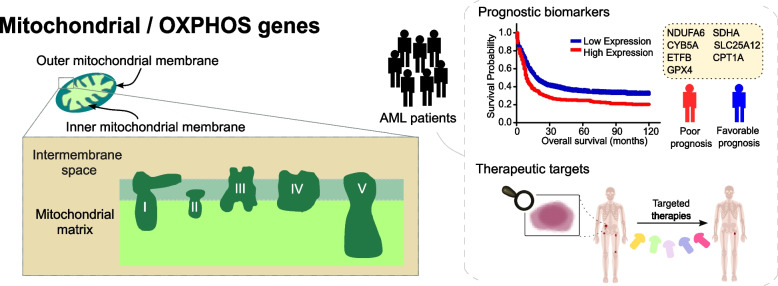
Abstract
Background
Despite significant advances in comprehending its tumorigenic role, the prognostic and therapeutic potential of targeting oxidative phosphorylation (OXPHOS) in acute myeloid leukemia (AML) remain obscure.
Methods
The prognostic value of ~ 200 mitochondrial/OXPHOS genes as candidate biomarkers was examined in AML patients over ~ 10 years follow-up using Kaplan–Meier and Cox regression analyses. Furthermore, the transcript levels of the assessed markers were inspected in healthy bone marrow tissues and the dependencies of AML cells on the assessed genes were examined.
Results
Elevated levels of NADH:ubiquinone oxidoreductase subunit A6 (NDUFA6), succinate dehydrogenase complex flavoprotein subunit A (SDHA), solute carrier family 25 member 12 (SLC25A12), electron transfer flavoprotein subunit beta (ETFB), carnitine palmitoyltransferase 1A (CPT1A) and glutathione peroxidase 4 (GPX4) were associated with poor overall survival of AML patients. SLC25A12, ETFB and CPT1A were overexpressed in AML compared to healthy tissues. Cytochrome B5 type A (CYB5A)high, SLC25A12high and GPX4high AML patients displayed higher levels of circulating and engrafted blasts compared to low-expressing cohorts. NPM1 and SRSF2 mutations were frequent in SDHAlow and CPT1Alow AML patients respectively. FLT3-ITD, NPM1 and IDH1 mutations were prevalent in CPT1Ahigh AML patients. FLT3-ITD AMLs were more dependent on OXPHOS.
Conclusions
This study identifies NDUFA6 and SDHA as novel companion prognostic biomarkers which might present a rational strategy for personalized therapy of AML patients.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12957-024-03581-5.
Keywords: Acute myeloid leukemia, Mitochondrial biomarker, Oxidative phosphorylation, Prognosis
Background
Acute myeloid leukemia (AML) is an aggressive type of blood cancer which is characterized by impaired differentiation and unlimited proliferation of AML blasts [1, 2]. Despite remarkable advances in treating other types of cancer, conventional chemotherapy remained the main standard of care therapy for AML patients [2–4]. With the advent of cutting-edge genomics technologies, genetic mutations were identified as fundamental players contributing to AML pathogenesis [1, 2, 5, 6]. Of note, multiple genetic lesions might co-exist in the same AML patient. Nucleophosmin 1 (NPM1) mutation is one of the common AML mutations which is associated with abnormal cytoplasmic localization of NPM1 [7]. Isocitrate dehydrogenase 1 (IDH1) and isocitrate dehydrogenase 2 (IDH2) mutations account for ~ 15–25% of AML mutations. Mutated IDH1 and IDH2 trigger the production of 2-hydroxyglutarate oncometabolite which is associated with defective hematopoietic differentiation [8]. Mutations in CCAAT enhancer binding protein A gene (CEBPA), a crucial transcription factor for the differentiation of granulocytes, are also among the frequent AML mutations. FMS-like tyrosine kinase 3 (FLT3) internal tandem duplication (FLT3-ITD) mutations and FLT3 tyrosine kinase domain (FLT3-TKD) mutations promote constitutive activation of FLT3 signaling which ultimately favour the proliferation of AML cells [5]. Over the last decades, several targeted agents as isocitrate dehydrogenase (IDH) inhibitors, fms-like tyrosine kinase 3 (FLT3) inhibitors, B-cell lymphoma 2 (BCL2) inhibitors, hypomethylating agents and menin inhibitors were developed and some were approved for AML therapy [5, 9–17].
Several studies underscored the role of oxidative phosphorylation (OXPHOS) in mediating the pathogenesis as well as resistance of diverse types of cancer including AML [18–21]. To meet their heightened demands of energy and biosynthetic precursors, cancer cells extensively metabolize substrates as glucose, glutamine and fatty acids to ultimately generate electrons which transfer across mitochondrial complexes [18]. Co-culturing AML cells with bone marrow derived stromal cells triggered OXPHOS and mitochondrial ATP synthesis in AML cells which exhibited chemoresistance [20]. Indeed, OXPHOS signature was enriched in cytarabine-resistant persisting AML cells [22]. Targeting mitochondrial protein synthesis or electron transfer enhanced the anti-leukemic activity of cytarabine [22]. Pharmacological inhibition of mitochondrial OXPHOS exhibited potent anticancer activities [23]. Of note, AML cells are vulnerable to inhibitors of the respiratory chain complexes as well as oxidative stressors [24]. Via inhibiting mitochondrial complex I, mubritinib elicited strong anti-AML effects in vitro and in vivo [25]. Tackling AML heterogeneity (including leukemic stem cells (LSCs) + more differentiated/mature AML blasts) is fundamental for eradicating AML [26–29]. Unlike hematopoietic stem cells (HSCs) which rely on glycolysis, LSCs are OXPHOS-dependent [30]. The imipridone ONC213 demonstrated potent anti-AML activity, particularly in LSCs, via inhibiting α-ketoglutarate dehydrogenase which impeded OXPHOS [31]. Durable remissions of AML patients who were treated with venetoclax (a selective BCL2 inhibitor) and azacitidine (a hypomethylating agent) were linked to inhibition of electron transport chain (ETC) complex II and hence OXPHOS suppression which preferentially targeted LSCs [32].
According to the NCI’s Cancer Stat Facts, the 5-year survival rate of AML patients is approximately 32%. Development of reliable and sensitive prognostic biomarkers is critically warranted for stratifying AML patients into low-risk and high-risk cohorts and guiding clinicians towards rational “tolerable and efficacious” therapeutic regimens. Diverse methods are proposed to assess mitochondrial oxygen consumption [33, 34]. Eight mitochondrial biomarkers predicted the progression and poor overall survival (OS) of gastric cancer patients [35]. More studies are needed to assess the prognostic potential of OXPHOS genes in AML patients. Indeed, this study aimed at systematically investigating the prognostic value of OXPHOS genes as candidate biomarkers to predict the clinical outcome of AML patients.
Materials and methods
Kaplan–Meier (K-M) survival analyses
To perform Kaplan–Meier (K-M) analysis on OXPHOS transcripts, the list of human genes encoding proteins involved in oxidative phosphorylation (HALLMARK_OXIDATIVE_PHOSPHORYLATION) was downloaded from GSEA MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/human/geneset/HALLMARK_OXIDATIVE_PHOSPHORYLATION.html.). Hazard-ratios (HR) for the overall survival (OS) and event-free survival (EFS) were calculated, with 120 months (~ 10 years) follow-up threshold at the best auto-selected cut-off on AML datasets (GSE1159, GSE6891, GSE8970, GSE12417 and GSE37642) and P-values were calculated using the log-rank test. K-M curves were also generated online by the K-M-plotter, using univariate analysis. False discovery rate (FDR) was computed using Benjamini–Hochberg method as described (https://kmplot.com/analysis/index.php?p=service&cancer=aml). Genes linked to poor prognosis (i.e. HR > 1) were selected for further evaluation given their therapeutic potential. For the filtered genes, multivariate Cox regression was exploited to validate the potential effect of clinical variables and gene expression on the survival of AML patients. K-M analyses were also restricted to AML patients with IDH1, IDH2, NPM1, FLT3-ITD, FLT3-TKD and CEBPA mutations. The most updated version of the database was exploited for this analysis [36].
Gene Expression Profiling Interactive Analysis (GEPIA)
Gene Expression Profiling Interactive Analysis (GEPIA) carries out OS analysis based on the gene expression in biospecimens obtained from AML patients [37]. In this study, we investigated the potential candidacy of mitochondrial / OXPHOS mediators as prognostic biomarkers for AML patients. To this end, HRs were calculated based on Cox proportional-hazards model at the median cut-off, and P-values were calculated using the logrank test [37].
cBioportal – OS analysis
To validate the results obtained from Km plotter and GEPIA database and inspect the effects of the expression of the indicated genes on the OS of AML patients, RNA-seq, clinical and survival data of BeatAML.2 dataset (OHSU, Cancer Cell, 2022) were downloaded from cBioPortal [38]. HR for the OS were calculated, with 120 months (~ 10 years) follow-up threshold at the best auto-selected cut-off and P-values were calculated using the log-rank test. The receiver operating characteristic (ROC) curves and all statistical computations were executed to evaluate the potential prognostic value of the indicated genes for the OS status (1:DECEASED or 0: LIVING) using timeROC package of R software (R.4.4.1). ROC curves comprise two parameters: true positive rate (sensitivity) and false positive rate (1-specificity) [39].
Protein expression analysis
The subcellular localization and expression of the assessed markers were further verified at the protein levels by exploiting the Human Protein Atlas database (https://www.proteinatlas.org/) as previously described [40].
Analysis of the expression levels of mitochondrial/OXPHOS genes in AML and healthy bone marrow-derived mononuclear cells
Exploiting Vizome database, the mRNA expression of the indicated mitochondrial/OXPHOS genes in biospecimens obtained from AML patients as well as that from healthy bone marrow-derived mononuclear cells (BMNCs) were downloaded from BeatAML.2 dataset [38].
Analysis of the percent of blasts in the peripheral blood and bone marrow in AML patients with low and high expression levels of mitochondrial/OXPHOS genes
Based on the mRNA expression levels of mitochondrial/OXPHOS genes (mRNA expression z-scores relative to all samples – log RNA-Seq RPKM) in AML patients (OHSU dataset, Cancer Cell, 2022) [38], cBioPortal tool was exploited to inspect the percent of circulating blasts as well as blasts in the bone marrow biospecimens obtained from AML patients with low (z score ≤ -1) and high (z score ≥ 1) expression levels of the assessed genes.
Analysis of the frequencies of genetic mutations in AML patients with low and high expression levels of mitochondrial/OXPHOS genes
Based on the mRNA expression levels of mitochondrial/OXPHOS genes (mRNA expression z-scores relative to all samples – log RNA-Seq RPKM), AML patients (OHSU dataset, Cancer Cell, 2022) were categorized into low (z score ≤ -1) and high (z score ≥ 1) expressing cohorts [38]. cBioPortal tool was then exploited to investigate the frequency of genetic mutations including missense mutations, inframe mutations, and truncating mutations in both cohorts of AML patients.
Functional enrichment analysis of mitochondrial/OXPHOS co-expressing genes in primary human AML biospecimens
Genome-wide transcriptomics datasets of AML patients were obtained from cBioportal database (OHSU, Cancer Cell 2022) [38, 41]. Statistically significant co-expressed genes with a correlation value > 0.5 and adjusted P value ≤ 0.05 were selected for pathway enrichment. Indeed, we generated lists of the enriched genes associated with the indicated genes (NDUFA6, SDHA, CYB5A, SLC25A12, ETFB and CPT1A) to be exploited for functional annotation and enrichment analysis of gene ontology—biological processes 2023 using Enrichr [42].
DepMap analysis of AML dependency on mitochondrial/OXPHOS genes
DepMap cancer dependency map identifies preferential dependencies of cancer cell lines [43, 44]. The dependency scores of human AML cell lines based on the indicated mitochondrial/OXPHOS genes were analysed using DepMap datasets of Sanger CRISPR Gene Effect (Project Score, Chronos) [43]. Lower Chronos score indicates a higher probability that the investigated gene is fundamental for that cancer cell line [43].
Results
Elevated levels of mitochondrial/OXPHOS genes correlate with poor prognosis of AML patients
Overall survival (OS) or the time from randomization to death is considered the gold standard endpoint in oncology clinical trials. To in silico test OXPHOS genes as candidate biomarkers for AML patients, we downloaded the list of human genes encoding proteins involved in OXPHOS (n = 200) from GSEA MSig database and exploited publically available transcriptional profiling datasets of AML patients (n = 734) with 10 years of follow-up using K-M plotter database [36]. Indeed, K-M analysis revealed that elevated transcript levels of NADH:ubiquinone oxidoreductase subunit A6 (NDUFA6), NADH:ubiquinone oxidoreductase subunit A8 (NDUFA8), NADH:ubiquinone oxidoreductase subunit A9 (NDUFA9), NADH:ubiquinone oxidoreductase subunit B5 (NDUFB5), NADH:ubiquinone oxidoreductase subunit B8 (NDUFB8), NADH:ubiquinone oxidoreductase subunit C1 (NDUFC1), NADH:ubiquinone oxidoreductase subunit S6 (NDUFS6), succinate dehydrogenase complex flavoprotein subunit A (SDHA), cytochrome B5 type A (CYB5A), solute carrier family 25 member 12 (SLC25A12), electron transfer flavoprotein subunit beta (ETFB), carnitine palmitoyltransferase 1A (CPT1A), phytanoyl-CoA 2-hydroxylase (PHYH), translocase of inner mitochondrial membrane 9 (TIMM9), cytochrome c oxidase subunit 7A2 (COX7A2), cytochrome c oxidase copper chaperone (COX11), cytochrome B5 reductase 3 (CYB5R3) and voltage dependent anion channel 2 (VDAC2) were significantly associated with poor OS in AML patients (log-rank P value < 0.05 and FDR = 1%) (Supplementary Table 1 and Fig. 1A-G). Notably, NDUFA6, SDHA, CYB5A, SLC25A12, ETFB and CPT1A remained significant when running a multivariate analysis including gender and treatment (untreated and chemotherapy treatment).
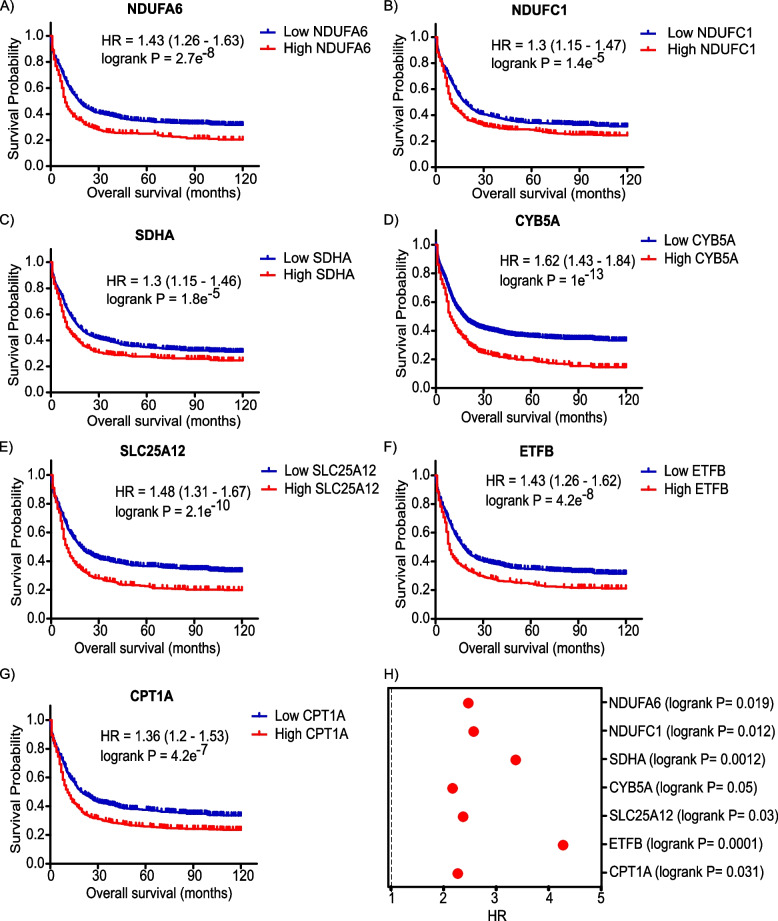
Fig. 1.
Elevated levels of mitochondrial/OXPHOS mediators correlate with poor prognosis of AML patients
A-G Kaplan-Meier survival plots of A NADH:ubiquinone oxidoreductase subunit A6 (NDUFA6), B NADH:ubiquinone oxidoreductase subunit C1 (NDUFC1), C succinate dehydrogenase complex flavoprotein subunit A (SDHA), D cytochrome B5 type A (CYB5A), E solute carrier family 25 member 12 (SLC25A12), F electron transfer flavoprotein subunit beta (ETFB) and G carnitine palmitoyltransferase 1A (CPT1A)) in AML patients with a follow-up threshold of 120 months based on KM plotter AML dataset. Samples were divided into low (blue) and high (red) expression cohorts for each gene. Hazard ratio (HR) and log-rank P values are illustrated in each plot. H HR and log-rank P values of the prognostic potential of indicated genes in AML patients based on Gene Expression Profiling Interactive Analysis (GEPIA) database
Event-free survival (EFS) is defined as the time from randomization to an event (as disease progression, treatment discontinuation or death). Yin and colleagues reported that EFS provided more precise assessment of the efficacy of drug therapy in AML patients [39]. Indeed, heightened levels of NDUFA6, NDUFC1, SDHA, CYB5A, SLC25A12, ETFB and CPT1A were associated with poor EFS in AML patients (Table 1), Next, we sought to validate the prognostic value of the above-mentioned mitochondrial/OXPHOS mediators for AML patients utilizing the Gene Expression Profiling Interactive Analysis (GEPIA) tool [37]. Genome-wide transcriptomics (RNA-Seq) datasets used by GEPIA is based on the UCSC Xena project [37]. GEPIA survival analysis confirmed that increased expression of NDUFA6, NDUFC1, SDHA, CYB5A, SLC25A12, ETFB and CPT1A significantly correlated with shorter OS of AML patients (Fig. 1H).
Table 1.
Hazard ratios (HR) and log-rank P values of the assessed biomarkers in AML patients (n = 525) for event-free survival (EFS). Follow up threshold: 120 months
| Gene | Hazard ratio (HR) | Log-rank P |
|---|---|---|
| NDUFA6 | 1.48 (1.18-1.85) | 0.00058 |
| NDUFC1 | 1.38 (1.1-1.73) | 0.005 |
| SDHA | 1.45 (1.14-1.84) | 0.0025 |
| CYB5A | 1.33 (1.05-1.69) | 0.0194 |
| SLC25A12 | 1.63 (1.29-2.07) | 4.6e-05 |
| ETFB | 1.37 (1.07-1.75) | 0.011 |
| CPT1A | 1.41 (1.13-1.77) | 0.0023 |
NDUFA6 NADH ubiquinone oxidoreductase subunit A6, NDUFC1 NADH ubiquinone oxidoreductase subunit C1, SDHA succinate dehydrogenase complex flavoprotein subunit A, CYB5A cytochrome B5 type A, SLC25A12 solute carrier family 25 member 12, ETFB electron transfer flavoprotein subunit beta, CPT1A carnitine palmitoyltransferase 1A, GPX4 glutathione peroxidase 4
Furthermore, we took advantage of the BeatAML2 dataset which comprises real-world cohort of ~ 940 biospecimens with genomic, transcriptomic, and clinical annotations obtained from young (< 45 years) and older patients with de novo, transformed, or therapy-related AMLs [38]. Notably, upregulated levels of NDUFA6, SDHA, SLC25A12, ETFB and CPT1A were significantly associated with worse OS in AML patients (Table 2).
Table 2.
Hazard ratios (HR) and log-rank P values of the assessed biomarkers in AML patients (BeatAML dataset, OHSU, Cancer Cell 2022) for overall survival (OS). Follow up threshold: 120 months
| Gene | Hazard ratio (HR) | Log-rank P |
|---|---|---|
| NDUFA6 | 1.26 (1.01-1.58) | 0.04 |
| SDHA | 1.4 (1.14-1.73) | 0.0016 |
| SLC25A12 | 1.27 (1.04-1.56) | 0.0198 |
| ETFB | 1.39 (1.13-1.7) | 0.0016 |
| CPT1A | 1.21 (1-1.48) | 0.053 |
| GPX4 | 1.3 (1.04-1.62) | 0.02 |
NDUFA6 NADH ubiquinone oxidoreductase subunit A6, SDHA succinate dehydrogenase complex flavoprotein subunit A, SLC25A12 solute carrier family 25 member 12, ETFB electron transfer flavoprotein subunit beta, CPT1A carnitine palmitoyltransferase 1A, GPX4 glutathione peroxidase 4
Next, we exploited the Human Protein Atlas database to inspect the subcellular localization and expression of the assessed markers (Supplemental Table 2). Indeed, NDUFA6 and NDUFC1 encodes for accessory subunits of the mitochondrial complex I which promotes the transfer of electrons from NADH to the ETC [23, 45]. SDHA encodes for a major catalytic subunit of mitochondrial complex II [46]. In the citric acid cycle, complex II promotes the oxidation of succinate to fumarate [46]. CYB5A is a flavoprotein reductase which catalyzes electron transfer from NADH to target substrate [47]. SLC25A12 functions as a mitochondrial aspartate/glutamate carrier [48, 49]. ETFB is an electron-transfer-flavoprotein which transfers electrons between flavoprotein dehydrogenases and flavoprotein ubiquinone oxidoreductase [50]. CPT1A is the first rate-limiting enzyme of fatty acid oxidation which promotes the mitochondrial uptake of fatty acids [51]. In line with our analysis, SLC25A12, ETFB and CPT1A were reported to predict the prognosis of AML patients [49, 52].
In addition, time-dependent ROC and area under time dependent ROC curve (AUC) analysis revealed that the AUC values of NDUFA6 were 0.56, 0.54 and 0.72 at 12, 48 and 100 months respectively (Figure.S1A). The AUC values of SDHA were 0.58, 0.56 and 0.56 whereas those of SLC25A12 were 0.50, 0.49 and 0.55 at 12,24 and 48 months respectively (Figure.S1B-C). ETFB had AUC values of 0.56, 0.56 and 0.59 whereas CPT1A had AUC values of 0.51, 0.49 and 0.53 at 1,2 and 4 years respectively (Figure.S1D-E).
Elevated levels of NDUFA6, SDHA, CYB5A, SLC25A12, ETFB and CPT1A are differentially associated with poor prognosis in AML patients with distinct mutations
Restricting K-M OS survival analyses to AML patients with distinct mutations revealed that upregulated levels of NDUFA6, SDHA and CPT1A were associated with unfavourable prognosis in mutated IDH1 AML patients (Table 3). Higher levels of NDUFA6 and CPT1A significantly correlated with poor OS of AML patients with IDH2 mutation. Elevated levels of NDUFA6, SDHA, CYB5A, SLC25A12, ETFB and CPT1A were associated with shorter OS of NPM1 and FLT3-ITD mutated AML patients (Table 3). Upsurged levels of SDHA, ETFB and CPT1A were linked to unfavourable prognosis of AML patients with FLT3-TKD mutation. Increased expression of SLC25A12 and CPT1A correlated with poor OS of AML patients with CEBPA mutation (Table 3).
Table 3.
Hazard ratios (HR) and log-rank P values of the assessed biomarkers associated with poor prognosis in patients with mutated AMLs
| Type of AML mutation (n) | Hazard ratio (HR) | Log-rank P |
|---|---|---|
|
IDH1 mutation (n = 51) |
NDUFA6: 3.28 (1.41–7.64) | 0.0037 |
| SDHA: 2.1 (1.02- 4.32) | 0.0405 | |
| CPT1A: 2.68 (1.32–5.43) | 0.0044 | |
|
IDH2 mutation (n = 57) |
NDUFA6: 2.45 (1.14–5.25) | 0.0178 |
| CPT1A: 2.48 (1.25–4.92) | 0.007 | |
|
NPM1 mutation (n = 217) |
NDUFA6:1.85 (1.18–2.88) | 0.0059 |
| SDHA: 1.67 (1.16.2.41) | 0.0057 | |
| CYB5A: 1.49 (1.05–2.1) | 0.0245 | |
| SLC25A12: 1.79 (1.18–2.73) | 0.0055 | |
| ETFB: 1.54 (1.07–2.21) | 0.0191 | |
| CPT1A: 1.45 (1.02–2.06) | 0.035 | |
|
FLT3-ITD mutation (n = 191) |
NDUFA6: 1.5 (1.07–2.1) | 0.0188 |
| SDHA: 2.59 (1.85–3.64) | 1e-08 | |
| CYB5A: 1.5 (1.06–2.13) | 0.0198 | |
|
SLC25A12: 1.58 (1.13–2.2) ETFB: 1.58 (1.12–2.23) |
0.0065 0.0079 |
|
| CPT1A: 2.09 (1.5–2 | 9.9e-06 | |
|
FLT3-TKD mutation (n = 79) |
SDHA: 2.17 (1.14–4.14) | 0.0163 |
| ETFB: 2.48 (1.31–4.7) | 0.0041 | |
| CPT1A: 3.41 (1.42–8.17) | 0.003 | |
|
CEBPA mutation (n = 52) |
SLC25A12: 4.48 (1.32–15.18) | 0.0084 |
| CPT1A: 2.57 (1.11–5.95) | 0.023 |
IDH1 isocitrate dehydrogenase 1, IDH2 isocitrate dehydrogenase 2, NPM1 nucleophosmin 1, ITD internal tandem duplication, TKD tyrosine kinase domain, CEBPA CCAAT/enhancer-binding protein alpha
SLC25A12, ETFB and CPT1A are significantly overexpressed in AML biospecimens compared to healthy bone marrow-derived mononuclear cells
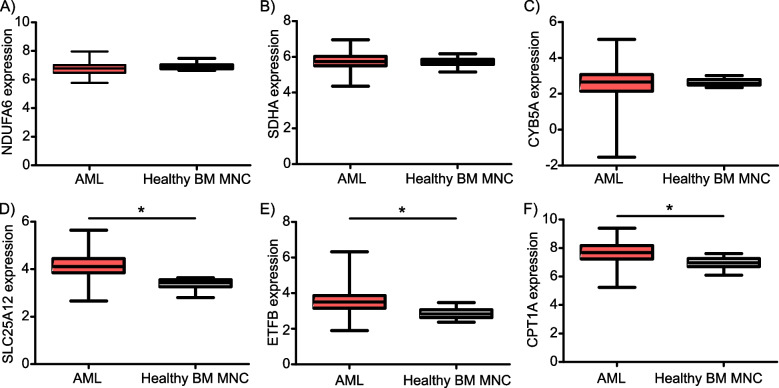
Next, we questioned whether the above-mentioned biomarkers are differentially expressed in primary human AML versus normal biospecimens. Exploiting BeatAML2 dataset revealed that there were no statistically significant differences in the mRNA levels of NDUFA6, SDHA, and CYB5A in biospecimens obtained from AML patients compared to healthy BMNCs (Fig. 2A-C). However, the transcript levels of SLC25A12, ETFB and CPT1A were significantly higher in AML compared to healthy BMNCs (Fig. 2D-F).
Fig. 2.
Solute carrier family 25 member 12 (SLC25A12), electron transfer flavoprotein subunit beta (ETFB) and carnitine palmitoyltransferase 1A (CPT1A) are significantly overexpressed in AML biospecimens compared to healthy bone marrow-derived mononuclear cells (BM MNC)
A-F Transcript levels of the indicated genes (A: NADH:ubiquinone oxidoreductase subunit A6 (NDUFA6), B: succinate dehydrogenase complex flavoprotein subunit A (SDHA), C: cytochrome B5 type A (CYB5A), D: solute carrier family 25 member 12 (SLC25A12), E: electron transfer flavoprotein subunit beta (ETFB) and F: carnitine palmitoyltransferase 1A (CPT1A)) in AML biospecimens as well as healthy bone marrow-derived mononuclear cells obtained from BeatAML.2 dataset (38). *: P value ≤ 0.05 compared to healthy bone marrow-derived mononuclear cells
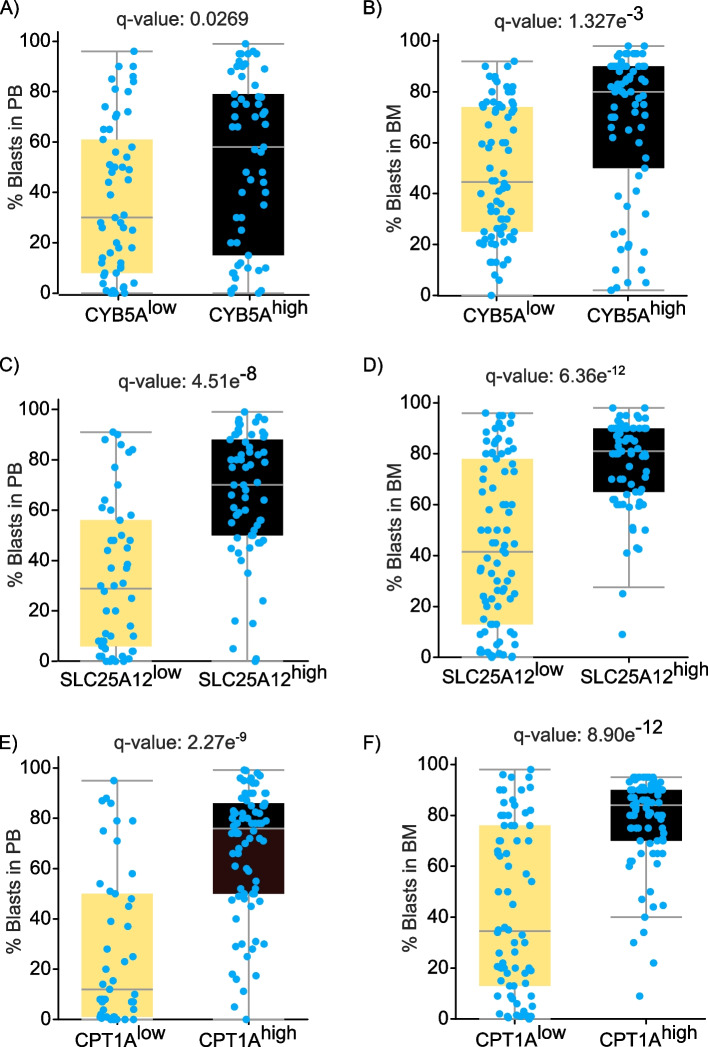
AML patients with higher levels of CYB5A, SLC25A12 or CPT1A had significantly higher levels of circulating as well as engrafted blasts compared to low-expressing cohorts
Monitoring the percent of blasts in the peripheral blood as well as bone marrow is critical for the diagnosis, prognosis, and therapy of AML patients [2, 53, 54]. Accordingly, we inspected whether AML patients with low and high levels of biomarkers as NDUFA6, SDHA, CYB5A, SLC25A12, ETFB and CPT1A have differential leukemic burden in their peripheral blood and bone marrow biospecimens. To this end, we categorized AML patients (BeatAML2, OHSU, Cancer 2022 dataset) according to the mRNA expression of the assessed genes (z-scores relative to all samples – log RNA-Seq RPKM) into low (z score ≤ -1) and high (z score ≥ 1) expressing cohorts. Statistical analyses of the clinical attributes of low- and high-expressing cohorts are summarized in Supplemental Table 3. Notably, CYB5Alow AML cohort had significantly lower percent of blasts in both the peripheral blood as well as bone marrow compared to CYB5Ahigh AML cohort (Fig. 3A-B). Similar findings were also noted in SLC25A12low versus SLC25A12high and in CPT1Alow versus CPT1Ahigh AML cohorts (Fig. 3C-F). It is worth mentioning that the current regimen and biospecimen type of SLC25A12low cohort (69% bone marrow aspirate and 31% peripheral blood) were statistically significant from that of SLC25A12high cohort (45.13% bone marrow aspirate, 7.01% leukapheresis and 47.79% peripheral blood) (Supplemental Table 3). Otherwise, the age at biospecimen collection, ethnicity category, sex, specimen type and current regimen were not statistically different among other low- and high-expressing cohorts (Supplemental Table 3).
Fig. 3.
The percent of AML blasts in the peripheral blood as well as the bone marrow of AML patients with low and high expression levels of the indicated genes
Based on the mRNA expression levels of the indicated genes (mRNA expression z-scores relative to all samples – log RNA-Seq RPKM) in AML patients (OHSU dataset, Cancer Cell, 2022) [38], cBioPortal tool was exploited to inspect the percent of circulating blasts as well as blasts in the bone marrow biospecimens of AML patients with low (z score ≤ -1) and high (z score ≥ 1) expression levels of the assessed genes. A, B The percent of circulating blasts (A) and blasts in the bone marrow (B) of cytochrome B5 type A (CYB5A)low and CYB5Ahigh AML patients (BeatAML.2 dataset [38]). C, D The percent of circulating blasts (C) and blasts in the bone marrow (D) of solute carrier family 25 member 12 (SLC25A12)low and SLC25A12high AML patients. E, F The percent of circulating blasts (E) and blasts in the bone marrow (F) of carnitine palmitoyltransferase 1A (CPT1A)low and CPT1Ahigh AML cohorts
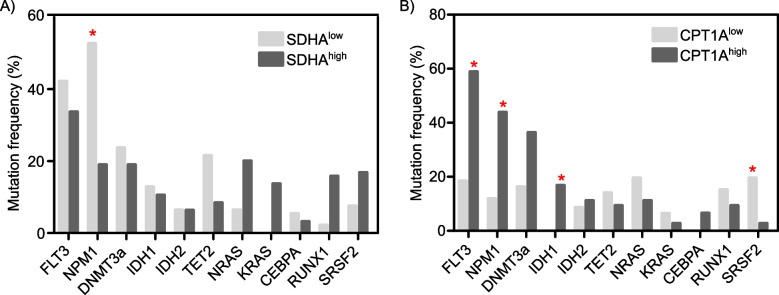
Prevalence of genetic mutations in AML patients with low and high levels of mitochondrial/OXPHOS genes
Given the prevalence of genetic mutations in AML, we examined the correlation between the transcriptional levels of the forementioned genes (low versus high expressing cohorts) and AML mutations using the cBioPortal tool [41]. Intriguingly, NPM1 and serine/arginine-rich splicing factor 2 (SRSF2) mutations were higher in SDHAlow and CPT1Alow AML cohorts respectively (Fig. 4A-B). In contrast, FLT3-ITD, NPM1 and IDH1 mutations were more common in CPT1Ahigh AML patients (Fig. 4B).
Fig. 4.
Frequency of genetic mutations in AML patients with low and high levels of the indicated genes
A, B Mutation frequencies of the indicated genes in AML patients with low and high expression levels of succinate dehydrogenase complex flavoprotein subunit A (SDHA) (A) and carnitine palmitoyltransferase 1A (CPT1A) (B). *: Adjust P value ≤ 0.05
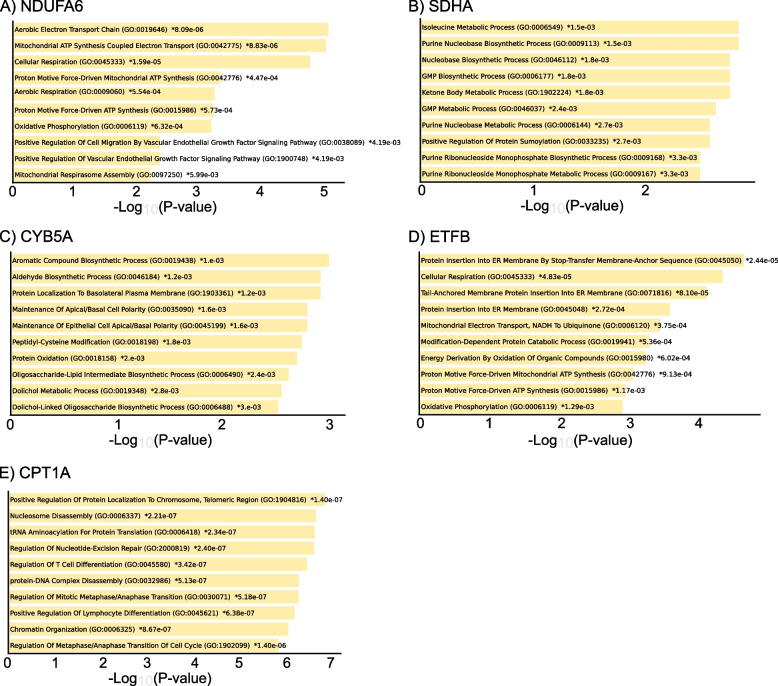
Functional prediction and pathway enrichment analysis of mitochondrial/OXPHOS genes in primary human AML biospecimens
We examined the genes which are co-expressed with mitochondrial/OXPHOS genes using the cBioPortal tool [38, 41]. The expression of NDUFA6 positively correlated with the upregulation of several genes including succinate dehydrogenase complex iron sulfur subunit B (SDHB), cysteine rich with EGF like domains 2 (CRELD2), adaptor related protein complex 2 subunit sigma 1 (AP2S1), HIG1 hypoxia inducible domain family member 1A (HIGD1A), cytochrome C oxidase subunit 7B (COX7B), proteasome maturation protein (POMP), myosin 1C (MYO1C), mitochondrial ribosomal protein S15 (MRPS15), NADH: ubiquinone oxidoreductase subunit B9 (NDUFB9), calmodulin binding transcription activator 1 (CAMTA1), G protein regulated inducer of neurite outgrowth 1 (GPRIN1) and BolA family member 3 (BOLA3). Gene Ontology (GO) enrichment analysis of NDUFA6 co-expressing genes showed the enrichment of distinct biological processes (BP) as mitochondrial ATP synthesis coupled electron transport, cellular respiration, oxidative phosphorylation, positive regulation of cell migration by vascular endothelial growth factor signalling pathway, positive regulation of vascular endothelial growth factor signalling pathway and mitochondrial respirasome assembly (Fig. 5A).
Fig. 5.
Gene ontology of the biological process of mitochondrial/OXPHOS co-expressed genes in AML patients
A-E) Bar chart of top enriched terms from the GO_Biological_Process_2023 gene set library which are significantly co-expressed with NADH: ubiquinone oxidoreductase subunit A6 (NDUFA6) (A), succinate dehydrogenase complex flavoprotein subunit A (SDHA) (B), cytochrome B5 type A (CYB5A) (C), electron transfer flavoprotein subunit beta (ETFB) (D) and carnitine palmitoyltransferase 1A (CPT1A) (E) in biospecimens obtained from AML patients (OHSU Cancer Cell 2022 dataset). The cut-off for Spearman’s correlation is > 0.5 and adjusted P value ≤ 0.05. The top 10 enriched terms for the input gene set are displayed based on the -log10 (P value), with the actual P value illustrated next to each term. The term at the top has the most significant overlap with the input query gene set
On the other hand, the expression of SDHA positively correlated with the expression of acetyl-CoA acetyltransferase 1 (ACAT1), SUMO1 activating enzyme subunit 1 (SAE1), ATP synthase membrane subunit C locus 3 (ATP5MC3), TNF receptor associated protein 1 (TRAP1), guanine monophosphate synthase (GMPS) and protein phosphatase 1 catalytic subunit gamma (PPP1CC). GO BP analysis of SDHA co-expressing genes highlighted the enrichment of isoleucine metabolic process, purine nucleibase biosynthetic process, ketone body metabolic process and positive regulation of protein sumoylation (Fig. 5B).
GO BP enrichment analysis of CYB5A co-expressed genes revealed their association with aromatic compound biosynthetic process, aldehyde biosynthetic process, protein localization to basolateral plasma membrane, maintenance of apical/basal cell polarity, peptidyl-cysteine modification, protein oxidation, oligosaccharide-lipid intermediate biosynthetic process, and dolichol metabolic process (Fig. 5C).
ETFB co-expressed genes were rather enriched with biological processes as protein insertion into ER membrane by stop-transfer membrane-anchor sequence, cellular respiration, mitochondrial electron transport, NADH to ubiquinone and energy derivation by oxidation of organic compounds (Fig. 5D).
GO enrichment analysis of CPT1A co-expressed genes revealed the enrichment of BP as positive regulation of protein localization to chromosome, telomeric region, nucleosome disassembly, tRNA aminoacylation for protein translation, regulation of nucleotide-excision repair, regulation of mitotic metaphase/anaphase transition and chromatin organization (Fig. 5E).
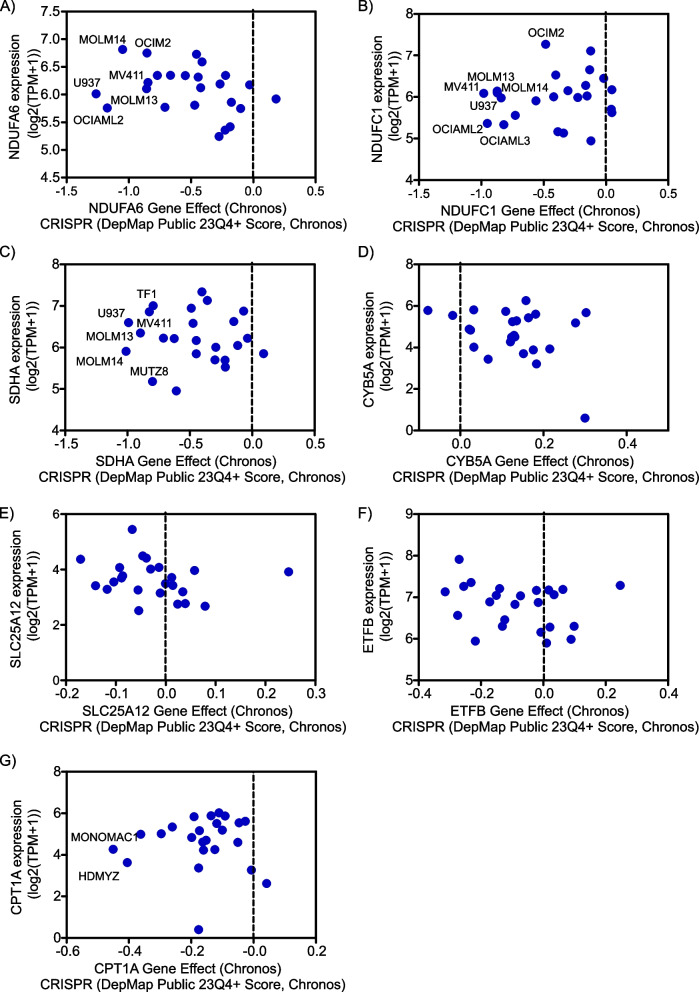
Differential dependency of AML cells on mitochondrial/OXPHOS genes
Based on the above-mentioned findings, we questioned whether human AML cells are dependent on the evaluated mitochondrial/OXPHOS genes. At genome-wide scale, the CRISPR–Cas9 system objectively identifies genes which are indispensable for the proliferation and survival of cancer cells including AML [44, 55]. To this end, we exploited the Cancer Dependency Map database (which comprises large-scale functional genomics profiling using CRISPR loss-of-function screens) to investigate the potential dependency of AML cells on mitochondrial/OXPHOS genes. Dempster and colleagues developed Chronos model which addressed several limitations associated with other models including sgRNA efficacy, variable screen quality and copy number bias [43]. Negative Chronos scores indicate slower growth of cancer cells [43].
Indeed, AML cells exhibited heterogeneous responses to CRISPR-mediated genetic pertubation of OXPHOS genes which encode for subunits of the OXPHOS complexes: I (NDUFA6 and NDUFC1) and II (SDHA) (Fig. 6A-C). Of note, AML cell lines with FLT3-ITD mutations (as MV4-11, MOLM13 and MOLM14) [5] were more vulnerable to the genetic depletion of NDUFA6, NDUFC1 and SDHA (Fig. 6A-C). In contrast, modest dependencies of AML cells were observed for CRISPR KO of CYB5A, SLC25A12, ETFB and CPT1A (Fig. 6D-G). Overall, these findings highlight the therapeutic potential of targeting mitochondrial/OXPHOS dependent/addicted AML cells.
Fig. 6.
Differential dependencies of AML cells on mitochondrial/OXPHOS genes
A-G Dot plot depicting the gene dependency effect (CRISPR, DepMap Public 23Q4+ Score, Chronos) of AML cells on the indicated mitochondrial/OXPHOS genes (A: NADH : ubiquinone oxidoreductase subunit A6 (NDUFA6), B: NADH : ubiquinone oxidoreductase subunit C1 (NDUFC1), C: succinate dehydrogenase complex flavoprotein subunit A (SDHA), D: cytochrome B5 type A (CYB5A), E: solute carrier family 25 member 12 (SLC25A12), F: electron transfer flavoprotein subunit beta (ETFB) and G: carnitine palmitoyltransferase 1A (CPT1A)) plotted on the X-axis versus their corresponding mRNA expression level (log2(TPM+1) plotted on the Y-axis
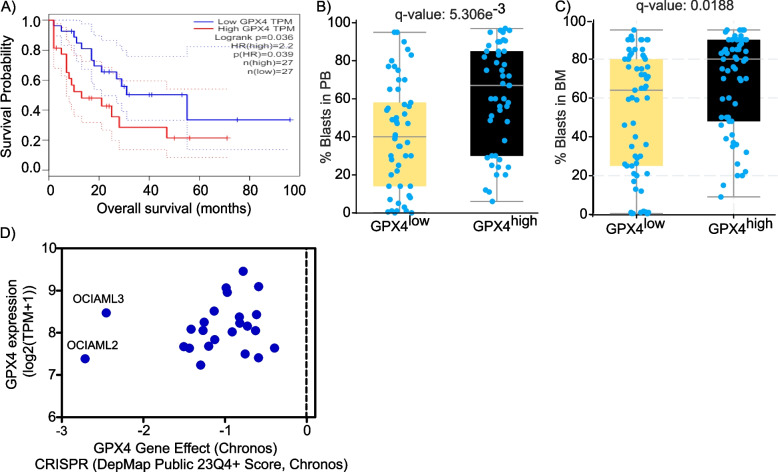
Heightened expression of glutathione peroxidase 4 (GPX4) is associated with higher percent of circulating and engrafted blasts of AML patients and DNMT3a mutant AML cells are dependent on GPX4
Glutathione peroxidase 4 (GPX4) is included in the geneset of HALLMARK_OXIDATIVE_PHOSPHORYLATION downloaded from GSEA MSigDB. As an antioxidant, GPX4 promotes the reduction of hydrogen peroxide, organic hydroperoxides and lipid hydroperoxides [56]. Km plotter database and GEPIA survival analyses showed controversial results in terms of the prognostic value of GPX4 expression in AML patients. Unlike Km plotter, the survival analyses of GEPIA and BeatAML.2 datasets demonstrated that elevated GPX4 levels are significantly associated with worse OS of AML patients (Fig. 7A and Supplemental Table 2). The AUC values of the ROC curve of GPX4 were 0.51, 0.52 and 0.62 at 2, 4 and 6 years respectively (Figure.S1F). GPX4high AML cohort had significantly higher percent of blasts in both the peripheral blood as well as bone marrow compared to GPX4low AML cohort (Fig. 7B-C).
Fig. 7.
Increased expression of glutathione peroxidase 4 (GPX4) is associated with higher percent of circulating and engrafted blasts as well as poor prognosis of AML patients and DNMT3a mutant AML cells are dependent on GPX4
A Kaplan–Meier survival curve depicting the hazard ratio (HR) and log-rank P values of the prognostic potential of GPX4 gene in AML patients based on Gene Expression Profiling Interactive Analysis (GEPIA) database. B, C The percent of circulating blasts (B) and blasts in the bone marrow (C) of GPX4low and GPX4high AML patients (BeatAML.2 dataset (38)). D Dot plot depicting the gene dependency effect (CRISPR, DepMap Public 23Q4 + Score, Chronos) of AML cells on GPX4 plotted on the X-axis versus their corresponding mRNA expression level (log2(TPM + 1) plotted on the Y-axis
Several patient-derived AML cell lines were evidently susceptible to CRISPR KO of GPX4 (Fig. 7D). Notably, OCIAML2 and OCIAML3 AML cell lines were preferentially responsive to the genetic ablation of GPX4. It is worth noting that OCI-AML2 and OCI-AML3 are the only known human AML cell lines with the DNMT3A mutation. Altogether, these findings underscore the therapeutic potential of targeting GPX4 for AML therapy.
Discussion
The present study underscored the prognostic value of mitochondrial/OXPHOS genes as NDUFA6, SDHA, SLC25A12, ETFB CPT1A and GPX4 in AML patients. Consistent with the present findings, breast cancer patients with high NDUFA6 levels had shorter OS [57]. Elevated levels of NDUFC1 also predicted unfavourable prognosis of hepatocellular carcinoma patients [32]. In line with this study, elevated levels of SLC25A12, ETFB and CPT1A were associated with poor prognosis of AML patients [49, 52]. High CPT1A levels were associated with worse OS of nasopharyngeal carcinoma patients post-radiotherapy [51]. Inspecting BeatAML.2 dataset, the present study also showed that CYB5Ahigh, SLC25A12high and CPT1Ahigh AML patients had significantly higher levels of circulating as well as engrafted blasts compared to CYB5Alow, SLC25A12low and CPT1Alow AML patients respectively. CYB5A reduces ferric hemoglobin to ferrous hemoglobin, which is required for stearoyl-CoA-desaturase activity [47]. Stearoyl coA desaturase converts saturated fatty acids into monounsaturated fatty acids. Intriguingly, high expression of stearoyl-coA-destaurase is associated with poor prognosis of gastric cancer patients [58].
More specifically, this study found that NPM1 W288Cfs*12 frameshift insertion mutations are prevalent in SDHAlow and CPT1Ahigh AML patients. NPM1 mutant insertions as W288Cfs*12 (tryptophan 288 to cysteine frameshift at exon 12) result in a change in the C-terminal domain (CTD) amino acid sequence which is required for nucleolar localization signalling [59]. The present study also shed light on the prevalence of SRSF2 P95H missense mutation in CPT1Alow AML patients which is located in the RNA binding domain of the SRSF2 splicing factor protein and leads to distorted RNA binding of SRFS2 (56). Likewise, the frequently occurring IDH1 R132H mutation in CPT1Alow AML patients is located in the catalytic site of the IDH1 protein which converts α-ketoglutarate to 2-hydroxyglutarate oncometabolite [1, 8, 60].
The present study also shed light on the heterogeneous responsiveness of AML cells to genetic depletion of OXPHOS genes as NDUFA6, NDUFC1 and SDHA. Knocking down NDUFC1 evidently impeded the proliferation, cell cycle progression, migration and invasion and triggered apoptotic cell death of hepatocellular carcinoma cells [32] Notably, FLT3-ITD+ AML cell lines (MV4-11, MOLM13 and MOLM14) [5] were more suseptible to genetic intereference with subunits of mitochondiral complex I (NDUFA6 and NDUFC1) and mitochondrial complex II (SDHA). Consistently, Baccelli and colleagues reported that AML addiction on mitochondrial complex I activity was strongly associated with the presence of NPM1 and FLT3-ITD mutations as well as those mutations affecting DNA methylation genes (as DNMT3a, IDH1, IDH2, and TET2) [25]. Intriguingly, IACS-010759 (an OXPHOS complex I inhibitor) synergized with quizartinib (a FLT3 inhibitor) on FLT3-ITD+ and FLT3-ITD− AMLs [61]. Despite its promising preclinical activity, Phase I study reported the narrow therapeutc index and dose-limiting toxicities of IACS-010759 when evaluated in patients with solid tumors as well as AML [62]. Erdem and colleagues demonstrated that FLT3-ITD+ AML patients had overactivated mitochondrial ETC complex II [57]. Although abrogating mitochondrial ETC complex II triggered apoptotic cell death, FLT3-ITD+ AMLs adapted by importing lactate to be exploited for mitochondrial respiration. Blocking lactate transport evidently augmented the anti-AML activity of ETC complex II inhibitors [57]. Blunting CPT1A sensitized nasopharyngeal cancer cells to radiotherapy via triggering mitochondrial apoptosis [51].
Compared with GPX4low AML patients, GPX4high cohort had higher percent of blasts both in the peripheral blood as well as bone marrow. Wei and colleagues showed that AML patients with high expression of GPX1, GPX3, GPX4, and GPX7 were associated with worse prognosis [58]. In accordance with the present findings, pharmacological as well as genetic suppression of GPX4 induced ferroptosis (a non-apoptotic cell death triggered by iron-dependent lipid peroxidation) in AML cells [58]. In this study, DNMT3a mutant AML cell lines (OCIAML2 and OCIAML3) were dependent on GPX4. Indeed, further mechanistic studies are needed to decipher the molecular basis underlying the preferential sensitivity of these AML cell lines and examine whether this is linked to DNMT3a mutation.
Altogether, the present study is a comprehensive and systematic bioinformatic analysis based on multiple databases and datasets of AML patients. However, further in vitro and in vivo validation of these findings is warranted.
Conclusions
To the best of our knowledge, this is the first study which systematically investigated the potential value of OXPHOS genes which could serve as prognostic biomarkers for AML patients. Moreover, this study highlighted differential dependencies of AML cells on NDUFA6, SDHA, CYB5A, SLC25A12, ETFB, and CPT1A which could rationally guide personalized therapy of AML patients. Nonetheless, caution is warranted to address the feasibility and safety profile of mitochondrial ETC complex I inhibitors before further advancing their preclinical and clinical development for AML therapy.
Supplementary Information
Supplementary Material 2: Figure S1. Area under the curve (AUC) of receiver operating characteristic (ROC) curves of the indicated genes in biospecimens obtained from AML patients (BeatAML.2 cBioportal dataset (OHSU, Cancer Cell, 2022). A-F) ROC curves and AUC of NADH:ubiquinone oxidoreductase subunit A6 (NDUFA6) (A), succinate dehydrogenase complex flavoprotein subunit A (SDHA) (B), solute carrier family 25 member 12 (SLC25A12) (C), electron transfer flavoprotein subunit beta (ETFB) (D), carnitine palmitoyltransferase 1A (CPT1A) (E) and glutathione peroxidase 4 (GPX4) (F) at the indicated time points of overall survival.
Acknowledgements
None.
Abbreviations
- COX7A2
Cytochrome c oxidase subunit 7A2
- COX11
Cytochrome c oxidase copper chaperone
- CPT1A
Carnitine palmitoyltransferase 1A
- CYB5A
Cytochrome B5 type A
- CYB5R3
Cytochrome B5 reductase 3
- ETC
Electron transport chain
- ETFB
Electron transfer flavoprotein subunit beta
- GPX4
Glutathione peroxidase 4
- GEPIA
Gene expression profiling interactive analysis
- NDUFA6
NADH:ubiquinone oxidoreductase subunit A6
- NDUFA8
NADH:ubiquinone oxidoreductase subunit A8
- NDUFA9
NADH:ubiquinone oxidoreductase subunit A9
- NDUFB5
NADH:ubiquinone oxidoreductase subunit B5
- NDUFB8
NADH:ubiquinone oxidoreductase subunit B8
- NDUFC1
NADH:ubiquinone oxidoreductase subunit C1
- NDUFS6
NADH:ubiquinone oxidoreductase subunit S6
- OS
Overall survival
- PHYH
Phytanoyl-CoA 2-hydroxylase
- SDHA
Succinate dehydrogenase complex flavoprotein subunit A
- SLC25A12
Solute carrier family 25 member 12
- TIMM9
Translocase of inner mitochondrial membrane 9
- VDAC2
Voltage dependent anion channel 2
Author's contributions
A.K.A. conceived, carried out data curation and bioinformatic analysis, prepared all the figures and wrote the main manuscript text.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abdel-Aziz AK. Advances in acute myeloid leukemia differentiation therapy: A critical review. Biochem Pharmacol [Internet]. 2023;215(July):115709. Available from: doi: 10.1016/j.bcp.2023.115709. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140(12):1345–77. [DOI] [PubMed] [Google Scholar]
- 3.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N Engl J Med. 2017;377(5):454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yee KW, Schittenhelm M, O'Farrell AM, et al. Synergistic effect of SU11248 with cytarabine or daunorubicin on FLT3 ITD-positive leukemic cells. Blood. 2004;104(13):4202–9. 10.1182/blood-2003-10-3381. [DOI] [PubMed]
- 5.Abdel-Aziz AK, Dokla EME, Saadeldin MK. FLT3 inhibitors and novel therapeutic strategies to reverse AML resistance: An updated comprehensive review. Crit Rev Oncol Hematol. 2023;191(April):104139. 10.1016/j.critrevonc.2023.104139. [DOI] [PubMed] [Google Scholar]
- 6.Prajapati S, Kumari N, Bhowmik D, Gupta R. Recent advancements in biomarkers, therapeutics, and associated challenges in acute myeloid leukemia. Ann Hematol. 2024. 10.1007/s00277-024-05963-x. [DOI] [PubMed]
- 7.Guan W, Zhou L, Li Y, Yang E, Liu Y, Lv N, et al. Profiling of somatic mutations and fusion genes in acute myeloid leukemia patients with FLT3-ITD or FLT3-TKD mutation at diagnosis reveals distinct evolutionary patterns. Exp Hematol Oncol. 2021;10(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel KP, Ravandi F, Ma D, Paladugu A, Barkoh BA, Medeiros LJ, et al. Acute myeloid leukemia with IDH1 or IDH2 mutation: requency and clinicopathologic features. Am J Clin Pathol. 2011;135(1):35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daver N, Perl AE, Maly J, Levis M, Ritchie E, Litzow M, et al. Venetoclax Plus Gilteritinib for FLT3 -Mutated Relapsed/Refractory Acute Myeloid Leukemia. J Clin Oncol. 2022;40(35):4048–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galanis A, Ma H, Rajkhowa T, Ramachandran A, Small D, Cortes J, et al. Crenolanib is a potent inhibitor of flt3 with activity against resistance-Conferring point mutants. Blood. 2014;123(1):94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Song Y, Liu D. Gilteritinib: A novel FLT3 inhibitor for acute myeloid leukemia. Biomark Res. 2019;7(1):104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes JE, Khaled S, Martinelli G, Perl AE, Ganguly S, Russell N, et al. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): a multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2019;20(7):984–97. [DOI] [PubMed] [Google Scholar]
- 13.DiNardo CD, Stein AS, Stein EM, Fathi AT, Frankfurt O, Schuh AC, et al. Mutant isocitrate dehydrogenase 1 inhibitor ivosidenib in combination with azacitidine for newly diagnosed acute myeloid leukemia. J Clin Oncol. 2021;39(1):57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salamero O, Montesinos P, Willekens C, Pérez-Simón JA, Pigneux A, Récher C, et al. First-in-Human Phase I Study of Iadademstat (ORY-1001): A First-in-Class Lysine-Specific Histone Demethylase 1A Inhibitor, in Relapsed or Refractory Acute Myeloid Leukemia. J Clin Oncol. 2020;38(36):4260–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Aziz AK, Minucci S. Comparing apples with oranges: Studying LSD1 inhibitors in cellular assays. Pharmacol Res. 2019:104345. 10.1016/j.phrs.2019.104345. [DOI] [PubMed]
- 16.Abdel-Aziz AK, Pallavicini I, Ceccacci E, Meroni G, Saadeldin MK, Varasi M, et al. Tuning mTORC1 activity dictates the response of acute myeloid leukemia to LSD1 inhibition. Haematologica. 2019 Sep 19;haematol.2019.224501. Available from: 10.3324/haematol.2019.224501. [DOI] [PMC free article] [PubMed]
- 17.Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8(3):277–84. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3624763/pdf/nihms412728.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Beauchamp L, Himonas E, Helgason GV. Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia. Leukemia. 2022;36(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Xu W, Zeng H, He Z, Lu X, Zuo D, et al. OXPHOS-dependent metabolic reprogramming prompts metastatic potential of breast cancer cells under osteogenic differentiation. Br J Cancer. 2020;123(11):1644–55. 10.1038/s41416-020-01040-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou D, Wang B, You R, Wang X, Liu J, Zhan W, et al. Stromal cells promote chemoresistance of acute myeloid leukemia cells via activation of the IL-6/STAT3/OXPHOS axis. Ann Transl Med. 2020;8(21):1346–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CL, Lin CY, Kung HJ. Targeting mitochondrial oxphos and their regulatory signals in prostate cancers. Int J Mol Sci. 2021;22(24):13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7(7):716–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galemou Yoga E, Parey K, Djurabekova A, Haapanen O, Siegmund K, Zwicker K, et al. Essential role of accessory subunit LYRM6 in the mechanism of mitochondrial complex I. Nat Commun. 2020;11(1):1–8. 10.1038/s41467-020-19778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood. 2015;125(13):2120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baccelli I, Gareau Y, Lehnertz B, Gingras S, Spinella JF, Corneau S, et al. Mubritinib targets the electron transport chain complex I and reveals the landscape of OXPHOS dependency in acute myeloid leukemia. Cancer Cell. 2019;36(1):84-99.e8. [DOI] [PubMed] [Google Scholar]
- 26.Laverdière I, Boileau M, Neumann AL, Frison H, Mitchell A, Ng SWK, et al. Leukemic stem cell signatures identify novel therapeutics targeting acute myeloid leukemia. Blood Cancer J. 2018;8(6):52. 10.1038/s41408-018-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baquero P, Dawson A, Mukhopadhyay A, Kuntz EM, Mitchell R, Olivares O, et al. Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia. 2019;33(4):981–94. 10.1038/s41375-018-0252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leiva M, Moretti S, Soilihi H, Pallavicini I, Peres L, Mercurio C, et al. Valproic acid induces differentiation and transient tumor regression, but spares leukemia-initiating activity in mouse models of APL. Leukemia. 2012;26(7):1630–7. [DOI] [PubMed] [Google Scholar]
- 29.Moretti S, Abdel-Aziz AK, Ceccacci E, Pallavicini I, Santoro F, de Thé H, et al. Co-targeting leukemia-initiating cells and leukemia bulk leads to disease eradication. Leukemia. 2022;36(5):1306–12. [DOI] [PubMed] [Google Scholar]
- 30.Peng M, Huang Y, Zhang L, Zhao X, Hou Y. Targeting mitochondrial oxidative phosphorylation eradicates acute myeloid leukemic stem cells. Front Oncol. 2022;12(April):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Y, Carter JL, Li X, Fukuda Y, Gray A, Lynch J, et al. The imipridone ONC213 targets α-Ketoglutarate dehydrogenase to induce mitochondrial stress and suppress oxidative phosphorylation in acute myeloid leukemia. Cancer Res. 2024;84(7):1084–100. 10.1158/0008-5472.CAN-23-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med. 2018;24:1859–66. 10.1038/s41591-018-0233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fovez Q, Laine W, Goursaud L, Berthon C, Germain N, Degand C, et al. Clinically relevant oxygraphic assay to assess mitochondrial energy metabolism in acute myeloid leukemia patients. Cancers (Basel). 2021;13(24):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helmy SWA, Abdel-Aziz AK, Dokla EME, Ahmed TE, Hatem Y, Abdel Rahman EA, et al. Novel sulfonamide-indolinone hybrids targeting mitochondrial respiration of breast cancer cells. Eur J Med Chem. 2024;268:1–18. 10.1016/j.ejmech.2024.116255. [DOI] [PubMed] [Google Scholar]
- 35.Sotgia F, Lisanti MP. Mitochondrial biomarkers predict tumor progression and poor overall survival in gastric cancers: Companion diagnostics for personalized medicine. Oncotarget. 2017;8(40):67117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gyorffy B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience. 2023;45(3):1889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottomly D, Long N, Schultz AR, Kurtz SE, Tognon CE, Johnson K, et al. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell. 2022;40(8):850-864.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh P, Patel M, Bhowmik D, Kumari N, Prajapati S, Gupta R. Identification of biomarkers affecting patient survival in cancers. World Acad Sci J. 2024;6(53). 10.3892/wasj.2024.268.
- 40.Patel M, Singh P, Gandupalli L, Reeshu G. Identification and evaluation of survival-associated common chemoresistant genes in cancer. Biomed Biotechnol Res J. 2024;8(3):320–7. [Google Scholar]
- 41.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(128):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dempster JM, Boyle I, Vazquez F, Root DE, Boehm JS, Hahn WC, et al. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021;22(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helmy SWA, Abdel-Aziz AK, Dokla EME, Ahmed TE, Hatem Y, Abdel Rahman EA, et al. Novel sulfonamide-indolinone hybrids targeting mitochondrial respiration of breast cancer cells. Eur J Med Chem. 2024;268(February):116255. 10.1016/j.ejmech.2024.116255. [DOI] [PubMed] [Google Scholar]
- 45.Han F, Liu J, Chu H, Cao D, Wu J, Fu H, et al. Knockdown of NDUFC1 inhibits cell proliferation, migration, and invasion of hepatocellular carcinoma. Front Oncol. 2022;12(September):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renkema GH, Wortmann SB, Smeets RJ, Venselaar H, Antoine M, Visser G, et al. SDHA mutations causing a multisystem mitochondrial disease: Novel mutations and genetic overlap with hereditary tumors. Eur J Hum Genet. 2015;23(2):202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall R, Yuan S, Wood K, Katona M, Straub AC. Cytochrome b5 reductases: Redox regulators of cell homeostasis. J Biol Chem. 2022;298(12):102654. 10.1016/j.jbc.2022.102654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aoki Y, Cortese S. Mitochondrial Aspartate/Glutamate Carrier SLC25A12 and Autism Spectrum Disorder: a Meta-Analysis. Mol Neurobiol. 2016;53(3):1579–88. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Yin C, Chen Q, Chen J, Ma Y, Wang L, et al. Prognostic Vvalue of SLC25A12 expression in patients with acute myeloid leukemia based on integrated analysis of multi-dimensional clinical data. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020;28(2):377–84. [DOI] [PubMed] [Google Scholar]
- 50.Henriques BJ, Katrine Jentoft Olsen R, Gomes CM, Bross P. Electron transfer flavoprotein and its role in mitochondrial energy metabolism in health and disease. Gene. 2021;776(July 2020):145407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan Z, Xiao L, Tang M, Bai F, Li J, Li L, et al. Targeting CPT1A-mediated fatty acid oxidation sensitizes nasopharyngeal carcinoma to radiation therapy. Theranostics. 2018;8(9):2329–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang N, Zhang X, Chen Q, Kantawong F, Wan S, Liu J, et al. Identification of a Mitochondria-Related Gene Signature to Predict the Prognosis in AML. Front Oncol. 2022;12(March):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wijnen NE, de Boer B, Klein K, Abrahamsson J, De Moerloose B, Zeller B, et al. Early Peripheral Blood Blast Clearance As a Prognostic Marker for Early Treatment Response in Pediatric Acute Myeloid Leukemia. Blood. 2023;142(Supplement 1):2950–2950. 10.1182/blood-2023-185597. [Google Scholar]
- 54.Yang CF, Gau JP, Hsiao LT, Hsu CY. Clinical significance of blast percentage assessed by bone marrow trephine biopsy and aspirate smear of myeloid malignancies. Pathology. 2021;53(6):740–5. [DOI] [PubMed] [Google Scholar]
- 55.Hou P, Wu C, Wang Y, Qi R, Bhavanasi D, Zuo Z, et al. A genome-wide CRISPR screen identifies genes critical for resistance to FLT3 inhibitor AC220. Cancer Res. 2017;77(16):4402–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bapat A, Keita N, Martelly W, Kang P, Seet C, Jacobsen JR, et al. Myeloid Disease Mutations of Splicing Factor SRSF2 Cause G2-M arrest and skewed differentiation of human hematopoietic stem and progenitor cells. Stem Cells. 2018;36(11):1663–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang B, Wu S, Zeng L, Tang Y, Luo L, Ouyang L, et al. Impact of NDUFAF6 on breast cancer prognosis: linking mitochondrial regulation to immune response and PD-L1 expression. Cancer Cell Int. 2024;24(1):1–21. 10.1186/s12935-024-03244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mrózek K, Kohlschmidt J, Blachly JS, Nicolet D, Carroll AJ, Archer KJ, et al. Outcome prediction by the 2022 European LeukemiaNet genetic-risk classification for adults with acute myeloid leukemia: an Alliance study. Leukemia. 2023;37(4):788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chin L, Wong CYG, Gill H. Targeting and Monitoring Acute Myeloid Leukaemia with Nucleophosmin-1 (NPM1) Mutation. Int J Mol Sci. 2023;24(4):3161. 10.3390/ijms24043161. [DOI] [PMC free article] [PubMed]
- 60.Chen D, Xia S, Wang M, Lin R, Li Y, Mao H, et al. Mutant and wild-type isocitrate dehydrogenase 1 share enhancing mechanisms involving distinct tyrosine kinase cascades in cancer. Cancer Discov. 2019;9(6):756–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu X, Han L, Busquets J, Collins M, Lodi A, Marszalek JR, et al. The Combined Treatment With the FLT3-Inhibitor AC220 and the Complex I Inhibitor IACS-010759 Synergistically Depletes Wt- and FLT3-Mutated Acute Myeloid Leukemia Cells. Front Oncol. 2021;11(August):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yap TA, Daver N, Mahendra M, Zhang J, Kamiya-Matsuoka C, Meric-Bernstam F, et al. Complex I inhibitor of oxidative phosphorylation in advanced solid tumors and acute myeloid leukemia: phase I trials. Nat Med. 2023;29(1):115–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 2: Figure S1. Area under the curve (AUC) of receiver operating characteristic (ROC) curves of the indicated genes in biospecimens obtained from AML patients (BeatAML.2 cBioportal dataset (OHSU, Cancer Cell, 2022). A-F) ROC curves and AUC of NADH:ubiquinone oxidoreductase subunit A6 (NDUFA6) (A), succinate dehydrogenase complex flavoprotein subunit A (SDHA) (B), solute carrier family 25 member 12 (SLC25A12) (C), electron transfer flavoprotein subunit beta (ETFB) (D), carnitine palmitoyltransferase 1A (CPT1A) (E) and glutathione peroxidase 4 (GPX4) (F) at the indicated time points of overall survival.
Data Availability Statement
No datasets were generated or analysed during the current study.