Abstract
For Enterobacteriaceae such as Salmonella spp. and Escherichia coli, no unified interpretive resistance criteria exist for streptomycin, an epidemiologically important antibiotic. As part of the National Antimicrobial Resistance Monitoring System, we had previously used a minimum inhibitory concentration of ≥64 μg mL−1 as an epidemiological cutoff value (ECV) to define non-wild-type isolates. To identify whether this ECV correlated with genetic determinants of resistance, we performed whole-genome sequencing of 463 Salmonella and E. coli isolates to identify streptomycin resistance genotypes. From this analysis, we found that using a streptomycin resistance breakpoint of ≥64 μg mL−1 classified over 20% of strains possessing aadA or strA/strB resistance genes as wild-type. Therefore, to improve the concordance between genotypic and phenotypic data, we propose reducing the phenotypic cutoff values to ≥32 μg mL−1 for both Salmonella and E. coli, to be used widely as ECVs to categorize non-wild-type isolates.
Keywords: Salmonella, E. coli, whole-genome sequencing, streptomycin, resistance, genotyping
One sentence summary:
We used whole-genome sequencing to identify streptomycin resistance genotypes of Salmonella and Escherichia coli, combining this information with susceptibility data to establish new streptomycin epidemiological cutoff values for these bacteria.
INTRODUCTION
Streptomycin was one of the first natural antibiotics discovered, and was introduced into clinical medicine in 1943. As an aminoglycoside, streptomycin works by binding to the bacterial 30S ribosomal subunit to disrupt protein synthesis. This effectively kills many types of bacteria, but human clinical use has been limited due to its toxicity (Bottger et al. 2001). However, streptomycin has been widely used to treat bacterial infections of plants and animals (Sundin and Bender 1996).
Streptomycin resistance has become prevalent among Enterobacteriaceae due to the expression of aminoglycoside modifying enzymes (Mingeot-Leclercq, Glupczynski and Tulkens 1999). In Escherichia coli and Salmonella, streptomycin resistance is often mediated by aadA genes, which are usually present on integrons and result in streptomycin adenylation (Hollingshead and Vapnek 1985). Other common genes include strA (aph(3′)-Ib) and strB (aph(6′)-Id), which are typically found together and produce phosphotransferases that modify streptomycin at different positions (Scholz et al. 1989). Streptomycin resistance genes are frequently associated with mobile genetic elements that disseminate multiple antimicrobial resistance genes in Enterobacteriaceae. As a result, streptomycin resistance has been used as an important epidemiological marker to indicate the likelihood of multidrug-resistance in pathogens (Scholz et al. 1989). For instance, Salmonella Typhimurium DT104, a significant human and animal pathogen, has several co-expressed resistance genes, yielding the resistance phenotype of ACSSuT (resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole and tetracycline) (Hsu et al. 2013). Tracking and identifying the origins and sources of such important multidrug-resistant pathogens is important for foodborne outbreak investigations, and can help reduce the burden of foodborne illness.
Although there is no clinical streptomycin resistance breakpoint for Enterobacteriaceae, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) utilizes an epidemiological cutoff value (ECV) of ≤16 μg mL−1 to identify wild-type strains of both Salmonella and E. coli (www.eucast.org). These ECVs are used to define the upper limit of the wild-type populations of each bacterium for streptomycin, and are intended to refer to organisms that are likely to be missing resistance mechanisms. However, EUCAST sets ECVs based solely on the distribution of minimum inhibitory concentrations (MICs) among populations of bacteria, not based on the presence of known resistance mechanisms in these strains.
The National Antimicrobial Resistance Monitoring System (NARMS) and others use ≥64 μg mL−1 to categorize streptomycin-resistant strains of Enterobacteriaceae (Zhao et al. 2006; Deckert et al. 2010). Several groups have attempted to establish streptomycin ECVs for E. coli and/or Salmonella based on genotypic data, but have had vastly different conclusions (Sunde and Norstrom 2005; Doran et al. 2006; Garcia-Migura et al. 2012). One potential source of these ambiguous results is that they relied on PCR tests to detect resistance genes, and thus may not have comprehensively evaluated their presence. Instead we used whole-genome sequencing (WGS) to identify streptomycin resistance genes to more rigorously evaluate the correlation between streptomycin resistance genotypes and phenotypes.
MATERIALS AND METHODS
WGS and analysis
WGS was performed on the Miseq platform for 337 Salmonella isolates of 36 serotypes (Table S1 Supporting Information) and 126 E. coli of 104 serotypes (Table S2 Supporting Information). Isolates were grown on blood agar plates at 35°C, and genomic DNA was extracted with a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) as per the manufacturer’s instructions. WGS was performed using the Miseq platform with v2 or v3 reagent kits (Illumina, San Diego, CA, USA). Assembly was performed de novo for each isolate with CLC Genomics Workbench version 7.5 using automated assembly parameters. All assemblies were of at least 30-fold coverage and contained fewer than 400 contigs. The strA/strB and aadA genes were identified using BlastX analysis with an in-house resistance gene database containing 44 aadA, 12 strA and 14 strB variants (Tyson et al. 2015). Salmonella serotypes were determined by traditional serology (FDA 2015), while E. coli serotypes were identified by sequence-based methods (Joensen et al. 2015).
Antimicrobial susceptibility testing
Streptomycin MICs were determined by broth microdilution using a Sensititre® system (Trek Diagnostic Systems, Cleveland, OH, USA) according to standardized protocols (CLSI 2015). Plate CMV3AGNF was used, containing streptomycin drug concentrations from 2 to 64 μg mL−1 in 2-fold increments. The ECOFFinder tool was used to calculate ECVs statistically based on MIC distributions (Turnidge, Kahlmeter and Kronvall 2006).
RESULTS AND DISCUSSION
A total of 337 Salmonella isolates of 36 serotypes from retail meats were subjected to WGS and antimicrobial susceptibility testing, and displayed a wide variety of streptomycin susceptibility phenotypes (Fig. 1A). Most isolates lacking known streptomycin resistance genes had MIC values ≤16 μg mL−1 (189/191, 99.0%), with two isolates having MICs of 32 μg mL−1 (Fig. 1A). As previously established, the presence of aadA genes was associated with decreased susceptibility to streptomycin (Hollingshead and Vapnek 1985), as a vast majority of isolates with aadA genes had MIC values of ≥32 μg mL−1 (89.5%) (Fig. 1A). Similar to previous results (Sunde and Norstrom 2005), isolates with strA/strB had elevated MICs relative to those with aadA genes, with most isolates (88.0%) having MICs of ≥64 μg mL−1. The combined expression of aadA and strA/strB further increased streptomycin MIC levels, as 93.8% of isolates with both types of genes had MIC values >64 μg mL−1 (Fig. 1A).
Figure 1.
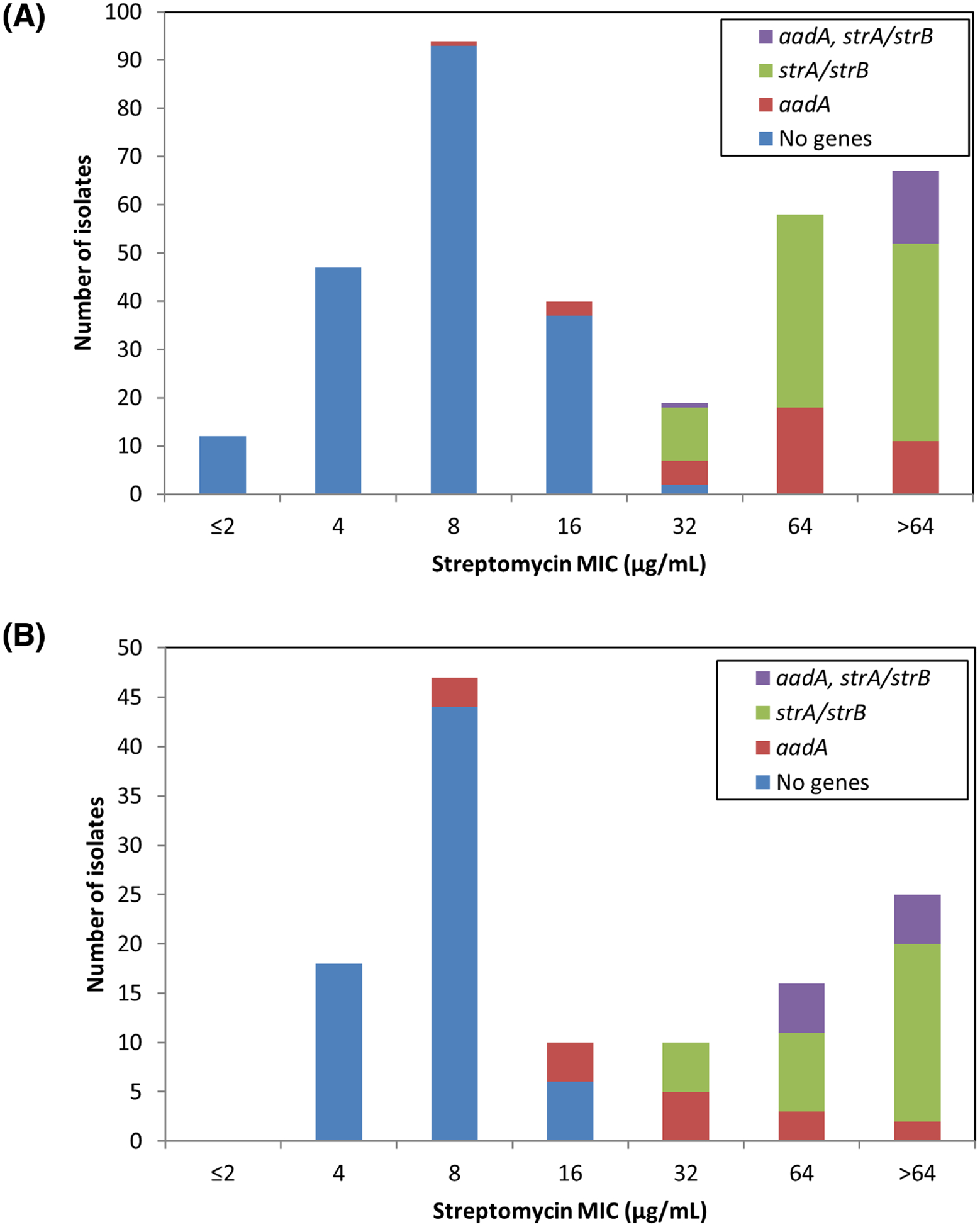
Association of streptomycin resistance genes and MIC. (A) Salmonella isolates were evaluated for susceptibility to streptomycin, and categorized based on those without known streptomycin resistance genes, with aadA genes, with strA/strB genes, or with both. Numbers in parentheses indicate the number of isolates with each genotype. (B) The streptomycin MIC distribution for E. coli is similarly depicted.
Genotypic and phenotypic data generally agreed for strains carrying strA/strB with the original NARMS ECV of ≥64 μg mL−1, although this was not the case for isolates with aadA genes. At the ≥64 μg mL−1 ECV, genotypes predicted all resistance phenotypes, since all isolates that were phenotypically resistant at this threshold possessed resistance genes. However, many isolates with resistance genes had MICs below this level, resulting in an overall correlation between genotypes and phenotypes of 93.8% (Table 1). Reducing the threshold to ≥32 μg mL−1 resulted in increasing genotype–phenotype correlations to 98.2% (Table 1), so we suggest establishing the Salmonella ECV at ≥32 μg mL−1 to denote non-wild-type strains.
Table 1.
Genotype–phenotype correlation for Salmonella at different streptomycin cutoff values.
| Phenotype:WT | Phenotype:NWT | |||||
|---|---|---|---|---|---|---|
| ECV (μg mL−1) | Genotype:R | Genotype:S | Genotype:R | Genotype:S | Correlation (%) | |
| ≥64 | 21 | 191 | 125 | 0 | 93.8 (316/337) | |
| ≥32 | 4 | 189 | 142 | 2 | 98.2 (331/337) | |
| ≥16 | 1 | 152 | 145 | 39 | 88.1 (297/337) | |
| ≥8 | 0 | 59 | 146 | 132 | 60.8 (205/337) | |
WT refers to wild-type, and NWT refers to non-wild-type.
From animal sources, 126 E. coli isolates of 104 serotypes were selected for WGS and antimicrobial susceptibility testing (Table S2 Supporting Information). Among the 68 isolates without any streptomycin resistance genes, all had MIC values from 4 to 16 μg mL−1 (Fig. 1B), similar to what was observed with Salmonella (Fig. 1A). Isolates with aadA genes had a broad range of streptomycin MIC values, from 8 to >64 μg mL−1. Although most had elevated MIC levels relative to isolates lacking resistance mechanisms, three isolates had MICs of 8 μg mL−1. These three isolates appeared to be highly related, as each was of the same serotype and possessed identical class 1 integrons containing aadA genes. A different isolate, N34557PS, had the same gene cassette yet had a streptomycin MIC of >64 μg mL−1. This suggests that additional unknown genetic factors may be present that disrupt aadA activity or otherwise influence streptomycin susceptibility in the three isolates with MICs of 8 μg mL−1. As observed with Salmonella, E. coli isolates with strA/strB genes had higher streptomycin MICs than those expressing aadA (Fig. 1B). The additional presence of aadA genes further increased MICs, as all isolates with both aadA and strA/strB genes had MICs ≥64 μg mL−1, the original resistance breakpoint. Although most isolates with resistance genes had MIC values of ≥64 μg mL−1, a decreased ECV resulted in better agreement between resistance genotypes and phenotypes (Table 2). In fact, decreasing the resistance threshold to ≥32 μg mL−1 raised the correlation from 86.5% to 94.4%. Reducing the ECV further, to ≥16 μg mL−1, resulted in a correlation of 92.9%, which is only slightly lower than that achieved with the ≥32 μg mL−1 cutoff (Table 2). This is because several isolates both with and without streptomycin resistance mechanisms had MICs of 16 μg mL−1 (Fig. 1B). We chose ≤16 μg mL−1 to designate the wild-type population since 16 μg mL−1 fits the definition of the upper limit of the isolates lacking resistance mechanisms. It is also only one dilution away from the most common MIC for isolates lacking mechanisms (8 μg mL−1), and therefore within the error range of broth microdilution for isolates with that MIC (Steward et al. 1999).
Table 2.
Genotype–phenotype correlation for E. coli at different streptomycin cutoff values.
| Phenotype:WT | Phenotype:NWT | ||||
|---|---|---|---|---|---|
| ECV (μg mL) | Genotype:R | Genotype:S | Genotype:R | Genotype:S | Correlation (%) |
| ≥64 | 17 | 68 | 41 | 0 | 86.5 (109/126) |
| ≥32 | 7 | 68 | 51 | 0 | 94.4 (119/126) |
| ≥16 | 3 | 62 | 55 | 6 | 92.9 (117/126) |
| ≥8 | 0 | 18 | 58 | 50 | 60.3 (76/126) |
WT refers to wild-type, and NWT refers to non-wild-type.
The ECVs determined by genotypic methods were compared to those from using ECOFFinder, a statistics-based method that identifies ECVs based on MIC distributions (Turnidge, Kahlmeter and Kronvall 2006). Based on this analysis, a Salmonella ECV of ≤32 μg mL−1 and E. coli ECV of ≤16 μg mL−1 would refer to wild-type strains, using the typical 97.5% wild-type value cutoff (Fig. 2). The E. coli ECV agrees with our results, whereas the Salmonella ECV from ECOFFinder is higher, suggesting that statistical methods do not always concur with conclusions based on resistance genotyping, which we argue is more valuable in determining the true wild-type population of bacteria. Our ability to use WGS-based techniques to dramatically improve the genotype–phenotype correlations underscores the importance of identifying appropriate ECVs to more accurately report emerging trends in antimicrobial resistance.
Figure 2.
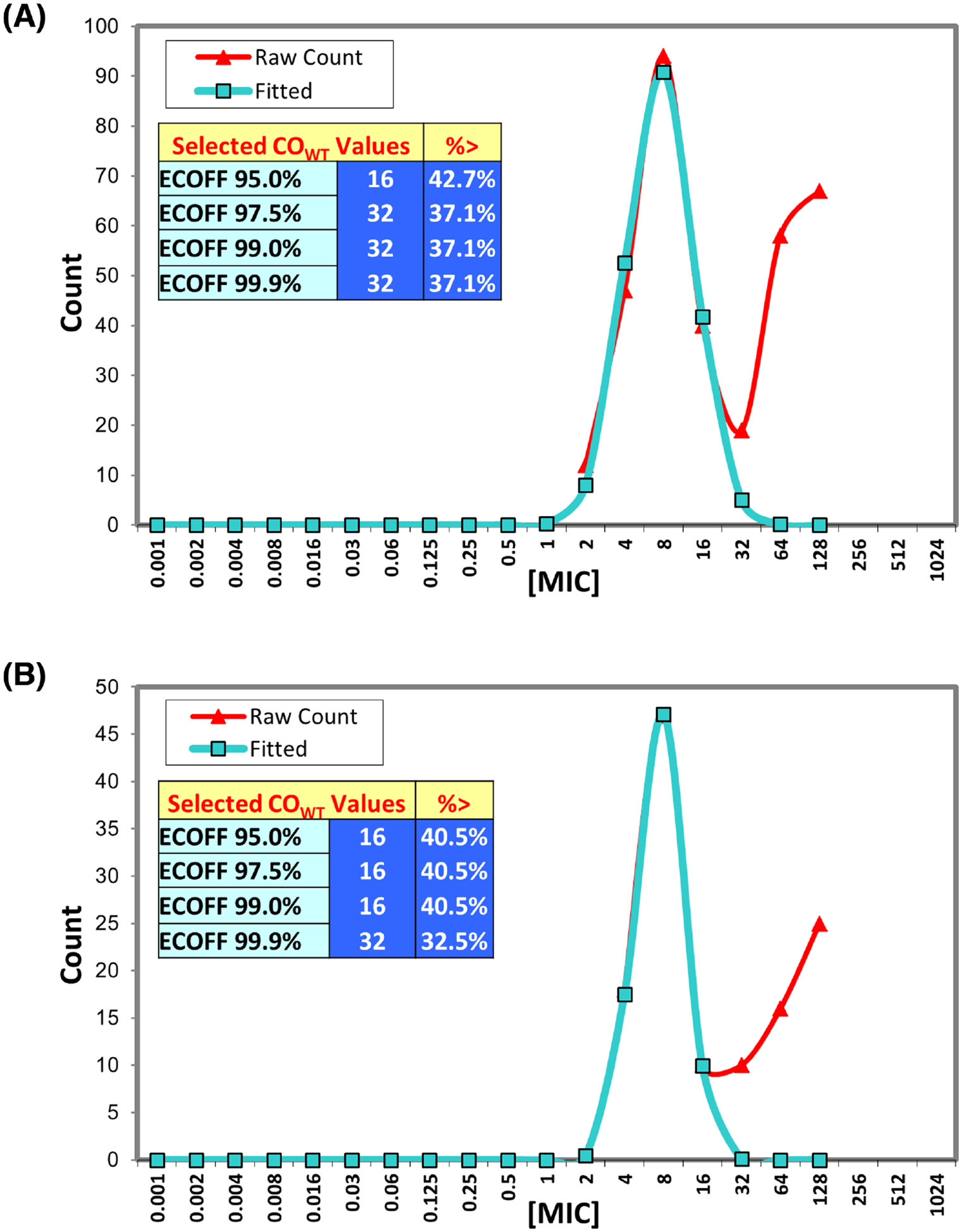
Using ECOFFinder to determine ECVs. (A) Salmonella MIC distributions were input into the ECOFFinder tool (Turnidge, Kahlmeter and Kronvall 2006) to determine potential ECVs. (B) Escherichia coli MIC distributions were similarly input into ECOFFinder.
CONCLUSIONS
In this work, we investigated the correlation between genotypic and phenotypic resistance to streptomycin for E. coli and Salmonella. To create a better concordance between genotypic and phenotypic data as well as unify streptomycin resistance reporting, we suggest that susceptibility testing results of ≥32 μg mL−1 for both Salmonella and E. coli be used to denote non-wild-type strains. These results contrast with some conclusions made by other groups, likely due to their reliance on PCR-based techniques to identify streptomycin resistance genes (Garcia-Migura et al. 2012). In contrast, our use of WGS resulted in a more unbiased ability to broadly identify resistance determinants, confirming the robustness of this technique in identifying important genes. Overall, our data demonstrate the power of WGS in predicting resistance phenotypes and should complement existing methods to expand our knowledge of antimicrobial resistance mechanisms.
Supplementary Material
Footnotes
DISCLAIMER
The views expressed in this article are those of the author(s) and do not necessarily reflect the official policy of the Department of Health and Human Services, the US Food and Drug Administration or the US Government. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Food and Drug Administration.
SUPPLEMENTARY DATA
Conflict of interest. None declared.
REFERENCES
- Bottger EC, Springer B, Prammananan T et al. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep 2001;2:318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—tenth edition. CLSI Document M07-A10. Wayne, PA, USA: Clinical and Laboratory Safety Institute, 2015. [Google Scholar]
- Deckert A, Gow S, Rosengren L et al. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) farm program: results from finisher pig surveillance. Zoonoses Public Hlth 2010;57 (Suppl 1):71–84. [DOI] [PubMed] [Google Scholar]
- Doran G, NiChulain M, DeLappe N et al. Interpreting streptomycin susceptibility test results for Salmonella enterica serovar Typhimurium. Int J Antimicrob Ag 2006;27:538–40. [DOI] [PubMed] [Google Scholar]
- FDA. National Antimicrobial Resistance Monitoring System (NARMS). 2012 Retail Meat Report. Rockville, MD: U.S. Department of Health and Human Services, Food and Drug Administration, 2015. [Google Scholar]
- Garcia-Migura L, Sunde M, Karlsmose S et al. Establishing streptomycin epidemiological cut-off values for Salmonella and Escherichia coli. Microb Drug Resist 2012;18:88–93. [DOI] [PubMed] [Google Scholar]
- Hollingshead S, Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyl-transferase. Plasmid 1985;13:17–30. [DOI] [PubMed] [Google Scholar]
- Hsu YM, Tang CY, Lin H et al. Comparative study of class 1 integron, ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline (ACSSuT) and fluoroquinolone resistance in various Salmonella serovars from humans and animals. Comp Immunol Microb 2013;36:9–16. [DOI] [PubMed] [Google Scholar]
- Joensen KG, Tetzschner AM, Iguchi A et al. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 2015;53:2410–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. Amino-glycosides: activity and resistance. Antimicrob Agents Ch 1999;43:727–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz P, Haring V, Wittmann-Liebold B et al. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 1989;75:271–88. [DOI] [PubMed] [Google Scholar]
- Steward CD, Stocker SA, Swenson JM et al. Comparison of agar dilution, disk diffusion, MicroScan, and Vitek antimicrobial susceptibility testing methods to broth microdilution for detection of fluoroquinolone-resistant isolates of the family Enterobacteriaceae. J Clin Microbiol 1999;37:544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde M, Norstrom M. The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J Antimicrob Chemoth 2005;56:87–90. [DOI] [PubMed] [Google Scholar]
- Sundin GW, Bender CL. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol Ecol 1996;5:133–43. [DOI] [PubMed] [Google Scholar]
- Turnidge J, Kahlmeter G, Kronvall G. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infec 2006;12:418–25. [DOI] [PubMed] [Google Scholar]
- Tyson GH, McDermott PF, Li C et al. WGS accurately predicts antimicrobial resistance in Escherichia coli. J Antimicrob Chemoth 2015;70:2763–9. [DOI] [PubMed] [Google Scholar]
- Zhao S, McDermott PF, Friedman S et al. Antimicrobial resistance and genetic relatedness among Salmonella from retail foods of animal origin: NARMS retail meat surveillance. Foodborne Pathog Dis 2006;3:106–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


