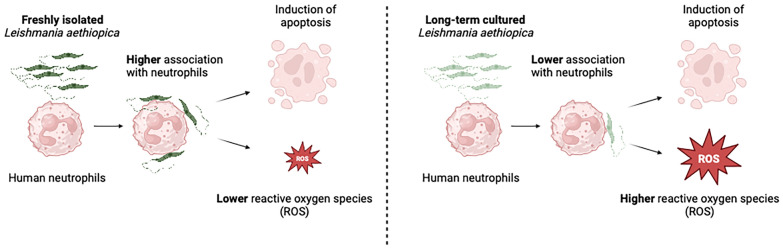
Abstract
Background
In Ethiopia, cutaneous leishmaniasis is mainly caused by Leishmania (L.) aethiopica parasites and presents in three main clinical forms. It is still not clear if the host immune response plays a role in the development of these different presentations. Since neutrophils are likely to be one of the first immune cells present at the site of the sand fly bite, we set up an in vitro model of infection of neutrophils with L. aethiopica and assessed some of the main neutrophil effector functions: association with and internalisation of parasites, apoptosis and ROS production. We used three freshly isolated clinical isolates and one isolate that has been kept in culture for decades.
Results
Our results showed by flow cytometry that all four L. aethiopica isolates had the ability to associate with neutrophils. The three clinical isolates of L. aethiopica associated more efficiently with neutrophils than the long-term cultured L. aethiopica. At 18 h, two distinct populations of neutrophils were identified that associated with L. aethiopica, CD15high and CD15low neutrophils. Confocal microscopy demonstrated that all isolates can be internalised. Our results also showed that all parasites induced apoptosis in L. aethiopica-associated neutrophils. Moreover, our results showed that after 2 h, L. aethiopica-associated neutrophils upregulated their production of ROS, but to a greater extent with the long-term cultured L. aethiopica. After 18 h of incubation, CD15lowparasite+ showed an impaired ability to produce ROS compared to CD15highparasite+.
Conclusions
Using this in vitro model, our results show that different L. aethiopica parasite isolates, most notably long-term cultured parasites, had differential effects on neutrophil effector functions.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-024-06489-x.
Keywords: Leishmania aethiopica, Neutrophils, ROS, Phagocytosis, Apoptosis
Background
Cutaneous leishmaniasis (CL) is caused by over 20 different species of Leishmania parasites, which are transmitted to their mammalian hosts during the blood meal of infected sand fly vectors. CL, the most common form of leishmaniasis, is endemic in at least 90 different countries. In 2022, over 205,000 cases were reported [1].
In Ethiopia, CL is mainly caused by Leishmania aethiopica [2] and presents in three main clinical forms: diffuse cutaneous leishmaniasis (DCL), characterised by numerous non-ulcerating nodules; mucocutaneous leishmaniasis (MCL), where the lesions affect the mucosa of the nose and/or mouth; and localised cutaneous leishmaniasis (LCL), characterised by small lesions that progress to ulcers. Whereas LCL usually heals spontaneously, it is not the case for DCL and MCL; both forms are difficult to treat, and relapses are frequent [3].
The mechanisms responsible for the development of these different clinical presentations of CL are not clearly understood. In a recent study, we showed that chemokine and cytokine levels in plasma as well as parasite genetic factors were not associated with different clinical presentations of CL [4]. However, only a small number of parasites isolated from DCL and MCL lesions were sequenced, which might explain why we did not identify individual genetic variants significantly associated with disease presentation. Other factors such as endosymbiont RNA viruses that are harboured by some species of Leishmania parasites (Leishmania RNA virus, LRV) can be an important virulence factor, as they cause a more severe form of CL, such as MCL [5]. LRV was also identified in five out of 11 isolates of L. aethiopica; the disease manifestations were not identified [6]. Host genetics has also been shown to play a role in disease development. For examples, there was an association between polymorphisms in IFN-γ and IL-4 between self-healing and chronic CL caused by Leishmania major [7]. Polymorphism located in the IL10 promoter was associated with increased risk of developing lesions in patients infected with Leishmania braziliensis [8]. However, in a recent study of 2066 CL cases caused by L. braziliensis and 2046 controls, genome-wide significance was not found [9]; yet, IFNG-AS1 was of particular interest as a non-coding antisense RNA, as it has been shown to promote IFN-γ secretion in T cells and NK cells [9]. The immune response can also influence CL development [10]. In contrast to the experimental models of CL [11–13], there is no distinct T helper (Th)1 or Th2 profile in human CL. de Mesquita et al. showed higher plasma levels of cytokines such as IFNγ, IL-1β, IL-6, IL-12p70, TNFα, IL-17, IL-1RA, IL-4, IL5, IL10, IL-13, IL-2, IL-7, IL-9, and IL-15; chemokines such as eotaxin, IL-8, IP-10, MCP-1, MIP-1α, and MIP1β; and G-CSF and GM-CSF [14] in patients infected with L. guyanensis. In another study with L. braziliensis-infected patients, antigen-stimulated PBMCs produced high levels of interferon (IFN)-γ and interleukin (IL)-4 during active disease; however, after healing, the levels of IFN-γ were maintained, but those of IL-4 were low [15]. Da-Cruz et al. showed that antigen-specific productions of IFNγ and IL-5 by PBMCs from L. braziliensis patients with mucosal leishmaniasis (ML) were elevated compared to patients with CL and that no IL-4 was detectable in CL patients, but low levels were present in ML patients [16]. In DCL patients, PBMCs are unable to mount an efficient immune response [17, 18].
During any infection, neutrophils are key cells of the innate immune response that are quickly recruited following pathogen entry. They possess an array of pathogen recognition molecules, such as Toll-like receptors (TLRs), Fcγ receptor, first and third complement receptor (CR1 and CR3) and mannose receptor [19, 20]. Once engaged, neutrophils can phagocytose and kill microbes by releasing enzymes such as myeloperoxidase and elastase in the phagosome; they can degranulate and release toxic molecules, reactive oxygen species and neutrophil extracellular traps (NETs), which may kill the pathogens in the microenvironment, as well as produce cytokines and chemokines that will promote the recruitment of other immune cells and shape the adaptive immune response [21, 22].
Most of our knowledge of neutrophil effector functions during leishmaniasis is derived from mouse models. Two-photon microscopy showed that neutrophils are quickly recruited to the site of sand fly bites [23, 24]. Leishmania amazonensis can be killed by NETs, but other species such L. donovani, L. infantum and L. mexicana can survive within NETs. Leishmania mexicana can even multiply inside neutrophils (summarised in [25, 26]). Use of neutropenic Genista mice or depletion of neutrophils with monoclonal antibodies at the time of Leishmania infection showed that neutrophils can contribute to exacerbation or control of the infection, depending on several factors, such as the route of infection, genetic background of the mice and parasite strains (summarised in [25]).
In humans, it has been well documented that neutrophils are quickly recruited to the site of inflammation [27, 28]. Since the sand fly bite results in the formation of a pool of blood, neutrophils will be present at the site of infection and further attracted in numbers; they are therefore likely to be one of the first innate immune cells to interact with Leishmania parasites.
The interactions between neutrophils and live L. aethiopica parasites have not been characterised, and it is not possible to study these interactions by using a mouse model since injection of L. aethiopica in mice does not cause symptoms [29, 30]. Therefore, the availability of an in vitro cellular model of L. aethiopica infection might be useful in identifying differences in neutrophil effector functions in response to the parasites causing different clinical forms of CL.
Here, we set up an in vitro model of infection of neutrophils with L. aethiopica to measure some of the main effector functions of neutrophils and compare these responses between infections with freshly isolated clinical isolates of L. aethiopica and long-term cultured L. aethiopica.
Methods
Sample collection
Three millilitres of blood was collected in heparin tubes from healthy non-endemic controls and was processed immediately after collection: following density gradient centrifugation on Histopaque-1077 (Sigma-Aldrich, Gillingham, UK), neutrophils were isolated from the erythrocyte fraction by dextran sulphate sedimentation, as described in [31], resuspended in Roswell Park Memorial Institute Medium (RPMI) containing 5% heat-inactivated foetal bovine serum (FBS) (Sigma-Aldrich, Gillingham, UK) (complete RPMI, cRPMI), 50 IU/ml penicillin and 50 mg/ml streptomycin (Merck, Darmstadt, Germany) and immediately used for flow cytometry. Neutrophil (as defined by CD15+ cells) purity was > 95% (gating strategy is shown in Figure S1) and their viability, as determined by 7-aminoactinomycin D (AAD) (Biolegend, London, UK), was > 99.0% (Figure S1).
Leishmania parasites
We have previously described a cohort of CL patients recruited in Nefas Mewcha, Gayint, Northern Ethiopia, from January 2019 to January 2022 [2]. Here, we used three L. aethiopica clinical isolates (L. aethiopica 1, 2 and 3) from the lesions of three different LCL patients recruited in the study described in [2]. To confirm that these isolates were L. aethiopica, we mapped RNA-seq reads from the isolates to the reference genome for L. aethiopica [32] obtained from TriTrypdb using STAR v2.7.0 [33] and then used samtools v1.17 [34] to call the consensus sequence of these mapped reads over the heat shock protein (HSP) 70 locus (LAEL147_000511500), which has been widely used to speciate Leishmania isolates. We compared these reconstructed sequences with previously obtained sequences from a range of Leishmania species (principally from [35]) using mafft v7.45 [36] to align the sequences, trimAl v2.0 [37] (with flag-strictplus), and then building a phylogeny using raxmlHPC v8.2.12 [38] (a single tree search under a GTR+gamma model of nucleotide substitution). This analysis confirmed that the isolates used here have sequences identical to L. aethiopica (accession FN395019) and differing by a single SNP from two other L. aethiopica isolates (FN395020 and FN395021). Other species formed separate groups on a phylogeny based on these sequences.
Another isolate of L. aethiopica (MHOM/ET/72/L100) [39, 40] was also used. Although it is not known precisely how long and how many times it had been kept frozen and in culture, it is known that it was isolated over 40 years ago [29]; this long-term cultured L. aethiopica isolate was therefore identified as L. aethiopica “laboratory” (L. aethiopica lab). A large stock of frozen stationary phase L. aethiopica 1, 2 and 3 and lab were prepared for further analysis. Once thawed, the parasites were used for a maximum of 3 weeks.
The following culture medium was used to keep the parasites in culture: M199 medium with 25 mM HEPES, 0.2 μM folic acid, 1 mM hemin, 1 mM adenine, 800 μM Biopterin, 50 IU/ml penicillin, 50 mg/ml streptomycin and 10% FBS (Sigma-Aldrich, Gillingham, UK) and the parasites were incubated at 26 °C. The parasites were passaged twice a week in new parasite medium.
Metacyclic promastigotes were isolated via agglutination with peanut agglutinin (PNA) (Merck, Darmstadt, Germany) as described previously [41] and were stained using CellTrace™ Far Red dye (FR) (Invitrogen, Loughborough, UK), using a 1 µM FR solution. Following a 20-min incubation at room temperature, the suspension was diluted 1 in 10 in cRPMI medium and incubated for a further 5 min at room temperature to quench any free dye remaining in the solution. At the end of the incubation, parasites were washed and resuspended in cRPMI medium and used for further experiments.
Neutrophil effector functions
Flow cytometry
Human neutrophils (1 × 105 cells/ml) were co-cultured with 1 × 106 cells/ml FR labelled L. aethiopica isolates for 2 h, at 37 °C, 5% CO2, in cRPMI. Cells were then washed twice with phosphate-buffered saline (PBS, Sigma-Aldrich, Gillingham, UK), and cells to be used for the 2 h incubation were labelled with CD15eFluor 450 (clone MMA) (eBioscience, Santa Clara, CA, USA) for 20 min at 4 °C before being washed and immediately used for flow cytometry analysis. Cells for the 18 h incubation were resuspended in cRPMI and incubated for a further 16 h at 37 °C, 5% CO2, washed twice with PBS and processed as indicated above for the 2 h incubation.
Association of parasites with neutrophils
The % of association between neutrophils [stained with anti-human CD15eFluor 450 (clone MMA), eBioscience, Santa Clara, CA, USA] and FR labelled L. aethiopica was assessed by flow cytometry.
Confocal microscopy
After 2 and 18 h of incubation, cells were transferred to poly-l-lysine (0.01% solution, Sigma-Aldrich, Gillingham, UK) coated coverslips and were incubated at room temperature. After 30 min, the cells were washed twice with PBS and fixed with 2% (w/v) paraformaldehyde (Sigma-Aldrich, Gillingham, UK) for 20 min. Cells were then washed three times with PBS and incubated with CD15 (C3D-1) mouse anti-human monoclonal antibody (Thermo Fisher Scientific, Loughborough, UK) overnight at 4 °C, followed by anti-IgG (H+L) highly cross-adsorbed secondary antibody (Alexa Fluor 555) (Thermo Fisher Scientific, Loughborough, UK). After 1 h of incubation in the dark, the coverslips were washed three times with PBS and placed onto a slide containing 50 µl mounting media (VECTASHIELD mounting media, Vector Laboratories, Newark, CA, USA). Slides were visualised under a ZEISS LSM 880 confocal laser scanning microscope (Zeiss, Cambourne, UK) under 60× magnification; 1.00 Airy unit (1AU) pinhole size was used. Image acquisition was done using Zen black software and the 3D z-stack orthogonal images were analysed by Zen 3.3 (blue edition) software.
Apoptosis
The PE-Annexin V/7-amino-actinomycin D (7-AAD) apoptosis detection kit (BioLegend, Greenwood, UK) was used to detect apoptosis according to the manufacturer’s protocol. Briefly, following the 2 and 18 h incubations with the parasites and labelling of neutrophils with CD15 antibody as described above, cells were washed and re-suspended with 100 µl Annexin V binding buffer, and 5 µl PE-Annexin V and 5 µl 7AAD (7-amino-actinomycin D) were added to the cell suspension. After 15 min of incubation in the dark at room temperature, a further 400 µl Annexin V binding buffer was added and the cells were immediately analysed by flow cytometry.
ROS detection assay
ROS-ID™ Total ROS detection kit (Enzo Life Sciences, Farmingdale, NY, USA) was used to evaluate the production of ROS by neutrophils according to the manufacturer’s protocol. Briefly, following the 2 and 18 h incubations with the parasites and labelling of neutrophils with CD15 antibody as described above, cells were washed; 25 nM ROS detection solution in 500 µl PBS was added to the cells, which were incubated for 30 min at 37 °C, 5% CO2, and immediately used for flow cytometry analysis.
Flow cytometry acquisition was performed using an LSRII (BD Biosciences, Wokingham, UK) and data were analysed using Summit v4.3 software (Beckman Coulter, Brea, CA, USA).
Statistical analysis
Data were evaluated for statistical differences as specified in the legend of each figure, using GraphPad Prism 10 (San Diego, CA, USA). The following tests were used: Mann-Whitney and Kruskal-Wallis. Results are expressed as mean ± SD. Differences were considered statistically significant at P < 0.05. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Results
Association of aethiopica parasites with neutrophils
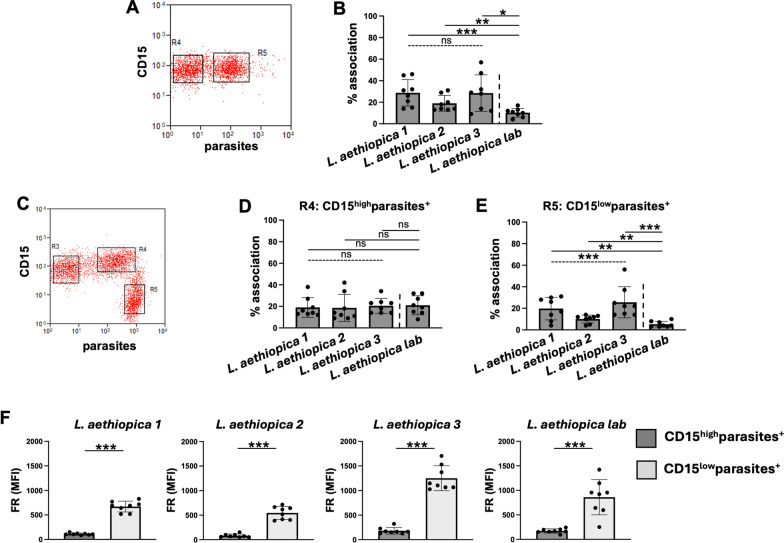
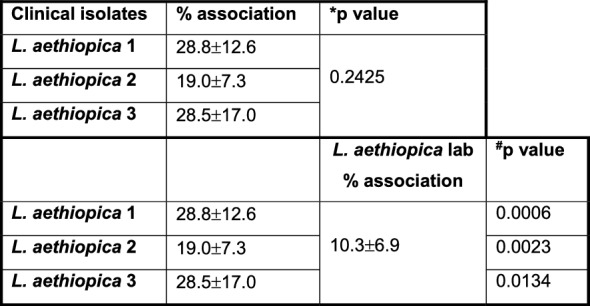
We first assessed by flow cytometry whether the three clinical and laboratory isolates of L. aethiopica can associate with neutrophils after 2 h of co-incubation; we chose a 2 h incubation as after 2 h the percentages of association between neutrophils and parasites were plateauing. Results presented in Fig. 1A, B show that L. aethiopica can associate with neutrophils and that the percentages of the L. aethiopica lab associated with neutrophils were significantly lower than for the three clinical isolates (Fig. 1B and Table 1). There were no significant differences among the three clinical isolates (Fig. 1B and Table 1).
Fig. 1.
Association of neutrophils with different isolates of Leishmania aethiopica; 1 × 105 cells/ml neutrophils were co-cultured with 1 × 106 cells/ml FR labelled L. aethiopica isolates for 2 h (A, B) and 18 h (C–F). The percentages of neutrophils associated with L. aethiopica were determined by flow cytometry. A Dot plot showing neutrophils unassociated (gate R4) and associated (gate R5) with L. aethiopica. B % of neutrophils associated with the four different isolates of L. aethiopica. C Dot plot showing the three different population of neutrophils: CD15intermediate (int)parasite− (gate R3), CD15highparasite+ (gate R4) and CD15lowparasite+ (gate R5). D % of CD15highparasite+ associated with the four different isolates of L. aethiopica. E % of CD15lowparasite+ associated with the four different isolates of L. aethiopica. F Comparison in FR MFI between CD15highparasite+ and CD15lowparasite+ for each L. aethiopica. Data are presented as scatter plot with bar (mean with standard deviation), with each dot representing the value for one experiment. Statistical differences were determined using Kruskal-Wallis (dotted line) and Mann-Whitney (solid line) tests
Table 1.
Association of neutrophils with Leishmania aethiopica following 2 h of co-incubation
1 × 105 cells/ml neutrophils were co-cultured with 1 × 106 cells/ml FR labelled L. aethiopica isolates for 2 h. The percentages of neutrophils associated with L. aethiopica were determined by flow cytometry. Statistical differences were determined using Kruskal-Wallis (*) and Mann-Whitney (#) tests
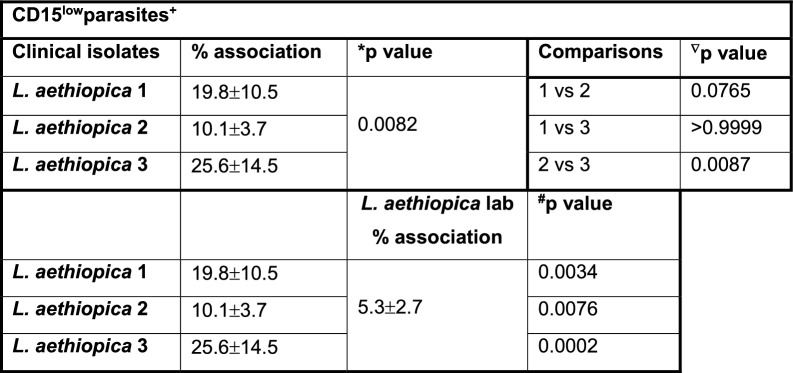
Depending on culture conditions, such as low levels of glucose or oxygen, neutrophils can have a short life span in vitro [42, 43]. However, for co-culture of neutrophils with Leishmania parasites, it has been shown to delays apoptosis for up to 24 h [44], suggesting that it prolongs neutrophil survival. Here, we first assessed whether their ability to associate with parasites was maintained after 18 h of co-culture. As shown in Fig. 1C, there were two distinct populations of neutrophils associated with the parasites: CD15high (gate R4) and CD15low (gate R5). This was the case for all four isolates of L. aethiopica (data not shown). There were no significant differences between different isolates in the percentages of parasites associated with neutrophils in the CD15highparasites+ (Fig. 1D and Table 2). However, the percentages of CD15low cells associated with L. aethiopica 2 were significantly lower compared to L. aethiopica 3 (Fig. 1E and Table 3). Furthermore, the percentages of the three clinical isolates associated with CD15low cells were all significantly higher compared to L. aethiopica lab (Fig. 1E and Table 3). Since there were differences in FR intensities among the four parasite isolates (Figure S2), it was not possible to compare the intensity of association between the neutrophils and different isolates. However, it was possible to compare the CD15high and the CD15low populations for each parasite isolate. As shown in Fig. 1F, the median fluorescent intensity (MFI) of the FR labelled parasites were always significantly higher in the CD15low population, suggesting that more parasites were associated with the CD15low population.
Table 2.
Comparison of the association of CD15high neutrophils with the different Leishmania aethiopica isolates after 18 h
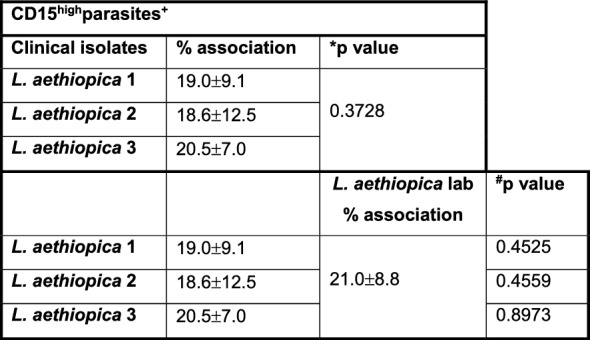
1 × 105 cells/ml neutrophils were co-cultured with 1 × 106 cells/ml FR labelled L. aethiopica isolates for 18 h. The percentages of CD15high neutrophils associated with L. aethiopica were determined by flow cytometry. Statistical differences were determined using Kruskal-Wallis (*) and Mann-Whitney (#) tests
Table 3.
Comparison of the association of CD15low neutrophils with the different Leishmania aethiopica isolates after 18 h
1 × 105 cells/ml neutrophils were co-cultured with 1 × 106 cells/ml FR labelled L. aethiopica isolates for 18 h. The percentages of neutrophils associated with L. aethiopica were determined by flow cytometry. Statistical differences were determined using Kruskal–Wallis (*), Dunn’s multiple comparison (∇) and Mann–Whitney (#) tests
Internalisation of L. aethiopica by neutrophils
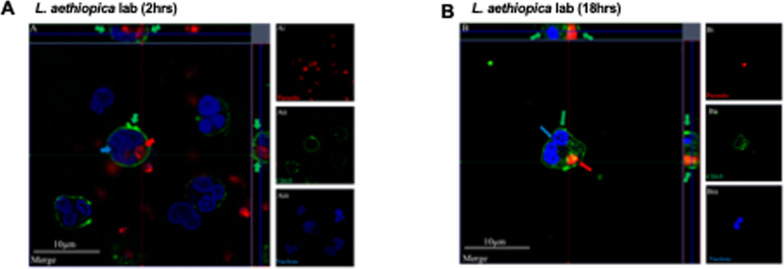
Next, confocal microscopy was used to demonstrate that L. aethiopica is internalised by neutrophils and does not only associate, as shown by flow cytometry. Results presented in Fig. 2 (L. aethiopica lab) and Figures S3 (L. aethiopica 1, 2 and 3) show that L. aethiopica parasites were internalised within neutrophils following 2 and 18 h of incubation. The top and right panels, both delineated by a grey line, show the horizontal and vertical view of the z-stack. Both show that the internalised parasites are surrounded by CD15+ neutrophil membrane.
Fig. 2.
Internalisation of Leishmania aethiopica by neutrophils; 1 × 105 cells/ml neutrophils were co-cultured with 1 × 106 cells/ml FR labelled L. aethiopica lab for 2 h (A) and 18 h (B) and cells were labelled as described in “Methods”. The red arrows point to the parasite (FR), the green arrows to the CD15 (Alexa Fluor 555) and the blue arrow to the nucleus (DAPI). These are representative images of at least three independent experiments
Neutrophil apoptosis
To determine how co-culture of neutrophils with L. aethiopica impacts their ability to undergo apoptosis, we measured the percentages of apoptotic cells [as defined by Annexin V+ 7-AAD− (early apoptosis) or Annexin V+ 7-AAD+ (late apoptosis) neutrophils]. The gating strategy is shown in Figure S4. As shown in Figure S4D, E, most gated apoptotic neutrophils were in the early stage of apoptosis. Following 2 h of incubation, the percentages of apoptotic cells were systematically increased in neutrophils associated or not with all L. aethiopica isolates compared to neutrophils alone (Table S1). Of note, the % of apoptotic neutrophils were significantly higher in neutrophils associated with the three clinical isolates compared to unassociated neutrophils; this was however not the case with L. aethiopica lab (Table S2).
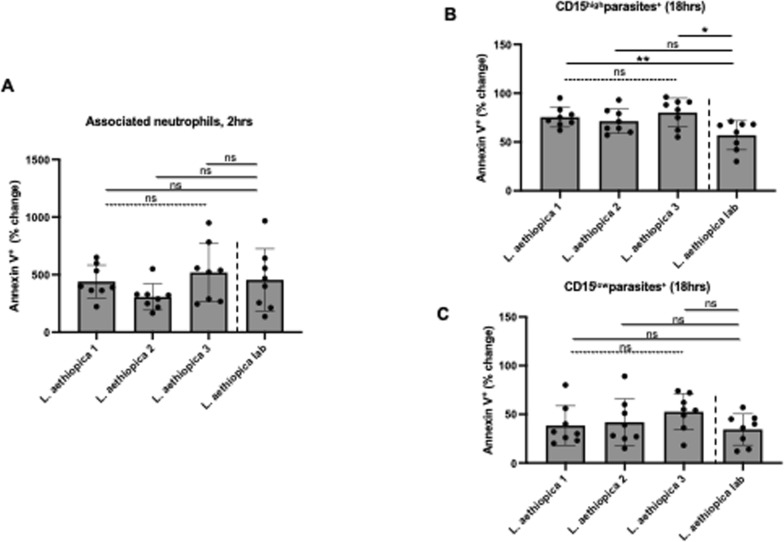
To be able to compare the % of neutrophils undergoing apoptosis following incubation with the different parasite isolates, % changes in apoptosis were assessed between baseline (neutrophils incubated without parasites) and those neutrophils co-incubated with L. aethiopica. Results show that after 2 h of incubation, there was no significant difference between any isolates in % change in apoptosis by neutrophils associated (Fig. 3A) or not (data not shown) with L. aethiopica between all isolates.
Fig. 3.
% change in apoptotic neutrophils between the different parasite isolates; 1 × 105 cells/ml neutrophils were co-cultured with 1 × 106 cells/ml FR labelled L. aethiopica isolates for 2 h (A) and 18 h (B, C). The % change was measured by deducting the % apoptotic (as defined by Annexin V+7-AAD−) CD15high neutrophils co-cultured with the parasites from the % of apoptotic neutrophils cultured in the absence of parasites. Data are presented as scatter plot with bar (mean with standard deviation), with each dot representing the value for one experiment. Statistical differences were determined using Kruskal-Wallis (dotted line) and Mann-Whitney (solid line) tests
After 18 h of incubation (gating strategy shown in Figure S5), most gated apoptotic neutrophils were in the early stage of apoptosis (Figure S5). Results presented in Table S3 show that for all isolates, the % Annexin V+ 7-AAD− neutrophils were similar between all four parasite isolates in CD15intparasites− and lower in CD15highparasites+ and CD15lowparasites+ compared to baseline. When comparing the % Annexin V+ 7-AAD− neutrophils among the three populations of neutrophils, it was always highest in the CD15intparasites+ and lowest in the CD15lowparasites+ (Table S4).
There were no significant differences in % change in the frequency of apoptotic cells among the four different isolates in CD15intparasite− (data not shown). In the CD15highparasites+, the % change was significantly higher with L. aethiopica 1 and 3, but not 2, compared to L. aethiopica lab (Fig. 3B and Table 4). However, there were no significant differences in % change between any parasites isolates in the CD15lowparasites+ (Fig. 3C).
Table 4.
% change in apoptotic CD15high neutrophils between the different parasite isolates after 18 h
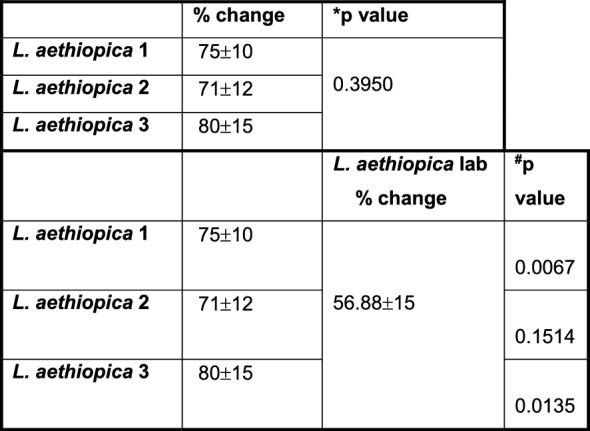
1 × 105 cells/ml neutrophils were cultured in the presence or the absence of 1 × 106 cells/ml FR labelled L. aethiopica isolates for 18 h. The % change was measured by deducting the % apoptotic (as defined by Annexin V+7AAD−) CD15high neutrophils co-cultured with the parasites from the % of apoptotic neutrophils cultured in the absence of parasites. Statistical differences were determined using Kruskal-Wallis (*) and Mann-Whitney (#) tests
ROS production by L. aethiopica-associated neutrophils
Next, we assessed the ability of neutrophils to upregulate ROS following co-incubation with L. aethiopica (gating strategy shown in Figure S6). Following 2 h of incubation, ROS production (MFI) was systematically significantly increased in L. aethiopica-associated neutrophils, but it was not significantly higher in unassociated neutrophils, except for a borderline difference with L. aethiopica 2 (P = 0.0433, Table S5). Of note, the levels of ROS production (MFI) were significantly higher in L. aethiopica-associated neutrophils than in unassociated ones (Table S6).
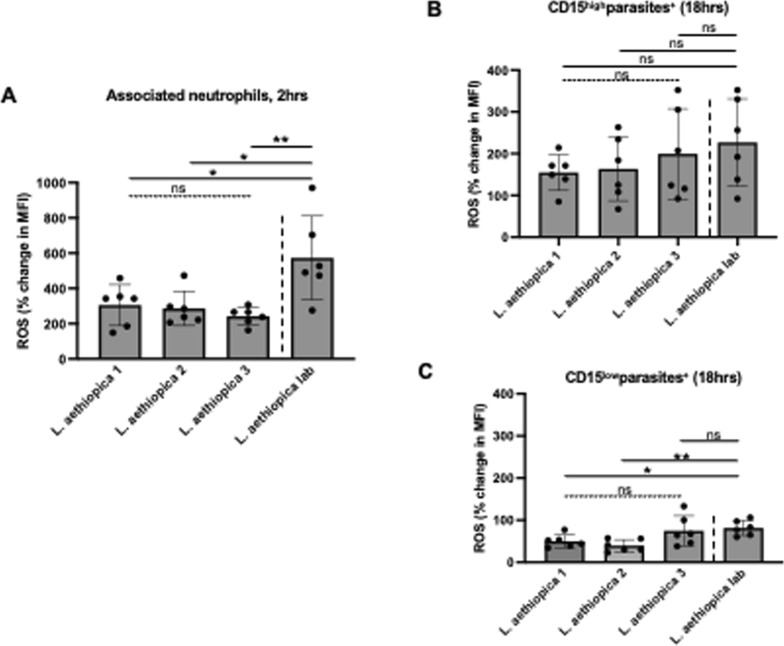
To be able to compare the levels of ROS production by neutrophils between the different parasite isolates, % changes in ROS production were assessed between baseline (neutrophils incubated without parasites) and those neutrophils co-incubated with L. aethiopica. Results shown in Fig. 4A and Table 5 show that after 2 h of incubation, there was no significant difference in % change in ROS production by neutrophils associated with L. aethiopica among the three clinical isolates. However, it was significantly higher for L. aethiopica lab compared to the clinical isolates (Fig. 4A and Table 5). There was no significant difference in ROS production among all four isolates in the L. aethiopica-unassociated neutrophils (data not shown).
Fig. 4.
% change in ROS MFI in neutrophils between the different parasite isolates; 1 × 105 cells/ml neutrophils were co-cultured with 1 × 106 cells/ml FR labelled L. aethiopica isolates for 2 h (A) and 18 h (B, C). The % change was measured by deducting the ROS MFI in the neutrophils co-cultured with the parasites from the ROS MFI in the neutrophils cultured in the absence of parasites. Data are presented as scatter plot with bar (mean with standard deviation), with each dot representing the value for one experiment. Statistical differences were determined using Kruskal-Wallis (dotted line) and Mann-Whitney (solid line) tests
Table 5.
% change in ROS MFI in neutrophils between the different parasite isolates after 2 h
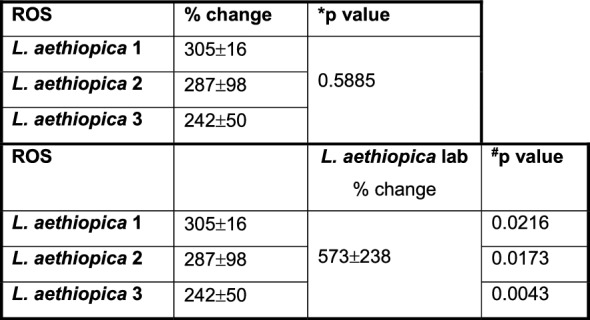
1 × 105 cells/ml neutrophils were cultured in the presence or the absence of 1 × 106 cells/ml FR labelled L. aethiopica isolates for 2 h. The % change was measured by deducting the ROS MFI in the neutrophils co-cultured with the parasites from the ROS MFI in the neutrophils cultured in the absence of parasites. Statistical differences were determined using Kruskal-Wallis (*) and Mann-Whitney (#) tests
After 18 h of incubation (gating strategy shown in Figure S7), results presented in Table S7 show that ROS MFI was similar when comparing baseline with CD15intparasites− and CD15highparasites+ neutrophils for all four L. aethiopica isolates. However, ROS MFI was significantly lower in CD15lowparasites+ compared to baseline for all three clinical isolates, but not L. aethiopica lab (Table S7). When comparing ROS production between the three populations of neutrophils, the CD15highparasites+ always produced the highest levels of ROS compared to CD15lowparasites+ (Table S8). There were no significant differences in % change among the four different isolates in CD15highparasite+ (Fig. 4B) and CD15intparasite− (data not shown). However, the % changes in ROS production in CD15lowparasite+ neutrophils were lower with L. aethiopica 1 and 2, but not 3, compared to L. aethiopica lab (Fig. 4C and Table 6).
Table 6.
% change in ROS MFI in CD15low neutrophils between the different parasite isolates after 18 h
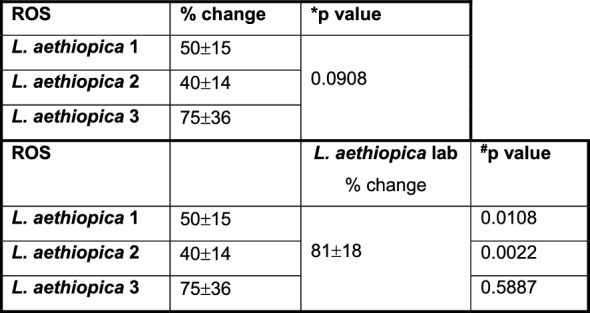
1 × 105 cells/ml neutrophils were cultured in the presence or the absence of 1 × 106 cells/ml FR labelled L. aethiopica isolates for 18 h. The % change was measured by deducting the ROS MFI in the CD15low neutrophils co-cultured with the parasites from the ROS MFI in the CD15low neutrophils cultured in the absence of parasites. Statistical differences were determined using Kruskal-Wallis (*) and Mann-Whitney (#) tests
Discussion
Here we set up an in vitro model of infection of human neutrophils with L. aethiopica and show that parasites can be phagocytosed by neutrophils and that association of neutrophils with L. aethiopica modulates apoptosis and ROS production. Our results also show that L. aethiopica lab associated less and induced more ROS production compared to freshly isolated L. aethiopica. We also identify some differences in the percentages of associated parasites among the three clinical isolates. Many studies imply that association as shown by flow cytometry equates to phagocytosis. However, by using flow cytometry alone, it is not possible to differentiate cells that are bound to neutrophils from those that have been internalised [45]. Techniques such as imaging flow cytometry or confocal microscopy need to be used to validate internalisation [45].
Little is known about the mechanisms influencing the different clinical presentations of cutaneous lesions caused by L. aethiopica. It has been previously suggested that differences in parasites are associated with the different clinical manifestations [46–48]. A later study suggested that the genetic variability did not correlate with the different manifestations [49]. In agreement with these results, in the largest study to date analysing genetic variations between L. aethiopica isolated from different lesions, we found no individual genetic variants were significantly associated with disease presentation [4]. It is also possible that Leishmania RNA viruses play a role in the development of disease manifestations [5, 6, 50].
Here, we used three L. aethiopica parasites freshly isolated from patients with localised cutaneous leishmaniasis (LCL) and one isolate that had been kept in culture for decades; the history of the number of in vitro passages and its origin have been poorly characterised [29]. Leishmania aethiopica parasites cannot be passaged in vivo [29, 30] to maintain virulence [51–53]. Therefore, to minimise variations due to the time in culture, once the parasites were growing from the cultured skin scrapings from CL patients, they were immediately frozen and shipped to the UK. Large stocks of parasites were produced and frozen, and once thawed, the parasites were kept in culture for a maximum of 3 weeks before a new tube was thawed. It has been shown that the metacyclogenesis of parasites is key in determining their virulence [54]. Therefore, to maximise our control over the stage of parasites used in these experiments, parasites were grown to a stationary phase and metacyclic parasites were purified using PNA.
Long-term in vitro culture has been associated with the loss of virulence of Leishmania parasites in both phagocytic cells, as shown by fewer amastigotes per cells [51–53], and animal models, as shown by a lower parasite burden or smaller lesions [53, 55, 56]. Several virulence factors have been identified in Leishmania parasites [57]. In particular, reduced expressions of lipophosphoglycan (LPG) and glycoprotein (GP) 63 have been shown to result from long-term in vitro culture [58, 59]. Both these molecules are important in the phagocytosis of Leishmania parasites [19, 60]. This might therefore explain why L. aethiopica lab associated significantly less with neutrophils than with the freshly isolated L. aethiopica parasites.
Interestingly, following 18 h of incubation, in addition to a population of neutrophils that did not associate with L. aethiopica, two other populations of neutrophils associated with L. aethiopica were identified based on the expression levels of CD15. These results show that following co-incubation of neutrophils with L. aethiopica, at least three populations of neutrophils can be identified that have different abilities to associate with the parasites. The heterogeneity of neutrophils is now well recognised [22, 61, 62]. scRNAseq analysis of circulating neutrophils showed three major populations [63]. Another study showed a high level of heterogeneity of neutrophils following phenotypic characterisation of peripheral neutrophils in healthy individuals and compared to patients with different pathological conditions [64]. We also found differences in the percentages of associated CD15low neutrophils among the three clinical isolates at 18 h; these results suggest that even though all three clinical isolates were obtained from lesions of LCL patients, there still might be differences between these parasites.
Infection of neutrophils by L. major and L. infantum has been shown to result in delayed apoptosis over time, suggesting that the parasites prolong the survival of neutrophils, thereby allowing the intracellular parasites to survive longer [44, 65]. Our results show that after 2 h of incubation, there were increased percentages of apoptotic associated and unassociated neutrophils compared to baseline. This was in contrast to 18 h, when there were significantly lower percentages of apoptotic associated neutrophils (CD15high and CD15low), but not unassociated neutrophils (CD15int), compared to baseline. The study by van Zandbergen et al. showed that apoptotic neutrophils can be phagocytosed by macrophages and that the phagocytosed parasites are then able to survive and multiply in these macrophages [66]. It is therefore possible that it will also be the case with L. aethiopica and that apoptotic neutrophils will be taken up by phagocytic cells, monocytes in the blood and macrophages if they enter tissues. It has been previously shown that compared to uninfected neutrophils, L. major-infected neutrophils have an increased ability to take up noninfected apoptotic cells [67]; this was associated with the downregulation of ROS production and better survival of parasites in the neutrophils. Thus, the high percentages of unassociated neutrophils identified in our study might contribute to better survival of the intracellular parasites. Whereas it has been shown that Leishmania-infected neutrophils undergo apoptosis [44, 65–67], there is little information about unassociated neutrophils undergoing apoptosis. They might become apoptotic because of a transient contact with Leishmania parasites, induction of ROS or production of cytokines such as TNFα (summarised in [68]).
In our study, after 18 h, there were fewer apoptotic cells in associated neutrophils compared to baseline. This contradicts the study by Oualha et al., as they show an increase in the percentages of apoptotic neutrophils following co-incubation with L. major and L. infantum compared to baseline. This might be due to differences in the experimental conditions, such as different parasite strains, a tenfold higher number of neutrophils co-cultured with parasites and differences in the stage of the parasites, as in this study, they did not use PNA to isolate the metacyclic parasites [65].
It has been previously shown that infection of neutrophils with different parasite species such as L. infantum [69, 70], L. braziliensis [71] and L. donovani [72] results in upregulation of ROS. One study showed higher levels of ROS by neutrophils in response to L. aethiopica; however, lysate and not live L. aethiopica were used in this study [73]. In our study, ROS was upregulated in neutrophils following 2 h of co-culture with all parasite isolates. ROS was also increased in unassociated neutrophils; this is likely caused by transient contact with the parasites during the 2 h co-culture. In contrast, after 18 h, ROS was similar to baseline levels in CD15intparasites− and in CD15highparasites+ neutrophils and significantly lower in CD15lowparasites+. The latter subpopulation was also the population that had the highest % of associated parasites, suggesting that the parasites might manipulate the neutrophils to limit the levels of ROS production to survive more efficiently. In support of these data, ROS had already been shown to be reduced following exposure of L. major-infected neutrophils to apoptotic cells [67]. Furthermore, a study by Mollinedo et al. has shown that L. major- and L. donovani-containing phagosomes do not fuse with specific and tertiary granules, thereby preventing the production of ROS [74]. Al Tuwaijri et al. have also shown that different preparations of L. major parasites reduced the respiratory burst by neutrophils [75].
Of note, L. aethiopica lab induced higher levels of ROS at both 2 and 18 h compared to the clinical isolates. Both LPG [76] and GP63 [77] have been shown to inhibit the oxidative burst. As discussed above, the extensive time in culture might have resulted in lower expression levels of LPG and GP63 on L. aethiopica lab and thereby impacted the levels of ROS production.
Conclusions
In this study we characterised, for the first time to our knowledge, neutrophil effector functions in response to different isolates of L. aethiopica. Our results show that care should be taken when using parasites that have been kept in culture for a long time and highlight that a more standardised way to isolate primary cells and infectious metacyclic parasites should be used.
In the absence of a mouse model, in vitro infection of phagocytic cells might identify different functional profiles that could shed light on the different presentation of lesions caused by L. aethiopica.
Supplementary Information
Acknowledgements
The authors are thankful to staff of Nefas Mewcha Hospital for their enthusiastic collaboration.
Author contributions
Conception of the study: EA, GG, PK. Acquisition of the data: EA, ECC, EY, YT, PK. Analysis: EA, ECC, YT, SS, JAC, GG, PK. Interpretation of the data: EA, ECC, YT, SS, JAC, GG, PK. Draft of the manuscript: EA, PK. Revision of the manuscript: EA, ECC, EY, YT, SS, JAC, GG, PK.
Funding
This research is jointly funded by the UK Medical Research Council (MRC) and the Foreign Commonwealth and Development Office (FCDO) under the MRC/FCDO Concordat agreement (MR/R021600/1) (EY, JAC, PK) and “Applied Biosciences for Health” Marie Sklodowska-Curie grant 801604 (EA, GG).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Declarations
Ethics approval and consent to participate
Ethical approval was obtained from the Faculty Ethic Committee University of Greenwich (FES-FREC-20-01.04.08.CA). Informed written consent was obtained from each participant.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
G. Getti and P. Kropf share last authorship.
Contributor Information
G. Getti, Email: g.t.m.getti@greenwich.ac.uk
P. Kropf, Email: p.kropf@imperial.ac.uk
References
- 1.Ruiz-Postigo JA, Jain S, Madjou S, Virrey Agua JF, Maia-Elkhoury AN, Valadas S, et al. Global leishmaniasis surveillance, 2022: assessing trends over the past 10 years. Wkly Epidemiol Rec. 2023;40:471–87. [Google Scholar]
- 2.Yizengaw E, Gashaw B, Yimer M, Takele Y, Nibret E, Yismaw G, et al. Demographic characteristics and clinical features of patients presenting with different forms of cutaneous leishmaniasis, in Lay Gayint, northern Ethiopia. PLoS Negl Trop Dis. 2024;18:e0012409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Henten S, Adriaensen W, Fikre H, Akuffo H, Diro E, Hailu A, et al. Cutaneous leishmaniasis due to Leishmania aethiopica. EClinicalMedicine. 2018;6:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yizengaw E, Takele Y, Franssen SU, Gashaw B, Yimer M, Abera A, et al. Parasite genetic variation and systemic immune responses are not associated with different clinical presentations of cutaneous leishmaniasis caused by Leishmania aethiopica. BioRxiv. 2024. 10.1101/2024.04.19.590259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Carvalho RVH, Lima-Junior DS, da Silva MVG, Dilucca M, Rodrigues TS, Horta CV, et al. Leishmania RNA virus exacerbates leishmaniasis by subverting innate immunity via TLR3-mediated NLRP3 inflammasome inhibition. Nat Commun. 2019;10:5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zangger H, Hailu A, Desponds C, Lye LF, Akopyants NS, Dobson DE, et al. Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis. 2014;8:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamali-Sarvestani E, Rasouli M, Mortazavi H, Gharesi-Fard B. Cytokine gene polymorphisms and susceptibility to cutaneous leishmaniasis in Iranian patients. Cytokine. 2006;35:159–65. [DOI] [PubMed] [Google Scholar]
- 8.Salhi A, Rodrigues V Jr, Santoro F, Dessein H, Romano A, Castellano LR, et al. Immunological and genetic evidence for a crucial role of IL-10 in cutaneous lesions in humans infected with Leishmania braziliensis. J Immunol. 2008;180:6139–48. [DOI] [PubMed] [Google Scholar]
- 9.Blackwell JM, Fakiola M, Castellucci LC. Human genetics of leishmania infections. Hum Genet. 2020;139:813–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novais FO, Amorim CF, Scott P. Host-directed therapies for cutaneous leishmaniasis. Front Immunol. 2021;12:660183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sacks DL, Noben-Trauth N. The immunology of susceptibility and resistance to Leishmania major in mice. Nat Rev Immunol. 2002;2:845–58. [DOI] [PubMed] [Google Scholar]
- 12.Kaye P, Scott P. Leishmaniasis: complexity at the host–pathogen interface. Nat Rev Microbiol. 2011;9:604–15. [DOI] [PubMed] [Google Scholar]
- 13.Rossi M, Fasel N. The criminal association of Leishmania parasites and viruses. Curr Opin Microbiol. 2018;46:65–72. [DOI] [PubMed] [Google Scholar]
- 14.de Mesquita TGR, Junior J, da Silva LDO, Silva GAV, de Araujo FJ, Pinheiro SK, et al. Distinct plasma chemokines and cytokines signatures in Leishmania guyanensis-infected patients with cutaneous leishmaniasis. Front Immunol. 2022;13:974051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho SG, Da-Cruz AM, Bertho AL, Santiago MA, De-Luca P. Immunologic patterns associated with cure in human American cutaneous leishmaniasis. Braz J Med Biol Res. 1998;31:139–42. [DOI] [PubMed] [Google Scholar]
- 16.Da-Cruz AM, Bittar R, Mattos M, Oliveira-Neto MP, Nogueira R, Pinho-Ribeiro V, et al. T-cell-mediated immune responses in patients with cutaneous or mucosal leishmaniasis: long-term evaluation after therapy. Clin Diagn Lab Immunol. 2002;9:251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Convit J, Pinardi ME, Rondon AJ. Diffuse cutaneous leishmaniasis: a disease due to an immunological defect of the host. Trans R Soc Trop Med Hyg. 1972;66:603–10. [DOI] [PubMed] [Google Scholar]
- 18.Machado GU, Prates FV, Machado PRL. Disseminated leishmaniasis: clinical, pathogenic, and therapeutic aspects. An Bras Dermatol. 2019;94:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno N, Wilson ME. Receptor-mediated phagocytosis of Leishmania: implications for intracellular survival. Trends Parasitol. 2012;28:335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossi M, Fasel N. How to master the host immune system? Leishmania parasites have the solutions! Int Immunol. 2018;30:103–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burn GL, Foti A, Marsman G, Patel DF, Zychlinsky A. The neutrophil. Immunity. 2021;54:1377–91. [DOI] [PubMed] [Google Scholar]
- 22.Aroca-Crevillen A, Vicanolo T, Ovadia S, Hidalgo A. Neutrophils in physiology and pathology. Annu Rev Pathol. 2024;19:227–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters NC, Egen JG, Secundino N, Debrabant A, Kimblin N, Kamhawi S, et al. In vivo imaging reveals an essential role for neutrophils in leishmaniasis transmitted by sand flies. Science. 2008;321:970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurrell BP, Schuster S, Grun E, Coutaz M, Williams RA, Held W, et al. Rapid sequestration of Leishmania mexicana by neutrophils contributes to the development of chronic lesion. PLoS Pathog. 2015;11:e1004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurrell BP, Regli IB, Tacchini-Cottier F. Different Leishmania species drive distinct neutrophil functions. Trends Parasitol. 2016;32:392–401. [DOI] [PubMed] [Google Scholar]
- 26.Regli IB, Passelli K, Hurrell BP, Tacchini-Cottier F. Survival mechanisms used by some Leishmania species to escape neutrophil killing. Front Immunol. 2017;8:1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–75. [DOI] [PubMed] [Google Scholar]
- 28.Di Domizio J, Belkhodja C, Chenuet P, Fries A, Murray T, Mondejar PM, et al. The commensal skin microbiota triggers type I IFN-dependent innate repair responses in injured skin. Nat Immunol. 2020;21:1034–45. [DOI] [PubMed] [Google Scholar]
- 29.Humber DP, Hetherington CM, Atlaw T, Eriso F. Leishmania aethiopica: infections in laboratory animals. Exp Parasitol. 1989;68:155–9. [DOI] [PubMed] [Google Scholar]
- 30.Akuffo HO, Walford C, Nilsen R. The pathogenesis of Leishmania aethiopica infection in BALB/c mice. Scand J Immunol. 1990;32:103–10. [DOI] [PubMed] [Google Scholar]
- 31.Cloke T, Garvery L, Choi BS, Abebe T, Hailu A, Hancock M, et al. Increased arginase activity correlates with disease severity in HIV seropositive patients. J Infect Dis. 2010;202:374–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren WC, Akopyants NS, Dobson DE, Hertz-Fowler C, Lye LF, Myler PJ, et al. Genome assemblies across the diverse evolutionary spectrum of Leishmania protozoan parasites. Microbiol Resour Announc. 2021;10:e0054521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G. Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol. 2010;10:238–45. [DOI] [PubMed] [Google Scholar]
- 36.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beverley SM, Ismach RB, Pratt DM. Evolution of the genus Leishmania as revealed by comparisons of nuclear DNA restriction fragment patterns. Proc Natl Acad Sci USA. 1987;84:484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Getti GT, Aslam SN, Humber DP, Stevenson PC, Cheke RA. The effect of cicerfuran, an arylbenzofuran from Cicer bijugum, and related benzofurans and stilbenes on Leishmania aethiopica, L. tropica and L. major. Planta Med. 2006;72:907–11. [DOI] [PubMed] [Google Scholar]
- 41.Sacks DL, Hieny S, Sher A. Identification of cell surface carbohydrate and antigenic changes between noninfective and infective developmental stages of Leishmania major promastigotes. J Immunol. 1985;135:564–9. [PubMed] [Google Scholar]
- 42.Scheel-Toellner D, Wang K, Craddock R, Webb PR, McGettrick HM, Assi LK, et al. Reactive oxygen species limit neutrophil life span by activating death receptor signaling. Blood. 2004;104:2557–64. [DOI] [PubMed] [Google Scholar]
- 43.Blanter M, Gouwy M, Struyf S. Studying neutrophil function in vitro: cell models and environmental factors. J Inflamm Res. 2021;14:141–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aga E, Katschinski DM, van Zandbergen G, Laufs H, Hansen B, Müller K, et al. Inhibition of spontaneous apoptosis of neutrophil granulocytes by the intracellular parasite Leishmania major. J Immunol. 2002;169:898–905. [DOI] [PubMed] [Google Scholar]
- 45.Smirnov A, Solga MD, Lannigan J, Criss AK. An improved method for differentiating cell-bound from internalized particles by imaging flow cytometry. J Immunol Methods. 2015;423:60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akuffo H, Schurr E, Andersson G, Yamaneberhan T, Britton S. Responsiveness in diffuse versus local cutaneous leishmaniasis is due to parasite differences. Scand J Immunol. 1987;26:717–21. [DOI] [PubMed] [Google Scholar]
- 47.Akuffo HO, Fehniger TE, Britton S. Differential recognition of Leishmania aethiopica antigens by lymphocytes from patients with local and diffuse cutaneous leishmaniasis. Evidence for antigen-induced immune suppression. J Immunol. 1988;141:2461–6. [PubMed] [Google Scholar]
- 48.Akuffo H, Maasho K, Blostedt M, Hojeberg B, Britton S, Bakhiet M. Leishmania aethiopica derived from diffuse leishmaniasis patients preferentially induce mRNA for interleukin-10 while those from localized leishmaniasis patients induce interferon-gamma. J Infect Dis. 1997;175:737–41. [DOI] [PubMed] [Google Scholar]
- 49.Schönian G, Akuffo H, Lewin S, Maasho K, Nylen S, Pratlong F, et al. Genetic variability within the species Leishmania aethiopica does not correlate with clinical variations of cutaneous leishmaniasis. Mol Biochem Parasitol. 2000;239:239–48. [DOI] [PubMed] [Google Scholar]
- 50.Shita EY, Semegn EN, Wubetu GY, Abitew AM, Andualem BG, Alemneh MG. Prevalence of Leishmania RNA virus in Leishmania parasites in patients with tegumentary leishmaniasis: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2022;16:e0010427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grimm F, Brun R, Jenni L. Promastigote infectivity in Leishmania infantum. Parasitol Res. 1991;77:185–91. [DOI] [PubMed] [Google Scholar]
- 52.Magalhaes RD, Duarte MC, Mattos EC, Martins VT, Lage PS, Chavez-Fumagalli MA, et al. Identification of differentially expressed proteins from Leishmania amazonensis associated with the loss of virulence of the parasites. PLoS Negl Trop Dis. 2014;8:e2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinha R, Mathumalar C, Raghwan, Das S, Shadab M, et al. Genome plasticity in cultured Leishmania donovani: comparison of early and late passages. Front Microbiol. 2018;9:1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DaSilva R, Sacks DL. Metacyclogenesis is a major determinant of Leishmania promastigote virulence and attenuation. Infect Immun. 1987;55:2802–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segovia M, Artero JM, Mellado E, Chance ML. Effects of long-term in vitro cultivation on the virulence of cloned lines of Leishmania major promastigotes. Ann Trop Med Parasitol. 1992;86:347–54. [DOI] [PubMed] [Google Scholar]
- 56.Moreira D, Santarem N, Loureiro I, Tavares J, Silva AM, Amorim AM, et al. Impact of continuous axenic cultivation in Leishmania infantum virulence. PLoS Negl Trop Dis. 2012;6:e1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumari D, Mahajan S, Kour P, Singh K. Virulence factors of Leishmania parasite: their paramount importance in unraveling novel vaccine candidates and therapeutic targets. Life Sci. 2022;306:120829. [DOI] [PubMed] [Google Scholar]
- 58.Mukhopadhyay S, Sen P, Majumder HK, Roy S. Reduced expression of lipophosphoglycan (LPG) and kinetoplastid membrane protein (KMP)-11 in Leishmania donovani promastigotes in axenic culture. J Parasitol. 1998;84:644–7. [PubMed] [Google Scholar]
- 59.Ali KS, Rees RC, Terrell-Nield C, Ali SA. Virulence loss and amastigote transformation failure determine host cell responses to Leishmania mexicana. Parasite Immunol. 2013;35:441–56. [DOI] [PubMed] [Google Scholar]
- 60.Kaushal RS, Naik N, Prajapati M, Rane S, Raulji H, Afu NF, et al. Leishmania species: a narrative review on surface proteins with structural aspects involved in host–pathogen interaction. Chem Biol Drug Des. 2023;102:332–56. [DOI] [PubMed] [Google Scholar]
- 61.Ng LG, Ostuni R, Hidalgo A. Heterogeneity of neutrophils. Nat Rev Immunol. 2019;19:255–65. [DOI] [PubMed] [Google Scholar]
- 62.Hedrick CC, Malanchi I. Neutrophils in cancer: heterogeneous and multifaceted. Nat Rev Immunol. 2022;22:173–87. [DOI] [PubMed] [Google Scholar]
- 63.Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su J, et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat Immunol. 2020;21:1119–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montaldo E, Lusito E, Bianchessi V, Caronni N, Scala S, Basso-Ricci L, et al. Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress. Nat Immunol. 2022;23:1470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oualha R, Barhoumi M, Marzouki S, Harigua-Souiai E, Ben Ahmed M, Guizani I. Infection of human neutrophils with Leishmania infantum or Leishmania major strains triggers activation and differential cytokines release. Front Cell Infect Microbiol. 2019;9:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Zandbergen G, Klinger M, Mueller A, Dannenberg S, Gebert A, Solbach W, et al. Cutting edge: neutrophil granulocyte serves as a vector for Leishmania entry into macrophages. J Immunol. 2004;173:6521–5. [DOI] [PubMed] [Google Scholar]
- 67.Salei N, Hellberg L, Kohl J, Laskay T. Enhanced survival of Leishmania major in neutrophil granulocytes in the presence of apoptotic cells. PLoS ONE. 2017;12:e0171850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Figueroa E, Alvarez-Carrasco P, Ortega E, Maldonado-Bernal C. Neutrophils: many ways to die. Front Immunol. 2021;12:631821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quintela-Carvalho G, Luz NF, Celes FS, Zanette DL, Andrade D, Menezes D, et al. Heme drives oxidative stress-associated cell death in human neutrophils infected with Leishmania infantum. Front Immunol. 2017;8:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quintela-Carvalho G, Goicochea AMC, Mancur-Santos V, Viana SM, Luz YDS, Dias BRS, et al. Leishmania infantum defective in lipophosphoglycan biosynthesis interferes with activation of human neutrophils. Front Cell Infect Microbiol. 2022;12:788196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conceicao J, Davis R, Carneiro PP, Giudice A, Muniz AC, Wilson ME, et al. Characterization of neutrophil function in human cutaneous leishmaniasis caused by Leishmania braziliensis. PLoS Negl Trop Dis. 2016;10:e0004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yizengaw E, Getahun M, Tajebe F, Cruz Cervera E, Adem E, Mesfin G, et al. Visceral leishmaniasis patients display altered composition and maturity of neutrophils as well as impaired neutrophil effector functions. Front Immunol. 2016;7:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chanyalew M, Abebe M, Endale B, Girma S, Tasew G, Bobosha K, et al. Enhanced activation of blood neutrophils and monocytes in patients with Ethiopian localized cutaneous leishmaniasis in response to Leishmania aethiopica neutrophil activation in Ethiopian cutaneous leishmaniasis. Acta Trop. 2021;220:105967. [DOI] [PubMed] [Google Scholar]
- 74.Mollinedo F, Janssen H, de la Iglesia-Vicente J, Villa-Pulgarin JA, Calafat J. Selective fusion of azurophilic granules with Leishmania-containing phagosomes in human neutrophils. J Biol Chem. 2010;285:34528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Al Tuwaijri AS, Al Mofleh IA, Mahmoud AA. Effect of Leishmania major on human polymorphonuclear leucocyte function in vitro. J Med Microbiol. 1990;32:189–93. [DOI] [PubMed] [Google Scholar]
- 76.Lodge R, Diallo TO, Descoteaux A. Leishmania donovani lipophosphoglycan blocks NADPH oxidase assembly at the phagosome membrane. Cell Microbiol. 2006;8:1922–31. [DOI] [PubMed] [Google Scholar]
- 77.Shio MT, Christian JG, Jung JY, Chang KP, Olivier M. PKC/ROS-mediated NLRP3 inflammasome activation is attenuated by Leishmania zinc-metalloprotease during infection. PLoS Negl Trop Dis. 2015;9:e0003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.