Abstract
Objectives: The study aims to characterize BRCA1/2 mutations in Pakistani gastric cancer (GC) patients, identifying unique pathogenic variants and evaluating their potential as diagnostic biomarkers, while also exploring therapeutic avenues for personalized treatment strategies. Methodology: In this study, we investigated the role of Breast Cancer gene 1 (BRCA1) and Breast Cancer gene 2 (BRCA2) mutations in Pakistani GC patients and their functional implications using Next-Generation Sequencing (NGS). Results: Through NGS, we identified a total of 19 mutations in BRCA1 and 11 mutations in BRCA2, all with high mutation quality scores. In silico analysis revealed one pathogenic mutation in BRCA1 and one in BRCA2, indicating a potential link to disease development. Notably, two pathogenic mutations (BRCA1 p.Ala1823Ser and BRCA2 p.Gln92fs) were exclusively observed in the Pakistani GC population, suggesting unique genetic markers. Further examination utilizing The Cancer Genome Atlas (TCGA) data confirmed the absence of these mutations in non-Pakistani GC samples, emphasizing their specificity. Sanger sequencing validated these findings. Real Time Quantitative PCR (RT-qPCR) analysis indicated significantly decreased BRCA1/2 gene expression in samples harboring pathogenic mutations, suggesting their potential as biomarkers for GC diagnosis. Immunohistochemistry corroborated this by showing reduced protein expression levels in mutated samples. Enrichment analysis highlighted associations of BRCA1/2 genes with DNA damage response pathways and cancer-related processes. DrugBank exploration revealed potential therapeutic agents targeting mutated BRCA1/2, such as Cisplatin and Estradiol, offering new avenues for treatment. Conclusion: Our study identifies novel BRCA1/2 mutations specific to Pakistani GC patients, suggesting a distinct genetic susceptibility profile. These findings not only contribute to understanding GC pathogenesis but also hold implications for personalized treatment strategies in this population.
Keywords: BRCA genes, next generation sequencing, diagnosis, treatment
Introduction
Gastric cancer (GC), also termed stomach cancer, stands as a formidable global health challenge, significantly impacting morbidity and mortality rates [1,2]. It ranks fifth among the most prevalent cancers and is the third leading cause of cancer-related deaths worldwide, underscoring the urgency to delve into its molecular intricacies [3-6]. Recent strides in genomic research have revolutionized our comprehension of cancer, including GC, by illuminating the genetic alterations propelling tumorigenesis and affecting treatment responses [7-9]. One avenue of focus in this exploration is mutations within the BRCA1 and BRCA2 genes, initially associated with hereditary breast and ovarian cancer but increasingly implicated in various cancer types, including GC [10-12].
GC manifests as a heterogeneous disease with diverse histological subtypes, including intestinal, diffuse, and mixed forms [13,14]. Its etiology is multifaceted, involving a complex interplay of genetic and environmental factors. While extensive research has scrutinized environmental influences like diet, smoking, and Helicobacter pylori infection in gastric carcinogenesis, genetic factors are gaining recognition as pivotal contributors to its development [15-17]. Although somatic mutations in key driver genes such as TP53, APC, and CDH1 are well-documented in GC, the significance of BRCA1 and BRCA2 mutations in gastric tumorigenesis is increasingly acknowledged [11,18,19]. Mutations in these genes can disrupt DNA repair mechanisms, induce genome instability, and heighten susceptibility to cancer.
The prevalence of BRCA1 and BRCA2 mutations in GC exhibits variability across populations and geographic regions [20,21]. Studies report diverse mutation frequencies, suggesting that the contribution of BRCA1 and BRCA2 to GC susceptibility is intricate and potentially influenced by genetic and environmental factors [22-25]. Comprehending the prevalence and functional repercussions of these mutations is pivotal for tailoring personalized treatment strategies and identifying candidates for targeted therapies.
This comprehensive research endeavor aims to scrutinize the prevalence, spectrum, and clinical implications of BRCA1 and BRCA2 mutations in GC patients. Leveraging advanced Next-Generation Sequencing (NGS) technology, we conducted mutational analysis of these genes in a GC cohort. By delineating the mutational landscape of BRCA1 and BRCA2 in GC, our objective is to elucidate their potential roles as molecular biomarkers and therapeutic targets.
Methodology
Collection of the GC samples
Our study involved the meticulous collection of 42 GC tissue samples from patients undergoing surgical resection at the District Headquarter (DHQ), Teaching Hospital, Dera Ismail Khan, KPK, a distinguished healthcare institution in Pakistan. The procurement of these tissue specimens adhered strictly to Helsinki ethical guidelines and protocols [26], ensuring the safeguarding of patients’ rights and privacy. Prior to initiating this research, ethical clearance was diligently obtained from PMAS-Arid Agriculture University, Rawalpindi, Pakistan, underscoring our dedication to conducting the study with the highest standards of integrity and adherence to ethical principles.
The study’s inclusion criteria for GC patients comprised individuals with a confirmed diagnosis of GC, as histologically verified by biopsy reports, who demonstrated willingness to participate in the research. Exclusion criteria encompassed patients with a history of other malignancies, those undergoing treatment for GC, and individuals unable or unwilling to provide informed consent. Additionally, patients with incomplete medical records or insufficient tissue samples for genetic analysis were excluded from the study. Detailed clinical information of the included patients is provided in Table 1.
Table 1.
An overview of gastric cancer (GC) patient’s characteristics in the present study
| Sr. no | Characteristics | Sample count (n) |
|---|---|---|
| 1 | Sex | |
| Male | 38 | |
| Female | 4 | |
| 2 | Age | |
| >60 | 2 | |
| <60 | 40 | |
| 3 | Treatment | |
| Pre-treatment | 42 | |
| Post-treatment | 0 | |
Nucleic acid extraction
In the present study, we employed a highly reliable kit-based method for DNA and RNA extraction. For DNA extraction, the GeneJET Genomic DNA Purification Kit (cat # K0721, Thermo Fisher) was used, while for RNA extraction, the GeneJET Genomic RNA Purification Kit (cat # K0732, Thermo Fisher) was utilized. After the extraction of DNA and RNA, it was crucial to verify the genetic material’s purity. To do so, the A260/A280 absorbance ratio, measured at 260 nm and 280 nm, was employed as a criterion. Samples with an A260/A280 ratio falling within the range of 1.8 to 2.0 were selected for subsequent downstream analyses.
NGS analysis
The genomic DNA isolated was appropriately diluted to achieve the required concentration for polymerase chain reaction (PCR)-based library preparation. Targeted amplification of the coding regions and splicing sites of BRCA1 (NM 007294) and BRCA2 (NM 000059) genes was carried out using the AmpliSeq for Illumina BRCA Panel. The uniquely indexed libraries specific to the BRCA panel were then prepared following the guidelines outlined in the AmpliSeq for Illumina BRCA Panel Reference Guide. Subsequently, paired-end sequencing by synthesis was performed using the MiSeq sequencer from Illumina, located in San Diego, CA, USA.
The base quality and amplicon coverage of the raw sequencing reads were analyzed using the local run manager of the MiSeq sequencer. Following this, the cleaned reads, meeting the quality threshold of a phred score greater than 30, were aligned to the human reference genome hg19/GRCh37. This alignment was accomplished using the BWA-MEM Whole-Genome Aligner, version 0.7.9a-isis-1.0.2. Subsequently, mismatched calls were identified as mutations using the Pisces Variant Caller, version 5.2.9.23. These genetic mutations were then characterized and annotated with the assistance of the Illumina Annotation Engine, version 2.0.11-0-g7fb24a09.
Interpretation of the mutations
The interpretation of genetic mutations adhered to guidelines established by the American College of Medical Genetics (ACMG) and the Association for Molecular Pathology (AMP) [27]. To classify these mutations into categories such as pathogenic, likely pathogenic, variants of uncertain significance (VUS), likely benign, or benign, a combination of in-silico prediction tools and curated external databases were utilized. Consistent with results from in-silico prediction software, including SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and Mutation Taster (http://www.mutationtaster.org/), the functional impact of these variants on the BRCA1 and BRCA2 protein products was assessed. Additionally, the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) was consulted to evaluate the clinical significance of the respective mutations.
Frequencies of the observed BRCA1/2 mutations in the gnomAD database
The gnomAD database stands as an invaluable asset within the realm of genomics and genetics [28]. It consolidates and integrates an extensive array of genomic data sourced from diverse populations, serving as a comprehensive repository of genetic variation information. Encompassing data from both exome and whole-genome sequencing, gnomAD provides a robust platform for exploring genetic variants, including single nucleotide polymorphisms (SNPs) and insertions/deletions (indels). Researchers and clinicians alike rely on gnomAD to gauge the frequency and distribution of genetic variants across human populations, facilitating the interpretation of genomic findings across various domains, spanning from rare disease diagnosis to population genetics. In this study, we leveraged the gnomAD platform to evaluate the prevalence of identified BRCA1/2 mutations among Asian individuals diagnosed with gastric cancer.
Analysis of the observed BRCA1/2 mutations in TCGA
The cBioPortal is a pivotal resource in the field of cancer research, providing a user-friendly platform for the exploration and analysis of large-scale cancer genomics datasets [29]. Researchers worldwide rely on the cBioPortal to gain insights into the genomic alterations and molecular profiles of various cancer types. This valuable tool offers interactive visualizations, enabling the study of genetic mutations, copy number variations, and gene expression patterns in the context of cancer. In the present study, cBioPortal database was utilized to check the presence of the observed BRCA1/2 mutations in the GC TCGA dataset.
Sanger sequencing
A subset of clinical gastric GC samples, including seven samples displaying pathogenic mutations, underwent sequencing for the complete coding regions using Sanger sequencing. The sequencing process utilized the ABI PRISM DyeDeoxy Terminator Cycle Sequencing Kit and an ABI 3100 Genetic Analyzer from Applied Biosystems in accordance with the methodology outlined by Coppa et al. [30]. The reference sequence employed for BRCA1 was NM_007294.3, and for BRCA2, it was NM_000059.3.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
Initially, the synthesis of first-strand cDNA was carried out using the cDNA Synthesis SuperMix from TransGen Biotech Co., Ltd., following the manufacturer’s recommended protocols. Subsequently, RT-qPCR was conducted using the TB Green® Premix EX Taq™ II (Takara Bio, Inc.) on an Applied Biosystems 7900 Real-Time PCR System (Thermo Fisher Scientific, Inc.). The RT-qPCR protocol involved an initial denaturation step at 95°C for 30 seconds, followed by 40 cycles of denaturation at 95°C for 5 seconds and annealing/extension at 55°C for 34 seconds. A final extension step was performed at 72°C for 5 minutes, followed by a 15-minute extension at 72°C. The expression levels of RNA were determined using the comparative 2-ΔΔCq method [31], with GAPDH expression as the internal RNA standard. The primers used for RT-qPCR are given below. All reactions were performed in triplicate.
GAPDH-F: 5’-ACCCACTCCTCCACCTTTGAC-3’, GAPDH-R: 5’-CTGTTGCTGTAGCCAAATTCG-3’; BRCA1-F: 5’-CTGAAGACTGCTCAGGGCTATC-3’, BRCA1-R: 5’-AGGGTAGCTGTTAGAAGGCTGG-3’; BRCA2-F: 5’-GGCTTCAAAAAGCACTCCAGATG-3’, BRCA2-R: 5’-GGATTCTGTATCTCTTGACGTTCC-3’.
Receiver operating characteristic (ROC) curve
Using the RT-qPCR expression data, we constructed ROC curves for BRCA1/2 expression levels utilizing the SRPLOT web resource available at https://bioinformatics.com.cn/srplot.
Immunohistochemistry (IHC) analysis
The tissue sections underwent deparaffinization, and for antigen retrieval, they were subjected to heat treatment in an EDTA (ethylenediaminetetraacetic acid) solution at pH 8.0. Protein expression levels of the mutated genes in GC tissue samples were evaluated using 4-μm-thick sections obtained from formalin-fixed, paraffin-embedded (FFPE) specimens. Monoclonal antibodies targeting BRCA1 (EPR19433, abcam) and BRCA2 (EPR23442-43, abcam) were utilized, and staining was carried out using the Ventana BenchMark XT staining system (Roche, Tokyo, Japan). In this analysis, non-pathogenic mutated tissue samples were used as the comparative reference. The assessment of tumor positivity was determined by a pathologist based on the presence of nuclear staining in tumor tissue or negativity in the absence of nuclear staining. Protein expression was evaluated, taking into account staining intensity.
Gene enrichment analysis
Metascape is a powerful and user-friendly bioinformatics tool designed for functional enrichment analysis [32]. It streamlines the interpretation of large-scale omics data by identifying enriched biological processes, pathways, and molecular functions. Metascape leverages a comprehensive gene annotation database, making it an invaluable resource for researchers seeking to unravel the biological significance of their datasets. In the present study, we used the Metascape tool to perform gene enrichment analysis of BRCA1/2 genes.
Results
NGS analysis and mutation detection
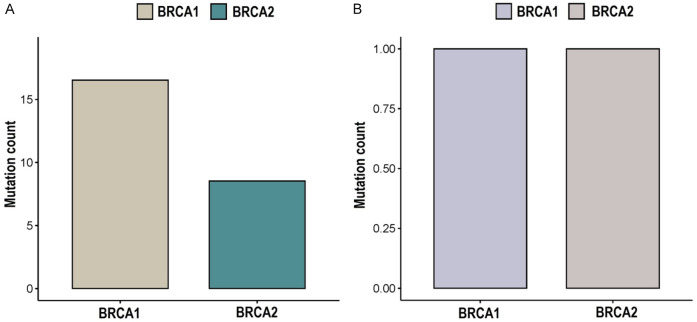
In this investigation, a cohort of 42 GC patients was studied, consisting of 38 males and 4 females (Table 1). The age of these individuals ranged from 18 to 68 years, with an average age of 37 years. Through comprehensive genetic analysis, we detected a total of 19 mutations in the BRCA1 gene and 11 mutations in the BRCA2 gene among the GC samples (Table 2). Notably, all of these identified mutations exhibited a high mutation quality score of 100. The sequencing data demonstrated an impressive coverage rate of 99.1%, and the average Quality score (Q30) reached a notable 98%. In-depth in silico analysis and interpretation based on ClinVar data revealed that within the BRCA1 gene, there were 1 pathogenic mutation (6%) and 15 benign mutations (94%) among the subjects (Figure 1 and Table 2). Similarly, within the BRCA2 gene, the analysis uncovered 1 pathogenic mutation (13%) and 7 benign mutations (87%) within the studied cohort (Figure 1 and Table 2).
Table 2.
Incidence and categorization of mutations detected in the BRCA1/2 genes among GC patients
| Sr. no | Gene | NM:c.DNA | Protein | Nature (ClinVar) | Nature (In silico analysis) | No. patients |
|---|---|---|---|---|---|---|
| 1 | BRCA1 | NM_007294.4:c.5467G>T | p.Ala1823Ser | Pathogenic | DC | 7 |
| 2 | NM_007294.4:c.5158A>G | p.Thr1720Ala | Benign | Non-DC | 15 | |
| 3 | NM_007294.4:c.5117G>C | p.Gly1706Ala | Benign | Non-DC | 9 | |
| 4 | NM_007294.4:c.5044G>A | p.Glu1682Lys | Benign | Non-DC | 9 | |
| 5 | NM_007294.4:c.5024C>T | p.Thr1675Ile | Benign | Non-DC | 11 | |
| 6 | NM_007294.4:c.4985T>C | p.Phe1662Ser | Benign | Non-DC | 10 | |
| 7 | NM_007294.4:c.4956G>A | p.Met1652Ile | Benign | Non-DC | 14 | |
| 8 | NM_007294.4:c.4955T>C | p.Met1652Thr | Benign | Non-DC | 19 | |
| 9 | NM_007294.4:c.4913A>T | p.Glu1638Val | Benign | Non-DC | 14 | |
| 10 | NM_007294.4:c.4910C>T | p.Pro1637Leu | Benign | Non-DC | 11 | |
| 11 | NM_007294.4:c.4840C>T | p.Pro1614Ser | Benign | Non-DC | 4 | |
| 12 | NM_007294.4:c.4837A>G | p.Ser1613Gly | Benign | Non-DC | 12 | |
| 13 | NM_007294.4:c.4816A>G | p.Lys1606Glu | Benign | Non-DC | 14 | |
| 14 | NM_007294.4:c.4729T>C | p.Ser1577Pro | Benign | Non-DC | 12 | |
| 15 | NM_007294.4:c.4691T>C | p.Leu1564Pro | Benign | Non-DC | 17 | |
| 16 | NM_007294.4:c.4682C>T | p.Thr1561Ile | Benign | Non-DC | 5 | |
| 1 | BRCA2 | NM_000059.4:c.275_276insCCAT | p.Gln92fs | Pathogenic | DC | 7 |
| 2 | NM_000059.4:c.1662T>G | p.Cys554Trp | Benign | Non-DC | 18 | |
| 3 | NM_000059.4:c.1744A>C | p.Thr582Pro | Benign | Non-DC | 21 | |
| 4 | NM_000059.4:c.1786G>C | p.Asp596His | Benign | Non-DC | 12 | |
| 5 | NM_000059.4:c.1792A>G | p.Thr598Ala | Benign | Non-DC | 13 | |
| 6 | NM_000059.4:c.1796C>T | p.Ser599Phe | Benign | Non-DC | 11 | |
| 7 | NM_000059.4:c.1798T>C | p.Tyr600His | Benign | Non-DC | 9 | |
| 8 | NM_000059.4:c.2350A>G | p.Met784Val | Benign | Non-DC | 17 |
DC = Disease causing.
Figure 1.
This figure illustrates a comprehensive summary of mutations identified in both the overall and pathogenic categories within the BRCA1/2 genes among gastric cancer (GC) patients utilizing Whole Exome Sequencing (WES). A. Provides an overview of the total count of detected mutations in BRCA1/2 genes among GC patients. B. Specifically highlights the count of pathogenic mutations in BRCA1/2 genes among GC patients.
Of particular interest is the observation that among the seven GC patients outlined in Table 1, we identified two pathogenic mutations, encompassing one mutations in BRCA1 (p.Ala1823Ser) and other one in the BRCA2 gene (p.Gln92fs). Given the direct association of pathogenic mutations with disease development, the subsequent phase of our investigation focused on scrutinizing the prevalence and functional implications of these pathogenic mutations among GC patients.
Frequencies of the observed BRCA1/2 pathogenic mutations in gnomAD database
Examination of pathogenic mutations in the BRCA1 gene (p.Ala1823Ser) and in the BRCA2 gene (p.Gln92fs). As listed in Table 2, across the gnomeAD database yields significant insights. Remarkably, these specific mutations have not been previously documented among Asian GC patients, consistently displaying a frequency of 0 in this database. This intriguing observation not only underscores the rarity of these mutations within the Asian GC population but also suggests a distinctive prevalence pattern unique to the Pakistani population in the context of gastric GC.
Analysis of the observed BRCA1/2 mutations in TCGA
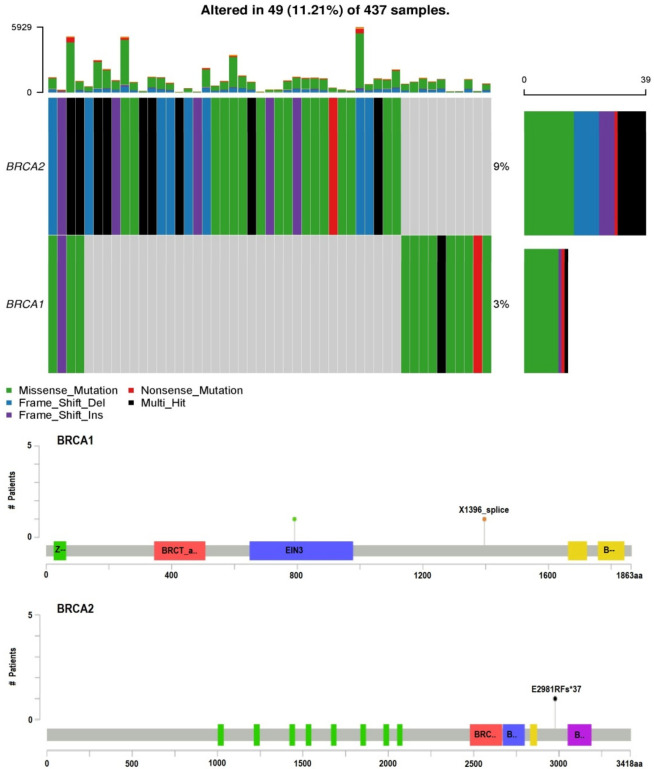
In the subsequent phase of this study, an extensive mutational exploration of the BRCA1/2 genes was conducted within GC samples obtained from the TCGA dataset, utilizing the cBioPortal platform. The primary objective was to identify potential genetic variations and assess their prevalence across the TCGA dataset. The findings of this analysis revealed a distinct trend: the pathogenic mutations in BRCA1 gene (p.Ala1823Ser) and in the BRCA2 gene (p.Gln92fs) that were identified in GC patients of Pakistani origin were notably absent within the TCGA GC samples (Figure 2). This observation further highlights the exceptional nature of these particular pathogenic mutations within the context of GC among individuals of Pakistani descent. In sum, these mutations appear to represent distinct genetic markers associated with susceptibility to GC in the Pakistani population.
Figure 2.
This figure displays visual representations employing oncoplots and lollipop plots to illustrate the observed BRCA1/2 mutations within The Cancer Genome Atlas (TCGA) gastric cancer (GC) patient cohort. The two rows depict the percentage of samples featuring BRCA1/2 mutations, and the accompanying lollipop plots emphasize the protein-level amino acid alterations resulting from these mutations.
Sanger sequencing
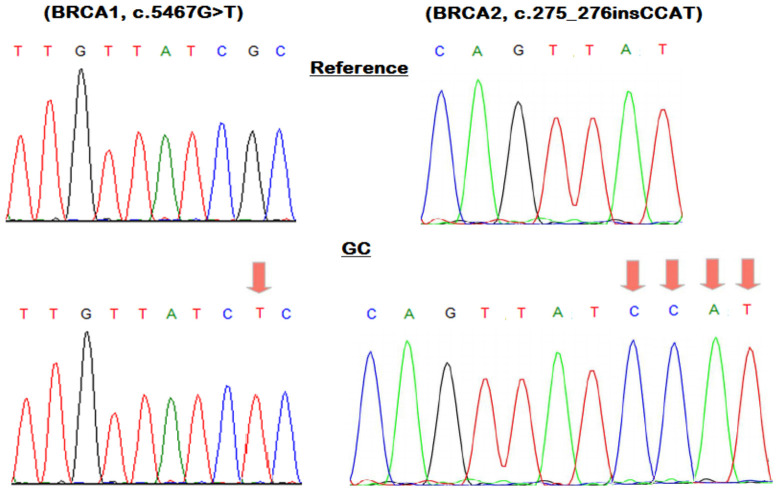
Sanger sequencing was conducted for the entire coding regions of BRCA1 and BRCA2 in seven GC samples that exhibited pathogenic mutations: BRCA1 (c.5467G>T) and BRCA2 (c.275_276insCCAT). As anticipated in representative images, Sanger sequencing validation affirmed the presence of the identified BRCA1 and BRCA2 pathogenic variants previously detected by NGS (Figure 3).
Figure 3.
This figure showcases a representative DNA sequencing chromatogram capturing the BRCA1/2 pathogenic mutations observed in gastric cancer (GC) samples.
RT-qPCR-based expression analysis of BRCA1/2 genes
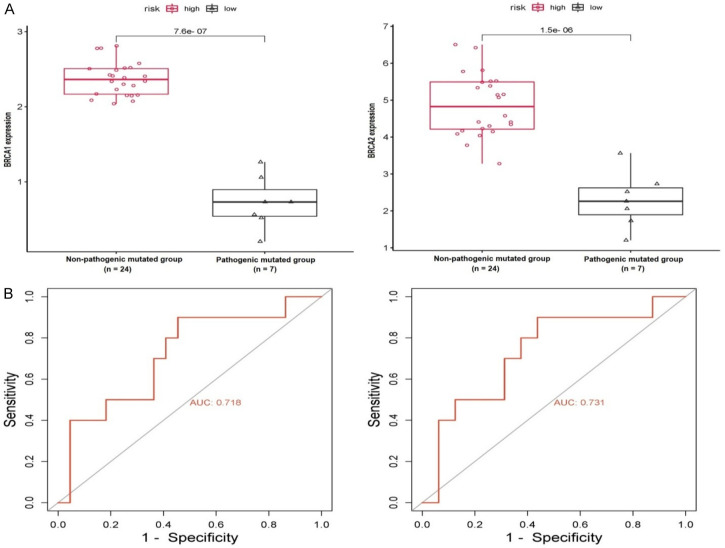
The evaluation of BRCA1/2 gene expression was carried out using RT-qPCR in two distinct subsets of GC samples. One subset consisted of samples harboring pathogenic mutations in in BRCA1 gene (p.Ala1823Ser) and in the BRCA2 gene (p.Gln92fs), while the other subset comprised samples without such mutations, representing the non-pathogenic mutation group. Upon analyzing the results of the RT-qPCR analysis, a noticeable trend became evident: the expression levels of BRCA1/2 genes exhibited a significant decrease within the GC samples containing BRCA1/2 pathogenic mutations, in clear contrast to the non-pathogenic mutation group (Figure 4A). This intriguing finding suggests a potential association between BRCA1/2 mutations and the heightened expression of these genes within the context of GC.
Figure 4.
This figure presents an evaluation of relative expression and receiver operating characteristics (ROC) curve analyses for BRCA1/2 genes, comparing groups of gastric cancer (GC) samples with pathogenic mutations and those without. (A) Illustrates the analysis of relative expression for BRCA1/2 genes using RT-qPCR, while (B) displays ROC curves derived from RT-qPCR expression data for BRCA1/2 genes. The classification threshold was set at a significance level of P<0.05.
Moreover, the ROC curves derived from the RT-qPCR expression data of BRCA1/2 genes offered further confirmation of their precision and sensitivity as prospective biomarkers (see Figure 4B). These curves illustrated the genes’ capacity (Figure 4B) to effectively distinguish conditions, underscoring their value as dependable biomarkers for GC diagnosis.
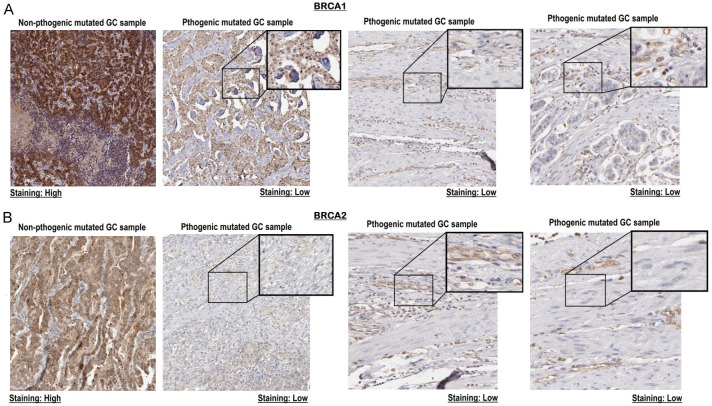
IHC-based expression analysis of BRCA1/2 proteins
We conducted an IHC analysis to assess the proteomic expression of BRCA1/2 proteins in GC tissue samples. Specifically, we examined one tissue sample with pathogenic mutations in BRCA1 gene (p.Ala1823Ser) and in the BRCA2 gene (p.Gln92fs), and another tissue sample without any pathogenic mutations. The goal was to investigate potential disparities in protein expression between these two categories of samples.
Upon scrutinizing the staining results, a significant pattern became evident (Figure 5). The GC tissue samples with pathogenic mutations displayed notably lower levels of BRCA1/2 protein expression when compared to their counterparts lacking these mutations (Figure 5). This finding suggests a possible correlation between the presence of pathogenic mutations in the BRCA1/2 genes and down-regulation in the expression of these proteins within the context of GC.
Figure 5.
This figure presents the proteomic expression analysis of BRCA1/2 proteins, conducted through immunohistochemistry (IHC), to compare gastric cancer (GC) samples with pathogenic mutations against those without. (A) Presents IHC images of the BRCA1 protein in GC tissues samples, and (B) presents IHC images of the BRCA2 protein in GC tissues samples. Variations in expression were evaluated by examining staining intensities.
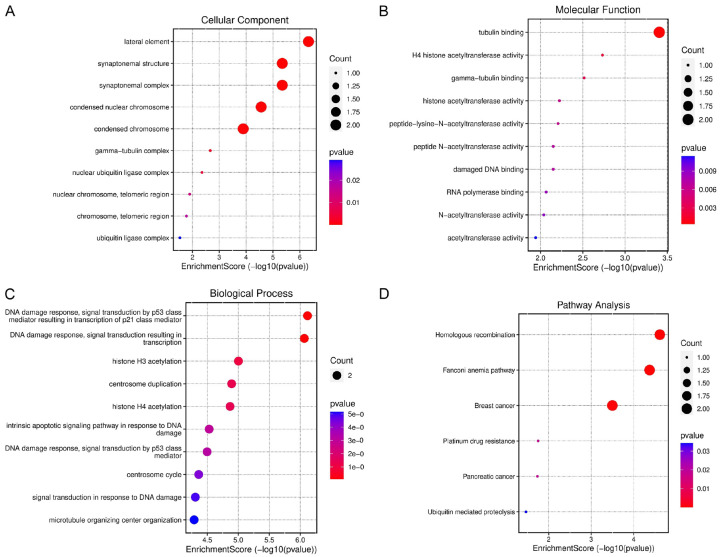
Enrichment analysis of BRCA1/2 genes
Next, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. Within the cellular component (CC) analysis, BRCA1/2 genes exhibited enrichments in terms such as “lateral element”, “synaptonemal structure”, “synaptonemal complex”, “condensed nuclear chromosome”, and “nuclear chromosome” among others (Figure 6A). In the case of molecular function (MF), they displayed enrichments in terms like “tubulin binding”, “H4 histone acetyltransferase activity”, “gamma tubulin binding”, and “acetyltransferase activity” (Figure 6B). Furthermore, within biological processes (BP), these genes were associated with terms including “DNA damage response”, “signal transduction by p53 class mediator resulting in transcription of P21 class mediator”, “DNA damage response”, “signal transduction resulting in transcription”, and “histone H3-acetylation” (Figure 6C). Additionally, in the KEGG pathway analysis, they were enriched in pathways such as “homologous recombination”, “Fanconi anemia pathway”, “breast cancer “and” platinum drug resistance in cancer” (Figure 6D).
Figure 6.
This figure illustrates the outcomes of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses conducted on BRCA1/2 genes via Metascape. (A) Displays Cellular Component (CC) terms related to BRCA1/2 genes, (B) Highlights Molecular Function (MF) terms, (C) Showcases Biological Process (BP) terms, and (D) Outlines Kyoto Encyclopedia of Genes and Genomes (KEGG) terms associated with BRCA1/2 genes. A significance level of P<0.05 served as the cutoff criterion.
BRCA1/2-associated drugs
In our comprehensive investigation, we made use of the DrugBank database to systematically explore therapeutic possibilities aimed at modulating the expression of mutated BRCA1 and BRCA2. Through our meticulous analysis, Cisplatin, Estradiol, Tretinoin, Genistein, Acetaminophen, and Quercetin (Table 3) emerged as compounds with significant potential to effectively up-regulate the expression levels of BRCA1/2 genes. This discovery holds substantial promise for innovative therapeutic approaches within the context of GC.
Table 3.
DrugBank-based BRCA1/2 associated drugs
| Sr. No | Hub gene | Drug name | Effect | Reference | Group |
|---|---|---|---|---|---|
| 1 | BRCA1 | Cisplatin | Increase expression of BRCA1 mRNA | A22234 | Approved |
| Estradiol | A21155 | ||||
| Tretinoin | A24464 | ||||
| Genistein | A22773 | ||||
| 2 | BRCA2 | Acetaminophen | Increase expression of BRCA2 mRNA | A20418 | Approved |
| Quercetin | A21498 | ||||
| Estradiol | A21155 | ||||
| Resveratrol | A23885 | ||||
| Metribolone | A23234 | ||||
| Genistein | A22773 |
mRNA = Messenger RNA.
Discussion
Gastric cancer (GC) continues to pose a significant global health challenge due to its complex etiology and diverse genetic alterations [33]. GC is the fifth most prevalent cancer and the third primary contributor to cancer-related fatalities on a global scale. This emphasizes the critical need for a deeper exploration into the molecular foundations of this disease [34,35].
In this study, we aimed to elucidate the role of BRCA1 and BRCA2 mutations in Pakistani GC patients and assess their functional consequences. The findings of this study shed light on the potential implications of these mutations in the context of GC development and progression. Comprehensive analysis in the current study involved the use of NGS to scrutinize BRCA1/2 genes in a cohort of 31 Pakistani GC patients. This approach revealed the presence of pathogenic mutations in BRCA1 gene (p.Ala1823Ser) and in the BRCA2 gene (p.Gln92fs) along with various benign mutations. The pathogenic mutations are of particular significance as they have been well-documented for their association with hereditary breast and ovarian cancers [36-38]. To the best of our knowledge, this study marks the first documentation of such pathogenic mutations in GC within the Pakistani population.
To explore the functional consequences of these observed pathogenic mutations, we conducted expression analyses of BRCA1 and BRCA2 genes at both mRNA and protein levels. The results yielded a striking and consistent pattern - GC samples harboring pathogenic mutations in these genes displayed a down-regulation of BRCA1/2 gene expression as compared other GC samples which do not harbor pathogenic mutations in those genes. Earlier studies have also indicated that pathogenic mutations typically result in the disruption and impaired function of the mutated gene in cancer patients [39-41].
The observed down-regulation of the mutated BRCA1 gene (p.Ala1823Ser) and BRCA2 gene (p.Gln92fs) in the current study is noteworthy for several reasons. Firstly, it underscores the impact of BRCA1/2 mutations beyond their canonical role in DNA repair [42,43]. These mutations appear to exert an influence on gene expression, potentially contributing to the molecular alterations associated with GC. Secondly, the down-regulation was observed not only at the mRNA level but also at the protein level, indicating a multifaceted effect on the cellular processes regulated by BRCA1/2. Prior investigations have also suggested that the decrease in protein-level expression of BRCA1/2 genes leads to the disruption of crucial cellular processes related to DNA repair [44,45]. These BRCA1/2 down-regulation-based finding underscores the potential clinical relevance of the pathogenic mutations in the context of GC. The observed down-regulation of BRCA1/2 genes in GC samples with pathogenic mutations has far-reaching implications for the management of GC patients. It suggests a potential role for these mutations as biomarkers with diagnostic and therapeutic significance. Down-regulation of BRCA1/2 genes in Pakistani GC patients may contribute to genomic instability and altered DNA repair mechanisms, which could impact disease progression and response to therapy. Therefore, targeted therapies that exploit the underlying biology of these mutations may offer new avenues for treatment, potentially enhancing treatment response and patient outcomes. In addition, we have investigated potential drugs (Cisplatin, Estradiol, Tretinoin, Genistein, Acetaminophen, and Quercetin) from the DrugBank database that have the capability to enhance the expression of BRCA1/2 genes when used in the treatment of GC patients.
Finally, pathway analysis revealed the involvement of BRCA1/2 genes in various divers’ pathways, including “homologous recombination”, “Fanconi anemia pathway”, “breast cancer “and” platinum drug resistance in cancer”. The dysregulation of these pathways is already well document in various cancers [46-48].
While our study provides compelling evidence of the functional consequences of BRCA1/2 mutations in GC, it is not without limitations. The sample size, although informative, is relatively small, and further validation in larger cohorts is essential. Additionally, in-depth mechanistic studies are warranted to elucidate the precise molecular pathways through which these mutations impact gene expression and GC progression.
Conclusion
In conclusion, our study highlights the presence of pathogenic mutations in BRCA1 and BRCA2 genes in Pakistani GC patients and underscores their functional consequences in terms of down-regulated gene expression. These findings open new avenues for research into the molecular underpinnings of GC and the development of targeted therapies for patients with these mutations. Further investigation is needed to harness the full clinical potential of these discoveries and improve the management of GC in affected populations.
Acknowledgements
This work was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2024R294), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. The authors extend their appreciation to the Researchers supporting project number (RSP2024R190), King Saud University, Riyadh, Saudi Arabia.
Disclosure of conflict of interest
None.
References
- 1.Schwarz RE. Current status of management of malignant disease: current management of gastric cancer. J Gastrointest Surg. 2015;19:782–788. doi: 10.1007/s11605-014-2707-x. [DOI] [PubMed] [Google Scholar]
- 2.Berretta S, Berretta M, Fiorica F, Di Francia R, Magistri P, Bertola G, Fisichella R, Canzonieri V, Tarantino G, Di Benedetto F. Multimodal approach of advanced gastric cancer: based therapeutic algorithm. Eur Rev Med Pharmacol Sci. 2016;20:4018–4031. [PubMed] [Google Scholar]
- 3.McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664–674. doi: 10.1038/nrgastro.2014.143. [DOI] [PubMed] [Google Scholar]
- 4.Dicken BJ, Bigam DL, Cass C, Mackey JR, Joy AA, Hamilton SM. Gastric adenocarcinoma: review and considerations for future directions. Ann Surg. 2005;241:27–39. doi: 10.1097/01.sla.0000149300.28588.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ullah L, Hameed Y, Ejaz S, Raashid A, Iqbal J, Ullah I, Ejaz SA. Detection of novel infiltrating ductal carcinoma-associated BReast CAncer gene 2 mutations which alter the deoxyribonucleic acid-binding ability of BReast CAncer gene 2 protein. J Cancer Res Ther. 2020;16:1402–1407. doi: 10.4103/jcrt.JCRT_861_19. [DOI] [PubMed] [Google Scholar]
- 6.Yasir M, Nawaz A, Ghazanfar S, Okla MK, Chaudhary A, Al WH, Ajmal MN, AbdElgawad H, Ahmad Z, Abbas F, Wadood A, Manzoor Z, Akhtar N, Din M, Hameed Y, Imran M. Anti-bacterial activity of essential oils against multidrug-resistant foodborne pathogens isolated from raw milk. Braz J Biol. 2022;84:e259449. doi: 10.1590/1519-6984.259449. [DOI] [PubMed] [Google Scholar]
- 7.Comen EA, Bowman RL, Kleppe M. Underlying causes and therapeutic targeting of the inflammatory tumor microenvironment. Front Cell Dev Biol. 2018;6:56. doi: 10.3389/fcell.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9:77–102. doi: 10.1007/s13167-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Sahar AM, Li C, Chaudhary A, Yousaf I, Saeedah MA, Mubarak A, Haris M, Nawaz M, Reem MA, Ramadan FA, Mostafa AAM, Feng W, Hameed Y. A detailed multi-omics analysis of GNB2 gene in human cancers. Braz J Biol. 2022;84:e260169. doi: 10.1590/1519-6984.260169. [DOI] [PubMed] [Google Scholar]
- 10.Cavanagh H, Rogers KM. The role of BRCA1 and BRCA2 mutations in prostate, pancreatic and stomach cancers. Hered Cancer Clin Pract. 2015;13:16. doi: 10.1186/s13053-015-0038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckley KH, Niccum BA, Maxwell KN, Katona BW. Gastric cancer risk and pathogenesis in BRCA1 and BRCA2 carriers. Cancers (Basel) 2022;14:5953. doi: 10.3390/cancers14235953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalil T, Okla MK, Al-Qahtani WH, Ali F, Zahra M, Shakeela Q, Ahmed S, Akhtar N, AbdElgawad H, Asif R, Hameed Y, Adetunji CO, Farid A, Ghazanfar S. Tracing probiotic producing bacterial species from gut of buffalo (Bubalus bubalis), South-East-Asia. Braz J Biol. 2022;84:e259094. doi: 10.1590/1519-6984.259094. [DOI] [PubMed] [Google Scholar]
- 13.Tatematsu M, Tsukamoto T, Inada K. Stem cells and gastric cancer: role of gastric and intestinal mixed intestinal metaplasia. Cancer Sci. 2003;94:135–141. doi: 10.1111/j.1349-7006.2003.tb01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, Chan KH, Chan AS, Tsui WY, Ho SL, Chan AK, Man JL, Foglizzo V, Ng MK, Chan AS, Ching YP, Cheng GH, Xie T, Fernandez J, Li VS, Clevers H, Rejto PA, Mao M, Leung SY. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 15.Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42:211–217. doi: 10.1016/j.gtc.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 17.Hameed Y, Ejaz S. TP53 lacks tetramerization and N-terminal domains due to novel inactivating mutations detected in leukemia patients. J Cancer Res Ther. 2021;17:931–937. doi: 10.4103/jcrt.JCRT_536_19. [DOI] [PubMed] [Google Scholar]
- 18.Zhang ZZ, Liu YJ, Yin XL, Zhan P, Gu Y, Ni XZ. Loss of BRCA1 expression leads to worse survival in patients with gastric carcinoma. World J Gastroenterol. 2013;19:1968–74. doi: 10.3748/wjg.v19.i12.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semba S, Yokozaki H, Yasui W, Tahara E. Frequent microsatellite instability and loss of heterozygosity in the region including BRCA1 (17q21) in young patients with gastric cancer. Int J Oncol. 1998;12:1245–1251. doi: 10.3892/ijo.12.6.1245. [DOI] [PubMed] [Google Scholar]
- 20.Blay P, Santamaría I, Pitiot AS, Luque M, Alvarado MG, Lastra A, Fernández Y, Paredes Á, Freije JM, Balbín M. Mutational analysis of BRCA1 and BRCA2 in hereditary breast and ovarian cancer families from Asturias (Northern Spain) BMC Cancer. 2013;13:243. doi: 10.1186/1471-2407-13-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Fan I, Tang J, Li S, Zhang S, Shaw PA, Narod SA. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98:1694–1706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 22.Iarmarcovai G, Bonassi S, Botta A, Baan RA, Orsiere T. Genetic polymorphisms and micronucleus formation: a review of the literature. Mutat Res. 2008;658:215–233. doi: 10.1016/j.mrrev.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Migliore L, Coppedè F. Genetic and environmental factors in cancer and neurodegenerative diseases. Mutat Res. 2002;512:135–153. doi: 10.1016/s1383-5742(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 24.Khan M, Hameed Y. Discovery of novel six genes-based cervical cancer-associated biomarkers that are capable to break the heterogeneity barrier and applicable at the global level. J Cancer Res Ther. 2023;9000:2023. [Google Scholar]
- 25.Usman M, Hameed Y, Ahmad M. Does human papillomavirus cause human colorectal cancer? Applying Bradford Hill criteria postulates. Ecancermedicalscience. 2020;14:1107. doi: 10.3332/ecancer.2020.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.General Assembly of the World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. J Am Coll Dent. 2014;81:14–18. [PubMed] [Google Scholar]
- 27.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudmundsson S, Singer-Berk M, Watts NA, Phu W, Goodrich JK, Solomonson M Genome Aggregation Database Consortium. Rehm HL, MacArthur DG, O’Donnell-Luria A. Variant interpretation using population databases: lessons from gnomAD. Hum Mutat. 2022;43:1012–1030. doi: 10.1002/humu.24309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coppa A, Nicolussi A, D’Inzeo S, Capalbo C, Belardinilli F, Colicchia V, Petroni M, Zani M, Ferraro S, Rinaldi C, Buffone A, Bartolazzi A, Screpanti I, Ottini L, Giannini G. Optimizing the identification of risk-relevant mutations by multigene panel testing in selected hereditary breast/ovarian cancer families. Cancer Med. 2018;7:46–55. doi: 10.1002/cam4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng L, Wang L, Ajani J, Xie K. Molecular basis of gastric cancer development and progression. Gastric Cancer. 2004;7:61–77. doi: 10.1007/s10120-004-0277-4. [DOI] [PubMed] [Google Scholar]
- 34.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 35.Ahmad M, Khan M, Asif R, Sial N, Abid U, Shamim T, Hameed Z, Iqbal MJ, Sarfraz U, Saeed H, Asghar Z, Akram M, Ullah Q, Younas QA, Rauf L, Hadi A, Maryam S, Hameed Y, Khan MR, Tariq E, Saeed S. Expression characteristics and significant diagnostic and prognostic values of ANLN in human cancers. Int J Gen Med. 2022;15:1957–1972. [Google Scholar]
- 36.Sekine M, Nagata H, Tsuji S, Hirai Y, Fujimoto S, Hatae M, Kobayashi I, Fujii T, Nagata I, Ushijima K, Obata K, Suzuki M, Yoshinaga M, Umesaki N, Satoh S, Enomoto T, Motoyama S, Tanaka K Japanese Familial Ovarian Cancer Study Group. Mutational analysis of BRCA1 and BRCA2 and clinicopathologic analysis of ovarian cancer in 82 ovarian cancer families: two common founder mutations of BRCA1 in Japanese population. Clin Cancer Res. 2001;7:3144–3150. [PubMed] [Google Scholar]
- 37.Suszynska M, Ratajska M, Kozlowski P. BRIP1, RAD51C, and RAD51D mutations are associated with high susceptibility to ovarian cancer: mutation prevalence and precise risk estimates based on a pooled analysis of ~30,000 cases. J Ovarian Res. 2020;13:50. doi: 10.1186/s13048-020-00654-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu W, Li H, Hameed Y, Abdel-Maksoud MA, Almutairi SM, Mubarak A, Aufy M, Alturaiki W, Alshalani AJ, Mahmoud AM, Li C. Elucidating the clinical and immunological value of m6A regulator-mediated methylation modification patterns in adrenocortical carcinoma. Oncol Res. 2023;31:819–831. doi: 10.32604/or.2023.029414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Backwell L, Marsh JA. Diverse molecular mechanisms underlying pathogenic protein mutations: beyond the loss-of-function paradigm. Annu Rev Genomics Hum Genet. 2022;23:475–498. doi: 10.1146/annurev-genom-111221-103208. [DOI] [PubMed] [Google Scholar]
- 40.Ansari S, Gantuya B, Tuan VP, Yamaoka Y. Diffuse gastric cancer: a summary of analogous contributing factors for its molecular pathogenicity. Int J Mol Sci. 2018;19:2424. doi: 10.3390/ijms19082424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Usman M, Hameed Y. GNB1, a novel diagnostic and prognostic potential biomarker of head and neck and liver hepatocellular carcinoma. J Cancer Res Ther. 2023;9000:2023. [Google Scholar]
- 42.Nathansen J, Meyer F, Müller L, Schmitz M, Borgmann K, Dubrovska A. Beyond the double-strand breaks: the role of DNA repair proteins in cancer stem-cell regulation. Cancers (Basel) 2021;13:4818. doi: 10.3390/cancers13194818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleury H, Carmona E, Morin VG, Meunier L, Masson JY, Tonin PN, Provencher D, Mes-Masson AM. Cumulative defects in DNA repair pathways drive the PARP inhibitor response in high-grade serous epithelial ovarian cancer cell lines. Oncotarget. 2017;8:40152–40168. doi: 10.18632/oncotarget.10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosen EM, Fan S, Pestell RG, Goldberg ID. BRCA1 gene in breast cancer. J Cell Physiol. 2003;196:19–41. doi: 10.1002/jcp.10257. [DOI] [PubMed] [Google Scholar]
- 45.Vodicka P, Andera L, Opattova A, Vodickova L. The interactions of DNA repair, telomere homeostasis, and p53 mutational status in solid cancers: risk, prognosis, and prediction. Cancers (Basel) 2021;13:479. doi: 10.3390/cancers13030479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leung W, Baxley RM, Moldovan GL, Bielinsky AK. Mechanisms of DNA damage tolerance: post-translational regulation of PCNA. Genes (Basel) 2018;10:10. doi: 10.3390/genes10010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovalchuk I. Roles of RAD18 in DNA replication and post-replication repair (PRR). In: Genome Stability. Elsevier; 2021. pp. 275–292. [Google Scholar]
- 48.Vaziri C, Tateishi S, Mutter-Rottmayer E, Gao Y. Roles of RAD18 in DNA replication and postreplication repair. In: Genome Stability. Elsevier; 2016. pp. 257–273. [Google Scholar]