Abstract
Background: Previous investigations into the correlation between tumor necrosis factor-α (TNF-α) gene polymorphism and susceptibility to angina have produced a range of results. This study aims to clarify the potential significance of TNF-α polymorphism as a contributing factor to the onset of angina. Methods: A comprehensive review of all corresponding literature was conducted across several major databases, covering their records from inception through to March 2024. Data were synthesized using meta-analysis techniques. Results: Notable associations were identified in various genetic comparisons: allele contrast (A vs. G: OR=0.77, 95% CI=0.70-0.86, P<0.001), homozygote comparison (AA vs. GG: OR=3.35, 95% CI=2.51-4.48, P<0.001), heterozygote comparison (GA vs. GG: OR=1.69, 95% CI=1.19-2.40, P=0.003), recessive genetic model (AA vs. GG/GA: OR=2.40, 95% CI=1.91-3.03, P<0.001), and dominant genetic model (AA/GA vs. GG: OR=1.84, 95% CI=1.27-2.65, P=0.001). Furthermore, ethnicity-based subgroup analyzes revealed that these associations were especially significant within the Asian population and unstable angina, with all genetic models demonstrating statistical significance (P<0.05). Conclusions: TNF-α gene is significantly associated with an increased risk of angina, particularly among the Asian population and unstable angina. This finding underscores the importance of considering genetic factors in the assessment and management of angina risk.
Keywords: Angina, TNF-α, gene polymorphism, meta-analysis
Introduction
Coronary Atherosclerotic Heart Disease (CHD), also known as coronary heart disease, is primarily caused by the narrowing of blood vessels due to atherosclerosis. This condition manifests itself through various clinical characteristics, including stable or unstable angina pectoris, heart failure, and arrhythmias [1]. Various risk factors contribute to coronary heart disease, such as hypertension, diabetes, elevated blood cholesterol levels, smoking, and poor dietary habits. CHD, particularly angina pectoris, is distinguished by its high rates of morbidity and mortality, which require diverse treatment strategies and prognoses based on different risk stratifications [2]. Furthermore, in elderly patients, the association of coronary heart disease with other chronic diseases such as hypertension and diabetes complicate clinical management and treatment, emphasizing the importance of early diagnosis and accurate disease evaluation to improve patient outcomes.
Coronary angiography (CAG) is recognized as the gold standard clinically for diagnosing coronary heart disease, capable of qualitatively and quantitatively assessing angina. However, its invasive nature makes it unsuitable for widespread screening purposes. Furthermore, the propensity to overtreat patients diagnosed via CAG, coupled with an increased incidence of postoperative adverse cardiovascular events, not only increases treatment costs, but also imposes a significant economic burden on patients. Consequently, there is an urgent need for reliable indicators capable of accurately predicting the risk of angina. With the deepening of the research on the pathogenesis of coronary heart disease, more and more research has been done on cytokines involved in the inflammatory response, and their functions have been given more and more attention. High plasma tumor necrosis factor-α (TNF-α) level in patients with coronary heart disease is associated with a higher risk of recurrent cardiovascular accidents [3,4]. TNF-α can directly damage vascular endothelial cells, increase their permeability, blood cholesterol easily penetrates the intima and deposits in the blood vessels, promoting the adhesion of monocytes to endothelial cells, promoting the migration and proliferation of vascular smooth muscle cells (VSMC), and participating in the occurrence and development of acute coronary syndrome [5,6]. TNF-α can also induce endothelial cells to release endothelin, causing endothelial damage and coronary artery spasms, thereby promoting platelet aggregation and thrombosis. TNF-α has an obvious procoagulant effect on endothelial cells [7]. It changes the anticoagulant activity of endothelial cells by stimulating the synthesis of thrombin, and stimulates the synthesis of platelet-activating factor to promote thrombosis. Inflammatory factors play an important role in the occurrence and development of atherosclerosis. TNF-α is an important predictor of the severity of coronary heart disease. The inflammatory marker TNF-α is significantly elevated in the acute stage of coronary artery disease [8].
In recent years, extensive literature has highlighted the impact of gene polymorphisms on the risk of developing angina pectoris. Tumor Necrosis Factor α (TNF-α), a multifunctional pro-inflammatory cytokine, is quite crucial in multiple important processes, including apoptosis, lipid metabolism, and the progression of chronic inflammation and malignancies. Polymorphisms within the TNF-α promoter region, especially at the -308 locus, which can undergo a conversion from G to A, have been extensively investigated. Such single nucleotide polymorphism changes could influence the gene’s transcription level, potentially affecting disease pathogenesis.
The findings of previous reports have been markedly inconsistent. Therefore, this study seeks to investigate the relationship through a comprehensive meta-analysis. This effort aims to provide valuable information for the prevention and treatment of angina, contributing to a more nuanced understanding of its genetic roots.
Materials and methods
Data collection
A systematic search was carried out by two independent authors in a selection of major databases. This search spanned from the inception of each database up to March 2024, ensuring a thorough inclusion of relevant literature. The keywords used for this extensive search encompassed “polymorphism” or “polymorphisms”, “angina”, “TNF-α”, and “tumor necrosis factor-α”, with a focus on the literature published in PubMed to maximize the scope of our research.
Inclusion and exclusion criteria
The inclusion criteria were precisely defined: (a) Case-control studies investigating the relationship between TNF-α rs1800629 -308 G/A polymorphism and the risk of angina; other types of studies were not included, including randomized controlled trials, cohort studies, reviews and case report. (b) Studies providing sufficient data. (c) Studies investigating the population with angina. Studies were not enrolled if they investigated other cardiovascular diseases including myocardial infarction, ischemic heart disease, stroke, cardiomyopathy, rheumatic heart disease, hypertensive heart disease, and endocarditis. Exclusion criteria were set to omit (a) studies not adhering to a case-control design; (b) studies lacking adequate data for the computation of OR and 95% CI computation; and (c) studies where the primary subjects were animals, ensuring that the focus remained on human-related research.
Data extraction and methodological quality assessment
The data and pertinent information were meticulously extracted and evaluated by the first and second authors, strictly adhering to the established inclusion and exclusion criteria. Essential elements extracted included the name of the literature, genotyping methods, control sources, and Newcastle-Ottawa Scale (NOS) scores. All studies included in the present meta-analysis follow the criteria for angina with authoritative references. In instances of disagreement between the two authors, a discussion was initiated with the corresponding author to reach a consensus. Given the nature of this meta-analysis, which involves the aggregation of existing data, the requirement for ethics approval and patient consent was deemed unnecessary.
Statistical analysis
To determine the distribution differences among TNF-α polymorphism genotypes and alleles of the TNF-polymorphism, the chi-square test (X2) based Q test was used. If P<0.1 or I2>50% of the heterogeneity test, suggesting the existence of heterogeneity, a random-effects model (the DerSimonian and Laird method) was applied to count the summary ORs; otherwise, If P≥0.1 and I2≤50% of the heterogeneity test, suggesting no existence of heterogeneity, the fixed-effects model (the Mantel-Haenszel method) was applied. Hardy-Weinberg equilibrium (HWE) for control groups was assessed using the chi-square test to ensure the integrity of the baseline distribution. Odds ratios (OR) and 95% confidence intervals (CI) were performed using logistic regression modeling, providing a measure of the strength of the association between TNF-α polymorphism and angina risk. Heterogeneity among included studies was evaluated using the Q-statistics and I-2 statistics, which guided the decision to use a fixed-effects or random-effects model based on the degree of heterogeneity observed. The meta-regression and subgroup analysis were applied to find the source of heterogeneity. The characteristics were enrolled as covariates in the meta-regression including genotyping methods (TaqMan vs. PCR-RFLP), ethnicity (Caucasian vs. Asian), source of controls (HB vs. PB), quality scores (High-quality vs. Low-quality) and HWE conformity (Yes vs. No). Sensitivity analysis and the evaluation of publication bias were performed following protocols established in previous meta-analyses [9-22]. Stata 15.0 and PRISMA 2009 checklist were used, ensuring a rigorous and transparent reporting of our meta-analysis methodology and the whole meta-analysis was based on the planned population, intervention, comparison, and outcome (PICO) elements. The study has been registered with the INPLASY (registration number: INPLASY202470120). The present study was implemented by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
Results
Overview of study selection
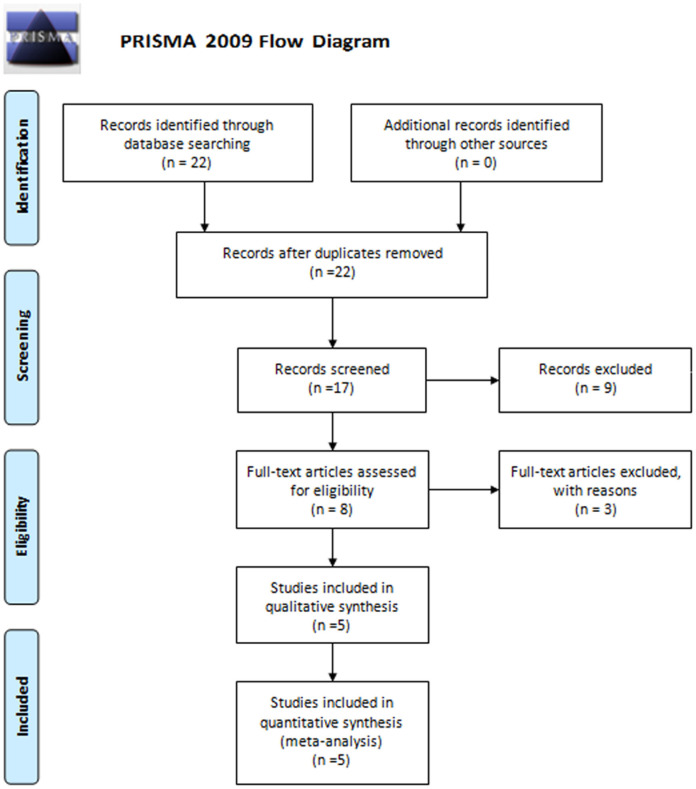
The selection process is depicted through a PRISMA 2009 flow diagram (Figure 1). Detailed information on all the included literature is presented in Table 1. In total, five studies were selected for inclusion in this meta-analysis [23-27].
Figure 1.
Flow diagram of study selection for the TNF-α polymorphism on angina risk. TNF-α: tumor necrosis factor-α.
Table 1.
General information of eligible studies enrolled in the meta-analysis
| Literature | Ethnics (Country) | Genotyping method | Control Origin | type | Matching standard | HWE conformity | NOS |
|---|---|---|---|---|---|---|---|
| Virginie (2003a) | Caucasian (France) | PCR-RFLP | PB | SA | Age, sex, ethnicity | Yes | 8 |
| Virginie (2003b) | Hispanics (France) | PCR-RFLP | PB | UA | Age, sex, ethnicity | Yes | 8 |
| Athanassios (2008a) | African (Greece) | PCR-SSP | HB | SA | Age, sex, ethnicity | Yes | 7 |
| Athanassios (2008b) | Caucasian (Greece) | PCR-SSP | HB | UA | Age, sex, ethnicity | Yes | 7 |
| Liu (2009a) | Asian (China) | PCR-RFLP | PB | SA | Age, sex, ethnicity | Yes | 9 |
| Liu (2009b) | Asian (China) | PCR-RFLP | PB | UA | Age, sex, ethnicity | Yes | 9 |
| Shevchenko (2010a) | Caucasian (Russia) | PCR-RFLP | HB | SA | Age, sex, ethnicity | Yes | 8 |
| Shevchenko (2010b) | African (Russia) | PCR-RFLP | HB | UA | Age, sex, ethnicity | Yes | 8 |
| Abdulfattah (2023a) | Asian (Iraqi) | TaqMan | PB | SA | Age, sex, ethnicity | Yes | 9 |
| Abdulfattah (2023b) | Asian (Iraqi) | TaqMan | HB | UA | Age, sex, ethnicity | Yes | 9 |
PB, Population-based; HB, Hospital-based; HWE, Hardy-Weinberg equilibrium; NOS, Newcastle-Ottawa scale.
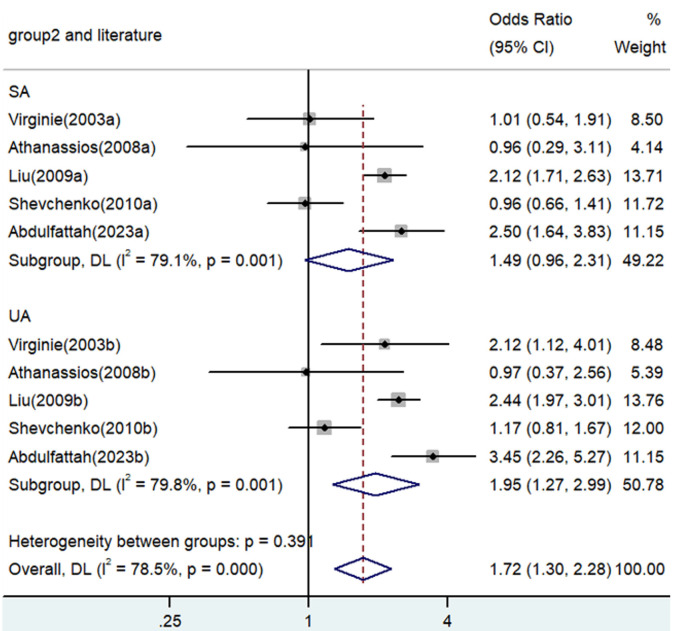
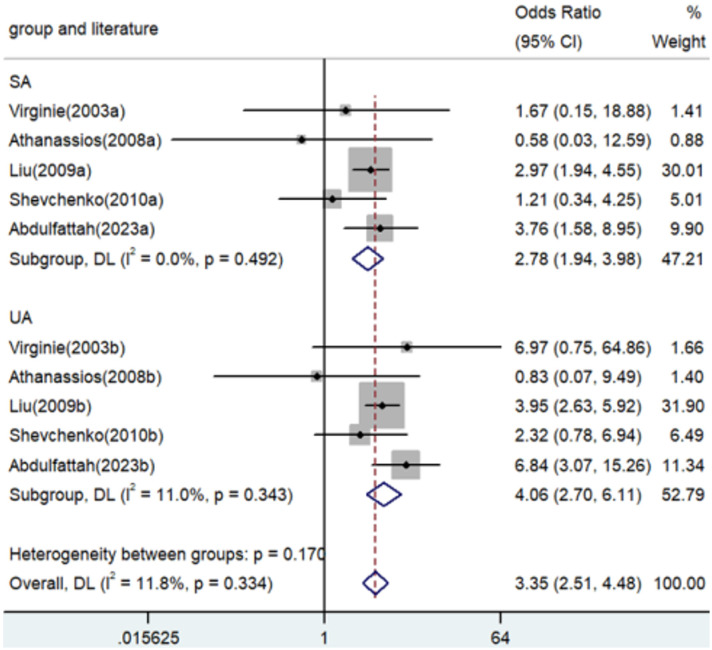
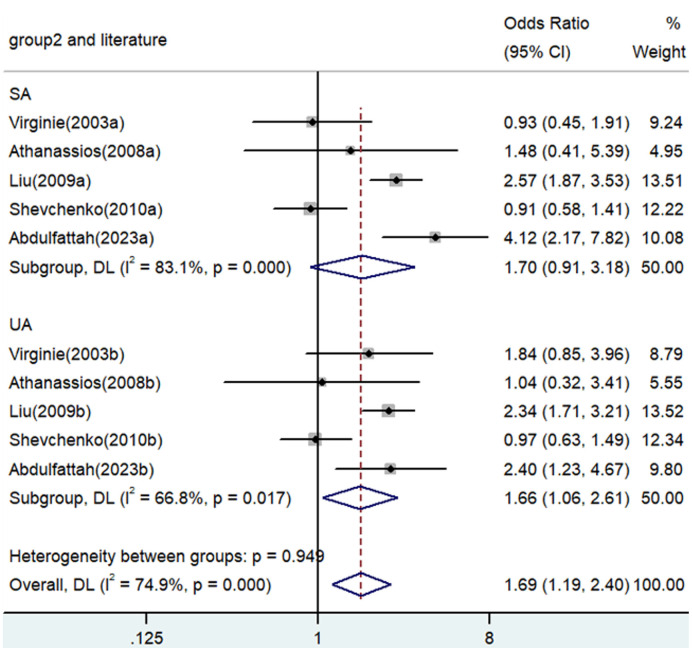
Association between TNF-α polymorphism and risk of angina
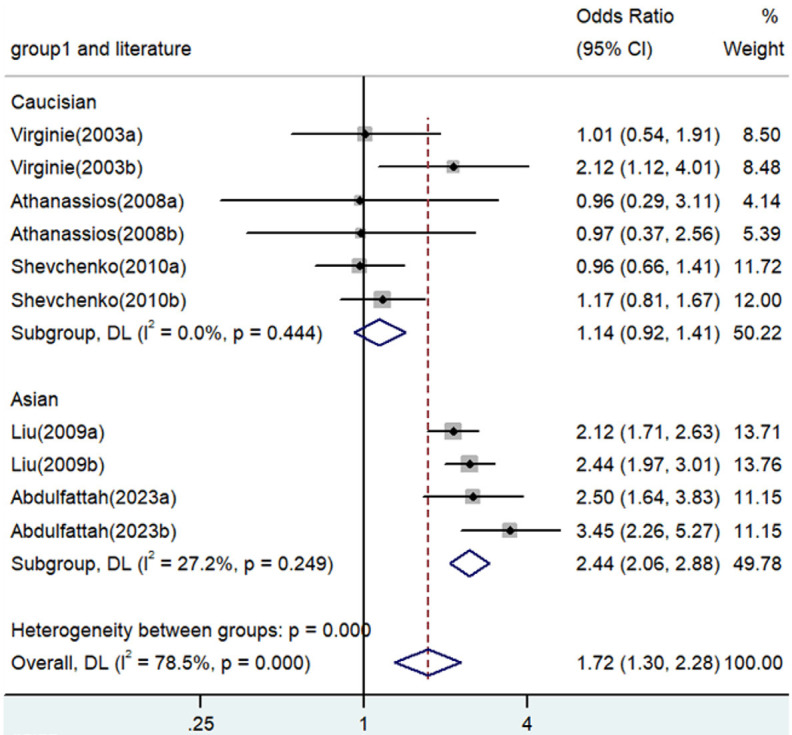
Significant associations between TNF-α polymorphisms and angina risk were identified in various genetic contrasts (all P<0.05) (Figures 2, 3, 4, 5 and 6). Furthermore, subgroup analyzes based on ethnicity and type of angina revealed that these associations were especially pronounced within the Asian population and unstable angina (Figures 2, 3, 4, 5, 6, 7, 8, 9, 10 and 11), with all genetic models showing significant associations (P<0.05), as detailed in Tables 2 and 3.
Figure 2.
Forest plot of the associations between TNF-α polymorphism and angina risk through allele contrast (A vs. G). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 3.
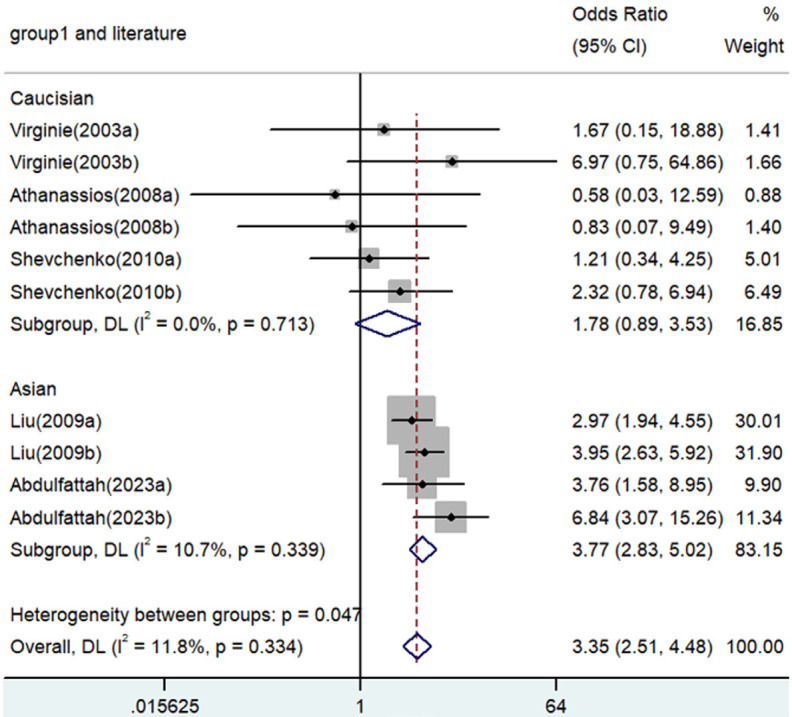
Forest plot of associations between TNF-α polymorphism and angina risk through homozygote comparison (AA vs. GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 4.
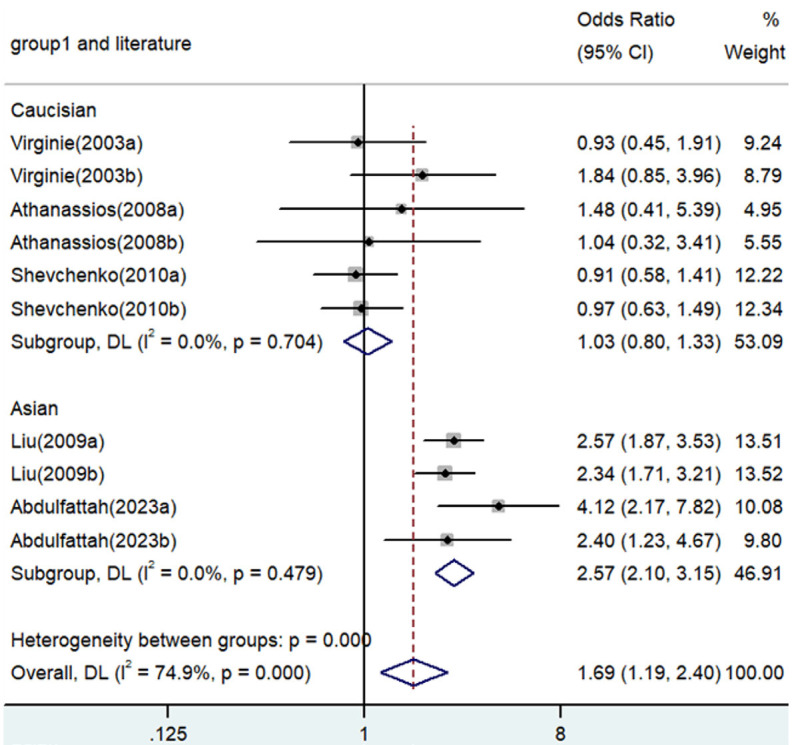
Forest plot for the associations between TNF-α polymorphism and angina risk through a heterozygous comparison (GA vs. GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 5.
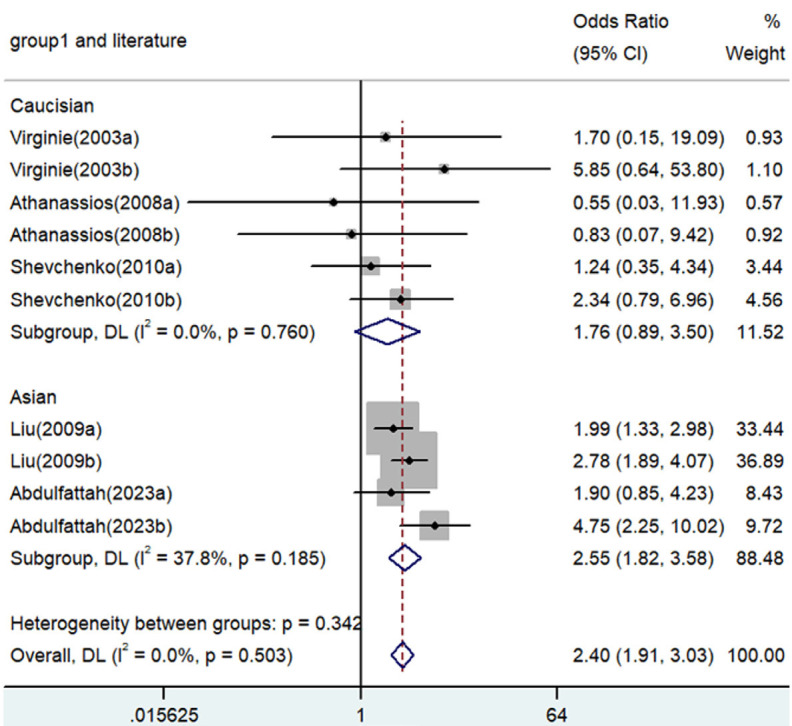
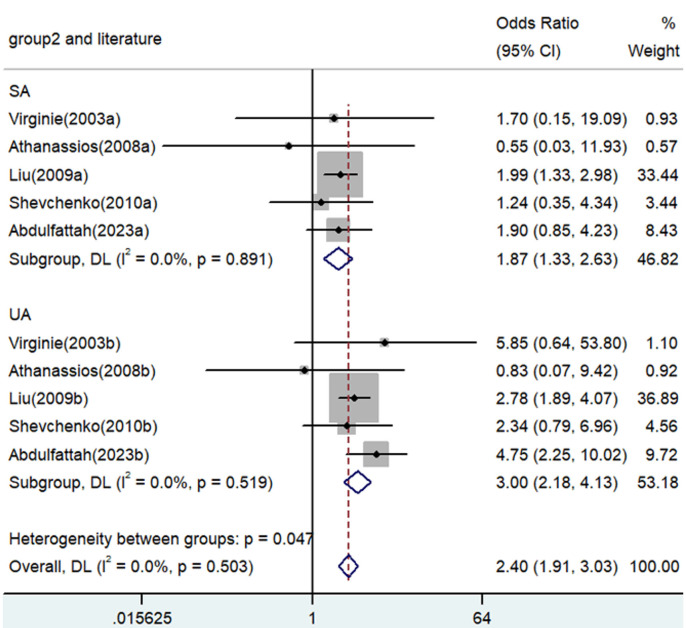
Forest plot for the associations between TNF-α polymorphism and angina risk through the recessive genetic model (AA vs. GA/GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 6.
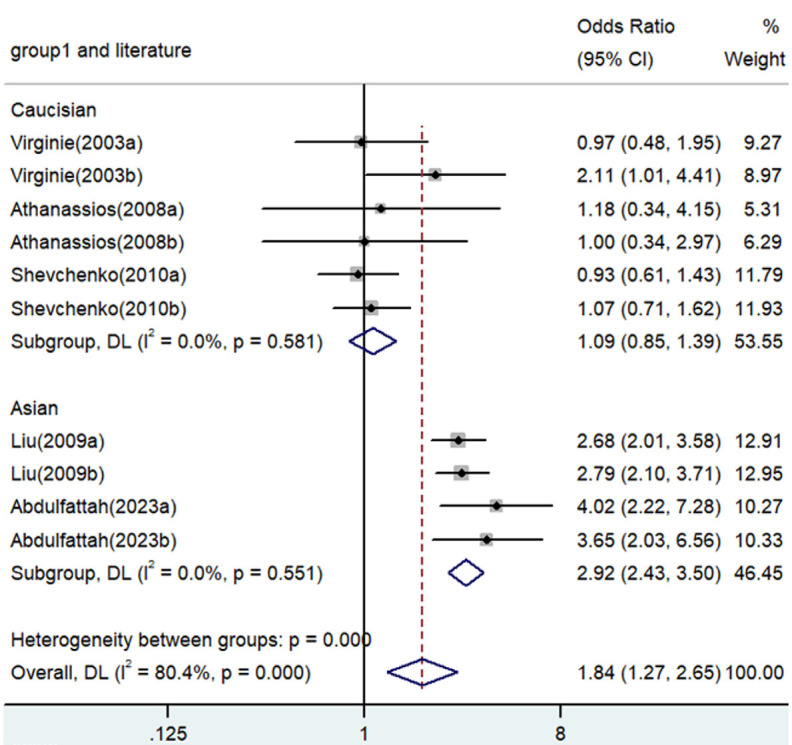
Forest plot for the associations between TNF-α polymorphism and angina risk through the dominant genetic model (AA/GA vs. GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 7.
Forest plot for the associations between TNF-α polymorphism and angina risk through the dominant genetic model based on type of angina (AA/GA vs. GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 8.
Forest plot for the associations between TNF-α polymorphism and angina risk through the dominant genetic model based on type of angina (AA/GA vs. GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 9.
Forest plot for the associations between TNF-α polymorphism and angina risk through the dominant genetic model based on type of angina (AA/GA vs. GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 10.
Forest plot for the associations between TNF-α polymorphism and angina risk through the dominant genetic model based on type of angina (AA/GA vs. GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Figure 11.
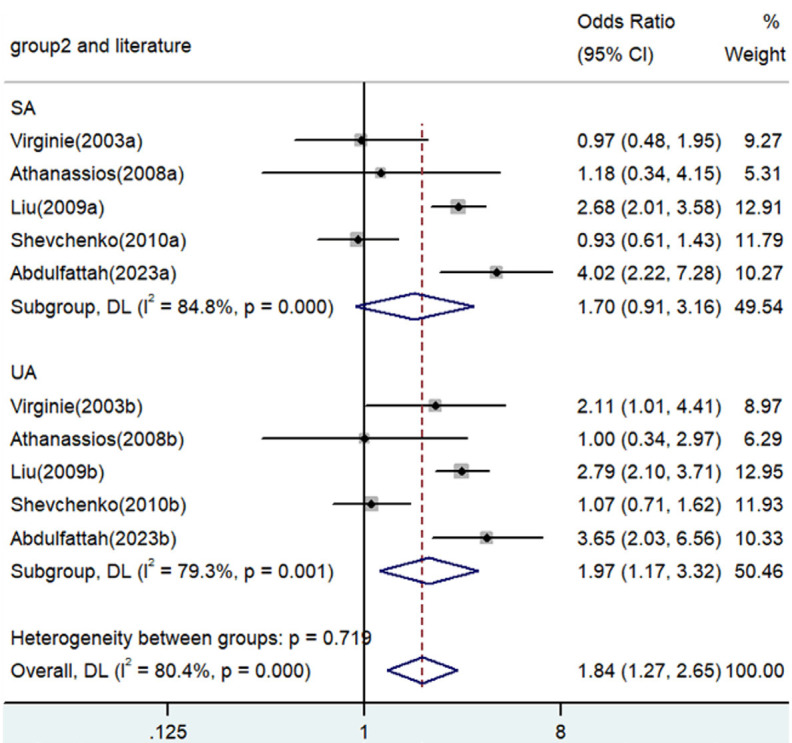
Forest plot for the associations between TNF-α polymorphism and angina risk through the dominant genetic model based on type of angina (AA/GA vs. GG). OR, odds ratio; CI, confidence interval. TNF-α: tumor necrosis factor-α.
Table 2.
Quality assessment of case-control studies according to the Newcastle-Ottawa scale
| Literature | Selection of enrolled study subjects | Between-group comparability | Exposure outcomes and factors | Total |
|---|---|---|---|---|
| Virginie (2003a) | 2 | 3 | 3 | 8 |
| Virginie (2003b) | 2 | 3 | 3 | 8 |
| Athanassios (2008a) | 2 | 3 | 2 | 7 |
| Athanassios (2008b) | 2 | 3 | 2 | 7 |
| Liu (2009a) | 3 | 3 | 3 | 9 |
| Liu (2009b) | 3 | 3 | 3 | 9 |
| Shevchenko (2010a) | 3 | 3 | 2 | 8 |
| Shevchenko (2010b) | 3 | 3 | 2 | 8 |
| Abdulfattah (2023a) | 3 | 3 | 3 | 9 |
| Abdulfattah (2023b) | 3 | 3 | 3 | 9 |
| Average | 2.6 | 3.0 | 2.6 | 8.2 |
Table 3.
Meta-analysis of the TNF-α polymorphism and angina risk
| Comparison | Population | N | Test of association | Mode | Test of heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| OR | 95% CI | P | χ2 | P | I2 | ||||
| A versus. G | Overall | 10 | 1.72 | 1.30-2.28 | <0.001 | Random | 41.81 | 0 | 78.5 |
| Caucasian | 6 | 1.14 | 0.92-1.41 | 0.243 | Fixed | 4.77 | 0.444 | 0 | |
| Asian | 4 | 2.44 | 2.06-2.88 | <0.001 | Fixed | 4.12 | 0.249 | 27.2 | |
| SA | 5 | 1.49 | 0.96-2.31 | 0.076 | Random | 19.18 | 0.001 | 79.1 | |
| UA | 5 | 1.95 | 1.27-2.99 | 0.002 | Random | 19.81 | 0.001 | 79.8 | |
| AA versus. GG | Overall | 10 | 3.35 | 2.51-4.48 | <0.001 | Fixed | 10.12 | 0.334 | 11.8 |
| Caucasian | 6 | 1.78 | 0.89-3.53 | 0.102 | Fixed | 2.91 | 0.713 | 0 | |
| Asian | 4 | 3.77 | 2.83-5.02 | <0.001 | Fixed | 3.36 | 0.339 | 10.7 | |
| SA | 5 | 2.78 | 1.94-3.98 | <0.001 | Fixed | 3.41 | 0.492 | 0 | |
| UA | 5 | 4.06 | 2.70-6.11 | <0.001 | Fixed | 4.49 | 0.343 | 11.0 | |
| GA versus. GG | Overall | 10 | 1.69 | 1.19-2.40 | 0.003 | Random | 35.86 | 0 | 74.9 |
| Caucasian | 6 | 1.03 | 0.80-1.33 | 0.811 | Fixed | 2.98 | 0.704 | 0 | |
| Asian | 4 | 2.57 | 2.10-3.15 | <0.001 | Fixed | 2.48 | 0.479 | 0 | |
| SA | 5 | 1.70 | 0.91-3.18 | 0.094 | Random | 23.70 | 0 | 83.1 | |
| UA | 5 | 1.66 | 1.06-2.61 | 0.027 | Random | 12.05 | 0.017 | 66.8 | |
| AA versus. GA/GG | Overall | 10 | 2.40 | 1.91-3.03 | <0.001 | Fixed | 8.32 | 0.503 | 0 |
| Caucasian | 6 | 1.76 | 0.89-3.50 | 0.105 | Fixed | 2.61 | 0.760 | 0 | |
| Asian | 4 | 2.55 | 1.82-3.58 | <0.001 | Fixed | 4.82 | 0.185 | 37.8 | |
| SA | 5 | 1.87 | 1.33-2.63 | <0.001 | Fixed | 1.12 | 0.891 | 0 | |
| UA | 5 | 3.00 | 2.18-4.13 | <0.001 | Fixed | 3.24 | 0.519 | 0 | |
| AA/GA versus. GG | Overall | 10 | 1.84 | 1.27-2.65 | 0.001 | Random | 46.02 | 0 | 80.4 |
| Caucasian | 6 | 1.09 | 0.85-1.39 | 0.494 | Fixed | 3.78 | 0.581 | 0 | |
| Asian | 4 | 2.92 | 2.43-3.50 | <0.001 | Fixed | 2.10 | 0.551 | 0 | |
| SA | 5 | 1.70 | 0.91-3.16 | 0.096 | Random | 26.34 | 0 | 84.8 | |
| UA | 5 | 1.97 | 1.17-3.12 | 0.011 | Random | 19.28 | 0.001 | 79.3 | |
SA, stable angina; UA, unstable angina; OR, odds ratio; CI, confidence interval; TNF-α, tumor necrosis factor-α.
Evaluation of between-studies heterogeneity
Analysis of the overall population revealed significant heterogeneity in three genetic models. To identify the source of heterogeneity, subgroup analyzes were performed on the basis of ethnicity and type of angina. The results of this subgroup analyze identified ethnicities as the main source of heterogeneity.
Sensitivity analysis and publication bias
Stability was achieved through sensitivity analysis, which involved excluding each study one by one. Overall, the findings remained consistent, underscoring the robustness of our meta-analysis. Additionally, a funnel plot could not find any evident asymmetry, and Egger’s test confirmed the reliability of these findings (P=0.760), indicating that there was no significant publication bias.
Discussion
Stable angina pectoris, characterized by a significant reduction in blood supply to the myocardium due to coronary artery stenosis, leads to rapid ischemia and hypoxia under increased myocardial load. This condition manifests clinically as chest tightness and pain, which typically subsides after rest or sublingual administration of nitroglycerin, a hallmark of coronary heart disease. The 2020 China Cardiovascular Disease and Health Report indicates an increasing trend in the prevalence and mortality of coronary heart disease within the country. Specifically, in 2019, mortality rates in urban and rural areas of China were reported to be 121.59/100,000 and 130.14/100,000, respectively, with a higher incidence in men than in women. This trend underscores the increasing economic and health burden of this condition on society. Chronic stable angina pectoris, characterized by consistent symptoms without significant fluctuation in the frequency of episodes or pain intensity, is the result of temporary myocardial ischemia. Prolongation of this disease without effective treatment can lead to complications such as arrhythmia, heart failure, and acute coronary syndrome. In particular, unstable angina pectoris, which bridges stable angina and acute myocardial infarction, has a higher annual mortality rate, highlighting the critical need for its prevention and management.
This study has produced positive findings in the general population, identifying ethnicity as a primary source of heterogeneity. We have discovered that the TNF-α gene significantly increases the risk of angina, especially among the Asian population, despite the fact that some heterogeneity was observed in the results for unstable angina (UA) and stable angina (SA), which could potentially compromise the reliability of these findings. To our knowledge, this meta-analysis is the first exploration of the association between TNF-α gene polymorphisms and angina risk, highlighting a significant correlation within the Asian demographic. TNF-α is a kind of cytokine with various biological activities, mainly involved in anti-infection, immune regulation and enhancement of monocyte activity, and it plays an important role in the immune defense system of the body. TNF-α is mainly secreted by macrophages, and promotes its own synthesis in an autocrine way, concentrated in the lesion site, and various infections and external stimuli can cause the increase of TNF-α expression. In physiological state or activated state TNF-α can inhibit collagen gene expression in vascular smooth muscle cells and can cause plaque instability. TNF-α can promote the expression of proto-oncogenes, produce platelet growth factors, make the coagulation and anticoagulation system unbalanced, promote the formation of thrombosis, and cause the occurrence of angina pectoris. In addition, TNF-α gene polymorphisms may increase or reduce TNF-α expression, which in turn affects the probability or risk of angina.
This study has a few areas for improvement. Expanding the diversity of the included studies to comprise more diverse ethnic groups, such as African and other populations would potentially enhance the comprehensiveness and applicability of our meta-analysis. Furthermore, the impact of several confounding factors remains assessed due to the paucity of data, coupled with the relatively small overall sample size. Thirdly, Mendelian randomization has not been applied in the present meta-analysis. Mendelian randomization of SNPs is a randomization method based on single nucleotide polymorphisms used to conduct causal inference in genetic association studies and genomics studies. Based on Mendelian laws of genetics, the genetic information in a parent’s genome is distributed in a random manner among the offspring. In SNP mendelian randomization, researchers use known associations between SNPS and a particular trait or disease to randomly group individuals according to their genotype. In future studies, we will model the effects of randomization on a disease or specific trait to infer a causal relationship between this SNP and that disease or trait. Lastly, we only investigate the -308 locus of the TNF-α gene. Although the -308 locus is the most popular and the most studied of all the sites, we are still committed to investigating more sites in future studies, to make our results more convincing, so as to better understand the etiology and risk of angina.
In conclusion, our analysis suggests a notable association between TNF-α gene polymorphisms and an increased risk of angina within the Asian population and unstable angina. Further research is essential to validate these findings and explore the underlying mechanisms of this association, potentially paving the way for improved diagnostic and therapeutic strategies for angina in diverse populations.
Acknowledgements
We thank everyone in our experimental group for helping us in providing valuable feedback, advice, and comments.
Disclosure of conflict of interest
None.
References
- 1.Li H, Sun K, Zhao R, Hu J, Hao Z, Wang F, Lu Y, Liu F, Zhang Y. Inflammatory biomarkers of coronary heart disease. Front Biosci (Schol Ed) 2018;10:185–196. doi: 10.2741/s508. [DOI] [PubMed] [Google Scholar]
- 2.Dong Y, Chen H, Gao J, Liu Y, Li J, Wang J. Molecular machinery and interplay of apoptosis and autophagy in coronary heart disease. J Mol Cell Cardiol. 2019;136:27–41. doi: 10.1016/j.yjmcc.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y, Wang L, Zhan Y, Zhang Z, Chen D, Xiang Y, Xie C. The expression of SAH, IL-1beta, Hcy, TNF-alpha and BDNF in coronary heart disease and its relationship with the severity of coronary stenosis. BMC Cardiovasc Disord. 2022;22:101. doi: 10.1186/s12872-021-02388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mir R, Elfaki I, Jha CK, Javid J, Babakr AT, Banu S, Mir MM, Jamwal D, Khullar N, Alzahrani KJ, Chahal SMS. Biological and clinical implications of TNF-alpha promoter and CYP1B1 gene variations in coronary artery disease susceptibility. Cardiovasc Hematol Disord Drug Targets. 2021;21:266–277. doi: 10.2174/1871529X22666211221151830. [DOI] [PubMed] [Google Scholar]
- 5.Gagat M, Zielinska W, Mikolajczyk K, Zabrzynski J, Krajewski A, Klimaszewska-Wisniewska A, Grzanka D, Grzanka A. CRISPR-based activation of endogenous expression of TPM1 inhibits inflammatory response of primary human coronary artery endothelial and smooth muscle cells induced by recombinant human tumor necrosis factor alpha. Front Cell Dev Biol. 2021;9:668032. doi: 10.3389/fcell.2021.668032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uddin MS, Kabir MT, Jakaria M, Mamun AA, Niaz K, Amran MS, Barreto GE, Ashraf GM. Endothelial PPARgamma is crucial for averting age-related vascular dysfunction by stalling oxidative stress and ROCK. Neurotox Res. 2019;36:583–601. doi: 10.1007/s12640-019-00047-5. [DOI] [PubMed] [Google Scholar]
- 7.Gao H, Zhang Q, Chen J, Cooper DKC, Hara H, Chen P, Wei L, Zhao Y, Xu J, Li Z, Cai Z, Luan S, Mou L. Porcine IL-6, IL-1beta, and TNF-alpha regulate the expression of pro-inflammatory-related genes and tissue factor in human umbilical vein endothelial cells. Xenotransplantation. 2018;25:e12408. doi: 10.1111/xen.12408. [DOI] [PubMed] [Google Scholar]
- 8.Sweigert PJ, Bansal VK, Hoppensteadt DA, Saluk JL, Syed DA, Fareed J. Inflammatory and metabolic syndrome biomarker analysis of vascular outcomes in end-stage renal disease. Int J Angiol. 2017;26:43–48. doi: 10.1055/s-0036-1593409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Wang Z, Yan Y, Li P, Yang Z, Qin L, Mo W. XRCC3 C18067T polymorphism contributes a decreased risk to both basal cell carcinoma and squamous cell carcinoma: evidence from a meta-analysis. PLoS One. 2014;9:e84195. doi: 10.1371/journal.pone.0084195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin LY, Chen X, Li P, Yang Z, Mo WN. Association between the XRCC3 Thr241Met polymorphism and cervical cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 2014;14:6703–6707. doi: 10.7314/apjcp.2013.14.11.6703. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Chen X, Liu B, Li S, Liu M, Xue H. Quantitative assessment of the associations between DNA repair gene XRCC3 Thr241Met polymorphism and gastric cancer. Tumour Biol. 2014;35:1589–1598. doi: 10.1007/s13277-013-1219-8. [DOI] [PubMed] [Google Scholar]
- 12.Yan Y, Chen X, Li T, Li M, Liang H. Association of OGG1 Ser326Cys polymorphism and pancreatic cancer susceptibility: evidence from a meta-analysis. Tumour Biol. 2014;35:2397–2402. doi: 10.1007/s13277-013-1317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Yan Y, Li P, Yang Z, Qin L, Mo W. Association of GSTP1 -313A/G polymorphisms and endometriosis risk: a meta-analysis of case-control studies. Eur J Obstet Gynecol Reprod Biol. 2013;171:362–367. doi: 10.1016/j.ejogrb.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Mo W, Peng Q, Su X. Lack of association between Fas rs180082 polymorphism and risk of cervical cancer: an update by meta-analysis. BMC Med Genet. 2013;14:71. doi: 10.1186/1471-2350-14-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Si D, Yao Y, Chen X, Qiu J. Ethnicity-stratified analysis of the association between P53 rs1042522 polymorphism and women HPV infection: a meta-analysis. Microb Pathog. 2021;161:105099. doi: 10.1016/j.micpath.2021.105099. [DOI] [PubMed] [Google Scholar]
- 16.Niu K, Chen X, Lu Y. COL3A1 rs1800255 polymorphism is associated with pelvic organ prolapse susceptibility in caucasian individuals: evidence from a meta-analysis. PLoS One. 2021;16:e0250943. doi: 10.1371/journal.pone.0250943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin X, Wu Y, Yin S, Chen X, Zhang Y. Association between the IL-10 and IL-6 polymorphisms and brucellosis susceptibility: a meta-analysis. BMC Med Genet. 2020;21:63. doi: 10.1186/s12881-020-01006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin X, Yin S, Zhang Y, Chen X. Association between TLR2 Arg677Trp polymorphism and tuberculosis susceptibility: a meta-analysis. Microb Pathog. 2020;144:104173. doi: 10.1016/j.micpath.2020.104173. [DOI] [PubMed] [Google Scholar]
- 19.Yuanyuan G, Xue Y, Yachao L, Xiao F, Xu C. Association between IL-18 -607 C/A polymorphism and the risk of prostate cancer: a meta-analysis of case-control studies. Asian Pac J Cancer Prev. 2019;20:1595–1602. doi: 10.31557/APJCP.2019.20.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin X, Yin S, Zhang Y, Chen X. Quantitative assessment of the association between IL-10 -592 A/C polymorphism and Kawasaki disease risk in Chinese population: evidence from a meta-analysis. Cardiol Young. 2018;28:811–815. doi: 10.1017/S1047951118000380. [DOI] [PubMed] [Google Scholar]
- 21.Jin X, Yin S, Zhang Y, Chen X. Association between TLR2 + 2477G/A polymorphism and bacterial meningitis: a meta-analysis. Epidemiol Infect. 2018;146:642–647. doi: 10.1017/S0950268818000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Jiang M, Zhao RK, Gu GH. Quantitative assessment of the association between ABC polymorphisms and osteosarcoma response: a meta-analysis. Asian Pac J Cancer Prev. 2015;16:4659–4664. doi: 10.7314/apjcp.2015.16.11.4659. [DOI] [PubMed] [Google Scholar]
- 23.Abdulfattah SY, Samawi FT. Estimating the role of single-nucleotide polymorphism (rs1800629)-308 G/A of TNF-alpha gene as genetic marker associated with angina pectoris in a sample of Iraqi patients. J Genet Eng Biotechnol. 2023;21:2. doi: 10.1186/s43141-022-00454-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernard V, Pillois X, Dubus I, Benchimol D, Labouyrie JP, Couffinhal T, Coste P, Bonnet J. The -308 G/A tumor necrosis factor-alpha gene dimorphism: a risk factor for unstable angina. Clin Chem Lab Med. 2003;41:511–516. doi: 10.1515/CCLM.2003.077. [DOI] [PubMed] [Google Scholar]
- 25.Manginas A, Tsiavou A, Chaidaroglou A, Giamouzis G, Degiannis D, Panagiotakos D, Cokkinos DV. Inflammatory cytokine gene variants in coronary artery disease patients in Greece. Coron Artery Dis. 2008;19:575–582. doi: 10.1097/MCA.0b013e32831286e8. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Jin W, Lu L, Chen QJ, Shen WF. Association between the -1031T/C polymorphism in tumor necrosis factor-alpha gene and unstable angina. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26:571–574. doi: 10.3760/cma.j.issn.1003-9406.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 27.Shevchenko AV, Golovanova OV, Konenkov VI, Tolkacheva OM, Romashchenko AG, Maksimov VN, Voevoda MI. Analysis of polymorphism of three positions of promoter region of TNF- gene in patients with ischemic heart disease, unstable angina and myocardial infarction. Kardiologiia. 2010;50:9–14. [PubMed] [Google Scholar]