ABSTRACT
Domibacillus sp. 8LH is a whitish bacterium isolated from the pools of the Cuatro Cienegas Basin (CCB) in the state of Coahuila and belongs to the Bacillaceae family. It grows in circular colonies of about 6 mm in diameter and is capable of forming biofilms. This strain was identified because, in previous experiments in our laboratory, it presented altruistic interactions when co-cultured with bacteria of the genus Bacillus that participate in the nitrogen cycle. This altruistic behavior confers to this Domibacillus strain a potential use for the construction of bacterial consortia with diverse biotechnological applications.
KEYWORDS: Cuatro Ciénegas, genome, Domibacillus
ANNOUNCEMENT
During the experimental stage of the project, we studied the interactions of bacteria from two different sites. We found an interesting phenotype present in one of the bacteria, “Strain 8.” During the experimental phase, “strain 8” helped the growth and development of different bacteria involved in the nitrogen cycle (Martinez-Perez,E. , Zaragoza-Fernandez,A. , Marquez-Cianci, L. , Rosas-Barrera,M. , Aguilera-Najera,D. ,Tapia-Lopez,R. ,Rodriguez-Cruz,U.E. , Souza-Saldivar,V. and Eguiarte,L.E. unpublished data). This phenotype was present regardless of the origin of the bacteria, the Cuenca de Cuatro Cienegas (CCB), or two mangrove sites in Topolobambo, Sinaloa. This bacterial strain was extracted from the lagoon of Los Hundidos within CCB; an oasis located in the state of Coahuila, Mexico.
The evidence obtained in this work indicates that the sequenced strain is within the phylum Bacillota, class Bacilli, order Bacilliales, family Bacillaceae, and belongs to the genus Domibacillus. This genus is composed of nine species at the time of writing. The strain Domibacillus sp. LH8 has unique characteristics described so far in the genus, presenting a cooperative behavior in the presence of Bacillaceae strains that possess genes within the nitrogen cycle.
MATERIALS AND METHODS
Water samples were taken from the Los Hundidos lagoon (geographic coordinates 26° 52' 11.84" N; 102° 01' 12.60" W), located in the Pozas Rojas system in CCB. Surface water samples were collected in sterilized bottles and kept in a cold cooler until use.
In the laboratory, the water was filtered on sterile GF/F filters (0.2 mm nominal pore size; Whatman) using a Millipore filtration device. Each GF/F filter was introduced into marine media plates (Difco & BBL/BD Diagnostics, 1984), and incubated at 30°C for 3 days. After achieving independent colony growth, they were inoculated individually on the same media under similar conditions for 1 day. After morphological and phenotypical characterization of a single colony of strain, 8LH was grown in liquid medium, one portion of this culture was used for glycerol storage at −70°C and the other portion was used for glycerol storage at −70°C. Samples were collected under SEMARNAT scientific permit SGPA/DGVS/04225/21.
DNA was extracted from an axenic culture with the DNAeasy Blood & Tissue QIAGEN’s DNA Library extraction kit (according to the manufacturer instructions), DNA quality was checked by electrophoresis, and libraries were prepared with the Illumina DNA LIBRARY PREP 2, the DNA fragments were done by tagmentation. Libraries of 600 bp were sequenced using the Illumina Miseq platform with 2 × 150 bp paired libraries using MiSeq Reagents Kit v2. A total of 2,714,126 reads paired-end were obtained. We performed a quality analysis of the raw reads using the FASTQC program (1). To perform the de novo assembly we used three different software tools; VELVET (2), SPADES (3), and MEGAHIT (4) in order to compare the obtained results and select the best one. The comparison was made using the QUAST (5) bioinformatic program. Finally, The genome was annotated using PROKKA (6).
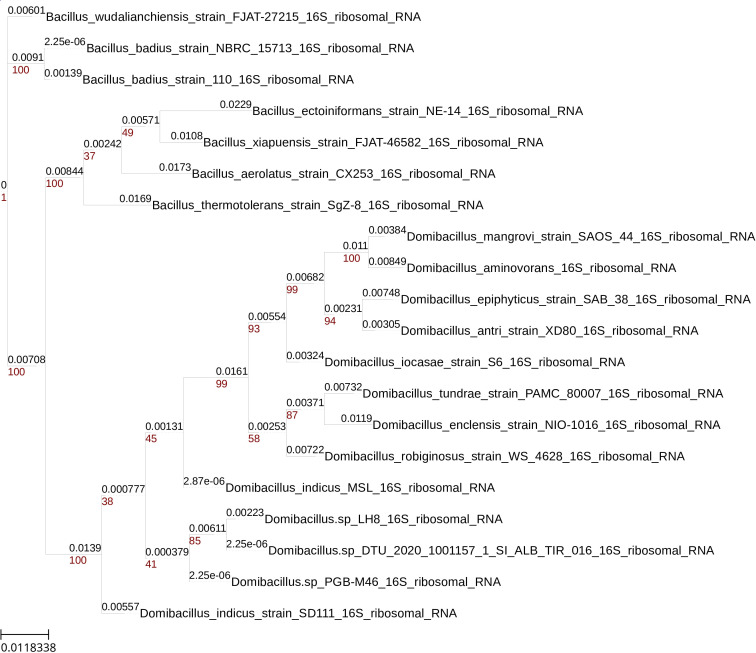
The sequences of the 16S rRNA subunit, previously obtained with Barrnap software, were used for taxonomic identification. The sequences were compared with sequences from the NCBI, SILVA(7), and RDP databases using Blast. Next, the closest sequences were aligned using MUSCLE (8). The result of this alignment was used to infer a maximum likelihood phylogenetic tree by nucleotide substitution model selection using the IQ-TREE (9, 10) web service (available at http://www.iqtree.org/). As an additional method, to improve the accuracy of our results, we used the tools ANIclustermap and FastANI (Yoshiyama Y, 2022; 11, 12), to measure the ANI of our genome with the rest of the genomes of the genus used with the phylogenetic tree (Fig. 1).
Fig 1.
Phylogenetic tree constructed with IQ-Tree Phylogenetic tree inferred with the maximum likelihood method using IQ-Tree. The 16S sequences of organisms of the genera Domibacillus and Bacillus.(13–20).
RESULTS AND DISCUSSION
The best assembly obtained was obtained with SPADES. The final genome has a final length of approximately 5.0 MB, an average GC content of 47% whole genome, and an N50 of 137.12 kb consisting of nine scaffolds whose coverage is above 35 and 4,710 genes according to the results reported by Prokka. Using data obtained from 16S rRNA sequences and SILVA and RDP data, we found matches between different species of the genera Domibacillus and Bacillus. To confirm our results, we used the fastANI program. According to fastANI the new strain shares 95.8% nucleotide identity with Domibacillus sp. DTU, coinciding with results obtained with 16S sequences.
This work contributes to the understanding of the taxonomic classification of bacteria, adds to the growing body of literature on the genus Domibacillus, and expands our knowledge of the diversity and distribution of this group of bacteria.
Software
All the programs used in this methodology were performed using the default parameters unless otherwise stated in the methodology.
Fastqc (version 0.12.0)
VELVET (version 1.2.10 )
SPADES (version 3.15.5 )
MEGAHIT (version 1.2.9 )
QUAST (version 5.2 )
MUSCLE (version 3.8.425 )
Barrnap (version 3 )
FastANI (version 1.33)
ANIclustermap (version 1.3 )
IQ-TREE (version 1.6.12)
PROKKA (version 1.14.5)
PGAP (version 6.6 )
ACKNOWLEDGMENTS
We are greatly thankful for the specialized technical support Drs. Morena Avitia Cao Romero and Marco Tulio Solano De la Cruz.
Pappit IN204822 to V.S.
Contributor Information
Valeria Souza, Email: souza@unam.mx.
David A. Baltrus, The University of Arizona, Tucson, Arizona, USA
DATA AVAILABILITY
Complementary images of the results presented in this work are available at Figshare DOI:https://doi.org/10.6084/m9.figshare.25371538.v2.(24–30) Both raw reads and assembled data were submitted on the NCBI platform to the following Bioproject: PRJNA1013342. The genome assembly is registered with the ID: JAVJNG000000000. The raw read data are registered in the SRA: SRR25930181. The public version of the genome was annotated using PGAP (21–23), while this work was performed using a Prokka annotation.
REFERENCES
- 1. Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 2. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- 4. Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 5. Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 7. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–6. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90k prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimoyama Y. 2022. ANIclustermap: a tool for drawing ANI clustermap between All-vs-All microbial genomes. Available from: https://github.com/moshi4/ANIclustermap
- 13. Verma A, Ojha AK, Pal Y, Kumari P, Schumann P, Gruber-Vodicka H, Dastager SG, Natarajan RK, Mayilraj S, Krishnamurthi S. 2017. An investigation into the taxonomy of “B.aminovorans” and its reclassification to the genus Domibacillus as Domibacillus aminovorans sp. nov. Syst Appl Microbiol 40:458–467. doi: 10.1016/j.syapm.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 14. Sharma A, Dhar SK, Prakash O, Vemuluri VR, Thite V, Shouche YS. 2014. Description of Domibacillus indicus sp. nov., isolated from ocean sediments and emended description of the genus Domibacillus. Int J Syst Evol Microbiol. 64:3010–3015. doi: 10.1099/ijs.0.064295-0 [DOI] [PubMed] [Google Scholar]
- 15. Verma A, Ojha AK, Dastager SG, Natarajan R, Mayilraj S, Krishnamurthi S. 2017. Domibacillus mangrovi sp. nov. and Domibacillus epiphyticus sp. nov., isolated from marine habitats of the central west coast of India. Int J Syst Evol Microbiol 67:3063–3070. doi: 10.1099/ijsem.0.002085 [DOI] [PubMed] [Google Scholar]
- 16. Xu D, Wang L, Wang G, Zheng S. 2016. Domibacillus antri sp. nov., isolated from the soil of a cave. Int J Syst Evol Microbiol 66:2502–2508. doi: 10.1099/ijsem.0.001080 [DOI] [PubMed] [Google Scholar]
- 17. Gyeong HR, Baek K, Hwang CY, Park KH, Kim HM, Lee HK, Lee YK. 2015. Domibacillus tundrae sp. nov., isolated from active layer soil of tussock tundra in Alaska, and emended description of the genus Domibacillus. Int J Syst Evol Microbiol 65:3407–3412. doi: 10.1099/ijsem.0.000429 [DOI] [PubMed] [Google Scholar]
- 18. Sun Q-L, Sun L. 2016. Description of Domibacillus iocasae sp. nov., isolated from deep-sea sediment, and emended description of the genus Domibacillus. Int J Syst Evol Microbiol 66:982–987. doi: 10.1099/ijsem.0.000823 [DOI] [PubMed] [Google Scholar]
- 19. Seiler H, Wenning M, Scherer S. 2013. Domibacillus robiginosus gen. nov., sp. nov., isolated from a pharmaceutical clean room. Int J Syst Evol Microbiol 63:2054–2061. doi: 10.1099/ijs.0.044396-0 [DOI] [PubMed] [Google Scholar]
- 20. Sonalkar VV, Mawlankar R, Krishnamurthi S, Tang S-K, Dastager SG. 2014. Domibacillus enclensis sp. nov., isolated from marine sediment, and emended description of the genus Domibacillus. Int J Syst Evol Microbiol 64:4098–4102. doi: 10.1099/ijs.0.068924-0 [DOI] [PubMed] [Google Scholar]
- 21. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C, Marchler-Bauer A, Thibaud-Nissen F. 2021. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res 49:D1020–D1028. doi: 10.1093/nar/gkaa1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O’Neill K, Li W, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng C, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD. 2018. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46:D851–D860. doi: 10.1093/nar/gkx1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2019. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. doi: 10.1101/602110 [DOI] [PMC free article] [PubMed]
- 25. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C-Y, Graham M, Van Domselaar G, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res 51:W484–W492. doi: 10.1093/nar/gkad326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33:1635–1638. doi: 10.1093/molbev/msw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic,prokaryotic, and viral genomes. Mol Biol Evol 38:4647–4654. doi: 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sahbou A-E, Iraqi D, Mentag R, Khayi S. 2022. BuscoPhylo: a webserver for Busco-based phylogenomic analysis for non-specialists. Sci Rep 12:17352. doi: 10.1038/s41598-022-22461-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Complementary images of the results presented in this work are available at Figshare DOI:https://doi.org/10.6084/m9.figshare.25371538.v2.(24–30) Both raw reads and assembled data were submitted on the NCBI platform to the following Bioproject: PRJNA1013342. The genome assembly is registered with the ID: JAVJNG000000000. The raw read data are registered in the SRA: SRR25930181. The public version of the genome was annotated using PGAP (21–23), while this work was performed using a Prokka annotation.