ABSTRACT
A patient with fever presented to the referral infectious disease hospital in Kathmandu, Nepal. Metagenomic sequencing of the patient’s serum recovered a near-complete genome of human immunodeficiency virus-1 (HIV-1), distinct from previous HIV-1 genomes from Nepal in GenBank. It shared 92.48% nucleotide identity with an HIV-1 subtype C isolate from India.
KEYWORDS: HIV, fever of unknown origin, WGS
ANNOUNCEMENT
Human immunodeficiency virus-1 (HIV-1) is a member of the genus Lentivirus in the family Retroviridae. GenBank contains only a limited number of near-complete HIV-1 genome sequences from Nepal. The most recent was reported in 2020 from samples collected in 2015.
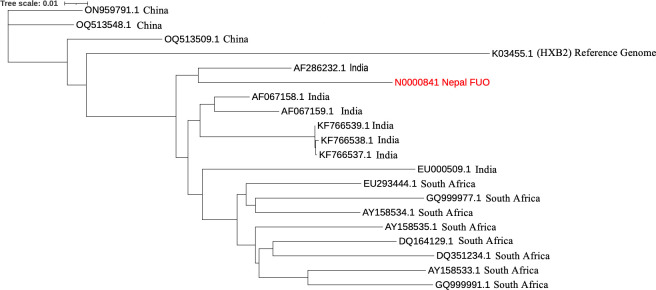
A 51-year-old male presented with fever of unknown origin (FUO) at Sukraraj Tropical and Infectious Disease Hospital (STIDH), Kathmandu, Nepal, in June 2023. Total nucleic acids were extracted from the patient’s serum using an Invitrogen Pure Link Viral RNA/DNA Mini Kit (Thermo Fisher Scientific) following the instruction manual and eluted in 50 µL RNase-free water. cDNA was obtained by reverse transcription using M-MLV Reverse Transcriptase (Promega) with a 31-base random primer (TACCGTAGAGCTGCTANNNNNNNNNNNNNNN). This was followed by second-strand synthesis with Sequenase Enzyme (Applied Biosystems, Thermo Fisher Scientific) and PCR amplification using a primer consisting of the first 16 bases of the reverse transcription primer (TACCGTAGAGCTGCTA) (1, 2). A library was constructed using the Illumina DNA Prep kit and sequenced (2 × 150 bp paired-end) on the Illumina NextSeq 500. A total number of 5,019,323 reads were obtained. Next generation sequencing (NGS) data were analyzed for the presence of viral sequences using the cloud-based platform CZ ID (3). Many HIV reads were detected; therefore, MEGAHIT v.1.2.9 (4) was used to assemble the raw reads, generating multiple contigs, of which one, 8,792 nucleotides long contig (N0000841 Nepal FUO) with an average read depth of 7×, mapped to HIV-1. The average length of the individual HIV sequence reads was 150 nucleotides. The GC content of the contig was 41.47%. The contig shared 92.48% identity with the HIV-1 subtype C isolate reported from India (accession # AF067158.1) via NCBI blastn (Date of access: 5 June 2024). VAPiD (5) v.1.6.7 was used for annotation with AF067158.1 as the reference genome. A maximum likelihood phylogenetic tree was generated with the 18 whole genome sequences of HIV-1 with highest identity % blastn scores downloaded from NCBI (accessed on 5 June 2024) with 1,000 bootstrap replications using W-IQ-TREE (6) and visualized using iTOL (7) (Fig. 1). The multiple sequence alignment was performed using Clustal Omega (8). Default parameters were used except where otherwise noted. There were 465 publicly accessible GenBank HIV-1 genome sequences (including 14 partial genomes of >8,000 bp) from Nepal (accessed on 5 June 2024) using search phrase “HIV Nepal.” Among Nepalese HIV sequences, online NCBI blastn algorithm pipeline with default parameters demonstrated the highest identity (91.91%) to HIV-1 isolate NP20 collected in 2015 (accession # MK493077.1). This study informs our understanding of the current status of the HIV-1 genome diversity in the Nepalese population.
Fig 1.
Maximum likelihood phylogenetic tree of the Nepalese HIV-1 isolate reported here (N0000841) with 18 complete genome sequences with the highest percentage identity and a reference genome. N0000841 lies in the HIV-1 clade reported from India.
ACKNOWLEDGMENTS
This study was approved by Nepal Health Research Council, Nepal (approval # 274-2020) and the Human Research Protection Office of Washington University in St. Louis, MO, USA (approval # 202004087). This project was supported by National Institute of Health grant U01AI151810.
Contributor Information
Krishna Das Manandhar, Email: krishna.manandhar@gmail.com.
Simon Roux, DOE Joint Genome Institute, Berkeley, California, USA.
DATA AVAILABILITY
The genome assembly (N0000841 Nepal FUO) has been deposited in GenBank, accession # PP879189. The version described in this paper is the first version PP879189.1. The raw reads have been submitted to SRA: SRS22420834 under BioProject PRJNA1148996 with BioSample: SAMN43226821.
REFERENCES
- 1. Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR. 2015. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21:1228–1234. doi: 10.1038/nm.3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang D, Urisman A, Liu Y-T, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, Latreille JP, Wilson RK, Ganem D, DeRisi JL. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol 1:E2. doi: 10.1371/journal.pbio.0000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kalantar KL, Carvalho T, de Bourcy CFA, Dimitrov B, Dingle G, Egger R, Han J, Holmes OB, Juan Y-F, King R, et al. 2020. IDseq-an open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Gigascience 9:giaa111. doi: 10.1093/gigascience/giaa111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 5. Shean RC, Makhsous N, Stoddard GD, Lin MJ, Greninger AL. 2019. VAPiD: a lightweight cross-platform viral annotation pipeline and identification tool to facilitate virus genome submissions to NCBI GenBank. BMC Bioinformatics 20:48. doi: 10.1186/s12859-019-2606-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Letunic I, Bork P. 2021. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome assembly (N0000841 Nepal FUO) has been deposited in GenBank, accession # PP879189. The version described in this paper is the first version PP879189.1. The raw reads have been submitted to SRA: SRS22420834 under BioProject PRJNA1148996 with BioSample: SAMN43226821.