Abstract
Biliary atresia (BA), resulting from abnormal development of the liver’s internal or external bile ducts, can lead to liver damage and potentially fatal cirrhosis. Type I cystic biliary atresia is a relatively uncommon, but clinically significant variant of BA. It is critical to develop experimental models of BA to examine the etiology and pathogenesis, which remain elusive, and to develop future therapeutics. Here, we have successfully generated a panel of human induced pluripotent stem cells (hiPSCs) from five Japanese patients carrying type I cystic BA. These hiPSC lines exhibited characteristics of self-renewal and pluripotency. These cells held normal karyotypes mostly, but one of them carried hemizygous deletions, the clinical significance of which is unknown yet. Whole genome sequence analysis indicated that some of the mutations or single nucleotide polymorphisms (SNPs) commonly found in these patients are related to hepatobiliary abnormality. Given the limited understanding of the molecular pathogenesis of cystic BA, attributed to unknown factors of genetic and environmental causes, these cellular resources will be instrumental in replicating disease phenotypes and in advancing novel therapies for this disease.
Keywords: Human induced pluripotent stem cells, Biliary atresia (BA), Self-renewal, Pluripotency, Whole genome sequencing
Introduction
Biliary atresia (BA) is an obstructive fibrotic cholangiopathy of the intrahepatic and extrahepatic bile ducts that causes pathological jaundice and liver failure in early infancy. Even if surgical removal of extrahepatic bile duct remnants and subsequent hepatoenterostomy are successful, most patients are likely to present with progressive liver dysfunction. It is the most common indication for pediatric liver transplantation [1]. Further improvement in outcome will require a greater understanding of the mechanisms of biliary injury and liver fibrosis. There are currently no convincing theories regarding the cause of BA, including abnormal organ development, viral infection, genetic predisposition, and maternal immunity. Thus, to elucidate the pathogenesis of BA and to develop alternative therapeutics, sophisticated disease models are required.
Animal models of BA have been generated with several methods. Heterozygous deletion of the SRY- related HMG-box 17 (Sox17) gene in mice exhibits a condition like BA [2–4]. Pkd1l1-deficient mice and zebrafish exhibited bile duct hypertrophy, reduced biliary drainage, and liver fibrosis [5–7]. Mice infected with rotavirus displayed symptoms similar to human biliary atresia, with bile duct obstruction, bile duct proliferation, and liver inflammation with fibrosis [8]. Also, a plant toxin, biliatresone, has been shown to induce BA-like symptoms in several animal models [9–11]. Neonatal BALB/C mice injected with biliatresone developed clinical signs of biliary obstruction, and dysplasia or the absence of extrahepatic biliary tract lumen, which confirmed the occurrence of BA [12]. In another study, neonatal C57BL/6 J mice exposed to biliatresone exhibited clinical symptoms of BA, such as jaundice, twisted and enlarged extraphepatic bile ducts, and cholestasis [13]. Also, the pups of mice treated with low-dose biliatresone during pregnancy have altered bile acid and immune profiles similar to those observed in BA patients, even in the absence of significant histological changes [14]. These studies demonstrate that biliatresone-treated mice are useful for modeling BA and may serve as a tool for exploring therapeutic interventions. However, it is unclear whether these models reflect the natural pathogenesis of human BA or not [15, 16].
Research on samples and data from BA patients is only possible after the onset of the disease [17, 18]. Recent studies have focused on the potential of BA-specific human liver organoids as research models. Biliary organoids derived from liver biopsies of BA patients revealed molecular and functional evidences of delayed epithelial development in BA patients [19]. Human liver organoids treated with a synthetic immunostimulant that is structurally similar to a double-stranded RNA (dsRNA) found in the reovirus and rotavirus, polyinosinic:polycytidylic acid (poly (I:C)), exhibited morphology and genetic signature highly compatible to organoids developed from BA liver samples [20]. Also, human liver organoids treated with biliatresone exhibited a decrease in cell–cell tight junctions, polarity changes, increased epithelial permeability, and loss of cilia and cilia function in cholangiocytes [21]. These studies demonstrate that human liver organoids from BA patients or chemical treatments are excellent disease models to recapitulate the symptoms of BA. In contrast, building an experimental model that can monitor the bile duct damage process during development will lead to further elucidation of the pathogenesis of BA. Patient-derived human induced pluripotent stem cells (hiPSCs) can be promising tools to recapitulate the pathogenesis during development. Indeed, only one pioneering study reported that BA-specific hiPSCs showed deficiency in biliary differentiation along with increased fibrosis, the two key disease features of BA to date [22]. To expand this report to the general conclusions of the modeling of BA, more hiPSC samples are required since the genetic cause of BA is unknown, and the ethnicity and geographical distribution of the epidemiology of BA may be variable [23, 24].
Here, we have established type I cystic BA-specific hiPSC lines from five Japanese female patients and performed whole genome sequencing (WGS) on these samples. Type I cystic BA is a relatively uncommon, but clinically significant variant of BA. These hiPSC lines derived from type I cystic BA patients should contribute to the development of pathological models and the elucidation of the molecular pathogenesis of cystic BA.
Results and discussion
We collected PBMCs (peripheral blood mononuclear cells) from five patients of type I cystic BA patients. The general clinical information of these patients is shown in Table 1. All the patients were diagnosed soon after birth and performed Kasai surgery operation. These patients were not associated with Alagille syndrome or Hajdu–Cheney syndrome because they did not develop any symptoms other than hepatobiliary systems. Also, morphologies of the common bile duct in these patients show cystic dilatation, which differs from hypoplasia observed in Alagille syndrome patients. No liver transplantation was performed on these patients until the collection of PBMCs. The hiPSC lines derived from these cystic BA patients (cBA-hiPSCs) were generated using episomal plasmid vectors from these PBMCs [25, 26].
Table 1.
Summary of general clinical information of cystic type I BA patients recruited for this study
| Patient number | HiPSC line name | Age at blood collection (years) | Sex | Ethnicity | Age at Kasai surgery operation (days) |
|---|---|---|---|---|---|
| 1 | HiPS-cBA1 | 3 | Female | Asia | 59 |
| 2 | HiPS-cBA2 | 6 | Female | Asia | 64 |
| 3 | HiPS-cBA3 | 25 | Female | Asia | 62 |
| 4 | HiPS-cBA4 | 28 | Female | Asia | 37 |
| 5 | HiPS-cBA5 | 5 | Female | Asia | 28 |
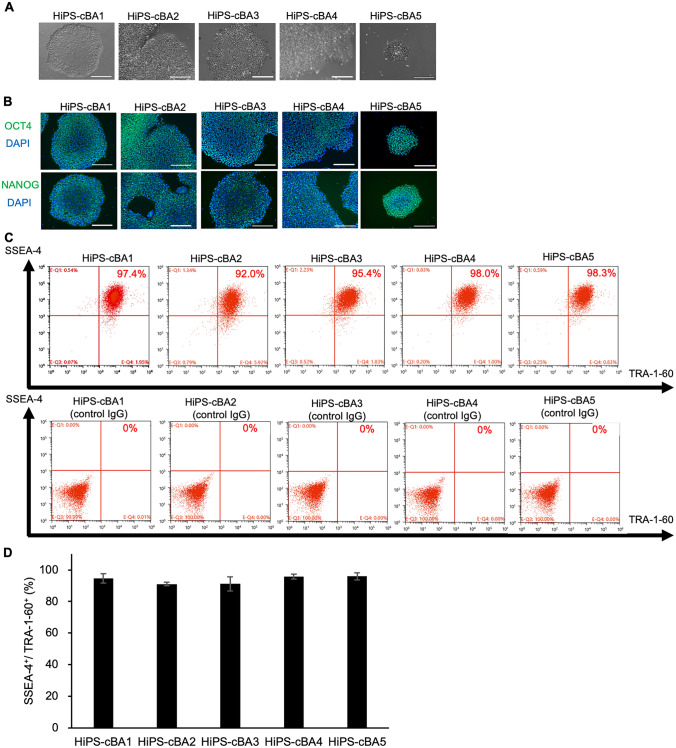
The generated cBA-hiPSC lines were cultured in an undifferentiated state and characterized to confirm self-renewal capacity and pluripotency. All the cBA-hiPSC lines formed typical human embryonic stem cell (hESC)-like colonies under feeder-free culture conditions (Fig. 1A). The expression of the self-renewal markers, OCT3/4 and NANOG, was detected in these cBA-hiPSC lines with immunocytochemistry (Fig. 1B). The expression of cell surface markers, SSEA-4 and TRA-1-60, was also detected with flow cytometry (Fig. 1C). All the cBA-hiPSC lines contained cells more than 90% positive for SSEA-4 and TRA-1-60 (Fig. 1D).
Fig. 1.
Self-renewal marker expression of hiPSC lines derived from cystic BA patients. A Morphologies of BA-specific hiPSCs taken with phase contrast microscopy. Scale bars, 200 µm. B Immunocytochemistry of OCT3/4 and NANOG on cBA-hiPSCs. Scale bars, 200 µm. C Flow cytometry analysis of SSEA-4 and TRA-1-60 on cBA-hiPSCs. Top panels: samples with anti-SSEA1 and anti-TRA-1-60 antibodies. Bottom panels: samples with control IgG. D The ratio of positive cells for SSEA-4 and TRA-1-60. The bar graph shows mean + standard errors (SE), n = 3 (biological samples)
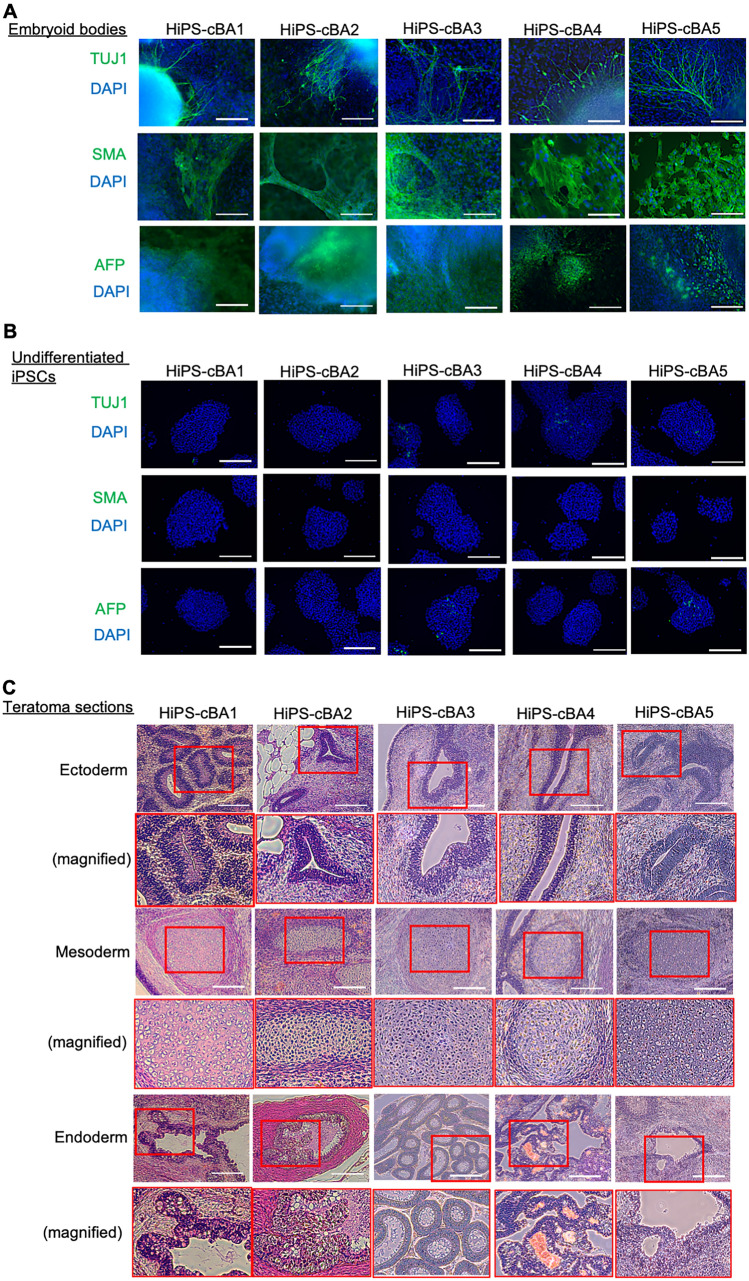
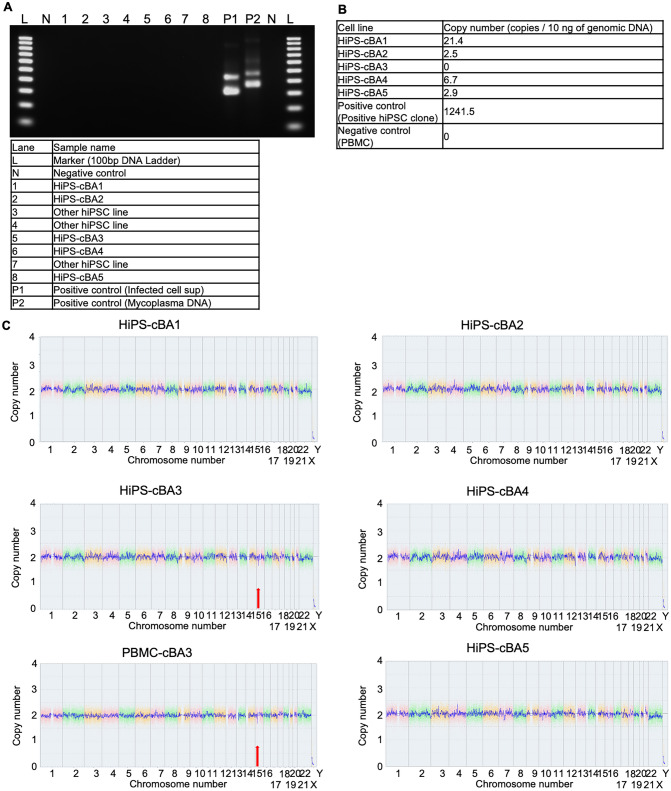
The pluripotency of these cBA-hiPSC lines was assessed using embryoid body (EB) formation and teratoma formation assays. Immunocytochemistry for an ectodermal marker, TUJ1 (TUBB3; tubulin, beta 3 class III), a mesodermal marker, SMA (smooth muscle actin), and an endodermal marker, AFP (α-fetoprotein), demonstrated that EBs generated from these cBA-hiPSC lines contained differentiated cells from all three germ layers (Fig. 2A and B). Teratomas derived from these lines contained tissues from all three germ layers, such as neuroepithelium or melanocyte (ectoderm), cartilage (mesoderm), and gastrointestinal-like structures (endoderm) (Fig. 2C). All cBA-hiPSC lines tested negative for mycoplasma contamination (Fig. 3A). The episomal plasmid vectors used for generating hiPSCs were confirmed to be absent or little in the genomic DNA of all cBA-hiPSC lines by qPCR targeting of the EBNA1 sequence (Fig. 3B). CNV (copy number variation) microarray analysis revealed that these lines had normal karyotypes, except for a heterozygous deletion of 16p12.2, which includes the METTL9, IGSF6, and OTOA genes in HIPS-cBA3 (Fig. 3C). This deletion was also found in the original PBMCs from the same patient. This large deletion is considered potentially pathogenic, especially for causing hearing loss when both OTOA alleles are disrupted [27, 28]. However, whether this deletion is linked to biliary atresia remains uncertain.
Fig. 2.
Pluripotency of hiPSC lines derived from cystic BA patients. A Immunocytochemistry of TUJ1, SMA, and AFP on embryoid bodies (EBs) differentiated from BA-specific hiPSCs. Scale bars, 200 µm. B Immunocytochemistry of TUJ1, SMA, and AFP on undifferentiated cBA-hiPSCs. Scale bars, 200 µm. C Sections with hematoxylin and eosin (HE) staining on teratomas derived from cBA-hiPSCs. Neural rosettes, cartilages, or intestine-like secretion tissues are indicated as derivatives of ectoderm, mesoderm, or endoderm, respectively. Red rectangles are magnified regions. Scale bars, 200 µm
Fig. 3.
Characterization of hiPSC lines derived from cystic BA patients. A Mycoplasma test using PCR on conditioned medium samples. B Episomal vector test using PCR on genomic DNA samples. C Copy number analysis from CGH array on genomic DNA samples. Red arrows indicate heterozygous deletion of the 16p12.2 region
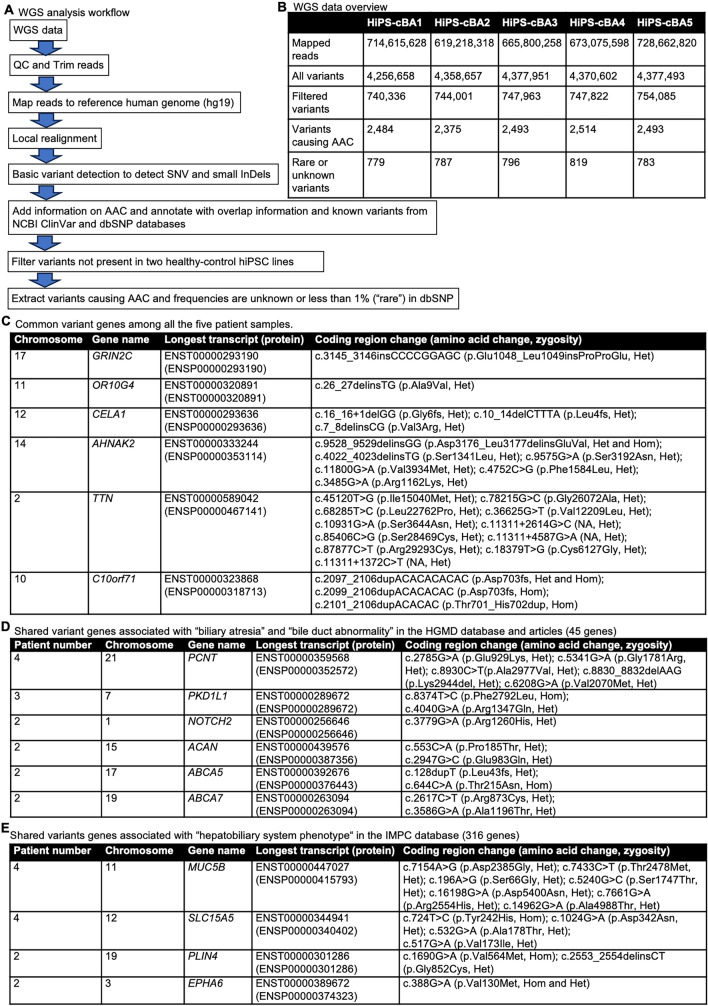
WGS was performed on these cBA-hiPSC lines. The workflow of the analysis is shown in Fig. 4A. From each sample, 740,000–750,000 variants were obtained from 610 to 730 million mapped reads (Fig. 4B). After filtering variants with two healthy control hiPSC samples and selecting those causing amino acid changes (AAC) with a frequency of less than 1% (considered "rare") or unknown, approximately 800 variants were identified in each sample. Common variant genes found in all patient samples included GRIN2C, OR10G4, CELA1, AHNAK2, TTN, and C10orf71; however, no published studies have linked these genes to hepatobiliary abnormality (Fig. 4C). Next, we extracted variant genes shared by two or more patients and cross-referenced them with previous studies and the Human Gene Mutation Database (HGMD) [29] to identify those potentially associated with bile duct abnormalities. In this category, PCNT (pericentin), PKD1L1 (polycystic kidney disease 1-like 1), ACAN (aggrecan), ABCA5 (ATP-binding cassette subfamily A member 5), ABCA7, and NOTCH2 were identified (Fig. 4D). PCNT was identified as a de novo mutation in BA patients, and PCNT knockout zebrafish exhibited reduced biliary flow [30]. Variants in PKD1L1 genes were also identified to be associated with BA in several studies [31, 32]. Pkd1l1-deficient mice and zebrafish showed abnormal phenotypes related to BA [5–7]. NOTCH2 is primarily a gene responsible for Alagille or Hajdu–Cheney syndrome, but may also be implicated in BA etiology [32]. ACAN, ABCA5, and ABCA7 have not been specifically linked to BA, but further research would be needed to clarify their roles. We also extracted variant genes shared by two or more patients and cross-referenced them with genes associated with "hepatobiliary system phenotypes" from the International Mouse Phenotyping Consortium (IMPC) [33]. In this category, MUC5B (mucin 5B), SLC15A5 (solute carrier family 15 member 5), PLIN4 (perilipin 4), and EPHA6 were identified (Fig. 4E). MUC5B is specifically expressed in gallbladder [34, 35], and homozygous Muc5b-deficient mice have smaller livers (https://www.mousephenotype.org/data/genes/MGI:1921430). SLC15A5 is a relatively uncharacterized gene, but homozygous knockout mice exhibit enlarged gallbladders, a sign of cholecystitis, according to the IMPC database (https://www.mousephenotype.org/data/genes/MGI:3607714). Knockout mice for PLIN4 or EPHA6 show abnormal liver morphology (https://www.mousephenotype.org/data/genes/MGI:1929709, https://www.mousephenotype.org/data/genes/MGI:108034). In summary, we identified interesting variants in genes potentially related to BA or hepatobiliary abnormality in these cBA-hiPSCs, although the impact of these variants on protein functionality remains unclear. These findings suggest that BA is not a monogenic disease but rather polygenic, or it could be associated with viral perinatal infections, toxins, and immune dysregulation. Notably, the PCNT and PKD1L1 genes are involved in cilia formation, and cilia dysfunction may be a key molecular mechanism underlying BA pathogenesis [5, 21].
Fig. 4.
Whole genome sequencing (WGS) analysis of cystic BA patients. A Workflow of WGS analysis. QC: quality check, hg19: human genome assembly, Genome Reference Consortium (GRC) h37, SNV: single nucleotide variant, InDels: insertions or deletions, AAC: amino acid changes, and SNP: single nucleotide polymorphism. B Summary of WGS and variant numbers in the analysis of each sample. C Common variant genes among all the five patient samples. Chromosome numbers, gene names, longest transcript and its proteins in Ensembl format, coding region change with amino acid change, and zygosity are shown. D Shared variant genes associated with “biliary atresia” and “bile duct abnormality” in the HGMD database and articles (45 genes). Patient numbers, chromosome numbers, gene names, longest transcript and its proteins in Ensembl format, coding region change with amino acid change, and zygosity are shown. E Shared variant genes associated with “hepatobiliary system phenotype" in the HGMD database and articles (316 genes). Patient numbers, chromosome numbers, gene names, longest transcript and its proteins in Ensembl format, coding region change with amino acid change, and zygosity are shown
In this study, we successfully generated five cBA-hiPSC lines with WGS information. These hiPSC lines derived from type I cystic biliary atresia patients should contribute to the development of pathological models and the elucidation of the molecular pathogenesis, since the genetic and/or environmental factors to cause BA have not been fully elucidated.
Materials and methods
Establishment and culture of hiPSCs
The hiPSCs were generated from PBMCs obtained from BA patients using episomal plasmid vectors to express OCT4, SOX2, KLF4, L-MYC, LIN28, mp53DD, and EBNA1 (Epi5 episomal iPSC reprogramming kit; A15960, Thermo Fisher Scientific) [25, 26]. The resulting cBA-hiPSCs were designated as HiPS-cBA1 (BRCi028-A in hPSCreg), HiPS-cBA2 (BRCi029-A in hPSCreg), HiPS-cBA3 (BRCi032-A in hPSCreg), HiPS-cBA4 (BRCi033-A in hPSCreg), and HiPS-cBA5 (BRCi035-A in hPSCreg). For seeding cBA-hiPSCs at every passage, StemFit AK02N medium (Ajinomoto, Tokyo, Japan; mixed with supplements B and C) supplemented with 10 μM Y-27632 (Wako, Osaka, Japan) and 0.25 μg/cm2 purified laminin-511 fragment (iMatrix-511 silk; Matrixome, Osaka, Japan) was prepared [36]. The plating density was 2,500 cells per cm2. The cBA-hiPSCs were single-cell passaged every 6–8 days using 0.5 × TrypLE Select (TrypLE Select (Gibco, Thermo Fisher, Waltham, MA) diluted 1:1 with 0.5 mM EDTA solution) or just 0.5 mM EDTA solution (Nacalai Tesque, Kyoto, Japan). The medium was changed every other day with StemFit AK02N medium (Ajinomoto; mixed with supplements B and C) from the next day of the passage.
Quantification of remaining EBNA1 sequence in genomic DNA
Genomic DNA was extracted from cBA-hiPSCs at passage numbers 5–15 using DNeasy Blood & Tissue (Qiagen, Venlo, NL). Genomic DNA samples (10 ng) were used as a template to detect the remaining episomal vectors using quantitative PCR (QPCR). The template DNA samples were mixed with Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) and primer sets. QPCR was performed with QuantStudio 3 real-time PCR system (Thermo Fisher Scientific) with default conditions. Copy numbers were calculated from the standard curves of several amounts of synthesized DNA oligos’ corresponding EBNA1 sequence. The primer sets for EBNA1 are listed in Table 2.
Table 2.
Reagent information
| Antibodies used for immunocytochemistry/flow cytometry/western blot | ||||
|---|---|---|---|---|
| Antibody | Dilution | Company cat # | RRID | |
| Pluripotency marker | Goat anti-OCT3/4 | 1:200 | R and D Systems, Cat# AF1759 | RRID:AB_354975 |
| Pluripotency marker | Rabbit anti-NANOG | 1:500 | ReproCELL Incorporated, Cat# RCAB004P-F, | RRID:AB_1560380 |
| Pluripotency marker | DyLight 550 Mouse anti-SSEA-4 | 1:125 | Stemgent, Cat# 09–0097 | RRID:AB_2784538 |
| Pluripotency marker | Alexa Fluor 488 Mouse anti-TRA-1–60 | 1:125 | BioLegend, Cat# 330,613 | RRID:AB_2295395 |
| Differentiation marker (Ectoderm) | Mouse anti-TUJ1 | 1:250 | R and D Systems, Cat# MAB1195 | RRID:AB_357520 |
| Differentiation marker (Mesoderm) | Mouse anti-SMA | 1:250 | R and D Systems, Cat# MAB1420 | RRID:AB_262054 |
| Differentiation marker (Endoderm) | Mouse anti-AFP | 1:200 | R and D Systems, Cat# MAB1368 | RRID:AB_357658 |
| Secondary antibody for immunocytochemistry | Donkey anti-goat IgG Alexa Flour 546 | 1:200 | Thermo Fisher Scientific, Cat# A-11056 | RRID:AB_2534103 |
| Secondary antibody for immunocytochemistry | Goat anti-Rabbit IgG Alexa Flour 555 | 1:500 | Thermo Fisher Scientific, Cat# A-21428 | RRID:AB_2535849 |
| Secondary antibody for immunocytochemistry | Donkey anti-mouse IgG Alexa Fluor 488 | 1:500 | Thermo Fisher Scientific, Cat# A-21202 | RRID:AB_141607 |
| Primers | ||||
| Target | Size of band | Forward/reverse primer (5′-3′) | ||
| Mycoplasma detection | Nested-PCR, 1st step PCR (MCGpF11/MCGpR1) | 350–850 bp |
ACACCATGGGAG(C/T)TGGTAAT/ CTTC(A/T)TCGACTT(C/T)CAGACCCAAGGCAT |
|
| Mycoplasma detection | Nested-PCR, 2nd step PCR (R16–2/MCGpR21) | 200–750 bp |
GTG(C/G)GG(A/C)TGGATCACCTCCT/ GCATCCACCA(A/T)A(A/T)AC(C/T)CTT |
|
| Episomal vector detection | EBNA1 (genomic qPCR) | 61 bp | ATCAGGGCCAAGACATAGAGATG / GCCAATGCAACTTGGACGTT | |
Immunocytochemistry and flow cytometry
Immunocytochemistry and flow cytometry were performed following our previous studies [37–42] at passage number 5–15. For immunocytochemistry, cells were fixed with 4% paraformaldehyde (Wako) and washed with phosphate-buffered saline (PBS). They were permeabilized with 0.1% Triton X-100 (Wako) in PBS and blocked with 1% bovine serum albumin (BSA; Wako) in PBS. These cell samples were incubated with primary antibodies at 4 ºC overnight. After washing these samples three times with PBS, they were incubated with secondary antibodies at room temperature for 1 h. Following three additional PBS washes, the nuclei were stained using Fluoro-KEEPER Antifade Reagent Non-Hardening Type with DAPI (Nacalai Tesque). Images of the cells were captured using a BZ-X800 fluorescence microscope (Keyence). For flow cytometry, the cells were dissociated using Accutase (Nacalai Tesque), resuspended in a custom buffer (0.5% EDTA in PBS supplemented with 1% FBS), and incubated for 1 h with antibodies. The stained cells were analyzed using an SH800S cell sorter and its accompanying software (Sony). The primary and secondary antibodies used in this study are listed in Table 2.
In vitro three-germ-layer differentiation assay by forming embryoid bodies (EBs)
EB formation assay was performed with our previous protocol with minor modifications [37–42]. Briefly, 1.2 × 106 cells were suspended in StemFit AK02N (supplemented with 12 μL of 10 mM Y-27632 solution) into a V-shape 96-well plate. From day 2, these cell aggregates were cultured in DMEM high glucose (Nacalai Tesque) supplemented with 10% fetal bovine serum (Biosera) (EB medium) for 8 days and plated on 0.1w/v% gelatin solution (WAKO)-coated plate in the EB medium for another 8 days. On day 16, these samples were fixed for immunocytochemistry.
In vivo three-germ-layer differentiation assay by forming teratomas
All animal experiments were approved by the Animal Experimentation Committee at the RIKEN Tsukuba Institute and performed according to the committee’s guiding principles and the “Guide for the Care and Use of Laboratory Animals” published by the National Institutes of Health. Teratomas were formed following our previous studies [37–42]. Briefly, 1.0 × 106 cells were suspended in 50% Matrigel solution (Matrigel (Corning): Stem Fit AK02N medium (Ajinomoto) with 10 µM Y-27632 (Wako) = 1:1) and injected into NSG mice. Teratomas were collected after around 3 months. Differentiation was validated by hematoxylin–eosin (HE) staining (performed at GenoStaff, Tokyo, Japan).
Virtual karyotyping (CNV/CGH microarray)
Virtual karyotyping assays were performed following our previous study [37–42]. Genomic DNA samples were extracted from hiPSCs at passage numbers 5–15 or PBMCs with DNeasy Blood & Tissue (Qiagen, Venlo, NL). Virtual karyotyping was performed with a GeneChip Scanner 3000 (Thermo Fisher Scientific) using KaryoStat Assay Arrays (Thermo Fisher Scientific) and analyzed with Chromosomal Analysis Suite (ChAS) software (Thermo Fisher Scientific).
Mycoplasma tests
Indirect DNA fluorescent staining and nested PCR were performed on cBA-hiPSC samples. The cBA-hiPSC culture medium was tested for mycoplasma contamination by staining with bisBenzimide H 33258 (Sigma-Aldrich) after 5–6 days of co-culture with VERO cells (RCB0001, RIKEN BRC Cell Bank), which served as mycoplasma infection indicator cells. DNA samples were extracted and analyzed using nested PCR at passage number 10. For the PCR, AmpliTaq Gold 360 DNA Polymerase (Thermo Fisher Scientific) was used. The same thermocycling conditions were applied for both PCRs: an initial denaturation at 95 °C for 10 min, followed by 30 cycles of thermocycling (30 s at 95 °C, 2 min at 55 °C, and 2 min at 72 °C) with a final extension at 72 °C for 5 min, and a hold at 4 °C. The PCR products were analyzed via electrophoresis on 2% agarose gel and stained with ethidium bromide. The primers used for the PCR are listed in Table 2.
WGS analysis
Genomic DNA samples from these cBA-hiPSCs were used for whole genome sequence analysis. These genomic DNA samples were fragmented using an ultrasonicator. The fragmented DNA is end-repaired and an A (adenine) is added to the 3ʼ end, and the entire long adapter sequence is ligated. Purification and size selection are then performed. PCR-free library was generated with Illumina DNA PCR-free Prep for these samples. Whole genome sequencing was used to produce around 600 M reads (~ 90 Gb data) from 150 × 2 paired-end (PE150) reads for each sample with Illumina NovaSeq X Plus 25B. Sequence data were analyzed using CLC genomics workbench, version 20. With this software, “quality check (QC) for sequence reads”, “trim reads”, “map reads to reference”, “local realignment”, “basic variant detection”, “amino acid changes”, “annotate with overlap information”, “annotate from known variants”, “filter with control samples”, and “export variant list in csv form” were performed on each sample. Exported variant lists were analyzed with Excel for Mac (ver. 16.89.1) using “autofilter” and “countif” functions. As healthy control hiPSCs, 1383D6 (HPS1006 in RIKEN cell bank) [36] and HiPS-NB1RGB (HPS5067) [43] lines were used.
Acknowledgements
We would like to express our sincere gratitude to Ms. Yasuko Hemmi and Dr. Tamami Wakabayashi for their technical support, and to Ms. Kumiko Omori for her administrative support.
Funding
This research was supported in part by grants from a RIKEN internal research budget, a Dokkyo International Medical Education and Research Foundation Prize, and AMED (23bm1423025h0001).
Data availability
The WGS data are available in the NBDC human database as research ID: hum0486. The authors confirm that the other data supporting the findings of this study are available within the article.
Declarations
Conflicts of interest
There was no conflict of interest for the works of this manuscript.
Ethics approval
The generation and use of human iPSCs were approved by the Ethics Committee of RIKEN BioResource Research Center and the Ethics Committee of the medical hospital of the Medical Department of University of Tokyo, Dokkyo Medical University, and the RIKEN BioResource Research Center (approved No. 2020075G and Tsukuba 29–3).
Informed consent
Formal informed consent was obtained from all the patients who participated in this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ningxin Ge and Kan Suzuki equally contributed.
Contributor Information
Kan Suzuki, Email: kan0228suzuki@gmail.com.
Yohei Hayashi, Email: yohei.hayashi@riken.jp.
References
- 1.Ando H, Inomata Y, Iwanaka T, et al. Clinical practice guidelines for biliary atresia in Japan: a secondary publication of the abbreviated version translated into English. J Hepatobiliary Pancreat Sci. 2021;28:55–61. [DOI] [PubMed] [Google Scholar]
- 2.Uemura M, Higashi M, Pattarapanawan M, et al. Gallbladder wall abnormality in biliary atresia of mouse Sox17(+/-) neonates and human infants. Dis Model Mech. 2020. 10.1242/dmm.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higashiyama H, Ozawa A, Sumitomo H, et al. Embryonic cholecystitis and defective gallbladder contraction in the Sox17-haploinsufficient mouse model of biliary atresia. Development. 2017;144:1906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uemura M, Ozawa A, Nagata T, et al. Sox17 haploinsufficiency results in perinatal biliary atresia and hepatitis in C57BL/6 background mice. Development. 2013;140:639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim YZ, Zhu M, Wang Y, et al. Pkd1l1-deficiency drives biliary atresia through ciliary dysfunction in biliary epithelial cells. J Hepatol. 2024. 10.1016/j.jhep.2024.02.031. [DOI] [PubMed] [Google Scholar]
- 6.Hellen DJ, Bennett A, Malla S, et al. Liver-restricted deletion of the biliary atresia candidate gene Pkd1l1 causes bile duct dysmorphogenesis and ciliopathy. Hepatology. 2023;77:1274–86. [DOI] [PubMed] [Google Scholar]
- 7.Ali RQ, Meyer-Miner A, David-Rachel M, et al. Loss of zebrafish pkd1l1 causes biliary defects that have implications for biliary atresia splenic malformation. Dis Model Mech. 2023. 10.1242/dmm.049326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riepenhoff-Talty M, Schaekel K, Clark HF, et al. Group A rotaviruses produce extrahepatic biliary obstruction in orally inoculated newborn mice. Pediatr Res. 1993;33:394–9. [DOI] [PubMed] [Google Scholar]
- 9.Koo KA, Lorent K, Gong W, et al. Biliatresone, a reactive natural toxin from dysphania glomulifera and D. littoralis: discovery of the toxic moiety 1,2-Diaryl-2-propenone. Chem Res Toxicol. 2015;28:1519–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patman G. Biliary tract: newly identified biliatresone causes biliary atresia. Nat Rev Gastroenterol Hepatol. 2015;12:369. [DOI] [PubMed] [Google Scholar]
- 11.Waisbourd-Zinman O, Koh H, Tsai S, et al. The toxin biliatresone causes mouse extrahepatic cholangiocyte damage and fibrosis through decreased glutathione and SOX17. Hepatology. 2016;64:880–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Wang J, Zhan Y, et al. The synthetic toxin biliatresone causes biliary atresia in mice. Lab Invest. 2020;100:1425–35. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt HC, Hagens J, Schuppert P, et al. Biliatresone induces cholangiopathy in C57BL/6J neonates. Sci Rep. 2023;13:10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta K, Xu JP, Diamond T, et al. Low-dose biliatresone treatment of pregnant mice causes subclinical biliary disease in their offspring: evidence for a spectrum of neonatal injury. PLoS One. 2024;19: e0301824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen C, Madadi-Sanjani O. Role of viruses in biliary atresia: news from mice and men. Innov Surg Sci. 2018;3:101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tam PKH, Yiu RS, Lendahl U, Andersson ER. Cholangiopathies—towards a molecular understanding. EBioMedicine. 2018;35:381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe E, Kawashima Y, Suda W, et al. Discovery of candidate stool biomarker proteins for biliary atresia using proteome analysis by data-independent acquisition mass spectrometry. Proteomes. 2020. 10.3390/proteomes8040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konishi KI, Mizuochi T, Takei H, et al. A Japanese prospective multicenter study of urinary oxysterols in biliary atresia. Sci Rep. 2021;11:4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amarachintha SP, Mourya R, Ayabe H, et al. Biliary organoids uncover delayed epithelial development and barrier function in biliary atresia. Hepatology. 2022;75:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung PH, Babu RO, Wu Z, Wong KK, Tam PK, Lui VC. Developing biliary atresia-like model by treating human liver organoids with polyinosinic: polycytidylic acid (Poly (I:C)). Curr Issues Mol Biol. 2022;44:644–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hai-Bing Y, Sivasankaran MS, Ottakandathil BR, et al. Environmental toxin biliatresone-induced biliary atresia-like abnormal cilia and bile duct cell development of human liver organoids. Toxins (Basel). 2024. 10.3390/toxins16030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian L, Ye Z, Kafka K, et al. Biliary atresia relevant human induced pluripotent stem cells recapitulate key disease features in a dish. J Pediatr Gastroenterol Nutr. 2019;68:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boonstra K, Beuers U, Ponsioen CY. Epidemiology of primary sclerosing cholangitis and primary biliary cirrhosis: a systematic review. J Hepatol. 2012;56:1181–8. [DOI] [PubMed] [Google Scholar]
- 24.Girard M, Jannot AS, Besnard M, Jacquemin E, Henrion-Caude A. Biliary atresia: does ethnicity matter? J Hepatol. 2012;57:700–1. [DOI] [PubMed] [Google Scholar]
- 25.Okita K, Matsumura Y, Sato Y, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–12. [DOI] [PubMed] [Google Scholar]
- 26.Okita K, Yamakawa T, Matsumura Y, et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–66. [DOI] [PubMed] [Google Scholar]
- 27.Tassano E, Ronchetto P, Calcagno A, et al. ‘Distal 16p12.2 microdeletion’ in a patient with autosomal recessive deafness-22. J Genet. 2019. 10.1007/s12041-019-1107-0. [PubMed] [Google Scholar]
- 28.Sugiyama K, Moteki H, Kitajiri SI, et al. Mid-frequency hearing loss is characteristic clinical feature of OTOA-associated hearing loss. Genes (Basel). 2019. 10.3390/genes10090715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenson PD, Mort M, Ball EV, et al. The human gene mutation database (HGMD((R))): optimizing its use in a clinical diagnostic or research setting. Hum Genet. 2020;139:1197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam WY, Tang CS, So MT, et al. Identification of a wide spectrum of ciliary gene mutations in nonsyndromic biliary atresia patients implicates ciliary dysfunction as a novel disease mechanism. EBioMedicine. 2021;71: 103530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berauer JP, Mezina AI, Okou DT, et al. Identification of polycystic kidney disease 1 like 1 gene variants in children with biliary atresia splenic malformation syndrome. Hepatology. 2019;70:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sok P, Sabo A, Almli LM, et al. Exome-wide assessment of isolated biliary atresia: a report from the national birth defects prevention study using child-parent trios and a case-control design to identify novel rare variants. Am J Med Genet A. 2023;191:1546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groza T, Gomez FL, Mashhadi HH, et al. The International mouse phenotyping consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 2023;51:D1038–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandenhaute B, Buisine MP, Debailleul V, et al. Mucin gene expression in biliary epithelial cells. J Hepatol. 1997;27:1057–66. [DOI] [PubMed] [Google Scholar]
- 35.Fagerberg L, Hallstrom BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa M, Taniguchi Y, Senda S, et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep. 2014;4:3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bershteyn M, Hayashi Y, Desachy G, et al. Cell-autonomous correction of ring chromosomes in human induced pluripotent stem cells. Nature. 2014;507:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi Y, Hsiao EC, Sami S, et al. BMP-SMAD-ID promotes reprogramming to pluripotency by inhibiting p16/INK4A-dependent senescence. Proc Natl Acad Sci U S A. 2016;113:13057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arai Y, Takami M, An Y, et al. Generation of two human induced pluripotent stem cell lines derived from two juvenile nephronophthisis patients with NPHP1 deletion. Stem Cell Res. 2020;45: 101815. [DOI] [PubMed] [Google Scholar]
- 40.Kuramochi Y, Awaya T, Matsuo-Takasaki M, et al. Generation of two human induced pluripotent stem cell lines derived from two X-linked adrenoleukodystrophy patients with ABCD1 mutations. Stem Cell Res. 2021;53: 102337. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu T, Matsuo-Takasaki M, Luijkx D, et al. Generation of human induced pluripotent stem cell lines derived from four DiGeorge syndrome patients with 22q11.2 deletion. Stem Cell Res. 2022;61:102744. [DOI] [PubMed] [Google Scholar]
- 42.Song D, Takahashi G, Zheng YW, et al. Retinoids rescue ceruloplasmin secretion and alleviate oxidative stress in Wilson’s disease-specific hepatocytes. Hum Mol Genet. 2022;31:3652–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borisova E, Nishimura K, An Y, et al. Structurally-discovered KLF4 variants accelerate and stabilize reprogramming to pluripotency. iScience. 2022;25:103525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The WGS data are available in the NBDC human database as research ID: hum0486. The authors confirm that the other data supporting the findings of this study are available within the article.