Abstract
Background
Pediatric cow milk allergies (CMA) can occur in immunoglobulin (Ig) E and non-IgE-mediated forms. Unlike IgE-mediated allergies, the mechanisms of disease pathogenesis in non-IgE-mediated food allergy and an association with microbiome has not been well established. Previous studies have identified the presence of altered humoral responses to gut bacteria in IgE mediated allergies. Here, we analyzed IgA, IgE and IgG responses to gut bacteria in subjects with either IgE or non-IgE mediated CMA to identify relative proportions of Ig-coated bacteria and characterize unique disease specific microbial signatures.
Methods
Multi-parametric flow cytometry analysis was used to identify IgA, IgE and IgG responses to gut bacteria in CMA patients. Cell sorting of Ig coated gut bacteria was subsequently performed followed by high throughput 16S rRNA gene sequencing and specific patterns of humoral responses to gut bacteria assessed in each study group.
Results
We identified significant alterations in IgA and IgG gut bacterial coating patterns in CMA subjects. Proportions of IgA-coated bacteria were decreased in IgE mediated CMA subjects without atopic dermatitis (ALL) and non-IgE mediated CMA subjects (ENP), compared to healthy controls (CON). In comparison, IgG-coated bacteria was significantly elevated in CMA subjects with atopic dermatitis (AD). Alpha and beta diversities displayed significant differences in IgA-, IgE-, and IgG-coated bacteria in AD and ENP groups. Significant differences in bacteria coated by IgA, IgE and IgG were detected at Phyla, Genus and Species levels and associated bacterial dysbiosis in IgE and non-IgE mediated allergies were identified. Linear discriminant analysis (LDA) effect size (LEFse) revealed unique disease associated bacterial signatures, including several pathogenic bacteria namely Bacteroides fragilis, Ruminococcus gnavus, Eubacterium dolichum, Fusobacterium, Clostridium neonatale and Robinsoniella peoriensis. Receiver operating characteristic curve analysis confirmed the efficiency of using the bacterial signatures identified as biomarkers for disease.
Conclusions
Altered IgA and IgG responses to gut bacteria were identified in CMA subjects. The disease-specific responses were associated with alterations in bacterial diversity and concomitant dysbiosis of Ig-coated bacteria in IgE-mediated and non-IgE-mediated CMA pediatric subjects. The identification of pathogenic bacteria uniquely associated with different classes of allergic disease indicates a role of these bacteria in driving disease-specific pathological phenotypes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05850-z.
Keywords: IgA, IgE, IgG, Gut bacteria, Immunoglobulin-coating, Cow milk allergy, Microbial dysbiosis
Background
Cow milk allergy (CMA) is the most prevalent food allergy amongst young children, manifesting as either an IgE mediated or non-IgE mediated allergic response [1]. CMA typically presents within the first year of life and amongst infants at one year of age, rates of CMA have been estimated to be between 0.5% and 3% [2], with the occurrence of CMA showing significant increases over the past three decades [1]. Studies looking at CMA resolution have shown a variety of outcomes, with one study showing 47% resolution by age 4 to 5 [3], whereas others have noted a continued prevalence of 57% CMA reactive children at age 10 [4]. In comparison, the incidence of CMA in the adult population is considerably lower with a recent study reporting a rate of 1.9%, of which 77.3% of these adults had first experienced CMA during early childhood [5]. Previous studies have shown that dysbiosis in the gut microbiome plays a significant role in the development of pediatric CMA [6, 7], as the gut microbiota has a pivotal role in the maintenance of homeostasis and in regulating immune responses at the intestinal barrier [8]. Several factors, including the mode of delivery [9], use of antibiotics [10] and breast feeding versus formula feeding [11–13], can contribute to dysbiosis of the gut microbiota, which in turn, leads to the predisposition of the development of atopic disease [14]. Reduced prenatal and postnatal microbial exposure is associated with a predisposition to the development of IgE mediated allergic diseases [15–18]. Under homeostatic conditions, the gut microbiota forms a symbiotic relationship with the host through reciprocal interactions between the intestinal mucosal surface, and the host immune system [19]. The development of a diverse gut microbiome helps in shaping immune tolerance to symbiotic bacteria and allows both the induction of protective responses against pathogens and maintenance of regulatory pathways [20, 21]. The presence of a diverse microbiome results in the immune system becoming tolerized to innocuous antigens from commensal bacteria and potential food allergens, through the induction of an IgA mediated antibody response, in association with a corresponding complement of regulatory T cells [14]. However, perturbations to the composition of the microbiome can cause microbial dysbiosis, leading to abnormal immune responses, inflammation and allergic diseases [20, 22].
In contrast to IgE mediated allergic diseases, the mechanisms associated with disease pathogenesis in non-IgE mediated food allergy are not yet well described and a link with microbial dysbiosis has not been definitively established [23, 24]. A previous study on non-IgE mediated CMA identified overlapping signatures in the dysbiosis exhibited by IgE and non-IgE mediated CMA children [25] and a second recent study identified a significant decrease in fecal Bifidobacterium to be a potential biomarker associated with onset of non-IgE mediated CMA. In general, non-IgE-mediated food allergies primarily affect the gastrointestinal system and are characterized by enteropathy and colitis-based symptoms in disorders including; food protein-induced allergic enteropathy (FPE), food protein-induced enterocolitis syndrome (FPIES) and food protein-induced allergic proctocolitis (FPIAP) [26]. Non-IgE mediated allergies are thought to be mediated by cell driven adaptive and innate immune mechanism [27] based on the delayed onset of symptoms following ingestion of allergens in combination with the absence of an IgE mediated response [28].
The immunoglobulins IgA, IgG and IgE are produced as a result of class switching events following activation and differentiation of naive IgM-bearing B cells, in response to an antigenic or allergenic stimulation [29]. While IgA is predominantly secreted across mucosal barriers and in breast milk and functions as an immune modulator at mucosal surfaces, IgG antibodies are most highly abundant in lymph and blood and offer broad systemic protection [29]. IgE is found at the lowest concentration, due to the tight regulation of IgE class switching events and sequestration at cell surfaces via binding to high and low FcE receptors [29]. Although class switching in favor of IgE occurs in the nasal [30] and bronchial mucosa [31] of allergic patients, the specific sites and role of IgE production in the gastrointestinal tract are still poorly characterized [29].
Previous studies have shown that IgA and IgG antibodies can bind to fecal microbes in murine samples, as well as in patient samples with inflammatory bowel disease (IBD) [32, 33]. In the context of IBD, IgG coating was observed mainly in subjects with IBD and was largely absent in healthy controls [34]. Studies have shown that IgA secreted at mucosal surfaces coats a fraction of the intestinal microbiota through both T-cell dependent and independent pathways, where the alternate pathways were shown to produce IgA that bound distinct subsets of microbes [35]. Further, IgA has been shown to maintain the composition of the microbiota and decreased concentrations of intestinal IgA have been correlated with lower levels of bacterial diversity [36] and high levels of intestinal IgA have been associated with a reduced risk of developing IgE mediated allergic disease [37]. Further, a reduction in levels of IgA coated bacteria and an increase in IgE coated bacteria was observed in food allergic patients [38]. Food allergy has also been associated with increased anti-commensal IgE-coating in the presence of Akkermansia muciniphila in fiber-deprived mice [39].
The association of Ig-coated bacteria (IgA, IgE and IgG) with disease has not previously been studied in the context of IgE vs. non-IgE mediated allergic disease. In this study, we recruited children with both IgE and non-IgE mediated CMA and identified the presence of differential patterns of bacterial Ig coating amongst disease groups, leading to the identification of altered Ig-coated microbial signatures in CMA subjects, which uniquely distinguished the specific allergic groups studied.
Methods
Ethics approval and consent to participate
The study obtained written parental consent from all participants before enrollment. The study protocol (1500751) was approved by the Institutional Review Board (IRB) at Sidra Medicine, Doha, Qatar.
Study design and patient recruitment
Recruitment of study subjects was conducted following Institutional Review board (Sidra Medicine, Doha, Qatar) approval, #1706011686. Informed consent was obtained from the parents/guardians of the study subjects. Pediatric subjects, aged between 1 and 4 years, with clinical symptoms of CMA, were recruited to the study and were grouped into IgE and non-IgE mediated allergic groups based on the clinical manifestations and the time of onset allergic symptoms. Age-matched healthy subjects served as the control group. A total of 38 study subjects were recruited and categorized into four groups; Control population with no history of food allergies (CON = 13), IgE mediated CMA (ALL = 8), IgE mediated CMA with Atopic Dermatitis (AD = 10), non-IgE mediated CMA with Enteropathy (ENP = 7), all patient characteristics are included in Table 1. Study exclusion criteria were, use of oral steroids, recent use of antibiotics, atopic dermatitis without sensitivity to milk, early onset inflammatory bowel diseases not related to milk ingestion, celiac disease and chronic diarrhea due to other immune defects. From all the study subjects, data was collected from a questionnaire which provided information regarding the familial history of allergy and environmental exposures.
Table 1.
Demographics of study subjects
| Patient demographics | CON | ALL | AD | ENP | p ALL vs. CON | p AD vs. CON | p ENP vs. CON |
|---|---|---|---|---|---|---|---|
| N | 13 | 8 | 10 | 7 | N/A | N/A | N/A |
| Male (%)ª | 38 | 75 | 60 | 43 | ns | ns | ns |
| Caesarean delivery (%)ª | 38 | 25 | 50 | 29 | ns | ns | ns |
| Gestational age, weeks (mean, SD)º | 37.92, 2.10 | 38.88, 1.13 | 39.10, 1.97 | 39.14, 0.69 | ns | ns | ns |
| Birth weight, kg (mean, SD)º | 2.76, 0.56 | 2.88, 0.45 | 3.25, 0.45 | 3.23, 0.69 | ns | ns | ns |
| Age of Onset, months (mean, SD)º | N/A | 5.12, 3.39 | 4.2, 2.6 | 3.85, 4.4 | N/A | N/A | N/A |
| Breast Feeding 0–6 months (%)ª | 92.31 | 100.00 | 90.00 | 100.00 | ns | ns | ns |
| Formula Feeding 0–6 months (%)ª | 30.70 | 12.50 | 40.00 | 28.50 | ns | ns | ns |
| Solid Food Introduction, months (mean, SD)º | 5.46, 1.71 | 5.87, 0.35 | 6.9, 1.91 | 7.28, 4.78 | ns | ns | ns |
| Playgroup at 6 months (%)ª | 7.69 | 12.50 | 10.00 | 28.57 | ns | ns | ns |
| Maternal antibiotic use during pregnancy (%)ª | 30.77 | 50.00 | 10.00 | 14.29 | ns | ns | ns |
| Exposure to cigarette smoke at home (%)ª | 15.38 | 12.50 | 0.00 | 0.00 | ns | ns | ns |
| Vitamin D deficiency at birth (%)ª | 76.92 | 87.50 | 70.00 | 85.71 | ns | ns | ns |
| Direct familial allergy history-parents/siblings (%)ª | 15.38 | 50.00 | 80.00 | 71.43 | ns | ** | * |
| Indirect familial allergy history-grandparents (%)ª | 0.00 | 37.50 | 40.00 | 0.00 | * | * | ns |
ªChi square used as statistical test
ºKruskal Wallis with Dunn’s multiple comparison used as statistical test
ImmunoCAPS assay for total and cow-milk specific IgE
Serum measurements of total IgE levels and total cow milk protein, casein-specific, β-lactoglobulin specific and α-lactalbumin specific IgE (sIgE), were performed using a fluoroenzyme immunoassay auto-analyzer, the ImmunoCAP 100 platform (ThermoFisher), according to the manufacturer’s guidelines [40] at the Hematology, Immunology and Transfusion Division, Sidra Medicine, Doha, Qatar.
Stool sample collection
Stool samples were collected in collection tubes and were frozen immediately at − 20 °C after collection and subsequently stored at − 80 °C until processing. The frozen stool samples were thawed immediately before the bacterial isolation.
Bacterial isolation, flow cytometry and sorting
Frozen stool samples were thawed on ice and 100 mg of stool sample was homogenized in 1X DPBS at a concentration of 100 mg/ml and centrifuged at 50 g for 15 min at 4 °C to remove large particles in the stool. Supernatants were recovered and washed twice at 8000 g for 5 min at 4 °C with 1 ml 1X DPBS. Following an additional wash, bacterial pellets were resuspended and incubated for 20 min at 4 °C in 1X DPBS containing 20% mouse serum (Sigma-Aldrich, Cat# M5905). Bacteria were then stained in a mixture of 1:20 IgA-APC (Clone: IS11-8E10, Miltenyi Biotec, Cat# 130-113-472), 1:20 IgE-PE (Clone: MB10-5C4, Miltenyi Biotec, Cat# 130-119-899) and 1:20 IgG-PerCP Vio700 (Clone: IS11-3B2.2.3, Miltenyi Biotec, Cat# 130-119-880) and Hoechst 33342 (10 μg/ml, BD Pharmingen, Cat# 561908) for 1 h on ice. Unstained and Hoescht only controls for each sample were also prepared. The samples were then washed in FACS buffer (1X DPBS + 1%FBS + 2 Mm EDTA) and acquired on BD FACS Aria III. IgA, IgE and IgG single positive samples were acquired based on gating established from unstained and Hoescht only samples, in addition to Fluorescent Minus One (FMO) controls for each antibody. All samples were subsequently analyzed using FlowJo (Treestar) software.
Bacterial DNA extraction
Total DNA from the sorted mono-Ig coated bacterial samples was extracted using QIAamp Fast DNA stool Mini Kit (QIAGEN, Cat# 51604), according to manufacturer’s instructions. The extracted DNA were stored at − 20 °C. The DNA integrity and concentration was measured using Qubit™ dsDNA Quantitation High Sensitivity Assay kit before the sequencing was performed.
16S rRNA gene amplification and sequencing
From the total isolated DNA, the V3–V4 regions of the 16S rRNA gene was amplified using Illumina Nextera XT library preparation Kit (Illumina, Cat# FC-131-1002,) and specific primers for V3-V4 region amplification:
Forward 5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG
Reverse 5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC
PCR amplification was done using equal starting DNA concentration. The amplified PCR products were purified using the Illumina 16S metagenomic sequencing library preparation protocol as per the instructions using AMPure XP magnetic beads (Beckman Coulter, Cat# A63881). The dual Index PCR was performed using the same protocol and purified as per the instructions. High-throughput sequencing was performed on an Illumina MiSeq2 × 300 platform in accordance with the manufacturer’s instructions.
Sequence analysis and taxonomic identification
The sequenced data was demultiplexed using the MiSeq Control Software (MCS), processed, and analyzed as described in the previous work using QIIME v1.9.0 pipeline [41]. Operational taxonomic units (OTUs) were generated by aligning against the Greengenes database (Version 13_8) with a confidence threshold of 97% [42]. Alpha diversity measures (Chao1, Observed, Shannon and Simpon indices) were calculated using “Phyloseq”, an R package [43]. Beta diversity indices were shown as principal coordinate analysis and R package ‘‘MicroEco’’ was used to perform the differences in the Bray–Curtis distance matrix between the study groups [44]. Differentially abundant taxa between the studied groups were identified using Linear Discriminant Analysis Effect Size (LEfSe) tool [45]. Microbial dysbiosis was measured using the package “dysbiosisR” and shared OTUs/taxa between the study groups were performed using the packages “MicrEco” and “Microbiota Process” [46, 47].
Statistical analysis
Statistical tests were performed using GraphPad PRISM software 10.2.3 (GraphPad Software, La Jolla, CA, USA) and data are shown as mean ± SEM. Shapiro-Wilks normality analysis was used to determine whether the data was normally distributed. Quantitative variables were compared using Kruskal Wallis-Dunn’s post-hoc analysis, one-way or two-way ANOVA-Tukey post-hoc analysis and one-way ANOVA-Dunnet post-hoc analysis as indicated for each figure panel. Statistical significance is defined as *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001. Data are represented as mean ± SEM.
Results
Altered IgA coating within gut bacterial populations in IgE and non-IgE mediated cow milk allergic pediatric subjects
To study the differential immunoglobulin responses generated against gut resident bacteria in allergic subjects, we utilised a flow cytometry-based antibody detection assay to allow for the identification and multiparametric analysis of Ig bound bacteria and subsequent sorting. Using multicolor flow cytometry, we performed parallel phenotyping of IgA, IgE and IgG coated bacteria in control (n = 13) and CMA subjects (n = 25). Allergic subjects were grouped into IgE mediated CMA without atopic dermatitis (ALL = 8), IgE mediated CMA with atopic dermatitis (AD = 10) and non-IgE mediated CMA with enteropathy (ENP = 7) groups (Fig. 1A). The demographics of the study subjects are detailed in Table 1. There were no significant differences in the gender, mode of delivery, gestational age, breastfeeding/formula feeding, solid food introduction, playgroup, maternal use of antibiotics and exposure to cigarette smoke, between the controls and those infants with allergic manifestations (Table 1). However, significant differences in both direct and indirect familial histories of allergy were detected (Table 1). ImmunoCAPs testing was conducted to determine levels of total IgE, total cow milk protein-, casein-, β-lactoglobulin- and α-lactalbumin-specific-IgE (sIgE). Our data showed increased levels of IgE and sIgE in the IgE mediated CMA groups (ALL and AD), whereas basal levels were detected in both CON and ENP groups (Table 2).
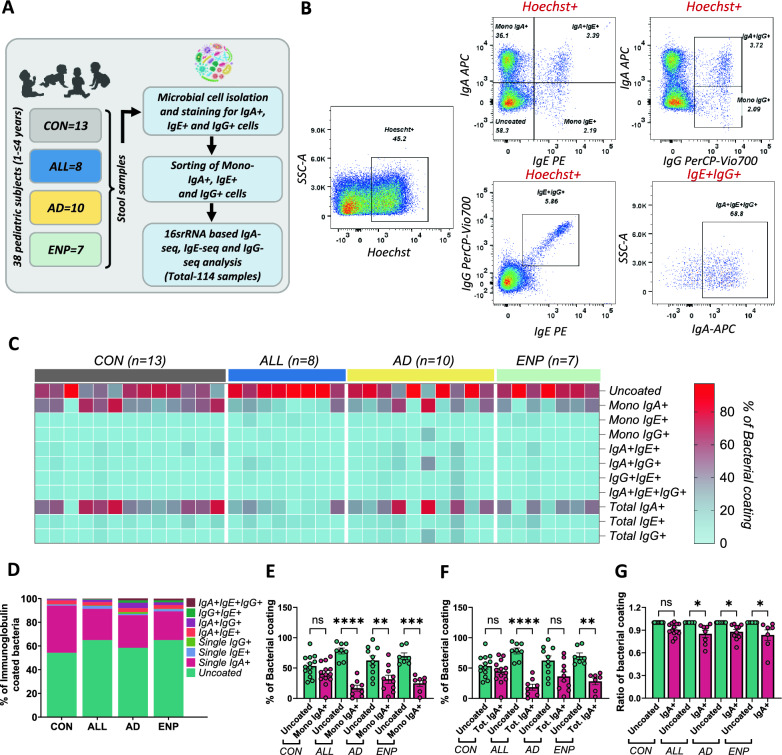
Fig. 1.
Altered IgA coating in cow milk allergic subjects. A Study outline. B Gating strategy used for identifying bacterial coating in the study subjects. C Heat-map showing the differential coating of bacterial populations in the study subjects. D Stacked plot for differential bacterial coating among the study groups. E and F Quantitative analysis of differential distribution of IgA + (E) and total IgA + (F) alongside uncoated bacteria within the study groups. G Quantitative analysis of the ratio of mono-IgA + to total IgA + coated bacteria compared to the normalised level of uncoated bacteria within the study groups. Quantitative variables were compared using one-way ANOVA and Tukey post-hoc analysis. Statistical significance is defined as *p < 0.05, **p < 0.01 and ***p < 0.001. Data are represented as mean ± SEM. See also Supplementary Figure S1 for fluorescence minus one and background staining controls
Table 2.
Comparative analysis of plasma IgE levels in study subjects for total and cow-milk allergen-specific IgE
| Plasma IgE levels (kUAD/L) | CON | ALL | AD | ENP | p ALL vs. CON | p AD vs. CON | p ENP vs. CON | p ALL vs. AD | p ALL vs. ENP | p AD vs. ENP |
|---|---|---|---|---|---|---|---|---|---|---|
| IgE Total (mean, SD)º | 24.38, 24.75 | 117.8, 99.51 | 956.4, 1101 | 17.26, 14.6 | ns | ** | ns | ns | * | ** |
| sIgE CMP (F2) (mean, SD)º | 0.14, 0.12 | 9.18, 8.95 | 111.6, 270.4 | 0.21, 0.35 | * | ** | ns | ns | ** | ** |
| sIgE Casein (F78) (mean, SD)º | 0.08, 0.07 | 6.55, 9.16 | 102.2, 255.4 | 0.08, 0.05 | * | * | ns | ns | ns | ns |
| sIgE β-lactoglobulin (F77) (mean, SD)º | 0.06, 0.06 | 2.13, 2.28 | 8.17, 11.91 | 0.06, 0.08 | * | ** | ns | ns | * | ** |
| sIgE α-lactalbumin (F76) (mean, SD)º | 0.06, 0.09 | 3.01, 2.02 | 5.84, 7.26 | 0.14, 0.33 | * | ns | ns | ns | ** | * |
ºKruskal Wallis with Dunn’s multiple comparison used as statistical test
IgA, IgE, and IgG-coated bacterial cells extracted from fecal samples, were analyzed by flow cytometry according to the gating strategy outlined (Fig. 1B and Supplementary Figure S1). Initial examination of the bacterial Ig coating revealed that a substantial proportion of bacteria remained uncoated, with no IgA, IgE or IgG binding detected (Fig. 1C and D). Amongst the Ig-coated bacteria, we identified a predominance of IgA coating across all subject groups (Fig. 1C and D). In contrast, IgE and IgG coating were observed at lower levels in all four study groups (Fig. 1C and D). We identified that Ig coating of bacteria occurred in multiple configurations as single, double and triple positive bacteria were observed across the study populations. Single- and double-coated bacteria were generally observed at higher frequencies, whereas triple-coated bacteria were observed at the lowest frequency and were absent in a proportion of the samples (Fig. 1C and D).
Previous studies have identified the association of reduced fecal IgA with food sensitization [48] and a decreased level of IgA coated bacteria in individuals with allergic conditions [21]. To investigate the patterns of overall IgA coating within our CMA study groups, we compared the percentages of mono-coated IgA + (Fig. 1E) and total IgA + (Fig. 1F) bacteria against uncoated bacteria. Here, we observed a decrease in the proportion of bacteria bound by IgA compared to uncoated bacteria present, with significant reduction in the mono-IgA + coated bacteria in all the three allergic disease states (Fig. 1E), while the total IgA + showed reduction of IgA + coated bacteria in ALL and ENP groups (Fig. 1F). In contrast, we observed levels of both mono-IgA + coated and total IgA + coated bacteria to be similar to those of uncoated bacteria in our control group (Fig. 1E and F), potentially indicating the presence of an altered IgA bound microbial abundance and composition amongst the CMA groups. To further assess changes in differential IgA coating, we quantified the fraction of mono-IgA + to total IgA + coated bacteria and compared the proportion of bacterial IgA coating against the normalized levels of uncoated bacterial proportions in each study group. Here, we identified that the bacterial IgA coating significantly decreased in ALL, AD and ENP allergy groups, while there was no significant differences in the CON group (Fig. 1G), further indicating that, a reduction of bacterial IgA coating occurs in both IgE and non-IgE mediated pediatric CMA subjects.
Differential patterns of Ig coating in CMA subjects
To identify the differential patterns of Ig coating present among the study groups, we quantified and compared the levels unbound and bound bacteria across the study groups. On analyzing the proportions of mono-IgA, IgE and IgG-coated bacteria, we identified a decrease in the percentages of IgA coating in all allergic groups with a significant decrease observed in the ALL group and the ENP group as compared with controls (Fig. 2A). Although reported by previous studies for IgE mediated allergic disease [21, 38], a decrease in bacterial IgA coating in a non-IgE-mediated allergic disease has not been previously identified. Although we observed variations in mono-IgE and mono-IgG coated bacteria, this did not yield a statistically significant result (Fig. 2B and C). Patterns of total IgA-coated bacteria showed decreased percentages in all allergy groups, with a significant decrease of total IgA-bound bacteria detected in the ALL and ENP groups (Fig. 2D). We also observed a significant increase in the total IgG bound bacteria in the AD group (Fig. 2F). However, levels of total IgE-coated bacteria, did not show any significant differences between the control and allergic subjects (Fig. 2E). Analysis of the proportion of uncoated bacteria revealed an increase in the percentage of unbound bacteria in allergic subjects, but only the ALL group showed a significant increase as compared to controls (Fig. 2G).
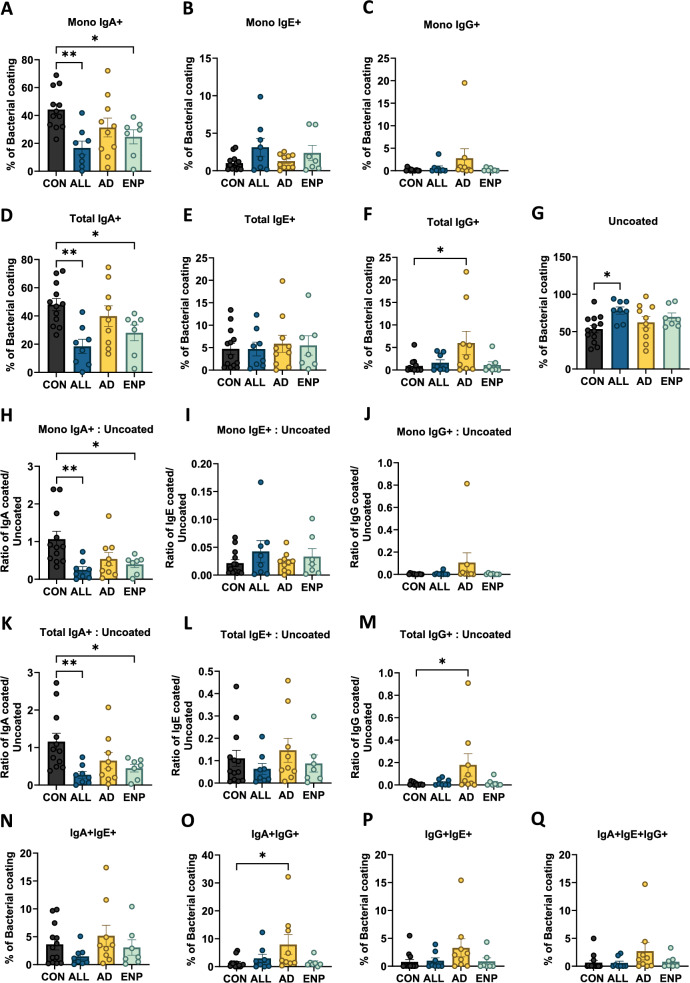
Fig. 2.
Differential bacterial coating of IgA, IgE and IgG in cow milk allergic subjects. A, B Comparative analysis of mono-Ig coated and total Ig coated bacterial populations with respect to control subjects. C Quantification of uncoated bacteria across the study subjects. D and E Comparative analysis of the ratio of mono-Ig coated (D) and total Ig coated (E) bacteria to uncoated bacteria in the study subjects. F Comparison analysis of dual IgA + IgE +, IgA + IgG + and IgE + IgG + coated bacteria across the study groups. F Quantification of triple coated (IgA + IgE + IgG +) bacteria across the study subjects. Quantitative variables were compared using one-way ANOVA and Dunnet post-hoc analysis. Statistical significance is defined as *p < 0.05, **p < 0.01 and ***p < 0.001. Data are represented as mean ± SEM
Further, we analyzed the ratios of mono and total IgA, IgE and IgG-coated bacteria to uncoated bacteria present (Fig. 2H–M). We observed a significant decrease in mono and total IgA-coated bacteria in ALL and ENP groups (Fig. 2H and K), and a significant increase in the ratio of total IgG-bound bacteria in the AD group (Fig. 2M). Next, we looked at the proportions of IgA + IgE +, IgA + IgG +, IgE + IgG + and IgA + IgE + IgG + coated bacteria present (Figs. 2N and Q). Interestingly, we identified a significant increase in the proportion of dual bound IgA + IgG + bound bacteria in the AD group compared to the CON group (Fig. 2O). Together, these findings indicate the presence of significant alterations in the proportions of Ig-coated bacteria in the IgE and non-IgE mediated CMA groups. Additionally, no increases in levels of IgE-coated bacteria were detected in the subjects with IgE-mediated disease, despite the presence of significantly higher levels of IgE in their plasma (Table 2).
Immunoglobulin coating reveals alterations in levels of coated fecal bacterial OTUs
To identify whether unique Ig coated bacterial profiles are associated with the different disease groups, we sorted bacterial populations that were mono-IgA, mono-IgE or mono-IgG-coated and performed 16S rRNA gene sequencing. After quality control, a total of 9,955,536 sequences (median: 102,614) were used for OTU generation. Following filtering and alignment, a total of 8126 OTUs were generated, with an average of 87,329 reads per sample.
We initially examined the patterns of unique and common OTUs identified as being bound by IgA, IgE or IgG (Fig. 3A–D). In CON, ALL and AD patients. Here, the relative Ig-recognized OTU set size present, indicated a pattern of Ig recognition where IgA > IgE > IgG (Fig. 3A–C), whereas for ENP patients, the set size pattern showed IgE > IgA > IgG (Fig. 3D). Interestingly, in all subject groups, we identified bacterial OTUs that were recognized by more than one of each of the mono-Ig subsets studied. Here, these shared OTUs recognized all three Ig subsets and constituted the largest OTU groups observed in all four of the study groups (Fig. 3A–D). This indicated that different bacterial OTUs belonging to the same taxonomic level could be individually coated by IgA, IgE and IgG.
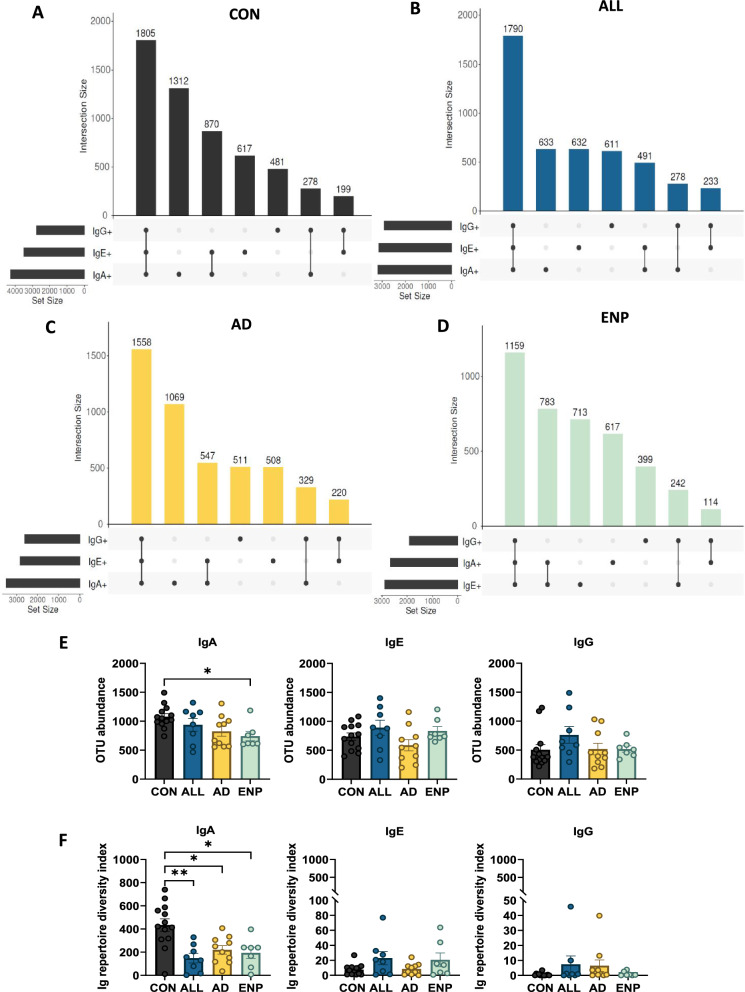
Fig. 3.
Immunoglobulin coating reveals alterations in levels of coated fecal bacterial OTUs. A–D Upset plots showing bacterial taxa across differentially coated subsets within A CON B ALL, C AD and D ENP groups. E Comparative analysis of total detected bacterial OTUs in the study groups. F Ig repertoire diversity index analysis as a measure of the percentage of mono-Ig bound bacteria together with that of the total OTU counts individually for IgA, IgE and IgG coated bacteria [Ig repertoire diversity index = (% of Mono-Ig coating) × (OTU abundance)]. Quantitative variables were compared using Kruskal–Wallis and Dunn’s post-hoc analysis. Statistical significance is defined as *p < 0.05 and **p < 0.01. Data are represented as mean ± SEM
The OTUs recognized individually by any two Ig subsets displayed a pattern where OTUs common to IgA + IgE + > IgA + IgG + > IgE + IgG + in CON, ALL and AD, whereas in the ENP group, the pattern observed was IgA + IgE + > IgE + IgG + > IgA + IgG + (Fig. 3A–D).
Further, as an estimate of the Ig-bound repertoire diversity present for each study subset, we first analyzed the total OTU counts in each study group for IgA, IgE and IgG subsets and identified a significantly lower number of IgA bound OTUs in the ENP group as compared to the CON group (Fig. 3E). We then used the number of OTUs present in individual study subjects’ samples to calculate the comparative proportion of bacteria bound by unique Ig’s as determined by flow cytometry to generate an Ig repertoire diversity index for IgA, IgE and IgG. This analysis showed a significant decrease in the repertoire diversity index for the IgA subset in all three CMA groups (Fig. 3F). No significant differences were observed in the diversity index calculated among the study groups for the IgE and IgG bound subsets (Fig. 3F).
Loss of alpha diversity and altered beta diversity in cow milk allergic subjects
The alpha diversity of the Ig coated bacteria was assessed to determine the species richness and evenness amongst the different allergic conditions. Alpha diversity was measured using Observed, Chao1, Shannon and Simpson indexes (Figs. 4A–C). Observed index reflecting the community richness by counting the unique OTUs showed a significant decrease in the alpha diversity of IgA coated bacteria; between controls and ENP patients (Fig. 4A). No significant difference was observed between controls and ALL or AD patients. Similarly, Chao1 index that estimates the total number of taxa while reflecting the rare OTUs showed a significant decrease of diversity in the ENP patients compared to controls. No significant difference was reflected in the Shannon or Simpson indices (Fig. 4A). In contrast, IgE-coated bacteria showed no significant differences in the alpha diversity in any of the allergic groups compared to controls (Fig. 4B). Among IgG-coated bacteria, the AD group showed decreased alpha diversity using the Shannon and Simpson indices (Fig. 4C).
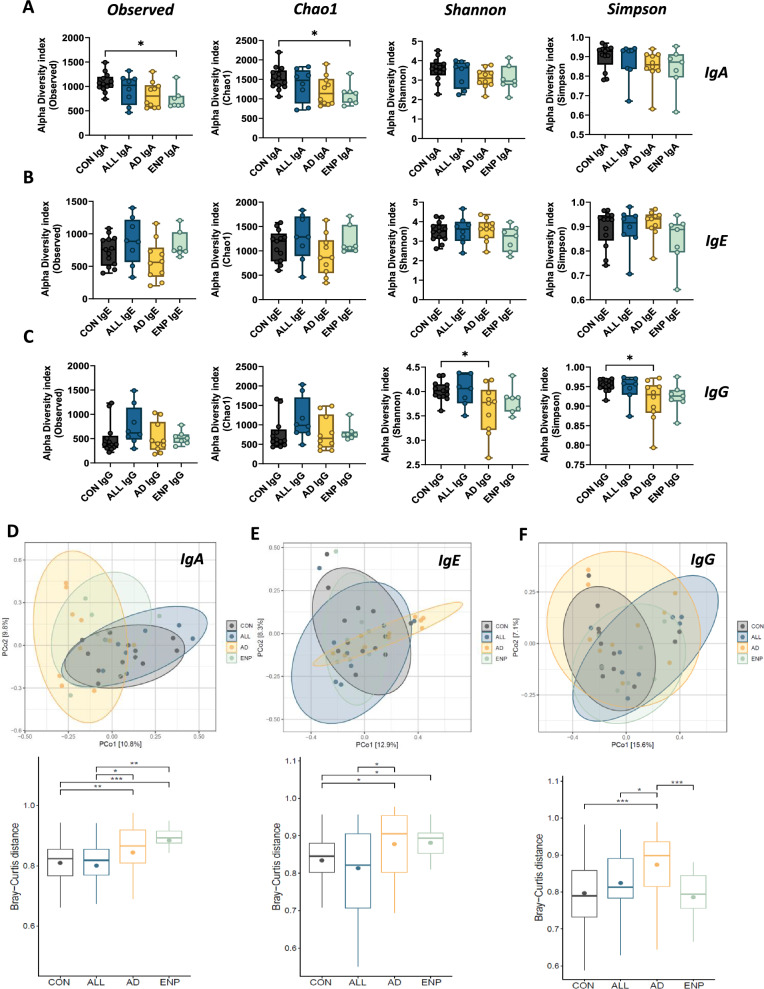
Fig. 4.
Alpha and beta diversity in cow milk allergic pediatric subjects. A Comparison of alpha diversity in IgA, IgE and IgG groups using Observed, Chao1, Shannon and Simpson indices. B, C, D Principal co-ordinate analysis plots of IgA (B), IgE (C) and IgG (D) coated bacteria (above) and box plots for the same (below). Quantitative variables were compared using one-way ANOVA and Dunnet post-hoc analysis for alpha diversity and Wilcoxon test for beta diversity. Statistical significance is defined as *p < 0.05, **p < 0.01 and ***p < 0.001. Data are represented as mean ± SEM
Next, we examined the beta diversity using the Bray–Curtis distance matrix which revealed that the AD group was significantly distant from the CON and ALL groups in IgA (Fig. 4D), IgE (Fig. 4E), and IgG-coated (Fig. 4F) bacterial groups. The ENP group was significantly distant from the CON group in IgA and IgE bacterial coating (Fig. 4D and E). The IgA coated ENP group also showed significant dissimilarity from the ALL group (Fig. 4D). The AD group showed significant dissimilarity of IgG coating, distinct from CON, ALL and ENP groups (Fig. 4F). Taken together, the above results identified that AD and ENP groups showed a loss of alpha diversity in IgA and IgG subsets and also displayed significant dissimilarity of beta diversity in IgA subset (AD and ENP) and IgG subset (AD) compared to controls.
Dysbiosis is associated with differentially coated bacterial subsets in cow milk allergy
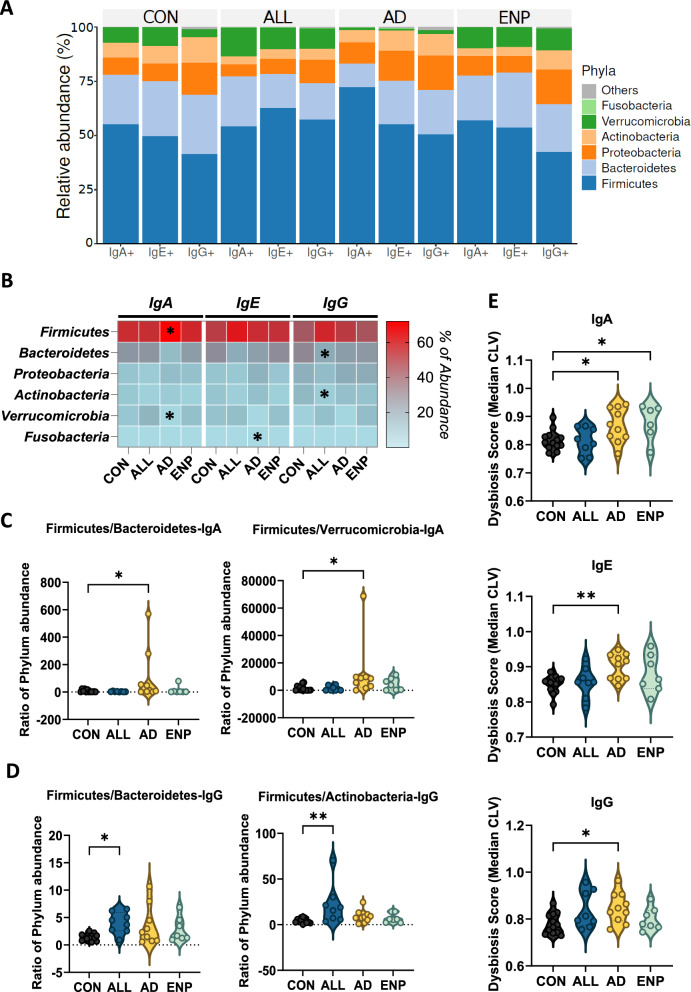
We next assessed the bacterial communities coated by IgA, IgE or IgG and identified that the majority of the coated bacteria belonged to the Firmicutes and Bacteroidetes phyla followed by Proteobacteria, Actinobacteria, Verruocomicrobia and Fusobacteria in the order of abundance (Fig. 5A). Analysis of the differences in the phyla abundance showed that in the IgA-coated AD group, Firmicutes were highly abundant (Fig. 5B and Supplementary Figure S2A), whereas Verrucomicrobia was present at a lower abundance as compared to the CON group (Fig. 5B and Supplementary Figure S2D). In the IgE coated subset, Fusobacteria was higher in AD group, compared to controls (Fig. 5B and Supplementary Figure S2F). While, in the IgG subset, ALL subjects had a significantly lower abundance of Bacteroidetes (Fig. 5B and Supplementary Figure S2B) and Actinobacteria in comparison to controls (Fig. 5B and Supplementary Figure S2E).
Fig. 5.
Dysbiosis is associated with differential bacterial Ig coating in CMA subjects. A Most abundant phyla among the mono- IgA, IgE and IgG coated bacteria. B Heat-map showing the significant differential distribution of phyla recognition patterns in IgA, IgE and IgG subsets. Each subset has been compared with the control group in the respective subsets. C, D Quantitative analysis of dysbiosis at phyla level analyzing Firmicutes/Bacteroidetes and Firmicutes/Verrucomicrobia in IgA coated (C) and Firmicutes/Bacteroidetes and Firmicutes/Actinobacteria in IgG coated bacteria (D). E Quantitative analysis of dysbiosis using Median CLV dysbiosis score in IgA, IgE and IgG coated bacteria. Quantitative variables were compared using Kruskal–Wallis and Dunn’s post-hoc analysis for panels B–D and one-way ANOVA and Dunnet post-hoc analysis for panel E. Statistical significance is defined as *p < 0.05, **p < 0.01. See also Additional file 1, Supplementary Figure S2. Data are represented as mean ± SEM
To assess the dysbiosis associated with the bacterial coating across the different allergic groups, we calculated the ratios of Firmicutes in comparison to the other major phyla identified across the Ig-coated subsets. In the IgA-coated bacterial subset, we identified an increase in the ratio of Firmicutes to Bacteroidetes in the AD group as compared to the CON group (Fig. 5C). The ratio of Firmicutes to Verruocomicrobia, increased in the AD group compared to controls in the IgA-coated bacterial subset (Fig. 5C). IgG-bound bacteria showed an increase in the ratio of Firmicutes to Bacteroidetes and an increase in Firmicutes to Actinobacteria in the ALL group (Fig. 5D). These results align with a previous study which reported an enrichment of Firmicutes and decrease of Bacteroidetes, Proteobacteria and Actinobacteria in subjects with food allergies [49].
The levels of dysbiosis associated with bacterial subsets at the OTU level were then evaluated using median CLV score, as described previously [47]. Based on this scoring analysis, we identified bacterial dysbiosis to be associated with IgA, IgE and IgG-bound bacteria in AD subjects, as compared to controls (Fig. 5E). IgA-coated bacteria in ENP subjects also displayed a significant level of dysbiosis compared to controls (Fig. 5E).
Preferential coating of bacteria of Ig’s differs by cow milk allergic phenotype
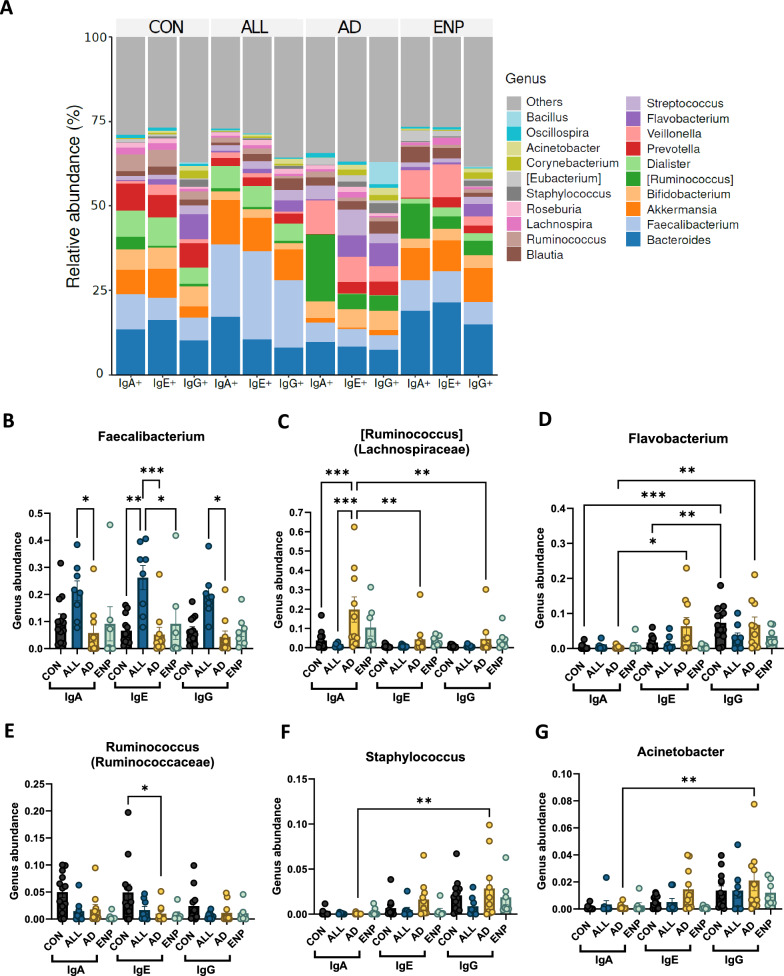
Next, we explored the bacterial composition at the genus level, to identify their differential abundance among CMA patient subsets. The top 25 abundant bacterial genera across all three subsets—IgA, IgE, and IgG were primarily identified (Fig. 6A). For each individual subset of IgA, IgE and IgG-coated bacteria, the top-most abundant 25 genera are shown in Supplementary Figure S3 A-C. Although Bacteroides was identified as the dominant genus within the three Ig subsets (Fig. 6A and Supplementary Figure S3 A-C), we did not identify any significant differences in the binding of Bacteroides to IgA, IgE or IgG between the study groups (Supplementary Figure S4). Interestingly, the second most abundant genus, Faecalibacterium (Fig. 6A and Supplementary Figure S3 A-C) was found predominantly coated by IgA, IgE and IgG in the ALL group (Fig. 6A and B). Faecalibacterium displayed the highest significant differences within the IgE bound subset, with significantly higher coating in the ALL group compared to CON, AD and ENP groups (Fig. 6B). In addition, within IgA and IgG-coated subsets, Faecalibacterium showed significantly higher binding in the ALL group compared to AD group (Fig. 6B).
Fig. 6.
Preferential bacterial coating of most abundant bacteria in CMA subjects. A Top 25 most abundant bacterial genera across mono- IgA, IgE and IgG coated bacteria. B–G Quantitative analysis of differential abundance of IgA, IgE and IgG coated B Faecalibacterium, C Ruminococcus from Lachnospiraceae D Flavobacterium E Ruminococcus from Ruminococcaceae F Staphylococcus and G Acinetobacter. Quantitative variables were compared using two-way ANOVA and Tukey post-hoc analysis. Statistical significance is defined as *p < 0.05, **p < 0.01 and ***p < 0.001. Data are represented as mean ± SEM. (See also Supplementary Figure S3 and Supplementary Figure S4)
Ruminococcus genus from the family Lachnospiraceae was significantly enriched within the IgA coated bacteria from the AD group, as compared to IgA-coated Ruminococcus in CON and ALL groups (Fig. 6C). In addition, the abundance of IgA-coated Ruminococcus was also significantly greater than IgE and IgG coated Ruminococcus (Fig. 6C). Flavobacterium showed significantly higher rates of IgE and IgG coating than IgA coating in the AD group (Fig. 6D). Control subjects also exhibited a higher level of IgG-coated Flavobacterium, as compared to IgA and IgE coated bacteria (Fig. 6C). Additionally, a second Ruminococcus genus from the Ruminococcaceae family exhibited significant decreases in IgE coating in the AD group in comparison to the control group (Fig. 6E), and Staphylococcus and Acinetobacter were found to be predominantly IgG-coated in the AD group (Fig. 6F and G). Akkermansia, Bifidobacterium, Veillonella, Dialister, Prevotella, Streptococcus, Blautia, Lachnospira, Roseburia and Eubacterium did not show significant changes in their preferences of binding to IgA, IgE or IgG (Supplementary Figure S4). However, Corynebacterium showed significant difference between IgA and IgG bound bacteria in the control group (Supplementary Figure S4).
Identification of uniquely dominant bacterial targets of Ig response in CMA groups
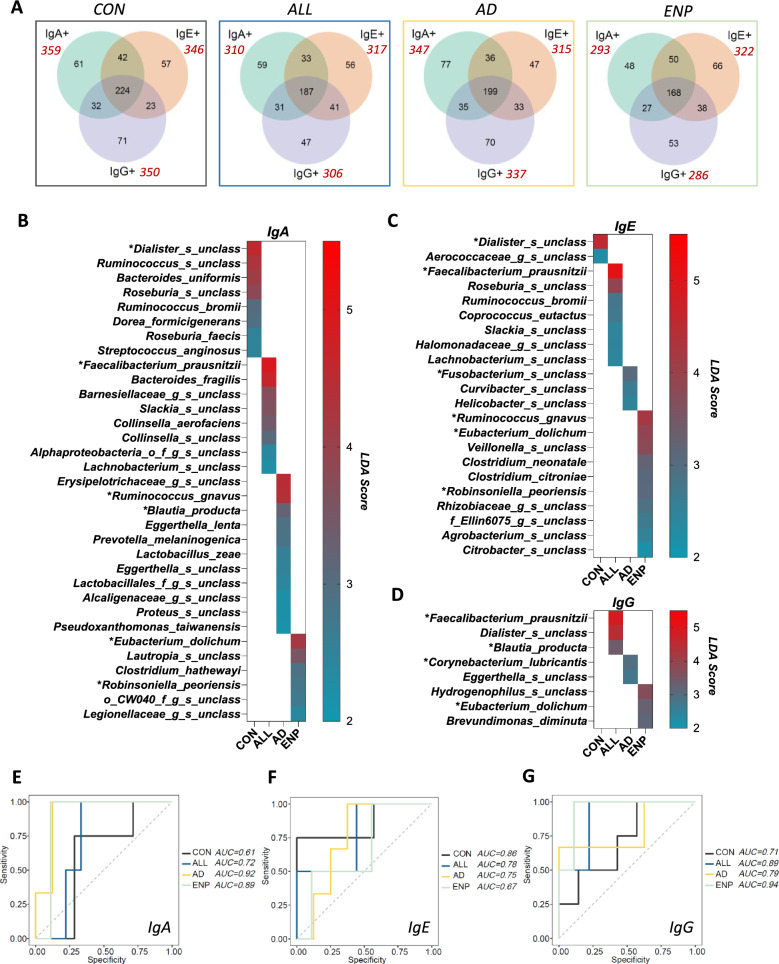
To further identify the bacterial targets of Ig’s at the species level, we identified the unique and common bacterial species bound by IgA, IgE and IgG subsets present in the study groups (Fig. 7A). Subsequently we characterized the most abundant bacterial targets bound by specific Ig’s across the study groups, LEfSe analysis was conducted at the species level, allowing the determination of the bacterial species that were most predominantly coated by an individual Ig in each of the different study groups (Fig. 7B–D). The most abundant uniquely IgA coated bacterial subsets within the CON group were Dialister, Ruminococcus, Bacteroides uniformis, Roseburia, Ruminococcus bromii, Dorea formicigenerans, Roseburia faecis and Streptococcus anginosus (Fig. 7B). Roseburia species, Ruminococcus bromii have been previously identified to be highly abundant in non-allergic conditions [50]. Interestingly, in the CON group, we identified predominance of only two bacterial species in the IgE coated subset, namely, Dialister and an unclassified genus from the Aerococcaceae family, and none in the IgG-coated subset (Fig. 7C and D).
Fig. 7.
Identification of uniquely dominant bacterial targets of Ig response in CMA groups. A Unique and common bacterial species present in IgA, IgE and IgG subsets among the study groups. Total number of bacterial species bound by each Ig subset in the different study groups are shown in red. B–D LEFse analysis showing uniquely abundant bacteria with LDA score above 2 within B IgA, C IgE and D IgG coated bacteria. E–G ROC curve analysis showing the AUC analysis for E IgA, F IgE and G IgG coated bacteria
Faecalibacterium prausnitzii was identified as coated by IgA, IgE and IgG in the ALL group (Fig. 7B–D). We also determined that Eubacterium dolichum was predominantly coated by IgA, IgE and IgG in the ENP group (Fig. 7B–D). Ruminococcus gnavus was found to be predominantly coated with IgA in the AD group and with IgE in the ENP group (Fig. 7B and C). Bacteroides fragilis, a species from Barneselliaceae family, Slackia, Collinsella aerofaciens, a second Collinsella species, a species from phyla Alphaproteobacteria and Lachnobacterium were all found to be most abundant bacteria among the IgA coated bacteria in the ALL group (Fig. 7B). In addition to Ruminococcus gnavus, the IgA coated AD group showed predominance of a species from Erysipelotrichaceae, Blautia producta, Eggerthella lenta, Prevotella melaninogenica, Lactobacillus zeae, an unclassified Eggerthella species, two unclassified species from order Lactobacillales and family Alcaligenaceae, Proteus and Pseudoxanthomonas taiwanensis (Fig. 7B). The AD group displayed the highest number of IgA coated species identified, as compared to the rest of the subject groups (Fig. 7B). In addition to the presence of Eubacterium dolichum, in the IgA coated ENP group, we identified the presence of Lautropia, Clostridium hathawayei, Robinsonella peoriensis, two unclassified species from order CW040 and a member of the Legionellaceae family (Fig. 7B).
Among the IgE-coated bacteria, we identified the highest number of uniquely coated bacterial OTUs (Fig. 3D) and highest number of uniquely bound bacterial species (Fig. 7A) present in the ENP group. While Roseburia, Ruminococcus bromi, Coprococcus eutactus, Slackia, a species from Halomonadaceae family and Lachnobacterium were found to be highly IgE coated in the ALL group, Fusobacterium, Curvibacter and Helicobacter were most abundant in the IgE coated AD group samples (Fig. 7C). In the ENP group, following Ruminococcus gnavus, Eubacterium dolichum was found to be predominantly IgE-coated followed by Veillonella, Clostridium neonatale, Clostridium citroniae, Robinsoniella peoriensis, two unclassified species from Rhizobiaceae and Ellin6075, Agrobacterium and Citrobacter (Fig. 7C).
Bacterial IgG coating in the ALL group was found to be highest for Dialister and Blautia producta, both of which followed Faecalibacterium prausnitzii (Fig. 7D). Corynebacterium lubricantis and Eggerthella were predominantly IgG coated in the AD group (Fig. 7D). The elevated proportion of IgG bound bacteria observed in the AD group could potentially have been driven by the presence of these pathogenic bacteria. Hydrogenophilus and Brevundimonas diminuta, in addition to Eubacterium dolichum were found to be highly IgG coated in the ENP group (Fig. 7D).
To understand the specificity of the biomarkers identified in each of the allergic groups and Ig subsets, the area under the ROC curve (AUC) analysis for IgA, IgE and IgG subsets was performed, using the bacterial biomarkers identified from the LEFse analysis in Fig. 7B–D. Analysis of the AUC values for the IgA-coated subset generated values of 0.61, 0.72, 0.92 and 0.89 for CON, ALL, AD and ENP groups respectively (Fig. 7E). For the IgE-coated CON, ALL, AD and ENP groups, AUC values were 0.86, 0.78, 0.75 and 0.67 respectively (Fig. 7F). The IgG-coated subset gave AUC values of 0.71, 0.89, 0.79 and 0.94 respectively for the CON, ALL, AD and ENP groups (Fig. 7G). As such, these results indicate that the Ig-specific binding of the identified microbes (Fig. 7B–D) are reliable biomarkers associated with IgE and non-IgE mediated cow milk allergy.
Discussion
In this study, we sought to identify the presence of differential Ig-specific bacterial coating patterns of IgA, IgE and IgG to gut bacteria in pediatric CMA subjects and to discern whether there are disease specific patterns of humoral responses through the application of Ig based bacterial sorting and subsequent 16S rRNA gene sequencing. A thorough understanding of the patterns of antibody responses generated against specific gut bacteria may aid in understanding the functional relevance of Ig’s in allergic disease and could potentially be developed as biomarkers for the severity of allergy or as a therapeutic target for modulating responses in specific disease settings. It has been previously shown that elevated intestinal IgA levels are protective against allergic diseases [37]. Moreover, IgA has been identified by several groups as an important factor for the maintenance of intestinal homeostasis, by inhibiting bacterial virulence and through shaping gut microbial composition to promote colonization by symbiotic bacteria [51–53]. The microbiota in the gut can also steer IgA production [54] which controls bacterial translocation and enables neutralization of bacteria toxins at the mucosal surface [55]. Our preliminary results identified a significantly decreased proportion of unique and total IgA bound bacteria in both IgE and non-IgE mediated CMA conditions by pairwise comparison in control and allergic subjects. Furthermore, quantification of the unique and total IgA coating in allergic subjects in comparison to non-allergic control subjects identified a significant decrease in bacterial IgA coating for IgE mediated ALL and non-IgE mediated ENP groups. An elevated level of mono-IgG and total IgG bacterial coating was identified as a unique feature for the IgE mediated AD group. AD subjects also exhibited a significant increase in IgA + IgG + and IgE + IgG + dual coated bacteria. Notably, the IgE coating of bacteria in the gut, did not show any differences between CON and the IgE mediated allergic groups, despite the presence of significantly greater levels of IgE in plasma for the ALL and AD groups (Table 2). Although the presence of IgE in the gut has previously been reported to be positively corelated between plasma and fecal samples in patients with atopic allergic disease [56] and those with parasitic infections [57, 58], our results indicate that the levels of plasma IgE present, may not directly correspond with the degree of bacterial IgE coating in the gut. Moreover, these studies have either focused on total levels of free IgE [57] or on the presence of allergen specific IgE and not specifically on gut bacteria coated by IgE [59]. Additionally, it has been shown that IgE could act as an antigen/allergen transepithelial transporter in combination with CD23 indicating it may play a distinct role in gut associated humoral responses [60]. To our knowledge the only prior study to directly assess IgE binding of bacteria in the human gut was a recent report by Abdel-Gadir et al., where, in contrast to our findings, they identified increased levels of bacterial IgE coating in IgE mediated food allergic subjects [38]. The disparity in these findings may potentially be due to differences in study populations including their younger age of recruitment, higher levels of multi-sensitization, or other clinical factors. Additionally, it should be noted that in the murine model employed by Abdel-Gadir et al. they observed that allergic sensitization with OVA and staphylococcal enterotoxin B, did not lead to an increase in the levels of IgE coated bacteria present in wild type animals. An additional study by Parrish et al. [39] also assessed IgE binding to gut microbes in a murine model. Here, increased levels of IgE coated bacteria were found to be present after five weeks on a fiber free diet, however this difference was only detectable in fecal content and was not detectable in ileal content. Taken together, these data provide an initial insight into how altered humoral immune responses to gut bacteria induced by the onset of food allergic conditions may be important in shaping the course of disease. However, many questions still remain unanswered as to the exact role of IgE mediated immune responses in the gut, especially in relation to potential microbiome specific responses, how IgE-producing memory B cells or plasma cells are generated, and whether IgE production is localized to the gut [61, 62] or is transported from the plasma [63].
The overall decrease in alpha diversity and dysbiosis we identified in association with the CMA subjects could potentially be due to the altered levels of IgA and IgG detected in these subjects. Alternatively, the increased prevalence of pathogenic bacteria identified under allergic conditions including, Bacteroides fragilis, Collinsella aerofaciens (IgA-ALL), Ruminococcus gnavus (IgA-AD), Clostridium hathewayi, Robinsonella peoriensis (IgA-ENP), could have contributed to decreases in IgA levels in these CMA subjects, through previously identified mechanisms such as the proteolytic cleavage of IgA which results in a decreased proportion of IgA bound bacteria [64, 65]. We also observed an increase in IgG bound bacteria in the AD group, possibly due to the increased abundance of pathogenic bacteria identified in this group, namely, Staphylococcus, Acinetobacter and Corynebacterium.
Following analysis of the 20 most abundant genera bound by IgA, IgE or IgG, we identified significant differences in the bacterial coating of Faecalibacterium prausnitzii, Ruminococcus gnavus, Flavobacterium, Ruminococcus bromii, Staphylococcus and Acinetobacter among the CMA groups. Our study found Staphylococcus and Corynebacterium to be more highly recognized by IgG, which is consistent with the established roles of Staphylococcus [66] and Corynebacterium [67] in atopic dermatitis. Moreover, atopic dermatitis has been reported to precede the development of food allergy [68] and skin colonization with Staphylococcus aureus has been shown to increase the susceptibility to food allergy [69]. Our study indicates that Staphylococcus aureus is also more prevalent in the gut of IgE mediated CMA subjects presenting with atopic dermatitis, which was observed to be predominantly bound by IgG suggesting the induction of an inflammatory response. We also identified an increased abundance of IgG coated Acinetobacter in the AD group (Fig. 6G). Acinetobacter was in one instance shown to be associated with protection against allergic sensitization and inflammation [70]. However, it has also been alternately reported that Acinetobacter baumannii, Acinetobacter lwoffii and Acinetobacter haemolyticus are associated with an increased incidence of AD [71].
Faecalibacterium prausnitzii has been previously found to be abundant in food and respiratory allergic conditions [50] and has also been shown to be strongly associated with atopic dermatitis [72]. Our observation strengthens these previous studies which have identified an increased abundance of Faecalibacterium prausnitzii in allergic conditions. Likewise, our results show Eubacterium dolichum to be highly bound by IgA, IgE and IgG in the ENP group, aligning with a previous study on Eubacterium dolichum which reported an increased bacterial burden in a mouse model of food allergy [73]. Eubacterium dolichum, hence could be a potential biomarker for non-IgE mediated cow milk allergy. Robinsoniella peoriensis, identified as highly IgA and IgE coated in the ENP group, could also be a potential biomarker for determining a predisposition to non-IgE mediated cow milk allergy. Further, Ruminococcus gnavus has been previously shown to be associated with increased allergic conditions in infants [74] and children [50] and a pathogenic role for Ruminococcus gnavus has also been found in association with diarrhea-predominant irritable bowel syndrome, through increasing serotonin biosynthesis [75]. Bacteroides fragilis was identified as highly coated by IgA in the ALL group (Fig. 7A) and although a previous study has reported the role of Bacteroides fragilis in the maintenance of tolerance [76], its increased abundance has also been associated with peanut and nut-allergic patients [77].
Conclusions
Although IgE mediated allergic diseases have been previously studied in association with the microbiome, non-IgE mediated food allergies are not yet well described, and their microbial association has not been clearly established. Our results have identified significant differences in how the bacterial Ig coating with respect to IgA, IgE and IgG varies in pediatric IgE and non-IgE mediated cow milk allergic conditions. This is the first-time bacterial Ig coating has been studied in the context of a non-IgE mediated allergy. Here, we report a previously unidentified association between decreased IgA coating and non-IgE mediated CMA. We also identify an elevated IgG binding in subjects presenting with atopic dermatitis associated with IgE mediated CMA. Moreover, we show the existence of specific gut bacterial signatures that are preferentially bound by IgA, IgE and IgG in both IgE and non-IgE mediated CMA conditions. Hence, the results from our study, provide further evidence that alterations in humoral responses that result in dysbiosis may be an underlying cause of allergic disease; raising the prospect for developing new insights into the clinical diagnosis of non-IgE mediated allergic conditions, and the design of future therapeutic interventions, by targeting the bacterial species identified here.
Supplementary Information
Acknowledgements
We thank all study participants and their parents for their involvement in this study.
Abbreviations
- CMA
Cow milk allergy
- Ig
Immunoglobulin
- CON
Healthy controls
- ALL
IgE mediated CMA subjects without atopic dermatitis
- AD
IgE mediated CMA subjects with atopic dermatitis
- ENP
Non-IgE mediated CMA subjects
- FPE
Food protein-induced allergic enteropathy
- FPIES
Food protein-induced enterocolitis syndrome
- FPIAP
Food protein-induced allergic proctocolitis
- IBD
Inflammatory bowel disease
- OTU
Operational taxonomic unit
- LEFSe
Linear discriminant analysis effect size
Author contributions
All listed authors contributed to this study. TA, FB and NVP contributed to the study design. TA, FB and SM contributed to study execution. AA, ME and MA enrolled patients and recorded clinical information. FB performed the consenting, collection of questionnaires and coordinated sample collection. TA performed flow cytometry assays, bacterial sorting and analysis, and DNA isolation from sorted Ig coated bacterial cells. JC and GG contributed to design and optimal sorting of Ig coated bacterial cells. SM performed 16S rRNA gene sequencing and analysis. TA and NVP wrote the manuscript. TA, SM and NVP contributed to data analysis. TA, SM, JC, SAK and NVP engaged in study interpretation and critical analysis. All authors reviewed the manuscript.
Funding
This work was supported by funding from Sidra Medicine Internal Research Fund to Dr. Nicholas van Panhuys.
Availability of data and materials
16S rRNA gene sequencing analysis data are deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA1127830 [(Run Selector:: NCBI (nih.gov), SRP520730-SRA-NCBI (nih.gov)].
Declarations
Ethics approval and consent to participate
The study obtained written parental consent from all participants before enrollment. The study protocol (1500751) was approved by the Institutional Review Board (IRB) at Sidra Medicine, Doha, Qatar
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial or non-financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Savage J, Johns CB. Food allergy: epidemiology and natural history. Immunol Allergy Clin North Am. 2015;35(1):45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flom JD, Sicherer SH. Epidemiology of cow’s milk allergy. Nutrients. 2019;11(5):1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elizur A, et al. Natural course and risk factors for persistence of IgE-mediated cow’s milk allergy. J Pediatr. 2012;161(3):482–7. [DOI] [PubMed] [Google Scholar]
- 4.Santos A, Dias A, Pinheiro JA. Predictive factors for the persistence of cow’s milk allergy. Pediatr Allergy Immunol. 2010;21(8):1127–34. [DOI] [PubMed] [Google Scholar]
- 5.Gupta RS, et al. Prevalence and severity of food allergies among US adults. JAMA Netw Open. 2019;2(1): e185630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriki D, et al. The role of the gut microbiome in cow’s milk allergy: a clinical approach. Nutrients. 2022;14(21):4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrickx DM, et al. Assessment of infant outgrowth of cow’s milk allergy in relation to the faecal microbiome and metaproteome. Sci Rep. 2023;13(1):12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantazi AC, et al. Relationship between gut microbiota and allergies in children: a literature review. Nutrients. 2023;15(11):2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biasucci G, et al. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(Suppl 1):13–5. [DOI] [PubMed] [Google Scholar]
- 10.Yassour M, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8(343):343ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, et al. Association between breastmilk microbiota and food allergy in infants. Front Cell Infect Microbiol. 2021;11: 770913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heine RG. Food allergy prevention and treatment by targeted nutrition. Ann Nutr Metab. 2018;72(Suppl 3):33–45. [DOI] [PubMed] [Google Scholar]
- 13.Oddy WH. Breastfeeding, childhood asthma, and allergic disease. Ann Nutr Metab. 2017;70(Suppl 2):26–36. [DOI] [PubMed] [Google Scholar]
- 14.Augustine T, et al. Microbial dysbiosis tunes the immune response towards allergic disease outcomes. Clin Rev Allergy Immunol. 2023;65(1):43–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ege MJ, et al. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol. 2006;117(4):817–23. [DOI] [PubMed] [Google Scholar]
- 16.Fall T, et al. Early exposure to dogs and farm animals and the risk of childhood asthma. JAMA Pediatr. 2015;169(11): e153219. [DOI] [PubMed] [Google Scholar]
- 17.Ege MJ, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364(8):701–9. [DOI] [PubMed] [Google Scholar]
- 18.Sbihi H, et al. Thinking bigger: how early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy. 2019;74(11):2103–15. [DOI] [PubMed] [Google Scholar]
- 19.Maynard CL, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dzidic M, et al. Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. J Allergy Clin Immunol. 2017;139(3):1017-1025.e14. [DOI] [PubMed] [Google Scholar]
- 22.Johnson CC, Ownby DR. The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl Res. 2017;179:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caparros E, et al. Intestinal microbiota is modified in pediatric food protein-induced enterocolitis syndrome. J Allergy Clin Immunol Glob. 2022;1(4):217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mennini M, et al. Microbiota in non-IgE-mediated food allergy. Curr Opin Allergy Clin Immunol. 2020;20(3):323–8. [DOI] [PubMed] [Google Scholar]
- 25.Berni Canani R, et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow’s milk allergy. Sci Rep. 2018;8(1):12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Iede M, et al. Perspectives on non-IgE-mediated gastrointestinal food allergy in pediatrics: a review of current evidence and guidelines. J Asthma Allergy. 2023;16:279–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goswami R, et al. Systemic innate immune activation in food protein-induced enterocolitis syndrome. J Allergy Clin Immunol. 2017;139(6):1885–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berin MC. Immunopathophysiology of food protein-induced enterocolitis syndrome. J Allergy Clin Immunol. 2015;135(5):1108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamji MH, et al. The role of allergen-specific IgE, IgG and IgA in allergic disease. Allergy. 2021;76(12):3627–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fokkens WJ, Vinke JG, KleinJan A. Local IgE production in the nasal mucosa: a review. Am J Rhinol. 2000;14(5):299–303. [DOI] [PubMed] [Google Scholar]
- 31.Ying S, et al. Local expression of epsilon germline gene transcripts and RNA for the epsilon heavy chain of IgE in the bronchial mucosa in atopic and nonatopic asthma. J Allergy Clin Immunol. 2001;107(4):686–92. [DOI] [PubMed] [Google Scholar]
- 32.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158(5):1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin R, et al. Clinical significance of soluble immunoglobulins A and G and their coated bacteria in feces of patients with inflammatory bowel disease. J Transl Med. 2018;16(1):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masu Y, et al. Immunoglobulin subtype-coated bacteria are correlated with the disease activity of inflammatory bowel disease. Sci Rep. 2021;11(1):16672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunker JJ, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43(3):541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catanzaro JR, et al. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep. 2019;9(1):13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kukkonen K, et al. High intestinal IgA associates with reduced risk of IgE-associated allergic diseases. Pediatr Allergy Immunol. 2010;21(1 Pt 1):67–73. [DOI] [PubMed] [Google Scholar]
- 38.Abdel-Gadir A, et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat Med. 2019;25(7):1164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parrish A, et al. Akkermansia muciniphila exacerbates food allergy in fibre-deprived mice. Nat Microbiol. 2023;8(10):1863–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scala E, et al. Comparison of the performance of Skin Prick and ISAC Tests in the diagnosis of allergy. Eur Ann Allergy Clin Immunol. 2020;52(6):258–67. [DOI] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8(4): e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C, et al. microeco: an R package for data mining in microbial community ecology. FEMS Microbiol Ecol. 2021;97(2):fiaa255. [DOI] [PubMed] [Google Scholar]
- 45.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shetty SA, et al. dysbiosisR: an R package for calculating microbiome dysbiosis measures. 2022.
- 47.Lloyd-Price J, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smeekens JM, et al. Fecal IgA, antigen absorption, and gut microbiome composition are associated with food antigen sensitization in genetically susceptible mice. Front Immunol. 2020;11: 599637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ling Z, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80(8):2546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Filippis F, et al. Specific gut microbiome signatures and the associated pro-inflamatory functions are linked to pediatric allergy and acquisition of immune tolerance. Nat Commun. 2021;12(1):5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakajima A, et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med. 2018;215(8):2019–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okai S, et al. High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol. 2016;1(9):16103. [DOI] [PubMed] [Google Scholar]
- 53.Sterlin D, et al. Human IgA binds a diverse array of commensal bacteria. J Exp Med. 2020; 217(3). [DOI] [PMC free article] [PubMed]
- 54.Moreau MC, et al. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun. 1978;21(2):532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tezuka H, Ohteki T. Regulation of IgA production by intestinal dendritic cells and related cells. Front Immunol. 2019;10:1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiu CY, et al. Cross-talk between airway and gut microbiome links to IgE responses to house dust mites in childhood airway allergies. Sci Rep. 2020;10(1):13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andre F, et al. IgE in stools as indicator of food sensitization. Allergy. 1995;50(4):328–33. [DOI] [PubMed] [Google Scholar]
- 58.Kolmannskog S. Immunoglobulin E in feces of children with intestinal Ascaris lumbricoides infestation. Int Arch Allergy Appl Immunol. 1986;80(4):417–23. [DOI] [PubMed] [Google Scholar]
- 59.Sereme Y, et al. Multiplex specific IgE profiling in neonatal stool of preterms predicts IgE-mediated disease. Allergies. 2023;3(1):58–71. [Google Scholar]
- 60.Tu Y, Perdue MH. CD23-mediated transport of IgE/immune complexes across human intestinal epithelium: role of p38 MAPK. Am J Physiol Gastrointest Liver Physiol. 2006;291(3):G532–8. [DOI] [PubMed] [Google Scholar]
- 61.Kolmannskog S, Haneberg B. Immunoglobulin E in feces from children with allergy. Evidence of local production of IgE in the gut. Int Arch Allergy Appl Immunol. 1985;76(2):133–7. [DOI] [PubMed] [Google Scholar]
- 62.Iweala OI, Burks AW. IgE producers in the gut expand the gut’s role in food allergy. Nat Rev Gastroenterol Hepatol. 2020;17(7):384–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tu Y, et al. CD23-mediated IgE transport across human intestinal epithelium: inhibition by blocking sites of translation or binding. Gastroenterology. 2005;129(3):928–40. [DOI] [PubMed] [Google Scholar]
- 64.Janoff EN, et al. Pneumococcal IgA1 protease subverts specific protection by human IgA1. Mucosal Immunol. 2014;7(2):249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Sousa-Pereira P, Woof JM. IgA: structure, function, and developability. Antibodies (Basel). 2019;8(4):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baker BS. The role of microorganisms in atopic dermatitis. Clin Exp Immunol. 2006;144(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paller AS, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(1):26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leung DY, Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment approaches. J Allergy Clin Immunol. 2014;134(4):769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones AL, Curran-Everett D, Leung DYM. Food allergy is associated with Staphylococcus aureus colonization in children with atopic dermatitis. J Allergy Clin Immunol. 2016;137(4):1247-1248.e3. [DOI] [PubMed] [Google Scholar]
- 70.Fyhrquist N, et al. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J Allergy Clin Immunol. 2014;134(6):1301-1309.e11. [DOI] [PubMed] [Google Scholar]
- 71.Paramita DA, Khairina A, Lubis NZ. Bacterial colonization in atopic dermatitis. Bali Med J. 2022;11(3):1924–9. [Google Scholar]
- 72.Song H, et al. Faecalibacterium prausnitzii subspecies–level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016;137(3):852–60. [DOI] [PubMed] [Google Scholar]
- 73.Liu S, et al. Herbal Formula-3 ameliorates OVA-induced food allergy in mice may via modulating the gut microbiota. Am J Transl Res. 2019;11(9):5812–23. [PMC free article] [PubMed] [Google Scholar]
- 74.Chua HH, et al. Intestinal dysbiosis featuring abundance of ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology. 2018;154(1):154–67. [DOI] [PubMed] [Google Scholar]
- 75.Zhai L, et al. Ruminococcus gnavus plays a pathogenic role in diarrhea-predominant irritable bowel syndrome by increasing serotonin biosynthesis. Cell Host Microbe. 2023;31(1):33-44.e5. [DOI] [PubMed] [Google Scholar]
- 76.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blázquez AB, Berin MC. Microbiome and food allergy. Transl Res. 2017;179:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene sequencing analysis data are deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA1127830 [(Run Selector:: NCBI (nih.gov), SRP520730-SRA-NCBI (nih.gov)].