Abstract
Mycobacteria is a diverse genus that includes both innocuous environmental species and serious pathogens like Mycobacterium tuberculosis, Mycobacterium leprae, and Mycobacterium ulcerans, the causative agents of tuberculosis, leprosy, and Buruli ulcer, respectively. This study focuses on Mycobacterium marinum, a closely related species known for its larger genome and ability to infect ectothermic species and cooler human extremities. Utilizing whole-genome sequencing, we conducted a comprehensive pan-genome analysis of 100 M. marinum strains, exploring genetic diversity and its impact on pathogenesis and host specificity. Our findings highlight significant genomic diversity, with clear distinctions in core, dispensable, and unique genes among the isolates. Phylogenetic analysis revealed a broad distribution of genetic lineages, challenging previous classifications into distinct clusters. Additionally, we examined the synteny and diversity of the virulence factor CpnT, noting a wide range of C-terminal domain variations across strains, which points to potential adaptations in pathogenic mechanisms. This study enhances our understanding of M. marinum's genomic architecture and its evolutionary relationship with other mycobacterial pathogens, providing insights that could inform disease control strategies for M. tuberculosis and other mycobacteria.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75228-0.
Keywords: Mycobacterium marinum, Pan genome, Evolution, Virulence genes
Subject terms: Bacterial genetics, Bacterial genomics
Introduction
Mycobacteria represents a distinct genus of microbes, which has membership ranging from ubiquitous and saprophytic species to deadly pathogens affecting a large population of the world. The important human pathogens within this genus are M. tuberculosis, M. leprae and M. ulcerans that are the causative agents of tuberculosis, leprae and Bruruli ulcer, respectively. Members of the M. tuberculosis complex have a conserved genome structure without recent events of horizontal gene transfer (HGT)1,2. This means that this pathogen is only able to evolve through gene mutation (point mutations, deletion and duplication) and subsequent clonal expansion of successful adaptations. Consequently, members of the M. tuberculosis complex show niche-adaptation and reductive evolution3. As compared to M. tuberculosis, the genomes of M. leprae and M. ulcerans show an even more drastic reductive genome evolution: both these pathogens contain a large number of pseudogenes, 16% and 27% respectively4,5. The current situation suggests that these three mycobacterial pathogens have likely evolved through HGT events6,7.
Interestingly, these three pathogens are not only relatively closely related to each other, but also to facultative pathogens such as Mycobacterium marinum. This pathogen is capable of causing infections in a wide range of cold-blooded species without clear evidence of host adaptation. M. marinum is also able to infect cooler parts of the human body, such as the extremities, leading to conditions commonly referred to as fish tank granuloma8. M. marinum has a larger genome as compared to the three human pathogens9,10, reflecting its status as a facultative pathogen that is also able to thrive in temperate and tropical water9. Comparative genomics analysis of M. marinum isolates revealed that the strains could be categorized into two clusters, which significantly influence the pathogenicity of these species11,12. Notably, representative strains of the first cluster, primarily composed of M. marinum strains isolated from humans, caused an acutely lethal disease in zebrafish. In contrast, strains of the second cluster are not usually linked to human infections and induced a chronic progressive disease in animals characterized by granuloma formation12. However, there is still limited information available regarding the evolutionary relationship and genomic diversity of M. marinum strains, and there has been relatively little focus on genomic studies and strain-specific genes in M. marinum to date.
Whole-genome sequencing (WGS) has enabled the determination of a pan-genome through comparative genomic analysis of multiple strains, providing an alternative approach to address various microbiological questions related to outbreak, evolution, antibiotic resistance, pathogenicity, and transmission13. Analysis of multiple genomes of individuals from the same species have indeed shown considerable intra-species diversity, primarily stemming from variances in the gene and transposable element repertoire among strains14. The pan-genome encompasses the entire gene collection of a particular species, including core genes shared by all strains, dispensable genes shared by two or more strains, and unique genes (singletons) specific to certain strains. This concept is valuable in characterizing bacterial species, as many species exhibit significant differences in gene contents15. Core genes are responsible for the primary phenotypes and fundamental biological processes, while dispensable and unique genes may participate in additional metabolic pathways, such as adaptation to specific hosts, virulence, antibiotic resistance, and other functions that confer selective advantages over other species16. Pan-genome analysis has been widely used in microbiology and has provided significant insights into pathogenesis, bacterial evolution, drug resistance, host specialization, HGT, and the identification of potential vaccine candidates against bacterial infections17–20.
This study is a comprehensive analysis of the pan-genome of M. marinum. The research focused on analyzing the genomic diversity of all M. marinum isolates, which comprised 60 from an online database (Supplementary Table S6) and 40 strains gathered in this study from humans and cold-blooded animals in various geographical areas. This analysis aimed to identify differences and similarities that could affect pathogenesis and host specificity. The approach included identifying core genes shared among all isolates (core genome), genes present in some but not all strains (dispensable genome), and strain-specific genes15. This research aims to enhance understanding of host-specific differences, pathogenicity, and evolutionary relationships, with potential applications in improving specific control measures for M. tuberculosis strain types.
Results
Phylogenetic analysis
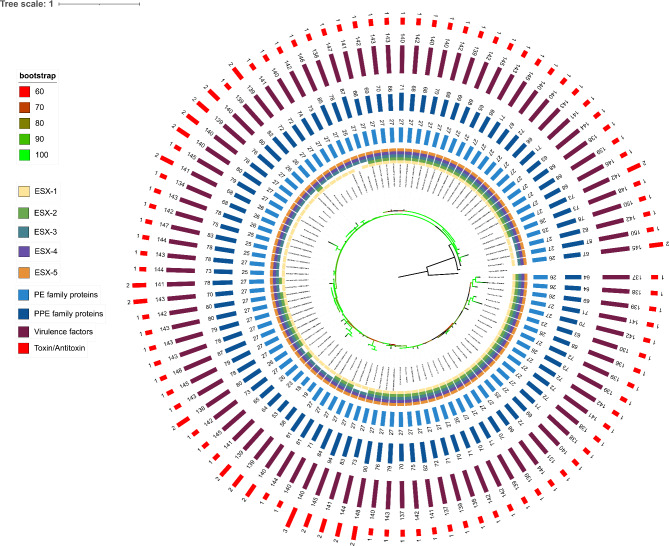
Synteny analysis revealed that the genomes exhibit a relative high degree of collinearity (Supplementary Fig. S1). We constructed a phylogenetic tree as descripted in the methods section, which included 100 M. marinum strains with Mycobacterium basiliense serving as the outgroup (Fig. 1). Based on the average nucleotide identity (ANI) heatmap, these 100 strains were divided into two distinct groups (Supplementary Fig. S2a), consistent with previous reports11,12 either based on virulence or ANI similarity. The phylogenetic analysis indicated that the strains could be divided into 2 lineages, with the so-called M lineage including 51 strains and the Aronson lineage including 49 strains. However, only one of these groups, the M lineage, forms a monophyletic clade. The other group, or Aronson lineage, although nucleotide-wise similar, comprise sister branches to the established clade, M lineage, and form a paraphyletic group (Supplementary Fig. S2b). This finding challenges the interpretations from current studies that consider both groups as separate clusters.
Fig. 1.
Phylogenetic tree of M. marinum strains. This figure illustrates the phylogenetic relationships among M. marinum strains analyzed in this study. The tree is represented in a circular layout with several concentric rings, each denoting different genetic characteristics of these strains. Starting from the innermost ring to the outermost, the annotations include: presence and variation of the ESX-1, ESX-2, ESX-3, ESX-4 and ESX-5 locus, distribution of PE, PPE family proteins, virulence factors and toxin/antitoxin systems. Each ring uses a color code to indicate the presence, absence, or type of each feature, providing a comprehensive overview of the genomic attributes that contribute to the phylogenetic placement of each strain. The different numbers of each strain indicate the total occurrences of different traits.
Type VII secretion systems in M. marinum
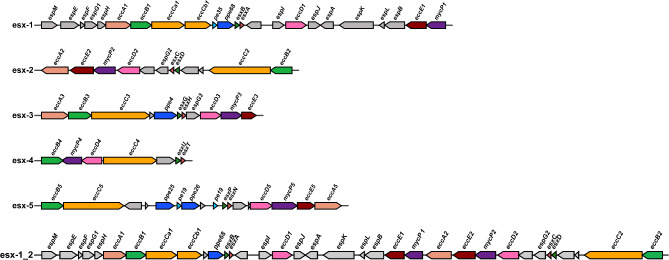
Type VII secretion systems (T7SSs) are essential for the secretion of effector proteins in mycobacteria. Mycobacteria, whether fast-growing or slow-growing, have five paralogous ESX loci, ranging from ESX-1 to ESX-5. However, the number of ESX systems encoded varies among different mycobacterial strains21. Previous studies have highlighted four membrane-associated ESX conserved components (EccB, EccC, EccD, and EccE), which establish a channel across the cytoplasmic membrane. The eccE gene is absent in one of the ESX loci (esx-4). Additionally, a conserved membrane-bound mycosin protease, MycP, serves to stabilize the core membrane complex. These core components of the T7SS were used as anchors for identifying the loci, leading to some findings.
In this study, the completeness of the ESX-1 to ESX-5 loci across 100 M. marinum strains was assessed, with the detailed results documented in Supplementary Table S1: ESX-3, ESX-4, and ESX-5 each in 100 strains. The universal presence of ESX-3, ESX-4, and ESX-5, underscores the critical role these systems play in the survival of M. marinum. The ESX-1 was complete in 99 strains, whereas ESX-2 was only present in 68 strains.
Manual verification was employed to assess locus completeness, especially in cases where core components appeared fragmented, which was predominantly due to assembly fragmentation as shown in Supplementary Fig. S3. For instance, the M. marinum H_15151 strain was missing eccCa1 from esx-1; however, the nucleotide sequences upstream the eccCb1 gene shared 98.98% identity with eccCa1 from the M strain, suggesting that fragmented assembly probably led to the partial loss of core components, hence the locus ESX-1 is considered as complete in this strain.
Notably, only one strain, DE4381 (GenBank accession number: GCA_003431585.1), exhibited a bonafide incomplete ESX-1 locus. Previous studies indicate that strain DE4381 isolated from saltwater fish, also known as “1218S” which is a smooth colony variant of 1218R, carries a deletion that spans ten genes within the esx-1 locus, including eccCa1, eccCb1, and eight other regional genes (espF, espG1, espH, eccA1, eccB1, pe35, ppe68, esxB), while duplicated esx-1 region (referred to as esx-6) include these genes of eccB1, pe35, ppe68 and esxB11. M. marinum ESX-1 locus contains proteins analogous to M. tuberculosis's EspI and EspJ, but their sequence identity and coverage are low. Although we consider them as espI and espJ based on the synteny of the locus, further evidence is needed to confirm this identification.
Most variation was observed for the ESX-2 locus. Similar as in M. tuberculosis, the esx-2 locus is closely linked to the esx-1 locus, with esx-1 located upstream of esx-2, as depicted in Fig. 2. Manual examination of the 32 strains lacking esx-2 locus showed that the region of difference is relatively conserved compared to strain 050,016 (GenBank accession number: GCA_028594325.1), who possesses a complete esx-2 locus and essential components are present on the same contig in the sequencing data (Supplementary Fig. S4). Notably, strains sharing the same deletion in esx-2 do not cluster into a single haplogroup on the phylogenetic tree (Fig. 1), which implies a horizontal event of this deletion followed by proliferation of the descendants. ESX-2 is the most obscure type VII system in mycobacteria, thus far no conditions were found that induced ESX-2 secretion and no specific phenotypic functions have been linked to this system.
Fig. 2.
Gene clusters of the ESX-1 to ESX-5 systems in M. marinum. The principal components of the ESX secretion systems include eccA, eccB, eccC, eccD, eccE, and mycP, with the exception that the esx-4 locus does not contain eccE4. The loci from ESX-1 to ESX-5 are depicted in various colors representing distinct protein families; regions colored in gray denote proteins from the Esp family or other regional proteins. The esx-1_2 locus represents the situation where the esx-1 gene cluster is contiguous with the esx-2 cluster.
The distribution of PE and PPE family proteins in M. marinum
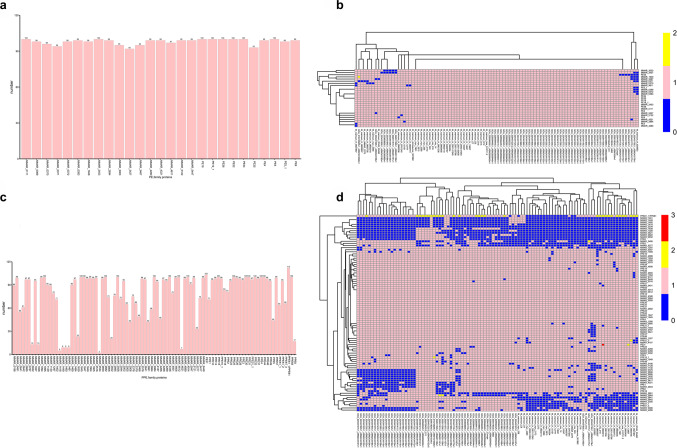
Next to the type VII secretion systems, we also examined their most abundant substrates, the PE and PPE family proteins. Our analysis of PE/PPE family proteins in M. marinum highlights their unique presence in mycobacteria. Analysis of 100 M. marinum strains revealed 27 types of PE family proteins, with little variance in their quantities across strains (Fig. 3a). Eight types of PE family proteins were consistently present in every strain (PE5, PE34, PE32, PE26, PE19_1, PE19, MMAR_2453, MMAR_0111), while PE35 was found in 93 strains (Fig. 3b, Supplementary Table S2). Conversely, significant diversity was observed in PPE family proteins, with distinct variations in abundance among different strains. Notably, PPE61_1 is a paralog of PPE61, which shares 96.16% identity with PPE61. PPE61/PPE61_1were the most prevalent in 100 strains, and PPE61 is a glycoprotein associated with drug efflux pumps22 (Fig. 3c, Supplementary Table S3). Seventeen types of PPE family proteins could be identified in each M. marinum strain, highlighting their widespread presence and potential functional significance (Fig. 3d, Supplementary Table S3).
Fig. 3.
Distribution of PE/PPE family proteins across 100 strains of M. marinum. The total counts of each (a) PE and (c) PPE family protein, respectively, with the protein names listed on the horizontal axes and the total counts on the vertical axes. Heatmaps showing the distribution patterns of (b) PE and (d) PPE family proteins across the strains, with strain names on the horizontal axes and protein names on the vertical axes.
Toxin/antitoxin systems
TA systems are typically composed of two components that include a stable toxin and a relatively unstable antitoxin. There are 79 pairs TA systems spanning the entire genome in M. tuberculosis H37Rv genome23. We used the 79 pairs of TA proteins as anchor to find the homologous proteins in M. marinum and have found that only 8 types of TA pairs (UCAT5, UCAT1, UCAT6, VapBC49, HigAB3, VapBC14, UCAT3, HigAB2) were present in M. marinum strains (Supplementary Fig. S5a, Supplementary Fig. S5b, Supplementary Table S4) with different abundance, for example UCAT5, which is uncharacterized, is universally present in all of the strains, whereas VapBC49 is present in only 3 strains. These 3 strains form a monophyletic group in terms of phylogeny (Fig. 1).
Distribution of virulence factors in M. marinum
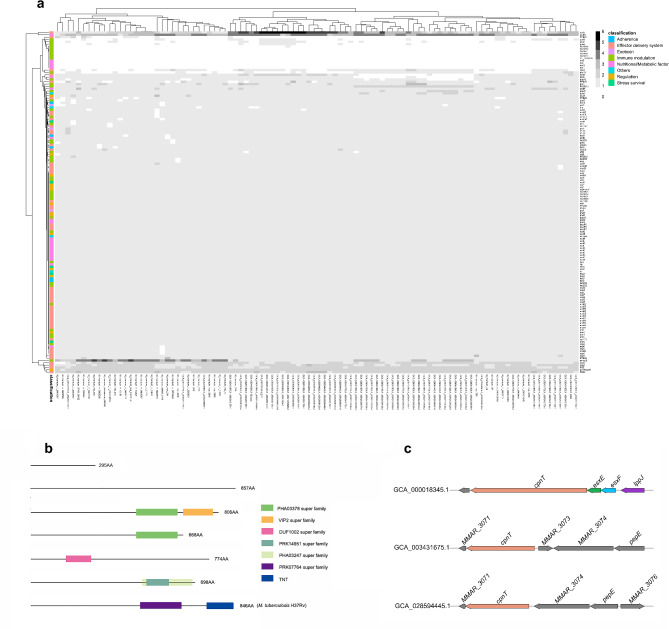
Based on the VFDB database (http://www.mgc.ac.cn/VFs/), all virulence factors related to Mycobacterium were downloaded, mainly from M. tuberculosis, which are categorized into eight primary groups including adherence, effector delivery system, exotoxin, immune modulation, nutritional/metabolic factor, stress survival, regulation and others. While the identities of EspI and EspJ from M. tuberculosis and M. marinum are low, we used EspI and EspJ from M. marinum to perform homologous clustering. Nine virulence factors could not be found in strains of M. marinum, including narG, narH, narI, narJ, PE_PGRS30, Rv2954c, Rv2956, Rv2957 and Rv2958c. The most abundant virulence factor is mbtM, with 342 genes in all strains, indicating that many strains have multiple copies of this virulence factor. (Fig. 4a, Supplementary Table S5).
Fig. 4.
Distribution of virulence factors and conserved domains of CpnT in 100 strains of M. marinum. (a) Heatmap displays the presence of 146 distinct virulence factors from M. tuberculosis across each M. marinum strain, with strains labeled along the horizontal axis and virulence factors listed on the vertical axis. (b) The variability in the conserved domains of the CpnT protein within M. marinum, where rectangles of different colors denote the various conserved domains. The different lengths of the line represent the different lengths of the CpnT amino acids. (c) The three synteny types of the cpnT gene.
CpnT is the only extracellular toxin of M. tuberculosis24. It is a two-domain protein, with each domain having distinct roles: the N-terminal domain is required for secretion by the type VII secretion systems. In addition, this N-terminal domain has been indicated to act as a transport pore in the mycobacterial outer membrane. The C-terminal domain contains the toxin activity and hydrolyzes the coenzyme NAD in the cytosol of infected host cells, leading to cell death25,26.
From previous genome analysis, we know that the C-terminal is radically different between M. tuberculosis strains and the M. marinum strain M. The diversity of the C-terminal domain of CpnT across different mycobacterial species was investigated using a homology search based on the N-terminal domain (Fig. 4b). This approach successfully identified seven distinct variants of the CpnT protein. Notably, a significant proportion of these variants (73.91%) lack any known Pfam domains, suggesting uncharacterized functional aspects or novel protein interactions. Among the characterized domains, the C-terminal regions were found to include domains from several super-families: PHA03378 (EBNA-3B), VIP2 (actin-ADP-ribosylating toxin), DUF1002 (unknown function), PRK14951 (DNA polymerase III subunits gamma and tau), and PHA03247 (large tegument protein UL36). These were primarily identified in M. marinum. Additionally, domains from PRK07764 (DNA polymerase III subunits gamma and tau) and TNT (inducing necrotic cell death) superfamilies were observed specifically in M. tuberculosis H37Rv. The presence of these diverse domains highlights potential functional diversities and adaptations of CpnT related to mycobacterial species-specific pathogenic mechanisms and cellular processes.
We have also explored the synteny patterns surrounding the cpnT within the genomes. This analysis revealed that cpnT is consistently preceded by three genes: esxE, esxF and lppJ in the majority of isolates, similar to the situation in M. tuberculosis. There are 4 cpnT genes present in a different location where adjunct to MMAR_3073, MMAR_3074 and pepE, and 5 cpnT homologues were adjunct to MMAR_3074, pepE and MMAR_3076.(Fig. 4c) The presence of the cpnT gene in a locus surrounded by genes coding for a short-chain membrane-associated dehydrogenase, a conserved hypothetical reductase, and regulatory proteins, rather than in the locus with secretion-related genes, suggests a different regulatory or functional context for cpnT in this specific arrangement. Such genomic contexts can offer insights into the diverse roles that cpnT might play in different Mycobacterium species or under varying environmental pressures.
Pan-genome analysis
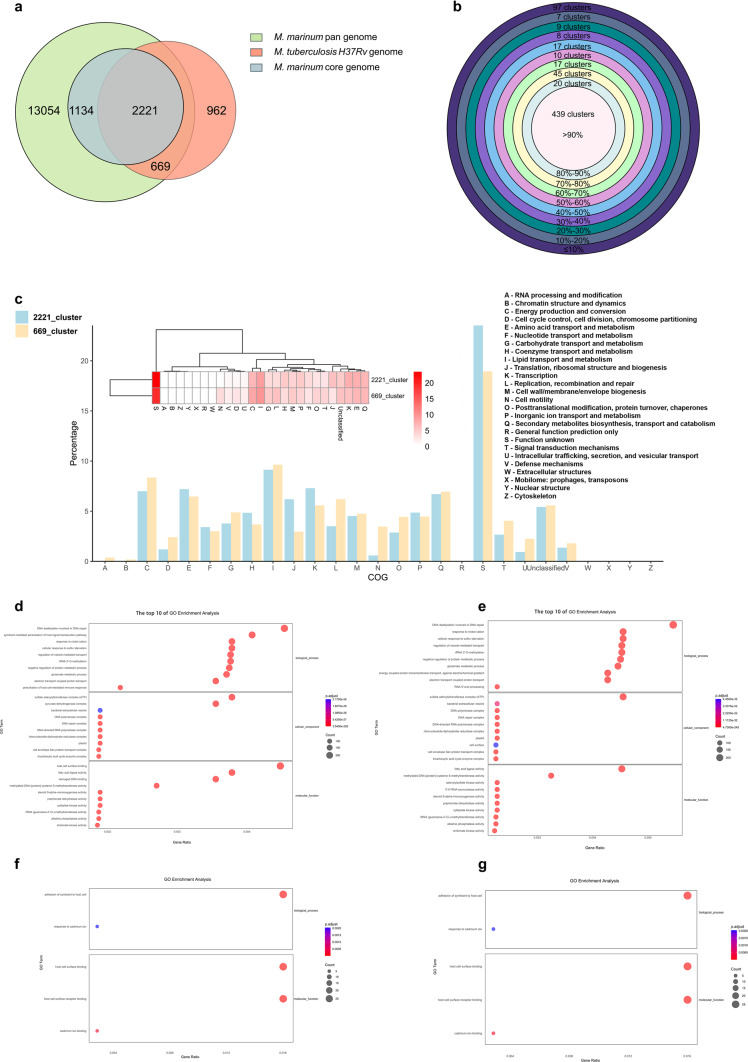
There are currently seven mainly recognized M. tuberculosis lineages (L1–L7), with two lineages (L2, L4) that have been well represented in taxonomic and phylogeographic evaluations. The representative strains of Lineage 1 to Lineage 7 are respectively N0072 strain, N0031 strain, N0054 strain, H37Rv strain, N1272 strain, N0091 strain and N3913 strain27. We conducted a cluster analysis of 100 strains of M. marinum and each representative strains of 7 lineages of M. tuberculosis, respectively. The analysis revealed that the clustering patterns across these distinct groups were not significantly different, as illustrated in the accompanying figure, mainly because gene variation in M. tuberculosis is very low (supplementary Fig. S6). In this study, we conducted an extensive analysis of orthologous proteins between 100 strains of M. marinum and M. tuberculosis H37Rv using CD-HIT. Our results identified 17,078 clusters that included genes from M. marinum and 3,852 clusters that incorporated genes from M. tuberculosis H37Rv. The presence of 17,078 clusters containing M. marinum genes suggests a robust diversity in this bacterial, possibly aiding its survival in diverse environmental conditions compared to the more host-restricted M. tuberculosis.
Notably, 3,355 clusters were characterized as core gene clusters for M. marinum, indicating essential functions preserved across all strains studied, and 2,221 clusters represented the intersection between the core gene clusters of M. marinum and M. tuberculosis H37Rv, which points to fundamental mycobacterial survival and virulence mechanisms that are conserved across these two species. Furthermore, an analysis of the pan-genomes (defined as genes present in less than 99% of strains) revealed 669 intersecting clusters between the less conserved genes of M. marinum and the genes of M. tuberculosis H37Rv (Fig. 5a). This indicates that M. tuberculosis and M. marinum are more related than previously anticipated. Among these genes that are present in M. tuberculosis and only present in a small subset of M. marinum strains that are the usual suspects, such as genes coding for putative transposases and prophage-related genes. However, interestingly, there are also several PPE and PE_PGRS genes. Possibly, these genes have played roles in niche-specific adaptations or immune evasion strategies.
Fig. 5.
Pangenome analysis of M. marinum. (a) The homology clusters of core genome and pan genome of 100 M. marinum strains and M. tuberculosis H37Rv. (b) The intersection(669_cluster) between pan genome of M. marinum and M. tuberculosis H37Rv was divided into 10 grades. (c) The functional annotation of 2221_cluster that is the intersection between M. marinum core genome and M. tuberculosis H37Rv and 669_cluster that is the intersection between M. marinum pan genome and M. tuberculosis H37Rv. (d) The top 10 of GO enrichment analysis of all genes of M. marinum of 669_cluster. The abscissa is the enrichment factor, indicating the ratio of the gene proportion annotated to the pathway in the all genes to that annotated to the pathway in all 100 M. marinum genes. The larger the enrichment factor, the more significant the enrichment level of genes in this pathway. (e) The top 10 of GO enrichment analysis of more than 90% strains of 669_cluster. (f) The GO enrichment analysis of 50%-90% strains of 669_cluster. (g) The GO enrichment analysis of less than 50% strains of 669_cluster.
To delve deeper into the genetic architecture, these 669 intersecting clusters were subdivided into 10 groups based on genes prevalence across strains, with the majority (65.62% or 439 clusters) being present in more than 90% of the strains (Fig. 5b). Within these groups, the cluster known as "2221_cluster," which comprises core genes of M. marinum, was found to be particularly significant in functional group J, more so than the "669_cluster," which consists of pan genes of M. marinum. Conversely, the “669_cluster” was more significant in functional group N than the “2221_cluster” (Fig. 5c).
The 669_cluster was further categorized based on the proportion of strains they appeared in into four grades: all clusters, > 90%, 50%-90%, and < 50%. GO enrichment analysis was performed to annotate the biological process (BP), cellular component (CC) and molecular function (MF) associated with these genes in different categories. Consistently, the top one of BP, CC and MF for 669_cluster are DNA dealkylation involved in DNA repair, sulfate adenylyltransferase complex (ATP) and host cell surface binding (Fig. 5d). The top one of BP, CC and MF for clusters present in more than 90% of strains included DNA dealkylation involved in DNA repair, sulfate adenylyltransferase complex (ATP) and fatty acid ligase activity (Fig. 5e). Clusters found in 50%-90% of strains and clusters present in less than 50% of strains showed enrichment for BP in the adhesion of symbiont to host cell and response to cadmium ion (Fig. 5f,g). This detailed categorization and analysis illuminate the diverse functional landscapes within these mycobacterial species.
Discussion
Our phylogenetic analysis of M. marinum suggests a notable divergence from previously established classifications into two distinct clades11,12. A clade is defined as a group of organisms that includes an ancestor and all its descendants, and is distinct from other organisms that do not share the same ancestry. Our results depict a single large monophyletic group (Supplementary Fig. S2b), which is M lineage, and the Aronson lineage does not form a monophyletic group. This observation challenges the existing paradigm that categorizes the species into two separate clusters. One plausible explanation for this discrepancy is insufficient sampling. The representation of genetic diversity within M. marinum is critical for accurately resolving its phylogenetic structure. In this study, even though this is the largest repository of M. marinum genomes thus far, the current limited number of strains may have skewed the phylogenetic tree, leading to the appearance of a more homogenous group rather than distinct clades.
The significance of T7SSs as secretion systems for effector proteins among mycobacterial species has led to the evolution of all five systems (ESX-1 to ESX-5) and their secretion components. Our findings indicate that the genes coding for three out of these five systems are conserved across all studied strains—ESX-3, ESX-4, and ESX-5—while the ESX-1 was present in 99 strains and ESX-2 in only 62 strains (refer to Fig. 1). This variability aligns with previous research, highlighting the diversity of these systems in mycobacteria21.
Moreover, the consistent presence of ESX-3, ESX-4, and ESX-5 in all strains, along with ESX-1 in 99% of these strains under investigation, emphasizes the crucial role these systems play in M. marinum's survival. In our study, we observed the absence of eccCa1 and eccCb1 genes in the DE4381 strain due to a deletion in the esx-1 locus. This absence shows the significance of this secretion system for virulence, as supported by previous findings in M. tubercuolosis DK9897 strain. This strain was unable to disrupt the phagosomal membrane, and the reinsertion of EccCa1-EccCb1-PE35 proteins resulted in the activation of specific EsxA T-cell responses, enhancing strain virulence28. Interestingly, among the 100 strains examined, 32 strains were uniquely found to lack ESX-2 locus. Despite the region of difference being relatively conserved compared to strain 050,016 (GenBank accession number: GCA_028594325.1), these strains did not exhibit the same haplogroup cluster in the phylogenetic tree (refer to Fig. 1). This observation suggests a wide distribution of genetic lineages, posing a challenge to prior classifications of M. marinum into distinct clades.
The examination of PE/PPE family proteins across 100 M. marinum strains unveiled minor variations in PE family proteins, whereas significant diversity was noted in PPE family proteins among these strains (Fig. 3a,c). Notably, seventeen proteins within the PPE family were conserved across all M. marinum strains, underscoring their potential implications for pathogenicity and host interaction (Fig. 3d). This observation aligns with recent research indicating that genetic variations in pe/ppe genes contribute to the diversity observed in M. tuberculosis lineages29. There are some reports about the functions of PE/PPE family proteins. PPE37 play a role in iron homeostasis through functions in heme–iron acquisition30. PE5–PPE4 are important for the iron-acquisition function of ESX-331. These analyses indicate that part of these PE/PPE proteins fulfill more conserved functions, such as nutrient import (PE/PPE proteins mediate nutrient transport across the outer membrane of M. tuberculosis)32, whereas others play roles in strain adaptation and niche differentiation, which could include virulence related functions. Furthermore, our analysis revealed a lower prevalence of TA pair systems in the M. marinum genomes (8 pairs) compared to that in the M. tuberculosis genomes (79 pairs) (Supplementary Fig. S5a, Supplementary Table S4). Interestingly, only three strains falling under the same monophyletic group harbored the VapBC49 systems, indicating a recent evolutionary divergence of these systems in M. marinum. In line with this, these genes adjacent to the VapBC49 operon are coding for some HGT-associated proteins, such as virion-associated phage protein, putative transposase for insertion sequence.
Additionally, the association between TA systems and the host–pathogen interface suggests a novel prospective role beyond their involvement in growth under various stress conditions33. These findings shed light on the evolutionary relationship of this gene group with pathogenicity. Pathogens and hosts have co-evolved, with hosts developing new defense mechanisms targeting specific gene products, prompting pathogens to develop new virulence factors. Analysis of these M. marinum genomes revealed 342 mbtM in greater abundance than other types virulence factors, while nine virulence factors were absent from all studied strains (Fig. 4a, Supplementary Table S5). The exploration and understanding of these undiscovered virulence factors are crucial for further understanding the pathogenic mechanisms of M. marinum, to provide new ideas for the pathogenesis of M. tuberculosis.
Our study elucidated a wide variation in the C-terminal domain functions of the CpnT protein, with 73.91% of these variants lacking Pfam domains, suggesting uncharacterized functional and novel protein interactions (Fig. 4b). Furthermore, we examined the Pfam domain TNT, which is associated with the inducing necrotic cell death superfamilies. Previous research has demonstrated that this domain can lead to the killing of macrophages by hydrolyzing NAD34. The presence of the cpnT within a locus flanked by genes such as a short-chain membrane-associated dehydrogenase, a conserved hypothetical reductase, and regulatory proteins, rather than a linkage with secretion associated genes, implies a distinct regulatory or functional context for cpnT. This suggests evolutionary adaptations or gene rearrangements that enable cpnT to contribute to cellular functions. This notion is supported by recent findings indicating that CpnT secretion relies on intact ESX-1, ESX-4, and ESX-5 systems during M. tuberculosis infection25. These genomic contexts offer insights into understanding the diverse functions that cpnT could serve across various Mycobacterium species or in response to different environmental challenges.
Our analysis aimed to identify orthologous proteins between 100 strains of M. marinum and M. tuberculosis H37Rv using CD-HIT. Results revealed 17,078 and 3,852 clusters from M. marinum and M. tuberculosis, respectively, incorporating genes from M. tuberculosis H37Rv (Fig. 5a). The extensive number of clusters (17,078 clusters) containing M. marinum genes reveals robust diversity within this bacterium, supporting its ability to survive in diverse environmental conditions compared to the more restricted host range of M. tuberculosis. Furthermore, our findings indicated 3,355 clusters comprising core genes of M. marinum and some genes of M. tuberculosis H37Rv. This suggests genetic conformity among M. marinum strains, as these genes encompass essential functions related to fundamental survival across all studied strains. Deep analysis of the genetic architecture of the 669 intersecting clusters based on gene prevalence across strains revealed most of them present in more than 90% of the strains, indicating distinct genomic diversity in M. marinum (Fig. 5b,c). This finding is supported by previous studies on 81 strains, which revealed extensive genomic diversity within M. marinum11. Notably, the "2221_cluster," comprised core genes of M. marinum involved in translational and ribosomal structure and biogenesis, crucial for adaptation to variant environmental conditions (Fig. 5c). Annotation enrichment analysis of molecular pathways associated with genes in different categories indicated diverse functions. The top one of BP, CC and MF for 669_cluster are DNA dealkylation involved in DNA repair, sulfate adenylyltransferase complex (ATP) and host cell surface binding (Fig. 5d). The top one of BP, CC and MF for clusters present in more than 90% of strains included DNA dealkylation involved in DNA repair, sulfate adenylyltransferase complex (ATP) and fatty cid ligase activity (Fig. 5e). Clusters found in 50%-90% of strains and clusters present in less than 50% of strains showed enrichment for BP in the adhesion of symbiont to host cell and response to cadmium ion (Fig. 5f,g). Collectively, these findings, in line with previous results, suggest a higher number of mutational hotspots in the M. marinum genome relative to other mycobacteria such as M. tuberculosis11, potentially explaining the diverse host range of this species.
Our study contributes to a deeper understanding of the genomic architecture of M. marinum and its evolutionary connections with other mycobacterial pathogens. By elucidating the correlations between genomic diversity and the pathogenesis of M. marinum strains, particularly in response to environmental variations, our findings offer valuable insights. These insights may inform the development of disease control strategies not only for M. tuberculosis but also for other mycobacteria species.
Methods
Culturing, DNA isolation and sequencing of bacteria
M. marinum isolates included in this study were obtained from the collections of the National Institute of Public Health and the Environment (RIVM) (Bilthoven, The Netherlands), the Institute of Aquaculture (Stirling, United Kingdom), and the Institute for Animal Science and Health (CIDC) (Lelystad, The Netherlands). Bacteria were grown at 30 °C on Middlebrook 7H10 agar plates supplemented with 10% OADC (oleic acid–albumin–dextrose–catalase, BD Biosciences), 0.2% Tween and 1 mg of D-arabinose per ml to decrease clumping of cells, or in shaking cultures in Middlebrook 7H9 liquid medium supplemented with 10% ADC (albumin–dextrose–catalase, BD Biosciences) and 0.05% Tween80. Genomic DNA was prepared using the bead beater–phenol/chloroform extraction method. Paired-end genomic DNA libraries were prepared using TruSeq DNA Sample Preparation Kits V2 (Illumina Inc, San Diego, CA, USA) and sequenced on an Illumina HiSeq2500 as described before10.
Genome assembly, annotation and data acquisition
PhiX reads trimal, adapter removal, quality control, and error correction of the raw sequence reads were done using BBDuk (https://jgi.doe.gov/data-and-tools/software-tools/bbtools/bb-tools-user-guide/bbduk-guide/) with the paraments “k = 25, minlen = 25, mink = 11 and hdist = 1”. A total of 43 M. marinum genomes obtained in this study underwent de novo assembly using SPAdes (v3.12.0)35 with the default settings. Sixty genome assemblies were obtained from National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/), the detailed information of the strains can be found in Supplementary Table S6. All genomes underwent quality control using CheckM (v1.2.2)36. One hundred assemblies with completeness higher than 97% and contamination lower than 2% were used for downstream analysis. Annotations of all assemblies were conducted with Prokka (v 1.11)37. Additionally, ProgressiveMauve (v20150226)38 was employed to carry out multiple genome alignments of the 40 M. marinum assemblies sequenced in this study that passed quality control.
Phylogenetic analysis
Marker genes extracted from the genome assemblies were aligned with PhyloPhlAn (v3.0.67)39. Subsequently, IQ-TREE (v2.1.4)40 was utilized for construct the phylogenetic tree with Maximum-likelihood, setting the ultrafast bootstrap parameter to 10,000, with all other parameters left at their default settings and VT + F + R2 being the best-fit model. The resulting tree was visualized with iTol (v6.9; https://itol.embl.de/)41. Mycobacterium basiliense served as the outgroup in the phylogeny construction. Average Nucleotide Identity (ANI) values among different M. marinum strains were calculated using fastANI (v1.1)42 with default parameters.
Comparative genomics analysis of M. marinum
Protein sequences of the Type VII Secretion Systems (T7SSs) in M. marinum were extracted from strain M. marinum M (GenBank accession number: GCA_000018345.1), and the homologues genes were identified in 100 M. marinum strains using Proteinortho (v6.0.29)43. The presence of these five core components, including eccB, eccC, eccD, eccE, and mycP, is essential for the formation of the corresponding ESX loci except for ESX-4, which lacks eccE in its locus44. In this study, a locus is considered complete if it encompasses all five essential genes, with the exception of ESX-4. If the number of core components for a particular locus falls below the expected count (fewer than 5 genes), a manual check is conducted to determine if this deficiency is due to assembly fragmentation. Additionally, a BLASTn analysis of the contig boundaries is performed to verify if the incomplete locus results from issues in the assembly process.
Loci of various types were visualized using R studio (v4.2.2), with ggplot2 (v3.4.4, https://ggplot2.tidyverse.org) and gggenes (v0.5.1, https://wilkox.org/gggenes/) as the necessary installed packages. A manual comparison of strains lacking esx-2 with the reference strain (GenBank accession number: GCA_028594325.1) was conducted using ACT (v18.2.0) to identify the location of the esx-2 deletion. Additionally, proteins from the PE/PPE families, known virulence factors, and various toxin/antitoxin (T/A) systems like VapBC and MazEF, that previously identified in M. tuberculosis H37Rv, were retrieved and homology searches were done across the same set of M. marinum strains for comparison.
Pan-genome analysis
To investigate the core genome and relationships among M. marinum assemblies and M. tuberculosis, homologous clustering approach was utilized. This analysis was carried out using CD-HIT (v4.8.1)45, with settings that included a sequence identity threshold of 60% and length difference cutoff of 60%. The genes present in ≥ 99% strains of M. marinum were designated core genes. Cluster of orthologous groups were assigned using eggNOG-mapper (v2.1.12)46 to determine functional differences between the core and dispensable genes. The GO annotation was used to annotate the genes from M. marinum assemblies, serving as the database for the enrichment analysis. The package clusterProfiler (4.6.2)47 from R was utilized to perform gene set pathway enrichment analysis for different gene clusters.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We extend our sincere thanks to the colleagues from Bioinformatics Laboratory and Bioinformatics Platform at The First Hospital of Jilin University for their valuable discussions and suggestions, which have significantly enhanced the quality of this manuscript. We also wish to thank the KAUST Core Lab for providing the sequencing services as well as the support from medical writers, proof-readers and editors.
Author contributions
AMA conceptualized the study. AMA, QG, AP and WB obtained the funding and supervised the work. AMA, SA and RU isolated, cultured bacterial strains and prepared the libraries. MZ and QG performed the data analysis, and MZ, QG, MA, and AMA prepared the initial draft of the manuscript, followed by edits from LS, WB, QG, and AMA. All authors have commented on various sections of the manuscript, which were finally curated and incorporated in the final version by AMA and QG.
Funding
Whole genome sequencing was supported by the baseline funding of AP at King Abdullah University of Science and Technology (KAUST). Bioinformatics analysis was supported by the Research start-up funds of Prof. Qingtian Guan, grant number 04045970001, The Bethune Project of Jilin University 2024B20 of Lei Song and Science and Technology Development Project of Changchun City of Lei Song, grant number 23YQ10. Work at AMA’s Laboratory is supported by the QU internal Grant (QUCG-CMED-21/22-2).
Data availability
Genome assemblies of isolates have been deposited in the European Nucleotide Archive (ENA) database (https://www.ebi.ac.uk/ena/browser/home) under BioProject accession number PRJEB76167, BioSample numbers SAMEA115663384 to SAMEA115663423.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qingtian Guan, Email: Qingtian_guan@jlu.edu.cn.
Abdallah M. Abdallah, Email: Abdallah.musa@qu.edu.qa
References
- 1.Hoffmann, C., Leis, A., Niederweis, M., Plitzko, J. M. & Engelhardt, H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA105, 3963–3967. 10.1073/pnas.0709530105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith, N. H., Gordon, S. V., de la Rua-Domenech, R., Clifton-Hadley, R. S. & Hewinson, R. G. Bottlenecks and broomsticks: The molecular evolution of Mycobacterium bovis. Nat. Rev. Microbiol.4, 670–681. 10.1038/nrmicro1472 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Veyrier, F. J., Dufort, A. & Behr, M. A. The rise and fall of the Mycobacterium tuberculosis genome. Trends Microbiol.19, 156–161. 10.1016/j.tim.2010.12.008 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T. et al. Massive gene decay in the leprosy bacillus. Nature409, 1007–1011. 10.1038/35059006 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Stinear, T. P. et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res.17, 192–200. 10.1101/gr.5942807 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becq, J. et al. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol. Biol. Evol.24, 1861–1871. 10.1093/molbev/msm111 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Veyrier, F., Pletzer, D., Turenne, C. & Behr, M. A. Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol. Biol9, 196. 10.1186/1471-2148-9-196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ucko, M. & Colorni, A. Mycobacterium marinum infections in fish and humans in Israel. J. Clin. Microbiol.43, 892–895. 10.1128/jcm.43.2.892-895.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stinear, T. P. et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res.18, 729–741. 10.1101/gr.075069.107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weerdenburg, E. M. et al. Genome-wide transposon mutagenesis indicates that Mycobacterium marinum customizes its virulence mechanisms for survival and replication in different hosts. Infect. Immun.83, 1778–1788. 10.1128/iai.03050-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, S. et al. Extensive genomic diversity among Mycobacterium marinum strains revealed by whole genome sequencing. Sci. Rep.8, 12040. 10.1038/s41598-018-30152-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Sar, A. M. et al. Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infect. Immun.72, 6306–6312. 10.1128/iai.72.11.6306-6312.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uchiya, K. I. et al. Comparative genome analyses of Mycobacterium avium reveal genomic features of its subspecies and strains that cause progression of pulmonary disease. Sci. Rep.7, 39750. 10.1038/srep39750 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tettelin, H., Riley, D., Cattuto, C. & Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol.11, 472–477. 10.1016/j.mib.2008.09.006 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Tettelin, H. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA102, 13950–13955. 10.1073/pnas.0506758102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhari, N. M., Gupta, V. K. & Dutta, C. BPGA—An ultra-fast pan-genome analysis pipeline. Sci. Rep.6, 24373. 10.1038/srep24373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maione, D. et al. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science309, 148–150. 10.1126/science.1109869 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vernikos, G., Medini, D., Riley, D. R. & Tettelin, H. T. Ten years of pan-genome analyses. Curr. Opin. Microbiol.23, 148–154. 10.1016/j.mib.2014.11.016 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Dunn, B., Richter, C., Kvitek, D. J., Pugh, T. & Sherlock, G. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res.22, 908–924. 10.1101/gr.130310.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muzzi, A., Masignani, V. & Rappuoli, R. The pan-genome: Towards a knowledge-based discovery of novel targets for vaccines and antibacterials. Drug Discov. Today12, 429–439. 10.1016/j.drudis.2007.04.008 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Rivera-Calzada, A., Famelis, N., Llorca, O. & Geibel, S. Type VII secretion systems: Structure, functions and transport models. Nat. Rev. Microbiol.19, 567–584. 10.1038/s41579-021-00560-5 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Yari, S. et al. A potent subset of Mycobacterium tuberculosis glycoproteins as relevant candidates for vaccine and therapeutic target. Sci. Rep.13, 22194. 10.1038/s41598-023-49665-2 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, A., Venkataraman, B., Vasudevan, M., & Gopinath Bankar, K. Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep.7, 5868 (2017). 10.1038/s41598-017-06003-7. [DOI] [PMC free article] [PubMed]
- 24.Pajuelo, D. et al. Toxin secretion and trafficking by Mycobacterium tuberculosis. Nat. Commun.12, 6592. 10.1038/s41467-021-26925-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izquierdo Lafuente, B., Ummels, R., Kuijl, C., Bitter, W., and Speer, A. Mycobacterium tuberculosis Toxin CpnT Is an ESX-5 Substrate and Requires Three Type VII Secretion Systems for Intracellular Secretion. mBio (2021). 10.1128/mBio.02983-20. [DOI] [PMC free article] [PubMed]
- 26.Danilchanka, O. et al. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc. Natl. Acad. Sci. USA111, 6750–6755. 10.1073/pnas.1400136111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borrell, S. et al. Reference set of Mycobacterium tuberculosis clinical strains: A tool for research and product development. PLoS ONE14, e0214088. 10.1371/journal.pone.0214088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clemmensen, H. S. et al. An attenuated Mycobacterium tuberculosis clinical strain with a defect in ESX-1 secretion induces minimal host immune responses and pathology. Sci. Rep.7, 46666. 10.1038/srep46666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gómez-González, P. J. et al. Functional genetic variation in pe/ppe genes contributes to diversity in Mycobacterium tuberculosis lineages and potential interactions with the human host. Front. Microbiol.14, 1244319. 10.3389/fmicb.2023.1244319 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tullius, M. V., Nava, S., & Horwitz, M. A. PPE37 is essential for mycobacterium tuberculosis heme-iron acquisition (HIA), and a defective PPE37 in Mycobacterium bovis BCG prevents HIA. Infect. Immun. (2019). 10.1128/iai.00540-18. [DOI] [PMC free article] [PubMed]
- 31.Tufariello, J. M. et al. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc. Natl. Acad. Sci. USA113, E348-357. 10.1073/pnas.1523321113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Q. et al. PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science367, 1147–1151. 10.1126/science.aav5912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan, S. et al. Toxin-antitoxin system of mycobacterium tuberculosis: Roles beyond stress sensor and growth regulator. Tuberculosis (Edinb)143, 102395. 10.1016/j.tube.2023.102395 (2023). [DOI] [PubMed] [Google Scholar]
- 34.Sun, J. et al. The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD. Nat. Struct. Mol. Biol.22, 672–678. 10.1038/nsmb.3064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A. & Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinf.70, e102. 10.1002/cpbi.102 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res.25, 1043–1055. 10.1101/gr.186072.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics30, 2068–2069. 10.1093/bioinformatics/btu153 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Darling, A. C., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res.14, 1394–1403. 10.1101/gr.2289704 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asnicar, F., et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 30. Nat. Commun.11, 2500 (2020). 10.1038/s41467-020-16366-7. [DOI] [PMC free article] [PubMed]
- 40.Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol.32, 268–274. 10.1093/molbev/msu300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293-w296. 10.1093/nar/gkab301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goris, J. et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol.57, 81–91. 10.1099/ijs.0.64483-0 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Lechner, M. et al. Proteinortho: Detection of (co-)orthologs in large-scale analysis. BMC Bioinf.12, 124. 10.1186/1471-2105-12-124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Y., et al. Crosstalk between the ancestral type VII secretion system ESX-4 and other T7SS in Mycobacterium marinum. iScience25, 103585 (2022). 10.1016/j.isci.2021.103585. [DOI] [PMC free article] [PubMed]
- 45.Li, W. & Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics22, 1658–1659. 10.1093/bioinformatics/btl158 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol.38, 5825–5829. 10.1093/molbev/msab293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics16, 284–287. 10.1089/omi.2011.0118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies of isolates have been deposited in the European Nucleotide Archive (ENA) database (https://www.ebi.ac.uk/ena/browser/home) under BioProject accession number PRJEB76167, BioSample numbers SAMEA115663384 to SAMEA115663423.