Abstract
Nuclear metabolism and DNA damage response are intertwined processes, but the precise molecular links remain elusive. Here, we explore this crosstalk using triple-negative breast cancer (TNBC) as a model, a subtype often prone to DNA damage accumulation. We show that the de novo purine synthesis enzyme IMPDH2 is enriched on chromatin in TNBC compared to other subtypes. IMPDH2 chromatin localization is DNA damage dependent, and IMPDH2 repression leads to DNA damage accumulation. On chromatin, IMPDH2 interacts with and modulates PARP1 activity by controlling the nuclear availability of NAD+ to fine-tune the DNA damage response. However, when IMPDH2 is restricted to the nucleus, it depletes nuclear NAD+, leading to PARP1 cleavage and cell death. Our study identifies a non-canonical nuclear role for IMPDH2, acting as a convergence point of nuclear metabolism and DNA damage response.
Subject terms: DNA damage and repair, Cancer metabolism, Breast cancer
Metabolism plays an important role in response to DNA damage, however the underlying mechanisms are less clear. Here, the authors identify a non-canonical role of IMPDH2 wherein it is recruited to the chromatin following DNA damage and mediates PARP1-dependent DNA damage repair via the regulation of nuclear NAD+ levels.
Introduction
Oncogenic signals orchestrate a metabolic rewiring that boosts metabolic processes to meet the highly biosynthetic demands of cancer cells1. Intermediate metabolites generated by the activity of metabolic enzymes can also diffuse into the nucleus and alter epigenetics as they act as essential cofactors for the covalent modification of histones and DNA2,3. More recently, metabolic enzymes have been detected in the nucleus, challenging the old paradigm that metabolites passively diffuse through nuclear pores, paving the path for a new area of research focused on nuclear metabolism4. Examples of nuclear metabolic activities include the folate enzyme Methylenetetrahydrofolate dehydrogenase, cyclohydrolase, and formyltetrahydrofolate synthetase 1 (MTHFD1) that interacts with the epigenetic reader Bromodomain and extra-terminal domain 4 (BRD4) to activate transcriptional programs that drive cancer progression5; the nuclear production of adenosine monophosphate (ATP) by Nucleoside diphosphate linked moiety X (NUDX5) that sustains chromatin remodeling in response to hormones6; the localization of nucleotide synthesis enzymes at the replication fork7; and the nuclear translocation of metabolic enzymes providing Acetyl-CoA8–10 and S-Adenosyl methionine11 for epigenetic regulation. Metabolic enzymes localized in the nucleus have been recently shown to participate in the DNA damage response (DDR). For instance, the nuclear fraction of ATP-citrate lyase (ACLY) is phosphorylated in response to DNA damage to provide acetyl-CoA for histone acetylation near sites of double-strand breaks (DSBs) to promote DNA repair by Homologous Recombination (HR)10. Recently, we also showed that the mitochondrial detoxifying enzyme Peroxiredoxin 1 (PRDX1) relocates to the nucleus in response to DNA damage to assist in nuclear Reactive Oxygen Species (ROS) scavenging and promote DNA damage repair12.
Cancer cells often have defects in DNA damage response routes, leading to increased mutational burden and genomic instability that contribute to cancer progression. However, defective DNA damage response is a double-edged sword. Excessive DNA damage signaling can result in cell death, which offers a window of opportunity for therapeutic intervention. HR-deficient cancer cells, for example, are selectively sensitive to treatment with DNA damage response inhibitors like Poly[ADP-ribose] polymerase (PARP) inhibitors13, which act by trapping PARP complexes at sites of single-strand breaks. This eventually leads to the formation of improperly repaired DSBs, resulting in cell death. PARP inhibitors are approved for the treatment of HR-defective tumor types for ovarian, pancreatic, prostate, and breast cancer14. Presently, PARP inhibition is the primary targeted therapy approved for TNBC15, the most aggressive subtype of breast cancer for which treatment options have limited efficacy beyond traditional chemotherapy.
The fate of a cell following DNA damage largely depends on the level of activation of PARP1. When DNA damage is excessive, the hyperactivation of PARP1 leads to either programmed cell death (apoptosis, or parthanatos) or necrosis, instead of promoting DNA repair16–18. The choice of “how to die” depends primarily on the level of residual energy the cell has after attempting to repair the damage. In cells with severely damaged DNA, PARP-1 hyperactivity can lead to elevated nicotinamide adenine dinucleotide (NAD+) consumption, resulting in energy depletion and necrosis19,20. This cascade can be blocked by caspase-mediated cleavage of PARP1, which instead triggers apoptosis. However, it is unknown how the cell senses NAD+ levels to regulate this process. One hypothesis could be that NAD+-consuming enzymes can control PARP1 activity by utilizing NAD+ thereby limiting the PARP1 activation window. In this regard, sirtuins, a class of NAD+-dependent deacetylases, have been hypothesized as modulators of PARP121, and their activation renders cells more susceptible to apoptosis22. However, sirtuins irreversibly consume NAD+, thus contributing to NAD+ depletion and energy exhaustion rather than a fine-tuned modulation of PARP1 activity.
In this work, we identify Inosine monophosphate dehydrogenase 2 (IMPDH2), a metabolic enzyme known for its role in catalyzing the rate-limiting step in the de novo biosynthesis of guanine nucleotides, as a chromatin partner of PARP1. Upon induction of DNA damage, IMPDH2 is recruited to chromatin, where it interacts with PARP1. This interaction modulates NAD+ availability, thus controlling PARP1 activity.
Results
IMPDH2 chromatin localization increases in triple-negative breast cancer
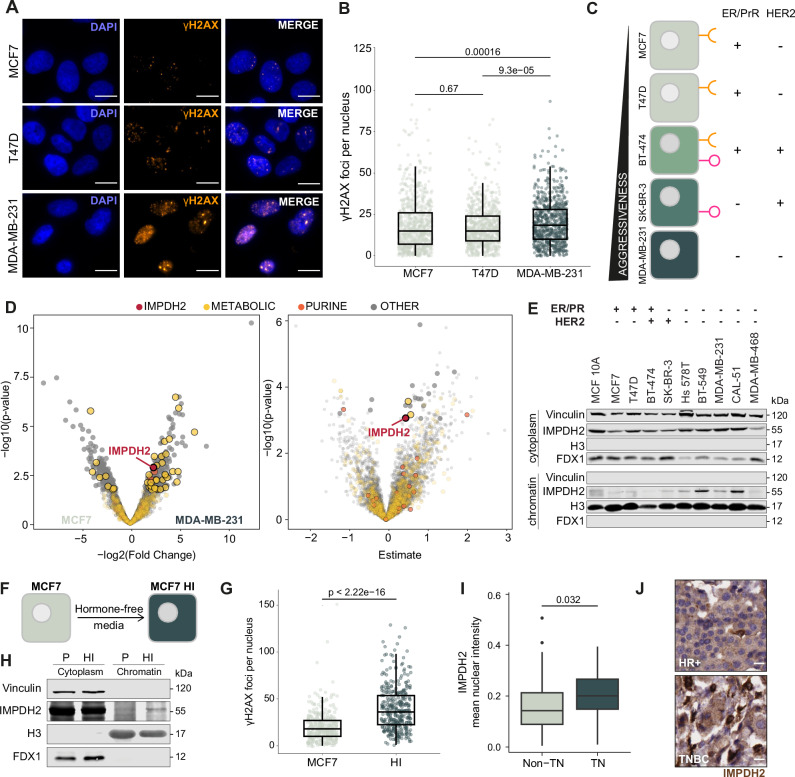
TNBC cells are more prone to accumulate DNA damage than other breast cancer cells23–25. To verify whether this could also be observed in breast cancer cell lines, endogenous DNA damage levels were compared in MCF7 and T47D cells, which are commonly used as Estrogen /Progesterone Receptor positive (ER/PrR+) breast cancer models, and MDA-MB-231 cells as TNBC model. High-throughput immunofluorescence (HT-IF) imaging revealed that MDA-MB-231 cells had significantly higher levels of ɣH2AX, a common DNA damage marker, than MCF7 and T47D cells, confirming that the TNBC model had increased levels of endogenous DNA damage (Fig. 1A, B). Since DNA damage repair inherently involves chromatin and is metabolically demanding26, we wondered whether the differences in endogenous DNA damage observed in different breast cancer cells might be associated with changes in chromatin-bound metabolic activities. We extracted the proteins associated with the chromatin (hereafter, the chromatome) from five breast cancer cell lines belonging to the four major molecular subtypes, which are MCF7 and T47D (ER/PrR+), BT-474 [ER/PrR+ and human epidermal growth factor receptor 2 positive (HER2+)], SK-BR-3 (HER2+), and MDA-MB-231 (TNBC) (Fig. 1C). Chromatome extract purity was first checked by western blot using Histone 3 (H3) as a chromatin marker and Vinculin as a cytoplasmic marker (Supplementary Fig. 1A). Subsequently, we performed Mass Spectrometry (MS) analysis of the chromatomes (Supplementary Data 1). Following signal normalization (Supplementary Fig. 1B), we conducted compartment enrichment analysis and observed that the chromatome fraction was enriched in chromatin and nuclear proteins, and depleted of proteins belonging to other compartments (Supplementary Fig. 1C), validating the subcellular fractionation. Gene Ontology (GO) analysis revealed terms related to the nuclear environment (Supplementary Fig. 1D), further confirming the quality of our dataset. Moreover, we observed good separation between the different cell lines and robust clustering of the replicates. MDA-MB-231 and T47D cell lines had a distinct composition of chromatin-associated proteins compared to MCF7, BT-474 and SK-BR-3, which clustered closer together (Supplementary Fig. 1E). We compared the chromatome of the TNBC cell line MDA-MB-231 with those of the other breast cancer cell lines. We observed that the enzyme IMPDH2, which regulates guanosine monophosphate (GMP) synthesis27, was enriched on chromatin in MDA-MB-231 cells when compared to MCF7 (Fig. 1D, left panel), BT-474, SK-BR-3 and T47D cells (Supplementary Fig. 1F and Supplementary Data 2), suggesting a higher requirement for its chromatin localization in the TNBC cells. Breast cancer aggressiveness usually increases from ER+ to TNBC (Fig. 1C). To identify proteins with increased chromatin abundance along with breast cancer aggressiveness, we performed regression analysis across the cell lines. This analysis indicated that IMPDH2 chromatin abundance correlates with breast cancer cell aggressiveness (Fig. 1D, right panel and Supplementary Data 3). Chromatome western blot analysis of several breast cancer cell lines confirmed that IMPDH2 was more abundant on chromatin in cell lines belonging to the TNBC subtype (Hs 578T, BT-549, MDA-MB-231, CAL-51, MDA-MB-468) (Fig. 1E). ER+ breast cancer cells can transform into TNBC-like cells by bypassing ER signaling dependency for growth28. MCF7 cells cultured without estrogen led to the selection of cells capable of growth in an ER-independent manner, thus mimicking this transition to a more malignant phenotype characterized by the progression from hormone-dependent to hormone-independent (HI) growth (Fig. 1F). The resulting MCF7 HI population displayed a heterogeneous phenotype, with some cells acquiring proliferative capacity and others remaining quiescent. By HT-IF, cells were separated based on their integrated DAPI intensity (DAPI signal multiplied by nucleus area) to enrich only for proliferating cells in G2/M. The quantification of γH2AX foci demonstrated that MCF7 HI proliferating cells exhibited higher levels of DNA damage than parental MCF7 cells (Fig. 1G). Moreover, MCF7 HI cells showed increased levels of IMPDH2 within the chromatin compartment (Fig. 1H). Furthermore, we performed immunohistochemistry using a commercial tissue microarray (TMA) containing 114 human samples of different breast cancer subtypes and grades obtained from patients. When analyzing the results based on tumor grade, we observed that the nuclear localization of IMPDH2, as well as its nuclear signal intensity, were significantly higher in grades 2 and 3 than in grade 1, confirming the correlation with IMPDH2 nuclear localization and tumor aggressiveness (Supplementary Fig. 1G, H). A comparison of TNBC samples with those of the other breast cancer subtypes revealed that TNBC samples had significantly higher levels of nuclear IMPDH2 (Fig. 1I, J), confirming our observation with cell lines.
Fig. 1. IMPDH2 is a chromatin-associated metabolic enzyme.
Representative pictures (A) and quantification (B) of ɣH2AX foci (ɣH2AX orange, DAPI blue; scale bar 15 μm) in breast cancer cell lines representing the different subtypes of breast cancer (MCF7, n = 1282; T47D, n = 1169; MDA-MB-231, n = 748; unpaired two-tailed Wilcoxon test). C Schematic overview of the breast cancer cell lines representing the different subtypes of breast cancer. D Volcano plots of changes in protein abundance on chromatin in MDA-MB-231 vs MCF7 (left panel) and linear regression coefficient of protein chromatin intensity against cell line aggressiveness (right panel); IMPDH2 highlighted in red, purine synthesis and metabolic pathways colored in orange and yellow, respectively; linear regression with multiple comparison adjustment (FDR). E Western blot detection of IMPDH2 protein on cytosolic and chromatin fractions of breast cancer cell lines representing the different subtypes of breast cancer; experiment performed twice with similar results. F Schematic representation of the generation of hormone independent cells (HI) from parental (P) MCF7 cells. G Quantification of ɣH2AX foci in MCF7 and HI cells, enriched for G2/M cell cycle (sampling for n = 363 cells in three technical replicates, unpaired two-tailed Wilcoxon test). H Western blot detection of IMPDH2 protein on cytosolic and chromatin fractions of MCF7 P and HI cells; experiment performed twice with similar results. Quantification of IMPDH2 nuclear signal (I) and representative images (scale bar 15 μm) (J) of immunohistochemical detection of IMPDH2 protein on non_TN and TN samples from a breast cancer tissue microarray (non-TN, n = 66; TN, n = 25; unpaired two-tailed Wilcoxon test). All box plots indicate the median value (central line), interquartile range IQR (box boundaries), and up to 1.5*IQR beyond the box boundaries (whiskers). Source data are provided as a Source Data file.
Overall, our results indicate that IMPDH2 is increased on chromatin in TNBC cell lines and patient-derived samples. Given that TNBC cells are more prone to accumulate high levels of DNA damage, we hypothesize that the chromatin-associated function of IMPDH2 may be related to the regulation of the DNA damage response.
Repression of IMPDH2 leads to DNA damage accumulation beyond replication stress
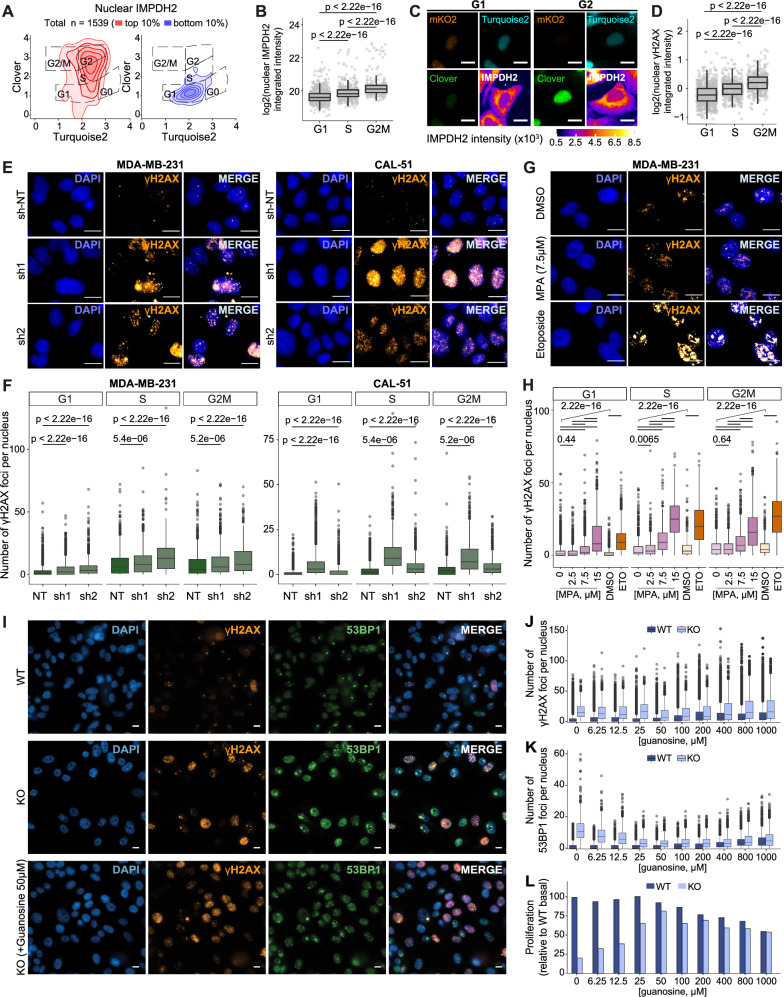
During the G2 phase of the cell cycle, cells check for faithful DNA replication during the S phase and correct any possible errors29. To unveil a possible connection between DNA damage repair and nuclear IMPDH2 requirements, we used an in-house optimized U2OS FUCCI system12 that allows cell cycle tracking and performed IMPDH2 HT-IF to quantify its subcellular distribution during cell cycle progression. We observed that, while the cytoplasmic amount of IMPDH2 changed slightly across the cell cycle (Supplementary Fig. 2A), IMPDH2 accumulated in the nucleus from late S to the end of the G2 phase (Fig. 2A–C), and cells also displayed increased levels of DNA damage, as shown by the quantification of γH2AX in HT-IF (Fig. 2D). To investigate whether the downregulation of IMPDH2 could potentially cause DNA damage accumulation, we utilized two shRNAs to reduce IMPDH2 expression in MDA-MB-231 and CAL-51 cells (Supplementary Fig. 2B). IMPDH2 downregulation partially decreased cell growth in both cell lines (Supplementary Fig. 2C, D). The ɣH2AX HT-IF analysis revealed a significant increase in DNA damage in IMPDH2 knock-down cells, particularly during the S to G2M phases of the cell cycle (Fig. 2E, F and Supplementary Fig. 2E). We then aimed to determine whether the catalytic activity of IMPDH2 was necessary to maintain DNA damage under control. To address this, we administered mycophenolic acid (MPA) - an IMPDH2 inhibitor - to MDA-MB-231 cells for 72 h, using concentrations of 2.5, 7.5, and 15 μM. MPA treatment caused a dose-dependent increase of IMPDH2 protein levels (Supplementary Fig. 2F) and reduced viability at the highest concentration used (15 μM) (Supplementary Fig. 2G). HT-IF showed that there was a significant increase in ɣH2AX signal in MPA treated cells already at 7.5 μM (Fig. 2G, H and Supplementary Fig. 2H) and 15 μM induced DNA damage levels comparable to etoposide -a Topoisomerase 2 (TOP2A) inhibitor that induces double strand breaks30,31 -suggesting that the enzymatic activity of IMPDH2 is required to keep DNA damage in check. We also detected increased levels of phosphorylated Ataxia Telangiectasia and Rad3-related protein (pATR), a master regulator of DDR, in cells treated with 7.5 µM MPA for 72 h, although no differences were detected in phosphorylated Cell Cycle Checkpoint Kinase 1 (pChk1), one of ATR downstream substrates32 (Supplementary Fig. 2I). To unravel whether IMPDH2 has a role in the control of DNA damage, we produced IMPDH2 knockout (KO) cells. The KO was achieved by targeting the IMPDH2 locus with the CRISPR-Cas9 system in MDA-MB-231 cells and confirmed by western blot analysis (Supplementary Fig. 2J). Total loss of IMPDH2 in MDA-MB-231 cells led to increased DNA damage, detected by accumulation of ɣH2AX, Tumor Protein P53 Binding Protein 1 (53BP1) (Fig. 2I) and pATR (Supplementary Fig. 2K), in line with previous results obtained by IMPDH2 partial loss (knock-down) or inhibition (MPA treatment). To formally assess the contribution of the catalytic activity of IMPDH2 to the DNA damage response, we reconstituted IMPDH2-KO cells with the wild-type version of IMPDH2 (WT) or with a catalytic dead mutant (CD) in which the active site residue Cys331 is mutated to Ala (C331A)33 (Supplementary Fig. 2L). The IMPDH2-CD reconstitution was defective in rescuing ɣH2AX accumulation as compared to the WT, indicating a role of the catalytic activity of IMPDH2 in the DNA damage response, which agreed with the results obtained by IMPDH2 inhibition with MPA (Supplementary Fig. 2M). IMPDH2 KO cells displayed proliferative defects (Supplementary Fig. 2N, O), as the depletion of IMPDH2 renders cells auxotrophic to guanosine34. Supplementing KO cells with 400 μM of guanosine34 partially recovered their proliferation, while it had a detrimental effect on IMPDH2 wild type (WT) MDA-MB-231, most probably due to a cell cycle defect induced by nucleotide imbalance35 (Supplementary Fig. 2N, O). By HT-IF, we observed that KO cells had increased nuclei area even if cultured with guanosine (Supplementary Fig. 2P, Q), in agreement with a G2-phase arrest, which was not fully rescued by guanosine supplementation (Supplementary Fig. 2R). We reasoned that the limited recovery of KO cells in the presence of guanosine supplementation might be due to an overaccumulation of DNA damage caused by the absence of IMPDH2, which goes beyond the restoration of guanosine levels. To distinguish between replication stress induced by nucleotide imbalance35 and double-strand breaks, we performed HT-IF of Replication Protein A 70 kDa subunit (RPA70) and ɣH2AX. RPA accumulates in response to replication stress and binds to single-stranded DNA (ssDNA), which is converted into DSBs if the damage cannot be promptly repaired36,37. Following guanosine withdrawal RPA foci increased over time in IMPDH2 KO cells (Supplementary Fig. 2S), suggesting a progressive increase of replication stress upon guanosine deprivation. In contrast, ɣH2AX foci were constantly high and only slightly increased at the latest time point (Supplementary Fig. 2T), indicating that lack of IMPDH2 induces DNA damage independent of replication stress. Furthermore, guanosine titration indicated that none of the concentrations tested was able to completely prevent ɣH2AX (Fig. 2I, J) and 53BP1 accumulation (Fig. 2I, K) or rescue proliferation (Fig. 2L), corroborating that IMPDH2 has a role in the control of DNA damage beyond guanosine synthesis.
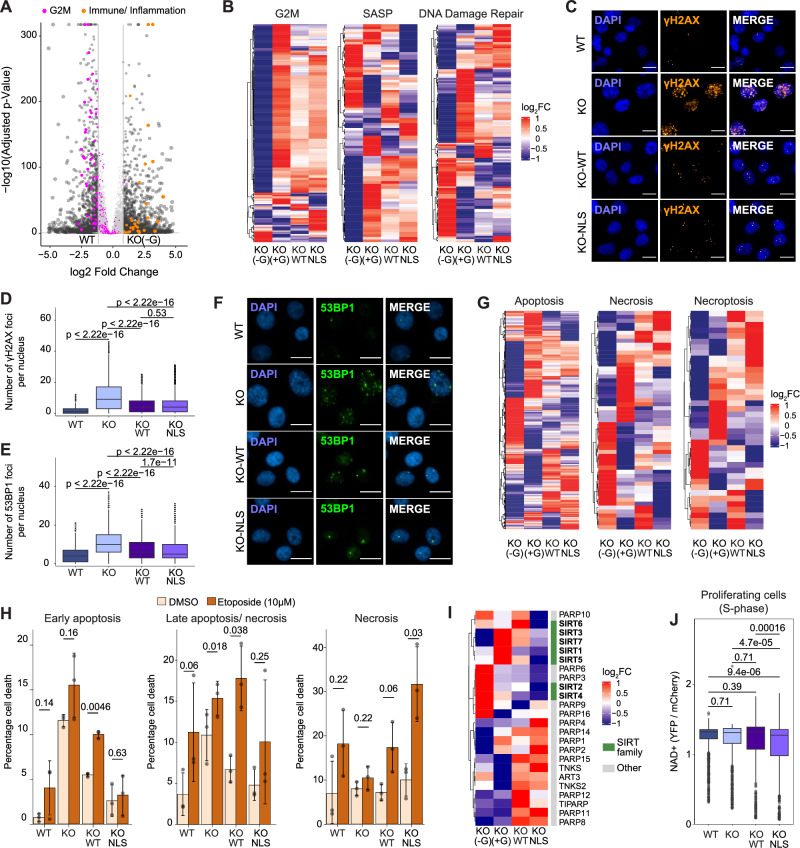
Fig. 2. IMPDH2 downregulation or pharmacological inhibition leads to DNA damage accumulation and G2/M arrest.
A Cell cycle profile of U2OS cells using the adapted FUCCI system, with the scaled density of nuclear IMPDH2 high (top 10%, left panel) and IMPDH2 low (bottom 10%, right panel) expressing cells. B Quantification of IMPDH2 nuclear integrated intensity across the different cell cycle phases (nG1 = 635, nS = 418, nG2M = 243; unpaired two-tailed Wilcoxon test). C Representative pictures of cells in G1 or G2M phases of the cell cycle (scale bar 25 μm). D Quantification of ɣH2AX nuclear integrated intensity across the different cell cycle phases (nG1 = 751, nS = 387, nG2M = 375; unpaired two-tailed Wilcoxon test). Representative pictures (E) and quantification (F) of ɣH2AX foci (ɣH2AX orange, DAPI blue; scale bar 15 μm) in MDA-MB-231 (NT, n = 6081; sh1, n = 5534; sh2, n = 2916) and CAL-51 (NT, n = 16171; sh1, n = 7916; sh2, n = 15196) cell lines 72 h after knockdown of IMPDH2; unpaired two-tailed Wilcoxon test. Representative pictures (G) and quantification (H) of ɣH2AX foci (ɣH2AX orange, DAPI blue; scale bar 15 μm) in MDA-MB-231 cells treated with increasing amounts of MPA (72 h) or etoposide control (3 h) (MPA 0, n = 6086; MPA 2.5 μM, n = 6005, MPA 7.5 μM, n = 5018, MPA 15 μM, n = 3005; DMSO, n = 2996; ETO, n = 5230; unpaired two-tailed Wilcoxon test). Guanosine titration in MDA-MB 231 WT and IMPDH2 KO cells (WT: n0 = 10683, n6.25 = 10079, n12.5 = 10368, n25 = 10784, n50 = 9917, n100 = 9275, n200 = 8232, n400 = 7834, n800 = 7326, n1000 = 5874; KO: n0 = 2161, n6.25 = 3444, n12.5 = 4147, n25 = 7014, n50 = 8741, n100 = 7009, n200 = 7446, n400 = 6388, n800 = 6268, n1000 = 5807) with representative pictures (I) (DAPI blue, ɣH2AX orange, 53BP1 green; scale bar 10 μm) and quantification of ɣH2AX foci (J), 53BP1 foci (K) and nuclei counting (L) with the percentage relative to WT nuclei count without guanosine. All box plots indicate the median value (central line), interquartile range IQR (box boundaries) and up to 1.5*IQR beyond the box boundaries (whiskers). Source data are provided as a Source Data file.
Taken together, these results indicate that IMPDH2 nuclear localization increases during the G2/M phase of the cell cycle in which the DNA damage checkpoint operates, and its catalytic activity is involved in keeping DNA damage under control.
IMPDH2 localizes in the nucleus in response to DNA damage
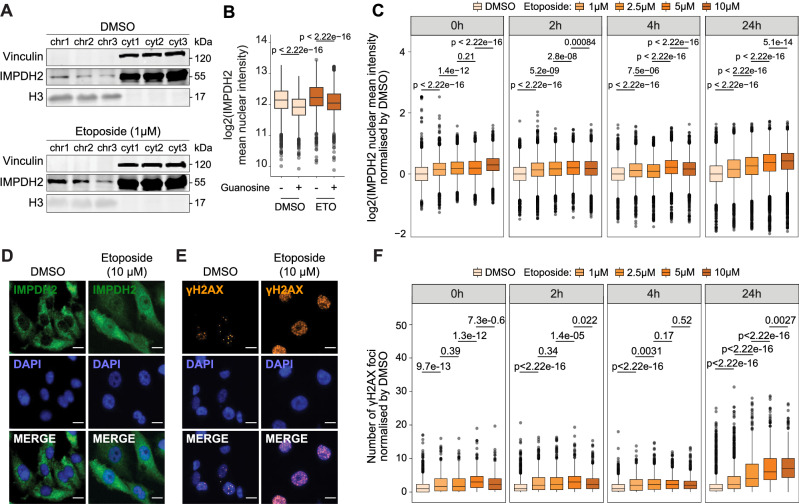
We recently published a dataset of chromatome-MS and metabolomics profiling of U2OS cells, which were treated with etoposide (1 μM, 3 h) to induce DNA damage, and released in etoposide free media to allow DNA damage repair12. By analyzing the data focusing on the enrichment of chromatin-associated metabolic enzymes, we observed that IMPDH2 was the most enriched metabolic enzyme on chromatin, and overall, one of the most enriched proteins on chromatin, following 24 h etoposide release (Supplementary Fig. 3A and Supplementary Data 4). Other metabolic enzymes of the purine pathway were also detected on chromatin after 24 h etoposide release, in particular the Trifunctional Purine Biosynthetic Protein Adenosine-3 (GART) and the Nucleoside Diphosphate Kinase family NME proteins - NME1, NME2, NME3- which are nucleoside diphosphate kinases (NDPKs) that catalyze the transfer of a ɣ-phosphate group to nucleoside diphosphates, mainly GDP38, suggesting a local increase of GTP availability in response to DNA damage. Metabolomics profiling showed a progressive increase of triphosphate nucleosides and a concomitant decrease in the monophosphate-nucleoside forms over time after release (Supplementary Fig. 3B). Of note, inosine monophosphate (IMP), the substrate of IMPDH2, decreased over release time while the levels of guanosine triphosphate increased, suggesting de novo biosynthesis of guanosine nucleotides. We performed a western blot analysis of the chromatome using MDA-MB-231 cells, replicating the conditions of the U2OS chromatome-MS and metabolomics experiments. Results confirmed that IMPDH2 increased on chromatin in etoposide-treated MDA-MB-231 cells after 24 h of release (Fig. 3A). We hypothesized that IMPDH2 may act as a nucleotide sensor that localizes to the nucleus due to the increasing nucleotide demand generated by the DNA damage repair. Supporting this hypothesis, guanosine supplementation reduced IMPDH2 nuclear levels in DMSO-treated cells and decreased IMPDH2 nuclear localization in etoposide-treated cells, as quantified by HT-IF (Fig. 3B). To investigate the role of IMPDH2 in response to DNA damage induction, we challenged MDA-MB-231 cells with increasing concentration of etoposide to analyze with HT-IF the dynamics of ɣH2AX accumulation and IMPDH2 nuclear recruitment either immediately after the treatment (0 h) or following 2, 4, or 24 h release. Results showed a dose-dependent increase of IMPDH2 in the nucleus following etoposide treatment (Fig. 3C, D), which persisted until 24 h post-release. Similar results were observed when treating CAL-51 cells with increasing concentrations of etoposide (Supplementary Fig. 3C). ɣH2AX followed the same behavior as nuclear IMPDH2, remaining elevated in MDA-MB-231 cells treated with high concentrations of etoposide even after 24 h of release (Fig. 3E, F), indicating a failure to repair damage. Interestingly, both IMPDH2 and ɣH2AX showed a similar dynamic of accumulation during the S/G2M phases of the cell cycle (Supplementary Fig. 3D, E), suggesting that increasing DNA damage stimulates IMPDH2 nuclear localization. Finally, we inhibited ATR and Ataxia Telangiectasia Mutated (ATM) to understand whether their activity in the DNA damage response signal transduction was required for IMPDH2 nuclear translocation. We observed that none of these conditions impeded IMPDH2 nuclear increase upon etoposide treatment. However, ATR inhibition, (but not ATM), completely abrogated ɣH2AX increase upon etoposide treatment, suggesting that the nuclear increase of IMPDH2 depends on DNA damage rather than on ɣH2AX increase (Supplementary Fig. 3F, G).
Fig. 3. IMPDH2 increases on chromatin in a DNA damage-dependent manner.
A Western blot of MDA-MB-231 cells cytosolic (cyt) and chromatin (chr) fractions in the control condition (DMSO) and after 3 h of etoposide treatment and 24 h of release. As cytosolic and nuclear markers, Vinculin and H3 were used, respectively (n = 3). B Quantification of nuclear IMPDH2 signal intensity in MDA-MB-231 cells treated with DMSO or etoposide in the presence (+G) or absence of guanosine supplementation (50 μM); 1 h post-supplementation with guanosine cells were exposed to 10 µM etoposide for 3 h followed by a 24 h recovery period (DMSO: nDMSO = 3011, nGuanosine = 6849; Etoposide: nDMSO = 3431, nGuanosine = 4259, outliers removed, 3 SD; unpaired two-tailed Wilcoxon test). Immunofluorescence of nuclear IMPDH2 and mean intensity quantification normalized by DMSO in MDA-MB-231 cells (DMSO, n0h = 7700; n2h = 6999; n4h = 7372; n24h = 13846; 1 μM, n0h = 7018; n2h = 6816; n4h = 7362; n24h = 10647; 2.5 μM, n0h = 6520; n2h = 7310; n4h = 8409; n24h = 9746; 5 μM, n0h = 6397; n2h = 7029; n4h = 7685; n24h = 8412; 10 μM, n0h = 6635; n2h = 7019; n4h = 6864; n24h = 8770) treated with increasing concentrations of etoposide after treatment or 2, 4 and 24h post-release (n = 3 biological replicates, unpaired two-tailed Wilcoxon test) (C), representative pictures of nuclear IMPDH2 (green) and DAPI (blue) in DMSO control condition and 10 μM etoposide treatment after 24 h of release; non-confocal mode, scale bar 15μm (D). Immunofluorescence of nuclear ɣH2AX, representative pictures (orange) and DAPI (blue) in DMSO control condition and 10 μM etoposide treatment after 24 h of release; non-confocal mode, scale bar 15 μm (E) and foci quantification in MDA-MB-231 cells (DMSO, n0h = 884; n2h = 1070; n4h = 1323; n24h = 2174; 1 μM, n0h = 642; n2h = 1322; n4h = 1270; n24h = 1927; 2.5 μM, n0h = 815; n2h = 1322; n4h = 1532; n24h = 1222; 5 μM, n0h = 651; n2h = 1382; n4h = 2212; n24h = 966; 10 μM, n0h = 657; n2h = 1140; n4h = 1182; n24h = 1226) treated with increasing concentrations of etoposide after treatment or 2, 4 and 24h post-release (n = 3 biological replicates, unpaired two-tailed Wilcoxon test) (F). All box plots indicate the median value (central line), interquartile range IQR (box boundaries), and up to 1.5*IQR beyond the box boundaries (whiskers). Source data are provided as a Source Data file.
These data indicated that IMPDH2 localizes to the nucleus in a manner dependent on DNA damage, with this localization being contingent on guanosine demand.
IMPDH2 interacts with PARP1 on chromatin to quench the DNA damage response
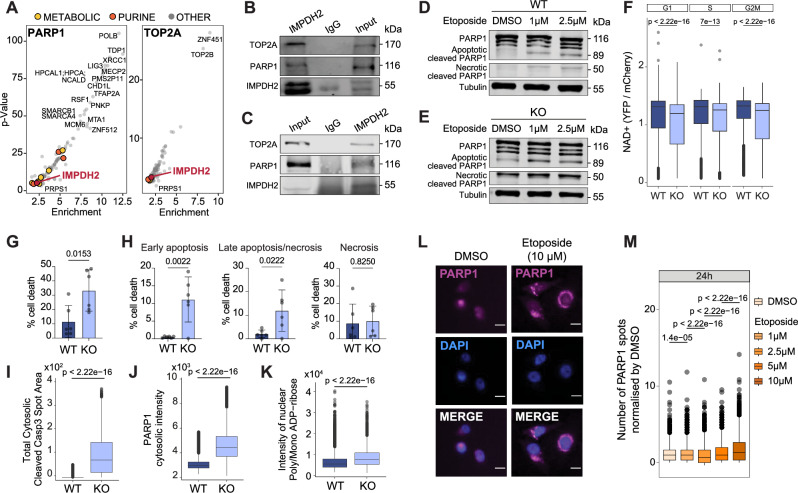
Next, we investigated how IMPDH2 might regulate DNA damage levels. In the OpenCell resource39, IMPDH2 scored as a common interactor between PARP1 and TOP2A (Fig. 4A and Supplementary Data 5). This evidence strengthened our hypothesis of a possible role of chromatin-associated IMPDH2 in the DNA damage response, given that PARP1 and TOP2A are chromatin master regulators of the DNA damage repair process. We performed an IMPDH2 pull-down in MDA-MB-231 chromatome extracts and confirmed that IMPDH2 can interact with PARP1 and TOP2A on chromatin (Fig. 4B). IMPDH2 pull-down in nuclear extracts from the Hs 578T TNBC cell line further validated this interaction (Fig. 4C). DNA damage response is a dynamic process; the initial stage involves the activation of the DNA damage cascade, followed by the participation of intermediate factors involved in the physical repair of damaged DNA, and ends with the late factors which stop the signaling cascade returning the cell into the normal state. PARP1 is one of the earliest factors in the DNA damage repair process, and its recruitment to the DNA damage foci occurs within minutes of the damage induction40. During the process of DNA damage, PARP1 is the primary consumer of energy20. The poly ADP-ribosylation (PARylation) reaction, which is initiated by PARP1 to recruit downstream DNA damage repair factors, requires NAD+ as a substrate. The degree of PARP1 activation serves as a regulatory mechanism that modulates the energy expenditure during the DNA damage repair process. The strength of the DNA damage determines the extent of PARP activation, which in turn affects the availability of NAD+41. When NAD+ is depleted due to extensive DNA damage the cell undergoes necrosis and PARP1 is cleaved by cathepsins into fragments of approximately 50 kDa. In contrast, when apoptosis occurs, PARP1 is inactivated by Caspase 3 cleavage, resulting in the generation of a small fragment of approximately 24 kDa that remains on chromatin and a larger fragment of 89 kDa that translocates to the cytosol. The fragmentation and cytoplasmic translocation of PARP1 are hallmarks of DNA damage-mediated cellular apoptosis16,42. We characterized the PARP1 fragmentation profile of either MDA-MB-231 WT or KO cells 24 h post etoposide release (either 1 or 2.5 μM). WT MDA-MB-231 cells exhibited a modest, dose-dependent increase in the PARP1 apoptotic fragment (89 kDa) with minimal necrosis. This indicated that the cells could regulate their NAD+ levels and either survived or underwent apoptosis. KO MDA-MB-231 cells, however, displayed clear apoptotic (89 kDa) and necrotic (50 kDa) fragments even in the absence of etoposide, which increased upon etoposide treatment (Fig. 4D, E). These data suggest that the lack of IMPDH2 provokes a lethal loss of the control of cellular NAD+ levels. Indeed, using a nuclear NAD+ sensor43, we observed that IMPDH2 KO cells displayed a reduction in nuclear NAD+, in support of our hypothesis (Fig. 4F). Furthermore, IMPDH2 KO cells in the absence of guanosine supplementation showed increased cell death when compared to WT cells (Fig. 4G) both in early apoptosis and late apoptosis/necrosis, as assessed by AnnexinV/DAPI staining (Fig. 4H) with increased levels of cytosolic cleaved Caspase-3 and PARP1 (Fig. 4I, J). In addition, IMPDH2 loss led to increased nuclear poly/mono-ADP-ribose levels, reflecting increased PARP1 activity in these cells (Fig. 4K). In the context of etoposide dose-response settings, we observed that PARP1 cytoplasmic translocation increased considerably 24 h after etoposide 10 μM treatment in WT MDA-MB-231 (Fig. 4L, M and Supplementary Fig. 4A) and CAL51 cells (Supplementary Fig. 4B), which also induced the highest proliferative defect (Supplementary Fig. 4C, D). This was concomitant with the maximum increase of nuclear IMPDH2 (Fig. 3C, D), suggesting a causal relationship between PARP1 cytoplasmic translocation and IMPDH2 nuclear localization. The enzymatic reaction performed by IMPDH2 reduces NAD+, the substrate required for PARP1 activation41. We hypothesized that the binding of IMPDH2 to PARP1 on chromatin might create a local NAD+-deprived environment that stops PARP1 activity, preventing cellular energy deprivation. We reasoned that if IMPDH2 controls PARP1 activity, IMPDH2 inhibition may expand the activation capacity of PARP1, thus decreasing cell sensitivity to PARP1 inhibitors. Thus, we treated MDA-MB-231 cells with MPA (IMPDH2 inhibitor) and Olaparib (PARP1 inhibitor) either alone or in combination. We observed that MPA and Olaparib behaved antagonistically; cells were more resistant to Olaparib treatment when MPA was used in combination (Supplementary Fig. 4E), further corroborating our hypothesis. Importantly, the combination of MPA with Carboplatin, a DNA cross-linking agent that does not directly target PARP1, did not show the same effect (Supplementary Fig. 4F).
Fig. 4. IMPDH2 interacts with PARP1 on chromatin for fine-tuned modulation of DDR.
A IMPDH2 shown in red, scored as an interactor of PARP1 and TOP2A DNA damage effectors (from OpenCell resource). Purine pathway enzymes are shown in orange, rest of metabolic enzymes in yellow. B Western blot of the chromatin fraction from MDA-MB-231 cells of IMPDH2 and IgG immunoprecipitation showing PARP1 and TOP2A pull-down after 3 h of 1 μM etoposide treatment and 24 h release, experiment performed twice with similar results. C Western blot of the nuclear-enriched fraction from Hs 578T cells of IMPDH2 and IgG immunoprecipitation showing IMPDH2, PARP1 and TOP2A, experiment performed twice with similar results. Western blot of MDA-MB-231 cells WT (D) and KO (E) showing full PARP1 and apoptotic and necrotic PARP1 cleaved, experiment performed twice; tubulin was used as a loading control. F Quantification of nuclear NAD+ as YFP/mCherry ratio across cell cycle phases in WT and KO cells cultured in the presence of guanosine 400 μM (WT: nG1 = 2316, nS = 1348, nG2/M = 1761; KO: nG1 = 2747, nS = 1359, nG2/M = 1282; outliers removed, 3 SD; unpaired two-tailed Wilcoxon test). Measurement of total cell death (G) and early apoptosis, late apoptosis/necrosis (H) in MDA-MB-231 WT and KO cells without guanosine supplementation for 96 h (WT: n = 6; KO: n = 6; unpaired two-tailed t-test); mean values +/− SD. Quantification of total cytosolic cleaved Casp3 Spot Area (I) (nWT= 79460, nKO= 7453; outliers removed, 3 SD; unpaired two-tailed Wilcoxon test), PARP1 cytosolic intensity (J) (nWT= 41640 nKO= 2297; outliers removed, 3 SD; unpaired two-tailed Wilcoxon test) and nuclear poly/mono ADP-ribose signal intensity (K) (nWT = 32600, nKO = 364; unpaired two-tailed Wilcoxon test) in MDA-MB 231 WT and KO cells without guanosine supplementation for 96 h. Immunofluorescence of PARP1 representative pictures (magenta) and DAPI (blue) in DMSO control and 10 μM etoposide treatment after 24 h of release; non-confocal mode, scale bar 15μm (L) and number of PARP1 spots quantification in MDA-MB-231 cells (DMSO, n = 3748; n1μM = 3165; n2.5 μM = 3763; n5μM = 2898; n10μM = 3740; unpaired two-tailed Wilcoxon test) (M). All box plots indicate the median value (central line), interquartile range IQR (box boundaries), and up to 1.5*IQR beyond the box boundaries (whiskers). Source data are provided as a Source Data file.
Taken together, these results suggest that nuclear IMPDH2 controls PARP1 activity, preventing cell death.
Forcing nuclear IMPDH2 localization impairs nuclear NAD+ balance
To ascertain the contribution of IMPDH2 subcellular localization to DNA damage repair and cell fitness, we reconstituted KO MDA-MB-231 cells with either the WT form of IMPDH2 (KO-WT) or its nuclear-restricted form tagged with a 3x nuclear localization sequence (KO-NLS) (Supplementary Fig. 5A). Next, we did a transcriptomic analysis, which compared WT and KO cells, in the presence (+G) or absence (-G) of guanosine supplementation, with KO-WT and KO-NLS. The replicates demonstrated robust clustering, as evidenced by the PCA plot (Supplementary Fig. 5B). The greatest segregation was observed between WT cells and KO cells without guanosine supplementation, which exhibited a considerable number of differentially regulated genes (Fig. 5A, Supplementary Data 6). Gene ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome analyses once again showed that KO cells in the absence of guanosine exhibited proliferation defects (Supplementary Fig. 5C–E). Furthermore, Differential Gene Expression (DGE) analysis revealed that KO cells with guanosine supplementation as well as both KO-WT and KO-NLS reconstitutions were able to partially restore proliferation (Fig. 5B, G2M, left panel). This indicated that IMPDH2 catalytic activity is equally efficient in sustaining guanosine synthesis independently of the enzyme compartmentalization, as confirmed by the proliferation assays (Supplementary Fig. 5F, G). The lack of a complete proliferative recovery in the presence of guanosine was not due to downregulation of nucleoside transporters in the KO cells (Supplementary Fig. 5H), indicating that KO cells underwent a phenotypic rewiring beyond the imbalance of nucleotides. KO cells exhibited increased immune-related signaling (Supplementary Fig. 5D), which was indicative of a senescence-associated secretory phenotype (SASP). DGE analysis showed that the expression of SASP associated genes was strongly impacted by the loss of IMPDH2 and only partially recovered when KO cells were cultured in presence of guanosine or reconstituted with WT- or NLS-IMPDH2 (Fig. 5B, SASP, middle panel). Moreover, IMPDH2 KO dramatically altered the expression of genes involved in DNA damage signaling, and neither guanosine supplementation nor reconstitution with either the WT or NLS form of IMPDH2 was able to restore the expression of these genes to baseline levels (Fig. 5B, DNA damage repair, right panel). The quantification of ɣH2AX and 53BP1 in KO-WT and KO-NLS cells confirmed the partial recovery of the DNA damage induced by IMPDH2 loss (Fig. 5C–F). Consequently, we focused on cell death signatures to ascertain whether we could identify differences between conditions. DGE analysis of apoptosis, necrosis and necroptosis showed partial rescue in guanosine-treated cells and cells reconstituted with WT or NLS-IMPDH2. Moreover, each of the conditions showed a different gene expression profile for each of the cell death signatures analyzed (Fig. 5G). With AnnexinV/DAPI staining and FACS analysis we observed that neither WT nor NLS reconstitution was able to completely rescue cell death. Moreover, after two days of 10 µM etoposide treatment such differences were exacerbated and while WT reconstituted cells exhibited a pro-apoptotic phenotype, NLS reconstituted cells were more prone to necrosis, suggesting a state of enhanced energy deprivation (Fig. 5H). Therefore, we sought to identify alterations in NAD+ metabolism that could explain such energy unbalance. First, we tested whether KO-NLS cells would be more sensitive to the Nicotinamide Phosphoribosyltransferase (NAMPT) inhibitor FK86644, which blocks the NAD+ salvage pathway. Instead, we observed that KO-WT cells were more sensitive to NAMPT inhibition than KO-NLS cells (Supplementary Fig. 5I). By examining NAMPT expression in our RNAseq data, we realized that KO-WT cells express lower levels of NAMPT (Supplementary Fig. 5J), which could be the reason for their hypersensitivity compared to KO-NLS cells. Therefore, we analyzed the expression of NAD+-consuming enzymes and found that NLS cells considerably downregulated the expression of the sirtuin family (SIRT1-7) (Fig. 5I, Supplementary Data 14), which together with the PARP family are the major NAD+ consumers45. These data may indicate that forcing IMPDH2 nuclear localization requires cells to limit nuclear NAD+ consumption by other enzymes to avoid nuclear energy depletion. Indeed, using a nuclear NAD+ sensor43, we observed that KO-NLS cells had lower nuclear NAD+ basal levels than the other conditions, supporting our hypothesis (Fig. 5J).
Fig. 5. Forcing nuclear IMPDH2 localization impacts on the transcriptome profile and nuclear NAD+ balance.
A Volcano plot of differentially expressed genes in KO-G vs WT condition (−1>log2FC > 1, FDR < 0.05); for differential expression analysis, the p-value was calculated by Wald’s Test and adjusted for multiple hypothesis by Benjamini-Hochberg criteria (FDR). Highlighted in magenta genes related to hallmark G2M checkpoint signature (MSigDB, M5901), in ochre to immune/inflammation pathways (KEGG hsa04672, hsa05323, hsa04940, hsa05332, hsa05330, hsa04658, hsa05310, hsa05150, hsa05320, hsa05321). B Heatmap of differentially expressed genes relative to WT from G2M checkpoint (Supplementary Data 7), SASP (Supplementary Data 8), and DNA repair hallmark (Supplementary Data 9), extracted from msigdbr R package. Representative pictures (C) and quantification (D) of ɣH2AX foci (ɣH2AX orange, DAPI blue; scale bar 15 μm) in MDA-MB-231 WT, KO, KO-WT and KO-NLS without guanosine supplementation for 96 h (WT, n = 28604; KO, n = 2707; KO-WT, n = 13572; KO-NLS, n = 14449; unpaired two-tailed Wilcoxon test). Quantification (E) and representative pictures (53BP1 green, DAPI blue; scale bar 15 μm) (F) of 53BP1 foci in WT, KO, KO-WT and KO-NLS without guanosine supplementation for 96 h (WT, n = 7642; KO, n = 1983; KO-WT, n = 4707; KO-NLS, n = 6548; unpaired two-tailed Wilcoxon test). G Heatmap of differentially expressed genes relative to WT from Apoptotic (GO:0006915, Supplementary Data 10), Necrosis (msigdbr, Supplementary Data 11), and Necroptotic Process (GO:0070266, Supplementary Data 12), GO terms from GO.db R package. H Cell death measurements in WT, KO, KO-WT, and KO-NLS treated with DMSO or etoposide 10 μM for 48 h (n = 3 biological replicates, paired two-tailed t-test); mean values +/− SD. I Heatmap of differentially expressed genes relative to WT from the NAD + ADP-ribosyltransferase activity (GO:0003950) expanded to include SIRT1-7 family (Supplementary Data 13). J Quantification of nuclear NAD+ as YFP/mCherry ratio for WT, KO with guanosine 50 μM supplementation, KO-WT and KO-NLS in S-phase enriched cells (WT, n = 1149; KO, n = 1069; KO-WT, n = 578; KO-NLS, n = 645, unpaired two-tailed Wilcoxon test); outliers removed to improve visualization. All box plots indicate the median value (central line), interquartile range IQR (box boundaries), and up to 1.5*IQR beyond the box boundaries (whiskers). Source data are provided as a Source Data file.
The results of our transcriptomic analysis indicated that the reconstitution of IMPDH2 is not sufficient to rescue the phenotype induced by IMPDH2 KO. This suggests that an imbalance in IMPDH2 expression levels (Supplementary Fig. 5K) or localization is detrimental to cell fitness, the DNA damage response, and overall NAD+ usage.
Forcing nuclear IMPDH2 localization leads to PARP1 apoptotic cleavage
Given the observed differences in NAD+ balancing in KO-WT and KO-NLS cells, we sought to explore the differential response of these reconstituted cells in the presence of DNA damage. First, we applied a mild etoposide treatment (2.5 µM) for 96 h and observed an increased PARP1 apoptotic cleaved fragment (89 kDa) in WT, KO-WT, and KO-NLS cells with minimal necrosis (PARP1 necrotic fragment, 50 kDa), indicating that they can efficiently trigger apoptosis in presence of DNA damage. However, KO-NLS reconstituted cells showed a visible PARP1 apoptotic fragment even in the absence of etoposide (Fig. 6A). Thus, we performed a shorter treatment with etoposide (5 µM, 3 h) and released the cells for 24 h. We observed that in this situation WT and KO-WT cells proficiently recovered from the DNA damage and 24 h after etoposide release the PARP1 apoptotic cleaved fragment was practically absent. On the contrary, KO-NLS cells showed apoptotic PARP1 cleavage even in the absence of etoposide that persisted 24 h post etoposide release (Fig. 6B). Therefore, we performed HT-IF to quantify PARP1 cytoplasmic translocation in KO-NLS and KO-WT. We observed that the KO-NLS cells exhibited augmented PARP1 cytosolic localization even in the absence of etoposide (Fig. 6C, D), in agreement with the observed PARP1 cleaved baseline status of these cells observed by western blot (Fig. 6A, B). Etoposide treatment augmented PARP1 cytosolic localization in all the tested conditions, with a higher increase observed when IMPDH2 was forced into the nucleus (Fig. 6C, D), which is consistent with decreased NAD+ availability. Similarly, KO-NLS cells exhibited elevated cleaved Caspase 3 signal in the absence of etoposide, when compared to KO-WT cells. Etoposide treatment resulted in an increase in cleaved Caspase 3 in both reconstituted conditions. However, the degree of increase was higher in KO-NLS cells (Fig. 6E, F). We thus sought to ascertain whether the augmented cytosolic PARP1 and cleaved Caspase 3 levels observed in the KO-NLS cells were attributable to impaired PARP1 functionality. To this end, we performed nuclear poly/mono-ADP-ribose quantification via HT-IF. We observed a reduction in poly/mono-ADP-ribose levels in the nucleus of KO-NLS cells compared to KO-WT cells (Fig. 6G), which is consistent with a role for nuclear IMPDH2 in reducing PARP1 activity. We reasoned that if nuclear IMPDH2 limits PARP1 activity by restricting NAD+ availability, NAD+ supplementation might be able to rescue poly/mono-ADP-ribose levels in these cells. Corroborating the functional connection between nuclear IMPDH2, the nuclear NAD+ pool availability and PARP1, NAD+ supplementation partially rescued nuclear poly/mono-ADP-ribose levels in KO-NLS cells (Fig. 6H). Furthermore, nuclear NAD+ levels were quantified in KO-WT and KO-NLS cells following treatment with etoposide at an early (2 h) and late (24 h) time point. In KO-WT cells, NAD+ levels decreased immediately after etoposide treatment, indicating proper PARP1 functionality, and recovered by 24 h after treatment. On the contrary, KO-NLS cells showed lower basal levels of nuclear NAD+ and did not further decrease after etoposide treatment, suggesting that when IMPDH2 is forced into the nucleus PARP1 is unable to further consume NAD+ in response to DNA damage induction (Fig. 6I, J), in agreement with the observed reduced PARP1 activity.
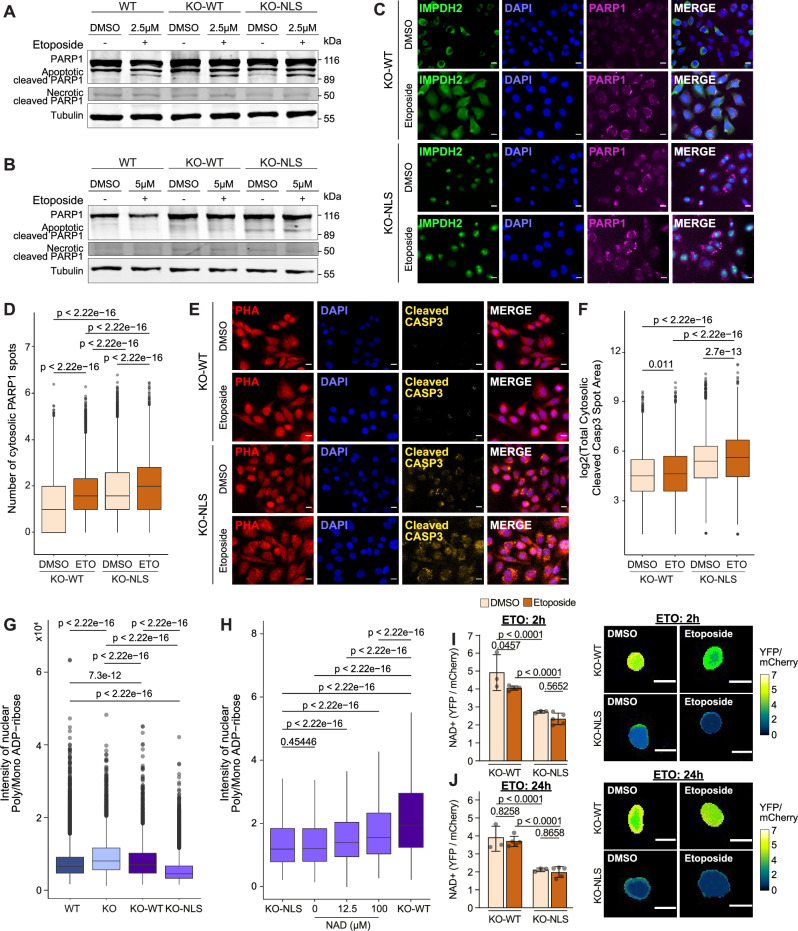
Fig. 6. Forcing nuclear IMPDH2 localization leads to a reduction of nuclear poly/mono ADP-ribose levels that can be rescued by NAD+ supplementation.
A, B Western blot analysis of PARP1 fragmentation in WT, KO-WT, and KO-NLS treated with DMSO or etoposide without release and after 24 h release (experiments performed twice). Tubulin is used as loading control. Immunofluorescence pictures of IMPDH2 (green), DAPI (blue), and PARP1 (magenta) from DMSO and 10 μM 24 h etoposide treatment, scale bar 15μm (C), and quantification of cytosolic PARP1 spots in KO-WT and KO-NLS cells normalized by DMSO (DMSO, nKO-WT = 75011; nKO-NLS = 125677, n10μM KO-WT = 45449; n10μM KO-NLS = 75939). Unpaired two-tailed Wilcoxon test, (n = 3 and 3 technical replicates per condition) (D). Immunofluorescence pictures of Phalloidin (red), cleaved Caspase3 (yellow) and DAPI (blue) from DMSO and 10μM 24 h etoposide treatment, scale bar 15 μm (E), and quantification of total cytosolic cleaved Caspase spot area in KO-WT and KO-NLS cells (DMSO, nKO-WT = 50568; nKO-NLS = 82455, n10μM KO-WT = 447736; n10μM KO-NLS = 80561). Unpaired two-tailed Wilcoxon test, (n = 3 and 3 technical replicates per condition) (F). G Quantification of nuclear poly/mono ADP-ribose signal intensity in MDA-MB-231 WT, KO, KO-WT, KO-NLS cells in the absence of guanosine supplementation for 96 h (nWT = 32590, nKO= 4575, nKO-WT = 3798, nKO-NLS = 15868) (n = 3 biological replicates, unpaired two-tailed Wilcoxon test). H Quantification of nuclear poly/mono ADP-ribose signal intensity for a titration with increasing NAD+ concentrations in KO-WT and KO-NLS cells (nKO-NLS = 15868, n0 = 10376, n12.25 = 2194, n100 = 2965, nKO-WT = 3798; unpaired two-tailed Wilcoxon test); outliers removed, and constantly added to improve visualization. Quantification of intranuclear NAD+ as YFP/mCherry ratio for KO-WT and KO-NLS cells with representative images (scale bar 15μm) after DMSO or 10 μM etoposide treatment for 2 h (I) and 24 h (J). Two-way Anova with either Tukey’s or Sidak multiple comparisons depending on intra or inter-comparison. For each timepoint 3 biological replicates were performed for the DMSO condition and 5 biological replicates for the ETO condition. All box plots indicate median value (central line), interquartile range IQR (box boundaries), and up to 1.5*IQR beyond the box boundaries (whiskers). Source data are provided as a Source Data file.
The results of this study indicate that nuclear IMPDH2 is necessary for the proper functioning of the DNA damage response. However, increasing IMPDH2 nuclear levels results in a reduction of NAD+ levels, which in turn affects the PARP1 activity and predisposes cells to death.
Discussion
The DNA damage response is a critical cellular process in which cells are placed under severe metabolic stress. The consumption of NAD+ by PARP1 activation can lead to cell death if not kept under control. However, how PARP1 activation is regulated remains largely unknown, and a better understanding of this process may lead to the improvement of treatment strategies based on PARP1 inhibition, such as those for stroke46,47, Alzheimer’s disease48,49, or cancer14,50.
Our study shows that the metabolic enzyme IMPDH2 localizes to the nucleus upon guanosine demand in a DNA damage-dependent manner. IMPDH2 is upregulated in a variety of tumor types51–54, drives aberrant nucleolar activity in a model of glioblastoma55, and promotes tumor progression and metastasis in prostate cancer56. Furthermore, the presence of IMPDH2 has been detected in the nucleus of Drosophila cells after DNA replication or under oxidative stress, where it acts as a transcriptional repressor57. However, its nuclear-associated functionality in human cells remained largely unknown. Here we show that nuclear IMPDH2 is required for cells to balance their response to DNA damage by keeping nuclear NAD+ availability and PARP1 activation under control.
First, we identified that IMPDH2 is enriched on chromatin in TNBC cell lines, TNBC patient samples, and in advanced breast cancers (Fig. 1), conditions typically characterized by high levels of DNA damage23,24,58. Furthermore, we demonstrated that IMPDH2 accumulates in the nucleus during the S phase of the cell cycle, consistent with a role in controlling DNA damage after DNA replication, and that its repression leads to accumulation of DNA damage and proliferative defects that persist despite guanosine supplementation (Fig. 2). In addition, we showed that IMPDH2 localizes to the nucleus in a manner that is dependent on DNA damage and guanosine demand (Fig. 3). On chromatin, IMPDH2 interacts with PARP1 (Fig. 4), and the highest IMPDH2 nuclear localization coincides with PARP1 cleavage and cytoplasmic translocation. These results suggest that nuclear IMPDH2 can control cell fate during DNA damage by interacting with PARP1 and regulating its activation levels. The localization and expression levels of IMPDH2 are relevant for cell fitness, DNA damage repair, and the balance of nuclear NAD+ levels (Fig. 5). Indeed, by forcing IMPDH2 into the nucleus, we observed that even in the absence of exogenous DNA damage, PARP1 is cleaved and translocated to the cytoplasm as a result of decreased nuclear NAD+ levels (Fig. 6).
Overall, this study breaks new ground by demonstrating that the presence of a metabolic enzyme in the nucleus can limit the availability of local substrates and thus influence cell fate. Given the relevance of PARP1 in the treatment of various pathologies, it is possible that the presence of nuclear IMPDH2 could be used as a biomarker to stratify patients who will respond better or worse to PARP1 inhibition. Moreover, the involvement of nuclear IMPDH2 in the regulation of the DNA damage response may extend beyond the regulation of PARP1 activity. Indeed, nuclear IMPDH2 also interacts with TOP2A (Fig. 4A–C), which is another master regulator of chromatin functions and the DNA damage response. Given the catalytic activity of IMPDH2, which favors GTP production at the expense of ATP, and the fact that TOP2A requires ATP for its activity, it is plausible that this interaction tunes TOP2A activity in a manner analogous to what we have demonstrated here for PARP1.
Methods
Cell culture
MCF7 (ATCC #HTB-22), T47D (ATCC #HTB-133), BT-474 (ATCC #HTB-20), SK-BR-3 (ATCC #HTB-30), Hs 578T (ATCC #HTB-126), BT-549 (ATCC #HTB-122), MDA-MB-231 (ATCC #HTB-26), CAL-51 (DSMZ #ACC 302), MDA-MB-468 (ATCC #HTB-132) and U-2OS (ATCC; #HTB-96) were cultured in DMEM media (Gibco; #11966025) supplemented with 10% Fetal Bovine Serum (FBS, Gibco; #10270106) and 1% penicillin/streptomycin (Gibco; #15140122) at 37 °C and 5% CO2. For MCF 10A (ATCC #CRL-10317), DMEM/F12 media (Gibco; #11320-033) was used supplemented with 5% Horse Serum, 20 ng/mL EGF, 0.5 g/mL hydrocortisone, 10 g/mL insulin, 100 ng/μL cholera toxin and 1% penicillin/streptomycin. To obtain the MCF7 hormone-independent cell line, MCF7 cells were cultured in hormone-deprived media for at least 4 weeks. Specifically, were cultured in phenol-red free media (Opti-MEM™ I Reduced Serum Medium, no phenol red, Thermo Scientific, #11058021) and charcoal FBS (FBS, Charcoal Stripped, sterile-filtered, suitable for cell culture, Sigma-Aldrich, #F6765-500mL). Cells were trypsinized using Trypsin-EDTA (Trypsin-EDTA (0.25%), phenol red, Thermo Scientific, #25200072). Cells used in this manuscript arise from the CRG-PRBB cell line collection and have been previously authenticated. Mycoplasma contamination was ruled out by qPCR every two weeks. All cell lines used were of the female sex.
Compounds
Mycophenolic acid (MPA, MedChemExpress #HY-B0421), etoposide (ETO, MedChemExpress #HY-13629), NAMPT inhibitor (FK866, MedChem Express, # HY-50876), guanosine (Merck, # G6264-5G), ATM inhibitor (KU-55933, Dismed, #HY-12016), ATR inhibitor ((S)-AZD6738, MedChem Express, #HY-19323A) and Olaparib (MedChemExpress #HY-10162) were dissolved in DMSO, Carboplatin (MedChemExpress # HY-17393) and NAD+ (Thermo Fisher Scientific #J62337.03) were dissolved in H2O.
Plasmids generation
The plasmids, primers, and gene blocks (IDT) used in this study are listed in Tables 1–3 respectively. To generate the plasmids needed for CRISPR-Cas9 KO/knock-in, the Brand and Winter protocol was followed59. To obtain the cutting vectors sgIMPDH2ex1_GW223 and sgIMPDH2ex7_GW223, primers with sense and antisense sgRNA sequences (primers 1-4) were designed to generate the sgRNA. The cutting vector GW223_pX330A_sgX_sgPITCh (2 μg) was digested with BbsI-HF (New England Biolab; #R3539) in Cutsmart Buffer (New England Biolab; #B6004) for 1 h, dephosphorylated with Shrimp Alkaline Phosphatase (rSAP) (New England Biolabs; #M0371) and gel purified with the QIAquick PCR & Gel Cleanup Kit (Qiagen; #28506). Sense and antisense oligos were annealed with T4 Polynucleotide Kinase (PNK) (New England Biolab; #M0201) and ligated with the digested cutting plasmid with T4 DNA ligase (New England Biolab; #M0202). The ligated fragments were transformed into DH5α E. coli competent cells (Thermo Fisher Scientific; #18265017) and single colonies were analyzed with Sanger sequencing (Eurofins) to select positive clones (primer 5). The repair vectors HYG_GW209 and BSD_GW209 were generated in several steps. Two GeneBlocks (LHA-hyg-bsd and RHA-hyg-bsd) were designed and cloned into GW209_pCRIS-PITChv2-C-dTAG-Puro (BRD4) to generate the corresponding homology arms in exon 1 and exon 7. The SV40pr-Hygromicin fragment was amplified from GW209_pCRIS-PITChv2-C-dTAG-Puro (BRD4) with primers 6–7 using the Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific; #F530). After purification, the PCR product and the SV40pr-Blasticidine geneblock were cloned separated into GW209_pCRIS- PITChv2-C-dTAG-Puro (BRD4) (2 μg) digested with MluI-HF (New England Biolabs; #R3198) in Cutsmart Buffer for 1 h, dephosphorylated with rSAP and gel purified with the QIAquick PCR & Gel Cleanup Kit. After, the PCR product and the gene blocks were cloned into the digested repair vector using the Gibson reaction approach for 3 h at 50 °C, followed by DH5α E. coli cell transformation. Single clones were Sanger sequenced (primers 8-10 for HYG_GW209, primers 8, 10, 11 for BSD_GW209). To obtain the lentiviral plasmids to generate the reconstituted IMPDH2 versions (KO-WT, KO-NLS), different strategies were followed. For the KO-WT version, two oligos containing the 3xHA (Primers 12, 13) were annealed as described above, amplified with Primers 14 and 15 using the Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific; #F530) and purified with QIAquick PCR & Gel Cleanup Kit (Qiagen; #28506). A geneblock corresponding to wild-type IMPDH2 was ordered (WT-IMPDH2-GB) and was cloned via Gibson assembly with the 3xHA PCR product in GO-HYG plasmid prior cut with NheI (New England Biolabs; # R3131S) and PmeI (New England Biolabs; # R0560S) for 3 h at 50 °C, followed by DH5α E. coli cells transformation. Single clones were Sanger sequenced (Primers 16–18). For the KO-NLS version, a geneblock to introduce the 3xNLS (C-MYC NLS:cctgctgctaagagagtgaaactggat, Nucleoplasmin NLS: aagcggcccgctgctactaagaaggctggtcaggctaagaagaagaag, SV40 NLS: gatccgaagaagaagcgaaaggtc) was amplified with Primers 19 and 20 using the Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific; #F530). The KO-WT vector was cut with AscI (New England Biolabs; # R0558S) and NheI. Both products were cleaned with QIAquick PCR & Gel Cleanup Kit and ligated via Gibson reaction for 3 h at 50 °C followed by DH5α E. coli cells transformation. Single clones were Sanger sequenced (Primer 16). The hygromycin gene was substituted by Puromycin and for that, the KO-WT, and KO-NLS were cut with KpnI (New England Biolabs; # R3142S) and PsiI (New England Biolabs; # R0657) and gel purified with QIAquick PCR & Gel Cleanup Kit (Qiagen; #28506). The sv40PR-puro fragment was amplified using Primers 21 and 22 using the Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific; #F530) and the resulting purified fragment was cloned via ligation into KO-WT and KO-NLS with T4 DNA ligase (New England Biolab; #M0202). The ligated fragments were transformed into DH5α E. coli competent cells (Thermo Fisher Scientific; #18265017) and single colonies were analyzed with Sanger sequencing to select positive clones (Primers 23 and 24). In order to generate the catalytic dead version of IMPDH2 harboring the mutation C331A, the KO-WT-LV vector was cut with SbfI-HF (New England Biolab; #R3642) and BamHI-HF (New England Biolab; #R3136) and the backbone gel purified with QIAquick PCR & Gel Cleanup Kit. A geneblock containing the mutation was cloned via Gibson assembly for 3 h at 50 °C, followed by DH5α E. coli cells transformation. Single clones were Sanger sequenced with primer 25. For IMPDH2 knockdown, shRNAs targeting IMPDH2 (Sigma-Aldrich TRCN0000026534, sh1; Sigma-Aldrich TRCN0000026591, sh2) or non-mammalian genes (Sigma-Aldrich SHC002, Non-Targeting, NT) were used. To generate the 3xNLS- pLV-FiNad sensor plasmid, the pLV-FiNad vector was cut with KpnI and BamHI (New England Biolabs; # R3136S) enzymes in Cutsmart Buffer for 1 h and gel purified with QIAquick PCR & Gel Cleanup Kit (Qiagen; #28506). A gene block corresponding to the 3xNLS fragment (3xNLS-FN) was amplified with primers 26 and 27 using the Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific; #F530). The product was cloned in the cut vector via Gibson reaction for 4 h at 50 °C and transformed into DH5α E. coli competent cells (Thermo Fisher Scientific; #18265017) and single colonies were analyzed with Sanger sequencing to select positive clones (Primers 28 and 29).
Table 2.
Primers
| Primer 1 | GACGACGGACTCACAGCACA |
| Primer 2 | TGTGCTGTGAGTCCGTCGTC |
| Primer 3 | GGACAGACCTGAAGAAGAAT |
| Primer 4 | ATTCTTCTTCAGGTCTGTCC |
| Primer 5 | GCTGGCCTTTTGCTCACATG |
| Primer 6 | CAAAGCATGCATCTCAATTAGTCAGCAACCAGGTGTGGAAAGTCCCCAGG |
| Primer 7 | GGTTTGTCCAAACTCATCAATGTATCTTAAATAGGGACTACCCACTAGCCTCCAAAGATG |
| Primer 8 | GGGTCATTAGTTCATAGCCC |
| Primer 9 | CTCTATCAGAGCTTGGTTGACGG |
| Primer 10 | CCCACTGTCCTTTCCTAATA |
| Primer 11 | AAGGACAGTGATGGACAGCCG |
| Primer 12 | CGTCAGATCCGCTAGCGCCACCATGGGGCAACAAGGATACCCGTATGATGTTCCGGATTACGCTGGCTACCCATACGACGTCCCAGACTACGCTGGCTACCCATACGACGTCCCAGACTACGCTCAAGGAGTGCG |
| Primer 13 | CGCACTCCTTGAGCGTAGTCTGGGACGTCGTATGGGTAGCCAGCGTAGTCTGGGACGTCGTATGGGTAGCCAGCGTAATCCGGAACATCATACGGGTATCCTTGTTGCCCCATGGTGGCGCTAGCGGATCTGACG |
| Primer 14 | CTGGTTTAGTGAACCGTCAGATCCGCTAGCGC |
| Primer 15 | GACTACGCTCAAGGAGTGCGGCCGCGGGGGCGCGCCGCCGACTACCTGATTAGTGGGGGCACGTCCTACGTGCCAGACGACGGA |
| Primer 16 | GCAGAGCTGGTTTAGTGAACCG |
| Primer 17 | CAGGGAAATTCCATCTTCCAGATCAATATGA |
| Primer 18 | CTAGGTACAATTCGATATCAAGCTTATCG |
| Primer 19 | GAGCTGGTTTAGTGAACCGTCAGA |
| Primer 20 | GGCTACCCATACGACGTCCCAGACTACGCTGGCTACCCATACGACGTCCCAGACTACGCTCAAGGAGTGCGGCCGCGGGGGCGCGCCGCCGACTACCTGATTAGT |
| Primer 21 | GGGTTTTCCAGTCACACCTCAGGTACCTTTAAGACCAATGACTTACAAGGCAG |
| Primer 22 | CCCCAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTT |
| Primer 23 | CGCGCAGCAACAGATGGA |
| Primer 24 | TCGTAGAAGGGGAGGTTGCGGG |
| Primer 25 | CGATAAGCTTGATATCGAATTGTACCTAG |
| Primer 26 | TGGCTAGCATGGCGGATCATCCATGCCTGCTGCTAAGAGAGTGAAACTGG |
| Primer 27 | CCTCCTCGCCCTTGCTCACCATGCCGACCTTTCGCTTCTTCTTCGGATCC |
| Primer 28 | TACGACTCACTATAGGGAGACCCAAG |
| Primer 29 | CTCTGGCTAACTAGAGAACCCAC |
Table 1.
Plasmids
| GW223_pX330A_sgX_sgPITCh | Brand et al.59 |
| sgIMPDH2ex1_GW223 | This study |
| sgIMPDH2ex7_GW223 | This study |
| HYG_GW209 | This study |
| BSD_GW209 | This study |
| GW209_pCRIS-PITChv2-C-dTAG-Puro (BRD4) | Brand et al.59 |
| GO-Hyg | Moretton et al.12 |
| KO-WT-LV | This study |
| KO-NLS-LV | This study |
| TRCN0000026534 | Sigma-Aldrich |
| TRCN0000026591 | Sigma-Aldrich |
| SHC002 | Sigma-Aldrich |
| pLV-FiNad | Zou et al.43 |
| 3xNLS- pLV-FiNad | Adapted for this study from Zou et al.43 |
Table 3.
GeneBlocks
| LHA-hyg-bsd | CGCGGTCCTCGGAGACACGCGGCGGTGTCCTGTGTTGGCCATGGCCGACTACCTGATTAGTGGGGGCACGTCCTACGTGCCAGAC |
| RHA-hyg-bsd | GGGACTACCCACTAGCCTCCAAAGATGCCAAGAAACAGCTGCTGTGTGGGGCAGCCATTGGCACTCATGAGGATGACAAGT |
| SV40pr-Blasticidine | CAAAGCATGCATCTCAATTAGTCAGCAACCAGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTCTCCGCCCCATGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCTGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAAAGCTCCCGGGAGCTTGTATATCCATTTTCGGATCTGATCAAGAGACAGGATGAGGATCGTTTCGCATGGCCAAGCCTTTGTCTCAAGAAGAATCCACCCTCATTGAAAGAGCAACGGCTACAATCAACAGCATCCCCATCTCTGAAGACTACAGCGTCGCCAGCGCAGCTCTCTCTAGCGACGGCCGCATCTTCACTGGTGTCAATGTATATCATTTTACTGGGGGACCTTGTGCAGAACTCGTGGTGCTGGGCACTGCTGCTGCTGCGGCAGCTGGCAACCTGACTTGTATCGTCGCGATCGGAAATGAGAACAGGGGCATCTTGAGCCCCTGCGGACGGTGCCGACAGGTGCTTCTCGATCTGCATCCTGGGATCAAAGCCATAGTGAAGGACAGTGATGGACAGCCGACGGCAGTTGGGATTCGTGAATTGCTGCCCTCTGGTTATGTGTGGGAGGGCTAAGCGGGACTCTGGGGTTCGAAATGACCGACCAAGCGACGCCCAACCTGCCATCACGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGGTTGGGCTTCGGAATCGTTTTCCGGGACGCCGGCTGGATGATCCTCCAGCGCGGGGATCTCATGCTGGAGTTCTTCGCCCACCCCAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTAAATAGGGACTACCCACTAGCCTCCAAAGATGCCAAGAAAC |
| WT-IMPDH2-GB | GCCGACTACCTGATTAGTGGGGGCACGTCCTACGTGCCAGACGACGGACTCACAGCACAGCAGCTCTTCAACTGCGGAGACGGCCTCACCTACAATGACTTTCTCATTCTCCCTGGGTACATCGACTTCACTGCAGACCAGGTGGACCTGACTTCTGCTCTGACCAAGAAAATCACTCTTAAGACCCCACTGGTTTCCTCTCCCATGGACACAGTCACAGAGGCTGGGATGGCCATAGCAATGGCGCTTACAGGCGGTATTGGCTTCATCCACCACAACTGTACACCTGAATTCCAGGCCAATGAAGTTCGGAAAGTGAAGAAATATGAACAGGGATTCATCACAGACCCTGTGGTCCTCAGCCCCAAGGATCGCGTGCGGGATGTTTTTGAGGCCAAGGCCCGGCATGGTTTCTGCGGTATCCCAATCACAGACACAGGCCGGATGGGGAGCCGCTTGGTGGGCATCATCTCCTCCAGGGACATTGATTTTCTCAAAGAGGAGGAACATGACTGTTTCTTGGAAGAGATAATGACAAAGAGGGAAGACTTGGTGGTAGCCCCTGCAGGCATCACACTGAAGGAGGCAAATGAAATTCTGCAGCGCAGCAAGAAGGGAAAGTTGCCCATTGTAAATGAAGATGATGAGCTTGTGGCCATCATTGCCCGGACAGACCTGAAGAAGAATCGGGACTACCCACTAGCCTCCAAAGATGCCAAGAAACAGCTGCTGTGTGGGGCAGCCATTGGCACTCATGAGGATGACAAGTATAGGCTGGACTTGCTCGCCCAGGCTGGTGTGGATGTAGTGGTTTTGGACTCTTCCCAGGGAAATTCCATCTTCCAGATCAATATGATCAAGTACATCAAAGACAAATACCCTAATCTCCAAGTCATTGGAGGCAATGTGGTCACTGCTGCCCAGGCCAAGAACCTCATTGATGCAGGTGTGGATGCCCTGCGGGTGGGCATGGGAAGTGGCTCCATCTGCATTACGCAGGAAGTGCTGGCCTGTGGGCGGCCCCAAGCAACAGCAGTGTACAAGGTGTCAGAGTATGCACGGCGCTTTGGTGTTCCGGTCATTGCTGATGGAGGAATCCAAAATGTGGGTCATATTGCGAAAGCCTTGGCCCTTGGGGCCTCCACAGTCATGATGGGCTCTCTCCTGGCTGCCACCACTGAGGCCCCTGGTGAATACTTCTTTTCCGATGGGATCCGGCTAAAGAAATATCGCGGTATGGGTTCTCTCGATGCCATGGACAAGCACCTCAGCAGCCAGAACAGATATTTCAGTGAAGCTGACAAAATCAAAGTGGCCCAGGGAGTGTCTGGTGCTGTGCAGGACAAAGGGTCAATCCACAAATTTGTCCCTTACCTGATTGCTGGCATCCAACACTCATGCCAGGACATTGGTGCCAAGAGCTTGACCCAAGTCCGAGCCATGATGTACTCTGGGGAGCTTAAGTTTGAGAAGAGAACGTCCTCAGCCCAGGTGGAAGGTGGCGTCCATAGCCTCCATTCGTATGAGAAGCGGCTTTTCTGAAAACGAATTCGTCGAGGGACCTA |
| 3xNLS | GAGCTGGTTTAGTGAACCGTCAGATCCGCTAGCGCCACCATGCCTGCTGCTAAGAGAGTGAAACTGGATAAGCGGCCCGCTGCTACTAAGAAGGCTGGTCAGGCTAAGAAGAAGAAGGATCCGAAGAAGAAGCGAAAGGTCGGGCAACAAGGATACCCGTATGATGTTCCGGATTACGCTGGCTACCCATACGACGTCCC |
| CD | GGGAAGACTTGGTGGTAGCCCCTGCAGGCATCACACTGAAGGAGGCAAATGAAATTCTGCAGCGCAGCAAGAAGGGAAAGTTGCCCATTGTAAATGAAGATGATGAGCTTGTGGCCATCATTGCCCGGACAGACCTGAAGAAGAATCGGGACTACCCACTAGCCTCCAAAGATGCCAAGAAACAGCTGCTGTGTGGGGCAGCCATTGGCACTCATGAGGATGACAAGTATAGGCTGGACTTGCTCGCCCAGGCTGGTGTGGATGTAGTGGTTTTGGACTCTTCCCAGGGAAATTCCATCTTCCAGATCAATATGATCAAGTACATCAAAGACAAATACCCTAATCTCCAAGTCATTGGAGGCAATGTGGTCACTGCTGCCCAGGCCAAGAACCTCATTGATGCAGGTGTGGATGCCCTGCGGGTGGGCATGGGAAGTGGCTCCATCGCCATTACGCAGGAAGTGCTGGCCTGTGGGCGGCCCCAAGCAACAGCAGTGTACAAGGTGTCAGAGTATGCACGGCGCTTTGGTGTTCCGGTCATTGCTGATGGAGGAATCCAAAATGTGGGTCATATTGCGAAAGCCTTGGCCCTTGGGGCCTCCACAGTCATGATGGGCTCTCTCCTGGCTGCCACCACTGAGGCCCCTGGTGAATACTTCTTTTCCGATGGGATCCGGCTAAAGAAATATCGCGGTATGGG |
| 3xNLS-FN | CCTGCTGCTAAGAGAGTGAAACTGGATAAGCGGCCCGCTGCTACTAAGAAGGCTGGTCAGGCTAAGAAGAAGAAGGATCCGAAGAAGAAGCGAAAGGTC |
Lentiviral production
Lentiviral particles were produced in HEK293T cells seeded in 150 mm plates using a standard Polyethylenimine (PEI)-mediated transfection method (Polysciences; #23966-1). In brief, 5.5 μg of pCMV-dR8_91 and 4.2 μg of pVSV-G packaging plasmids were mixed with 8.4 μg of plasmid of interest in 1 mL of Opti-MEM (Gibco; #11058021). In parallel, 54.6 l of PEI (1 mg/mL) were mixed with 900 μL of Opti-MEM. After 5 min, the two mixtures were combined, incubated for 20 min to allow complex formation, and added dropwise to the cells in serum-free media. After 6 h, the media was changed to regular media. Virus-containing supernatant was collected 48 h after and lentiviral transduction was performed in a 6-well plate format using 1 mL virus-containing supernatant per well in the presence of polybrene (10 μg/mL). Transduced cells were selected with puromycin (MDA-MB-231, 2 μg/mL; CAL-51, 10 μg/mL).
Generation of IMPDH2 KO cells
MDA-MB-231 cells were nucleofected using the Lonza Amaxa Kit V (Lonza, #VCA-1003) and Amaxa Nucleofector (Lonza) following the MDA-MB-231 protocol. Briefly, 1^106 trypsinized cells were resuspended in supplement nucleofector solution and nucleofected using the X-013 program. Specifically, for the hygromycin and blasticidin knock-in, cells were nucleofected with 4 μg of the sgIMPDH2ex1_GW223 and sgIMPDH2ex7_GW223 cutting vectors, which contain Cas9, and 4 μg of the HYG_GW209 and BSD_GW209 repair vectors, using the intron-tagging strategy described by Serebrenik et al.60. Cells were supplemented with Guanosine (Sigma-Aldrich #G6264) at 400 μM concentration and after two days of nucleofection cells were treated with hygromycin (Sigma-Aldrich; #H3274) and blasticidine (Sigma-Aldrich; #15205) at 250 μg/mL and 6 μg/mL, respectively for 7 days. The KO population was confirmed by western blot with primary rabbit monoclonal antibody Anti-IMPDH2 [EPR8364(B)] (Abcam #ab131158; 1:1000).
Whole cell extracts
Whole-cell extracts were obtained by lysing with 2%SDS lysis buffer (2%SDS, 50 mM Tris-HCl pH 7.4, 10% glycerol) and boiling at 95 °C for 5 min, followed by quantification in a NanoDrop spectrophotometer using the Protein A280 mode. For detection of phosphoproteins, cells were lysed in Radioimmunoprecipitation Assay Buffer (RIPA) buffer supplemented with proteinase inhibitors (Roche; #4693132001) and phosphatase inhibitors (Roche; 4906845001) for 20 min at 4 °C followed by centrifugation at 18400 g for 15 min at 4 °C. Protein concentration was quantified with Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; #PIER23225) according to the manufacturer’s instructions.
Proliferation assays
Cell viability
Cell viability was assessed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay in a 96-well plate format. At the desired time point, MTT reagent was added to the wells diluted in serum-free media at a final concentration of 0.5 mg/mL. After 3 h of incubation at 37 °C/5% CO2, media was aspirated and crystals were dissolved by the addition of isopropanol. After 5 min shaking, A565nm was measured in a TECAN Infinite M200 Plate Reader.
Growth curves
For growth rate determination, a crystal violet assay was performed in a 12-well plate format. At the desired time points, plates were washed with PBS, fixed with 10% neutral buffered formalin (Sigma-Aldrich #HT501128) for 10 min/RT followed by washing with PBS. For crystal violet staining, 0.1% Crystal Violet (SIGMA, C-3886) in 20% methanol was added to the plates for 15 min/shaking/RT. Plates were then washed twice with water, allowed to air-dry, and solubilized by the addition of 10% acetic acid. A590nm was measured in a TECAN Infinite M200 Plate Reader.
Cell cycle analysis
Cells were trypsinized and incubated in suspension in DMEM media for 45 min at 37 °C, at 1 million cells/mL with 5 μg/mL Hoechst 33342 (Life Technologies, # H3570) final concentration. Hoechst signal from 10000 cells was measured with LSRFortessa Cell Analyzer (BD Biosciences) and the results were analyzed with FlowJo_V10.
Cell death assays
Cells were treated as indicated and stained with fluorescent conjugates of Annexin V (Alexa Fluor® 647 Annexin V, 640912, BioLegend) and DAPI (62248, Thermo Fisher) and analyzed on a flow cytometry LSRFortessa instrument (BD, Franklin Lakes, NJ, USA). Viable cells are Annexin V and DAPI negative. Cell death is expressed as 100%-viable cells, and it is divided into early (Annexin V positive), late apoptosis/late necrosis (Annexin V and DAPI positive), and necrosis (DAPI positive).
Immunofluorescence
Immunofluorescence was performed in a 96-well plate format using black/clear bottom plates (PhenoPlate, PerkinElmer #6055508). At the desired experimental point, plates were washed with PBS and fixed with 4% formaldehyde in PBS [Formaldehyde 16% (w/v) methanol-free, Pierce™ # 28908] during 15 min/RT. Plates were then washed twice with PBS and the permeabilization step was performed by the addition of 0.2% Triton X-100 in PBS during 20 min/RT. After two PBS washes, the blocking step was performed with 5% bovine serum albumin (BSA, lyophilized powder, Sigma-Aldrich, #A2153-50G) in PBS for 1 h/RT, followed by incubation with primary antibodies diluted in 0.5% BSA in PBS for 2 h/RT. Plates were then rinsed with 0.05% Tween 20 in PBS, washed with PBS and incubated with secondary antibodies [Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488, (Invitrogen #A-21206); Goat anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 (Invitrogen #A-21424), Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (Thermo Scientific #A-21244)] diluted 1:1000 in 0.5% BSA in PBS for 1 h/RT in the dark.
Plates were then rinsed with 0.05% Tween 20 in PBS, washed with PBS, and incubated with DAPI at 1 μg/mL (4′,6-diamidino-2-phenylindole, Sigma-Aldrich #MBD0015) or Hoechst 33342 (Life Technologies, # H3570) for 7 min/RT, followed by two PBS washes. Images were acquired with an Operetta High Content Screening System (PerkinElmer) and analyzed using the Harmony software (version 4.9).
Primary antibodies used were Anti-IMPDH2 rabbit monoclonal antibody [EPR8364(B)] (Abcam #ab131158; 1:200), HA-Tag (C29F4) rabbit monoclonal antibody (Cell Signaling Technology #3724; 1:500), phospho-Histone H2A.X (Ser139) mouse monoclonal antibody, clone JBW301 (Millipore #05-636-I; 1:500), PARP1 Monoclonal antibody (Proteintech 66520-1-Ig; 1:400), Cleaved Caspase-3 (Asp175) (Cell Signaling Technology #9661; 1:400), PARP1 Monoclonal Antibody (C.384.8), (Thermo-Fisher #MA5-15031; 1:800), poly/mono-ADP-ribose (D9P7Z) rabbit monoclonal antibody (WERFEN ESPAÑA; S.A.U.; #89190S, 1:3000), RPA70/RPA1 Antibody (Cell Signaling Technology #2267; 1:50), Anti 53BP1 rabbit (Novus Biologicals #NB100-304; 1:5000).
To mark cytoplasm when needed, conjugated phalloidin was used (Phalloidin543, Biotium #00043).
Immunohistochemistry
Tissue microarray (TMA) BR1141a (TissueArray.Com) was used for immunohistochemical detection of IMPDH2 protein. The slide was placed in the oven at 65 °C/ON, deparaffinized, and rehydrated. The antigen retrieval step was performed by boiling sodium citrate buffer (pH = 6.0) in a pressure cooker, followed by incubation in 3% H2O2 in PBS for 15 min for inactivation of endogenous peroxidase. Blocking and permeabilization step was performed with 1%BSA, and 0.3% Triton-X100 in PBS during 1 h/RT, followed by incubation with Anti-IMPDH2 rabbit monoclonal antibody [EPR8364(B)] (Abcam #ab131158) diluted 1:250 in 1%BSA in PBS ON/4 °C. Following PBS washes, TMA was incubated with ImmPRESS (Peroxidase) Polymer Anti-Rabbit IgG Reagent (Vector Laboratories #MP-7451) for 2 h/RT, PBS washed and stained with 3,3‐diaminobenzidine (DAB, Agilent Dako #K346711-2) for 1 min. The slide was then counterstained with hematoxylin 20% for 2 min, dehydrated, and mounted. Images were obtained with a VENTANA DP 200 slide scanner and analyzed with QuPath software61.
Chromatome fractionation
Cells were lysed in 1.5% CHAPS (3-cholamidopropyl dimethylammonium 1-propane sulfonate) in PBS for 20 min to break the cytosolic membrane and centrifuged for 5 min at 720 g at 4 °C. The supernatant was harvested as the cytosolic fraction. The nuclear pellet was resuspended in Cytoplasmic Lysis Buffer (IGEPAL 0.1%, NaCl 150 mM, Tris-HCl 10 mM pH 7 in H2O), placed on the top of a Sucrose Gradient Buffer (NaCl 150 mM, sucrose 25%, Tris-HCl 10 mM pH 7 in H2O) and centrifuged for 5 min at 1200 g at 4 °C. Purified nuclei were then washed 3 times by resuspending in Nuclei Washing Buffer (EDTA 1 mM, IGEPAL 0.1% in PBS) and centrifuged for 5 min at 1200 g at 4 °C. Then, the washed nuclear pellet was resuspended in Nuclei Resuspension Buffer (EDTA 1 mM, NaCl 75 mM, 50% sucrose, Tris-HCl 20 mM pH 8 in H2O) and the nuclear membrane was lysed by adding Nuclei Lysis Buffer (EDTA 0.2 mM, HEPES 20 mM pH 7.5, IGEPAL 0.1%, NaCl 300 mM in H2O), vortexing and incubating for 2 min. After centrifugation for 2 min at 16000 g at 4 °C, the resulting chromatin was resuspended in Benzonase Digestion Buffer (15 mM HEPES pH 7.5, 0.1% IGEPAL, TPCK 5 μg/mL) and sonicated on a Bioruptor Pico (Diagenode) for 15 cycles 30 sec ON/30 sec OFF in 1.5 mL Diagenode tubes (Diagenode; #C30010016). Finally, sonicated chromatin was digested with benzonase enzyme (VWR; #706643; 2.5U) for 30 min at room temperature, and the resulting sample was harvested as chromatome fraction. All the steps were performed on ice and all buffers were supplemented with proteinase inhibitors (Roche; #4693132001). Cytosolic and chromatome extracts were quantified with Pierce BCA Protein Assay Kit (Thermo Scientific; #PIER23225).
IMPDH2 Pull-down
Protein A Dynabeads (Thermo Scientific; #10001D) were incubated for 4 h on a rotating wheel at 4 °C with primary antibodies IMPDH2 (Proteintech; # 12948-1-AP; 5 μg) or negative control IgG (Sigma-Aldrich; #I5006; 5 μg). Then, antibody-bound beads were incubated with 2 mg chromatome extracts overnight on a rotating wheel at 4 °C. The complexes were then washed three times with Nuclei Wash Buffer (EDTA 1 mM, IGEPAL 0.1% in PBS). For nuclear fraction pull-down, cells were washed twice with cold PBS and lysed using Low Salt Buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 0,1% IGEPAL, 10% Glycerol) and centrifuged for 15 min at 800 g at 4 °C. The supernatant was discarded as the cytosolic fraction. The nuclear pellet was resuspended in High Salt Buffer (20 mM HEPES pH 7.4, 350 mM NaCl, 1 mM MgCl2, 0.5% Triton X-100, 10% glycerol). After 10 min incubation, 15 cycles of sonication (30 sec ON/30 sec OFF) were performed on a Bioruptor Pico (Diagenode) in 1.5 mL Diagenode tubes (Diagenode; #C30010016). The sonicated chromatin was digested with benzonase enzyme (VWR; #706643; 2.5U) for 30 min. Then, the sample is centrifuged at 15700 g at 4 °C for 10 min. The supernatant corresponds to the nuclear protein extract and Balance Buffer (20 mM HEPES pH 7.4, 1 mM MgCl2, 10 mM KCl) is added to reach 150 mM NaCl concentration. Protein concentration is quantified with Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; #PIER23225) according to the manufacturer’s instructions and measured with TECAN Infinite M200 Plate Reader. All the steps were performed on ice and all buffers were supplemented with proteinase inhibitors (Roche; #4693132001). 1000 μg of nuclear protein extract was incubated overnight on a rotating wheel at 4 °C with primary antibodies IMPDH2 (Proteintech; # 12948-1-AP; 5 μg) or negative control IgG (Sigma-Aldrich; #I5006; 5 μg). Protein A Dynabeads (Thermo Fisher Scientific; #10001D) were washed three times with Washing Buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.5% Triton X-100, 10% glycerol). Then, 50 uL beads were added to the protein-antibody extract for 2 h on a rotating wheel at 4 °C. The beads were washed three times with Washing Buffer and complexed eluted with 2× Laemmli Sample Buffer (Bio-Rad; #1610747) after boiling at 95 °C for 5 min. All the steps were performed on ice and the Washing Buffer was supplemented with proteinase inhibitors (Roche; #4693132001).
Western Blot
Protein samples were separated by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) method and transferred to a 0.45 μm nitrocellulose membrane (AmershamTM #10600002) for immunoblotting. Membranes were blocked with 5% non-fat milk (skim milk, Merck, # 70166-500 G) in TBS-Tween 0.05% (Tween® 20, for molecular biology, viscous liquid, Sigma-Aldrich, # P9416-100mL) and blotted with primary antibodies. Fluorescence-conjugated secondary antibodies Alexa Fluor Plus 800 goat anti-rabbit IgG (Thermo Fisher Scientific #A32735; 1:10000) and Alexa Fluor™ 680 goat anti-mouse IgG (Thermo Fisher Scientific #A21058; 1:10000) were used for signal detection with Odyssey CLx Imaging System (LI-COR Biosciences).
Primary antibodies used were Anti-IMPDH2 rabbit monoclonal antibody [EPR8364(B)] (Abcam #ab131158; 1:1000), HA-Tag (C29F4) rabbit monoclonal antibody (Cell Signaling Technology #3724; 1:1000), α-Tubulin mouse monoclonal (Sigma-Aldrich #T9026; 1:1000), GAPDH (D4C6R) mouse monoclonal antibody (Cell Signaling Technology #97166; 1:1000), Vinculin (E1E9V) XP® rabbit monoclonal antibody (Cell Signaling Technology #13901; 1:1000), FDX1 rabbit polyclonal antibody (Thermo Fisher Scientific #PA5-59653; 1:1000), Histone H3 (1B1B2) mouse monoclonal antibody (Cell Signaling Technology #14269; 1:10000), Phospho-Chk1 (Ser345) (133D3) Rabbit mAb (Cell Signaling Technology #2348; 1:1000), Chk1 (2G1D5) Mouse mAb (Cell Signaling Technology #2360; 1:1000), Phospho-ATR (Ser428) Antibody #2853, ATR (C-1) antibody (Santa Cruz Technologies # sc-515173; 1:500), Topo IIa mouse monoclonal antibody (Santa Cruz Technologies # sc-365916; 1:1000), Anti-PARP1 rabbit polyclonal antibody (ProteinTech 13371-1-AP; 1:1000), Cleaved Caspase-3 (Asp175) (Cell Signaling Technology #9661; 1:1000).
All the uncut full western blot presented in main and Supplementary Figs. can be found in the Source Data and Supplementary Information files, respectively.
3xNLS-FiNad Sensor
3xNLS-FiNad vector was introduced in MDA-MB-231 cells by transfection with TransIT®-BrCa Transfection Reagent (Mirus Bio, # MIR 5500) or nucleofection. Transfection with TransIT was performed in a 6-well plate format according to manufacturer’s instructions and then cells were transferred to 96-well plate format for Operetta acquisition. For nucleofection, the Lonza Amaxa Kit V (Lonza, #VCA-1003) and Amaxa Nucleofector (Lonza) were used following the standard MDA-MB-231 protocol. In summary, 1^106 KO-WT and KO-NLS trypsinized cells were resuspended in supplement nucleofector solution and nucleofected using the X-013 program with 4 μg of the 3xNLS-FiNad vector. Cells were seeded in 96-well plate format using black/clear bottom plates (PhenoPlate, PerkinElmer #6055508). Cells were treated with DMSO or 10μM etoposide for 2 and 24 h. For both time points, cells were incubated with Hoechst 33342 (Life Technologies #H3570) for 10 min and washed twice with PBS To obtain the dual-excitation ratio, images were acquired with an Operetta High Content Screening System (PerkinElmer) using a 488 nm excitation laser and 500 to 550 nm emission range, and a 561 nm excitation laser and a 570 to 630 nm emission range. The analysis of the YFP and mCherry ratio was obtained with the Harmony software (version 4.9).
FUCCI cells generation
To generate a stable U-2OS cell line with a Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) system, cells were transduced with viral particles containing the vectors pLL3.7m-mTurquoise2-SLBP(18-126)-Neomycin and pLL3.7m-Clover-Geminin(1-110)-IRES-mKO2-Cdt(30-120)-Hygromycin in the presence of polybrene (Sigma-Aldrich; #TR1003G; 10 μg/mL). Since these vectors contained neomycin and hygromycin resistance cassettes, respectively, 24 h after transduction, the media was replaced with fresh media containing 200 μg/mL Geneticin (Thermo Scientific; #10131035) or 150 μg/mL hygromycin (Sigma-Aldrich; #H3274), respectively. Antibiotic selection lasted 4–7 days. Transduced cells were further selected through FACS sorting (BD Influx) to keep cells that showed proper activation and degradation of the FUCCI system. The FUCCI system used in this study is an adaptation of FUCCI462 to show the 3 cell cycle-regulated fusion proteins Clover-Geminin, SLBP-Turquoise2, and Cdt1-mKO2. FUCCI cells were used for immunofluorescence as previously described. The top 10% with the highest and lowest IMPDH2 staining in the nucleus or the cytosol were selected.
RNA-sequencing
Sample preparation
RNA was extracted using the PureLink RNA mini kit (Thermo Fisher Scientific; #12183018 A) at day 4 from WT (MDA-MB-231 wild-type cells reconstituted with empty vector), KO -G (IMPDH2 KO reconstituted with empty vector cultured in the absence of guanosine supplementation), KO + G (IMPDH2 KO reconstituted with empty vector cultured in the presence of 400 μM guanosine supplementation), KO-WT (IMPDH2 KO reconstituted with wild-type IMPDH2) and KO-NLS (IMPDH2 KO reconstituted with a nuclear-specific version of IMPDH2). Libraries were prepared using the TruSeq stranded mRNA Library Prep (ref. 20020595, Illumina) according to the manufacturer’s protocol, to convert total RNA into a library of template molecules of known strand origin and suitable for subsequent cluster generation and DNA sequencing. Briefly, 500 ng of total RNA was used for polyA‐mRNA selection using Oligo‐dT beads, and two rounds of purification were performed. During the second elution of the mRNA, this was fragmented under elevated temperature and primed with random hexamers for cDNA synthesis, which was performed using reverse transcriptase (SuperScript II; ref. 18064‐014, Invitrogen). Then, the second strand cDNA was synthesized, incorporating dUTP in place of dTTP and generating blunt‐ended ds cDNA. A single ‘A’ nucleotide was added to the 3’ ends of the blunt fragments (A‐tailing) and immediately afterward the Truseq adapter was ligated. Finally, PCR selectively enriched those DNA fragments that had adapter molecules on both ends. The PCR was performed using Unique Dual Indexes and the master mix provided with the kit. All purification steps were performed using AgenCourt AMPure XP beads (ref. A63882, Beckman Coulter). Final libraries were analyzed using Bioanalyzer DNA 1000 or Fragment Analyzer Standard Sensitivity (ref. 5067‐1504 or ref. DNF‐473, Agilent) to estimate the quantity and validate the size distribution, and were then quantified by qPCR using the KAPA Library Quantification Kit KK4835 (ref. 07960204001, Roche). Libraries were sequenced 1 * 51 + 10 + 10 bp on Illumina’s NextSeq2000.
Data analysis
51 base-pairs, single-end reads were filtered for low-quality reads (>20) and adapters were removed by utilizing TrimGalore (v. 0.6.10). Salmon index (hg19 reference genome) with decoys and k-mer length 19, as well as gene-level quantification was done with Salmon (v 1.10.0) software63. All subsequent analyses were carried out in the R programming environment (v 4.4). Transcript abundance at the gene level was estimated with the tximport package64, followed by differential expression between conditions versus wild-type (3 replicates each) with Deseq265. All the results shown in heatmaps are based on log-fold shrinkage with apeglm package66. The criteria for significance in Gene Ontology (GO) Term and KEGG pathway following the differential expression analysis were set at a minimum log-fold change of 1 and an adjusted p-value less than 0.05 (Benjamini-Hochberg correction). Gene annotation was done using the EnsDb.Hsapiens.v86. GO term and KEGG enrichment analysis were performed with ClusterProfiler67,68 package with default parameters and the universe defined as all the genes with more than 10 reads across all conditions. Redundant terms were removed from the enrichment64 results using the GOSemSim69,70 package, employing the simplify function. Heat maps were constructed with ComplexHeatmap package71,72 with Pearson distance and Ward.D linkage, and values are all based on shrunk log-fold change of treatment versus wild-type for better visualization.
Mass Spectrometry (MS)-based analysis of chromatin-associated proteome
For mass spectrometry-based analysis, three biological replicates were prepared per condition.
Chromatome extraction
The cellular pellet was resuspended in 1 mL of SB buffer (HEPES 10 mM, KCl 10 mM, MgCl2 1.5 mM, sucrose 0.1%, and proteinase inhibitor cocktail (Sigma 4693159001). After a 30 min incubation at 4 °C, the tubes were centrifuged for 3 min at 2000 g at 4 °C. The supernatant was stored for further analysis as it comprises the cytoplasmic fraction of cells and was used as a control. The pellet containing the nuclei of the cells was resuspended in 0.5 mL of SB buffer and then ultracentrifuged on a sucrose gradient over SC buffer (HEPES 10 mM, KCl 10 mM, MgCl2 1.5 mM, Sucrose 2.1 M and proteinase inhibitors. Ultracentrifugation was performed at 53,000 rpm for 3 h at 4 °C with reduced acceleration and deceleration of 5 out of 10 in a TLA100.3 rotor and an Optima TableTop Ultracentrifuge (Beckman Coulter). After the ultracentrifugation, the supernatant containing the nuclear proteins was discarded while the pellet containing the chromatin and its associated proteins was resuspended on 0.5 mL of buffer SB and later centrifuged at 20000 g for 45 min at 4 °C to remove the rest of the sucrose gradient. Two washing steps were then performed by using a buffer composed of HEPES 10 mM and proteinase inhibitors (Sigma 4693159001). The pellet contains chromatin and its associated proteins. To release the chromatin-associated proteins, a sonication procedure was performed followed by a DNA digestion step. For the sonication, the chromatin pellet was resuspended in 0.25 mL of Benzonaze digestion buffer [HEPES 15 mM, EDTA 1 mM, EGTA 1 mM, 5 mg/mL TPCK, 1% of NP40 and proteinase inhibitors (Sigma 4693159001)]. We used a BioRuptor Pico sonication device from Diagenode, specially designed for chromatin, DNA, and RNA shearing. Chromatin was sheared through 15 cycles of 30 sec of sonication followed by 45 sec of pause. DNA was then degraded through the enzymatic action of 1μL of Benzonaze (Millipore 70664) and RNA was removed with the addition of the RNAse A at a 1:1000 dilution (Thermo EN0531). DNA and RNA digestions were performed during 30–40 min incubation at 4 °C in a rotary wheel. At this point, proteins associated with chromatin were released into the solution. The final chromatome sample from 40 million cells (253.5 μL) was then split into three equal samples. Sample 1 was kept in its native state and immediately stored at −80 °C for further use. Samples 2 and 3 were treated to denature proteins either with 6 M urea or with SDS 2%. Both samples were also stored at −80 °C until used.
Sample preparation for MS
Samples (20 μg) were reduced with dithiothreitol (100 mM, 37 °C, 60 min) and alkylated in the dark with iodoacetamide (5μmol, 25 °C, 20 min). The resulting protein extract was washed with 2 M urea with 100 mM Tris-HCl and then with 50 mM ammonium bicarbonate for digestion with endoproteinase LysC (1:10 w:w, 37 °C, o/n, Wako, cat #129-02541) and then for trypsin digestion (1:10 w:w, 37 °C, 8 h, Promega cat #V5113) following Wiśniewski et al73 FASP procedure. After digestion, the peptide mix was acidified with formic acid and desalted with a MicroSpin C18 column (The Nest Group, Inc.) before LC-MS/MS analysis.
Chromatographic and mass spectrometric analysis
Samples were analyzed using an Orbitrap Eclipse mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) coupled with an EASY-nLC 1200 (Thermo Fisher Scientific, San Jose, CA, USA). Peptides were loaded directly onto the analytical column and were separated in a water-acetonitrile 90-min gradient by reversed-phase chromatography using a 50-cm column with an inner diameter of 75μm, packed with 2μm C18 particles spectrometer (Thermo Scientific, San Jose, CA, USA). The mass spectrometer was operated in data-dependent acquisition (DDA) mode and full MS scans with 1 micro scan at a resolution of 120,000 were used over a mass range of m/z 350–1500 with detection in the Orbitrap mass analyzer. Auto gain control MS1 was set to 1E5 and charge state filtering disqualifying singly charged peptides was activated. In each cycle of data-dependent acquisition analysis, following each survey scan, the most intense ions above a threshold ion count of 10000 were selected for fragmentation. The number of selected precursor ions for fragmentation was determined by the “Top Speed” acquisition algorithm and a dynamic exclusion of 60 s. Fragment ion spectra were produced via high-energy collision dissociation (HCD) at a normalized collision energy of 28% and they were acquired in the ion trap mass analyzer. AGC MS2 was set to 1E4, and an isolation window of 1.6 m/z and a maximum injection time of 200 ms were used. All data were acquired with Xcalibur software v4.1.31.9. Digested bovine serum albumin (New England Biolabs cat #P8108S) was analyzed between each sample to avoid sample carryover and to assure the stability of the instrument and QCloud has been used to control instrument longitudinal performance during the project. Acquired spectra were analyzed using the Proteome Discoverer software suite (v2.4, Thermo Fisher Scientific) and the Mascot search engine74 (v2.6, Matrix Science). The data were searched against a Swiss-Prot human database (April 2019) plus a list of common contaminants and all the corresponding decoy entries75. For peptide identification a precursor ion mass tolerance of 7 ppm was used for the MS1 level, trypsin was chosen as the enzyme, and up to three missed cleavages were allowed. The fragment ion mass tolerance was set to 0.5 Da for MS2 spectra. Oxidation of methionine and N-terminal protein acetylation were used as variable modifications whereas carbamidomethylation on cysteines was set as a fixed modification. False discovery rate (FDR) in peptide identification was set to a maximum of 5%. Peptide quantification data were retrieved from the “Precursor ions quantifier” node from Proteome Discoverer (v2.4) using 2 ppm mass tolerance for the peptide extracted ion current (XIC). The obtained values were used to calculate protein fold changes and their corresponding adjusted p-values.
Bioinformatic analysis
For the comparison of the protein lists retrieved by mass spectrometry among the samples, we used the Panther Classification System (http://pantherdb.org/) following the protocol described in Thomas et al.76. Protein lists were classified according to their molecular function, biological process, and metabolic pathways among other criteria. Classifications were visualized by pie charts generated also through the Panther Classification System.
Data processing
Chromatin data were normalized using the normalize_vsn and median_normalisation functions from the DEP77 and proDA78 packages, respectively. The rest of the pipeline was followed according to the DEP package, with the inclusion of impute.mi function for protein-imputation from the imp4p package79. Q-value FDR control was implemented through the qvalue R package80. Known subcellular localizations for proteins were obtained from the pRoloc R package81, and the normalization of proteins to their expected whole-cell extract (WCE) levels for untreated breast cancer cell lines was performed through the ProteomicRuler in Perseus. The WCE protein expression was obtained from the CCLE proteomics dataset82. Analysis was facilitated by the tidyverse83 collection of packages. Chromatin protein abundance association of cancer cell line aggressiveness was performed using the lm function.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The authors would like to thank the Genomics Unit at the CRG for assistance with the sequencing, the CRG Proteomics Facility (Barcelona, Spain) for the chromatome MS analysis, the CRG Flow Cytometry Facility (Barcelona, Spain) for the cell cycle analysis, Dr. Ivan Perez-Nuñez from Dr. Toni Celià-Terrassa group (IMIM, Barcelona, Spain) for helping with the IHC analysis. The CRG/UPF Proteomics Unit is part of the Spanish Infrastructure for Omics Technologies (ICTS OmicsTech). We acknowledge the support of the Spanish Ministry of Science and Innovation through the Centro de Excelencia Severo Ochoa (CEX2020-001049-S, MCIN/AEI /10.13039/501100011033), and the Generalitat de Catalunya through the CERCA program. N.P.L. is supported by a Boehringer Ingelheim Fonds Ph.D. fellowship. L.P.R. is supported by a Fundación Francisco Cobo (Eduardo Gallego) postdoctoral fellowship. The Sdelci lab’s contributions to this study have received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 852343), and from the Spanish Plan Estatal grants (Ministerio de Ciencia e Innovación, PID2019-110598GA-I00/AEI/10.13039/501100011033 and PID2022-141740NB-I00 funded by MICIU /AEI /10.13039/501100011033 / FEDER, UE) and from Fundación BBVA under Beca Leonardo a Investigadores y Creadores Culturales 2022 (LEO22-2-2290-BBM-BAS-167).
Author contributions
L.E. and M.G.C. performed and/or analyzed most of the experiments shown in Figs. 1–6 and Supp. Figures 1–5. A.S. contributed to experimental assays of immunofluorescence and immunoprecipitation, data analysis, and data visualization. S.K. performed the chromatin proteome analysis, OpenCell resource data analysis, and analysis of enriched proteins on chromatin. A.G.Z. performed the FUCCI experiments and contributed to data analysis and visualization. C.A.V. contributed to knockdown and pharmacological inhibition experiments. L.G.L. and M.G. contributed to optimization and chromatin sample preparation for mass spectrometry analysis. I.S. performed RNA-seq data analysis and visualization. N.P.L. contributed to statistical analysis and data visualization R.G. contributed to data analysis and visualization. L.P.R. generated the hormone-independent cell line. E.D. performed TMA data analysis. M.B. and J.M. performed cell death assays and analyses. S.S. conceptualized and designed the project, supervised the study, and acquired the funding. S.S., M.G.C. and A.S. wrote the manuscript with contributions from all the authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Protein interactors of TOP2A and PARP1 were derived from the OpenCell project39. Data for IMPDH2 protein expression upon etoposide treatment and release were obtained from Moretton et al.12. The raw proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the PRIDE repository84 with the dataset identifier PXD043522. RNA-sequencing data generated in this study have been deposited in the NCBI GEO database under accession code GSE271269. High-throughput microscopy data are publicly available in the SdelciLab GitHub repository [https://github.com/SdelciLab/IMPDH2_chromatin]85. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Code availability
All the scripts used for this manuscript are publicly available in the GitHub repository [https://github.com/SdelciLab/IMPDH2_chromatin]85.
Competing interests
The author declares no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lorena Espinar, Marta Garcia-Cao.
Contributor Information
Marta Garcia-Cao, Email: marta.garciacao@crg.eu.
Sara Sdelci, Email: sara.sdelci@crg.eu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53877-z.
References
- 1.Pavlova, N. N., Zhu, J. & Thompson, C. B. The hallmarks of cancer metabolism: Still emerging. Cell Metab.34, 355–377 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etchegaray, J.-P. & Mostoslavsky, R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol. Cell62, 695–711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid, M. A., Dai, Z. & Locasale, J. W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol.19, 1298–1306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boon, R., Silveira, G. G. & Mostoslavsky, R. Nuclear metabolism and the regulation of the epigenome. Nat. Metab.2, 1190–1203 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Sdelci, S. et al. MTHFD1 interaction with BRD4 links folate metabolism to transcriptional regulation. Nat. Genet51, 990–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright, R. H. G. et al. ADP-ribose–derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science352, 1221–1225 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Murthy, S. & Reddy, G. P. Replitase: Complete machinery for DNA synthesis. J. Cell. Physiol.209, 711–717 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Takahashi, H., McCaffery, J. M., Irizarry, R. A. & Boeke, J. D. Nucleocytosolic Acetyl-Coenzyme A Synthetase is required for histone acetylation and global transcription. Mol. Cell23, 207–217 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Sutendra, G. et al. A Nuclear Pyruvate Dehydrogenase complex is important for the generation of Acetyl-CoA and Histone acetylation. Cell158, 84–97 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Sivanand, S. et al. Nuclear Acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell67, 252–265.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, S. et al. Serine and SAM responsive complex SESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol. Cell60, 408–421 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Moretton, A. et al. A metabolic map of the DNA damage response identifies PRDX1 in the control of nuclear ROS scavenging and aspartate availability. Mol. Syst. Biol.19, e11267 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Kamel, D., Gray, C., Walia, J. S. & Kumar, V. PARP inhibitor drugs in the treatment of breast, ovarian, prostate and pancreatic cancers: an update of clinical trials. CDT19, 21–37 (2018). [DOI] [PubMed]
- 15.Singh, D. D., Parveen, A. & Yadav, D. K. Role of PARP in TNBC: Mechanism of inhibition, clinical applications, and resistance. Biomedicines9, 1512 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaitanya, G. V., Alexander, J. S. & Babu, P. P. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal8, 31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, P. et al. Molecular mechanisms of Parthanatos and its role in diverse diseases. IJMS23, 7292 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mashimo, M. et al. The 89-kDa PARP1 cleavage fragment serves as a cytoplasmic PAR carrier to induce AIF-mediated apoptosis. J. Biol. Chem.296, 100046 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrabi, S. A. et al. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc. Natl Acad. Sci. USA111, 10209–10214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murata, M. M. et al. NAD+ consumption by PARP1 in response to DNA damage triggers metabolic shift critical for damaged cell survival. MBoC30, 2584–2597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantó, C., Sauve, A. A. & Bai, P. Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes. Mol. Asp. Med.34, 1168–1201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolthur-Seetharam, U., Dantzer, F., McBurney, M. W., Murcia, G. D. & Sassone-Corsi, P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle5, 873–877 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro, E. et al. Triple negative breast cancers have a reduced expression of DNA repair genes. PLoS ONE8, e66243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagelkerke, A. et al. Constitutive expression of γ-H2AX has prognostic relevance in triple negative breast cancer. Radiother. Oncol.101, 39–45 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Chopra, N. et al. Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat. Commun.11, 2662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moretton, A. & Loizou, J. I. Interplay between cellular metabolism and the DNA damage response in cancer. Cancers12, 2051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedstrom, L. IMP Dehydrogenase: Structure, mechanism, and inhibition. Chem. Rev.109, 2903–2928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heitz, F. et al. Differences in the receptor status between primary and recurrent breast cancer - the frequency of and the reasons for discordance. Oncology84, 319–325 (2013). [DOI] [PubMed] [Google Scholar]
- 29.O’Connell, M. J., Walworth, N. C. & Carr, A. M. The G2-phase DNA-damage checkpoint. Trends Cell Biol.10, 296–303 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Montecucco, A., Zanetta, F. & Biamonti, G. Molecular mechanisms of etoposide. EXCLI J.14, 95–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan, D. M., Latham, M. D. & Ross, W. E. Proliferation-dependent topoisomerase II content as a determinant of antineoplastic drug action in human, mouse, and Chinese hamster ovary cells. Cancer Res.47, 3973–3979 (1987). [PubMed] [Google Scholar]
- 32.Blackford, A. N. & Jackson, S. P. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell66, 801–817 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Futer, O. et al. A mutational analysis of the active site of human type II inosine 5′-monophosphate dehydrogenase. Biochim. et. Biophys. Acta (BBA) - Protein Struct. Mol. Enzymol.1594, 27–39 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Pelletier, J. et al. Nucleotide depletion reveals the impaired ribosome biogenesis checkpoint as a barrier against DNA damage. EMBO J.39, e103838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diehl, F. F. et al. Nucleotide imbalance decouples cell growth from cell proliferation. Nat. Cell Biol.24, 1252–1264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates, L. A. et al. A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA. Nat. Commun.9, 5447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whelan, D. R. et al. Super-resolution visualization of distinct stalled and broken replication fork structures. PLoS Genet16, e1009256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puts, G. S., Leonard, M. K., Pamidimukkala, N. V., Snyder, D. E. & Kaetzel, D. M. Nuclear functions of NME proteins. Lab. Investig.98, 211–218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho, N. H. et al. OpenCell: Endogenous tagging for the cartography of human cellular organization. Science375, eabi6983 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mortusewicz, O., Amé, J.-C., Schreiber, V. & Leonhardt, H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res.35, 7665–7675 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo, X. & Kraus, W. L. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev.26, 417–432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, Y., Dawson, V. L. & Dawson, T. M. Poly(ADP-ribose) signals to mitochondrial AIF: A key event in parthanatos. Exp. Neurol.218, 193–202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou, Y. et al. Illuminating NAD+ metabolism in live cells and in vivo using a genetically encoded fluorescent sensor. Dev. Cell53, 240–252.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasmann, M. & Schemainda, I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res63, 7436–7442 (2003). [PubMed] [Google Scholar]
- 45.Covarrubias, A. J., Perrone, R., Grozio, A. & Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol.22, 119–141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, S., Luo, W. & Wang, Y. Emerging role of PARP‐1 and PARthanatos in ischemic stroke. J. Neurochem.160, 74–87 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, J. et al. Delayed PARP-1 inhibition alleviates post-stroke inflammation in male versus female mice: differences and similarities. Front. Cell. Neurosci.14, 77 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martire, S., Mosca, L. & d’Erme, M. PARP-1 involvement in neurodegeneration: A focus on Alzheimer’s and Parkinson’s diseases. Mech. Ageing Dev.146–148, 53–64 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Mao, K. & Zhang, G. The role of PARP1 in neurodegenerative diseases and aging. FEBS J.289, 2013–2024 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Eikesdal, H. P. et al. Olaparib monotherapy as primary treatment in unselected triple negative breast cancer. Ann. Oncol.32, 240–249 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Wang, W. et al. Shikonin is a novel and selective IMPDH2 inhibitor that target triple‐negative breast cancer. Phytother. Res.35, 463–476 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Duan, S. et al. IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways. J. Exp. Clin. Cancer Res37, 304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian, Y., Zhang, J., Chen, L. & Zhang, X. The expression and prognostic role of IMPDH2 in ovarian cancer. Ann. Diagn. Pathol.46, 151511 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Xu, Y. et al. High expression of IMPDH2 is associated with aggressive features and poor prognosis of primary nasopharyngeal carcinoma. Sci. Rep.7, 745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kofuji, S. et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat. Cell Biol.21, 1003–1014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wieczorek, P., Bałut-Wieczorek, M., Jasinski, M., Szabłoński, W. & Antczak, A. Inosine monophosphate dehydrogenase 2 as a marker of aggressive and advanced prostate cancer. Cent. Eur. J. Urol.71, 399–403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozhevnikova, E. N. et al. Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol. Cell47, 133–139 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Shi, Y., Jin, J., Ji, W. & Guan, X. Therapeutic landscape in mutational triple negative breast cancer. Mol. Cancer17, 99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brand, M. & Winter, G. E. Locus-specific knock-in of a degradable tag for target validation studies. in Target Identification and Validation in Drug Discovery (eds. Moll, J. & Carotta, S.) vol. 1953 105–119 (Springer New York, New York, NY, 2019). [DOI] [PubMed]
- 60.Serebrenik, Y. V., Sansbury, S. E., Kumar, S. S., Henao-Mejia, J. & Shalem, O. Efficient and flexible tagging of endogenous genes by homology-independent intron targeting. Genome Res29, 1322–1328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep.7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bajar, B. T. et al. Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat. Methods13, 993–996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics35, 2084–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: J. Integr. Biol.16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, G. Gene ontology semantic similarity analysis using GOSemSim. in Stem Cell Transcriptional Networks (ed. Kidder, B. L.) vol. 2117 207–215 (Springer US, New York, NY, 2020) [DOI] [PubMed]
- 70.Yu, G. et al. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics26, 976–978 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Gu, Z. Complex heatmap visualization. iMeta1, e43 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wiśniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Perkins, D. N., Pappin, D. J. C., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis20, 3551–3567 (1999). [DOI] [PubMed] [Google Scholar]
- 75.Beer, L. A., Liu, P., Ky, B., Barnhart, K. T. & Speicher, D. W. Efficient quantitative comparisons of plasma proteomes using label-free analysis with MaxQuant. in Serum/Plasma Proteomics (eds. Greening, D. W. & Simpson, R. J.) vol. 1619 339–352 (Springer New York, New York, NY, 2017). [DOI] [PMC free article] [PubMed]
- 76.Mi, H. et al. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc.14, 703–721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, X. et al. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc.13, 530–550 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Ahlmann-Eltze, C. proDA: differential abundance analysis of label-free mass spectrometry data. Bioconductor R Package Version1, https://github.com/const-ae/proDA (2022).
- 79.Gianetto, Q. G., Wieczorek, S., Couté, Y. & Burger, T. APeptide-Level Multiple Imputation Strategy Accounting for the Different Natures of Missing Values in Proteomics Data. bioRxiv 2020.05.29.122770; 10.1101/2020.05.29.122770 (2020).
- 80.Storey, J. D., Bass, A. J., Dabney, A. & Robinson, D. _qvalue: Q-value estimation for false discovery rate (2022).
- 81.Thul, P. J. et al. A subcellular map of the human proteome. Science356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Wickham, H. et al. Welcome to the Tidyverse. JOSS4, 1686 (2019). [Google Scholar]
- 84.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Espinar, L. et al. Nuclear IMPDH2 controls the DNA damage response by modulating PARP1 activity. GitHub repository 10.5281/zenodo.13770471.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Protein interactors of TOP2A and PARP1 were derived from the OpenCell project39. Data for IMPDH2 protein expression upon etoposide treatment and release were obtained from Moretton et al.12. The raw proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the PRIDE repository84 with the dataset identifier PXD043522. RNA-sequencing data generated in this study have been deposited in the NCBI GEO database under accession code GSE271269. High-throughput microscopy data are publicly available in the SdelciLab GitHub repository [https://github.com/SdelciLab/IMPDH2_chromatin]85. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
All the scripts used for this manuscript are publicly available in the GitHub repository [https://github.com/SdelciLab/IMPDH2_chromatin]85.