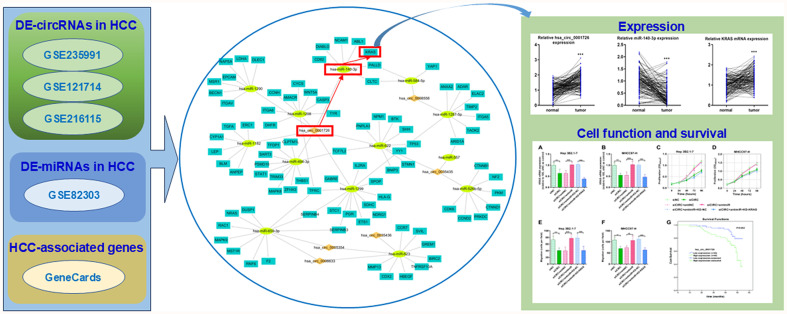
Abstract
Background and Aims
Hepatocellular carcinoma (HCC) is one of the most fatal malignancies. Epigenetic mechanisms have revealed that noncoding RNAs, such as microRNAs (miRNAs) and circular RNAs (circRNAs), are involved in HCC progression. This study aimed to construct a circRNA-miRNA-mRNA network in HCC and validate one axis within the network.
Methods
HCC-related transcriptome data were obtained from the Gene Expression Omnibus, and HCC-related genes were sourced from GeneCards to identify differentially expressed circRNAs and miRNAs. The targeting relationships between circRNA-miRNA and miRNA-mRNA interactions were predicted. The involvement of the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC was evaluated through cellular experiments and survival analyses.
Results
We identified six differentially expressed circRNAs in HCC, which were linked to 13 miRNAs and 88 mRNAs. A network containing 34 circRNA-miRNA pairs and 194 miRNA-mRNA pairs was constructed. Cell proliferation and migration assays confirmed the role of hsa_circ_0001726 in promoting HCC progression, possibly through the miR-140-3p/KRAS axis. Survival analysis verified that hsa_circ_0001726 was a prognostic factor for overall survival in patients with HCC. The hsa_circ_0001726/miR-140-3p/KRAS axis also mediates lenvatinib resistance in HCC cells.
Conclusions
The HCC circRNA/miRNA/mRNA network provides new insights into the post-transcriptional regulatory mechanism of HCC. The hsa_circ_0001726/miR-140-3p/KRAS axis is involved in HCC progression and lenvatinib resistance.
Keywords: Circular RNA, microRNA, Prognosis, Hepatocellular cancer, Bioinformatics, Tyrosine kinase inhibitor
Graphical abstract
Introduction
Population-based cancer incidence data from the Global Cancer Observatory indicate a rising trend in liver cancer incidence.1 Globally, 4.7% of the 19.3 million new cancers and 8.3% of cancer-related mortalities are associated with liver cancer. 2 Nearly half of all liver cancer cases and liver cancer deaths worldwide occur in China (45.3% and 47.1%, respectively). 3 The epidemiology of liver cancer is dominated by hepatocellular carcinoma (HCC), which accounts for about 70% of primary liver cancers. 4 HCC is one of the most fatal malignancies, usually diagnosed at advanced stages with very limited therapeutic options, leading to a dismal prognosis. 5 Current survival rates for liver cancers remain low (21%). 1 The therapeutic outcomes for patients with hepatobiliary cancer are unsatisfactory due to the bleak prognosis, leaving significant room for improvement.
The etiologies and geographical trends of HCC are complex. Molecular heterogeneity contributes to the complexity of HCC at different levels. 6 Recently, next-generation sequencing-based technologies have been widely used for multiomic and single-cell analyses. These advances have enabled the creation of high-resolution atlases of tumor genomes and epigenomes, accelerating our understanding of tumor spatial and temporal heterogeneity. 7 HCC involves a multitude of genetic and epigenetic aberrations. 8 A deeper understanding of cancer epigenetics in tissue samples and biopsies is increasingly valuable in predicting patient prognosis and treatment response in clinical settings. Epigenetic mechanisms have revealed that alterations in ncRNAs, such as miRNAs and circRNAs, promote uncontrolled cell growth, metastasis, and HCC progression. 9,10 These ncRNAs have also emerged as a critical area of research in HCC-targeting therapies. 11
In the present research, bioinformatics analyses were used to identify differentially expressed circRNAs, miRNAs, and HCC-related genes in HCC. The target miRNAs of the circRNAs and mRNAs of the miRNAs were predicted and intersected with the differentially expressed miRNAs and HCC-related genes to identify common targets. A network was subsequently established, and the hsa_circ_0001726/miR-140-3p/KRAS axis was further investigated and validated.
Methods
RNA expression data retrieval and processing
RNA-seq RNA expression data for HCC and adjacent noncancerous liver samples were obtained from the Gene Expression Omnibus (GEO). The GSE235991 dataset contains five high-invasive and five low-invasive HCC tissues for circRNA expression profiling. The GSE121714 and GSE216115 datasets provide circRNA microarray analysis data for HCC and nontumor tissues. GSE82303, a dataset of blood exosomal miRNA profiles, includes two subsets, GSE82301 and GSE82302, with 72 patients with normal livers and 32 patients with HCC.
Construction of the circRNA-miRNA-gene network
The GSE235991, GSE121714, and GSE216115 datasets were analyzed for differentially expressed circRNAs in HCC using the GEO2R online tool (logFC > 1; p < 0.05). The common circRNAs among these datasets were identified using a Venn diagram. Potential downstream miRNAs were collected from ENCORI, the Circular RNA Interactome, and the Cancer-Specific CircRNA database. The GSE82303 dataset was analyzed for differentially expressed miRNAs in HCC using GEO2R (logFC > 1; p < 0.05). The downstream miRNAs were then intersected with the differentially expressed miRNAs in HCC using VennPainter to obtain the HCC-related, circRNA-targeting miRNAs. The miRNA targets were obtained from TargetScan and the Encyclopedia of RNA Interactomes (ENCORI/starBase). HCC-associated genes were retrieved from GeneCards using the keyword “hepatocellular carcinoma” and filtered by a twofold mean relevance score. The overlapping genes from miRNA-targeting genes and HCC-associated genes were used to construct the circRNA-miRNA-gene network, which was visualized using Cytoscape software.
Gene Ontology (GO) and pathway enrichment analysis
The genes involved were subjected to enrichment analysis of GO terms for biological process, cellular component, molecular function, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using OmicShare (https://www.omicshare.com/tools/).
Patient material
HCC patients included in this study (n = 133) were enrolled at the 900TH Hospital of the Joint Logistics Support Force. These patients had no history of cancer-related therapy or other serious diseases. All patients provided written informed consent, and the study was approved by the institutional ethical review board. Patient information, including characteristics and clinicopathological variables, is described in Table 1, following current clinical definitions in China.
Table 1. Association of hsa_circ_0001726 expression with clinicopathological features in hepatocellular carcinoma (n = 133).
| Characteristics | Level | Low expression of hsa_circ_0001726 | High expression of hsa_circ_0001726 | p | Test |
|---|---|---|---|---|---|
| T stage | T1&T2 | 43 (32.33) | 31 (23.31) | 0.017 | Chi-Square |
| T3&T4 | 22 (16.54) | 37 (27.82) | |||
| N stage | N0 | 63 (47.37) | 63 (47.37) | 0.441 | Fisher’s Exact |
| N1 | 2 (1.50) | 5 (3.76) | |||
| M stage | M0 | 63 (47.37) | 65 (48.87) | 1.000 | Fisher’s Exact |
| M1 | 2 (1.50) | 3 (2.26) | |||
| BCLC stage | Stage I & Stage II | 50 (37.59) | 44 (33.08) | 0.122 | Chi-Square |
| Stage III & Stage IV | 15 (11.28) | 24 (18.05) | |||
| Gender | Female | 28 (21.05) | 25 (18.80) | 0.457 | Chi-Square |
| Male | 37 (27.82) | 43 (32.33) | |||
| Age | ≤61 | 34 (25.56) | 38 (28.57) | 0.679 | Chi-Square |
| >61 | 31 (23.31) | 30 (22.56) | |||
| BMI | ≤24 | 29 (21.80) | 35 (26.32) | 0.429 | |
| >24 | 36 (27.07) | 33 (24.81) | |||
| Histologic grade | G1 & G2 | 46 (34.59) | 41 (30.83) | 0.204 | Chi-Square |
| G3 & G4 | 19 (14.29) | 27 (20.30) | |||
| AFP | ≤356 ng/ml | 54 (40.60) | 53 (39.85) | 0.455 | Chi-Square |
| >356 ng/ml | 11 (8.27) | 15 (11.28) | |||
| Fibrosis Ishak score (vs.) | 0/1/2 | 49 (36.84) | 52 (39.10) | 0.884 | Chi-Square |
| 3/4/5/6 | 16 (12.03) | 16 (12.03) | |||
| OS event, n (%) | Alive | 55 (41.35) | 41 (30.83) | 0.002 | Chi-Square |
| Dead | 10 (7.52) | 27 (20.30) |
BCLC, Barcelona Clinic Liver Cancer; BMI, body mass index; AFP, alpha fetoprotein; OS, overall survival.
Cell culture
HCC cells were cultured in specific media (Gibco, Carlsbad, CA, USA) as recommended by the supplier (China Center for Type Culture Collection). For transfection experiments, the cells were preincubated with the indicated concentrations of inhibitors, mimics, or siRNAs for 24 h.
Establishment of lenvatinib-resistant cell lines
To determine the half-maximal inhibitory concentration (IC50) of lenvatinib, MHCC97-H, and Hep 3B2.1-7 cells were incubated with various concentrations of lenvatinib, and cell viability was measured after three days. For the establishment of lenvatinib-resistant cell lines, MHCC97-H and Hep 3B2.1-7 cells were incubated with lenvatinib at the approximate IC50 concentration, which was increased by 0.2 µmol/L per week for 6 months. These cells were termed MHCC97-H-LR and Hep 3B2.1-7-LR, respectively.
RNA isolation and real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Fresh tumor samples were collected and preserved in RNAlater (Qiagen, Hilden, Germany). Total RNA was extracted from tissues and cells using the RNeasy kit (Qiagen, Valencia, USA), and RNA concentrations were measured using an ND-100 Nanodrop spectrophotometer. cDNAs were obtained through reverse transcription using the PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa Bio, Japan). RT-qPCR was performed using RT-Taq/SYBR Green QPCR reagents (Invitrogen, Carlsbad, USA) on a Light-Cycler96 thermocycler (Roche, Basel, Switzerland). Relative expression values were calculated using the 2–ΔΔCT method.
Dual-luciferase reporter assay
The wt-circ plasmid contained the synthesized 3′-untranslated region (UTR) wild-type sequence of hsa_circ_0001726, while the mut-circ plasmid contained the mutant sequence, along with the Fluc coding sequence. The WT KRAS luciferase reporter plasmid was generated using the full-length KRAS 3′ UTR vector, whereas the MUT KRAS plasmid was generated with the mutant KRAS 3′ UTR vector. The cells were subjected to lysis in Passive Lysis Buffer (Progma, USA), and luciferase activity was measured following the Dual-Luciferase® Reporter 1000 Assay System protocol (Promega).
Cell proliferation assay
The proliferation of transfected or treated cells was evaluated using the BeyoClick EdU Cell Proliferation Kit (APExBIO, Houston, USA). Cells were seeded into 96-well plates at 40% confluence. The kit reagent was added to each well on different days (zero, one, two, three, and four days), followed by measurement of absorption at 450 nm two hours later.
In vitro Transwell migration
HCC cells were subjected to an assessment of in vitro Transwell migratory ability using HTS Transwell-96 Permeable Support with an 8.0 µm pore polyester membrane (Corning Life Sciences - Axygen, Union City, USA). Cells were pretreated with serum-free medium and added to the top wells of Transwell inserts. The bottom wells were filled with 600 µL of media containing 10% fetal bovine serum (Gibco, Grand Island, USA). After 24 h of incubation, cells in the bottom wells were harvested and counted.
Statistical analysis
For group comparisons, continuous variables were compared using the Mann-Whitney U test, while categorical variables were compared using the chi-square test or Fisher’s exact test. Logistic regression analysis was used for Kaplan-Meier analysis with categorical variables. Univariate and multivariate Cox regression analyses were performed to evaluate the odds ratios for associations between survival risk factors and events. Statistical significance was set at a two-sided p-value < 0.05.
Results
Differentially expressed circRNAs in HCC based on microarray data
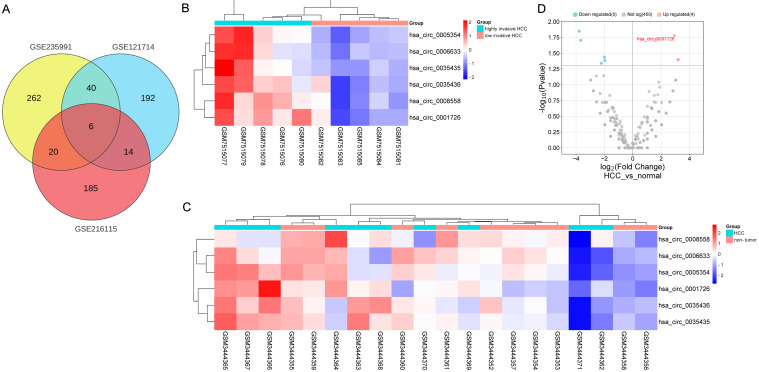
We retrieved the GSE235991, GSE121714, and GSE216115 datasets to identify differentially expressed circRNAs in HCC. A total of 328 differentially expressed circRNAs were identified from the GSE235991 dataset, 252 from the GSE121714 dataset, and 225 from the GSE216115 dataset. Among these differentially expressed circRNAs, hsa_circ_0001726, hsa_circ_0008558, hsa_circ_0005354, hsa_circ_0035436, hsa_circ_0035435, and hsa_circ_0006633 were common (Fig. 1A). The detailed expression levels of these six circRNAs are depicted in Figure 1B–D.
Fig. 1. Identification of differentially expressed circRNAs in hepatocellular carcinoma (HCC).
(A) Venn diagram of the intersection of differentially expressed circRNAs among the three datasets. (B, C) Heatmaps of six HCC-related differentially expressed circRNAs in the GSE235991 and GSE121714 datasets. (D) Volcano plot of HCC circRNAs in the GSE216115 dataset.
Differentially expressed miRNAs in HCC based on microarray data and circRNA target prediction
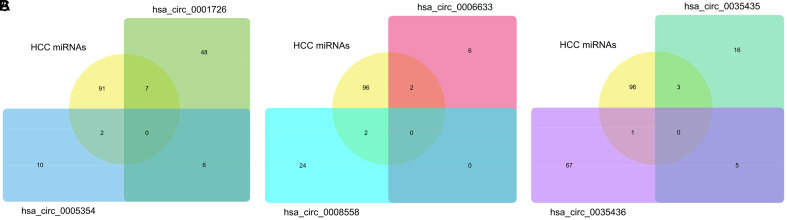
The GSE82303 dataset was retrieved and analyzed to identify 100 differentially expressed miRNAs in HCC. The prediction of downstream miRNAs was conducted via databases. Hsa-miR-1182, hsa-miR-1208, hsa-miR-1290, hsa-miR-1299, hsa-miR-140-3p, hsa-miR-494-3p, and hsa-miR-622 were identified as hsa_circ_0001726-targeting differentially expressed miRNAs in HCC, while hsa-miR-623 and hsa-miR-659-3p were identified as hsa_circ_0005354-targeting miRNAs in HCC (Fig. 2A). Hsa-miR-623 and hsa-miR-659-3p were also identified as hsa_circ_0006633-targeting miRNAs in HCC, and hsa-miR-1287-5p and hsa-miR-584-5p were identified as hsa_circ_0008558-targeting miRNAs in HCC (Fig. 2B). Three miRNAs—hsa-miR-557, hsa-miR-526b-5p, and hsa-miR-622—were observed to target differentially expressed miRNAs in HCC, while hsa-miR-623 was identified as the hsa_circ_0035436-targeting miRNA in HCC (Fig. 2C).
Fig. 2. Venn diagrams of the intersection of hsa_circ_0001726/hsa_circ_0005354-targeting miRNAs (A), hsa_circ_0008558/hsa_circ_0006633-targeting miRNAs (B), and hsa_circ_0035436/hsa_circ_0035435-targeting miRNAs with HCC-related differentially expressed miRNAs from GSE80303 (C).
HCC, hepatocellular carcinoma.
The circRNA-miRNA-mRNA regulatory network involved in HCC
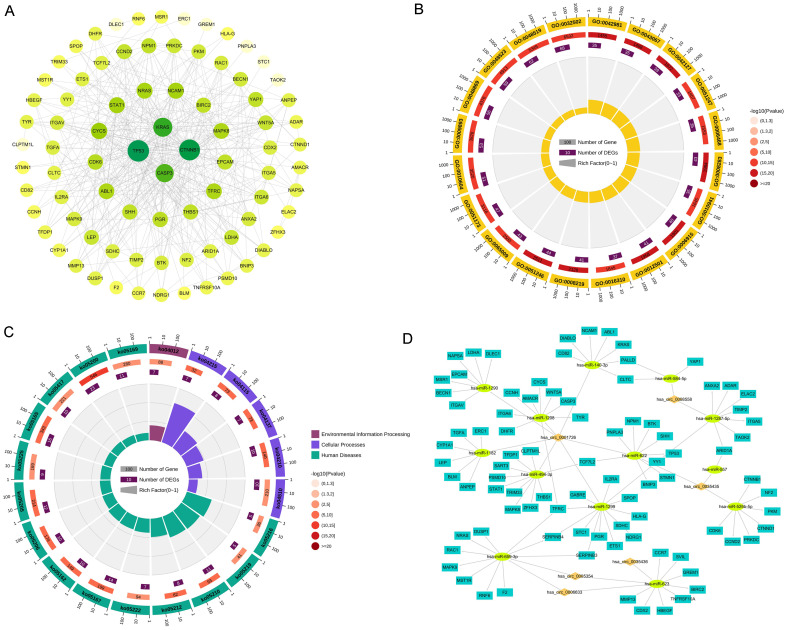
HCC-related genes were obtained from GeneCards and intersected with the miRNA target genes, resulting in a total of 99 genes. After protein-protein interaction prediction, disconnected nodes in the network were hidden, and the interaction network was analyzed via Cytoscape software, which revealed 82 nodes and 426 edges (Fig. 3A). The first six biological process terms and the first six GO terms enriched by the genes included GO:0008283 cell population proliferation, GO:0042127 regulation of cell population proliferation, GO:0012501 programmed cell death, GO:0006915 apoptotic process, GO:0042981 regulation of the apoptotic process, and GO:0043067 regulation of programmed cell death (Fig. 3B). Pathways in cancer, apoptosis-multiple species, microRNAs in cancer, the ErbB signaling pathway, and the hepatocellular carcinoma pathway were among the top 20 KEGG pathways (Fig. 3C). Finally, the circRNA-miRNA-gene regulatory network involved in HCC was constructed (Fig. 3D), consisting of 34 circRNA-miRNA pairs and 194 miRNA-mRNA pairs.
Fig. 3. Identification of differentially expressed mRNAs and construction of a network.
(A) Interaction of HCC-associated genes. (B) Circular diagram of Gene Ontology (GO) function annotation. (C) Circular diagram of Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment. (D) Interaction network consisting of 6 circRNAs, 13 miRNAs, and 88 mRNAs.
Validation of hsa_circ_0001726, miR-140-3p, and KRAS expression in HCC
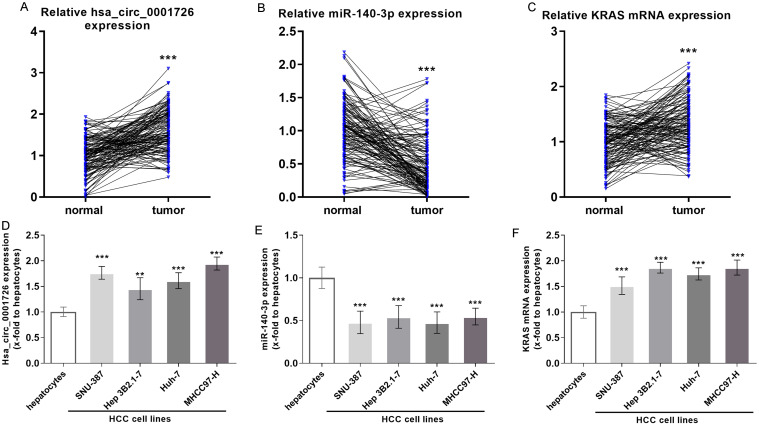
The microarray dataset analyses revealed the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC. Furthermore, using RT-qPCR, the expression of hsa_circ_0001726, miR-140-3p, and KRAS was evaluated in 133 HCC and matched adjacent normal tissues. We confirmed that hsa_circ_0001726 was significantly upregulated in HCC (Fig. 4A). miR-140-3p was downregulated in HCC (Fig. 4B), while KRAS mRNA was upregulated (Fig. 4C). Similar alterations were observed in HCC cells (Fig. 4D, F).
Fig. 4. Validation of hsa_circ_0001726, miR-140-3p, and KRAS expression in HCC patients.
Expression levels of hsa_circ_0001726 (A), miR-140-3p (B), and KRAS (C) in HCC tissue samples. Expression levels of hsa_circ_0001726 (D), miR-140-3p (E), and KRAS (F) in HCC cells. **p < 0.01, ***p < 0.001. HCC, hepatocellular carcinoma.
Validation of the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC
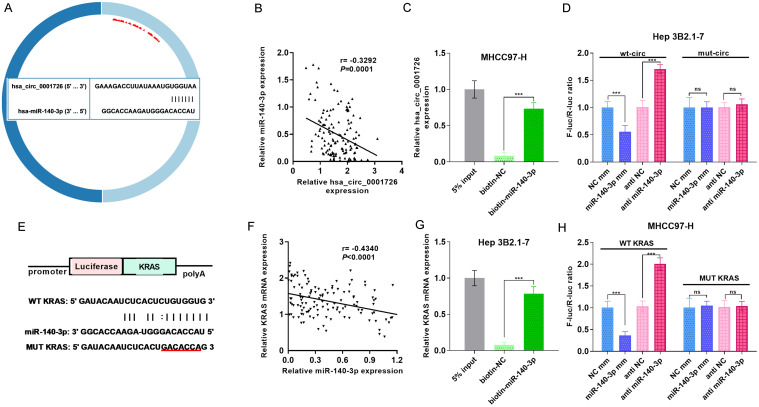
To further validate the relationships between hsa_circ_0001726 and miR-140-3p, as well as between miR-140-3p and KRAS, we performed dual-luciferase reporter assays and RNA pull-down analyses. Prediction revealed binding sites in hsa_circ_0001726 and miR-140-3p (Fig. 5A). Notably, hsa_circ_0001726 was negatively correlated with miR-140-3p (Fig. 5B). A pull-down experiment in MHCC-97-H cells revealed that hsa_circ_0001726 was more enriched in pull-down experiments involving miR-140-3p than in the negative control (Fig. 5C). A dual-luciferase reporter assay in Hep 3B2.1-7 cells confirmed these findings, revealing enhanced luciferase activity in miR-140-3p-inhibited cells and reduced activity in miR-140-3p-overexpressing cells (Fig. 5D). We also examined the relationship between miR-140-3p and KRAS (Fig. 5E–H).
Fig. 5. Validation of the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC.
(A) Predicted binding sites between hsa_circ_0001726 and miR-140-3p. (B) The correlation between the expression levels of hsa_circ_0001726 and miR-140-3p. (C) RNA pull-down assay for hsa_circ_0001726. (D) Dual-luciferase reporter assay for hsa_circ_0001726. (E) Predicted binding sites between miR-140-3p and KRAS. (F) The correlation between the expression levels of miR-140-3p and KRAS. (G) RNA pull-down assay for KRAS mRNA. (H) Dual-luciferase reporter assay for KRAS mRNA. ***p < 0.001. ns, not significant. HCC, hepatocellular carcinoma.
Role of the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC progression
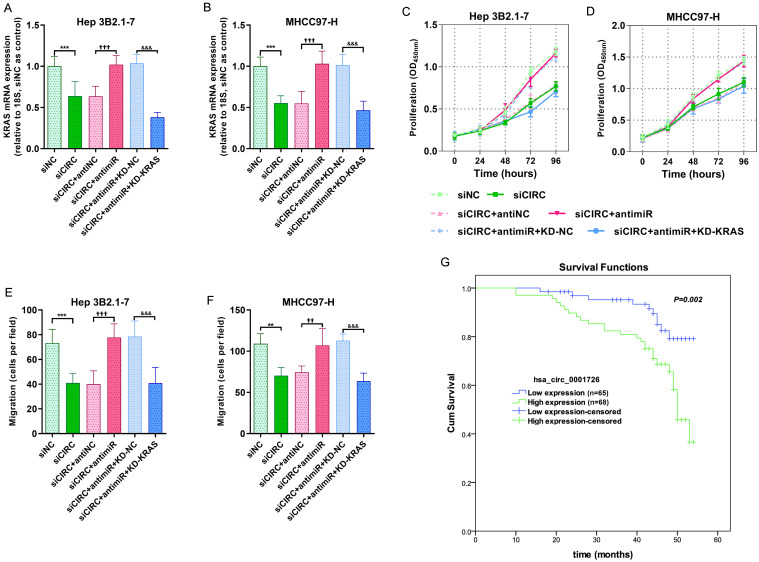
To investigate the role of the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC progression, we analyzed HCC cell proliferation and migration after modulating the axis. KRAS mRNA expression was significantly downregulated after hsa_circ_0001726 inhibition, but this effect was rescued by the miR-140-3p inhibitor and reversed by KRAS knockdown (Fig. 6A, B). Cell proliferation assays revealed that hsa_circ_0001726 knockdown inhibited cell growth, which was partly reversed by the miR-140-3p inhibitor and restored by KRAS knockdown (Fig. 6C, D). Similarly, hsa_circ_0001726 and KRAS knockdown inhibited HCC cell migration, while the miR-140-3p inhibitor promoted HCC cell migration (Fig. 6E, F). We next assessed the clinical role of hsa_circ_0001726 in the prognosis of HCC patients. hsa_circ_0001726 expression was significantly correlated with T stage and overall survival (Table 1). Cox regression analysis was used to calculate the risk of poor prognosis (hazard–risk ratio) for the high and low hsa_circ_0001726 populations. There was a 2.900-fold (95% CI: 1.402–5.998) increased risk of disease recurrence or death in high-hsa_circ_0001726-expressing patients compared with low-hsa_circ_0001726-expressing patients after univariate Cox proportional hazards analysis and a 2.402-fold (95% CI: 1.152–5.009) increased risk after multivariate analysis (Table 2). The cumulative survival of the enrolled HCC patients was estimated via Kaplan-Meier analyses, which revealed that HCC patients with high hsa_circ_0001726 levels had a lower mean survival rate than those with low hsa_circ_0001726 levels (Fig. 6G).
Fig. 6. Hsa_circ_0001726 promotes HCC progression via the miR-140-3p/KRAS axis.
(A, B) KRAS mRNA expression levels determined via RT-qPCR after transfection. (C, D) Proliferative capacity measured using CCK-8 assay. (E, F) Migratory ability measured by Transwell assay. (G) Comparison of overall survival curves for HCC patients based on hsa_circ_0001726 expression (log-rank test p = 0.002). **p < 0.01, ***p < 0.001, versus siNC. ††p < 0.01, †††p < 0.001, versus siCIRC+antiNC. &&&p < 0.001, versus siCIRC+antimiR+KD-NC. HCC, hepatocellular carcinoma.
Table 2. A multivariate and univariate Cox proportional hazards analysis of hsa_circ_0001726 expression in hepatocellular carcinoma overall survival.
| Characteristics | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| Hazard ratio (95% CI) | p-value | Hazard ratio (95% CI) | p-value | |
| Hsa_circ_0001726 | 2.900 (1.402–5.998) | 0.004 | 2.402 (1.152–5.009) | 0.019 |
| T stage (T3 & T4 vs. T1&T2) | 2.507 (1.275–4.9300 | 0.008 | 1.302 (0.547–3.101) | 0.551 |
| N stage (N1 vs. N0) | 2.220 (0.679–7.256) | 0.187 | ||
| M stage (M1 vs. M0) | 1.536 (0.367–6.424) | 0.556 | ||
| BCLC stage (Stage III & Stage IV vs Stage I & Stage II) | 2.435 (1.270–4.668) | 0.007 | 1.793 (0.787–4.081) | 0.164 |
| Gender (Male vs Female) | 1.169 (0.609–2.241) | 0.639 | ||
| Age (>61 vs ≤61) | 1.173 (0.611–2.250) | 0.631 | ||
| BMI (≤24 vs >24) | 1.517 (0.786–2.926) | 0.214 | ||
| Histologic grade (G3 & G4 vs G1 & G2) | 2.018 (1.055–3.860) | 0.034 | 1.756 (0.895–3.446) | 0.102 |
| AFP (>356 ng/ml vs ≤356 ng/ml) | 2.580 (1.274–5.224) | 0.008 | 2.247 (1.099–4.595) | 0.027 |
| Fibrosis Ishak score (3/4/5/6 vs. 0/1/2) | 1.464 (0.734–2.920) | 0.279 | ||
BCLC, Barcelona Clinic Liver Cancer; BMI, body mass index; AFP, alpha fetoprotein; OS, overall survival.
Role of the hsa_circ_0001726/miR-140-3p/KRAS axis in lenvatinib resistance
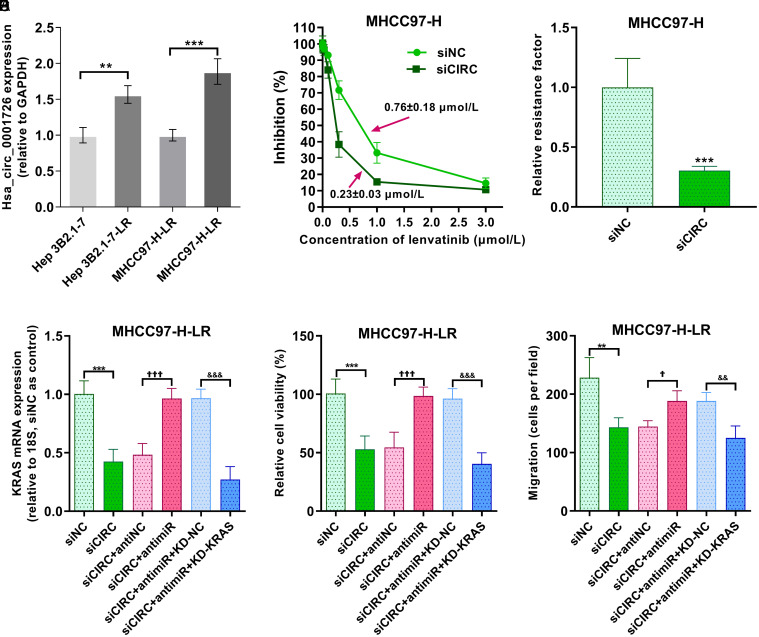
To examine whether the hsa_circ_0001726/miR-140-3p/KRAS axis influences lenvatinib resistance, lenvatinib-resistant Hep 3B2.1–7 and MHCC97-H cells were established. Hsa_circ_0001726 was upregulated in lenvatinib-resistant HCC cells, particularly in lenvatinib-resistant MHCC97-H cells (Fig. 7A). After treatment with varying concentrations of lenvatinib, hsa_circ_0001726-knockdown MHCC97-H cells exhibited a lower IC50 value for lenvatinib compared with negative controls (Fig. 7B). As shown in Figure 7C, compared with negative control cells, hsa_circ_0001726-silenced MHCC97-H cells were more sensitive to lenvatinib and presented a decreased relative resistance factor. KRAS mRNA expression was subsequently examined in transfected MHCC97-H cells via RT-qPCR, which confirmed that lenvatinib-resistant MHCC97-H cells were successfully transfected (Fig. 7D). The viability of hsa_circ_0001726-knockdown cells resistant to lenvatinib was lower than that of negative control-transfected cells resistant to lenvatinib, but the miR-140-3p inhibitor and KRAS siRNA reversed this effect (Fig. 7E). The migration of lenvatinib-resistant cells was measured via Transwell assay, revealing less migration in hsa_circ_0001726-knockdown and lenvatinib-resistant cells compared with negative control cells (Fig. 7F).
Fig. 7. Role of the hsa_circ_0001726/miR-140-3p/KRAS axis in lenvatinib resistance in HCC.
(A) Hsa_circ_0001726 expression in lenvatinib-resistant HCC cells. **p < 0.01, ***p < 0.001. (B) The IC50 of hsa_circ_0001726 in MHCC-97-H cells with or without hsa_circ_0001726 knockdown. (C) The relative resistance factor. ***p < 0.001. (D) KRAS mRNA expression level determined via RT-qPCR after transfection. (E) Proliferative capacity measured via CCK-8 assay. (F) Migratory ability measured via Transwell assay. **p < 0.01, ***p < 0.001, versus siNC. †p < 0.05, †††p < 0.001, versus siCIRC+antiNC. &&p < 0.01, &&&p < 0.001, versus siCIRC+antimiR+KD-NC. HCC, hepatocellular carcinoma.
Discussion
Advances in sequencing technologies have led to the discovery of ncRNAs, and increasing research has revealed their regulatory role in cellular processes and pathways in cancer. 12 It is challenging to uncover ncRNA functions in isolation. CircRNAs regulate the abundance of specific miRNAs through mechanisms known as sequestration; miRNAs, in turn, target the mRNAs of various genes through complementary combinations; and vice versa, each gene can be targeted by multiple miRNAs. Thus, circRNAs link associated miRNAs and genes into a complex regulatory network. 13 The highly complex characteristics of circRNA interactions support their regulatory roles in important cellular processes. In our study, we analyzed recent datasets to identify differentially expressed circRNAs in HCC. We combined dataset analysis and in silico target prediction approaches to construct a circRNA-miRNA-gene network in HCC patients. Additionally, cell line experiments were conducted to validate the role of the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC. We revealed that hsa_circ_0001726 promotes HCC progression and predicts HCC prognosis.
The decoy role of circRNAs is based on their ability to sequester miRNAs. 14 CircRNAs containing complementary sequences for miRNAs can function as sponges for several miRNAs. The hypothesis of a ceRNA network suggests that circRNAs regulate the expression of other genes and influence tumorigenesis and cancer progression. 15 Based on comprehensive analysis, Zhang et al. established a prognostic ceRNA network of 21 circRNAs, five miRNAs, and seven hub mRNAs for hepatocellular carcinoma. 16 Our experimental results revealed an HCC-related network consisting of six circRNAs, 13 miRNAs, and 88 mRNAs. Hsa_circ_0001726 has been identified as a back-spliced junction and a prognostic marker in esophageal squamous cell carcinoma. 17 Hsa_circ_0008558 modulates the maturation and exosomal dissemination of miR-17, enhancing colorectal carcinoma metastasis. 18 Hsa_circ_0008558 was shown to be downregulated in HCC. 19 Hsa_circ_0005354 has been linked to the risk of acute myeloid leukemia in pediatric patients. 20 Hsa_circ_0035436 exhibited significant differences between clear cell renal cell carcinoma cell lines and 293T cell lines. 21 Hsa_circ_0006633 is expressed at low levels in HCC patients, is associated with poor overall survival, and can sponge miR-545-3p to promote Smad7 expression, thereby inhibiting HCC progression. 22
For a long time after their discovery, ncRNAs were considered nonfunctional molecules or “transcriptional noise.” However, in recent years, ncRNAs have been recognized for their crucial biological roles as transcription and translation regulators in various diseases, including cancer. 23 This recognition has prompted scientists to explore their potential in clinical practice, considering them new targets or strategies for cancer treatment. We found that hsa_circ_0001726 promotes HCC by accelerating cell proliferation and migration, and its knockdown can inhibit HCC progression. A clear correlation between hsa_circ_0001726 expression and HCC T stage was observed. The predictive power of hsa_circ_0001726 for overall survival in HCC patients was also demonstrated. These findings suggest that hsa_circ_0001726, along with the axis it forms (hsa_circ_0001726/miR-140-3p/KRAS), has potential as a target for HCC therapy.
RAS is a member of the small GTPase family, and its gain-of-function mutations are among the leading causes of human oncogenesis. 24 However, attempts to develop RAS-targeted therapies have often failed. Therefore, the development of RAS inhibitors targeting upstream regulatory molecules offers a novel approach to overcoming anti-RAS challenges. 24 The KRAS family is encoded by KRAS genes in humans. The RAF/MEK/ERK pathway is the primary pathway involved in the anticancer effects of some promising anti-HCC tyrosine kinase inhibitors, such as lenvatinib. 25 In our study, we found that KRAS silencing inhibits HCC cell proliferation and migration, consistent with previous results. 26 Furthermore, we validated the role of the hsa_circ_0001726/miR-140-3p/KRAS axis in lenvatinib resistance in HCC. Our results showed that downregulation of hsa_circ_0001726 increases miR-140-3p expression and decreases KRAS mRNA expression in lenvatinib-resistant HCC cells, thereby inhibiting HCC progression. These results support our hypothesis that hsa_circ_0001726 is involved in lenvatinib resistance in HCC. However, this study lacks more in-depth mechanistic investigations of the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC and lenvatinib resistance.
Conclusions
This study established an HCC-related circRNA-miRNA-mRNA regulatory network and identified hsa_circ_0001726 as a novel potential biomarker for HCC prognosis. Additionally, we experimentally revealed the involvement of the hsa_circ_0001726/miR-140-3p/KRAS axis in HCC progression and lenvatinib resistance.
Ethical statement
All analyses were approved by the 900TH Hospital of Joint Logistics Support Force Institutional Ethical Review Board (No. 2023016) and were conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent.
Data sharing statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77(6):1598–1606. doi: 10.1016/j.jhep.2022.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massarweh NN, El-Serag HB. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control. 2017;24(3):1073274817729245. doi: 10.1177/1073274817729245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konyn P, Ahmed A, Kim D. Current epidemiology in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2021;15(11):1295–1307. doi: 10.1080/17474124.2021.1991792. [DOI] [PubMed] [Google Scholar]
- 6.Llovet JM, Pinyol R, Kelley RK, El-Khoueiry A, Reeves HL, Wang XW, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer. 2022;3(4):386–401. doi: 10.1038/s43018-022-00357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suresh A, Dhanasekaran R. Implications of genetic heterogeneity in hepatocellular cancer. Adv Cancer Res. 2022;156:103–135. doi: 10.1016/bs.acr.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagaraju GP, Dariya B, Kasa P, Peela S, El-Rayes BF. Epigenetics in hepatocellular carcinoma. Semin Cancer Biol. 2022;86(Pt 3):622–632. doi: 10.1016/j.semcancer.2021.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Meng H, Niu R, Huang C, Li J. Circular RNA as a Novel Biomarker and Therapeutic Target for HCC. Cells. 2022;11(12):1948. doi: 10.3390/cells11121948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv Y, Wang Z, Yuan K. Role of Noncoding RNAs in the Tumor Immune Microenvironment of Hepatocellular Carcinoma. J Clin Transl Hepatol. 2023;11(3):682–694. doi: 10.14218/JCTH.2022.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474(24):4219–4251. doi: 10.1042/BCJ20170079. [DOI] [PubMed] [Google Scholar]
- 12.Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65(4):625–639. doi: 10.1042/EBC20200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu B, Jia W, Feng Y, Wang J, Wang J, Zhu D, et al. Exosome-transported circHDAC1_004 Promotes Proliferation, Migration, and Angiogenesis of Hepatocellular Carcinoma by the miR-361-3p/NACC1 Axis. J Clin Transl Hepatol. 2023;11(5):1079–1093. doi: 10.14218/JCTH.2022.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong G, Lin Y, Huang Z. Identification of a novel circRNA-miRNA-mRNA regulatory axis in hepatocellular carcinoma based on bioinformatics analysis. Sci Rep. 2023;13(1):3728. doi: 10.1038/s41598-023-30567-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Tao H, Li J, Zhang E, Liang H, Zhang B. Comprehensive analysis of the competing endogenous circRNA-lncRNA-miRNA-mRNA network and identification of a novel potential biomarker for hepatocellular carcinoma. Aging (Albany NY) 2021;13(12):15990–16008. doi: 10.18632/aging.203056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Zhu D, Zhao Z, Sun M, Wang F, Li W, et al. RNA sequencing reveals the expression profiles of circRNA and identifies a four-circRNA signature acts as a prognostic marker in esophageal squamous cell carcinoma. Cancer Cell Int. 2021;21(1):151. doi: 10.1186/s12935-021-01852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19(1):60. doi: 10.1186/s12943-020-01184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunagawa Y, Yamada S, Sonohara F, Kurimoto K, Tanaka N, Suzuki Y, et al. Genome-wide identification and characterization of circular RNA in resected hepatocellular carcinoma and background liver tissue. Sci Rep. 2021;11(1):6016. doi: 10.1038/s41598-021-85237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye F, Fan C, Peng M, Liu S, Dong J, Yang L, et al. Screening and validating circular RNAs that estimate disease risk and treatment response of pediatric acute myeloid leukemia: a microarray-based analyses and RT-qPCR validation. J Cancer Res Clin Oncol. 2023;149(13):11233–11245. doi: 10.1007/s00432-023-04879-9. [DOI] [PubMed] [Google Scholar]
- 21.Wei X, Dong Y, Chen X, Ren X, Li G, Wang Y, et al. Construction of circRNA-based ceRNA network to reveal the role of circRNAs in the progression and prognosis of metastatic clear cell renal cell carcinoma. Aging (Albany NY) 2020;12(23):24184–24207. doi: 10.18632/aging.104107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng KL, Diao N, Zhou ZW, Fang CK, Wang JN, Zhang Y, et al. CircFGGY Inhibits Cell Growth, Invasion and Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma via Regulating the miR-545-3p/Smad7 Axis. Front Cell Dev Biol. 2022;10:850708. doi: 10.3389/fcell.2022.850708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36. doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 24.Qiu Y, Wang Y, Chai Z, Ni D, Li X, Pu J, et al. Targeting RAS phosphorylation in cancer therapy: Mechanisms and modulators. Acta Pharm Sin B. 2021;11(11):3433–3446. doi: 10.1016/j.apsb.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Zhu P, Ye Z, Wang M, Yang H, Huang S, et al. YRDC Mediates the Resistance of Lenvatinib in Hepatocarcinoma Cells via Modulating the Translation of KRAS. Front Pharmacol. 2021;12:744578. doi: 10.3389/fphar.2021.744578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W, Deng B, Lin P, Liu C, Li B, Huang Q, et al. Ribosome profiling analysis identified a KRAS-interacting microprotein that represses oncogenic signaling in hepatocellular carcinoma cells. Sci China Life Sci. 2020;63(4):529–542. doi: 10.1007/s11427-019-9580-5. [DOI] [PubMed] [Google Scholar]