Abstract
Environmental osmolarity plays a crucial role in regulating the functions and behaviors of both host cells and pathogens. However, it remains unclear whether and how environmental osmotic stimuli modulate bacterial‒host interfacial adhesion. Using single-cell force spectroscopy, we revealed that the interfacial adhesion force depended nonlinearly on the osmotic prestimulation of host cells but not bacteria. Quantitatively, the adhesion force increased dramatically from 25.98 nN under isotonic conditions to 112.45 or 93.10 nN after the host cells were treated with the hypotonic or hypertonic solution. There was a strong correlation between the adhesion force and the number of host cells harboring adherent/internalized bacteria. We further revealed that enhanced overexpression levels of collagen XV and II were responsible for the increases in interfacial adhesion under hypotonic and hypertonic conditions, respectively. This work provides new opportunities for developing host-directed antibacterial strategies related to interfacial adhesion from a mechanobiological perspective.
Subject terms: Antimicrobials, Cellular microbiology, Cellular microbiology
Introduction
Osmotic stress is widespread in host environments because of the potential influence of many factors, such as diet1, physiology2, diseases3,4 and drugs5. For example, a 7-fold increase in osmolarity at lesion sites3 can be induced by cystic fibrosis in the airway. Osmotic pressure perturbations can be caused by not only changes in luminal contents6 but also conditions such as osmotic diarrhea7, inflammatory bowel disease8 and the use of osmotic drugs5 in the gut. Osmotic homeostasis profoundly regulates cell cycle arrest9, immunity10–12, migration and other cell functions and behaviors13. It has been reported that hypertonic osmotic pressure modulates vascular barrier function and proactively prevents infectious diseases from progressing to a severe stage14. There is growing evidence that hyperosmolality induced by a high-salt diet impairs antimicrobial neutrophil responses10, triggers autoimmune disease12, and enhances proinflammatory activity in macrophages11. Osmotic pressure also mediates bacterial growth15,16 and biofilm formation17,18. For example, mild osmotic stress can cause the gut microbiota S24-7 family to undergo abundant extinction in circumstances involving long-term changes5, whereas osmotic pressure gradients in extracellular matrices play crucial roles in controlling the growth, thickening and spreading of Bacillus subtilis biofilms17. However, it is currently unclear whether and how extracellular osmolarity modulates bacterial and host cell interactions.
Pathogenic bacteria‒host interactions and the resulting bacterial infections are likely to be life-threatening and seriously affect human health19,20. In essence, bacterial infection is a multistep process that allows bacteria to adhere to host cells and subsequently invade, colonize, and spread to other sites within the host21,22. Generally, interfacial contact and adhesion between bacteria and host cells is the first step for bacteria to infect host cells23, where bacterial pathogens have already evolved to express specific adhesins that adhere to host cells through proteins such as fibrinogen24, fibrin, and collagen25–27. Staphylococcus aureus (S. aureus), for example, expresses specific collagen-binding adhesins28,29, i.e., collagen adhesins (Cna), which contribute to bacterial adhesion to the host and subsequent bacterial virulence30–32. In recent years, several attempts have been made to characterize the interactions between bacteria and host cells via atomic force microscopy (AFM)33–35. Beaussart and coworkers reported that the interactions between Pseudomonas aeruginosa and A549 cells involve bacterial pili extension and the formation of membrane tethers from host cells34. Additionally, AFM revealed that S. aureus mutants lacking fibronectin-binding protein B (FnBPB) or coagulation factor B (ClfB) had a reduced ability to adhere to the skin of patients with atopic dermatitis (AD), which could enhance antimicrobial therapy in patients with AD35. To date, one of the major challenges in traditional bacterium-directed chemotherapies lies in the crisis of antibiotic resistance, which has led to increasing interest in designing antibacterial strategies associated with host microenvironments to increase antibiotic efficacy36–39. In fact, biomechanical microenvironments around host cells can play crucial roles in bacterial‒cell interactions, such as the binding of adhesins to host cell receptors40, bacterial infection of epithelial cells and the intracellular accumulation of antibiotics41. It is very likely that quantitative studies on the regulatory effects of biophysical/biomechanical microenvironments on bacterial‒cell interactions provide new ideas for the development of host-acting antibacterial therapies36,42,43.
Here, we focus on the interfacial adhesion interactions between bacterial pathogens and host cells regulated by osmotic homeostasis. With single-cell force spectroscopy (SCFS), we quantify the adhesion forces between a bacterium and a single host cell under different osmotic pressures. We subsequently reveal that host cells under isotonic conditions can more effectively resist bacterial infection by minimizing bacterial–cell adhesion forces. With the aid of RNA sequencing, we show that, as different collagen subtypes, collagen XV and II in host cells can be upregulated in hypotonic and hypertonic environments, respectively, which leads to an increase in interfacial adhesion forces and the subsequent exacerbation of infection. These findings imply that targeting different isoforms of collagen proteins expressed by host cells modulated by environmental osmolality may be a new host-directed antibacterial strategy.
Results
Hypotonic or hypertonic stimulation increases bacterial adhesion to host cells
The bacterial‒host interactions mediated by environmental osmolality are widespread in the intestinal microenvironment and play essential roles in regulating the physiological and pathological functions of the intestines1 (Fig. 1A). To mimic the interfacial interactions in vivo, we first constructed an in vitro model of bacteria‒cell interactions, where environmental osmolality was precisely controlled by adding a certain amount of deionized water or D-mannitol solution44,45 to regular cell culture medium (Supplementary Table 1). After exposing IEC-6 cells (rat small intestinal epithelial cell line-6) to hypotonic (0.5× and 0.75×), isotonic (1×), and hypertonic (1.5× and 2×) solutions for 3 h, we found that these osmotic stimuli did not exhibit obvious cytotoxicity (Supplementary Fig. 1A and Fig. 2), even though their volumes responded to changes in the environmental osmotic pressure (Supplementary Fig. 1B, C).
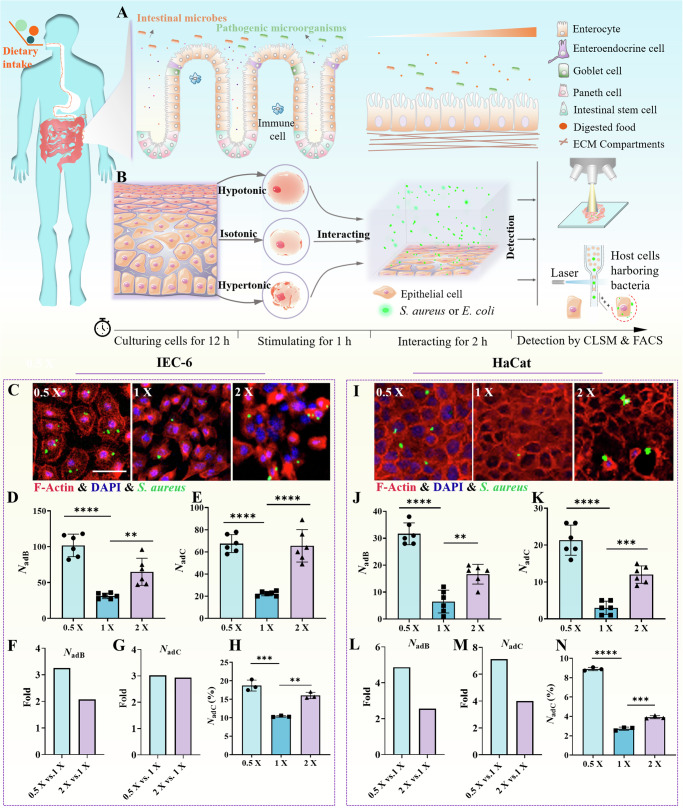
Fig. 1. Interactions between bacteria and host cells are regulated by environmental osmotic pressures.
A Schematic diagram of interactions between bacteria and cells in the intestinal microenvironment. B Schematic diagram of an in vitro model of bacterial interaction with epithelial cells under different osmotic pressures. The host cells were cultured for 12 h in regular culture medium and then stimulated for 1 h in a specific osmotic mixture. Then, they were allowed to interact with GFP-expressing S. aureus or E. coli for another 2 h and rinsed three times with Dulbecco’s phosphate-buffered saline (DPBS) solution. Finally, the osmoregulated bacterium‒cell interactions were quantified. C Images of GFP-expressing S. aureus adherent/internalized to IEC-6 cells after the host cells were prestimulated with hypotonic (0.5x), isotonic (1x), and hypertonic (2x) solutions, which were acquired with a laser scanning confocal microscope. The F-actin cytoskeleton was labeled in red by rhodamine phalloidin, and the nuclei were labeled in blue by DAPI. D and E Statistical results of the number of bacteria adherent/internalized to host cells (NadB) and the number of host cells harboring adherent/internalized bacteria (NadC) in microscopy images with an imaging field of view of 337 × 337 μm2, where the host cells were prestimulated with hypotonic (0.5x), isotonic (1x), and hypertonic (2x) solutions, respectively. F and G Fold changes in NadB and NadC under hypertonic and hypotonic conditions relative to the indices in the isotonic situation. (H) Statistical results of the number of host cells harboring adherent/internalized bacteria (NadC (%)) quantified through flow cytometry. I–N are similar to C–H, respectively, except that the host cells were replaced with the HaCat cell line. All the statistical data (mean ± SD) originated from at least three independent experiments for each specific condition. Statistical analyses based on one-way ANOVA were used in the experiments, and **, *** and **** denote P < 0.01, P < 0.001 and P < 0.0001, respectively. Scale bar: 50 μm.
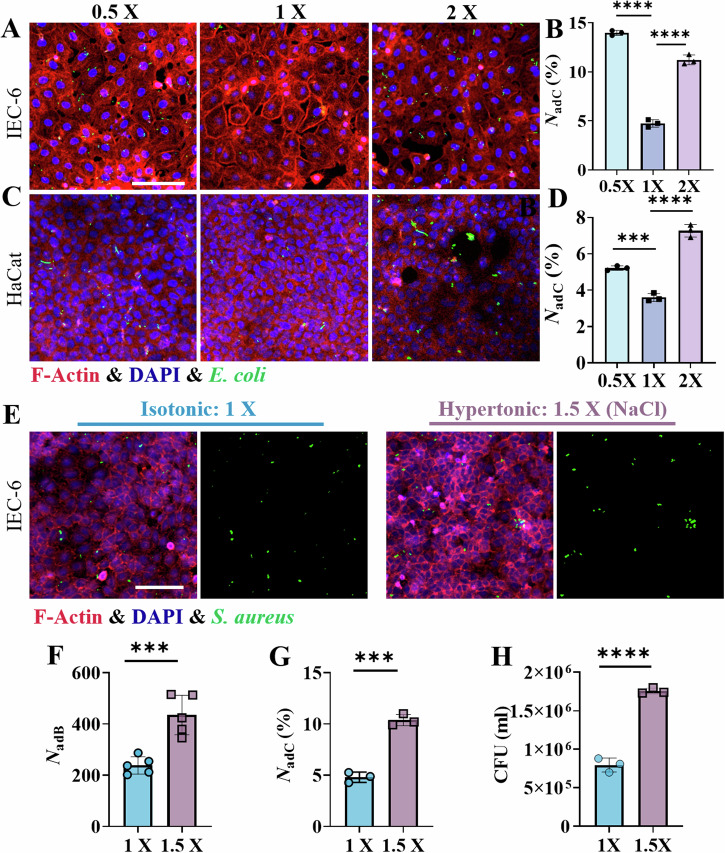
Fig. 2. Interactions between the gram-negative bacterium E. coli and host cells following prestimulation with hypotonic (0.5×), isotonic (1×) and hypertonic (2×) solutions, as well as the effects of different penetrants such as NaCl.
Typical images of GFP-expressing E. coli adherent/internalized to IEC-6 cells (A) and HaCat cells (C). Scale bar: 100 μm. Statistical results of the number of host cells (B: IEC-6; D: HaCat) harboring adherent/internalized bacteria, i.e., NadC (%), which was quantified by flow cytometry. E Typical fluorescence images of GFP-expressing S. aureus adherent/internalized to IEC-6 cells after the host cells were stimulated with an isotonic solution (1x, left) or a hypertonic solution prepared with NaCl (1.5x, right). Scale bar: 100 μm. F Statistical results of the number of bacteria adherent/internalized to host cells (NadB) with an imaging field of view of 337 × 337 μm2, where the host cells were stimulated with an isotonic solution (1x) or a hypertonic (1.5x) solution prepared with NaCl. G Statistical analysis of the number of host cells harboring adherent/internalized bacteria (NadC (%)) by flow cytometry. H Colony count-based statistical results of bacteria adherent/internalized to host cells that were prestimulated with isotonic (1x) and hypertonic (1.5x) solutions prepared with NaCl. All the statistical data are presented as the means ± SDs from at least three independent experiments for each specific condition, and two-sided unpaired t-tests were used in the analyses, with *** and **** indicating P < 0.001 and P < 0.0001, respectively.
Subsequently, we stimulated IEC-6 and HaCat (human keratinocyte) cell monolayers with the prepared hypotonic (0.5x and 0.75x), isotonic (1x), and hypertonic (1.5x and 2x) solutions for 1 h and then added S. aureus or E. coli to interact with the host cell monolayers for another 2 h at different osmotic pressures (Fig. 1B), where the number of bacteria added in the experiments was 100 times that of the host cells. Our experimental data showed that, in comparison with isotonic stimulation, both hypotonic and hypertonic stimulations of the host cells induced increases in the number of adherent bacteria on the host monolayers (Fig. 1C, I and Supplementary Fig. 3). When the prestimulation of host cells was changed from the isotonic condition to the hypotonic or hypertonic condition, both the number of bacteria adherent/internalized to host cells (NadB) and the number of host cells harboring adherent/internalized bacteria (NadC) increased significantly (Fig. 1D, E, J, K; Supplementary Fig. 3 and Supplementary Fig. 4). Quantitatively, after prestimulation of the host cell monolayers with hypotonic and hypertonic solutions, NadB increased by approximately 3.25-fold and approximately 2.07-fold, respectively, for the IEC-6 monolayers (Fig. 1F) and by approximately 4.87-fold and approximately 2.56-fold, respectively, for the HaCat monolayers (Fig. 1L). Moreover, NadC increased by approximately 3.02-fold and approximately 2.93-fold for the IEC-6 monolayers (Fig. 1G) and by approximately 7.11-fold and approximately 4.00-fold for the HaCat monolayers (Fig. 1M). Similar trends in the osmoregulated bacteria‒cell interactions were also further confirmed by flow cytometric analysis (Fig. 1H, N and Supplementary Fig. 5). Additionally, lysis of post-infected cells for colony counting resulted in increases in total and adherent bacteria with abnormal osmotic pressure (Supplementary Fig. 6A).
To assess the generality of this phenomenon, the gram-negative bacteria E. coli was used to interact with IEC-6 cells and HaCat cells, which also presented increasing interactions between bacteria and host cells in both hypo- and hyperosmotic environments (Fig. 2A–D and Supplementary Fig. 6B). The influence of the osmotic agents themselves was also considered in this work. When the hypertonic solution of D-mannitol was replaced with a hypertonic solution prepared with NaCl to stimulate the host cell monolayers, we found a similar experimental phenomenon in which the hypertonic stimulation of the host cells led to increases in the number of bacteria adherent/internalized to host cells and the number of host cells harboring adherent/internalized bacteria (Fig. 2E–H), which implied that these osmotic agents themselves had little effect on osmotically stimulated bacterial‒cell interactions. To further elucidate the impact of hypotonic stimulation on bacterial–host cell interactions, we performed a comparative analysis of bacterial‒host interactions under isotonic, isotonic & low-nutrient, hypotonic, and hypotonic conditions under normal nutrient conditions (Supplementary Table 2 and Supplementary Fig. 7). Our experimental findings indicated that hypotonic conditions led to increased bacterial‒host interactions, demonstrating the significant role of osmolarity in modulating bacterial‒host interactions.
Environmental osmotic pressures regulate bacterial–cell adhesion forces
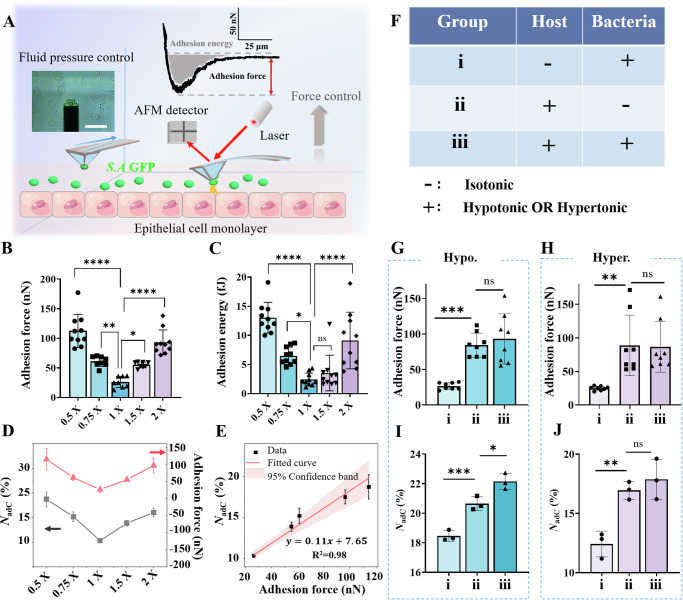
By means of SCFS based upon fluidic force microscopy (FluidFM)23, we next quantified the interfacial adhesion forces between the bacteria and the host cell monolayers in hypotonic (0.5× and 0.75×), isotonic (1×), and hypertonic (1.5× and 2×) solutions. We used a probe with a specified fluid channel to capture a single bacterium (S. aureus) and allowed it to gently approach the underlying host cell monolayer at a speed of 1 μm/s until they were in complete contact (Fig. 3A). After the probe with the bacterium was paused for 30 s to allow sufficient adhesive interaction, we retracted it to its initial position at the same speed of 1 μm/s (Video S1). In this way, we could quantitatively estimate the interfacial adhesion force and adhesion energy between the bacterium and the host cell monolayer. The mean adhesion force between the bacterium (S. aureus) and the IEC-6 cell monolayer increased from 25.98 nN (isotonic, 1×) to 61.1 nN (hypotonic, 0.75x) and 112.45 nN (hypotonic, 0.5×) or to 55.02 nN (hypertonic, 1.5×) and 93.1 nN (hypertonic, 2×) (Fig. 3B). These results indicated that either the hypertonic or hypotonic environment resulted in a significant increase in the interfacial adhesion force. A similar conclusion could be drawn from the measured data derived from the interfacial adhesion energy (Fig. 3C). There was a strong correlation between the measured interfacial adhesion force and NadC (%) under different osmotic stimulation prestimulations (Fig. 3D). More interestingly, there was an approximately linear relationship (i.e., NadC (%) = 0.11*Fad + 7.65), between the NadC (%), which was quantified by flow cytometry, and the measured interfacial adhesion force, which was within the range of 20–120 nN (Fig. 3E), implying that the interfacial adhesion forces exerted an important impact on the interaction between the bacteria and host cells. Furthermore, we investigated whether bacteria or host cells play a more dominant role in regulating their interfacial adhesion forces after osmotic stimulation. To this end, we employed hypotonic or hypertonic solutions to stimulate bacteria, host cells or both (Fig. 3F) and accordingly quantified the bacterium‒cell adhesion forces (Fig. 3G, H) and the number of host cells harboring adherent/internalized bacteria (NadC (%)) (Fig. 3I, J). Neither the interfacial adhesion forces nor NadC (%) increased significantly when only the bacteria themselves were treated with the hypotonic or hypertonic solutions, which had little effect on bacterial viability or growth (Supplementary Fig. 8), and adhesion (Supplementary Fig. 9). In contrast, whenever the host cells experienced hypotonic or hypertonic stimulation, both the interfacial adhesion force and NadC (%) increased significantly in our experiments (Fig. 3G–J). Likewise, the surface roughness and stiffness of the host cells were characterized by AFM after stimulation in either hypotonic or hypertonic environments. As shown in Supplementary Fig. 10, the indices of the osmotically stimulated cells did not significantly change. These findings suggest that the host cells undergoing osmotic stimulation have a more pronounced effect on the interfacial interactions than do the bacteria involved and that the host cells may respond to the surface components, thus affecting bacterial colonization.
Fig. 3. Quantification of interfacial adhesion forces between bacteria and host cell monolayers prestimulated with hypotonic (0.5× and 0.75×), isotonic (1×), and hypertonic (1.5× and 2×) solutions.
A Schematic diagram of the characterization of interfacial adhesion forces between bacteria and host cell monolayers using single-cell force spectroscopy (SCFS) based on fluid force microscopy (FluidFM). B Interfacial adhesion force and C adhesion energy between a single bacterium and the host cell monolayers that were prestimulated with hypotonic (0.5× and 0.75×), isotonic (1×), and hypertonic (1.5× and 2×) solutions. D Correlation between the interfacial adhesion force and NadC (%) quantified by flow cytometry in different osmotic solutions. The points and curve in red represent the data of the adhesion force, whereas those in black denote the data of NadC (%). E An approximately linear relationship was observed between NadC (%) and the quantified interfacial adhesion forces in the range of 20–120 nN. F List of experimental groups in which hypotonic or hypertonic solutions were used to stimulate bacteria (i), host cells (ii) or both (iii). G and H Comparisons of interfacial adhesion forces in the three experimental groups after stimulation with the hypotonic or hypertonic solutions, respectively. I and J Comparisons of NadC (%) in the three experimental groups after stimulation with the hypotonic or hypertonic solution, respectively. All the statistical data are presented as the mean ± SD from at least three independent experiments for each condition. Statistical analyses based upon one-way ANOVA were adopted, and *, **, *** and **** denote P < 0.05, P < 0.01, P < 0.001 and P < 0.0001, respectively.
RNA sequencing reveals distinct transcriptional profiles among hypo-, iso-, and hyperosmotic environments
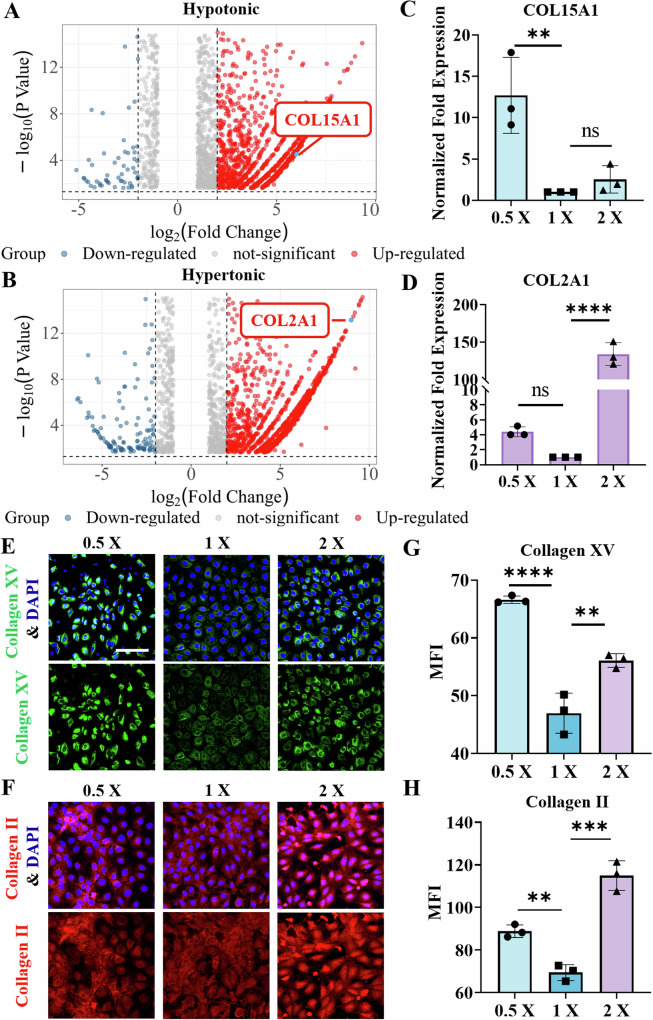
We employed a transcriptomic approach to explore the underlying reason for the significant increase in interfacial adhesion forces between the bacteria and the host cell monolayers after treatment with the hypotonic or hypertonic solutions. The RNA sequencing data revealed that the transcriptional level of the host cells significantly changed once the host cells were treated with the hypotonic or hypertonic solutions (Supplementary Fig. 11 and Supplementary Table 3). Notably, the expression levels of several genes related to different isoforms of collagen were increased (Fig. 4A, B). A more detailed list of the major differentially expressed genes is presented in Supplementary Tables 4 and 5.
Fig. 4. Transcriptomic alterations in IEC-6 cells after osmotic stimulation.
Volcano plots of differential gene expression in IEC-6 cells treated with (A) hypotonic (0.5×) and (B) hypertonic (2×) solutions. In these plots, the genes expressed at relatively high levels are represented by red dots, whereas the genes expressed by relatively low levels are denoted by blue dots. Additionally, the genes encoding COL15 A1 and COL2A1 are highlighted in the plots. C and D The relative expression levels of the COL15 A1 and COL2A1 genes were quantified by qPCR and normalized to that of GAPDH. Relative expression was calculated using the 2−∆ ∆ CT method. E and F Representative immunofluorescence images of collagen XV (green) and collagen II (red) after IEC-6 cells were treated with hypotonic or hypertonic solutions, respectively. The nuclei were labeled blue with DAPI. Scale bar: 100 μm. G and H The corresponding changes in the mean fluorescence intensity (MFI) of collagen XV (green) and collagen II (red) in IEC-6 cells treated with hypotonic or hypertonic solutions, respectively. The data were quantified with ImageJ software. All the statistical data are presented as the means ± SDs, and statistical analyses based on one-way ANOVA were employed, where **, *** and **** indicate P < 0.01, P < 0.001 and P < 0.0001, respectively.
In fact, collagen, a protein abundantly found in host cells26,46,47, serves as a primary target for colonization by numerous pathogens and is also closely linked to the formation of biofilms48,49. We subsequently investigated the impact of the osmoregulation of different subtypes of collagen on bacterial‒host cell interactions. Our qPCR analyses revealed that the expression of COL15A1 was indeed elevated in hypo-osmotic environments, whereas hyperosmotic environments led to an increase in the expression of COL2A1 (Fig. 4C, D, and Supplementary Fig. 12). The primers used in the qPCR assays are listed in Supplementary Table 6. Similarly, we confirmed the upregulated expression of these collagen isoforms at the protein level by immunofluorescence staining (Fig. 4E, F). Compared with those under isotonic conditions, the expression levels of both collagen isoforms under hypotonic or hypertonic conditions were increased (Fig. 4G, H). Specifically, the expression of collagen XV was significantly increased under hypotonic conditions (Fig. 4G), whereas that of collagen II was significantly increased under hypertonic conditions (Fig. 4H). The results of the immunoblot analysis were consistent with the immunofluorescence results (Supplementary Fig. 13).
Inhibition of collagen reduces infection in hyperosmotic and hypotonic environments
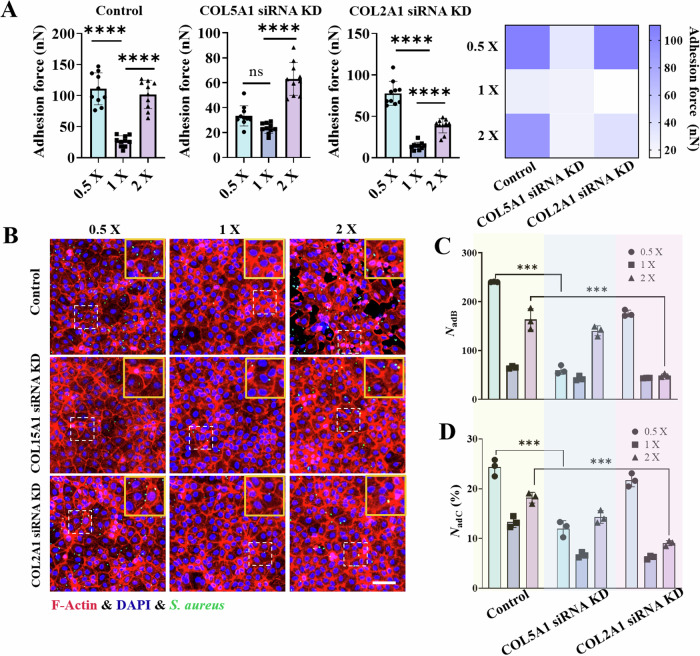
To validate the roles of COL15A1 and COL2A1 in the hypo- and hyperosmotic stress-regulated interactions between bacteria and host cells, we transfected siRNAs targeting COL15A1 or COL2A1 into IEC-6 cells (Supplementary Table 7), which effectively reduced the transcriptome and protein expression of the target genes (Supplementary Fig. 14). We subsequently measured the interfacial adhesion forces between a single bacterium (S. aureus) and IEC-6 cells treated with siRNAs targeting COL15A1 or COL2A1 (Fig. 5A). Compared with those in the control group, the interfacial adhesion forces significantly decreased after the COL15A1 siRNA-treated IEC-6 cells were pretreated with the hypotonic or hypertonic solutions (Fig. 5A). In particular, no significant difference in the interfacial adhesion forces was observed when these COL15A1 siRNA-treated cells were stimulated with hypotonic (0.5×) or isotonic (1×) solutions. These findings indicated that the overexpression of collagen XV triggered by hypotonic stimulation was responsible for the substantial increase in the interfacial adhesion forces under hypotonic conditions. Conversely, compared with those in the control group, the adhesion forces between the bacteria and the COL2A1 siRNA-treated IEC-6 cells significantly decreased (Fig. 5A). Specifically, a significant decrease in the interfacial adhesion forces was observed between the bacteria and the COL2A1 siRNA-treated IEC-6 cells after prestimulation with the hypertonic solution (2×). These findings suggest that the overexpression of collagen II induced by hypertonic stimulation plays an essential role in mediating interfacial adhesion interactions under hypertonic conditions. Moreover, both the number of bacteria adherent/internalized to host cells (NadB) and the number of host cells harboring adherent/internalized bacteria (NadC (%)) decreased significantly after the COL15A1 siRNA- and COL2A1 siRNA-treated cells were stimulated with hypotonic (0.5×) and hypertonic (2×) solutions, respectively (Fig. 5B, C, D), results that were in qualitatively consistent with the aforementioned measurement data of the interfacial adhesion forces.
Fig. 5. Interactions between bacteria (S. aureus) and IEC-6 cells whose genes encoding COL15A1 or COL2A1 were knocked down.
A Statistical analysis of the interfacial adhesion forces between a single bacterium and IEC-6 cells (control group) or between IEC-6 cells with COL15A1 or COL2A1 gene knockdown under different osmotic environments. B Representative fluorescence images of bacteria‒cell interactions showing GFP-expressing bacteria (green) adhering to and internalized by the underlying IEC‒6 cell monolayers. The nuclei are labeled blue, whereas the cytoskeletons are labeled red. C and D Comparisons of the number of bacteria adherent/internalized to host cells (NadB) and the number of host cells harboring adherent/internalized bacteria (NadC(%)) under hypotonic (0.5×), isotonic (1×) and hypertonic (2×) conditions, where COL15A1 siRNA KD and COL2A1 siRNA KD denote IEC-6 cells in which the gene encoding either COL15A1 or COL2A1 was knocked down based on the well-developed small interfering RNA technique. These data were counted under an imaging field of view of 500 × 500 μm2. All the statistical data are presented as the mean ± SD from at least three independent experiments for each experimental group. Statistical analyses were performed based on one-way ANOVA. *** and **** denote P < 0.001 and P < 0.0001, respectively. Scale bar: 100 μm.
Discussion
In essence, interfacial adhesion plays important roles in the infection and pathogenesis of various bacterial pathogens, such as Aeromonas veronii50, Helicobacter pylori51 and Pseudomonas aeruginosa48. Blocking bacterial adhesion is currently recognized as an effective antibacterial strategy52,53. For example, the adhesion protein FimH of uropathogenic E. coli interacts with M428439, which is a high-affinity inhibitory mannoside of host cells. The interaction weakens bacterial adhesion to the host cell surface via mannoside, leading to a reduction in the colonization of uropathogenic E. coli. These findings suggest a potential antiadhesion strategy as a novel antimicrobial therapy. The quantitative characterization of interfacial adhesion forces between bacteria and host cells based on SCFS paves the way for accurate exploration of the mechanobiological mechanisms of bacterial adhesion and subsequent infection.
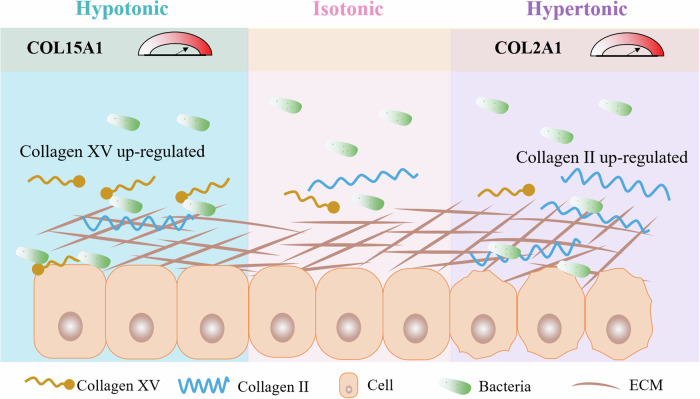
Overall, our study investigated the regulatory effect of environmental osmotic pressures on bacterial‒host interactions and their mechanobiological mechanism. We revealed that these osmotic pressures have the ability to remodel collagen subtypes and thus dominate mechanical adhesion between bacteria and host cells. Collagen subtype-dependent bacterial–host interfacial adhesion not only involves diverse bacterial–host cell interactions but also provides us with new strategies for developing host-specific antibacterial therapies from a mechanobiological perspective. Additionally, our findings highlight the importance of maintaining osmotic homeostasis to promote host cell defense against gram-positive and gram-negative bacteria. Specifically, we learned that hypotonicity and hypertonicity directly enhance bacterial adhesion to host cells via the overexpression of collagen XV or II triggered by environmental osmolarity, as illustrated in Fig. 6. In general, collagen XV is a nonfibrillar collagen with multiple breaks54,55 and is known to play crucial roles in antiangiogenic and antitumor functions56. In contrast, collagen II is commonly found in hyaline cartilage, which provides flexibility and support to bones and joints57. The osmotic pressure-induced overexpression of different subtypes of collagen and their key roles in bacteria‒cell interactions suggest that some siRNA-based targeted drugs or biomaterials can be designed to synergistically address bacterial infections in the future. Given the key roles of adhesion in both pathogenic and probiotic bacteria, it is imperative that future research prioritizes the regulation of adhesion across a broader spectrum of bacterial species58. This is particularly critical, as probiotics still face major challenges in effectively colonizing the gut to compete with pathogens and provide immunomodulatory benefits. One possible solution is to develop bioengineered probiotic strains to increase their adhesion to the host59. It is also worthwhile to further dissect the interactions between pathogenic bacteria and probiotics and their combined effects, as well as the adhesion ability of the microbiota under the regulation of environmental osmotic pressure. We have focused on the modulation of osmotic pressure in the host cells. However, the interactions between penetrants and bacteria (e.g., adhesins) should also be considered. The FimH antagonist (M4284)39 increases the osmotic pressure of the culture medium, demonstrating differing effects on the colonization of S. aureus and E. coli, respectively (see Supplementary Fig. 15 and Supplementary Table 8). The influence of drugs on osmotic pressure and their subsequent effects on pathogens should be comprehensively evaluated in future studies.
Fig. 6. Schematic representation of interfacial adhesion between bacteria and host cells regulated by environmental osmotic pressures.
In a hypotonic environment, the increase in type XV collagen in host cells enhances bacterial adhesion to the host. In contrast, in a hypertonic environment, the increase in type II collagen in the host cells also enhances the adhesion of bacteria to the host.
Methods
Mammalian cell culture
Rat small intestinal epithelial cells (IEC-6, ATCC CRL-1592) and human immortalized keratinocyte cells (HaCat, Cell Bank of the Chinese Academy of Sciences SCSP-5091) were cultured at 37 °C under 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin (P&S). Prior to the experiments, the cells were cultured in culture medium without penicillin/streptomycin for approximately 12 h until they reached confluence.
Bacterial strains and culture
The gram-positive bacterium Staphylococcus aureus (S. aureus, ATCC 29213) and the gram-negative bacterium Escherichia coli (E. coli, ATCC 25922) were used in the present study. These strains were transfected with the Psc19-GFP (erythromycin resistant) plasmid to express green fluorescent protein (GFP) for imaging and flow cytometric analysis. GFP-expressing S. aureus and E. coli were selected with the addition of 50 µg/ml erythromycin in Luria–Bertani (LB) broth or LB agar. Bacteria in the logarithmic growth phase were centrifuged at 5000 rpm and then resuspended in DPBS before performing the bacteria‒cell interaction experiments.
Preparation of solutions with different osmotic pressures
In the experiments, Dulbecco’s modified Eagle’s medium (DMEM) containing high glucose produced by HyClone (Cat No. SH30243.01) was used directly as the isotonic solution (1×), whose osmotic pressure was approximately 344 mOsm·kg−1, as measured with a freezing point osmometer (FASKE210). Hypotonic solutions of 0.5× and 0.75× were prepared by diluting the abovementioned isotonic solution (1×) with deionized (DI) water at ratios of 1:1 and 2:1 (v/v), respectively, and their osmotic pressures were found to be approximately 168 and 227 mOsm·kg−1. The hypertonic solutions of 1.5× and 2× were prepared by adding 30 mg/ml or 50 mg/ml D-mannitol to the isotonic solution (1×), respectively. The corresponding osmotic pressures were approximately 499 and 612 mOsm·kg−1 for the 1.5× and 2× solutions, respectively. Supplementary Table 1 lists the reagent formulas of the osmotic solutions with different osmotic pressures. Another hypertonic solution (1.5×) was also prepared by mixing 100 mM NaCl, 200 mM sorbitol or 200 mM glucose. Additionally, hypotonic or isotonic solutions with different nutrient levels (i.e., isotonic, isotonic & low nutrition, hypotonic and hypotonic & normal nutrition) were prepared as listed in Supplementary Table 2. M4284 (10 μM) and D-mannose (1 mM and 200 mM) were used to measure osmotic pressure and carry out interaction of bacteria-host cells.
Cell viability assay
Cell viability was quantitatively assessed using a live/dead assay kit (Beyotime, C2015l). Briefly, IEC-6 cells grown in cell culture dishes were incubated with various osmotic media for 3 h and then stained with calcein-AM and propidium iodide (PI) from the Live/Dead Assay Kit according to the manufacturer’s instructions. Finally, the live and dead cells were imaged with an inverted confocal laser scanning microscope (Nikon A1, Japan). The cytotoxic activity was also assessed using the CCK8 assay. Briefly, IEC-6 cells or HaCat cells (5 × 104 cells/well) were incubated in a 96-well plate for 12 h. Then, the solutions with different osmotic pressures were used to treat IEC-6 cells or HaCat cells for an additional 3 h. The CCK8 solution was then incubated with IEC-6 cells or HaCat cells for 1 h at 37 °C. Finally, the absorbance at 450 nm was measured using a plate reader (Thermo Fisher Scientific, Varioskan LUX).
Quantification of bacteria adherent to host monolayers
IEC-6 or HaCat cells were seeded in 6-well plates at a density of 3 × 105/well and grown in DMEM without P&S until they reached 80% confluence. The cells were then exposed to media with different osmotic pressures and incubated for 1 h. Subsequently, S. aureus or E. coli was added to interact with the cells for an additional 2 h in solutions with different osmotic pressures, where the number of GFP-labeled bacteria used in the experiments was 100 times greater than that of the mammalian cells. Afterward, we quantified the bacterium‒cell interactions by confocal laser scanning microscopy and flow cytometric analyses. To this end, the cells were first rinsed three times with DPBS solution, fixed with 4% paraformaldehyde (PFA) and stained with TRITC-conjugated phalloidin (Yeasen, 40734ES75) and DAPI (Beyotime, C1002) per the manufacturer’s instructions. The fluorescence images were acquired with an inverted confocal laser scanning microscope (Nikon A1, Japan) or a super-resolution microscope (AIRY, Polay-SIM, China) and analyzed with the well-developed software ImageJ. The function ‘analyze-analyze particles’ in ImageJ software was employed to determine the number of infected bacteria in the obtained images. To quantify the bacterium‒cell interaction by flow cytometry, we detached the cells that interacted with the bacteria from the cell culture dishes using 0.25% trypsin‒EDTA and resuspended them in DPBS supplemented with 2% bovine serum albumin (BSA) (BD, FACSverse). For each assay, we used at least 1 × 104 cells for flow cytometric analysis. Additionally, the cells that interacted with the bacteria were lysed with 1% Triton X-100 and serially diluted in a gradient for analysis by the flat colony counting method.
Characterization of interfacial adhesion forces
We employed single-cell force spectroscopy (SCFS) based on fluidic force microscopy (FluidFM, CytoSurge) to measure the interfacial adhesion forces between a single bacterium and the underlying host cell monolayers with different osmotic pressures. The spring constant of the FluidFM probe was 0.6 N/m, and the microfluidic channel aperture was 300 nm. A pressure controller was attached to the probe to apply a specific pressure to trap a single bacterium. In the experiment, the probe with a single bacterium gently approached the underlying host cell monolayer at a speed of 1 μm/s. After the bacterium was fully in contact with the host cell monolayer, the probe was paused for 30 s to allow sufficient adhesive interaction between them. The probe together with the bacterium was then retracted to its initial position at the same speed of 1 μm/s. In the experiments, individual bacteria were reacquired for each test and randomly selected at different locations in the epithelial monolayers. During the retreat process of the probe, the curve of the adhesion force versus probe displacement was recorded accordingly, and the interfacial adhesion force and adhesion energy were accordingly quantified with the help of CytoSurge software. All the experiments were performed in an incubator containing 5% CO2 and 90% humidity at 37 °C.
Adhesion between bacteria and substrates and bacterial viability testing
To assess the influence of environmental osmotic pressure on bacteria, we performed SYTO/PI dead-live double-staining tests and colony counting experiments on bacteria exposed to different osmotic pressures for 6 h. Subsequently, we examined the growth curves of bacteria grown under different osmotic conditions for 12 h. Furthermore, we detected the ability of bacteria to adhere to collagen-modified substrates under different osmotic pressure environments. To this end, we plated ~105 bacteria/ml on collagen-modified glass at different osmolarities and allowed them to adhere for 6 h before imaging. In the adhesion tests, bacteria were exposed to a specific osmotic pressure for 6 h before their adhesion was assessed. Additionally, we used a heat-treated group as a comparison to detect changes in interfacial adhesion forces, where the bacteria were heat-treated at 95 °C.
Assays of the surface roughness and stiffness of host cells
An atomic force microscope (AFM, Asylum Research MFP-3D) was utilized to characterize the surface roughness and stiffness of the host cells. Prior to the AFM-based experiments, IEC-6 cells were exposed to hypotonic, isotonic, or hypertonic solutions for 1 h. The force‒displacement data were subsequently obtained by indenting live IEC-6 cells under hypotonic or hypertonic conditions using a 5 μm sphere-tipped probe (Bruker, NP-O10) with a spring constant of 0.03 N/m, where the approach velocity was 1 μm/s and the setpoint force was 5 nN. The Young’s moduli of the host cells were evaluated by fitting the measured force‒displacement curves based on the classical Hertz model. The surface roughness of the host cells was measured in solutions with different osmolarities with another AFM (Bruker, Biomic Force Microscope), where PeakForce tapping mode was adopted and the spring constant of the pyramid probe was 0.03 N/m (Bruker, MLCT-BIO). The surface roughness of the host cells was finally analyzed using NanoScope Analysis software (Bruker AXS Corporation, Santa Barbara, CA, USA).
RNA isolation and RNA sequencing
IEC-6 cells were seeded in 6-well plates at a density of 3 × 105 cells/well and grown for 24 h. After stimulation with media with different osmotic pressures for 3 h, the cells were washed three times with PBS. Then, the cells were lysed using TRNzol buffer (TIANGEN, cat #DP424) and harvested in an RNase-free tube. Next, total RNA was purified according to the manufacturer’s protocol (RNA Mini Kit, TIANGEN). All RNA sequencing (RNA-seq) analyses were performed using at least three biological replicates. The RNA libraries were prepared and sequenced on the Illumina HiSeq platform by GENEWIZ, Inc. (Suzhou, China). The obtained clean data were analyzed using HISAT2 and HTSEQ software. Differential expression analysis was performed using the DESeq2 Bioconductor software package. The dispersion and logarithmic fold changes were calculated using data-driven prior distributions. Genes with Padj values less than 0.05 were considered differentially expressed. GOSeq (v1.34.1) was adopted to identify gene ontology (GO) terms annotating a list of enriched genes with significant Padj values less than 0.05. The RNA-seq data used in this study are available in the NCBI SRA database under accession code PRJNA1069618.
Gene expression
We performed gene expression analysis by quantitative qPCR on IEC-6 cells grown to 80% confluence in 6-well plates after hypotonic or hyperosmotic stress. After 3 h of exposure to hypotonic or hypertonic stress, the medium was removed, and the cells were washed twice with DPBS. Then, the cells were lysed with cell lysis buffer, total RNA was purified following the manufacturer’s protocol (RNA Mini Kit, TIANGEN), and 1 μg of total RNA was used to perform reverse transcription using a reverse transcription kit (Takara, RR037A). The resulting cDNA was diluted 5-fold and then subjected to PCR with TB Green MIX (Takara, RR430S). Using the GAPDH gene as an internal control, we evaluated the expression levels of collagen genes in three independent replicates for each group. The 2-∆∆Ct method was used to determine the relative gene expression levels. The corresponding primers are listed in Supplementary Table 6.
Immunofluorescence imaging
Cells on glass substrates were stimulated with different osmotic pressures, fixed with 4% PFA for 15 min, and washed three times with PBS. The samples were blocked with 5% BSA in PBS containing 0.3% Triton X-100 for 1 h. Subsequently, they were incubated overnight at 4 °C with primary antibodies (rabbit anti-COL2A1 polyclonal antibody, Abbkine; rabbit anti-COL15A1 polyclonal antibody, Thermo Fisher) diluted 1:500 in PBS containing 5% BSA. After the primary antibodies were removed, the samples were washed three times with PBS, and fluorescent secondary antibodies (DyLight 680, goat anti-rabbit IgG, Abbkine; DyLight 488, goat anti-rabbit IgG, Abbkine) were added as appropriate. Next, the samples were diluted 1:500 in 2% BSA in PBS and incubated with secondary antibodies for 1 h. Afterward, the F-actin cytoskeleton and nuclei were stained with 0.2 μM TRITC phalloidin (Yeasen, 40734ES75) and 2 μg/ml DAPI (Beyotime, C1002) according to the manufacturer’s instructions. Finally, the samples were stored in 1 ml of PBS and imaged with an inverted confocal laser scanning microscope (Nikon A1, Japan).
Transfection of IEC-6 cells with siRNA
To knock down the COL2A1 and COL15A1 genes, we designed a double-stranded siRNA, which was synthesized by GenePharma Company (Suzhou). The sequences of COL2A1 and COL15A1 are listed in Supplementary Table 7. IEC-6 cells were seeded in 6-well plates and allowed to reach 60–70% confluence 24 h before transfection. Then, the siRNAs were transfected using Lipofectamine RNAiMAX at a final concentration of 50 nM following the manufacturer’s instructions. After 6–8 h of incubation, the medium containing the siRNA was replaced with complete medium containing serum. Protein and total RNA were extracted 24 h after transfection for Western blotting and qPCR experiments. The transfected cells were used within 24 h for adhesion assays and infection experiments.
Western blotting
To confirm the effect of siRNA knockdown, we transfected IEC-6 cells and then lysed them with RIPA buffer (Beyotime, P0013B) containing a protease inhibitor cocktail (phenylmethylsulfonyl fluoride, PMSF). The total cell lysates were separated by SDS‒PAGE and subsequently transferred onto a PVDF membrane (Millipore, 0.45 μm pore size), which was incubated with the corresponding antibodies (COL15A1 polyclonal antibody from Invitrogen (PA5-115179) and COL2A1 monoclonal antibody from Santa Cruz Biotechnology (sc-52658)) overnight at 4 °C. Immunodetection was performed using a western-light chemiluminescence detection system (Talant). Membrane blots of COL2A1 or COL15A1 were imaged in the range of 60–270 kDa, while those of GAPDH (36 kDa) were imaged in the range of 30–60 kDa.
Statistical analysis
Unless otherwise stated, the data are presented as the means ± SDs. One-way analysis of variance (ANOVA) was used for multiple-group comparisons. Intergroup comparisons were analyzed by Student’s t-test (two-tailed). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. All of the statistical analyses were conducted with at least three independent samples.
Supplementary information
Acknowledgements
The authors thank the Core Facilities of the School of Life Sciences and the National Center for Protein Sciences at Peking University. This work was supported by the National Natural Science Foundation of China (Grant nos. 12372175, 12402215 and 12402360), the Beijing Natural Science Foundation (Grant no. 7244515), the Tianjin Science and Technology Plan Project (Grant no. 22ZYQYSY00030), and the Tianjin Key Medical Discipline (Specialty) Construction Project (Grant no. TJYXZDXK-062B). Graphics software and pictures Illustrations were made using Microsoft Office PowerPoint under Peking University licenses.
Author contributions
H.X. and J.H. designed research; H.X., Y.F., Y.D., Y.H., Y.J. and S.Q. did experiments; H.X., X.D., L.S., X.L., and K.H. analyzed data; and H.X. and J.H. wrote the paper. All authors reviewed the manuscript.
Data availability
The datasets supporting the findings of this study are available at Supplementary Data 1. Additional raw data from this study is available from the corresponding author on reasonable request. The RNA-seq data used in this study are available in the NCBI SRA database under the accession code PRJNA1069618.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00600-x.
References
- 1.Ichiki, T. et al. Sensory representation and detection mechanisms of gut osmolality change. Nature602, 468–474 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Jafari, N. V. & Rohn, J. L. The urothelium: a multi-faceted barrier against a harsh environment. Mucosal Immunol.15, 1127–1142 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato, T. et al. Mucus concentration–dependent biophysical abnormalities unify submucosal gland and superficial airway dysfunction in cystic fibrosis. Sci. Adv.8, 12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pflugfelder, S. C. & De Paiva, C. S. The pathophysiology of dry eye disease. Ophthalmology124, S4–S13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tropini, C. et al. Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell173, 1742–1754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overduin, J., Tylee, T. S., Frayo, R. S. & Cummings, D. E. Hyperosmolarity in the small intestine contributes to postprandial ghrelin suppression. Am. J. Physiol. Gastrointest. Liver Physiol.306, 1108–1116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilleri, M. Diagnosis and treatment of irritable bowel syndrome. J. Am. Med. Assoc.325, 865 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Chang, J. T. & Longo, D. L. Pathophysiology of inflammatory bowel diseases. N. Engl. J. Med.383, 2652–2664 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Thiemicke, A. & Neuert, G. Kinetics of osmotic stress regulate a cell fate switch of cell survival. Sci. Adv.7, eabe1122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jobin, K. et al. A high-salt diet compromises antibacterial neutrophil responses through hormonal perturbation. Sci. Transl. Med.12, eaay3850 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Eddie lp, W. K. & Medzhitov, R. Macrophages monitor tissue osmolarity and induce inflammatory response through NLRP3 and NLRC4 inflammasome activation. Nat. Commun.6, 6931 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinewietfeld, M. et al. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature496, 518–522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, H. P. et al. The nuclear piston activates mechanosensitive ion channels to generate cell migration paths in confining microenvironments. Sci. Adv.7, eabd4058 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang, J. H. et al. Mechanobiological adaptation to hyperosmolarity enhances barrier function in human vascular microphysiological system. Adv. Sci.10, 2206384 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojas, E., Theriot, J. A. & Huang, K. C. Response of Escherichia coli growth rate to osmotic shock. PNAS111, 7807–7812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rojas, E. R., Huang, K. C. & Theriot, J. A. Homeostatic cell growth is accomplished mechanically through membrane tension inhibition of cell-wall synthesis. Cell Syst.5, 578–590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seminara, A. et al. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. PNAS109, 1116–1121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan, J., Nadell, C. D., Stone, H. A., Wingreen, N. S. & Bassler, B. L. Extracellular-matrix-mediated osmotic pressure drives Vibrio cholerae biofilm expansion and cheater exclusion. Nat. Commun.8, 327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuipulotu, D., Mathur, A., Ngo, C. & Man, S. M. Bacillus cereus: epidemiology, virulence factors, and host–pathogen interactions. Trends Microbiol.29, 458–471 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Howden, B. P. et al. Staphylococcus aureus host interactions and adaptation. Nat. Rev. Microbiol.21, 380–395 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dufrêne, Y. F. Sticky microbes: forces in microbial cell adhesion. Trends Microbiol.23, 376–382 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Camiello, V., W.Peterson, B., Van der Mei, H. C. & Busscher, H. Role of adhesion forces in mechanosensitive channel gating in Staphylococcus aureus adhering to surfeces. Npj Biofilms Microbiol.6, 31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng, Y. et al. Geometric constraint-triggered collagen expression mediates bacterial-host adhesion. Nat. Commun.14, 8165 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khateb, H. et al. The role of nanoscale distribution of fibronectin in the adhesion of Staphylococcus aureus studied by protein patterning and DNA-PAINT. ACS Nano16, 10392–10403 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster, T. J. The MSCRAMM Family of cell-wall-anchored surface proteins of Gram-positive Cocci. Trends Microbiol.27, 927–941 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Foster, T. J., Geoghegan, J. A., Ganesh, V. K. & Höök, M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol.12, 49–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, C. et al. Catch bond-mediated adhesion drives Staphylococcus aureus host cell invasion. Nano Lett.23, 5297–5306 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Arora, S., Gordon, J. & Hook, M. Collagen binding proteins of Gram-positive pathogens. Front. Microbiol.12, 628798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Álvarez, S., Leiva-Sabadini, C., Schuh, C. M. A. P. & Aguayo, S. Bacterial adhesion to collagens: implications for biofilm formation and disease progression in the oral cavity. Crit. Rev. Microbiol.48, 83–95 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Valotteau, C. et al. Single-cell and single-molecule analysis unravels the multifunctionality of the Staphylococcus aureus collagen-binding protein Cna. ACS Nano11, 2160–2170 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Beaussart, A., Feuillie, C. & El-Kirat-Chatel, S. The microbial adhesive arsenal deciphered by atomic force microscopy. Nanoscale12, 23885–23896 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Sullan, R. M. A. et al. Single-cell force spectroscopy of pili-mediated adhesion. Nanoscale6, 1134–1143 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Bowen, W. R., Fenton, A. S., Lovitt, R. W. & Wright, C. J. The measurement of Bacillus mycoides spore adhesion using atomic force microscopy, simple counting methods, and a spinning disk technique. Biotechnol. Bioeng.79, 170–179 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Beaussart, A. et al. Nanoscale adhesion forces of Pseudomonas aeruginosa type IV pili. ACS Nano8, 10723–10733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towell, A. M. et al. Staphylococcus aureus binds to the N-terminal region of corneodesmosin to adhere to the stratum corneum in atopic dermatitis. PNAS118, e2014444118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, W. P. J., Wucher, B. R., Nadell, C. D. & Foster, K. R. Bacterial defences: mechanisms, evolution and antimicrobial resistance. Nat. Rev. Microbiol.21, 519–534 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Lallow, E. O. et al. Molecular distribution in intradermal injection for transfer and delivery of therapeutics. Front. Drug Deliv.3, 1095181 (2023). [Google Scholar]
- 38.Shaw, T. D. et al. Mesenchymal stromal cells: an antimicrobial and host-directed therapy for complex infectious diseases. Clin. Microbiol. Rev.34, e00064–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, X., Wu, Y., Mao, C., Shen, J. & Zhu, K. Host-acting antibacterial compounds combat cytosolic bacteria. Trends Microbiol.30, 761–777 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Pierrat, X., Wong, J. P. H., Al-Mayyah, Z. & Persat, A. The mammalian membrane microenvironment regulates the sequential attachment of bacteria to host cells. mBio12, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, X. et al. Extracellular matrix stiffness modulates host-bacteria interactions and antibiotic therapy of bacterial internalization. Biomaterials277, 121098 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Yao, T. et al. Response mechanisms to acid stress promote LF82 replication in macrophages. Front. Cell Infect. Microbiol.13, 1255083 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia, T. et al. The phosphate-induced small RNA EsrL promotes E. coli virulence, biofilm formation, and intestinal colonization. Sci. Signal.16, eabm0488 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Nadesalingam, A., Chen, J. H. K., Farahvash, A. & Khan, M. A. Hypertonic saline suppresses NADPH oxidase-dependent neutrophil extracellular trap formation and promotes apoptosis. Front. Immunol.9, 359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta, K. et al. Actomyosin contractility drives bile regurgitation as an early response during obstructive cholestasis. J. Hepatol.66, 1231–1240 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Herman-Bausier, P. et al. Mechanical strength and inhibition of the Staphylococcus aureus collagen-binding protein Cna. mBio7, e01529–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zong, Y. et al. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J.24, 4224–4236 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laventie, B.-J. et al. A surface-induced asymmetric program promotes tissue colonization by Pseudomonas aeruginosa. Cell Host Microbe25, 140–152 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Green, L. R. et al. Cooperative role for tetraspanins in adhesin-mediated attachment of bacterial species to human epithelial cells. Infect. Immun.79, 2241–2249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, Y. et al. Autoinducer-2 promotes adherence of Aeromonas veronii through facilitating the expression of MSHA type IV pili genes mediated by c-di-GMP. Appl. Environ. Microb.89, e00819–e00823 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Javaheri, A. et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol.2, 16189 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Flores-Mireles, A. L., Pinkner, J. S., Caparon, M. G. & Hultgren, S. J. EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice. Sci. Transl. Med.6, 257ra127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss, G. L. et al. Architecture and function of human uromodulin filaments in urinary tract infections. Science369, 1005–1010 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Adams, J., Gatseva, A., Sin Yuan, Y., Brezzo, G. & Van Agtmael, T. Basement membrane collagens and disease mechanisms. Essays Biochem.63, 297–312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Izzi, V. et al. Exploring the roles of MACIT and multiplexin collagens in stem cells and cancer. Semin. Cancer Biol.62, 134–148 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Cui, Y. et al. Clusterin suppresses invasion and metastasis of testicular seminoma by upregulating COL15a1. Mol. Ther. Nucl. Acids26, 1336–1350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bretaud, S., Nauroy, P., Malbouyres, M. & Ruggiero, F. Fishing for collagen function: about development, regeneration and disease. Semin. Cell Dev. Biol.89, 100–108 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Gorreja, F. & Walker, W. A. The potential role of adherence factors in probiotic function in the gastrointestinal tract of adults and pediatrics: a narrative review of experimental and human studies. Gut Microbes14, 2149214 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lourenço, M. et al. The gut environment regulates bacterial gene expression which modulates susceptibility to bacteriophage infection. Cell Host Microbe30, 556–569 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the findings of this study are available at Supplementary Data 1. Additional raw data from this study is available from the corresponding author on reasonable request. The RNA-seq data used in this study are available in the NCBI SRA database under the accession code PRJNA1069618.