Abstract
The tripartite ancestral structure is a recently proposed model for the genetic origin of modern Japanese, comprising indigenous Jomon hunter-gatherers and two additional continental ancestors from Northeast Asia and East Asia. To investigate the impact of the tripartite structure on genetic and phenotypic variation today, we conducted biobank-scale analyses by merging Biobank Japan (BBJ; n = 171,287) with ancient Japanese and Eurasian genomes (n = 22). We demonstrate the applicability of the tripartite model to Japanese populations throughout the archipelago, with an extremely strong correlation between Jomon ancestry and genomic variation among individuals. We also find that the genetic legacy of Jomon ancestry underlies an elevated body mass index (BMI). Genome-wide association analysis with rigorous adjustments for geographical and ancestral substructures identifies 132 variants that are informative for predicting individual Jomon ancestry. This prediction model is validated using independent Japanese cohorts (Nagahama cohort, n = 2993; the second cohort of BBJ, n = 72,695). We further confirm the phenotypic association between Jomon ancestry and BMI using East Asian individuals from UK Biobank (n = 2286). Our extensive analysis of ancient and modern genomes, involving over 250,000 participants, provides valuable insights into the genetic legacy of ancient hunter-gatherers in contemporary populations.
Subject terms: Evolutionary biology, Genetic variation, Evolutionary genetics, Genome informatics, Risk factors
Using biobank data from over 250,000 participants, the authors present a detailed picture of regional and individual variation in the tripartite ancestral structure of modern Japanese populations and demonstrate its impact on elevated body mass index.
Introduction
Anatomically modern humans, who originated in Africa, began a global dispersal 50–60 thousand years ago (kya) through a series of migrations, settlements, and admixture1,2. The arrival in East Asia can be traced back to at least 40–50 kya, as they gradually spread across the region3. A crucial event in human history was the encounter between indigenous hunter-gatherers and immigrant farmers, subsequently leading to significant shifts in lifestyle4. While this transition to farming occurred on a global scale, its timing and process varied from one region to another; the agricultural revolution in continental East Eurasia dates back to around 10 kya5.
Archeological evidence suggests that humans occupied the Japanese archipelago, an insular region of East Eurasia, as early as 38 kya, during the Paleolithic period6. Still, our understanding of their ancestral connection to modern populations is limited due to the scarcity of ancient DNA data7. Among the well-studied ancestral groups in Japan are the Jomon, a cultural group of hunter-gatherer-fishers who inhabited the archipelago as far back as 16.5 kya8,9. The Jomon are notable for their pioneering use of pottery, which is among the earliest instances in the world10. The Jomon period lasted until ~3 kya, when immigrants from the continent introduced rice cultivation during the Yayoi period that spanned from 3 to 1.7 kya. This agricultural revolution prompted sociopolitical developments, leading to the establishment of the Japanese state in the Kofun period, which began around 1.7 kya and endured for 200–300 years11.
A long-standing model of the origin of Japanese populations is a dual-ancestral structure12,13. This model states that modern Japanese people are a mixture of indigenous Jomon hunter-gatherers from Southeast Asia and immigrant farmers from Northeast Asia. However, a recent ancient DNA study provides compelling evidence that the genetic origin of Japanese populations consists of three distinct ancestors (i.e., tripartite ancestral structure): (1) ancient hunter-gatherer Jomon, (2) Northeast Asian ancestry introduced during the agrarian phase, the Yayoi period, and (3) East Asian ancestry brought in the state formation phase, the Kofun period14. This tripartite structure, which was established during the Kofun period, persists in contemporary populations14–16. Japanese populations are well characterized with their north-to-south gradient of genomic variation17–19. However, the applicability of the tripartite model and variability of the three distinct ancestral components throughout the archipelago remains unknown due to limitations in samples and geographic representation of modern individuals used in previous studies14,15. It is thus crucial to model this ancestral structure using comprehensive, population-level genomic data.
Recent advancements in paleogenomics have made it possible not only to identify diverse genetic ancestors20–23, both locally and globally, but also to uncover their impact on health and disease in contemporary populations, such as the exacerbation and resistance of coronavirus disease 2019 through Neanderthal-introgressed genes24,25 or the genetic predisposition to multiple sclerosis brought by Steppe ancestry26. However, the extent to which the human past has shaped phenotypic variation today remains poorly understood, especially in non-European contexts27. Here, we focus on the Japanese archipelago, where the hunting-gathering Jomon period lasted for more than 10,000 years in the insular environment, and where the genetic remnants of this ancient hunter-gatherer ancestry persist in present-day populations.
In this study, we present an integrated analysis of ancient human genomes (n = 22) and modern Japanese genomes, utilizing the first cohort of Biobank Japan (BBJ, n = 171,287), one of the largest population-based cohorts in a non-European population, encompassing participants from all regions of the archipelago. Our approaches involve five key steps: First, we evaluate the applicability of the tripartite ancestral model to different subpopulations defined by their geography or genetic clusters. Second, we quantify the impact of Jomon ancestry on phenotypic variation among Japanese individuals. Third, we identify Jomon-related genetic variants by employing a genome-wide association study (GWAS) method, with robust adjustment for geographic and genetic substructures, and control for genomic inflation of test statistics. Fourth, we demonstrate the predictive power of Jomon-related variants in estimating an individual’s Jomon ancestry using independent Japanese cohorts (the Nagahama cohort n = 2993 and the second cohort of BBJ n = 72,695). Finally, we apply the Jomon predictive model to East Asian (EAS) individuals within UK Biobank and replicate the phenotypic impact of Jomon ancestry (UKB EAS, n = 2286).
Our study provides a comprehensive understanding of the genetic legacy of ancient hunter-gatherers in contemporary descendants throughout the Japanese archipelago and highlights its impact on phenotypic variation today.
Results
Inference for the tripartite ancestral structure in Biobank Japan dataset
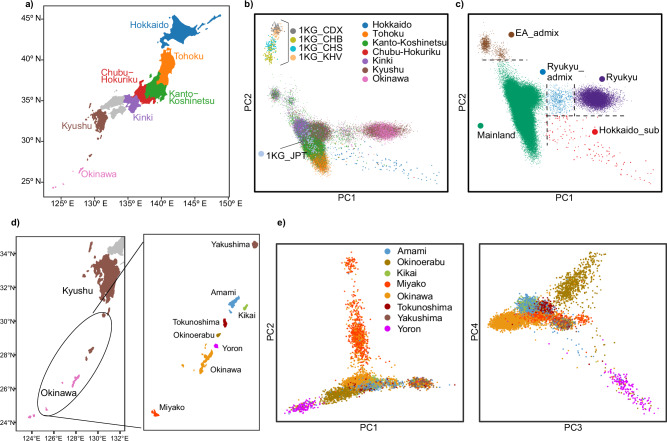
To assess the fit of the tripartite model in contemporary populations, we used BBJ GWAS data, comprising participants from hospitals across seven geographic regions in Japan28,29 (Fig. 1a and Supplementary Data 1). The total number of participants is 171,287, with regional distribution from northeast to southwest throughout the archipelago as follows: Hokkaido = 7955, Tohoku = 11,013, Kanto-Koshinetsu = 94,981, Chubu-Hokuriku = 9489, Kinki = 25,200, Kyushu = 15,962, and Okinawa = 5804. Our PCA defines distinct clusters (Fig. 1b), with a separation of the Ryukyu cluster, primarily including individuals from Okinawa, from the rest of the populations as reported in previous studies17,18. There are also regional clusters observable from Tohoku, Kanto-Koshinetsu, Kinki, and Kyushu respectively. Based on the PCA results, we define five distinct genetic clusters in the Japanese populations: EastAsian_admix (EA_admix; n = 1019), Mainland (n = 159,642), Ryukyu_admix (n = 640), Ryukyu (n = 9847), and Hokkaido_sub (n = 139) (Fig. 1c). Population stratification is further evident even within the Ryukyu Islands, reflecting their geographic affinities in this local insular context (Fig. 1d, e, Yakushima; n = 431, Amami; n = 1531, Kikai; n = 561, Okinoerabu; n = 845, Tokunoshima; n = 476, Yoron; n = 167, Okinawa; n = 4795, Miyako; n = 827).
Fig. 1. Population structure of Biobank Japan.
a Seven geographic regions, each represented by a different color, indicate the locations where participants were registered at local hospitals. b A scatter plot of PCA for BBJ participants with 1KG EAS samples. The colors of the BBJ participants correspond to those used in (a). c Clustering results of BBJ participants based on PCA. d Geographic locations of the Ryukyu Islands. e Scatter plots of PCA for BBJ participants from the Ryukyu Islands. The individuals are colored differently according to the locations of their registered hospitals. In (a, d) the map of Japan is drawn using the R package “jpndistrict” (https://github.com/uribo/jpndistrict). PC Principal component, 1KG 1000 Genomes Project, JPT Japanese in Tokyo, CDX Chinese Dai in Xishuangbanna, CHB Han Chinese in Beijing, CHS Han Chinese South, KHV Kinh in Ho Chi Minh City, NEA Northeast Asian, EA East Asian.
We then merged this diverse set of Japanese individuals with ancient genomic data from Japan and the Eurasian continent14. To find the sites that are present in both the array-typing genome data from BBJ and the pseudo-diploid data from ancient humans, we converted the highly accurate BBJ imputed dosage data (Rsq ≥ 0.7) into genotype data (details in “Methods”). This conversion resulted in 2,038,260 shared variants (n = 171,287 of BBJ and n = 22 of ancient genomes). We subsequently applied qpAdm in AdmixTools30,31, based on a set of source populations defined in a previous study14 (see Supplemental Data 2), to evaluate the fit of admixture models and to estimate admixture proportions at both population and individual levels in the biobank. To comprehensively capture the geographic and genetic diversity of Japanese populations, we fitted the tripartite model to geographically-defined populations throughout the Japanese archipelago and the Ryukyu Islands (Fig. 1b, e), five genetically-defined populations (Fig. 1c), as well as the entire BBJ dataset. This analysis demonstrates that the tripartite structure provides a better fit for all populations, both at broad and local scales, when compared to all possible dual-ancestral structure models (Supplementary Data 3). The only exception is EA_admix, where the population can be sufficiently explained by a two-way admixture between Northeast Asian and East Asian ancestry.
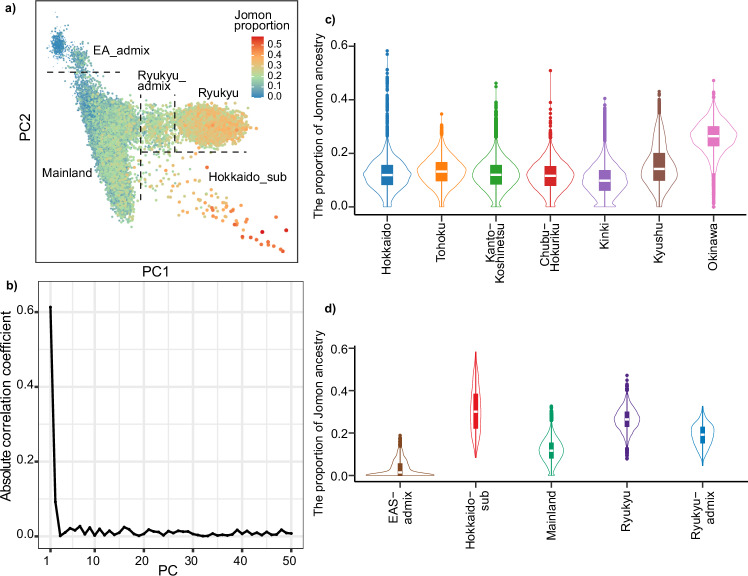
In the whole BBJ dataset, the proportions of the three distinct ancestral components closely align with those reported in the previous study (Jomon: 12.4%, Northeast Asia: 21.2%, and East Asia: 66.4%)14. However, Jomon ancestry exhibits regional variation, ranging from 9.8% in Kinki to 26.1% in Okinawa (Fig. 2a). Within the Ryukyu Islands, there is an elevated level of Jomon ancestry, with the highest proportion observed on Yoron Island (Fig. 2b). Jomon ancestry is even higher in one of the genetically-defined populations, Hokkaido_sub (31.6%, Fig. 2c and Supplementary Data 3), which primarily includes a subset of individuals from Hokkaido. In contrast, EA_admix, possibly representing continental individuals from East Asia, has very little Jomon ancestry; this may explain why the admixture model without Jomon ancestry is preferred. The Mainland cluster mirrors the proportion of the entire BBJ as it includes the majority of the samples in the data (159,642 out of 171,287). Even when we separate individuals from this cluster based on their geographic origins (i.e., where the sample collection took place), this proportion remains relatively consistent across different regions (Supplementary Fig. 1). The Ryukyu cluster represents the ancestral composition from Okinawa, while the proportions in the Ryukyu_admix cluster fall between those of the Mainland and Ryukyu clusters (Supplementary Data 3).
Fig. 2. Variation in tripartite ancestry structures across groups of Biobank Japan participants.
Bar plots show the proportions of three distinct ancestors. a Populations are defined either as a group of the entire BBJ samples or by the regions where participants were registered (Hokkaido = 7955, Tohoku = 11,013, Kanto-Koshinetsu = 94,981, Chubu-Hokuriku = 9489, Kinki = 25,200, Kyushu = 15,962, and Okinawa = 5804). b Individuals recruited from the Ryukyu Islands are grouped by their specific islands (Yakushima = 431, Amami = 1531, Kikai = 561, Okinoerabu = 845, Tokunoshima = 476, Yoron = 167, Okinawa = 4795, and Miyako = 827). c The BBJ samples are split into five different insular populations based on their PCA clusters shown in Fig. 1 (Mainland = 159,642, Hokkaido_sub = 139, Ryukyu = 9847, Ryukyu_admix = 640, and EA_admix = 1019). Proportions in the bar plots represent the mean values of three ancestral components estimated using qpAdm, with error bars indicating standard errors. Values at the top of each bar indicate the tail probabilities of the tripartite model for each group. NEA Northeast Asian, EA East Asian.
Next, we asked whether Jomon ancestry is uniquely present in the Japanese populations or whether this ancestry is also observable in continental populations using f4-statistics with the form of f4(Mbuti, Jomon; Han, X). Our target populations include those in the Simon Genome Diversity Project (SGDP) panel, the 1000 Genomes Project (1KG), and the subpopulations within BBJ. As shown in previous studies8,9,14, there is no extra genetic affinity between the Jomon and any of the populations tested, except for Ulchi in the SGDP or the East Asians (EAS) within the 1KG (Supplementary Fig. 2). Among the 1KG EAS population, only Japanese in Tokyo (JPT) show a significant affinity to the Jomon. Within the BBJ participants, the Hokkaido_sub and Ryukyu subpopulations exhibit an extremely strong affinity with the Jomon as consistent with the higher Jomon ancestry in our admixture modeling (Fig. 1e), as well as previous studies8,9.
Overall, our analysis provides detailed pictures of regional variation in the tripartite ancestral structures throughout the archipelago.
Variation in the tripartite structure among individuals
Our admixture modeling reveals that two subpopulations, Ryukyu and Hokkaido_sub, have higher proportions of Jomon ancestry compared to the other subpopulations (Fig. 2). To visualize the genetic distance between ancient and present-day populations, we projected the ancient or modern individuals, who represent the three distinct ancestors underlying the tripartite structure of modern Japanese populations, along with additional ancient Japanese individuals (i.e., Yayoi and Kofun) onto the PCA plots. While the Kofun are included within variation in the present-day populations, the Jomon are clustered in a position extending from the Ryukyu and Hokkaido_sub subpopulations. The two individuals from the Yayoi period, who are morphologically considered as the Jomon32 but genetically admixed between the Jomon and continental ancestry14, are positioned between the Jomon and the present-day populations (Supplementary Fig. 3a).
Given that the population-based admixture modeling only represents an averaged pattern in a group of individuals, we then apply the tripartite model to each participant in BBJ individually. This model fits 154,339 out of the 171,287 individuals (90.1%), with varying proportions of three ancestral components. The proportions of these three ancestors are negatively correlated with each other (Supplementary Fig. 3b, c), supporting a previously proposed scenario that two continental ancestors, Northeast Asian and East Asian, are likely to have arrived in the archipelago independently14. Approximately 5% of the individuals did not conform to the tripartite model (8932 individuals), showing a better fit for a two-way admixture involving either Jomon and East Asian ancestry or Northeast Asian and East Asian ancestry. However, 28 individuals were excluded from subsequent analysis due to insufficient support for the dual ancestry models over the tripartite model, as indicated by nested p < 0.05. There was also a minor exception, where a model of East Asian ancestry alone, rather than any dual structure models, was preferred in ten individuals (Supplementary Fig. 4). It’s important to note that none of the models could adequately fit the remaining individuals, comprising nearly 5% (7962 individuals). This is likely due to the use of a tail probability cut-off of 5%, which accounts for the inherent variability in the data, as a defined proportion of all tests would be expected to deviate from the models tested.
By incorporating the proportion of Jomon ancestry onto the PCA plots, it becomes evident that there is a noticeable gradient in the Jomon proportion along the first and second principal components (Fig. 3a). Indeed, the Jomon proportion has a remarkably strong correlation with the first two principal components (Fig. 3b; |R| = 0.61 and 0.09 for PC1 and PC2, respectively; see Supplementary Data 4). Similar patterns of correlation are also observable between Northeast Asian or East Asian ancestry and the PCs; however, the strength of correlation is not as pronounced as that observed for Jomon (|R| = 0.14 for Northeast Asian and PC1; |R| = 0.26 for East Asian and PC1). The correlation between Jomon ancestry and PCs is still evident even upon focusing solely on individuals from the Mainland or Ryukyu cluster (Supplementary Fig. 5). These findings strongly indicate that Jomon ancestry has a critical role in shaping genomic variation among Japanese individuals at the level of PCA. Furthermore, these individual-based estimates not only mirror the population-based patterns in terms of their means but also highlight significant variation in Jomon ancestry across the Japanese archipelago (Fig. 3c, d).
Fig. 3. Genetic legacy of Jomon ancestry throughout Japanese populations.
Proportions of Jomon ancestry are estimated through admixture modeling under models optimal for each individual (n = 163,243). a Projection of the Jomon proportions onto the PCA plots. b Absolute values of the correlation coefficients between the Jomon proportions and PCs. The correlation coefficient is calculated using Pearson’s method. c Boxplots representing variation in the Jomon proportion across BBJ participants within each of their registered regions. d Boxplots representing variation in the Jomon proportion across the participants within each of different genetic clusters defined by PCA. In (c, d), boxes denote the interquartile range (IQR) and the median is shown as white horizontal bars; whiskers extend to 1.5 times the IQR; outliers are shown as individual points. PC Principal component, EA East Asian.
Impacts of Jomon ancestry on phenotypic variation in Japanese populations
Next, we explored whether or not Jomon ancestry has any phenotypic impact on present-day populations. Among 163,243 individuals from BBJ, who were successfully modeled for their genetic ancestry, the average proportion of the Jomon component is 12.5 ± 6.3% (mean ± SD). There is no substantial difference in the Jomon proportion between ages or between genders (Supplementary Fig. 6).
We then tested associations of Jomon ancestry with 80 different complex traits by making robust adjustments for genetic and geographic subpopulations (see “Methods” and Supplementary Data 5). To account for type I error, we set a threshold of statistical significance at P < 0.05/80 = 6.3 × 10−4, based on the Bonferroni correction. We confirm that this threshold was calibrated by simulating 10 dummy heritable phenotypes as negative controls (“Methods”). Our analysis with all BBJ participants identifies significant associations with an increase in body mass index (BMI) (Fig. 4a and Supplementary Data 6; Beta = 0.012, Standard error [SE] = 0.003, P = 3.0 × 10−5). However, it is important to note that regional disparity, such as the higher BMI in Okinawa compared to the other regions33, may potentially confound this association, even when adjusting for geographic factors as covariates. To address this concern, we confined our analysis to individuals included in the Mainland cluster (n = 152,148; see Fig. 1c). The association with BMI remains statistically significant (Fig. 4b and Supplementary Data 7; Beta = 0.012, SE = 0.003, P = 7.9 × 10−5). We further confirmed the significance of the association with BMI regardless of sex or age (see Supplementary Fig. 7).
Fig. 4. Associations of Jomon ancestry with 80 complex traits.
Associations are tested using a generalized linear model in (a) the entire BBJ participants (n = 163,243) and (b) the participants only in the Mainland cluster (n = 152,148). The quantitative traits are modeled by linear regression, while the binary traits are analyzed with logistic regression. The direction of the triangle corresponds to the sign of the beta coefficient for each trait. The gray dashed line represents the statistical significance based on the Bonferroni correction (P < 0.05/80 = 6.3 × 10−4). Control indicates 10 dummy phenotypes. All traits labeled on the plots have nominal significance with P < 0.05. Details of statistical test results are presented in Supplementary Data 6 and 7. P-values are computed by linear regression or logistic regression. All statistical tests are two-sided and unadjusted for multiple comparisons. BMI Body mass index, BW Body weight, LVM Left ventricular mass, E/A E/A ratio, RBC Red blood cell count, MCH Mean corpuscular volume, LDLC low-density lipoprotein cholesterol.
While it may be argued that our approach is conservative, we prioritize addressing any potential inflation of association signals stemming from population stratification34. Therefore, it is crucial to account for its effect by incorporating PCs as covariates, despite the stratification originating from variation in Jomon ancestry across individuals to some degree. To mitigate this issue, we also employed an alternative methodology where phenotypes underwent correction by regressing their measures with all covariates, including PCs, before being tested for associations with Jomon ancestry. We observed a significant association in BMI, while none of the other traits reached statistical significance (Supplementary Fig. 8, and Supplementary Data 8 and 9). Taking all results together, our analyses consistently demonstrate the robustness of the BMI signals, regardless of the methods used for correcting population stratification (see Fig. 4 and Supplementary Fig. 8).
To assess the impact of Jomon ancestry on BMI, we incorporated the proportion of Jomon ancestry as a covariate in our GWAS for BMI (Supplementary Fig. 9a and Supplementary Data 11). While the majority of lead SNPs remain consistent, we observed a reduction in the effect of Jomon ancestry on polygenic scores (PGS) of BMI (Supplementary Fig. 9b). It is worth noting that this effect persists to some extent, indicating a functional correlation between Jomon ancestry and BMI-PGS. Still, the PGS tends to be inflated when Jomon ancestry is not accounted for in GWAS (Supplementary Fig. 9c), with the degree of inflation weakly correlating with the proportion of Jomon ancestry (Supplementary Fig. 9d). These findings suggest that the effect sizes estimated from GWAS without including Jomon ancestry as a covariate may be biased due to residual confounding.
Our analysis also includes height, which has been shown to exhibit a north-to-south gradient in Japanese populations35. The association of Jomon ancestry with the decrease of height is observable only if PCs are not accounted for in the test (Supplementary Fig. 10 and Supplementary Data 10), indicating that this association can be confounded by population stratification. Overall, these results suggest that the genetic legacy of the ancient hunter-gatherer Jomon significantly influences BMI across populations today, regardless of geographic differences, which may consequently contribute to an increased risk of obesity. Additionally, we tested the effect of the use of the Jomon proportion on the predictive power of the polygenic score (PGS) for BMI (see “Method”). The incremental predictive performance is limited with a magnitude of −2.8 × 10−3.
Identification of genetic variants underlying variation in Jomon ancestry
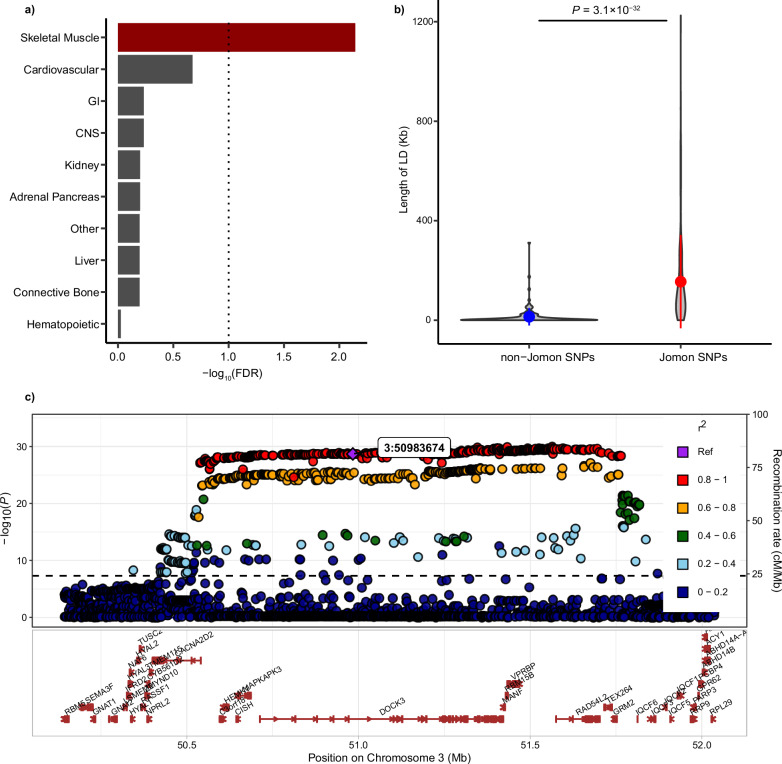
We performed a genome-wide investigation of variants associated with variation in Jomon ancestry among individuals. In this analysis, an individual proportion of Jomon ancestry is considered as a proxy phenotype. However, a conventional null hypothesis, as used in standard genotype-phenotype association studies, is not directly applicable to this phenotype since it is inferred from the genomic data. To ensure robust correction for genomic and geographic subpopulations and to control for genomic inflation of the test statistics, we adopted a double genomic control correction method and a mixed linear model (MLM) approach that includes PCs as covariates36. Furthermore, we conducted a meta-analysis of the association tests for individuals within the Mainland and Ryukyu clusters separately, as the Jomon proportions significantly differ between these groups (Fig. 3d). To gain biological insights into the genetic signals enriched for Jomon ancestry, we conducted stratified linkage disequilibrium score regression (S-LDSC) on the genome-wide meta-analyzed result. Our S-LDSC identified a significant enrichment of the heritability in skeletal muscle cells across major cell groups (Fig. 5a).
Fig. 5. Functional and genomic characterization of Jomon-related marker SNPs.
a Stratified LD score regression analysis is conducted with major 10 cell type groups. False discovery rate (FDR) is calculated using the Benjamini-Hochberg method. A dotted vertical line represents the FDR of 0.1. b Violin plots compare the lengths of regions in strong LD with each of 132 Jomon-related SNPs (r2 > 0.8) to those of 132 frequency-matched non-Jomon-related SNPs. Dots represent the mean lengths, with bars showing the standard deviations. A p-value (3.1 × 10−32) is calculated by the Wilcoxon rank sum test, reflecting a two-sided test. c A locus plot highlights a Jomon-related SNP, which is shown as a purple diamond in the upper plot, that is linked with the longest haplotype among all 132 variants. A dashed line defines a statistical significance as a p-value of 5.0 × 10−8. Lower plots represent the protein-coding genes, based on GENCODE, that are present in the highlighted region. The P-value is computed by inverse variance weighted meta-analysis using METAL. All statistical tests are two-sided and unadjusted for multiple comparisons. GI Gastrointestinal, CNS Central nerve system, LD Linkage disequilibrium, SNP Single nucleotide polymorphism.
Based on our association analysis, we classify a variant as Jomon-related if it has a positive Z-score and reaches a genome-wide significance (P < 5.0 × 10−8) after rigorous control measures. This results in 132 independent variants identified from LD clumping (details in “Methods”; Table 1 and Supplementary Data 12). To explore evolutionary contexts of these Jomon-related variants, we examined haplotype structures of the genomic regions containing Jomon-related variants. Contrasting these haplotype structures with those of allele frequency-matched non-Jomon-related variants (i.e., variants with P > 0.05), we find that the Jomon-related variants exhibit significantly longer haplotypes than non-Jomon-related variants (P = 3.1 × 10−32) (Fig. 5b, c and Supplementary Data 12). This strongly supports a Jomon origin for these extended haplotypes, supported by strong linkage disequilibrium throughout the genome, possibly due to the high genetic homogeneity and small effective population size (~1000) within the Jomon population14,37. Furthermore, the persistence of these long Jomon-derived haplotypes in the contemporary populations can be attributed to the relatively recent admixture with the continental ancestors14, coupled with insufficient time for recombination to break apart the haplotypes. Leveraging selection scan results based on singleton density score (SDS)17, which detects selection over approximately the past 100 generations38, we further observe an enrichment of selection signals in the Jomon-derived haplotypes (P = 0.008). These results support the idea that the Jomon-related variants can serve as markers for quantifying Jomon ancestry, tag Jomon-derived haplotypes, and potentially have been subject to recent selective pressures.
Table 1.
List of the top 10 Jomon-related variants ranked by statistical significance in BBJ
| Frequency of alternative allele | FST between JPT and EAS | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBJ | 1KG populations | Jomon | |||||||||||||||||
| rsID | Chr | Pos (hg19) | Ref/Alt | Loci | Nearest gene | P | PheWAS | Mainland | Ryukyu | EAS | AFR | AMR | EUR | SAS | CDX | CHB | CHS | KHV | |
| rs28729170 | 1 | 188,505,272 | C/A | 1q31 | PLA2G4A | 8.1 × 10−33 | - | 0.78 | 0.83 | 0.72 | 0.74 | 0.66 | 0.79 | 0.85 | 0.86 | 0.01 | 0.03 | 0.03 | 0.00 |
| rs13017060 | 2 | 15,641,500 | A/C | 2p24 | NBAS | 1.7 × 10-32 | BMI, Body weight | 0.29 | 0.38 | 0.16 | 0.20 | 0.52 | 0.63 | 0.41 | 0.86 | 0.10 | 0.03 | 0.13 | 0.04 |
| rs2645158 | 19 | 31,885,791 | A/G | 19q12 | TSHZ3 | 5.4 × 10−30 | T2D | 0.38 | 0.36 | 0.42 | 0.82 | 0.58 | 0.66 | 0.62 | - | 0.00 | 0.00 | 0.00 | 0.01 |
| rs6446239 | 3 | 50,983,674 | G/C | 3p21 | DOCK3 | 2.0 × 10−29 | Height, Body weight | 0.68 | 0.73 | 0.65 | 0.62 | 0.71 | 0.97 | 0.95 | 0.83 | 0.01 | 0.02 | 0.00 | 0.02 |
| rs1026980 | 6 | 47,624,988 | C/T | 6p12 | ADGRF2 | 3.8 × 10−26 | - | 0.34 | 0.39 | 0.31 | 0.54 | 0.47 | 0.68 | 0.72 | - | 0.00 | 0.00 | 0.00 | 0.00 |
| rs2057165 | 6 | 64,966,049 | T/A | 6q12 | EYS | 3.5 × 1025 | - | 0.50 | 0.53 | 0.50 | 0.71 | 0.66 | 0.71 | 0.73 | 0.56 | 0.02 | 0.09 | 0.03 | 0.04 |
| rs4302225r | 2 | 84,050,402 | C/T | 2p11 | SUCLG1 | 2.8 × 10−23 | - | 0.50 | 0.54 | 0.50 | 0.39 | 0.58 | 0.72 | 0.62 | - | 0.00 | 0.00 | 0.00 | 0.00 |
| rs4981864 | 14 | 32,086,838 | G/T | 14q12 | NUBPL | 2.1 × 10−24 | - | 0.55 | 0.56 | 0.61 | 0.19 | 0.54 | 0.58 | 0.53 | 0.83 | 0.05 | 0.00 | 0.00 | 0.00 |
| rs629577 | 10 | 38,335,178 | A/G | 10p11 | ZNF33A | 3.4 × 10−22 | - | 0.49 | 0.55 | 0.42 | 0.46 | 0.48 | 0.51 | 0.74 | - | 0.00 | 0.01 | 0.00 | 0.02 |
| rs6097031 | 20 | 51,188,901 | G/C | 20 | ZFP64 | 5.2 × 10−21 | - | 0.35 | 0.46 | 0.32 | 0.88 | 0.35 | 0.30 | 0.41 | 0.91 | 0.00 | 0.00 | 0.01 | 0.01 |
All 132 Jomon-related variants are listed in Supplementary Data 11, with their FST values shown in Supplementary Data 12. The P-value is computed by inverse variance weighted meta-analysis using METAL. All statistical tests are two-sided and unadjusted for multiple comparisons. Nearest gene names are described in Italic.
Ref/Alt Reference/Alternative allele (effect allele is alternative allele), Alt freq. the alternative allele frequency, BBJ Biobank Japan, 1KG 1000 Genomes Project, T2D type 2 diabetes.
These Jomon-related variants are significantly more frequent in JPT compared to the other East Asian populations within the 1KG dataset (Table 1, Supplementary Fig. 11, and Supplementary Data 12). Even within the BBJ population, the Ryukyu group exhibits higher frequencies of these variants than the mainland group, supporting their strong linkage with the Jomon-derived segments. We measured the fixation index (FST) for the 132 Jomon-related variants in order to assess their specificity in the Japanese population. While there is no major difference in FST across the East Asian populations, rs536618 at 4p12 and rs2871660 at 1q31 show relatively high FST values in the Japanese population (Supplementary Figs. 12 and 13; Supplementary Data 13). Notably, the top variant, rs13017060, is located in the intron region of the NBAS gene, exhibiting pleiotropic effects on the increase of BMI and body weight in modern Japanese populations29. It also serves as an expression quantitative trait locus (eQTL) for NBAS in heart muscle, as evidenced by GTEx data39. Moreover, the same is associated with increased leg fat mass in the UK Biobank40,41. These findings provide further evidence supporting the significant association between Jomon ancestry and BMI.
Validation of the tripartite model and Jomon-related variants using independent Japanese cohorts
To validate our admixture modeling of the tripartite structure, we utilized two independent Japanese cohorts, the Nagahama cohort and the second cohort of BBJ (BBJ-2nd). The Nagahama cohort includes participants from Nagahama city, Shiga Prefecture, within the Kinki area of Japan (comprising Nagahama A with n = 1549 and Nagahama B with n = 1444)42. BBJ-2nd is an additional independent cohort, which is distinct from the first BBJ cohort, representing various regions across the Japanese archipelago (n = 72,695).
Our validation process consists of two different approaches: (i) estimating the proportion of Jomon ancestry at the individual level and (ii) deriving the Jomon ancestry prediction score as a form of PGS. The PCA of BBJ-2nd confirms the significant correlations of Jomon ancestry with the first two PCs (Fig. 6a; Supplementary Data 4), as observed in the first BBJ cohort (Fig. 3a). We then derived the prediction scores of Jomon ancestry using 132 Jomon-related variants identified from the first BBJ cohort. Under the robust adjustment for population stratification, the variance of Jomon ancestry explained by the prediction scores are as follows: 014, 0.12, and 0.13 in Nagahama A, Nagahama B, and BBJ-2nd respectively (PNagahama A = 2.5 × 10−50, PNagahama B = 1.3 × 10−40, and PBBJ-2nd < 1.0 × 10−300). Notably, the observed Jomon proportions gradually increase in line with the prediction scores across the different cohorts when the scores are grouped into deciles (Fig. 6b and Supplementary Fig. 14).
Fig. 6. Validation of the genetic legacy of Jomon ancestry and the predictive power of 132 Jomon-related variants using an independent cohort of BBJ-2nd.
a The PCA plots include all individuals from BBJ-2nd (n = 68,632), with their respective Jomon proportions overlaid. b The Jomon component prediction scores measured from all individuals within the cohort (n = 72,695) are split into the deciles. The boxplot shows the distribution of observed Jomon proportions in each decile of the predicted scores. R2 indicates the squared value of the Pearson’s correlation coefficient between the prediction score and the residuals of the Jomon proportion regressed out with 10 PCs. P-value reflects a two-sided test. In (b), boxes denote the interquartile range (IQR) and the median is shown as white horizontal bars; whiskers extend to 1.5 times the IQR; outliers are shown as individual points. PC Principal component.
The predictive power of Jomon-related variants shows an increase when the p-value cut-off is relaxed from 5.0 × 10−8 to 1.0 × 10−3, resulting in the variance of 0.27 (Nagahama A), 0.26 (Nagahama B), and 0.24 (BBJ-2nd) respectively for Jomon ancestry explained by genetic variants (Supplementary Fig. 15). However, it is crucial to note that this clumping and thresholding method has been demonstrated to potentially overfit the data, particularly when relaxing the p-value cut-offs43. Therefore, we adhere to a p-value cut-off of 5 × 10−8 for predicting Jomon ancestry in subsequent analysis.
Replication of the association between Jomon ancestry and BMI using East Asian individuals from UK Biobank
We sought to replicate our finding on the link between Jomon ancestry and the increase of BMI using a completely independent cohort. Our focus is on East Asian individuals within UK Biobank (UKB EAS). We first selected the individuals of UKB EAS based on the PCA plots (n = 2286; Supplementary Fig. 16). The f4-test, in the form of f4(Mbuti, Jomon; Han, UKB EAS), shows that the Jomon are symmetrically related to Han and UKB EAS (Z = 0.93). We then split UKB EAS into different groups based on their self-reported ethnic backgrounds and performed f4-test for each group individually (Supplementary Fig. 17). This analysis identifies Ethnic Group (EG) 6 (i.e., the self-reported Other ethnic population, n = 541) that shows a higher genetic affinity to Jomon than Han with a Z-score of 5.4. None of the other EGs support a significant affinity with Jomon, including EG5, consisting of self-reported Chinese individuals (n = 1375). These results suggest that participants in EG6 carry Jomon ancestry and are a suitable subpopulation for our replication analysis.
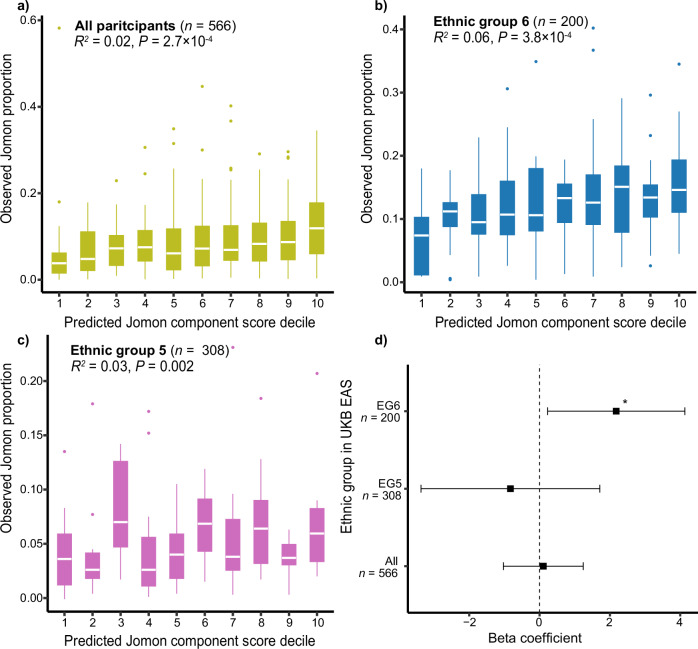
By fitting the admixture models to the UKB EAS cohort (n = 2286), we were able to quantify Jomon ancestry for 566 individuals. The predictive power of the scores based on 132 Jomon-related variants account for ~2% of the total variance in the observed Jomon proportion (R2 = 0.02 and P = 2.7 × 10−4; Fig. 7a). This power substantially increases when only focusing on EG 6 within UKB EAS (R2 = 0.06 and P = 3.8 × 10−4; Fig. 7b). In contrast, this prediction becomes less effective in EG5 (R2 = 0.03, and P = 0.002 Fig. 7c), aligning with the apparent scarcity of Jomon ancestry in this group (Supplementary Fig. 17).
Fig. 7. Prediction of Jomon ancestry and replication of phenotypic association using UKB EAS.
Jomon component prediction scores are split into deciles. The boxplot shows the distribution of observed Jomon proportions in a given decile of the predicted scores based on (a) all UKB EAS participants (n = 566), (b) EG6 (self-reported Other ethnic population; n = 200), and (c) EG5 (self-reported Chinese population; n = 308). The EG number is referred from Data-coding of UKB. d The forest plots represent the effect size of Jomon ancestry on BMI in UKB EAS, EG5, and EG6. Squares indicate the point estimates, while error bars indicate 95% confidence intervals. Asterisks represent P < 0.05 (P = 0.03 in EG6). In (a, b, c), boxes denote the interquartile range (IQR) and the median is shown as white horizontal bars; whiskers extend to 1.5 times the IQR; outliers are shown as individual points. R2 indicates the squared value of the Pearson’s correlation coefficient between the prediction score and the residuals of the Jomon proportion regressed out with 10 PCs. P-values reflect two-sided tests. In (d), P-values are computed by linear regression. All statistical tests are two-sided and unadjusted for multiple comparisons.
Finally, we tested the effect of Jomon ancestry on BMI in UKB EAS. There is no significant association either in the entire UKB EAS or EG5 of UKB EAS (Fig. 7d; Beta = 0.11, SE = 0.58, P = 0.86 in the entire UKB EA; Beta = −0.83, SE = 1.30, P = 0.52 in EG5). However, when focusing exclusively on EG6, which exhibits a higher Jomon genetic influence, we identify a significant association between Jomon ancestry and an increase in BMI (Beta = 2.2, SE = 0.99, P = 0.03).
Overall, these results highlight the potential influence of Jomon ancestry on the risk of obesity in contemporary populations regardless of differences in their living environments, as observed between UK and Japan.
Discussion
Our study provides comprehensive insights into the genetic legacy of ancient hunter-gatherers in the Japanese archipelago by combining ancient human genomes from Japan and continental East Asia with modern genomic data of more than 250,000 Japanese individuals. Leveraging this extensive biobank-scale dataset, we demonstrate that the recently proposed model for the genetic origin of modern Japanese populations, known as the tripartite ancestry structure14, widely and consistently fits better than the dual structure model across the Japanese archipelago. The composition of the three distinct ancestors varies among different geographic and genetic subpopulations within Japan. Among these ancestral components, the ancient hunter-gatherer Jomon stands out as the most influential in increasing BMI in contemporary individuals, as well as in shaping genomic variation, both at the individual and population levels. Our novel GWAS approach, coupled with rigorous controls for genomic inflation factors, effectively identifies genetic variants associated with the Jomon ancestry that modern Japanese possess. We have then pinpointed a set of 132 independent variants as genetic markers for predicting an individual’s Jomon ancestry. The predictive power of these markers is validated with independent cohorts of Japanese populations. Furthermore, we successfully replicate the phenotypic impact of Jomon ancestry by studying a cohort of individuals who have Jomon components as their genetic ancestor, residing in the United Kingdom.
Our analysis presents the first in-depth characterization of the tripartite structure across the entire archipelago. It unveils substantial variation in the proportion of Jomon ancestry, mirroring the genetic ancestry continuum observed in present-day populations. This proportion is extremely high in individuals from the Ryukyu and Hokkaido_sub clusters, as indicated in previous studies8,9. The continental ancestor (i.e., Northeast Asian and East Asian) is also observable in individuals from Okinawa. However, it is important to note that these continental components were not directly introduced from the continent but by immigrants from the main islands of Japan, who already possessed the tripartite ancestor15. This migration has been estimated to have occurred around the eleventh century AD, marking the end of the prehistoric period in the area. Until this transition, it is widely accepted that people with Jomon-like genetic characteristics continued to inhabit the region for at least several thousand years44. Therefore, the elevated levels of Jomon ancestry in Okinawa can be attributed to this historical event.
In contrast to the Ryukyu or Hokkaido_sub clusters, the Kinki region exhibits a relatively low level of Jomon ancestry. Historical records indicate the enduring presence of governmental center in this area, implying more frequent interactions with people from the Asian continent than in other regions45. Our biobank-scale analysis provides a refined picture of the genetic makeup of people throughout the Japanese archipelago both at broad and local scales.
The finding on the significant association between increased Jomon ancestry and the increase in BMI aligns with the observations that people from Okinawa have not only elevated levels of BMI but also a higher propensity for obesity compared to those from the main island33. The obesogenic environment in present-day populations is certainly a substantial contributor to the ongoing obesity epidemics. Nonetheless, we identify this phenotypic impact of Jomon ancestry in the populations from Japan, as well as the UK, where individuals are exposed to varying degrees of obesogenic environments. Therefore, the ancient hunter-gatherer ancestry plays a key role in increasing BMI in Japan, which could also be linked to the disparities in obesity prevalence among Asian populations residing in Western countries46. By emphasizing the importance of incorporating Jomon ancestry as a confounding factor in GWAS for BMI, this analysis provides a proof-of-concept for research that bridges our human past with current health challenges.
Given that the set of genetic variants associated with the proportion of Jomon ancestry likely tags genomic segments descended from the Jomon, they could serve as ancestral informative markers (AIMs), offering a convenient way to predict an individual’s Jomon ancestry. We demonstrate the predictive power of these markers by leveraging three additional validation cohorts. Furthermore, the Jomon-related variants are significantly enriched with active functions in skeletal muscle cells, some of which are also associated with increased BMI, body weight, and height. These findings suggest potential adaptations related to the high physical activity required for hunting and gathering lifestyles47, as supported by selection scans in the Jomon showing that the top variants are associated with increased bone mineral density37. However, the genetic legacy of this selection may now pose a risk factor for elevated levels of BMI through the interaction with modern environments. Our study provides evidence of how the past action of natural selection has shaped present-day disease risks, primarily due to rapid changes in diet and societal lifestyle. It is important to note that further analysis with post-Jomon genomes (e.g., Yayoi or Kofun genomes) is crucial to uncover how Jomon segments have been inherited by descendants as lifestyles transformed from hunting and gathering to farming.
Nonetheless, our research has several caveats. First, we employed modern genomic data converted from imputed genome-wide array data. Consequently, our modern data remains ascertained, and the use of large-scale whole-genome sequence data could offer an unbiased set of variants for analysis. Regarding ancient genomic data, all Jomon data used in this study are shotgun-sequenced, rather than capture-sequenced8,9,14,48. Still, most of the data are of low-coverage and were analyzed as pseudo-haploid data alongside the modern dataset. This may potentially restrict our ability to establish associations between modern and ancient genomes at a finer resolution49. However, the emerging genotype imputation for ancient genomes may provide an innovative solution to enhance the depth of genotypic profiles derived from such low-coverage data37,50,51.
In summary, our integrated analysis of modern and ancient genomes unveils the genetic legacy of ancient hunter-gatherers in populations today and its impact on phenotypic variation, shedding light on the significance of understanding a person’s genetic ancestry not only for tracing their genetic origins but also for controlling confounding effects in GWAS. The field of ancient genomics is rapidly evolving, and future research that encompasses a diverse range of ancient humans across various time periods and geographic locations will be essential in providing a more comprehensive understanding of the extent to which the human past has shaped genomic and phenotypic variation in contemporary populations.
Methods
Biobank Japan (BBJ) dataset
We used 171,287 individuals from the first cohort of BBJ, which enrolled participants between 2003 and 2007. BBJ is a hospital-based genome cohort that included participants affected by at least one of 47 diseases from 12 medical institutes located in seven regions in Japan28. All participants provided written informed consent, which was approved by the ethical committees of the Institute of Medical Science, the University of Tokyo. This study was approved by the ethical committees of Osaka University Graduate School of Medicine and Graduate School of Medicine, the University of Tokyo.
We excluded (i) individuals with lower call rates (<99%), (ii) closely related individuals with genetic relatedness ≥0.178 calculated from a genetic related matrix (GRM) by GCTA (version 1.93.3beta2), or (iii) individuals were excluded if they were positioned far from the Japanese cluster defined within the 1000 Genomes Project (1KG) dataset in PCA plots using PLINK2 (v2.00a2.3). Based on where participants were registered, we defined seven geographically-defined populations for the Japanese archipelago (i.e., Hokkaido, Tohoku, Kanto-Koshinetsu, Chubu-Hokuriku, Kinki, Kyushu, and Okinawa, which are ordered from northeast to southwest Japan) and eight populations for the Ryukyu Islands (i.e., Yakushima, Amami, Kikai, Okinoerabu, Tokunoshima, Yoron, Okinawa, and Miyako). The five subpopulations were identified from visual inspection of the PCA plots (i.e., Mainland, Ryukyu, Ryukyu_admix, EA_admix, and Hokkaido_sub).
BBJ GWAS data were genotyped using the Illumina HumanOmniExpressExome BeadChip or a combination of the Illumina HumanOmniExpress and HumanExome BeadChips. The quality control of the genotypes has been described elsewhere29,52. In brief, we excluded variants with the following criteria: (i) call rate <99%, (ii) P value for Hardy-Weinberg equilibrium (HWE) < 1.0 × 10−6, (iii) number of heterozygotes <5, and (iv) a concordance rate <99.5% or a non-reference concordance rate between GWAS array and whole genome sequencing. The genotype data were phased by Eagle v2 and imputed with WGS merged the 1000 Genomes Project Phase3v5 (n = 2504) and BBJ1K WGS (n = 1037) using Mimimac3 software (2.0.1).
Data merging between modern and ancient genomic data
We used ancient genomes from the Japanese archipelago and East Eurasian continent, which had been previously compiled and integrated with the Simons Genome Diversity Project (SGDP) panel (SGDP_Ancient)14. This SGDP_Ancinet dataset includes 14 ancient Japanese that were all shotgun sequenced. Under the stringent quality control (e.g., ancient DNA damage or low-coverage data), the ancient genomes (n = 22) were pseudo-diploid called for total 3,867,366 sites that were transversion only and a minor allele frequency of 1%14. To further merge ancient genome data with the BBJ genotype data, we converted the accurately imputed dosage data of BBJ (Rsq ≥ 0.7) to genotype data using PLINK2 with the following options: fill-missing-from-dosage and hard-call-threshold 0.499. Then, we extracted sites that are present in both SGDP_Ancient and BBJ genotype data using PLINK (v1.90b4.4), resulting in the final merged dataset containing a total of 2,038,260 sites.
Admixture modeling and f4 tests
We applied qpAdm in AdmixTools (version 7.0.2)30 to the final merged data. In line with a previous study14, our analysis only used transversion sites with global minor allele frequencies of 1%, coupled with the option of “allsnps: NO.” In qpAdm, we set the left and right populations as the source of admixture and reference populations, respectively. We then selected nine Eurasians as a right population; Sardinian (n = 3), Kusunda (n = 2), Papuan (n = 14), Dai (n = 4), Ami (n = 2), Naxi (n = 3)53, Tianyuan (n = 1)3,54, Chokhopani (n = 1)3,54, and Mal’ta (n = 1)55. As the left population, we set three ancient populations: Jomon (n = 12)8,9,14,48, Northeast Asian (n = 2; WLR_BA_o and HMMH_MN, Bronze Age and Middle Neolithic individuals from the West Liao River basin)56, and Han (n = 3; the SGDP panel, details in Supplementary Data 2)14.
We evaluated whether the tripartite structure provides a better fit to the data than any other possible scenarios at population levels based on p-values for nested models with a cut-off of 0.05. These alternative models include dual structure models involving any combination of Jomon, Northeast Asian, and East Asian ancestry, as well as single ancestry models. When conducting admixture modeling at individual levels, we considered the tripartite model to be a good fit if (i) a tail probability was >0.05 and/or (ii) estimates of admixture fractions were feasible (i.e., >0.0 and <1.0). The correlation of admixture proportions of the three distinct ancestors was calculated using Pearson’s method. For individuals who did not support the tripartite model due to the tail probability being below 5%, we sought to identify an alternative dual ancestry model with the highest tail probability, subsequently confirmed through nested p > 0.05 by comparing the tripartite and dual structure models. If a single ancestry model was plausible, we further assessed whether a specific ancestor alone can adequately explain the individual using the nested p-values.
The f4-statistics were measured in the form of f4(Mbuti, Jomon; Han, X) using qpDstat with the f4 mode in AdmixTools. We used the BBJ and the populations in the 1KG (n = 2504) or the SGDP as target populations (X in the form).
Curation of phenotypes in Biobank Japan
BBJ collected clinical conditions, laboratory data, and behavioral habit information for all participants through interviews and reviews of medical records using a standardized questionnaire. We selected 80 traits (3 anthropometrics, 55 biomarkers, 2 behavioral habits, 2 reproductive traits, and 18 diseases). We utilized the data for individuals over the age of 18 years, but only included the drinking and smoking traits for those over 20 years. For quantitative biomarkers, we applied the same processing and quality control methods as previously reported (Supplementary Data 4)19,29. In brief, we used the laboratory values measured during the participants’ first visit to the recruitment center and adjusted the values based on the type of medication used. We then applied a rank-based inverse normal transformation to normalize biomarker traits. Behavioral traits, including drinking and smoking history (ever versus never drinking and ever versus never smoking), were analyzed as binary phenotypes57,58. Reproductive traits were coded as age at menarche and age at menopause59. We excluded individuals if their age at menarche was below 10 or above 20 years, or if their age at menopause was below 40 or above 60 years. Patients with myocardial infarction, stable angina, and unstable angina were reclassified as having coronary artery disease (CAD)60. 18 diseases were selected from a group of target diseases in BBJ, with sufficient numbers of cases and controls (Arrhythmia, Asthma, Cataract, CAD, Dyslipidemia, Ischemic stroke, Congestive heart failure, Osteoporosis, Glaucoma, Chronic hepatitis C, Colorectal cancer, Gastric cancer, Pollinosis, Urolithiasis, Rheumatoid arthritis, Prostate cancer, Breast cancer, and Type 2 diabetes). Individuals who were not affected by a particular disease under study were treated as controls.
In addition, we set a dummy phenotype as a negative control. We simulated 10 phenotypes with predefined heritability (h2 = 0.5) from 10,000 causal variants randomly sampled from BBJ GWAS data using the GCTA GWAS simulation method61. The values were normalized by applying rank-based inverse normal transformation.
Phenotypic impact of Jomon ancestry in Japanese populations
Associations between Jomon ancestry and traits were tested using a glm() function implemented in R software (version 4.1.0). We applied a linear regression model to quantitative traits and a logistic regression model to diseases and behavioral habits, with adjustment for covariates. The proportion of Jomon ancestry was normalized using the rank-based inverse normal transformation method. As covariates, we included sex, age, age squared, top 20 PCs, 45 disease statuses, geographic regions, PCA clusters, and trait specific covariates listed in Supplementary Data 5. For BMI, we further tested the association with Jomon ancestry, stratified by sex and age groups, with the age threshold set at 65 years old, which reflects a mean age among the BBJ participants.
To address potential multicollinearity issues between Jomon ancestry and PCs, we employed an alternative approach. Initially, we conducted regression analysis of the quantitative trait measurements on all covariates, including PCs. Subsequently, we tested the associations between Jomon ancestry and the residuals derived from this regression using a linear regression model.
Assessment of genome-wide estimation and polygenic prediction with and without Jomon ancestry
We investigated the influence of the Jomon proportions on GWAS for BMI and the accuracy of BMI PGS. We performed BMI GWAS on all BBJ participants using a generalized MLM approach of GCTA-fastGWA62. We employed a sparse GRM constructed with variants subject to minimal LD pruning. Covariates in the original BMI GWAS included sex, age, age squared, the top 20 PCs, and 45 disease statuses. To evaluate the impact of Jomon ancestry, we introduced Jomon proportions as an additional covariate and compared the results with those obtained from the original GWAS.
To compute PGS, we adopted a five-fold leave-one-group-out GWAS method due to the absence of independent external reference for GWASs or genotype data with Japanese ancestry19,63. In brief, we initially split the BBJ individuals randomly into five subsets. We then performed BMI GWAS on samples, excluding the subset under investigation (i.e., target subset), using GCTA-fastGWA. To estimate the posterior effects of SNPs from GWAS summary data, we utilized PRS-CS-auto (version Jun 4, 2021) with the HapMap3 LD reference panel of 1KG EAS64,65. These posteriors of effect sizes were estimated both from the original GWAS and from the GWAS with the covariate of Jomon ancestry. Finally, we applied the PLINK2 score function to compute PGS for individuals within the target subset, comparing those scores with and without the Jomon ancestry covariate.
The incremental prediction performance of the Jomon ancestry was quantified with the difference in R2 as follows:
| 1 |
where R2PGS is the R2 of PGS modeled by trait ~PGS + covariates (sex, age, age squared, the top 20 PCs, 45 disease status, geographic regions, and PCA clusters), while R2PGS+Jomon is the R2 of the same PGS model but with the additional inclusion of the Jomon proportion.
Identification of Jomon-related variants
To detect genetic variants associated with Jomon ancestry, we conducted a genome-wide association analysis between the Jomon proportions and 6,861,976 biallelic variants (MAF ≥ 1% and Rsq ≥ 0.7). To strictly control the genomic inflation factor of test statistics, we adopted the following correction methods: (i) MLM-based approach using GCTA-fastGWA with the adjustment of covariates: age, age squared, sex, the top 20 PCs, 45 disease status, geographic regions, and PCA clusters; (ii) fixed-effect meta-analysis of Mainland summary data including individuals from the Mainland and EA_admix clusters (n = 151,075) and of Ryukyu summary data including individuals from the Ryukyu, Ryukyu admix, and Hokkaido_sub clusters (n = 10,080) using METAL (version 2020-05-05); and (iii) double genomic control correction method using METAL36. We then computed a Z score for each variant by considering the sign of the beta coefficient and the associated p-value.
To identify independent variants, we performed LD clumping on those variants with positive Z scores using BBJ1K and 1KG EAS as reference populations, with the following PLINK parameters: p1 = 1, p2 = 1, r2 = 0.01, kb=2000. Jomon-related variants (total 132 variants) were defined as those that are independent, not located in the HLA loci, and satisfy a genome-wide significance (P < 5.0 × 10−8). We calculated the FST value for the Jomon-related variants among the EAS populations in 1KG using Hudson’s FST implemented in PLINK266. Variants with p-values greater than 0.05 were classified as non-Jomon-related. We then identified 132 non-Jomon-related variants matching allele frequencies of the Jomon-related variants using a nearest neighbor matching method, contrasting the Jomon-related and non-Jomon-related variants by examining their frequency differences between the Japanese and continental East Asian populations, as well as their haplotypic lengths with LD greater than 0.8. The enrichment of selection signals in the Jomon-related variants was tested based on Z-scores of SDSs estimated in a previous study17. The sum of squared values of rank-based normalized Z-scores were compared to a chi-squared distribution with the degree of freedom equal to the number of available variants.
Stratified LD score regression was applied to the meta-analyzed summary data of the Jomon proportion, which estimated cell group enrichment with the recommended baseline LD model67. For LD score regression, we adopted the HapMap3 SNPs, excluding those within the HLA region, and used pre-computed LD scores among the 1KG EAS populations, which were obtained from the LDSC software website68. We investigated the pleiotropic effects of Jomon-related variants using BioBank Japan PheWeb for the Japanese population (https://pheweb.jp/) and Open Targets Genetics for Europeans (https://genetics.opentargets.org/). We also evaluated eQTL effects of the variants using GTEx (https://gtexportal.org/home/).
Validation with independent Japanese cohorts
As independent replication cohorts, we used the Nagahama cohort study and the second BBJ cohort (BBJ-2nd). The Nagahama cohort study is a community-based and recruited participants from Nagahama City, Shiga Prefecture, Japan42. The cohort was genotyped using six different genotyping arrays. We then selected two platforms (Nagahama A; Illumina Human610-Quad Beadchip, and Nagahama B; Illumina HumanOmni2.5-4v1 Beadchip) with a large number of samples. We excluded individuals with low call rates, high heterozygosity rates, closely related individuals, and PCA outliers from the EAS populations (n = 1591 in Nagahama A and n = 1444 in Nagahama B)69. We also excluded variants with (i) call rate <0.98, (ii) MAF < 1%, and (iii) HWE P < 1.0 × 10−6. Genotype data were phased by Eagle v2 and imputed with the reference panel from the 1000 Genomes Project Phase3v5 and BBJ1K using Mimimac3. As described above, we converted the high-quality imputed dosage data to genotype data and merged them with ancient genome data. We obtained 1,982,989 shared variants in Nagahama A and 2,109,225 shared variants in Nagahama B.
The BBJ-2nd is an additional cohort of independent participants from the first cohort of BBJ. The BBJ-2nd consisted of ~80,000 individuals collected between 2013 and 2018. The subjects were genotyped using an Illumina Asian Screening Array chip. As in the first cohort, we applied stringent QC filters to both participants and SNPs as described elsewhere70,71. Briefly, we excluded individuals with a low call rate (<0.98), closely related individuals (King’s kinship index ≥ 0.0884), and outliers from the EAS cluster in the PCA with the samples of the HapMap3 project (n = 72,695). We also excluded variants with call rate <0.99, minor allele count <5, HWE P < 1.0 × 10−10 and >0.05 of allele frequency difference when compared with the Japanese WGS reference panels. The genotype data was phased by SHAPEIT (version 4.2.1) and imputed with the reference panel from the 1000 Genomes Project Phase3v5 and BBJ1K using Mimimac4 (version 1.0.1). Converting the high-quality imputed dosage data (Rsq ≥ 0.7) to genotype data, we merged the ancient genome data to obtain 1,940,657 shared variants.
We estimated proportions of Jomon ancestry for individuals in each cohort by using qpAdm in the same way as the first cohort of BBJ. We derived a score for predicting Jomon ancestry based on 132 independent Jomon-related variants by using the PLINK2 score option, which is defined as Jomon component prediction score. After regressing the Jomon proportion out with 10 PCs as covariates, we estimated the variance explained by the scaled Jomon component prediction score using Pearson’s correlation. In each cohort, principal components were computed using the projection method on the PCs of the first cohort of BBJ. To evaluate the power of the Jomon component prediction score, we employed a clumping and thresholding approach. We built the Jomon component score for BBJ-2nd individuals using genetic variants meeting the following p-value thresholds: 5 × 10−8, 5 × 10−7, 1 × 10−6, 1 × 10−5, 1 × 10−4, 1 × 10−3, 0.01, 0.05, 0.1. To assess the proportion of variance explained by the Jomon component score, we calculated the adjusted R2 from a full model that included the score and all covariates, compared to a null model without the score.
Replication analysis using East Asians individuals in UK Biobank
As an independent source of an East Asian (EAS) population, we focused on EAS individuals from UK Biobank (UKB), which is a population-based cohort that recruited 500,000 individuals between 40 and 69 years old from across the United Kingdom72. EAS individuals were extracted by visual inspection on the PCA plots including 1KG populations as the reference for EAS ancestry (n = 2286; Supplementary Fig. 11). UKB was genotyped with either the Applied Biosystems UK BiLEVE Axiom Array or the Applied Biosystems UKB Axiom Array. The genotypes were imputed using the Haplotype Reference Consortium, UK10K, and the 1000 Genomes Phase 3 reference panel using IMPUTE472. The detailed characteristics of the cohort have been described elsewhere72. For UKB EAS individuals, we converted the variants with INFO score ≥0.8 to genotype data and obtained 3,610,183 sites shared with ancient genome data.
The genetic affinity between the Jomon and UKB EAS or between the Jomon and each of different ethnic background groups within UKB EAS was tested using f4-statistics with the form of f4(Mbuti, Jomon; Han, X). The breakdown of self-reported ethnic backgrounds at the participants’ initial visit (Data-Field 21000) is as follows: Data-coding 1 for White (n = 1), Data-coding 1001 for British (n = 1), Data-coding 2003 for White and Asian (n = 2), Data-coding 2004 for Any other mixed background (n = 3), Data-coding 3 for Asian or Asian British (n = 2), Data-coding 3004 for Any other Asian background (n = 300), Data-coding 5 for Chinese (n = 1375), and Data-coding 6 for Other EG (n = 541).
We estimated the Jomon proportion for individuals in each cohort by using qpAdm. In cases where individuals did not support the tripartite ancestry structure, we applied the same procedure used for BBJ to identify a plausible model with a two-way admixture, where Jomon served as one of the source ancestors. Jomon component prediction scores based on 132 independent Jomon-related variants were calculated using PLINK2.
To assess the association between Jomon ancestry and BMI in UKB EAS, we computed the mean BMI for participants with measurements taken two or three times. The association tests were adjusted for sex, age, age squared, ascertainment center information, batch information, and ethnic backgrounds. This approach was also applied to EG5 (i.e., self-reported Chinese, n = 1375) and 6 (i.e., self-reported Other ethnic group, n = 541) with the same covariates, except for ethnic backgrounds.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank all participants and investigators of the Biobank Japan Project, Nagahama cohort study, and UK Biobank. We would like to express our deepest gratitude to our co-author T.T., who passed away in July of 2024, for his invaluable contribution to BioBank Japan Project. K.Y. was supported by AMED (JP223fa627002) and Center for Advanced Modality and DDS (CAMaD), Osaka University. K. Suzuki., T.Y. and T.K. were supported AMED (JP20km0405202, JP23tm0424218). T.G was supported by SAKIGAKE Project in Kanazawa University, MEXT KAKENHI Grant Number (20H05822), JSPS KAKENHI Grant Number (21H04358). S.Nakagome. was supported by JSPS KANENHI (22H02711) and Wellcome Trust ISSF Award. Y.O. was supported by JSPS KAKENHI (22H00476), and AMED (JP23km0405211, JP23km0405217, JP23ek0109594, JP23ek0410113, JP23kk0305022, JP223fa627002, JP223fa627010, JP233fa627011, JP23zf0127008, JP23tm0524002), JST Moonshot R&D (JPMJMS2021, JPMJMS2024), Takeda Science Foundation, Bioinformatics Initiative of Osaka University Graduate School of Medicine, Institute for Open and Transdisciplinary Research Initiatives, Center for Infectious Disease Education and Research (CiDER), and Center for Advanced Modality and DDS (CAMaD), Osaka University.
Author contributions
K.Y., S.Nakagome., and Y.O. designed this study and wrote the manuscript. K.Y., S.Namba., K.Sonehara., S.S., and N.P.C. conducted data curation and bioinformatics analysis. K. Suzuki., S.H., S.K., H.A., K. Matsuura., Y.M., Y.F., T.T., K. Matsuda., T.G., T.Y., T.K., and S.Nakagome. collected the samples. S.Nakagome. and Y.O. supervised this study.
Peer review
Peer review information
Nature Communications thanks William Barrie and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The summary statistics from the genome-wide association analysis for Jomon ancestry have been deposited in the National Bioscience Database Center (NBDC) Human Database (https://humandbs.dbcls.jp/en/) under accession code hum0197 (https://humandbs.dbcls.jp/en/hum0197-latest). The genotype data of BBJ are available from the NBDC Human Database (research ID: hum0014 and hum0311). The genotype data of the Nagahama cohort study are available from the NBDC Human Database (research ID: hum0012). UKB analysis was conducted using application number 47821 (https://www.ukbiobank.ac.uk/). All ancient genomic data used in this study were previously compiled14.
Code availability
We used publicly available software for analysis. The software used is described in the “Methods” section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Shigeki Nakagome, Email: nakagoms@tcd.ie.
Yukinori Okada, Email: yokada@sg.med.osaka-u.ac.jp.
the Biobank Japan Project:
Shinichi Higashiue, Shuzo Kobayashi, Yuji Yamanashi, Yoichi Furukawa, Takayuki Morisaki, Yoshinori Murakami, Yoichiro Kamatani, Kaori Muto, Akiko Nagai, Yusuke Nakamura, Wataru Obara, Ken Yamaji, Kazuhisa Takahashi, Satoshi Asai, Yasuo Takahashi, Hiroki Yamaguchi, Yasunobu Nagata, Satoshi Wakita, Chikako Nito, Yu-ki Iwasaki, Shigeo Murayama, Kozo Yoshimori, Yoshio Miki, Daisuke Obata, Masahiko Higashiyama, Akihide Masumoto, Yoshinobu Koga, Yukihiro Koretsune, Koichi Matsuda, and Yukinori Okada
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54052-0.
References
- 1.Bergström, A., Stringer, C., Hajdinjak, M., Scerri, E. M. L. & Skoglund, P. Origins of modern human ancestry. Nature590, 229–237 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang, M. A. et al. 40,000-year-old individual from Asia provides insight into early population structure in Eurasia. Curr. Biol.27, 3202–3208.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazaridis, I. et al. Genomic insights into the origin of farming in the ancient Near East. Nature536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, C.-C. et al. Genomic insights into the formation of human populations in East Asia. Nature591, 413–419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakazawa, Y. On the Pleistocene population history in the Japanese Archipelago. Curr. Anthr.58, S539–S552 (2017). [Google Scholar]
- 7.Mizuno, F. et al. Population dynamics in the Japanese Archipelago since the Pleistocene revealed by the complete mitochondrial genome sequences. Sci. Rep.11, 12018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gakuhari, T. et al. Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations. Commun. Biol.3, 437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanzawa-Kiriyama, H. et al. Late Jomon male and female genome sequences from the Funadomari site in Hokkaido, Japan. Anthropol. Sci.127, 83–108 (2019). [Google Scholar]
- 10.Habu, J. Ancient Jomon of Japan. (Cambridge University Press, 2004).
- 11.Mizoguchi, K. The Archaeology of Japan: From the Earliest Rice Farming Villages to the Rise of the State. (Cambridge University Press, 2013). 10.1017/cbo9781139034265.
- 12.Hanihara, K. Dual structure model for the population history of the Japanese. Jpn. rev.2, 1–33 (1991). [Google Scholar]
- 13.Hudson, M. J., Nakagome, S. & Whitman, J. B. The evolving Japanese: the dual structure hypothesis at 30. Evol. Hum. Sci.2, e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooke, N. P. et al. Ancient genomics reveals tripartite origins of Japanese populations. Sci. Adv.7, eabh2419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooke, N. P. et al. Genomic insights into a tripartite ancestry in the Southern Ryukyu Islands. Evol. Hum. Sci.5, e23 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbeets, M. et al. Triangulation supports agricultural spread of the Transeurasian languages. Nature599, 616–621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada, Y. et al. Deep whole-genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat. Commun.9, 1631–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakaue, S. et al. Dimensionality reduction reveals fine-scale structure in the Japanese population with consequences for polygenic risk prediction. Nat. Commun.11, 1569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto, K. et al. Genetic footprints of assortative mating in the Japanese population. Nat. Hum. Behav.7, 65–73 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skov, L. et al. The nature of Neanderthal introgression revealed by 27,566 Icelandic genomes. Nature582, 78–83 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Sankararaman, S. et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature507, 354–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dannemann, M. & Kelso, J. The contribution of Neanderthals to phenotypic variation in modern humans. Am. J. Hum. Genet.101, 578–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, L., Wolf, A. B., Fu, W., Li, L. & Akey, J. M. Identifying and interpreting apparent Neanderthal ancestry in African individuals. Cell180, 677–687.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Zeberg, H. & Pääbo, S. A genomic region associated with protection against severe COVID-19 is inherited from Neandertals. Proc. Natl Acad. Sci. USA118, e2026309118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeberg, H. & Pääbo, S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature587, 610–612 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Barrie, W. et al. Elevated genetic risk for multiple sclerosis emerged in steppe pastoralist populations. Nature625, 321–328 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marnetto, D. et al. Ancestral genomic contributions to complex traits in contemporary Europeans. Curr. Biol.32, 1412–1419.e3 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Nagai, A. et al. Overview of the BioBank Japan Project: study design and profile. J. Epidemiol.27, S2–S8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet.53, 1415–1424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patterson, N. et al. Ancient admixture in human history. Genetics192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaifu, Y., Sakaue, K. & Kono, R. T. Early Jomon and Yayoi human skeletal remains from Shimomotoyama Rock Shelter, Sasebo, Nagasaki prefecture, Japan. Anthropol. Sci.125, 25–38 (2017). [Google Scholar]
- 33.Matsushita, Y. et al. Overweight and obesity trends among Japanese adults: a 10-year follow-up of the JPHC Study. Int. J. Obes.32, 1861–1867 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Berg, J. J. et al. Reduced signal for polygenic adaptation of height in UK Biobank. eLife8, e39725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoya, M., Shimizu, H. & Higuchi, Y. Geographical distribution of adolescent body height with respect to effective day length in Japan: an ecological analysis. PLoS ONE7, e50994 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooke, N. P. et al. Genomic imputation of ancient Asian populations contrasts local adaptation in pre- and post-agricultural Japan. iScience27, 110050 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Field, Y. et al. Detection of human adaptation during the past 2000 years. Science354, 760–764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mountjoy, E. et al. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat. Genet.53, 1527–1533 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghoussaini, M. et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res49, D1311–D1320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imaizumi, A. et al. Genetic basis for plasma amino acid concentrations based on absolute quantification: a genome-wide association study in the Japanese population. Eur. J. Hum. Genet27, 621–630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi, S. W., Mak, T. S.-H. & O’Reilly, P. F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc.15, 2759–2772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamagiwa, K. Early human cultural and communal diversity in the Ryukyu islands. Okinawan J. Isl. Stud.3, 3–15 (2022). [Google Scholar]
- 45.Mizoguchi, K. Nodes and edges: a network approach to hierarchisation and state formation in Japan. J. Anthr. Archaeol.28, 14–26 (2009). [Google Scholar]
- 46.Gong, S., Wang, K., Li, Y., Zhou, Z. & Alamian, A. Ethnic group differences in obesity in Asian Americans in California, 2013–2014. BMC Public Heal21, 1589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pontzer, H., Wood, B. M. & Raichlen, D. A. Hunter‐gatherers as models in public health. Obes. Rev.19, 24–35 (2018). [DOI] [PubMed] [Google Scholar]
- 48.McColl, H. et al. The prehistoric peopling of Southeast Asia. Science361, 88–92 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Orlando, L. et al. Ancient DNA analysis. Nat. Rev. Methods Prim.1, 14 (2021). [Google Scholar]
- 50.Sousa da Mota, B. et al. Imputation of ancient human genomes. Nat. Commun.14, 3660 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cassidy, L. M. et al. A dynastic elite in monumental Neolithic society. Nature582, 384–388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akiyama, M. et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat. Commun.10, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mallick, S. et al. The Simons genome diversity project: 300 genomes from 142 diverse populations. Nature538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong, C. et al. Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe. Proc. Natl Acad. Sci. USA115, E11248–E11255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raghavan, M. et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature505, 87–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ning, C. et al. Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun.11, 2700 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matoba, N. et al. GWAS of smoking behaviour in 165,436 Japanese people reveals seven new loci and shared genetic architecture. Nat. Hum. Behav.3, 471–477 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Matoba, N. et al. GWAS of 165,084 Japanese individuals identified nine loci associated with dietary habits. Nat. Hum. Behav.4, 308–316 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Horikoshi, M. et al. Elucidating the genetic architecture of reproductive ageing in the Japanese population. Nat. Commun.9, 1977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishigaki, K. et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet.52, 669–679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet.88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang, L. et al. A resource-efficient tool for mixed model association analysis of large-scale data. Nat. Genet.51, 1749–1755 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Sakaue, S. et al. Trans-biobank analysis with 676,000 individuals elucidates the association of polygenic risk scores of complex traits with human lifespan. Nat. Med.26, 542–548 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun.10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, Y. et al. Global Biobank analyses provide lessons for developing polygenic risk scores across diverse cohorts. Cell Genom.3, 100241 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhatia, G., Patterson, N., Sankararaman, S. & Price, A. L. Estimating and interpreting FST: the impact of rare variants. Genome Res.23, 1514–1521 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet.47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet.47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yasumizu, Y. et al. Genome-wide natural selection signatures are linked to genetic risk of modern phenotypes in the Japanese population. Mol. Biol. Evol.37, 1306–1316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akiyama, Y. et al. Genome-wide association study identifies risk loci within the major histocompatibility complex region for Hunner-type interstitial cystitis. Cell Rep. Med.4, 101114 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Naito, T. et al. Genetic risk of primary aldosteronism and its contribution to hypertension: a cross-ancestry meta-analysis of genome-wide association studies. Circulation147, 1097–1109 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The summary statistics from the genome-wide association analysis for Jomon ancestry have been deposited in the National Bioscience Database Center (NBDC) Human Database (https://humandbs.dbcls.jp/en/) under accession code hum0197 (https://humandbs.dbcls.jp/en/hum0197-latest). The genotype data of BBJ are available from the NBDC Human Database (research ID: hum0014 and hum0311). The genotype data of the Nagahama cohort study are available from the NBDC Human Database (research ID: hum0012). UKB analysis was conducted using application number 47821 (https://www.ukbiobank.ac.uk/). All ancient genomic data used in this study were previously compiled14.
We used publicly available software for analysis. The software used is described in the “Methods” section.