ABSTRACT
We report the complete genome sequence of a rabies virus obtained by direct metagenomics from the cerebellum of a gold panner who died of unknown encephalitis in French Guiana. Phylogenetic analysis exhibited a close genetic relationship with vampire bat-related isolates, confirming the second case of human rabies identified in this territory.
KEYWORDS: rabies, French Guiana, complete genome, metagenomics, encephalitis, diagnosis, Illumina, next generation sequencing, bat, human
ANNOUNCEMENT
Rabies remains a major but still neglected tropical disease, with an estimated number of 59,000 fatal human cases worldwide each year (1). Rabies virus (RABV) is the principal etiological agent, belonging to the species Rabies lyssavirus within the genus Lyssavirus, family Rhabdoviridae (order Mononegavirales) (2). Apart from domestic dogs, other mammals such as bats can act as natural hosts and vectors. In French Guiana, an overseas territory localized in the northern part of South America, rabies transmitted by hematophagous bats (mainly the Desmodus rotundus species) remains endemic. Regular cases are detected in cattle, domestic carnivores, or even bats (3, 4). In 2008, the first confirmed human case of vampire rabies was detected (5). We describe here the RABV genome sequence of the second human Guyanese case, whose diagnosis was based on direct metagenomics.
A post-mortem cerebellum biopsy (lab ref 2024-6-10) was collected from a deceased patient presenting with encephalitis of unknown etiology in late February 2024 in Cayenne Hospital, French Guiana. This patient first developed symptoms while working on an illegal gold mining site in the Amazon rainforest. Nucleic acids were extracted with a DNA/RNA Microprep plus kit (Zymo Research). Total RNA was used for reverse transcription using random hexamer primers and the SuperScript IV First-Strand cDNA Synthesis kit (Thermofisher Scientific) and amplified with the Quantitect Whole Transcriptome kit (Qiagen). All steps were performed according to the manufacturer’s instructions. A sequencing-ready library was prepared using the TruSeq DNA PCR-Free prep kit (Illumina) and sequenced on a NextSeq2000 system (Illumina) generating nearly 14.4 million of 300-bp single-end length reads (88.2% of unique reads and 12.8% of duplicate reads).
Nearly 77 million contigs were generated and identified against the RVDB database (v25.0) using Microseek, retaining only contigs with a length of 100 nucleotides (6). Among them, a contig (9,010 nt) related to Lyssavirus rabies species was identified. A second dedicated workflow built on Galaxy@Pasteur (a Galaxy platform hosted by Institut Pasteur) was subsequently performed, combining cleaning, de novo assembly, and mapping (CLC Assembly Cell, Qiagen, v5.1.10) (7–10). A final consensus was generated with the nearly 8 million reads retained and manually edited using SnapGene (v7.2) before visualization using Tablet (11).
This complete RABV genome was 11,922 nucleotides in length, with 294.80× coverage on average and 18,078 (0.22%) mapped reads. The five canonical genes were present, encoding the nucleoprotein (N, 1,353 nt, 450 aa), phosphoprotein (P, 894 nt, 297 aa), matrix protein (M, 609 nt, 202 aa), glycoprotein (G, 1,575 nt, 524 aa), and RNA polymerase (L, 6,387 nt, 2,128 aa). The sequences of leader and trailer extremities were complete, with 58 and 69 nucleotides in length, respectively (checking done after alignment with genetically close and available complete genomes). The transcription initiation signal AACA and the transcription termination polyadenylation (TTP) sequence TGA7 were observed for all the genes, except for the G gene, which presented the AGA7 motif for TTP.
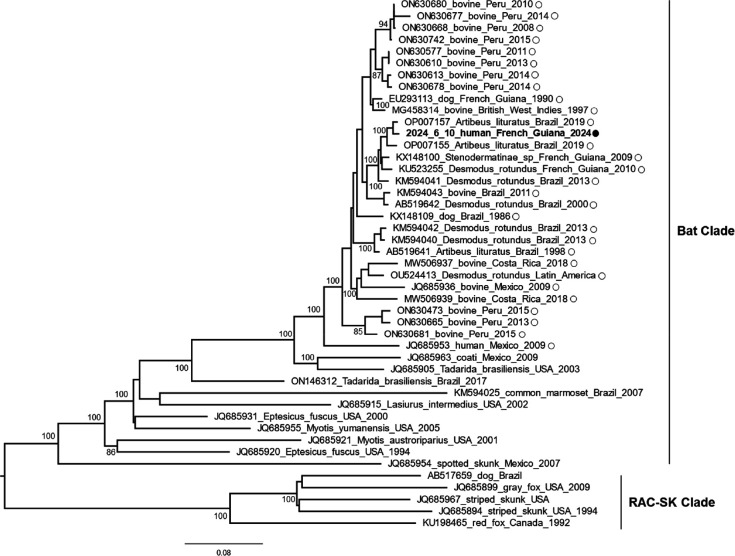
Maximum likelihood (ML) phylogenetic analysis was performed on the five concatenated open reading frames (ORFs) with different representative American strains using PhyML (12), and genetic analysis confirmed that the virus clustered together with vampire bat-related isolates within the Bat Clade (Fig. 1) (13).
Fig 1.
Phylogenetic analysis of the RABV strain (2024-6-10) obtained from the cerebellum of a deceased patient presenting with encephalitis of unknown etiology in French Guiana and different representative American strains. The tree was based on the multiple sequence alignment corresponding to the concatenation of the five ORFs (10,815–10,818 nt), performed with ClustalW (v2.1) (14) implemented in Galaxy@Pasteur (9). The tree was constructed using the ML approach based on the generalized time-reversible model proportion of invariable sites plus gamma-distributed rate heterogeneity (GTR+I+Γ4) utilizing Subtree Prune-and-Regraft (SPR) branch-swapping, as estimated in PhyML 3.0 (12) with Smart Model Selection, also implemented in Galaxy@Pasteur. The robustness of individual nodes was estimated using 100 bootstrap replicates. All tools were run with default parameters unless otherwise specified. The ML phylogenetic tree was visualized with FigTree (http://tree.bio.ed.ac.uk/). Only bootstrap values ≥80 are indicated for the major branches. The scale bar indicates nucleotide substitutions per site. Each sequence denomination contains the GenBank accession number, the nature of the infected species, and the country and year of isolation, where applicable. Rabies virus isolates associated with vampire bats are indicated by an open circle. The isolate (2024_6_10_RABV_GUY) described in this study is indicated in bold, with a black circle. RAC-SK, raccoon–skunk (rabies virus associated with the raccoon and skunk host animals).
ACKNOWLEDGMENTS
We thank the other members of the Laboratory for Urgent Response to Biological Threats (CIBU) for their technical help, especially Séverine Matheus for her network. We are also grateful to the Biomics Core Facility of Institut Pasteur for access to their facilities.
This work was funded through the convention with the Direction Générale de la Santé (for CIBU), with the support of the DURABLE EU4Health program under grant agreement 101102733 (for ERI unit).
Contributor Information
Laurent Dacheux, Email: laurent.dacheux@pasteur.fr.
Valérie Caro, Email: valerie.caro@pasteur.fr.
Jelle Matthijnssens, Katholieke Universiteit, Leuven, Belgium.
DATA AVAILABILITY
The complete genome sequence of the vampire bat-related rabies virus from French Guiana was deposited in GenBank under accession number PP764242 and BioProject accession number PRJNA1108161.
ETHICS APPROVAL
The lab investigations described in this article were carried out as part of the diagnosis activities of the Laboratory for Urgent Response to Biological Threats (CIBU). After the patient’s death, the relatives were informed about the use of the post-mortem cerebellum biopsy for genetic characterization of the rabies strain, after the diagnosis had been established, and consent was obtained.
REFERENCES
- 1. Hampson K, Coudeville L, Lembo T, Sambo M, Kieffer A, Attlan M, Barrat J, Blanton JD, Briggs DJ, Cleaveland S, et al. 2015. Estimating the global burden of endemic canine rabies. PLoS Negl Trop Dis 9:4. doi: 10.1371/journal.pntd.0003709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walker PJ, Freitas-Astúa J, Bejerman N, Blasdell KR, Breyta R, Dietzgen RG, Fooks AR, Kondo H, Kurath G, Shi PL, Stone M, Tesh DM, Tordo RB, Vasilakis N, Whitfield N. 2022. ICTV virus taxonomy profile: Rhabdoviridae 2022. J Gen Virol 103. doi: 10.1099/jgv.0.001689 [DOI] [PubMed] [Google Scholar]
- 3. Lavergne A, Darcissac E, Bourhy H, Tirera S, de Thoisy B, Lacoste V. 2016. Complete genome sequence of a vampire bat rabies virus from French Guiana. Genome Announc 4:e00188-16. doi: 10.1128/genomeA.00188-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Thoisy B, Bourhy H, Delaval M, Pontier D, Dacheux L, Darcissac E, Donato D, Guidez A, Larrous F, Lavenir R, Salmier A, Lacoste V, Lavergne A. 2016. Bioecological drivers of rabies virus circulation in a neotropical bat community. PLoS Negl Trop Dis 10:e0004378. doi: 10.1371/journal.pntd.0004378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meynard J-B, Flamand C, Dupuy C, Mahamat A, Eltges F, Queuche F, Renner J, Fontanella J-M, Hommel D, Dussart P, Grangier C, Djossou F, Dacheux L, Goudal M, Berger F, Ardillon V, Krieger N, Bourhy H, Spiegel A. 2012. First human rabies case in French Guiana, 2008: epidemiological investigation and control. PLoS Negl Trop Dis 6:e1537. doi: 10.1371/journal.pntd.0001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pérot P, Bigot T, Temmam S, Regnault B, Eloit M. 2022. Microseek: a protein-based metagenomic pipeline for virus diagnostic and discovery. Viruses 14:1990. doi: 10.3390/v14091990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luo D-S, Li B, Shen X-R, Jiang R-D, Zhu Y, Wu J, Fan Y, Bourhy H, Hu B, Ge X-Y, Shi Z-L, Dacheux L. 2021. Characterization of novel rhabdoviruses in Chinese bats. Viruses 13:64. doi: 10.3390/v13010064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pallandre L, Luo D, Feuvrier C, Lieffrig F, Pozet F, Dacheux L, Bigarré L. 2020. Revisiting the classification of percid perhabdoviruses using new full-length genomes. Viruses 12:649. doi: 10.3390/v12060649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mareuil F, D-A O, Ménager H. 2017. A public galaxy platform at pasteur used as an execution engine for web services. F1000Res 1030. doi: 10.7490/f1000research.1114334.1 [DOI] [Google Scholar]
- 10. Dacheux L, Dommergues L, Chouanibou Y, Doméon L, Schuler C, Bonas S, Luo D, Maufrais C, Cetre-Sossah C, Cardinale E, Bourhy H, Métras R. 2019. Co-circulation and characterization of novel African arboviruses (genus Ephemerovirus) in cattle, Mayotte island, Indian Ocean, 2017. Transbound Emerg Dis 66:2601–2604. doi: 10.1111/tbed.13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012 [DOI] [PubMed] [Google Scholar]
- 12. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 13. Troupin C, Dacheux L, Tanguy M, Sabeta C, Blanc H, Bouchier C, Vignuzzi M, Duchene S, Holmes EC, Bourhy H. 2016. Large-scale phylogenomic analysis reveals the complex evolutionary history of rabies virus in multiple carnivore hosts. PLOS Pathog 12:e1006041. doi: 10.1371/journal.ppat.1006041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of the vampire bat-related rabies virus from French Guiana was deposited in GenBank under accession number PP764242 and BioProject accession number PRJNA1108161.