Abstract
Purpose: This study aimed to explore the effectiveness of preoperative computed tomography (CT) radiomic models combined with clinical and CT imaging features for predicting spread through air spaces (STAS) in patients with T1 lung adenocarcinoma. Methods: The preoperative CT and clinical data of 219 patients with T1 invasive lung adenocarcinoma confirmed by surgery were retrospectively analyzed and randomly divided into training and test sets at a ratio of 7:3. Univariable and multivariable logistic analyses were performed on the clinical and CT manifestations to screen independent predictive factors for STAS (+), and a clinical model was constructed. Radiomic features were extracted from the tumor (T), peritumoral (P) and tumor-peritumoral (TP) regions to construct radiomic models (Model T, Model P and Model TP), and the optimal radiomic model was identified. A combined model was then built on the basis of the best radiomic score (Radscore) and clinically independent predictors. For each model, the effectiveness in predicting STAS (+) was assessed with receiver operating characteristic (ROC) curve analysis, including calculation of the area under the curve (AUC), and a nomogram was created. Calibration curve analysis was used to assess model calibration, and decision curve analysis (DCA) was used to evaluate the clinical value of the model. Results: Emphysema, the preoperative carcinoembryonic antigen (CEA) level, and the consolidation tumor ratio (CTR) were identified as independent predictors of STAS (+) (all P < 0.01). Model T was considered the optimal radiomic model. In the training set, the AUC of the combined model was greater than that of the clinical model (0.93 vs. 0.85, P < 0.01). However, no significant difference in the AUC was found between the combined model and Model T (0.93 vs. 0.92, P > 0.05). In the test set, the AUC of the combined model was greater than that of the clinical model (0.92 vs. 0.85, P < 0.05), but there was no significant difference compared to the AUC of Model T (0.92 vs. 0.90, P = 0.13). The AUC of Model T was greater than that of the clinical model in the training set (0.92 vs. 0.85, P < 0.01), but this difference was not significant in the test set (0.90 vs. 0.85, P = 0.35). The clinical model, radiomic Model T, and combined model all had high degrees of calibration. Finally, the clinical net benefit of the combined model was greater than that of the other two models with the threshold ranged from 0.10 to 0.40. Conclusion: The preoperative CT radiomics model combined with clinical and CT imaging features can effectively predict STAS in T1 invasive lung adenocarcinoma patients.
Keywords: Invasive lung adenocarcinoma, spread through air spaces, radiomic model
Introduction
The prognosis of lung adenocarcinoma, one of the most common types of non-small cell lung cancer [1], is closely related to early and accurate tumor detection and prediction. Especially in the early stage of lung adenocarcinoma, that is, stage T1, when the tumor size is small (≤ 3 cm) and has not invaded beyond the lung lobes, accurate prediction of tumor behaviors - especially the risk of spread through air spaces (STAS) - is crucial for guiding clinical treatment and assessing disease prognosis [2]. STAS refers to the spread of tumor cells along the airways, a phenomenon that has been proven to be an independent risk factor for postoperative recurrence and poor outcomes in patients with lung adenocarcinoma [3,4].
Although traditional clinical and imaging features play a role in predicting survival outcomes in patients with lung adenocarcinoma, these features often fail to fully reveal the biological behavior of the tumor. Previous research has indicated that the presence of STAS can be predicted through the evaluation of computed tomography (CT) scans [5]. This phenomenon has been linked with certain CT characteristics, such as solid nodules, centrally located low attenuation areas, poorly defined opacities, air bronchograms, and a high consolidation-to-tumor ratio. Nonetheless, these qualitative CT characteristics are subject to individual interpretation bias, which could result in misclassification errors. With the development of radiomics, computers are able to extract histological features from CT images, efficiently quantify image information, reduce the impact of subjective judgments, and enhance predictive accuracy [6-8]. By extracting high-dimensional quantitative features from conventional medical images, radiomics can reveal the microscopic heterogeneity of tumors and their environment, thereby providing more comprehensive biological information on tumor behavior [9-11]. Some studies have shown that radiomics can quantify tumor characteristics and predict STAS in patients with lung cancer. Jiang et al. [12] were the first to apply a radiomics-based random forest (RF) model based on CT images to predict STAS in lung adenocarcinoma, achieving an area under the curve (AUC) of 0.754 (with a sensitivity of 0.880 and specificity of 0.588). Chen et al. [13] developed a CT-based radiomic naïve Bayes model that also performed well in the presurgical prediction of STAS in stage I lung adenocarcinoma patients (external validation AUC = 0.69). Han et al. [14] applied the same radiomic approach to presurgical stage IA lung adenocarcinoma (LUAD) patients, achieving AUC values of 0.812 and 0.850 in the training and test sets, respectively, with logistic regression. These studies relied primarily on various radiomic features, including shape, intensity, and texture, which were then fed into the corresponding machine learning algorithms to predict STAS. However, these methods also focused only on regions within the tumor, which overlooks subtle changes in the peritumoral microenvironment. Conversely, our study considers the potential impact of the peritumoral area. Therefore, we performed radiomic analysis on tumor, peritumoral microenvironment, and the combined tumor and peritumoral microenvironment areas and then integrated clinical factors and CT characteristics to construct a joint prediction model. Through this method, we expect our model to provide patients with more precise treatment selection and prognosis assessment, particularly by playing a key role in the treatment decision-making process for early-stage lung adenocarcinoma.
Materials and methods
General information
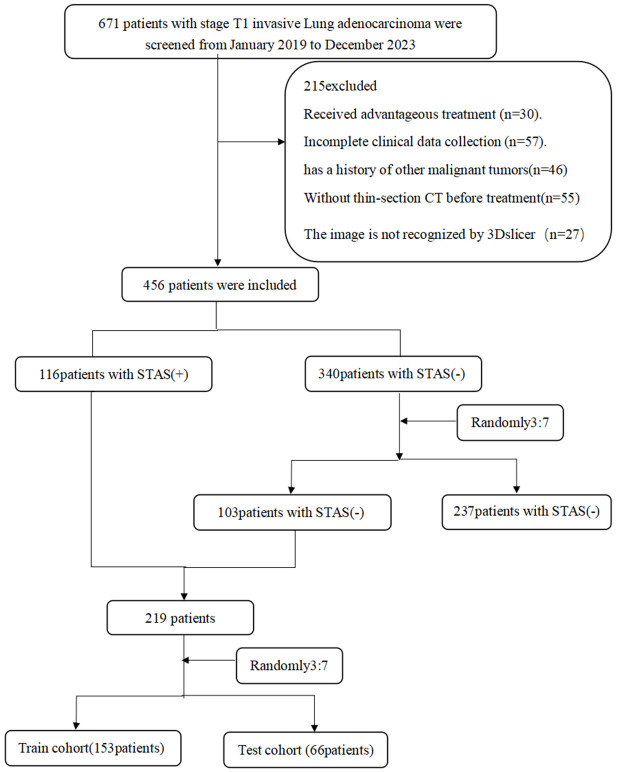
This study was conducted according to the tenets of the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of Huzhou First People’s Hospital, which waived the need for informed patient consent. A retrospective analysis was conducted on 219 patients with solitary or part-solitary T1-stage invasive lung adenocarcinoma diagnosed by postoperative pathology from January 2019 to October 2023. The cohort included 113 males and 106 females aged 29 to 83 years, with an average age of 64.9±7.1 years. Among them, 116 patients were STAS (+), and 103 patients were STAS (-). The patients were divided into training and test sets at a 7:3 ratio; ultimately, 154 patients were included in the training set (82 STAS (+) and 72 STAS (-)), and 65 patients were included in the test set (34 STAS (+) and 31 STAS (-)). The data in the training set and the test set were not significantly different (Table 1).
Table 1.
The clinical and radiological features of patients in the training set and the test set
| Characteristics | Test set (n = 65) | Training set (n = 154) | P value |
|---|---|---|---|
| Gender | 0.991 | ||
| Male n (%) | 33 (50.8) | 80 (51.9) | |
| Female n (%) | 32 (49.2) | 74 (48.1) | |
| Age | 64.9±9.58 | 64.5±9.15 | 0.362 |
| Lymph code | 0.11±0.31 | 0.12±0.32 | 0.576 |
| Emphysema | 0.34±0.47 | 0.36±0.48 | 0.217 |
| Smoke | 0.24±0.43 | 0.28±0.45 | 0.054 |
| CEA | 0.21±0.41 | 0.18±0.39 | 0.116 |
| CA125 | 0.05±0.22 | 0.05±0.22 | 0.858 |
| Diameter | 1.68±0.64 | 1.67±0.65 | 0.671 |
| Location | 2.84±1.49 | 2.89±1.53 | 0.409 |
| Lobulation sign | 0.69±0.47 | 0.66±0.48 | 0.106 |
| Bubble sign | 0.40±0.49 | 0.41±0.49 | 0.943 |
| Satellite lesion | 0.04±0.19 | 0.04±0.19 | 0.769 |
| Vascular convergence | 0.57±0.50 | 0.56±0.50 | 0.789 |
| Spiculation | 0.47±0.50 | 0.45±0.50 | 0.223 |
| Air bronchogram | 0.28±0.45 | 0.28±0.45 | 0.973 |
| Pleural indentation | 0.39±0.49 | 0.39±0.49 | 0.886 |
| CTR | 1.84±0.60 | 1.85±0.63 | 0.694 |
Inclusion criteria: (i) Maximum tumor diameter no greater than 3 cm on preoperative CT imaging, according to the 8th edition of the lung cancer TNM staging guidelines; (ii) CT imaging data obtained within one month before surgery; (iii) Pathological diagnosis of invasive lung adenocarcinoma.
Exclusion criteria: (i) Neoadjuvant therapy; (ii) Multiple pulmonary nodules on preoperative CT images; (iii) Past or current history of other malignant tumors; (iv) CT images that could not be fully read by 3D Slicer software; and (v) Distant metastasis (Figure 1).
Figure 1.
Flowchart of the patient selection procedure.
Clinical data: The clinical information of the patients was collected from the electronic medical records system of our hospital and included age, sex, smoking history, and CEA and CA125 levels.
Instruments and methods
Chest scans were performed using a German Siemens Definition AS 64-slice 128-slice spiral CT. The scanning range extended from the thoracic inlet to the diaphragm. The subject was placed in a supine position, then instructed to inhale deeply and hold their breath. The scanning parameters were as follows: tube voltage of 120 kV, tube current of 120 mA, window width of 1300-1500, window level of -600 to -700, pitch of 1.0, and rack rotation time of 0.33 seconds per 360 degrees. The lung window was reconstructed with the lung method, with a reconstruction thickness and slice spacing of 1.25 mm. The mediastinal window reconstruction thickness and slice spacing were 5 mm.
Image analysis
Traditional imaging features were independently evaluated by two experienced chest radiologists (with 5 and 10 years of experience in lung nodule diagnosis) who were blinded to the pathological and clinical information of the patients. When the two radiologists had inconsistent interpretations of the imaging features, a consensus was reached through discussion.
The evaluated imaging features included lesion location, composition (solid or part-solid), maximum diameter on three-dimensional imaging, margin (clear/blurred), lobulation (present/absent), spiculation (present/absent), cavitation (present/absent), the air bronchogram sign (present/absent), vessel convergence (present/absent), pleural retraction (present/absent), and peritumoral ground-glass opacities (present/absent). Lobulation was defined as the presence of at least 3 arc-shaped protrusions greater than 2 mm in height alternating with indentations, resulting in a lobed appearance, which may be related to uneven peripheral tumor growth and differentiation. The maximum diameter was measured on coronal and sagittal views after multiplanar reconstruction (MPR) from three orthogonal directions. Spiculation was defined as the presence of small, approximately 2 mm spike-like projections or linear opacities, in a fine linear or dense brush-like pattern, extending from the margin of the mass into the lung parenchyma, which may represent thickened interlobular septa containing coarse lymphatics or tumor-associated fibrotic scarring. Cavitation was defined as the presence of an area of extremely low density (< 5 mm) within the lesion. The air bronchogram sign was defined as visualization of an air-filled bronchus within the lesion. Vessel convergence was defined as the convergence of vessels toward the lesion or the direct continuation of vessels into the lesion. Pleural retraction was defined as the presence of single or multiple linear bands of varying thickness connecting the lesion to the pleura, with or without pleural puckering or indentation. The peritumoral ground-glass opacity sign was defined as the presence of a hazy, indistinct high-density area surrounding the lung tumor, with a density between that of the normal lung parenchyma and the solid tumor. The images were processed on a GE AW4.6 workstation equipped with Thoracic VCAR software, which facilitates a detailed analysis of the lung parenchyma. The threshold limits were carefully set, with an upper limit of 3072 HU and a lower limit of -1024 HU. The emphysema threshold was specifically set to -950 HU to accurately assess the presence and extent of emphysematous changes. The system automatically generates the lung air volume (LAV%) for the entire lungs and for specific segments, including the upper, middle, and lower lobes of the right lung and the upper and lower lobes of the left lung. This meticulous approach to image collection, feature assessment, and quantitative analysis ensures robustness and reproducibility in our radiological evaluations.
Pathological examination
On the basis of the 2015 World Health Organization classification of lung cancer and the study by Kadota et al., we developed the following criteria for defining STAS. Hematoxylin and eosin (H&E) staining was performed, and tumor cells were observed in airway spaces without substantial stromal invasion. Two pathologists simultaneously reassessed the included patients’ slides following H&E staining with a multiheaded microscope. In cases of disagreement or inconsistency, a consensus was reached through discussion. STAS was defined as the presence of tumor cells within the normal alveolar spaces adjacent to the main tumor, which appeared as microvascular clusters, small solid nests, or single cells. To avoid confusion with cells artificially displaced during tumor handling, at least three tumor sections were examined under the microscope, and the margins of the tumor were first identified at low magnification in each section. According to the presence or absence of STAS, the study population was divided into two groups: patients with STAS and patients without STAS.
Tumor image segmentation and feature extraction
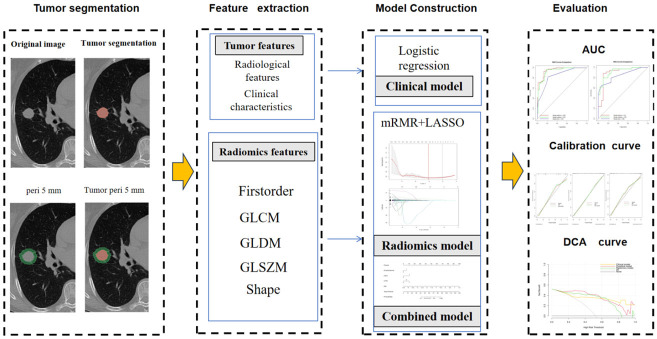
Tumor segmentation was manually performed with 3D Slicer (version 5.7.0, https://www.slicer.org) by a radiologist with 5 years of clinical experience who was blinded to the patients’ clinical and pathological information. The segmentation involved initially delineating the tumor ROI (ROIT) on the lung window images that contained the largest cross-sectional area of the tumor. The software then automatically expanded this boundary by 5 mm to obtain the tumor-peritumoral region ROI (ROITP); the area of the peritumoral region (ROIP) was then calculated by subtracting the area of ROIT from that of ROITP. The chest wall soft tissue, bones, and mediastinal overlaps within the 5 mm periphery of the tumor were then manually excluded. Image preprocessing included resampling all images to a voxel size of 1×1×1 mm to standardize the voxel spacing. Data normalization was achieved through z score standardization (zero-mean normalization).
Feature extraction was performed with the radiomics extension in 3D Slicer for each lesion, yielding a total of 899 features. These included first-order statistics, shape, gray level dependence matrix (GLDM), gray level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), gray level ize zone matrix (GLSZM), neighboring gray tone difference matrix (NGTDM), and wavelet-based features. These features allow the quantitative analysis of various dimensions of the tumor for characterizing its properties. To validate the extracted features, a second radiologist with 10 years of diagnostic imaging experience randomly selected the imaging data of 30 patients for ROI delineation and radiomic feature extraction.
Feature selection and model development
After the features were extracted from both the ROIT and ROIP with the above feature extraction methods, a feature fusion approach was employed to obtain the ROITP feature set. These features were then randomly divided into training and test sets at a 7:3 ratio. Prior to feature screening, the features of the training set were normalized and scaled to the same order of magnitude. Thereafter, feature screening was performed on the training set, which consisted of the following three steps:
(1) Mann-Whitney U test and feature screening: All radiomic features were subjected to the Mann-Whitney U test and feature screening. Only radiomic features with a p value less than 0.05 were retained.
(2) Spearman correlation analysis: For features with high repeatability, the Spearman rank correlation coefficient was calculated to assess the correlation between them. If the correlation coefficient between any two features was greater than 0.9, only one of the features was retained.
Greedy recursive deletion: To maximize the ability to describe the features, a greedy recursive deletion strategy was adopted, whereby the most redundant feature in the current set was deleted at each step.
(3) Least absolute shrinkage and selection operator (LASSO) regression: A LASSO model was then used to construct feature signatures from the exploratory dataset. On the basis of the regularization parameter λ, LASSO contracts all regression coefficients toward zero and sets the coefficients for many unrelated features exactly to zero. The optimal λ was determined via 10-fold cross-validation, identifying the value of λ that produced the smallest cross-validation error.
The features that retained nonzero coefficients after LASSO were used for regression model fitting and combined into an “imaging omics signature”. A logistic regression classifier was then applied to build radiomic models (Model T, Model TP, and Model P), calculate the radiomic score (Radscore), and select the best radiomic model.
Statistical analysis
Statistical analysis was performed with SPSS 25.0 software and R software (version 4.4.0).
Continuous variables are expressed as the means ± standard deviation (SD) and ranges, whereas qualitative variables are expressed as raw numbers, proportions and percentages. The normality of the continuous variables was tested with the Kolmogorov-Smirnov test. Differences between normally distributed continuous variables were analyzed with the t test, whereas those between nonnormally distributed continuous variables were analyzed with the Mann-Whitney U test. Qualitative variables were compared with the chi-square test. Univariable and multivariable logistic regression analyses were performed on the clinicopathological and CT features. The Spearman correlation coefficient (r) was subsequently used to represent correlations between each pair of features. Intraobserver and interobserver consistency were evaluated with the intraclass correlation coefficients (ICCs) via the absolute agreement method. ICCs > 0.75 indicated that there was good consistency between the two readers. Receiver operating characteristic (ROC) curve analysis was used to evaluate the prediction performance of each model. The DeLong test was applied to evaluate significant differences between the AUC values of the ROC curves. Calibration curve analysis was used to evaluate the degree of calibration of each model, and the clinical value of the models was evaluated with decision curve analysis (DCA). Statistical significance was established for a two-tailed P value of less than 0.05.
Results
Construction of the clinical model
Emphysema, the preoperative CEA level, and the consolidation-to-tumor ratio (CTR) were identified as independent clinical predictors of STAS (+) (all P < 0.01, Table 2). These variables were used to construct the clinical model.
Table 2.
Logistic regression analysis identified independent clinical and CT predictors for spread through air spaces (STAS) (+)
| Characteristics | Univariable logistic regression | Multivariable logistic regression | ||
|---|---|---|---|---|
|
|
|
|||
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Gender | 2.891 (1.678, 5.046) | < 0.001 | ||
| Age | 1.025 (0.997, 1.056) | 0.079 | ||
| Lymph code | 11.585 (3.290, 73.564) | 0.001 | ||
| Emphysema | 4.887 (2.636, 9.437) | < 0.001 | 4.729 (1.888, 12.667) | 0.001 |
| Smoke | 4.202 (2.115, 8.889) | < 0.001 | ||
| CEA | 1.071 (4.396, 32.188) | < 0.001 | 8.992 (2.707, 36.491) | < 0.001 |
| CA125 | 1.069 (0.312, 3.812) | 0.914 | ||
| Diameter | 4.417 (2.673, 7.646) | < 0.001 | ||
| Location | 1.173 (0.980, 1.408) | 0.083 | ||
| Lobulation sign | 3.004 (1.681, 5.483) | < 0.001 | ||
| Bubble sign | 0.805 (0.467, 1.385) | 0.435 | ||
| Satellite lesion | 1.501 (0.359, 7.469) | 0.584 | ||
| Vascular convergence | 0.984 (0.575, 1.683) | 0.954 | ||
| Spiculation | 2.852 (1.653, 4.987) | < 0.001 | ||
| Air bronchogram | 1.404 (0.775, 2.573) | 0.266 | ||
| Pleural indentation | 1.932 (1.115, 3.387) | 0.020 | ||
| CTR | 0.095 (0.041, 0.188) | < 0.001 | 0.185 (0.069, 0.449) | < 0.001 |
Construction of the radiomic models
Following assessment with the ICC, 899 features were retained from the ROIT, ROITP, and ROIP, respectively. Using the minimum redundancy maximum relevance (mRMR) method and LASSO, the optimal radiomic features were selected, resulting in the use of 8, 3, and 3 features for constructing Models T, TP, and P, respectively (Figure 2). Model T predicted STAS (+) in the training and test sets with AUCs of 0.91 and 0.90, respectively, which were slightly higher than those of Model TP (0.90 and 0.86) and Model P (0.86 and 0.88). However, the differences were not statistically significant (all P > 0.05). Model T was ultimately chosen as the optimal radiomic model (Table 3).
Figure 2.
Overall design flowchart of this study.
Table 3.
The efficacy of a radiomics model in predicting STAS (+)
| Model | AUC (95% CI) | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|
| Training set | ||||
| T | 0.912 (0.866, 0.958) | 0.800 | 0.913 | 0.892 |
| TP | 0.900 (0.850, 0.949) | 0.841 | 0.917 | 0.844 |
| P | 0.855 (0.832, 0.937) | 0.854 | 0.791 | 0.824 |
| Test set | ||||
| T | 0.898 (0.808, 0.987) | 0.935 | 0.882 | 0.892 |
| TP | 0.863 (0.759, 0.968) | 0.941 | 0.774 | 0.831 |
| P | 0.878 (0.791, 0.964) | 0.735 | 0.903 | 0.800 |
Construction of the combined model
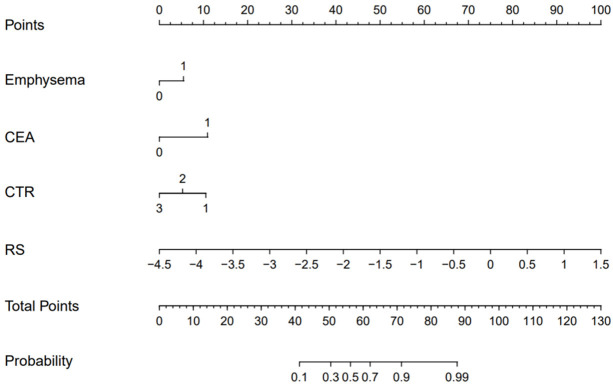
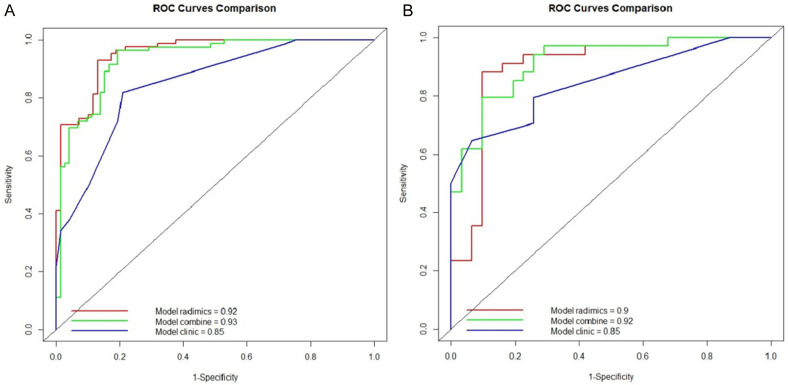
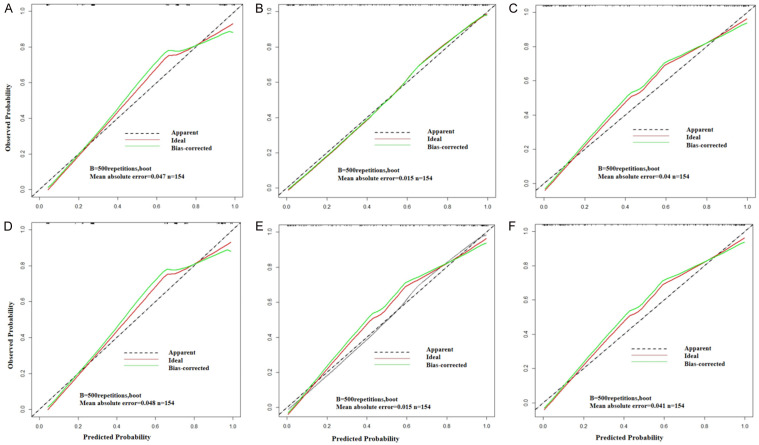
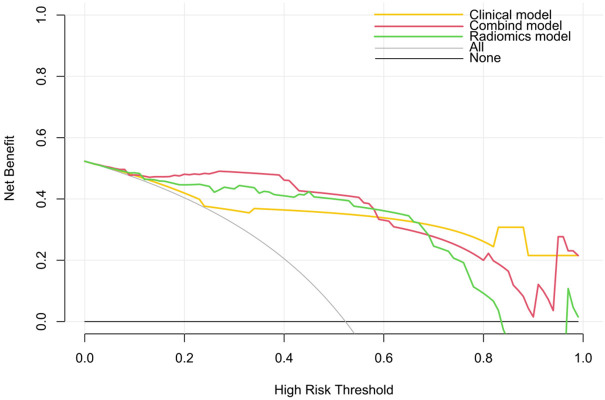
A combined model was constructed using the Radscore from Model T and the independent clinical predictors. Figure 3 presents the nomogram derived for this combined model. The AUC in predicting STAS (+) in the training set was greater for the combined model than that for the clinical model (0.93 vs. 0.85, Z = -3.99, P < 0.001), but the difference between the combined model and Model T was not significant (0.93 vs. 0.91, Z = -1.32, P = 0.19). In the test set, the AUC for the combined model was greater than that for the clinical model (0.93 vs. 0.86, Z = -0.93, P = 0.35) and Model T (0.93 vs. 0.90, Z = -1.51, P = 0.13), but the differences were not statistically significant. The differences in the AUCs between the clinical model and Model T were statistically significant in the training set (Z = -2.81, P < 0.01) but not in the test set (Z = -0.36, P > 0.05) (Figure 4 and Table 4). Calibration curve analysis indicated that the clinical model, Model T, and the combined model all exhibited high calibration (Figure 5). DCA demonstrated that, over a threshold range from 0.10 to 0.40, the combined model offered better clinical net benefits than the clinical and radiomic models, as illustrated in Figure 6.
Figure 3.
Nomogram for the combined model.
Figure 4.
Receiver operating characteristic (ROC) curves of the clinical model, Model T, and the combined model for predicting spread through air spaces (STAS) in the (A) training set and (B) testing set.
Table 4.
The efficacy of a various model in predicting STAS (+)
| Model | AUC (95% CI) | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|
| Training set | ||||
| T | 0.912 (0.866, 0.958) | 0.800 | 0.913 | 0.892 |
| Clinical | 0.854 (0.799, 0.910) | 0.812 | 0.768 | 0.792 |
| Combined | 0.927 (0.886, 0.967) | 0.847 | 0.884 | 0.864 |
| Test set | ||||
| T | 0.898 (0.808, 0.987) | 0.935 | 0.882 | 0.892 |
| Clinical | 0.858 (0.772, 0.944) | 0.806 | 0.794 | 0.800 |
| Combined | 0.930 (0.861, 0.999) | 0.935 | 0.853 | 0.892 |
Figure 5.
Calibration curves for the clinical model, Model T, and the combined model in the training and testing sets. A: Clinical model calibration curve in the training set; B: Model T calibration curve in the training set; C: Combined model calibration curve in the training set; D: Clinical model calibration curve in the testing set; E: Model T calibration curve in the testing set; F: Combined model calibration curve in the testing set.
Figure 6.
Decision curve analysis (DCA) plot depicting the clinical net benefits of each model.
Discussion
STAS significantly impacts the recurrence-free survival (RFS) and overall survival (OS) of lung cancer patients [15-17]. Precisely identifying the presence of this invasive characteristic before surgery can aid in choosing the most suitable surgical approach, ultimately improving patient outcomes and extending survival [18,19]. This study investigated the efficacy of preoperative CT-based radiomic models combined with clinical and CT imaging features in predicting the risk of STAS in patients with T1-stage invasive lung adenocarcinoma. The results demonstrated that the combined model, incorporating both radiomic features and traditional clinical features, had greater accuracy and clinical value in predicting STAS than the individual model components. Moreover, the developed nomogram converted the model’s complex regression equations into an easily interpretable visual format, simplifying the preoperative evaluation of STAS status.
Through univariable and multivariable logistic regression analyses, we identified emphysema, preoperative CEA level, and consolidation-to-tumor ratio (CTR) as independent predictors of STAS. These findings are consistent with the literature, indicating the important role of these clinical and imaging features in the prognosis of T1-stage lung adenocarcinoma. Unlike the findings of other studies, we also identified spiculation, the pleural indentation sign, and vascular convergence as predictors of STAS, although they were not ultimately found to be independent predictors. Multivariable logistic regression analysis verified that the CTR was an independent risk factor for predicting STAS, which aligns with findings from previous studies [20,21]. Compared with the tumor diameter and solid component diameter, the CTR provides a better representation of tumor aggressiveness. A higher CTR is associated with an increased probability of STAS, in agreement with clinical T staging, which emphasizes the diameter of the solid component over the total tumor diameter [22,23]. The present study also revealed a correlation between the presence of STAS and CEA level; 64.6% (75/116) of patients with CEA levels below 5 μg/L and 35.3% (41/116) of patients with CEA levels of 5 μg/L or higher were STAS (+), whereas 95.1% (98/103) of patients with CEA levels below 5 μg/L and 4.9% (5/103) of patients with CEA levels of 5 μg/L or higher were STAS (-). These findings indicate that patients with CEA levels of 5 μg/L or higher have an increased likelihood of being STAS (+), which is consistent with the conclusions of previous studies [24,25]. Notably, emphysema was also identified as an independent predictor of STAS, which is also consistent with the findings of previous studies [23].
However, relying solely on these clinical features may not fully capture the tumor’s biological behavior, underscoring the need to incorporate radiomic features. Radiomics has emerged as a pivotal area of research in medical imaging analysis, particularly in oncology. 3D Slicer has been widely used for radiomic analysis in various diseases [26-28]. However, no studies have used this program in delineating ROIs or the radiomic plugin for extracting radiomic features in predicting airway dissemination. In this study, the use of 3D Slicer (version 5.7.0) software for tumor segmentation and feature extraction demonstrated the powerful capabilities of radiomics in quantitative tumor characterization. By precisely segmenting the tumor and its peritumoral region and extracting a comprehensive set of features, we obtained 899 distinct radiomic features, including GLDM, shape, GLCM, GLRLM, GLSZM, NGTDM, first-order statistic, and wavelet-based features. These features enabled a detailed characterization of various tumor dimensions, such as shape, texture, and intensity distribution, providing rich information that could enhance the accuracy of tumor diagnosis. Moreover, this comprehensive analysis could be used to differentiate tumor type, evaluate disease prognosis, and predict treatment responses.
In this study, Model T yielded high AUC values in both the training and testing sets (0.91 and 0.90, respectively), significantly outperforming the model utilizing only clinical features (AUC of 0.85 in both sets). By resampling images to a voxel size of 1×1×1 mm and applying z score standardization, the researchers ensured the consistency and comparability of the imaging data. Additionally, the reliability of the extracted features was validated through repeat segmentation and feature extraction by two experienced radiologists, and the final features were selected on the basis of an ICC greater than 0.75, ensuring that the features were stable and consistent. Taken together, these findings indicate the substantial potential of radiomic features extracted by 3D Slicer in predicting STAS.
While Model T demonstrated excellent predictive ability on its own, the combined model, incorporating clinical features with Model T, achieved the highest AUC values in both the training and testing sets (0.93 and 0.92, respectively). Although the differences in the AUCs were not statistically significant compared with those of Model T alone, DCA revealed that the combined model yielded the greatest net clinical benefits, particularly within the threshold range of 0.10 to 0.40. These findings suggest that the combined model may offer greater practicality and reliability in clinical applications than the individual models. Calibration curve and decision curve analyses further validated the reliability and clinical value of the models. All the models exhibited a high degree of calibration, indicating consistent performance across the different datasets.
The robustness and accuracy of our radiomic model might have been affected by differences in CT scanning parameters (such as differences in contrast administration or the reconstruction kernels used) or by demographic differences within the dataset (such as the prevalence of STAS-positive tumors or type of CT lesions) [29]. These factors can impact the stability and reliability of the extracted radiomic features. Nonetheless, we assumed that these CT-related differences did not decisively impact our study’s outcomes. This assumption is supported by the results from our internal validation set, which was composed of the data from CT scans obtained with different scanners and protocols; in this validation set, the combined model demonstrated a greater AUC (0.878) than the clinical model (0.854) in preoperatively predicting STAS. This finding aligns with results from a previous study that assessed the performance of a radiomic model using data characterized by significant heterogeneity in terms of CT device manufacturer and type and CT protocol [30]. From the initial phases of devising the study design, it is crucial to avoid selection bias. The inclusion and exclusion criteria need to be clearly defined and strictly adhered to, and it is imperative that these criteria lead to an unbiased study sample that can be generalized beyond that of the study institution.
This study has several limitations. First, the relatively small sample size may affect the generalizability of the results. Second, given the retrospective nature of the study, there is potential for selection bias. Finally, the study was conducted at a single center. Future multicenter studies with larger sample sizes are needed to validate our findings.
Conclusions
In conclusion, preoperative CT-based radiomic models combined with clinical and CT imaging features can effectively predict the risk of STAS in T1-stage invasive lung adenocarcinoma. The combined model demonstrated excellent predictive accuracy and clinical utility, suggesting its potential for broader application in clinical practice. Improved risk assessment through the use of such models can allow clinicians to devise more precise treatment plans, thereby improving patient outcomes.
Acknowledgements
We would like to thank everyone who contributed to this article.
Disclosure of conflict of interest
None.
References
- 1.Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, Wender RC. Cancer screening in the United States, 2019: a review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2019;69:184–210. doi: 10.3322/caac.21557. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, Ishikawa Y, Kerr KM, Noguchi M, Pelosi G, Powell CA, Tsao MS, Wistuba I WHO Panel. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 3.Yanagawa N, Shiono S, Endo M, Ogata SY. Tumor spread through air spaces is a useful predictor of recurrence and prognosis in stage I lung squamous cell carcinoma, but not in stage II and III. Lung Cancer. 2018;120:14–21. doi: 10.1016/j.lungcan.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 4.Aly RG, Rekhtman N, Li X, Takahashi Y, Eguchi T, Tan KS, Rudin CM, Adusumilli PS, Travis WD. Spread through air spaces (STAS) is prognostic in atypical carcinoid, large cell neuroendocrine carcinoma, and small cell carcinoma of the lung. J Thorac Oncol. 2019;14:1583–1593. doi: 10.1016/j.jtho.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyokawa G, Yamada Y, Tagawa T, Kamitani T, Yamasaki Y, Shimokawa M, Oda Y, Maehara Y. Computed tomography features of resected lung adenocarcinomas with spread through air spaces. J Thorac Cardiovasc Surg. 2018;156:1670–1676. e4. doi: 10.1016/j.jtcvs.2018.04.126. [DOI] [PubMed] [Google Scholar]
- 6.Lin MW, Chen LW, Yang SM, Hsieh MS, Ou DX, Lee YH, Chen JS, Chang YC, Chen CM. CT-based deep-learning model for spread-through-air-spaces prediction in ground glass-predominant lung adenocarcinoma. Ann Surg Oncol. 2024;31:1536–1545. doi: 10.1245/s10434-023-14565-2. [DOI] [PubMed] [Google Scholar]
- 7.Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology. 2019;290:290–297. doi: 10.1148/radiol.2018181352. [DOI] [PubMed] [Google Scholar]
- 8.Zeng C, Zhang W, Liu M, Liu J, Zheng Q, Li J, Wang Z, Sun G. Efficacy of radiomics model based on the concept of gross tumor volume and clinical target volume in predicting occult lymph node metastasis in non-small cell lung cancer. Front Oncol. 2023;13:1096364. doi: 10.3389/fonc.2023.1096364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Liu X, Jiang C, Kang W, Pan Y, Tang X, Luo Y, Gong J. CT-based super-resolution deep learning models with attention mechanisms for predicting spread through air spaces of solid or part-solid lung adenocarcinoma. Acad Radiol. 2024;31:2601–2609. doi: 10.1016/j.acra.2023.12.034. [DOI] [PubMed] [Google Scholar]
- 10.Zhou H, Bai HX, Jiao Z, Cui B, Wu J, Zheng H, Yang H, Liao W. Deep learning-based radiomic nomogram to predict risk categorization of thymic epithelial tumors: a multicenter study. Eur J Radiol. 2023;168:111136. doi: 10.1016/j.ejrad.2023.111136. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Wu X, Fang T, Ge Z, Liu J, Wu Q, Zhou L, Shen J, Zhou C. Prognostic impact of tumor spread through air spaces for T2aN0 stage IB non-small cell lung cancer. Cancer Med. 2023;12:15246–15255. doi: 10.1002/cam4.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang C, Luo Y, Yuan J, You S, Chen Z, Wu M, Wang G, Gong J. CT-based radiomics and machine learning to predict spread through air space in lung adenocarcinoma. Eur Radiol. 2020;30:4050–4057. doi: 10.1007/s00330-020-06694-z. [DOI] [PubMed] [Google Scholar]
- 13.Chen D, She Y, Wang T, Xie H, Li J, Jiang G, Chen Y, Zhang L, Xie D, Chen C. Radiomics-based prediction for tumour spread through air spaces in stage I lung adenocarcinoma using machine learning. Eur J Cardiothorac Surg. 2020;58:51–58. doi: 10.1093/ejcts/ezaa011. [DOI] [PubMed] [Google Scholar]
- 14.Han X, Fan J, Zheng Y, Ding C, Zhang X, Zhang K, Wang N, Jia X, Li Y, Liu J, Zheng J, Shi H. The value of CT-based radiomics for predicting spread through air spaces in stage IA lung adenocarcinoma. Front Oncol. 2022;12:757389. doi: 10.3389/fonc.2022.757389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv Y, Li S, Liu Z, Ren Z, Zhao J, Tao G, Zheng Z, Han Y, Ye B. Impact of surgery and adjuvant chemotherapy on the survival of stage I lung adenocarcinoma patients with tumor spread through air spaces. Lung Cancer. 2023;177:51–58. doi: 10.1016/j.lungcan.2023.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Ding Y, Li J, Li X, Xu M, Geng H, Sun D. Impact of preoperative biopsy on tumor spread through air spaces in stage I non-small cell lung cancer: a propensity score-matched study. BMC Pulm Med. 2022;22:293. doi: 10.1186/s12890-022-02090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyokawa G, Yamada Y, Tagawa T, Oda Y. Significance of spread through air spaces in early-stage lung adenocarcinomas undergoing limited resection. Thorac Cancer. 2018;9:1255–1261. doi: 10.1111/1759-7714.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez Moreno JC, Aljamal AA, Bahmad HF, Febres-Aldana CA, Rassaei N, Recine M, Poppiti R. Correlation between spread through air spaces (STAS) and other clinicopathological parameters in lung cancer. Pathol Res Pract. 2021;220:153376. doi: 10.1016/j.prp.2021.153376. [DOI] [PubMed] [Google Scholar]
- 19.Mino-Kenudson M. Significance of tumor spread through air spaces (STAS) in lung cancer from the pathologist perspective. Transl Lung Cancer Res. 2020;9:847–859. doi: 10.21037/tlcr.2020.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Lyu D, Zhang D, Hu L, Wu J, Tu W, Xiao Y, Fan L, Liu S. Nomogram based on clinical characteristics and radiological features for the preoperative prediction of spread through air spaces in patients with clinical stage IA non-small cell lung cancer: a multicenter study. Diagn Interv Radiol. 2023;29:771–785. doi: 10.4274/dir.2023.232404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Y, Zhao S, Liu X, Ren J, Li J, Zhang W, Xu M, Sun D. The value of frozen section diagnosis of tumor spread through air spaces in small-sized (≤ 2 cm) non-small cell lung cancer. World J Surg Oncol. 2023;21:195. doi: 10.1186/s12957-023-03092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Lyu D, Cheng C, Zhou T, Tu W, Xiao Y, Zuo C, Fan L, Liu S. Preoperative nomogram for predicting spread through air spaces in clinical-stage IA non-small cell lung cancer using 18F-fluorodeoxyglucose positron emission tomography/computed tomography. J Cancer Res Clin Oncol. 2024;150:185. doi: 10.1007/s00432-024-05674-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Travis WD, Asamura H, Bankier AA, Beasley MB, Detterbeck F, Flieder DB, Goo JM, MacMahon H, Naidich D, Nicholson AG, Powell CA, Prokop M, Rami-Porta R, Rusch V, van Schil P, Yatabe Y International Association for the Study of Lung Cancer Staging and Prognostic Factors Committee and Advisory Board Members. The IASLC Lung Cancer Staging Project: proposals for coding T categories for subsolid nodules and assessment of tumor size in part-solid tumors in the forthcoming eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2016;11:1204–1223. doi: 10.1016/j.jtho.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 24.Shimomura M, Miyagawa-Hayashino A, Omatsu I, Asai Y, Ishihara S, Okada S, Konishi E, Teramukai S, Inoue M. Spread through air spaces is a powerful prognostic predictor in patients with completely resected pathological stage I lung adenocarcinoma. Lung Cancer. 2022;174:165–171. doi: 10.1016/j.lungcan.2022.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Shou H, Wen H, Wang X, Wang H, Lu C, Gu J, Xu F, Zhu Q, Wang L, Ge D. An individual nomogram can reliably predict tumor spread through air spaces in non-small-cell lung cancer. BMC Pulm Med. 2022;22:209. doi: 10.1186/s12890-022-02002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colen RR, Rolfo C, Ak M, Ayoub M, Ahmed S, Elshafeey N, Mamindla P, Zinn PO, Ng C, Vikram R, Bakas S, Peterson CB, Rodon Ahnert J, Subbiah V, Karp DD, Stephen B, Hajjar J, Naing A. Radiomics analysis for predicting pembrolizumab response in patients with advanced rare cancers. J Immunother Cancer. 2021;9:e001752. doi: 10.1136/jitc-2020-001752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Kim SH, Kim Y, Park J, Park GE, Kang BJ. Radiomics nomogram: prediction of 2-year disease-free survival in young age breast cancer. Cancers (Basel) 2022;14:4461. doi: 10.3390/cancers14184461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homps M, Soyer P, Coriat R, Dermine S, Pellat A, Fuks D, Marchese U, Terris B, Groussin L, Dohan A, Barat M. A preoperative computed tomography radiomics model to predict disease-free survival in patients with pancreatic neuroendocrine tumors. Eur J Endocrinol. 2023;189:476–484. doi: 10.1093/ejendo/lvad130. [DOI] [PubMed] [Google Scholar]
- 29.Van Calster B, Steyerberg EW, Wynants L, van Smeden M. There is no such thing as a validated prediction model. BMC Med. 2023;21:70. doi: 10.1186/s12916-023-02779-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bassi M, Russomando A, Vannucci J, Ciardiello A, Dolciami M, Ricci P, Pernazza A, D’Amati G, Mancini Terracciano C, Faccini R, Mantovani S, Venuta F, Voena C, Anile M. Role of radiomics in predicting lung cancer spread through air spaces in a heterogeneous dataset. Transl Lung Cancer Res. 2022;11:560–571. doi: 10.21037/tlcr-21-895. [DOI] [PMC free article] [PubMed] [Google Scholar]