Abstract
Human astrovirus (HAstV) is a global cause of gastroenteritis in infants, the elderly, and the immunocompromised. However, the molecular mechanisms that control its susceptibility are not fully understood, as the functional receptor used by the virus has yet to be identified. Here, a genome‐wide CRISPR‐Cas9 library screen in Caco2 cells revealed that the neonatal Fc receptor (FcRn) can function as a receptor for classical HAstV (Mamastrovirus genotype 1). Deletion of FCGRT or B2M, which encode subunits of FcRn, rendered Caco2 cells and intestinal organoid cells resistant to HAstV infection. We also showed that human FcRn expression renders non‐susceptible cells permissive to viral infection and that FcRn binds directly to the HAstV spike protein. Therefore, our findings provide insight into the entry mechanism of HAstV into susceptible cells. We anticipate that this information can be used to develop new therapies targeting human astroviruses, providing new strategies to treat this global health issue.
Keywords: Astrovirus, FcRn, HAstV, Neonatal Fc receptor
Knocking out of neonatal Fc receptor in Caco2 cells inhibited classical Human Astrovirus infection.
1. INTRODUCTION
Astroviruses (family Astroviridae), nonenveloped viruses with a positive‐sense single‐stranded RNA genome, cause gastroenteritis in humans and animals worldwide. Astroviruses are classified into two groups based on their capsid amino acid sequences: Avastrovirus, which infects birds, and Mamastrovirus, which infects mammals. Mamastrovirus consists of 19 groups, including the human‐infecting Mamastrovirus genotype 1 (HAstV serotypes 1–8), 6 (MLB1 and 2), 8 (VA2, 4, and 5), and 9 (VA1 and 3) (de Benedictis et al., 2011).
Classical‐type human astroviruses (e.g., Mamastrovirus genotype 1), unlike novel‐type astroviruses (e.g., Mamastrovirus genotype 6, 8, and 9), require capsid processing by trypsin to infect cells. Several human and primate cell lines, especially those derived from the gastrointestinal tract, are susceptible to astroviruses (Brinker et al., 2000). Recently, astroviruses were shown to replicate in a human intestine‐derived organoid (Kolawole et al., 2019). The VA (Brown et al., 2015; Frémond et al., 2015; Król et al., 2021; Lum et al., 2016; Naccache et al., 2015; Quan et al., 2010), MLB (Sato et al., 2016), and classical HAstV (Koukou et al., 2019) have been detected in the central nervous system of immunocompromised patients. Cases of viremia involving MLBs (Cordey et al., 2016; Cordey et al., 2018; Holtz et al., 2011; Lau et al., 2017; Sato et al., 2016; Wylie et al., 2012) or classical HAstVs (van der Doef et al., 2016; Wunderli et al., 2011; Zanella et al., 2021) have also been reported in humans. However, the mechanism underlying viral spread from the intestinal tract or other primary infection sites remains unknown.
Astrovirus genome contains four open reading frames (ORFs): ORF1a, ORF1b, ORF2, and ORFx. ORF1a and ORF1a/b encode nsP1a and nsP1ab polyproteins, respectively, which are further divided into a protease, VPg and RdRp, as well as several proteins of unknown function. ORF2 encodes a capsid protein precursor, VP90 (M1 to E787), while ORFx overlaps with ORF2 in a shifted reading frame and encodes an XP protein involved in virus assembly and/or release (Lulla & Firth, 2020). VP90 is cleaved by caspase to form VP70 (M1 to D657). Extracellular trypsin‐mediated processing leads to the formation of a mature capsid consisting of the core, VP34 (M1 to S411) and spike proteins, VP27 (T394 to R648), along with the removal of some spike proteins, VP25 (S424 to R648). As antibodies targeting the spike are neutralizing (Bass & Upadhyayula, 1997; Bogdanoff et al., 2017), VP27 is believed to contain the receptor binding domain. Trypsin treatment significantly alters the structure of HAstV (Dryden et al., 2012), potentially facilitating receptor interaction.
During cellular infection, HAstV is believed to initially bind to a surface‐expressed polysaccharide through the VP27 region, which is conserved among serotypes (Dong et al., 2011). Viral entry into the cell may then be facilitated through clathrin‐mediated endocytosis, escape from the late endosomes (Méndez et al., 2014), and potentially involve uncoating by protein disulfide isomerase A4 (PDIA4) to release its genome (Aguilar‐Hernández et al., 2020). However, a functional receptor for HAstV has not been identified, and the entry mechanism has not been fully elucidated.
Here, we identified the neonatal Fc receptor (FcRn) as a receptor of HAstV using genome‐wide CRISPR‐Cas9 screening. Although FcRn was directly bound to a spike protein, depleting FcRn did not impair HAstV binding to the cell surface. However, HAstV infection was dramatically prevented in FcRn‐deficient cells, indicating that FcRn plays a role in viral uptake. Our findings suggest that FcRn is an important factor for HAstV infection and provides critical insight into the mechanism underlying HAstV entry.
2. RESULTS
2.1. Identification of host genes essential for HAstV infection
Caco2 cells derived from human colon adenocarcinoma are susceptible to HAstV and are frequently used in various studies on HAstV (Willcocks et al., 1990). HAstV typically does not induce death in Caco2 cells (Hargest et al., 2021), but apoptosis can be induced at higher multiplicities of infection (MOIs) (Guix et al., 2004). For multiple cycle of infections, infected Caco2 cells were cultured with trypsin to facilitate the maturation of newly shed virus. To conduct CRISPR knockout screenings (Sanjana et al., 2014) to identify host factors involved in HAstV replication in Caco2 cells, it was necessary to keep the cells on plates under trypsin‐containing conditions. First, Caco2 cells were plated on collagen‐coated plates to prevent detachment by trypsin for several days. Subsequently, highly susceptible and cytopathic effect‐prone Caco2 cells were cloned (Figure S1a,b).
A single‐guide RNA (sgRNA) library pool was transduced into Cas9‐expressing Caco2 (Caco2/Cas9) cells. The cells were then exposed to HAstV4 and incubated in a trypsin‐containing medium for 2 days, and incubated further for propagation of cells that had escaped infection (Figure 1a). This infection–amplification step was repeated five times in the first round and then twice in the second round. Genomic DNA was extracted from the escaped cells, and the sgRNA regions were amplified and sequenced using next‐generation sequencing (NGS). The results suggested that the genes FCGRT and B2M could be involved in susceptibility to HAstV4a (Figure 1b). These genes encode the α (or heavy) chain (encoded by FCGRT) and β2 microglobulin (β2m; encoded by B2M) of the neonatal Fc receptor, a non‐classical Major Histocompatibility Complex (MHC) class I molecule.
FIGURE 1.
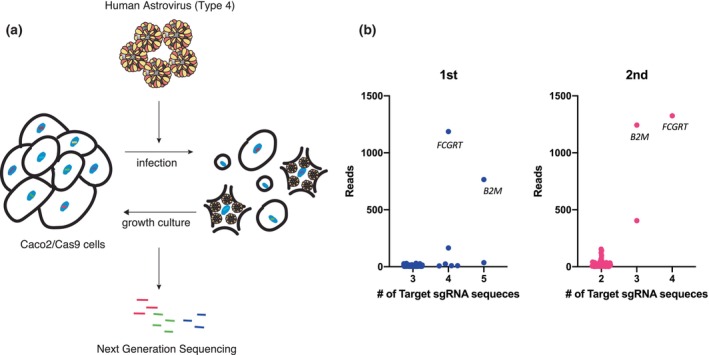
CRISPR screening revealed that FCGRT and B2M were involved in HAstV infection. (a) Schema of the screening method for cells resistant to HAstV infection. Caco2 cells transduced with a CRISPR‐Cas9 library pool were infected with HAstV4 (MOI = 5) multiple times until almost no surviving cells were visible. The genomic DNA of the surviving cells was purified, and determined for sgRNA sequences via next‐generation sequencing (NGS). (b) NGS analysis revealed knockout genes enriched in HAstV‐resistant cells. The library contained multiple gRNAs for each target gene, and the X‐axis indicates the number of sgRNA detected for each gene via NGS. Y‐axis represents the total read counts per gene. Each dot represents single sgRNA detected by NGS analysis and multiple sgRNAs targeting FCGRT and B2M were observed in the first (blue) and second (pink) rounds.
To determine the role of these gene products in HAstV infection in Caco2 cells, we established FCGRT‐ or B2M‐knockout Caco2 cells, as well as FCGRT‐B2M double‐knockout (FB KO) Caco2 cells. Knockout of FCGRT (F KO) and/or B2M (B KO) was confirmed via western blotting (Figure 2a). HAstV4 RNA was significantly amplified in infected parental Caco2/Cas9 cells, but showed much less amplification in F KO, B KO, or FB KO Caco2 cells (Figure 2b). When HAstV1, the most prevalent among classical HAstV (Vu et al., 2017), was introduced to the knockout cells, viral RNA amplification was again reduced in the knockout cell lines. To confirm that the loss of susceptibility was caused by the gene knockouts, each gene was reintroduced to cells using lentivirus encoding sgRNA‐resistant FCGRT or B2M. The addition of FCGRT and B2M restored the HAstV RNA levels to that of the parental cells (Figure 2a,b). Additionally, the susceptibilities of FB KO Caco2 cells to HAstVs 2, 3, 5, 6, 7, and 8 serotypes were similarly decreased compared to parental cells (Figure 2c). The extracellular viral titer of HAstV 1 or 4 in the supernatant of infected FB KO cells showed a significant decrease (Figure 2d). These findings suggest that FcRn is essential for HAstV infection in Caco2 cells.
FIGURE 2.
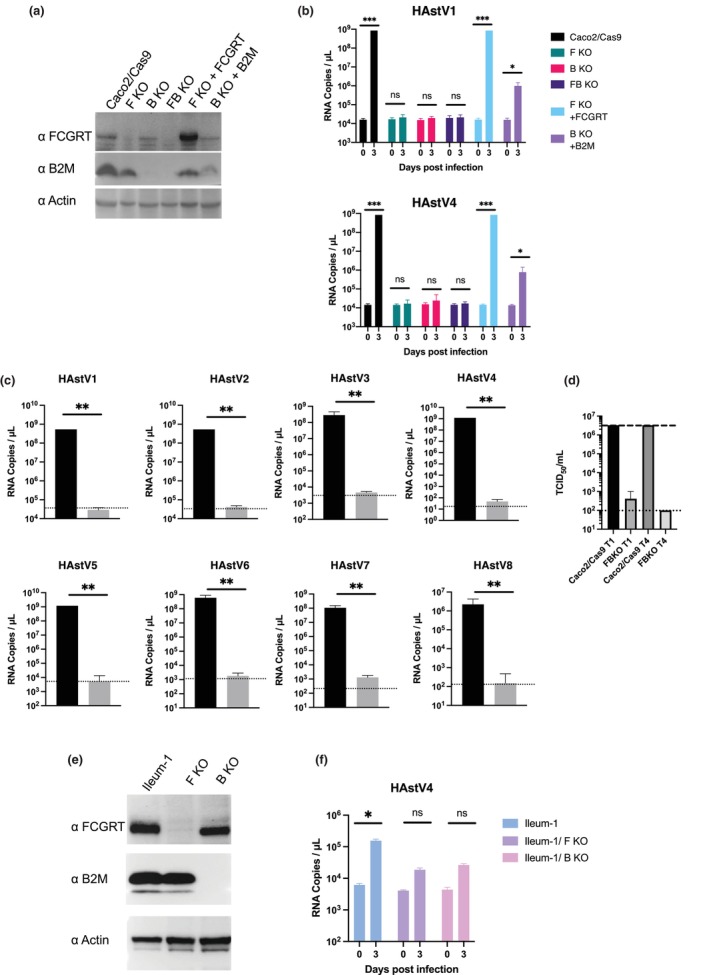
Knocking out FCGRT or B2M prevents human astrovirus infection of Caco2 cells and an ileum‐derived human intestinal organoid (Ileum‐1). (a) Detection of FCGRT and B2M in Caco2 cells. Caco2/Cas9 knockout cells targeted for FCGRT (F KO), B2M (B KO), and both (FB KO), and cells rescued for FCGRT (F KO + FCGRT) and for B2M (B KO + B2M) were detected for FCGRT and B2M by Western blot. Actin was used as a loading control. (b) Knockout of FCGRT or B2M inhibited HAstV infection in Caco2/Cas9 cells. The cells were incubated with HAstV1 or 4 (MOI 0.05) for 1 h, washed twice, and incubated in a fresh medium containing 5 μg/ml trypsin. Culture supernatants were collected immediately after adding fresh medium (Day 0) and at 3 days post‐infection (day 3). RNA copies in the culture supernatant were determined via qPCR. Each data bar represents the geometric mean for eight wells of inoculated cells. Error bars denote SD. Each experiment was performed three times, and representative data are shown in this figure. Significance was determined by the Mann–Whitney test (***p < 0.0005; *p < 0.05; ns, not significant). (c) Knocking out FCGRT and B2M (FB KO) inhibited infection by all serotypes of classical HAstV (Mammastrovirus 1) in Caco2/Cas9 cells. Caco2/Cas9 (black) or FB KO Caco2 (gray) cells were incubated in a culture medium containing trypsin‐activated HAstV for 1 h. After washing with the medium three times, the cells were cultured with 5 μg/ml trypsin for 3 days at 37°C. The amount of genomic RNA of each virus was determined by qPCR. Each data bar represents the geometric mean for eight wells of inoculated cells. Error bars denote SD. Each experiment was performed two or more times, and representative data are shown in this figure. Significance was determined by the Mann–Whitney test (**p < 0.005). (d) The median tissue culture infectious dose (TCID50) of progeny virus of HAstV1 (T1) or HAstV4 (T4) produced from Caco2/Cas9 and FB KO cells at 3 days post‐inoculation was determined. Each data bar represents the geometric mean for six wells of Caco2/Cas9 or FBKO cells. Error bars denote SD. The dashed or dotted line indicates the upper or lower limit of TCID50. (E) Detection of FCGRT and B2M in corresponding knockout Ileum‐1 cells. Actin was used as a loading control. (F) Each gene‐knockout Ileum‐1 monolayer was inoculated with culture medium containing trypsin‐activated HAstV4. After washing twice with basal culture medium, cells were cultured at 37°C for 3 days. Genomic RNA copies of each virus were determined by qPCR. Each data bar represents the geometric mean for four wells of inoculated monolayers. Error bars denote genomic SD. Each experiment was performed twice, and representative data are shown in this figure. Significance was determined by the Mann–Whitney test (*p < 0.05; ns, not significant).
2.2. Astrovirus susceptibility is reduced by FcRn deletion in human intestinal organoids
The primary target of HAstV is the intestinal epithelium, and astrovirus infections have recently been established in organoids derived from the small intestine or colon (Kolawole et al., 2019). Therefore, we investigated whether FcRn has played a role in HAstV susceptibility in a human intestinal organoid derived from normal human ileum (Ileum‐1). In FCGRT‐ or B2M‐knockout Ileum‐1, the respective expression of FCGRT or B2M expression was not observed (Figure 2e). HAstV4, activated by trypsin treatment, was inoculated into the Ileum‐1 monolayer. The increase in HAstV4 RNA level was similar to the human enteric organoid (Kolawole et al., 2019). However, since Ileum‐1 could not be cultured with trypsin, the newly produced virus could not be activated for infection, resulting in lower HAstV amplification compared to Caco2/Cas9 cells. Nonetheless, the HAstV4 RNA level was significantly reduced in FCGRT‐knockout Ileum‐1 (Figure 2f).
Since enterovirus B also uses FcRn as a receptor (Morosky et al., 2019; Zhao et al., 2019), the replication of other gastroenteritis viruses—GII.4‐type human norovirus (HuNoV) and GI.1‐type human sapovirus (HuSaV)—was also studied in the knockout Ileum‐1. A fecal specimen containing GII.4‐type HuNoV was inoculated into Ileum‐1, and progeny production in the culture supernatant was determined 3 days post‐infection. As previously shown (Haga et al., 2020), deletion of the fucosyltransferase‐2 gene (FUT2) from Ileum‐1 greatly reduced Ulex europaeus agglutinin‐1 (UEA‐1) binding to the cellular surface and the susceptibility to GII.4‐type HuNoV (Figure S2a,b). Infection of GI.1‐type HuSaV to HuTu80 cells (Takagi et al., 2020) and human intestinal organoids (Euller‐Nicolas et al., 2023) is known to require sodium glycocholate (GlyCA), which was included in the culture medium (Figure S2c). In contrast to the results observed for HAstV4, neither FCGRT nor B2M deletion affected susceptibility to HuNoV and HuSaV infections (Figure S2d,e). The data suggest that FcRn is not universally used in intestinal virus infection, though it is used by enterovirus B and HAstV.
2.3. Exogenous expression of FCGRT and B2M renders unsusceptible cells permissive to HAstV
Susceptibility to HAstV varies among different cell lines (Brinker et al., 2000). To further investigate the role of FcRn in HAstV susceptibility, we compared FcRn expression and susceptibility in various cell lines. First, we compared HAstV1 and HAstV4 susceptibility and FCGRT or B2M mRNA expression in different human cell lines using data from the Human Protein Atlas (www.proteinatlas.org) (Table S1). In cell lines with lower FCGRT mRNA expression compared to Caco2 cells, susceptibility to HAstV was also lower. For example, cells from the large intestine (HCT116 and HCT15), cells from the small intestine (HuTu80), and HeLa cells were significantly less susceptible than Caco2 cells (Brinker et al., 2000). FCGRT protein expression in these cell lines was lower than in Caco2/Cas9 cells, whereas B2M levels were higher or comparable in HCT116, HuTu80, and Hela cells, but lower in HCT15 cells (Figure 3a). Next, we investigated whether susceptibility to HAstV could change when exogenous FCGRT was transduced into these cell lines (Figure 3b). Parental HeLa cells and HuTu80 cells exhibited low susceptibility to HAstV1 and marginal susceptibility to HAstV4, whereas HCT15 and HCT116 cells showed even lower susceptibility to both strains (Figure 3c), consistent with previous findings (Brinker et al., 2000). Transduction of FCGRT into HCT116, HuTu80, and HeLa cells, where endogenous B2M was present, increased susceptibility to HAstV1 and HAstV4. However, this effect was not observed in HCT15 cells. Further transduction of B2M into HCT15/FCGRT cells (HCT15/FCGRT/B2M) improved replication of HAstV1 and HAstV4 (Figure 3c), similar to results observed in B2M knock out Caco2/Cas9 cells abolished HAstV susceptibility (Figure 2b). Transduction of B2M into HCT116 cells had little effect on increasing HAstV susceptibility (Figure 3c). Apparently, endogenous B2M in HCT116 cells was sufficient enough to form functional FcRn and support HAstV infection when FCGRT was transduced.
FIGURE 3.
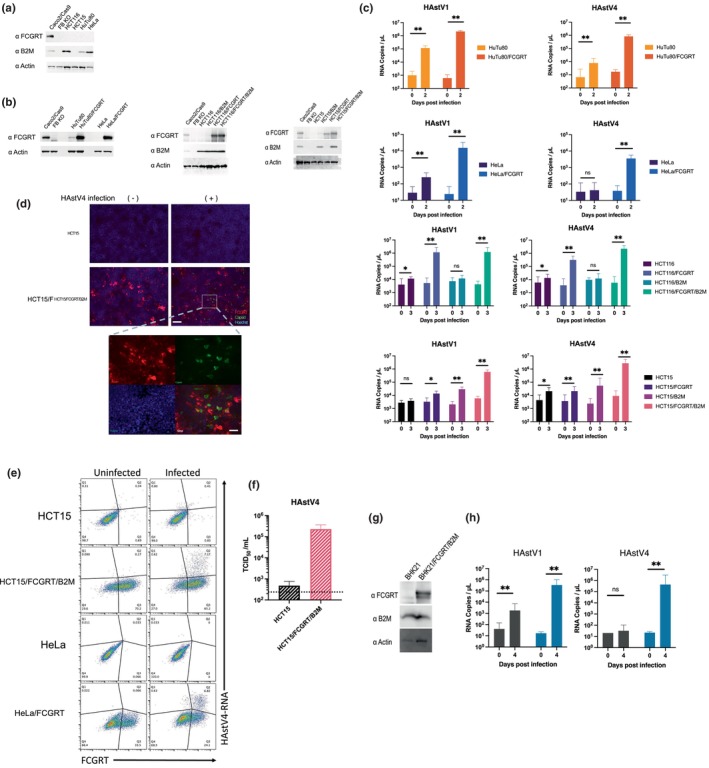
The exogenous expression of FCGRT and B2M renders cell lines susceptible to HAstV. (a) HCT116, HCT15, HuTu80, and HeLa cells exhibited lower FCGRT expression compared to Caco2/Cas9 cells. Additionally, HCT15 cells showed undetectable B2M expression, similar to FB KO cells. (b) FCGRT was transduced into each cell line using a lentiviral vector, while B2M was transduced into HCT116 and HCT15 cells. Stable expression of FCGRT and B2M was detected using Western blotting with actin serving as a loading control. (c) Established cell lines were inoculated in a culture medium containing trypsin‐activated HAstV1 or HAstV4 (MOI 1) for 1 h. After washing with medium three times, cells were cultured without trypsin at 37°C for 2 or 3 days. Genomic RNA copies of each virus were determined by qPCR. Each experiment was performed twice, and representative data are shown in this figure. Each data bar represents the geometric mean of six technical replicates in each experiment. Error bars denote SD. Significance was determined by the Mann–Whitney test (***p < 0.0005; *p < 0.05; ns, not significant). (d) HCT15 cells and HCT15/FCGRT/B2M cells were infected with HAstV4 (MOI 10) and incubated for 24 h. Cells were fixed and treated with anti‐FCGRT antibodies (red) before permeabilization. Then cells were treated with anti‐HAstV capsid antibodies (green). Scale bar on the upper panel is 150 μm. The lower panel is an expanded image of the area indicated with a white square on the image of HAstV‐infected HCT15/FCGRT/B2M cells. Scale bar on the lower panel is 50 μm. (e) FCGRT‐expressing cells were permissive to HAstV. HCT15, HCT15/FCGRT/B2M, HeLa, and HeLa/FCRT cells were infected with HAstV4 (MOI 10) and incubated for 24 h. Cells were fixed for PrimeFlow analysis and then treated with anti‐FCGRT antibody and subsequently with fluorescent secondary antibody. The X‐axis indicates FCGRT expression and the Y‐axis indicates HAstV genome RNA replication in the cells. (f) Median tissue culture infectious dose (TCID50) of progeny virus produced from HCT15 or HCT15/FCGRT/B2M cells at 3 days post‐inoculation was determined. Each data bar represents the geometric mean of three wells of HCT15 or HCT15/FCGRT/B2M cells. Error bars denote SD.
To determine the causal relationship between FCGRT expression and viral susceptibility, we first confirmed that viral capsid protein expression was observed in cells expressing FCGRT on the cellular surface after HAstV4 infection of HCT15 or HCT15/FCGRT/B2M. The cells were fixed and treated with anti‐FCGRT antibodies before permeabilization to detect FCGRT protein on the cell surface. They were then permeabilized to detect the capsid protein of infected cells using anti‐HAstV capsid antibodies. FCGRT expression was clearly observed on the cellular surface of HCT15/FCGRT/B2M, and the capsid protein was also detected only in HCT15/FCGRT/B2M infected with HAstV4 (Figure 3d). However, not all HCT15/FCGRT/B2M cells expressed FCGRT on their surface, and some cells expressing the capsid protein did not show detectable FCGRT surface expression. Less than 10% of HCT15/FCGRT/B2M cells showed detectable FCGRT expression on the cell surface (Figure S3a). We inferred that FcRn was shuttled between the cellular surface and the cytoplasm because FcRn is known to be involved in the uptake and shedding of antibodies and albumin (He et al., 2008; Pyzik et al., 2019). The number of FCGRT‐positive cells increased after the permeabilizing treatment of HCT15/FCGRT/B2M (Figure S3b). Next, to further confirm HAstV replication in FCGRT‐expressing cells, we used PrimeFlow, an in situ hybridization assay that detects specific RNA, including viral RNA. This method has been applied to detect viral RNAs in cells infected with human immunodeficiency virus (Bertram et al., 2019; Rao et al., 2021), Zika virus (McDonald et al., 2018), and yellow fever virus (Sinigaglia et al., 2018). Specific probes designed to hybridize to genomic RNA regions of HAstV4 were used to detect infected cells in HCT15/FCGRT/B2M and HeLa/FCGRT cells, as well as their parental cells. By using anti‐FCGRT antibody to mark cells expressing detectable FCGRT, we sorted the cells harboring both viral RNA and FCGRT using fluorescence‐activated cell sorting (FACS) (Figure 3e). It was evident that cells harboring the PrimeFlow signal were also positive for FCGRT, strongly suggesting that FCGRT expression plays a crucial role in HAstV infection in the cell.
Finally, the supernatant from HAstV4‐infected HCT15 or HCT15/FCGRT/B2M cell cultures was examined for the presence of infectious progeny virus by the median tissue culture infectious dose (TCID50) using Caco2/Cas9 cells (Figure 3f). The progeny virus produced by HCT15/FCGRT/B2M cells was infectious to Caco2/Cas9 cells and showed apparent CPE. These findings indicated that FCGRT and B2M expression were highly associated with permissiveness to HAstV.
Next, exogenous FCGRT and B2M transduction was conducted in non‐human cells, specifically Madin–Darby canine kidney (MDCK) cells, which are non‐susceptible to HAstV infection (Brinker et al., 2000). Although a significant expression of both FCGRT and B2M was detectable upon transduction by western blotting (Figure S4a), and the presence of FCGRT on the cell surface was apparent (Figure S4b), viral RNA amplification after HAstV1/4 infection was minimal (Figure S4c). Occasional increases in viral RNA were detected, but a clear reproducibility of viral amplification was not achieved, suggesting that FcRn is not the sole determinant for HAstV propagation in MDCK cells. In BHK21 (Syrian hamster kidney) cells, HAstV1 showed marginal replication, while HAstV4 did not. However, transduction of FGGRT and B2M increased susceptibility to both HAstV1 and HAstV4 (Figure 3g,h). These findings suggested that FcRn was important for HAstV susceptibility in non‐human cells, but there may be other cellular factors essential for HAstV replication to gain permissiveness, such as the maturation of the HAstV capsid, which requires a host enzyme to cleave it intracellularly (Aguilera‐Flores et al., 2022; Méndez et al., 2004).
2.4. FcRn is involved in the early phase of infection
To investigate the role of FcRn during the viral life cycle, we asked whether FcRn is involved in viral attachment to cells. Caco2/Cas9 cells were inoculated with activated HAstV, and the extent of bound virus was evaluated by the number of viral RNA copies. When comparing the surface‐bound HAstV1 or HAstV4 between Caco2/Cas9 cells that expressed or did not express FcRn, no significant difference was found in the number of bound viruses (Figure 4a). Next, we treated Caco2/Cas9 cells with an anti‐FCGRT antibody before virus infection to block the interaction between FcRn and HAstV. The results showed that the anti‐FCGRT antibody dose‐dependently inhibited viral infection, as judged from the reduced viral amplification (Figure 4b). Since antibodies are ligands for FcRn, cells were also treated with an isotype control antibody, but this treatment did not inhibit HAstV infection. These findings indicated that cell‐surface binding of HAstV appeared independent of FcRn, but HAstV infection was highly dependent on FcRn expression on the cell surface.
FIGURE 4.
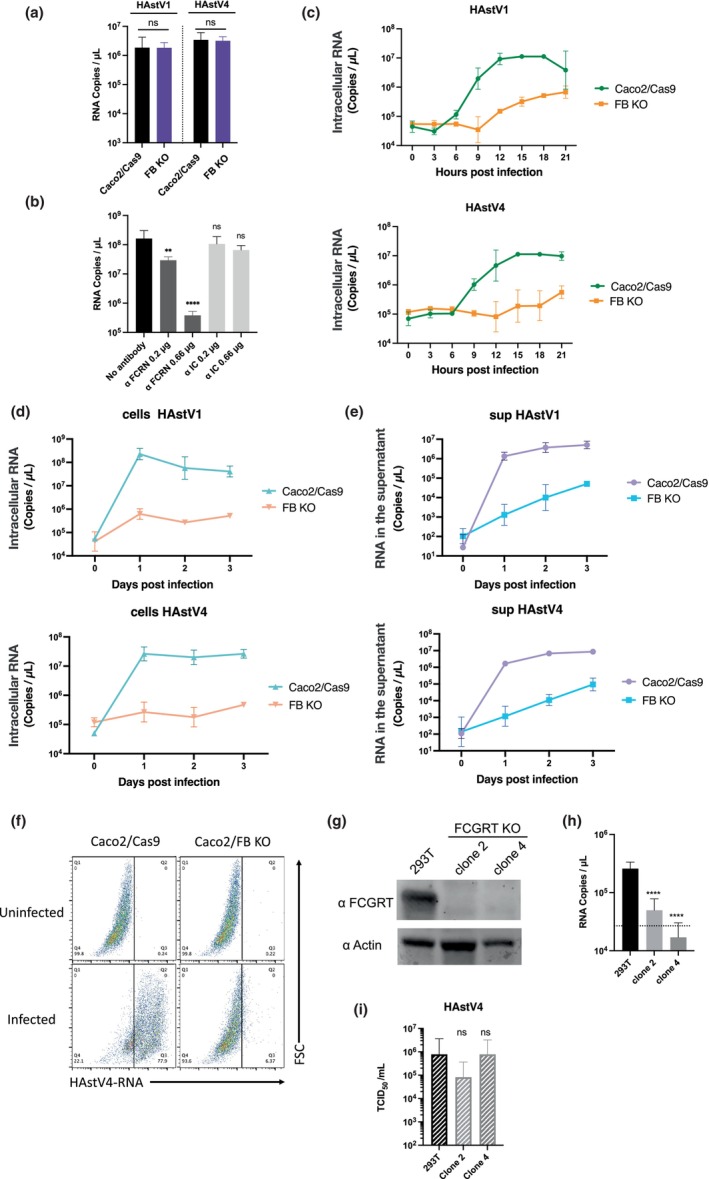
FcRn is involved in the early stages of human astrovirus replication. (a) Caco2/Cas9 cells were incubated with activated HAstV (MOI 50) at 4°C for 1 h. After washing with MEM (−) three times, RNA was extracted from the cells with the bound virus, and copies were quantified by qPCR. Each data bar represents the geometric mean of four wells. Error bars denote SD. Significance was determined by the Mann–Whitney test (ns, not significant). (b) Anti‐FCRN antibody blocked HAstV1 infection. Caco2 cells were cultured with 0.66 or 0.2 μg/ml of anti‐FCRN antibody (NBP1‐89128: Novus) or 2, 0.66, or 0.2 μg/ml rabbit isotype control IgG (ab37415: Abcam) for 1 h before infection. The Caco2 cells were incubated with the activated virus (MOI 0.005) and anti‐FCRN at the indicated concentrations for 1 h. The cells were washed three times with fresh medium and cultured for 3 days with each antibody at the indicated concentrations. Each data point represents the geometric mean of six wells. Significance was determined by Dunn's multiple comparisons test (****p < 0.0001; **p < 0.005; ns, not significant). (c) Trypsin‐activated HAstV1 or HAstV4 was inoculated into the cells (MOI 1). At each time point, the culture medium was aspirated from the wells, and the cells were washed twice with PBS. RNA was extracted, and copies were quantified by qPCR. Each point represents the mean of five wells. Error bars denote SD. (d, e) Viral RNA copies in cells and culture supernatant were reduced in FcRn‐KO cells. Trypsin‐activated HAstV1 or −4 was inoculated into the cells (MOI 1). Each day, culture medium (c) and cells (d) were collected separately. The cells were washed with PBS twice, and RNA was extracted. RNA copies in the culture medium and cells were quantified by qPCR. Each data point represents the mean of four wells. Error bars denote SD. (f) Infected cells were detected using PrimeFlow technology. Caco2 cells were exposed to trypsin‐activated HAstV4 (MOI 10) and incubated in a trypsin‐free medium for 24 h. Cells were detached from the wells and were treated with a HAstV‐specific probe according to the manufacturer's protocol. Probe‐hybridized cells were analyzed using a FACSMelody cell sorter (BD Biosciences) and FlowJo software (BD Biosciences). (g, h) Knocking out FCGRT prevents HAstV4 infection in 293T cells. (g) Detection of FCGRT in 293T cells and FCGRT‐KO clones. Actin was used as a loading control. (H) HAstV4 was incubated with trypsin to activate it. After activation, viruses were treated with FBS to inactivate trypsin and were inoculated into cells. After 1 h incubation at 37°C and subsequent washing with the medium (twice), the cells were cultured at 37°C for 3 days. The fold increase of genomic RNA copies determined by qPCR was calculated by dividing RNA copies at 3 days from 1 day. Each data bar represents the geometric mean of four wells of infected cells. Error bars denote SD. Each experiment was performed twice, and representative data are shown in this figure. Significance was determined by Dunn's multiple comparisons test (**p < 0.005; *p < 0.05; ns, not significant). (i) Viral RNA extracted from HAstV4 was transfected into 293T cells and FCGRT‐KO clones. After 72 h, the culture supernatant was serially diluted and inoculated into Caco2/Cas9 cells. After 5 days of incubation, the median tissue culture infectious dose (TCID50) was determined. Each data bar represents the geometric mean of four wells of transfected 293T cells or FCGRT‐KO clones. Each experiment was performed three times, and representative data are shown in this figure. Error bars denote SD. Significance was determined by the Dunn's multiple.
While HAstV RNA levels in FB KO Caco2 cells were completely inhibited compared to parental cells at 3 days post‐infection at an MOI of 0.05 (Figure 2b), a slight increase in viral RNA replication was observed in FB KO Caco2 cells over 21 h post‐infection at a high MOI (Figure 4c). In contrast, a significant increase in viral RNA was observed in parental Caco2/Cas9 cells at 9 h post‐infection (Figure 4c). Extending the observation to 3 days post‐infection, there was no apparent increase in viral RNA replication in FB KO cells (Figure 4d). However, a small but steady increase in extracellular viral RNA levels was observed in FB KO Caco2 cells (Figure 4e), which was not evident in the previous results (Figure 2b,c). In contrast, extracellular viral RNA produced from Caco2/Cas9 cells increased to approximately 104 times on the first‐day post‐infection (Figure 4e). The results confirmed that FCGRT/B2M was important for initiating viral RNA replication in the early phase of infection, but also suggested that an infectious route independent of FcRn could contribute to the infection. This apparent inconsistency could likely be due to the high titer at inoculation in that experiment (Figure 4e). Since virus replication was observed even in FB KO cells, we further investigated whether the proportion of infected cells was different in Caco2/Cas9 cells and FB KO cells by detecting replicated viral RNA with PrimeFlow RNA assay. Upon infection with trypsin‐activated HAstV4 at a MOI of 10, the proportion of infected cells at 24 h post‐infection was 77.9% for Caco2/Cas9 and 6.37% for FB KO cells (Figure 4f), suggesting that some FB KO cells were still permissive to HAstV at a higher MOI. Although the implication of FcRn is not clear, extracellular vesicles were reported to facilitate human astrovirus infection probably via an unconventional HAstV infectious pathway (Baez‐Navarro et al., 2022).
Since FcRn can mediate the endocytosis of antibodies and their release (He et al., 2008; Pyzik et al., 2019), FcRn could be involved in viral release from cells. To investigate this, the extent of viral release was examined using two 293T/FCGRT‐KO cell lines and their parental cells (Figure 4g). It is known that transfection of viral RNA into cells can yield infectious virions (Geigenmüller et al., 1997). Therefore, HAstV4 genomic RNA extracted from virions was transfected into the cells, and the infectious virions produced by each cell line was examined using TCID50. The 293T cells have a slight susceptibility to HAstV (Brinker et al., 2000), and FCGRT depletion abolished this susceptibility (Figure 4g,h). However, there was no significant difference in the levels of infectious virion shedding among cell lines after HAstV4 genomic RNA transfection (Figure 4i). Thus, our results indicated that FcRn is involved in the early stages of viral replication. Additionally, the results suggested the presence of another infection route, which is FcRn‐independent, in Caco2/Cas9 cells.
2.5. FcRn directly binds to Mamastrovirus 1
We have demonstrated a causal relationship between FcRn and the susceptibility of HAstV in the early stages of infection. To evaluate whether FcRn directly interacts with HAstV and acts as a HAstV receptor, we investigated the localization of FcRn in cell lines. FcRn is involved in transporting albumin and IgG into and out of cells and is expressed on the cell surface (Pyzik et al., 2019). While others have observed FcRn expression on the surface of Caco2 cells (Hornby et al., 2014), we were unable to detect endogenous FcRn expression in Caco2/Cas9 cells using available antibodies against FCGRT. Instead, we used HeLa/FCGRT and HCT15/FCGRT/B2M cells to detect surface FCGRT localization. Transduction of FCGRT into these cells increased FCGRT detection in over half of the cell population (Figure 3d,e). However, cell surface FCGRT expression was limited in fewer cells, as shown by applying anti‐FCGRT antibodies to cells without permeabilization (Figure S3). Since the population of FcRn‐supplemented HeLa and HCT15 cells infected with HAstV was significantly lower than those expressing FCGRT (Figure 3d,e), the results support the notion that infection occurs in cells expressing FcRn on the cell surface.
An enzyme‐linked immunosorbent assay (ELISA) was performed to examine the direct interaction between recombinant human FcRn (r‐hFcRn) protein and HAstV4. Immature HAstV4 was prepared in the absence of trypsin and purified by isopycnic CsCl‐gradient centrifugation (Figure S5). Purified and serially diluted HAstV4 was immobilized on the ELISA plate, and the wells were treated with or without trypsin. Trypsin treatment reduced HAstV4 detection (Figure 5a), potentially by removing some of the spike protein, VP25, from virus particles. Next, the immobilized HAstV4 were treated with r‐hFcRn. The r‐hFcRn bound to immature HAstV4, and the amount of r‐hFcRn binding to mature HAstV4 was reduced, while recombinant mouse FcRn (r‐mFcRn) did not bind to immobilized HAstV4 (Figure 5b). Since maturation of HAstV by trypsin treatment reduced r‐hFcRn binding to HAstV, r‐hFcRn was predicted to bind to the spike region of the capsid protein. Therefore, we produced recombinant spike protein (r‐VP25) using Escherichia coli and confirmed that r‐hFcRn could directly bind to r‐VP25. Since recombinant VP27 (T394 to P645) which has interacting domain to core capsid VP34, was insoluble when expressing by Escherichia coli, recombinant VP25 (r‐VP25: G429 to P645) was created by removing the N‐terminus of VP27 and was used for evaluating direct interaction to r‐FcRn. Although the homology of the spike region among serotypes was 42–75% (Figure S6), FcRn knockout in Caco2/Cas9 cells abolished the susceptibility of all serotypes of HAstV. We then succeeded in producing soluble type 1, 4, 5, 6, and 7 r‐VP25 and demonstrated that r‐hFcRn could dose‐dependently bind to each type of r‐VP25 (Figures 5c and S8). Finally, we estimated the docking site in silico. The binding sites of FcRn to IgG or serum albumin are well documented (Pyzik et al., 2019; Sockolosky & Szoka, 2015). As the isotype control IgG did not interfere with HAstV1 infection (Figure 4b) and albumin, which was contained in the blocking buffer, did not interfere the binding between the spike protein and FcRn (Figures 4c and 5b), the binding site of FcRn to the spike would be different from the binding region to these proteins. We then simulated the HAstV spike‐hFcRn docking model using the Dock application in the Molecular Operating Environment (MOE). The obtained docking model suggested that the binding site of FcRn to the spike (Figure 5d) was distinctly different from the binding site to IgG or albumin (PDB ID: 4N0U, shown in Figure S9) (Oganesyan et al., 2014). These findings suggest that human FcRn directly binds to the spike region, which is conserved among serotypes, and acts as a functional receptor of HAstV.
FIGURE 5.
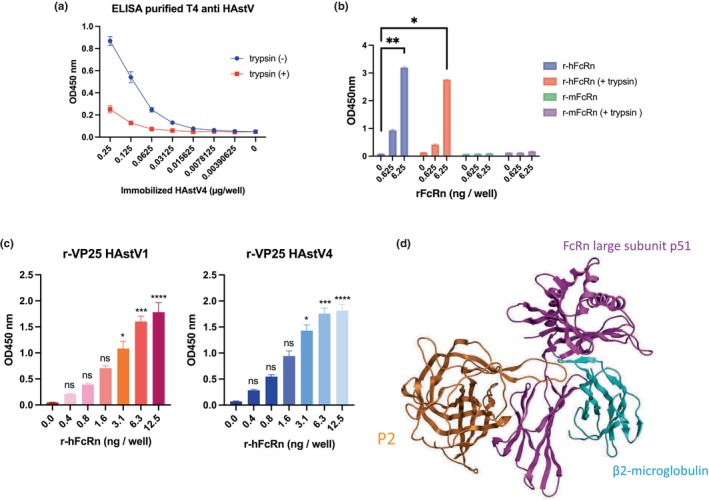
FcRn directly binds to spike protein of human astrovirus. (a) Immobilized HAstV was detected by ELISA. Massively produced HAstV4 without trypsin was purified, and serially diluted HAstV4 was immobilized on the plate. Wells were treated with or without trypsin (10 μg/ml) and viruses were detected by anti‐HAstV capsid antibody (8E7) and subsequent HRP‐conjugated anti‐mouse IgG. Each data point represents the mean of three wells. Error bars denote SD. (b) Recombinant FcRn bound to HAstV. Purified HAstV4 (250 ng) was immobilized on the plate and incubated with or without trypsin (10 μg/ml). His‐tagged recombinant human (h) or mouse (m) FcRn bound to viruses was detected by HRP‐conjugated anti‐His antibody (D291‐7:MLB). Each data point represents the mean of three wells. Error bars denote SD. (c) Recombinant human FcRn (r‐hFcRn) bound to type 1 or 4 of recombinant VP25. Each His‐tagged VP25 was expressed in BL21. Purified His‐tagged VP25 (2.5 μg) was immobilized and incubated with diluted r‐hFcRn. The r‐hFcRn bound to VP25 was detected by an anti‐FcRn antibody. Data are representative of two independent experiments. Each data point represents the mean of three wells. Error bars denote SD. Significance was determined by Dunn's multiple comparisons test (**p < 0.005; *p < 0.05; ns, not significant). (d) Docking model of the HAstV1 spike protein (P2) (PDB: 5EWO) and the human FcRn (PDB: 6FGB). The docking simulation was performed by using the Dock application in MOE software. The orange ribbon is the spike protein, the magenta ribbon is FcRn large subunit (FCGRT), and the cyan ribbon is β2‐macroglobulin (B2M).
3. DISCUSSION
Here, we demonstrated that depleting FcRn in Caco2 cells and a human intestinal organoid inhibited HAstV infection, while exogenous FcRn expression rendered several human cell lines susceptible to HAstV. Additionally, we found that FcRn played no role in HuNoV or HuSaV infection. We also showed that FcRn is directly bound to the HAstV spike protein, suggesting that FcRn acted as a receptor for HAstV.
Our genome wide gene screen revealed that genes encode FcRn heterodimer are essential for HAstV infection. However, the host factors previously reported (Aguilar‐Hernández et al., 2020; Méndez et al., 2014) to be involved in HAstV infection were not detected. It is possible that the deletion of these factors could be detrimental to cell survival or reduce HAstV progeny production, rather than preventing HAstV‐induced cell death.
FcRn is categorized as a non‐classical MHC class I molecule. It is highly expressed in infants' intestines, where it functions to transfer maternal IgG from milk to the intestines (Rodewald & Kraehenbuhl, 1984; Simister & Rees, 1985). IgG binds to FcRn at the apical membrane of the intestines and is then released at the basolateral membrane in a process regulated by pH (He et al., 2008). FcRn is expressed not only in the intestinal epithelium of neonatal rodents but also ubiquitously in various adult tissues (Pyzik et al., 2019). It is expressed in the epithelial cells of the intestinal tract, kidney, placenta, and liver, as well as in hematopoietic cells, vascular endothelia, and at the blood–brain barrier, working to recycle IgG or albumin by transporting them into and then out of cells (Pyzik et al., 2019). The primary target of HAstV is the intestines, and infection can cause gastroenteritis and, less frequently, encephalitis in immunocompromised patients. In cases where HAstV is detected in encephalitis of immunocompromised patients, VA1 (Mamastrovirus 9) (Brown et al., 2015; Król et al., 2021; Lum et al., 2016; Naccache et al., 2015; Quan et al., 2010) is the most frequently detected, while MLB1(Mamastrovirus 6) (Sato et al., 2016) and HAstV1 and 4 (Mamastrovirus 1) (Koukou et al., 2019; Wunderli et al., 2011) have also been detected. Despite the spike region having about a 40% identity between HAstV1 and HAstV4 (Figure S6), FcRn directly bound to the spike protein (VP25) of both types of HAstV1, 4, 5, 6, and 7 (Figures 5c and S7). We also showed that FcRn KO cells reduced susceptibility to classical HAstVs (Figure 2c), suggesting that FcRn works as a receptor for classical HAstVs. Indeed, in a previous study, VA1 infected primary astrocytes and immortalized neuronal cell lines, but minimal HAstV4 infection was observed (Janowski et al., 2019). According to the Human Protein Atlas (www.proteinatlas.org) (Berglund et al., 2008), FcRn is expressed in vascular endothelial cells in the brain and in some neuronal cell lines, implying that HAstV‐spread causing viremia and infection to the central nervous system could involve FcRn‐mediated viral entry.
Since FcRn can bind to both mature and immature HAstV4, HAstV tropism to enteric tissues would not be determined by HAstV maturation by trypsin. Although FcRn expression is not restricted to enteric tissues, the localization of FcRn in different tissues has not been fully revealed; therefore, FcRn localization to the cell surface could be another factor in determining HAstV tropism. Since only minor populations of exogenous FCGRT‐transduced cells expressed FCGRT on the cell surface (Figures 3d,e and S3), there must be a mechanism to regulate the localization of FcRn expression. We have not tested if FcRn would also bind to VP34. Since FcRn binding interface to antibodies and albumin encompassing significantly large surface area, its binding to virion may not confined to VP25 but could extend to VP34. Further experiment is needed to define binding region of FcRn to viral particles.
Cleavage of capsid proteins by trypsin, which induces efficient HAstV infection, exposes the spike protein VP27 and allows neutralizing monoclonal antibodies against HAstV to bind to VP27 (Bass & Upadhyayula, 1997; Bogdanoff et al., 2017). Neutralizing antibodies may block the binding of VP27 to FcRn, as we showed that FcRn directly binds to VP27. After HAstV binds to FcRn, HAstV may be taken into the cell via the clathrin heavy chain (CHC) pathway. Studies have shown that a CHC inhibitor or CHC siRNA reduces HAstV8 infectivity (Méndez et al., 2014), and FcRn‐mediated transcytosis of IgG is also associated with clathrin (He et al., 2008). Taken together, these findings support the idea that HAstV binds to surface‐expressed FcRn and enters the cell via clathrin‐mediated endocytosis. Once inside, FcRn may play a role in viral uncoating because it functions pH‐dependently as an uncoating receptor for enterovirus B (Zhao et al., 2019). Our experiments on FB KO Caco2 cells infected with higher viral titers (Figure 4c,f) also suggest that independent FcRn‐mediated infection routes exist. Recently, extracellular vesicles were reported to facilitate human astrovirus infection without the need for trypsin activation (Baez‐Navarro et al., 2022). The role of FcRn during viral RNA replication was not examined in this study. We found that transfection of virion‐derived viral genomic RNA could generate infectious particles (Figure 4i), but not efficient enough to monitor viral RNA replication. DNA‐based reverse genetics system shown to recapitulate viral RNA replication could be applicable if FcRn would involve the viral replication process (Chapellier et al., 2015). Although FcRn is known for releasing IgG or albumin from the cytoplasm and enhancing human immunodeficiency virus‐1 trafficking via Env‐specific IgG binding to FcRn in genital tract epithelial cells (Gupta et al., 2013), FcRn was not associated with HAstV shedding.
In this study, we demonstrated that FcRn serves as a receptor for HAstV (Mamastrovirus 1 and 9). Our data offer new insights into the mechanism underlying FcRn‐mediated HAstV entry. Targeting FcRn could be a potential strategy for developing anti‐HAstV therapeutics for patients with gastroenteritis or encephalitis.
4. EXPERIMENTAL PROCEDURES
4.1. Cells and viruses
Caco2 cells (American Type Culture Collection, ATCC: HTB‐37) were cultured in minimum essential medium (MEM; Nacalai Tesque) containing 10% fetal bovine serum (FBS), 1% nonessential amino acids (Nacalai Tesque, 06344‐56), 1% sodium pyruvate (Gibco, 11360‐070), and 1% antibiotic–antimycotic (Gibco, 15240‐062). HeLa cells (ATCC: CCL‐2) and MDCK cells (ATCC: CCL‐34) were cultured in Dulbecco's modified Eagle's medium (DMEM; Nacalai Tesque) containing 10% FBS and 1× penicillin–streptomycin (Gibco, 15140‐122). HuTu80 cells (ATCC: HTB‐40) were cultured in Iscove's Modified Dulbecco's Medium (Sigma‐Aldrich) containing 5% FBS, GlutaMAX (Gibco), and 1× penicillin–streptomycin. HCT15 cells (ATCC: CCL‐225) were cultured in RPMI medium (Nacalai Tesque) containing 10% FBS and 1× penicillin–streptomycin. HCT116 cells (ATCC: CCL‐247) were cultured in McCoy's 5a Medium (Gibco) containing 10% FBS and 1× penicillin–streptomycin. BHK21 cells (ATCC: CCL‐10) were cultured in MEM containing 10% FBS and 1× penicillin–streptomycin. Ileum‐1 was prepared as previously described (Sugimoto et al., 2021) and was established at the Keio University School of Medicine. Ileum‐1 was maintained as described in (Fujii et al., 2015) with the following modifications: Ileum‐1 was embedded in Matrigel matrix (Corning) and cultured in IntestiCult Organoid Growth Medium (Veritas). HAstV1 (VR‐1936), HAstV2 (VR‐1943), HAstV3 (VR‐1944), HAstV5 (VR‐1947), HAstV6 (VR‐1948), and HAstV7 (VR‐1949) were provided by the ATCC, and HAstV4 was a kind gift from Dr. Mitsuaki Oseto (Hachiya et al., 1999). HAstV8 was kindly gifted from Dr. Albert Bosch.
4.2. Screening of astrovirus susceptibility genes
The Cas9‐expressing lentiviral vector (lentiCas9‐Blast, Addgene) and the Human GeCKOv2 CRISPR knockout pooled library (Sanjana et al., 2014) were provided by Addgene (# 1000000049). Cas9 was induced in Caco2 cells and selected with 5 μg/ml of blasticidin‐S (Takara). The GeCKO2 library was then induced in the Caco2/Cas9 cells and selected with 5 μg/ml of puromycin (Takara). Transduced cells were incubated with MEM (−) plus type 4 HAstV (MOI 5). After 2 days, the culture medium was replaced with MEM (+), and the cells were incubated for 6 h, washed three times with PBS, and then incubated with MEM (−) plus HAstV. These infection–growth cycles were repeated five times in the first round and twice in the second round until almost no surviving cells were visible in Caco2/Cas9 cells. To facilitate screenings, the 17 clones of Caco2/Cas9 cells were tested for susceptibility of HAstV1 and HAstV4 by detecting infected cells using immunofluorescence. Six clones showing high proportion of infected cells were pooled and used to generate genome‐wide gene knockout library. The library‐induced Caco2/Cas9 cells, that escaped HAstV infection were grown in MEM (+), and genomic DNA was then extracted. Integration of the sgRNA sequence into genomic DNA was identified using PCR with the following primers: 5′‐GACTATCATATGCTTACCGTAAC‐3′ and 5′‐AAAAAGCACCGACTCGGTGCCAC‐3′. Amplified fragments were sequenced via next‐generation sequencing as previously described (Haga et al., 2016).
4.3. Astrovirus infection and quantitative real‐time PCR (qPCR)
Each serotype of HAstV was treated with 10 μg/ml of trypsin type IX‐S (trypsin from porcine pancreas Type IX‐S: SIGMA, T0303‐1G) at 37°C for 1 h. Cells were cultured on a collagen I‐coated 96‐well plate (Corning), inoculated with the virus, and incubated at 37°C for 1 h. The multiplicity of infection (MOI) was adjusted for each experiment. After washing with MEM containing 1% nonessential amino acids, 1% sodium pyruvate, 20 mM HEPES (pH 7.5), and 1% antibiotic–antimycotic solution (MEM [−]), the cells were incubated in MEM (−) or MEM (−) containing 5 μg/ml trypsin IX‐S. The supernatant was collected immediately after adding the medium, labeled as “Day 0.”
4.4. Quantitative real‐time PCR (qPCR) for quantification of Astrovirus genomic copies
Viral RNA in the supernatant was quantified using a Norovirus G1/G2 high‐speed probe detection kit (Toyobo) with primers and probes specific to astrovirus. For classical HAstVs, the forward primer sequence was 5′‐ACTGCDAAGCAGCTTCGTGA‐3′, and the reverse primer sequence was 5′‐CTTGCTAGCCATCRCACTTCT‐3′. The probe sequence was 5′‐ROX‐CACWGAAGAGCAACTCCATCGCAT‐BHQ2‐3′. These primers and probes target the boundary of ORF1 and ORF2 of the HAstV genome. The reaction time and temperature are as follows: pre‐incubation at 42°C for 4 min, then 95°C for 10 s, a two‐step reaction of denaturing at 95°C for 1 s, and annealing‐extension at 50°C for 15 s repeated for 45 cycles.
4.5. HAstV4 infection in Ileum‐1
HAstV4 was incubated with 10 μg/ml of trypsin type IX‐S (Trypsin from porcine pancreas type IX‐S: SIGMA, T0303‐1G) for 1 h, and then FBS was added at 1/10th of the virus solution to inactivate trypsin. The activated HAstV4 was inoculated onto monolayer Ileum‐1 cells and incubated for 1 h. After inoculation, the cells were washed three times with 200 μl of basal culture medium, and then 200 μl of ENRA medium (described in Data S1) was added. At 3 days post‐infection, the cell culture supernatant was collected, and RNA copies were quantified by qPCR as described above. The Light Cycler 480 (Roche) was used for the qPCR reaction.
4.6. Quantification of virus binding on the cell surface and viral RNA replication in the cells
For quantification of the bound virus, cells were incubated with activated HAstV (MOI 50) at 4°C for 1 h. After washing with MEM (−) three times, RNA was extracted using a NucleoSpin 8 kit (Macherey‐Nagel). To quantify viral RNA replication in the cells, HAstV was first treated with trypsin at 37°C for 1 h to activate it, and then trypsin was inactivated by adding FBS and cells were inoculated with HAstV (MOI 1). At each time point (Figure 4d–f), the supernatant and cells were collected separately. The cells were washed with phosphate‐buffered saline (PBS) after aspirating the supernatant. RNA from the cells was extracted using a NucleoSpin 8 kit (Macherey‐Nagel).
4.7. Blocking assay with anti‐FcRn antibody
Caco2/Cas9 cells were cultured with either 0.66 or 0.2 μg/ml of anti‐FcRn antibody (NBP1‐89128: Novus) or 0.66, or 0.2 μg/ml of rabbit isotype control IgG (ab37415: Abcam) for 1 h before infection. Subsequently, the Caco2 cells were incubated with the activated virus (MOI 0.005) along with 0.66, or 0.2 μg/ml of each antibody for 1 h. The cells were then washed with MEM (−) three times and cultured for 3 days with 0.66, or 0.2 μg/ml of each antibody. The HAstV genomic RNA copies in the culture supernatant at 3 days post‐infection were quantified by qPCR.
4.8. Immature HAstV4 preparation for ELISA
Caco2/Cas9 cells were plated in six T225 flasks. The cells were then activated with trypsin and infected with HAstV4 (MOI 1) for 1 h at 37°C. After infection, the cells were washed with fresh medium and incubated for 2 days in MEM (−). The viruses in the collected culture supernatant (180 ml) were centrifuged through a 30% (w/v) sucrose cushion for 2 h at 124,000×g using a Beckman SW32 Ti rotor. They were further purified by isopycnic CsCl‐gradient centrifugation in FBS‐free MEM (0.44 g/ml) for 24 h at 150,000×g using a Beckman SW55 Ti rotor. After centrifugation, the viruses, visible as a white band with a density of approximately 1.35 g/ml, were collected. This solution was diluted 10 times in FBS‐free MEM, and the viruses were centrifuged again at 150,000×g for 3 h. The precipitates were suspended in MEM (−) and the protein concentration was determined using a Qubit (Thermo). A virus solution (5 ng) was treated with 10 μg/ml of trypsin at 37°C for 1 h and then mixed with Laemmli's SDS/PAGE sample buffer. The lysates were separated on a 5–20% polyacrylamide gel (ePAGEL; ATTO), and the gel was stained with Bio‐Safe CBB G‐250 (Bio‐Rad).
4.9. ELISA for quantifying immobilized HAstV4
Purified HAstV4 was immobilized at 0.25 to 0.00390625 μg per well (three times serially diluted with PBS) on a 384‐well plate (Thermo Fisher Scientific). The immobilized HAstV4 was treated with or without trypsin (10 μg/ml) and washed with PBS. After blocking with Block Ace solution (Waken B Tech), the wells were incubated with anti‐HAstV capsid (8E7) mouse monoclonal IgG (1:1000, Santa Cruz Biotechnology) as a primary antibody, followed by HRP‐conjugated anti‐mouse IgG antibody (1:5000 dilution, Cell Signaling) as a secondary antibody. KPL SureBlue Reserve TMB Microwell Peroxidase Substrate (Sera Care) was used as a substrate, and the absorbance at 450 nm was detected using an Ensight multimode plate reader (PerkinElmer).
4.10. ELISA for binding of recombinant FcRn and immobilized HAstV4
Purified HAstV4 was immobilized (0.25 μg/well) on a 384‐well plate (Thermo Fisher Scientific). The immobilized HAstV4 was then treated with or without trypsin (10 μg/ml) followed by a wash with PBS. After blocking with Block Ace solution (Waken B Tech), the wells were incubated with His‐tagged recombinant human FcRn (r‐hFcRn) or mouse FcRn (r‐mFcRn) (Sino Biological), which were serially diluted 10 times with PBS (6.25–0.0625 ng/well). To detect the binding of rFcRn to HAstV4, the wells were treated with HRP‐conjugated anti‐His antibody (1:5000 dilution, MBL D291‐7). The KPL SureBlue Reserve TMB Microwell Peroxidase Substrate (Sera Care) was used as a substrate, and the absorbance at 450 nm was measured using an Ensight multimode plate reader (PerkinElmer).
4.11. ELISA for binding of recombinant FcRn and recombinant VP25
Recombinant VP25 (r‐VP25) (diluted to 50 μg/ml) was immobilized on a 96‐well plate (Thermo Fisher Scientific). After blocking with 1% bovine serum albumin/PBS (Sigma‐Aldrich), recombinant human FcRn (r‐hFcRn) (Sino Biological) was incubated on the plate at 37°C for 1 h. After washing, the wells were treated with anti‐FcRn mouse monoclonal IgG (1:1000 dilution, ab228975, Abcam) as the primary antibody, followed by HRP‐conjugated anti‐mouse IgG antibody (1:5000 dilution, Cell Signaling) as the secondary antibody. The KPL SureBlue Reserve TMB Microwell Peroxidase Substrate (Sera Care) was used as a substrate, and the absorbance at 450 nm was detected using an Ensight multimode microplate reader (PerkinElmer). Each type of immobilized VP25 was directly detected by HRP‐conjugated anti‐His antibody (1:5000 dilution, MBL D291‐7).
4.12. Exogenous expression of FCGRT and B2M
Complementary DNA of FCGRT or B2M was obtained from Caco2 cells and cloned into the lentivirus vector‐encoded plasmids pLVSIN‐puro or pLVSIN‐neo, respectively. These constructs were then transduced into various cell lines. The cDNA sequence for each gene can be found in the Supplementary Material. Transduced cells were selected using 2 μg/ml puromycin or 400 μg/ml G‐418 (Nacalai Tesque). CRISPR‐resistant mutations were introduced for FCGRT (5′‐ACGGCGAAATTTGCGCTCAA‐3′) and B2M (5′‐ACTCACGTTGTATGGCTTCC‐3′) by modifying the gRNA‐targeted FCGRT sequences: (5′‐ACCGCCAAGTTCGCGCTGAA‐3′) and B2M sequence: (5′‐ACTCACGCTGGATAGCCTCC‐3′). Mutants were generated using either pLVSIN‐puro (FCGRT) or pLVSIN‐neo (B2M) and a Prime Star Max (Takara) mutagenesis kit. The selection marker of the resistant FCGRT vector was replaced with hygromycin.
4.13. The PrimeFlow RNA assay
The PrimeFlow RNA assay (Thermo Fisher Scientific) was performed according to the manufacturer's instructions. Caco2/Cas9, FB‐KO, HCT15, HCT15/FCGRT/B2M, HeLa or HeLa/FCGRT cells were infected with HAstV4 (MOI 10) and harvested after 2 days using enzyme‐free, PBS‐based cell dissociation buffer (Thermo Fisher Scientific). HAstV4‐specific probes were designed for the whole genomic region based on the information provided on the company's website (https://www.thermofisher.com/jp/ja/home/life‐science/cell‐analysis/flow‐cytometry/flow‐cytometry‐assays‐reagents/rna‐detection‐flow‐cytometry.html) (Thermo Fisher Scientific). After hybridizing the probe for the PrimeFlow assay, the cells were treated with Block Ace solution (Waken B Tech) for blocking, followed by anti‐FcRn/FCGRT antibody (Novus Biologicals) as the primary antibody and Alexa Fluor 647 donkey anti‐rabbit IgG (Thermo Fisher Scientific) as the secondary antibody. The cells were then analyzed using a FACSMelody cell sorter (BD Biosciences) and FlowJo software (BD Biosciences).
4.14. Flow cytometric analysis
The expression of FCGRT was analyzed using FACS. Cells were detached from the plate with a cell dissociation buffer (Thermo Fisher Scientific). The cells were fixed with 4% paraformaldehyde and blocked with Block Ace solution (Waken B Tech). Subsequently, 0.05% Triton‐X solution was used for permeabilization. The anti‐FcRn/FCGRT antibody (Novus Biologicals) was then incubated with the cells for 30 min on ice. Following incubation, the cells were washed and incubated with Alexa Fluor 647 donkey anti‐rabbit IgG (Thermo Fisher Scientific) for an additional 30 min on ice. The antibodies were diluted in Block Ace solution (Waken B Tech), and the washing steps were carried out in ice‐cold PBS without magnesium or calcium salts (PBS (−)). Finally, the cells were analyzed using a FACSMelody cell sorter (BD Biosciences) and FlowJo software (BD Biosciences).
4.15. Viral RNA transfection
The RNA was extracted from HAstV4 that had been cultured with Caco2 cells under trypsin‐containing conditions using a collagen‐I‐coated T75 flask (Iwaki). The virus in the culture supernatant was collected by ultracentrifugation (150,000×g, 2 h, 4°C) using a SW55Ti rotor (Beckman Coulter), and the genomic RNA was purified using a High Pure Viral RNA Kit (Roche) according to the manufacturer's protocol. The 293T or FCGRT‐KO 293T cells were plated on a 12‐well plate 1 day before transfection, and the culture medium was replaced with DMEM without FBS just before transfection. The cells were transfected with 100 ng of purified RNA with Polyethylenimine “Max” (Polysciences) as a transfection reagent and cultured for 48 h. The culture supernatants were concentrated by ultracentrifugation and serially diluted to determine TCID50 using Caco2/Cas9 cells.
4.16. Immunofluorescence image
The cells were infected with HAstV4 (MOI 10) and incubated in the absence of trypsin for 24 h. Then, cells were fixed with 4% paraformaldehyde and treated with Block Ace solution (Waken B Tech) for blocking. Rabbit anti‐FCGRT IgG (1000× dilution, ab228975, Abcam) was then added. After washing, the cells were permeabilized with 0.05% Triton‐X solution and treated with Block Ace solution for blocking again. After blocking, mouse anti‐HAstV capsid IgG (1:100 dilution at 37°C for 1 h, 8E7, Santa Cruz Biotechnology) was added. Subsequently, Alexa Fluor 568 conjugated anti‐rabbit IgG (1:1000 dilution, Thermo Fisher Scientific) and Alexa Fluor 488 conjugated anti‐mouse IgG (1:1000 dilution, Thermo Fisher Scientific) were added as secondary antibodies, and treated with Hoechst 33342 stain (Thermo Fisher Scientific). Images were captured using a BZ‐X 800 fluorescence microscope (Keyence).
4.17. In silico docking model of the spike protein and FcRn
The docking model of the spike protein and the FcRn extracellular domain was constructed using the Dock application in the Molecular Operating Environment (MOE). The docking simulation utilized the crystal structure of the spike protein of HAstV1 at a resolution of 0.95 Å (PDB ID code 5EWO) and the crystal structure of the human FcRn extracellular domain at a resolution of 2.90 Å (PDB ID code 6FGB).
4.18. Statistical analysis
All statistical analyses were conducted using GraphPad Prism version 8.0 (GraphPad Software). The significance difference test between the two groups was determined by the Mann–Whitney test. The significance difference test between multiple groups was determined by Dunn's multiple comparisons test. p values <0.05 were considered statistically significant.
AUTHOR CONTRIBUTIONS
Kei Haga: Conceptualization; data curation; funding acquisition; writing – original draft. Takashi Tokui: Data curation; writing – original draft. Kana Miyamoto: Data curation. Reiko Takai‐Todaka: Data curation; writing – original draft. Shiori Kudo: Data curation. Azusa Ishikawa: Data curation. Ryoka Ishiyama: Data curation. Akiko Kato: Data curation. Masaru Yokoyama: Data curation. Kazuhiko Katayama: Conceptualization; funding acquisition; project administration; supervision. Akira Nakanishi: Conceptualization; data curation; funding acquisition; writing – original draft.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Supporting information
DATA S1. Supporting Information.
ACKNOWLEDGMENTS
Prof. Toshiro Sato, Keio University School of Medicine, kindly provided the human intestinal organoid (Ileum‐1). The Electron micrograph was captured by Prof. Kazuyoshi Murata, National Institute for Physiological Sciences. This work is supported in part by funding from the Japan Agency for Medical Research and Development (AMED) (Akira Nakanishi and Kazuhiko Katayama) and the JSPS KAKENHI Grant‐in‐Aid for Scientific Research (C) (Kei Haga). We would like to thank Editage (www.editage.com) for English language editing.
Haga, K. , Tokui, T. , Miyamoto, K. , Takai‐Todaka, R. , Kudo, S. , Ishikawa, A. , Ishiyama, R. , Kato, A. , Yokoyama, M. , Katayama, K. , & Nakanishi, A. (2024). Neonatal Fc receptor is a functional receptor for classical human astrovirus. Genes to Cells, 29(11), 983–1001. 10.1111/gtc.13160
Kei Haga, Takashi Tokui, and Kana Miyamoto have contributed equally to this study.
Communicated by: Atsushi Miyawaki.
Contributor Information
Kazuhiko Katayama, Email: katayama@lisci.kitasato-u.ac.jp.
Akira Nakanishi, Email: nakanishi@waka.kindai.ac.jp.
REFERENCES
- Aguilar‐Hernández, N. , Meyer, L. , López, S. , DuBois, R. M. , & Arias, C. F. (2020). Protein disulfide isomerase A4 is involved in genome uncoating during human astrovirus cell entry. Viruses, 13(1), 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera‐Flores, C. , Lopez, T. , Zamudio, F. , Sandoval‐Jaime, C. , Perez, E. I. , Lopez, S. , DuBois, R. , & Arias, C. F. (2022). The capsid precursor protein of astrovirus VA1 is proteolytically processed intracellularly. Journal of Virology, 96, e0066522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez‐Navarro, C. , Quevedo, I. R. , Lopez, S. , Arias, C. F. , & Isa, P. (2022). The association of human astrovirus with extracellular vesicles facilitates cell infection and protects the virus from neutralizing antibodies. Journal of Virology, 96, e0084822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, D. M. , & Upadhyayula, U. (1997). Characterization of human serotype 1 astrovirus‐neutralizing epitopes. Journal of Virology, 71, 8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund, L. , Björling, E. , Oksvold, P. , Fagerberg, L. , Asplund, A. , al‐Khalili Szigyarto, C. , Persson, A. , Ottosson, J. , Wernérus, H. , Nilsson, P. , Lundberg, E. , Sivertsson, Å. , Navani, S. , Wester, K. , Kampf, C. , Hober, S. , Pontén, F. , & Uhlén, M. (2008). A genecentric human protein atlas for expression profiles based on antibodies. Molecular & Cellular Proteomics, 7, 2019–2027. [DOI] [PubMed] [Google Scholar]
- Bertram, K. M. , Botting, R. A. , Baharlou, H. , Rhodes, J. W. , Rana, H. , Graham, J. D. , Patrick, E. , Fletcher, J. , Plasto, T. M. , Truong, N. R. , Royle, C. , Doyle, C. M. , Tong, O. , Nasr, N. , Barnouti, L. , Kohout, M. P. , Brooks, A. J. , Wines, M. P. , Haertsch, P. , … Harman, A. N. (2019). Identification of HIV transmitting CD11c+ human epidermal dendritic cells. Nature Communications, 10, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanoff, W. A. , Campos, J. , Perez, E. I. , Yin, L. , Alexander, D. L. , & DuBois, R. M. (2017). Structure of a human Astrovirus capsid‐antibody complex and mechanistic insights into virus neutralization. Journal of Virology, 91(2), e01859–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker, J. P. , Blacklow, N. R. , & Herrmann, J. E. (2000). Human astrovirus isolation and propagation in multiple cell lines. Archives of Virology, 145, 1847–1856. [DOI] [PubMed] [Google Scholar]
- Brown, J. R. , Morfopoulou, S. , Hubb, J. , Emmett, W. A. , Ip, W. , Shah, D. , Brooks, T. , Paine, S. M. L. , Anderson, G. , Virasami, A. , Tong, C. Y. W. , Clark, D. A. , Plagnol, V. , Jacques, T. S. , Qasim, W. , Hubank, M. , & Breuer, J. (2015). Astrovirus VA1/HMO‐C: An increasingly recognized neurotropic pathogen in immunocompromised patients. Clinical Infectious Diseases, 60, 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapellier, B. , Tange, S. , Tasaki, H. , Yoshida, K. , Zhou, Y. , Sakon, N. , Katayama, K. , & Nakanishi, A. (2015). Examination of a plasmid‐based reverse genetics system for human astrovirus. Microbiology and Immunology, 59, 586–596. [DOI] [PubMed] [Google Scholar]
- Cordey, S. , Hartley, M. A. , Keitel, K. , Laubscher, F. , Brito, F. , Junier, T. , Kagoro, F. , Samaka, J. , Masimba, J. , Said, Z. , Temba, H. , Mlaganile, T. , Docquier, M. , Fellay, J. , Kaiser, L. , & D'Acremont, V. (2018). Detection of novel astroviruses MLB1 and MLB2 in the sera of febrile Tanzanian children. Emerging Microbes & Infections, 7, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordey, S. , Vu, D. L. , Schibler, M. , L'Huillier, A. G. , Brito, F. , Docquier, M. , Posfay‐Barbe, K. M. , Petty, T. J. , Turin, L. , Zdobnov, E. M. , & Kaiser, L. (2016). Astrovirus MLB2, a new gastroenteric virus associated with meningitis and disseminated infection. Emerging Infectious Diseases, 22, 846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Benedictis, P. , Schultz‐Cherry, S. , Burnham, A. , & Cattoli, G. (2011). Astrovirus infections in humans and animals: Molecular biology, genetic diversity, and interspecies transmissions. Infection, Genetics and Evolution, 11, 1529–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, J. , Dong, L. , Méndez, E. , & Tao, Y. (2011). Crystal structure of the human astrovirus capsid spike. Proceedings of the National Academy of Sciences of the United States of America, 108, 12681–12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden, K. A. , Tihova, M. , Nowotny, N. , Matsui, S. M. , Mendez, E. , & Yeager, M. (2012). Immature and mature human astrovirus: Structure, conformational changes, and similarities to hepatitis E virus. Journal of Molecular Biology, 422, 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euller‐Nicolas, G. , Le Mennec, C. , Schaeffer, J. , Zeng, X. L. , Ettayebi, K. , Atmar, R. L. , Le Guyader, F. S. , Estes, M. K. , & Desdouits, M. (2023). Human sapovirus replication in human intestinal enteroids. Journal of Virology, 97, e0038323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frémond, M. L. , Pérot, P. , Muth, E. , Cros, G. , Dumarest, M. , Mahlaoui, N. , Seilhean, D. , Desguerre, I. , Hébert, C. , Corre‐Catelin, N. , Neven, B. , Lecuit, M. , Blanche, S. , Picard, C. , & Eloit, M. (2015). Next‐generation sequencing for diagnosis and tailored therapy: A case report of astrovirus‐associated progressive encephalitis. J Pediatric Infect Dis Soc, 4, e53–e57. [DOI] [PubMed] [Google Scholar]
- Fujii, M. , Matano, M. , Nanki, K. , & Sato, T. (2015). Efficient genetic engineering of human intestinal organoids using electroporation. Nature Protocols, 10, 1474–1485. [DOI] [PubMed] [Google Scholar]
- Geigenmüller, U. , Ginzton, N. H. , & Matsui, S. M. (1997). Construction of a genome‐length cDNA clone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. Journal of Virology, 71, 1713–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guix, S. , Bosch, A. , Ribes, E. , Dora Martínez, L. , & Pintó, R. M. (2004). Apoptosis in astrovirus‐infected CaCo‐2 cells. Virology, 319, 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S. , Gach, J. S. , Becerra, J. C. , Phan, T. B. , Pudney, J. , Moldoveanu, Z. , Joseph, S. B. , Landucci, G. , Supnet, M. J. , Ping, L. H. , Corti, D. , Moldt, B. , Hel, Z. , Lanzavecchia, A. , Ruprecht, R. M. , Burton, D. R. , Mestecky, J. , Anderson, D. J. , & Forthal, D. N. (2013). The neonatal fc receptor (FcRn) enhances human immunodeficiency virus type 1 (HIV‐1) transcytosis across epithelial cells. PLoS Pathogens, 9, e1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya, M. , Matsui, M. , Sanogo, M. , Oseto, M. , Morooka, K. , & Ushijima, H. (1999). Genetic variation in the capsid region of human astrovirus serotype 4 isolated in Japan. Microbiology and Immunology, 43, 1067–1070. [DOI] [PubMed] [Google Scholar]
- Haga, K. , Ettayebi, K. , Tenge, V. R. , Karandikar, U. C. , Lewis, M. A. , Lin, S. C. , Neill, F. H. , Ayyar, B. V. , Zeng, X. L. , Larson, G. , Ramani, S. , Atmar, R. L. , & Estes, M. K. (2020). Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor‐dependent human norovirus infection. mBio, 11(2), e00251–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga, K. , Fujimoto, A. , Takai‐Todaka, R. , Miki, M. , Doan, Y. H. , Murakami, K. , Yokoyama, M. , Murata, K. , Nakanishi, A. , & Katayama, K. (2016). Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proceedings of the National Academy of Sciences of the United States of America, 113, E6248–E6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargest, V. , Davis, A. E. , Tan, S. , Cortez, V. , & Schultz‐Cherry, S. (2021). Human astroviruses: A tale of two strains. Viruses, 13(3), 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, W. , Ladinsky, M. S. , Huey‐Tubman, K. E. , Jensen, G. J. , McIntosh, J. R. , & Björkman, P. J. (2008). FcRn‐mediated antibody transport across epithelial cells revealed by electron tomography. Nature, 455, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz, L. R. , Wylie, K. M. , Sodergren, E. , Jiang, Y. , Franz, C. J. , Weinstock, G. M. , Storch, G. A. , & Wang, D. (2011). Astrovirus MLB2 viremia in febrile child. Emerging Infectious Diseases, 17, 2050–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby, P. J. , Cooper, P. R. , Kliwinski, C. , Ragwan, E. , Mabus, J. R. , Harman, B. , Thompson, S. , Kauffman, A. L. , Yan, Z. , Tam, S. H. , Dorai, H. , Powers, G. D. , & Giles‐Komar, J. (2014). Human and non‐human primate intestinal FcRn expression and immunoglobulin G transcytosis. Pharmaceutical Research, 31, 908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski, A. B. , Klein, R. S. , & Wang, D. (2019). Differential in vitro infection of neural cells by Astroviruses. mBio, 10(4), e01455–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolawole, A. O. , Mirabelli, C. , Hill, D. R. , Svoboda, S. A. , Janowski, A. B. , Passalacqua, K. D. , Rodriguez, B. N. , Dame, M. K. , Freiden, P. , Berger, R. P. , Vu, D. L. , Hosmillo, M. , O'Riordan, M. X. D. , Schultz‐Cherry, S. , Guix, S. , Spence, J. R. , Wang, D. , & Wobus, C. E. (2019). Astrovirus replication in human intestinal enteroids reveals multi‐cellular tropism and an intricate host innate immune landscape. PLoS Pathogens, 15, e1008057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukou, G. , Niendorf, S. , Hornei, B. , Schlump, J.‐U. , Jenke, A. C. , & Jacobsen, S. (2019). Human astrovirus infection associated with encephalitis in an immunocompetent child: A case report. Journal of Medical Case Reports, 13, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Król, L. , Turkiewicz, D. , Nordborg, K. , Englund, E. , Stenberg, L. , Karlsson Lindsjö, O. , Lind Karlberg, M. , & Pronk, C. J. (2021). Astrovirus VA1/HMO encephalitis after allogeneic hematopoietic cell transplantation: Significant role of immune competence in virus control. Pediatric Blood & Cancer, 68, e29286. [DOI] [PubMed] [Google Scholar]
- Lau, P. , Cordey, S. , Brito, F. , Tirefort, D. , Petty, T. J. , Turin, L. , Guichebaron, A. , Docquier, M. , Zdobnov, E. M. , Waldvogel‐Abramowski, S. , Lecompte, T. , Kaiser, L. , & Preynat‐Seauve, O. (2017). Metagenomics analysis of red blood cell and fresh‐frozen plasma units. Transfusion, 57, 1787–1800. [DOI] [PubMed] [Google Scholar]
- Lulla, V. , & Firth, A. E. (2020). A hidden gene in astroviruses encodes a viroporin. Nature Communications, 11, 4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum, S. H. , Turner, A. , Guiver, M. , Bonney, D. , Martland, T. , Davies, E. , Newbould, M. , Brown, J. , Morfopoulou, S. , Breuer, J. , & Wynn, R. (2016). An emerging opportunistic infection: Fatal astrovirus (VA1/HMO‐C) encephalitis in a pediatric stem cell transplant recipient. Transplant Infectious Disease, 18, 960–964. [DOI] [PubMed] [Google Scholar]
- McDonald, E. M. , Duggal, N. K. , Ritter, J. M. , & Brault, A. C. (2018). Infection of epididymal epithelial cells and leukocytes drives seminal shedding of Zika virus in a mouse model. PLoS Neglected Tropical Diseases, 12, e0006691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez, E. , Muñoz‐Yañez, C. , Sánchez‐San Martín, C. , Aguirre‐Crespo, G. , Baños‐Lara Mdel, R. , Gutierrez, M. , Espinosa, R. , Acevedo, Y. , Arias, C. F. , & López, S. (2014). Characterization of human astrovirus cell entry. Journal of Virology, 88, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez, E. , Salas‐Ocampo, E. , & Arias, C. F. (2004). Caspases mediate processing of the capsid precursor and cell release of human astroviruses. Journal of Virology, 78, 8601–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosky, S. , Wells, A. I. , Lemon, K. , Evans, A. S. , Schamus, S. , Bakkenist, C. J. , & Coyne, C. B. (2019). The neonatal Fc receptor is a pan‐echovirus receptor. Proceedings of the National Academy of Sciences of the United States of America, 116, 3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache, S. N. , Peggs, K. S. , Mattes, F. M. , Phadke, R. , Garson, J. A. , Grant, P. , Samayoa, E. , Federman, S. , Miller, S. , Lunn, M. P. , Gant, V. , & Chiu, C. Y. (2015). Diagnosis of Neuroinvasive Astrovirus infection in an immunocompromised adult with encephalitis by unbiased next‐generation sequencing. Clinical Infectious Diseases, 60, 919–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan, V. , Damschroder, M. M. , Cook, K. E. , Li, Q. , Gao, C. , Wu, H. , & Dall'Acqua, W. F. (2014). Structural insights into neonatal Fc receptor‐based recycling mechanisms. The Journal of Biological Chemistry, 289, 7812–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyzik, M. , Sand, K. M. K. , Hubbard, J. J. , Andersen, J. T. , Sandlie, I. , & Blumberg, R. S. (2019). The neonatal Fc receptor (FcRn): A misnomer? Frontiers in Immunology, 10, 1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan, P. L. , Wagner, T. A. , Briese, T. , Torgerson, T. R. , Hornig, M. , Tashmukhamedova, A. , Firth, C. , Palacios, G. , Baisre‐de‐Leon, A. , Paddock, C. D. , Hutchison, S. K. , Egholm, M. , Zaki, S. R. , Goldman, J. E. , Ochs, H. D. , & Lipkin, W. I. (2010). Astrovirus encephalitis in boy with X‐linked agammaglobulinemia. Emerging Infectious Diseases, 16, 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, S. , Lungu, C. , Crespo, R. , Steijaert, T. H. , Gorska, A. , Palstra, R. J. , Prins, H. A. B. , van Ijcken, W. , Mueller, Y. M. , van Kampen, J. J. A. , Verbon, A. , Katsikis, P. D. , Boucher, C. A. B. , Rokx, C. , Gruters, R. A. , & Mahmoudi, T. (2021). Selective cell death in HIV‐1‐infected cells by DDX3 inhibitors leads to depletion of the inducible reservoir. Nature Communications, 12, 2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald, R. , & Kraehenbuhl, J. P. (1984). Receptor‐mediated transport of IgG. The Journal of Cell Biology, 99, 159s–164s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana, N. E. , Shalem, O. , & Zhang, F. (2014). Improved vectors and genome‐wide libraries for CRISPR screening. Nature Methods, 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M. , Kuroda, M. , Kasai, M. , Matsui, H. , Fukuyama, T. , Katano, H. , & Tanaka‐Taya, K. (2016). Acute encephalopathy in an immunocompromised boy with astrovirus‐MLB1 infection detected by next generation sequencing. Journal of Clinical Virology, 78, 66–70. [DOI] [PubMed] [Google Scholar]
- Simister, N. E. , & Rees, A. R. (1985). Isolation and characterization of an fc receptor from neonatal rat small intestine. European Journal of Immunology, 15, 733–738. [DOI] [PubMed] [Google Scholar]
- Sinigaglia, L. , Gracias, S. , Décembre, E. , Fritz, M. , Bruni, D. , Smith, N. , Herbeuval, J.‐P. , Martin, A. , Dreux, M. , Tangy, F. , & Jouvenet, N. (2018). Immature particles and capsid‐free viral RNA produced by yellow fever virus‐infected cells stimulate plasmacytoid dendritic cells to secrete interferons. Scientific Reports, 8, 10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockolosky, J. T. , & Szoka, F. C. (2015). The neonatal fc receptor, FcRn, as a target for drug delivery and therapy. Advanced Drug Delivery Reviews, 91, 109–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, S. , Kobayashi, E. , Fujii, M. , Ohta, Y. , Arai, K. , Matano, M. , Ishikawa, K. , Miyamoto, K. , Toshimitsu, K. , Takahashi, S. , Nanki, K. , Hakamata, Y. , Kanai, T. , & Sato, T. (2021). An organoid‐based organ‐repurposing approach to treat short bowel syndrome. Nature, 592, 99–104. [DOI] [PubMed] [Google Scholar]
- Takagi, H. , Oka, T. , Shimoike, T. , Saito, H. , Kobayashi, T. , Takahashi, T. , Tatsumi, C. , Kataoka, M. , Wang, Q. , Saif, L. J. , & Noda, M. (2020). Human sapovirus propagation in human cell lines supplemented with bile acids. Proceedings of the National Academy of Sciences, 117, 32078–32085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Doef, H. P. J. , Bathoorn, E. , van der Linden, M. P. M. , Wolfs, T. F. W. , Minderhoud, A. L. C. , Bierings, M. B. , Wensing, A. M. J. , & Lindemans, C. A. (2016). Astrovirus outbreak at a pediatric hematology and hematopoietic stem cell transplant unit despite strict hygiene rules. Bone Marrow Transplantation, 51, 747–750. [DOI] [PubMed] [Google Scholar]
- Vu, D. L. , Bosch, A. , Pintó, R. M. , & Guix, S. (2017). Epidemiology of classic and novel human astrovirus: Gastroenteritis and beyond. Viruses, 9(2), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks, M. M. , Carter, M. J. , Laidler, F. R. , & Madeley, C. R. (1990). Growth and characterisation of human faecal astrovirus in a continuous cell line. Archives of Virology, 113, 73–81. [DOI] [PubMed] [Google Scholar]
- Wunderli, W. , Meerbach, A. , Guengoer, T. , Berger, C. , Greiner, O. , Caduff, R. , Trkola, A. , Bossart, W. , Gerlach, D. , Schibler, M. , Cordey, S. , McKee, T. A. , Van Belle, S. , Kaiser, L. , & Tapparel, C. (2011). Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One, 6, e27483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie, K. M. , Mihindukulasuriya, K. A. , Sodergren, E. , Weinstock, G. M. , & Storch, G. A. (2012). Sequence analysis of the human virome in febrile and afebrile children. PLoS One, 7, e27735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanella, M. C. , Cordey, S. , Laubscher, F. , Docquier, M. , Vieille, G. , Van Delden, C. , Braunersreuther, V. , Ta, M. K. , Lobrinus, J. A. , Masouridi‐Levrat, S. , Chalandon, Y. , Kaiser, L. , & Vu, D. L. (2021). Unmasking viral sequences by metagenomic next‐generation sequencing in adult human blood samples during steroid‐refractory/dependent graft‐versus‐host disease. Microbiome, 9, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , Zhang, G. , Liu, S. , Chen, X. , Peng, R. , Dai, L. , Qu, X. , Li, S. , Song, H. , Gao, Z. , Yuan, P. , Liu, Z. , Li, C. , Shang, Z. , Li, Y. , Zhang, M. , Qi, J. , Wang, H. , du, N. , … Gao, G. F. (2019). Human neonatal fc receptor is the cellular Uncoating receptor for enterovirus B. Cell, 177, 1553–1565.e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DATA S1. Supporting Information.


