ABSTRACT
Two ascomycetes, Neofusicoccum sp. and Xylaria sp., were isolated from healthy leaves of the tropical gymnosperms Zamia pseudoparasitica (Z2) and Zamia nana (Z50) from Panama. The two draft genomes possess a broad predicted repertoire of carbohydrate-degrading CAZymes, peptidases, and secondary metabolites, with more secondary metabolite clusters in the Xylaria isolate.
KEYWORDS: cycad, endophyte, pathogen, ascomycetes, secondary metabolites, Zamia
ANNOUNCEMENT
Neofusicoccum and Xylaria are two common endophytic fungi (1, 2) isolated from two endemic cycad species from Panama. Cycads are the most endangered group of plants—nearly 72% of the 375 species have a critical IUCN status. The main threats are deforestation and poaching. To our knowledge, these are the two first fungal genomes isolated from cycads.
The two cultures were sampled from Zamia pseudoparasitica (Z2) and Zamia nana (Z50) from El Copé (8°40′12.12″N, 80°36′13.26″W) and El Valle de Antón (8°37′18.32.52″N, 80° 7′13.9548″W), respectively, in Central Panamá. Briefly, middle sections of leaf samples were cut into 50 2 × 2 mm2 fragments and surface sterilized by placing them in a small strainer that was submerged and shaken constantly while they were passed through a disinfection battery using a 70% ethanol wash for 2 min, 1% sodium hypochlorite for 3 min, and sterile distilled water for 1 min. The fragments were seeded on large Petri dishes (90 × 14 mm) containing solid potato dextrose agar (PDA) and incubated at 24°C–26°C (ambient light) for approximately 1 week to allow fungal growth to emerge. To isolate pure cultures, a fragment of mycelium was taken from each cultivar, transferred to a test tube with inclined PDA, and grown for nearly 2 months using sterile tweezers. Cultures have been deposited in the collection of Department of Microbiology, Universidad de Panama.
Genomic DNA was extracted using a cetyltrimethylammonium bromide (3) method (obtaining up to 120 ng in 11.7 µL). The genomic DNA was used for library synthesis using a KAPA HyperPlus Kit (Roche), according to the manufacturer’s instructions. The library was quantified and sequenced on an Illumina MiSeq 150-bp paired-end run (300 cycles, v2 kit) at the Smithsonian Tropical Research Institute (Panamá). DNA reads were cleaned and trimmed using Trimmomatic version 0.36 (4) (-phred33), read quality was assessed using FastQC version 0.11.8 (5), and de novo assembled using SPADes version 3.14.1 (6). Genome quality and coverage were assessed using Minimap 2.1.0 (7). Fungal identity was verified using BUSCO version 5.0.0 (8), BLAST version 2.9.0+ (9), and BlobTools version 1.1 (10). After selecting only ascomycete contigs and verifying their taxonomic identity using BLAST, BUSCO was used to estimate the completeness of the filtered assemblies.
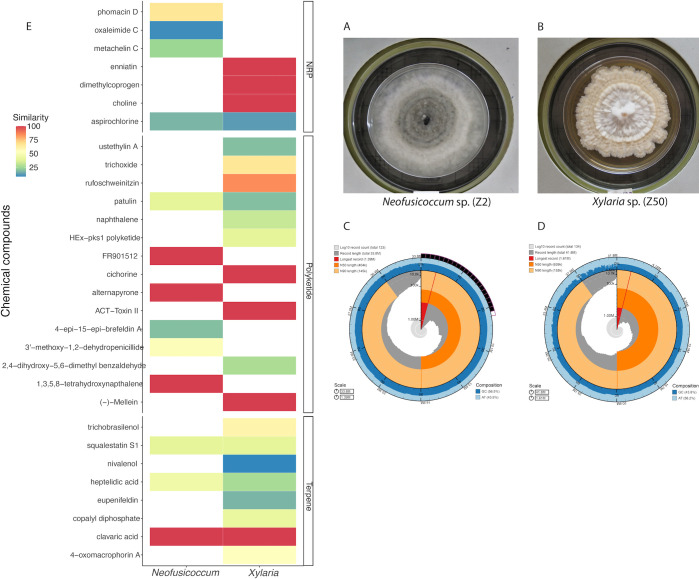
We then used the Funannotate version 1.8.12 pipeline (11) to mask repeats, predict, annotate, and compare the genomes. We used the “funannotate predict” command to train and run three ab initio gene predictors—AUGUSTUS version 3.3.2 (12), GlimmerHMM version 3.4 (13), and SNAP v2006-07-28 (14). Functional prediction of the gene models was performed using InterProScan version 5.57-90.0 (15) with mapping to Gene Ontology (GO) terms, eggNOG-mapper version 2 (16), the Clusters of Orthologous Groups of proteins (17), Pfam domains, the Carbohydrate-Active Enzyme database [CAZY (18)], the secreted protein database [MEROPSv12 (19)], and InterProScan version 5.57-90.0 (15) for fungal transcription factors. We explored the richness of secondary metabolite gene clusters (SMGCs) using antiSMASH version 6.1.1 (20). The relaxed search was conducted on scaffolds and annotated genes (from the funannotate output “annotate results”) using the online settings knownClusterBlast, ClusterBlast, SubClusterBlast, ActiveSiteFinder, Cluster Pfam analysis, and Pfam-based GO term annotation. The genome statistics for each strain are indicated in Fig. 1; Table 1.
Fig 1.
Morphological and genomic features of fungal genomes. (A) Culture of Neofusicoccum sp. (Z2). (B) Culture of Xylaria sp. (Z50). (C) Snail plot indicating general features of the genomes, such as N50, scaffold length, and GC content of Neofusicoccum sp. (Z2). (D) Snail plot indicating general features of the genomes such as N50, scaffold length, and GC content of Xylaria sp. (Z50). (E) Secondary metabolite gene clusters (SMGCs) predicted from antiSMASH analyses for both genomes, highlighting non-ribosomal peptides (NRPs), polyketides, and terpenes. A complete annotation of the SMGCs can be found at https://github.com/jcarlosvillarreal/fungal_cycad_genomes_Panama.
TABLE 1.
Genome statistics for fungal isolates from Zamia pseudoparasitica (Z2) and Zamia nana (Z50) from Panama
| Parameter | Neofusicoccum parvum (Z2) | Xylaria sp. (Z50) |
|---|---|---|
| No. of clean reads | 5,599,532 | 6,504,418 |
| Total genome size (bp) | 33,764,537 | 41,770,564 |
| Largest scaffold | 1,389,236 | 1,612,932 |
| Number of scaffolds | 123 | 134 |
| N50 (bp) | 403,681 | 638,724 |
| Coverage (×) | 62 | 64 |
| GC content (%) | 56.53 | 43.83 |
| No. of genes | 9,753 | 10,245 |
| No. of proteins | 9,634 | 10,030 |
| No. of tRNAs | 119 | 215 |
| Completeness (%) (BUSCO) | 92,5 | 84 |
| Number of secondary metabolite gene clusters |
50 | 95 |
| Number of CAZY enzymes | 450 | 446 |
| Number of secreted peptidases | 343 | 344 |
| Accession no. | JBAWJY000000000 | JBAWJU00000000 |
| SRA | SRX22736318 | SRX22949399 |
| BioSample | SAMN38641487 | SAMN38693397 |
ACKNOWLEDGMENTS
We thank STRI, Universidad de Panamá, the program Canada Research Chair, Canada Foundation for Innovation #36781 and #39135, and SENACYT for providing funding. Thanks to Maycol Madrid for field assistance, Adriel Sierra Pinilla for help with figures, Cely González and Eyda Gomez for help at the Naos Laboratory (STRI), and to Dr. Hernán D. Capador–Barreto.
This project was funded by SENACYT No. 12-2018-4-FID16-237 to K.S. and J.C.V. and the Simons Foundation No. 429440 (W. Wcislo). All collection permits were issued by Ministerio de Ambiente, Panamá, no. SE/P-10-2020.
Contributor Information
Juan Carlos Villarreal Aguilar, Email: jcvil9@ulaval.ca.
Jason E. Stajich, University of California Riverside, Riverside, California, USA
DATA AVAILABILITY
The Whole Genome Shotgun project has been deposited in GenBank under the accession no. JBAWJY000000000 (Neofusicoccum sp. Z2) and JBAWJU00000000 (Xylaria sp. Z50). The Project number is PRJNA1048497 and the SRA accession numbers for the raw MiSeq data are SRX22736318 (Z2) and SRX22949399 (Z50). Annotated versions of the genomes can be found in the https://doi.org/10.5281/zenodo.12521997 and github: https://github.com/jcarlosvillarreal/fungal_cycad_genomes_Panama.
REFERENCES
- 1. Rodriguez RJ, White JF Jr, Arnold AE, Redman RS. 2009. Fungal endophytes: diversity and functional roles. New Phytol 182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x [DOI] [PubMed] [Google Scholar]
- 2. U’Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE. 2012. Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am J Bot 99:898–914. doi: 10.3732/ajb.1100459 [DOI] [PubMed] [Google Scholar]
- 3. Doyle J, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 752:11–15. [Google Scholar]
- 4. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Online. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 6. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 9. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laetsch DR, Blaxter ML. 2017. BlobTools: interrogation of genome assemblies. F1000Res 6:1287. doi: 10.12688/f1000research.12232.1 [DOI] [Google Scholar]
- 11. Palmer JM, Stajich J. 2020. Funannotate v1.8.1: eukaryotic genome annotation
- 12. Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34:W435–9. doi: 10.1093/nar/gkl200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Majoros WH, Pertea M, Salzberg SL. 2004. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20:2878–2879. doi: 10.1093/bioinformatics/bth315 [DOI] [PubMed] [Google Scholar]
- 14. Korf I. 2004. Gene finding in novel genomes. BMC Bioinform 5:59. doi: 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38:5825–5829. doi: 10.1093/molbev/msab293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. 2021. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res 49:D274–D281. doi: 10.1093/nar/gkaa1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drula E, Garron ML, Dogan S, Lombard V, Henrissat B, Terrapon N. 2022. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res 50:D571–D577. doi: 10.1093/nar/gkab1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rawlings ND, Barrett AJ, Bateman A. 2012. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40:D343–50. doi: 10.1093/nar/gkr987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. 2021. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35. doi: 10.1093/nar/gkab335 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Whole Genome Shotgun project has been deposited in GenBank under the accession no. JBAWJY000000000 (Neofusicoccum sp. Z2) and JBAWJU00000000 (Xylaria sp. Z50). The Project number is PRJNA1048497 and the SRA accession numbers for the raw MiSeq data are SRX22736318 (Z2) and SRX22949399 (Z50). Annotated versions of the genomes can be found in the https://doi.org/10.5281/zenodo.12521997 and github: https://github.com/jcarlosvillarreal/fungal_cycad_genomes_Panama.