Abstract
Background: Morinda officinalis saponins (MOS), a traditional Chinese medicine extracted from M. officinalis roots, have been used as a health supplement. Existing evidence suggests that extracts from this plant can be used for osteoporosis treatment. However, the molecular mechanisms underlying the anti-osteoporotic effects of M. officinalis remain poorly understood. Methods and Results: In this study, we investigated the osteogenesis-promoting effects of MOS on human umbilical cord-derived mesenchymal stem cells (HUC-MSCs). Alkaline phosphatase staining, alizarin red staining, and quantitative reverse transcription-PCR demonstrated that MOS promoted the osteogenic differentiation of HUC-MSCs in a concentration-dependent manner. RNA sequencing results showed that the expression of key osteogenic differentiation-related genes, including BMP4, as well as the activity of transforming growth factor-β and calcium signaling pathways increased following MOS treatment. Furthermore, treatment with the bone morphogenetic protein (BMP) antagonist Noggin reversed the MOS-induced pro-osteogenic differentiation effects and the upregulation of osteoblast-specific markers. Conclusions: Overall, the results indicate that MOS can partially promote osteogenic differentiation of HUC-MSCs by regulating the BMP-SMAD signaling pathway. These findings indicate the potential utility of MOS as a therapeutic agent for osteoporosis, particularly in the context of stem cell therapy.
Keywords: Morinda officinalis saponins, human umbilical cord-derived mesenchymal stem cells, osteogenic differentiation, RNA sequencing, stem cell therapy
Introduction
Mesenchymal stem cells (MSCs), which are capable of osteogenic, chondrogenic, and adipogenic lineage commitment, are multipotent cells obtained from adipose tissue, bone marrow, umbilical cord, and Wharton’s jelly (WJ) [1-3]. Owing to their functions, such as tissue repair, hematopoietic support, and immunomodulation, MSCs are considered ideal seed cells for different treatments [4]. They exhibit promising potential for the clinical treatment of multi-organ injuries owing to their low immunogenicity, extensive and convenient availability, sufficient quantity, robust differentiation ability, rapid proliferation, comprehensive application prospects, and absence of ethical controversy [3,5-7]. Human umbilical cord-derived mesenchymal stem cells (HUC-MSCs), which are isolated from the human umbilical cord, have the ability to differentiate into various cell types, rendering them a promising candidate for cell-based therapy [8]. HUC-MSCs are currently used in the treatment of various diseases, such as osteoarthritis, diabetes and related complications, systemic lupus erythematosus, and viral infections [9]. Moreover, HUC-MSCs can be used as a noninvasive treatment for COVID-19 [10]. These clinical applications suggest their promising role in regenerative medicine in the future.
Morinda officinalis, a dicotyledonous plant widely grown in tropical and subtropical regions of the world, is one of the most well-known Chinese herbs. Recognized for over 2000 years in Northeast Asia, its roots have the ability to bolster the immune system and fortify the bones and kidneys, as documented in the Chinese pharmacopeia [11]. The herb has been commonly used to treat rheumatoid arthritis, impotent dermatitis, and menstrual disorders [12,13]. Recent research has unveiled various pharmacological effects of M. officinalis, including immunomodulation, anti-tumor, anti-aging, and anti-fatigue properties [14,15]. M. officinalis root extracts can promote bone formation in vivo and exhibit beneficial effects in the prevention and treatment of osteoporosis [13,16]. Morinda officinalis saponins (MOS) are important active ingredients of M. officinalis and are currently mainly used as nutraceuticals. Despite numerous experimental studies demonstrating the beneficial effects of M. officinalis extracts, particularly polysaccharides and oligosaccharides, on the promotion of osteoblast proliferation and activity, induction of osteogenic differentiation in bone marrow MSCs, and bone metabolism [17-19], the effects of MOS on HUC-MSCs remain unexplored. In this study, we investigated the effects of MOS on the proliferation and osteogenic differentiation of HUC-MSCs and the resulting transcriptomic changes.
Materials and methods
Media and reagents
MOS was purchased from Xi’an Yatu Biotechnology Co., Ltd. (Xi’an, China), stored in sealed ziplock bags in a dry environment, and preserved at -20°C after dissolving in water. HUC-MSCs were supplied by Biotech & Biomedicine Group., Ltd. (Shenyang, China). Dulbecco’s modified Eagle’s medium (DMEM), α-minimum essential medium (MEM), and penicillin (10000 Units/mL)/streptomycin (10000 μg/mL) (P/S) were purchased from Gibco (Grand Island, NY, USA). Fetal bovine serum (FBS) was sourced from ExCell Company (Shanghai, China), while L-glutamine (200 mM) was purchased from Sangon Biotech (Shanghai, China). The BCIP/NBT alkaline phosphatase (ALP) color development kit was purchased from Beyotime Biotechnology (Shanghai, China), and the cell counting kit-8 (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan). Alizarin red staining kit was procured from Beijing Solarbio Science & Technology Co., Ltd. Additionally, β-glycerophosphate disodium salt hydrate and ascorbic acid were acquired from Sigma-Aldrich (St. Louis, MO, USA). Recombinant human bone morphogenetic protein 4 (BMP4) was purchased from PeproTech (Rocky Hill, NJ, USA), and Noggin was obtained from MedChem Express (Monmouth Junction, NJ, USA).
Cell culture
HUC-MSCs were cultured and maintained in DMEM complete medium (DMEM supplemented with 10% FBS, 1% P/S, and 1% L-glutamine) in a 5% CO2 humidified incubator at 37°C until 80% confluence was reached. HUC-MSCs from passages 4 to 6 were used for all experiments, and the complete medium was replaced every two days.
Morphological characteristics and cytotoxicity test
HUC-MSCs (5 × 105 cells/well, passages 6) were seeded onto 12-well plates and incubated for 24 h. Subsequently, the experimental groups were treated with six different concentrations of MOS (0, 4, 20, 100, 500, and 2500 μg/mL) for 1, 6, 24, and 48 h. Cell morphology was observed and morphological data were collected using an inverted microscope (S80-SLIDER, Leica Microsystems, Wetzlar, Germany). Cytotoxic effects of MOS on HUC-MSCs were determined using the CCK-8 kit. Briefly, HUC-MSCs (5 × 103 cells/well, passage 6) were seeded in 96-well plates and incubated for 24 h, followed by culturing with different MOS concentrations (0, 5, 25, 50, 100, and 200 μg/mL) for 24, 48, 72, and 96 h, with three parallel control wells in each group. Subsequently, 10 μL CCK-8 reagent was added to each well, the plates were incubated for 3 h in a 5% CO2 humidified incubator at 37°C, and the absorbance was measured at 450 nm.
Osteoblast differentiation assay
For in vitro HUC-MSC differentiation, passage 5 HUC-MSCs were plated at a density of 1.0 × 105 cells/well in 12-well plates. Cells were cultured in complete medium until they reached a confluence of 80%; subsequent replacement with phosphate-buffered saline (PBS) preceded the introduction of osteogenic differentiation medium. Osteogenic media (OM), comprising α-MEM, 5% FBS, 1% P/S, 100 μg/mL ascorbic acid, 5 mM β-glycerophosphate disodium salt hydrate, and varying concentrations of MOS (5, 25, 50, and 100 μg/mL), were used for differentiation. The media were replaced every two days, and three biological replicates were set for each group.
ALP and alizarin red staining assays
At the initial stage of osteogenic differentiation of HUC-MSCs (seven days), cells were stained using an ALP staining kit. After medium removal, the cells were washed with PBS, fixed in 4% paraformaldehyde for 15 min, and washed thrice with PBS. Subsequently, the cells were stained with an ALP staining solution and incubated in the dark at 30°C for 12 h. To investigate the effect of MOS on mineralization, alizarin red staining was performed at the late stage of osteogenic differentiation (21 days). After staining with a 0.2% alizarin red solution for 30 min at room temperature, the reaction was terminated by washing the HUC-MSCs gently with ultrapure water; the cells were then observed under a microscope (DS-Ri2, Nikon, Tokyo, Japan). The positive cell area was quantified using ImageJ v1.52 (National Institutes of Health, Bethesda, MD, USA).
Quantitative reverse transcription-PCR (RT-qPCR)
Total RNA was extracted from HUC-MSCs using TRIzol reagent (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Briefly, passage 5 HUC-MSCs were plated at a density of 1.0 × 105 cells per well in six-well plates, and each well received 500 μL TRIzol reagent after five days of osteogenic differentiation induction. After centrifugation for 10 min at 12000 × g and 4°C, the supernatants were collected for subsequent RNA isolation. cDNA was synthesized using the Prime-Script RT Reagent Kit (TaKaRa Biotech, Tokyo, Japan). The mRNA levels of osteogenic differentiation markers (RUNX2, OCN, and OPN) were determined via RT-qPCR using SYBR Premix Ex Taq (TaKaRa Biotech) and Bio-Rad CFX 96 real-time PCR detection system (Hercules, CA, USA) with QuantStudioTM Design and Analysis Software v1.4.3 (Thermo Fisher Scientific, Waltham, MA, USA). All primer sequences for the tested genes are listed in Table 1. The cycling conditions were as follows: denaturation at 95°C for 1 min, followed by 40 cycles at 95°C for 15 s, 60°C for 15 s, and 72°C for 45 s. The successful generation of individual amplicons was confirmed through melting curve analysis. Gene expression levels were determined using the 2-ΔΔCq method and normalized to GAPDH levels [20].
Table 1.
Primer sequences used for quantitative reverse transcription-PCR-based analysis of gene expression
| Gene name | Accession No. | Primer Sequence (5’ to 3’) |
|---|---|---|
| Runx2 | NM_001024630.4 | F: GACAAGCACAAGTAAATCATTGAACTACAG |
| R: GTAAGGCTGGTTGGTTAAGAATCTCTG | ||
| Ocn | NM_198406.3 | F: ACCAGGTAATGCCAGTTTGC |
| R: CCCCCTCTAGCCTAGGACC | ||
| Opn | NM_001040058.2 | F: CTGAAACCCACAGCCACA |
| R: TGTGGAATTCACGGCTGA | ||
| Gapdh | NM_002046.7 | F: AGCCACATCGCTCAGACAC |
| R: GCCCAATACGACCAAATCC |
RNA sequencing (RNA-seq) analysis
The cells were sub-cultured in a 6-cm dish once the viability of HUC-MSCs revived in a 10 cm dish reached 90%. HUC-MSCs were cultured with 25 μg/mL MOS for 48 h; a normal control group (complete medium without MOS) was also established, with three parallel wells in each group. The cell pellets were washed, collected, and sent to Shenzhen Promegene Technology Co., Ltd. (Shenzhen, China) for sequencing. All purified libraries were sequenced on DNBSEQ-T7RS using Geneplus to acquire 150 bp paired-end sequence reads. Raw RNA-seq data were deposited in the NCBI Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/PRJNA843361).
Western blotting
HUC-MSCs were inoculated into six-well plates and subsequently cultured with OM+BMP4, OM+BMP4+Noggin, OM+Noggin, OM+MOS, OM+MOS+Noggin, or OM medium for 24 h. The cells were lysed with the standard radioimmunoprecipitation assay lysis buffer supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), and the extracted proteins were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. Following electrophoresis, the resolved proteins were transferred onto polyvinylidene difluoride membranes, which were blocked with 5% non-fat milk and incubated with primary antibodies against p-SMAD1/5/9 (Cell Signaling Cat# 13820) and beta-actin (Sigma-Aldrich Cat# A1978). Thereafter, the membranes were washed and probed with an anti-rabbit secondary antibody (R&D Systems). Finally, the membranes were examined using an enhanced chemiluminescence detection system (Beyotime Cat# P0018AS) and visualized with a Bio-Rad imaging system (Hercules, CA, USA).
Statistical analysis
Cell experiments were repeated three times. All values are presented as mean ± standard deviation. Significant differences were determined using a one-way analysis of variance followed by Dunnett’s or Tukey’s multiple comparison test. GraphPad Prism 9.0 (GraphPad Software Inc., La Jolla, CA, USA) was used to perform data analysis and statistical tests, while edgeR package v3.26.8 (Bioconductor, Roswell Park Cancer Institute, Buffalo, NY, USA) was used to identify differentially expressed genes (DEGs). DEGs were evaluated based on the following criteria: (i) more than 1.41-fold change in gene expression and (ii) P < 0.05. The fold change in the expression of each gene was calculated by comparing the standardized read counts of MOS-treated cells with those of the non-treated cells (fold change = standardized read counts of MOS-treated cells/standardized read counts of non-treated cells). Log2(fold change) was used for convenience, where |Log2(fold change)| > 1 indicated at least a twofold change. *P-values < 0.05 were considered statistically significant.
Results
MOS administration dose for HUC-MSCs
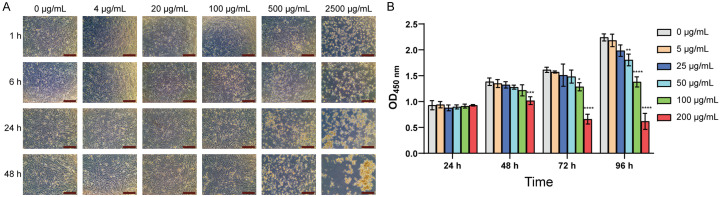
The cytotoxicity of various MOS concentrations to HUC-MSCs was tested to determine the appropriate concentration for this study. An analysis of the cells under a microscope revealed that cell death was evident after 1 h when MOS concentration exceeded 500 μg/mL, indicating significant toxicity to HUC-MSCs at this concentration (Figure 1A). CCK-8 assay results showed that the proliferation of HUC-MSCs was not affected by low MOS concentrations (5 and 25 μg/mL). However, MOS concentrations of 50 and 100 μg/mL were toxic to HUC-MSCs, leading to the inhibition of cell proliferation (Figure 1B). Therefore, the 200 μg/mL concentration of MOS was defined as the sublethal concentration.
Figure 1.
MOS affects HUC-MSC proliferation. A. The cells grew within 48 h of treatment with a particular MOS concentration (magnification: × 40, scale bar: 500 μm). B. CCK-8 assay results showing cell viability within 96 h of treatment with different MOS concentrations. P < 0.05; all values are presented as mean ± SD; n = 3 (vs. 0 μg/mL group; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Significant differences compared with those in the 0 μg/mL group were analyzed using a one-way ANOVA with Dunnett’s post-hoc test. MOS: Morinda officinalis saponins; HUC-MSCs: human umbilical cord-derived mesenchymal stem cells; CCK-8: cell counting kit-8; SD: standard deviation.
MOS promoted the osteogenic differentiation and mineralization of HUC-MSCs
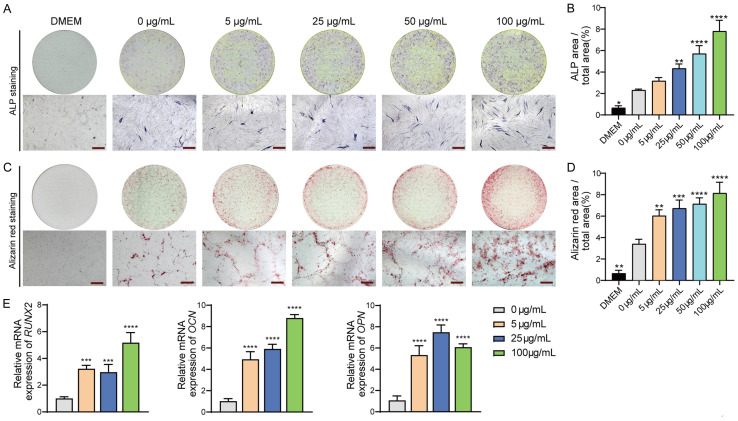
The ALP staining assay (Figure 2A, 2B) showed that MOS treatment resulted in an increase in the number of ALP-positive cells in a dose-dependent manner after seven days of osteogenic differentiation. Alizarin red staining was used to investigate the effects of MOS on calcium nodule formation after 21 days (Figure 2C, 2D). Notably, the number of calcium nodules significantly increased following treatment with MOS and was positively correlated with MOS concentration. A comparative analysis of ALP and alizarin red staining revealed a significant positive effect of MOS on the osteogenic differentiation of HUC-MSCs. The effect of MOS on HUC-MSCs was more pronounced at a concentration of 100 μg/mL. The expression of the osteogenic differentiation markers RUNX2, OCN, and OPN in the MOS-treated group was significantly increased (P < 0.05) compared with that in the control group (Figure 2E).
Figure 2.
MOS promotes HUC-MSC osteogenic differentiation and mineralization in a concentration-dependent manner. A and B. HUC-MSCs were cultured in DMEM complete medium and osteogenic differentiation medium with and without MOS for seven days, respectively. ALP staining assay was performed, and the staining area was quantified using ImageJ. C, D. HUC-MSCs were cultured for 21 days. Alizarin red staining results; the stained area was quantified using ImageJ; the orange-red dots represent calcium nodules. E. Expression of osteogenic markers. Data are presented as mean ± SD; n = 3 (vs. 0 μg/mL group; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Significant differences compared with those in the 0 μg/mL group were analyzed using one-way ANOVA with Dunnett’s post-hoc test. Magnification: × 40, scale bar: 500 μm. MOS: Morinda officinalis saponins; HUC-MSCs: human umbilical cord-derived mesenchymal stem cells; DMEM: Dulbecco’s modified eagle medium; ALP: alkaline phosphatase; SD: standard deviation.
Potential mechanisms underlying the effects of MOS on the osteogenic differentiation of HUC-MSCs based on RNA-seq analysis
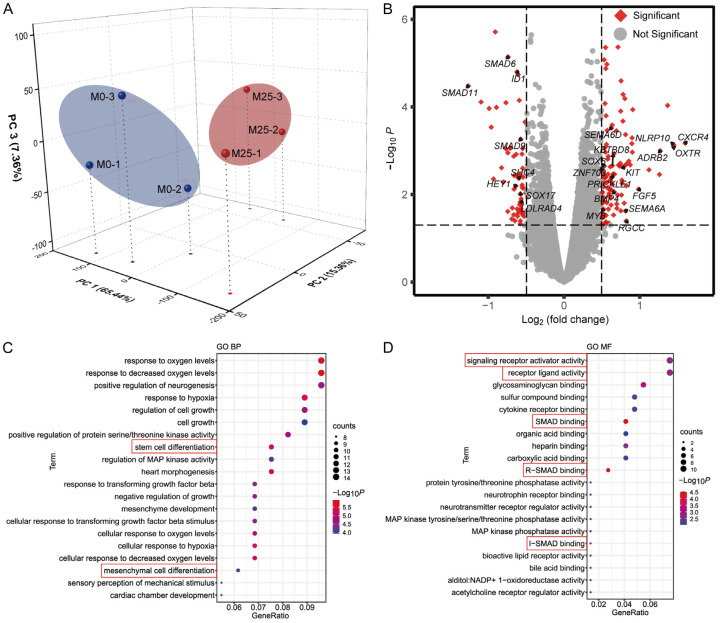
To further investigate the potential mechanisms underlying MOS-induced osteogenic differentiation in HUC-MSCs, we analyzed the gene expression profiles of HUC-MSCs cultured with and without MOS (25 μg/mL) using RNA-seq. To determine significant differences between the MOS-treated (M25) and non-treated (M0) samples, we performed 3D principal component analysis (PCA) to reduce dimensionality and evaluate the independence of each group (Figure 3A). The PCA results showed that M25 and M0 were divided into two distinct groups. The cumulative contribution rates for each principal component on the vertical axis were 65.44%, 15.38%, and 7.36% for PC1, PC2, and PC3, respectively, indicating that the three principal components were sufficient for distinguishing the two groups.
Figure 3.
RNA sequencing analysis results. A. The 3D principal component analysis of six experimental samples was visualized based on MOS-treated (M25, red) and non-treated (M0, blue) samples. B. A volcano plot showing differential gene expression and annotations of significantly upregulated or downregulated differentially expressed genes (red). C, D. GO enrichment analysis of terms in the biological process (BP) and molecular function (MF) categories; the ontologies represent the top 20 significantly enriched terms in each category. MOS: Morinda officinalis saponins; GO: Gene Ontology.
Next, we compared differences in gene expression between the control and MOS-treated groups using edgeR. A total of 165 DEGs were identified based on a threshold of (false discovery rate |log2(fold change)| > 0.5 and P < 0.05), with 104 and 61 DEGs being upregulated and downregulated, respectively, in the MOS-treated group. Volcano plots showing the number of DEGs identified after screening and the genes associated with osteogenic differentiation are shown in Figure 3B. The expression levels of osteogenic differentiation genes based on the RNA-seq data are summarized in Tables S1 and S2.
To further investigate the functional states of HUC-MSCs and potential molecular regulators after MOS treatment, the DEGs were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. DEGs were significantly enriched in biological process (BP) terms (Figure 3C), stem cell and mesenchymal cell differentiation. In the context of stem cell differentiation, specific terms exhibited significant enrichment, with upregulated genes (BMP4, EDN1, KBTBD8, KIT, MYB, PRICKLE1, SEMA6A, SEMA6D, and SOX6) and downregulated genes (SHC4 and SOX17) being involved. The term for mesenchymal cell differentiation included both upregulated genes (BMP4, EDN1, KBTBD8, RGCC, SEMA6A, SEMA6D, and ZNF703) and downregulated genes (HEY1 and LDLRAD4). The most significantly enriched molecular function (MF) terms included R-SMAD binding, SMAD binding, I-SMAD binding, signaling receptor activator activity, and receptor ligand activity (Figure 3D). Collectively, the results suggest that the transforming growth factor (TGF)-β/BMP-SMAD signaling pathway plays a pivotal role in the induction of osteogenic differentiation by MOS in HUC-MSCs.
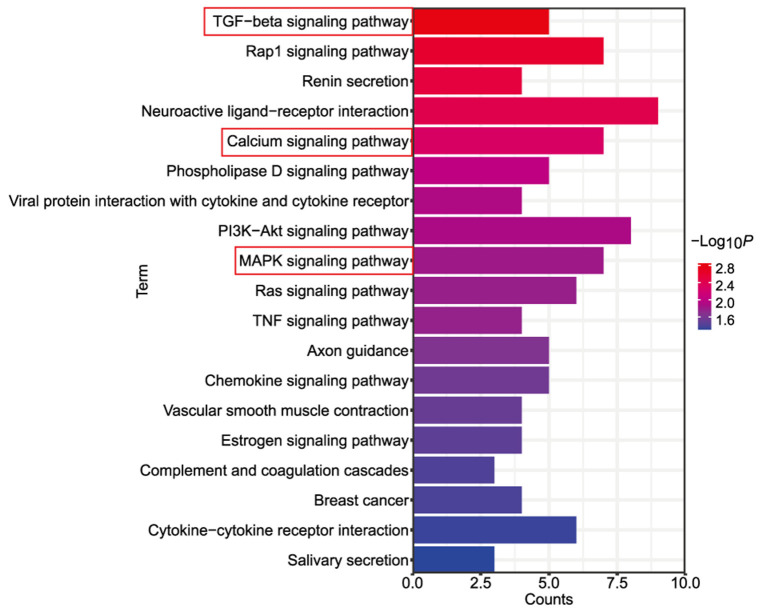
A total of 72 genes were mapped onto signaling pathways, as determined using the KEGG pathway enrichment analysis. The TGF-β, calcium, and mitogen-activated protein kinase (MAPK) signaling pathways were among the 19 pathways that were significantly enriched (Figure 4). Notably, the TGF-β signaling pathway emerged as an essential pathway, aligning with the enriched biological terms for SMAD mentioned above.
Figure 4.
KEGG pathway enrichment analysis results. The top 19 significantly enriched pathways were ranked by gene count. Three of the pathways, namely, the TGF-β, calcium, and MAPK signaling pathways, were closely associated with osteogenic differentiation. KEGG: Kyoto Encyclopedia of Genes and Genomes; TGF-β: transforming growth factor-β; MAPK: mitogen-activated protein kinase.
MOS promoted osteogenic differentiation via BMP-SMAD signaling
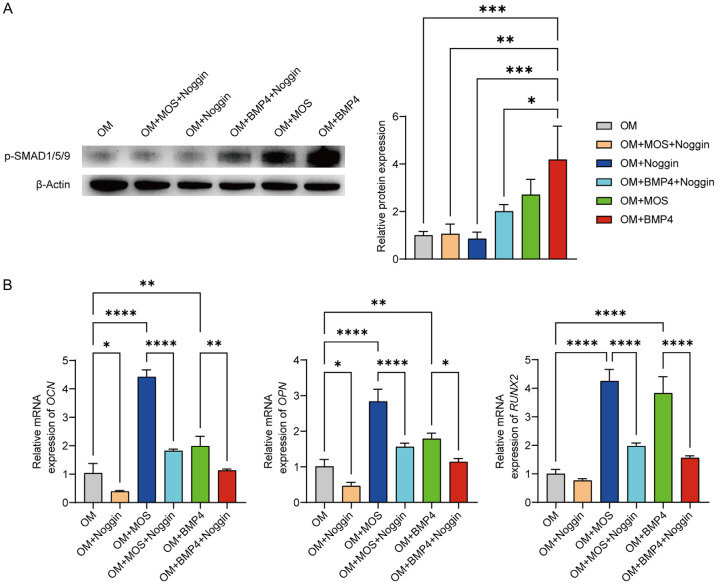
To investigate the mechanisms underlying the MOS-induced osteogenic differentiation of HUC-MSCs, we treated cells with the BMP pathway inhibitor Noggin, in combination with MOS, and used BMP4 as the positive control. Western blotting was used to analyze the changes in p-SMAD1/5/9 protein expression levels (Figure 5A), while RT-qPCR was used to quantify the mRNA expression of the osteogenic markers OCN, OPN, and RUNX2 (Figure 5B). Western blot analysis showed increased p-SMAD1/5/9 protein expression levels in the OM+MOS group and that the addition of Noggin reversed this increase. In addition, RT-qPCR results showed a significant increase in the expression of osteogenesis-specific genes in the OM+MOS group; however, the expression of these genes was significantly reduced in the OM+MOS+Noggin group. These results confirmed the influence of MOS on the BMP-SMAD signaling pathway.
Figure 5.
MOS-induced osteogenic differentiation of HUC-MSCs via the BMP-SMAD signaling pathway. A. HUC-MSCs were cultured for 24 h. Changes in p-SMAD1/5/9 protein expression levels were detected using western blotting. p-SMAD1/5/9 protein expression in HUC-MSCs was quantified using ImageJ and normalized to β-actin expression. B. RT-qPCR analysis of mRNA levels in HUC-MSCs treated for six days. Data are presented as mean ± SD; n = 3 (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Significant differences were analyzed using a one-way ANOVA with Tukey’s post-hoc test. MOS: Morinda officinalis saponins; HUC-MSCs: human umbilical cord-derived mesenchymal stem cells; RT-qPCR: quantitative reverse transcription-PCR; SD: standard deviation.
Discussion
The clinical application of MSCs depends on their ability to differentiate into multiple cell types, including osteoblasts, adipocytes, and chondrocytes, and their potential for self-renewal [3,21]. HUC-MSCs are widely used in experimental studies to analyze in vitro osteogenic differentiation potential [22,23]. Numerous studies have shown that some traditional herbal medicines can significantly improve the osteogenic differentiation ability of MSCs during osteogenic differentiation [22]. Previous studies have demonstrated that certain carbohydrate components, such as polysaccharides and oligosaccharides of M. officinalis, are crucial for the prevention and treatment of osteoporosis [18,24-26]. However, no experimental study has determined whether MOS can promote the osteogenic differentiation of HUC-MSCs. Furthermore, little is known regarding the molecular mechanisms through which MOS promote osteogenic differentiation.
The directed differentiation of MSCs is regulated by several intracellular signaling pathways and transcription factors [27]. BMP4, a member of the TGF-β superfamily, is a growth factor that actively contributes to bone formation [28]. Multiple members of the TGF-β signaling family can regulate the differentiation of MSCs into specific cells by influencing the activity and expression of key transcription factors [29-31]. Except for BMP3, all other BMPs generally promote the osteogenic differentiation of MSCs [32-34]. In this study, BMP4 was significantly upregulated after MOS treatment, which is consistent with our experimental results regarding the induction of osteogenic differentiation in HUC-MSCs by MOS. In addition, the expression of CXCR4 [35], FGF5 [36], and ADRB2 [37], which promote the osteogenic differentiation of MSCs, significantly increased. Notably, apart from the expression of positive regulators of osteogenic differentiation, the expression of SMAD6 [38] and ID1 [39], which are known to negatively regulate the osteogenic differentiation of MSCs, was reduced after MOS treatment. Previous studies have shown that the differentiation of MSCs into osteoblasts is accompanied by the expression of Ca2+-binding proteins [40], and calcium signaling is activated during MSC osteogenic differentiation [41]. The MAPK signaling pathway is a major signal transducer that regulates the osteogenic differentiation of MSCs [42]; suppression of the MAPK signaling pathway leads to a significant reduction in the expression of osteogenesis-related genes [43]. MAPK is the only active signaling molecule involved in the regulation of the three MSC differentiation lineages (osteogenic, adipogenic, and chondrogenic lineages) [44]. In the present study, the TGF-β, calcium, and MAPK signaling pathways were significantly enriched at the transcriptome level after MOS treatment.
The TGF-β superfamily comprises TGF-β, BMP, activins, and related proteins [45]. In the canonical SMAD pathway, BMP2 and BMP4 ligands can phosphorylate the downstream mediators SMAD1, SMAD5, and SMAD9 by binding to the BMP receptor complex at the cell surface and then forming a complex with SMAD4, which is translocated into the nucleus to activate osteogenic genes to stimulate osteoclast differentiation [46,47]. Therefore, we selected BMP4 as a positive control for the promotion of osteoclast differentiation through BMP signaling. Noggin’s primary structure consists of an acidic amino-terminal and a cysteine-rich carboxy-terminal region. This clip snakes around the BMP ligand and occludes the surfaces of the growth factor from binding to both the BMP types I and II receptors, thereby acting as an efficient antagonist of BMP signaling [48]. Our results showed that Noggin significantly reduced the effects of MOS on osteogenic differentiation and osteogenesis-related gene expression. These findings suggest that MOS can promote osteogenic differentiation through the BMP-SMAD signaling pathway.
Although our study elucidated the osteogenic effects of MOS on HUC-MSCs, it is essential to acknowledge that in vitro findings may not fully replicate the complex in vivo microenvironment. Future investigations incorporating animal models and clinical trials are warranted to validate the translational potential of MOS in osteoporosis treatment.
Conclusion
Our study provides a molecular basis that enhances our understanding of the mechanisms through which MOS promote the osteogenic differentiation of HUC-MSCs, thereby providing novel insights into the application of MOS in MSC-based treatments for bone diseases. Overall, MOS can partially promote the osteogenic differentiation of HUC-MSCs through the BMP-SMAD signaling pathway, which suggests that MOS are promising agents for the prevention and adjunct treatment of osteoporosis using stem cell therapy. Future studies should explore the long-term effects and safety profiles of MOS in vivo and address questions related to optimal dosage, potential side effects, and sustained efficacy. Additionally, investigating the synergistic effects of MOS in combination with other osteogenic agents or pharmaceuticals could enhance the understanding of their therapeutic applications in bone regeneration and osteoporosis management.
Acknowledgements
This work was supported by Jiangxi Natural Science Foundation for distinguished young scholars (20212ACB216001), Startup Foundation for Advanced Talents (QD201910), Jiangxi key research and development program (20203BBG73063), Jiangxi Double Thousand Plan (jxsq2019101064), Graduate Innovation Special Fund project of Jiangxi Education Department (YC2021-S804), Liaoning Revitalization Talents Program (XLYC2002027), Construction of Liaoning technological innovation center (1590826279052), Central government funds for guiding local scientific and Technological Development (2021JH6/10500225), Scientific Research Fund of Education Department of Liaoning Province in 2021 (LJKZ0464), and Young Scientists Nurturing Program from Department of Education of Liaoning Province (LQ2020022).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Hass R, Kasper C, Böhm S, Jacobs R. Different populations and sources of human mesenchymal stem cells (MSC): a comparison of adult and neonatal tissue-derived MSC. Cell Commun Signal. 2011;9:12. doi: 10.1186/1478-811X-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almalki SG, Agrawal DK. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation. 2016;92:41–51. doi: 10.1016/j.diff.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 4.Zhang LB, He M. Effect of mesenchymal stromal (stem) cell (MSC) transplantation in asthmatic animal models: a systematic review and meta-analysis. Pulm Pharmacol Ther. 2019;54:39–52. doi: 10.1016/j.pupt.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Harrell CR, Markovic BS, Fellabaum C, Arsenijevic A, Volarevic V. Mesenchymal stem cell-based therapy of osteoarthritis: current knowledge and future perspectives. Biomed Pharmacother. 2019;109:2318–2326. doi: 10.1016/j.biopha.2018.11.099. [DOI] [PubMed] [Google Scholar]
- 6.Han B, Zhou L, Guan Q, da Roza G, Wang H, Du C. In vitro expansion and characterization of mesenchymal stromal cells from peritoneal dialysis effluent in a human protein medium. Stem Cells Int. 2018;2018:5868745. doi: 10.1155/2018/5868745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Liu G, Wang X, Hu Y, Luo H, Ye L, Feng Z, Li C, Kuang M, Zhang L, Zhou Y, Qi X. hUC-MSCs: evaluation of acute and long-term routine toxicity testing in mice and rats. Cytotechnology. 2022;74:17–29. doi: 10.1007/s10616-021-00502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T, Xia M, Gao Y, Chen Y, Xu Y. Human umbilical cord mesenchymal stem cells: an overview of their potential in cell-based therapy. Expert Opin Biol Ther. 2015;15:1293–1306. doi: 10.1517/14712598.2015.1051528. [DOI] [PubMed] [Google Scholar]
- 10.Shu L, Niu C, Li R, Huang T, Wang Y, Huang M, Ji N, Zheng Y, Chen X, Shi L, Wu M, Deng K, Wei J, Wang X, Cao Y, Yan J, Feng G. Treatment of severe COVID-19 with human umbilical cord mesenchymal stem cells. Stem Cell Res Ther. 2020;11:361. doi: 10.1186/s13287-020-01875-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, Peng Q, Xu Y, Xu H, Wan Y, Li Z, Qiu Y, Xia W, Guo Z, Li H, Jin H, Hu B. Morinda officinalis oligosaccharides ameliorate depressive-like behaviors in poststroke rats through upregulating GLUT3 to improve synaptic activity. FASEB J. 2020;34:13376–13395. doi: 10.1096/fj.201902546RR. [DOI] [PubMed] [Google Scholar]
- 12.Yang YJ, Shu HY, Min ZD. Anthraquinones isolated from Morinda officinalis and Damnacanthus indicus. Yao Xue Xue Bao. 1992;27:358–364. [PubMed] [Google Scholar]
- 13.Wu YB, Zheng CJ, Qin LP, Sun LN, Han T, Jiao L, Zhang QY, Wu JZ. Antiosteoporotic activity of anthraquinones from Morinda officinalis on osteoblasts and osteoclasts. Molecules. 2009;14:573–583. doi: 10.3390/molecules14010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Xu H, Xu Y, Lu G, Peng Q, Chen J, Bi R, Li J, Chen S, Li H, Jin H, Hu B. Morinda officinalis oligosaccharides alleviate depressive-like behaviors in post-stroke rats via suppressing NLRP3 inflammasome to inhibit hippocampal inflammation. CNS Neurosci Ther. 2021;27:1570–1586. doi: 10.1111/cns.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan C, Huang D, Shen X, Qin N, Jiang K, Zhang D, Zhang Q. Identification and characterization of a polysaccharide from the roots of Morinda officinalis, as an inducer of bone formation by up-regulation of target gene expression. Int J Biol Macromol. 2019;133:446–456. doi: 10.1016/j.ijbiomac.2019.04.084. [DOI] [PubMed] [Google Scholar]
- 16.Seo BI, Ku SK, Cha EM, Park JH, Kim JD, Choi HY, Lee HS. Effect of Mornidae Radix extracts on experimental osteoporosis in sciatic neurectomized mice. Phytother Res. 2005;19:231–238. doi: 10.1002/ptr.1683. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JH, Xin HL, Xu YM, Shen Y, He YQ, Hsien-Yeh, Lin B, Song HT, Juan-Liu, Yang HY, Qin LP, Zhang QY, Du J. Morinda officinalis How. - A comprehensive review of traditional uses, phytochemistry and pharmacology. J Ethnopharmacol. 2018;213:230–255. doi: 10.1016/j.jep.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 18.He YQ, Zhang Q, Shen Y, Han T, Zhang QL, Zhang JH, Lin B, Song HT, Hsu HY, Qin LP, Xin HL, Zhang QY. Rubiadin-1-methyl ether from Morinda officinalis How. Inhibits osteoclastogenesis through blocking RANKL-induced NF-κB pathway. Biochem Biophys Res Commun. 2018;506:927–931. doi: 10.1016/j.bbrc.2018.10.100. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Zhao X, Wang W. lncRNA and mRNA sequencing of the left testis in experimental varicocele rats treated with Morinda officinalis polysaccharide. Exp Ther Med. 2021;22:1136. doi: 10.3892/etm.2021.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 22.He S, Yang S, Zhang Y, Li X, Gao D, Zhong Y, Cao L, Ma H, Liu Y, Li G, Peng S, Shuai C. LncRNA ODIR1 inhibits osteogenic differentiation of hUC-MSCs through the FBXO25/H2BK120ub/H3K4me3/OSX axis. Cell Death Dis. 2019;10:947. doi: 10.1038/s41419-019-2148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrijantini N, Hartono P. Phenotype characteristics and osteogenic differentiation potential of human mesenchymal stem cells derived from Amnion Membrane (HAMSCs) and Umbilical Cord (HUC-MSCs) Acta Inform Med. 2019;27:72–77. doi: 10.5455/aim.2019.27.72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Zhang S, Jiang K, Li T, Yan C. Bioassay-guided isolation and evaluation of anti-osteoporotic polysaccharides from Morinda officinalis. J Ethnopharmacol. 2020;261:113113. doi: 10.1016/j.jep.2020.113113. [DOI] [PubMed] [Google Scholar]
- 25.Xia T, Dong X, Lin L, Jiang Y, Ma X, Xin H, Zhang Q, Qin L. Metabolomics profiling provides valuable insights into the underlying mechanisms of Morinda officinalis on protecting glucocorticoid-induced osteoporosis. J Pharm Biomed Anal. 2019;166:336–346. doi: 10.1016/j.jpba.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Jiang K, Huang D, Zhang D, Wang X, Cao H, Zhang Q, Yan C. Investigation of inulins from the roots of Morinda officinalis for potential therapeutic application as anti-osteoporosis agent. Int J Biol Macromol. 2018;120:170–179. doi: 10.1016/j.ijbiomac.2018.08.082. [DOI] [PubMed] [Google Scholar]
- 27.Sun Q, Liu S, Feng J, Kang Y, Zhou Y, Guo S. Current status of MicroRNAs that target the Wnt signaling pathway in regulation of osteogenesis and bone metabolism: a review. Med Sci Monit. 2021;27:e929510. doi: 10.12659/MSM.929510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seeherman HJ, Berasi SP, Brown CT, Martinez RX, Juo ZS, Jelinsky S, Cain MJ, Grode J, Tumelty KE, Bohner M, Grinberg O, Orr N, Shoseyov O, Eyckmans J, Chen C, Morales PR, Wilson CG, Vanderploeg EJ, Wozney JM. A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix. Sci Transl Med. 2019;11:eaar4953. doi: 10.1126/scitranslmed.aar4953. [DOI] [PubMed] [Google Scholar]
- 29.Minina E, Wenzel HM, Kreschel C, Karp S, Gaffield W, McMahon AP, Vortkamp A. BMP and Ihh/PTHrP signaling interact to coordinate chondrocyte proliferation and differentiation. Development. 2001;128:4523–4534. doi: 10.1242/dev.128.22.4523. [DOI] [PubMed] [Google Scholar]
- 30.Oshimori N, Fuchs E. The harmonies played by TGF-β in stem cell biology. Cell Stem Cell. 2012;11:751–764. doi: 10.1016/j.stem.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grafe I, Alexander S, Peterson JR, Snider TN, Levi B, Lee B, Mishina Y. TGF-β family signaling in mesenchymal differentiation. Cold Spring Harb Perspect Biol. 2018;10:a022202. doi: 10.1101/cshperspect.a022202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokabu S, Gamer L, Cox K, Lowery J, Tsuji K, Raz R, Economides A, Katagiri T, Rosen V. BMP3 suppresses osteoblast differentiation of bone marrow stromal cells via interaction with Acvr2b. Mol Endocrinol. 2012;26:87–94. doi: 10.1210/me.2011-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen D, Ji X, Harris MA, Feng JQ, Karsenty G, Celeste AJ, Rosen V, Mundy GR, Harris SE. Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J Cell Biol. 1998;142:295–305. doi: 10.1083/jcb.142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu DD, Zhang JC, Zhang Q, Wang SX, Yang MS. TGF-β/BMP signaling pathway is involved in cerium-promoted osteogenic differentiation of mesenchymal stem cells. J Cell Biochem. 2013;114:1105–1114. doi: 10.1002/jcb.24451. [DOI] [PubMed] [Google Scholar]
- 35.Gong ZM, Tang ZY, Sun XL. LncRNA PRNCR1 regulates CXCR4 expression to affect osteogenic differentiation and contribute to osteolysis after hip replacement. Gene. 2018;673:251–261. doi: 10.1016/j.gene.2018.05.043. [DOI] [PubMed] [Google Scholar]
- 36.Park GC, Song JS, Park HY, Shin SC, Jang JY, Lee JC, Wang SG, Lee BJ, Jung JS. Role of fibroblast growth factor-5 on the proliferation of human tonsil-derived mesenchymal stem cells. Stem Cells Dev. 2016;25:1149–1160. doi: 10.1089/scd.2016.0061. [DOI] [PubMed] [Google Scholar]
- 37.Alves Barreto AE, Balera Brito VG, Patrocinio MS, Ballassoni BB, Tfaile Frasnelli SC, Penha Oliveira SH. β1-adrenergic receptor but not β2 mediates osteogenic differentiation of bone marrow mesenchymal stem cells in normotensive and hypertensive rats. Eur J Pharmacol. 2021;911:174515. doi: 10.1016/j.ejphar.2021.174515. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Zheng GF, Xu XF. MicroRNA-186 improves fracture healing through activating the bone morphogenetic protein signalling pathway by inhibiting SMAD6 in a mouse model of femoral fracture: an animal study. Bone Joint Res. 2019;8:550–562. doi: 10.1302/2046-3758.811.BJR-2018-0251.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Y, Kang Q, Luo Q, Jiang W, Si W, Liu BA, Luu HH, Park JK, Li X, Luo J, Montag AG, Haydon RC, He TC. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279:32941–32949. doi: 10.1074/jbc.M403344200. [DOI] [PubMed] [Google Scholar]
- 40.Carlier A, Chai YC, Moesen M, Theys T, Schrooten J, Van Oosterwyck H, Geris L. Designing optimal calcium phosphate scaffold-cell combinations using an integrative model-based approach. Acta Biomater. 2011;7:3573–3585. doi: 10.1016/j.actbio.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Viti F, Landini M, Mezzelani A, Petecchia L, Milanesi L, Scaglione S. Osteogenic differentiation of MSC through calcium signaling activation: transcriptomics and functional analysis. PLoS One. 2016;11:e0148173. doi: 10.1371/journal.pone.0148173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol. 2007;176:709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Zhuang Y, Ouyang N, Yu H. Cytochalasin D promotes osteogenic differentiation of MC3T3-E1 cells via p38-MAPK signaling pathway. Curr Mol Med. 2019;20:79–88. doi: 10.2174/1566524019666191007104816. [DOI] [PubMed] [Google Scholar]
- 44.Mei Y, Bian C, Li J, Du Z, Zhou H, Yang Z, Zhao RC. miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J Cell Biochem. 2013;114:1374–1384. doi: 10.1002/jcb.24479. [DOI] [PubMed] [Google Scholar]
- 45.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 46.Chen G, Deng C, Li YP. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012;8:272–288. doi: 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu DD, Zhang CY, Liu Y, Li J, Wang YX, Zheng SG. RUNX2 regulates osteoblast differentiation via the BMP4 signaling pathway. J Dent Res. 2022;101:1227–1237. doi: 10.1177/00220345221093518. [DOI] [PubMed] [Google Scholar]
- 48.Krause C, Guzman A, Knaus P. Noggin. Int J Biochem Cell Biol. 2011;43:478–481. doi: 10.1016/j.biocel.2011.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.