osmotic balance, ROS detoxification, and ion transport. For example, WRKY8 regulates ion homeostasis by managing antiporters like NHX and interacting with jasmonic and salicylic acid pathways. Finally, aquaporins such as PIP1 and PIP2 are critical for water transport and maintaining osmotic balance under salinity stress, though chronic salt exposure reduces their activity.127 Ultimately, these interconnected pathways help plants become more resistant to salinity and other environmental stresses.
2. Breeding and genetic engineering approaches for enhanced tolerance
Methods of improvement breeding and gene manipulative techniques. The biosynthesis of plants modified in terms of gene TFs, using transformation with vectors carrying genes encoding respective TFs, has already been reported to become tolerant to salt. This is important for integrating salt-sensory pathways with the large repertoire of genes involved in plant adaptation to salinity.128, 129 These genes play a cardinal role in plant abiotic stress tolerance, involving diverse challenging elements like salinity! Several studies were reported for drug-resistant plants with modified TF gene regulation that resulted in increased salt tolerance supremacy in them. These may be expressed and responsible for plant stress responses.130 Earlier studies in different plant species have demonstrated the synergistic role played by TFs and promoter regions on gene expression which is imperative for salt stress tolerance. For instance, transgenic Oryza sativa lines with heightened expression of OsDREB2A exhibited enhanced tolerance to salinity to their wildtype counterparts.131 Furthermore, transgenic Arabidopsis plants expressing TF genes displayed improved growth under salt, drought, and temperature.132 In another study, overexpression of the HhBREB2 gene, a member of the AP2/EREBP TF family isolated from Halimodendron halodendron, bolstered salt and drought tolerance in Arabidopsis, underscoring its significance in salinity-associated signalling.76 MYB-type TFs, exemplified by OsMYB2 in rice, have also demonstrated efficacy in enhancing resistance to various stresses such as salinity, low temperature and dehydration.133 Moreover, abiotic stress responses, such as salinity and drought, are mediated by the action of No Apical Meristem (NAC) proteins, initially characterized for their roles in plant development. In oil crops, such as soybean and rapeseed, the overexpression of NAC genes has shown promise in enhancing tolerance to both salt and drought stresses. For instance, the transformation of soybeans with stress-responsive NAC genes, like GmNAC20, has demonstrated improved growth under saline conditions.134 Similarly, overexpression of NAC genes in oilseed rape has led to increased tolerance to salt stress, higher crop yield, and improved water-use efficiency.135 These responses include maintaining plant growth, reducing water potential, and regulating ion transport, particularly Na+ and Cl−, which play a significant role in plant stress tolerance. While Cl− is necessary at low concentrations, its toxicity at higher levels can intensify stress, further underlining the importance of ion transporters and aquaporin channels as key targets for enhancing oil crop resilience.
Upon uptake by outer root cells, Na+ and Cl− ions traverse to the root xylem before reaching the stem, where they can be kept in between apoplastic spaces. Some salt-tolerant species excrete these ions through specialized structures like glands or bladders. Salt tolerance mechanisms encompass various strategies, including the synthesis of osmoprotectants, compatible solutes, and antioxidants to mitigate ion-induced stress. However, their production incurs high energy costs, potentially impacting crop yields.136 Under salt stress, mainly restricts Na+ and Cl− uptake/transport while keeping homeostasis of beneficial ions like K+, required for normal growth.137 The plasma membrane’s transportation process of Na+/H+ antiporters and intracellular compartmentalization take place to exclude ions from cells.138 Moreover, other regulatory networks like the SOS system and PM H+-ATPases H+/pyrophosphatases for ion regulation/exclusion contribute to this process. Moving forward, clever genetic editing approaches toward ion transport components with less negative fold-effects on development might be prospects for improved salt tolerance in crops. Because of such complexity in the determination of plant micronutrient requirements and speciation during uptake, it is essential to match these data with specific ion uptake systems through understanding tissue- or developmental stage-specific expression patterns and by careful genetic engineering. Continued investigation in this field is highly important to clarify the individual functions of transporters and develop strategies for enhanced salt-stress resistance.139
2.1. Ion transport and salt stress tolerance
Salt stress may cause changes in intracellular levels of Na+ concentration mediated by various proteins or pathways; for example, the plasma membrane proton pumps (H+-ATPases) contribute to maintaining plant salt tolerance by transporting ions from cells. The principal H+ ATPase pumps aid in creating electrochemical potential gradients that are crucial for multiple physiological functions. These processes, such as regulating stomatal pores, phloem loading, and cell enlargement, are mediated by different signalling cascades of ABA to facilitate nutrient uptake through roots. Transgenic strategies targeting H+ pump genes indicate significant applications in improving the salt stress tolerance of various plant species. Researchers try to adjust the expression levels or activities of these pumps, hoping that they will facilitate ion transport and prevent cellular ionic imbalances, thereby increasing biomass under salinity. This mechanism, awaiting deeper molecular clarification (the H+ pump-related salt stress tolerance), could easily be utilized to produce alkali-resilient crops.
2.2. Na+ transport in chloroplasts and salt stress adaptation
The transport of Na + in chloroplasts is essential for photosynthesis and tolerance to salinity stress. Import of Na(+) into chloroplasts provides the driving force for the production of phosphoenolpyruvate (PEP), an essential substrate in photosynthetic carbon dioxide fixation. This process is performed by a Na+/pyruvate symporter, BASS2, which co-transports Na+ and pyruvate into the chloroplasts. Meanwhile, sodium leaves the chloroplasts by a sodium transporter, NHD1. Chloroplast Na + transport and its relation to photosynthesis in higher plants play a crucial role.140Na+ is also moved into chloroplasts to support the synthesis of PEP required for the fixation of CO2 in plants during photosynthesis. Knockout mutants of vacuolar Na+/H+ antiporter NHD1 in Arabidopsis thaliana that were impaired for Na+ export also showed down-regulation of photosynthesis, resulting in reduced tolerance under salt stress.141 his further underscores the significance of Na + homeostasis inside chloroplasts for plant performance. Halophytes (salt-tolerant plants) overcome stomatal limitations by employing CO2-concentrating mechanisms in saline environments. In addition, it has been shown that the influx of Na+ in the chloroplast stroma might be needed for grana formation, which could explain why halophytes simply increase the number of chloroplasts per cell.142 Additionally, tonoplast-localized K+ channels such as TPK1 and TPK3 are involved in mediating the intracellular pools of K+/Na+ ratios.143 Deciphering the fine molecular machinery on how transporters mediate cations and anions movement across chloroplast envelope membranes as well as thylakoids, keeping a balance toward PSII activities under salt stress conditions would be vital for unveiling plant response to salinity stress at the molecular level and breeding salt-tolerant crops.144 Potassium (K+) transporters are essential for plant metabolism since their function is linked with intracellular K + balance and salinity-related stress. PM-localized H + pumps (proton ATPase) maintain the membrane potential that allows influx transport of K through voltage-gated channels and exclusion Na via cytoplasmic-membrane localized Na+/H-exchangers like SOS1 which also aide in nutrient uptake as seen with sulfate transporter SULTR4;2 loss-of-function mutant roots enhance sodium deposits but are impaired in sulfur absorption1.145
Calcium channels with increased K+/Na+ selectivity, found in salt-tolerant plants like Thellungiella salsuginea, contribute to improved salt tolerance by reducing Na + uptake.146 Targeting these channels to restrict Na+ uptake can benefit K+/Na+ homeostasis without affecting the negative membrane potential, allowing selective K+ uptake via inward-rectifying K+ channels.147 Other membrane proteins from the high-affinity K+ transporter (HKT) family may be able to facilitate the the influx of Na+ ions into root cells.148 Genetic modification of HKT1 (now HKT2;1) can enhance K+/Na+ selectivity and salt tolerance.149 Halophytes can tolerate salt by maintaining high K+ uptake while restricting Na+ influx into root cells.150 Despite genetic modifications of multiple ion transport components to enhance salinity resistance.151 Targeting these systems alone or in combination to improve salt tolerance remains a significant challenge in plant biotechnology.
2.3. RNA interference approaches to produce salt-tolerant plants
RNA interference (RNAi) is a powerful tool for targeted silencing or modifying gene expression in plants, including for improving salt tolerance. This process involves small RNA molecules, such as microRNAs (miRNAs), which can precisely silence targeted genes without affecting the expression of other genes. miRNAs are produced from double-stranded RNAs cleaved by enzymes like DICER or DCL, leading to RNAi.152 They are key players in epigenetic regulation of gene expression and imprinting, genome stability and heritable maintenance within plants, or can act as a defence against biotic or abiotic stresses. Modifying specific microRNAs (miRNAs) or their targets presents a novel method for engineering salinity tolerance in crops.153
2.4. Qtlomics of salt tolerance
Quantitative Trait Loci (QTL) have significantly advanced marker development and crop breeding efforts, particularly for traits related to salt tolerance. This progress has been facilitated by extensive physiological and molecular studies on stress tolerance mechanisms, as well as the availability of information on stress-specific and shared adaptation mechanisms. Several important QTLs associated with salt tolerance have been identified in various crops such as rice, cotton, soybean, and maize.154 One of the most significant QTLs is Saltol, which is linked to Na+/K+ homeostasis under salt stress. Saltol has been introduced into elite rice cultivars, resulting in improved seedling-stage salt stress tolerance and superior agronomic performance.155 In addition to Saltol, other QTLs have also been identified and used in breeding programs aimed at developing salt-tolerant crop varieties. For example, SSR markers have been utilized to find biomarkers associated with salt tolerance in cotton and cucumber.156 In maize, QTLs for salt tolerance have been mapped using high-density SNP markers, leading to the identification of candidate genes involved in ion homeostasis.157 In soybeans, the salt tolerance-associated gene GmSALT3 was isolated using fine mapping methods. This gene, encoding a cation/H+ exchanger family transporter, was shown to play a role in salt tolerance and is localized to the endoplasmic reticulum.158 Advances in genome sequencing and accessibility to genome datasets have further accelerated progress in molecular breeding for salt tolerance.
2.5. Alternative splicing and salt stress tolerance
Alternative splicing, a process where multiple mature mRNA variants are produced from a single gene, has emerged as a significant regulatory mechanism in plants, particularly during stress responses such as salt stress. This phenomenon generates diverse transcriptomic and proteomic profiles, as different splice variants can yield proteins with altered structures, functions, and cellular localizations.159 Several splicing factors have been identified in plants, and their roles in stress responses are increasingly being recognized. For instance, splicing factors like Sm-like conserved protein 5 (LSm5) and PRP31 have been linked to stress response and pre-mRNA splicing regulation in Arabidopsis thaliana. Under cold stress, PRP31 plays a critical role in modulating the expression of cold-responsive genes by regulating pre-mRNA splicing.160 Alternative splicing can also produce premature termination codons (PTCs) in transcripts, leading to their degradation or the production of truncated proteins. Examples include the dehydration-responsive element-binding (DREB) protein 2B in rice, where alternative splicing generates isoforms that promote stress tolerance under drought and high-temperature conditions.161 Heat stress has also been shown to induce alternative splicing in plants, with studies demonstrating that splice isoforms of heat-shock transcription factor A2 (HSFA2) in Arabidopsis and rice play distinct roles in stress response.162 In grapevines, high-temperature stress leads to intron retention events, indicating a key post-transcriptional regulatory mechanism.163 Furthermore, spliceosomal proteins like multiprotein bridging factor 1c and U1A have been implicated in alternative RNA splicing regulation and stress response in plants.164 Exploring the halobiome, which encompasses halophilic and halotolerant organisms such as bacteria, algae, fungi, and halophytes, holds promise for identifying genes associated with salt tolerance. Halophytes, in particular, have evolved various structural and developmental adaptations to thrive in hypersaline environments, including succulence, leaf abscission, and the presence of salt-secreting structures such as salt glands or salt hairs. These plants employ cellular mechanisms to manage salt stress, such as reducing Na+ influx, compartmentalizing Na+ ions in vacuoles, or eliminating Na+ ions through plasma membrane anti-porters.165 Genes isolated from halophytes or their homologs have been successfully used to enhance salt tolerance in crop plants. Cis-regulatory elements and promoter sequences of stress-inducible genes in halophytic plants have shown strong expression under saline conditions, providing effective tools for transgene expression. Genes encoding TFs, enzymatic and non-enzymatic antioxidants, and ion transporters from halophytes have been utilized to improve salt tolerance in non-halophytic plants.166 Moreover, genes from non-plant halobionts, such as osmoprotectant synthesis enzymes and antioxidative enzymes, have been transferred into plants, resulting in enhanced salt tolerance.167, 168 For instance, genes involved in ectoine synthesis from the halophilic bacterium Halomonas elongata and glycine betaine synthesis from the halotolerant cyanobacterium Aphanothece halophytica have been expressed in transgenic plants, leading to improved growth under saline conditions.169 In addition to transgenic approaches, whole-genome and transcriptome analyses of halotolerant organisms have provided valuable insights into candidate genes for crop improvement. Sequencing efforts have elucidated the genomes of various halotolerant organisms, offering potential targets for transformation studies.
3. Genome editing in plants to reduce soil salinity stress
Genome editing (GE) technologies, including CRISPR-Cas9, have revolutionized plant breeding by enabling precise modifications in crop genomes. While GE has been extensively applied in various crop plants, its use in developing salt-tolerant plants has been relatively limited.170 Nonetheless, recent studies have demonstrated the potential of GE to enhance salt tolerance by targeting specific genes associated with salt stress response pathways. One successful application of GE involved editing the ARGOS8 maize genomic sequence in maize using CRISPR-Cas9, resulting in maize lines with higher and ubiquitous expression levels of gene 4.171 Given the complexity of salt tolerance, which involves multiple genes and pathways, multiplex GE techniques have been proposed to target multiple genes simultaneously. This approach allows for the induction of specific changes in salt-responsive pathways, potentially leading to more effective enhancement of salt tolerance. Base editors, such as cytidine deaminases, offer advantages for inducing transition mutations in plants, including the introduction of nonsense mutations to disrupt gene function.172, 173 The CRISPR/Cpf1 system has also shown promise for gene editing, particularly in AT-rich regions of the genome. Cpf1 nucleases have demonstrated the ability to introduce accurate gene insertions and indel mutations in the rice genome, offering another tool for precise GE in plants174. As research in this area continues to advance, GE approaches are likely to play an increasingly important role in crop improvement efforts aimed at enhancing salt tolerance and ensuring food security in saline environments.175 Progress in genomic techniques, like next-gen sequencing genome-wide association studies (GWAS), and functional genomics, have provided valuable insights into the complex regulatory networks underlying salinity stress responses in plants.176 Integration of genetic, genomic, and molecular approaches offers significant potential for accelerating the breeding of salt-tolerant crops to mitigate the impact of salinity stress on agricultural productivity and food security. Understanding the molecular basis and pathways related to salinity tolerance is essential for the development of salt-tolerant crop varieties capable of sustaining productivity under saline conditions. For instance, there are also mechanisms of adapting to salinity stress that are known as ABA-dependent and ABA-independent pathways.
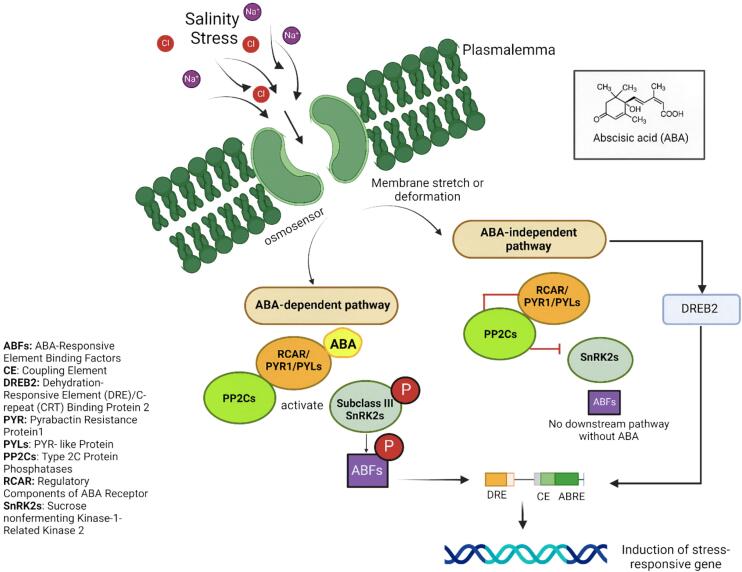
4. Aba-dependent vs ABA-Independent pathways
ABA-dependent pathways involve the direct action of ABA in signalling processes when adapting to stress. In these pathways, ABA binds to its receptors (PYR/PYL/RCAR), which then inhibit protein phosphatase 2Cs (PP2Cs). This inhibition activates kinase 2 protein (SnRK2), which phosphorylates subsequent targets involved in stress responses, such as TFs (e.g., ABI5) and ion channels (eg: SLAC1). ABA-dependent pathways are crucial for processes like stomatal closure, seed dormancy and germination inhibition during stress conditions.177 On the other hand, ABA-independent pathways do not rely directly on the presence of ABA. These pathways involve other signalling molecules and mechanisms, such as osmotic stress sensors, that can activate similar downstream components, including SnRK2 kinases, but through different initial triggers. This pathway helps plants respond to stress independently of ABA, ensuring a more robust and versatile stress response system.178 Both pathways are illustrated in Fig. 2 below. Understanding both pathways provides insight into how plants integrate multiple signals to survive and adapt to challenging environments, which is crucial for developing stress-tolerant crop varieties (Fig. 3).
Fig. 3.
Transduction of osmotic signal of salinity stress through ABA-dependent and ABA-independent pathways.
4.1. Omics approaches to salinity stress tolerance
Panomic strategies are those approaches that can combine multi-omics data (genomics, transcriptome, proteome, and metabolome) as a hope for the future of agriculture improvement.179 Such an integrative way of information can provide a holistic comprehension of biological systems and help identify essential molecular agents involved with plants under abiotic stresses.180 A number of existing tools and techniques have been further combined with ontologies to cater to the multi-omics data. Generated multi-omics data must be deposited in omics repositories and supported by visualization platforms nowadays, in addition to the tremendous results from diverse high-throughput screening methods.181 In this review, the promise of panomics is exemplified by integrated systems analysis using multi-omic data and functional annotation together with knowledge-guided pathway analyses.182 This strategy allows even more accurate phenotyping through careful pattern identification in a complex by using machine learning, to identify uncharacterized mechanisms that contribute positively to abiotic stress tolerance. These data also contribute to facilitating the building of less-random networks (related candidate genes, proteins), molecules (metabolites), and biomarkers associated with relevant traits for crop improvement based on multi-omics approaches. This can further be utilized to improve tolerance and susceptibility in practical plant species under targeted breeding or genetic engineering programs.183
5. Insights from genomics, transcriptomics, and metabolomics studies
Salinity stress poses a significant threat to global agricultural productivity, affecting numerous crop species worldwide. Understanding the molecular mechanisms underlying salinity stress tolerance is essential for developing resilient crop varieties capable of thriving in saline environments. In recent years, the integration of genomics, transcriptomics, and metabolomics approaches has provided valuable insights into the complex regulatory networks and biochemical pathways involved in plant responses to salinity stress.184 Salinity stress, resulting from the accumulation of soluble salts in soil or irrigation water, represents a major environmental constraint to agricultural productivity worldwide. High soil salinity levels can adversely affect plant growth, development, and productivity, leading to significant economic losses in crop production. The complex nature of salinity stress tolerance involves intricate interactions among various physiological, biochemical, and molecular processes within plants. Understanding these mechanisms is crucial for devising effective strategies to enhance crop resilience to salinity stress. Multi-omics approaches, including genomics, transcriptomics, and metabolomics, offer valuable tools for dissecting the molecular mechanisms of salinity stress tolerance in plants. Integration of omics data enables a systems-level understanding of stress response networks and facilitates the identification of potential targets for crop improvement.185
5.1. Genomics insights into salinity stress tolerance
Advancements in genomic technologies have enabled the identification of genetic determinants associated with salinity stress tolerance in plants.186 Genome-wide association studies (GWAS), quantitative trait loci (QTL) mapping, and comparative genomics analyses have revealed genetic variations underlying salinity tolerance traits across diverse plant species. Candidate genes involved in ion homeostasis, osmotic regulation, and stress signalling pathways have been identified through genomic approaches, providing valuable targets for genetic engineering and breeding programs aimed at improving salinity stress tolerance in crops.187, 188
5.2. Transcriptomics profiling of salinity stress response
Transcriptomic studies have provided comprehensive insights into the dynamic changes in gene expression patterns during plant responses to salinity stress. Differential gene expression analyses have uncovered key transcriptional regulators, stress-responsive genes, and signalling pathways implicated in salinity stress adaptation.189 Moreover, transcriptomic profiling has elucidated the crosstalk between different stress response pathways and the regulatory networks governing plant acclimation to salinity stress.190 Integration of transcriptomic data with other omics layers has facilitated the identification of candidate genes and regulatory elements modulating salinity stress tolerance in plants.191
6. Metabolomics approaches for understanding salinity stress adaptation
Metabolomics analyses offer a holistic view of plant metabolism under salinity stress conditions, providing insights into the metabolic pathways and biochemical processes involved in stress tolerance mechanisms. Metabolite profiling studies have revealed changes in primary and secondary metabolite levels in response to salinity stress, highlighting metabolic adjustments crucial for osmotic regulation, antioxidant defence, and energy metabolism.192 Integration of metabolomics data with genomics and transcriptomics datasets has enabled the identification of metabolic signatures associated with salinity stress tolerance, offering potential biomarkers for crop breeding programs.193
6.1. Integrative multi-omics approaches for unraveling plant tolerance
Integration of genomics, transcriptomics, and metabolomics data holds promise for elucidating the complex interplay between genetic, transcriptional, and metabolic networks underlying salinity stress tolerance in plants.194 Multi-omics approaches enable the identification of key molecular players, regulatory pathways, and metabolic signatures associated with stress adaptation, providing a comprehensive understanding of plant responses to salinity stress.195 Challenges in data integration, computational analysis, and validation of omics-derived findings necessitate collaborative efforts among researchers and the development of robust analytical frameworks for effective multi-omics studies in plant stress biology.
7. Future directions and prospects
7.1. Challenges in applying research to field conditions
The complex biology of plant tolerance to salt poses a massive challenge to the practical implementation of research discoveries in the field. Because several interacting genes and regulatory systems rule this feature, it is challenging to anticipate the behaviour of selectively grown or genetically engineered plants outside of laboratory settings. Field circumstances are very different from controlled laboratory surroundings because of the variations in soil composition, temperature, and biotic variables. The effective implementation of study findings is difficult due to this disparity. The application of salinity tolerance techniques might also be hampered by socioeconomic factors and legislative barriers, such as the approval of genetically modified organisms (GMOs). Novel technologies provide encouraging opportunities to enhance the development of crops that can withstand salt. GE techniques like CRISPR-Cas9 allow for precise alterations in genes associated with salinity tolerance, enabling the swift development of adaptable crop varieties. These technologies can target multiple genes simultaneously, bypassing the lengthy traditional breeding cycles. Moreover, multi-omics approaches—encompassing genomics, transcriptomics, proteomics, and metabolomics—provide a holistic understanding of the molecular mechanisms underlying salinity tolerance. Advances in high-throughput phenotyping and AI-driven data analysis further enhance the ability to identify key traits and predict the performance of new varieties under saline conditions. Advancing research in salinity tolerance hinges on global collaboration and effective data sharing. Partnerships among researchers, institutions, and countries enable the pooling of knowledge, resources, and technologies, accelerating scientific advancements. International consortia can standardize research methodologies, share extensive datasets, and conduct multi-location trials to evaluate the performance of salinity-tolerant crops in diverse environments. Open-access databases and repositories for omics data, supported by sophisticated bioinformatics tools, enhance the accessibility and utility of research findings, fostering transparency and reproducibility in scientific research.
8. Conclusion
In conclusion, tackling the challenges of salinity stress in agriculture requires an integrated approach that combines cutting-edge technologies, collaborative efforts, and practical field applications. Continued innovation and research in this field are vital for ensuring global food security and developing sustainable agricultural practices capable of mitigating the adverse effects of soil salinization.
9. Consent for publication
All authors agreed to publish this review.
CRediT authorship contribution statement
Sarah Alrajeh: Writing – original draft. Muhammad Naveed Khan: Writing – original draft. Aidhya Irhash Putra: Writing – original draft. Dhafar N. Al-ugaili: Investigation, Data curation. Khalid H. Alobaidi: Visualization, Validation. Othman Al Dossary: Supervision, Project administration. Jameel R. Al-Obaidi: Writing – review & editing, Supervision, Conceptualization. Azi Azeyanty Jamaludin: Methodology, Investigation. Mohammed Yahya Allawi: Resources. Bilal Salim Al-Taie: Writing – original draft, Methodology. Norafizah Abdul Rahman: Writing – original draft, Methodology. Norasfaliza Rahmad: Writing – original draft, Methodology.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The author would like to express sincere thanks to the Faculty of Science and Mathematics (FSM) and the Research Management and Innovation Center (RMIC) at Universiti Pendidikan Sultan Idris for their invaluable support and assistance in conducting this study.
References
- 1.Candellone E., Aleta A., Ferraz de Arruda H., Meijaard E., Moreno Y. Characteristics of the vegetable oil debate in social-media and its implications for sustainability. Communications Earth & Environment. 2024;5:391. doi: 10.1038/s43247-024-01545-x. [DOI] [Google Scholar]
- 2.Adeleke B.S., Babalola O.O. Oilseed crop sunflower (Helianthus annuus) as a source of food: nutritional and health benefits. Food Sci Nutr. 2020;8(9):4666–4684. doi: 10.1002/fsn3.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slimani A, Oufdou K, Meddich A. Combining intercropping and co-inoculation of AMF and PGPR mitigate salinity in barley and alfalfa by improving photosynthetic capacity, nutrient acquisition, and redox potential. Plant Biosystems - An International Journal Dealing with all Aspects of Plant Biology.1-10. doi:10.1080/11263504.2024.2392573.
- 4.Shanker A., Venkateswarlu B. Abiotic stress in plants: mechanisms and adaptations. BoD–Books on Demand. 2011 [Google Scholar]
- 5.Glenn EP, Brown JJ, Khan MJ. Mechanisms of salt tolerance in higher plants. Mechanisms of environmental stress resistance in plants. 2022:83-110. Doi: 10.1201/9780203747803.
- 6.Gu H., Wang Y., Xie H., et al. Drought stress triggers proteomic changes involving lignin, flavonoids and fatty acids in tea plants. Sci Rep. 2020;10(1):15504. doi: 10.1038/s41598-020-72596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolatabadi N., Toorchi M., Valizadeh M., Bandehagh A. The proteome response of salt-sensitive rapeseed (Brassica napus L.) genotype to salt stress. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 2019;47(1):17–23. doi: 10.15835/NBHA47111133. [DOI] [Google Scholar]
- 8.Meena S., Deb S., Samtani H., Khurana P. Dissecting the molecular function of Triticum aestivum STI family members under heat stress. Front Genet. 2020;11:873. doi: 10.3389/fgene.2020.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arif Y., Singh P., Siddiqui H., Bajguz A., Hayat S. Salinity induced physiological and biochemical changes in plants: an omic approach towards salt stress tolerance. Plant Physiol Biochem. 2020;156:64–77. doi: 10.1016/j.plaphy.2020.08.042. [DOI] [PubMed] [Google Scholar]
- 10.Das P, Nutan K, Singla-Pareek S, Pareek A. Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Review. Frontiers in Plant Science. 2015-September-09 2015;6Doi: 10.3389/fpls.2015.00712. [DOI] [PMC free article] [PubMed]
- 11.Wani S.H., Kumar V., Khare T., et al. Engineering salinity tolerance in plants: progress and prospects. Planta. 2020;251:76. doi: 10.1007/s00425-020-03366-6. [DOI] [PubMed] [Google Scholar]
- 12.Slimani A., Ait-El-Mokhtar M., Ben-Laouane R., et al. Molecular and systems biology approaches for harnessing the symbiotic interaction in mycorrhizal symbiosis for grain and oil crop cultivation. Int J Mol Sci. 2024;25(2) doi: 10.3390/ijms25020912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muthuramalingam P., Jeyasri R., Rakkammal K., et al. Multi-Omics and integrative approach towards understanding salinity tolerance in rice: a review. Biology. 2022;11(7):1022. doi: 10.3390/biology11071022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madeira C., Costa P.M. Proteomics in systems toxicology. Adv Protein Chem Struct Biol. 2021;127:55–91. doi: 10.1016/bs.apcsb.2021.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Kausar R, Komatsu S. Proteomic Approaches to Uncover Salt Stress Response Mechanisms in Crops. Int J Mol Sci. Dec 28 2022;24(1)doi:10.3390/ijms24010518. [DOI] [PMC free article] [PubMed]
- 16.Anupama A., Bhugra S., Lall B., Chaudhury S., Chugh A. Morphological, transcriptomic and proteomic responses of contrasting rice genotypes towards drought stress. Environ Exp Bot. 2019;166 doi: 10.1016/j.envexpbot.2019.06.008. [DOI] [Google Scholar]
- 17.Yin S., Liu L., Gan W. The roles of post-translational modifications on mTOR signaling. Int J Mol Sci. 2021;22(4):1784. doi: 10.3390/ijms22041784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai C., Miao L., Zhang Y., Zhang L., Li H., Zhang S. An enzyme response-regulated colorimetric assay for pattern recognition sensing application using biomimetic inorganic-protein hybrid nanoflowers. Chem Eng J. 2022;431 doi: 10.1016/j.cej.2021.134107. [DOI] [Google Scholar]
- 19.Sivanich M.K., Gu T.-J., Tabang D.N., Li L. Recent advances in isobaric labeling and applications in quantitative proteomics. Proteomics. 2022;22(19–20) doi: 10.1002/pmic.202100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupree E.J., Jayathirtha M., Yorkey H., Mihasan M., Petre B.A., Darie C.C. A critical review of bottom-up proteomics: the good, the bad, and the future of this field. Proteomes. 2020;8(3) doi: 10.3390/proteomes8030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy Choudhury A., Roy S.K., Trivedi P., et al. Label-free proteomics approach reveals candidate proteins in rice (Oryza sativa L.) important for ACC deaminase producing bacteria-mediated tolerance against salt stress. Environ Microbiol. 2022;24(8):3612–3624. doi: 10.1111/1462-2920.15937. [DOI] [PubMed] [Google Scholar]
- 22.Meng X, Liu S, Dong T, et al. Comparative Transcriptome and proteome analysis of salt-tolerant and salt-sensitive sweet potato and overexpression of IbNAC7 confers salt tolerance in arabidopsis. Original research. Frontiers in Plant Science. 2020-August-27 2020;11Doi: 10.3389/fpls.2020.572540. [DOI] [PMC free article] [PubMed]
- 23.Rehman H.M., Chen S., Zhang S., et al. Membrane proteomic profiling of soybean leaf and root tissues uncovers salt-stress-responsive membrane proteins. Int J Mol Sci. 2022;23(21):13270. doi: 10.3390/ijms232113270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan J, Li Z, Wang Q, et al. Phosphoproteomic profiling reveals early salt-responsive mechanisms in two foxtail millet cultivars. Original Research. Frontiers in Plant Science. 2021-September-20 2021;12Doi: 10.3389/fpls.2021.712257. [DOI] [PMC free article] [PubMed]
- 25.Mansour M.M.F., Hassan F.A.S. How salt stress-responsive proteins regulate plant adaptation to saline conditions. Plant Mol Biol. 2022;108(3):175–224. doi: 10.1007/s11103-021-01232-x. [DOI] [PubMed] [Google Scholar]
- 26.Hasanuzzaman M., Raihan M.R.H., Masud A.A.C., et al. Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int J Mol Sci. 2021;22(17):9326. doi: 10.5772/895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paliwal S., Tripathi M.K., Tiwari S., et al. Molecular advances to combat different biotic and abiotic stresses in linseed (Linum usitatissimum L.): a comprehensive review. Genes. 2023;14:1461. doi: 10.3390/genes14071461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saleem S., Mushtaq N.-U., Shah W.-H., Rasool A., Hakeem K.-R., Rehman R.-U. Morpho-physiological, biochemical and molecular adaptation of millets to abiotic stresses: a review. Phyton-International Journal of Experimental Botany. 2021;90(5):1363–1385. doi: 10.32604/phyton.2021.014826. [DOI] [Google Scholar]
- 29.Guo M, Wang X-S, Guo H-D, et al. Tomato salt tolerance mechanisms and their potential applications for fighting salinity: a review. Review. Frontiers in Plant Science. 2022-September-14 2022;13Doi: 10.3389/fpls.2022.949541. [DOI] [PMC free article] [PubMed]
- 30.Bhatt D., Nath M., Sharma M., Bhatt M.D., Bisht D.S., Butani N.V. Role of growth regulators and phytohormones in overcoming environmental stress. Protective Chemical Agents in the Amelioration of Plant Abiotic Stress. 2020:254–279. [Google Scholar]
- 31.Zhao C., Zhang H., Song C., Zhu J.-K., Shabala S. Mechanisms of plant responses and adaptation to soil salinity. The Innovation. 2020;1 doi: 10.1016/j.xinn.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan P., Liu F., Li R., Wang S., Luo J. Chloroplasts— beyond energy capture and carbon fixation: tuning of photosynthesis in response to chilling stress. Int J Mol Sci. 2019;20(20):5046. doi: 10.3390/ijms20205046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Obaidi J.R., Rahmad N., Hanafi N.M., Halabi M.F., Al-Soqeer A.A. Comparative proteomic analysis of male and female plants in Jojoba (Simmondsia chinensis) leaves revealed changes in proteins involved in photosynthesis, metabolism, energy, and biotic and abiotic stresses. Acta Physiol Plant. 2017;39:179. doi: 10.1007/s11738-017-2485-7. [DOI] [Google Scholar]
- 34.Sharma S., Joshi J., Kataria S., et al. In: Plant Life under Changing Environment. Tripathi D.K., Pratap Singh V., Chauhan D.K., editors. Academic Press; 2020. Chapter 27 - Regulation of the Calvin cycle under abiotic stresses: an overview; pp. 681–717. [Google Scholar]
- 35.Zahra N., Hafeez M.B., Ghaffar A., et al. Plant photosynthesis under heat stress: effects and management. Environ Exp Bot. 2023;206 doi: 10.1016/j.envexpbot.2022.105178. [DOI] [Google Scholar]
- 36.Sarwar M., Anjum S., Ali Q., Alam M.W., Haider M.S., Mehboob W. Triacontanol modulates salt stress tolerance in cucumber by altering the physiological and biochemical status of plant cells. Sci Rep. 2021;1 doi: 10.1038/s41598-021-04174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan X., Cao X., Zhou H., et al. Carbon dioxide fertilization effect on plant growth under soil water stress associates with changes in stomatal traits, leaf photosynthesis, and foliar nitrogen of bell pepper (Capsicum annuum L.) Environ Exp Bot. 2020;179 doi: 10.1016/j.envexpbot.2020.104203. [DOI] [Google Scholar]
- 38.Hannachi S, Steppe K, Eloudi M, Mechi L, Bahrini I, Van Labeke M-C. Salt Stress induced changes in photosynthesis and metabolic profiles of one tolerant (‘Bonica’) and one sensitive (‘Black Beauty’) eggplant cultivars (Solanum melongena L.). Plants. 2022;11(5):590. Doi: 10.3390/plants11050590. [DOI] [PMC free article] [PubMed]
- 39.Salim Akhter M., Noreen S., Mahmood S., et al. Influence of salinity stress on PSII in barley (Hordeum vulgare L.) genotypes, probed by chlorophyll-a fluorescence. Journal of King Saud University - Science. 2021;33 doi: 10.1016/j.jksus.2020.101239. [DOI] [Google Scholar]
- 40.Zahra N., Al Hinai M.S., Hafeez M.B., et al. Regulation of photosynthesis under salt stress and associated tolerance mechanisms. Plant Physiol Biochem. 2022;178:55–69. doi: 10.1016/j.plaphy.2022.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Peng Z., Wang Y., Geng G., et al. Comparative analysis of physiological, enzymatic, and transcriptomic responses revealed mechanisms of salt tolerance and recovery in tritipyrum. Front Plant Sci. 2022;12 doi: 10.3389/fpls.2021.800081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiba R., Urano Y., Noguchi N. α-Tocopherol suppresses 24(S)-hydroxycholesterol-induced cell death via inhibition of endoplasmic reticulum membrane disruption. Steroids. 2023;189 doi: 10.1016/j.steroids.2022.109136. [DOI] [PubMed] [Google Scholar]
- 43.Liu J., Lin Y., Lin H., Lin M., Fan Z. Impacts of exogenous ROS scavenger ascorbic acid on the storability and quality attributes of fresh longan fruit. Food Chemistry: x. 2021;12 doi: 10.1016/j.fochx.2021.100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dewanjee S., Bhattacharjee N., Chakraborty P., Bhattacharjee S. In: Carotenoids: Structure and Function in the Human Body. Zia-Ul-Haq M., Dewanjee S., Riaz M., editors. Springer International Publishing; 2021. Carotenoids as Antioxidants; pp. 447–473. [Google Scholar]
- 45.Hossain A., Pamanick B., Venugopalan V.K., et al. In: Emerging Plant Growth Regulators in Agriculture. Aftab T., Naeem M., editors. Academic Press; 2022. Chapter 1 - Emerging roles of plant growth regulators for plants adaptation to abiotic stress–induced oxidative stress; pp. 1–72. [Google Scholar]
- 46.Kumar A, Singh S, Gaurav AK, Srivastava S, Verma JP. Plant Growth-Promoting Bacteria: Biological Tools for the Mitigation of Salinity Stress in Plants. Review. Frontiers in Microbiology. 2020-July-07 2020;11Doi: 10.3389/fmicb.2020.01216. [DOI] [PMC free article] [PubMed]
- 47.Liu B., Peng X., Han L., Hou L., Li B. Effects of exogenous spermidine on root metabolism of cucumber seedlings under salt stress by GC-MS. Agronomy. 2020;10(4):459. doi: 10.3390/agronomy10040459. [DOI] [Google Scholar]
- 48.Chin C.F., Teoh E.Y., Chee M.J.Y., Al-Obaidi J.R., Rahmad N., Lawson T. Comparative proteomic analysis on fruit ripening processes in two varieties of tropical mango (Mangifera indica) Protein J. 2019;38(6):704–715. doi: 10.1007/s10930-019-09868-x. [DOI] [PubMed] [Google Scholar]
- 49.Amin I., Rasool S., Mir M.A., Wani W., Masoodi K.Z., Ahmad P. Ion homeostasis for salinity tolerance in plants: a molecular approach. Physiol Plant. 2021;171(4):578–594. doi: 10.1111/ppl.13185. [DOI] [PubMed] [Google Scholar]
- 50.Hussain S., Hussain S., Ali B., et al. Recent progress in understanding salinity tolerance in plants: story of Na+/K+ balance and beyond. Plant Physiol Biochem. 2021;160:239–256. doi: 10.1016/j.plaphy.2021.01.029. [DOI] [PubMed] [Google Scholar]
- 51.Lindberg S., Premkumar A. Ion changes and signaling under salt stress in wheat and other important crops. Plants. 2024;13(1):46. doi: 10.3390/plants13010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hussain S., Khalid M.F., Sohail M., et al. In: Approaches to the Remediation of Inorganic Pollutants. Hasanuzzaman M., editor. Springer Singapore; 2021. Role of Transporters in Accumulating Salt Ions by Halophytes; pp. 11–40. [Google Scholar]
- 53.Chakraborty K., Mondal S., Bhaduri D., Mohanty A., Paul A. In: Plant Nutrition and Food Security in the Era of Climate Change. Kumar V., Srivastava A.K., Suprasanna P., editors. Academic Press; 2022. Chapter 11 - Interplay between sodium and chloride decides the plant’s fate under salt and drought stress conditions; pp. 271–314. [Google Scholar]
- 54.Basu S., Kumar A., Benazir I., Kumar G. Reassessing the role of ion homeostasis for improving salinity tolerance in crop plants. Physiol Plant. 2021;171(4):502–519. doi: 10.1111/ppl.13112. [DOI] [PubMed] [Google Scholar]
- 55.Balasubramaniam T., Shen G., Esmaeili N., Zhang H. Plants’ response mechanisms to salinity stress. Plants. 2023;12(12):2253. doi: 10.3390/plants12122253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rasouli F., Kiani-Pouya A., Zhang H., Shabala S. In: Biology and Biotechnology of Quinoa: Super Grain for Food Security. Varma A., editor. Springer Singapore; 2021. Mechanisms of Salinity Tolerance in Quinoa; pp. 221–242. [Google Scholar]
- 57.Ali Q., Shahid S., Nazar N., et al. In: Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives II: Mechanisms of Adaptation and Stress Amelioration. Hasanuzzaman M., editor. Springer Singapore; 2020. Use of Phytohormones in Conferring Tolerance to Environmental Stress; pp. 245–355. [Google Scholar]
- 58.Al-Obaidi JR, Yahya Allawi M, Salim Al-Taie B, et al. The environmental, economic, and social development impact of desertification in Iraq: a review on desertification control measures and mitigation strategies. Environ Monit Assess. May 20 2022;194(6):440. doi:10.1007/s10661-022-10102-y. [DOI] [PubMed]
- 59.Mann A., Lata C., Kumar N., Kumar A., Kumar A., Sheoran P. Halophytes as new model plant species for salt tolerance strategies. Frontiers in Plant Science. 2023;14 doi: 10.3389/fpls.2023.1137211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haider S., Iqbal J., Naseer S., et al. Molecular mechanisms of plant tolerance to heat stress: current landscape and future perspectives. Plant Cell Rep. 2021;40(12):2247–2271. doi: 10.1007/s00299-021-02696-3. [DOI] [PubMed] [Google Scholar]
- 61.Al-Obaidi J.R. Proteomics of edible mushrooms: a mini-review. Electrophoresis. 2016;37(10):1257–1263. doi: 10.1002/elps.201600031. [DOI] [PubMed] [Google Scholar]
- 62.Razi K., Muneer S. Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit Rev Biotechnol. 2021;41(5):669–691. doi: 10.1080/07388551.2021.1874280. [DOI] [PubMed] [Google Scholar]
- 63.Slimani A., Raklami A., Benmrid B., Oufdou K., Meddich A. Salt-tolerant plant growth-promoting rhizobacteria mitigate salinity in barley by improving photosynthetic capacity, antioxidant activity, and soil fertility. Biologia. 2023;78(12):3367–3379. doi: 10.1007/s11756-023-01541-0. [DOI] [Google Scholar]
- 64.Slimani A., Ait-El-Mokhtar M., Ben-Laouane R., et al. Signals and machinery for mycorrhizae and cereal and oilseed interactions towards improved tolerance to environmental stresses. Plants. 2024;13(6) doi: 10.3390/plants13060826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mustafa G, Komatsu S. Plant proteomic research for improvement of food crops under stresses: a review. https://doi.org/10.1039/D1MO00151E. Molecular Omics. 2021;17(6):860-880. Doi: 10.1039/D1MO00151E. [DOI] [PubMed]
- 66.Kchouk S., Melsen L.A., Walker D.W., Oel P.R.V. A geography of drought indices : mismatch between indicators of drought and its impacts on water and food securities. Nat Hazards Earth Syst Sci. 2022;22(2):323–344. doi: 10.5194/nhess-22-323-2022. [DOI] [Google Scholar]
- 67.Aslam M.M., Rashid M.A.R., Siddiqui M.A., et al. Recent insights into signaling responses to cope drought stress in rice. Rice Sci. 2022;29(2):105–117. doi: 10.1016/j.rsci.2021.08.001. [DOI] [Google Scholar]
- 68.Choudhary J.R., Get S., Tripathi A., et al. In: Augmenting Crop Productivity in Stress Environment. Ansari S.A., Ansari M.I., Husen A., editors. Springer Nature Singapore; 2022. Breeding Efforts for Crop Productivity in Abiotic Stress Environment; pp. 63–103. [Google Scholar]
- 69.Yang Y, Saand MA, Huang L, et al. Applications of Multi-Omics Technologies for Crop Improvement. Review. Frontiers in Plant Science. 2021-September-03 2021;12Doi: 10.3389/fpls.2021.563953. [DOI] [PMC free article] [PubMed]
- 70.Rao A., Barkley D., França G.S., Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596(7871):211–220. doi: 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lill J.R., Mathews W.R., Rose C.M., Schirle M. Proteomics in the pharmaceutical and biotechnology industry: a look to the next decade. Expert Rev Proteomics. 2021;18(7):503–526. doi: 10.1080/14789450.2021.1962300. [DOI] [PubMed] [Google Scholar]
- 72.Chen Y., Wang Y., Yang J., Zhou W., Dai S. Exploring the diversity of plant proteome. J Integr Plant Biol. 2021;63(7):1197–1210. doi: 10.1111/jipb.13087. [DOI] [PubMed] [Google Scholar]
- 73.Bhushan S, Sharma D, Kaul S, Dhar MK, Sharma M. Recent Strategies to Engineer Alkaloid Biosynthesis in Medicinal Plants. Medicinal Plants: Their Response to Abiotic Stress. Springer; 2023:391-416.
- 74.Mahapatra K., Mukherjee A., Suyal S., Dar M.A., Bhagavatula L., Datta S. Regulation of chloroplast biogenesis, development, and signaling by endogenous and exogenous cues. Physiol Mol Biol Plants. 2024;30(2):167–183. doi: 10.1007/s12298-024-01427-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Christopher J.A., Stadler C., Martin C.E., et al. Subcellular proteomics. Nature Reviews Methods Primers. 2021;1:32. doi: 10.1038/s43586-021-00029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma M., Sidhu A.K., Samota M.K., Gupta M., Koli P., Choudhary M. Post-translational modifications in histones and their role in abiotic stress tolerance in plants. Proteomes. 2023;11(4):38. doi: 10.3390/proteomes11040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaya C., Uğurlar F., Adamakis I.-D.-S. Molecular mechanisms of CBL-CIPK signaling pathway in plant abiotic stress tolerance and hormone crosstalk. Int J Mol Sci. 2024;25:5043. doi: 10.3390/ijms25095043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muleya V., Lois L.M., Chahtane H., Thomas L., Chiapello M., Marondedze C. (De)Activation (Ir)reversibly or degradation: dynamics of post-translational protein modifications in plants. Life. 2022;12(2):324. doi: 10.3390/life12020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Z., Zhu L., Zhao F., et al. Plant salinity stress response and nano-enabled plant salt tolerance. Frontiers in Plant Science. 2022;13 doi: 10.3389/fpls.2022.843994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh M., Nara U., Kumar A., Choudhary A., Singh H., Thapa S. Salinity tolerance mechanisms and their breeding implications. J Genet Eng Biotechnol. 2021;19:173. doi: 10.1186/s43141-021-00274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gul Z., Tang Z.-H., Arif M., Ye Z. An insight into abiotic stress and influx tolerance mechanisms in plants to cope in saline environments. Biology. 2022;11(4):597. doi: 10.3390/biology11040597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Athar H-u-R, Zulfiqar F, Moosa A, et al. Salt stress proteins in plants: An overview. Review. Frontiers in Plant Science. 2022-December-16 2022;13Doi: 10.3389/fpls.2022.999058. [DOI] [PMC free article] [PubMed]
- 83.Chowdhury S., Basu A., Kundu S. Overexpression of a new osmotin-like protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. original research. Front Plant Sci. 2017:8. doi: 10.3389/fpls.2017.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bashir M.A., Silvestri C., Ahmad T., et al. Osmotin: a cationic protein leads to improve biotic and abiotic stress tolerance in plants. Plants. 2020;9(8):992. doi: 10.3390/plants9080992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ozturk M., Turkyilmaz Unal B., García-Caparrós P., Khursheed A., Gul A., Hasanuzzaman M. Osmoregulation and its actions during the drought stress in plants. Physiol Plant. 2021;172(2):1321–1335. doi: 10.1111/ppl.13297. [DOI] [PubMed] [Google Scholar]
- 86.Rathor P., Borza T., Bahmani R., et al. Expression of a heat shock protein 70 from the brown alga ectocarpus sp. imparts salinity stress tolerance in arabidopsis thaliana. J Appl Phycol. 2023;35(2):803–819. doi: 10.1007/s10811-022-02897-7. [DOI] [Google Scholar]
- 87.López A.M.Q., dos Santos Silva A.L. Proteomics and bioremediation using prokaryotes. Genomics Approach to Bioremediation. 2023:485–502. [Google Scholar]
- 88.Raggi L, Caproni L, Ciancaleoni S, D’Amato R, Businelli D, Negri V. Investigating the genetic basis of salt-tolerance in common bean: a genome-wide association study at the early vegetative stage. Scientific Reports. 2024/03/04 2024;14(1):5315. Doi: 10.1038/s41598-024-55403-z. [DOI] [PMC free article] [PubMed]
- 89.Daldoul S., Hanzouli F., Hamdi Z., et al. The root transcriptome dynamics reveals new valuable insights in the salt-resilience mechanism of wild grapevine (Vitis vinifera subsp. sylvestris). Original Research. Front Plant Sci. 2022;13 doi: 10.3389/fpls.2022.1077710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Waters E.R., Vierling E. Plant small heat shock proteins – evolutionary and functional diversity. New Phytol. 2020;227(1):24–37. doi: 10.1111/nph.16536. [DOI] [PubMed] [Google Scholar]
- 91.Halder T., Choudhary M., Liu H., Chen Y., Yan G., Siddique K.H.M. Wheat proteomics for abiotic stress tolerance and root system architecture: current status and future prospects. Proteomes. 2022;10(2):17. doi: 10.3390/proteomes10020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian J.-j., Zhang J.-m., Yu E.-m., et al. Identification and analysis of lipid droplet-related proteome in the adipose tissue of grass carp (Ctenopharyngodon idella) under fed and starved conditions. Comp Biochem Physiol D: Genomics Proteomics. 2020;36 doi: 10.1016/j.cbd.2020.100710. [DOI] [PubMed] [Google Scholar]
- 93.Raza A., Ashraf F., Zou X., Zhang X., Tosif H. In: Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives i: General Consequences and Plant Responses. Hasanuzzaman M., editor. Springer Singapore; 2020. Plant Adaptation and Tolerance to Environmental Stresses: Mechanisms and Perspectives; pp. 117–145. [Google Scholar]
- 94.Hussain H., Mustafa Kamal M., Al-Obaidi J.R., Hamdin N.E., Ngaini Z., Mohd-Yusuf Y. Proteomics of sago palm towards identifying contributory proteins in stress-tolerant cultivar. Protein J. 2020;39(1):62–72. doi: 10.1007/s10930-019-09878-9. [DOI] [PubMed] [Google Scholar]
- 95.Cui Y., Zeng X., Xiong Q., et al. Combining quantitative trait locus and co-expression analysis allowed identification of new candidates for oil accumulation in rapeseed. J Exp Bot. 2020;72(5):1649–1660. doi: 10.1093/jxb/eraa563. [DOI] [PubMed] [Google Scholar]
- 96.Yan M., Zheng L., Li B., Shen R., Lan P. Comparative proteomics reveals new insights into the endosperm responses to drought, salinity and submergence in germinating wheat seeds. Plant Mol Biol. 2021;105(3):287–302. doi: 10.1007/s11103-020-01087-8. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y., Li D., Zhou R., et al. A collection of transcriptomic and proteomic datasets from sesame in response to salt stress. Data Brief. 2020;32 doi: 10.1016/j.dib.2020.106096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nie L., Feng J., Fan P., et al. Comparative proteomics of root plasma membrane proteins reveals the involvement of calcium signalling in NaCl-facilitated nitrate uptake in Salicornia europaea. J Exp Bot. 2015;66(15):4497–4510. doi: 10.1093/jxb/erv216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luo J., Tang S., Peng X., et al. Elucidation of cross-talk and specificity of early response mechanisms to salt and PEG-simulated drought stresses in brassica napus using comparative proteomic analysis. PLoS One. 2015;10(10):e0138974. doi: 10.1371/journal.pone.0138974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y., Wei M., Liu A., et al. Comparative proteomic analysis of two sesame genotypes with contrasting salinity tolerance in response to salt stress. J Proteomics. 2019;201:73–83. doi: 10.1016/j.jprot.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 101.Kholghi M., Toorchi M., Bandehagh A., et al. Comparative proteomic analysis of salt-responsive proteins in canola roots by 2-DE and MALDI-TOF MS. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2019;1867(3):227–236. doi: 10.1016/j.bbapap.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 102.Shaki F., Ebrahimzadeh Maboud H., Niknam V. Differential proteomics: Effect of growth regulators on salt stress responses in safflower seedlings. Pestic Biochem Physiol. 2020;164:149–155. doi: 10.1016/j.pestbp.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 103.Wang Y., Peng X., Salvato F., et al. Salt-adaptive strategies in oil seed crop Ricinus communis early seedlings (cotyledon vs. true leaf) revealed from proteomics analysis. Ecotoxicol Environ Saf. 2019;171:12–25. doi: 10.1016/j.ecoenv.2018.12.046. [DOI] [PubMed] [Google Scholar]
- 104.Bittencourt C.B., Carvalho da Silva T.L., Rodrigues Neto J.C., et al. Insights from a multi-omics integration (MOI) study in oil palm (Elaeis guineensis Jacq.) response to abiotic stresses: part one—salinity. Plants. 2022;11:1755. doi: 10.3390/plants11131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ben Abdallah M., Trupiano D., Polzella A., et al. Unraveling physiological, biochemical and molecular mechanisms involved in olive (Olea europaea L. cv. Chétoui) tolerance to drought and salt stresses. J Plant Physiol. 2018;220:83–95. doi: 10.1016/j.jplph.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 106.Banaei-Asl F., Bandehagh A., Uliaei E.D., et al. Proteomic analysis of canola root inoculated with bacteria under salt stress. J Proteomics. 2015;124:88–111. doi: 10.1016/j.jprot.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 107.Banaei-Asl F., Farajzadeh D., Bandehagh A., Komatsu S. Comprehensive proteomic analysis of canola leaf inoculated with a plant growth-promoting bacterium, Pseudomonas fluorescens, under salt stress. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2016;1864(9):1222–1236. doi: 10.1016/j.bbapap.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 108.Javed M., Zafar Z.U., Ashraf M. Leaf proteome analysis signified that photosynthesis and antioxidants are key indicators of salinity tolerance in canola (Brassica napus L.) Pak J Bot. 2019;51(6):1955–1968. doi: 10.30848/PJB2019-6(38). [DOI] [Google Scholar]
- 109.Çulha Erdal Ş., Eyidoğan F., Ekmekçi Y. Comparative physiological and proteomic analysis of cultivated and wild safflower response to drought stress and re-watering. Physiol Mol Biol Plants. 2021;27(2):281–295. doi: 10.1007/s12298-021-00934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jia H., Shao M., He Y., Guan R., Chu P., Jiang H. Proteome dynamics and physiological responses to short-term salt stress in brassica napus leaves. PLOS One. 2015;10(12):e0144808. doi: 10.1371/journal.pone.0144808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alkharabsheh H.M., Seleiman M.F., Hewedy O.A., et al. Field crop responses and management strategies to mitigate soil salinity in modern agriculture: a review. Agronomy. 2021;11(11):2299. doi: 10.3390/agronomy11112299. [DOI] [Google Scholar]
- 112.Wang X., Yao Q., Zhang D.-m., et al. Effects of acute salinity stress on osmoregulation, antioxidant capacity and physiological metabolism of female Chinese mitten crabs (Eriocheir sinensis) Aquaculture. 2022;552 doi: 10.1016/j.aquaculture.2022.737989. [DOI] [Google Scholar]
- 113.Sugumar T., Shen G., Smith J., Zhang H. Creating Climate-resilient crops by increasing drought, heat, and salt tolerance. Plants. 2024;13(9):1238. doi: 10.3390/plants13091238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chakraborty K., Basak N., Bhaduri D., et al. In: Plant Nutrients and Abiotic Stress Tolerance. Hasanuzzaman M., Fujita M., Oku H., Nahar K., Hawrylak-Nowak B., editors. Springer Singapore; 2018. Ionic Basis of Salt Tolerance in Plants: Nutrient Homeostasis and Oxidative Stress Tolerance; pp. 325–362. [Google Scholar]
- 115.Hasanuzzaman M., Fujita M. Plant responses and tolerance to salt stress: physiological and molecular interventions. Int J Mol Sci. 2022;23(9):4810. doi: 10.3390/ijms23094810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Al-Obaidi J.R., Jamaludin A.A., Rahman N.A., Ahmad-Kamil E.I. How plants respond to heavy metal contamination: a narrative review of proteomic studies and phytoremediation applications. Planta. 2024;259:103. doi: 10.1007/s00425-024-04378-2. [DOI] [PubMed] [Google Scholar]
- 117.Prusty A., Panchal A., Singh R.K., Prasad M. Major transcription factor families at the nexus of regulating abiotic stress response in millets: a comprehensive review. Planta. 2024;259:118. doi: 10.1007/s00425-024-04394-2. [DOI] [PubMed] [Google Scholar]
- 118.Altaf M.A., Mandal S., Behera B., et al. Salinity stress tolerance in solanaceous crops: current understanding and its prospects in genome editing. J Plant Growth Regul. 2023;42(7):4020–4036. doi: 10.1007/s00344-022-10890-0. [DOI] [Google Scholar]
- 119.Shah W.H., Rasool A., Saleem S., et al. Understanding the integrated pathways and mechanisms of transporters, protein kinases, and transcription factors in plants under salt stress. International Journal of Genomics. 2021;2021 doi: 10.1155/2021/5578727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Venkataraman G., Shabala S., Véry A.-A., et al. To exclude or to accumulate? Revealing the role of the sodium HKT1;5 transporter in plant adaptive responses to varying soil salinity. Plant Physiol Biochem. 2021;169:333–342. doi: 10.1016/j.plaphy.2021.11.030. [DOI] [PubMed] [Google Scholar]
- 121.Nadeem M., Ali M., Kubra G., et al. In: Climate Change and Food Security with Emphasis on Wheat. Ozturk M., Gul A., editors. Academic Press; 2020. Chapter 6 - Role of osmoprotectants in salinity tolerance in wheat; pp. 93–106. [Google Scholar]
- 122.Aazami M.A., Rasouli F., Ebrahimzadeh A. Oxidative damage, antioxidant mechanism and gene expression in tomato responding to salinity stress under in vitro conditions and application of iron and zinc oxide nanoparticles on callus induction and plant regeneration. BMC Plant Biol. 2021;21:597. doi: 10.1186/s12870-021-03379-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu Y., Yu M., Zhang S., et al. Transcriptomic identification of wheat AP2/ERF transcription factors and functional characterization of TaERF-6-3A in response to drought and salinity stresses. Int J Mol Sci. 2022;23(6):3272. doi: 10.3390/ijms23063272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu H., Tang X., Zhang N., Li S., Si H. Role of bZIP transcription factors in plant salt stress. Int J Mol Sci. 2023;24(9):7893. doi: 10.3390/ijms24097893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Punia H., Tokas J., Malik A., et al. Genome-wide transcriptome profiling, characterization, and functional identification of NAC transcription factors in sorghum under salt stress. Antioxidants. 2021;10(10):1605. doi: 10.3390/antiox10101605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang M., Yu Z., Zeng D., et al. Transcriptome and metabolome reveal salt-stress responses of leaf tissues from dendrobium officinale. Biomolecules. 2021;11(5):736. doi: 10.3390/biom11050736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jia J., Liang Y., Gou T., et al. The expression response of plasma membrane aquaporins to salt stress in tomato plants. Environ Exp Bot. 2020;178 doi: 10.1016/j.envexpbot.2020.104190. [DOI] [Google Scholar]
- 128.Ge X., Du J., Zhang L., Qu G., Hu J. PeCLH2 gene positively regulate salt tolerance in transgenic populus alba × populus glandulosa. Genes. 2023;14(3) doi: 10.3390/genes14030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shelake RM, Kadam US, Kumar R, Pramanik D, Singh AK, Kim JY. Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives. Plant Commun. Nov 14 2022;3(6):100417. doi:10.1016/j.xplc.2022.100417. [DOI] [PMC free article] [PubMed]
- 130.Ritonga F.N., Ngatia J.N., Wang Y., Khoso M.A., Farooq U., Chen S. AP2/ERF, an important cold stress-related transcription factor family in plants: a review. Physiol Mol Biol Plants. 2021;27(9):1953–1968. doi: 10.1007/s12298-021-01061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ganguly M., Roychoudhury A., Sengupta D.N., Datta S.K., Datta K. Independent overexpression of OsRab16A and AtDREB1A exhibit enhanced drought tolerance in transgenic aromatic rice variety Pusa Sugandhi 2. J Plant Biochem Biotechnol. 2020;29(3):503–517. doi: 10.1007/s13562-020-00565-w. [DOI] [Google Scholar]
- 132.Chen D., Fu L., Su T., et al. N6-methyladenosine methylation analysis reveals transcriptome-wide expression response to salt stress in rice roots. Environ Exp Bot. 2022;201 doi: 10.1016/j.envexpbot.2022.104945. [DOI] [Google Scholar]
- 133.Liu X., Wu D., Shan T., et al. The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol Biol. 2020;103(4):545–560. doi: 10.1007/s11103-020-01010-1. [DOI] [PubMed] [Google Scholar]
- 134.Yang C, Huang Y, Lv P, et al. NAC Transcription Factor GmNAC12 Improved Drought Stress Tolerance in Soybean. Int J Mol Sci. Oct 10 2022;23(19)doi:10.3390/ijms231912029. [DOI] [PMC free article] [PubMed]
- 135.Dai R, Zhan N, Geng R, et al. Progress on Salt Tolerance in Brassica napus. Plants (Basel). Jul 21 2024;13(14)doi:10.3390/plants13141990. [DOI] [PMC free article] [PubMed]
- 136.Goswami S.K., Kashyap A.S., Kumar R., Gujjar R.S., Singh A., Manzar N. Harnessing rhizospheric microbes for eco-friendly and sustainable crop production in saline environments. Curr Microbiol. 2023;81:14. doi: 10.1007/s00284-023-03538-z. [DOI] [PubMed] [Google Scholar]
- 137.Malakar P., Chattopadhyay D. Adaptation of plants to salt stress: the role of the ion transporters. J Plant Biochem Biotechnol. 2021;30(4):668–683. doi: 10.1007/s13562-021-00741-6. [DOI] [Google Scholar]
- 138.Xie Q, Zhou Y, Jiang X. Structure, function, and regulation of the plasma membrane Na+/H+ antiporter salt overly sensitive 1 in plants. Review. Frontiers in Plant Science. 2022-March-31 2022;13Doi: 10.3389/fpls.2022.866265. [DOI] [PMC free article] [PubMed]
- 139.Afzal M., Hindawi S.E.S., Alghamdi S.S., et al. Potential breeding strategies for improving salt tolerance in crop plants. J Plant Growth Regul. 2023;42(6):3365–3387. doi: 10.1007/s00344-022-10797-w. [DOI] [Google Scholar]
- 140.Gadelha C.G., Coutinho Í.A.C., Pinheiro S.K.P., et al. Sodium uptake and transport regulation, and photosynthetic efficiency maintenance as the basis of differential salt tolerance in rice cultivars. Environ Exp Bot. 2021;192 doi: 10.1016/j.envexpbot.2021.104654. [DOI] [Google Scholar]
- 141.Tsujii M., Tanudjaja E., Uozumi N. Diverse physiological functions of cation proton antiporters across bacteria and plant cells. Int J Mol Sci. 2020;21(12):4566. doi: 10.3390/ijms21124566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Suo J., Zhao Q., David L., Chen S., Dai S. Salinity response in chloroplasts: insights from gene characterization. Int J Mol Sci. 2017;18(5):1011. doi: 10.3390/ijms18051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ali S., Tyagi A., Park S., Bae H. Understanding the mechanobiology of phytoacoustics through molecular Lens: mechanisms and future perspectives. J Adv Res. 2023 doi: 10.1016/j.jare.2023.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Farooq N., Khan M.O., Ahmed M.Z., et al. Salt-induced modulation of ion transport and PSII photoprotection determine the salinity tolerance of amphidiploid brassicas. Plants. 2023;12(14):2590. doi: 10.3390/plants12142590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sardans J., Peñuelas J. Potassium control of plant functions: ecological and agricultural implications. Plants. 2021;10(2):419. doi: 10.3390/plants10020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Han Y., Yin S., Huang L., et al. A sodium transporter HvHKT1;1 confers salt tolerance in barley via regulating tissue and cell ion homeostasis. Plant Cell Physiol. 2018;59(10):1976–1989. doi: 10.1093/pcp/pcy116. [DOI] [PubMed] [Google Scholar]
- 147.Raddatz N, Morales de los Ríos L, Lindahl M, Quintero FJ, Pardo JM. Coordinated Transport of Nitrate, Potassium, and Sodium. Review. Frontiers in Plant Science. 2020-March-06 2020;11Doi: 10.3389/fpls.2020.00247. [DOI] [PMC free article] [PubMed]
- 148.Ali A., Raddatz N., Pardo J.M., Yun D.-J. HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species. Physiol Plant. 2021;171(4):546–558. doi: 10.1111/ppl.13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gao L.-W., Yang S.-L., Wei S.-W., Huang D.-F., Zhang Y.-D. Supportive role of the Na+ transporter CmHKT1;1 from Cucumis melo in transgenic Arabidopsis salt tolerance through improved K+/Na+ balance. Plant Mol Biol. 2020;103(4):561–580. doi: 10.1007/s11103-020-01011-0. [DOI] [PubMed] [Google Scholar]
- 150.Hafeez M.B., Raza A., Zahra N., et al. In: Handbook of Bioremediation. Hasanuzzaman M., Prasad M.N.V., editors. Academic Press; 2021. Chapter 22 - Gene regulation in halophytes in conferring salt tolerance; pp. 341–370. [Google Scholar]
- 151.Wu D., Saleem M., He T., He G. The mechanism of metal homeostasis in plants: a new view on the synergistic regulation pathway of membrane proteins, lipids and metal ions. Membranes. 2021;11(12):984. doi: 10.3390/membranes11120984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yuan S., Zhao J., Li Z., et al. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Horticulture. Research. 2019;6 doi: 10.1038/s41438-019-0130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Waheed S., Anwar M., Saleem M.A., Wu J., Tayyab M., Hu Z. The critical role of small RNAs in regulating plant innate immunity. Biomolecules. 2021;11(2):184. doi: 10.3390/biom11020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xu P., Guo Q., Meng S., et al. Genome-wide association analysis reveals genetic variations and candidate genes associated with salt tolerance related traits in Gossypium hirsutum. BMC Genomics. 2021;22:26. doi: 10.1186/s12864-020-07321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lokeshkumar B.M., Krishnamurthy S.L., Rathor S., et al. Morphophysiological diversity and haplotype analysis of saltol QTL region in diverse rice landraces for salinity tolerance. Rice Sci. 2023;30(4):306–320. doi: 10.1016/j.rsci.2023.02.001. [DOI] [Google Scholar]
- 156.Shehzad M, Ditta A, Cai X, et al. Identification of salt stress-tolerant candidate genes in the BC2F2 population at the seedling stages of G. hirsutum and G. darwinii using NGS-based bulked segregant analysis. Original Research. Frontiers in Plant Science. 2023-July-03 2023;14Doi: 10.3389/fpls.2023.1125805. [DOI] [PMC free article] [PubMed]
- 157.Cao Y., Zhou X., Song H., Zhang M., Jiang C. Advances in deciphering salt tolerance mechanism in maize. The Crop Journal. 2023;11(4):1001–1010. doi: 10.1016/j.cj.2022.12.004. [DOI] [Google Scholar]
- 158.Cho K.-H., Kim M.Y., Kwon H., Yang X., Lee S.-H. Novel QTL identification and candidate gene analysis for enhancing salt tolerance in soybean (Glycine max (L.) Merr.) Plant Sci. 2021;313 doi: 10.1016/j.plantsci.2021.111085. [DOI] [PubMed] [Google Scholar]
- 159.Taha M.S., Haghighi F., Stefanski A., et al. Novel FMRP interaction networks linked to cellular stress. FEBS J. 2021;288(3):837–860. doi: 10.1111/febs.15443. [DOI] [PubMed] [Google Scholar]
- 160.Butt H., Bazin J., Prasad K.V.S.K., et al. The rice serine/arginine splicing factor RS33 regulates Pre-mRNA splicing during abiotic stress responses. Cells. 2022;11(11):1796. doi: 10.3390/cells11111796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Niu X., Luo T., Zhao H., Su Y., Ji W., Li H. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene. 2020;740 doi: 10.1016/j.gene.2020.144514. [DOI] [PubMed] [Google Scholar]
- 162.Rosenkranz RRE, Ullrich S, Löchli K, Simm S, Fragkostefanakis S. Relevance and Regulation of Alternative Splicing in Plant Heat Stress Response: Current Understanding and Future Directions. Review. Frontiers in Plant Science. 2022-June-23 2022;13Doi: 10.3389/fpls.2022.911277. [DOI] [PMC free article] [PubMed]
- 163.Ju Y.-l., Min Z., Zhang Y., Zhang K.-k., Liu M. Fang Y-l. Transcriptome profiling provide new insights into the molecular mechanism of grapevine response to heat, drought, and combined stress. Sci Hortic. 2021 doi: 10.1016/j.scienta.2021.110076. [DOI] [Google Scholar]
- 164.Angarola B.L., Anczuków O. Splicing alterations in healthy aging and disease. WIREs RNA. 2021;12(4):e1643. doi: 10.1002/wrna.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nizam A., Meera S.P., Kumar A. Genetic and molecular mechanisms underlying mangrove adaptations to intertidal environments. iScience. 2022;25 doi: 10.1016/j.isci.2021.103547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sengupta S., Pehlivan N., Mangu V., Rajasekaran K., Baisakh N. Characterization of a stress-enhanced promoter from the grass halophyte, spartina alterniflora L. Biology. 2022;11(12):1828. doi: 10.3390/biology11121828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zulfiqar F., Akram N.A., Ashraf M. Osmoprotection in plants under abiotic stresses: new insights into a classical phenomenon. Planta. 2019;251:3. doi: 10.1007/s00425-019-03293-1. [DOI] [PubMed] [Google Scholar]
- 168.Ejaz S., Fahad S., Anjum M.A., et al. In: Sustainable Agriculture Reviews 39. Lichtfouse E., editor. Springer International Publishing; 2020. Role of Osmolytes in the Mechanisms of Antioxidant Defense of Plants; pp. 95–117. [Google Scholar]
- 169.Khare T., Jamla M., Mathur V., Kumar V. Exploring halobiome resources for developing salt-tolerant crops: a perspective review. J Plant Growth Regul. 2024 doi: 10.1007/s00344-024-11266-2. [DOI] [Google Scholar]
- 170.Sami A., Xue Z., Tazein S., et al. CRISPR–Cas9-based genetic engineering for crop improvement under drought stress. Bioengineered. 2021;12(1):5814–5829. doi: 10.1080/21655979.2021.1969831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li N, Zeng Y, Wang M, et al. Characterization of phage resistance and their impacts on bacterial fitness in Pseudomonas aeruginosa. Microbiology Spectrum. 2022;10(5):e02072-22. Doi: 10.1128/spectrum.02072-22. [DOI] [PMC free article] [PubMed]
- 172.León-Félix J, Villicaña C. The impact of quorum sensing on the modulation of phage-host interactions. J Bacteriol. 2021;203(11):10.1128/jb. 00687-20. Doi: 10.1128/jb.00687-20. [DOI] [PMC free article] [PubMed]
- 173.Molla K.A., Sretenovic S., Bansal K.C., Qi Y. Precise plant genome editing using base editors and prime editors. Nat Plants. 2021;7(9):1166–1187. doi: 10.1038/s41477-021-00991-1. [DOI] [PubMed] [Google Scholar]
- 174.Bandyopadhyay A., Kancharla N., Javalkote V.S., Dasgupta S., Brutnell T.P. CRISPR-Cas12a (Cpf1): a versatile tool in the plant genome editing tool box for agricultural advancement. Review. Frontiers in Plant Science. 2020 doi: 10.3389/fpls.2020.584151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Atta K, Mondal S, Gorai S, et al. Impacts of salinity stress on crop plants: improving salt tolerance through genetic and molecular dissection. Review. Frontiers in Plant Science. 2023-September-15 2023;14Doi: 10.3389/fpls.2023.1241736. [DOI] [PMC free article] [PubMed]
- 176.Billah M., Aktar S., Brestic M., et al. Progressive genomic approaches to explore drought-and salt-induced oxidative stress responses in plants under changing climate. Plants. 2021;10(9):1910. doi: 10.3390/plants10091910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hasan M.M., Liu X.-D., Waseem M., et al. ABA activated SnRK2 kinases: an emerging role in plant growth and physiology. Plant Signal Behav. 2022;17 doi: 10.1080/15592324.2022.2071024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yoshida T., Mogami J., Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 179.Hina A., Abbasi A., Arshad M., et al. Utilization of multi-omics approaches for crop improvement. OMICs-based Techniques for Global Food Security. 2024:91–121. [Google Scholar]
- 180.Pandey P., Srivastava S., Pandey A.K., Dubey R.S. In: Plant Life under Changing Environment. Tripathi D.K., Pratap Singh V., Chauhan D.K., editors. Academic Press; 2020. Chapter 22 - Abiotic-stress tolerance in plants-system biology approach; pp. 577–609. [Google Scholar]
- 181.Eicher T., Kinnebrew G., Patt A., et al. Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites. 2020;10(5):202. doi: 10.3390/metabo10050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mahmood U., Li X., Fan Y., et al. Multi-omics revolution to promote plant breeding efficiency. Systematic Review. Frontiers Plant Sci. 2022 doi: 10.3389/fpls.2022.1062952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Derbyshire M.C., Batley J., Edwards D. Use of multiple ‘omics techniques to accelerate the breeding of abiotic stress tolerant crops. Current Plant Biol. 2022;32 doi: 10.1016/j.cpb.2022.100262. [DOI] [Google Scholar]
- 184.Ullah M.A., Abdullah-Zawawi M.-R., Zainal-Abidin R.-A., Sukiran N.L., Uddin M.I., Zainal Z. A review of integrative omic approaches for understanding rice salt response mechanisms. Plants. 2022;11(11):1430. doi: 10.3390/plants11111430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yadav A.K., Shree B., Lakhwani D., Singh A.K. In: Integrative Omics. Gupta M.K., Katara P., Mondal S., Singh R.L., editors. Academic Press; 2024. Chapter 18 - Omics technologies for crop improvement; pp. 293–313. [Google Scholar]
- 186.Desa S., Saidin S., Azeyanty J.A., et al. In: New Frontiers in Plant-Environment Interactions: Innovative Technologies and Developments. Aftab T., editor. Springer Nature Switzerland; 2023. Plant-Environment Interactions: Proteomics, Metabolomics and Genetic Engineering Perspective; pp. 15–51. [Google Scholar]
- 187.Su H, Ma D, Zhu H, Liu Z, Gao F. Transcriptomic response to three osmotic stresses in gills of hybrid tilapia (Oreochromis mossambicus female × O. urolepis hornorum male). BMC Genomics. 2020/01/31 2020;21(1):110. Doi: 10.1186/s12864-020-6512-5. [DOI] [PMC free article] [PubMed]
- 188.Kumar N., Soren K.R., Bharadwaj C., et al. Genome-wide transcriptome analysis and physiological variation modulates gene regulatory networks acclimating salinity tolerance in chickpea. Environ Exp Bot. 2021;187 doi: 10.1016/j.envexpbot.2021.104478. [DOI] [Google Scholar]
- 189.Shi P., Gu M. Transcriptome analysis and differential gene expression profiling of two contrasting quinoa genotypes in response to salt stress. BMC Plant Biol. 2020;20:568. doi: 10.1186/s12870-020-02753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang W., Pang J., Zhang F., Sun L., Yang L., Siddique K.H.M. Transcriptomic and metabolomics-based analysis of key biological pathways reveals the role of lipid metabolism in response to salt stress in the root system of Brassica napus. Plant Growth Regul. 2022;97(1):127–141. doi: 10.1007/s10725-021-00788-4. [DOI] [Google Scholar]
- 191.Geng L., Zhang W., Zou T., et al. Integrating linkage mapping and comparative transcriptome analysis for discovering candidate genes associated with salt tolerance in rice. Original Research. Frontiers Plant Sci. 2023:14. doi: 10.3389/fpls.2023.1065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sarri E., Termentzi A., Abraham E.M., et al. Salinity stress alters the secondary metabolic profile of M. sativa, M. arborea and their hybrid (Alborea) Int J Mol Sci. 2021;22:4882. doi: 10.3390/ijms22094882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hamany Djande C.Y., Pretorius C., Tugizimana F., Piater L.A., Dubery I.A. Metabolomics: a tool for cultivar phenotyping and investigation of grain crops. Agronomy. 2020;10(6):831. doi: 10.3390/agronomy10060831. [DOI] [Google Scholar]
- 194.Razzaq M.K., Aleem M., Mansoor S., et al. Omics and CRISPR-Cas9 approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int J Mol Sci. 2021;22(3):1292. doi: 10.3390/ijms22031292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Iqbal Z., Iqbal M.S., Khan M.I.R., Ansari M.I. Toward integrated multi-omics intervention: rice trait improvement and stress management. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.741419. [DOI] [PMC free article] [PubMed] [Google Scholar]