Abstract
Schizophrenia is a disabling mental disorder that affects 1% of people over their lifetime. The etiology and mechanism of schizophrenia are very complex, and many genes are involved in many different signaling pathways in the etiology of this disease. According to recent studies, one of the important mechanisms altered in this disorder is the regulation of immune system and the inflammation mechanism. In the present study, we evaluated the peripheral blood expression pattern of four lncRNAs and three protein-coding genes in the treatment- naïve patients, and medicated patients compared with sex and age-matched controls. In the medicated-patients, expression levels of IFNG, IL18RAP, AC007278.2 were significantly up-regulated (P < 0.05); and the expression level of IFNG-AS1-001 was significantly down-regulated compared to healthy controls (P < 0.05). However, levels of IL18R1, AC007278.3 and IFNG-AS1-003 were not different between these groups. In the treatment-naïve patients, IFNG, IL18R1, IL18RAP, IFNG-AS1-001, AC007278.2, and AC007278.3 were significantly up-regulated compared to controls. On the other hand, IFNG-AS1-003 was significantly down-regulated in the treatment-naïve patients compared to controls. Based on the Spearman correlation matrix, there was a significant correlation between genes in the treatment-naïve patients. We also showed the high sensitivity and specificity of IFNG-AS1-003, IFNG, IL18R1, and AC007278.3 in the identification of treatment-naïve patients from controls. The current study contributes further evidence to the understanding of the role of lncRNAs in the pathogenesis of schizophrenia. Future research is necessary to establish the validity of lncRNAs as peripheral markers for this condition.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78220-w.
Keywords: Schizophrenia, IFNG, IL18R, IL18RAP, IFNG-AS1-001
Subject terms: Genetics, Diseases
Introduction
Schizophrenia is one of the most disabling mental disorders that affects about 1% of people over their lifetime1. The prevalence of this disease is approximately 0.3 to 0.7% worldwide. The disease usually occurs sporadically. Its symptoms usually appear in adulthood and early adolescence, leading to severe disability and high level of stress1. This disorder is determined by the existence of various symptoms, including positive symptoms (hallucinations, delusions, unusual behaviors, unusual speech, abnormal thought, and movement disorder), negative symptoms (insensibility, lack of delight, attention impairment, and sociality withdrawal), and cognitive symptoms (imperfection in executive function and precision, and agnosia)2. The etiology of schizophrenia has not been completely identified3. However, it is clear that mechanisms of schizophrenia are very complex, and many environmental and genetic factors play a pivotal role in causing this disease. There is massive evidence indicating that changes in gene expression in the immune system contribute to the pathogenesis of this disorder4,5. Dysregulation of the immune system and its intricate interplay with the nervous system might play a part in the etiology and physiological mechanisms of schizophrenia6. The reciprocal relationship between the immune system and the brain has sparked a rising curiosity regarding the involvement of the immune system in neuropsychiatric disorders. Notably, the atypical blood lymphocyte parameters, such as the levels of total T lymphocytes and T helper cells, have specifically attracted attention7.
Numerous studies have shown that inflammation and immunity play significant roles in the development of symptoms associated with schizophrenia8,9. These studies have provided compelling evidence that systemic inflammation can have a profound impact on the brain, resulting in alterations in mood, cognition, and behavior10.
Long non-coding RNAs are a subset of non-coding transcripts and have a length of more than 200 nucleotides11. Studies have demonstrated that some lncRNAs are effective in controlling the behavior of immune cells and immune responses12, including the differentiation and activity of T and B cells, macrophages, and NK cells. Several lncRNAs play pivotal roles in cell function and participate in the pathogenesis and development of various diseases such as cancer13and especially neuropsychiatric disorders and neurodegenerative diseases such as Alzheimer’s disease14, Parkinson’s disease15, major depressive disorder16, autism spectrum disorders17and multiple sclerosis18.
Based on the functional roles in the regulation of immune responses, we selected four lncRNAs, namely IFNG-AS1-001, IFNG-AS1-003, AC007278.2, and AC007278.3; and three protein-coding genes, namely IFNG, IL18R1, and IL18RAP to assess their expression in peripheral blood of treatment-naive and medicated schizophrenia patients compared with matched healthy controls. IFNGhas been suggested to partake in the pathogenesis of schizophrenia19,20. Methylation of the IFNG locus is regulated by IFNG-AS1-00121. IFNG-AS1-003 gene is also located on the same chromosome as the IFNG22, possibly contributing to regulation of this gene. Similarly, IL-18is involved in the pathologic events seen in this disorder23. This cytokine exerts its effect through its receptor being encoded by IL18R1 and IL18RAPgenes24. Two functionally related lncRNAs, namely AC007278.2 and AC007278.3 are located on chromosome 2 inside the introns of IL18R1 and IL18RAP, respectively.
This study provides new insights into the altered expression of protein-coding and non-coding genes related to the immune system and proposes them as contributors in the pathogenesis of schizophrenia and as novel biomarkers for the diagnosis of schizophrenia.
Materials and methods
Study participants
The present study was performed on 50 medicated and 25 treatment-naive schizophrenia patients, and 50 sex and age-matched healthy controls. Cases were recruited from Razi hospital, Tabriz, Iran. The fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-V)25 was applied in the diagnostic process. The participants with alcohol drinking and substance abuse or cigarette smoking were excluded from the study. Healthy controls were selected from those were referred to the health centers of Tabriz University of Medical Sciences. The Mini-International Neuropsychiatric Interview was used for the assessment of healthy controls. Exclusion criteria were the existence of any systemic disorder, psychiatric condition, pregnancy, or a history of psychopathology in a first-degree biological relative. The study protocol was approved by the Shahroud University of Medical Sciences Ethical Committee (IR.SHMU.REC.1398.113). Informed consent forms were signed by all participants/or their guardians.
RNA extraction and cDNA synthesis
In the first step, 5 ml of the peripheral blood was gathered in K2-EDTA-containing tubes. The RNA of all samples was extracted by using the Hybrid-R blood RNA extraction Kit (Gene All, Seoul, Korea). The Quality of the extracted RNAs was verified by 1% agarose gel electrophoresis, and to eliminate any genomic DNA contamination, samples were treated with DNase I (Thermo scientific, Deutschland, Germany). The quantity of RNA was appraised by Nanodrop equipment (Thermo Scientific, MA, USA). The extracted RNA was subsequently converted to cDNA by using the High-Capacity cDNA Reverse Transcription FIRE Script RT cDNA Synthesis Kit (Solis Bio Dyne, Estonia).
Primer design
Primer design was accomplished using the NCBI Primer designing tool and verified through blasting in the nucleotide BLAST database to confirm specific binding to target sequences. The primer sequences are detailed in Table 1.
Table 1.
Primers sequences.
| Primers | Sequence 5’– 3’ | Tm (C°) | |
|---|---|---|---|
| IFNG-AS1-001 | F | ACATACTTCCACCAGAGA | 62 |
| R | TTCCACAACACTATCAACT | ||
| IFNG-AS1-003 | F | CCAGCACCATAATTCCAG | 60 |
| R | GAAGACATGAGCACTGAC | ||
| AC007278.2 | F | ATCATCTGTATGCTGTCTAAC | 60 |
| R | AACCATATAATGAGGCTGTC | ||
| AC007278.3 | F | TTGAAGAGGAGATTAAGTATTAGG | 64 |
| R | TCGATCATCTTCACATTCACATC | ||
| IFNG | F | GTTCTCTTGGCTGTTACTG | 58 |
| R | CATTATCCGCTACATCTGAAT | ||
| IL18R1 | F | GAGAAACATTTTGGGTATAAGTTATG | 62 |
| R | CTCTATCAGTGAGTGGATTTC | ||
| IL18RAP | F | AACACTCTACTCTGGCAAA | 62 |
| R | ATCCTTTAATTCGCTCTCCT | ||
| UBC | F | GGATTTGGGTCGCAGTTCTTG | 60 |
| R | TGCCTTGACATTCTCGATGG | ||
| YWHAZ | F | ACTTTTGGTACATTGTGGCTTC | 60 |
| R | CCGCCAGGACAAACCAGTA | ||
qRT-PCR
Expression levels of genes were assessed in all participants using the RealQ Plus 2 × PCR Master Mix Green with high ROX (Amplicon, Odense, Denmark). Cycling reactions were carried out in Step One Plus Real-Time PCR equipment (Applied Biosystems, Foster City, CA, USA). After evaluation of Ct values of UBC and YWHAZ, we used UBC gene as the reference gene based on its constant expression in the peripheral blood of schizophrenia patients. The stable expression of UBC was also confirmed by NormFinder software (https://www.moma.dk/software/normfinder).
Statistical analysis
Relative expression levels of genes were measured in all samples. The Ln [Efficiency^ΔΔCT] method considers the transcript levels of UBC as normalizer. The Shapiro-Wilk test was performed to evaluate the normality of the data. One-way ANOVA and Bonferroni’s multiple comparisons test were used for comparison of expression data between study groups. Correlations between expressions of genes were valued by calculation of Spearman correlation coefficients. Data was analyzed using the GraphPad Prism 8.0.0 software. The diagnostic power of the transcript levels of genes was measured by depicting receiver operating characteristic (ROC) curves.
Results
General data of patients and controls
A total of 50 medicated patients, 25 treatment-naïve patients, and 50 healthy controls were recruited for the current case-control study. No significant difference was found between the age and sex ratios of cases and controls. Demographic data of the participants in the study are summarized in Table 2.
Table 2.
Demographic data of patients and controls.
| Study groups | Parameters | Values | ||
|---|---|---|---|---|
| Treatment-naive patients | Gender (number, %) | Male | 20 (80%) | |
| Female | 5 (20%) | |||
| Age (Years, mean ± SD) | Male | 27 ± 7.33 | ||
| Female | 24 ± 3.71 | |||
| Family History (number, %) | Yes | 15 (60%) | ||
| No | 10 (40%) | |||
| Education (%) | Illiterate | 40% | ||
| School | 32% | |||
| High School | 24% | |||
| University | 4% | |||
| PANNS (mean ± SD) | 72.52 ± 8.47 | |||
| BPRS (mean ± SD) | 51.16 ± 4.69 | |||
| Positive symptoms score (mean ± SD) | 17.12 ± 3.52 | |||
| Negative symptoms score (mean ± SD) | 16.84 ± 4.32 | |||
| CRP (mg/L) (mean ± SD) | 11.32 ± 3.23 | |||
| Medicated Patients | Gender (number, %) | Male | 42 (84%) | |
| Female | 8 (16%) | |||
| Age (Years, mean ± SD) | Male | 42.26 ± 8.42 | ||
| Female | 37.33 ± 11.5 | |||
| Age at onset (Years, mean ± SD) | Male | 28.36 ± 6 | ||
| Female | 29 ± 3.6 | |||
| Duration (Years, mean ± SD) | Male | 14.45 ± 6.8 | ||
| Female | 8 ± 8.18 | |||
| Family History (number, %) | Yes | 27 (54%) | ||
| No | 23 (46%) | |||
| Education (%) | Illiterate | 20% | ||
| School | 30% | |||
| High School | 44% | |||
| University | 6% | |||
| PANNS (mean ± SD) | 58.12 ± 7.87 | |||
| BPRS (mean ± SD) | 33.62 ± 6.33 | |||
| Positive symptoms score (mean ± SD) | 12.86 ± 3.38 | |||
| Negative symptoms score (mean ± SD) | 10.68 ± 2.85 | |||
| CRP (mg/L) (mean ± SD) | 5.88 ± 2.22 | |||
| Clozapine dose (mg/day) (mean ± SD) | 172 ± 82.16 | |||
| Controls | Gender (number, %) | Male | 42 (84%) | |
| Female | 8 (16%) | |||
| Age (Years, mean ± SD (range)) | Male | 39.5 ± 8.81 | ||
| Female | 41 ± 9.51 | |||
| Education (%) | Illiterate | 0 | ||
| School | 10% | |||
| High School | 48% | |||
| University | 42% | |||
Expression assays
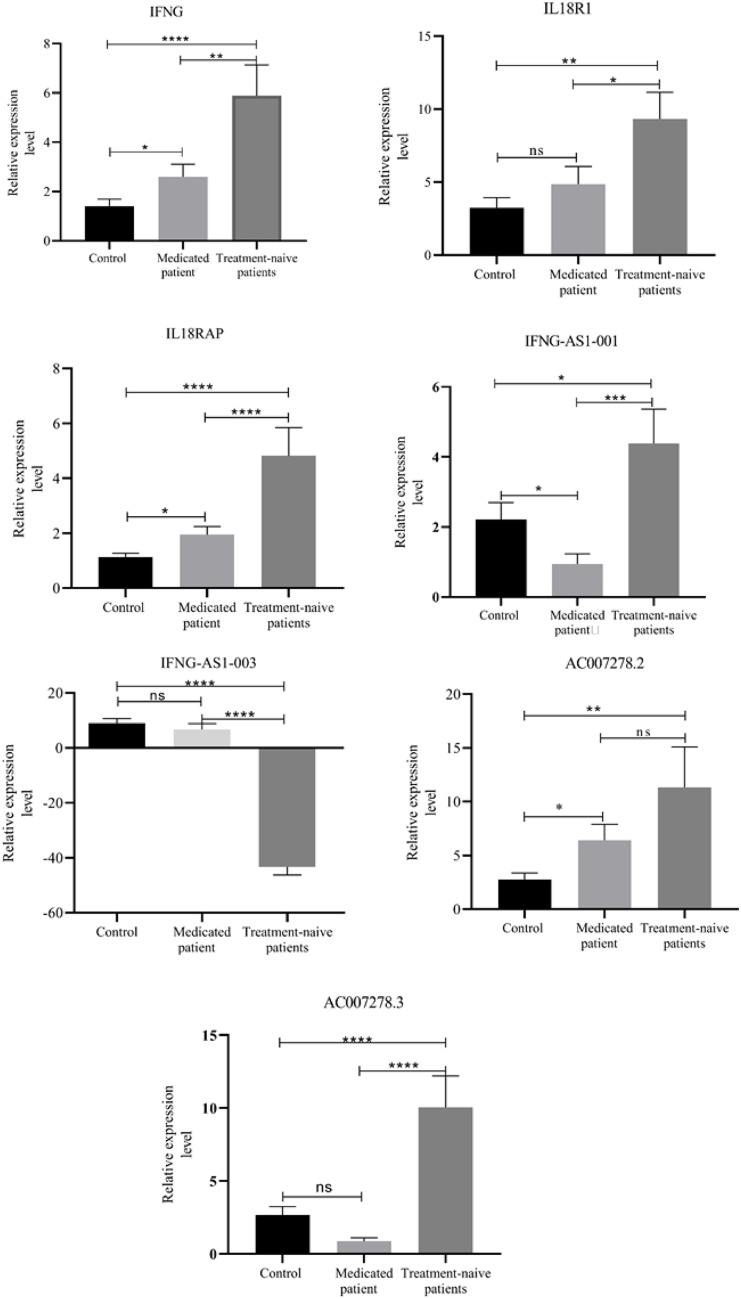
Relative expression of genes was compared between medicated patients, treatment-naïve patients, and healthy controls. Expression level of IFNG-AS1-001 was significantly downregulated in medicated-patient (P < 0.05); while expression levels of IFNG, IL18RAP, and AC007278.2 were significantly upregulated in medicated patients compared to controls (P < 0.05). Expression levels of IFNG-AS1-003, AC007278.3 and IL18R1 were not significantly different between this group of patients and healthy controls (Fig. 1).
Figure 1.
Expressions levels of genes in treatment-naïve and medicated patients compared to healthy controls. Expression levels of genes were calculated using the (Efficiency^-ΔΔCT) method. ns: not significant, P < 0.05(*), P < 0.01(**), P < 0.001(***), P ≤ 0.0001(****).
Expression levels of IFNG, IFNG-AS1-001, IL18R1, IL18RAP, AC007278.2, and AC007278.3 were significantly upregulated in treatment-naïve patients compared to controls (P < 0.0001). On the other hand, expression level of IFNG-AS1-003 was significantly downregulated in treatment-naïve patients compared to controls (P < 0.0001) (Fig. 1).
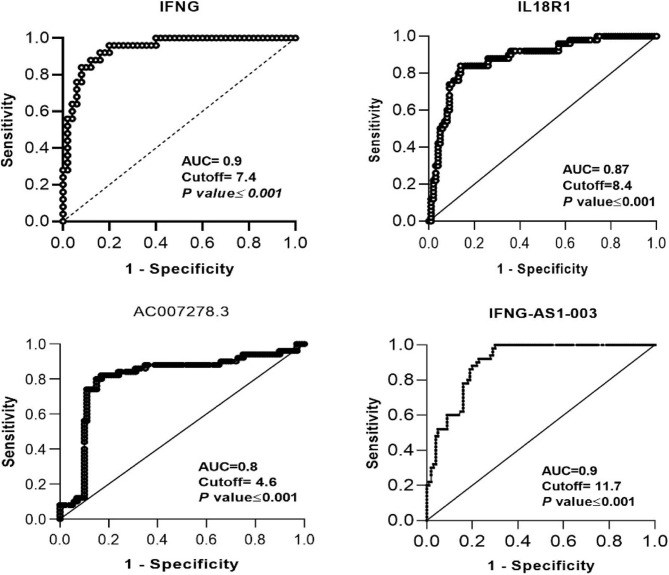
ROC curve analysis
In the present study, we evaluated the diagnostic power of transcript quantities of IFNG, IL18R1, IL18RAP, IFNG-AS1-003, AC007278.2, and AC007278.3 in identifying between the treatment-naïve patients and controls by depicting ROC curve (Table 3).
Table 3.
Result of ROC curve analysis between treatment-naïve patients and controls.
| Gene | Specificity | Sensitivity | AUC | Cutoff | p-value |
|---|---|---|---|---|---|
| IFNG-AS1-003 | 0.94 | 0.7 | 0.9 | 11.7 | < 0.001 |
| IL18R1 | 0.9 | 0.84 | 0.879 | 8.4 | < 0.001 |
| IFNG | 0.9 | 0.8 | 0.94 | 7.4 | < 0.001 |
| AC007278.3 | 0.82 | 0.84 | 0.8 | 4.66 | < 0.001 |
Based on the area under cover (AUC) value, INFG-AS1-003, IFNG, IL18R1, and AC007278.3 had powerful diagnostic power (AUC = 0.90, 0.9, 0.87, and 0.80, P < 0.0001) (Fig. 2).
Figure 2.
The diagnostic power of transcript quantities of IFNG, IL18R1, IFNG-AS1-003, and AC007278.3 in identifying between treatment-naïve patients and controls.
Additionally, we assessed the diagnostic power of all differentially expressed genes in 3 in identifying between treatment-naïve patients and controls (Fig. 3). Transcript levels of these genes could separate these groups with AUC = 0.93, sensitivity = 0.96 and specifiicity = 0.73.
Figure 3.
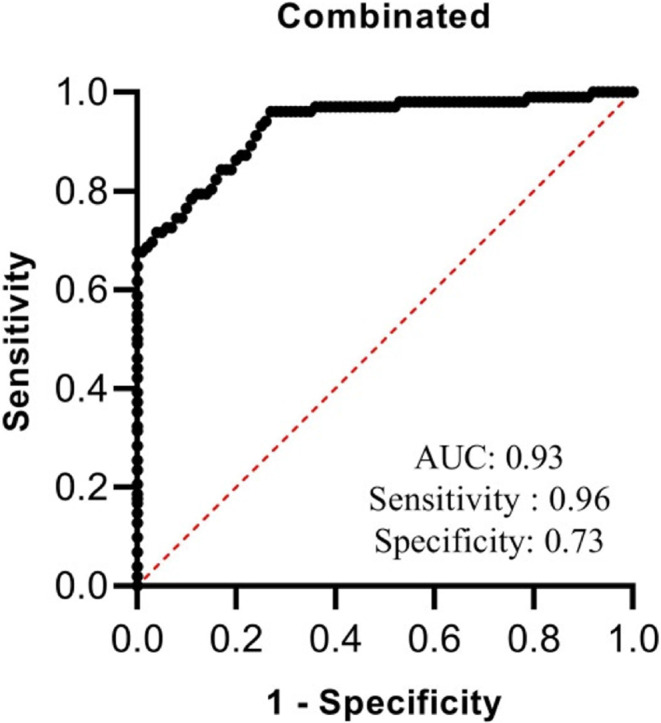
The diagnostic power of combination of transcript quantities of IFNG, IL18R1, IFNG-AS1-003, and AC007278.3 in identifying between treatment-naïve patients and controls.
Correlation matrix analysis
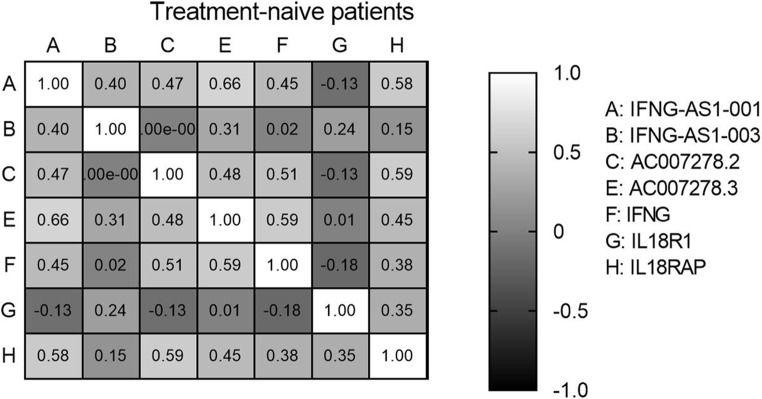
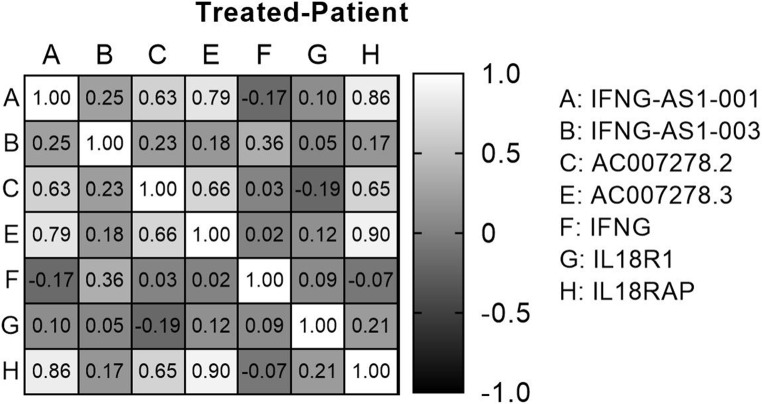
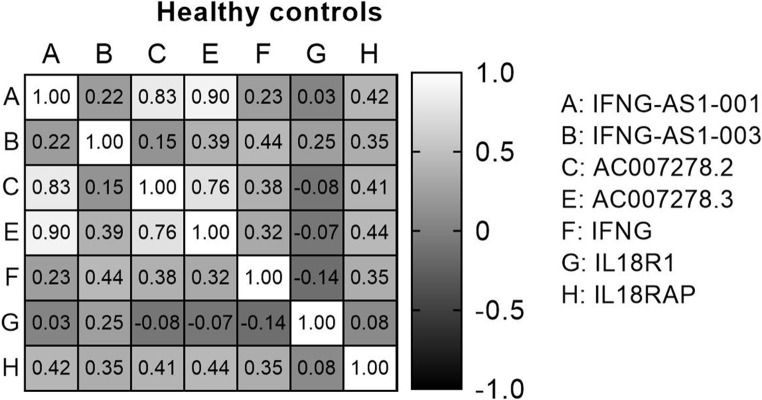
The analysis of the Spearman correlation matrix among genes in all subjects allows us to examine the connections between genes, as evidenced by their correlation coefficients and corresponding p-values. Significant positive and negative correlations between genes were observed in all three groups of medicated patients, treatment-naïve patients, and healthy controls, which indicated their expression relationship in the disease (Figs. 4, 5 and 6).
Figure 4.
Spearman correlation between genes in treatment-naive patients.
Figure 5.
Spearman correlation between genes in medicated patients.
Figure 6.
Spearman correlation between genes in healthy controls.
Finally, we assessed correlation between expression of genes and clinicopathological scores (Tables 4 and 5). In the medicated patients, we found positive correlations between expression of IL18RAP and BPRS, expression of IFNG-AS1-001 and both PANNS and negative symptoms score, and expression of AC007278.2 and PANNS. Moreover, inverse correlations were detected between CRP levels and expression levels of both IFNG-AS1-001 and AC007278.3.
Table 4.
Spearman correlation matrix analysis between genes and schizophrenia assessments criteria in the medicated patients.
| Genes | Parameters | PANNS | BPRS | Positive symptoms | Negative symptoms | CRP | Clozapine dose |
|---|---|---|---|---|---|---|---|
| IFNG | r |
0.017 0.905 |
0.048 0.739 |
0.042 0.771 |
0.151 0.296 |
-0.118 0.421 |
− 0.166 0.248 |
| P value | |||||||
| IL18R1 | r |
-0.182 0.206 |
-0.259 0.069 |
-0.174 0.227 |
0.201 0.161 |
-0.142 0.331 |
− 0.212 0.139 |
| P value | |||||||
| IL18RAP | r |
0.249 0.081 |
0.341* 0.015 |
0.175 0.225 |
0.029 0.843 |
0.051 0.729 |
0.144 0.318 |
| P value | |||||||
| IFNG-AS1-001 | r |
0.280* 0.049 |
0.080 0.579 |
0.130 0.370 |
0.309* 0.029 |
-0.287* 0.045 |
0.073 0.613 |
| P value | |||||||
| IFNG-AS1-003 | r |
0.162 0.261 |
0.073 0.616 |
0.090 0.533 |
0.081 0.574 |
-0.135 0.354 |
0.031 0.083 |
| P value | |||||||
| AC007278.2 | r |
0.3* 0.034 |
0.165 0.252 |
0.241 0.092 |
0.209 0.145 |
-0.161 0.270 |
0.69 0.632 |
| P value | |||||||
| AC007278.3 | r |
0.220 0.125 |
0.099 0.495 |
0.138 0.339 |
0.275 0.053 |
-0.319* 0.026 |
0.19 0.89 |
| P value |
Table 5.
Spearman correlation matrix analysis between genes and schizophrenia assessments criteria in treatment-naive patients.
| Genes | Parameters | PANNS | BPRS | Positive symptoms | Negative symptoms | CRP |
|---|---|---|---|---|---|---|
| IFNG | r |
-0.157 0.454 |
-0.108 0.606 |
0.219 0.293 |
-0.265 0.201 |
0.319 0.128 |
| P value | ||||||
| IL18R1 | r |
0.209 0.317 |
0.01 0.962 |
-0.265 0.201 |
0.092 0.662 |
-0.069 0.748 |
| P value | ||||||
| IL18RAP | r |
-0.053 0.801 |
-0.355 0.082 |
-0.162 0.438 |
0.267 0.197 |
-0.066 0.758 |
| P value | ||||||
| IFNG-AS1-001 | r |
-0.209 0.316 |
-0.557 0.004 |
-0.719 0.000 |
0.221 0.289 |
0.162 0.449 |
| P value | ||||||
| IFNG-AS1-003 | r |
0.248 0.232 |
0.197 0.346 |
-0.205 0.327 |
0.387 0.056 |
0.18 0.4 |
| P value | ||||||
| AC007278.2 | r |
0.021 0.92 |
0.14 0.505 |
0.147 0.484 |
-0.105 0.618 |
-0.175 0.415 |
| P value | ||||||
| AC007278.3 | r |
-0.072 0.731 |
-0.057 0.786 |
-0.231 0.268 |
0.367 0.071 |
-0.132 0.54 |
| P value |
Among treatment-naïve patients, expression of IFNG-AS1-001 was inversely correlated with BPRS and positive symptoms.
Discussion
After intensive research, it is now clear that a gene or protein cannot explain such a complex disease as schizophrenia. Schizophrenia is a multifactorial disease in which a large number of genes and different cellular signaling pathways are involved in its initiation and development. One of the important pathways involved in the development of schizophrenia is the inflammatory pathway26. Cytokines have a critical role in initiating and maintaining immune responses. They can easily cross the blood-brain barrier and act as a major mediator between the brain and the immune system27. Many studies showed that there is a significant alteration in the levels of inflammatory cytokines in the blood of schizophrenia patients compared to healthy controls28.
The aim of this study was to evaluate the expression level of IFNG, IL18R1, IL18RAP, and IFNG-AS1-001, IFNG-AS1-003, AC007278.2, and AC007278.3 in treatment-naïve schizophrenic patients who were in the acute phase of the disease as well as patients who were in the remission phase of the disease (medicated) compared to matched controls.
Interleukin 18 (IL-18) is one of the multifunctional cytokines that is structurally similar to the IL-1 family and is one of the main factors inducing the secretion of IFNG from T-helper cells29. IL-18 has a role in many psychiatric disorders. IL-18 signaling has been found to be interrupted in the central amygdala in an animal model of post-traumatic stress and alcohol use disorder30. This cytokine has a possible role in the modulation of the hypothalamic–pituitary–adrenal axis and might mediate the CNS dependent impacts on the susceptibility to related disorders31. Moreover, in many neurodegenerative and inflammatory diseases, the expression of IL-18 receptor (IL18R) and also the expression of IL18were increased32. The IL18R has two subunits, including IL18R1 (IL18α) and IL18RAP or (IL18ß)24. Two functionally related lncRNAs, namely AC007278.2 and AC007278.3 are located on chromosome 2 inside the introns of IL18R1 and IL18RAP, respectively. IL-18 is secreted by macrophage-like cells and plays a pivotal role in the response of helper T cells33. IL18R is widely present in neurons. Thus, the IL-18 that is synthesized in the CNS, can affect neuronal growth, differentiation, and apoptosis34. This interleukin exerts its inflammatory role by increasing IFNG production from T cells and NK cells. Expression level of IL18in the blood of patients with schizophrenia has been shown to be higher than controls23. This finding has also been confirmed by Szabo et al., the serum level of IL18 significantly increased in schizophrenia patients compared to HCs35.
Luo et al. have shown that the concentration of IL-18 in the serum of patients is not significantly altered between treatment-naïve and medicated patients, and the concentration level of IL-18 is increased in both groups compared to healthy controls36. Thus, one can infer that antipsychotic treatment does not change the concentration of IL-18. This finding was also verified in our study.
In the present study, we also evaluated the expression level of IL-18 receptor subunits, namely IL18R1 and IL18RAP. The findings from the earlier research indicate that the binding of IL-18 to IL18R1 induces a sequence of processes that activates multiple signaling pathways, such as NF-kB and MAPK pathways, resulting in the generation of inflammatory cytokines and chemokines37. Therefore, based on previous studies, there is a direct relationship between the level of IL-18 and IL18R, IL18RAP. In the present study, we showed that the expression levels of IL18R1 and IL18RAP were higher in both treatment-naive and medicated groups compared to healthy controls. We did not observe any significant expression changes between medicated and treatment-naive patients, which indicates that the use of antipsychotic drugs does not have a significant effect on the expression of IL-18 receptor subunits. In the ROC curve analysis of IL18R1, the sensitivity of 84% and the specificity of 90% indicated that it can be used as a suitable diagnostic biomarker to identify treatment-naïve patients from healthy with a expression cutoff of 8.4, which is statistically significant.
Another subunit of the IL18 receptor is IL18RAP. The expression of IL18RAP was significantly increased in treatment-naïve and medicated patients.
In line with our results, Xu et al. have analyzed GWAS and replicated the results in an independent cohort of schizophrenia patients. They showed association signals within IL18R1 and IL18RAP genes, with the most significant marker being IL18R1rs1035130. They have also reported altered levels of IL-18 and IL18R1 in schizophrenia patients compared with controls38.
We suggest that by increasing the expression of IL-18 receptor subunits, the expression of IL-18 also increases, which causes an increase in the amount of IFNG. IL-18 and IFNG are closely related cytokines that play important roles in the immune response. IL-18 is known to enhance the production of IFNG, and both cytokines can synergistically amplify immune responses39. In Spearman’s correlation matrix analysis, there was no significant relationship between the expression of IL18R1 and IL18RAP in any of study subgroups.
We found positive correlation between expression of IL18RAP and BPRS in the medicated patients. In a previous study, elevation of serum concentration of IL-18 and a certain polymorphisms within IL-18gene have been reported to be positively associated with the PANSS general psychopathology subscore and the PANSS total score40. However, there was no data about correlation between expression of IL-18 receptor subunits and mentioned scores.
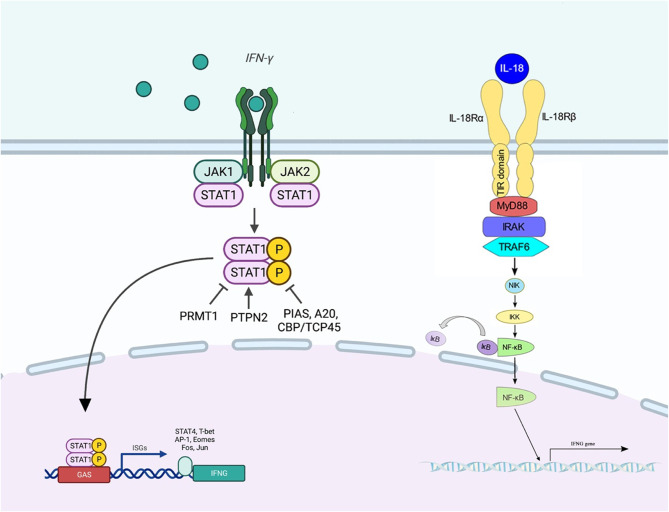
IFNG is a soluble cytokine of the type II class of IFNs41. It is a key activator of macrophages and inducer of MHC ΙΙ molecule expression42. Abnormal expression of IFNGhas been associated with numerous autoinflammatory and autoimmune diseases43. This cytokine is synthesized by T helper cells (particularly, Th1 cells), cytotoxic T cells, macrophages, mucosal epithelial cells, and NK cells. Moreover, it acts as a crucial autocrine signal for professional antigen-presenting cells (APCs) in early innate immune responses and a key paracrine signal in the adaptive immune response. Notably, expression of this molecule is induced by a number of cytokines, including IL-18 and type I IFN44. It is a cytokine of T helper 145and regulates the presentation of antigens and the division and differentiation of lymphocytes. Meta-analyses of cytokine changes in schizophrenia have shown that IFNG can be a marker of disease diagnosis. The IFNG secretion is controlled by APCs, IL12 and IL1846. When the IFNG ligand binds to the receptor, the receptor is dimerized and causes JAK1 and JAK2 to come close to each other, leading to the phosphorylation and activation of STAT1. Activated STAT1 causes activation of STAT4. Activated STAT4 goes to the nucleus and increases the expression of the IFNGgene47. Figure 7 shows the relationships between IL-18, IFNG, JAK1, JAK2, STAT1, and STAT4.
Figure 7.
The relationships between IL18, IFNG, JAK1, JAK2, STAT1, and STAT4. When the IFNG ligand binds to the receptor, the receptor is dimerized. Then, JAK1 and JAK2 become close to each other. This leads to the phosphorylation and activation of STAT1. Activated STAT1 causes activation of STAT4. This cascade of events leads to up-regulation of IFNG.
IFNGhas been shown to be significantly decreased in the medicated schizophrenia patients compared to healthy controls, while its expression has been up-regulated in the treatment-naïve patients48. Another study has shown that the expression level of this gene is higher in schizophrenia patients compared with controls49. In the medicated patients, we found positive correlations between expression of IFNG-AS1-001 and both PANNS and negative symptoms score, and expression of AC007278.2 and PANNS. Moreover, inverse correlations were detected between CRP levels and expression levels of both IFNG-AS1-001 and AC007278.3. However, among treatment-naïve patients, expression of IFNG-AS1-001 was inversely correlated with BPRS and positive symptoms. Thus, it can be inferred that medication affects correlation between mentioned genes and clinicopathological scores.
IL-18 and IFNG, two closely related cytokines, have a significant impact on the immune response as they often interact with each other. Both IL-18 and IFNG are classified as pro-inflammatory cytokines and play crucial roles in regulating immune responses, particularly in the context of defending the host against infections and managing the inflammatory response. One of the key aspects of their interaction is that IL-18 can stimulate the production of IFNG from various immune cells, such as T cells and NK cells. In response, IFNG can further enhance the expression of IL-18 receptors on immune cells. This creates a positive feedback loop within the immune response, where IL-18 and IFNG mutually reinforce each other’s effects. This interaction between IL-18 and IFNG serves to amplify the inflammatory response, leading to a more robust immune defense against pathogens. By working together, IL-18 and IFNG contribute to the coordination and regulation of immune responses, ultimately aiding in the protection of the host organism50. Based on previous studies, the elevated IL18R1 and IL18RAP expression can positively up-regulate the expression level of IFNG. In the present study, we showed that the expression level of IFNG in treatment-naïve patients significantly increased, and on the other hand, the level of IFNG expression in medicated patients compared decreased compared to controls. Based on the ROC curve analysis, IFNG had the highest AUC among the genes, at 0.94, and sensitivity of 0.8, specificity of 0.9 which significantly indicate its diagnostic ability (P < 0.0001).
IFNG-AS1, which is an intergenic lncRNAs, is located on the same chromosome as the IFNG gene, and is known as Tmevpg1; NEST51. IFNG-AS1has a number of variants52, and in the present study, we investigated the expression level of two variants, including IFNG-AS-001 and IFNG-AS-003. The expression level of the IFNG-AS1-001 variant decreased twofold in the medicated patients compared with controls, while its expression level was higher in the treatment-naïve patients. Most notably, while the expression level of the IFNG-AS-003 gene was similar between the medicated patients and controls, its expression was significantly lower in the treatment-naïve patients compared to controls.
Expression level of the IFNG-AS1-001 gene was decreased in the medicated patients compared with controls. IFNG-AS1-001 contributes to the methylation of the IFNGlocus and decreases its expression by binding to WDR5, a scaffolding protein in the H3K4 methyltransferase complex21. Thus, down-regulation of IFNG-AS1-001 is expected to result in the up-regulation of IFNG. IFNG-AS1-003 gene is also located on the same chromosome as the IFNG22. The observed downregulation of the IFNG-AS1-003variant in the current study is consistent with the previous studies that were conducted on other diseases such as Hashimoto’s disease53. In fact, AC007278.2 and IFNG-AS1-001 have been shown to be up-regulated in the relapsing phase in multiple sclerosis patients, while IFNG-AS1-003has been up-regulated in the remitting phase compared with relapsing phase52.
Expression levels of AC007278.2 and AC007278.3 were higher in the treatment-naïve patient compared to the controls. These lncRNAs are directly related with the increase of IL18R1 and IL18RAP genes. These genes are located on the positive strand of chromosome 2q12, and inside the IL18R1gene intron and play an important role in the expression and differentiation of T helper1 cells54. It has been disclosed that the expression level of AC007278.2 and AC007278.3 lncRNAs is aligned with the IL18R1 and IL18RAP gene expression. Up-regulation of the mentioned lncRNAs has been correlated with over-expression of IL18R1 and IL18RAP genes52. The increase in the expression of IL18R1 and IL18RAgenes causes the activation of STAT4 and IL-2, as well as activation of histone acetyltransferase and DNA methylase complex. Finally, IL18R1 and IL18RAP cause the differentiation of T helper1 cells, and this differentiation causes the production of pro-inflammatory cytokines from T helper1 cells, which is one of the main causes of autoimmune diseases55. Therefore, it is likely that the expression level of AC007278.2 and AC007278.3 is directly related to the level of expression of genes coding for the IL-18 receptor protein, and the use of antipsychotic drugs may reduce their levels. Based on the ROC curve analysis, AC007278.3 had the sensitivity of 84% and the specificity of 82% with AUC = 80%, thus it can be used as a suitable diagnostic biomarker to identify treatment-naïve patients from controls with an expression cutoff of 4.66.
Finally, assessment of pairwise correlation in the patients group showed that there was a significant direct relationship between genes. In both medicated and treatment-naïve patients, IFNG-AS1-001 showed significantly strong positive correlations with AC007278.2, AC007278.3, and IL18RAP. Moreover, AC007278.2 had a significant correlation with IFNG-AS1-001, AC007278.3 and IL18RAP in both groups. However, the correlations between AC007278.2/IFNG and AC007278.3/ IFNG were only significant in the treatment-naïve patients. On the other hand, the correlation between and IFNG and IFNG-AS1-003 was only significant among medicated patients. Thus, it can be inferred from the correlation data that medication affects the correlation between IFNG and other genes.
Conclusion
In summary, this study showed changes in the expression of a number of lncRNA genes and related protein-coding genes in the blood of patients with schizophrenia. Notably, IFNG, IFNG-AS1-003, IL18R1, and AC007278.3 genes were found to be suitable biomarkers in this disorder. In order to confirm the biomarker power of genes and gene expression changes in the acute phase and remission of the disease, more studies in a larger statistical population are suggested. Our study had some limitations including lack of assessment of expression of mRNA coding genes at protein level and lack of functional studies.
Supplementary Information
Acknowledgements
This study was founded by the Shahroud University of Medical Science, Iran. The present study was supported by Shahroud University of medical sciences as a MSc Thesis. We hereby acknowledge the research deputy for grant No 9870
Research involving human participants and/or animals
The study was carried out according to the principles of the 1964 Declaration of Helsinki. Each enrolled subject provided a written informed consent, and all study data were obtained and elaborated in accordance with our institutional ethical committee regulations. The study protocol was approved by the Shahroud University of Medical Sciences Ethical Committee (IR.SHMU.REC.1398.113).
Informed consent
Informed consent was obtained from patients.
Consent to participate
Not applicable.
Author contributions
Kamran Javidi and Shima Rahmani performed the majority of experiments and data analysis. Faezeh Mehdizadeh, Fatemeh Manafzadeh, Seyed Gholamreza Noor Azar and Shahrokh Aghayan contributed to the experiments and interpreted the results. Faezeh Mehdizadeh wrote the manuscript and helped with the experiments. Behzad Baradaran and Soudeh Ghafouri-Fard revised the manuscript critically for important intellectual content. Asghar Shayannia designed and conducted the project.
Data availability
The datasets generated and/or analysed during the current study are available in the NCBI repository: IFNG (NC_000012.12), IL18R (NC_058098.1), IL18RAP (NC_000002.12), IFNG-AS1(NC_000012.12).
Declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Asghar Shayannia, Email: a.shayannia@gmail.com.
Soudeh Ghafouri-Fard, Email: s.ghafourifard@sbmu.ac.ir.
References
- 1.McCutcheon, R. A., Marques, T. R. & Howes, O. D. Schizophrenia—an overview. JAMA Psychiatry. 77 (2), 201–210 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Correll, C. U. & Schooler, N. R. Negative symptoms in Schizophrenia: a review and clinical guide for Recognition, Assessment, and treatment. Neuropsychiatr Dis. Treat.16, 519–534 (2020). PubMed PMID: 32110026. Pubmed Central PMCID: PMC7041437. Epub 2020/02/29. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn, R. S. On the origins of schizophrenia. Am. J. Psychiatry. 177 (4), 291–297 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Ermakov, E. A., Melamud, M. M., Buneva, V. N. & Ivanova, S. A. Immune System abnormalities in Schizophrenia: an integrative view and translational perspectives. Front. Psychiatry. 13, 880568 (2022). PubMed PMID: 35546942. Pubmed Central PMCID: PMC9082498. Epub 2022/05/14. eng. [DOI] [PMC free article] [PubMed]
- 5.Müller, N. & Schwarz, M. J. Immune System and Schizophrenia. Curr. Immunol. Rev.6 (3), 213–220 (2010). PubMed PMID: 21057585. Pubmed Central PMCID: PMC2971548. Epub 2010/11/09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pouget, J. G. The emerging immunogenetic architecture of schizophrenia. Schizophr. Bull.44 (5), 993–1004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ermakov, E. A., Melamud, M. M., Buneva, V. N. & Ivanova, S. A. Immune system abnormalities in schizophrenia: an integrative view and translational perspectives. Front. Psychiatry. 13, 880568 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller, N., Weidinger, E., Leitner, B. & Schwarz, M. J. The role of inflammation in schizophrenia. Front. Neurosci.9, 372 (2015). PubMed PMID: 26539073. Pubmed Central PMCID: PMC4612505. Epub 2015/11/06. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallée, A. Neuroinflammation in Schizophrenia: the key role of the WNT/β-Catenin pathway. Int. J. Mol. Sci. 23 (5). 10.3390/ijms23052810 (2022). PubMed PMID: 35269952. Pubmed Central PMCID: PMC8910888. Epub 2022/03/11. eng. [DOI] [PMC free article] [PubMed]
- 10.Birnbaum, R. & Weinberger, D. R. A genetics perspective on the role of the (neuro) immune system in schizophrenia. Schizophr. Res.217, 105–113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bridges, M. C., Daulagala, A. C. & Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol.220 (2), e202009045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peltier, D. C., Roberts, A. & Reddy, P. LNCing RNA to immunity. Trends Immunol.43 (6), 478–495 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, M-C., Ni, J-J., Cui, W-Y., Wang, B-Y. & Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. cancer Res.9 (7), 1354 (2019). [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, N., Tie, C., Yu, B., Zhang, W. & Wan, J. Identifying lncRNA–miRNA–mRNA networks to investigate Alzheimer’s disease pathogenesis and therapy strategy. Aging (Albany NY). 12 (3), 2897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai, L-J. et al. LncRNA MALAT1 facilitates inflammasome activation via epigenetic suppression of Nrf2 in Parkinson’s disease. Mol. Brain. 13 (1), 1–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui, X. et al. Long non-coding RNA: potential diagnostic and therapeutic biomarker for major depressive disorder. Med. Sci. Monitor: Int. Med. J. Experimental Clin. Res.22, 5240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parikshak, N. N. et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 540 (7633), 423–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teimuri, S. et al. Integrative analysis of lncRNAs in Th17 cell lineage to discover new potential biomarkers and therapeutic targets in autoimmune diseases. Mol. Therapy-Nucleic Acids. 12, 393–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arolt, V. et al. Production of interferon-gamma in families with multiple occurrence of schizophrenia. Psychiatry Res.66 (2–3), 145–152 (1997). PubMed PMID: 9075278. Epub 1997/02/07. eng. [DOI] [PubMed] [Google Scholar]
- 20.Paul-Samojedny, M. et al. Association Study of Interferon Gamma (IFN-γ) + 874T/A gene polymorphism in patients with paranoid Schizophrenia. J. Mol. Neurosci.43(3), 309–315 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Collier, S. P., Henderson, M. A., Tossberg, J. T. & Aune, T. M. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J. Immunol.193 (8), 3959–3965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng, H. et al. Elevated expression of the long noncoding RNA IFNG-AS1 in the peripheral blood from patients with rheumatoid arthritis. J. Immunol. Res.2020, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Syed, A. A. S., He, L., Shi, Y. & Mahmood, S. Elevated levels of IL-18 associated with schizophrenia and first episode psychosis: a systematic review and meta‐analysis. Early. Interv. Psychiat.15 (4), 896–905 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Kaplanski, G. Interleukin-18: Biological properties and role in disease pathogenesis. Immunol. Rev.281 (1), 138–153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.American Psychiatric Association D & American Psychiatric Association, D. Diagnostic and Statistical Manual of Mental Disorders: DSM-5 (American psychiatric association Washington, DC, 2013). [Google Scholar]
- 26.Müller, N. Inflammation in schizophrenia: pathogenetic aspects and therapeutic considerations. Schizophr. Bull.44 (5), 973–982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Upthegrove, R., Khandaker, G.M. Cytokines, Oxidative Stress and Cellular Markers of Inflammation in Schizophrenia. In: Khandaker, G., Meyer, U., Jones, P. (eds) Neuroinflammation and Schizophrenia. Current Topics in Behavioral Neurosciences, vol 44. Springer, Cham. 10.1007/7854_2018_88 (2019) [DOI] [PubMed]
- 28.Lee, E. E., Hong, S., Martin, A. S., Eyler, L. T. & Jeste, D. V. Inflammation in schizophrenia: cytokine levels and their relationships to demographic and clinical variables. Am. J. Geriatric Psychiatry. 25 (1), 50–61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasuda, K., Nakanishi, K. & Tsutsui, H. Interleukin-18 in health and disease. Int. J. Mol. Sci.20 (3), 649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borgonetti, V. et al. IL-18 signaling in the Rat Central Amygdala is disrupted in a Comorbid Model of post-traumatic stress and Alcohol Use Disorder. Cells.12 (15). 10.3390/cells12151943 (2023). PubMed PMID: 37566022. Pubmed Central PMCID: PMC10416956. Epub 2023/08/11. eng. [DOI] [PMC free article] [PubMed]
- 31.Sugama, S. & Conti, B. Interleukin-18 and stress. Brain Res. Rev.58(1), 85–95 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Piancone, F., La Rosa, F., Marventano, I., Saresella, M. & Clerici, M. The role of the inflammasome in neurodegenerative diseases. Molecules. 26 (4), 953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang, D. et al. Differential regulation of transcription factor T-bet induction during NK cell development and T helper-1 cell differentiation. Immunity. 55 (4), 639–655 (2022). e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, D. et al. Interleukin-18 from neurons and microglia mediates depressive behaviors in mice with post-stroke depression. Brain. Behav. Immun.88, 411–420 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Szabo, A. et al. Increased circulating IL-18 levels in severe mental disorders indicate systemic inflammasome activation. Brain. Behav. Immun.99, 299–306 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Luo, Y., He, H., Zhang, J., Ou, Y. & Fan, N. Changes in serum TNF-α, IL-18, and IL-6 concentrations in patients with chronic schizophrenia at admission and at discharge. Compr. Psychiatr.90, 82–87 (2019). PubMed PMID: 30782515. Epub 2019/02/21. eng. [DOI] [PubMed] [Google Scholar]
- 37.Rex, D. et al. A comprehensive pathway map of IL-18-mediated signalling. J. cell. Communication Signal.14 (2), 257–266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Y. et al. Potential involvement of the interleukin-18 pathway in schizophrenia. J. Psychiatr. Res.74, 10–6 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Meda Spaccamela, V. et al. High levels of IL-18 and IFN-γ in chronically inflamed tissue in chronic granulomatous disease. Front. Immunol.10, 2236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, X. Y. et al. Serum IL-18 level, clinical symptoms and IL-18-607A/C polymorphism among chronic patients with schizophrenia in a Chinese Han population. Psychoneuroendocrinology68, 140–147 (2016). [DOI] [PubMed]
- 41.Burke, J. D. & Young, H. A. (eds) IFN-γ: A Cytokine at the Right time, is in the Right Place. Seminars in Immunology (Elsevier, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Axelrod, M. L., Cook, R. S., Johnson, D. B. & Balko, J. M. Biological consequences of MHC-II expression by tumor cells in cancer. Clin. Cancer Res.25 (8), 2392–2402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matta, B., Song, S., Li, D. & Barnes, B. J. Interferon regulatory factor signaling in autoimmune disease. Cytokine. 98, 15–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirlekar, B. & Pylayeva-Gupta, Y. IL-12 family cytokines in cancer and immunotherapy. Cancers. 13 (2), 167 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paibomesai, M. A. Epigenetic Influences on Bovine T-helper 1 and T-helper 2 Cytokines (interferon-gamma and Interleukin-4) in high and low Immune Responders around the (University of Guelph, 2017).
- 46.Zhu, Q. & Kanneganti, T-D. Cutting edge: distinct regulatory mechanisms control proinflammatory cytokines IL-18 and IL-1β. J. Immunol.198 (11), 4210–4215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Philips, R. L. et al. The JAK-STAT pathway at 30: much learned, much more to do. Cell. 185 (21), 3857–3876 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghafelehbashi, H., Pahlevan Kakhki, M., Kular, L., Moghbelinejad, S. & Ghafelehbashi, S. Decreased expression of IFNG-AS 1, IFNG and IL‐1B inflammatory genes in Medicated Schizophrenia and bipolar patients. Scand. J. Immunol.86 (6), 479–485 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Lorestani, R., Boozarpour, S., Alijanpour, S. & Ahangar, L. Evaluation of IFN-γ and T-bet expression levels as possible molecular markers of Schizophrenia. J. Cell. Mol. Res.11 (2), 82–89 (2020). [Google Scholar]
- 50.Nakanishi, K. Unique action of interleukin-18 on T cells and other immune cells. Front. Immunol.9, 358681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lodde, V. et al. Long noncoding RNAs and circular RNAs in autoimmune diseases. Biomolecules. 10 (7), 1044 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hosseini, A. et al. LncRNAs associated with multiple sclerosis expressed in the Th1 cell lineage. J. Cell. Physiol.234 (12), 22153–22162 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Peng, H. et al. The long noncoding RNA IFNG-AS1 promotes T helper type 1 cells response in patients with Hashimoto’s thyroiditis. Sci. Rep.5 (1), 17702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You, Y. et al. Integrated transcriptome profiling revealed that elevated long non-coding RNA-AC007278. 2 expression repressed CCR7 transcription in systemic lupus erythematosus. Front. Immunol.12, 615859 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hedl, M., Zheng, S. & Abraham, C. The IL18RAP region disease polymorphism decreases IL-18RAP/IL-18R1/IL-1R1 expression and signaling through innate receptor–initiated pathways. J. Immunol.192(12), 5924–5932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the NCBI repository: IFNG (NC_000012.12), IL18R (NC_058098.1), IL18RAP (NC_000002.12), IFNG-AS1(NC_000012.12).