Abstract
Non-homologous chromosomal contacts (NHCCs) between different chromosomes participate considerably in gene and genome regulation. Due to analytical challenges, NHCCs are currently considered as singular, stochastic events, and their extent and fundamental principles across cell types remain controversial. We develop a supervised and unsupervised learning algorithm, termed Signature, to call NHCCs in Hi-C datasets to advance our understanding of genome topology. Signature reveals 40,282 NHCCs and their properties across 62 Hi-C datasets of 53 diploid human cell types. Genomic regions of NHCCs are gene-dense, highly expressed, and harbor genes for cell-specific and sex-specific functions. Extensive inter-telomeric and inter-centromeric clustering occurs across cell types [Rabl’s configuration] and 61 NHCCs are consistently found at the nuclear speckles. These constitutive ‘anchor loci’ facilitate an axis of genome activity whilst cell-type-specific NHCCs act in discrete hubs. Our results suggest that non-random chromosome positioning is supported by constitutive NHCCs that shape genome topology along an off-centered spatial gradient of genome activity.
Subject terms: Machine learning, Nuclear organization, Genome
The authors develop a supervised and unsupervised learning algorithm Signature. Machine learning and network model analysis of Hi-C datasets across 62 2n genomes suggest that inter-chromosomal contacts demarcate genome topology along a spatial gradient of genome activity.
Introduction
Chromosomal interactions between different chromosomes (termed here: non-homologous chromosomal contacts [NHCCs]) have been shown to contribute to genome topology1–5. It is well established that NHCCs are critical for several biological processes, such as coalescing olfactory receptor genes to orchestrate their expression in multi-chromosomal hubs6,7, and forming the nucleolus through spatial proximity between acrocentric chromosomes8,9. Further underscoring the importance of NHCCs is their reorganization in human disease10,11.
Two commonly used approaches for investigating NHCCs are imaging and chromatin capture, both of which are limited in determining NHCCs. Specifically, imaging is not scalable to genome-wide approaches and chromosome conformation capture (i.e., proximity ligation-based Hi-C) as the most widely used technique to study 3D genome organization mainly focuses on analyzing intra-chromosomal contacts12,13. Moreover, both methodologies often caused discordant results when studying NHCCs that do not complement one another5,14,15. Importantly, Hi-C datasets contain ‘trans-reads’, but current computational and statistical analysis has limitations in confidently determining true NHCCs. Hence, NHCCs have been considered as stochastic, singular events5,16, that are not readily detectable in Hi-C data17,18. The non-ligation-based methodologies, such as SPRITE9, imaging approaches17,19–23, TSA-Seq24, and HiPore-C25 have assayed single cell types and although they determined NHCCs around the nuclear speckles and nucleoli9,25, their depth is not comparable to Hi-C. In summary, while some examples of NHCCs are well established and critical for cellular processes, we still lack a comprehensive view of the fundamental principles of NHCCs. This is owing to analytical Hi-C limitations where a robust statistical framework is required to confidently determine true NHCCs above background noise. Here, we developed a machine learning method assessing the Spatially Interacting GeNomic ArchitecTURE (Signature) towards a comprehensive and systematic detection of NHCCs, their extent across cell types, and their putative impact on non-random chromosome positioning. Signature is explicitly designed to examine intra- and inter-chromosomal interactions in Hi-C datasets (including Omni-C, capture Hi-C, and Micro-C26), without technical intricacy, further resources, and time to perform orthogonal approaches (i.e., GAM27, SPRITE9, HiPore-C25), which is advantageous for the field.
To derive the fundamental properties of NHCCs, we took a genome-wide approach across 53 diploid (2n) cell types. Specifically, Signature overcomes previous analytical limitations by supervised and unsupervised learning (Community Detection28) and tests all genomic contacts relative to the entire genomic context. Using Signature, we confidently determined multi-dimensional NHCCs in Hi-C data to provide a global view of how and where NHCCs contribute to genome function. NHCCs occur in genomic regions that are highly gene-dense, transcribed, and bound by transcription factors and that harbor genes related to cell-type function and sex. We uncover 61 constitutive NHCCs as ‘topological anchors’ at the speckles that occur across cell types and sexes and that maintain a spatial genome gradient along an axis of activity. Cell-type-specific NHCCs connect to the main gradient, but act in discrete spatial hubs. Collectively, we reveal that NHCCs are prevalent and deterministic in the inter-chromosomal topology across human 2n genomes.
Results
The Signature pipeline determines NHCCs in Hi-C datasets
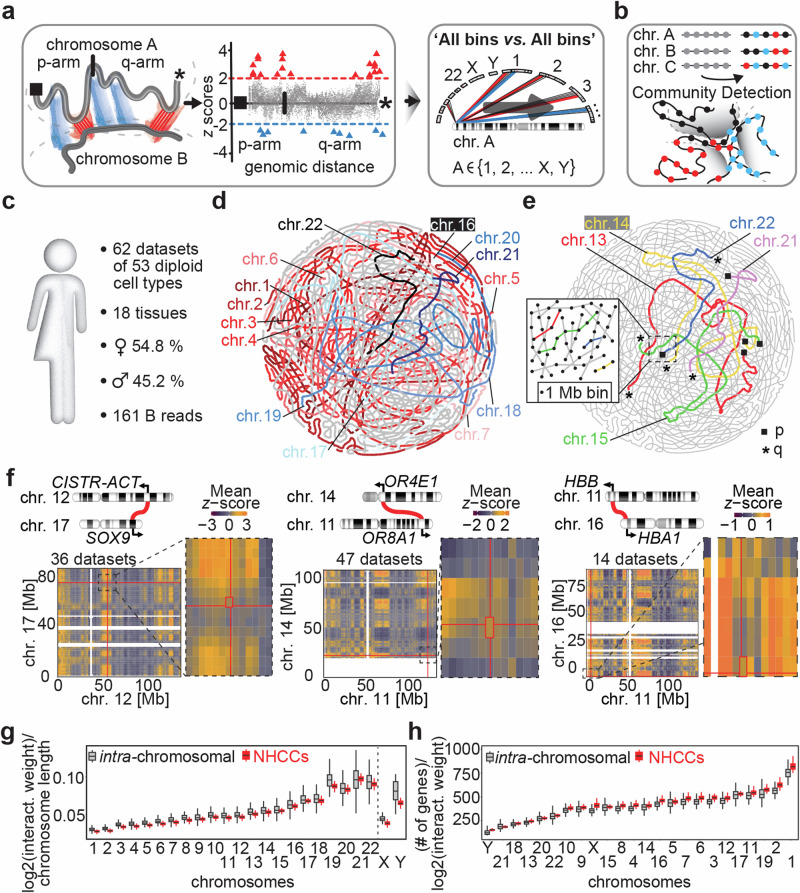
Classic Hi-C analysis identifies significant intra-chromosomal interactions by taking the linear genomic distance between two interaction anchors into account29. In contrast, distance as a concept for NHCCs is not the same. Thus, to define true NHCCs where loci of different chromosomes are in spatial proximity, our model is fitted against all genomic coordinates and evaluates inter-chromosomal Hi-C interaction weights between chromosomes. Specifically, we developed a non-parametric supervised learning approach (Local Weighted Polynomial Regression [LWPR]) that systematically assesses relationships between all loci on all chromosomes. LWPR queries each chromosomal position against all other genomic regions (1 megabase [Mb] bins) in an ‘All bins vs. All bins’ approach, which has not been accomplished for Hi-C analysis before (Fig. 1a, Supplementary Fig. 1a-e). The aim is to identify genomic regions with significantly increasing or decreasing interaction weights, either implying spatial proximity (NHCCs) or separation (non-interacting regions) in relation to the entire genomic background (Fig. 1a). To determine the best local regression fit, we cross-validated the span parameter. Thus, LWPR offers local insights into the spatial relationships of non-linear inter-chromosomal genome topology. Upon detecting how frequent an NHCC is observed versus all expected ‘locus-to-locus’ contacts, Signature evaluates if there is a significant interaction (or no interaction) and determines either a bona fide NHCC or non-interacting region by significant z-score-transformed p- and q-values (Fig. 1a, Supplementary Fig. 1f-n).
Fig. 1. Signature confidently maps NHCCs.
a Supervised learning in Signature. Scheme depicts genomic regions between different chromosomes that are evaluated for their significantly interacting (red triangle = positive z scores) and non-interacting bins (blue triangle = negative z scores). All genomic regions are queried against all other regions in an ‘All vs. All’ approach. Right: example of interaction weights with significance cutoffs (dashed lines) between two chromosomes. b Unsupervised learning by Community Detection (CD) in Signature. CD groups clusters of similar properties (i.e., interaction weights of intra-chromosomal interactions and NHCCs) in communities (black, red, blue). c Features of the body map analyzed with Signature. 161 billion (B) mapped reads derived from 62 datasets generated a compendium of 2n genomic interactions across human cell types, separated by sex. d Consecutive bins of each chromosome are strung together to generate the chromosomal outlines and to visualize CD-approximated genome topology across 62 Hi-C datasets. Large chromosomes 1-7 (red & pink) and small, gene-dense chromosomes 16-22 (blue & black) are highlighted. e Acrocentric chromosomes 13-15, 21, and 22 are colored in genome topology map. Telomeric p- arms and q-arms are shown as black squares or asterisks, respectively. Enlargement depicts how CD strung bins together to generate chromosomal outlines. f Ideograms depict reported NHCCs tested by Signature. Each heatmap represents a pair of interacting chromosomes. Mean z-scores are shown and red lines indicate genomic positions of reported loci (shown in Mb = megabases). Enlargements highlight region of interest, each cell is a 1 Mb bin. Unmapped regions such as acrocentric p arm of chromosome 14 are shown in white. g Interaction density per megabase of intra-chromosomal interactions (gray) and NHCCs (red) per chromosome (n = 62 Hi-C datasets). Box limits represent upper and lower quartiles. Central boxplot line represents the median and whiskers represent 1.5x IQR. h Same as panel g but for number of genes per chromosome (n = 62 Hi-C datasets).
Hi-C analysis is restricted to pairwise contacts, however, 3D genome organization results in multi-way interactions9,25,27,30,31. To overcome this limitation, we further added Community Detection (CD, unsupervised learning)28,32 to Signature. This feature complements supervised learning (LWPR) and visualizes spatial peculiarities of where NHCCs impact genome topology. CD can reveal multi-dimensionality of Hi-C data with either dense or loose NHCC associations, because it clusters bins based on their structures and interaction weights to deconvolute complex networks (Fig. 1b)32. Together, Signature includes supervised and unsupervised machine learning to identify NHCCs genome-wide and across cell types.
Benchmarking. Our incomplete understanding of NHCCs is mostly derived from probing single aneuploid cell lines. In order to resolve the inter-chromosomal topology of human 2n genomes, we analyzed solely diploid and near-diploid genomes to avoid aneuploidy bias. Specifically, we identified 62 Hi-C datasets (Supplementary Data 1) derived from 53 diploid cell types, devoid of aneuploidy, and comprising unprecedented ˜161 billion reads (Fig. 1c). Using Signature, we observed a total of 40,282 (q < 0.05) NHCCs (740,835 [p < 0.05], Supplementary Data 2). Notably, Signature cannot only identify NHCCs but also those genomic regions that have a significant depletion of interactions. Signature identified 186,429 (q < 0.05) significantly non-interacting regions (2,068,828 [p < 0.05], Supplementary Data 3), and 120,106 (q < 0.05) significant intra-chromosomal interactions at 50 kb genomic resolution (31,604,799 [p < 0.05], submitted to GEO, Methods, Supplementary Fig. 1j).
Next, we used CD to establish genome topology maps across the 62 datasets. Using intra- and inter-chromosomal interaction weights, and restricting the number of possible communities to 46, as this entity resembles diploid human genomes, CD recapitulated the known radial chromosomal arrangement33 (Fig. 1d, Supplementary Fig. 2a), and the nucleolus formation by acrocentric chromosomes8 (Fig. 1e, Supplementary Movie 1). These results indicate that CD can indeed probe and visualize spatial genome structure. Signature showed high detection rates and its methodology did not adversely affect interaction patterns (Supplementary Figs. 2b,d, 3a, b).
To further evaluate Signature’s performance, we compared it to orthogonal reference sets. First, we compared Signature to MERFISH20-identified NHCCs and determined that NHCCs occurred significantly more often at close spatial distances than non-interacting regions (Mann–Whitney test p = 0.014, Supplementary Fig. 3c), consistent with the mean NHCC distance (˜280 nm) that we defined by CRISPR live-cell imaging17. Signature identified speckle-associated NHCCs that had been determined by HiCAN34 and SPRITE9 (90.9% and 13.3% recall rate, respectively), and nucleolar-associated NHCCs (42.4% and 74.6% recall rate, Supplementary Fig. 3d, e). Importantly, Signature has high concordance with other methods for known topological features, but determines interactions that the comparable methods did not identify (Supplementary Fig. 3d, e). Genes annotated at Signature NHCCs that overlapped with either HiCAN or SPRITE NHCCs related to reported biological function, such as splicing at the speckles35, in a GO term analysis36. Moreover, Signature successfully validated reported NHCCs in up to 47 Hi-C datasets between CISTR-ACT & SOX910, between human olfactory receptors7, and hemoglobin genes37 (Fig. 1f), and FIRRE & YPEL438 (Supplementary Fig. 3f). The benchmarking suggests that Signature can confidently detect NHCCs in existing Hi-C data and uncovers an abundance of yet unexplored NHCCs which supports studying genome topology.
NHCCs are non-random
The numerous discovered NHCCs across cell types provide the opportunity to determine inherent properties of NHCCs. Therefore, we started exploring global features of all chromosomal contacts across chromosomes and cell types. First, intra-chromosomal contacts dominated in comparison to NHCCs (Supplementary Fig. 3g, h), which is expected due to the in situ Hi-C protocol39,40 and prevailed intra-chromosomal proximity17. Second, NHCC interaction density inversely related to chromosome size across all cell types (Fig. 1g), whilst NHCCs positively associated with intra-chromosomal contacts and chromosome length, and number of genes per chromosome (Pearson correlations r = 0.99 and r = 0.82, respectively, Supplementary Fig. 3 h). Overall, NHCCs seemed to be less dependent on the number of genes per chromosome (Fig. 1h). Notably, gene-poor chromosome 21 showed the highest interaction density of NHCCs per Mb (Fig. 1g), indicating that gene number may not be the sole determinant for NHCCs. However, the most gene-poor chromosome 18 had significantly fewer NHCCs than the gene-densest chromosome 19 (Mann-Whitney p < 2.2 × 10−16, Fig. 1h, Supplementary Fig. 3i), confirming earlier results13. In a global interaction matrix of all 62 Hi-C datasets, we observed more recurrent and non-random NHCCs among smaller chromosomes than between large ones (Fig. 2a). To investigate if NHCCs depend on intra-chromosomal interactions, we performed genome-wide correlations of averaged cis and trans interaction weights per bin for all 62 datasets. Moreover, using Hi-C datasets with the highest and lowest number of NHCCs, we tested if more NHCCs occur at the expense of intra-chromosomal contacts. We found weak negative correlations for both experiments (Pearson’s R = −0.18 and −0.22, p < 2.2 × 10−16, Supplementary Fig. 3j), that suggest that bins involved in NHCCs are mostly spatially separated from intra-chromosomal contacts. Collectively, the recurrent and deterministic NHCC patterns across chromosomes and cell types in our body map align with the paradigm of evolutionarily conserved chromosome positioning in a radial pattern33.
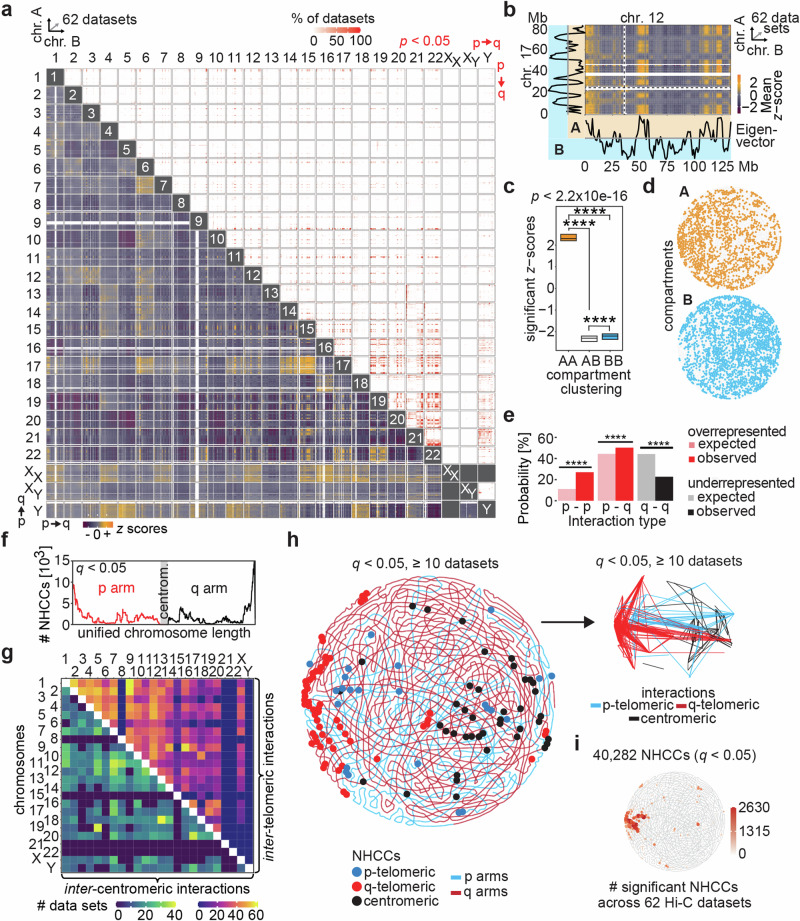
Fig. 2. Deterministic NHCC patterns and inter-telomeric and inter-centromeric NHCCs.
a Genome-wide matrix of interacting chromosomes with separated female gonosomes (XX), and male gonosomes (XY and Y). Lower left depicts mean z-scores (unmapped regions = white), and upper right depicts significant NHCCs (p < 0.05); red gradient = percent detection range across 62 datasets. b Interaction heatmap of chromosomes 12 and 17 with mean z-scores of 62 Hi-C datasets, in comparison to average eigendecomposition of each chromosome across 62 datasets. A compartment = blue; B compartment = orange; centromeres = dashed lines; unmapped regions = white. c Quantification of compartment clustering (AA: n = 3.2 × 106, AB: n = 1.64 × 106, or BB: n = 4.25 × 106) of significant positive and negative z-scores across 62 datasets. Asterisks depict significance determined by Mann-Whitney testing (p < 2.2 × 10−16, two sided). Central boxplot line represents the median, and box limits represent upper and lower quartiles. d CD genome topology map of genomic compartments (A = blue, B = orange) in across 62 datasets. e. Binomial testing (p–q: p = 3.94 × 10−138, p–p: p = 4.94 × 10−324, q-q: p = 4.94 × 10−324, two-sided) of expected and observed probabilities (%) of p–p, p–q, and q–q interactions based on chromosomal length in 62 Hi-C datasets; overrepresented = red; underrepresented = blue; n = 24 conditions per group (22 autosomes and two gonosomes). f Distribution of all 40,282 NHCCs (q < 0.05) along length of a unified chromosome. g Heatmap shows distribution of significant inter-telomeric and inter-centromeric NHCCs for each chromosomal pair across 62 Hi-C datasets. h CD-approximation of common inter-telomeric (q < 0.05, >10 datasets) and inter-centromeric NHCCs with chromosomal p- and q-arms shown in blue and red, respectively. On the right, linear interactions of NHCCs are shown. i Distribution of 40,282 NHCCs in genome topology map.
Inter-chromosomal compartment size. Mapping NHCCs indicated that they pertain to larger inter-chromosomal domains. Specifically, NHCCs spread across 1.84 Mb on average, whilst significantly non-interacting regions comprise 3.38 Mb (q < 0.05, Supplementary Fig. 3k-l), which partially explains why locus-specific NHCCs (kilobase range) were previously not readily detected in Hi-C14,16,17,41.
As reported before4,9,20,25, Signature also detected the association of homotypic A/A compartments with NHCCs, further benchmarking its accuracy to detect known topological features. Heterotypic A/B and homotypic B/B compartmentalization correlated with significantly non-interacting regions (Mann-Whitney test p < 2.2 × 10−16, Fig. 2b, c, Supplementary Data 4), indicating clear segregation of NHCCs from other genomic regions. Interestingly, genome topology maps showed off-centered inversely organized compartments, where B was as expected more in the periphery (Fig. 2d).
P/q arms. We next asked if NHCCs are biased by chromosomal structure (p and q arms) which remains unexplored. Significantly more p–p and p–q than q–q contacts occurred (binomial testing p < 3.94 × 10−138, Fig. 2e, Supplementary Fig. 3m), underscoring the deterministic nature of chromosome positioning and NHCCs across cell types. Chromosome types (meta-centric, sub-metacentric, acrocentric) did not affect NHCCs (Supplementary Fig. 3n). Interestingly, despite the heterogeneity of our body map, pervasive NHCCs in A compartments occurred in deterministic patterns especially among small gene-dense chromosomes with widespread p–p and p–q contacts.
Inter-chromosomal topology involves many inter-centromeric and inter-telomeric contacts
The structural feature of clustered telomeric and centromeric interactions on opposite ends of the nucleus (termed Rabl’s configuration [1885]42) is observed in yeast43, drosophila44, plants45, and mammals3,46, and has been proposed to reduce chromosomal entanglements42,47. Rabl’s configuration may support the mitotic heritability of global chromosome positioning15, but its existence in human genomes is unclear48,49. When analyzing mapped regions flanking the centromeres and sub-telomeric regions, Signature identified thousands of significant contacts of p-arms and particularly of q-arms across all cell types (Fig. 2f, Supplementary Fig. 3o). These NHCCs occurred significantly more often than expected, and more frequently than telomeric-centromeric NHCCs (Binomial testing empirical p = 0, Supplementary Fig. 3p). Remarkably, chromosomes 8, 15, 21, 22, and Y showed either no or sparse inter-telomeric and inter-centromeric NHCCs (Fig. 2g), similar to HiPore-C results25. Overall, these results suggest that Rabl’s configuration is a predominant feature of human genome topology and can be detected with Signature.
We next asked if the same inter-telomeric and inter-centromeric NHCCs commonly occur among cell types, which may support the idea of NHCCs as general determinants of genome structure. Upon interrogating and visualizing common NHCCs (q < 0.05, ≧ 10 datasets, 393 bins = 12.73% of hg38, Supplementary Data 6) in genome topology maps, we observed clustering and clear demarcations according to Rabl’s configuration (Fig. 2h). Specifically, off-centered common q-telomeric NHCCs occurred on one side of the CD-approximation, whilst p-telomeric NHCCs were diffused (Fig. 2h). Inter-centromeric NHCCs depicted more distributed interaction patterns distal to q-telomeric NHCCs (Fig. 2h). Notably, all 40,282 NHCCs (q < 0.05) were off-centered and asymmetrically organized (Fig. 2i, Supplementary Data 5), in congruence with the A compartment organization (Fig. 2d), and the highest density of common q-telomeric NHCCs (Fig. 2h). Our findings indicate that NHCCs across cell types are not randomly distributed across the entire genome and common inter-telomeric and inter-centromeric NHCCs contribute to the inherent genome structure.
NHCCs and genomic features demarcate genome topology along a spatial genome gradient
Thus far, Signature has identified common NHCCs and their off-centered pattern. Now, we further explore their collective fundamental properties. First, we focused on gene regulation, since transcription rates can define the size of a chromosomal territory50 which may influence inter-chromosomal topology. Common NHCCs (≧ 10 datasets, q < 0.05, Supplementary Data 6) harbored 23.47% of all genes (GENCODE V4251) and their gene density was up to 2.1-fold higher than expected (1 Mb bins, Fisher’s exact test p = 9.31 × 10−36, Fig. 3a). Gene expression across all 62 datasets was significantly higher for common NHCCs when compared to GTEx52 (Mann-Whitney test, range: p = 2.79 × 10−15 - p = 6.11 × 10−52, Fig. 3b). The same was true when we compared tissue-specific gene expression profiles from GTex samples matching our Hi-C data by showing that NHCC regions harbor the most active genes (Supplementary Fig. 4a). Similarly, we compared transcription factor (TF) binding from ChIPseq experiments53 in cell types that matched our tissue-specific unique NHCCs. We found more TF binding at NHCCs than globally (Mann-Whitney test range: p = 2.39 × 10−6–p = 1.15 × 10−71, Supplementary Fig. 4b). Gene expression of either of the two genomic regions that are involved in NHCCs was similar (Supplementary Fig. 4c), proposing that NHCC formation is independent of the counterpart’s expression level.
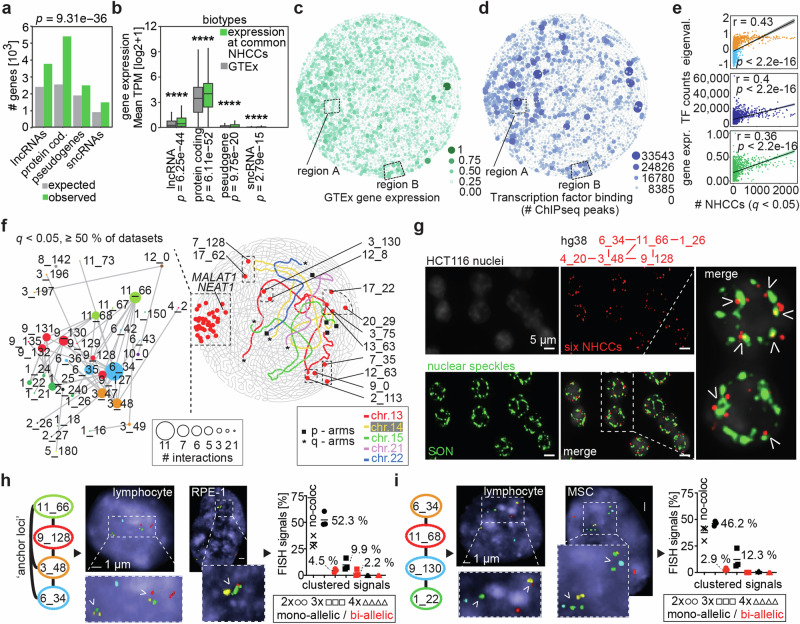
Fig. 3. Features of a spatial genome gradient.
a Number of observed genes at common NHCCs (q < 0.05, ≧ 10 datasets, n = 15,096 genes) in comparison to expected numbers of genes, separated in four biotypes. Fisher’s exact test determined significance (p = 9.31 × 10−36, two-sided). b Gene expression at common NHCCs (q < 0.05, ≧10 datasets) in comparison to GTEx52 (n = 56,201 genes). Mean TPM (log2 + 1) of four biotypes is shown; asterisks denote significance determined by Mann–Whitney testing (lncRNAs: [long non-coding RNAs]: p = 6.25 × 10-44, protein-coding genes: 6.11 × 10−52, pseudogenes: p = 9.75 × 10−20, sncRNAs [short nuclear RNAs]: p = 2.79 × 10−15, two-sided). Box limits represent upper and lower quartiles. Central boxplot line represents the median, whiskers represent 1.5x IQR. c Scaled average gene expression of GTex52 per 1 Mb bin across genome topology. Dashed boxes indicate regions with either an anticorrelated pattern of expression and TF binding (region A) or a correlated pattern (region B). d Summed TF peaks per 1 Mb bin are shown across genome topology. e Pearson correlations of the bin frequency involved in 40,282 NHCCs with compartments, gene expression (GTEx52), and TF binding53. Correlation coefficients r and p-values are depicted. f Genome topology map of constitutive NHCCs (>50% of datasets, red dots) and acrocentric chromosomes 13-15, 21, and 22 (colored). Telomeric acrocentric p-arm and q-arm NHCCs are shown as black squares or asterisks, respectively. Zoom-in shows community of constitutive NHCCs (red) with multi-way interactions (dot size). Genomic locations are annotated as 6_34 = chromosome 6, megabase 34–35, hg38). g Oligopainting of six ‘anchor loci’ (red, hg38: [chr_Mb]) with speckle marker SON (green) in HCT116 cells (n = 2, each biological replicate > 300 nuclei); arrows depict either co-localization or close spatial proximity; scale bars = 5 µm. h–i. FISH of ‘anchor loci’ ([chr_Mb]) in lymphocytes, RPE-1 and MSCs [each ˜100 nuclei]). White arrows exemplify clustered signals; scale bars = 1 µm. Plots show quantification of no-colocalizations, mono-allelic (black), and bi-allelic (red) signal frequencies of double (2x), triple (3x), and quadruple (4x) clustered signals of NHCCs. Means (%) and datapoints of three analyzed cell lines are shown.
The visualization of gene expression and TF binding results in genome topology maps (Fig. 3c, d) revealed again off-centered asymmetric patterns that overlapped with the archetypes of A compartments (Fig. 2d), common q-telomeric NHCCs (Fig. 2h), and all NHCCs (Figs. 2i). These genomic features are positively correlated with NHCCs (Fig. 3e). Remarkably, expression and TF binding were highly correlated (r = 0.6945), although not in a consistent pattern across the genome. Locally, we found regions with anticorrelated expression and TF binding pattern in the genome topology map. For example, region A (r = −0.161) harbors many bound TFs, whilst its genes are lowly expressed and relate to HDAC-deacetylated histones (log10[q] = −67.13, Fig. 3c, d), indicating a local enrichment of TFs. In contrast, other regions showed high correlation between expression and TF binding, such as region B where genes related to sensory perception function (r = 0.849, Fig. 3c-d). The combination of Signature results with other genomic features in genome topology maps, such as expression and ChIPseq data may help to reveal higher-order subnuclear structures and their function that have not been determined yet. Collectively, NHCCs form an off-centered asymmetric genome gradient where especially highly gene-dense and expressed regions interact. The spatial gradient along an axis of activity may either form because of TF accumulation, transcription, and RNA or these features are a consequence of chromatin flexibility and diffusibility of DNA and subnuclear organization.
A constitutive ‘topological anchor community’ converges at nuclear speckles
Given the spatial gradient of genome activity, we further explored if a coalescence of constitutive NHCCs supports the genome gradient’s activity and may explain constant principles of genome organization, such as sub-nuclear organization9, and the mitotic heritability of the genome1,15. Remarkably, LWPR determined 61 constitutive NHCCs in ≧50% of the 62 datasets (q < 0.05), of which 56 overlapped with the off-centered pattern identified by CD (overlap 91.8% [56/61], Fig. 3f, permutation testing empirical p = 0, Supplementary Fig. 5a, Supplementary Data 7 and Supplementary Movie 2). This ‘topological anchor community’ was proximal to q-telomeric NHCCs (Fig. 2h), converged with patterns of genomic features (Figs. 2d, 3c, d), and had even higher expression levels than remaining NHCCs (Mann–Whitney test p = 3.31 × 10−11, Supplementary Fig. 5b), and higher mean gene density (47.21 genes/Mb vs. 23.04 genes/Mb for genome). The involved loci showed up to 11 multi-way interactions with consecutive bins, and harbored chromosomal regions involved in sub-nuclear organization (MALAT154 and NEAT155), in chromatin-associated functions (KMT5B, KDM2A, HMGA, ABL1, etc.), and in RNA-binding and transcription (Fig. 3f).
Recently, others described that paraspeckle formation with NEAT1 is biased towards one side of the nucleus56, which aligns with our genome topology maps. Thus, we proposed that the ‘topological anchors’ may be part of the nuclear speckles35 where high gene density and expression contribute to the formation of these subnuclear organelles9,57. We tested this idea by oligopainting58 of six ‘anchor loci’ and confirmed them, their multi-way interactions, and their significant overlap with speckle marker SON59 (68.8%, n = 2, each > 300 nuclei, permutation testing empirical p = 0, Fig. 3g, Supplementary Fig. 5c). Moreover, we performed two multi-color FISH approaches, each with four different ‘anchor loci’ in lymphocytes, RPE-1, and MSCs (each > 100 nuclei). We measured either single alleles or bi-allelic proximities and found that at least two ‘anchor loci’ were proximal in 52.3% of cells on average (Fig. 3h). Three anchors interacted in 12.3% of cells (Fig. 3i), and even bi-allelic proximity occurred in several cells of the three investigated cell types (Supplementary Fig. 5d). The imaging of proximal ‘anchor loci’ validated Signature results, of both supervised and unsupervised learning approaches, and showed that constitutive NHCCs are in proximity to nuclear speckles.
Cell-type-specific NHCCs connect with the ‘topological anchor community’
Cell-type-specific NHCCs exemplified a relation to function7,60,61. Inspired by this finding, we next investigated the wide range of NHCC frequencies (8–65.3%) across our compendium of cell types (Supplementary Fig. 3g). We found that mature tissues (i.e., aorta, cortex, hippocampus, lung, thymus, ventricles, etc.) had more NHCCs than proliferating cell types (i.e., H9-hESCs, IMR90, RPE1, etc.) with more intra-chromosomal contacts (Fig. 4a, Supplementary Fig. 5e, f). To test if terminal differentiation and mitosis-related genome re-organization can affect NHCC extent and formation, we performed Omni-C across human chondrogenesis in organoids, where mesenchymal stem cells (MSCs) terminally differentiate into hypertrophic chondrocytes with reduced proliferation62,63 (Fig. 4b, Supplementary Fig. 5g, h). In the chondrogenic time-course, pre- and hypertrophic chondrocytes showed significantly more NHCCs across most chromosomes in comparison to MSCs and chondrogenic precursors (ANOVA p < 0.0001, Fig. 4c, d). This suggests that reduced genome re-organization and terminal differentiation facilitate NHCCs, probably by cell-type-specific transcriptional programs that evolve post-mitosis in a spatiotemporal manner.
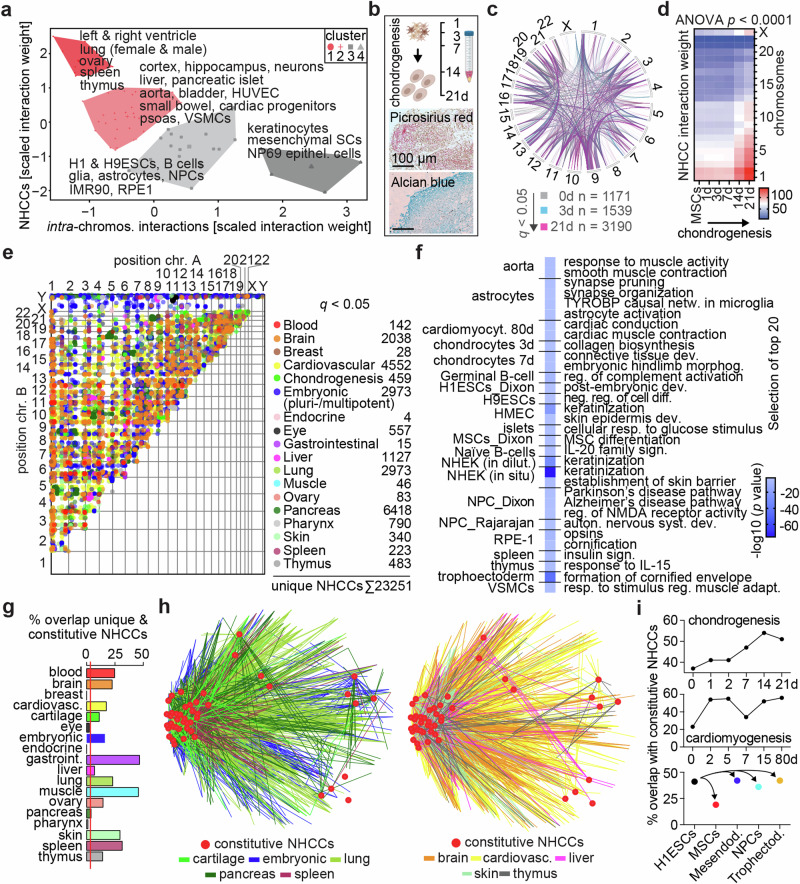
Fig. 4. Cell-type-specific NHCCs connect with the main spatial genome gradient.
a K-Means clustering of NHCCs and intra-chromosomal interactions. Labels specify datasets of underlying cluster. b Time course of chondrogenic differentiation (0–21 d) in 3D organoids. Picrosirius Red and Alcian Blue stainings show extra-cellular matrix deposition in sections of MSC-derived cartilage (day 21). Scale bars = 100 µm. c Number of NHCCs (q < 0.05) across chondrogenesis. MSCs (0 d), chondrogenic precursors (3 d) and hypertrophic chondrocytes (21 d) are shown (opacity 60%). Chromosome Y was omitted from circos plot as SCRC-4000 (MSCs) cells are of female origin. d NHCC interaction weights across chromosomes during chondrogenic differentiation. Two-way ANOVA determined significance (p < 0.0001, two biological replicates). e Scatterplot of unique (cell/tissue-type-specific) NHCCs (q < 0.05, opacity 50%) grouped into tissues that reflect the dataset origins. Number of observations are noted. f Selected examples of top 20 GO-terms of unique (cell-type-specific) NHCCs are listed per cell/tissue-type together with -log10(p values); one-sided, uncorrected. g Percent overlap of constitutive with tissue-specific NHCCs. Red line indicates expected probability (3.27%). h Shared interactions of constitutive ‘anchor loci’ (red) and unique NHCCs grouped into tissues (multi-colored lines, see Supplementary Data 1). i. Percent overlap of constitutive NHCCs in (top) chondrogenesis, (middle) cardiomyogenesis64, and (bottom) pluri- and multi-potent cells/tissues derived from H1-ESCs65.
Next, we determined NHCC variation among cell and tissue types. Unexpectedly, 57.7% of significant NHCCs (23,251/40,282; q < 0.05) were unique among the 62 datasets (Fig. 4e, Supplementary Fig. 6a, Supplementary Data 8), which is significantly higher than random selection (randomization test empirical p = 0, Supplementary Fig. 6b). Notably, genes at these unique NHCCs and also at significantly non-interacting regions related to meaningful biological functions by GO-term analysis36 (Fig. 4f, Supplementary Fig. 6c).
The high fraction of unique NHCCs motivated us to evaluate their relationship to constitutive NHCCs and the main spatial genome gradient. Most of the analyzed tissues (14/18) had a significant overlap of unique NHCCs with constitutive NHCCs (Fig. 4g). Visualizing these shared interactions showed that the main genome gradient maintains global structure by connecting constitutive and cell-/tissue-specific NHCCs (Fig. 4h). Notably, topological anchor loci increased in stem cell differentiations across chondrogenesis and cardiomyogenesis64, whilst H1-ESC-derived germ layers65 shared similar numbers of constitutive NHCCs (Fig. 4i). This high overlap between pluri-/multi-potent and differentiated cell types with constitutive NHCCs (mean 69.9%) indicates that topological anchors exist as prevalent structural feature in all cell states. Moreover, a subset of 10 cardiovascular Hi-C datasets recapitulated the spatial genome gradient with 96.7% of the constitutive NHCCs and showed asymmetric gene expression (Supplementary Fig. 7a, b), revealing that the genome gradient holds true in smaller subsamples. Collectively, our findings indicate that constitutive NHCCs globally connect and support an off-centered genome structure with clustered q-telomeric and centromeric NHCCs across cell types, and tissue-specific NHCCs dispersed in discrete hubs, which expands recent observations9,20,24,25.
Sex-specific NHCCs relate to sexual dimorphic features
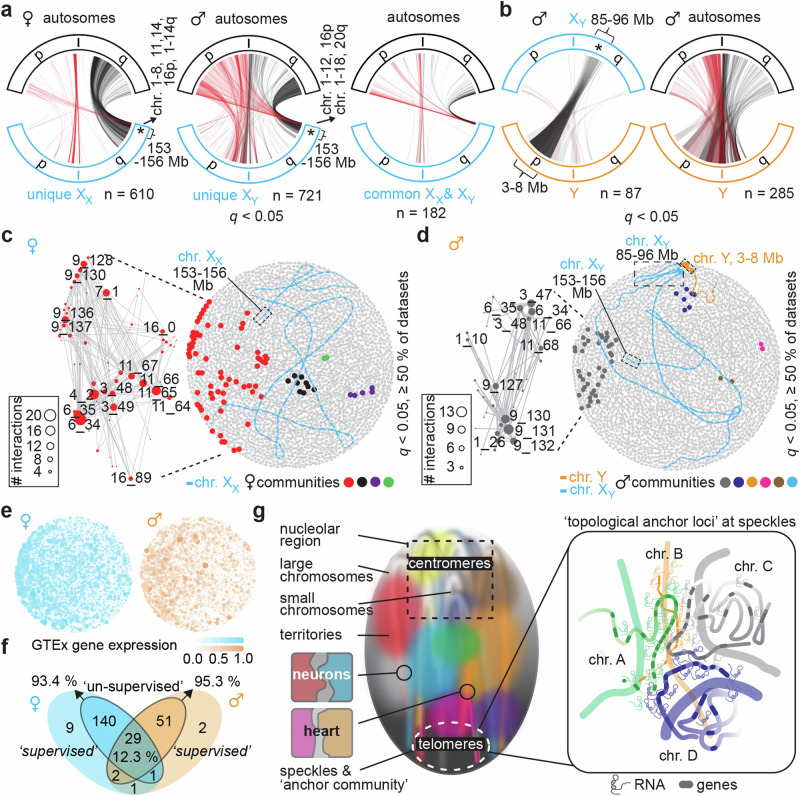
In the context of the spatial genome gradient and discrete function-related hubs, it is interesting to consider sex-specific gonosome-autosome NHCCs, which remain completely unknown. To do this, we first separated female (54.8%) and male (45.2%) datasets; although it is important to note that we did not analyze identical cell types for each sex, and then visualized all gonosome-autosome interactions separately. We found more unique than common NHCCs of the X chromosomes in both sexes (Fig. 5a, Supplementary Data 9-11). Genes annotated at XX-autosome contacts were enriched for mitochondrial function and cardiolipin metabolism (Supplementary Fig. 7c), whilst XY-autosome contacts related to lipid catabolism, carbohydrate metabolism, and skeletal muscle development (Supplementary Fig. 7d), which are reported sexual dimorphic processes66–68. Remarkably, male and female datasets showed an anchor locus on a Xq-telomeric region (~153–156 Mb, hg38), interacting with sub-telomeric regions of >50% of autosomes (both p and q arms, Fig. 5a).
Fig. 5. NHCCs relate to sex-specific features.
a Alluvial-style plots show unique gonosome-autosome NHCCs (q < 0.05) for chromosome X in (left) female datasets or in (center) male datasets, and (right) common gonosome-autosome NHCCs for chromosome X among male and female datasets. n = number of interactions. Interactions are colored based on autosome’s origin where p-arm = red, and q-arm = black (opacity 10%). XX = chromosome X from female data, XY = chromosome X from male data. X “chromosomes” are outlined in blue, Y “chromosomes” are outlined in orange, “autosomes” are outlined in black. Asterisks describe genomic regions of hg38 in megabases (Mb). b Same as panel a but for (left) X-Y gonosome-gonosome NHCCs and (right) Y gonosome-autosome NHCCs in male datasets. c Constitutive female NHCCs (>50% of datasets) are shown in genome topology map with XX (blue) and female communities (colored datapoints). Dashed boxes highlight Xq-telomeric anchor region. d Same as panel c, but for male datasets. In addition to Xq-telomeric anchor region, dashed boxes highlight constitutive XY-Y contacts. e Scaled average sex-specific gene expression of GTex52 per 1 Mb bin across (left) female and (right) male genome topology (high [intense color] vs. low expression [white]). f Overlap of supervised and unsupervised Signature results separated by sex. g Working model of the inter-chromosomal topology across human cell types. Constitutive NHCCs coalesce at the speckles and demarcate together with inter-centromeric and inter-telomeric interactions a major spatial genome gradient. Cell-type-specific NHCCs locate to discrete hubs distal to the main axis of genome activity (i.e., NHCCs in neurons, heart). Constitutive NHCCs happen in euchromatin and possess more genes with higher expression in comparison to non-interacting regions.
In male cell types, two genomic regions of XY-Y NHCCs (XY ~ 85-96 Mb; Y ~ 3-8 Mb) overlapped with a pseudoautosomal region of an X-chromosome-transposed region69, and centromeric XY-Y NHCCs matched with unique male trait associations70, i.e., hemoglobin concentration, body fat percentage, etc. (Fig. 5b, Supplementary Data 12, 13). Genes of XY-Y NHCCs related to fat differentiation and sterol signaling (Supplementary Fig. 7e, f)71,72.
We then evaluated constitutive male and female anchor loci (>50% of datasets, q < 0.05) in relation to the identified gonosome contacts. Sex-specific constitutive NHCCs were off-centered and asymmetrically organized with similar regions of chromosomes 3, 6, 9, and 11 (harboring MALAT1 and NEAT1) across both sexes (Fig. 5c, d). Remarkably, the topology maps recapitulated spatial proximity of the Xq-telomeric anchor region to constitutive NHCCs and the XY-Y NHCCs (Fig. 5c, d). To address gene expression among sexes in relation to the main spatial gradient, we plotted sex-separated GTEx expression data in genome topology maps. The spatial gradient along an axis of expression exists in each sex (Fig. 5e). High similarity between supervised and unsupervised learning (female: 93.4%; male: 95.3%) validated replicability of Signature (Fig. 5f).
Collectively, we find evidence that sex-specific NHCCs relate to sexual dimorphic processes and despite sex-biases, genome topology in both sexes involves the spatial genome gradient of activity and an Xq-telomeric anchor region close to constitutive NHCCs.
Discussion
Signature represents an applicable tool for multiscale and integrative Hi-C analysis of all genomic contacts in healthy and in disease states. Using Signature revealed an extensive repertoire of NHCCs that occur genome-wide and that harbor intrinsic properties for genome structure and gene regulation.
Hi-C assays genome organization as a mean across cells in all mitotic stages and our heterogenous body map reflects a spectrum from proliferating to terminally differentiated cell types and tissues. Nevertheless, our findings imply that many NHCCs are deterministic and that constitutive NHCCs together with gene-regulatory features are involved in an off-centered asymmetric spatial genome gradient that is common across many cell types and both sexes. Cell-type-specific NHCCs on the other hand locate to discrete function-related spatial environments distal to the main genome gradient (Fig. 5g).
The transmission of structural and regulatory features of genome organization is thought to involve a unified model of helical coils that allows interphase chromosomes to retain Rabl’s configuration, to build chromosomal territories and to fit into the nucleus73. This fits with our observation that Rabl’s configuration is congruent with the spatial genome gradient of activity observed in our analysis. Despite recent findings suggesting that condensin II prevents inter-centromeric clustering and negatively influences Rabl’s configuration in the human genome49, Rabl’s configuration and chromosomal territories may not be mutually exclusive. Rather, they both may coexist to structure a flexible genome architecture that can react to external stimuli and undergo mitotic re-organization. Inter-centromeric and inter-telomeric interactions are striking structural features in multiple organisms, where chromosomal interaction patterns are influenced by relative sizes of the involved chromosomes. Small chromosomes crowded in a boscage of all chromosomes tend to make more contacts74. This is consistent with our findings, especially in terminally differentiated cells with reduced mitosis-related genome re-organization.
The combination of supervised and unsupervised learning in Signature unveiled multi-chromosomal NHCCs, especially on chromosomes 6, 9, and 11. Local gene density and transcription on chromosome 11 (as one of the gene-densest and disease-richest chromosomes) influence its nuclear organization75. It harbors 40% of olfactory receptors76 which are involved in NHCCs7, and highly and ubiquitously expressed lncRNAs (i.e., MALAT1 and NEAT1) that organize speckles54,55. Moreover, this further validates that topological features determined by Signature are validated by known structures. Whether or not the largest autosomal heterochromatin block and sequence duplications on chromosome 977 that may be caused by NHCCs, are involved in the formation of the ‘anchor community’ remains to be determined. The largest tRNA cluster on chromosome 6 strongly interacts with NHCCs in HiPore-C25, and its MHC locus78 often loops away from its chromosomal territory upon transcription79, which likely supports NHCCs. Altogether, high concentrations of genes, RNAs, and TFs support NHCCs and the spatial genome gradient. This fits well with the findings that NHCCs can demarcate nuclear compartments together with expressed lncRNAs that remain proximal to their loci to enrich for otherwise diffusible molecules (i.e., RNA-binding proteins, TFs, etc.)10,38,80,81. However, euchromatin’s natural flexibility juxtaposed with organized heterochromatin formation may also support the diffusibility of DNA and the gradient formation and its NHCCs. Altogether, the reciprocal interplay between fine-scale genome organization of DNA, transcription, and RNA shapes the genomic architecture82,83.
Signature revealed clear evidence that constitutive NHCCs, as ‘topological anchor loci’ synergistically shape genome topology along a spatial gradient to maintain non-random chromosome positioning. Especially, p- and q-telomeric NHCCs coalesce regions of highest gene density and expression at the nuclear speckles (Fig. 5g). To which extent certain thresholds of RNA and protein interactors are required, and if RNA and DNA in phase-separated hubs as components of genome structure contribute to NHCC formation by clinging chromosomes together, remains to be addressed. Further exploring the biological functions of non-interacting regions, such as cis gene-regulatory hubs and intra-chromosomal organization would be interesting, as well as the extent of transient factors, such as transcriptional activity and cell cycle stages, that influence deterministic NHCC formation. Combining Signature outputs with additional genomic features (i.e., expression and ChIPseq data, methylation and acetylation pattern, recombination frequencies, etc.) in genome topology maps may complement Signature-derived data interpretation and identify yet uncharacterized subnuclear structures.
Signature uncovered rules of shared and separated spatial environments in the human genome. We defined NHCCs and their properties and underscore that NHCCs represent an important regulatory structure for genome topology.
Methods
Datasets
All studies were performed under the regulation of the SickKids Research Ethics Board (Study: #1000080135). Upon completion of required data access agreements, datasets were downloaded from the The European Genome-phenome Archive (EGA) for EGAS0000100476384, and from the NIMH Repository & Genomics Resource, a centralized national biorepository for genetic studies of psychiatric disorders for NPCs, glia, and neurons85. These data were generated as part of the PsychENCODE Consortium, supported by: U01DA048279, U01MH103339, U01MH103340, U01MH103346, U01MH103365, U01MH103392, U01MH116438, U01MH116441, U01MH116442, U01MH116488, U01MH116489, U01MH116492, U01MH122590, U01MH122591, U01MH122592, U01MH122849, U01MH122678, U01MH122681, U01MH116487, U01MH122509, R01MH094714, R01MH105472, R01MH105898, R01MH109677, R01MH109715, R01MH110905, R01MH110920, R01MH110921, R01MH110926, R01MH110927, R01MH110928, R01MH111721, R01MH117291, R01MH117292, R01MH117293, R21MH102791, R21MH103877, R21MH105853, R21MH105881, R21MH109956, R56MH114899, R56MH114901, R56MH114911, R01MH125516, and P50MH106934 awarded to: Alexej Abyzov, Nadav Ahituv, Schahram Akbarian, Alexander Arguello, Lora Bingaman, Kristin Brennand, Andrew Chess, Gregory Cooper, Gregory Crawford, Stella Dracheva, Peggy Farnham, Mark Gerstein, Daniel Geschwind, Fernando Goes, Vahram Haroutunian, Thomas M. Hyde, Andrew Jaffe, Peng Jin, Manolis Kellis, Joel Kleinman, James A. Knowles, Arnold Kriegstein, Chunyu Liu, Keri Martinowich, Eran Mukamel, Richard Myers, Charles Nemeroff, Mette Peters, Dalila Pinto, Katherine Pollard, Kerry Ressler, Panos Roussos, Stephan Sanders, Nenad Sestan, Pamela Sklar, Nick Sokol, Matthew State, Jason Stein, Patrick Sullivan, Flora Vaccarino, Stephen Warren, Daniel Weinberger, Sherman Weissman, Zhiping Weng, Kevin White, A. Jeremy Willsey, Hyejung Won, and Peter Zandi. Details on the origin of the other datasets can be found in the Supplementary Data 1 which also includes Pubmed IDs of the underlying papers.
Mapping of Hi-C and Omni-C data
We processed Hi-C/Omni-C data according to the 4D Nucleome recommendation86. Specifically, we mapped reads to the GRCh38 version 32 of the human reference genome using bwa (version 0.7.17; mem)87. We determined mapping statistics using samtools (version 1.5; view, flagstat)88, and filtered for valid Hi-C and Omni-C alignments by using pairtools (version 0.3.0; parse, sort, dedup, select ‘(pair_type == “UU”) or (pair_type == “UR”) or (pair_type == “RU”)’, split, select ‘True’) (https://github.com/open2c/pairtools). Indexing of the resulting pairs was done with pairix (version 0.3.7) (https://github.com/4dn-dcic/pairix). We normalized the data and applied binning for genomic resolutions (cis = 50 kb, NHCCs = 1 Mb) by using Cooler89. We ensured that pseudo-chromosome Y information was removed from female cooler data (refer to section “Sex determination of cell types”). Omni-C biological replicates of chondrogenic differentiation showed high reproducibility in Pearson’s correlations (normalized cooler output), done in R. For downstream analyses, sequences obtained from replicates were pooled separately (pairtools merge) and balanced (cooler balance89) to serve as a combined dataset per cell type. For all datasets, we aggregated genomic bins for 50 kb in cis and 1 Mb in trans into a Hi-C contact matrices and performed out-of-core matrix balancing using cooler (version 0.8.11; cload pairix, balance, dump)89.
Supervised learning
We employed a Local Weighted Polynomial Regression (LWPR) model90 to model the relationship between Hi-C/Omni-C interaction weights and genomic position (inter-chromosomal, in trans = 1 Mb bins). Beginning at each chromosome’s start, LWPR continuously measures locally in 1 Mb bins which other chromosomal regions (1 Mb bins) are in proximity in relation to the entire genomic background with all possible interactions. We applied LWPR with the same setting on all datasets.
LWPR is facile to use and processes Hi-C data in short run-times. For example, Signature’s parallel computing approach analyzes ˜3.87 B reads in ˜3 h with 38 CPUs (IntelR XeonR E5-2670 @ 2.60 GHz) and 4 GB RAM on a SLURM system. A step-by-step documentation to use Signature can be found on Github (https://github.com/MaassLab/Signature).
We considered linear genomic distance (intra-chromosomal, in cis = 50 kb bins) as previously described29, and added our cross-validation approach. In our supervised learning framework, we treated interaction weight as the response variable and genomic position as the predictor in our regression model. To do this, we linearly represented the anchor chromosome (Fig. 1a), and assigned each bin as an independent predictor variable, with the relationship formulated as:
| 1 |
Here, and denote the dependent (interaction weight) and independent (genomic position) variables of the LWPR model, respectively. represents the coefficient parameter, estimated through local polynomial fitting, and indicates the error term. For every bin , the LWPR assigns different weights to the data points in the vicinity of .
Central to LWPR is the determination of coefficients that minimize the weighted least square error of the fitted polynomial model, capturing the - relationship in the local neighborhood of each :
| 2 |
In this equation, represents the weight assigned to the data point (, ) based on its distance from , and stands for the number of data points. In our study, we employed nonparametric estimation91 to solve Eq. 2. We utilized four-fold stratified cross-validation to determine the optimal width of the span window (, i.e., smoothing parameter) for each bin, considering different percentages of neighboring interactions. Cross-validation was utilized to assess the performance and generalization of our predictive model92. For the NHCC analysis (inter-chromosomal, 1 Mb resolution), we evaluated five potential values for the dynamic span parameter, encompassing approximately the interactions in 4, 5, 6, 7, and 8 bins, to form the local neighborhood used for the LWPR fitting. To maintain consistency across different genomic resolutions, we multiplied the previous range by 20 (1 Mb/50 kb) for the 50 kb (cis) analysis, resulting in widths of 80, 100, 120, 140 and 160. Independently of the sequencing depth of a Hi-C dataset, LWPR recognizes the trends of interaction weights.
Our LWPR model calculates weighted average and weighted standard deviation for each genomic bin (LOESS fitted values), addressing distance signal bias (in cis), and considering interacting bins (NHCCs, in trans) genome wide. We transformed Hi-C/Omni-C signals into z-scores using the formula Zi=xi-/, where Zi represents the z-score and xi denotes the interaction weight of interaction i. Additionally, and indicate the LOESS fitted values for each genomic bin (distance in cis), respectively. Positive z-scores denote more frequent contacts than expected, while negative z-scores indicate fewer interactions than expected29. For each queried interaction in ‘All vs. All’, we obtained two z-scores. Depending on the chosen anchor (i.e., predictor variable - the chromosome used to calculate weighted mean and standard deviation for interacting bins across chromosomes), we selected the more conservative z-score that was closer to zero. The log2-transformed interaction weights of Hi-C data conform to a normal distribution (Supplementary Fig. 1). Consequently, z-scores follow a standard normal distribution, so we also extracted corresponding p-values from the normal distribution function. Finally, to mitigate the risk of false positive results, we performed the Benjamini-Hochberg method to control the false discovery rate. We identified interactions with positive z-scores and q values < 0.05 as significant results indicating highly interacting loci, while negative z-scores with q values < 0.05 represent significantly non-interacting loci. We further extracted annotated genes that mapped within the interacting bins and performed GO-term analysis as described below.
Unsupervised learning
We used Community Detection (CD) to investigate hubs (communities) of genomic interactions. CD operates on a (weighted) graph comprising nodes and edges, aiming to identify non-overlapping node clusters based on the network structure28. The most common approach for detecting communities in networks involves maximizing the function modularity by heuristic optimization algorithms to group nodes of the input network into clusters. Particularly, the Combo algorithm93 has shown higher relative success in maximizing modularity94. Similar to applying K-Means for clustering independent vector data to find clusters of similar datapoints in an unsupervised way, we employed Combo for CD to generate clusters from interdependent network data. Chromosomal interactions are interdependent data that cannot be clustered using vector clustering, justifying our use of CD for network clustering. We used interaction weights (both intra-chromosomal and inter-chromosomal) as CD input to generate clusters of interacting loci through weighted modularity maximization.
We restricted the number of possible communities to 46, as this entity resembles both maternal and paternal alleles of diploid human genomes. To attain a robust 3D genome structure, averting mapping noises, we computed the mean interaction weights from our 62 datasets for each mapped interaction. This treated each genomic locus as a node within a graph, enabling us to cluster all loci in the genome-wide graph based on their interactions into 46 distinct communities. We applied CD using the pycombo package in python 3.8 (using the parameters modularity_resolution = 1.4, max_communities = 46). We then used the outcome of CD to visualize estimations of genome topology by clustering interactive bins and linking consecutive bins in a string together to generate chromosome outlines.
To determine the overlap between results of CD and LWPR, we used permutation testing that involved 10,000 iterations of randomly selected NHCCs to examine the number of iterations in which both bins are in the same community. To visualize a maximum of 46 chromosomes (two gonosomes [female XX / male XY] and 22 pairs of autosomes = 24 possible chromosomes) in human diploid cells, we set the number of possible communities to 46. This resembles the human genome and allows each community to include only one chromosome’s bin if there is an intra-chromosomal domain structure isolated from the rest of the genome. For the visualization of the genome topology, we used the Gephi95 software. To reflect the results of CD and to ensure clear separation between bins from different communities, we first optimized the visualization process. We excluded inter-community interactions and plotted all bins as nodes using the ForceAtlas-296 visualization layout, based on intra-community Hi-C interaction weights. This step ensured that bins within each community are visualized close together and separated from other communities, in turn facilitating the visualization of each community in the genome topology map. The distribution across the topology map as a ‘mock nucleus’ resulted in 46 distinct communities, where bins with higher interaction weights in each community were placed closer together to better visualize their interactions. Next, we added the physical connections between consecutive bins as edges in the network and optimized the network layout using the Fruchterman-Reingold layout algorithm (parameters: area = 5000, gravity = 5, speed = 10). This ensured that bins within the same community remained close together and the structural connections between consecutive bins across chromosomes were maintained. In the final genome topology estimation, consecutive bins were positioned next to each other to outline of the chromosomal structures. Moreover, bins within the same community were as close to each other as the physical constraints allowed. For the 3D visualizations, the Helios web software97 was employed to showcase the 3D architectural estimation derived from the CD results.
Genome-wide significant NHCCs
To assess Signature’s capacity to map inter-chromosomal interactions, we checked reported NHCCs with 1 Mb resolution across all 62 datasets. We extracted any interaction involving both chromosomes of four reported NHCCs (i.e., chromosome 12 and 17 interactions for CISTR-ACT & SOX910) from our genome-wide result and plotted the average z-score of all datasets with a positive z-score for that reported NHCC. To address NHCCs genome-wide, we generated a global matrix, displaying NHCCs for all chromosome pairs across all datasets. We extracted interactions between each pair of chromosomes and plotted the average z-score of each interaction across the 62 datasets in a heatmap. To identify significant NHCCs across cell types (positive z-score with either p < 0.05 or q < 0.05), we calculated the percentage of datasets with significant interactions and plotted the results for the entire genome.
NHCCs vs. intra-chromosomal interactions
To assess the relative proportions of NHCCs and intra-chromosomal interactions in 62 diploid (2n) datasets, we divided all normalized Hi-C interaction weights (from cooler) into cis and trans-chromosomal interactions for each dataset.
MERFISH
We extracted loci of the MERFISH approach20 that matched both bins of our Signature-identified NHCCs and non-interacting regions (q < 0.05) in two Hi-C datasets of IMR90 cells (Supplementary Data 1). To analyze these contacts, we computed the spatial distance between two genomic regions using the Euclidean metric.
HiCAN
We intersected the top 100 speckle-associated and nucleolus-associated bins identified by the HiCAN34 approach with our Signature-identified NHCCs (q < 0.05). Cell lines were matched to 2x GM12878, HMEC, HUVEC, 2x IMR90, 2x NHEK, and 2x teloHAEC datasets (Supplementary Data 1). HiCAN bins were converted from 500 Kb to 1 Mb for direct comparison to Signature.
SPRITE
Genomic bins associated with nucleolar and active (“speckle”) hubs defined by SPRITE9 were intersected with our Signature-identified NHCCs (q < 0.05) in two datasets of GM12878 cells.
Eigendecomposition for compartment analysis
We utilized the eigendecomposition method implemented in the cooltools package98, and computed mean eigenvalues of each bin to generate the eigenvectors for the analysis of genomic compartments. Values above zero indicate A compartments (active), while those below zero were designated as B compartments (inactive).
Domain definition for NHCCs and non-interacting regions
We considered four different scenarios (I-IV) to determine the average size of NHCC and non-interacting domains (Supplementary Fig. 3k). I: consecutive bins of chromosome A, with a maximum of one bin as gap in between, interact with the same bin on chromosome B. This builds domains on chromosomes A and B. Individual interacting bins were considered once to measure average NHCC sizes. II: consecutive bins of chromosome A, with one bin as gap in between, interact with different single bins on chromosome B. This builds a domain on chromosome A only. III: consecutive bins of chromosome A, with one bin as gap in between, interact with similar setup on chromosome B. This builds domains on chromosomes A and B. IV: consecutive bins of chromosome A, with one bin as gap in between, interact with different bins on two different chromosomes B and C. This builds a domain on chromosome A only.
Binomial testing of p and q arm interactions
To investigate whether the structure of human chromosomes (p and q arms) influence chromosomal interactions and positioning, we focused on the interaction weights of p-p, q-q, and p-q arm contacts and took the length of each chromosomal arm into account. First, we assessed the impact of p- and q- arms on chromosomal contacts. We grouped all genome-wide interactions into p-p, p-q, and q-q categories and extracted the total interaction weights in each group. We then used the total length of p- and q- arms to calculate the probability of each interaction type as follow:
and,
| 3 |
Here , , and represent the probability of p-p, p-q, and q-q interactions, respectively, calculated based on the chromosomal arm length. The function L(.) represents length in megabases. Accordingly, the variables L(p), L(q), and L(c) represent the length of the respective underlying p-arm, q-arm, and the entire chromosome (p-arm plus q-arm).
Next, to assess significant over-representations and under-representations between the expected and actual NHCCs for each interaction type, we conducted binomial testing99. The expected probabilities, actual interaction weights, and total interaction weights of each interaction type were considered as hypothesized probability of success, number of trials, and number of successes in the binomial test respectively.
We then calculated the probability of each interaction type based on the p- and q-arm lengths for each chromosome (anchor chromosome), separately. The calculation was done using Eq. (3):
| 4 |
X denotes the arm of the anchor chromosome (indicated by anch index) and Y denotes the arm of all other chromosomes except anchor chromosome (indicated by targ index).
Using equation (4), we computed the probability of four possible interaction types (, , , and ) for all chromosomes. By aggregating the data from all 62 datasets, we extracted the interaction weights for each interaction type. Subsequently, we conducted the binomial testing (as described above) to determine statistically significant discrepancies between the observed and expected interaction weights for each interaction type.
Unified chromosome length
To visualize the distribution of identified NHCCs across all chromosomes along one single linear chromosome, we mapped NHCCs onto a unified representation. The calculation was done using Eq. 5:
| 5 |
represents the coordinates (start and end) of a bin of the selected chromosome, and represents their corresponding coordinates on the unified linear chromosome. Here, and denote the length of the selected chromosome and unified chromosome, respectively. To ensure a comprehensive representation, we included a gap size of 4.2% of the unified chromosome length between the p and q arms, which accounted for the average unmapped read percentage in the centromeric regions of all chromosomes.
Telomeric–centromeric interactions
To explore potential patterns in chromosomal interactions and positioning of telomeric and centromeric regions, we focused on specific segments of mapped sequences, excluding repetitive sequences. We selected 5% of mapped regions 5’ and 5% 3’ of each chromosome as subtelomeric regions (t), and 5% of mapped regions from both 5’ and 3’ of the centromeres (c; identified as unmapped regions in Hi-C). We excluded p arms of acrocentric chromosomes, as well as other regions where more than 2.5% of chromosomal sequence in respective regions were unmapped (Supplemental Fig. 3p). Utilizing a modified version of Eq. (3), tailored for centromeric and telomeric regions, we computed the expected probability of mapped regions of each interaction type (c-c, c-t, and t-t). Following a methodology analogous to testing the p- / q-arm interactions (see binomial testing of p and q arm interactions), we conducted a binomial testing to evaluate interactions of telomeric and centromeric regions, aiming to identify instances of either over- or under-representation for each interaction type.
Gene expression analysis using GTEx
We explored gene expression levels of genes among the common significant NHCCs (q < 0.05, ≧ 10 datasets, 393 bins = 12.73% of hg38 genome) genome-wide using GTEx gene IDs52. We used normalized gene expression from GTEx V8 with transcript per million (TPM) values for 56,201 genes. Since GTEx expression profiles are count data with non-negative values, we applied a log2-transformation after adding 1 to the expression levels to avert a log(0) numerical error.
First, we compared the mean TPM of genes annotated in our common NHCC bins with all genes in GTEx across four different biotypes using Mann-Whitney testing. We assigned the biotypes using GENCODE V42. Next, we extracted the number of genes for each biotype from the bins identified by Signature. To assess statistical significance, we calculated the expected number of genes for each biotype in the same bins using Eq. 6:
| 6 |
and represent the expected number and total number of genes in biotype , respectively. Additionally, and denote the number of bins identified by Signature (393) and the total number of bins in hg38 genome (3088), respectively. We then performed a Fisher’s exact test to determine if there is a significant difference between the actual and expected number of genes for each biotype. To investigate tissue-specific expression levels of the genes identified in Signature’s unique NHCCs, we separated the corresponding cell-types/samples from GTEx and from our Hi-C datasets to generate eight matched groups. We then examined whether there is a significant difference between average TPM of all genes and our tissue-specific genes using Mann-Whitney tests.
To visualize topological gene expression by CD, the GTEx catalogue was filtered to 18 Signature-matched cell-types and the mean TPM was calculated for every bin observed across the 62 Hi-C datasets (2813 = 91.1% of hg38 genome). This was repeated for sex-specific bins (F = 2786, M = 2802), filtering GTex catalogue to corresponding samples with annotated sex.
Transcription factor binding (ChIPseq)
We explored transcription factor binding in our Signature-identified NHCCs (q < 0.05) using the ChIP Atlas53 (“TF and others” track, human hg38, statistical significance threshold 50). Eight tissue groups were matched between the ChIPseq data and Signature’s Hi-C data. For each tissue group individually, we summed the peak counts for each bin identified in the NHCCs and compared this with the summed peak counts for every bin observed across the 62 Hi-C datasets. The statistical significance was examined using a Mann-Whitney test. To visualize topological transcription factor binding by CD, the summed peak counts across all matched tissues were determined for every possible bin observed across the 62 Hi-C datasets (n = 2813).
Tissue culture
hTERT-immortalized female adipose-derived primary human mesenchymal stem cells (ASC52telo [SCRC-4000; RRID:CVCL_U602], ATCC) were maintained in basal MSC media (PCS-500-030, ATCC), supplemented with 2% FBS (ThermoFisher), 5 ng/ml recombinant human FGF basic (R&D Systems 233-FB-010), 5 ng/ml recombinant human FGF acidic (R&D Systems 232-FA-025), 5 ng/ml recombinant human EGF (R&D Systems 236-EG-200), 2.4 mM L-Alanyl-L-Glutamine (ThermoFisher), and 0.2 mg/ml Geneticin (G418, ThermoFisher). TeloHAEC (SCRC-4052, RRID:CVCL_Z065, ATCC) were maintained in vascular cell basal medium (ATCC PCS-100-030) with VEGF (ATCC PCS-100-041). hTERT-RPE-1 (RRID:CVCL_4388, ATCC) were cultivated in DMEM:F12 (ThermoFisher) with 10% FBS (Canadian origin, ThermoFisher). HCT116 cells (RRID:CVCL_0291) were cultivated according to ATCC recommendations. All cells were maintained at sub-confluent conditions, maintained at 37 C with 5% CO2 and were passaged every 3-4 days. Mycoplasma testing was performed every eight weeks with LookOut Mycoplasma PCR detection kit (Sigma-Aldrich).
Oligopainting with immunofluorescence of speckle marker SON
We designed probes the recently described optimized Oligopaint protocol23. Briefly, we designed probes with 80 bases of homology to 1-Mb genomic targets, with an average probe density of 3.5 probes per kb, and directly labeled with Alexa 555. We performed two replicates of FISH and immunofluorescence experiments. Specifically, HCT116 cells were settled on fibronectin-coated slides for 2 h and fixed in 4% formaldehyde for 10 min. We then permeabilized cells in 0.5% Triton-PBS for 15 min, followed by adding 25 µl of hybridization mix, consisting of 2 pmol of each probe, 10% dextran sulfate, 2x SSCT (0.3 M NaCl, 0.03 M sodium citrate and 0.1% Tween 20), 50% formamide, 4% polyvinylsulfonic acid (PVSA)), 5.6 mM dNTPs and 10 μg RNase A, onto each slide, sealing under a coverslip, followed by denaturation at 92 oC for 5 min. Slides were then incubated overnight at 37 oC. On the next day, we washed slides in 2x SSCT at 60 oC for 15 min and twice at RT for 10 min. Slides were then blocked for 30 min in a 0.05% Tween-PBS (PBST) solution containing 1% bovine serum albumin (BSA). For speckle labeling, we incubated 1:500 dilution of anti-SON antibody (HPA023535, Sigma) overnight with each slide at 4 oC, washed the next day three times in 1% PBST alone for 5 min each, incubated in anti-rabbit Alexa 488 secondary (111-545-003, Jackson) at room temperature for 1 h, followed by an additional three washes in 1% for 5 min each. After mounting slides in SlowFade Gold Antifade (Invitrogen), we acquired images of > 300 nuclei in each replicate similar to Luppino et al. 100, using a Leica widefield microscope with a 1.4 NA ×63 oil-immersion objective (Leica) and Andor iXonμltra emCCD camera, then deconvoluted with Huygens Essential v20.04.03 (Scientific Volume Imaging), using the CMLE algorithm and signal:noise ratio of 40, and then analyzed using TANGO101 to identify and make measurements between the FISH spots and speckles.
Permutation analysis. We determined significance of clustering by comparing the observed clustering of genomic regions at speckles to an “expected” null distribution of clustering, generated by 10,000 permutations of the data. Specifically, each permutation maintained the observed number of genome-speckle associations per cell, but randomized which genomic regions interacted with which speckle. The maximum number of spots at any speckle was determined and averaged across all randomized cells, and this process was repeated for each permutation to generate a distribution of random clustering to compare against the observed clustering.
Multicolor FISH
We performed molecular cytogenetic studies on PHA-stimulated peripheral blood lymphocytes, MSCs (SCRC-4000, ATCC), and hTERT RPE-1 (ATCC) according to standard protocols102. For locus-specific labeling, DNA from bacterial artificial chromosome (BAC) probes from BACPAC Chori were used; their DNA was extracted, amplified and labeled by DOP-PCR (standard protocols). Zeiss Axioplan 2 and Axio Imager.Z2 fluorescence microscopes (Carl-Zeiss, Jena, Germany) equipped with appropriate filter sets to discriminate between a maximum of four different fluorochromes.
For FISH, we labeled RP11-661K21 (6p21.31; hg38:33,873,549-34,061,469) and RP11-57C19 (9q34.11 ~ 34.12; hg38:130,605,158-130,778,620) directly with DEAC, RP11-545E17 (9q34.11; hg38:128,699,415-128,866,078) and RP11-569N5 (11q13.2 ~ 13.3; hg38:68,548,288-68,727,966) with Spectrum Orange, RP11-142G8 (11q13.2; hg38:66,352,870-66,511,335) and RP11-466J21 (1p36.12; hg38:22,393,597-22,470,578) indirectly with Digoxigenin-Fluorescein, and RP11-24C3 (3p21.31; hg38:48,340,573-48,500,067) and RP3-349A12 (6p21.31; hg38:34,842,772-34,975,718) indirectly with Biotin-Cy5; DAPI was used as counterstain. We combined the eight probes in two independent four color FISH experiments (set1: RP11-661K21, RP11-545E17, RP11-142G8, RP11-24C3; set2: RP11-57C19, RP3-349A12, RP11-569N5, RP11-466J21). Digital images were captured using an IMAC S30 CCD camera and MetaSystems (Isis) software (Altlussheim, Germany). We used the same acquisition setting to image ˜100 nuclei from the same experiment, on the same slide, under the same microscopic conditions. To provide the best signal-to-noise ratios of every image, we used the Isis-standardized background control algorithm to allow quantitative analysis of the BAC signals. Images were post-processed by increasing the contrast of each acquired channel. We analyzed the arrangements of the BAC signals to one another within the same cell in a cell-by-cell manner for identifying the relative chromosomal positioning to each other. We refer to clustered signals where either signals of different channels directly overlapped (co-localized) or where signals were in close proximity. To distinguish close proximal signals from non-colocalized signals, we measured signal distances and put them in relation to the corresponding nucleus diameter. Then, we calculated means across all cells (˜100) analyzed in each experiment. We grouped the signals into groups of no colocalization, two-color, three-color, and four-color clustered signals and distinguished between mono-allelic and bi-allelic signals.
K-means and hierarchical clustering
We performed K-Means clustering with four clusters (evaluated using the elbow method) to gain insight into the distinct characteristics distinguishing cell types based on their cis contacts and NHCCs. We standardized the interaction weights by scaling them to a mean of zero and a standard deviation of one. Moreover, we examined interaction patterns across each chromosome using hierarchical clustering on NHCCs and intra-chromosomal interactions separately.
Chondrogenic differentiation
We performed chondrogenic differentiation in micromass pellet cultures for up to 21 days. We supplemented DMEM (4.5 g/L glucose) with 1 IU/mL heparin sulfate, 5% FBS, 1× ITS-X, 100 U/mL penicillin, 100 μg/mL streptomycin, 100 nM dexamethasone, 50 μM l-ascorbic-2-phosphate, 100 ng/μL recombinant human IGF-1, 10 ng/mL recombinant human TGF-β1 and 1 mM sodium pyruvate. We fixed chondrogenic pellets overnight with 4% paraformaldehyde, and paraffin-embedded tissue was sectioned for Picrosirius Red and Alcian blue staining. We exchanged the medium every two days for a total period of 21 days.
Immunohistochemistry of chondrogenic differentiations
Day 21 chondrogenic pellet cultures were fixed in 4% paraformaldehyde for 24 hr at 4 °C. Samples were embedded in paraffin, cut into five micrometer sections, deparaffinized, hydrated and then subjected to either Alcian Blue or Picrosirius Red staining. For Alcian Blue staining of mucins, sections were soaked in 3% acetic acid for 3 minutes, then in 1% Alcian Blue in 3% acetic acid for 30 minutes at room temperature and subsequently washed with distilled water. We stained collagen fibers with Picrosirius Red according to standard procedures in collaboration with the SickKids Pathology core.
RNA extraction, cDNA preparation, RT-qPCR
Chondrogenic identity was verified by measuring relative expression of chondrogenic markers (COL2A1, COL10A1, PTHLH, and SOX9). Total RNA was extracted from cells undergoing differentiation at day 0, 3, 7, 10, 14, and 21, using the phenol-chloroform extraction method according to standard protocols. Residual genomic DNA was removed using DNase I digestion (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized using SuperScript III First Strand Synthesis System (Invitrogen). qRT-PCRs were performed using PowerUp SYBR Master Mix (Applied Biosystems) analyzed by using the 2(-ΔΔCt) method. Relative expression was calculated using RPL13A as the housekeeping gene.
Omni-C
Hi-C13 and Omni-C (randomly digested chromatin, Dovetail Genomics, CA, USA) analyze genomic interactions. We generated Omni-C data of two independent MSC-derived chondrogenic differentiations (time course: 0, 1, 3, 7, 14, and 21 days). We FA-crosslinked samples for 10 minutes and pelleted them for 5 minutes with 2000 g. Pellets were washed in 800 µL wash buffer (100 mM NaCl, Tris pH8.0, 0.05% Tween-20) until fully resuspended. We removed supernatants after 5 minutes of centrifugation at 2000g and repeated the latter two steps. Cell pellets were frozen at -80 °C for library preparation. Dovetail Genomics, CA, USA prepared three Omni-C libraries (technical replicates) of each independent replicate.
Tissue-specific NHCCs
Of all 40,282 interactions, we singled out those NHCCs that displayed significance (q < 0.05) exclusively in a particular dataset and labeled them as ‘unique NHCCs’ (23,251). In order to assess the proportion of these unique interactions, we applied random selection to a simulation of NHCC numbers. For each dataset, we randomly selected the same number of NHCCs as presented in the dataset. For example, dataset aorta_Leung showed 2210 NHCCs. Thus, we randomly chose 2210 out of the total number of NHCCs without replacement. This process was repeated for all datasets, and then iterated 10000 times. In each iteration, we calculated the ratio of uniqueness (the number of NHCCs that was selected in only one dataset). Our actual unique NHCC ratio was 0.577, whereas the highest ratio observed in the 10,000 iterations was 0.162 (empirical p-value = 0).
Enrichment analysis
We performed functional GO-term analysis of genes with Metascape36, using GENCODE-annotated genes103 of 1 Mb bins as background. Default was q < 0.05 in chondrocytes 3 d and 7 d, germinal B-cells, HMEC, naïve B-cells, NPC_Rajarajan, and VSMCs 21 d. Due to the high number of genes in bins of unique NHCCs in other datasets, we adjusted the FDR cutoff to limit the number of genes (q < 0.025 NHEK in dilution; q < 0.01 aorta, astrocytes_cerebellum, cardiomyocytes 80 d, H9ESCs, MSCs_Dixon, NHEK in situ, NPC_Dixon, spleen, trophectoderm; q < 0.005 thymus; q < 0.001 H1ESCs_Dixon, islets, RPE-1).
Sex determination of cell types
Among the 62 analyzed datasets, 16 lacked information about the sample’s sex. To address this issue, we utilized logistic regression104 on the SAM files of 46 datasets with known sex to train a classifier. This classifier was trained on the proportion of gonosome-mapped reads and the mapped reads from the autosomes. This relies on the assumption that 2n diploid genomes with male (XY) gonosomes will produce 50% fewer mapped reads for the X chromosome than female (XX) genomes. We then split the datasets with sex labels (SAM files of 46 datasets) randomly into two sets: 75% as the training set and the remaining 25% as the validation set. The proportions of male and female labels were maintained in both sets. The sex determination classifier achieved 100% accuracy on the validation set, confirming the approach’s reliability. We then applied the trained classifier to predict the sex of 16 datasets without sex labels. To analyze sex-specific NHCCs across all datasets, we applied an FDR cutoff of q < 0.05 and converted genomic positions of sex-specific NHCCs into unified chromosome lengths (see unified chromosome length) to plot them in circos plots105.
Statistics and reproducibility
No statistical method was used to predetermine sample size and data was excluded upon outlier analysis. All statistical tests conducted were two-sided, unless stated otherwise. If multiple tests were carried out on the same data, error rates were corrected for multiple testing using Bonferroni correction or as stated in the results and methods. For statistical analysis, we used distinct samples in R version 4.2.1 (or as stated in the methods), Python version 3.8, and GraphPad Prism version 10.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The authors thank J. L. Rinn for comments on the manuscript, and the Hi-C data contributors of the Aifantis, Tsirigos, Martin-Subero, Ren, Akbarian, Lieberman-Aiden, Dekker, Lettre, Neuveut, Stitzel, Zhao, Fullwood, and Khavari labs for their work. We thank The Centre for Applied Genomics and the Research IT department (Andrew Maksymowsky), The Hospital for Sick Children, Toronto, Canada for assistance with high-throughput sequencing, and computation; and Filipi Nascimento Silva, Indiana University for Helios web support. We thank Dovetail Genomics (Drs. Marco Blanchette and Lisa Munding), LLC, 100 Enterprise Way, Suite A101, Scotts Valley, CA 95066, USA, for generating Omni-C libraries and for the collaborative support. DFL was supported by an Ontario Genomics-CANSSI Ontario Postdoctoral Fellowship in Genome Data Science, JWLB was supported by a CGS-D fellowship, and MM & SA received Restracomp fellowships from the SickKids Research Institute. This project was supported by Canada’s New Frontiers in Research Fund (NFRFE-2018-01305), the NSERC Discovery Grant (RGPIN-2020-04180), and the Canadian Institutes of Health Research (CIHR PJT 173542 [PGM]). PGM holds a Canada Research Chair Tier 2 in Non-coding Disease Mechanisms.
Author contributions
Conceptualization and Funding Acquisition: P.G.M.; Methodology and Formal Analysis: M.M., J.J.C., S.C.N., B.J.M., D.F.L., V.M., M.S., A.R.B., J.O.N., S.E.G., S.A., J.W.L.B., K.D., Sameen A, P.G.M.; Software: M.M., J.J.C., B.J.M., D.F.L., M.S., S.E.G., S.A.; Investigation: M.M., J.J.C., B.J.M., D.F.L., M.S.; Resources: T.L., M.D.W., A.S., E.F.J., A.W., P.G.M.; Validation: S.C.N., T.L., E.F.J., A.W.; Writing: P.G.M. & M.M.; Review and Editing all authors; Supervision: P.G.M.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The datasets generated in this study have been deposited in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/; accession numbers: GSE217358, GSM7757606, GSM7757607, GSM7757608, GSM7757609, GSM7757610, GSE242273). Source data are provided with this paper.
Code availability
The code of Signature (LWPR & Community detection), its documentation, and a demo of how to utilize Signature, as well as computational analysis required for graphical visualization are available at https://github.com/MaassLab/Signature106. Further custom code to reanalyze the data reported in this project is available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Inclusion, diversity, and ethics statement
We support inclusive, diverse, ethical and equitable conduct of research.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53983-y.
References
- 1.Cremer, T. & Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol.2, a003889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickmore, W. A. & van Steensel, B. Genome architecture: domain organization of interphase chromosomes. Cell152, 1270–1284 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branco, M. R. & Pombo, A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol.4, e138 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maass, P. G., Barutcu, A. R. & Rinn, J. L. Interchromosomal interactions: A genomic love story of kissing chromosomes. J. Cell Biol.218, 27–38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomvardas, S. et al. Interchromosomal interactions and olfactory receptor choice. Cell126, 403–413 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Monahan, K., Horta, A. & Lomvardas, S. LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature565, 448–453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McStay, B. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev.30, 1598–1610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinodoz, S. A. et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell174, 744–757.e24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maass, P. G. et al. A misplaced lncRNA causes brachydactyly in humans. J. Clin. Invest.122, 3990–4002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engreitz, J. M., Agarwala, V. & Mirny, L. A. Three-dimensional genome architecture influences partner selection for chromosomal translocations in human disease. PLoS ONE7, e44196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dekker, J. et al. Spatial and temporal organization of the genome: Current state and future aims of the 4D nucleome project. Mol. Cell83, 2624–2640 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker, J. Mapping the 3D genome: aiming for consilience. Nat. Rev. Mol. cell Biol.17, 741–742 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Payne, A. C. et al. In situ genome sequencing resolves DNA sequence and structure in intact biological samples. Science371, eaay3446 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bashkirova, E. & Lomvardas, S. Olfactory receptor genes make the case for inter-chromosomal interactions. Curr. Opin. Genet Dev.55, 106–113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maass, P. G., Barutcu, A. R., Weiner, C. L. & Rinn, J. L. Inter-chromosomal contact properties in live-cell imaging and in Hi-C. Mol. Cell69, 1039–1045 e1033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, R., Zhou, T. & Ma, J. Multiscale and integrative single-cell Hi-C analysis with Higashi. Nat. Biotechnol.40, 254–261 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maass, P. G. et al. Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING). Nat. Struct. Mol. Biol.25, 176–184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su, J. H., Zheng, P., Kinrot, S. S., Bintu, B. & Zhuang, X. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell182, 1641–1659 e1626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen, H. Q. et al. 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nat. Methods17, 822–832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takei, Y. et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature590, 344–350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, D. S. et al. High-throughput Oligopaint screen identifies druggable 3D genome regulators. Nature620, 209–217 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, Y. et al. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol.217, 4025–4048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong, J. Y. et al. High-throughput Pore-C reveals the single-allele topology and cell type-specificity of 3D genome folding. Nat. Commun.14, 1250 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krietenstein, N. et al. Ultrastructural details of mammalian chromosome architecture. Mol. Cell78, 554–565 e557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beagrie, R. A. et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature543, 519–524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fortunato, S. & Newman, M. E. J. 20 years of network community detection. Nat. Phys.18, 848–850 (2022). [Google Scholar]
- 29.Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature489, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beagrie, R. A. et al. Multiplex-GAM: genome-wide identification of chromatin contacts yields insights overlooked by Hi-C. Nat. Methods20, 1037–1047 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dotson, G. A. et al. Deciphering multi-way interactions in the human genome. Nat. Commun.13, 5498 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, W. et al. To embed or not: network embedding as a paradigm in computational biology. Front. Genet.10, 381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanabe, H. et al. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl Acad. Sci. USA99, 4424–4429 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joo, J. et al. Probabilistic establishment of speckle-associated inter-chromosomal interactions. Nucleic Acids Res.51, 5377–5395 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spector, D. L. & Lamond, A. I. Nuclear speckles. Cold Spring Harb. Perspect. Biol.3, a000646 (2011). [DOI] [PMC free article] [PubMed]
- 36.Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun.10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoenfelder, S. et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet.42, 53–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hacisuleyman, E. et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol.21, 198–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagano, T. et al. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol.16, 175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fudenberg, G. & Imakaev, M. FISH-ing for captured contacts: towards reconciling FISH and 3C. Nat. Methods14, 673–678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cremer, T. et al. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum. Genet.60, 46–56 (1982). [DOI] [PubMed] [Google Scholar]
- 43.Kim, S. et al. The dynamic three-dimensional organization of the diploid yeast genome. eLife6, e23623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hochstrasser, M., Mathog, D., Gruenbaum, Y., Saumweber, H. & Sedat, J. W. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J. Cell Biol.102, 112–123 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dogan, E. S. & Liu, C. Three-dimensional chromatin packing and positioning of plant genomes. Nat. Plants4, 521–529 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Zhang, C. et al. tagHi-C reveals 3d chromatin architecture dynamics during mouse hematopoiesis. Cell Rep.32, 108206 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Pouokam, M. et al. The Rabl configuration limits topological entanglement of chromosomes in budding yeast. Sci. Rep.9, 6795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parada, L. & Misteli, T. Chromosome positioning in the interphase nucleus. Trends Cell Biol.12, 425–432 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Hoencamp, C. et al. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science372, 984–989 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croft, J. A. et al. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol.145, 1119–1131 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frankish, A. et al. Gencode 2021. Nucleic Acids Res.49, D916–D923 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Consortium, G. T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou, Z., Ohta, T., Miura, F. & Oki, S. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res.50, W175–W182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hutchinson, J. N. et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics8, 39 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clemson, C. M. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell33, 717–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Todorovski, V. et al. Confined environments induce polarized paraspeckle condensates. Commun. Biol.6, 145 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen, Y. et al. TSA-seq mapping of nuclear genome organization. bioRxiv10.1101/307892 (2018).
- 58.Nguyen, S. C. & Joyce, E. F. Programmable chromosome painting with oligopaints. Methods Mol. Biol.2038, 167–180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ilik, I. A. et al. SON and SRRM2 are essential for nuclear speckle formation. eLife9, e60579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chandrasekaran, S. et al. Neuron-specific chromosomal megadomain organization is adaptive to recent retrotransposon expansions. Nat. Commun.12, 7243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Espeso-Gil, S. et al. Environmental enrichment induces epigenomic and genome organization changes relevant for cognition. Front. Mol. Neurosci.14, 664912 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldring, M. B., Tsuchimochi, K. & Ijiri, K. The control of chondrogenesis. J. Cell Biochem97, 33–44 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Hallett, S. A., Ono, W. & Ono, N. The hypertrophic chondrocyte: to be or not to be. Histol. Histopathol.36, 1021–1036 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y. et al. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet.51, 1380–1388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Demarest, T. G. & McCarthy, M. M. Sex differences in mitochondrial (dys)function: Implications for neuroprotection. J. Bioenerg. Biomembr.47, 173–188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rubin, J. B. et al. Sex differences in cancer mechanisms. Biol. Sex. Differ.11, 17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Acaz-Fonseca, E., Ortiz-Rodriguez, A., Lopez-Rodriguez, A. B., Garcia-Segura, L. M. & Astiz, M. Developmental sex differences in the metabolism of cardiolipin in mouse cerebral cortex mitochondria. Sci. Rep.7, 43878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Veerappa, A. M., Padakannaya, P. & Ramachandra, N. B. Copy number variation-based polymorphism in a new pseudoautosomal region 3 (PAR3) of a human X-chromosome-transposed region (XTR) in the Y chromosome. Funct. Integr. Genomics13, 285–293 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Sidorenko, J. et al. The effect of X-linked dosage compensation on complex trait variation. Nat. Commun.10, 3009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaikaew, K., Grefhorst, A. & Visser, J. A. Sex differences in brown adipose tissue function: sex hormones, glucocorticoids, and their crosstalk. Front Endocrinol. (Lausanne)12, 652444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hatori, M. et al. Light-dependent and circadian clock-regulated activation of sterol regulatory element-binding protein, X-box-binding protein 1, and heat shock factor pathways. Proc. Natl Acad. Sci. USA108, 4864–4869 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sedat, J. et al. A proposed unified interphase nucleus chromosome structure: preliminary preponderance of evidence. Proc. Natl Acad. Sci. USA119, e2119101119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duan, Z. et al. A three-dimensional model of the yeast genome. Nature465, 363–367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahy, N. L., Perry, P. E. & Bickmore, W. A. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol.159, 753–763 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor, T. D. et al. Human chromosome 11 DNA sequence and analysis including novel gene identification. Nature440, 497–500 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Humphray, S. J. et al. DNA sequence and analysis of human chromosome 9. Nature429, 369–374 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mungall, A. J. et al. The DNA sequence and analysis of human chromosome 6. Nature425, 805–811 (2003). [DOI] [PubMed] [Google Scholar]
- 79.Volpi, E. V. et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. cell Sci.113, 1565–1576 (2000). [DOI] [PubMed] [Google Scholar]
- 80.Quinodoz, S. A. et al. RNA promotes the formation of spatial compartments in the nucleus. Cell184, 5775–5790 e5730 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Unfried, J. P. & Ulitsky, I. Substoichiometric action of long noncoding RNAs. Nat. Cell Biol.24, 608–615 (2022). [DOI] [PubMed] [Google Scholar]
- 82.van Steensel, B. & Furlong, E. E. M. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. cell Biol.20, 327–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barutcu, A. R., Blencowe, B. J. & Rinn, J. L. Differential contribution of steady-state RNA and active transcription in chromatin organization. EMBO Rep.20, e48068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vilarrasa-Blasi, R. et al. Dynamics of genome architecture and chromatin function during human B cell differentiation and neoplastic transformation. Nat. Commun.12, 651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rajarajan, P. et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science362, eaat4311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dekker, J. et al. The 4D nucleome project. Nature549, 219–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abdennur, N. & Mirny, L. A. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics36, 311–316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Atkeson, C. G., Moore, A. W. & Schaal, S. in Lazy Learning (ed Aha, D. W.) 75-113 (Springer Netherlands, 1997).
- 91.Scrucca, L. Model-based SIR for dimension reduction. Comput. Stat. Data Anal.55, 3010–3026 (2011). [Google Scholar]
- 92.Mokhtaridoost, M., Maass, P. G. & Gonen, M. Identifying tissue- and cohort-specific RNA regulatory modules in cancer cells using multitask learning. Cancers (Basel)14, 4939 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sobolevsky, S., Campari, R., Belyi, A. & Ratti, C. General optimization technique for high-quality community detection in complex networks. Phys. Rev. E, Stat. Nonlinear, Soft Matter Phys.90, 012811 (2014). [DOI] [PubMed] [Google Scholar]
- 94.Aref, S., Mostajabdaveh, M. & Chheda, H. Heuristic modularity maximization algorithms for community detection rarely return an optimal partition or anything similar. In Computational Science – ICCS 2023 612–626 (Springer, 2023).
- 95.Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media3, 361–362 (2009). [Google Scholar]
- 96.Jacomy, M., Venturini, T., Heymann, S. & Bastian, M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE9, e98679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.GeitHub. Helios-Web (Version 0.7.9) (GitHub, 2023) http://heliosweb.io.
- 98.Open2C et al. Cooltools: enabling high-resolution Hi-C analysis in Python. bioRxiv10.1101/2022.10.31.514564 (2022). [DOI] [PMC free article] [PubMed]
- 99.Hollander, M., Wolfe, D. A. & Chicken, E. Nonparametric Statistical Methods 3rd edn. (Wiley, 2013).
- 100.Luppino, J. M. et al. Co-depletion of NIPBL and WAPL balance cohesin activity to correct gene misexpression. PLoS Genet.18, e1010528 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ollion, J., Cochennec, J., Loll, F., Escude, C. & Boudier, T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics29, 1840–1841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.R. S. Verma, A. B. Human Chromosomes—Manual of Basic Techniques. (Pergamon Press, 1989).
- 103.Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res.47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hastie, T., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning. 2nd edn. (Springer, 2009).
- 105.Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Mokhtaridoost, M. et al. Signature10.5281/zenodo.13873973 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The datasets generated in this study have been deposited in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/; accession numbers: GSE217358, GSM7757606, GSM7757607, GSM7757608, GSM7757609, GSM7757610, GSE242273). Source data are provided with this paper.
The code of Signature (LWPR & Community detection), its documentation, and a demo of how to utilize Signature, as well as computational analysis required for graphical visualization are available at https://github.com/MaassLab/Signature106. Further custom code to reanalyze the data reported in this project is available from the corresponding author on reasonable request.