Abstract
Although the metabolism of yolk lipids such as docosahexaenoic acid (DHA) is pivotal for embryonic development, the underlying mechanism remains elusive. Here we find that the zebrafish hydroxysteroid (17-β) dehydrogenase 12a (hsd17b12a), which encodes an intestinal epithelial-specific enzyme, is essential for the biosynthesis of long-chain polyunsaturated fatty acids in primitive intestine of larval fish. The deficiency of hsd17b12a leads to severe developmental defects in the primitive intestine and exocrine pancreas. Mechanistically, hsd17b12a deficiency interrupts DHA synthesis from essential fatty acids derived from yolk-deposited triglycerides, and consequently disrupts the intestinal DHA-phosphatidic acid (PA)-phosphatidylglycerol (PG) axis. This ultimately results in developmental defects of digestive organs, primarily driven by ferroptosis. Our findings indicate that the DHA-PA-PG axis in the primitive intestine facilitates the uptake of yolk lipids and promotes the expansion of digestive organs, thereby uncovering a mechanism through which DHA regulates embryonic development.
Subject terms: Organogenesis, Cell death, Development
Lipid utilization from the yolk sac plays a crucial role in embryonic development. Here, Chen et al. generate hsd17b12a mutant zebrafish and discover that a docosahexaenoic acid (DHA)-phosphatidic acid (PA)-phosphatidylglycerol (PG) (DHA-PA-PG) axis facilitates the uptake of yolk lipids and promotes the expansion of digestive organs.
Introduction
The yolk sac stores a large amount of maternal material accumulated during oogenesis, and serves as a nutrient source for embryonic development in both viviparous and oviparous vertebrates1. The metabolism of maternal materials plays an important role in embryonic development2. Unlike viviparous embryos, oviparous embryos depend entirely on yolk nutrients, highlighting the importance of the absorption and utilization of yolk nutrients for their embryonic development3. Zebrafish, as oviparous organisms, depend solely on a single yolk for nutrition until the endogenous-to-exogenous nutrient source transition (eeNST), which mainly occurs at 5 days post-fertilization (5 dpf)4. In zebrafish, the yolk syncytial layer (YSL) hydrolyzes lipids to release fatty acids and produces lipoproteins that export lipids to the developing embryos until exogenous nutrients are absorbed5. Previous studies also suggest that the primitive intestine is involved in the absorption of yolk lipids during embryonic development. For instance, lipid droplets were detected in the intestinal epithelial cells of sea bass during the lecithotrophic period6, and yolk-deposited fatty acids could be transported to the primitive intestine, and participate in lipid synthesis of zebrafish7. Dietary phosphatidylcholines (PCs) have been shown to accelerate the metabolism of triglycerides (TAGs) in the intestinal epithelium8. Nevertheless, the regulation of yolk lipid absorption by the primitive intestine has yet to be investigated.
Phospholipids (PLs), including phosphatidic acids (PAs), phosphatidylglycerols (PGs), and PCs, can be distinguished based on the variations in the sn-1 and sn-2-linked fatty acid (FA) composition9. Saturated fatty acids (SFAs) and monounsaturated fatty acids (MUFAs) are preferably linked to the sn-1 position of PLs, whereas long-chain poly-unsaturated fatty acids (LC-PUFAs) are linked to the sn-2 position, therefore the composition of side-chain fatty acids is closely related to the function of PLs10. It was reported that the 3H-palmitate injected into the yolk could be involved in PL metabolism of larvae at 3 dpf7. Yolk cells are also active in lipid metabolism during the development of zebrafish embryos11. During embryonic development of zebrafish prior to exogenous feeding, there is a gradual increase in the total amount of omega-3 (n-3) LC-PUFAs12, suggesting that zebrafish embryos are capable of LC-PUFA biosynthesis from maternally deposited PUFAs. However, LC-PUFAs cannot be synthesized de novo due to the genetic absence of ∆12 and ∆15 desaturase coding genes13, necessitating the conversion of linoleic acid (LA) and α-linolenic acid (ALA) from the yolk into LC-PUFAs via fatty acid elongation. Therefore, disruption of the LC-PUFA elongation could be a promising approach to investigate the role of LC-PUFA biosynthesis pathway in the utilization of maternally-deposited yolk lipids.
Hydroxysteroid (17β) dehydrogenases (Hsd17b) are enzymes that catalyze the conversion of 17-ketosteroids into 17β-hydroxysteroids14. Hsd17b12, a member of this family, serves as a 3-ketoacyl-CoA reductase and is involved in the biosynthesizing of LC-PUFAs in mammals15. HSD17B12 is strongly expressed in organs involved in lipid metabolism, including the liver, kidneys, skeletal muscle, and placenta, during embryonic development16. Hsd17b12-knockout mice die at the embryonic stage due to severe disruptions in organogenesis17. Moreover, liver-specific knockout of Hsd17b12 in adult mice resulted in weight loss, reduced food and water intake, hepatic steatosis and accumulation of lipids with side chains shorter than 18-carbon fatty acids18. Nevertheless, it remains largely unknown how Hsd17b12 regulates nutrient absorption and organ development.
In this study, we generated a mutant of the zebrafish hsd17b12a, an orthologue of mammalian Hsd17b12, and used the hsd17b12a mutant model to study the role of yolk-deposited lipid in early development. We show that the contents of DHA, and DHA-derived phosphatidic acid (PA) and PA-originated phosphatidylglycerol (PG), mainly synthesized in the primitive intestine, are severely decreased after disruption of hsd17b12a, which is specifically expressed in the intestine. The disruption of DHA-PA-PG axis results in intestinal ferroptosis and developmental defects of digestive organs in the mutants. In conclusion, our study uncovers an intestinal DHA-PA-PG axis that promotes the expansion of digestive organs through the absorption and utilization of maternally-deposited yolk lipids during embryonic development.
Results
Loss of hsd17b12a leads to defective LC-PUFA synthesis and embryonic lethality
The absorption and utilization of lipids are crucial for development. After the endogenous-to-exogenous nutrient source transition (eeNST) of zebrafish at 5 dpf, lipid absorption primarily occurs in the intestine, while lipid metabolism is predominantly regulated by the liver3. To further investigate the lipid dynamics before eeNST, we re-analyzed the data from Fraher et al.11, and found that PL levels increase in the body while decreasing in the yolk (Supplementary Fig. 1a), indicating dynamic changes in lipid composition within the yolk and body before 5 dpf. By re-analyzing a bulk-seq data for the liver and primitive intestine at 5 dpf19, we identified a large number of differentially expressed genes (DEGs) between intestine and liver (Supplementary Fig. 1b). Notably, we found an enrichment of glycerolipid metabolism in the liver and glycerophospholipid metabolism in the primitive intestine (Fig. 1a, b), underscoring the importance of the primitive intestine in glycerophospholipid metabolism before eeNST. In contrast, a bulk-seq analysis of the liver and intestine in adult zebrafish showed that the liver becomes the primary site for glycerophospholipid metabolism (Supplementary Tables 1 and 2). Therefore, these results suggest that the primitive intestine might play an important role in supplying PLs necessary for embryonic development.
Fig. 1. LC-PUFA synthesis facilitates the embryo-to-larval transition.
a KEGG pathway analysis of genes highly expressed in intestine compared to liver at 5 dpf (p-value < 0.05). b KEGG pathway analysis of genes highly expressed in liver compared to intestine at 5 dpf (p-value < 0.05). c The heatmap illustrates enzyme expression related to long-chain polyunsaturated fatty acid (LC-PUFA) synthesis in the liver and intestine of WT at 5 dpf, showing p-values and effect sizes (n = 3) d Schematic of the knock-in strategy for inserting 5×Myc-P2A-mCherry into the hsd17b12a locus using CRISPR/Cas9. e The mCherry signal, confirmed not to be autofluorescence, co-localizes with the EGFP signal in the intestine-specific transgenic fish Tg(ET33J1:EGFP) at 5 dpf (n > 3). Scale bar, 200 μm. f Immunofluorescence staining of the intestine in hsd17b12aKI/KI using a Myc-tag antibody at 5 dpf (n = 3). Nuclei stained with DAPI. Scale bar, 200 μm. g Immunofluorescence staining of the intestine in hsd17b12aKI/KI using Myc-tag and ERp72 (ER) antibodies at 5 dpf (n = 3). Nuclei stained with DAPI. Scale bar, 200 μm. h Expression of genes related to LC-PUFA synthesis in WT at 4 dpf was detected using section in situ hybridization (n = 3). Scale bar, 50 μm. i Overexpression of hsd17b12a rescued the swim bladder inflation defect (n > 3). The arrows indicate the location of the swim bladder. Scale bar, 500 μm. j Survival curves were plotted for WT, hsd17b12a+/−, hsd17b12a−/−, hsd17b12a−/− with hsd17b12a mRNA, and Tg(CMV:hsd17b12a)/hsd17b12a−/−, respectively. k Expression of genes related to LC-PUFA synthesis in hsd17b12a−/− at 4 dpf was detected using section in situ hybridization (n = 3). Scale bar, 50 μm. l Expression of genes related to LC-PUFA synthesis in the primitive intestine was quantified by RT-PCR in WT and hsd17b12a−/− at 4 dpf. m Differential analysis of relative amounts of total FAs (fatty acids) in hsd17b12a−/− larvae compared to WT at 4 dpf. Data in l, m are represented as mean ± S.D., using a two-tailed Student’s t test; each point represents an independent biological sample (n = 3). Source data are provided as a Source Data file.
PLs are incorporated with LC-PUFAs which are synthesized via a series of LC-PUFA synthases20. Analysis of enzyme gene expression related to LC-PUFA synthesis revealed elevated levels of hsd17b12a in the primitive intestine compared to the liver (Fig. 1c). Whole-mount in situ hybridization (WISH) analysis revealed that hsd17b12a is specifically expressed in the primitive intestine at 4 dpf and 5 dpf (Supplementary Fig. 1c, d). To determine the cellular localization of Hsd17b12a, we generated a knock-in zebrafish line, hsd17b12aKI/KI, using a recently developed induced primordial germ cell transplantation (iPGCT) and CRISPR/Cas9-mediated knock-in technology21. This knock-in strategy introduced a Myc-tagged Hsd17b12a protein and an mCherry reporter, linked by a P2A peptide, into the hsd17b12a locus (Fig. 1d). In the hsd17b12aKI/KI line, mCherry exhibited no autofluorescence and co-localized with EGFP in intestinal epithelial cells of Tg(ET33J1:EGFP)22 (Fig. 1e; Supplementary Fig. 1e). Swim bladder inflation and embryo survival remained unaffected in both hsd17b12aKI/KI and hsd17b12aKI/− lines (Supplementary Fig. 1f–h), confirming that the knock-in did not impair function of endogenous Hsd17b12a protein. Immunofluorescence analysis using a Myc-tag antibody on primitive intestinal sections of hsd17b12aKI/KI embryos demonstrated specific expression of Hsd17b12a in intestinal epithelial cells (Fig. 1f), and the signals mainly co-localized with the endoplasmic reticulum (ER) marker ERp72 (Fig. 1g). We further analyzed the expression of the enzyme coding genes responsible for LC-PUFA synthesis, and found that these genes, such as elovl1b, elovl7a, hacd4, and elovl2, are specifically expressed in intestinal epithelial cells in WT embryos at 4 dpf (Fig. 1h). These results indicate that the primitive intestine specifically expresses hsd17b12a and other LC-PUFA synthesizing enzyme genes, which may enable the intestine to synthesize LC-PUFAs.
To investigate the impact of primitive intestinal LC-PUFA synthesis on yolk lipid absorption and utilization, we used CRISPR/Cas9 to generate hsd17b12a mutants with disrupted NADB domains (Supplementary Fig. 1i, j). There was a significant down-regulation of hsd17b12a in mutants compared to wild-type (WT), with no compensatory upregulation of hsd17b12b (Supplementary Fig. 1k, l). Notable phenotypes in mutants included uninflated swim bladders (Fig. 1i; Supplementary Fig. 1m) and embryonic lethality before 8 dpf (Fig. 1j), and injection of hsd17b12a mRNA or transgenic overexpression of hsd17b12a rescued these phenotypes (Fig. 1i, j; Supplementary Fig. 1n–p). However, the mRNA-rescued mutants still exhibited developmental defects, such as short stature, due to the limited duration of hsd17b12a mRNA activity (Supplementary Fig. 1n). In the mutants, expression of the LC-PUFA synthesizing enzyme genes was absent in the primitive intestine as revealed by WISH analysis (Fig. 1h, k; Supplementary Fig. 1q) and RT-PCR further confirmed the results (Fig. 1l; Supplementary Fig. 1r). These indicate a defective synthesis ability in the mutant intestine, which led to decreased LC-PUFAs levels and increased levels of essential fatty acids (C18:2 and C18:3) (Fig. 1m). Therefore, the hsd17b12a mutants serve as a model to investigate the role of primitive intestinal LC-PUFA synthesis in yolk lipid absorption and utilization.
Digestive organ expansion is defective in hsd17b12a mutant
Given the specific expression of hsd17b12a in the primitive intestine and the observation that its mutation leads to uninflated swim bladders derived from the endoderm23, we investigated whether this mutation affects the development of other endodermal organs. RNA sequencing (RNA-seq) was performed to compare gene expression between mutants and WT embryos at 3 dpf and 4 dpf, and the DEGs were analyzed (Supplementary Fig. 2a, b). Specific markers were used to analyze the development of endodermal organs, liver markers (fabp10a, cp, and tfa)19; exocrine pancreas markers (trypsin, cpa5, and ctrl)24; endocrine pancreas markers (gcga for α-cells, ins for β-cells, and sst1.1 for δ-cells)25; intestine markers (fabp2 for enterocytes, agr2 for goblet cells, cldn15la for epithelial cells, and neurod1 for enteroendocrine)19; and swim bladder markers (pbx1a for epithelium cells, acta2 for mesenchyme cells, and elovl1a for mesothelium cells)23. Endodermal organ development progresses through two stages: anlage budding and organ expansion26–28. Bulk-seq analysis showed no significant changes in the expression of endodermal organ markers at 3 dpf (Supplementary Fig. 2c), but did show variations in some markers, particularly those associated with the exocrine pancreas, at 4 dpf (Supplementary Fig. 2c). This suggests that developmental defects in other endodermal organs in mutants primarily emerge during the organ expansion stage, rather than the anlage budding stage.
To confirm these developmental defects, we performed WISH using specific probes for various endodermal organs. WISH results indicated that hsd17b12a depletion does not affect the cell fate of swim bladders (Supplementary Fig. 2d). The alterations in exocrine pancreatic and hepatic signals observed via WISH are consistent with findings from bulk-seq analysis. At 3 dpf, there were no significant differences in the areas of the exocrine pancreas and liver between the mutants and WT (Fig. 2a, d–f). At 4 dpf, the areas of the exocrine pancreas and liver in mutants were significantly smaller compared to those in WT, with the most pronounced difference observed in exocrine pancreas (Fig. 2b, d–f). At 5 dpf, the exocrine pancreas and liver areas in mutants remained smaller than in WT (Fig. 2c; Fig. 2d–f). Further examination of endocrine pancreatic α-cells, β-cells, and δ-cells showed that only the δ-cell areas were significantly reduced in the mutants at 4 dpf (Fig. 2a–c; Fig. 2d–f; Supplementary Fig. 2e–g). Despite the known role of the endocrine pancreas in glucose metabolism29, glucose metabolism was not significantly affected by the hsd17b12a mutation (Supplementary Fig. 2h). Thus, the loss of hsd17b12a primarily impacts the expansion of the exocrine pancreas and liver.
Fig. 2. Disrupting Hsd17b12a results in organ expansion defects.
a–c The development of the endocrine pancreas, exocrine pancreas, and liver was examined using WISH (whole-mount in situ hybridization) at 3 dpf, 4 dpf, and 5 dpf in both WT and hsd17b12a−/− (n > 3). Scale bar, 200 μm. d–f The areas of β-cells (ins), exocrine pancreas (trypsin), and liver (fabp10a) were measured in WT and hsd17b12a−/− embryos at 3 dpf, 4 dpf and 5 dpf. g Fluorescence images of exocrine pancreas and liver in WT, hsd17b12a−/−, and Tg (CMV: hsd17b12a)/hsd17b12a−/− at 5 dpf (n > 3). Scale bar, 200 μm. h The areas of images of the liver and exocrine pancreas were quantified in WT, hsd17b12a−/− and Tg(CMV:hsd17b12a)/hsd17b12a−/−. Data in d–f, h are represented as mean ± S.D., using a two-tailed Student’s t test; each point represents an independent ±biological sample (n > 3). Source data are provided as a Source Data file.
Furthermore, a transgenic line Tg(fabp10:RFP;ela3l:EGFP), which labels liver and exocrine pancreas30, was used to analyze mutant and WT embryos. At 4 dpf and 5 dpf, the liver and exocrine pancreas were significantly smaller in mutants compared to WT or heterozygotes (Supplementary Fig. 2i–n). Transgenic overexpression of hsd17b12a restored expansion of the liver and exocrine pancreas in mutants (Fig. 2g, h; Supplementary Fig. 2o), confirming that hsd17b12a disruption specifically impairs digestive organ expansion. We further observed a down-regulation of genes encoding secreted digestive enzymes produced by acinar cells (Supplementary Fig. 2p)24, which are crucial for nutrient absorption in the intestine31. Impaired nutrient uptake likely leads to delayed embryonic development, evidenced by the shorter standard length of mutants compared to WT at 4 dpf (Supplementary Fig. 2q). These findings indicate that mutations in hsd17b12a affect digestive function.
To rule out the potential impact of maternal Hsd17b12a protein on organ expansion, we analyzed the knock-in-positive and -negative embryos from hsd17b12aKI/− females crossed with WT males (Supplementary Fig. 2r). Western blot (WB) analysis demonstrated that maternal Hsd17b12a protein persists up to 3 dpf but disappeared at 4 dpf (Supplementary Fig. 2s, t). However, a significant contribution from zygotic expression of Hsd17b12a was observed from 3 dpf (Supplementary Fig. 2s, t), indicating the defects observed at 4 dpf are mainly due to the zygotic loss of hsd17b12a. To further confirm this, we designed a morpholino (MO) to inhibit hsd17b12a translation by targeting the 5’-UTR and ATG sequence (Supplementary Fig. 3a). Injection of the hsd17b12a_MO at the 1-cell stage resulted in an uninflated swim bladder in WT embryos (Supplementary Fig. 3b, c) and decreased mCherry fluorescence in hsd17b12aKI/KI embryos (Supplementary Fig. 3d). Conversely, heat shock overexpression of hsd17b12a, which is not targeted by the MO (Supplementary Fig. 3a), rescued the uninflated swim bladder phenotype in hsd17b12a knockdown embryos (Supplementary Fig. 3e). These results confirm that the hsd17b12a_MO specifically inhibits hsd17b12a translation and causes defects in organ expansion and yolk absorption (Supplementary Fig. 3f), mimicking the zygotic mutants. Overall, these results collectively highlight the critical role of Hsd17b12a expressed in the primitive intestine for proper organ expansion during embryonic development.
Hsd17b12a promotes yolk lipid absorption and utilization
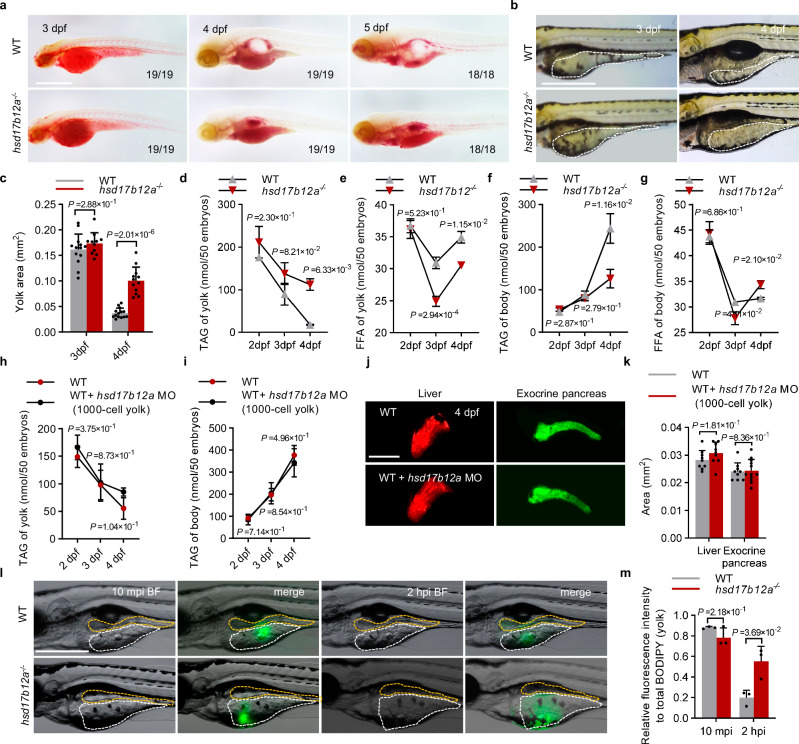
Vertebrates, including zebrafish, cannot synthesize LC-PUFAs de novo13, and rely on dietary linoleic acid (LA) and α-linolenic acid (ALA) for in vivo biosynthesis of LC-PUFAs via fatty acid elongation. In zebrafish, the yolk acts as a source of nutrients, and we have shown that hsd17b12a plays an important role in synthesizing LCFAs in the primitive intestine. We then examined the absorption and utilization of yolk lipids in hsd17b12a mutants. Oil Red O (ORO) staining showed that hsd17b12a mutants at 3–5 dpf had darker red ORO staining signals in the yolk, indicating an accumulation of TAGs compared with WT embryos (Fig. 3a). Further analysis showed that the size of the yolk region of the mutants was not different from that of the WT embryos at 3 dpf, but was significantly larger after 4 dpf (Fig. 3b, c). Previous studies have shown that altered lipid accumulation under yolk platelet activation in the YSL can affect its opacity32–34. However, the hsd17b12a mutant embryos showed no significant changes in YSL transparency (Supplementary Fig. 4a, b), suggesting that Hsd17b12a is not required for yolk platelet activation. The total amount of TAGs was significantly higher in the hsd17b12a mutant than in the WT, but the total amount of FFA was significantly lower (Supplementary Fig. 4c–e). Overexpression of hsd17b12a reduced TAG accumulation in hsd17b12a mutant (Supplementary Fig. 4f). The above results suggest that the utilization of TAGs may be blocked in the yolk of the hsd17b12a mutant.
Fig. 3. Malabsorption of yolk lipids results in expansion defects.
a ORO (Oil Red O) staining was performed on embryos of WT and hsd17b12a−/− at 3 dpf, 4 dpf, and 5 dpf (n > 3). Scale bar, 0.5 mm. b Images of yolk in WT and hsd17b12a−/− at 3 dpf and 4 dpf. The white dotted line indicates the yolk region (n > 3). Scale bar, 1 mm. c Quantification of the area of the yolk in WT and hsd17b12a−/− at 3 dpf and 4 dpf. d–g The body and yolk of WT and hsd17b12a−/− were isolated at 2 dpf, 3 dpf, and 4 dpf, and the TAG (triglyceride) and FFA (free fatty acid) levels in the body and yolk were detected. h, i TAG content in the yolk and body of embryos with suppressed expression of YSL hsd17b12a was measured at 2 dpf, 3 dpf, and 4 dpf. j, k Fluorescence imaging of the liver and exocrine pancreas at 4 dpf in WT and WT with suppressed YSL hsd17b12a expression (n > 3), and quantification of the liver and exocrine pancreas. Scale bar, 500 μm. l Assessment of lipid transport by injection of BODIPY FL C12 (green) into the yolk in WT and hsd17b12a−/− at 4 dpf (n = 3). Images were captured at 10 mpi (10 min post-injection) and 2 hpi (2 h post-injection). The orange dotted line indicates intestinal lumen. The white dotted line indicates the region of yolk. Scale bar, 0.5 mm. m Relative analysis of the fluorescence intensity in the yolk compared to the total fluorescence (BODIPY) intensity (0 mpi) in the yolk at 10 mpi and 2 hpi. Data in d–i are presented as mean ± S.D., using a two-tailed Student’s t-test; n ≥ 3. Data in c, k, m are represented as mean ± S.D., using a two-tailed Student’s t test; each point represents an independent biological sample (n ≥ 3). Source data are provided as a Source Data file.
Subsequently, we analyzed the dynamic changes in the TAG and FFA levels in yolk or body over time, after the embryonic body and yolk were dissected. Compared to the WT, the hsd17b12a mutant had a higher content of TAGs in the yolk and lower content of TAGs in the body from 4 dpf (Fig. 3d, f). From 3 dpf to 4 dpf, although both the mutant and WT embryos showed similar dynamic changes of FFA levels in yolk and body, we noted that the hsd17b12a mutant had significantly lower FFA content in the yolk and significantly higher FFA content in the body (Fig. 3e, g). This suggests that the utilization of TAGs is slower in the mutants than in the WT from 4 dpf.
To further investigate the effects of Hsd17b12a on yolk TAG utilization, we examined the expression of apolipoprotein (apo) genes5, microsomal triglyceride transfer protein (MTP)32, and CD3632,35, which are essential for the transport of yolk lipids from YSL into the body. Our results suggest that apo genes, mtp and cd36 were downregulated in mutants (Supplementary Fig. 4g–j). To exclude the possibility that YSL-specific expression of hsd17b12a affects yolk lipid utilization, we injected hsd17b12a_MO into the YSL at the 1000-cell stage to block YSL-expression of Hsd17b12a according to a previous study (Supplementary Fig. 5a)36. We found that YSL-specific inhibition of Hsd17b12a did not affect swim bladder inflation, yolk utilization, and larval growth (Supplementary Fig. 5b–f), TAG contents in both the yolk and body from 2 dpf to 4 dpf (Fig. 3h, i), and the expansion of exocrine pancreas and liver (Fig. 3j, k). Therefore, the impaired yolk lipid utilization and defective organ expansion in mutants are not due to Hsd17b12a function in the YSL. To evaluate defects in yolk lipid transport and utilization, we used fluorescently labeled fatty acids (BODIPY FL C12)7. At 4 dpf, BODIPY FL C12 was transported from the yolk to the primitive intestine within 2 h post-injection (hpi) in WT embryos, but remained within the yolk in mutants (Fig. 3l, m). All these indicate that Hsd17b12a-mediated LC-PUFA synthesis in the primitive intestine promotes the absorption and utilization of yolk lipids by the body.
Primitive intestine is defective in hsd17b12a mutant
To elucidate the mechanisms underlying impaired yolk lipid absorption and utilization in the primitive intestine of hsd17b12a mutants, we conducted a detailed analysis of intestinal morphology. The development of the primitive intestine was analyzed by WISH using the intestine-specific marker fabp219. At 3 dpf, the shape of the primitive intestine in mutants was similar to that in WT, although the fabp2 signal was more intense in mutants (Fig. 4a). By 4 dpf and 5 dpf, mutants exhibited reduced curvature and width of the primitive intestine compared to WT (Fig. 4a). Fluorescence labeling revealed impaired expansion of the primitive intestine and pronounced yolk accumulation in mutants (Fig. 4b). These findings suggest that intestinal development is impaired in mutants.
Fig. 4. The primitive intestinal structures exhibit defects.
a Intestinal development was examined using whole-mount in situ hybridization in both WT and hsd17b12a−/− at 3 dpf, 4 dpf, and 5 dpf (n > 3). Scale bar, 200 μm. b Intestinal fluorescence imaging of WT and hsd17b12a−/− (n = 5). The white arrows indicate the yolk region. Scale bar, 200 μm. c HE staining of paraffin sections of the intestine in hsd17b12a−/− and WT at 4 dpf and 5 dpf (n = 5). Scale bar, 100 μm. d The thickness of the primitive intestine was quantified in WT and hsd17b12a−/− at 4 dpf and 5 dpf. e Confocal imaging of the intestine in WT and hsd17b12a−/− at 4 dpf (n = 5). The white dotted line indicates the intestinal lumen. Scale bar, 100 μm. f, g Transmission electron microscopy micrographs (TEM) (n > 3) and quantification of the intestinal microvilli length in WT and hsd17b12a−/− at 4 dpf. Scale bar, 0.5 μm. h, i TEM micrographs (n = 3) and quantification of the intestinal tight junction length in WT and hsd17b12a−/− at 4 dpf. The white arrows indicate the tight junctions. Scale bar, 0.5 μm. j Immunofluorescence staining of proteins associated with tight junction formation in the intestine of WT and hsd17b12a−/− at 4 dpf (n = 3). Scale bar, 50μm. k Tg(CMV:hsd17b12a) rescued the expansion defects of primitive intestine in hsd17b12a−/− (n ≥ 3). Scale bar, 0.5 mm. l The area of the primitive intestine was quantified by the signal of fabp2 in WT, hsd17b12a−/− and Tg(CMV:hsd17b12a)/hsd17b12a−/−. Data in d, j, i, l are represented as mean ± S.D., using a two-tailed Student’s t test; each point represents an independent biological sample (n ≥ 3). Source data are provided as a Source Data file.
Zebrafish intestinal epithelial cells complete differentiation and remodeling to form a functional primitive intestine at 4 dpf4, therefore we analyzed primitive intestine of the mutant and WT embryos at 4 dpf and/or 5 dpf in different aspects. Hematoxylin-eosin (HE) staining indicated that primitive intestine in mutants appeared thinner with less pronounced folds compared to WT at 4 dpf and 5 dpf (Fig. 4c, d). Confocal microscopy showed that the mutants had fewer microvilli in the intestine lumen than the WT (Fig. 4e), and transmission electron microscopy (TEM) analysis further confirmed that microvilli in mutants were significantly shorter than those in WT (Fig. 4f, g). It is known that tight junctions are crucial for forming the intestinal epithelial barrier and maintaining intestinal function37, we further investigated tight junctions in the mutants and found that the length of tight junctions was significantly reduced compared to WT (Fig. 4h, i). Although Rab13, which regulates tight junction assembly by recruiting claudin proteins38, was not affected in the mutants, tight junction-forming proteins, such as Claudin-4 and Claudin-3, were absent in mutants (Fig. 4j), indicating a severe defect of tight junctions in the mutant intestinal epithelial cells. Finally, we showed that overexpression of hsd17b12a rescued the intestinal expansion defects in mutants (Fig. 4k, l). Therefore, disruption of hsd17b12a leads to defective intestinal structure, which is a major contributor to impaired intestinal function.
Ferroptosis causes defects in digestive organ expansion
To investigate the mechanisms underlying defects in digestive organ expansion, KEGG pathway analysis was conducted on embryos at 3 dpf and 4 dpf. At 3 dpf, enriched pathways included fatty acid synthesis and metabolism (Fig. 5a; Supplementary Data 1). By 4 dpf, additional enriched pathways involved digestion and absorption of fats and proteins, pancreatic secretion, tight junctions, apoptosis, ferroptosis and glycerophospholipid metabolism (Fig. 5b; Supplementary Data 2). These findings suggest that impaired fatty acid metabolism at 3 dpf further affects pathways crucial for nutrient digestion and absorption at 4 dpf.
Fig. 5. Ferroptosis hinders the digestive organ expansion.
a, b KEGG pathway analysis of differentially expressed genes in hsd17b12a−/− compared to WT at 3 dpf and 4 dpf (p-value < 0.05). c Single-cell sequencing analysis of the liver, intestine, and pancreas in hsd17b12a−/− and WT at 4 dpf. The “Liver” label indicates liver-origin cells, the “Pancreas” label indicates pancreas-origin cells, and the “Intestine” label indicates intestinal-origin cells. d The proportions of each cell type in hsd17b12a−/− and WT. e KEGG pathway analysis of differentially expressed genes in the intestine of hsd17b12a−/− compared to WT (p-value < 0.05). f Detection of ROS (reactive oxygen species) in WT and hsd17b12a−/− at 4 dpf (n = 6). The white arrowheads indicate the intestine. Scale bar, 0.5 mm. g MDA (malondialdehyde) levels were measured in WT and hsd17b12a−/− at 4 dpf. h RT-qPCR of acsl4a and acsl4b in WT and hsd17b12a−/− at 3 dpf and 4 dpf. i, j Assessment and quantification of intracellular Fe2+ levels in larvae using FerroOrange probe in WT and hsd17b12a−/− at 4 dpf (n > 3). k, l Evaluation of GPX4 protein levels in WT and hsd17b12a−/− by western blot and quantification of GPX4 relative to GAPDH (n = 3). m Immunofluorescence analysis of GPX4 protein in the primitive intestine of WT and hsd17b12a−/− at 4 dpf (n = 3). The white arrows indicate the intestinal epithelia. Scale bar: 25 μm. n Fluorescence imaging of the liver and exocrine pancreas in hsd17b12a−/− treated with the Acsl4 inhibitor Rosi (rosiglitazone) (n > 3). o Measurement of the exocrine pancreas and liver areas in both untreated and Rosi-treated hsd17b12a−/−. Scale bar: 250 μm. p, q Fluorescence imaging and quantification of the exocrine pancreas and liver areas in WT, hsd17b12a−/−, and Fer-1 (Ferrostatin-1)-treated hsd17b12a−/− at 5 dpf (n > 3). Scale bar: 200 μm. r RT-PCR of fabp10a and trypsin at 5 dpf after treatment with Fer-1. Data in g, h, j, l, o, q, r are represented as mean ± S.D., using a two-tailed Student’s t test; each point represents an independent biological sample (n ≥ 3). Source data are provided as a Source Data file.
To further elucidate the digestive organ expansion defects, single-cell RNA sequencing (scRNA-seq) analysis was performed on the intestine, liver, and exocrine pancreas of both mutants and WT at 4 dpf. UMAP clustering identified sixteen cell types in the integrated data, which were annotated according to specific markers (Fig. 5c; Supplementary Fig. 6a). Importantly, hsd17b12a expression was restricted to intestinal epithelial cells and enteroendocrine cells (Supplementary Fig. 6b). Compared to WT, the mutants showed increased cell fractions in the intestine and decreased cell fractions in hepatocytes, pancreatic exocrine cells, and immune cells (Fig. 5d). Subsequent analysis revealed downregulation of ribosome and amino acid metabolism pathways in the intestine. Notably, upregulation of the ferroptosis pathway, an iron-dependent form of cell death characterized by lipid peroxidation39, was found in mutant intestinal cells, but not in hepatocytes-1 and pancreatic exocrine cells (Fig. 5e; Supplementary Fig. 6c–e), indicating that hsd17b12a deficiency leads to ferroptosis in the primitive intestine.
We further analyzed ferroptosis in mutants at 3 and 4 dpf. The results showed that the levels of reactive oxygen species (ROS) and malondialdehyde (MDA), which are produced by lipid peroxidation39, were elevated in mutants compared to WT at 4 dpf (Fig. 5f, g). Expression of acsl4b, a marker of ferroptosis sensitivity39, was upregulated in mutants compared to WT at 3 and 4 dpf (Fig. 5h). Additionally, Fe2+ levels were also increased in the liver and intestine of mutants relative to WT (Fig. 5i, j). Gpx4 protein levels, which inhibit ferroptosis39, were reduced in the intestine of mutants compared to WT(Fig. 5k–m). These findings confirm that loss of hsd17b12a leads to ferroptosis in the primitive intestine.
To determine whether the expansion defects in the digestive organs are attributable to ferroptosis in the primitive intestine, we conducted rescue experiments using two ferroptosis inhibitors, rosiglitazone (Rosi), which inhibits Acsl4 activity40, and a potent inhibitor of ferroptosis, ferrostatin-1 (Fer-1)41,42. Interestingly, Rosi treatment rescued the expansion defect in the exocrine pancreas of mutants (Fig. 5n, o); and Fer-1 treatment partially rescued hepatic and exocrine pancreatic expansion defects in mutants (Fig. 5p–r). These results suggest that ferroptosis of primitive intestine in hsd17b12a mutant is responsible for the defects of liver and exocrine pancreas expansion.
DHA-PA-PG axis regulates ferroptosis and digestive organ expansion
To investigate whether the cell fate of primitive intestine was affected by hsd17b12a depletion, we re-clustered intestinal cells from the scRNA-seq data for further analysis. There are six distinct subclusters within intestinal cells, based on cell-type-specific markers (Fig. 6a; Supplementary Fig. 7a). By analyzing cell trajectories originating from stem cells43, we concluded that enterocytes can differentiate into brush border cells or intestinal epithelial cells (Supplementary Fig. 7b). There are no significant differences in cell differentiation trajectories in the liver, pancreas, and intestine of mutants compared to WT (Supplementary Fig. 7c). In the mutants, the proportions of intestinal epithelial cells and brush border cells were dramatically decreased, while the proportions of enterocytes and stem cells were greatly increased (Fig. 6b). Specifically, hsd17b12a is prominently expressed in enterocytes and intestinal epithelial cells (Fig. 6c), suggesting that depletion of hsd17b12a strongly affects the differentiation from enterocytes to intestinal epithelial cells. We then tried to screen transcriptional factors that are specifically expressed in hsd17b12a-positive intestinal epithelial cells. The results showed that two transcriptional factor genes, osr1, which is suggested to play a role in digestive organ development44, and tfcp2l1, previously shown to regulate lipid metabolism45, were specifically expressed in hsd17b12a-positive intestinal epithelial cells (Supplementary Fig. 7d–f). Interestingly, the hsd17b12a mutants displayed pectoral fin malformations (Supplementary Fig. 7g), which mimic those observed in the osr1-knockdown embryos46. We then conducted KEGG pathway analysis with the hsd17b12a-positive intestinal epithelial cells, and the results suggest that these cells play an important role in lipid metabolism (Fig. 6c, d), such as glycerophospholipid metabolism and fatty acid degradation. These findings suggest that the loss of hsd17b12a leads to a cell fate loss of intestinal epithelial cells, which may strongly affect glycerophospholipid metabolism.
Fig. 6. PG inhibits ferroptosis and promotes organ expansion.
a UMAP analysis classified the intestines of hsd17b12a−/− and WT. b Proportional statistics of the six intestinal cell types in hsd17b12a−/− and WT. c Violin plots showing hsd17b12a expression profiles in six intestinal cell types. d Identification of enriched pathways for highly expressed genes in intestinal epithelial cells (p-value < 0.05). e Heatmap depicting changes in lipid components in WT and hsd17b12a−/− larvae at 4 dpf (n = 3). f Differential analysis of free LC-PUFAs (long-chain polyunsaturated fatty acids) in hsd17b12a−/− larvae compared to WT at 4 dpf. g Differential analysis of TAGs (triglycerides) containing C18:2 or C18:3 in hsd17b12a−/− larvae compared to WT at 4 dpf. h, i Differential analysis of PA (phosphatidic acid) and PG (phosphatidylglycerol) in hsd17b12a−/− larvae compared to WT at 4 dpf. j, k RT-PCR analysis of acsl4b gene expression at 5 dpf after DHA (docosahexaenoic acid), PA, and PG immersion treatments. l RT-PCR analysis of fabp2, fabp10a, and trypsin expression in WT, hsd17b12a−/−, and DHA-immersed hsd17b12a−/− at 5 dpf. m, n Images and quantification of the exocrine pancreas areas in WT, hsd17b12a−/−, and DHA-immersed hsd17b12a−/− at 5 dpf (n = 3). Scale bar, 200 μm. o Overexpression of pgs1 rescued exocrine pancreatic expansion defects and facilitated yolk utilization in hsd17b12a−/− (n > 3). Scale bar, 250 μm. p, q Quantification of exocrine pancreas area and yolk area at 4 dpf. Yolk area highlighted with a white dotted line. r RT-PCR analysis of acsl4b gene expression at 4 dpf following pgs1 overexpression. s Model of the DHA-PA-PG axis in the primitive intestine regulating digestive organ expansion. In hsd17b12a−/−, the LC-PUFA synthesis is blocked, leading to TAG accumulation in the yolk, decreased LC-PUFA and LC-PUFA-containing phospholipids, further resulting in ferroptosis and defective digestive organ expansion. The red arrows indicate increased content, while the blue arrows indicate decreased content. Data in f–l, n, p, q, r are represented as mean ± S.D., using a two-tailed Student’s t test; each point represents an independent biological sample (n ≥ 3). Source data are provided as a Source Data file.
To investigate how glycerophospholipid metabolism regulates yolk lipid absorption and organ expansion, we conducted lipidomic analysis on the mutants and WT embryos at 4 dpf. There were reduced levels of free fatty acid (FFA), phosphatidic acid (PA), and phosphatidylglycerol (PG) in mutants compared to WT (Fig. 6e; Supplementary Data 3). It is known that PA is essential for glycerophospholipid metabolism, such as producing PG through the cytidine diphosphate (CDP)-DAG pathway (Supplementary Fig. 8a)47. We found that genes involved in glycerophospholipid metabolism, such as agpat2 and lpin2, were highly expressed in hsd17b12a-positive intestinal epithelial cells (Supplementary Fig. 8b). Further analysis of RNA-seq results suggests that most genes involved in glycerophospholipid metabolism, except lpin1, were downregulated in the intestines of mutants compared to WT (Supplementary Fig. 8c, d), which is in accordance with the decreased level of PG in the mutants. These results suggest that disruption of Hsd17b12a strongly impairs PG synthesis.
To investigate how LC-PUFA synthesis defects affect PG synthesis, we analyzed the LC-PUFA contents in mutants and WT. It was shown that all LC-PUFA contents were decreased in mutants compared to WT, with the most pronounced decrease observed in DHA (C22:6) (Fig. 6f). Although free LA (C18:2) and ALA (C18:3) contents were also decreased in mutants, the contents of TAGs containing LA and ALA in their side chains were increased (Fig. 6f, g). The contents of PA and PG containing LC-PUFAs were significantly decreased in mutants compared to WT (Fig. 6h, i). Thus, LC-PUFAs synthesized by hsd17b12a, such as DHA, are essential for PA and PG synthesis, through a DHA-PA-PG axis. Given that suppression of ferroptosis rescues organ expansion defects, we investigated whether disruption of the DHA-PA-PG axis contributes to ferroptosis. Treatment of mutants with DHA, PA, or PG significantly reduced acsl4b expression (Fig. 6j, k), suggesting an inhibition of ferroptosis. RT-PCR analysis demonstrated that DHA treatment improved marker gene expression in the intestine, liver, and exocrine pancreas of mutants (Fig. 6l). Live imaging further indicated partial restoration of the exocrine pancreas expansion defect (Fig. 6m, n). Therefore, our results indicate that the DHA-PA-PG axis is crucial for digestive organ expansion.
AGPAT2 and CDS1/2 interact to form functional complexes that facilitate PA metabolism through the CDP-DAG pathway47. In the mutant, both agpat2, which converts LPA to PA, and pgs1, which converts PA to PG, are specifically expressed in the primitive intestine and are downregulated compared to the WT (Supplementary Fig. 8c, d). However, overexpression of pgs1, but not agpat2, partially rescued exocrine pancreas expansion defects and yolk malabsorption in the hsd17b12a-knockdown embryos (Supplementary Fig. 8e–g). Furthermore, overexpression of pgs1 in the mutants rescued defects in exocrine pancreas expansion and yolk malabsorption (Fig. 6o–q), and also slightly suppressed ferroptosis, as indicated by decreased acsl4b expression and increased levels of Gpx4 protein (Fig. 6r; Supplementary Fig. 8h, i). To further confirm that the DHA-PA-PG axis influences organ expansion via ferroptosis, we used four gRNAs to knockout agpat2 and pgs1 and observe the resulting phenotype in F0 zebrafish48. Knockout of agpat2 and pgs1 resulted in impaired expansion of liver and exocrine pancreas, as well as reduced yolk absorption, while overexpression of these genes rescued knockout phenotypes (Supplementary Fig. 8j–o). Thus, the DHA-PA-PG axis promotes yolk lipid absorption and organ expansion, whereas its disruption leads to ferroptosis and defects in organ expansion (Fig. 6s).
Discussion
The yolk sac has garnered considerable attention in recent research due to its crucial role in nutrient absorption. Transcriptomic and single-cell omics studies in humans and mice have highlighted its significance, particularly in nutrient transport and embryonic development49,50. Disruption in yolk sac function, such as defects in vitelline duct closure or impaired nutrient absorption, can lead to developmental abnormalities like Meckel’s diverticulum51. Our study identifies the primitive intestine as a primary site for synthesizing LC-PUFAs and phospholipids (PLs), demonstrating that the Hsd17b12a-regulated DHA-PA-PG axis is crucial for yolk sac lipid absorption and digestive organ expansion. The discovery of these regulatory mechanisms is crucial for our understanding of early embryonic development during eeNST, and may provide insights into therapeutic strategies for gastrointestinal diseases related to nutrient absorption disorders.
Previous studies have shown the role of protein and glucose metabolism in digestive organ growth. For example, disruption of mTORC1 signaling or ribosome biosynthesis leads to hypoplasia of the digestive organs due to impairing cell proliferation52,53, and glucose transporters are required for maintaining nucleotide synthesis and for controlling liver outgrowth54. Nevertheless, the contribution of lipid metabolism to the digestive organ development has been elusive. Lipid metabolism is highly active during embryonic development, both in the yolk and within the embryo11. Despite its importance, the regulation of yolk sac lipid utilization by the embryo is not well understood. Our research reveals that the loss of Hsd17b12a disrupts DHA, PA, and PG synthesis, namely a DHA-PA-PG axis, in the primitive intestine, impairing yolk sac lipid absorption and digestive organ expansion.
In hsd17b12a mutants, a significant imbalance in cell fate occurs in the primitive intestine, characterized by the near absence of brush border cells and hsd17b12a-positive epithelial cells, alongside an increase in enterocytes. This imbalance underscores the pivotal role of hsd17b12a-positive epithelial cells in glycerophospholipid metabolism, influencing intestinal cell differentiation. Blocking the DHA-PA-PG axis or the glycerophospholipid metabolic pathway triggers ferroptosis in the primitive intestine. Inhibiting ferroptosis rescues the expansion defect in mutants, emphasizing the significant role of ferroptosis homeostasis in digestive organ development. PG, a negatively charged membrane phospholipid, facilitates material transport55, and maintains mitochondrial homeostasis56. Our study indicates that PG deficiency leads to ferroptosis and developmental defects in the digestive organs.
In viviparous vertebrates, placental nutrient sensing and hormone release regulate fetal nutrient supply, which is crucial for development57,58. The hormone-like peptide IGF2, produced by placental endocrine cells, modulates maternal glucose and lipid metabolism to support fetal growth59. Oviparous vertebrates rely exclusively on yolk nutrients for embryonic development, while mechanisms governing yolk nutrient release by embryos remain poorly understood. Our findings reveal that Hsd17b12a-mediated LC-PUFA synthesis in the primitive intestine stimulates the release and absorption of yolk lipids, promoting the expansion of digestive organs. These insights enhance our understanding of nutrient absorption mechanisms and could inform future research into related developmental and metabolic disorders.
Methods
Ethics statement
All animal experiments were conducted according to the standard animal guidelines approved by the Animal Care Committee of the University of Chinese Academy of Sciences and the Institute of Hydrobiology, Chinese Academy of Sciences.
Zebrafish
The zebrafish used in this study were maintained at the Chinese Zebrafish Resource Center of the National Aquatic Biological Resource Center (CZRC-NABRC, Wuhan, China, http://zfish.cn). The following transgenic lines were utilized: Tg(fabp10a:dsRed; ela3l:EGFP)60, and Tg(ET33J1:EGFP)22. To investigate the effects of hsd17b12a overexpression, Tg(CMV:mCherry;CMV:hsd17b12a) and Tg(hsp70:hsd17b12a-P2A-GFP) were generated using Tol2 transposase. The corresponding plasmid and Tol2 mRNA were co-injected into one-cell stage WT embryos.
Knockout and knock-in at the hsd17b12a locus
The hsd17b12a knockout mutants were generated using CRISPR/Cas9. The sgRNA sequence for targeting hsd17b12a was 5’-GGATATGGGTGCTGGGAA
AC-3’, designed with the CRISPR-scan tool61. To identify homozygous mutants, sequencing and PCR with specific primers were employed. For the knock-in procedure, a 5xMyc-P2A-mCherry cassette was introduced into the hsd17b12a genomic locus using a recently developed induced primordial germ cell transplantation (iPGCT) and CRISPR/Cas9-mediated knock-in technology21. The gRNA sequence for the knock-in vector and the endogenous target was 5’-GGGGTTTCAAGCCGTAGAC-3’. All microinjections were conducted at the one-cell stage embryos. Primers sequences used in these procedures are listed in Supplementary Tables 3 and 4.
Morpholino Oligonucleotide (MO) design and injection
The hsd17b12a morpholino oligonucleotide (hsd17b12a_MO) was designed by Gene Tools (https://www.gene-tools.com/). The sequence targeting both the 5’-UTR and the cDNA sequence with ATG is ACGTTAAACGACTCCATGTCTAC GG. To knock down hsd17b12a expression in whole embryos, MO was injected at the one-cell stage. For specific targeting of hsd17b12a expression in the yolk syncytial layer (YSL), MO was injected at the 1000-cell stage36. Injected embryos were subsequently incubated at 28 °C until the desired developmental stages were reached.
mRNA synthesis and yolk injection
hsd17b12a cDNAs were transcribed in vitro using the mMESSAGE mMACHINETM SP6 Transcription Kit (Invitrogen, Cat. AM1340) following the provided protocol. The transcribed mRNAs were injected into one-cell stage embryos. Survival curves for WT, mutants and mutants expressing hsd17b12a were generated by recording survival rates over a period of fifteen consecutive days.
Treatment of embryos with DHA, PA, PG, Rosi, and Fer-1
Docosahexaenoic acid (DHA, Sigma-Aldrich, Cat. D2534) powder was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 100 mM, and embryos were treated with 50 μM from 2 dpf to 4 dpf. DMSO treatment was served as a control. Phosphatidic acid (PA, Sigma-Aldrich, Cat. P9511), and phosphatidylglycerol (PG, Sigma-Aldrich, Cat. P8318) powders were dissolved in chloroform at a concentration of 100 mM, and embryos were treated with 50 μM from 2 dpf to 4 dpf. Chloroform treatment was served as a control. Ferrostatin-1 (Fer-1, Sigma-Aldrich, Cat. SML0583) powder was dissolved in DMSO at a concentration of 200 mM, and embryos were treated with 20 μM from 3 dpf to 4 dpf. DMSO treatment was served as a control. Rosiglitazone (Rosi, aladdin, Cat. 122320-73-4) powder was dissolved in DMSO at a concentration of 200 mM, and embryos were treated with 1 μM from 2 dpf to 4 dpf. DMSO treatment was served as a control.
Quantitative RT-PCR
Total RNA was isolated from zebrafish embryos of WT and hsd17b12a−/− at 4 dpf and 5 dpf using the FastPure Cell/tissue Total RNA Isolation Kit (Vazyme, Cat. RC101-01) according to the protocol provided with the Kit. cDNA was synthesized from the isolated RNA using HiScript lll All-in-one RT SuperMix Perfect for RT-PCR (Vazyme, Cat. R333) following the provided protocol. Quantitative RT-PCR was performed with Taq Pro Universal SYBR RT-PCR Master Mix (Vazyme, Cat. Q712) and SYBRGreen Supermix (BioRad, Cat. 172–5124) on a BioRad CFX96 Real-Time PCR Detection System. Expression values were calculated using the 2−∆∆CT method and normalized to β-actin. The expression in control sample was further normalized to 1. The primers used for RT-PCR are listed in Supplementary Table 5.
Analysis of yolk syncytial layer (YSL) opacity
To analyze yolk syncytial layer (YSL) opacity34, embryo images were obtained using transmitted light microscopy. The opacity of YSL the was quantified based on the gray value of the eyes in the images. The analysis was performed at 2 days post-fertilization (dpf), 3 dpf, and 4 dpf for both hsd17b12a mutants and WT embryos. Gray values were measured using ImageJ software to determine differences in opacity between groups.
Detection of Fe2+ levels and glucose levels
Fe2+ levels were detected using FerroOrange staining (DOJINDO, Cat. F374)62. Embryos were incubated with FerroOrange at a concentration of 10 μM for 1 h at 37 °C. After staining the embryos were imaged using a fluorescence microscope. Finally, the fluorescence intensity of the liver and intestine was measured using ImageJ. Free glucose levels in whole larvae were measured using a Glucose Kit (Jiancheng, Nanjing, China, Cat. A154-1-1), according to the protocol provided with the kit. Glucose concentration was quantified relative to the number of embryos.
Measurement of triacylglycerol and free fatty acids
Yolks and bodies of embryos were dissected and measured independently. The tissues were homogenized and centrifuged at 600 × g for 10 min. The supernatant was stored and analyzed using the Nonesterified Free Fatty Acids (FFAs) Assay Kit (Jiancheng, Nanjing, China, Cat.A042-2-1) and the Tissue Triglyceride Assay Kit (Jiancheng, Nanjing, China, Cat.A110-1-1), according to the protocol provided with the Kits. Lipid amounts were normalized to the number of embryos.
Detection of ROS levels and MDA levels
Lipid peroxidation was assessed in WT and mutant using a Reactive oxygen species Assay Kit (Jiancheng, Nanjing, China, E004-1-1) at 4 dpf, according to the protocol provided with the Kit. Malondialdehyde (MDA) levels, indicative of lipid peroxidation, were measured using a Microscale Malondialdehyde (MDA) Assay Kit (Jiancheng, Nanjing, China, A003-1-2) at 4 dpf, according to the protocol provided with the Kit.
Oil Red O staining and BODIPY-labeled fatty acid injection
WT and hsd17b12a−/− embryos at 3–5 dpf were fixed with 4% paraformaldehyde and stained with 0.3% Oil Red O solution63. BODIPYTM FL C12 (Invitrogen, Cat. D3822) were injected into the yolk of 4 dpf embryos and fluorescence intensity of BODIPY in the yolk was measured at 10 min post-injection (mpi) and 2 h post-injection (hpi) compared to the initial fluorescence intensity (0 mpi)7. Images were captured using a fluorescence microscope, and the fluorescence intensity was quantified using ImageJ software.
In situ hybridization
Digoxigenin (DIG)-labeled oligonucleotides were synthesized using T7 polymerase and PCR-amplified sequences of genes of interest. The primers used for synthesizing these probes are listed in Supplementary Table 6. Whole-mount in situ hybridization (WISH) was performed on WT and hsd17b12a−/− embryos at different developmental stages according to the protocol64. In situ hybridization on frozen section was performed65. In brief, larvae were embedded in OCT compound and sectioned at 12-μm thickness. Sections were air-dried, fixed in 4% PFA for 15 min, and then washed three times with 1x PBS for 5 min each. DIG-labeled probes were hybridized at 70 °C overnight. DIG antibodies were subsequently incubated at room temperature in the dark to visualize the hybridization signals.
Immunofluorescence on sections
Immunofluorescence of frozen intestinal sections was performed on WT and hsd17b12a−/− primitive intestine66. Sections were incubated with primary antibodies overnight at 4 °C. and subsequently incubated with secondary antibodies and DAPI overnight at 4 °C. The following primary antibodies were used: mouse anti-Myc tag (Cell Signal Technology, Cat. 2276, 1:8000), rabbit anti-Gpx4 (HUABIO, Cat. ET1706-45, 1:100), rabbit anti-Rab13 (SAB, Cat. 46176-1, 1:200), rabbit anti-Claudin-3 (Invitrogen, Cat. 18-7340, 1:200), mouse anti-ERp72 (HUABIO, Cat. EM1701-95, 1:50) and mouse anti-Claudin-4 (Invitrogen, Cat. 32-9400, 1:200). Secondary antibodies included anti-rabbit Alexa Fluor 680 and 488 and anti-mouse Alexa Fluor 488. Fluorescent images were captured using a Leica SP8 confocal microscope.
Western blotting
Protein extraction was performed on 50 larvae per sample from WT and hsd17b12a−/− embryos at 4 dpf, and western blotting (WB) was conducted67. The following primary antibodies were used for WB: mouse anti-GAPDH (DIA-AN, Cat. 2058, 1:3000) and rabbit anti-Gpx4 (HUABIO, Cat. ET1706-45, 1:100). Protein bands were detected using ECL western blotting detection reagents (Millipore, Cat. WBKLS0100, Billerica, Massachusetts) and visualized with the ChemicDoc MP imaging system (BioRad).
Histological analysis and transmission electron microscopy
Histomorphological analysis of WT and hsd17b12a−/− embryos at 4 dpf and 5 dpf was conducted using paraffin embedding and HE staining. For transmission electron microscopy (TEM), larvae were embedded in Epon812 resin after dehydration. Ultrathin sections (70–80 nm) were prepared using an ultrathin slicer. The sections were then examined using a TEM for detailed structural analysis.
Gas chromatography-mass spectrometer (GC-MS) analyzes
Fatty acids (FAs) of WT or hsd17b12a−/− 50 larvae were quantified using gas chromatography-mass spectrometer (GC-MS). Total lipids were extracted from dried larvae, and TAGs were separated. The FA content in TAGs was then analyzed using GC-MS.
Liquid chromatography–mass spectrometry (LC–MS) analyses
Lipids were extracted from approximately 30 mg of WT and hsd17b12a−/− embryos (n = 3 replicates) at 4 dpf using a modified version of Bligh and Dyer’s method68. Briefly, the tissues were homogenized in 750 µL of chloroform:methanol:MilliQ H2O (3:6:1) (v/v/v). The homogenate was then incubated at 200 × g for 1 h at 4 °C. At the end of the incubation, 350 µL of deionized water and 250 µL of chloroform were added to induce phase separation. The samples were then centrifuged and the lower organic phase containing the lipids was extracted into a clean tube. Lipid extraction was repeated by adding 450 µL of chloroform to the remaining aqueous phase. The lipid extracts were pooled into a single tube and dried in the SpeedVac under OH mode. Samples were stored at −80 °C until further analysis.
Lipidomic analyses were conducted at LipidALL Technologies using a Shimadzu Nexera 20-AD coupled with a Sciex QTRAP 6500 PLUS69. Sample reconstitution was performed in isotopic mixed standards using an Exion UPLC-QTRAP 6500 Plus (Sciex) liquid chromatography-mass spectrometry (LC-MS) system. All analyses were conducted in electrospray ionization (ESI) mode under the following conditions: curtain gas = 20, ion spray voltage = 5500 V, temperature = 400 °C, ion source gas 1 = 35, and ion source gas 2 = 35. Separation of individual lipid classes of polar lipids by normal phase (NP)-HPLC was carried out using a TUP-HB silica column (i.d.150 × 2.1 mm, 3 µm) with the following conditions:mobile phase A (chloroform:methanol: ammonium hydroxide, 89.5:10:0.5) and mobile phase B (chloroform:methanol: ammonium hydroxide:water, 55:39:0.5:5.5). Multiple reaction monitoring (MRM) transitions were established for comparative analysis of various polar lipids. Individual lipid species were quantified by referencing spiked internal standards. d9-PC32:0(16:0/16:0), d9-PC36:1p (18:0p/18:1), d7-PE33:1(15:0/18:1), d31-PS(d31-16:0/18:1), d7-PA33:1(15:0/18:1), d7-PG33:1(15:0/18:1), d7-PI33:1(15:0/18:1), C17-SL, d5-CL72:8(18:2)4, Cer d18:1/15:0-d7, d9-SM d18:1/18:1, d7-LPC18:1, d7-LPE18:1, C17-LPI, C17-LPA, C17-LPS, and C17-LPG, which were obtained from Avanti Polar Lipids. GM3-d18:1/18:0-d3 was purchased from Matreya LLC. Free fatty acids were quantified using d31-16:0 (Sigma-Aldrich) and d8-20:4 (Cayman Chemicals).
Glycerol lipids, including diacylglycerols (DAG) and triacylglycerols (TAG), were quantified using a modified version of reverse phase HPLC/MRM. Separation of neutral lipids was achieved on a Phenomenex Kinetex-C18 column (i.d.4.6 ×100 mm, 2.6 µm) using an isocratic mobile phase containing chloroform:methanol:0.1 M ammonium acetate 100:100:4 (v/v/v) at a flow rate of 300 µL/min for 10 min. Levels of short-, medium-, and long-chain TAGs were calculated by referencing spiked internal standards of TAG(14:0)3-d5, TAG(16:0)3-d5 and TAG(18:0)3-d5 obtained from CDN Isotopes, respectively. DAGs were quantified using d5-DAG17:0/17:0 and d5-DAG18:1/18:1 as internal standard (Avanti Polar Lipids). Quantification was achieved using the isotope internal standard based on neutral loss MS/MS techniques.
RNA-seq and data analysis
For embryo samples, 50 embryos at 3 dpf and 4 dpf were collected as a single sample. Total RNA from the samples was extracted using RNAprep Pure Micro Kit (TIANGEN, Cat. DP420) according to the provided protocol in the Kit. Genomic DNA was removed using DNase I (Vazyme). RNA-seq data for the intestine and liver at 5 dpf were obtained from the National Genomics Data Centre (CRA005219 and CRA005220, https://ngdc.cncb.ac.cn/gsa)19. RNA-seq transcriptome library construction and bioinformatics analysis were performed following standard protocols70.
Generating single-cell suspensions for single cell RNA-seq
Embryos at 4 dpf were first treated with 1x PBS (pH 7.4) containing an anesthetic to stabilize the embryos during dissection. Using two syringes with fine needles, the needle in the left hand was inserted into the swim bladder to stabilize the embryo, while the needle in the right hand was used to gently scraped and remove the tissue around the yolk. Subsequently, the skin surrounding the hindgut was peeled away. After draining the yolk and removing the skin around the hindgut, the needle in the right hand was positioned at the junction of the foregut and esophagus through an abdominal incision, and the junction was gently severed. The organs, including the intestine, liver, and pancreas, were then removed together and separated using the needle. All dissections were performed under a light microscope to ensure precision.
The liver, intestine, and pancreas tissues from WT (n = 100) and hsd17b12a−/− (n = 200) zebrafish were placed in 2 mL low-adsorption EP tubes containing 1 mL of L-15 (Sigma-Aldrich) and cut into pieces. Next, 0.25% trypsin (Biological Industries), 400 U/mL collagenase a (Aladdin), and 0.05% DNaseI (Roche) were added to 1 mL L-15, followed by incubation at 32 °C for 2–3 h in a water bath. Once the large tissue masses had disappeared, the resulting cell suspension was filtered sequentially through 70 μm and 40 μm cell strainers to remove small tissue debris. The reaction was terminated by adding 10% fetal bovine serum (FBS). Cells were washed three times with Dulbecco’s phosphate-buffered saline (DPBS) containing 0.04% bovine serum albumin (BSA) and resuspended in 100–200 μL of the same buffer. For viability assessment, 10 μL of the cell suspension was mixed with 10 μL of AOPI dye, then applied to a Countstar cell plate and analyzed using the Countstar instrument to determine viable cell concentration, viability, diameter, and aggregation rate.
Single-cell suspensions were processed using the 10X Genomics Chromium Next GEM Single Cell 3ʹ GEM, Library & Gel Bead Kit v3.1. Libraries were constructed according to the Chromium Next GEM Single Cell 3’ Reagent Kits v3.1 User Guide. Quality control of libraries involved quantification using a Nanodrop spectrophotometer, agarose gel electrophoresis to check for degradation and contamination, and analysis with the Agilent 2100 Bioanalyzer for library integrity and quantification. Paired-end sequencing with 150 bp read length on each end was performed on the Illumina HiSeq platform.
Processing and analysis of single-cell RNA-seq data
To analyze RNA-seq data from single cells, Cell Ranger (10X Genomics, version 7.1.0) was employed to process Illumina sequencing data and map reads to the reference genome of zebrafish (Ensembl GRCz11.108). The output matrices from Cell Ranger were analyzed using the Seurat R package (version 4.3.0) in RStudio. To reduce the number of low-quality cells and doublets, cells with fewer than 200 unique molecular identifiers (UMIs), more than 3000 UMIs, or greater than 10% mitochondrial reads were excluded. The Seurat package was used to normalize the expression values, cluster all cells in the integrated dataset at a resolution of 0.2, and cluster the intestinal cells in the integrated dataset at a resolution of 0.1. Following the identification of marker genes for each cluster using the ‘FindMarkers’ function in Seurat, the clusters were annotated based on known cell type markers in zebrafish.
Reagents and antibodies
See Supplementary Table 7.
Statistical analysis
WT, hsd17b12a+/−, and hsd17b12a−/− zebrafish embryos were obtained from incross of hsd17b12a+/− males and females and the genotypes of offspring were identified before all the experiments. Early embryos and WISH samples were imaged using a fluorescence stereomicroscope (Axio Zoom.V16, Zeiss), chemical stained sections were imaged using a microscope (Scope.A1, Zeiss), immunofluorescence sections were imaged using a confocal microscope (SP8, Leica). The replicate numbers or sample numbers are identified in the figures, figure legends or text. The diagram in Fig. 6s was created using Adobe Illustrator 2020, and the diagrams in Supplementary Fig. 3b and Supplementary Fig. 5a were drawn using Microsoft Office PowerPoint. Images were processed using Image J software (version 1.8.0), and data were analyzed using R software (version 4.3.0), or GraphPad Prism 8.0 software. All data are presented as mean values ± S.D.. Unpaired two-tailed Student’s t test was used to calculate the P values.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Source data
Acknowledgements
We thank Dr. Jingjing Zhang at Guangdong Medical University for providing the antibodies (Rab13, Claudin3, and Claudin4), and the Tg(ET33J1:EGFP) reporter fish. We would like to thank Kuoyu Li and Luyuan Pan at the China Zebrafish Resource Center, National Aquatic Biological Resource Center (CZRC, NABRC, http://zfish.cn) for raising the zebrafish, Zhixian Qiao, Xiaocui Chai, Fang Zhou, Yuan Xiao, and Zhenfei Xing at the Analysis and Testing Center, Institute of Hydrobiology, Chinese Academy of Sciences, for their assistance with experiments. This work was funded by the National Natural Science Foundation of China (32025037), Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB0730300), National Key R&D Program of China (2023YFD2401603), Ministry of Agriculture and Rural Affairs (NK2022010207), Natural Science Foundation of Wuhan, Science and Technology Special Fund of Hainan Province (ZDYF2024XDNY256), and the Key Laboratory of Breeding Biotechnology and Sustainable Aquaculture (CAS) to Y.S., and by the National Natural Science Foundation of China (grant No 32273134) and the Youth Innovation Promotion Association CAS (grant No. 2023353) to M.H. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Z.C. designed the research, performed experiments, analyzed data, and wrote the manuscript. M.H., H.W., X.L., R.Q., D.Y., X.Z., J.Z., and Q.Z. provided assistance with experiments. P.H. provided assistance with single-cell RNA-seq analyses. G.S. provided assistance with lipidomic analyses. Y.S. designed the research, analyzed data, wrote and revised the manuscript.
Peer review
Peer review information
Nature Communications thanks Yann Gibert, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The sequence data that support the findings of this study have been deposited in the Genome Sequence Archive at the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (CRA018363 [https://ngdc.cncb.ac.cn/gsa/search?searchTerm=CRA018363], CRA018331, and CRA018335) are publicly accessible at https://ngdc.cncb.ac.cn/gsa. The lipidomics data are provided in Supplementary Data 3. Public bulk RNA-seq data are available in the GSA database under the accession numbers CRA005219 and CRA005220. The remaining data are available within the Article, Supplementary Information or Source data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54258-2.
References
- 1.Ross, C. & Boroviak, T. E. Origin and function of the yolk sac in primate embryogenesis. Nat. Commun.11, 3760 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao, J. et al. Metabolic remodelling during early mouse embryo development. Nat. Metab.3, 1372–1384 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Quinlivan, V. H. & Farber, S. A. Lipid Uptake, Metabolism, and Transport in the Larval Zebrafish. Front. Endocrinol.8, 319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verbueken, E., et al. From mRNA Expression of Drug Disposition Genes to In Vivo Assessment of CYP-Mediated Biotransformation during Zebrafish Embryonic and Larval Development. Int. J. Mol. Sci.19, 3976 (2018). [DOI] [PMC free article] [PubMed]
- 5.Otis, J. P. et al. Zebrafish as a model for apolipoprotein biology: comprehensive expression analysis and a role for ApoA-IV in regulating food intake. Dis. Model Mech.8, 295–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García, M. P., Elbal, H. M. T. L. M. T. & Agulleiro, B. Development of the digestive tract of sea bass(Dicentrarchus labrax L). Light and electron microscopic studies. Anat. Embryol.204, 39–57 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Miyares, R. L., de Rezende, V. B. & Farber, S. A. Zebrafish yolk lipid processing: a tractable tool for the study of vertebrate lipid transport and metabolism. Dis. Model Mech.7, 915–927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saele, O., Rod, K. E. L., Quinlivan, V. H., Li, S. & Farber, S. A. A novel system to quantify intestinal lipid digestion and transport. Biochim Biophys. Acta Mol. Cell Biol. Lipids1863, 948–957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, B. & Tontonoz, P. Phospholipid Remodeling in Physiology and Disease. Annu. Rev. Physiol.81, 165–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, Q. et al. The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3. Nat. Commun.12, 6869 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraher, D. et al. Zebrafish Embryonic Lipidomic Analysis Reveals that the Yolk Cell Is Metabolically Active in Processing Lipid. Cell Rep.14, 1317–1329 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Monroig, O., Rotllant, J., Sanchez, E., Cerda-Reverter, J. M. & Tocher, D. R. Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim. Biophys. Acta1791, 1093–1101 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Pang, S. C. et al. Double Transgenesis of Humanized fat1 and fat2 Genes Promotes Omega-3 Polyunsaturated Fatty Acids Synthesis in a Zebrafish Model. Mar. Biotechnol.16, 580–593 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Kemilainen, H. et al. The Hydroxysteroid (17beta) Dehydrogenase Family Gene HSD17B12 Is Involved in the Prostaglandin Synthesis Pathway, the Ovarian Function, and Regulation of Fertility. Endocrinology157, 3719–3730 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Moon, Y. A. & Horton, J. D. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J. Biol. Chem.278, 7335–7343 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Sakurai, N. et al. Systemic distribution and tissue localizations of human 17beta-hydroxysteroid dehydrogenase type 12. J. Steroid Biochem. Mol. Biol.99, 174–181 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Rantakari, P. et al. Hydroxysteroid (17{beta}) dehydrogenase 12 is essential for mouse organogenesis and embryonic survival. Endocrinology151, 1893–1901 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Heikela, H. et al. Hydroxysteroid (17beta) dehydrogenase 12 is essential for metabolic homeostasis in adult mice. Am. J. Physiol. Endocrinol. Metab.319, E494–E508 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Gao, Y., et al. Unraveling Differential Transcriptomes and Cell Types in Zebrafish Larvae Intestine and Liver. Cells11, 3290 (2022). [DOI] [PMC free article] [PubMed]
- 20.Lands, W. E. M. Metabolism of Glycerolipides: A Comparison of Lecithin and Triglyceride Synthesis. J. Biol. Chem.231, 883–888 (1958). [PubMed] [Google Scholar]
- 21.Wang, X. et al. Induced formation of primordial germ cells from zebrafish blastomeres by germplasm factors. Nat. Commun.14, 7918 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, C. et al. Zebrafish hhex-null mutant develops an intrahepatic intestinal tube due to de-repression of cdx1b and pdx1. J. Mol. Cell Biol.11, 448–462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winata, C. L. et al. Development of zebrafish swimbladder: The requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev. Biol.331, 222–236 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Hostelley, T. L., et al. Exocrine pancreas proteases regulate β-cell proliferation in zebrafish ciliopathy models and in murine systems. Biol Open10, bio046839 (2021). [DOI] [PMC free article] [PubMed]
- 25.Singh, S. P., et al. A single-cell atlas of de novo β-cell regeneration reveals the contribution of hybrid β/δ-cells to diabetes recovery in zebrafish. Development149, dev199853 (2022). [DOI] [PubMed]
- 26.Field, H. A., Ober, E. A., Roeser, T. & Stainier, D. Y. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol.253, 279–290 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Field, H. A., Dong, P. D., Beis, D. & Stainier, D. Y. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev. Biol.261, 197–208 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Ng, A. N. et al. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev. Biol.286, 114–135 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Gut, P. et al. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat. Chem. Biol.9, 97–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong, P. D. et al. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat. Genet39, 397–402 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Brannon, P. M. Adaptation of the exocrine pancreas to diet. Annu Rev. Nutr.10, 85–105 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Wilson, M. H. et al. A point mutation decouples the lipid transfer activities of microsomal triglyceride transfer protein. PLoS Genet16, e1008941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avraham-Davidi, I. et al. ApoB-containing lipoproteins regulate angiogenesis by modulating expression of VEGF receptor 1. Nat. Med.18, 967–973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Templehof, H., Moshe, N., Avraham-Davidi, I., Yaniv, K. Zebrafish mutants provide insights into Apolipoprotein B functions during embryonic development and pathological conditions. JCI Insight6, e130399 (2021). [DOI] [PMC free article] [PubMed]
- 35.Pepino, M. Y., Kuda, O., Samovski, D. & Abumrad, N. A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev. Nutr.34, 281–303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakaguchi, T., Kikuchi, Y., Kuroiwa, A., Takeda, H. & Stainier, D. Y. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development133, 4063–4072 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Horowitz, A., Chanez-Paredes, S. D., Haest, X. & Turner, J. R. Paracellular permeability and tight junction regulation in gut health and disease. Nat. Rev. Gastroenterol. Hepatol.20, 417–432 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Köhler, K., Louvard, D. & Zahraoui, A. Rab13 regulates PKA signaling during tight junction assembly. J. Cell Biol.165, 175–180 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endale, H. T., Tesfaye, W. & Mengstie, T. A. ROS induced lipid peroxidation and their role in ferroptosis. Front Cell Dev. Biol.11, 1226044 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Askari, B. et al. Rosiglitazone inhibits acyl-CoA synthetase activity and fatty acid partitioning to diacylglycerol and triacylglycerol via a peroxisome proliferator-activated receptor-gamma-independent mechanism in human arterial smooth muscle cells and macrophages. Diabetes56, 1143–1152 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, X., et al. Biosynthetic deficiency of docosahexaenoic acid causes nonalcoholic fatty liver disease and ferroptosis-mediated hepatocyte injury. J. Biological Chem., 300, 107405 (2024). [DOI] [PMC free article] [PubMed]
- 42.Miotto, G. et al. Insight into the mechanism of ferroptosis inhibition by ferrostatin-1. Redox Biol.28, 101328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peron, M., et al. The stem-like Stat3-responsive cells of zebrafish intestine are Wnt/β-catenin dependent. Development147, dev188987 (2020). [DOI] [PMC free article] [PubMed]
- 44.Han, L. et al. Osr1 functions downstream of Hedgehog pathway to regulate foregut development. Dev. Biol.427, 72–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan, H. et al. Fatty acid oxidation is required for embryonic stem cell survival during metabolic stress. EMBO Rep.22, e52122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neto, A., Mercader, N. & Gomez-Skarmeta, J. L. The Osr1 and Osr2 genes act in the pronephric anlage downstream of retinoic acid signaling and upstream of Wnt2b to maintain pectoral fin development. Development139, 301–311 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Mak, H. Y. et al. AGPAT2 interaction with CDP-diacylglycerol synthases promotes the flux of fatty acids through the CDP-diacylglycerol pathway. Nat. Commun.12, 6877 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, R. S. et al. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev. Cell46, 112–125.e114 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Goh, I. et al. Yolk sac cell atlas reveals multiorgan functions during human early development. Science381, eadd7564 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cindrova-Davies, T. et al. RNA-seq reveals conservation of function among the yolk sacs of human, mouse, and chicken. Proc. Natl Acad. Sci. USA114, E4753–e4761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bass, L. & Wershil, B. K. Anatomy, histology, embryology, and developmental anomalies of the small and large intestine. In Sleisenger and Fordtran’s gastrointestinal and liver disease 10th ed Philadelphia, PA: Saunders, 1649 (Elsevier Inc, 2016).
- 52.He, J. et al. Ribosome biogenesis protein Urb1 acts downstream of mTOR complex 1 to modulate digestive organ development in zebrafish. J. Genet Genomics44, 567–576 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Li, Y. F. et al. Mycn regulates intestinal development through ribosomal biogenesis in a zebrafish model of Feingold syndrome 1. PLoS Biol.20, e3001856 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cox, A. G., et al. Yap regulates glucose utilization and sustains nucleotide synthesis to enable organ growth. EMBO J.37, e100294 (2018). [DOI] [PMC free article] [PubMed]
- 55.Susila, H. et al. Florigen sequestration in cellular membranes modulates temperature-responsive flowering. Science373, 1137–1142 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Zhang, X. et al. Defective Phosphatidylglycerol Remodeling Causes Hepatopathy, Linking Mitochondrial Dysfunction to Hepatosteatosis. Cell. Mol. Gastroenterol. Hepatol.7, 763–781 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Díaz, P., Powell, T. L. & Jansson, T. The role of placental nutrient sensing in maternal-fetal resource allocation. Biol. Reprod.91, 82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Napso, T., Yong, H. E. J., Lopez-Tello, J. & Sferruzzi-Perri, A. N. The Role of Placental Hormones in Mediating Maternal Adaptations to Support Pregnancy and Lactation. Front. Physiol.9, 1091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez-Tello, J. et al. Fetal manipulation of maternal metabolism is a critical function of the imprinted Igf2 gene. Cell Metab.35, 1195–1208.e1196 (2023). [DOI] [PubMed] [Google Scholar]
- 60.Korzh, S. et al. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev. Biol.8, 84 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods12, 982–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li, Q. et al. Inhibition of SLC7A11-GPX4 signal pathway is involved in aconitine-induced ferroptosis in vivo and in vitro. J. Ethnopharmacol.303, 116029 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Qiu, Y. et al. Carboxyl ester lipase is highly conserved in utilizing maternal supplied lipids during early development of zebrafish and human. Biochim. Biophys. Acta Mol. Cell Biol. Lipids1865, 158663 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc.3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Q., et al. Zebrafish cyp11c1 Knockout Reveals the Roles of 11-ketotestosterone and Cortisol in Sexual Development and Reproduction. Endocrinology161, bqaa048 (2020). [DOI] [PubMed]
- 66.Wang, Y., et al. Cyp11a2 Is Essential for Oocyte Development and Spermatogonial Stem Cell Differentiation in Zebrafish. Endocrinology163, bqab258 (2022). [DOI] [PubMed]
- 67.Thierer, J. H., Ekker, S. C. & Farber, S. A. The LipoGlo reporter system for sensitive and specific monitoring of atherogenic lipoproteins. Nat. Commun.10, 3426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lam, S. M., et al. Quantitative Lipidomics and Spatial MS-Imaging Uncovered Neurological and Systemic Lipid Metabolic Pathways Underlying Troglomorphic Adaptations in Cave-Dwelling Fish. Mol. Biol. Evol.39, msac050 (2022). [DOI] [PMC free article] [PubMed]
- 69.Lam, S. M. et al. A multi-omics investigation of the composition and function of extracellular vesicles along the temporal trajectory of COVID-19. Nat. Metab.3, 909–922 (2021). [DOI] [PubMed] [Google Scholar]
- 70.Ye, D., et al. A landscape of differentiated biological processes involved in the initiation of sex differentiation in zebrafish. Water Biol. Security1, 100059 (2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
The sequence data that support the findings of this study have been deposited in the Genome Sequence Archive at the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (CRA018363 [https://ngdc.cncb.ac.cn/gsa/search?searchTerm=CRA018363], CRA018331, and CRA018335) are publicly accessible at https://ngdc.cncb.ac.cn/gsa. The lipidomics data are provided in Supplementary Data 3. Public bulk RNA-seq data are available in the GSA database under the accession numbers CRA005219 and CRA005220. The remaining data are available within the Article, Supplementary Information or Source data file. Source data are provided with this paper.