Abstract
Background
Clinically important lineages in Klebsiella, especially those expressing multi-drug resistance (MDR), pose severe threats to public health worldwide. They arose from the co-evolution of the vertically inherited core genome and horizontal gene transfers by plasmids, which has not been systematically explored.
Methods
We designed KleTy, which consists of dedicated typing schemes for both the core genome and plasmids in Klebsiella. We compared the performance of KleTy with many state-of-the-art pipelines using both simulated and real data.
Results
Employing KleTy, we genotyped 33,272 Klebsiella genomes, categorizing them into 1773 distinct populations and predicting the presence of 87,410 plasmids from 837 clusters (PCs). Notably, Klebsiella is the center of the plasmid-exchange network within Enterobacteriaceae. Our results associated the international emergence of prevalent Klebsiella populations with only four carbapenem-resistance (CR) PCs, two hypervirulent PCs, and two hvCR-PCs encoding both carbapenemase and hypervirulence. Furthermore, we observed the ongoing international emergence of blaNDM, accompanied by the replacement of the previously dominant population, blaKPC-encoding HC1360_8 (CC258), during 2003–2018, with the emerging blaNDM-encoding HC1360_3 (CC147) thereafter. Additionally, expansions of hypervirulent carbapenem-resistant Klebsiella pneumoniae (hvCRKP) were evidenced in both populations, driven by plasmids of MDR-hypervirulence convergences.
Conclusions
The study illuminates how the global genetic landscape of Klebsiella has been shaped by the co-evolution of both the core genome and the plasmids, underscoring the importance of surveillance and control of the dissemination of plasmids for curtailing the emergence of hvCRKPs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-024-01399-0.
Keywords: Distributed cgMLST, Plasmid, Klebsiella pneumoniae, Multi-drug resistance, Hypervirulence
Background
Nosocomial infections by carbapenem-resistant Klebsiella pneumoniae (CRKP) that also express multidrug resistance (MDR), first reported in the 1990s, now have a worldwide distribution [1]. In particular, the emergence of hypervirulent CRKPs (hvCRKPs) in China, India, and many other countries has drawn special attention [2] due to their association with the increased prevalence of bloodstream infections [3]. To investigate infections and transmission of K. pneumoniae, a wide range of typing techniques have been utilized [4], including serotyping, pulsed-field gel electrophoresis (PFGE), and multi-locus sequence typing (MLST). State-of-the-art pipelines, such as Kleborate [4], rely on MLST and serotyping to characterize Klebsiella strains. Recently, cgMLST schemes for the K. pneumoniae Species Complex (KpSC) have been developed and hosted by Institut Pasteur (629 genes) [5], which also designed cgLINcodes to infer population structures. Clinical use of the cgMLST scheme, however, has been restricted by the requirement to upload nucleotide sequences into central databases, which can be difficult for clinicians and/or epidemiologists [6].
Mobile genetic elements harboring antimicrobial-resistant genes (ARGs) and virulence factors (VFs) have been regarded as one of the major driving factors behind the emergence of epidemic lineages in K. pneumoniae [7]. Notably, the recent emergence of ST11 hvCRKP strains in China has been associated with the acquisition of virulence factors (VFs) in the CRKPs [8], calling for systematic analyses of the complex interplay between the plasmids and the bacterial hosts. In addition, K. pneumoniae has long been regarded as the hub for inter-species horizontal genetic transfers (HGTs) of plasmids, especially among Enterobacteriaceae [9]. Investigation of the contextual genetic diversity of plasmids in Klebsiella will facilitate our understanding and control of the inter-species spreading of ARGs, which has been frequently associated with the plasmids [1]. Currently, low resolution limits state-of-the-art techniques, such as replicon typing and MOB typing [10], thereby highlighting the need for new algorithms that systematically predict and catalog plasmid content.
Here we describe an automatic pipeline, KleTy, that enables unified genotyping of both the core genome and the plasmids in all Klebsiella species. KleTy consists of three modules: (1) a population assignment module based on a novel dcgMLST + HierCC scheme, (2) a plasmid prediction module based on a novel plasmid clustering (PC) scheme, and (3) an ARG/VF prediction module. We showed that KleTy outperformed state-of-the-art pipelines in both population assignments and plasmid predictions based on the prediction of 1773 natural populations and 87,410 plasmids in 33,272 Klebsiella genomes. This work expanded our understanding of the genetic diversity of plasmids in Klebsiella and the dynamics of predominant populations that are associated with the global elevation of hvCRKP over decades.
Implementation
Genome sequence collection
To capture the broadest possible diversity of Klebsiella populations, we established a comprehensive genomic dataset of Klebsiella that consists of a total of 33,272 assemblies and short reads retrieved from GenBank (as of Aug. 2022, Additional file 1: Table S1). All short reads were assembled into draft genomes using EToKi [11]. These genomes represent isolates collected between 1886 and 2022 from 94 different countries, with the majority from the Americas (27.8%, n = 9243), Europe (25.2%, n = 8383), and Asia (20.5%, n = 6815). The STs, ARG/VF profiles, and capsular types of each genome were predicted using Kleborate v2.3.2 [4], and the genes were predicted and annotated using Prokka [12].
Establishments of the dcgMLST + HierCC schemes
Construction of the dcgMLST scheme consisted of three stages (Fig. 1a). First, to reduce genetic redundancy due to the over-representation of genetically nearly identical strains in Klebsiella, we employed fastANI [13] to estimate the pairwise genetic distances of all 33,272 genomes and separated them into single-linkage clusters of ≥ 99.8% identities. One genome with the greatest N50 value was chosen for each cluster and subjected to quality checking using FetchMG [14]. We kept only 7269 representative genomes that carried ≥ 37/40 single-copy core genes (SCGs), including 5109 that shared ≥ 95% ANIs to the K. pneumoniae type strain (GCA_000281755). Furthermore, we repeated the procedures above on the representative genomes to a further sub-selected seed set of 1478 genomes of < 99.3% identities, which were used as the seeds for pan-genome estimation.
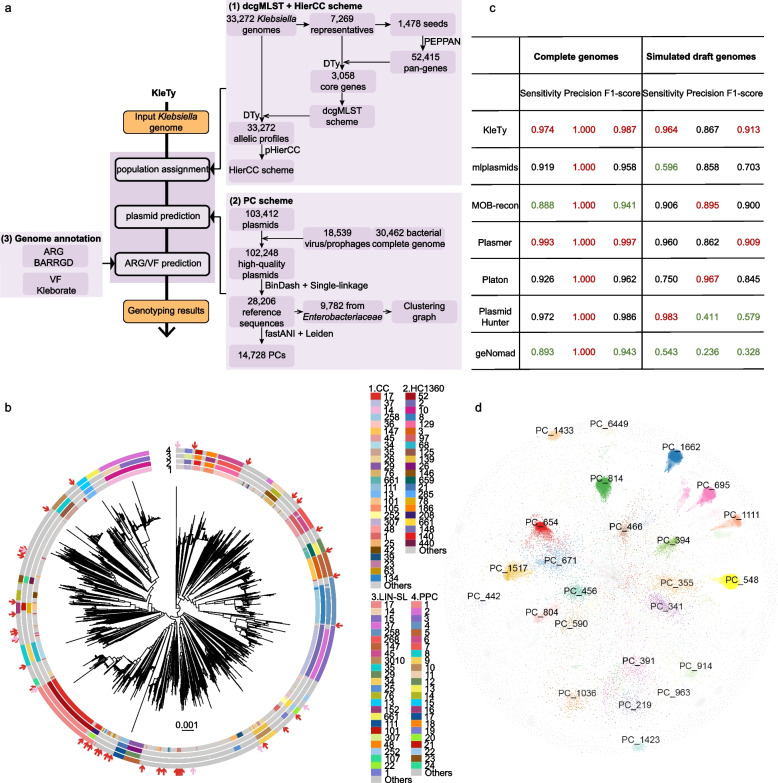
Fig. 1.
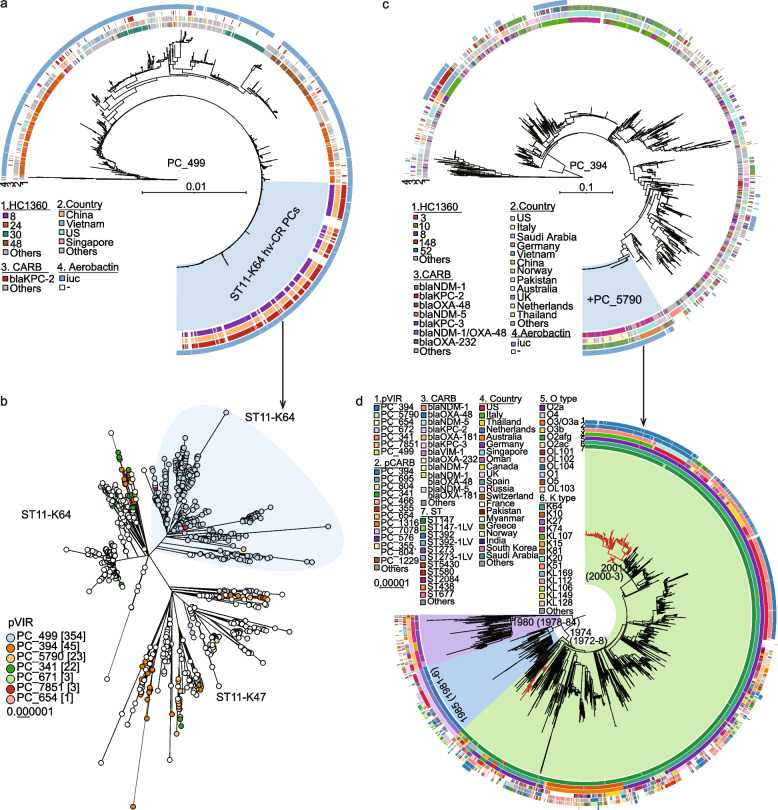
The workflow and evaluation results for the KleTy pipeline. a KleTy consists of three modules that (1) genotype each genome into a set of curated hierarchical clusters using the dcgMLST + HierCC scheme, (2) identify plasmids and assign each to one of the 14,728 clusters in the Plasmid Clustering (PC) scheme, and (3) predict ARGs and VFs based on curated gene sets. b Visualizing the HC1360 groups, clonal complexes (CCs), cgLINcodes sub-lineages (LIN-SLs), and PopPUNK clusters (PPCs) in the supertree of 5109 K. pneumoniae genomes summarized from 3058 gene trees (see the “Methods” section). The circular bars surrounding the tree from outer to inner showed the PPCs, LIN-SLs, HC1360s, and CCs of each genome (inset Keys). Arrows show the genomes inaccurately assigned to CC17 (red) or CC14 (pink). c Comparisons of the plasmid predictions by KleTy, mlplasmids, MOB-recon, Plasmer, Platon, PlasmidHunter, and geNomad on benchmark datasets of 1271 complete (columns 1–3) or draft (columns 4–6) Klebsiella genomes. The simulated draft genomes were assembled from simulated short reads by wgsim based on public complete genomes and made available at https://doi.org/10.5281/zenodo.12633486. The results with the two greatest or lowest values were highlighted in red or green, respectively. d The similarity network of 9782 reference plasmids from Enterobacteriaceae. Plasmids (nodes) from the top 30 most abundant PCs were color-coded
We applied PEPPAN [15] to predict a pan-genome of 52,415 genes based on the 1478 genomes in the seed set. Furthermore, we employed the EToKi MLSTdb module to identify and remove potential paralogs that shared over 80% amino acid similarity. A total of 42,061 pan genes were kept and used to build the whole-genome MLST (wgMLST) scheme for Klebsiella. We estimated the presence of genes in the wgMLST scheme in all 33,272 genomes using DTy (https://github.com/ADSGF203com/DTy) and selected a subset of 3058 core genes that (1) present in ≥ 95% of genomes, and (2) maintained intact open reading frames in > 94% of its alleles using EToKi cgMLST module. The distributed cgMLST scheme was built based on the core genes and made publicly available as part of the KleTy pipeline at https://github.com/zheminzhou/KleTy. DTy designated each allele in the dcgMLST based on the MD5 hash value of its sequence, rather than an arbitrary sequential integer from a central database in the traditional cgMLST scheme. This allowed each genome to be characterized as a collection of up to 3058 MD5 hash values, which each represented a unique core gene allelic sequence. All the genotyping results were also available in the KleTy repository.
Additionally, we hierarchically grouped the allelic profiles of all genomes into multi-level clusters using pHierCC [16] and evaluated the consistencies and cohesiveness of each cluster using the pHCCeval module, which is also in the pHierCC package. Briefly, pHCCeval estimated the similarity of all potential clustering thresholds pairwisely using the normalized mutual information (NMI) score, revealing “stable blocks” that yielded similar clusters sharing NMI of > 0.9. Furthermore, pHCCeval used silhouette scores to evaluate the cohesiveness of each cluster, identifying the optimal thresholds in these stable blocks for population inferences.
Comparison of HC1360s with clusters from other schemes
The clustering schemes were compared based on 5109 K. pneumoniae genomes in the representative set. First, the supertree of these genomes was estimated using a divide-and-conquer algorithm implemented in the cgMLSA package [17]. The CCs were estimated as clusters of single-locus variants of the 7-gene MLST profiles by the eBURST algorithm implemented in the goeBURST software [18]. Furthermore, we used PopPUNK to create a database of the 5109 genomes and used the bgmm model to separate them into 796 clusters. The command lines are:
poppunk –create-db –output < database_name > –r-files < query_list > –threads 20.
poppunk –fit-model bgmm –ref-db < database_name > –K 11.
Finally, we also uploaded all 5109 K. pneumoniae genomes to PathogenWatch (https://pathogen.watch) in batches of 1000 genomes each to obtain the cgLINcode predictions. Sub-lineage information was obtained for 5025 genomes, while the remaining 84 genomes were designed as “new.”
Scheme of plasmid clusters (PCs)
The complete sequences of 103,412 plasmids, spanning > 2400 species across the Tree of Life, were downloaded from GenBank (as of March 2023). Sequences that shared high similarity with bacterial chromosomes or viruses (≥ 95% identities and ≥ 60% coverages) were identified based on BLAST searches against all complete sequences of bacteria and viruses (https://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi), which were also downloaded from GenBank at the same time. A total of 102,248 high-quality plasmids were retained. We employed BinDash [19] to estimate the pair-wise genetic distances of the plasmids and grouped them into 28,206 single linkage clusters of ≥ 99% identities. The reference plasmid dataset was built by selecting one sequence of the greatest size for each cluster. Furthermore, we employed FastANI [13] with parameters of “–fragLen 1000 –minFraction 0.5” to calculate average nucleotide identity (ANI) between pairs of plasmids in the reference dataset and used the Leiden algorithm [20] to separate them into 14,728 plasmid clusters (PCs) with ANIs of ≥ 90% and alignment coverage of ≥ 50% (Fig. 1a). The similarity network of the plasmids and the resulting PCs were visualized using the Fruchterman Reingold layout algorithm [21] implemented in the Gephi software [22]. The resulting reference sequences, together with the associated host species and PC assignments, were all deposited in the KleTy repository (https://github.com/zheminzhou/KleTy/tree/main/db).
The plasmid prediction module in KleTy employs BLASTn [23] to align each Klebsiella genome onto the reference plasmids (Additional file 2: Fig. S1). To remove potential nonspecific matches, it removes any alignment with < 85% identity or < 400 bp length. KleTy evaluates each contig by its alignment coverages to reference plasmids and keeps only alignments that are located in contigs with ≥ 30% of its sequences similar to the corresponding plasmid. Furthermore, KleTy identifies the PCs iteratively using a greedy algorithm. In each iteration, a PC that has the greatest proportion of its sequences found in the assembly is selected, and all contigs with ≥ 50% coverage and ≥ 85% identity to the PC are removed from the following iterations. The program stops when no PCs had ≥ 50% of its sequences aligned by the contigs, and reports all the identified PCs as well as their associated contigs. Optionally, KleTy also explores potential PC fragments that were only partially aligned by the contigs. At this step, only contigs that had ≥ 50% of their sequences similar to a reference plasmid were kept. The iterative searching of PC fragments stops when no PCs had ≥ 30% of their sequences aligned by the contigs.
Gene annotation and network construction
KleTy predicts ARGs based on a modified version of Kleborate with its default parameters. Furthermore, KleTy employs BLASTn to predict gene encoding hypervirulence or resistance to biocides and metal by aligning the sequences to the reference genes hosted in Kleborate and AMRfinder, respectively, keeping only hits with ≥ 80% identities and ≥ 60% coverages. Furthermore, regions responsible for Inc, MOB, and MPF types were obtained by comparing to the reference sequences hosted in MOB-typer [10]. An HC1360 cluster was designated as multi-drug resistance if 50% or more of its genomes carry ARGs for at least three of the following drug categories: aminoglycosides, beta-lactams, carbapenems, ESBLs, fluoroquinolones, macrolides, phenicols, sulfonamides, tetracyclines, or trimethoprim. Furthermore, an HC1360 cluster was considered hypervirulence if ≥ 30% of its genomes carry both aerobactin (iuc) and salmochelin (iro) genes. Similarly, PCs with ≥ 20% of the carbapenemase genes, iuc gene, or both were designated carbapenem-resistant (CR-PCs), hypervirulent (hvPCs) PCs, or hvCR-PCs, respectively.
We visualized the association between bacterial hosts and the PCs in Enterobacteriaceae as a network, in which the nodes represent PCs or bacterial genera. Edges were drawn between a PC and bacteria if the PC was found in the corresponding genus. The resulting network was rendered and visualized using the OpenOrd layout algorithm [24] in Gephi.
Comparison of KleTy against other plasmid prediction tools
All 1271 complete Klebsiella genomes that were available in GenBank as of June 2023 were downloaded, encompassing 1273 chromosome sequences and 4796 plasmid sequences. Furthermore, for each complete genome, we simulated reads using wgsim with 55 × coverage and no error rate (“-N 1000000 -e 0 -1 150 -2 150”). SPAdes was used to perform de novo assembly of the simulated reads, and the plasmid- or chromosome-associated contigs were identified by aligning all contigs back to the corresponding complete genomes using BLASTn, with ≥ 98.5% identity and ≥ 80% coverage. A total of 1674 contigs (~ 1.3 per genome) that failed to align, or aligned to both chromosome and plasmids were excluded. Furthermore, contigs with a length smaller than 1000 bp were also removed because many of the prediction tools were not able to handle them. The final simulated dataset consists of 662,128 contigs from chromosomes and 64,981 contigs from plasmids, which are all deposited in a publically accessible repository (https://doi.org/10.5281/zenodo.12633486).
We evaluated the performance of KleTy in both the complete and draft assemblies and compared it with mlplasmids (v 2.1.0), MOB-recon (v 3.1.0), Plasmer (v 0.1), Platon (v 1.7), PlasmidHunter (v 1.2), and geNomad (v 1.7.1). The plasmid prediction tools were all run with the default parameters, using the following command lines:
-
Mlplasmids:
Mlplasmids < query_assembly > < output > 0.7 'Klebsiella pneumoniae'.
-
Mob-recon:
Mob_recon –infile < query_assembly > –outdir < output >
-
Plasmer:
Plasmer -g < query_assembly > -d /path/to/Plasmer/db/ -p < prefix > -o < output >
-
Platon:
Platon -t 4 -o < output > -p < prefix > < query_assembly >
-
PlasmidHunter:
Plasmidhunter -o < output > -i < query_assembly >
-
geNomad:
Genomad end-to-end –cleanup –splits 8 < query_assembly > < output > /path/to/genomad_db.
-
KleTy:
KleTy -q < query_assembly >
All predictions from each tool were also deposited in the aforementioned public repository and their precision, sensitivity, and F1-score were calculated using scikit-learn. Particularly, we also ran mlplasmids with a higher prob_threshold of 0.8, which was not reported here because it yielded worse predictions than the default prob_threshold in terms of the F1-score.
Phylogenetic analysis of the genomes
The minimum spanning tree of 33,272 Klebsiella genomes was constructed using the MSTreeV2 algorithm and visualized in GrapeTree [25]. The maximum-likelihood (ML) trees of HC1360_3 and HC1360_8 were calculated using the EToKi package [11]. Briefly, EToKi employs minimap2 [26] to align 1493 HC1360_3 genomes and 2530 HC1360_8 ST11 genomes onto the reference sequences of GCF_005944305 and GCF_011066505, respectively, to obtain a multi-sequence alignment. It then estimates ML trees based on the alignments using IQ-TREE [27]. Furthermore, we used RecHMM [28] to identify and remove DNA sketches that were imported by homologous recombination and estimated the tree again based on the remaining non-recombinant regions. All resulting trees were visualized online using either GrapeTree or iTOL v6 [29].
Phylogenetic analysis of the plasmids
Publically available, complete sequences of PC_499, PC_394, PC_456, and PC_1293 plasmids were retrieved from GenBank (Additional file 1: Table S2) and their alignment fractions to the reference plasmid (PC_499: AY378100; PC_394: OW969913; PC_456: CP031850; PC_1293: CP024041) were measured using BLASTn (Additional file 2: Fig. S2). Furthermore, contigs associated with each of these PCs, predicted by KleTy, were extracted from the corresponding Klebsiella assemblies. For each PC, we used “EToKi align” module to align all genomes onto the reference plasmid. Regions shared by ≥ 80% of the plasmids were maintained, accounting for 40–81% of the sequences in the reference. SNPs in these conserved regions were subjected to ML phylogeny, using the phylo module in EToKi, and the results were visualized in iTOL.
Inferences of population dynamics for HC1360_3
The spatiotemporal dynamics of the HC1360_3 population were estimated by TreeTime [30] based on the ML tree. We pruned all genomes without isolation dates from the tree, and ran TreeTime on the remaining tree with the parameters of “–keep-polytomies –confidence –covariation –time-marginal always –relax 0.5 0.5 –coalescent skyline –n-skyline 20” to obtain a date-calibrated tree. We then run TreeTime again on the dated tree in the “mugration” mode to estimate the ancestral international transmission of the bacteria with default parameters.
Statistical analysis
All statistical analyses were performed using R v4.2.2 or Python v3.8. The Normalised Mutual information score and the Silhouette score of the hierarchical clusters were calculated using scikit-learn implemented in pHCC_eval in the pHierCC package. The Cramer’s V statistic was used to test the strength of association between two categorical variables and computed using the rcompanion package in R. Both Pearson’s and Spearman’s correlations between the carbapenem resistance and geographic distributions were performed using the cor.test function in the stats package in R. Linear regression analysis was performed for the carbapenem resistance and geographic distribution of the HC1360s using the ggplot2 package (Method = lm) in R. The pairwise similarity of the four clustering schemes was assessed as the adjusted Rand index (ARI) using scikit-learn in Python. Furthermore, the Wallace index was also applied to measure the containment relationship of the three genome-based approaches. The precision, sensitivity, and F1-score for the results of each plasmid prediction pipeline were also calculated using scikit-learn. A p-value of < 0.01 was considered statistically significant in all tests.
Results
A unified genotyping scheme for chromosome and plasmid in Klebsiella
Here we establish KleTy, a tool that rapidly genotypes core genomes and plasmids in Klebsiella with three modules, including population assignment, plasmid prediction, and genome annotation (Fig. 1a). Based on KleTy, one can easily assign a given Klebsiella genome to a pre-defined population and identify clinically relevant genes and plasmids in ~ 60 s with eight threads.
Population assignment module based on dcgMLST + HierCC scheme
A total of 33,272 Klebsiella genomes were retrieved from public databases as of June 2022 (Additional file 1: Table S1). To reduce redundancy caused by genetically similar strains and simplify calculations, we first selected a seed set of 1478 sequences of < 99.3% average nucleotide identities (ANIs) and used them to estimate a pan-genome of 52,415 genes, including 3058 soft-core genes with ≥ 95% presence. These soft-core genes were employed to establish a distributed cgMLST (dcgMLST) scheme [6], an extension of the standard cgMLST scheme that allows de-centralized allelic designation using a hash-based algorithm (see the “Methods” section). Furthermore, we used pHierCC to evaluate a series of single-linkage clustering results designated after their allowed allelic differences from HC0, namely no observed allelic differences, to HC3057, which allowed different sequences for all but one core gene. A block of clustering levels from HC100 to HC1800 was found to be similar to each other based on their pairwise normalized mutual information scores (Additional file 2: Fig. S3), indicating gradual genetic diversifications [16]. Among them, HC1360 clusters (arrows in Additional file 2: Fig. S3b) had the greatest average silhouette score and exhibited high similarity to the clonal complexes (CCs) in the 7-gene MLST scheme, with an adjusted Rand index (ARI) of 0.967, likely representing natural populations in Klebsiella (Additional file 1: Table S3).
To evaluate the performance of population characterization, we selected a representative set of 5109 K. pneumoniae genomes with < 99.8% ANIs. A supertree of these strains was built by combining 3058 trees of their core genes using a divide-and-conquer strategy [17]. Furthermore, we calculated their CCs, HC1360s, and PopPUNK clusters (PPCs), and also obtained their cgLINcodes by uploading the genomes into PathogenWatch (https://pathogen.watch). The sub-lineage (LIN-SL) assignments in the cgLINcode were selected for the comparison because they exhibited the greatest silhouette score as proposed in Hennart et al. [5]. We obtained LIN-SL for 5025 strains, while the other 84 strains were designated as “NEW” without actual nomenclatures.
After mapping all four clustering results onto the supertree, we found that 16.8% of the CCs mistakenly generated paraphyletic groups (Fig. 1b, Additional file 1: Table S4). For example, CC17 was found in 45 monophyletic clades (red arrows in Fig. 1b, Figs. S2a, S2b) across the tree and CC14 was split into 10 clades (pink arrows in Fig. 1b, Figs. S2a, S2c). Notably, the representative set minimized the overrepresentation of strains from large populations of clinical relevance and thus amplified the inconsistencies between the CCs and the other three, genome-based clustering schemes. The CCs exhibited an ARI of 0.81 with the HC1360s, and slightly lower ARIs of 0.79 with the other two schemes. Nearly all of the CCs that were incompatible with HC1360s were also inconsistent with either LIN-SLs or PPCs or both (Table 1).
Table 1.
A summary of the correspondence between HC1360, CC, PPC, and LIN-SL
| Reference: query | Category | Count | Representative CCs |
|---|---|---|---|
| HC1360: CC | Consistent | 344 | |
| Incompatible | 468 | CC17 (n = 45); CC23 (n = 19); CC63 (n = 19); CC26 (n = 15); CC25 (n = 12); CC132 (n = 11); CC35 (n = 11); CC134 (n = 11); CC200 (n = 10); CC14 (n = 10) | |
| Merge | 74 | ||
| Split | 72 | ||
| LIN-SL: CC | Consistent | 386 | |
| Incompatible | 392 | CC17 (n = 44); CC23 (n = 19); CC63 (n = 18); CC26 (n = 14); CC25 (n = 12); CC14 (n = 11); CC200 (n = 11); CC35 (n = 10) | |
| Merge | 40 | ||
| Split | 135 | ||
| LIN-SL: HC1360 | Consistent | 599 | |
| Incompatible | 0 | ||
| Merge | 0 | ||
| Split | 242 | ||
| LIN-SL: PPC | Consistent | 648 | |
| Incompatible | 25 | ||
| Merge | 31 | ||
| Split | 157 | ||
| PPC: HC1360 | Consistent | 632 | |
| Incompatible | 30 | ||
| Merge | 52 | ||
| Split | 127 |
*consistent—identical clusters (comprise the same set of genomes); merge—multiple query clusters are merged into one cluster in the reference scheme; split—one query cluster is split into multiple clusters in the reference; incompatible—scenarios that cannot be assigned into any of the three categories above
Meanwhile, all genome-based approaches of HC1360, PopPUNK, and cgLINcode yielded clusters of good consistencies with the supertree (Fig. 1b). Particularly, HC1360 clusters exhibited high similarity (ARI = 0.90–0.91) with both PPCs and LIN-SLs, whereas the PPCs and LIN-SLs were less similar (ARI = 0.82). We attributed such differences to the varied sizes of the clusters. Broadly speaking, the PPCs were the largest, followed by HC1360s and LIN-SLs, evidenced by their directional Wallace indexes [31] of > 0.97 versus only 0.88–0.94 in the other direction. Notably, detailed investigations suggested that over 86% (599/693) of the HC1360s exhibited one-to-one correspondence with the LIN-SLs while the remaining 94 were split into 342 sub-clusters in the LIN-SLs (Additional file 1: Table S5). We also assessed the Klebsiella pneumoniae genomes in the representative set between different clustering methods (Additional file 2: Fig. S4). The genome-based schemes were generally comparable in clustering structure except for resolution differences. CCs varied more significantly. For instance, CC17 and CC14 were linked to numerous distinct HC1360s, LIN-SLs, and PPCs (Figs. S2b, S2c).
Plasmid prediction module based on pre-curated plasmid clusters (PCs)
We retrieved all 103,412 complete plasmids from over 2400 bacterial species in GenBank (as of March 2023) to capture their full genetic diversity. Using BinDash [19], we estimated the pairwise genetic distances of the plasmids and grouped them into single-linkage clusters (SLCs) with a distance threshold of < 0.01. One representative sequence from each cluster was selected and screened for potential mislabeling of chromosomal or viral DNAs (see the “Methods” section). Pairwise comparisons of the remaining 28,206 high-quality representatives were estimated using FastANI [13] (Additional file 2: Fig. S5) and subsequently grouped into plasmid clusters (PCs) using the Leiden algorithm [20] with a resolution of 0.01.
To optimize clustering, we tested various ANI and alignment fraction (AF) thresholds (Additional file 2: Fig. S6, Additional file 2: Fig. S7). At a fixed AF of 50%, the number of clusters decreased as ANI thresholds increased (Additional file 2: Fig. S7). Using the “elbow method,” we identified 90% ANI as optimal, yielding clusters 2000 fewer than those at 95% ANI but only slightly more than those at 85% and 80% ANI. This was also consistent with the fact that 82% of plasmid pairs with ≥ 50% AF had an ANI of ≥ 90%, while only 15% fell between 80 and 90% ANIs.
We also evaluated clustering with varying AFs and a fixed 90% ANI (Additional file 2: Fig. S6a). The number of PCs increased as AF thresholds rose, without a clear elbow point (Additional file 2: Fig. S6c). However, lower AF thresholds resulted in greater plasmid size variation within clusters, with some PCs showing larger standard deviations at AF < 50% (Additional file 2: Fig. S6b). Ultimately, we defined 14,728 plasmid clusters using 90% ANI and 50% AF (Fig. 1d, Additional file 1: Table S6; see the “Methods” section). A bimodal distribution emerged: the 20 most common PCs encompassed 23% of plasmids, while the majority of remaining PCs were rare, each containing 1–10 plasmids (Additional file 2: Fig. S8). This likely reflects the under-sampling of plasmid diversity in public databases, an issue further explored in Klebsiella.
Furthermore, the low-resolution Leiden algorithm produced clusters similar to SLCs, which suffered from the “chaining phenomenon,” where distant clusters might be merged due to a few close elements. To address this, we further divided each PC into complete-linkage clusters, designated as plasmid types (PTs), ensuring the plasmids within each PT were similar.
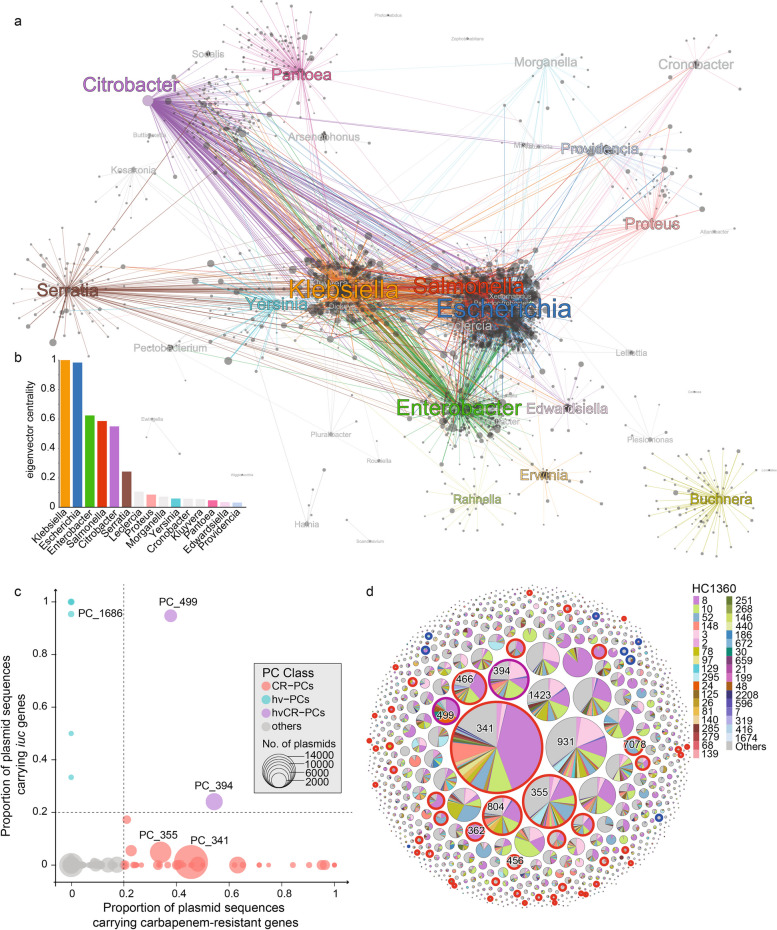
Based on the PCs, we constructed a plasmid-exchange network of Enterobacteriaceae by bridging each PC with its associated genera (Fig. 2a, Additional file 1: Table S7). For example, the PC_654, PC_671, PC_548, and PC_1517 contained over 90% Escherichia plasmids, while PCs such as PC_456, PC_466, and PC_219 spanned Escherichia, Salmonella, Enterobacter, and Citrobacter. Additionally, we found that 76% and 71% of the PCs harbored by Klebsiella and Escherichia were also present in other genera including Enterobacter and Salmonella. This makes Klebsiella and Escherichia the predominant centers of the plasmid-exchange network, with the greatest eigenvector centralities of 1.0 and 0.98, respectively (Fig. 2b). To minimize the impact of the unequal amount of public plasmids in each genus, we randomly downsampled the maximum number of plasmids each genus to 1000. With 100 random downsamplings, we found that Klebsiella and Escherichia still ranked at the top in terms of their eigenvector centralities (Additional file 1: Table S7).
Fig. 2.
The host association and genetic characteristics of the PCs. a A graph of the plasmid-exchange network in Enterobacteriaceae. Nodes show the plasmids (gray pots) and genera of the hosts (colored) and the colored edges show the presence of the plasmids in the corresponding hosts. b Histogram of the eigenvector centralities of genera in the network in part a. c Scatter plot of the average carriage of carbapenemase genes (X-axis) and the iuc gene (Y-axis) in each PC. Each circle represents a PC and is sized proportional to the number of associated plasmids and color-coded as in the Key. The two dashed lines show the criteria for assigning PCs as CR-PCs (≥ 0.2 carbapenemases), hvPCs (≥ 0.2 iuc), or hvCR-PCs. d Bubble plot of the HC1360 distribution for each PC. Each bubble indicates a PC and is sized relative to the associated plasmids. The piechart in each bubble shows the proportional presence of the PC in different HC1360s. The halos surrounding some bubbles indicate that these PCs belong to one of the CR-PCs (red), hvPCs (blue), or hvCR-PCs (purple), as defined in part c
We designed an algorithm as the plasmid module in KleTy for predicting PCs and compared its performance with six existing algorithms: mlplasmids [32], MOB-recon [10], Plasmer [33], Platon [34], PlasmidHunter [35], and recently published geNomad [36]. To this end, we retrieved all 1271 publically available complete Klebsiella genomes from GenBank (June 2023), containing 4796 plasmids (Additional file 1: Table S8). Furthermore, we also simulated 1271 draft assemblies, averaging 521 and 51 contigs for chromosomes and plasmids, respectively, by simulating and assembling the short reads from each complete genome. All tools performed reasonably well with complete genomes (F1-score of 0.940–0.997) (Fig. 1c), although MOB-recon and geNomad suffered from low sensitivities of ~ 0.89. In contrast, the performances varied in the draft genome dataset. KleTy ranked 2nd in sensitivity (0.964) and 3rd in precision (0.867), leading to the top F1-score of 0.913. Plasmer and MOB-recon also performed well with the draft assemblies, with an F1-score of 0.908 and 0.901, respectively. Particularly, Plasmer ranked 3rd in sensitivity (0.960) and MOB-recon ranked 2nd in precision (0.895). Platon and mlplasmids both had high precision of 0.967 and 0.857 but missed many true positives, resulting in a lower F1-score of 0.845 and 0.703, respectively. In contrast, PlasmidHunter ranked 1st in sensitivity (0.982) but had low precisions (0.411), resulting in an F1-score of 0.579. Finally, geNomad performed poorly in both sensitivity and precision, resulting in the lowest F1-score of 0.328, possibly because it was designed for metagenomic data rather than genomes.
ARG/VF prediction module
To evaluate the clinical importance of predicted populations and plasmids, we employed a third module that predicts ARGs and VFs based on a slightly modified version of Kleborate [4]. Additionally, we used the reference sequences hosted in AMRfinder [37] for predicting genes responsible for resistance to biocides and metals.
Genetic landscape of populations and plasmids in Klebsiella.
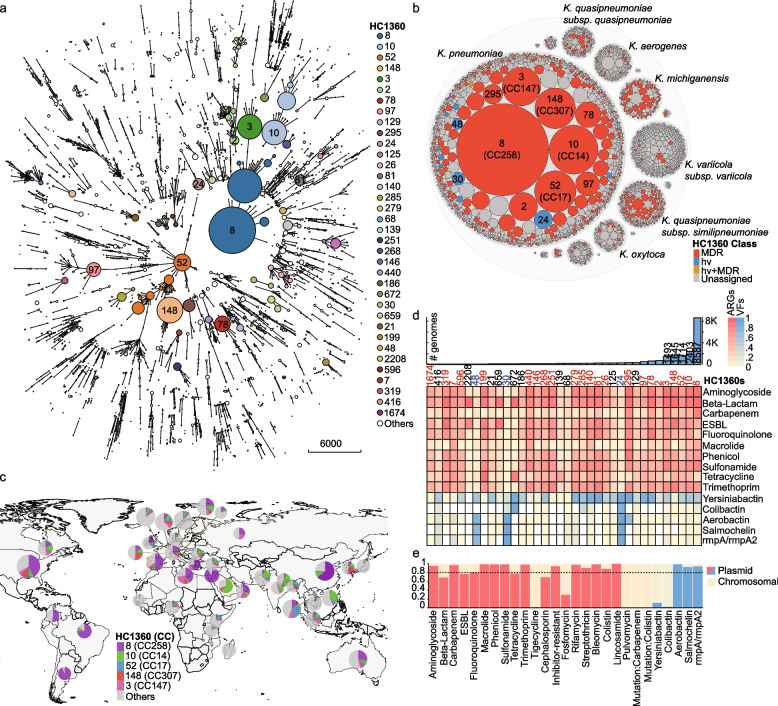
We identified 1773 HC1360 populations in Klebsiella based on all 33,272 publicly available genomes (Fig. 3a), making it slightly more divergent than Escherichia/Shigella, which has 1379 HC1100 populations [17]. Over 70% of the HC1360s fell within only three species of K. pneumoniae (43%, 757), Klebsiella variicola subsp. variicola (17%, 299), and Klebsiella quasipneumoniae subsp. similipneumoniae (11%, 193) (Additional file 1: Table S9). Notably, nearly half of the Klebsiella genomes were from one of the five predominant populations of HC1360_8 (CC258; 8587 strains), HC1360_10 (CC14; 2403), HC1360_52 (CC17; 1714), HC1360_148 (CC307; 1645), and HC1360_3 (CC147; 1493) (Fig. 3b). HC1360_8 (CC258) is the primary source of Klebsiella in China, Israel, the USA, and countries in South America and Europe; HC1360_10 (CC14) predominates in many countries around the Indian Ocean (Fig. 3c) and HC1360_52 (CC17) were prevalent in Thailand some other Southeast Asian countries. Furthermore, HC1360_148 (CC307) and HC1360_3 (CC147), two emerging populations, were found in almost all continents. HC1360_148 was prevalent in France, Korea, and the USA, while HC1360_3 was primarily from Italy, Libya, and Russia. None of the predominant HC1360s has been frequently found in North Europe or Africa, which might be attributed to their distinct epidemiological patterns [38]. Alternatively, variations in sampling frameworks—particularly in the selection of clinical, community, or environmental sources, or the focus on antimicrobial resistance—may have also influenced the results.
Fig. 3.
The population structure and geographical distribution of the Klebsiella genomes. a A minimum spanning tree of 33,272 Klebsiella genomes based on their dcgMLST profiles. Branches with < 400 allelic distances are collapsed. The HC1360 populations with ≥ 100 genomes are color-coded as in the Key and some are also labeled. Only branches with < 3035 allelic distances are shown by clarity. b Hierarchical bubble plot for the population structure of the Klebsiella genus. The bubbles indicate the genus, species, and HC1360 populations in Klebsiella. Some HC1360s are color-coded because the majority of the genomes in them are MDR (red), hypervirulence (blue), or both (orange). c Global distribution of the five major HC1360 populations in countries with ≥ 1000 genomes visualized using D3.js. d The heat plot of the prevalence of antimicrobial-resistant genes (ARGs; red) and hypervirulence factors (VFs; blue) in the top 36 HC1360 populations. The histogram on the top shows the numbers of genomes in the HC1360 populations. e The predicted percentages of each of the ARGs and VFs from the plasmids. The dotted line indicates 80% of plasmid origins
KleTy also predicted the presence of 87,410 plasmids from 837 PCs in Klebsiella (Additional file 1: Table S10). Of the identified PCs, 38% (314/837) were novel for Klebsiella, having been previously found only in other genera, such as Enterococcus, Escherichia, Staphylococcus, Acinetobacter, Bacillus, Neisseria, Enterobacter, and Salmonella (Additional file 1: Table S11). The addition of these plasmids reshaped the PC distribution in Klebsiella, substantially reducing the relative frequencies of rare PCs and increasing the prevalence of PCs associated with 11 to 500 plasmids, indicating improved plasmid sampling.
Additionally, KleTy assigned Incompatibility (Inc) types to ~ 78% (68,103/87,410) of the plasmids, with ColRNAI, IncFIB, and IncFII being the most frequent (Additional file 2: Fig. S9a). It also assigned 46,715 (53%) plasmids to MOB types, with MOBF, MOBP, and MOBH being the most frequent (Additional file 2: Fig. S9b). There was minimal association between typing schemes, with most PCs linking to multiple Inc and MOB types. In summary, our findings substantially expand our understanding of the genetic diversity in Klebsiella.
Emergence of MDR HC1360_8 (CC258) population with blaKPC-carrying plasmids.
Approximately 29% (524/1773) of the HC1360s from 13 Klebsiella species had > 50% of its genomes encoding resistance to ≥ 3 drug classes, and thus designed as MDR (Additional file 1: Table S12). Half (265/513) of these MDR HC1360s fell among K. pneumoniae, followed by K. quasipneumoniae subsp. similipneumoniae (82) and K. michiganensis (58) (Fig. 3b).
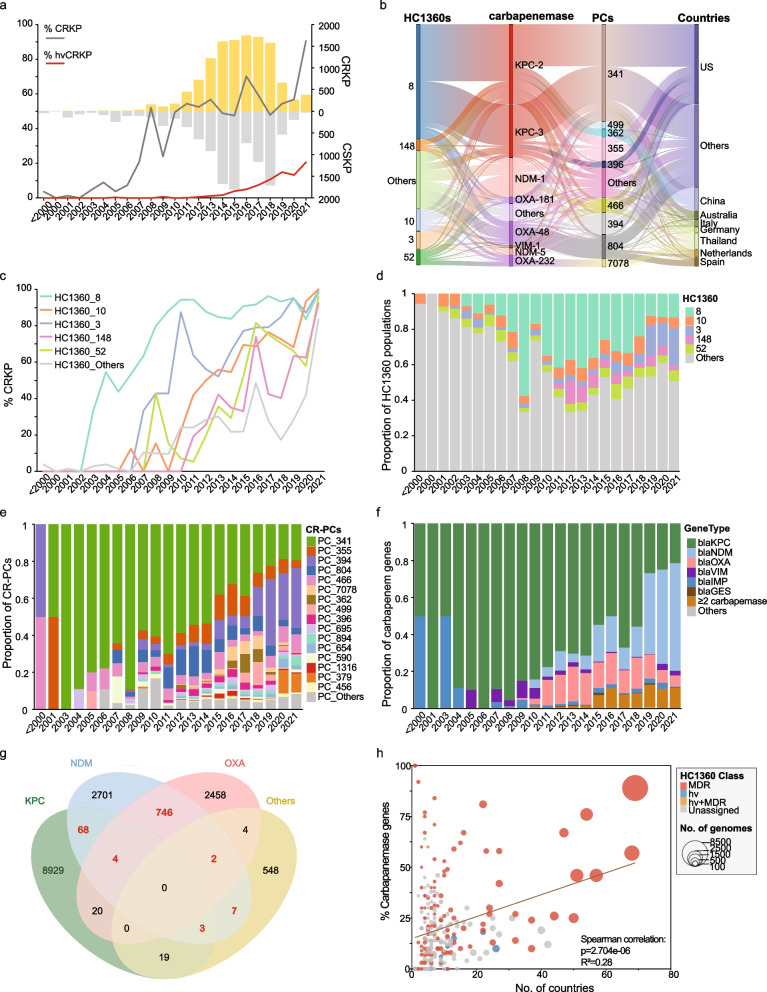
Nearly half (47%; 15,509/33,272) of the Klebsiella genomes exhibited carbapenem resistance (CRKP) due to the acquisition of carbapenemases (Fig. 4a). We found 50 carbapenem-resistant PCs (CR-PCs) that each has a ≥ 20% carbapenemase carriage rate (Fig. 2c). CR-PCs accounted for four of the six most abundant plasmids (Additional file 2: Fig. S9) and carbapenemases (Additional file 2: Fig. S10) in Klebsiella as previously reported [39], including PC_341 (37% blaKPC, 88% IncFIB/IncFII), PC_355 (17% blaKPC, 60% IncFIB/IncFII), PC_394 (34% blaNDM, 80% IncFIB/IncHI1B), and PC_804 (51% blaOXA, 99% IncL/M). Meanwhile, geographically restricted CR-PCs also exhibited almost exclusive association with only one category of carbapenemase (Fig. 4b), such as the blaKPC-2-encoding PC_362 and PC_499 in China and blaKPC-3-encoding PC_396 and PC_1671 in the US.
Fig. 4.
The prevalences and genetic characteristics of carbapenemase-carrying Klebsiella strains. a The annual frequencies of CRKP (yellow) and carbapenam-sensitive Klebsiella (CSKP; gray) in the 23,868 genomes with known isolation years. The relative percentages of the CRKP (dark gray) and hvCRKP (red) in each year are also shown as curves. All strains isolated before 2000 are assigned to one bin for clarity. b Sankey diagram shows the relationships between the HC1360 populations, carbapenemases, PCs, and the countries of the CRKPs. c The percentages of CRKPs per year for each of the five predominant HC1360 populations and the remaining. d–f The relative proportions of the predominant HC1360 populations (d), CR-PCs (e), and carbapenamases (f) per year. g Venn diagram of the carbapenemase profiles of the CRKPs. Each oval shows the carriages of one of the blaKPC, blaNDM, blaOXA, and other carbapenamases. The overlapping regions show genomes that each encode two or three carbapenemase categories simultaneously. The red numbers show the multi-carriage of both blaNDM and other carbapenamases. h The bubble plot of the country distributions (X-axis) and percentage of CRKPs (Y-axis) for each HC1360 population. The circles were sized relative to the number of genomes and color-coded according to the Key. The dark red line shows a positive correlation between the CRKP percentages and the country distributions of HC1360s by the linear regression, with Spearman’s correlation of R.2 = 0.28 (Pearson: 0.287), p = 2.704e − 6
In our genomic data, the earliest CRKP in HC1360_8 (CC258) was isolated in 2003. After the acquisition of a blaKPC-3-encoding PC_341 plasmid, HC1360_8 CRKP isolates were found to increase over time, reaching > 80% carriage in 2008 (Fig. 4c). Meanwhile, the isolation frequencies of HC1360_8 strains also increased, accounting for 58% of available Klebsiella genomes in 2008 (Fig. 4d). Early HC1360_8 isolates were predominantly ST258 from the USA, but by 2007 [40], blaKPC-2-producing ST11 strains from China also emerged as major contributors [8].
Two longitudinal monitoring projects, one in the USA (PRJNA288601, [41]) and the other in Australia (PRJNA529744, [42]), further documented the spread of these strains. The US study, covering eight states between 2013 and 2016, identified ST258 as the most prevalent strain (n = 207), representing 43.5% of all isolates. Nearly 99% of ST258 strains carried blaKPC, aligning with our data. In the Australian study, 361 strains were evaluated, with 48.3% carrying the blaKPC-2 plasmid and belonging to ST258. Both projects confirmed the dominance of HC1360_8 strains through 2018.
Other CRKP populations were also observed. For example, the blaOXA-48 carried by PC_804 accounted for ~ 12% of the isolates between 2011 and 2014 (Fig. 4b, e). However, the blaKPC-encoding HC1360_8 kept being the most prevalent CRKP population, accounting for ~ 30% of annual Klebsiella isolates until 2019 (Fig. 4d), when it was surpassed by the blaNDM-encoding HC1360_3 (CC147). Similar trends were also found in both the US and Australia, where HC1360_3 increased its frequencies in recent years (Additional file 2: Fig. S11).
Emergence of HC1360_3 (CC147) with plasmid-driven blaNDM-hypervirulence convergence.
HC1360_3 (CC147) was the most abundant population after 2019 and accounted for 13% to 21% of the Klebsiella strains (Fig. 4d), possibly associated with multiple reports of associated outbreaks [43]. It exhibited a diverse carbapenemase profile of blaNDM-1 (53%, 606/1138), blaNDM-5 (7%, 77), blaOXA-48 (10%, 111), and blaKPC-2 (6%, 68). We found an increase of blaNDM in HC1360_3 up to 89% (317/356) during its upsurge since 2019, indicating a possible association between blaNDM and the population expansion (Fig. 4f, Additional file 2: Fig. S12). In addition, the recent growth of blaNDM carriage rates in less common populations was observed. Over 70% of strains in HC1360s with < 1000 strains carry blaNDM after 2020, resulting in the general elevation of carbapenem resistance in Klebsiella to 90% by 2021. Moreover, blaNDM contributed to 94% (830/883) of multi-carbapenemase strains, which each carried two or more carbapenemases, further underscoring its complicated genetics (Fig. 4g).
Virulence also contributes to the epidemiology of Klebsiella. Hypervirulence in Klebsiella has been associated with the presence of five virulence loci, especially aerobactin (iuc), which encodes the siderophores aerobactin. The hvKPs and the CRKPs normally fall into different HC1360s (Fig. 3b, d) and are associated with the acquisition of different plasmids (Fig. 2d). However, some HC1360s have been reported to experience antimicrobial resistance (AMR) and virulence convergence, resulting in severe disease outbreaks [8, 44]. Using both iuc and carbapenemase genes as markers, we observed a steady increase of hvCRKP frequencies over time, from 4.5% before 2019 to 15.8% currently (Fig. 4a). This increase has been associated with convergence or conjugation of hv- and ARG-carrying plasmids [45].
Characteristics of hvCR PCs based on plasmid phylogenetic trees
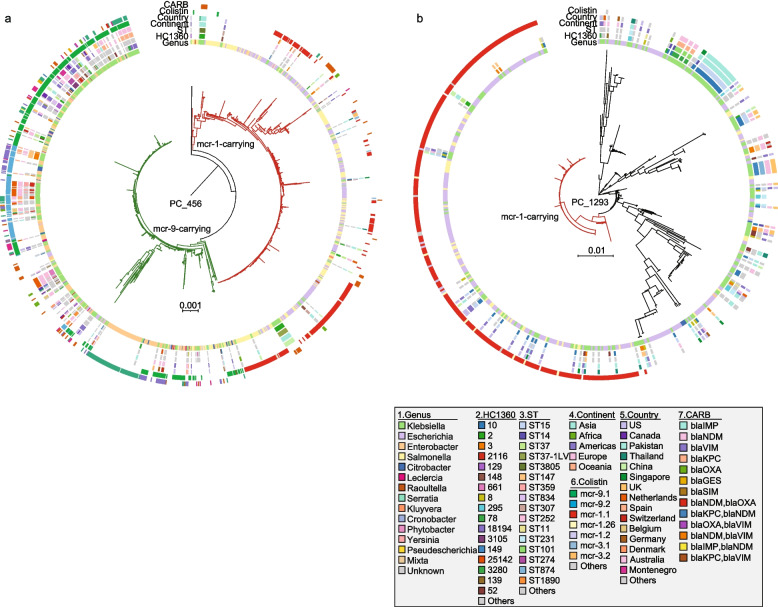
The PCs encompassed nearly all plasmids (99–100%) that shared a significant portion of conserved sequences (Additional file 2: Fig. S2), facilitating robust phylogenetic analysis. Using both predicted plasmids in Klebsiella and those from the reference dataset, we constructed phylogenetic trees for four PCs. The PC_499 tree was built based on 7755 SNPs in 178 KB core sequences (81% of the plasmid size), and the PC_394 tree was built based on 28,993 SNPs in 120 KB core sequences (40% of the plasmid size) (Fig. 5a, c). The PC_456 tree was built based on 11,903 SNPs in 149 KB core sequences (62% of the plasmid size), and the PC_1293 tree was built based on 2059 SNPs in 19 KB core sequences (57% of the plasmid size) (Fig. 6a, b).
Fig. 5.
The maximum-likelihood phylogeny of the major hvCRKP HC1360 populations and their associated hvCR-PCs. a The phylogeny of 361 public PC_499 plasmids and 1141 associated plasmids predicted from assemblies. The phylogeny was built based on 7755 SNPs from 178 KB of the conserved regions (shared by ≥ 80% of the plasmids), responsible for 81% of the reference plasmid (AY378100). The hvCR-PCs responsible for the majority of the hvCRKP in ST11-K64 in part B are highlighted in light blue. b The phylogeny of 1114 ST11 genomes that are from China or fall within the same clade of the Chinese strains. The shape in light blue highlights a cluster of ST11-K64 strains that are mostly hvCRKPs due to the acquisition of the hvCR PC_499 plasmids in part A. c The phylogeny of 378 public PC_394 plasmids and 2608 associated plasmids predicted from assemblies. The phylogeny was built based on 28,993 SNPs from 120 KB of the conserved regions (shared by ≥ 80% of the plasmids), responsible for 40% of the reference plasmid (OW969913). The hvCR-PCs responsible for the majority of the hvCRKPs in ST147 in part d resulted from a conjugation of the plasmids in PC_5790 and are highlighted in light blue. d The phylogeny of 1493 global HC1360_3 (CC147) genomes. The three MLST STs associated with HC1360_3 (CC147) are shown in colored arcs and the hvCRKP clades are highlighted in red, carrying the hvCR PC_394 plasmids in part c. The circular bars in parts a, c, and d show metadata associated with the plasmids or genomes, as in the Keys. Visualizations of the PC_499 and PC_394 plasmid trees are available in https://itol.embl.de/shared/2Lj8mfCZAiEmU
Fig. 6.
The maximum-likelihood phylogenies of mcr-carrying plasmids. a The phylogeny of 971 public PC_456 plasmids and 304 associated plasmids predicted from assemblies. The phylogeny was built based on 11,903 SNPs from 149 KB of the conserved regions (shared by ≥ 80% of the plasmids), responsible for 62% of the reference plasmid (CP031850). Red and green branches are each associated with mcr-1 or mcr-9 genes, respectively. b The phylogeny of 379 public PC_1293 plasmids and 149 associated plasmids predicted from assemblies. The phylogeny was built based on 2059 SNPs from 19 KB of the conserved regions (shared by ≥ 80% of the plasmids), responsible for 57% of the reference plasmid (CP024041). Red branches indicate plasmids carrying the mcr-1 gene. CARB: carbapenemase, ST: sequence type. Visualizations of the PC_456 and PC_1293 plasmid trees are available in https://itol.embl.de/shared/2Lj8mfCZAiEmU
From the phylogenetic trees, we found that two hvCR PCs had high levels of both iuc and carbapenemase genes: PC_499 (94.7% iuc, 37.8% carbapenemase, 89% IncFIB/IncHI1B) and PC_394 (24.1% iuc, 54.3% carbapenemase, 80% IncFIB/IncHI1B) (Fig. 2c, outer ring of Fig. 5 a/c). PC_499 resulted from the conjugation of pLVPK (also in PC_499), the most abundant hv-PC in Klebsiella, and the blaKPC-2-carrying plasmids in China (Fig. 5a). It has been exclusively associated with the recent emergence of the ST11-K64 hvCRKP clone in China and rarely found elsewhere. Through a phylogenetic analysis of 2530 ST11 genomes from China, we observed that > 90% of the ST11-K64 hvCRKPs fell into one monophyletic cluster that was associated with a cluster of PC_499 hvCR plasmids (Fig. 5b). The hvCRKPs were much fewer outside this cluster and were associated with acquisitions of multiple plasmids.
PC_394 has been associated with the currently emerging HC1360_3 (CC147) [46] and causing disease outbreaks internationally [43, 47] (Fig. 5c). It was formed by conjugation of the carbapenemase-carrying processor in PC_394 and the hypervirulence PC_5790. To investigate the population dynamics of HC1360_3 (CC147) and PC_394, we reconstructed a maximum-likelihood phylogeny based on 25,144 SNPs in the non-repetitive, non-recombinant core genome of 1493 international HC1360_3 strains (Fig. 5d) and estimated its date of origin and geographic transmission (Additional file 2: Fig. S13). The most recent common ancestor (MRCA) of HC1360_3 was estimated to be present before 1947 (CI95%: 1945–1954) in the USA, and it later diverged there into three lineages that broadly consisted of strains from ST147 (1244 strains), ST392 (139), and ST273 (90). The ST147 lineage likely emerged before 1974 (CI95%: 1972–1978) and was gradually transmitted into ≥ 48 countries. A transition of primary carbapenemase from blaOXA or blaKPC in early strains to blaNDM encoded by PC_394 or PC_695 after 2017 was observed (Fig. 5d). Furthermore, we predicted many AMR-virulence convergences along the phylogeny of PC_394, including one large cluster resulting from conjugation with PC_5790 plasmids (Fig. 5c). The resulting hvCR plasmids were independently acquired by HC1360_3 (CC147) to form hvCRKPs ≥ 9 times. Two of the resulting hvCRKP clusters have been circulating for ≥ 10 years, one responsible for repetitive infections in Russia [48], and the other for outbreaks in both Italy [43] and the USA [47].
Colistin resistance plasmids promote specific gene transmission across regions and hosts
Mobilized colistin resistance gene (mcr) resulted in reduced susceptibility of colistin, further limiting the treatment options [49]. We identified a total of 23 mcr-carrying PCs in Klebsiella. Most of these PCs were low in amount or had low mcr carriages, except for two, PC_456 and PC_1293, which accounted for 70% (205/292) of mcr in Klebsiella.
The IncHI2A PC_456 carries an average of eight ARGs per plasmid and was associated with two mcr variants, mcr-1 (6%) and mcr-9 (51%), each by plasmids from a different phylogenetic clade (Fig. 6a). The mcr-1-carrying clade consists of plasmids from Asian Klebsiella strains, as well as many from Salmonella and Escherichia. Meanwhile, the mcr-9-carrying clade consists of primarily Euroamerican strains, plus plasmids from Enterobacter, Citrobacter, and Escherichia. These findings reflected the presence of two dynamic plasmid pools in PC_456 each associated with different Enterobacteriaceae spp. In contrast, the IncX4 PC_1293 is rarely associated with ARGs other than mcr. It has been primarily found in Klebsiella, Escherichia, and Salmonella. All mcr-1-carrying plasmids fell into a genetically closely related cluster in the phylogeny, which accounts for 50% of the PC_1293 plasmids (Fig. 6b).
Discussion
Klebsiella populations that are clinically significant, particularly those that exhibit multi-drug resistance, arise from the co-evolution of the genetically conserved core genome and the highly variable, accessory genes that have been horizontally transmitted. Here we describe KleTy, an integrated tool that offers high-resolution genotyping solutions for both the core genome and plasmids based on the dcgMLST + HierCC and PC schemes, respectively.
Here we adopted a dcgMLST scheme for Klebsiella, which was proposed recently [6] to allow de-centralized cgMLST calculation by implementing MD5 hash for allele designations. Furthermore, based on the dcgMLST types, we separated Klebsiella genomes into HC1360 clusters that represent natural populations. These populations have been previously approximated as clonal complexes (CCs) in the legacy MLST scheme, sublineages in the cgLINcode scheme, and clusters in many standalone pipelines, such as PopPUNK. We showed that the genome-based approaches yielded more phylogenetic-compatible clusters than the CCs (Fig. 1b). Particularly, some renowned CCs, such as CC14 [50] and CC17 [51] which are associated with MDR infections globally, were found to be paraphyletic (Fig. S4), potentially leading to inaccurate biological interpretation. The HC1360 clusters had the greatest average ARI value when compared to other genome-based approaches and the CCs. Furthermore, the CC and PopPUNK clusters are unstable and may merge when adding new strains [52]. Both the HierCC and cgLINcode schemes overcome this problem by implementing an algorithm that ensures static clustering assignments [5, 16].
Plasmids have long been regarded as a “black hole” in phylogenetic research due to their highly variable gene contents and extensive HGTs across bacterial hosts [39]. KleTy managed to accurately predict plasmids and PCs based on a comprehensive reference dataset compiled from > 100 K existing plasmids. We demonstrated the superior performance of KleTy over six state-of-the-art pipelines of mlplasmids, MOB-recon, Plasmer, Platon, PlasmidHunter, and geNomad, in both complete and draft assemblies (Fig. 1c). Notably, all seven pipelines could be broadly separated by their algorithms into three categories: (1) similarity-based, which includes KleTy, MOB-recon, and Platon; (2) Kmer-based, which includes mlplasmids and Plasmer; (3) machine-learning based, which includes PlasmidHunter and geNomad. While the greatest sensitivity was found for results from PlasmidHunter, both machine-learning-based algorithms suffered from low precision of only 0.236–0.411 for the draft assemblies. Platon and mlplasmids, conversely, had high precision and low sensitivities of 0.596–0.750, which may be associated with the quality of their databases. Finally, KleTy, Plasmer, and MOB-recon all performed reasonably well, with balanced sensitivities and precisions (Fig. 1c). This indicates that the applications of machine-learning algorithms in detecting mobile genetic elements, such as plasmids, are still in need of further development.
We also used this massive PC dataset to investigate the dynamics of inter-species plasmid transfer among Enterobacteriaceae, which has been associated with the spread of ARGs [39] including carbapenemase and mcr genes (Fig. 2a). We demonstrated the pivotal role of Klebsiella and Escherichia in the plasmid-exchange network. These two species have been frequently associated with the worldwide dissemination of ARG-carrying plasmids, facilitating their transmission among other more virulent pathogens, such as Salmonella enterica and Vibrio cholerae [53, 54]. Moreover, we showed that the plasmid-exchange rate has been drastically underestimated due to a lack of understanding of the genetic landscape of plasmids. KleTy identified 314 new PCs that have not been previously reported in Klebsiella, reflecting HGTs from/to other species in Enterobacteriaceae or even other families. The inclusion of predicted plasmids reduced the frequency of rare PCs in Klebsiella, showing that next-generation sequencing data significantly improves plasmid sampling compared to those in GenBank. Expanding the use of the PC module to other bacterial species would allow a more systematic evaluation of plasmid exchange and its role in the cross-species spread of ARGs and other genes.
Notably, we found a lack of association between both Inc and MOB types and the PCs, consistent with previous reports [55]. Genes responsible for these traditional typings might have been horizontally transferred between different PCs, reflecting a new level of complexity. Intriguingly, among the 453 PCs that have been found between different species, 68% (308) have not been associated with MOB genes, which may have reflected a loss of MOB genes after the HGT. Alternatively, these plasmids might be hitchhikers that transferred together with the MOB-encoding plasmids in the same bacterial hosts [56].
Our analysis demonstrated the importance of chromosome-plasmid co-evolution in the formation of MDR epidemic lineages. Over 1/3 of the HC1360 populations in Klebsiella are MDR, mostly driven by plasmid-oriented ARGs (Fig. 3b, e). Notably, we identified 119 CRKP HC1360s that exhibited high levels (> 80%) of carbapenem resistance and demonstrated positive correlations (Fig. 4h) between the carbapenem resistance of the HC1360s and geographic distributions (P < 0.001).
It has also been clear that none of the chromosome, plasmid, or carbapenemase genes guarantees the success of a population. We found only weak associations between Klebsiella population and carbapenemase (Cramer’s V (CV): 0.37), carbapenemase and CR-PCs (CV: 0.29), and population and CR-PCs (CV: 0.45), indicating highly dynamic HGTs at all three levels. Even the five predominant populations are each associated with multiple carbapenemase genes and CR-PCs (Fig. 4b), and so are the five predominant PCs. Furthermore, while HC1360_8 (CC258) emerged after its acquisition of blaKPC-carrying PC_341, the same plasmid has also been found in 155 other HC1360s, most of which had very few clinical isolates. Similarly, blaNDM-carrying PC_394 was acquired by 99 HC1360s and resulted in the rise of only HC1360_3 (CC147) (Fig. 4). Moreover, blaNDM genes spread along in the “minority” HC1360s independent of the selective advantages in the hosts. All these findings demonstrate the influence of co-evolution in the emergence of Klebsiella populations: only those that had selective advantages in both chromosome and plasmids demonstrate prevalence.
The plasmid-mediated colistin resistance genes were first discovered in 2015 [42], posing a significant threat due to their potential for rapid spread of colistin resistance. We associated 70% of the mcr genes in Klebsiella to only two PCs of PC_456 (IncHI2A) and PC_1293 (IncX4) (Fig. 6). PC_456 exhibited a broad host range and carried both mcr-1 and mcr-9. In contrast, PC_1293 exclusively carried the mcr-1 genes and was found in a narrower range of hosts. These findings highlighted the fact that the characteristics of the plasmids pose a strong influence on the spreading potential and destiny of its associated ARGs.
The inclusion of over 30,000 strains in this study allowed a comprehensive overview of the population distribution worldwide. For example, substantial geographic specificities were found for HC1360_10 (CC14) and HC1360_52 (CC17), which were primarily found around the Indian Ocean and in Southeast Asia, respectively. Furthermore, our analysis revealed the global prevalence of the HC1360_8 (CC258) lineage during 2003–2018 (Fig. 4), in both the global dataset and two independent longitudinal surveillance projects [42, 57]. Carbapenem resistance in HC1360_8 was primarily attributed to blaKPC genes (87%), blaOXA (8%), and blaNDM (6%), each associated with a different panel of plasmids. Over 52% of blaOXA in HC1360_8 was associated with PC_804, which was also the primary source of blaOXA in other populations. Geographic specificity was found in HC1360_8 for blaKPC-carrying plasmids, which were PC_341 in Europe and the US and PC_499 and PC_362 in China. Finally, similar to other populations, blaNDM in HC1360_8 was associated with > 20 PCs with no clear geographical preference. These findings confirmed previous reports [58] and further revealed that HC1360_8 has not carried a different carbapenemase than other, less prevalent populations. Additionally, HC1360_148 (CC307) and HC1360_3 (CC147) were regarded as emerging populations recently, and we found that both populations have been reported across almost all continents, underscoring their threat to public health [59].
The recent emergence of hvCRKP due to AMR-virulence convergence raises major concerns due to the high bloodstream infection rates and limited antimicrobial treatment options [3]. While the overall frequencies of AMR-virulence convergences remained low (4.5%), substantially more hvCRKP strains have been sequenced in the past 5 years, up to ~ 20% (Fig. 4a). This could be partially attributed to sampling bias, while there was a general trend of two emerging populations: (1) blaKPC-2-carrying ST11-K64 hvCRKPs in HC1360_8 and (2) the blaNDM-carrying hvCRKPs in HC1360_3 (CC147). ST11-K64 hvCRKP strains have resulted from a convergence of classical virulence plasmid, pLVPK (PC_499) with CR-PCs [60]. It was first reported in 2017 in Zhejiang [8] and later also found in almost all provinces in China as part of an ongoing clonal replacement [61]. Our phylogenetic reconstruction of PC_499 (Fig. 5a) revealed that most (> 90%) of the blaKPC-virulence convergence in ST11-K64 hvCRKPs are associated with a narrow spectrum of PC_499 plasmids, indicating high genetic stability of the convergence. Notably, these hvCRKPs have been rarely found in other countries, indicating the presence of other, unknown factors that limit the spread of hvCRKP. High associations have been recognized between hvCRKPs and the virulence plasmid markers (iuc, iro, rmpA, rmpA2, peg-344) [62]. Here we selected iuc as the marker for hv-PCs because it has been well investigated and exhibited a direct association with sepsis, promoting the blood growth of bacteria by acquiring iron from transferrin [4].
Many of the hvCRKPs reported after 2019 belonged to HC1360_3 (CC147). This new hvCRKP group is of particular concern because both the bacterial hosts and the associated plasmids have been widely reported in Asia, Europe, and the Americas for decades before the AMR-virulence convergences. Furthermore, the HC1360_3 hvCRKPs have been causing disease outbreaks in Russia, Italy, and the USA [43, 47, 48]. Our results showed that all three outbreaks were associated with one genetically stable hvCR cluster in PC_369, and the resulting hvCRKPs likely have been endemic in each region for decades. These hvCRKPs likely have contributed to the potential emergence of HC1360_3 in the past 3 years. Effective research and controls are urgently needed for this previously underestimated population.
We acknowledge the limitations of our study. Sampling bias and incompleteness of the metadata may have limited our ability to accurately determine the prevalence of HC1360s and plasmids in specific regions. Specifically, many projects focused on clinically relevant isolates, particularly CRKP, contributing to a sampling bias in public databases and affecting our overall analysis. Additionally, the reference plasmid database may introduce bias, particularly for plasmids of highly variable or uncharacterized sequences. Broader samplings of both Klebsiella strains and plasmids from diverse sources are urgently needed to enhance our understanding of the genetic context of these pathogens and their associated ARGs.
Conclusions
In summary, we investigated the genetic landscape of Klebsiella, demonstrating the role of chromosome-plasmid interactions in facilitating the dissemination of antimicrobial resistance and virulence genes. We revealed two sequential global pandemic populations, HC1360_8 (CC258) which was primarily associated with blaKPC-carrying PC_341, and HC1360_3 (CC147) which has blaNDM-carrying PC_394. An ongoing expansion of carbapenemase-hypervirulence convergences was reported in both populations, underscoring the importance of understanding the association between plasmids and specific populations and genes, prompting monitoring of plasmids for effective prevention and control of serious infections caused by K. pneumoniae.
Availability and requirements
Project name: KleTy.
Project home page: https://github.com/zheminzhou/KleTy.
Operating system(s): Platform independent.
Programming language: Python.
Other requirements: None.
License: GNU General Public License v3.0.
Any restrictions to use by non-academics: None.
Supplementary Information
Additional file 1: Supplementary Tables 1–12.
Additional file 1: Supplementary Figs. 1–13.
Acknowledgements
We thank Dr. Karl Drlica for his critical reading and valuable comments.
Authors’ contributions
Conceptualization: ZZ, YH, and HL; resources: XL, HL, ZZ, and YG; methodology: ZZ, SL, SX, and YR; software: SL, GJ, and XL; writing-original draft preparation: HL and XL; writing-review and editing: HL, XL, JR, LZ, QJ, YH, WS, and ZZ; project administration: ZZ; all authors read and approved the final manuscript.
Funding
ZZ was supported by the National Natural Science Foundation of China (32170003, 32370099), the Natural Science Foundation of Jiangsu Province (BK20211311), the Provincial-level Talent Program for National Center of Technology Innovation for Biopharmaceuticals (NCTIB2024JS0101), Jiangsu Specially-appointed Professor Project, the Suzhou Top-Notch Talent Groups (ZXD2022003) and the Suzhou Science and Technology Innovations Project in Health Care (SKY2021013). YH was supported by the National Natural Science Foundation of China (32170032, 32370034), the National Major Youth Talent Project A, the Jiangsu Specially-appointed Professor Project, the Suzhou Innovation Leading Talent Project (ZXL2022456), and the Jiangsu Improvement Project of Science, Technology, and Education (CXZX202231). HL was supported by the National Natural Science Foundation of China (82202465). LZ was supported by the Graduate Research and Innovation Projects of Jiangsu Province (KYCX22_3187). LX was supported by National High-Level Hospital Clinical Research Funding (2022-PUMCH-A-114).
Data availability
The KleTy command-line toolbox is hosted on GitHub at https://github.com/zheminzhou/KleTy. A detailed README and test dataset are included. The entire set of 33,272 public Klebsiella genomes is available in GenBank, with the accession codes in Additional file 1: Table S1. The 1271 simulated draft assemblies for benchmarking plasmid prediction tools are available at https://doi.org/10.5281/zenodo.12633486.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
SX and YR are currently employed by Iotabiome Biotechnology Inc., a commercial entity. The company did not have any role in the design, execution, or interpretation of the research, nor did it influence the content or conclusions presented in this manuscript. All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Heng Li, Xiao Liu, Shengkai Li and Jie Rong contributed equally to this work.
Contributor Information
Wanping Sun, Email: sunwanping@suda.edu.cn.
Yuzhi Hong, Email: yzhong@suda.edu.cn.
Zhemin Zhou, Email: zmzhou@suda.edu.cn.
References
- 1.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–75. [DOI] [PubMed] [Google Scholar]
- 2.Lan P, Jiang Y, Zhou J, Yu Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26–34. [DOI] [PubMed] [Google Scholar]
- 3.Shankar C, Nabarro LE, Anandan S, Ravi R, Babu P, Munusamy E, et al. Extremely high mortality rates in patients with Carbapenem- resistant, hypermucoviscous klebsiella pneumoniae blood stream infections. J Assoc Physicians India. 2018;66:13–6. [PubMed] [Google Scholar]
- 4.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hennart M, Guglielmini J, Bridel S, Maiden MCJ, Jolley KA, Criscuolo A, et al. A dual barcoding approach to bacterial strain nomenclature: genomic taxonomy of klebsiella pneumoniae strains. Mol Biol Evol. 2022;39:msac135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong L, Zhang M, Sun L, Yang Y, Wang B, Yang H, et al. Distributed genotyping and clustering of Neisseria strains reveal continual emergence of epidemic meningococcus over a century. Nat Commun. 2023;14:7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durrant MG, Li MM, Siranosian BA, Montgomery SB, Bhatt AS. A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe. 2020;27:140–153.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. [DOI] [PubMed] [Google Scholar]
- 9.Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–9. [DOI] [PubMed] [Google Scholar]
- 10.Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microbial Genomics. 2018;4: e000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Z, Alikhan N-F, Mohamed K, Fan Y, the Agama Study Group, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- 13.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kultima JR, Sunagawa S, Li J, Chen W, Chen H, Mende DR, et al. MOCAT: a metagenomics assembly and gene prediction toolkit. PLoS ONE. 2012;7: e47656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z, Charlesworth J, Achtman M. Accurate reconstruction of bacterial pan- and core genomes with PEPPAN. Genome Res. 2020;30:1667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Z, Charlesworth J, Achtman M. HierCC: a multi-level clustering scheme for population assignments based on core genome MLST. Bioinformatics. 2021;37:3645–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Achtman M, Zhou Z, Charlesworth J, Baxter L. EnteroBase: hierarchical clustering of 100 000s of bacterial genomes into species/subspecies and populations. Philos Trans R Soc Lond B Biol Sci. 2022;377:20210240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francisco AP, Bugalho M, Ramirez M, Carriço JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics. 2009;10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X. BinDash, software for fast genome distance estimation on a typical personal laptop. Bioinformatics. 2019;35:671–3. [DOI] [PubMed] [Google Scholar]
- 20.Traag VA, Waltman L, Van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fruchterman TMJ, Reingold EM. Graph drawing by force-directed placement. Softw Pract Exp. 1991;21:1129–64. [Google Scholar]
- 22.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. ICWSM. 2009;3:361–2. [Google Scholar]
- 23.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin S, Brown WM, Klavans R, Boyack KW. OpenOrd: an open-source toolbox for large graph layout. Proc SPIE. 2011;7868: 786806. [Google Scholar]
- 25.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, McCann A, Weill F-X, Blin C, Nair S, Wain J, et al. Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc Natl Acad Sci U S A. 2014;111:12199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sagulenko P, Puller V, Neher RA. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace DL. A method for comparing two hierarchical clusterings: comment. J Am Stat Assoc. 1983;78:569. [Google Scholar]
- 32.Arredondo-Alonso S, Rogers MRC, Braat JC, Verschuuren TD, Top J, Corander J, et al. mlplasmids: a user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microbial Genomics. 2018;4: e000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Q, Gao S, Xiao B, He Z, Hu S. Plasmer: an accurate and sensitive bacterial plasmid prediction tool based on machine learning of shared k-mers and genomic features. Microbiol Spectr. 2023;11:e04645–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwengers O, Barth P, Falgenhauer L, Hain T, Chakraborty T, Goesmann A. Platon: identification and characterization of bacterial plasmid contigs in short-read draft assemblies exploiting protein sequence-based replicon distribution scores. Microbial Genomics. 2020;6:mgen000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian R, Imanian B. PlasmidHunter: Accurate and fast prediction of plasmid sequences using gene content profile and machine learning. preprint. 2023; 10.1101/2023.02.01.526640. [DOI] [PMC free article] [PubMed]
- 36.Camargo AP, Roux S, Schulz F, Babinski M, Xu Y, Hu B, et al. Identification of mobile genetic elements with geNomad. Nat Biotechnol. 2023. 10.1038/s41587-023-01953-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, et al. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. 2021;11:12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David S, Reuter S, Harris SR, Glasner C, the EuSCAPE Working Group, the ESGEM Study Group, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Che Y, Yang Y, Xu X, Břinda K, Polz MF, Hanage WP, et al. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc Natl Acad Sci U S A. 2021;118: e2008731118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei Z-Q, Du X-X, Yu Y-S, Shen P, Chen Y-G, Li L-J. Plasmid-Mediated KPC-2 in a Klebsiella pneumoniae Isolate from China. Antimicrob Agents Chemother. 2007;51:763–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutgring JD, Kent AG, Bowers JR, Jasso-Selles DE, Albrecht V, Stevens VA, et al. Comparison of carbapenem-susceptible and carbapenem-resistant Enterobacterales at nine sites in the USA, 2013–2016: a resource for antimicrobial resistance investigators. Microbial Genomics. 2023;9: 001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherry NL, Lane CR, Kwong JC, Schultz M, Sait M, Stevens K, et al. Genomics for molecular epidemiology and detecting transmission of carbapenemase-producing Enterobacterales in Victoria, Australia, 2012 to 2016. J Clin Microbiol. 2019;57:e00573–19. https://journals.asm.org/doi/full/10.1128/jcm.00573-19. [DOI] [PMC free article] [PubMed]
- 43.Di Pilato V, Henrici De Angelis L, Aiezza N, Baccani I, Niccolai C, Parisio EM, et al. Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. Lancet Microbe. 2022;3:e224–34. [DOI] [PubMed] [Google Scholar]
- 44.Pei N, Li Y, Liu C, Jian Z, Liang T, Zhong Y, et al. Large-scale genomic epidemiology of klebsiella pneumoniae identified clone divergence with hypervirulent plus antimicrobial-resistant characteristics causing within-ward strain transmissions. Microbiol Spectr. 2022;10:e02698–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie M, Yang X, Xu Q, Ye L, Chen K, Zheng Z, et al. Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun Biol. 2021;4:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Damjanova I, Toth A, Paszti J, Hajbel-Vekony G, Jakab M, Berta J, et al. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type -lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005–the new “MRSAs”? J Antimicrob Chemother. 2008;62:978–85. [DOI] [PubMed] [Google Scholar]
- 47.Lapp Z, Crawford R, Miles-Jay A, Pirani A, Trick WE, Weinstein RA, et al. Regional Spread of bla NDM-1-Containing Klebsiella pneumoniae ST147 in post-acute care facilities. Clin Infect Dis. 2021;73:1431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starkova P, Lazareva I, Avdeeva A, Sulian O, Likholetova D, Ageevets V, et al. Emergence of hybrid resistance and virulence plasmids harboring new Delhi Metallo-β-Lactamase in Klebsiella pneumoniae in Russia. Antibiotics. 2021;10: 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8. [DOI] [PubMed] [Google Scholar]
- 50.Matsumura Y, Peirano G, Motyl MR, Adams MD, Chen L, Kreiswirth B, et al. Global molecular epidemiology of IMP-producing enterobacteriaceae. Antimicrob Agents Chemother. 2017;61:e02729–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hetland MAK, Hawkey J, Bernhoff E, Bakksjø R-J, Kaspersen H, Rettedal SI, et al. Within–patient and global evolutionary dynamics of Klebsiella pneumoniae ST17. Microbial Genomics. 2023;9:mgen001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae Clonal Groups. Emerg Infect Dis. 2014;20:1812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meunier D. Florfenicol resistance in Salmonella enterica serovar Newport mediated by a plasmid related to R55 from Klebsiella pneumoniae. J Antimicrob Chemother. 2003;51:1007–9. [DOI] [PubMed] [Google Scholar]
- 54.Das B, Verma J, Kumar P, Ghosh A, Ramamurthy T. Antibiotic resistance in Vibrio cholerae: Understanding the ecology of resistance genes and mechanisms. Vaccine. 2020;38:A83–92. [DOI] [PubMed] [Google Scholar]
- 55.Redondo-Salvo S, Fernández-López R, Ruiz R, Vielva L, de Toro M, Rocha EPC, et al. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat Commun. 2020;11:3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coluzzi C, Garcillán-Barcia MP, De La Cruz F, Rocha EPC. Evolution of plasmid mobility: origin and fate of conjugative and nonconjugative plasmids. Mol Biol Evol. 2022;39: msac115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duffy N, Karlsson M, Reses HE, Campbell D, Daniels J, Stanton RA, et al. Epidemiology of extended-spectrum β-lactamase–producing Enterobacterales in five US sites participating in the Emerging Infections Program, 2017. 2022; [DOI] [PMC free article] [PubMed]
- 58.David S, Cohen V, Reuter S, Sheppard AE, Giani T, Parkhill J, et al. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2020;117:25043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18:344–59. [DOI] [PubMed] [Google Scholar]
- 60.Yang X, Sun Q, Li J, Jiang Y, Li Y, Lin J, et al. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes & Infections. 2022;11:841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Q, Jia X, Zhou M, Zhang H, Yang W, Kudinha T, et al. Emergence of ST11-K47 and ST11-K64 hypervirulent carbapenem-resistant Klebsiella pneumoniae in bacterial liver abscesses from China: a molecular, biological, and epidemiological study. Emerg Microb Infect. 2020;9:320–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J Clin Microbiol. 2018;56:e00776–00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Tables 1–12.
Additional file 1: Supplementary Figs. 1–13.
Data Availability Statement
The KleTy command-line toolbox is hosted on GitHub at https://github.com/zheminzhou/KleTy. A detailed README and test dataset are included. The entire set of 33,272 public Klebsiella genomes is available in GenBank, with the accession codes in Additional file 1: Table S1. The 1271 simulated draft assemblies for benchmarking plasmid prediction tools are available at https://doi.org/10.5281/zenodo.12633486.