Abstract
Campylobacter spp. commonly cause gastrointestinal illness in humans. Poultry meats have long been considered the predominant source of these infections, but few in-depth Campylobacter source attribution studies have been completed. We analyzed more than 1,300 Campylobacter isolates recovered from a number of animal and food sources, including dairy and beef cattle, pigs, poultry, and retail poultry meat, and compared them with Campylobacter isolates recovered from human clinical samples. Each isolate was subtyped using pulsed-field gel electrophoresis (PFGE) with SmaI and queried against the Centers for Disease Control and Prevention PulseNet database to identify human isolates with indistinguishable patterns. Half (49.5%) of the PFGE patterns from poultry animal and retail meat isolates were indistinguishable from patterns of at least one human isolate. Among the isolates from beef and dairy cows, 56.6 and 65.0%, respectively, of their PFGE patterns were indistinguishable from those of human isolates. Only a small portion of the PFGE patterns of Campylobacter isolated from pigs (9.5%) were found to have PFGE patterns in common with human isolates. These data imply that cattle may be larger contributors to Campylobacter infections than previously recognized and help further our understanding of potential sources of human campylobacteriosis.
Keywords: Campylobacter, Food animals, Pulsed-field gel electrophoresis, Source attribution
Campylobacter spp. are among the most common bacterial causes of foodborne illness in the United States, causing approximately 1 million illnesses each year (21), with approximately 90% of human infections attributed to Campylobacter jejuni (12). Most infections result in self-limiting gastroenteritis; however, some cases can progress to chronic sequelae such as Guillain-Barré syndrome, irritable bowel syndrome, and reactive arthritis (9). Therefore, identifying sources of Campylobacter is key to preventing its spread and ensuring a safe food supply.
Source attribution studies are critical to understanding the predominant sources of human illnesses and to determining where to direct resources to prevent the consumption of contaminated food. To perform source attribution studies, researchers need to discriminate and subtype bacterial strains to allow the direct comparison of isolates from human cases and source materials. Multilocus sequence typing has been used previously to differentiate Campylobacter isolates based on sequence polymorphisms in discrete genes (6). Although multilocus sequence typing data generally correlate well with data acquired using pulsed-field gel electrophoresis (PFGE) (29), PFGE has been found to have superior discriminatory power (20).
Food animals are significant reservoirs of Campylobacter. These bacteria have been found associated with pig, cattle, and poultry sources (3); however, most attribution studies have identified chickens as the major source of infection. In the United States, Campylobacter contaminates approximately 50% of retail chicken meat, with significantly less found in pork, ground turkey, and ground beef retail products (32). In Europe, as many as 80% of human Campylobacter infections have been thought to be associated with the consumption of chicken products (7), with some case-control studies in the United States supporting this conclusion (8). However, PFGE profile comparisons of Campylobacter isolates from retail poultry meats and humans have revealed significant differences (26), indicating the likelihood of additional Campylobacter reservoirs that contribute to human infection. In support of this, some studies have suggested that a large number of animal, food, and environmental reservoirs are important contributors to Campylobacter infection (30).
To better evaluate potential sources of human Campylobacter infections in the United States, we analyzed more than 1,300 Campylobacter isolates recovered from cecal samples of dairy and beef cattle, pigs, chickens, and turkeys, in addition to isolates recovered from retail poultry meats. Using PFGE to discriminate strains, we compared patterns from these isolates with those from human patients.
MATERIALS AND METHODS
Cecal sampling and Campylobacter isolation.
Cecal contents from pigs, cattle, chicken, and turkey animals were collected by U.S. Department of Agriculture, Food Safety and Inspection Service (USDA-FSIS) personnel from animals presented for slaughter at USDA-FSIS–inspected facilities throughout the United States in 2013 (28). Cecal samples were shipped to the Eastern USDA-FSIS laboratory, where samples were cultured for Campylobacter using published methods (15). Briefly, cecal samples suspended in buffered peptone water were enriched with Bolton enrichment broth for 48 h before samples were streaked onto Campylobacter-modified charcoal-cefoperazone-deoxycho-late agar plates for isolation. An attempt was made to obtain at least 300 isolates each from pig, dairy cattle, beef cattle, and chicken and turkey sources. All the pig and poultry isolates were analyzed, but the beef and dairy cattle isolates that were analyzed were randomly selected from a larger number of isolates. In the absence of the requisite number of poultry isolates, we added poultry retail meat isolates obtained in 2013 by the National Antimicrobial Resistance Monitoring System retail meat surveillance (28) to the analysis. Campylobacter speciation was performed using a PCR-based assay, as described in a previous article (31).
PFGE.
PFGE was performed per standardized protocols, in accordance with Centers for Disease Control and Prevention (CDC) PulseNet procedures (26). Briefly, Campylobacter genomic DNA was subjected to digestion by SmaI, electrophoresis, and GelRed 3X stain (Phenix Research, Candler, NC). The band patterns were analyzed using BioNumerics version 6.6 (Applied Maths, Austin, TX). We performed the analysis using 1.5% optimization and 1.5% band matching tolerance, with an unweighted pair group method with arithmetic mean–based cluster analysis. We compared the PFGE patterns of Campylobacter isolates to those from the CDC PulseNet database (24), which contains isolates obtained nationwide from human patients (10,320 isolates for C. jejuni and 413 for Campylobacter coli at time of analysis). Patterns were identified as indistinguishable using PFGE if they were identical according to this analysis. The same analysis criteria were also used in comparing the patterns from this study with each other. We used the program JVenn to display the number of PFGE patterns shared among sources (Fig. 1) (2).
FIGURE 1.
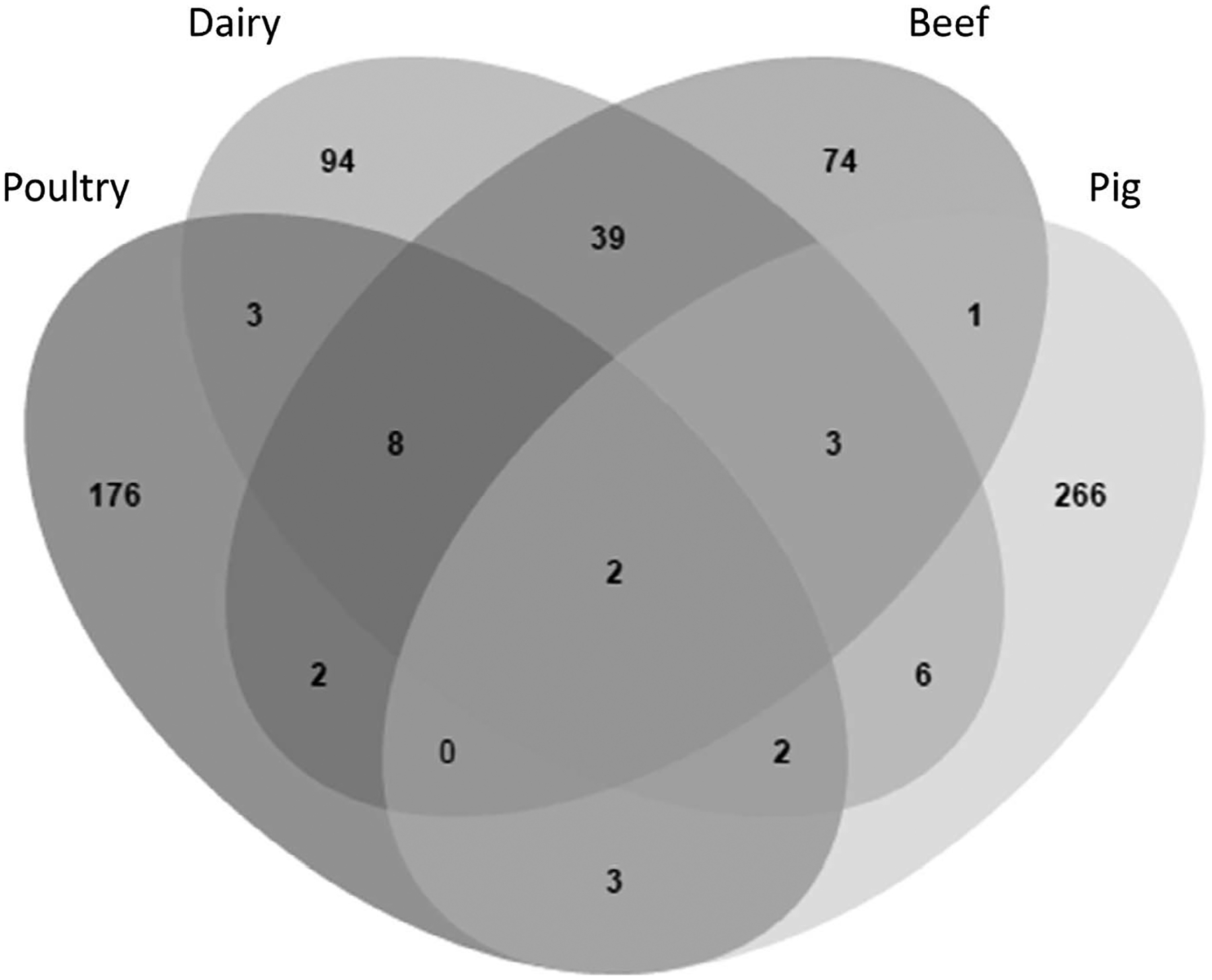
Diversity of PFGE patterns by source. The Venn diagram depicts the number of PFGE patterns that are unique and shared among sources. Poultry refers to patterns of isolates from both poultry animals and poultry retail meat.
Statistical analysis.
We analyzed the results using a chi-square test of independence (or Fisher’s exact test, where appropriate) to determine whether the differences were significant among sources in the percentage of indistinguishable patterns or isolates and in the percentage of isolates that were C. jejuni. We made additional pairwise comparisons using the Bonferroni correction to see which source accounted for more of the chi-square statistic. P values of less than 0.05 were denoted as significant.
RESULTS
To better understand the sources of the Campylobacter causing human infections, we selected 1,363 isolates from various sources for study. The goal was to compare at least 300 isolates each from the ceca of poultry, beef cattle, dairy cattle, and pigs with isolates from humans. All the poultry and pig isolates were selected, but the isolates we analyzed from beef and dairy cattle were randomly selected from different isolation dates to represent samples isolated from throughout the year because a larger number was available. The frequency of Campylobacter isolation was highest among the cecal samples collected from dairy cattle (42.6%), beef cattle (41.5%), and pigs (31.7%). In contrast, Campylobacter prevalence was lower among poultry, with only 21.8% of chickens and 9.5% of turkeys found to be positive. As a result, we obtained only 79 poultry animal isolates, so we collected an additional 230 isolates from poultry retail meats during the same time period to include in the analysis. Among the isolates selected for study, 721 (52.8%) were C. jejuni and 636 (46.7%) were C. coli. Five isolates were Campylobacter lari, and one was Campylobacter fetus. The frequency of isolation of each Campylobacter species differed greatly by source (Table 1). For instance, C. jejuni predominated among beef cattle (76.4%), dairy cattle (74.9%), and to a lesser extent, poultry animals and retail meat (62.8%). In contrast, C. coli constituted the vast majority of the isolates from pigs (97.8%), with only 1.6% of pig isolates being C. jejuni. The differences in frequency of C. jejuni isolated from the sources were significant for all sources (P < 0.05), except for beef and dairy cattle.
TABLE 1.
Campylobacter isolates with patterns indistinguishable from those from human patients based on sourcea
| Source | No. of isolates (C. jejuni/C. coli)b | % isolates indistinguishable | ||
|---|---|---|---|---|
| C. jejuni | C. coli | Overall | ||
| Beef cattle | 243/75 | 85.6 | 64.0 | 80.5 |
| Dairy cattle | 278/89 | 89.6 | 64.0 | 83.6 |
| Pigs | 6/357 | 66.7 | 12.6 | 13.7 |
| Poultry animals and retail meat | 194/115 | 74.2 | 47.0 | 64.1 |
| Total | 721/636 | 83.9 | 32.1 | 59.7 |
Patterns indistinguishable are indistinguishable from those of human isolates in the CDC PulseNet database.
Four C. lari were isolated from dairy cattle and one from pigs. One C. fetus was isolated from pigs. These six isolates were included in the overall data, with the five C. lari (but not the C. fetus) having patterns indistinguishable from those of human isolates.
To subtype strains and perform source attribution analysis, we performed PFGE on each isolate, comparing the patterns with each other as well as with those from human isolates in the CDC PulseNet database. There was considerable diversity in the PFGE patterns isolated from each source. For instance, among the 309 poultry animal and retail meat isolates, there were 196 distinct PFGE patterns (Table 2). Less diversity was observed among the isolates obtained from cattle, with a total of 129 and 157 PFGE patterns corresponding to the 318 beef and 371 dairy cattle isolates, respectively. The pig isolates had the greatest diversity of PFGE patterns, with 283 total patterns among 365 isolates.
TABLE 2.
Number of PFGE patterns from different sources indistinguishable from patterns of human isolatesa
| Source | Total no. of patterns | Overall % patterns indistinguishable | No. of common patterns indistinguishableb |
|---|---|---|---|
| Beef cattle | 129 | 56.6 | 23/23 |
| Dairy cattle | 157 | 65.0 | 34/34 |
| Pigs | 283 | 9.5 | 5/14 |
| Poultry animals and retail meat | 196 | 49.5 | 19/19 |
Patterns indistinguishable are indistinguishable from those of human isolates in the CDC PulseNet database.
Common patterns encompass three or more isolates from a particular source; this total is the denominator in each cell.
Despite the strain differences in Campylobacter from different sources, there were some PFGE patterns shared by multiple sources (Fig. 1). In total, 69 of 679 total patterns (10.2%) were shared among multiple sources, with just two patterns shared among all four sources (Fig. 1). Many patterns were shared between the isolates from beef and dairy cattle, with a total of 52 PFGE patterns in common. This contrasted with the poultry and pig isolates, which had relatively few patterns shared with either each other or with the cattle isolates (Fig. 1). Many isolates clustered into groups by isolation source; the representative PFGE patterns depicting these differences are shown in Figure 2.
FIGURE 2.
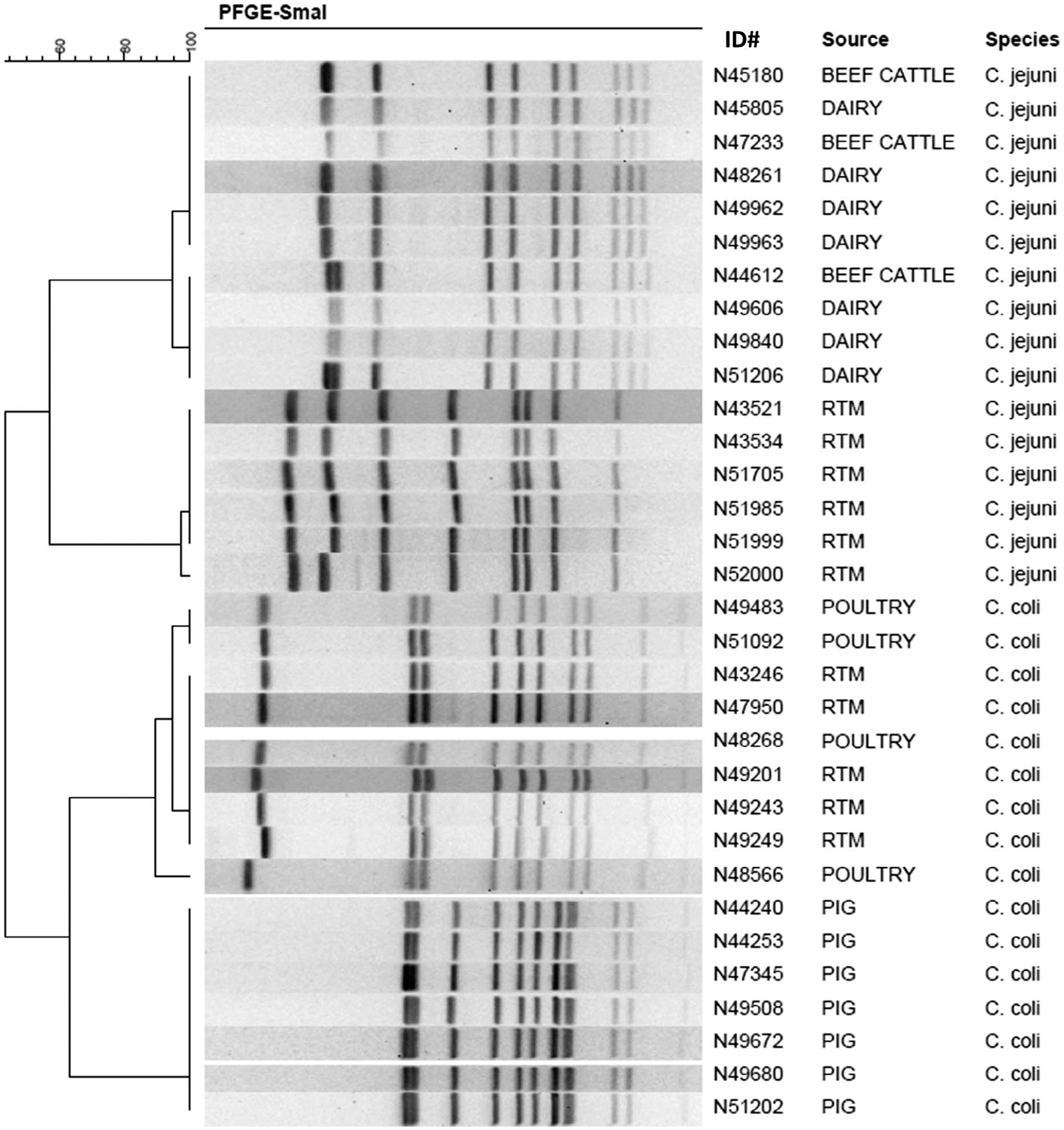
PFGE clusters from distinct sources. A dendrogram depicts the estimated relatedness of the PFGE patterns. The isolate identification number, isolate source, and Campylobacter species are depicted. RTM, retail poultry meat; poultry refers to isolates from poultry animal ceca.
Each of the PFGE patterns from the animal and poultry meat sources was compared with those present in the CDC PulseNet database, which includes patterns from human isolates obtained from both sporadic and outbreak infections. This analysis showed that 83.9% of C. jejuni and 32.1% of C. coli isolates had PFGE patterns that were indistinguishable from patterns from human isolates (Table 1). The difference between the two species was expected because C. coli is less common as a cause of human illness than C. jejuni (28). This is also reflected by the fact that, at the time of analysis, CDC PulseNet contained only about 400 C. coli human isolates with PFGE patterns, compared with more than 10,000 C. jejuni human isolates.
The different animal sources had substantial disparities when their PFGE patterns were compared to those from human isolates. Among the 196 PFGE patterns from poultry animal and retail meat isolates, 97 (49.5%) were indistinguishable from those of human isolates in the CDC PulseNet database (Table 2). Each of the 19 most common poultry patterns, containing at least three isolates per cluster, had corresponding patterns among human isolates. As a result, most Campylobacter isolates from poultry animals and retail meat (64.1%) had PFGE patterns indistinguishable from those of bacteria isolated from human patients (Table 1). In contrast, among the pig isolates only 9.5% of the PFGE patterns and 13.7% of all isolates had patterns indistinguishable from those of human isolates (Tables 1 and 2). Only 5 of the 14 common patterns (present in at least three pig strains) were indistinguishable from those of human isolates in the CDC PulseNet database. This suggests that pigs may be a less common source of Campylobacter causing human infections than poultry sources; however, it may also be attributable to the fact that nearly all the isolates from pigs were C. coli. For the cattle isolates, the majority of patterns were indistinguishable from those of the human isolates, comprising 56.6% of beef cattle and 65.0% of dairy cattle patterns (Table 2). Moreover, all the most common patterns from the isolates from each cattle source (23 patterns from beef cattle and 34 from dairy cattle sources) had patterns indistinguishable from those of human isolates. As a result, the beef and dairy cattle isolates had the highest correspondence with the human isolate PFGE profiles (80.5 and 83.6%, respectively) (Table 1). These percentages of isolates with indistinguishable patterns for the cattle isolates were significantly greater than those from any of the other sources (P < 0.0001), regardless of the Campylobacter species (Table 1).
One recent public health concern is the emergence of a Campylobacter clone associated with higher virulence, called clone SA (19). This clone, designated DBRS16.0008 (18), causes abortions in sheep as well as increased pathogenesis in mammalian hosts (4). The PFGE pattern corresponding to this clone was present in the CDC PulseNet database and was one of the most common PFGE patterns found in our collection, encompassing 37 total isolates: 1 from retail poultry meat, 2 from pigs, 14 from beef cattle, and 20 from dairy cattle (Fig. 3). Although these isolates are from only a single clonal group, these results suggest that some cattle isolates may be associated with greater pathogenesis in human hosts.
FIGURE 3.
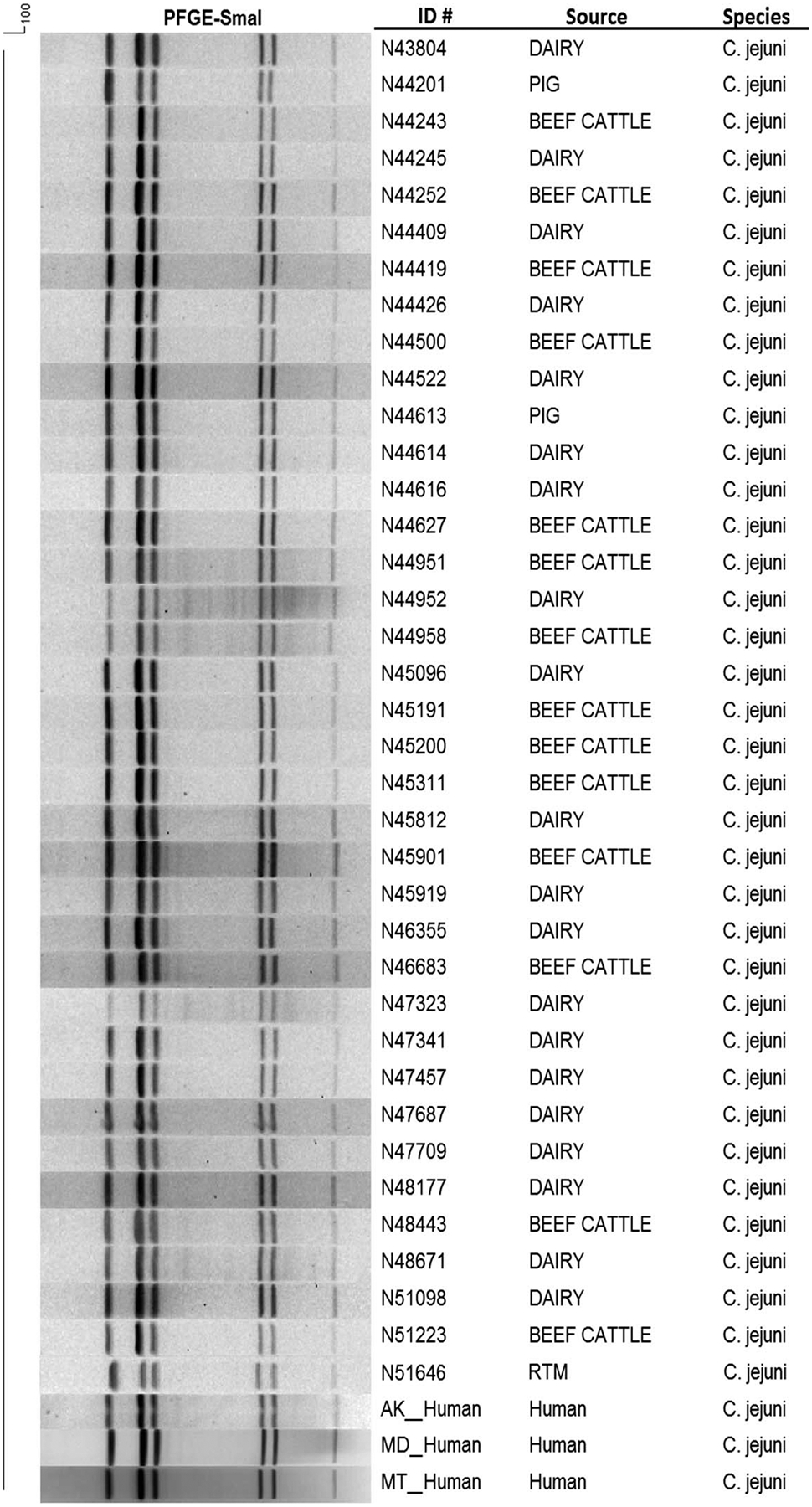
Depiction of a high-virulence Campylobacter SA clone (DBRS16.0008). Isolate identification number, source of isolate, and species are shown. Some human patterns were included for comparison. RTM, retail poultry meat.
DISCUSSION
Very few large-scale animal sampling studies of Campylobacter have been conducted for source attribution. We found that chickens, thought to be a major source of Campylobacter, had a relatively low prevalence of Campylobacter in their ceca despite the high contamination rate of poultry retail meats. The low cecal colonization rates were consistent with some previous studies (23), suggesting that cross-contamination from processing facilities may be responsible for the higher colonization in retail meats (13). In contrast to the low Campylobacter prevalence in poultry animals, we isolated much more Campylobacter from both cattle and pigs. Nevertheless, retail meat surveillance of ground beef and pork chops has found less than 1% of each meat type is contaminated with Campylobacter (27); therefore, these meat types presumably pose a lower risk for Campylobacter exposure. This could account for some of our results because only a minority of Campylobacter from pig ceca had patterns indistinguishable from those of human patients (9.5% of the patterns and 13.7% of the isolates). Over 80% of isolates from beef cattle ceca, however, had PFGE patterns indistinguishable from those of human patient isolates, despite the rare Campylobacter contamination of retail beef. This high frequency of indistinguishable patterns may be explained by the fact that beef cattle isolate PFGE patterns often clustered with those from dairy cattle, which may be sources of Campylobacter associated with the consumption of raw milk or dairy products (16).
Although chicken meat has long been considered the principal source of Campylobacter infections, other important sources have been identified based on outbreak investigations. In a joint U.S. Food and Drug Administration, CDC, and USDA retrospective epidemiological study (17), approximately 67% of Campylobacter outbreak infections were attributed to milk and dairy products. A more recent report (11) based on Campylobacter outbreaks from 1998 to 2012 estimated that 66% of outbreaks were attributable to the consumption of dairy products (90% credibility interval, 57 to 74%). The large proportion of cases associated with dairy products is mostly attributable to the consumption of raw milk because Campylobacter is effectively killed by pasteurization (1). Additionally, cattle may also be a source through water and environmental dissemination (5, 22). Despite the fact that these reports focused solely on Campylobacter outbreaks and not sporadic infections, these data indicate the importance of raw milk and other nonpoultry sources as significant contributors to Campylobacter infection.
The results of this study emphasize the continued relevance of PFGE in performing source attribution studies. Although there is some disagreement about whether PFGE is more discriminatory than multilocus sequence typing (20, 25), the vast amount of Campylobacter PFGE data from human clinical samples has proven valuable (14). Whole-genome sequencing is in the process of transforming outbreak detection and source attribution, although most outbreaks are still initially detected using PFGE, with subsequent whole-genome sequencing helping with trace-back analyses (10).
Despite the significant results of this study, there are some limitations to our analysis. For instance, the data are based solely on the presence of PFGE patterns from animal sources that are indistinguishable from human sources. This does not unambiguously assert that a given infection was caused by the consumption of a certain product because consumption data from the human disease cases was not available. Also, the poultry and pig isolates had a greater PFGE pattern diversity, making it potentially less likely for human sampling to result in as many pattern matches. In addition, for patterns shared by multiple sources, it is unclear which source was predominantly responsible for the human illnesses. The implementation of newer technologies, such as whole-genome sequencing, may provide sufficient resolution to perform these analyses. Additional case control studies focusing on exposures associated with human illnesses caused by specific PFGE patterns are necessary to strengthen the conclusions made in this study.
Several features of Campylobacter have impeded our ability to conduct thorough source attribution studies to date. Because most infections are sporadic, it is often not possible to identify the food vehicles causing the illness. The fastidious growth requirements of the genus also make it challenging to recover sufficient isolate numbers from food sources to make adequate comparisons. Applying common cecal culture methods resulted in high recovery rates and provided the number of isolates we needed to conduct the study reported here. As described previously, PFGE has been helpful for determining source attribution and investigating common source outbreaks of illness, and this study demonstrates that increased surveillance of Campylobacter from different sources can lead to new insights into the origins of human infections.
ACKNOWLEDGMENTS
We thank the CDC PulseNet Task Force for curating the PulseNet database. The views expressed in this article are those of the author(s) and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, or the U.S. Government. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration.
REFERENCES
- 1.American Academy of Pediatrics, Committee on Infectious Diseases and Committee on Nutrition. 2014. Consumption of raw or unpasteurized milk and milk products by pregnant women and children. Pediatrics 133:175–179. [DOI] [PubMed] [Google Scholar]
- 2.Bardou P, Mariette J, Escudie F, Djemiel C, and Klopp C. 2014. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boes J, Nersting L, Nielsen EM, Kranker S, Enoe C, Wachmann HC, and Baggesen DL. 2005. Prevalence and diversity of Campylobacter jejuni in pig herds on farms with and without cattle or poultry. J. Food Prot 68:722–727. [DOI] [PubMed] [Google Scholar]
- 4.Burrough ER, Sahin O, Plummer PJ, Zhang Q, and Yaeger MJ. 2009. Pathogenicity of an emergent, ovine abortifacient Campylobacter jejuni clone orally inoculated into pregnant guinea pigs. Am. J. Vet. Res 70:1269–1276. [DOI] [PubMed] [Google Scholar]
- 5.Clark CG, Price L, Ahmed R, Woodward DL, Melito PL, Rodgers FG, Jamieson F, Ciebin B, Li A, and Ellis A. 2003. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis 9:1232–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJ, Urwin R, and Maiden MC. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol 39:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.European Food Safety Authority (EFSA) Panel on Biological Hazards. 2010. Scientific opinion on quantification of the risk posed by broiler meat to human campylobacteriosis in the EU. EFSA J. 8:1437. doi: 10.2903/j.efsa.2010.1437. [DOI] [Google Scholar]
- 8.Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, Reddy S, Ahuja SD, Helfrick DL, Hardnett F, Carter M, Anderson B, Tauxe RV, and for the Emerging Infections Program FoodNet Working Group. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis 38(Suppl. 3):S285–S296. [DOI] [PubMed] [Google Scholar]
- 9.Gibney KB, O’Toole J, Sinclair M, and Leder K. 2014. Disease burden of selected gastrointestinal pathogens in Australia, 2010. Int. J. Infect. Dis 28:176–185. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann M, Luo Y, Monday SR, Gonzalez-Escalona N, Ottesen AR, Muruvanda T, Wang C, Kastanis G, Keys C, Janies D, Senturk IF, Catalyurek UV, Wang H, Hammack TS, Wolfgang WJ, Schoonmaker-Bopp D, Chu A, Myers R, Haendiges J, Evans PS, Meng J, Strain EA, Allard MW, and Brown EW. 2016. Tracing origins of the Salmonella Bareilly strain causing a food-borne outbreak in the United States. J. Infect. Dis 213(4):502–508. [DOI] [PubMed] [Google Scholar]
- 11.Interagency Food Safety Analytics Collaboration (IFSAC). 2015. Foodborne illness source attribution estimates for Salmonella, Escherichia coli O157 (E. coli O157), Listeria monocytogenes (Lm), and Campylobacter using outbreak surveillance data. Available at: https://www.cdc.gov/foodsafety/pdfs/ifsac-project-report-508c.pdf.
- 12.Janssen R, Krogfelt KA, Cawthraw SA, van Pelt W, Wagenaar JA, and Owen RJ. 2008. Host-pathogen interactions in Campylobacter infections: the host perspective. Clin. Microbiol. Rev 21:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keener KM, Bashor MP, Curtis PA, Sheldon BW, and Kathariou S. 2004. Comprehensive review of Campylobacter and poultry processing. Compr. Rev. Food Sci. Food Saf 3:105–116. [DOI] [PubMed] [Google Scholar]
- 14.Lehner A, Schneck C, Feierl G, Pless P, Deutz A, Brandl E, and Wagner M. 2000. Epidemiologic application of pulsed-field gel electrophoresis to an outbreak of Campylobacter jejuni in an Austrian youth centre. Epidemiol. Infect 125:13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Antimicrobial Resistance Monitoring System (NARMS). 2015. Manual of laboratory methods, 2nd ed. U.S. Food and Drug Administration, Washington, DC. [Google Scholar]
- 16.Orr KE, Lightfoot NF, Sisson PR, Harkis BA, Tweddle JL, Boyd P, Carroll A, Jackson CJ, Wareing DR, and Freeman R. 1995. Direct milk excretion of Campylobacter jejuni in a dairy cow causing cases of human enteritis. Epidemiol. Infect 114:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, and Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis 19:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahin O, Fitzgerald C, Stroika S, Zhao S, Sippy RJ, Kwan P, Plummer PJ, Han J, Yaeger MJ, and Zhang Q. 2012. Molecular evidence for zoonotic transmission of an emergent, highly pathogenic Campylobacter jejuni clone in the United States. J. Clin. Microbiol 50:680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahin O, Plummer PJ, Jordan DM, Sulaj K, Pereira S, Robbe-Austerman S, Wang L, Yaeger MJ, Hoffman LJ, and Zhang Q. 2008. Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. J. Clin. Microbiol 46:1663–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sails AD, Swaminathan B, and Fields PI. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol 41:4733–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, and Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis 17:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith KE, Stenzel SA, Bender JB, Wagstrom E, Soderlund D, Leano FT, Taylor CM, Belle-Isle PA, and Danila R. 2004. Outbreaks of enteric infections caused by multiple pathogens associated with calves at a farm day camp. Pediatr. Infect. Dis. J 23:1098–1104. [PubMed] [Google Scholar]
- 23.Stern NJ, Reiersen J, Lowman R, Bisaillon JR, Fridriksdottir V, Gunnarsson E, Hiett KL, and Campy-on-Ice C. 2005. Occurrence of Campylobacter spp. in cecal contents among commercial broilers in Iceland. Foodborne Pathog. Dis 2:82–89. [DOI] [PubMed] [Google Scholar]
- 24.Swaminathan B, Barrett TJ, Hunter SB, and Tauxe RV. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis 7:382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakur S, White DG, McDermott PF, Zhao S, Kroft B, Gebreyes W, Abbott J, Cullen P, English L, Carter P, and Harbottle H. 2009. Genotyping of Campylobacter coli isolated from humans and retail meats using multilocus sequence typing and pulsed-field gel electrophoresis. J. Appl. Microbiol 106:1722–1733. [DOI] [PubMed] [Google Scholar]
- 26.Thakur S, Zhao S, McDermott PF, Harbottle H, Abbott J, English L, Gebreyes WA, and White DG. 2010. Antimicrobial resistance, virulence, and genotypic profile comparison of Campylobacter jejuni and Campylobacter coli isolated from humans and retail meats. Foodborne Pathog. Dis 7:835–844. [DOI] [PubMed] [Google Scholar]
- 27.U.S. Food and Drug Administration. 2013. National Antimicrobial Resistance Monitoring System–Enteric Bacteria (NARMS): 2011 executive report. U.S. Department of Health and Human Services, Food and Drug Administration, Rockville, MD. [Google Scholar]
- 28.U.S. Food and Drug Administration. 2015. National Antimicrobial Resistance Monitoring System (NARMS): 2012–2013 Integrated Report. U.S. Department of Health and Human Services, Food and Drug Administration, Rockville, MD. [Google Scholar]
- 29.Wang X, Zhao S, Harbottle H, Tran T, Blickenstaff K, Abbott J, and Meng J. 2011. Antimicrobial resistance and molecular subtyping of Campylobacter jejuni and Campylobacter coli from retail meats. J. Food Prot 74:616–621. [DOI] [PubMed] [Google Scholar]
- 30.Whiley H, van den Akker B, Giglio S, and Bentham R. 2013. The role of environmental reservoirs in human campylobacteriosis. Int. J. Environ. Res. Public Health 10:5886–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao C, Ge B, De Villena J, Sudler R, Yeh E, Zhao S, White DG, Wagner D, and Meng J. 2001. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the greater Washington, D.C., area. Appl. Environ. Microbiol 67:5431–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Young SR, Tong E, Abbott JW, Womack N, Friedman SL, and McDermott PF. 2010. Antimicrobial resistance of Campylobacter isolates from retail meat in the United States between 2002 and 2007. Appl. Environ Microbiol 76:7949–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]


