Abstract
Recently, there have been reports worldwide of a multidrug-resistant, emergent Salmonella Infantis (ESI) clone with a large megaplasmid (pESI), often containing the extended-spectrum beta-lactamase gene blaCTX-M-65. This clone also has a gyrA mutation conferring fluoroquinolone resistance, further limiting treatment options. In the United States, this clone has also been found in poultry sources, indicating a likely source of human illnesses. We conducted short-read sequencing of Salmonella enterica isolated from retail meats as part of routine surveillance by the National Antimicrobial Resistance Monitoring System (NARMS). We analyzed the resulting data temporally and geographically to determine when and where the ESI clone has spread in the United States. We found the ESI clone was first found in retail meats in Tennessee in 2014, but by 2019 was throughout the United States and comprised 29% of all Salmonella isolated from retail chickens, and 7% from retail turkey. Of these isolates, 85.0% were within 20 single nucleotide polymorphisms (SNPs) of those causing human illnesses. Long-read sequencing data indicated substantial recombination in the pESI plasmid resulting in the presence of 0–10 resistance genes, despite all their chromosomes being within 31 SNPs of one another. This work demonstrates the rapid spread of this clone of Salmonella Infantis in poultry in the United States, with the potential for increased burden of human illness attributed to this multidrug-resistant pathogen.
Keywords: Salmonella enterica, antibiotic resistance, plasmids, poultry
Introduction
Salmonella enterica is a major cause of foodborne illness in the United States.1 Although most infections resolve on their own without need for treatment, severe infections may need antimicrobial therapy, usually with cephalosporins or fluoroquinolones.2 S. enterica serovar Infantis has been among the top 10 serotypes causing human illness in the United States every year since 2010.3 Poultry is the most common source of Salmonella Infantis,4 although it has also been found in swine and other sources.5 In the United States, the National Antimicrobial Resistance Monitoring System (NARMS) conducts routine surveillance for antimicrobial resistance in Salmonella and other foodborne pathogens, including in retail meats. Historically, we have found ~2–4% of retail meat Salmonella are of serovar Infantis, with most being pansusceptible to antimicrobials on the NARMS panel.6
In 2014, there was the first report of a multidrug-resistant, emergent Salmonella Infantis (ESI) in Israel, with subsequent findings in Italy, Japan, and Russia, among others.7–10 Subsequent reports and retrospective sequencing tracked the origins of this clone to South America.11,12 This clone carried a large plasmid ESI (pESI) that carried several antimicrobial resistance, metal, and virulence genes. This plasmid could transfer to other Salmonella and Escherichia coli and conferred increased adhesion and virulence in cell and mammalian models, likely owing to the presence of fimbrial genes.7 In 2017, we identified this clone of Salmonella Infantis in a retail chicken isolate in the United States as part of NARMS sampling.13 Additional surveillance identified this clone in poultry animals, and in several human cases of illness in the United States. Characterization of the pESI megaplasmid found it was ~300 kb and contained a variety of resistance genes.13 The most prominent of these was blaCTX-M-65, an extended-spectrum beta-lactamase (ESBL) gene, which confers resistance to third-generation cephalosporins.13 This was the first reported instance of blaCTX-M-65 in Salmonella from retail meats in the United States. Additional resistance genes were found in some isolates, including those conferring resistance to tetracycline, aminoglycosides, sulfonamides, phenicols, and fosfomycin, and all resistance genes were found on elements flanked by transposases or integrases.13 This clone also had a GyrA (D87Y) amino acid change conferring resistance to fluoroquinolones, a drug class not used in U.S. poultry production since 2005.14
An additional epidemiological study further characterized the rise of this Salmonella Infantis clone in the United States, where it accounted for nearly 10% of all Salmonella Infantis human cases by 2017.15 Initially most human illnesses were associated with international travel to South America, but domestically acquired cases began in 2014. These human isolates were highly related to those from chicken sources.15
Owing to the resistant nature of this pathogen and its contribution to human infections, we sought to determine whether this clone has spread further in the food supply. We used long-read DNA sequencing to understand the genetics underlying the evolution of the megaplasmid as a reference point for tracking its spread, as we attempt to further understand this important pathogenic clone.
Methods
Bacterial isolates and sequencing
As part of routine NARMS surveillance 3,934 Salmonella isolates were collected from retail meat sampling in 21 states from 2014 to 2019. A total of 453 of these were of the Salmonella Infantis ESI clone. Each of these isolates was sequenced by short-read sequencing on the Illumina MiSeq as previously described.16 In brief, genomic DNA was extracted with QIAmp 96 DNA QIAcube HT kits (Qiagen, Germantown, MD) on the automated QIAcube HT machine per the manufacturer’s instructions. Libraries were prepared with Nextera XT kits and sequenced with v3 reagent kits 2 × 300 bp on the Illumina MiSeq (Illumina, San Diego, CA). Illumina sequences were submitted to the sequence read archive (SRA) in National Center for Biotechnology Information (NCBI), where they were assembled by SKESA.17 SRA accessions are listed in Supplementary Table S1, with all sequences in BioProject PRJNA292661.
Data mining
The NCBI Isolates Browser (www.ncbi.nlm.nih.gov/pathogens/isolates) was analyzed for the ESI clone as of January 10, 2020. The isolates browser automatically assigns each sequence within 50 single nucleotide polymorphisms (SNPs) of one another into a single SNP cluster, generating phylogenetic trees based on the maximum compatibility method.18 This clone was identified by searching for the NCBI SNP cluster PDS000003955.782, which included 4,699 isolates. Isolates specific to NARMS retail meat sampling were identified from BioProject PRJNA292661.
Geographic analysis
We used the Geographic Information System (GIS) program ArcGIS 10.7 (Environmental Systems Research Institute, Redlands, CA) to input state-level metadata information and overlay geographic and temporal metadata onto a map of the United States. These included all 453 NARMS retail meat isolates with state and year information.
Long-read sequencing
Thirty-one isolates were selected for long-read sequencing, to represent isolates with diverse combinations of resistance genes. This sequencing was conducted as previously described using the 10-kb template preparation protocol using the Pacific Biosciences Sequel sequencer with Sequel sequencing kit v 3.0 (Pacific Biosciences, Menlo Park, CA).19 The long reads were assembled to contigs by HGAP4 or Microbial Assembly and contigs were circularized by Circlator.20,21 PacBio genome assemblies were submitted to GenBank with nucleotide accession numbers and BioSample numbers as listed in Supplementary Table S2. All sequences are under the same BioSamples as those associated with the short-read sequences and are in BioProject PRJNA292661.
Sequencing analysis
Resistance genes were identified in the Isolates Browser by using AMRFinder under default settings.22 The relatedness of retail meat ESI clones to human ESI clones was identified with the NCBI SNP tree, using information from the Min-diff column in the NCBI Isolates Browser.
SNP analysis from long-read sequencing data was conducted with kSNP version 3.1, and kmer size chosen by kChooser.23 A maximum likelihood tree was constructed by MEGA7.0 with support values estimated through 500 bootstraps.24 Plasmids were removed before conducting phylogenetic analysis. This tree included 31 ESI clones and three non-ESI Infantis as an outgroup.
Results
Prevalence of ESI clone
To have an all-encompassing definition of the ESI clone that was not limited by pulsed-field gel electrophoresis (PFGE) patterns or the presence of specific resistance genes such as blaCTX-M-65, we relied on phylogenetic analysis. We used the Isolates Browser housed at the NCBI website, which partitions sequences into distinct clusters within 50 SNPs. We analyzed the presence of isolates in the SNP cluster PDS000003955.782 in the Isolates Browser (https://www.ncbi.nlm.nih.gov/pathogens/isolates/#PDS000003955.782), containing 4,669 isolates at the time of analysis. We defined this cluster as isolates of the ESI clone, regardless of the presence of individual resistance genes.
In this SNP cluster, we found isolates in retail meats and humans as far back as 2009, with all from before 2012 isolated in Peru. There were no isolates from any NARMS sampling of U.S. retail meat before 2014, but by 2019, 29.2% of retail chicken Salmonella were of the ESI clone, and 6.7% of retail turkey isolates (Fig. 1). From 2014 to 2019, there were also two isolates from ground beef, among 67 total isolates from this source during this time. Further geographic analysis points to the rapid spread of the ESI clone. It was first identified in Tennessee and subsequently emerged in the northeastern United States before being found in all NARMS sampling states (Fig. 2). The largest number of retail meat isolates were in Maryland and Tennessee, although it is worth noting that NARMS does not sample in all 50 states.
FIG. 1.
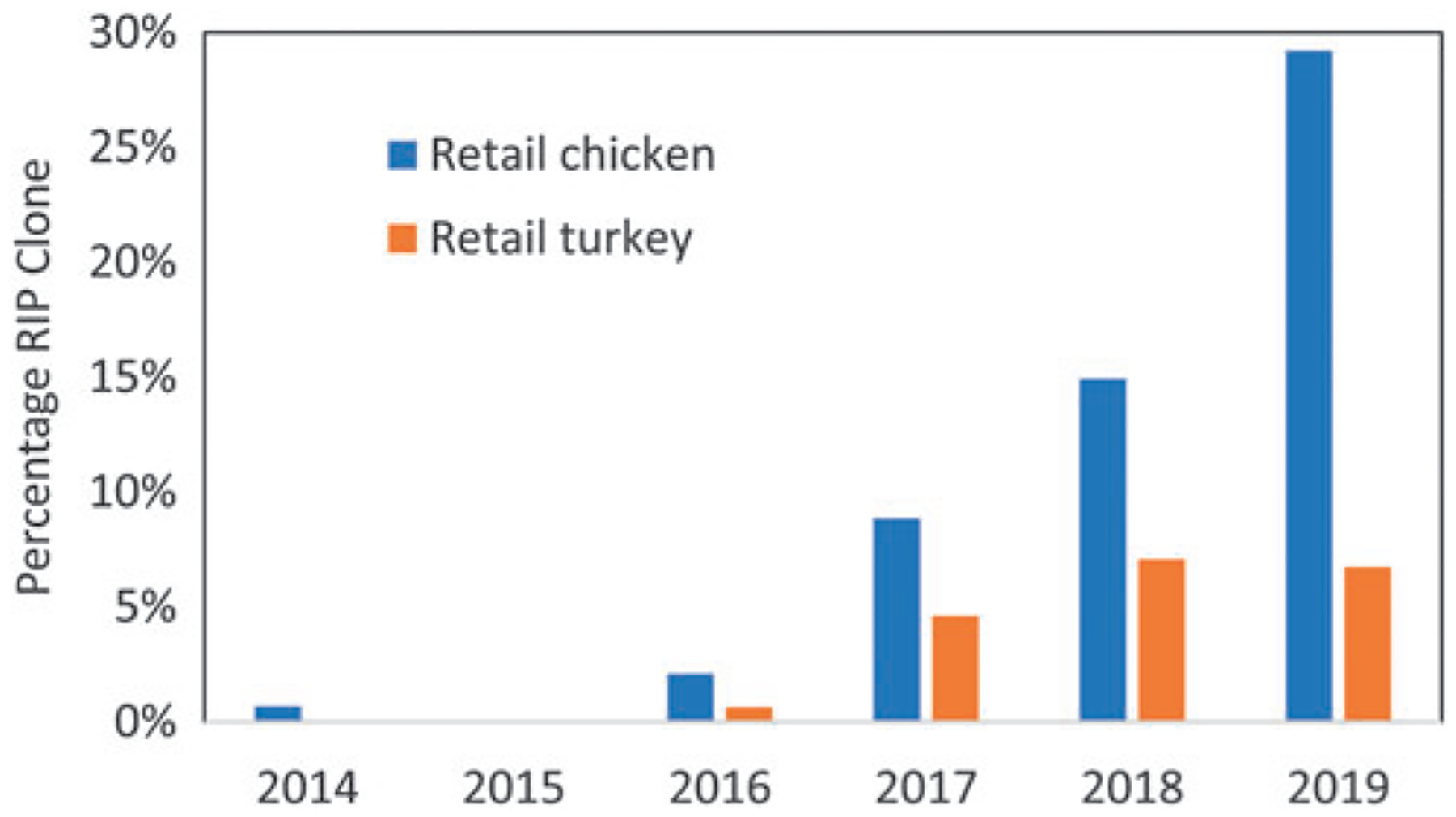
Prevalence of ESI clone from 2014 to 2019 from NARMS sampling among retail meat sources. ESI, emergent Salmonella Infantis; NARMS, National Antimicrobial Resistance Monitoring System.
FIG. 2.
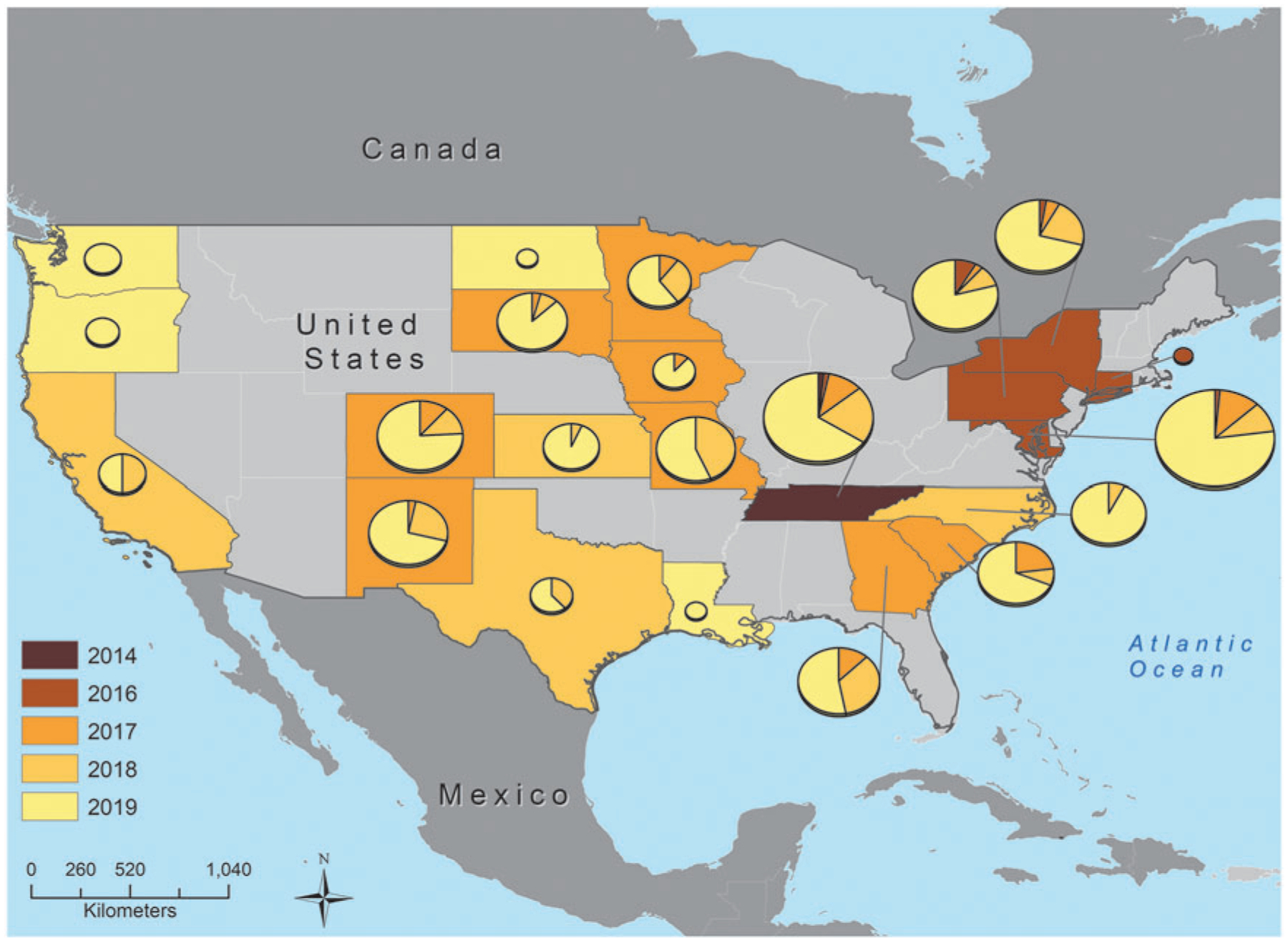
Geographic representation of ESI clone emergence. States are colored by the year the clone was first identified in retail meats, with pie charts representing the proportion of isolates from each year. The size of the pie chart is proportional to the number of isolates from each state. Note: not all states were part of surveillance for all years of this study, and states colored gray are not part of NARMS surveillance.
Resistance gene analysis and relatedness to human isolates
Among the 453 total isolates of the ESI clone collected from NARMS retail meat surveillance, there were 57 different combinations of resistance genes (Supplementary Table S1). Although most isolates had three to seven resistance genes (293/453, 64.7%), 32.9% had eight or more resistance genes, including 7.7% of isolates having 10 resistance genes (Fig. 3A). Five isolates had no resistance genes. The most common resistance genotype, in 19.2% of isolates (87/453), was with aadA1, sul1, and tet(A), which confer resistance to streptomycin, sulfisoxazole, and tetracycline, respectively. The most common pattern with blaCTX-M-65 was in 9.9% of isolates (45/453) and included aac(3)-IV, aadA1, aph(4)-Ia, blaCTX-M-65, floR, sul1, and tet(A), conferring resistance to various aminoglycosides, beta-lactams, chloramphenicol, sulfisoxazole, and tetracycline.
FIG. 3.
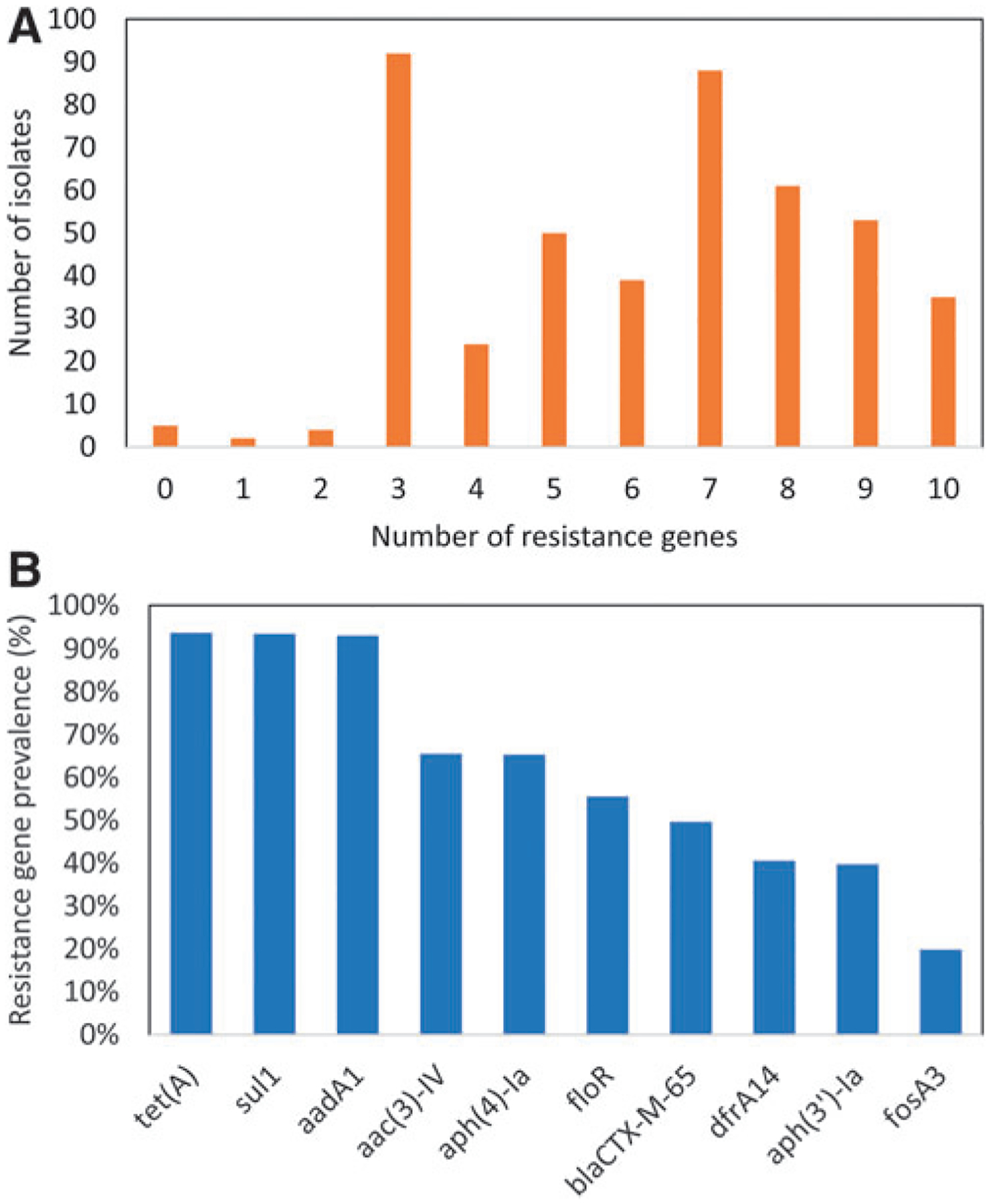
(A) Number of resistance genes among ESI clones, and (B) prevalence of each resistance gene among them.
Although the combinations of resistance genes varied widely among the isolates, they always comprised different combinations of a set of 10 resistance genes (Fig. 3B). These resistance genes are across several different drug classes, including tetracyclines (tet(A)), folate synthesis inhibitors (sul1, dfrA14), aminoglycosides (aadA1, aac(3)-IV, aph(4)-Ia, aph(3′)-Ia), beta-lactams (blaCTX-M-65), and phenicols (floR), and fosfomycin (fosA3). Each gene was in at least 20% of ESI clones. Based on GIS analysis, there was no clear geographic or temporal trend for the presence of specific resistance genes.
In the large ESI SNP cluster, there were 1,241 isolates from human illnesses collected as part of PulseNet surveillance, in BioProject PRJNA230403. Analysis from the NCBI Isolates Browser showed that 85.0% (385/453) of the retail meat isolates were within 20 SNPs of human cases, the threshold typically used for outbreak investigations.25 Furthermore, 62 of our retail meat isolates were within 5 SNPs of human isolates, with 45 having blaCTX-M-65 (72.6%). Only 49.7% of all the retail meat ESI clones had this gene.
Long-read sequencing
To characterize differences in plasmid content and resistance genes, we sequenced 31 isolates using PacBio long-read sequencing technologies. The 31 isolates were closely related and formed a separate clade from three Infantis from other NCBI SNP clusters, as expected (Fig. 4). This figure also depicts the complement of resistance genes in each isolate, illustrating the high level of recombination occurring within the pESI plasmid. All ESI clones were within 31 SNPs of each other, indicating a high degree of relatedness. Nevertheless, isolates in the same subclusters often had different combinations of resistance genes on their plasmids. This was particularly evident with closely related isolates such as N17S1234 and N17S1598, which were adjacent on the phylogenetic tree. Although some resistance genes were the same in both plasmids, N17S1234 had dfrA14 and aph(3′)-Ia, whereas N17S1598 had aadA1, sul1, and tet(A). Of interest, the isolate N19S0949 did not have any resistance genes, but still possessed the pESI plasmid, simply missing the regions containing resistance genes (Table 1). Isolate-level accession numbers and plasmid length and source information are given in Table 1.
FIG. 4.
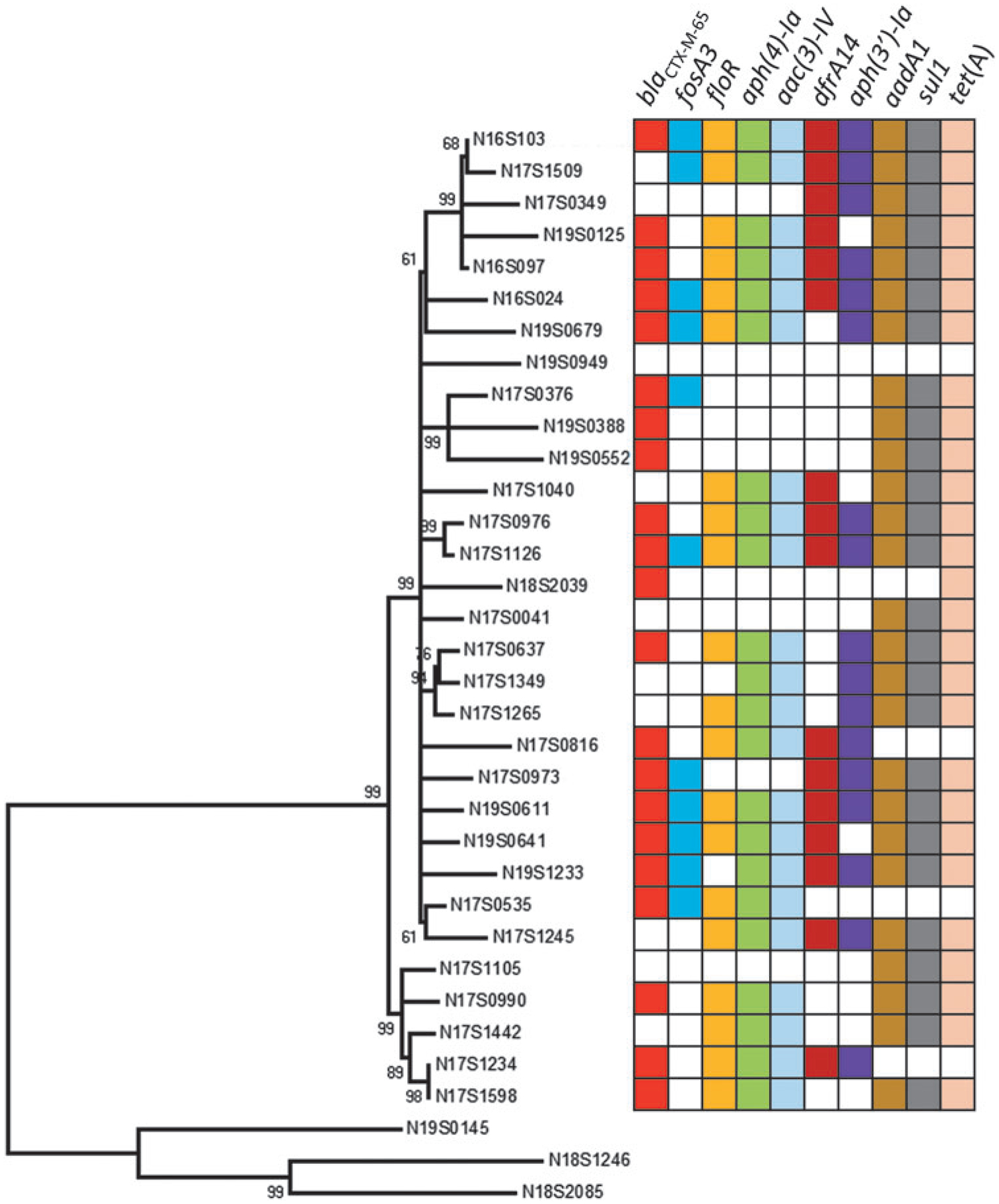
Phylogenetic tree of 31 ESI clones with closed genomes, including resistance gene information. Numbers in the figure are bootstrap values. The bottom three sequences are non-ESI clone Infantis included as an outgroup.
Table 1.
Plasmid Resistance Genes and Accession Numbers from Long-Read Sequencing of 31 Isolates
| NARMS ID | GenBank accession | Plasmid length | Source | Year | State | Plasmid genotype |
|---|---|---|---|---|---|---|
| N16S024 | CP052840 | 322,506 | Chicken | 2016 | CT | aac(3)-IV aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 floR fosA3 sul1 tet(A) |
| N16S097 | CP052838 | 318,524 | Chicken | 2016 | PA | aac(3)-IV aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 floR sul1 tet(A) |
| N16S103 | CP052836 | 322,451 | Chicken | 2016 | PA | aac(3)-IV aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 floR fosA3 sul1 tet(A) |
| N17S041 | CP052834 | 288,008 | Turkey | 2017 | GA | aadA1 sul1 tet(A) |
| N17S1040 | CP052832 | 306,079 | Chicken | 2017 | NY | aac(3)-IVa aadA1 aph(4)-Ia dfrA14 floR sul1 tet(A) |
| N17S1105 | CP052830 | 289,847 | Chicken | 2017 | SC | aadA1 sul1 tet(A) |
| N17S1126 | CP052828 | 322,509 | Turkey | 2017 | TN | aac(3)-IVa aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 floR fosA3 sul1 tet(A) |
| N17S1234 | CP051676 | 297,707 | Chicken | 2017 | CO | aac(3)-IVa aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 floR |
| N17S1245 | CP052826 | 299,741 | Chicken | 2017 | CO | aac(3)-IVa aadA1 aph(3’)-Ia aph(4)-Ia dfrA14 floR sul1 tet(A) |
| N17S1265 | CP052824 | 290,274 | Chicken | 2017 | CO | aac(3)-IVa aadA1 aph(3’)-Ia aph(4)-Ia floR sul1 tet(A) |
| N17S1349 | CP052822 | 298,238 | Chicken | 2017 | MD | aac(3)-IVa aadA1 aph(3’)-Ia aph(4)-Ia sul1 tet(A) |
| N17S1442 | CP052820 | 292,474 | Chicken | 2017 | SC | aac(3)-IVa aadA1 aph(4)-Ia floR sul1 tet(A) |
| N17S1509 | CP052818 | 306,853 | Chicken | 2017 | GA | aac(3)-IVa aadA1 aph(3’)-Ia aph(4)-Ia dfrA14 floR fosA3 sul1 tet(A) |
| N17S1598 | CP052816 | 303,467 | Chicken | 2017 | TN | aac(3)-IVa aadA1 aph(4)-Ia blaCTX-M-65 floR sul1 tet(A) |
| N17S349 | CP052814 | 300,799 | Turkey | 2017 | SC | aadA1 aph(3’)-Ia dfrA14 sul1 tet(A) |
| N17S376 | CP052812 | 293,162 | Chicken | 2017 | SD | aadA1 blaCTX-M-65 fosA3 sul1 tet(A) |
| N17S535 | CP052810 | 283,099 | Chicken | 2017 | IA | aac(3)-IVa aph(4)-Ia blaCTX-M-65 floR fosA3 |
| N17S637 | CP052808 | 312,913 | Chicken | 2017 | MN | aac(3)-IVa aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 floR sul1 tet(A) |
| N17S816 | CP052806 | 296,746 | Chicken | 2017 | PA | aac(3)-IVa aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 floR |
| N17S973 | CP052804 | 309,326 | Chicken | 2017 | MD | aadA1 aph(3’)-Ia blaCTX-M-65 dfrA14 fosA3 sul1 tet(A) |
| N17S976 | CP052802 | 319,023 | Turkey | 2017 | MD | aac(3)-IVa aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 floR sul1 tet(A) |
| N17S990 | CP052799 | 310,270 | Turkey | 2017 | MN | aac(3)-IVa aadA1 aph(4)-Ia blaCTX-M-65 floR sul1 tet(A) |
| N18S2039 | CP052797 | 168,393 | Turkey | 2018 | CA | None (blaCTX-M-65 tet(A) on chromosome) |
| N19S0125 | CP052795 | 317,309 | Chicken | 2019 | MD | aac(3)-IV aadA1 aph(4)-Ia blaCTX-M-65 dfrA14 floR sul1 tet(A) |
| N19S0388 | CP052793 | 290,416 | Chicken | 2019 | SD | aadA1 blaCTX-M-65 sul1 tet(A) |
| N19S0552 | CP052791 | 289,609 | Turkey | 2019 | TX | aadA1 blaCTX-M-65 sul1 tet(A) |
| N19S0611 | CP052788 | 322,462 | Turkey | 2019 | NC | aac(3)-IV aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 floR fosA3 sul1 tet(A) |
| N19S0641 | CP052786 | 321,524 | Chicken | 2019 | PA | aac(3)-IV aadA1 aph(4)-Ia blaCTX-M-65 dfrA14 floR fosA3 sul1 tet(A) |
| N19S0679 | CP052783 | 312,779 | Chicken | 2019 | NY | aac(3)-IV aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 floRfosA3 sul1 tet(A) |
| N19S0949 | CP052781 | 265,507 | Chicken | 2019 | TX | None |
| N19S1233 | CP052779 | 314,663 | Turkey | 2019 | TN | aac(3)-IV aadA1 aph(3’)-Ia aph(4)-Ia blaCTX-M-65 dfrA14 fosA3 sul1 tet(A) |
NARMS, National Antimicrobial Resistance Monitoring System.
In one isolate, N18S2039, the pESI plasmid was only 168 kb long, and had no resistance genes. The rest of the plasmid was integrated into the chromosome, including the region containing the blaCTX-M-65 and tet(A) genes. An 80-kb portion of this integrated plasmid aligned with >99% identity to the IncI1 plasmid pC193 from an E. coli in Bolivia.26 Because the pESI plasmid is thought to have come from South America, this E. coli plasmid may have been an original component that helped form the megaplasmid. The N18S2039 isolate was the only one with long-read sequencing data that had any chromosomal resistance genes.
Discussion
The emergence of the ESI clone of Salmonella Infantis is concerning, as its sudden rise in retail meats and contributions to human illnesses is combined with a propensity for multidrug-resistance. This is particularly a problem when a greater proportion of ESI clones closely related to those causing human infections have the ESBL gene blaCTX-M-65 than the overall proportion. Combined with fluoroquinolone resistance resulting from a gyrA mutation in the ESI clone, this can make infections caused by this clone difficult to treat, because fluoroquinolones and cephalosporins are the most commonly used drugs to treat severe salmonellosis.2
In this work, we used SNP clusters from NCBI’s Isolates Browser to identify the ESI clone, which has dramatically risen in prevalence in retail meats as it has spread across the United States. We believe the use of SNP trees, such as those produced in NCBI’s Isolates Browser, are an ideal method for identifying these emerging bacterial clones. This provides a more inclusive and forward-looking definition than methods such as PFGE clusters, which have previously excluded related isolates of the ESI clone.13 AMRFinder results from this approach also agreed with prior work finding two sites within the pESI plasmid with resistance genes, with blaCTX-M-65, fosA3, floR, aph(4)-Ia, aac(3)-VI, dfrA14, and aph(3)-Ia at one site and sul1, tet(A), and aadA1 at the other site.13
We believe long-read sequencing is an essential component of understanding the emergence and epidemiology of this clone, as it helped us determine the complement of resistance genes and their variability among highly related isolates. Long-read sequencing also identified an IncX4 plasmid with the blaTEM-1 gene in one isolate. This indicates the pESI plasmid was not restrictive in preventing acquisition or maintenance of additional plasmids, at least of this incompatibility type.
The unusual genotype and early epidemiological linkages highlight that the ESI clone originated outside the United States, likely in South America.15 After having arrived, the ESI clone has risen quickly and continuously, but why it has successfully spread in food animals and retail meats in the United States remains unknown. Since fluoroquinolone use was banned in poultry in 2005 and cephalosporin use has been restricted since 2012,14 the use of other antimicrobials may help select for the range of plasmid structures seen in the different isolates, including those with blaCTX-M-65. In fact, >90% of isolates had the tet(A) gene, conferring tetracycline resistance, and tetracyclines are the antimicrobial with highest annual sales in food animals in the United States.27 Thus, use of antimicrobials such as tetracycline has the potential to co-select for the continued presence of the pESI plasmid.
Another factor potentially contributing to the success of the ESI clone and its pESI plasmid are its association with virulence. Prior reports demonstrated that the pESI plasmid confers increased adhesion and invasion in cell culture models and increased virulence in a mouse model of infection.7 This adhesion and increased virulence are proposed to be mediated by two chapterone-usher fimbria gene clusters encoded by fea and ipf genes found on the pESI plasmid.7 Given that ESI clones without resistance genes are successfully spreading, it is likely that these other factors are also contributing to its increased dissemination. Why it has successfully spread in poultry sources, and not others such as cattle, where this clone has also been found,13 remains unknown.
Typing of the pESI plasmid is still difficult, given that it is a megaplasmid that appears to have arisen from multiple recombination events. PlasmidFinder classifies this as an IncFIB-type plasmid, as has been previously described.13,28 However, as a megaplasmid, it appears to have resulted from integration of an IncFIB and IncI1 plasmid. For isolate N18S2039, the IncI1 portion containing blaCTX-M-65 integrated into the chromosome, resulting in a much smaller IncFIB plasmid. A potential contribution from an IncP plasmid has also been postulated.7 Together, these findings indicate the difficulties in applying conventional plasmid typing methods to megaplasmids.
Despite the significance of our findings, there are some limitations to this study. For instance, not all states perform NARMS retail meat sampling, so our information on the prevalence and geographic distribution of the ESI clone is limited to ~20 states. In addition, for SNP cluster information we rely upon NCBI’s Isolates Browser, which includes sequences that are heavily biased toward those from the United States and United Kingdom, which have the most public data. Despite these limitations, robust NARMS sequencing efforts have included all retail meat Salmonella since sampling began in 2002. Although there may have been ESI clones in retail meats before 2014, when it was first detected, this probably would have been rare based on our sampling of thousands of retail meats for Salmonella each year.
In summary, the ESI clone has continued to increase among retail poultry in the United States, now accounting for almost one-third of all Salmonella isolates from retail chicken. Given the association of this clone and the pESI plasmid with multidrug-resistance and increased virulence, it is likely to have an increased burden on human health. There is the potential for similar spread in the European Union, where Salmonella Infantis is the number one Salmonella serotype from food animals, and number three from humans.29 The ESI clone, including the pESI plasmid and associated resistance and virulence genes, have been found in Europe and Asia,9,10 and accounted for a majority of pediatric Salmonella cases in a study in Peru.30
Furthermore, the potential spread of the pESI plasmid is of concern, as it has been successfully transferred to other enteric organisms, including other types of Salmonella.31 Thus, the ESI clone and its associated pESI plasmid pose a continued public health problem deserving of further attention and study.
Supplementary Material
Acknowledgments
NARMS acknowledges the contribution of its state and local public health laboratories and universities for bacterial isolation and some whole-genome sequencing. The authors also thank Andrea Presotto for helping with the geographic representation.
Funding Information
Funding was provided by the U.S. Food and Drug Administration as part of routine work.
Footnotes
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, or the U.S. Government. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration.
Disclosure Statement
No competing financial interests exist.
Supplementary Material
Supplementary Table S1
Supplementary Table S2
References
- 1.Tack DM, Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Scallan E, Lathrop S, Muse A, Ryan P, Smith K, Tobin-D’Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, and Geissler AL. 2019. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2015–2018. MMWR Morb. Mortal. Wkly. Rep 68:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K, Chan EWC, and Chen S. 2019. Evolution and transmission of a conjugative plasmid encoding both ciprofloxacin and ceftriaxone resistance in Salmonella. Emerg. Microbes Infect 8:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. 2018. National Salmonella Surveillance Annual Report, 2016. Atlanta, GA: Centers for Disease Control and Prevention. [Google Scholar]
- 4.Gymoese P, Kiil K, Torpdahl M, Osterlund MT, Sorensen G, Olsen JE,1 Nielsen EM, and Litrup E. 2019. WGS based study of the population structure of Salmonella enterica serovar Infantis. BMC Genomics 20:870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skarzynska M, Hoszowski A, Zajac M, Lalak A, Samcik I, Kwit R, and Wasyl D. 2017. Distribution of Salmonella Serovars along the food chain in Poland, 2010–2015. J. Vet. Res 61:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.NARMS. 2016. NARMS Integrated Report: 2014. The National Antimicrobial Resistance Monitoring System: Enteric Bacteria. Rockville, MD: U.S. Food and Drug Administration. [Google Scholar]
- 7.Aviv G, Tsyba K, Steck N, Salmon-Divon M, Cornelius A, Rahav G, Grassl GA, and Gal-Mor O. 2014. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ. Microbiol 16:977–994. [DOI] [PubMed] [Google Scholar]
- 8.Bogomazova AN, Gordeeva VD, Krylova EV, Soltynskaya IV, Davydova EE, Ivanova OE, and Komarov AA. 2020. Mega-plasmid found worldwide confers multiple antimicrobial resistance in Salmonella Infantis of broiler origin in Russia. Int. J. Food Microbiol 319:108497. [DOI] [PubMed] [Google Scholar]
- 9.Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, Tolli R, D’Incau M, Staffolani M, Di Giannatale E, Hendriksen RS, and Battisti A. 2015. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One 10:e0144802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama E, Ando N, Ohta T, Kanada A, Shiwa Y, Ishige T, Murakami K, Kikuchi T, and Murakami S. 2015. A novel subpopulation of Salmonella enterica serovar Infantis strains isolated from broiler chicken organs other than the gastrointestinal tract. Vet. Microbiol 175:312–318. [DOI] [PubMed] [Google Scholar]
- 11.Cartelle Gestal M, Zurita J, Paz YMA, Ortega-Paredes D, and Alcocer I. 2016. Characterization of a small outbreak of Salmonella enterica serovar Infantis that harbour CTX-M-65 in Ecuador. Braz. J. Infect. Dis 20: 406–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palma N, Pons MJ, Gomes C, Mateu J, Riveros M, Garcia W, Jacobs J, Garcia C, Ochoa TJ, and Ruiz J. 2017. Resistance to quinolones, cephalosporins and macrolides in Escherichia coli causing bacteraemia in Peruvian children. J. Glob. Antimicrob. Resist 11:28–33. [DOI] [PubMed] [Google Scholar]
- 13.Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, Li C, Morales C, Tyson GH, Mukherjee S, Brown AC, Green A, Wilson W, Dessai U, Abbott J, Joseph L, Haro J, Ayers S, McDermott PF, and Zhao S. 2017. Comparative analysis of extended-spectrum-beta-lactamase CTX-M-65-producing Salmonella enterica Serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother 61:e00488–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price LB, Lackey LG, Vailes R, and Silbergeld E. 2007. The persistence of fluoroquinolone-resistant Campylobacter in poultry production. Environ. Health Perspect 115:1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown AC, Chen JC, Watkins LKF, Campbell D, Folster JP, Tate H, Wasilenko J, Van Tubbergen C, and Friedman CR. 2018. CTX-M-65 extended-spectrum beta-lactamase-producing Salmonella enterica Serotype Infantis, United States. Emerg. Infect. Dis 24:2284–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S, Bodeis-Jones S, Kabera C, Gaines SA, Loneragan GH, Edrington TS, Torrence M, Harhay DM, and Zhao S. 2015. WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother 70:2763–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souvorov A, Agarwala R, and Lipman DJ. 2018. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherry JL 2017. A practical exact maximum compatibility algorithm for reconstruction of recent evolutionary history. BMC Bioinformatics 18:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyson GH, Li C, Ceric O, Reimschuessel R, Cole S, Peak L, and Rankin SC. 2019. Complete genome sequence of a carbapenem-resistant Escherichia coli isolate with bla NDM-5 from a dog in the United States. Microbiol. Resour. Announce 8:e00872–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, and Harris SR. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, and Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic. Acids. Res 44: 6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu CH, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, and Klimke W. 2019. Validating the AMRFinder Tool and Resistance Gene Database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother 63:e00483–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner SN, Slezak T, and Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Stecher G, and Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pightling AW, Pettengill JB, Luo Y, Baugher JD, Rand H, and Strain E. 2018. Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front Microbiol. 9:1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riccobono E, Di Pilato V, Di Maggio T, Revollo C, Bartoloni A, Pallecchi L, and Rossolini GM. 2015. Characterization of IncI1 sequence type 71 epidemic plasmid lineage responsible for the recent dissemination of CTX-M-65 extended-spectrum beta-lactamase in the Bolivian Chaco region. Antimicrob. Agents Chemother 59:5340–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.FDA. 2018. 2017 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals. Rockville, MD: U.S. Food and Drug Administration. [Google Scholar]
- 28.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, and Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother 58:3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen E, Rahav G, and Gal-Mor O. 2020. Genome sequence of an emerging Salmonella enterica serovar Infantis and genomic comparison with other S. Infantis strains. Genome Biol. Evol 12:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granda A, Riveros M, Martínez-Puchol S, Ocampo K, Laureano-Adame L, Corujo A, Reyes I, Ruiz J, and Ochoa TJ. 2019. Presence of extended-spectrum β-lactamase, CTX-M-65 in Salmonella enterica serovar Infantis isolated from children with diarrhea in Lima, Peru. J. Pediatr. Infect. Dis 14:194–200. [Google Scholar]
- 31.Aviv G, Rahav G, and Gal-Mor O. 2016. Horizontal transfer of the Salmonella enterica Serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. mBio 7:e01395–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


