Abstract
PURPOSE
Radium-223 improves overall survival (OS) and reduces skeletal events in patients with bone metastatic castration-resistant prostate cancer (CRPC), but relevant biomarkers are lacking. We evaluated automated bone scan index (aBSI) and circulating tumor cell (CTC) analyses as potential biomarkers of prognosis and activity.
PATIENTS AND METHODS
Patients with bone metastatic CRPC were enrolled on a prospective single-arm study of standard radium-223. 99mTc-MDP bone scan images at baseline, 2 months, and 6 months were quantitated using aBSI. CTCs at baseline, 1 month, and 2 months were enumerated and assessed for RNA expression of prostate cancer–specific genes using microfluidic enrichment followed by droplet digital polymerase chain reaction.
RESULTS
The median OS was 21.3 months in 22 patients. Lower baseline aBSI and minimal change in aBSI (<+0.7) from baseline to 2 months were each associated with better OS (P = .00341 and P = .0139, respectively). The higher baseline CTC count of ≥5 CTC/7.5 mL was associated with worse OS (median, 10.1 v 32.9 months; P = .00568). CTCs declined at 2 months in four of 15 patients with detectable baseline CTCs. Among individual genes in CTCs, baseline expression of the splice variant AR-V7 was significantly associated with worse OS (hazard ratio, 5.20 [95% CI, 1.657 to 16.31]; P = .00195). Baseline detectable AR-V7, higher aBSI, and CTC count ≥5 CTC/7.5 mL continued to have a significant independent negative impact on OS after controlling for prostate-specific antigen or alkaline phosphatase.
CONCLUSION
Quantitative bone scan assessment with aBSI and CTC analyses are prognostic markers in patients treated with radium-223. AR-V7 expression in CTCs is a particularly promising prognostic biomarker and warrants validation in larger cohorts.
INTRODUCTION
Radium-223 was US Food and Drug Administration (FDA)–approved in 2013 for the treatment of patients with castration-resistant prostate cancer (CRPC) with bone metastases and no known visceral metastases. In this clinical setting, radium-223 was shown in the ALSYMPCA trial to improve overall survival (OS),1 delay time to first symptomatic skeletal events,2 and improve quality of life as measured by patient-reported outcome (PRO) surveys.3 Despite these benefits and its widespread availability, prognostic and predictive biomarkers to guide the use of radium-223 are lacking. Prostate-specific antigen (PSA) responses are uncommon,4 and imaging assessment of bone metastases is limited.5
CONTEXT
Key Objective
Radium-223 can be an effective treatment for castration-resistant prostate cancer metastatic to bone, but biomarkers to guide its use are lacking. In this prospective study of patients receiving radium-223, automated 99mTc-MDP bone scan index (automated bone scan index [aBSI]) and circulating tumor cell (CTC) analyses were evaluated as potential biomarkers of prognosis and activity. To our knowledge, this is the first study to analyze CTC gene expression in this context.
Knowledge Generated
Lower baseline aBSI and minimal change in aBSI after 2 months of therapy were associated with better survival. High baseline CTC counts and AR-V7 expression in CTCs were found to be associated with poor survival after radium-223 therapy.
Relevance
If validated in larger cohorts, these biomarkers may be useful in guiding the use of radium-223. Stability of aBSI after 2 months identifies patients who are likely to benefit, whereas high levels of CTCs or AR-V7 at baseline suggest that other treatments should be considered.
In ALSYMPCA, serum PSA declines only occurred in 27% of patients and the mean percent PSA change at week 12 was an increase of 83.3%.4 A decrease in total alkaline phosphatase (tAP) at 12 weeks was observed in 87% of radium-223 patients and associated with better OS, but the role of tAP monitoring was less clear when not elevated at baseline. Imaging assessment of patients receiving radium-223, who have bone-predominant disease, has also been limited. On the basis of RECIST,6 blastic bone metastases typical of prostate cancer are not measurable. The Prostate Cancer Clinical Trials Working Group 3 (PCCTWG3) advises recording changes qualitatively.5 Radiographic assessment of this population is therefore limited.
Bone scan remains the most widely available imaging study for assessment of prostate cancer bone metastases. 68Ga and 18F prostate-specific membrane antigen positron emission tomography tracers have also become important imaging tools,7-10 but their serial use as biomarkers of progression or response is not yet established. Although bone scans to assess response to systemic therapies are qualitative in clinical practice, quantitative analysis offers a potential alternative. Automated bone scan index (aBSI) is a reproducible quantitative parameter, calculated using a computer-assisted diagnosis (CAD) system, that reflects percentage of total skeletal mass occupied by tumor.11 aBSI baseline value and its change during systemic therapy have been shown to be prognostic in several settings.12-18 Prospective studies in the setting of radium-223 have not been reported.
Circulating tumor cells (CTCs) represent a form of liquid biopsy. A CTC count of ≥5 per 7.5 mL blood using the CellSearch assay correlates with worse OS in patients with metastatic CRPC treated with different therapies,19 but this has not been prospectively demonstrated for radium-223. Beyond CellSearch, efficient microfluidic CTC isolation technology enables molecular analyses.20 A recent study using the microfluidic CTC-iChip established a droplet digital polymerase chain reaction (ddPCR) gene expression assay for quantitation of a composite digital CTCM score, on the basis of a panel of eight prostate-derived CTC transcripts, that predicts poor survival after abiraterone.21 In addition, expression of the androgen receptor (AR) splice variant AR-V7 in CTCs has been correlated with resistance to AR targeted therapies but not to docetaxel chemotherapy.22,23 AR-V7 has been shown to potentially mediate DNA repair and radiation resistance in vitro.24 It is unknown whether the presence of AR-V7 in CTCs correlates with response or prognosis in patients who receive radium-223.
We aimed to address these unmet needs through a single-arm prospective biomarker-driven study of radium-223 on its FDA-approved schedule for patients with bone metastatic CRPC who were candidates for standard-of-care radium-223.
PATIENTS AND METHODS
Participants
Participants were 18 years and older with confirmed prostate adenocarcinoma with bone-predominant metastases (≥two skeletal metastases with no lung, liver, or known brain metastases) and castration resistance. All were judged to have progressive disease sufficient to clinically justify standard-of-care radium-223 treatment. Notable exclusion criteria included previous treatment with a radionuclide, previous radiation to >25% of the bone marrow, fecal incontinence, and lymphadenopathy >6 cm and/or contributing to concurrent hydroureteronephrosis. The trial was Dana Farber/Harvard Cancer Center institutional review board–approved (protocol 14-075) and registered at ClinicalTrials.gov identifier: NCT02346526.
Treatment
This was a single-institution, single-arm prospective study in which all participants received radium-223 once monthly for up to six doses. The dose of radium-223 dichloride was 50-55 kBq/kg body weight as a bolus injection for up to six cycles. Participants were maintained on standard androgen deprivation therapy throughout. Study therapy was continued in the absence of clinical grounds for discontinuation (ie, clinical progression or unacceptable side effects as assessed by the treating investigator).
End Points
This was an exploratory biomarker study designed to assess imaging and circulatory biomarkers during treatment with radium-223. Bone scans were performed at pretreatment baseline, 2 months, and 6 months, and CTC analyses were performed at pretreatment baseline, 1 month, and 2 months (Fig 1A; Appendix Fig A1). OS was the clinical end point correlated with the exploratory biomarkers.
FIG 1.
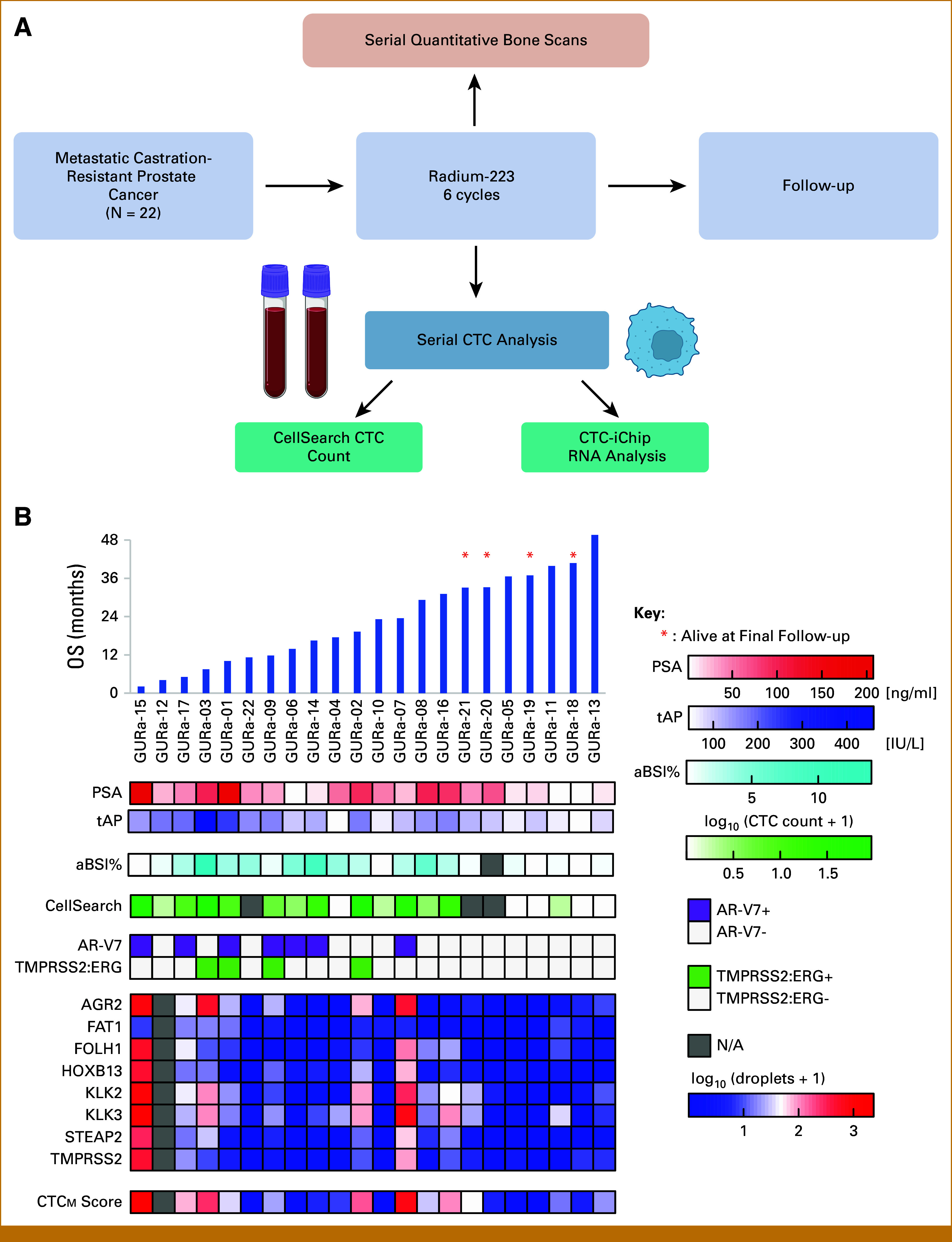
(A) Study schematic. Serial bone scans were obtained at pretreatment baseline, 2 months, and 6 months. Serial CTC analyses were performed using two different CTC methods at pretreatment baseline, 1 month, and 2 months. A detailed flow diagram of actual patient evaluations collected is shown in Appendix Figure A1. (B) Bar graph in the top panel shows patients in a clinically uniform prospective cohort (N = 22) ordered by OS from left to right (vertical blue bars). Four patients noted with red asterisks were alive at the time of final follow-up. Heatmap shows the level of signal of various assays for individual patients, including CTC expression of AR-V7, TMRPSS2:ERG, and prostate genes, CTCM score, CellSearch CTC count, and %aBSI. aBSI, automated bone scan index; CTC, circulating tumor cell; NA, not applicable; OS, overall survival; PSA, prostate-specific antigen; tAP, total alkaline phosphatase.
PRO Assessments
All participants were given validated PRO surveys at the time of study assessments at baseline and during therapy. These included the 5-level EuroQol (Eq-5D-3L) instrument and the MD Anderson Brief Pain Inventory (BPI). EuroQol included a Global Health Score. The BPI included a BPI Summation Pain Score.
aBSI Assessments
aBSI is a quantitative measure of the percentage of total skeletal mass occupied by tumor, calculated from 99mTc-MDP bone scan images using a CAD system.11 aBSI was calculated using aBSI v.3.6.1 software (EXINI Diagnostics AB, Sweden). As described by Ulmert et al,11 anatomic regions were segmented with abnormal hotspots automatically identified and classified as benign or metastatic. The mass fraction of the skeleton for each metastatic hot spot was determined, and the aBSI calculated as the sum of all such fractions. The hotspots were manually reviewed by a radiologist with fellowship training in nuclear medicine and molecular imaging before approval of the quantitative analysis. Assessments were at baseline, 2 months, and 6 months.
CTCs
We used two CTC assays: the commercially available CellSearch assay25 and the microfluidic CTC-iChip.26 RNA from enriched CTC samples (CTC-iChip product) was extracted. Genes expressed in CTCs (including eight prostate cancer–specific genes that comprise the CTCM score, AR-V7, and TMPRSS2:ERG) were assessed using ddPCR as previously described.21 Methods for CTC isolation, RNA extraction, cDNA synthesis, and ddPCR are presented in Appendix 1.
Statistical Analyses
OS curves were generated using the Kaplan-Meier method and compared using the log-rank test with the hazard ratio (HR) and 95% CI estimated by Cox proportional hazards regression. The changepoint method of Contal-O’Quigley27 was used to determine the optimal cut point for aBSI and CTCM scores in OS analysis. For the CellSearch CTC data, the threshold was set as five CTCs/7.5 mL on the basis of previous publications and FDA clearance.28 All genes were compared as ≤threshold versus >threshold. The threshold for each gene was mean + 2 standard deviation of the expression of that gene in healthy donors (data from the study by Miyamoto et al21). Thresholds for AGR2, FAT1, FOLH1, HOXB13, KLK2, KLK3, STEAP2 and TMPRSS2, AR-V7, and TMPRSS2:ERG were 2.96, 17.88, 0.78, 1.21, 0.19, 1.05, 2.07, 0.79, 0, and 0 (droplets/mL), respectively. The Wilcoxon rank-sum test was used to compare CTC counts in AR-V7– versus AR-V7+ patients.
Cox proportional hazards regression was used to control for established prognostic factors in multivariable analysis of promising biomarkers. In the clinical setting of radium-223 therapy, PSA, tAP, performance status (PS), and lactate dehydrogenase (LDH) have previously been reported to be prognostic.4 However, our ability to analyze PS was limited as PS was 0-1 in all patients within this cohort and LDH values were not available, and we therefore limited our analysis to PSA and tAP. To preserve power given the modest sample size, each candidate biomarker was analyzed in the presence of PSA or tAP separately, with two covariates in each multivariable model. PSA and tAP were analyzed as continuous variables, whereas AR-V7, CellSearch CTC count, CTCM score, baseline aBSI, and change in aBSI were dichotomized into binary variables on the basis of detection status, published cutoff, or optimal cut point determined in univariable analysis. As the study was not designed to achieve adequate power for multivariable analysis, P < .15 was used within an exploratory framework for the multivariable analyses to identify candidate biomarkers with the potential for independent prognostic value. For all univariable analyses, the conventional threshold of P < .05 was used for significance. All P values were reported on the basis of a two-sided hypothesis, and statistical analyses were performed using R, version 3.6.3 (Vienna, Austria).
RESULTS
Patient Characteristics and Clinical Prognostic Factors
Twenty-two men with bone metastatic CRPC were enrolled (September 2015-July 2019). Baseline clinical characteristics are detailed in Table 1. Sixty-four percent of participants had six or more bone metastases, and 23% had previous use of docetaxel. The median baseline PSA level was 38.5 ng/mL, and the median baseline tAP level was 117.5 ng/mL. Eighteen patients received at least three planned cycles of radium-223, and 14 of those received all six planned cycles. None of the on-study doses were delayed. The dosing weight range was 64.4-110.7 kg (median, 83.7 kg). Detailed schematics of the enrolled participants and completed assessments are included in Appendix Figure A1 and Figure 1A. Consistent with previous observations with radium-223 treatment,1 serum PSA rose in a majority of patients. PSA progression per PCCTWG35 was a frequent early event as 17 patients met this criterion within 2 months of baseline. The longitudinal behavior of PSA over time in each individual patient is shown in Appendix Figure A2.
TABLE 1.
Baseline Clinical Characteristics of the Cohort
| Characteristic | Radium-223 Cohort (N = 22) |
|---|---|
| Age, years | |
| Median (range) | 71 (49-88) |
| >75, No. (%) | 9 (41) |
| Race, No. (%) | |
| White | 18 (82) |
| Other | 4 (18) |
| Current use of bisphosphonates, No. (%) | |
| Yes | 4 (18) |
| No | 18 (82) |
| Any previous use of docetaxel, No. (%) | |
| Yes | 5 (23) |
| No | 17 (77) |
| ECOG performance status, No. (%) | |
| 0 | 10 (45) |
| 1 | 12 (55) |
| ≥2 | 0 (0) |
| Extent of disease, No. (%) | |
| <6 metastases | 8 (36) |
| 6-20 metastases | 7 (32) |
| >20 metastases | 7 (32) |
| Biochemical values, median (range) | |
| Total alkaline phosphatase, U/L | 117.5 (45-460) |
| PSA, ng/mL | 38.5 (1.1-207.4) |
| WBC, K/µL | 6.4 (4.1-9.1) |
Abbreviations: ECOG, Eastern Cooperative Oncology Group; PSA, prostate-specific antigen.
The median OS for the entire cohort was 21.3 months. Only four participants were alive longer than 30 months at the time of data cutoff (Appendix Table A1). Several baseline characteristics were evaluated for their prognostic value in this prospective cohort. A higher PSA at baseline was associated with worse survival (Fig 1B; median OS, 17.5 months for above median and 36.6 months for below median; log-rank P = .0402; HR, 2.96 [95% CI, 1.000 to 8.767]). Similarly, a higher tAP at baseline was associated with worse survival (Fig 1B; median OS, 11.2 months for above median and 39.9 months for below median; log-rank P = .000271; HR, 6.899 [95% CI, 2.125 to 22.40]). In addition to these biochemical prognostic factors, baseline Global Health Scores <95 (P = .00547; HR, 6.720 [95% CI, 1.462 to 30.90]) and baseline Summation Pain Scores ≥8 (P = .00139; HR, 6.374 [95% CI, 1.780 to 22.82]) were significantly associated with worse OS (Appendix Fig A3).
Quantitative Analysis of Bone Scan at Baseline and Over Time During Treatment Is Prognostic
Baseline aBSI was strongly prognostic (Figs 1B and 2A and 2B). When the study cohort was stratified relative to an optimized cut point of 0.9 (Methods section), men with a lower baseline aBSI value had longer survival (median OS, 39.9 months for aBSI <0.9 and 15.2 months for aBSI ≥0.9; P = .00341; HR, 5.874 [95% CI, 1.582 to 21.81]; Fig 2B). Moreover, the magnitude of change in aBSI from baseline to 2 months on treatment (the pre-specified primary study outcome measure) also significantly correlated with survival. OS was significantly longer for participants who had a relatively stable aBSI at 2 months (<+0.7 increase) compared with an increase of ≥+0.7 (median OS, 36.6 months v 17.9 months; P = .0139; HR, 4.146 [95% CI, 1.220 to 14.09]; Fig 2C). Among 12 patients with unfavorable prognosis baseline aBSI ≥0.9, only two had follow-up bone scan imaging with stability at 2 months (aBSI change <+0.7). If a patient exhibited stability at 2 months, he was likely to complete all six infusions (nine of 10 patients). Patients with 2-month progression were less likely to complete all six infusions (four completed, four discontinued).
FIG 2.
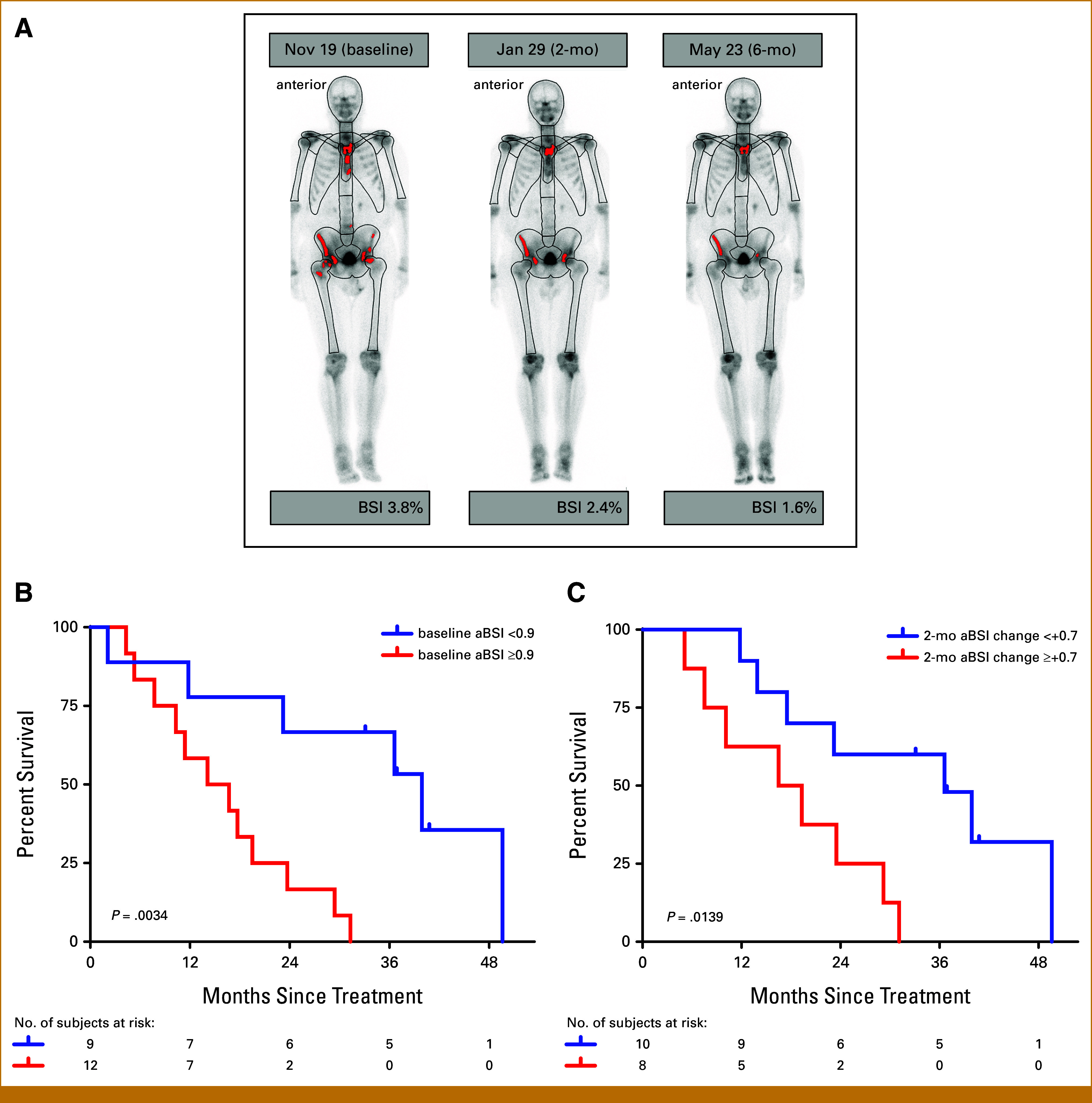
aBSI is prognostic for OS. (A) Time course of aBSI assessments in a single patient with metastatic prostate cancer at baseline (BSI, 3.8%), 2 months (aBSI, 2.4%), and 6 months (aBSI, 1.6%). (B) Kaplan-Meier estimates of OS by baseline aBSI relative to the optimized cut point of 0.9. (C) Kaplan-Meier estimates of OS by aBSI change of ≥+0.7 versus <+0.7 from baseline to 2 months. aBSI, automated bone scan index; OS, overall survival.
CTC Measurements at Baseline Are Prognostic in Patients Receiving Radium-223
We used two independent methods for CTC analysis (see the Methods section; Figs 1A and 1B). Using the previously described CellSearch CTC cutoff value of 5 CTCs/7.5 mL blood,19 patients with a lower baseline CTC count had a significantly longer OS (median OS, 32.9 months if <5 and 10.1 months if ≥5; P = .00568; HR, 4.328 [95% CI, 1.411 to 13.28]; Fig 1B and 3A). Similarly, with the CTC-iChip ddPCR gene expression assay, using an optimized cutoff CTCM score of 20 (see the Methods section), median OS was longer for those with lower baseline CTCM score (36.6 months for CTCM ≤20 and 15.6 months for CTCM >20; P = .0404; HR, 2.995 [95% CI, 0.9967 to 9.002]; Figs 1B and 3B).
FIG 3.
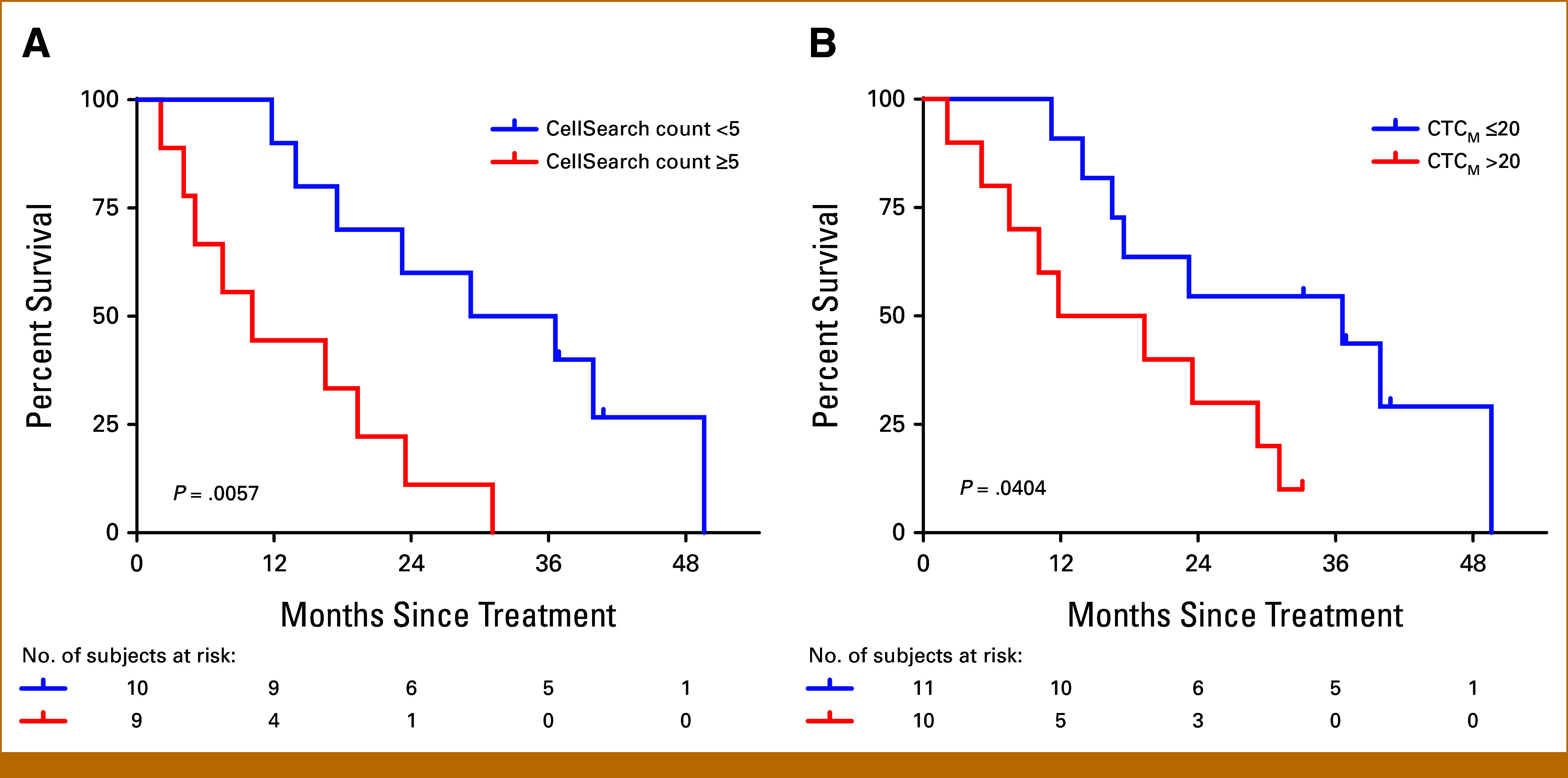
Higher baseline CellSearch count or CTCM score was associated with significantly worse OS. Kaplan-Meier estimates of OS by (A) baseline CellSearch CTC counts greater than or less than five CTCs/7.5 mL blood and (B) baseline CTCM score relative to the optimized cut point of 20. CTC, circulating tumor cell; OS, overall survival.
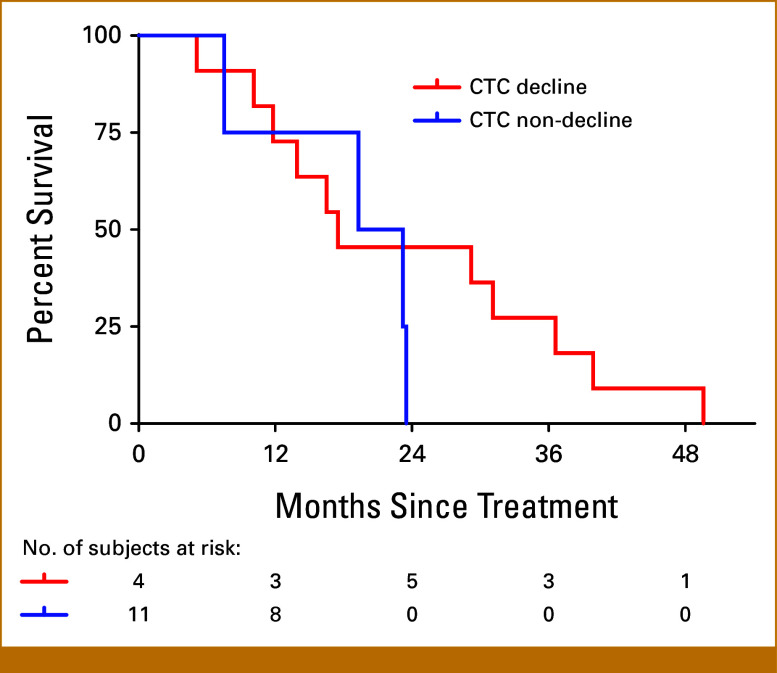
Longitudinal measurements of CellSearch CTC counts showed that a measurable decline in CTC counts during radium-223 therapy was relatively unusual (Appendix Fig A2). Although we had hypothesized that declining CTC counts would correlate with better OS, measurable declines in CTC counts were observed in only four of 15 patients who had detectable CTCs at baseline by the CellSearch assay. CTC decline did not correlate with OS (Appendix Fig A4).
Expression of Individual Genes Within CTCs at Baseline
We examined the prognostic value of baseline expression of individual genes within microfluidically isolated CTCs by ddPCR (Fig 1B). In univariable analyses, expression of classic AR-modulated genes such as KLK2, KLK3, and FOLH1 was not prognostic, but three transcripts (AR-V7, TMPRSS2:ERG, and STEAP2) were significantly associated with poorer OS (Table 2). Of these, AR-V7 stood out as most predictive of poor prognosis. The median OS was 11.8 months for patients with detectable baseline AR-V7 and 31.1 months for patients without detectable baseline AR-V7 (P = .00195; HR, 5.198 [95% CI, 1.657 to 16.31]; Fig 4A). To address the possibility that this finding was an artifact of the presence of CTC rather than a biologic difference, we evaluated for any association of AR-V7 expression with the CellSearch CTC assay at baseline. There was no significant association between CTC count and the presence of AR-V7 (Figs 4B and 4C).
TABLE 2.
Univariable Analysis of Genes Expressed in CTCs and HRs for OS
| Gene | HR (95% CI) | Log-Rank P |
|---|---|---|
| AR-V7 | 5.198 (1.657 to 16.31) | .0020* |
| TMPRSS2:ERG | 3.836 (1.087 to 13.54) | .0252** |
| AGR2 | 1.142 (0.420 to 3.104) | .794 |
| FAT1 | — | — |
| FOLH1 | 1.484 (0.506 to 4.351) | .469 |
| HOXB13 | 3.226 (0.901 to 11.55) | .058 |
| KLK2 | 1.002 (0.361 to 2.782) | .997 |
| KLK3 | 3.208 (0.727 to 14.16) | .104 |
| STEAP2 | 4.32 (1.420 to 13.15) | .0052** |
| TMPRSS2 | 1.474 (0.474 to 4.580) | .500 |
NOTE. HR for OS by expression of each individual gene, analyzed by the Cox proportional hazards regression model. P values are based on the log-rank test. All genes were compared as ≤threshold versus >threshold. The threshold for each individual gene was mean + 2 SD of the expression of that gene in healthy donors. All results for FAT1 in our cohort were less than the threshold.
Abbreviations: CTC, circulating tumor cell; HR, hazard ratio; OS, overall survival; SD, standard deviation.
*P < .01.
**P < .05.
FIG 4.
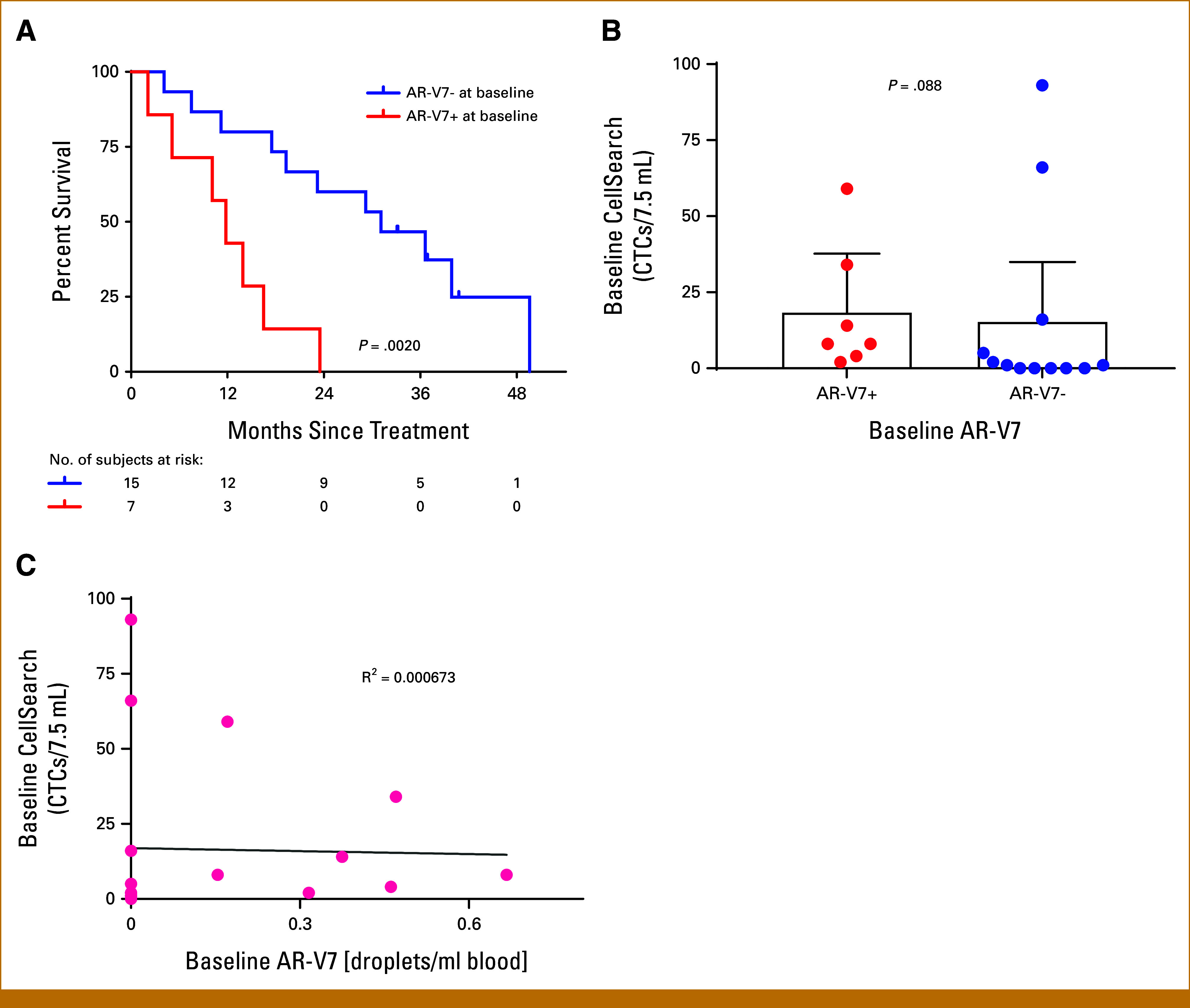
AR-V7 expression in CTCs at baseline is independently prognostic. (A) Kaplan-Meier estimates of OS by the AR-V7 signal in CTCs at baseline. (B) Box plot showing baseline CellSearch CTC counts by AR-V7 status. (C) Scatter plot of baseline CellSearch CTC counts versus baseline AR-V7 signal, showing no significant correlation between CTC count and the presence of AR-V7. CTC, circulating tumor cell; OS, overall survival.
Independent Prognostic Value of Biomarkers
Although our study was not powered for a multivariable analysis that includes multiple covariates in a single model, we used separate multivariable analyses of several promising biomarkers to examine their independent prognostic value when adjusted for established clinical prognostic factors (each model with two covariates). We selected for testing baseline CTCM score and aBSI on the basis of the above-described OS differences in univariable analysis. In addition, we selected baseline detectable AR-V7, baseline CellSearch count ≥5, and change in aBSI value from baseline to 2 months ≥0.7 as three biomarkers with the most potential clinical utility to predict a poor prognosis. AR-V7 expression was chosen given its biologic significance and prognostic value in other clinical settings29-31 and its potential role in mediating radiation resistance.24 Baseline CTC assessment by CellSearch was chosen because it is standardized and available. Change in aBSI at 2 months was chosen because it had the potential to be clinically useful to facilitate decision making at an early on-treatment timepoint.
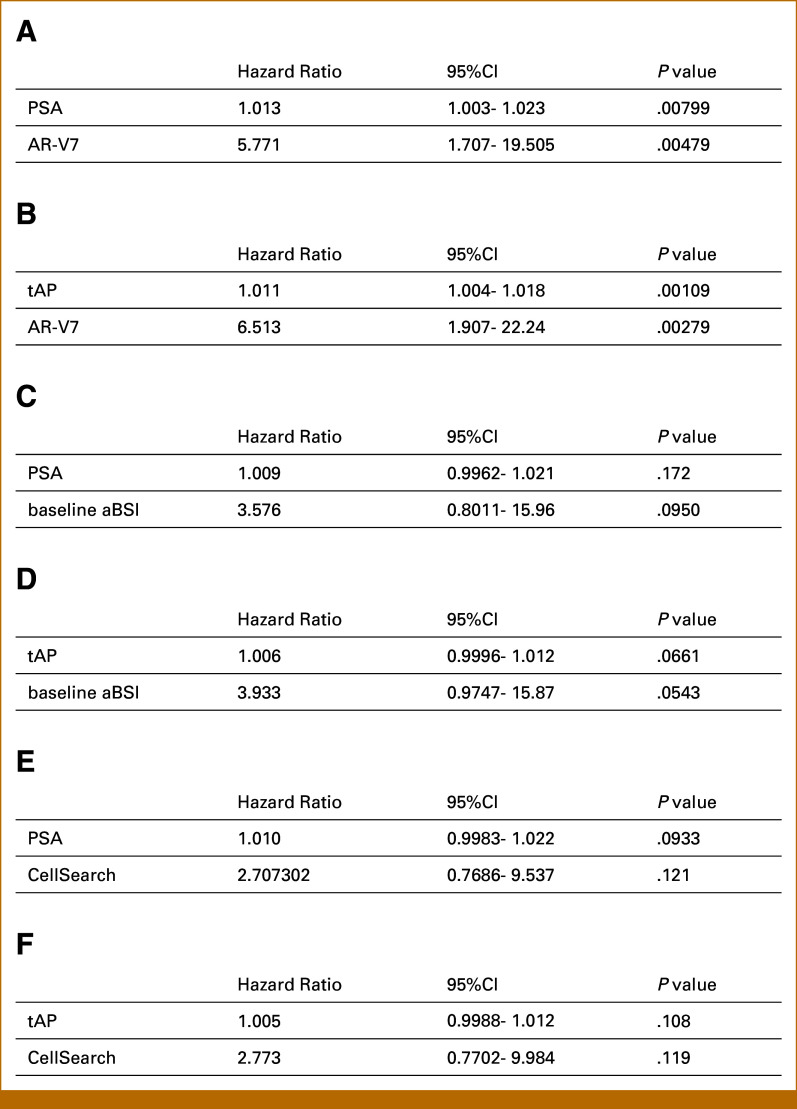
Of the five candidate biomarkers evaluated in these separate exploratory multivariable analyses, baseline AR-V7, baseline aBSI, and baseline CellSearch count retained independent prognostic value after controlling for known prognostic markers PSA or tAP (Appendix Fig A5). The adjusted HR for AR-V7 was 5.771 (95% CI, 1.707 to 19.50; P = .00479) when analyzed with PSA and 6.513 (95% CI, 1.907 to 22.24; P = .00279) when analyzed with tAP. The adjusted HR for baseline aBSI was 3.576 (95% CI, 0.8011 to 15.96; P = .0950) and 3.933 (95% CI, 0.9747 to 15.87; P = .0544), respectively. The adjusted HR for CellSearch count was 2.707 (95% CI, 0.7686 to 9.537; P = .121) and 2.773 (95% CI, 0.7702 to 9.984; P = .119), respectively.
DISCUSSION
The limitations of current standard imaging of prostate cancer within bone are problematic with the bone-predominant metastatic pattern of the radium-223 population. In this prospective biomarker study, quantitative aBSI provided prognostic information at baseline and identified a subset of patients after two infusions of radium-223 who were likely to complete all six infusions. The presence of CTCs at baseline by CellSearch was associated with inferior prognosis. Individual gene expression in CTCs as assessed by an RNA-based digital PCR assay showed that AR-V7 expression was a promising and independent baseline prognostic marker with radium-223.
Quantitative aBSI is an imaging methodology that is widely available and amenable to standardization. aBSI has previously been studied for its prognostic value and as an imaging biomarker of response.12-17,32 We found that baseline aBSI was a powerful prognostic factor. We also found that relative stability of aBSI at 2 months on treatment was a good prognostic factor even if baseline aBSI had been high. In the present cohort, those with stability at 2 months were likely to complete all six radium-223 infusions uneventfully (nine of 10). Given the dearth of informative on-treatment biomarkers in current clinical practice, the use of this 2-month reassessment strategy to identify patients likely to successfully complete all six treatments merits further study in larger multi-institutional cohorts.
Assessment of CTCs in the context of radium-223 is not currently standard clinical practice. One retrospective study focused on the enumeration of CTCs,33 and another evaluated the presence of the gamma-H2AX signal in CTCs as a marker of double-strand DNA breaks.34 To our knowledge, our study is the first to prospectively evaluate molecular RNA signatures in microfluidically isolated CTCs as potential biomarkers of prognosis with radium-223. The eight-gene CTCM panel was optimized to predict drug response and cancer progression in the setting of AR-targeted therapy,21 a clinical setting that is distinct from the non–AR-targeted radium-223. With radium-223, we found that baseline CTC expression levels of multiple AR-responsive genes (eg, KLK2, KLK3, AGR2) were not prognostic. Baseline CTC expression of STEAP2 was significantly associated with poorer prognosis, underscoring the need for further study of the potential importance of this transmembrane protein.35,36
AR-V7 stood out statistically as the CTC-expressed gene most predictive of OS after radium-223. To our knowledge, it has not been previously examined as a prognostic marker with radiopharmaceuticals. Our analysis suggests that AR-V7 is an independent negative prognostic factor in patients with mCRPC treated with radium-223, but this requires further validation in larger cohorts. AR-V7 has been shown to mediate DNA repair and radiation resistance in cell line models,24 providing mechanistic support for the observed association of AR-V7 positivity with poor prognosis despite radium-223.
This study has notable strengths, including its prospective design, clinically uniform cohort, and robust follow-up for OS. Quantitative analysis of bone scan has the advantage of broad availability of the imaging modality. An additional notable strength of the analyses is the integration of multiple types of assessments in a single well-characterized cohort. The depth of gene-by-gene molecular CTC analyses facilitated by the microfluidic CTC-iChip is novel in this context.
One limitation is the relatively small size of this single-institution cohort although it was adequately powered for univariable analysis. As some patients progressed or came off study for other reasons, increasingly fewer patients had data from later timepoints (Appendix Fig A6). Larger multi-institutional trials would be needed to detect independent prognostic value in multivariable analysis. Another limitation is the absence of uniform imaging for radiographic progression-free survival (rPFS) analysis after conclusion of radium-223. Data on post-radium rPFS would have been helpful to better discern the biologic significance of the observed association between AR-V7 and OS. Although aBSI is a well-described technique, application of our findings would require broader adoption of this methodology within clinical workflows. Finally, the AR-V7 RNA-based CTC assay used in this study is not widely available (eg, as compared with the protein-based Epic Sciences AR-V7 CTC assay22,23).
Clinical use of radium-223 is widespread a decade after its approval, but prognostic biomarkers and methods for on-treatment monitoring are limited. An additional study of quantitative aBSI at an early timepoint during radium-223 is needed. Promising baseline prognostic factors in this specific clinical setting include aBSI, the presence or absence of CTCs by CellSearch, and the presence or absence of detectable expression of AR-V7 in CTCs.
ACKNOWLEDGMENT
We are grateful to our patient participants and their families.
APPENDIX 1. SUPPLEMENTARY METHODS
Circulating Tumor Cell Isolation
At each prespecified time point (baseline, 1 months, and 2 months), 10 mL of blood was collected into a CellSave tube for CellSearch analysis and 10 mL of blood was collected into a separate EDTA tube for circulating tumor cell (CTC)-iChip analysis. CellSearch CTC enumeration was performed per the standard protocol25 at Brigham and Women's Hospital CTC Core Lab (Boston, MA). The CTC-iChip is a microfluidic device that isolates CTCs in an epitope-independent manner through negative selection and depletion of known blood components and including red blood cells and leukocytes, thus enriching CTCs that are intact and amenable to RNA expression analysis.26,27 This negative isolation technique does not tag the CTCs during their isolation, minimizing downstream bias. To maximize the recovery of viable CTCs with intact RNA, blood samples were processed within 4 hours of being collected from the patient. Blood samples were processed using the CTC-iChip as previously described.26 Briefly, whole blood samples were spiked with biotinylated antibodies against CD45 (R&D Systems, clone 2D1, Minneapolis, MN), CD66b (AbD Serotec, clone 80H3), and CD16 (Janssen Diagnostics, Titusville, FL), followed by incubation with Dynabeads MyOne Streptavidin T1 (Invitrogen, Waltham, MA) to achieve magnetic labeling of white blood cells. Blood was processed through the CTC-iChip to collect the enriched CTC product on ice. Enriched CTCs were centrifuged at 4,750 rpm, flash-frozen in the presence of RNAlater (Ambion, Waltham, MA), and stored at –80°C to preserve RNA integrity.
RNA Extraction and cDNA Synthesis
RNA from enriched CTC samples (CTC-iChip product) was extracted using the RNeasy Plus Micro Kit (Qiagen, Venlo, The Netherlands) or the AllPrep DNA/RNA Micro Kit (Qiagen, Venlo, The Netherlands). cDNA was generated from purified RNA using the SuperScript III First-Strand Synthesis System (Life Technologies, Waltham, MA).
Droplet Digital Polymerase Chain Reaction
Genes expressed in CTCs (including eight prostate cancer–specific genes that comprise the CTCM score, AR-V7, and TMPRSS2:ERG) were assessed using droplet digital polymerase chain reaction (ddPCR) as previously described using a Bio-Rad automated droplet generator and a droplet reader.21 Briefly, cDNA and primer/probe mixes were combined with ddPCR Supermix for Probes (Bio-Rad, Hercules, CA) in a 96-well plate, and droplets were automatically generated. Next, droplets were subjected to thermal cycling using a modified 45-cycle PCR that featured a 70°C step-down in between denaturation and annealing steps. After thermal cycling, droplets containing the transcript of interest were detected via fluorescence with the QX200 Droplet Reader System (Bio-Rad). The normalized CTCM score was calculated as described using the previously determined weighting coefficients.21
TABLE A1.
Patient-by-Patient Data on Systemic Therapies Before and After Radium-223
| Patient ID | Pre-223Ra Systemic | Cycles of 223Ra | Post-223Ra Systemic | OS, Days |
|---|---|---|---|---|
| 1 | Abiraterone | 6 | Docetaxel | 307 |
| 2 | Docetaxel | 6 | Abiraterone, enzalutamide | 587 |
| 3 | Abiraterone | 3 | Data not available | 228 |
| 4 | Abiraterone, enzalutamide | 6 | None | 533 |
| 5 | Cabozantinib, abiraterone | 6 | None | 1,115 |
| 6 | Abiraterone, docetaxel | 6 | None | 423 |
| 7 | Abiraterone | 6 | Enzalutamide + GS5829, docetaxel, abiraterone | 715 |
| 8 | Abiraterone | 2 | Docetaxel, cabazitaxel, enzalutamide | 888 |
| 9 | Abiraterone | 6 | Enzalutamide | 358 |
| 10 | Abiraterone, enzalutamide | 6 | Docetaxel, cabazitaxel, abiraterone | 706 |
| 11 | Abiraterone | 6 | PCM-075, docetaxel, enzalutamide | 1,214 |
| 12 | Enzalutamide | 1 | None | 125 |
| 13 | None | 6 | Abiraterone, enzalutamide | 1,511 |
| 14 | Abiraterone, docetaxel | 3 | Cabazitaxel | 502 |
| 15 | Abiraterone | 1 | None | 64 |
| 16 | Enzalutamide | 6 | Abiraterone (cabozantinib + atezolizumab) | 947 |
| 17 | Abiraterone | 3 | None | 156 |
| 18 | Abiraterone | 6 | Enzalutamide, docetaxel, 177Lu-PSMA-617 | Alive (1,243) |
| 19 | Docetaxel | 6 | Enzalutamide, abiraterone, cabazitaxel (REGN5678 + cemiplimab) | Alive (1,123) |
| 20 | Abiraterone, sipuleucel-T | 6 | Enzalutamide | Alive (1,011) |
| 21 | Abiraterone | 3 | Enzalutamide, docetaxel, 177Lu-PSMA-617 | Alive (1,008) |
| 22 | Abiraterone, enzalutamide | 2 | None | 342 |
NOTE. Data for each of the study participants are presented individually. Included systemic therapies are those known to prolong OS in the phase III study and those that were given on clinical trials. Four of the patients were alive at final data censorship; their survival duration as of that date is listed in parentheses. Systemic therapies used are a reflection of the timing of study conduct (eg, only late-enrolled and long-lived participants × two received 177Lu-PSMA-617 subsequent to radium-223).
Abbreviation: OS, overall survival.
FIG A1.
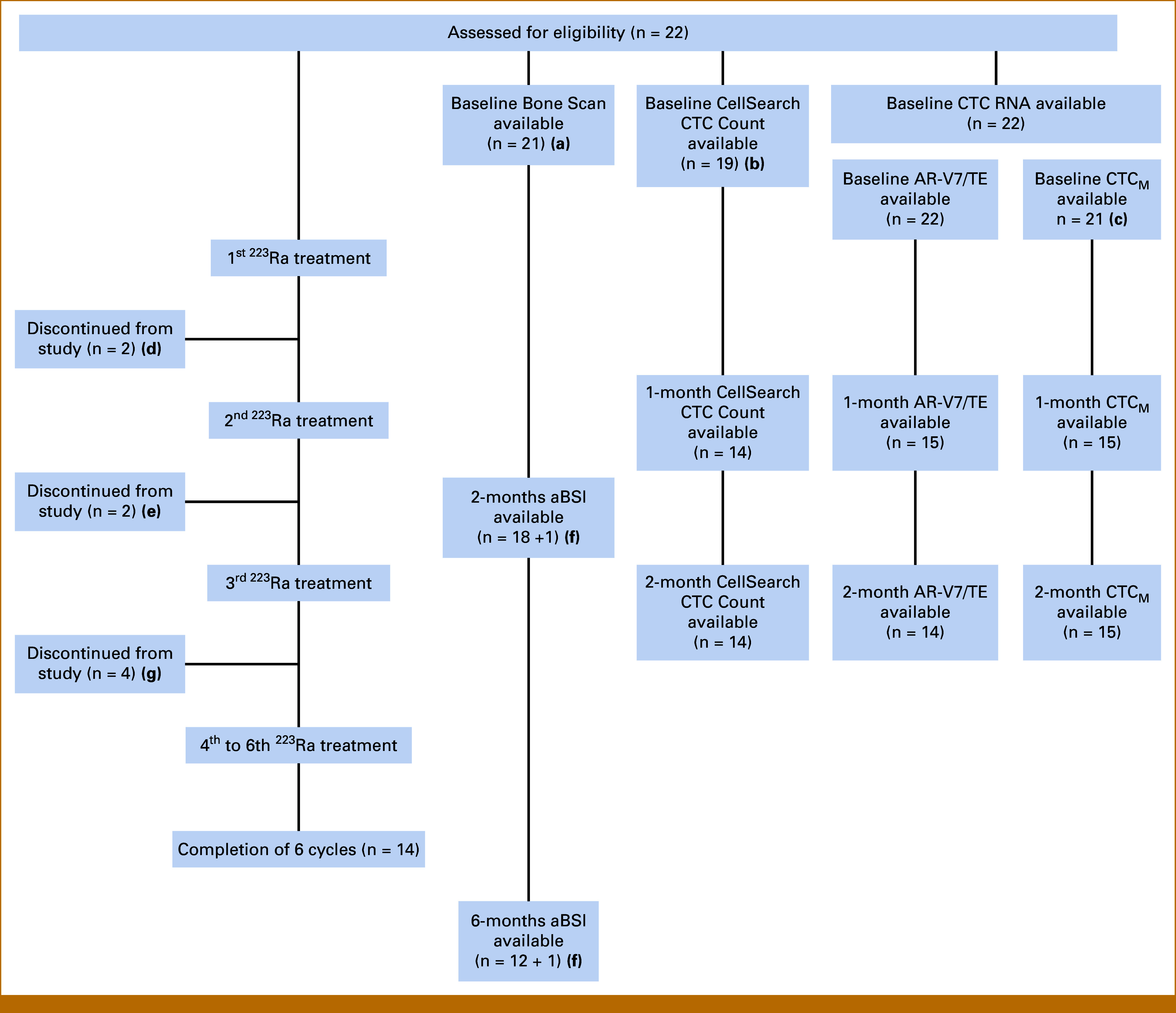
Detailed schematic description of the study. (A) One patient with the bone scan image not analyzable for aBSI; (B) laboratory operating CellSearch CTC enumeration assay closed after recruiting Pt 19; (C) one patient sample had a technical issue during processing; (D) patient decision to discontinue participation; (E) discontinuation from study because of disease progression: one patient discontinued before bone scan, and the other patient discontinued after bone scan; (F) one patient whose baseline image was not analyzable for aBSI, (G) disease progression. aBSI, automated bone scan index; CTC, circulating tumor cell; Pt, patient.
FIG A2.
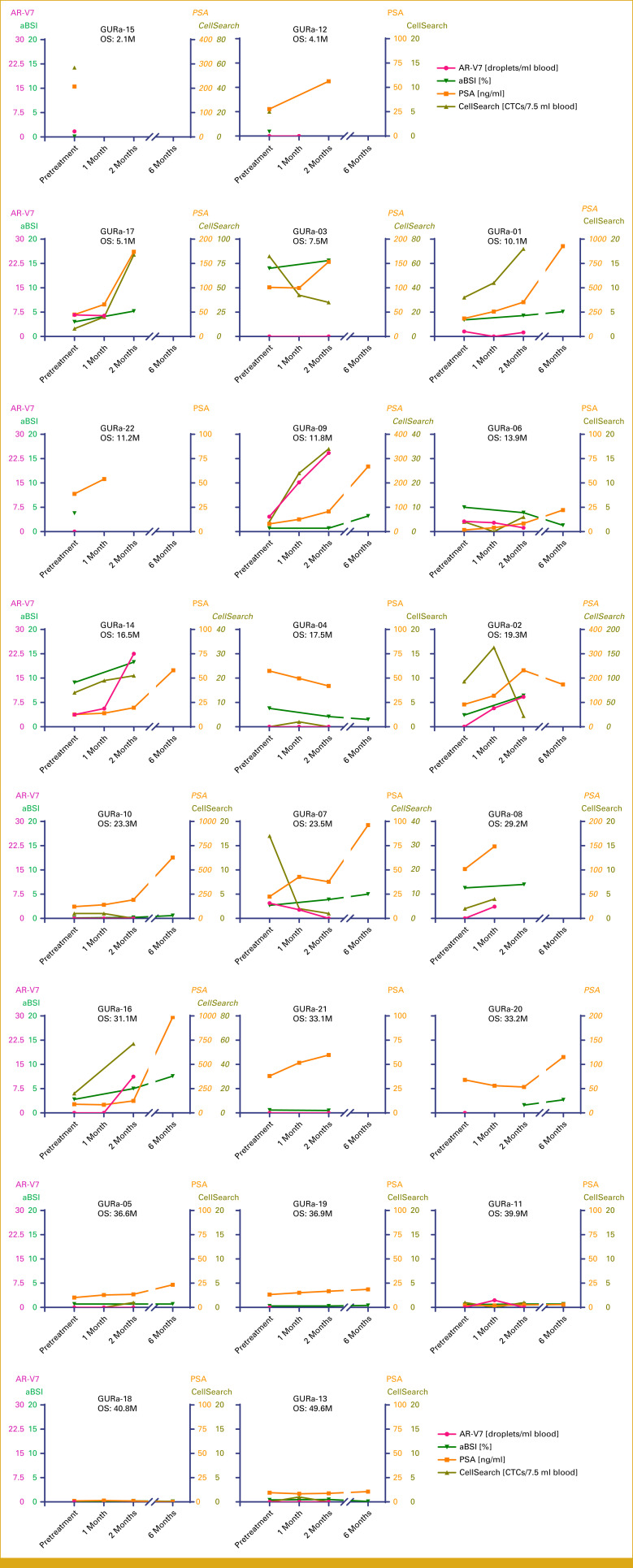
Plots showing longitudinal time courses of AR-V7, aBSI, PSA, and CellSearch CTC counts for each individual participant. Some PSA values and CellSearch CTC counts with high values were scaled differently and shown in italics. From top to bottom, left to right, these are aligned by the order in Figure 1B (ie, shortest survival to longest survival). aBSI, automated bone scan index; CTC, circulating tumor cell; OS, overall survival; PSA, prostate-specific antigen.
FIG A3.
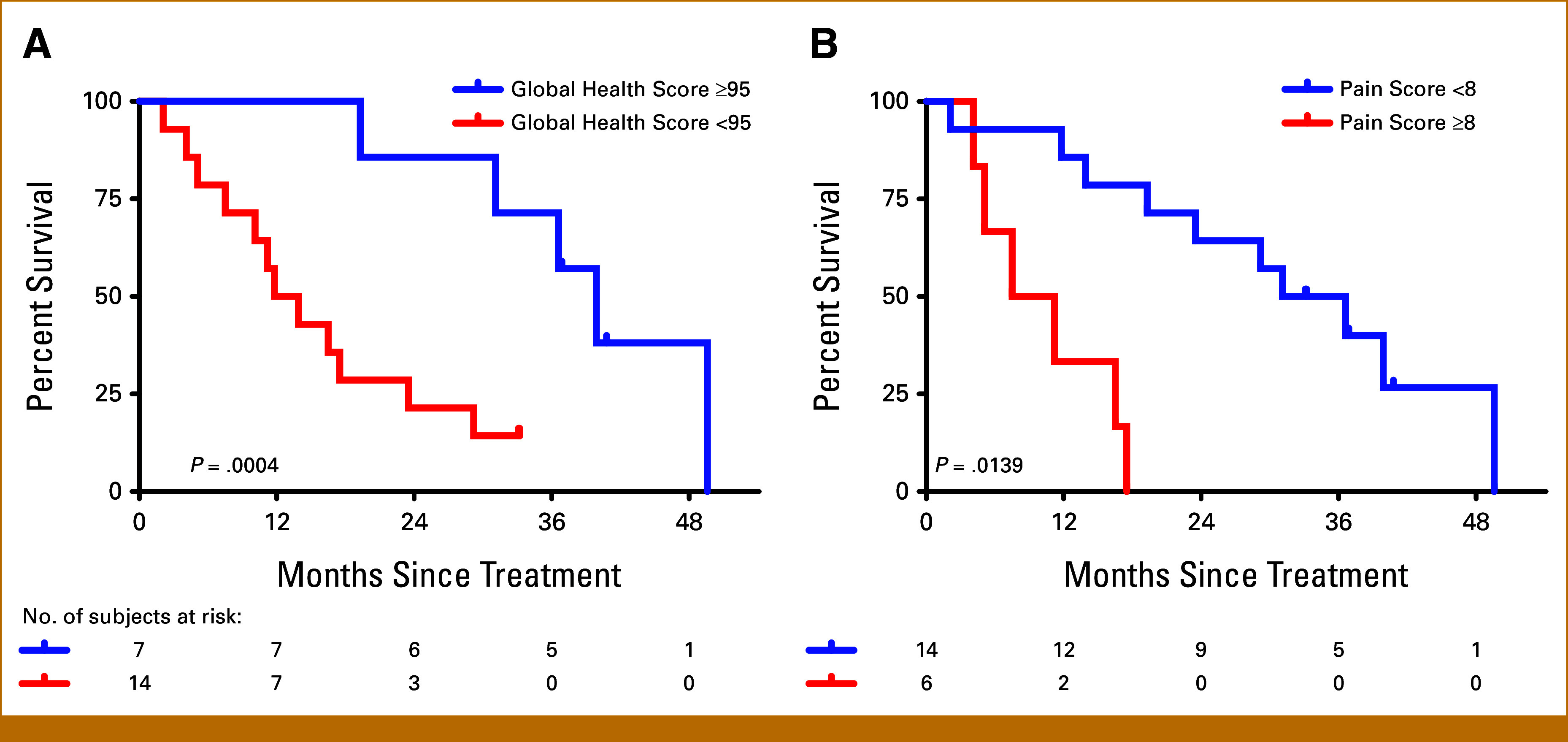
Kaplan-Meier estimates of overall survival by (A) baseline Global Health Score and (B) baseline Pain Score.
FIG A4.
Kaplan-Meier estimates of overall survival by CTC decline or nondecline. CTC, circulating tumor cell.
FIG A5.
Cox proportional hazards regression analyses of promising radium-223 biomarkers to examine for independent prognostic value when adjusted for PSA or tAP. Each separate multivariable model is limited to two covariates because of limitations of sample size. aBSI, automated bone scan index; HR, hazard ratio; PSA, prostate-specific antigen; tAP, total alkaline phosphatase.
FIG A6.
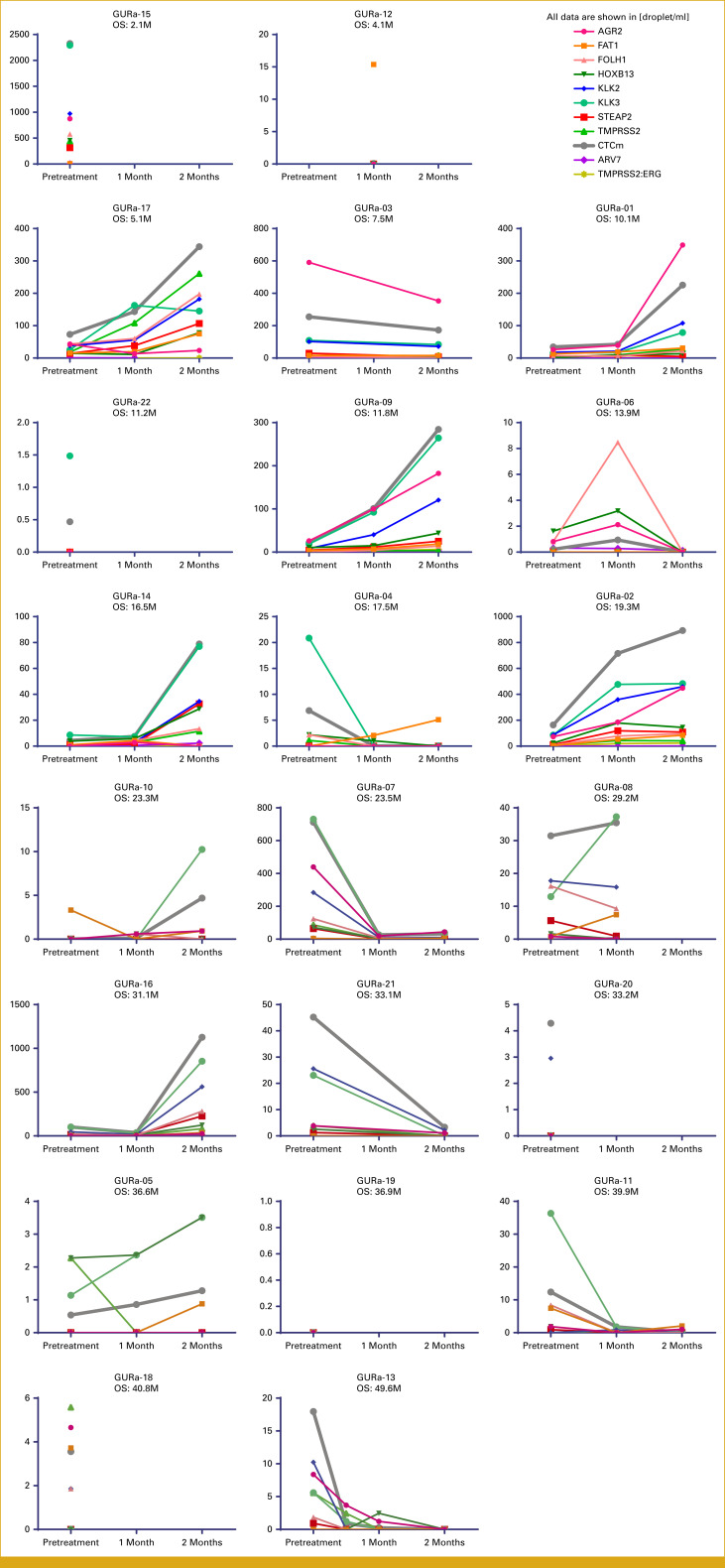
Plots showing longitudinal time courses of gene expression measured in CTCs for each individual participant. From top to bottom, left to right, these are aligned by the order in Figure 1B (ie, shortest survival to longest survival). CTC, circulating tumor cell; OS, overall survival.
EQUAL CONTRIBUTION
P.J.S. and K.O. contributed equally to this work.
SUPPORT
Supported through a sponsored research agreement from Bayer (P.J.S.), provision of software by Exini Diagnostics, and grants from the Prostate Cancer Foundation (D.T.M. and R.J.L.) and the NCI (U01CA268933 to M.T.).
AUTHOR CONTRIBUTIONS
Conception and design: Philip J. Saylor, Mehmet Toner, Daniel A. Haber, David T. Miyamoto, Matthew R. Smith
Financial support: Mehmet Toner, David T. Miyamoto
Administrative support: Philip J. Saylor, Mehmet Toner, David T. Miyamoto
Provision of study materials or patients: Philip J. Saylor, Daniel A. Haber, Richard J. Lee, Matthew R. Smith, Kara Olivier, Erika Meneely
Collection and assembly of data: Philip J. Saylor, Keisuke Otani, Rene Balza, Jacob Ukleja, Haley Pleskow, Rebecca Fisher, Erika Kusaka, Yukako S. Otani, Priscilla Oluwakemi Badusi, Erika Meneely, Kara Olivier, Alarice C. Lowe, David T. Miyamoto
Data analysis and interpretation: Philip J. Saylor, Keisuke Otani, Rene Balza, Jacob Ukleja, Haley Pleskow, Yukako S. Otani, Matthew R. Smith, Shyamala Maheswaran, Daniel A. Haber, Beow Y. Yeap, Richard J. Lee, David T. Miyamoto
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Philip J. Saylor
Research Funding: Bayer (Inst)
Rene Balza
Research Funding: International Skeletal Society (Inst)
Jacob Ukleja
Employment: Cleveland Clinic Florida, Novartis
Stock and Other Ownership Interests: Novartis
Matthew R. Smith
Consulting or Advisory Role: Bayer, Janssen Oncology, Amgen, Pfizer, Lilly, Novartis, Astellas Pharma, Ambrx
Research Funding: Janssen Oncology (Inst), Bayer (Inst), Lilly (Inst), ESSA (Inst), ORIC Pharmaceuticals (Inst)
Kara Olivier
Honoraria: Bayer
Mehmet Toner
Stock and Other Ownership Interests: AutoIVF, TellBio, General Fluidics, Sylvatica
Research Funding: AutoIVF and Sylvatica
Patents, Royalties, Other Intellectual Property: All my patents are owned by Massachusetts General Hospital, and some have been licensed to various companies and they are managed according to the institutional conflict rules
Shyamala Maheswaran
Stock and Other Ownership Interests: TellBio
Research Funding: We received funding from Radius Health to study that effect of Elacestrant, an estrogen receptor degrader, on metastatic breast cancer using CTCs as a model system
Patents, Royalties, Other Intellectual Property: Patents Awarded: (1) Title: Cadherins as Cancer Biomarkers (Patent Number: 10094837); Inventors: Shyamala Maheswaran, David Tsai Ting, Daniel A. Haber. (2) Title: Diagnosis and monitoring treatment of prostate cancer (Patent Number: 9417244); Inventors: Daniel A. Haber, Shyamala Maheswaran, David T. Miyamoto. (3) Title: Methods relating to circulating tumor cell clusters and the treatment of cancer (Patent Number: 10053692); Inventors: Nicola Aceto, Daniel A. Haber, Shyamala Maheswaran Patents Pending: (1) Title: Use of Müllerian inhibiting substance and interferons in treating tumors. Biomarkers of Cancer (Publication number: 20040151693); Inventors: Shyamala Maheswaran and Patricia K. Donahoe. (2) Title: Biomarkers of Cancer (Publication number: 20140031250); Inventors: Shyamala Maheswaran, David Tsai Ting, Daniel A. Haber. (3) Title: Biomarkers of Cancer (Publication number: 20140287956); Inventors: Shyamala Maheswaran, David Tsai Ting, Daniel A. Haber. (4) Title: Methods and assays relating to circulating tumor cells (Publication number: 20160312298); Inventors: David Tsai Ting, Daniel A. Haber, Shyamala Maheswaran. (5) Title: Targeting human satellite ii (HSATII; Publication number: 20170198288); Inventors: David Tsai Ting, Daniel A. Haber, Shyamala Maheswaran. (6) Title: Digital Analysis of Circulating Tumor Cells in Blood Samples (Publication number: 20180057889); Inventors: Daniel A. Haber, Ravi Kapur, Mehmet Toner, Shyamala Maheswaran, Xin Hong, David Tomoaki Miyamoto, Tanya Todorova, Sara Javaid. (7) Title: LNA-Based Mutant Enrichment Next-Generation Sequencing Assays (Publication number: 20180112259); Inventors: Tilak Sundaresan, Zongli Zheng, Daniel A. Haber, Shyamala Maheswaran, A. John Iafrete
Daniel A. Haber
Consulting or Advisory Role: ROME Therapeutics, TellBio
Research Funding: Novartis Institutes for BioMedical Research, Asteroid
Patents, Royalties, Other Intellectual Property: EGFR mutations to direct therapy in non–small-cell lung cancer, RNA-based molecular signatures of CTCs to direct breast cancer therapies, Wilms tumor gene WT1
Beow Y. Yeap
Stock and Other Ownership Interests: SISCAPA Assay Technologies
Consulting or Advisory Role: Guardant Health
Richard J. Lee
Consulting or Advisory Role: Janssen Oncology, Exelixis, Bayer, Blue Earth Diagnostics
Research Funding: Janssen
Expert Testimony: Boehringer Ingelheim
David T. Miyamoto
Research Funding: Cardiff Oncology (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Parker C, Nilsson S, Heinrich D, et al. : Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med 369:213-223, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Sartor O, Coleman R, Nilsson S, et al. : Effect of radium-223 dichloride on symptomatic skeletal events in patients with castration-resistant prostate cancer and bone metastases: Results from a phase 3, double-blind, randomised trial. Lancet Oncol 15:738-746, 2014 [DOI] [PubMed] [Google Scholar]
- 3.Nilsson S, Cislo P, Sartor O, et al. : Patient-reported quality-of-life analysis of radium-223 dichloride from the phase III ALSYMPCA study. Ann Oncol 27:868-874, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sartor O, Coleman RE, Nilsson S, et al. : An exploratory analysis of alkaline phosphatase, lactate dehydrogenase, and prostate-specific antigen dynamics in the phase 3 ALSYMPCA trial with radium-223. Ann Oncol 28:1090-1097, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scher HI, Morris MJ, Stadler WM, et al. : Trial design and objectives for castration-resistant prostate cancer: Updated recommendations from the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol 34:1402-1418, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenhauer EA, Therasse P, Bogaerts J, et al. : New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 45:228-247, 2009 [DOI] [PubMed] [Google Scholar]
- 7.Hofman MS, Lawrentschuk N, Francis RJ, et al. : Prostate-specific membrane antigen PET-CT in patients with high-risk prostate cancer before curative-intent surgery or radiotherapy (proPSMA): A prospective, randomised, multicentre study. Lancet 395:1208-1216, 2020 [DOI] [PubMed] [Google Scholar]
- 8.Calais J, Zhu S, Hirmas N, et al. : Phase 3 multicenter randomized trial of PSMA PET/CT prior to definitive radiation therapy for unfavorable intermediate-risk or high-risk prostate cancer [PSMA dRT]: Study protocol. BMC Cancer 21:512, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afshar-Oromieh A, Sattler LP, Mier W, et al. : The clinical impact of additional late PET/CT imaging with (68)Ga-PSMA-11 (HBED-CC) in the diagnosis of prostate cancer. J Nucl Med 58:750-755, 2017 [DOI] [PubMed] [Google Scholar]
- 10.Dietlein F, Kobe C, Neubauer S, et al. : PSA-stratified performance of (18)F- and (68)Ga-PSMA PET in patients with biochemical recurrence of prostate cancer. J Nucl Med 58:947-952, 2017 [DOI] [PubMed] [Google Scholar]
- 11.Ulmert D, Kaboteh R, Fox JJ, et al. : A novel automated platform for quantifying the extent of skeletal tumour involvement in prostate cancer patients using the Bone Scan Index. Eur Urol 62:78-84, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis ER, Jia X, Mezheritskiy IS, et al. : Bone scan index: A quantitative treatment response biomarker for castration-resistant metastatic prostate cancer. J Clin Oncol 30:519-524, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armstrong AJ, Kaboteh R, Carducci MA, et al. : Assessment of the bone scan index in a randomized placebo-controlled trial of tasquinimod in men with metastatic castration-resistant prostate cancer (mCRPC). Urol Oncol 32:1308-1316, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong AJ, Anand A, Edenbrandt L, et al. : Phase 3 assessment of the automated bone scan index as a prognostic imaging biomarker of overall survival in men with metastatic castration-resistant prostate cancer: A secondary analysis of a randomized clinical trial. JAMA Oncol 4:944-951, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uemura K, Miyoshi Y, Kawahara T, et al. : Prognostic value of an automated bone scan index for men with metastatic castration-resistant prostate cancer treated with cabazitaxel. BMC Cancer 18:501, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyoshi Y, Sakamoto S, Kawahara T, et al. : Correlation between automated bone scan index change after cabazitaxel and survival among men with castration-resistant prostate cancer. Urol Int 103:279-284, 2019 [DOI] [PubMed] [Google Scholar]
- 17.Mota JM, Armstrong AJ, Larson SM, et al. : Measuring the unmeasurable: Automated bone scan index as a quantitative endpoint in prostate cancer clinical trials. Prostate Cancer Prostatic Dis 22:522-530, 2019 [DOI] [PubMed] [Google Scholar]
- 18.McNamara MA, Oyekunle T, Chin BB, et al. : Patterns of response and progression in bone and soft tissue during and after treatment with radium-223 for metastatic castrate-resistant prostate cancer. Prostate 79:1106-1116, 2019 [DOI] [PubMed] [Google Scholar]
- 19.Scher HI, Heller G, Molina A, et al. : Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 33:1348-1355, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang WL, Pleskow HM, Miyamoto DT: Molecular analysis of circulating tumors cells: Biomarkers beyond enumeration. Adv Drug Deliv Rev 125:122-131, 2018 [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto DT, Lee RJ, Kalinich M, et al. : An RNA-based digital circulating tumor cell signature is predictive of drug response and early dissemination in prostate cancer. Cancer Discov 8:288-303, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scher HI, Graf RP, Schreiber NA, et al. : Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol 4:1179-1186, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armstrong AJ, Luo J, Nanus DM, et al. : Prospective multicenter study of circulating tumor cell AR-V7 and taxane versus hormonal treatment outcomes in metastatic castration-resistant prostate cancer. JCO Precis Oncol 4:2020, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y, Li R, Xu K, et al. : Androgen receptor variants mediate DNA repair after prostate cancer irradiation. Cancer Res 77:4745-4754, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scher HI, Jia X, de Bono JS, et al. : Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol 10:233-239, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozkumur E, Shah AM, Ciciliano JC, et al. : Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 5:179ra47, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Contal C, O'Quigley J: An application of changepoint methods in studying the effect of age on survival in breast cancer. Comput Stat Data Anal 30:253-270, 1999 [Google Scholar]
- 28.de Bono JS, Scher HI, Montgomery RB, et al. : Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 14:6302-6309, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Antonarakis ES, Lu C, Wang H, et al. : AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 371:1028-1038, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonarakis ES, Lu C, Chen Y, et al. : AR splice variant 7 (AR-V7) and response to taxanes in men with metastatic castration-resistant prostate cancer (mCRPC). J Clin Oncol 33, 2015. (suppl 7; abstr 138) [Google Scholar]
- 31.Scher HI, Lu D, Schreiber NA, et al. : Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol 2:1441-1449, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsui Y, Shiina H, Yamamoto Y, et al. : Prediction of survival benefit using an automated bone scan index in patients with castration-resistant prostate cancer. BJU Int 110:E628-E634, 2012 [DOI] [PubMed] [Google Scholar]
- 33.Carles J, Castellano D, Méndez-Vidal MJ, et al. : Circulating tumor cells as a biomarker of survival and response to radium-223 therapy: Experience in a cohort of patients with metastatic castration-resistant prostate cancer. Clin Genitourin Cancer 16:e1133-e1139, 2018 [DOI] [PubMed] [Google Scholar]
- 34.Chatzkel J, Mocha J, Smith J, et al. : Circulating tumor cells and γH2AX as biomarkers for responsiveness to radium-223 in advanced prostate cancer patients. Future Sci OA 6:FSO437, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burnell SEA, Spencer-Harty S, Howarth S, et al. : Utilisation of the STEAP protein family in a diagnostic setting may provide a more comprehensive prognosis of prostate cancer. PLoS One 14:e0220456, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burnell SEA, Spencer-Harty S, Howarth S, et al. : STEAP2 knockdown reduces the invasive potential of prostate cancer cells. Sci Rep 8:6252, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]